Sato et al. demonstrate that MYC expression renders MCF10A cells sensitive to down-regulation of UVSSA, as it alleviates stress during MYC-dependent transcription. UVSSA is a promising novel target for attacking MYC-addicted cancer cells.
Abstract
Cancer cells develop strong genetic dependencies, enabling survival under oncogenic stress. MYC is a key oncogene activated across most cancers, and identifying associated synthetic lethality or sickness can provide important clues about its activity and potential therapeutic strategies. On the basis of previously conducted genome-wide screenings in MCF10A cells expressing MYC fused to an estrogen receptor fragment, we identified UVSSA, a gene involved in transcription-coupled repair, whose knockdown or knockout decreased cell viability when combined with MYC expression. Synthetic sick interactions between MYC expression and UVSSA down-regulation correlated with ATM/CHK2 activation, suggesting increased genome instability. We show that the synthetic sick interaction is diminished by attenuating RNA polymerase II (RNAPII) activity; yet, it is independent of UV-induced damage repair, suggesting that UVSSA has a critical function in regulating RNAPII in the absence of exogenous DNA damage. Supporting this hypothesis, RNAPII ChIP-seq revealed that MYC-dependent increases in RNAPII promoter occupancy are reduced or abrogated by UVSSA knockdown, suggesting that UVSSA influences RNAPII dynamics during MYC-dependent transcription. Taken together, our data show that the UVSSA complex has a significant function in supporting MYC-dependent RNAPII dynamics and maintaining cell survival during MYC addiction. While the role of UVSSA in regulating RNAPII has been documented thus far only in the context of UV-induced DNA damage repair, we propose that its activity is also required to cope with transcriptional changes induced by oncogene activation.
Introduction
c-Myc (hereafter MYC) is one of the most frequently deregulated, overexpressed, or amplified proto-oncogenes in human cancers, ranging from hematopoietic to mesenchymal and epithelial tumors (Dang, 2012; Gabay et al., 2014; Rickman et al., 2018). Regulated MYC expression in response to mitogenic signals is essential in normal cells for controlling a wide range of processes, including proliferation, differentiation, and metabolism. Aberrant expression of MYC is sufficient to elicit multiple hallmarks of cancer, including uncontrolled cell division, as MYC is a rare bona fide proto-oncogene that does not require mutations to transform cells (Conacci-Sorrell et al., 2014).
Developing drugs against MYC has been challenging because MYC does not contain easily targetable domains, and MYC inhibition is lethal for normal dividing cells (McKeown and Bradner, 2014). As an alternative, targeting of cancer cells with aberrant MYC expression can be achieved by identifying genes or pathways that are specifically required for survival by cells expressing MYC at oncogenic levels. Such genes or pathways that are synthetic lethal (SL) with deregulated MYC expression (MYC-SL genes) provide valuable insight into MYC biology, as well as identify potential targets for attacking MYC-dependent cancer cells (Cermelli et al., 2014; Hsu et al., 2015; Lin et al., 2012a; Molenaar et al., 2009; Moser et al., 2012; Rottmann et al., 2005; Wang et al., 2004; Yang et al., 2010; Zhou et al., 2014; Sato et al., 2015). Notably, the striking variety of MYC-SL genes identified emphasizes how versatile and widespread the effects of deregulated MYC expression can be within a single cell, as well as across various cell types.
MYC is a member of the basic helix–loop–helix leucine zipper family of transcription factors and activates target genes through binding gene promoters harboring canonical E-box sequences with highest affinity (Dang, 2012). MYC also regulates gene expression in other ways beyond typical sequence-specific binding. Oncogenic MYC binds to and activates transcription at many genomic regions containing degenerate E-box sequences or even no E-boxes at all, as long as they are accessible (Fernandez et al., 2003; Orian et al., 2003; Zeller et al., 2006). MYC stimulates the activity of RNA polymerases (RNAPs) I and III in addition to RNAPII and increases the production of almost all types of RNA when expressed at high levels (Campbell and White, 2014). MYC also directly regulates global RNAPII promoter-proximal pause release (Adelman and Lis, 2012; Jonkers and Lis, 2015; Rahl et al., 2010). This genome-wide control of transcription is deregulated globally at gene promoters during oncogenic MYC expression (Rahl et al., 2010).
Thus, MYC has been proposed to function as a global transcription amplifier (Lin et al., 2012b; Nie et al., 2012). MYC likely incurs both local and global effects on transcription, depending on its abundance and cellular context. Indeed, MYC increases both de novo RNAPII binding to promoters and pause release at already active promoters (de Pretis et al., 2017). It is becoming increasingly clear that MYC’s role in cell proliferation and oncogenesis is not limited to controlling the expression of a subset of genes and that MYC’s influence on genome-wide transcription may be much more complex and global than originally thought (Baluapuri et al., 2020).
Apart from promoter-proximal pause release, RNAPII stalling and release have been extensively studied during transcription-coupled repair (TCR), or transcription-coupled nucleotide excision repair (TC-NER), a DNA repair pathway that supports efficient repair of bulky adducts or UV-induced lesions specifically on actively transcribed strands of DNA (Hanawalt and Spivak, 2008; Mellon et al., 1987; Spivak, 2016). The Cockayne syndrome A (CSA)/ERCC8 protein is recruited to stalled RNAPII at UV-induced lesions along with CSB/ERCC6, and germline mutations in CSA or CSB cause Cockayne syndrome (CS; Saijo, 2013; Vélez-Cruz and Egly, 2013; Spivak, 2016). CSA is part of an E3 ubiquitin ligase complex required for degradation of CSB following lesion repair and subsequent recovery of transcription (Groisman et al., 2006). CSA also recruits other TCR factors to RNAPII-stalled sites, including the UV-stimulated scaffold protein A–ubiquitin-specific–processing protease 7 (UVSSA–USP7) complex (Fei and Chen, 2012). UVSSA interacts with the USP7 deubiquitinase, and this complex is critical for the regeneration of hypophosphorylated RNAPII complexes required for de novo RNAPII loading, which is required for transcription recovery following TCR (Nakazawa et al., 2012; Schwertman et al., 2012; Zhang et al., 2012). UVSSA–USP7 also stabilizes CSB by preventing its premature proteasomal degradation (Fei and Chen, 2012; Higa et al., 2016; Nakazawa et al., 2012; Schwertman et al., 2012; Zhang et al., 2012). Intriguingly, germline inactivation of UVSSA results in UV-sensitive syndrome (UV-SS), a condition with much less severe symptoms than CS and other classical TCR disorders (Itoh et al., 1996; Ogi et al., 2013). Cells from UV-SS patients do not efficiently reinitiate transcription following UV irradiation, much like cells from CS patients; yet, UV-SS patient phenotypes are vastly milder, with no associated neurodegeneration or developmental defects (Itoh et al., 1996, 1995, 1994). Notably, while CSA and CSB have been reported to have roles outside of TCR, this has not yet been investigated for UVSSA.
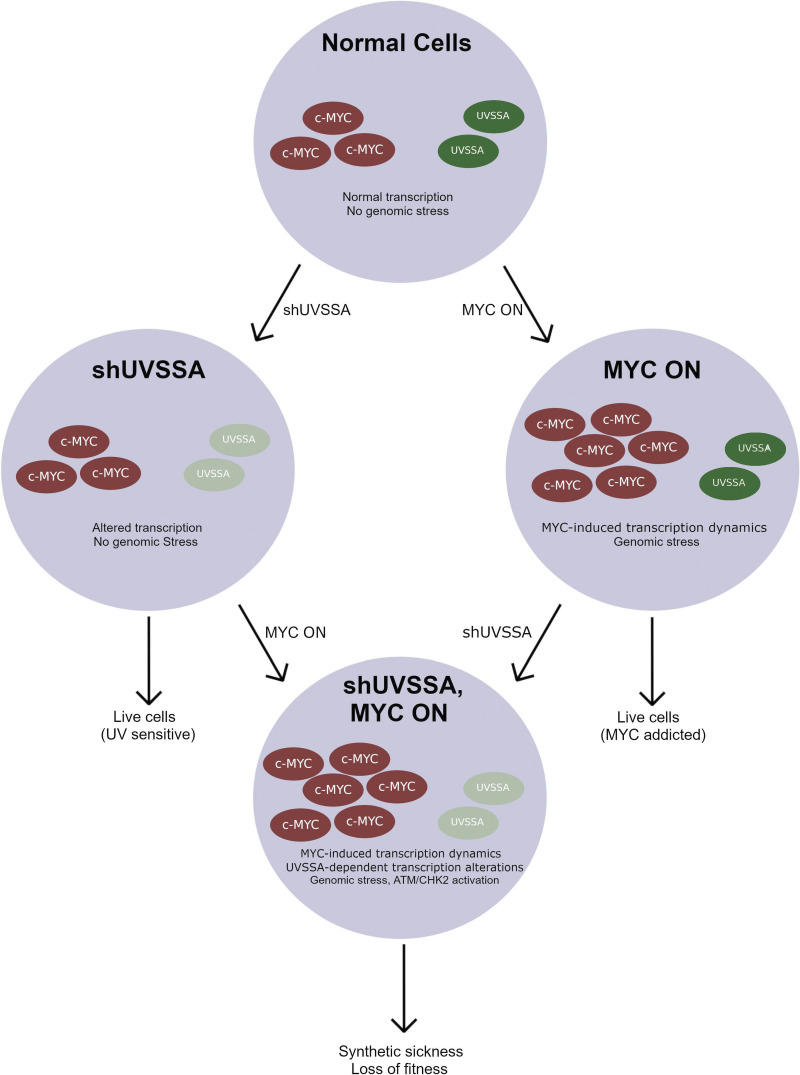
Here, we conducted a shRNA-based genome-wide screen to identify MYC-SL candidates followed by community enrichment protein analysis of the hits. One of the major protein communities we identified was highly enriched in DNA repair proteins and included UVSSA and CSA/ERCC8. Knockdown of UVSSA or CSA/ERCC8, or inhibition of USP7, decreased the viability of cells expressing a MYC- estrogen receptor fragment fusion protein (MYC-ER), validating our screen results. ATM/Checkpoint kinase 2 (CHK2) was activated upon MYC induction and UVSSA knockdown, suggesting genomic instability. Interestingly, attenuation of RNAPII activity with α-amanitin abrogated the synthetic sick (SS) effect, while reduction of DNA replication did not. This result suggested that MYC-induced transcription contributes to genome instability during MYC overexpression and UVSSA knockdown. Global RNAPII chromatin immunoprecipitation–sequencing (ChIP-seq) data showed that UVSSA knockdown greatly reduced the MYC-dependent increase in RNAPII occupancy at gene promoters and distribution of RNAPII on actively transcribing genes, implying that UVSSA may be required to support RNAPII stability or loading during MYC-induced transcription. Taken together, our results suggest that oncogene-dependent transcription stress is a vulnerability for MYC-deregulated cells and that the UVSSA complex has a previously unidentified function in protecting cells from this stress.
Results
Identification of MYC-SL/SS protein communities and candidate genes
To identify proteins involved in MYC-SL/SS interactions, we previously performed genome-wide screening in MCF10A-MYC-ER cells (Sato et al., 2015). In this shRNA-based screen, data were accrued after 4 wk of continuous MYC-ER expression at levels approximately twofold higher than control to mimic relatively low and steady MYC overexpression, present in many cancers addicted to MYC (Nesbit et al., 1999). As a result, MYC-SL/SS candidates that we recovered contained a mixture of acute SL targets as well as many SS targets that reduce the fitness of MYC-ER–expressing cells over time, in contrast to a more acute approach examining cell death within a shorter period of time with higher MYC expression.
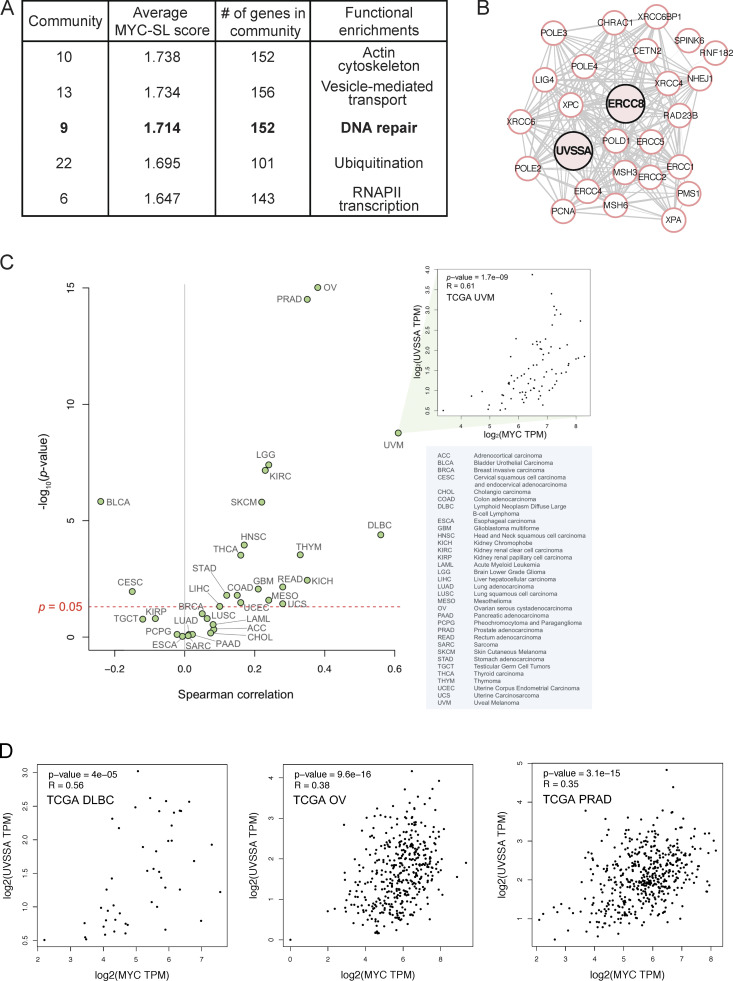
Here, we analyzed the data using a community enrichment approach based on protein–protein interaction networks. Proteins related to a common MYC-SL/SS biological process should present an enrichment of fold scores in the genome-wide screening data. Fold change increases from the shRNA screen were assigned to weigh nodes (proteins), then local network communities were detected and clustered. Resulting MYC-SL/SS communities were ranked according to the average MYC-SL/SS score of the included nodes. 18 MYC-SL/SS protein network communities were detected, covering a variety of cellular processes (cutoff, >100 nodes per community; Fig. 1 A; Table S1; see Materials and methods).
Figure 1.
MYC interacts with DNA repair protein community, and its expression correlates with UVSSA expression in a subset of tumor types. (A) The highest-scoring MYC-SL gene product communities. Gene product communities were analyzed using STRING interactions and ranked according to average MYC-SL scores of each node in the community. A network of gene products involved in DNA repair ranks third (bold). (B) Most densely interconnected nodes around UVSSA and ERCC8 within community 9. UVSSA and ERCC8 are emphasized in pink but do not represent special nodes. (C) Spearman correlation analysis of MYC and UVSSA expression in 33 TCGA cancer types (green dots) using GEPIA 2. Pop-out box shows an example scatterplot for a given TCGA cancer type with statistics shown. Blue box includes a legend listing abbreviations. (D) Scatterplots showing MYC and UVSSA expression in the diffuse large B cell lymphoma (DLBC), ovarian serous cystadenocarcinoma (OV), and prostate adenocarcinoma (PRAD) TCGA cancer samples with statistical values. Values measured in transcripts per million (TPM).
Notably, community 9 was ranked third for average MYC-SL/SS score and was highly enriched for DNA repair proteins. Within the 152 gene products in this community, NER (which encompasses both global NER and TC-NER in Kyoto Encyclopedia of Genes and Genomes pathways) ranked as the second most enriched pathway with high confidence (false discovery rate, 1.94e-35), after Fanconi anemia. We focused our attention on UVSSA (or KIAA1530), a gene in this dataset involved in TC-NER, since it has not previously been implicated in any process outside of UV damage or the repair of specific bulky adducts on DNA (Fig. 1, A and B; and Fig. S1). Spearman correlation analysis of MYC and UVSSA expression in 33 cancer types in The Cancer Genome Atlas (TCGA) using GEPIA 2 (Gene Expression Profiling Interactive Analysis) suggested strong correlation in at least a subset of cancers, including uveal melanoma and diffuse large B cell lymphoma, indicating that the relationship may be relevant to MYC-expressing cancer cells, at least in certain contexts (Fig. 1, C and D).
Figure S1.
MYC-SL community 9 network. All 152 nodes of community 9 are represented. UVSSA and ERCC8 are highlighted in pink but do not denote special nodes. Distance between nodes and thickness of gray links between nodes represent relative relationships between nodes, according to the STRING network. Analyzed in STRING, and image was generated by using Cytoscape.
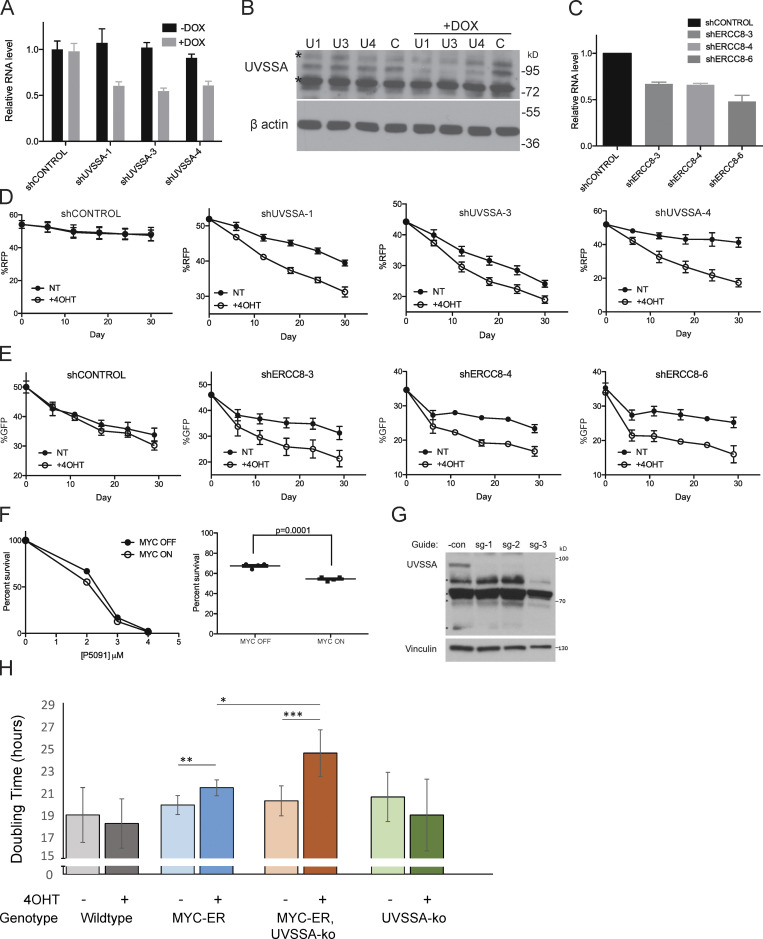
Importantly, two independent shRNA clones targeting UVSSA/KIAA1530 were recovered as MYC-SL/SS in the screen (log2 fold change [Log2FC], −2.39; P = 0.0216; and Log2FC, −2.35; P = 0.0983; Table S2). To validate this finding and confirm that UVSSA knockdown is SL or SS with MYC activation, we used the same MCF10A-MYC-ER cells from the original screen, expressing MYC-ER at approximately twofold above endogenous MYC. We down-regulated UVSSA expression using three unique shRNA clones targeting distinct sequences from the library shRNAs, achieving ∼40–50% knockdown of UVSSA mRNA levels (Fig. S2, A and B). To compare the fitness of shUVSSA cells to control cells, we employed a fluorescence-based competitive survival (FBCS) assay, a sensitive and internally controlled assay to detect loss of fitness over time. Cells with shUVSSA or shCONTROL coexpressing GFP or RFP were coplated with cells without shRNA in equal numbers, and the fraction of GFP/RFP-expressing cells was monitored over time. Cells with UVSSA down-regulation were markedly less viable with MYC activation over the course of 30 d (Figs. 2 A and S2 D). While the survival ratio of MYC on/MYC off cells remained approximately at 1.0 for shCONTROL cells, the ratio for shUVSSA-1 and shUVSSA-3 cells dropped to 0.81 and 0.78, respectively, and for shUVSSA-4, it dropped to 0.46 (P ≤ 0.001), validating that decreased UVSSA expression results in an SS phenotype with MYC-ER expression (Fig. 2 A). Notably, UV-related phenotypes dependent on shUVSSA-4 were restored by expressing shRNA-resistant wild-type UVSSA in previous studies, validating its specificity (Fei and Chen, 2012). To confirm the specificity of the synthetic sickness phenotype and rule out any potential off-target effects of UVSSA shRNA, we inactivated endogenous UVSSA via the CRISPR-based iSTOP technology (Billon et al., 2017) in MCF10A cells expressing MYC-ER and in control cells (Fig. S2 H). We assessed the impact of MYC expression in these lines by calculating the doubling time for a 6-d logarithmic growth period. As anticipated, MYC-ER activation increased cells’ doubling time. Notably, MYC activation in the UVSSA-knockout (UVSSA-KO) line triggered a significantly larger increase in doubling time than MYC-ER alone, confirming that the synthetic sickness observed with UVSSA knockdown was caused by loss of UVSSA (Fig. S2 H).
Figure S2.
Down-regulation, inhibition, or KO of UVSSA-USP7 or CSA/ERCC8 sensitizes MYC-overexpressing MCF10A cells. (A) qRT-PCR following knockdown of UVSSA. Three independent doxycycline (DOX)-inducible shRNA clones were used to produce stable UVSSA-knockdown cell lines. Black bars show UVSSA mRNA levels without shRNA induction, and gray bars show mRNA levels after 72 h of 1 µg/ml DOX treatment. Values were normalized to GAPDH. Error bars represent the SEM (n = 3). (B) Western blot following knockdown of UVSSA. Whole-cell lysates were prepared after 72 h of 1 µg/ml DOX treatment in stable cell lines with UVSSA knockdown and subjected to SDS-PAGE. Asterisks show nonspecific bands recognized by the UVSSA antibody. (C) qRT-PCR following knockdown of ERCC8. Three independent shRNA clones were used to produce stable ERCC8-knockdown cell lines. Values were normalized to GAPDH. Error bars represent the SEM (n = 3). (D) Representative raw data from UVSSA FBCS assays as seen in Fig. 2 A, comparing 4-hydroxy-tamoxifen (4OHT) treated and Not Treated (NT) conditions. Points denote RFP readings via FACS at the indicated time points. A shift in slope corresponds to sensitivity of each cell line to MYC-ER activation (treated with 200 nM 4OHT or vehicle). Error bars represent the SEM between technical replicates (n = 3). (E) Representative raw data from ERCC8 FBCS assays as seen in Fig. 2 B, comparing 4-hydroxy-tamoxifen (4OHT) treated and Not Treated (NT) conditions. Points denote GFP readings via FACS at the indicated time points. A shift in slope corresponds to the sensitivity of each cell line to MYC-ER activation (treated with 200 nM 4OHT or vehicle). Error bars represent the SEM between technical replicates (n = 3). (F) Clonogenicity assay following 24 h of treatment with varying doses of P5091 (treated with 200 nM 4OHT or vehicle). Percentage survival is based on colony numbers after 10 d of incubation. Left: A representative P5091 dose-dependent (0–4 µM) clonogenicity assay. Right: Clonogenicity difference between MYC on and MYC off cells after treatment with 2 µM P5091 for 24 h. P value was calculated from an unpaired t test (n = 4). (G) Western blot of UVSSA in iSTOP edited clonal lines. Cloned MCF10A lines edited by iSTOP using three different guides were lysed in radioimmunoprecipitation assay lysis buffer. The whole-cell lysates were then subjected to Western blot analysis using an anti-UVSSA antibody. The UVSSA-specific band is marked by an asterisk. Vinculin served as a loading control. *Nonspecific bands. (H) Doubling time for UVSSA-KO MCF10A cells. MCF10A cells with the indicated genotypes were grown with 4OHT (400 nM) or vehicle control for up to 6 d, and doubling time was calculated from cell counts, as described in the Materials and methods (n = 3, 4, 5, and 5, respectively). Statistical analysis was performed using Student’s t test, paired for intraline comparisons and unpaired for interline comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.005).
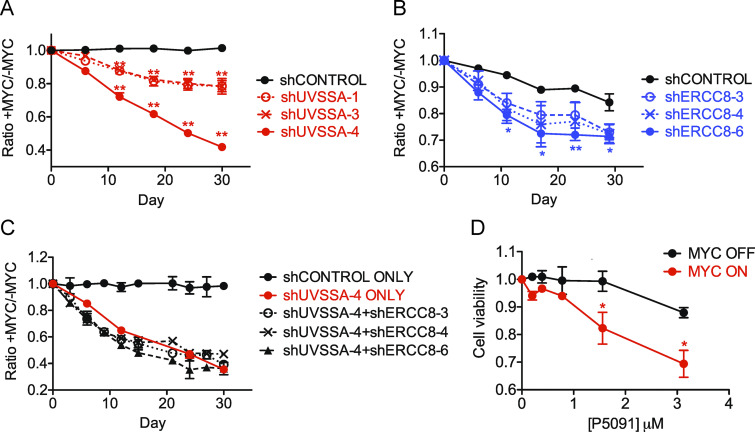
Figure 2.
Down-regulation of the UVSSA–USP7–ERCC8 pathway sensitizes MCF10A cells to MYC overexpression. (A) FBCS assay with three unique shRNA clones targeting UVSSA. Each curve represents the ratio of fluorescent signal between MYC on and MYC off conditions (treated with 200 nM 4OHT or vehicle) for each cell line determined by FACS at the indicated time points during the assay. A downward slope denotes sensitivity to MYC deregulation. Asterisks indicate significant difference from shCONTROL at each time point (**, P < 0.01). Error bars represent the SEM (n = 4). (B) FBCS assay of cell lines expressing three unique shRNAs against ERCC8. Asterisks indicate significant difference from shCONTROL at each time point (**, P < 0.01; *, P < 0.05). Error bars represent the SEM (n = 4). (C) FBCS assay of cell lines expressing shUVSSA-4 in combination with one of three shRNA clones against ERCC8. No significant difference was found between shUVSSA-4 alone and any of the double knockdowns. Error bars represent the SEM (n = 2). (D) MTS assay in the presence of USP7 inhibitor P5091. MCF10A-MYC-ER cells were subjected to MTS assays in MYC OFF or MYC ON conditions using indicated doses of P5091. Asterisks indicate significant difference from MYC off at each concentration (*, P < 0.05). Error bars represent the SEM (n = 3).
ERCC8, or CSA, which acts in a complex with UVSSA during TC-NER, was also identified as a high-confidence MYC-SL/SS candidate in the screen (Log2FC, −3.2; P = 0.03). To validate this result, three independent shRNA clones were used to modulate ERCC8 mRNA levels to 47–68% of control cells (Fig. S2 C). FBCS assays showed a 67–78% decrease in the MYC ON/MYC OFF survival ratio over the course of 18 d compared with control cells, validating ERCC8 as another MYC-SS gene (P ≤ 0.001; Figs. 2 B and S2 E).
Since UVSSA and ERCC8 associate within a protein complex in TCR, we next examined the effect of knocking down both UVSSA and ERCC8 on cell survival in MYC-ER cells. We generated double-knockdown cells using shRNA clone UVSSA-4 in combination with clone ERCC8-3, -4, or -6. The MYC ON/MYC OFF survival ratios over the course of 30 d dropped to 0.45, 0.49, and 0.41, respectively (P ≤ 0.001), not significantly different from UVSSA-4 alone (0.46; Fig. 2 C). This result suggests that UVSSA and ERCC8 likely act in a complex to yield the MYC-SS effect.
UVSSA is also in complex with USP7 during TCR. We asked whether inhibition of USP7 recapitulated the SS effect when combined with MYC-ER expression. Treatment of MCF10A-MYC-ER cells with P5091, a specific small-molecule inhibitor of USP7, sensitized MYC on cells approximately threefold over MYC off cells in 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays (Fig. 2 D). Clonogenic assays also revealed enhanced sensitivity of MYC-expressing cells to P5091 (S2F). These results support our hypothesis that down-regulating the UVSSA–USP7–ERCC8 complex is SS with MYC deregulation.
Down-regulation of UVSSA in MYC-ER cells results in ATM/CHK2 activation
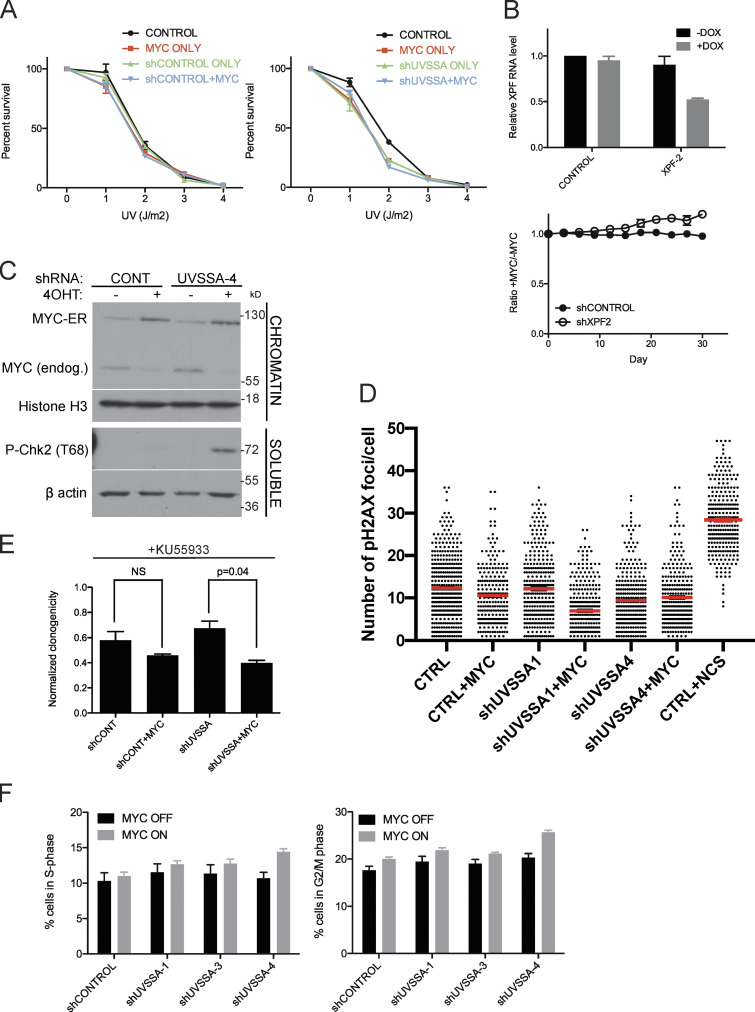
Since the only known function of UVSSA is in TCR, we assessed whether the SS effect upon UVSSA down-regulation in MYC-ER cells was due to increased sensitivity to UV damage. We treated shCONTROL or shUVSSA cells with increasing doses of UV-C in the presence or absence of MYC activation. MYC ON cells showed mild sensitivity to UV in both shCONTROL and shUVSSA cells, consistent with previous studies (Herold et al., 2002; Fig. S3 A). UVSSA knockdown alone also conferred UV sensitivity, as previously reported in UV-SS patient cells and knockdown cells (Nakazawa et al., 2012; Schwertman et al., 2012; Zhang et al., 2012). Importantly, the induction of MYC did not exacerbate the UV sensitivity of shUVSSA cells, suggesting the SS relationship between shUVSSA and MYC is independent of exogenous UV-induced DNA damage (Fig. S3 A). We then knocked down expression of XPF, a downstream NER endonuclease essential for UV repair, and performed an FBCS assay. XPF knockdown did not impair the survival of MYC on cells. Rather, MYC-ER induction appeared to moderately enhance survival of cells with XPF knockdown (Fig. S3 B). These results indicate that the MYC-SS effect of UVSSA knockdown is independent of its known role in UV-induced damage repair, suggesting a novel cellular role for UVSSA.
Figure S3.
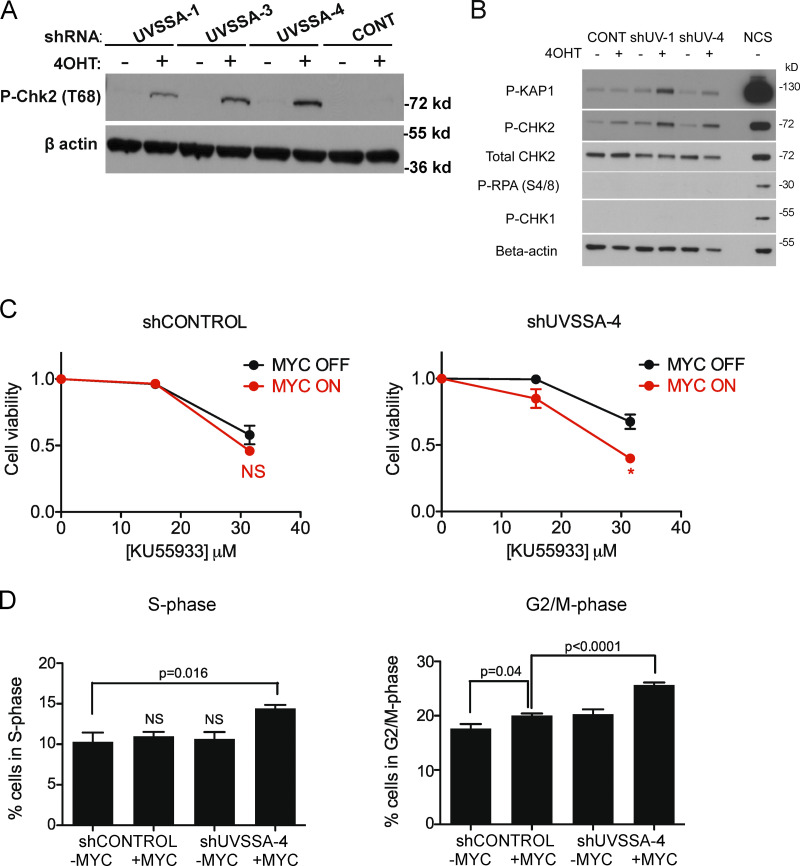
Down-regulation of UVSSA elicits a DNA damage response in MYC-deregulated cells. (A) Clonogenicity assay following UV treatment. shCONTROL cells (left) or shUVSSA-4 cells (right) were treated with vehicle or 1 µg/ml doxycycline (DOX), in combination with 200 nM 4OHT or vehicle, and subjected to increasing doses of UV-C radiation. Percentage survival is based on colony numbers after 10 d of incubation. Error bars represent the SEM (n = 3). (B) XPF knockdown and FBCS assays. XPF was down-regulated using DOX-inducible shRNA (XPF-2). qRT-PCR shows XPF mRNA levels without shRNA induction in black and mRNA levels after 72 h of 1 µg/ml DOX treatment in gray (top). Values were normalized to GAPDH, and error bars represent the SEM (n = 3). shCONTROL and shXPF cells were subjected to FBCS assays with 1 µg/ml DOX and 200 nM 4OHT or vehicle (bottom). Survival is indicated as a ratio of RFP signal between MYC on and MYC off conditions. Error bars represent the SEM (n = 3). (C) Western blot showing CHK2 activation in MYC-deregulated UVSSA knockdown cells. Indicated cell lines were treated with 200 nM 4OHT or vehicle and 1 µg/ml DOX every 3 d and harvested at day 9. Following cell fractionation, chromatin-bound and soluble cytoplasmic fractions were run on SDS-PAGE gels and subjected to Western blot analysis with the indicated antibodies. Histone H3 served as a loading control. (D) Quantification of H2AX foci following MYC induction. Indicated cell lines were treated with 200 nM 4OHT or vehicle and 1 µg/ml DOX for 3 d before fixing and immunostaining with H2AX antibodies. NCS was used as a positive control. Red lines indicate means. (E) Clonogenicity assays were performed for shCONTROL or shUVSSA-4 cells treated with 31.5 µM KU55933. Statistical analysis was performed using t tests. Error bars represent the SEM (n = 3). (F) Representative raw data for Fig. 3 D. Fractions of cells in S phase (left) or G2/M phase (right) were measured using FACS after 30 d of culturing cells with 1 µg/ml DOX and 200 nM 4OHT or vehicle. Error bars represent the SEM (n = 3).
MYC-deregulated cells show increased genomic stress. We asked whether UVSSA knockdown increases MYC-dependent genomic instability by monitoring the accumulation of various DNA damage markers. Phosphorylated CHK2 (T68), an indicator of ATM activation, was enriched in UVSSA-knockdown cells with MYC activation and peaked at 10 d after induction with 4-hydroxytamoxifen (4OHT; Figs. 3 A and S3 C). Accumulation of phosphorylated KAP1 (P-KAP1) was also observed, another specific target of ATM (Fig. 3 B). While ATM activation is often associated with the accumulation of DNA double-strand breaks (DSBs), we did not observe significant accumulation of P-CHK1 (S345) or P-RPA (S4/S8), in contrast to the rapid accumulation of these species following treatment with DSB-inducing agent neocarzinostatin (NCS; Fig. 3 B). The accumulation of phosphorylated H2A histone family member X (P-H2AX), another DSB marker and ATM target, was inconclusive by Western blot analysis; however, P-H2AX foci were markedly scarcer in all conditions compared with cells treated with NCS (Fig. S3 D). These results suggest that activation of ATR following DSB processing might be limited in cells with UVSSA knockdown and MYC-ER activation.
Figure 3.
Down-regulation of UVSSA elicits a DNA damage response in MYC-deregulated cells. (A) CHK2 activation in three independent shUVSSA cell lines following MYC induction. Indicated cell lines were treated with 200 nM 4OHT or vehicle and 1 µg/ml doxycycline every 3 d and harvested at day 9. Whole-cell lysates were subjected to Western blot analysis with anti–phospho-T68-CHK2 antibodies. β-Actin staining served as a loading control. (B) KAP1 phosphorylation in two independent shUVSSA cell lines following MYC induction. Indicated cell lines were treated with 200 nM 4OHT or vehicle and 1 µg/ml doxycycline every 3 d and harvested at day 9. Whole-cell lysates were subjected to Western blot analysis with indicated antibodies. β-Actin staining served as a loading control. NCS treatment served as a positive control for induced DNA DSBs. (C) MTS assays in the presence of ATM inhibitor KU55933. shCONTROL (left) or shUVSSA-4 (right) cells were subjected to standard MTS assays with increasing concentrations of KU55933. Asterisks indicate significant difference from MYC off at indicated concentration (*, P < 0.05). Error bars represent the SEM (n = 3). (D) Cell cycle distribution of shCONTROL or shUVSSA-4 cells in MYC on or MYC off conditions. Cells in S phase (left) or G2/M phase (right) were measured by propidium iodide staining after 30 d of culturing cells with 1 µg/ml doxycycline and 200 nM 4OHT or vehicle. P values are indicated where appropriate. Error bars represent the SEM (n = 3).
Since ATM directly activates and phosphorylates CHK2 and KAP1, we tested the sensitivity of MYC-induced UVSSA-knockdown cells to ATM inhibitor KU55933. While MYC induction alone did not confer increased sensitivity to KU55933, the combined knockdown of UVSSA with MYC induction increased the sensitivity of MCF10A cells to acute treatment with KU55933, suggesting that MYC-ER expression and UVSSA knockdown trigger an ATM-dependent checkpoint and prosurvival mechanism, consistent with previous results (Figs. 3 C and S3 E).
ATM-dependent checkpoint activation affects cell cycle progression. As expected, after 20 d of culturing UVSSA-knockdown cells in MYC OFF or MYC ON conditions, we observed a significant accumulation of cells in S and G2/M phase, indicating cell cycle slowing (Figs. 3 D and S3 F). Knockdown using clones UVSSA-1, -3, and -4 all resulted in an increase in S-phase cells compared with shCONTROL when combined with MYC induction (Figs. 3 D and S3 F). In MYC OFF conditions, there were no significant differences in the number of cells in S phase. The G2/M population was also increased with UVSSA knockdown and MYC-ER induction, consistent with checkpoint activation (Figs. 3 D and S3 F).
SS phenotype is dependent on RNAPII transcription
Because MYC regulates both DNA replication and transcription, we tested whether inhibiting DNA replication or transcription could alleviate the SS interaction between MYC-ER expression and UVSSA knockdown. In an FBCS assay, cells were either continuously treated with low doses of aphidicolin or α-amanitin in MYC ON or MYC OFF conditions (Fig. 4, A and B). Aphidicolin inhibits DNA replicative polymerases, whereas α-amanitin is a potent inhibitor of RNAPII. Treatment with α-amanitin (100 ng/ml) increased the MYC ON/MYC OFF survival ratio of UVSSA-knockdown cells to levels comparable to those of shCONTROL cells over the course of 14 d (Fig. 4 A, left; P ≤ 0.005). In contrast, after 26 d, the highest sublethal dose of aphidicolin (50 ng/ml) did not alter the MYC ON/MYC OFF ratio of UVSSA-knockdown cells (Fig. 4 B). Of note, the 100 ng/ml dose of α-amanitin was toxic to cells beyond 14 d. Nevertheless, the SS phenotype was transiently alleviated with α-amanitin, but not with aphidicolin. These data strongly suggest that MYC-dependent transcription is primarily responsible for the SS effect between MYC activation and UVSSA knockdown.
Figure 4.
SS effect between UVSSA knockdown and MYC induction depends on transcription. (A and B) FBCS assays in presence of α-amanitin or aphidicolin. shCONTROL or shUVSSA-4 cells were subjected to FBCS assays in the presence of 1 µg/ml doxycycline, 200 nM 4OHT or vehicle, and vehicle or indicated doses of α-amanitin (A) or aphidicolin (B) administered every 48–72 h over the course of the indicated number of days. Asterisks indicate significant difference of the data point with highest dose of drug from shUVSSA-4 only at the indicated time point (**, P < 0.01; *, P < 0.05). Error bars represent the SEM (n = 3). (C) FBCS assay in the presence of BRD4 inhibitor JQ1. MCF10A-MYC-ER cells expressing the indicated shRNA were subjected to FBCS assays with 200 nM 4OHT or vehicle, 1 µg/ml doxycycline, and 220 nM JQ1 or vehicle administered every 3 d. Asterisks indicate significant difference of the +JQ1 data point from its no JQ1 counterpart (in same color) at the denoted time points (**, P < 0.01; *, P < 0.05). Error bars represent the SEM (n = 3). (D) Quantification of P-CHK2 signal with or without transcription inhibitors by Western blot analysis. Signal quantification was internally normalized to β-actin signal, and P values were calculated using paired t tests. n = 13 for DMSO; n = 10 for JQ1; and n = 13 for triplotide conditions. (E) Effect of XPB inhibitor triptolide (TTL). Indicated cell lines were subjected to FBCS assays with 200 nM 4OHT or vehicle, 1 µg/ml doxycycline, and 56 pM TTL or vehicle administered every 3 d for 14 d. Survival ratio in each condition at day 14 is denoted in the graph. Error bars represent the SEM (n = 2). (F) Representative Western blot of markers of transcriptionally active chromatin. Whole-cell lysates were prepared as described in Materials and methods after the indicated cell lines were treated with 200 nM 4OHT or vehicle and 1 µg/ml doxycycline for 72 h and probed with the indicated antibodies. β-Actin served as the loading control. (G) Quantification of histone modification signals by Western blot analysis as in E. Each data point of H3K27Ac (left) or H3K4m31 (right) signal on Western blots was internally normalized to β-actin. P values were calculated using paired t tests.
To confirm whether the SS phenotype is dependent on transcription induced by MYC-ER, FBCS assays were employed in the presence or absence of (S)-(+)-tert-butyl-2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetate (JQ1), an inhibitor of the BET family of bromodomain proteins, most notably BRD4, that facilitate MYC-dependent transcription (Delmore et al., 2011). We observed that the decreased viability of MYC ON cells with UVSSA knockdown was partially alleviated by the addition of JQ1, as predicted (Fig. 4 C). JQ1 treatment also decreased P-CHK2 signal in MYC ON shUVSSA cells, consistent with transcription underlying ATM activation (Fig. 4 D). Notably, JQ1 also improved survival of MCF10A-MYC-ER cells without UVSSA knockdown, implying that MYC-dependent transcription is a stressor even for cells with unperturbed UVSSA expression.
Next, we examined the levels of post-translational histone modifications associated with transcriptionally active chromatin. We sought to assess whether transcription stress was reflected by an increase in histone marks following MYC activation and UVSSA knockdown. Monomethylated histone H3 lysine K4 (H3K4) and acetylated H3K27 levels mark regions of chromatin that contain primed or active enhancers and promoters, corresponding to elevated transcriptional activity (Gates et al., 2017). We observed increased levels of H3K4me1 and H3K27Ac when MYC-ER was activated in shCONTROL cells, consistent with MYC’s role in amplifying transcription (Fig. 4, F and G). Notably, H3K4me1 and H3K27Ac levels were further elevated when MYC-ER was combined with UVSSA knockdown, which could signify increased transcription, transcription-associated stress, or deregulation of signaling (Fig. 4 G). This result is consistent with our hypothesis that UVSSA knockdown in MYC-ER cells is associated with aberrant transcription.
Transcription factor II human (TFIIH) is essential for both RNAPII transcription and downstream DNA repair in global NER and TC-NER (Compe and Egly, 2012; Egly and Coin, 2011; Wang et al., 1994). Since TFIIH interacts with UVSSA during TC-NER, we assessed the impact of TFIIH inhibition on the SS interaction between MYC and UVSSA (Nakazawa et al., 2012; Okuda et al., 2017). Triptolide, a small-molecule inhibitor of XPB, a DNA helicase subunit of TFIIH, partially alleviated the survival defect upon MYC up-regulation and UVSSA knockdown (Fig. 4 E). Like JQ1, triptolide treatment also decreased P-CHK2 signal in MYC ON cells with UVSSA knockdown (Fig. 4 D). Consistent with our hypothesis, these results suggest that TFIIH-dependent RNAPII transcription contributes to the MYC-SS effect of UVSSA knockdown.
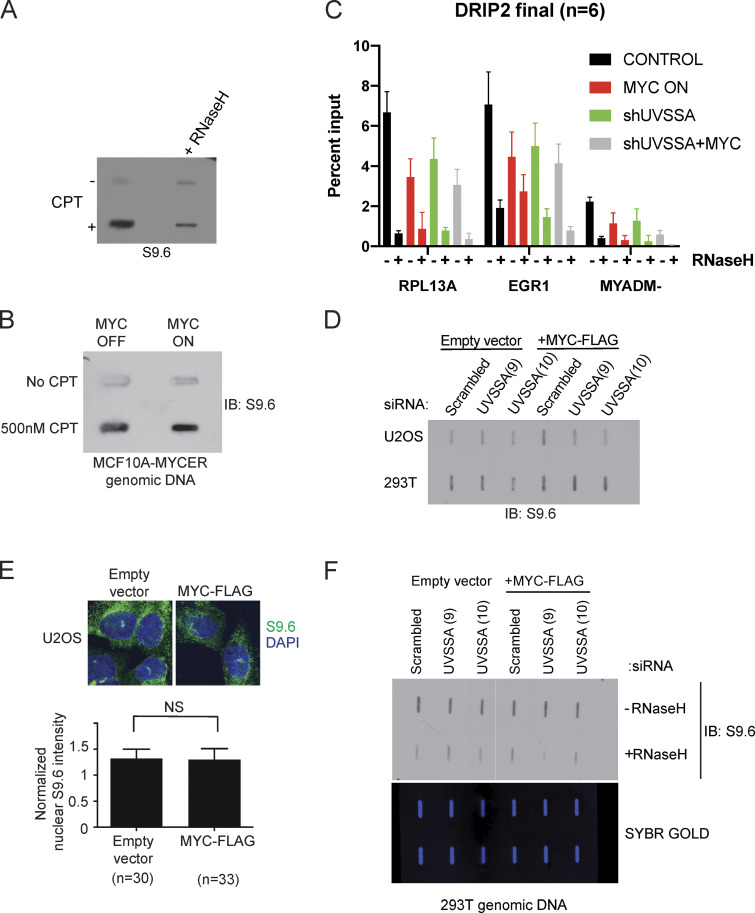
R-loops are RNA/DNA hybrid structures that occur at a steady state during normal transcription with distinct roles, including transcription control (Sanz et al., 2016; Skourti-Stathaki et al., 2011). R-loops can be a significant source of DNA damage relevant in oncogene-induced genome instability, as in the case of mutant HRAS (Kotsantis et al., 2016). Importantly, CSB, a member of the TCR pathway, participates in R-loop processing (Sollier et al., 2014). Increases or persistence of R-loops upon MYC induction and UVSSA knockdown during transcription could underlie the SS phenotype. To interrogate whether R-loops accumulate on chromatin upon MYC activation and/or UVSSA knockdown, we blotted whole genomic DNA using the S9.6 antibody, which can recognize RNA/DNA hybrids (Boguslawski et al., 1986; Phillips et al., 2013). As expected, we detected increased S9.6 signal upon camptothecin treatment, which was sensitive to treatment with RNase H1 (Fig. S5 A). However, we were unable to detect significant S9.6 signal using MCF10A-MYC-ER cells via slot blot analysis (Fig. S5 B). Next, we thought to monitor R-loops at specific sites known to be enriched in DNA/RNA hybrids. DNA/RNA hybrid immunoprecipitation (DRIP)–quantitative PCR (qPCR) revealed that MYC-ER expression, UVSSA knockdown, and their combination all decreased DRIP signal compared with control at two known R-loop rich loci (RPL13A and EGR1), consistent with our previous observations that R-loop signal does not increase (Fig. S5 C). Acute overexpression of MYC and siRNA knockdown of UVSSA in two different cell lines (U2OS or 293T) did not significantly alter S9.6 signal as detected by slot blot or S9.6 immunofluorescence analysis (Fig. S5, D–F). These results strongly suggest that the SS effect between MYC-ER and UVSSA knockdown is caused by genomic stress independent of R-loops.
Figure S5.
R-loops do not accumulate upon MYC activation or UVSSA knockdown. (A) Slot blot to examine S9.6 signal specificity. MCF10A-MYC-ER cells were treated with 500 nM camptothecin (CPT) or vehicle for 48 h. Genomic DNA was treated with vehicle or RNase H1 for 1 h, then purified and spotted onto a membrane and incubated with S9.6 antibody. (B) Slot blot to examine S9.6 signal following MYC induction. MCF10A-MYC-ER cells were simultaneously treated with 500 nM CPT or vehicle and 200 nM 4OHT or vehicle to induce MYC-ER and DNA damage for 48 h. Genomic DNA was purified and spotted onto a membrane and incubated with S9.6 antibody. (C) DRIP-qPCR signal upon MYC deregulation and UVSSA down-regulation. RPL13A and EGR1 are known R-loop rich loci, while MYADM- represents a negative control. Error bars represent SEM (n = 3). (D) Slot blot to examine S9.6 levels following MYC induction and UVSSA knockdown in various cell types. U2OS or 293T cells were transfected with siRNA against UVSSA and pcDNA-MYC-FLAG or empty vector for 48 h. Genomic DNA was isolated and subjected to slot blot analysis and staining with S9.6 antibody. (E) Immunofluorescence visualization of R-loops. U2OS cells were transfected with pcDNA-MYC-FLAG and subjected to immunofluorescence using S9.6 antibody after 48 h (top). The indicated number of cells was analyzed for S9.6 intensity in the nuclei (bottom). (F) Slot blot analysis of genomic DNA using S9.6 antibody. Purified genomic DNA harvested from 293T cells transfected with control siRNA or UVSSA siRNA (clones 9 and 10) and pcDNA3-MYC-FLAG or vector was harvested after 72 h. DNA was treated with RNase H1 or vehicle and spotted on a slot blot, and the membrane was incubated with S9.6 antibody (top). Subsequent to imaging, the membrane was stained with SYBR GOLD and visualized (bottom). IB, immunoblot.
MYC-driven changes in RNAPII occupancy and distribution are abrogated by UVSSA knockdown
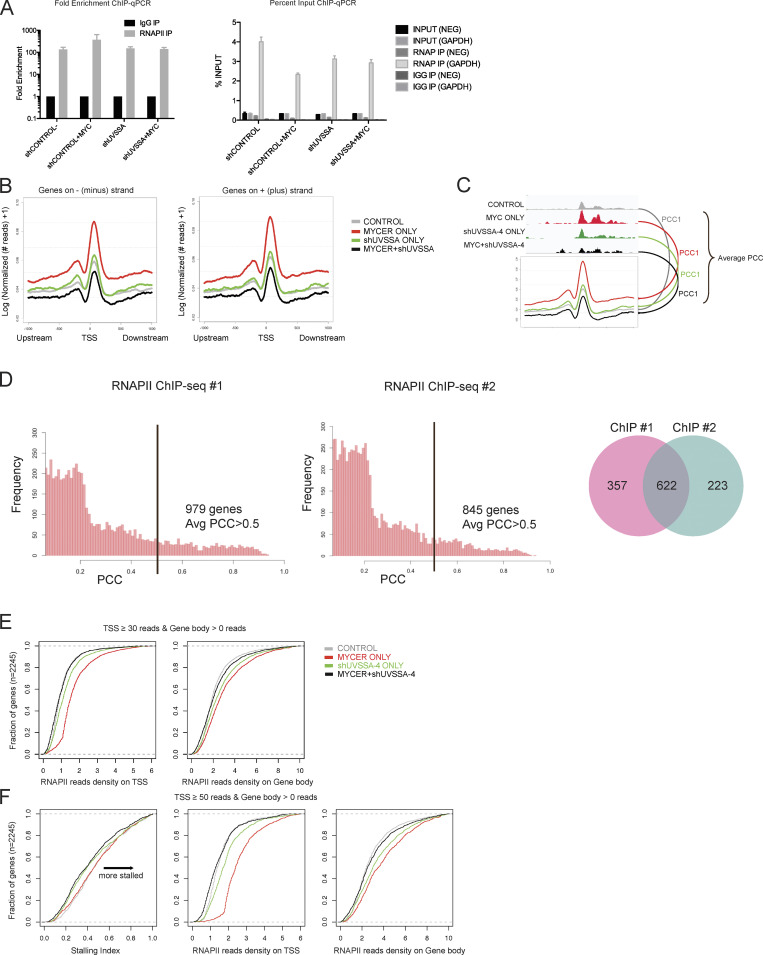
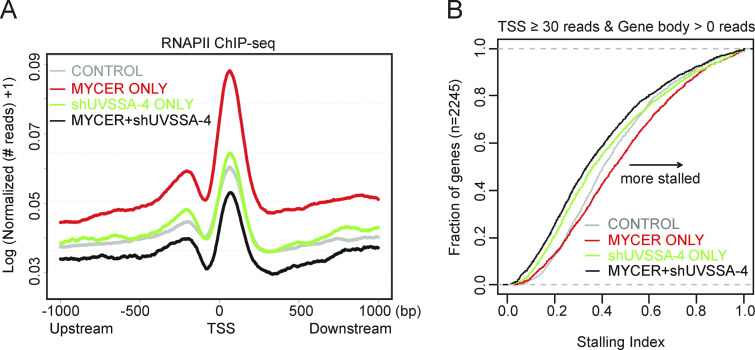
Given the role of UVSSA in RNAPII turnover during TCR, we hypothesized that UVSSA may be modulating RNAPII behavior during MYC-dependent transcription. We sought to compare genomic RNAPII occupancy in normal or MYC-deregulated conditions with or without UVSSA knockdown by performing genome-wide ChIP-seq using antibodies against RNAPII. ChIP signal from the N-20 antibody reflects all forms of RNAPII, regardless of the phosphorylation status. We first established that RNAPII signal was significantly enriched at the GAPDH promoter region but not in a promoter-negative intergenic region as confirmed by ChIP-qPCR (Fig. S4 A). Next, we examined RNAPII signal across all RNAPII-loaded transcription start sites (TSSs) genome-wide (Fig. 5 A). We observed a marked increase in RNAPII occupancy in the TSS when MYC-ER was activated, consistent with previous reports (Sabò et al., 2014; Fig. 5 A, red). UVSSA knockdown alone did not appear to significantly alter the RNAPII occupancy in TSS regions compared with control (Fig. 5 A, green). Notably, when MYC activation and UVSSA knockdown were combined, RNAPII signal in the TSS was drastically diminished to levels below control (Fig. 5 A, black). Genes on either DNA strand were equally affected (Fig. S4 B). Pearson correlation coefficient (PCC) analysis was used to identify individual gene loci that contributed most significantly to the differences (Fig. S4, C and D). Genes with a PCC score >0.5 overlapped significantly between duplicate ChIP-seq experiments, confirming our results (Fig. S4 D).
Figure S4.
RNAPII ChIP-seq quality control, correlation analysis of replicates, and calculation of SIs. (A) RNAPII ChIP-qPCR results shown by fold enrichment (left) or percentage input (right). GAPDH primers serve as a positive control for RNAPII enrichment, while NEG primers serve as a negative control. n = 4. Error bars represent SEM. (B) Normalized ChIP-seq footprints in a 2-kb window of genomic TSSs. Curves for genes on either the minus (left) or plus (right) strands were plotted, respectively. The y axis indicates logarithmic transformation of normalized read depths plus one per site. (C) Schematic for PCC analysis. For each gene, the PCC was calculated between the averaged ChIP-seq footprint curve and corresponding raw read depths around the TSS ±300 bp. PCCs were then averaged over all four conditions. (D) Distributions of averaged PCCs of genes from RNAPII ChIP-seq replicates 1 and 2. The black bar denotes a PCC of 0.5. Venn plot for genes associated with average PCC >0.5. (E) Empirical distribution of normalized read density on gene promoter (left) and gene body (right) regions. Genes with at least 30 reads on the promoter and at least 1 read in the gene body were selected for this analysis, and the cumulative SIs are denoted in Fig. 5 B. (F) Empirical distribution of RNAPII SIs (left) and normalized read density (center, right) for genes with at least 50 reads on the promoter and at least 1 read in the gene body. IP, immunoprecipitation.
Figure 5.
UVSSA knockdown alters MYC-driven RNAPII dynamics at TSSs and gene bodies. (A) Normalized PNAPII ChIP-seq footprints in a 2-kb window surrounding TSSs. The y axis indicates logarithmic transformation of normalized read depths plus one at each site. (B) Empirical distribution of RNAPII SIs. SIs were calculated as described in the text and plotted for all genes in which ⩾30 RNAPII reads were detected in the TSS window and ⩾1 reads were detected in the gene body (n = 2,245 genes).
Next, we assessed whether RNAPII distribution across genes is affected during MYC activation and UVSSA knockdown. We examined the RNAPII stalling index (SI) in each condition to compare the relative abundance of RNAPII peaks in promoter regions versus gene bodies. Of note, the SI reflects the ratio of distribution and not the absolute abundance of RNAPII peaks. Genes included in this analysis were those with at least 50 RNAPII reads in the TSS and all genes in the case of gene bodies. MYC activation alone increased the RNAPII SI compared with control, consistent with previous studies (Sabò et al., 2014; Fig. 5 B, red). UVSSA knockdown alone decreased the SI of over half of the genes examined compared with control, implying that fewer RNAPII peaks were distributed in TSS regions than in the corresponding gene bodies relative to control (Fig. 5 B, green). Remarkably, when MYC activation and UVSSA knockdown were combined, the SI was further decreased in almost the entire gene set, suggesting that the distribution of RNAPII peaks was shifted in favor of gene bodies in the majority of genes examined (Fig. 5 B, black; and Fig. S4, E and F). This result, in combination with the above data on TSS peaks, suggests that UVSSA affects RNAPII progression, turnover, or stability during normal transcription. This UVSSA function is critical for survival during MYC-induced transcription, giving UVSSA a novel role in RNAPII regulation in the absence of UV-induced DNA damage.
Discussion
Identifying genes or pathways specifically required for survival in MYC-deregulated cells, but dispensable for normal cells, has potential therapeutic value for many cancers. We focused on UVSSA as a MYC-SL/SS candidate, since two independent shUVSSA clones significantly scored as MYC-SL/SS in our genome-wide screen. Furthermore, UVSSA loss in humans is compatible with development and cell growth. UVSSA and CSA/ERCC8 were validated as MYC-SS genes using three shRNA clones possessing unique target sequences. Each shRNA decreased UVSSA or CSA/ERCC8 mRNA levels to various extents. Nevertheless, every shRNA clone that decreased UVSSA or CSA/ERCC8 mRNA levels impaired cell survival over time when combined with MYC-ER activation, establishing that the MYC-SS phenotype is specific to UVSSA or CSA/ERCC8 knockdown. Finally, KO of UVSSA by CRISPR exacerbated MYC-dependent increase in doubling time, confirming the specificity of the MYC-SS phenotype.
We show that the UVSSA complex has a novel role in supporting survival during MYC-induced transcription, likely involving alleviation of genomic stress. Specifically, this genetic interaction depends on active RNAPII transcription. It was unexpected that modulating DNA replication would not affect the MYC-SS phenotype, since MYC directly and indirectly stimulates DNA replication and induces DNA damage during S phase (Dominguez-Sola et al., 2007). We also expected elevated incidences of aphidicolin-sensitive transcription–replication conflicts in MYC-deregulated cells. However, aphidicolin was unable to dampen the SS phenotype, suggesting that loss of UVSSA affects a genomic process that is mostly independent of DNA replication during MYC activation.
Because α-amanitin was able to mitigate the SS phenotype, it is likely that transcription induced by MYC generates genomic stress. We failed to identify specific transcription-dependent toxic lesions that require UVSSA–USP7–ERCC8 for processing. We did not detect accumulation of R-loops, a potentially toxic intermediate of transcription (Fig. S5). Despite ATM/CHK2 activation, we did not detect evidence of DNA DSBs, at least compared with that from DSB-inducing agent NCS. This may reflect noncanonical ATM activation, which occurs in response to cellular or genomic stress without DSBs and warrants further investigation (Burgess and Misteli, 2015; Marteijn et al., 2017; Tresini et al., 2015). Besides R-loops, little is known about the contribution of transcription to DNA damage in mammalian cells (Callegari, 2016; D’Alessandro and d’Adda di Fagagna, 2016). In Escherichia coli and yeast, DNA damage can result from transcription-associated mutagenesis and recombination. However, it is not known whether analogous processes occur frequently in mammalian cells (Datta and Jinks-Robertson, 1995; Nickoloff, 1992). If they do, these structures may block RNAPII progression and signal for the recruitment of TCR complexes. Further studies are necessary to better understand the dynamics of DNA and RNA in actively transcribed regions under stress, especially with activated oncogenes.
Alternatively, it is possible that MYC-induced transcription does not generate specific DNA lesions. Deregulated or stressed RNAPII complexes may themselves act as obstacles or triggers that require the recruitment of specialized enzymes for removal or clearance. TCR is activated in response to stalled RNAPII at R-loops, as well as at 8-oxoguanine sites resulting from oxidative DNA damage, in addition to traditional UV adducts (Guo et al., 2013; Sollier et al., 2014). Given that UVSSA-USP7 participates in the regeneration of hypophosphorylated RNAPII complexes during TCR and that UVSSA interacts with stalled RNAPII, recruits TFIIH (van der Weegen et al., 2020), and promotes ubiquitination of RNAPII in TCR (Nakazawa et al., 2020), we hypothesize that this function may also be crucial under conditions of aberrant RNAPII regulation, such as during MYC overexpression, without a bulky adduct necessarily blocking the path. Whether endogenous UVSSA localizes to sites of active transcription remains to be tested.
Our ChIP-seq results suggest that UVSSA knockdown results in destabilization and/or eviction of RNAPII complexes from TSS regions of actively transcribing genes, especially when MYC is overexpressed. Whether the global landscape of RNAPII phosphorylation and/or progression rate is altered remains a question. Examination of the genome-wide distribution of differentially phosphorylated RNAPII complexes by ChIP-seq, as well as RNAPII progression using global run-on sequencing methods, is critical to provide further insight into the intricate changes in RNAPII dynamics that result from UVSSA knockdown in both MYC on and off backgrounds.
UV-SS and CS are both TCR-defective conditions; yet, they present with very different symptoms. These differences may arise from CSA and CSB having distinct roles outside of TCR, such as in transcription, oxidative DNA damage repair, and mitochondrial DNA damage repair (Groisman et al., 2006; Nicolai et al., 2015; Saijo, 2013; Vélez-Cruz and Egly, 2013). Although no such role outside of TCR has been reported for UVSSA, it has been established that the UVSSA–USP7–ERCC8 complex exists in a complex before UV damage and is found on chromatin before UV damage, and its abundance on chromatin does not increase after UV damage (Zhang et al., 2012). The interaction between the UVSSA–USP7–ERCC8 complex and CSB/ERCC6 is stimulated by UV damage; moreover, a recent study suggests that the interaction of UVSSA with RNAPII or TFIIH on chromatin is enhanced upon UV damage (van der Weegen et al., 2020). Interestingly, the UV-SS–causing UVSSA-C32R mutation only affects UVSSA’s interaction with TFIIH after UV damage and has no effect on the same interaction before UV damage (Nakazawa et al., 2012). Collectively, these results allow the possibility that the UVSSA–USP7–ERCC8 complex has roles outside of UV-induced damage repair, as we have uncovered in this study.
Our summarized results suggest that the UVSSA complex has a role in alleviating genomic stress during MYC-induced transcription (Fig. 6). Whether the UVSSA complex associates with RNAPII during normal transcription remains to be tested. However, our data show that UVSSA affects RNAPII dynamics in normal transcription to some degree. During MYC-dependent transcription, some silent genes are activated de novo; already active genes load additional RNAPII complexes due to MYC’s enhancer activity; and RNAPII pausing and release is stimulated. These changes in transcription trigger a requirement for the UVSSA complex. When UVSSA is down-regulated during MYC-ER induction, genomic stress is increased, likely as a result of impaired regulation of RNAPII stability or turnover, and results in loss of fitness over time.
Figure 6.
Summary and model of the role of UVSSA during oncogene-induced transcription stress. The UVSSA complex alleviates genomic stress during MYC-induced transcription. Whereas the role of UVSSA complex during normal RNAPII-associated transcription is not known, UVSSA is required for MYC-dependent transcription. UVSSA down-regulation during MYC-dependent transcription results in genomic stress, including aberrant RNAPII dynamics, transcription-dependent cell sickness, and activation of the ATM/CHK2 DNA damage response pathway.
Since many oncogenes regulate transcription, including MYC, KRAS, c-JUN, and nuclear factor-κB, it is tempting to speculate that transcription stress is not restricted to MYC and that, in turn, UVSSA could be a crucial factor in sustaining survival of cells that express a wide range of oncogenes that trigger transcription stress (Paccez and Zerbini, 2001). MYC expression is altered in many tumors; yet, it has been a particularly difficult protein to target directly with pharmacological agents, making synthetic lethality or sickness an attractive approach for MYC-dependent cancer therapy. Unlike many MYC-SL/SS candidates that have been identified in recent years, UVSSA is not an essential gene in normal cells. UV-SS patients with complete loss of function of UVSSA present with mild photosensitivity, but, importantly, they have no predisposition to cancer (Sarasin, 2012). Thus, targeting UVSSA could be a useful candidate for targeting MYC-dependent cancer cells.
Materials and methods
Cell culture
MCF10A cells were purchased from the American Type Culture Collection (ATCC; CRL-10317). MYC-ER was introduced into MCF10A cells retrovirally using pBabe-hygro-MYC-ER, and transduced clones were selected by the addition of 100 μg/ml Hygromycin B (Roche Holding AG). Clones were isolated by serial dilution. MCF10A cells were cultured in DMEM/F12 (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml EGF (PeproTech), 0.5 μg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml cholera toxin (Sigma-Aldrich), and 10 μg/ml insulin (Sigma-Aldrich), and antibiotics.U2OS (ATCC HTB-96) cells and 293T (ATCC CRL-3216) cells were cultured in DMEM (Invitrogen) with 10% FBS (Invitrogen) and antibiotics at 37°C and 5% CO2. All cell lines were originally purchased from ATCC. Mycoplasma contamination tests by PCR were administered periodically during culturing and showed negative results.
Production of lentivirus and shRNA
Glycerol stocks of lentiviral shRNA constructs (pGIPZ, pTRIPZ) were obtained from Thermo Scientific Open Biosystems. Clones used were UVSSA-1 (V3THS_398946), UVSSA-3 (V3THS_398945), UVSSA-4 (V2THS_139628), ERCC8-3 (V3LHS_332967), ERCC8-4 (V3LHS_332966), ERCC8-6 (V3LHS_404566), and XPF-2 (V3THS_356949). An empty vector was used as a negative control. Plasmids were isolated using the E.Z.N.A. Plasmid Miniprep kit (Omega Bio-Tek, Inc.) and packaged into lentivirus by transfecting (jetPEI; Polyplus-transfection S.A.) into 293T cells with pMD.G and pCMVR8.91. Viruses were collected in MCF10A media, filtered, and infected into MCF10A in the presence of 8 μg/ml polybrene (Sigma-Aldrich) and spin infected for 1 h at 1,000 rpm at RT. After 24 h of incubation, infected cells were selected by the addition of 2 μg/ml puromycin (Sigma-Aldrich).
shRNA screen
The pooled shRNA screen was performed using the Thermo Scientific Open Biosystems GIPZ Lentiviral Human shRNA-mir Library as previously described (Rodriguez-Barrueco et al., 2013). The library was packaged into lentivirus as described above, and MCF10A-MYC-ER cells were infected at 30% efficiency. Infected cells were selected with 2 μg/ml puromycin for two passages and divided into 10 independent populations. Five populations were treated with 200 nM 4OHT (every 48 h), and five were treated with vehicle. After 30 d, genomic DNA of surviving cells was isolated by lysing cells with DNA lysis buffer (1% SDS, 100 mM EDTA, pH 8.0, 50 mM Tris-HCl, pH 8, 100 nM NaCl), treating them with RNase A (Qiagen N.V.), and purifying DNA with phenol chloroform extraction and isopropanol precipitation. Genomic DNA isolated from surviving populations was subjected to PCR to recover the shRNA, using primers with barcoded adapter sequences compatible with the Illumina sequencing platform. The ∼450-bp PCR products were gel extracted and purified, pooled, then processed using the Illumina HiSeq 2000 system. Data analysis was then performed as previously described (Yu et al., 2013) to generate Log2FC values associated with each shRNA in the library between MYC ON and MYC OFF conditions. Briefly, HiSeq data were processed into FASTQ files and decoded to identify the shRNA represented by each read, and the number of reads for each shRNA was calculated. The data were then normalized to calculate shRNA representation in each sample. Quality assurance was performed at this step to identify outliers. Then, the differential representation of each shRNA was calculated by identifying enrichment or depletion of individual shRNAs between the start and end of the experiment. This enrichment or depletion was expressed as a Log2FC value.
Gene network community analysis
STRING interactions were adopted as the background biological network (https://string-db.org). Only the top 10% of highly confident interactions were adopted. Log2FC values from the raw shRNA screen data were used to weigh the nodes of the above network. Nodes with Log2FC scores >0 were removed. Nodes not covered in the raw shRNA screen data were also removed. If one gene was associated with multiple shRNAs in the screen library, the lowest Log2FC score was adopted. The R package igraph was used to cluster the node-weighted network. Specifically, we used the function cluster_infomap to detect local network communities with consideration of node weights (Rosvall and Bergstrom, 2008). After the clustering, each protein (node) was assigned to one community. Log2FC scores from the shRNA screen were converted to MYC-SL scores for this analysis by multiplying by a factor of −1. These converted MYC-SL scores were averaged for community members to give average MYC-SL values shown in Table S1.
FBCS assays
250,000 MCF10A-MYC-ER cells expressing stably integrated shRNA and 250,000 MCF10A-MYC-ER cells without shRNA were plated together. At the first passage, fluorescent cells were counted by live-cell flow cytometry and replated with either vehicle or 4OHT. At each subsequent passage, the remaining percentage of fluorescent cells was analyzed using flow cytometry. All cell sorting was performed with the BD FACSCalibur cell sorter (BD Biosciences). For experiments using α-amanitin (A2263; Sigma-Aldrich), aphidicolin (A0781; Sigma-Aldrich), or JQ1 (SML0974; Sigma-Aldrich), cells were supplemented every 72 h with the appropriate drug simultaneously with vehicle (100% ethanol) or 200 nM 4OHT (H7904; Sigma-Aldrich), and 1 μg/ml doxycycline (D9891; Sigma-Aldrich) or vehicle (PBS) for inducible shRNA lines, during the duration of the experiments.
mRNA isolation and quantitative RT-PCR (qRT-PCR)
RNA was isolated using the Nucleospin RNA II kit (Clontech Laboratories, Inc.). 1 µg of RNA was subjected to reverse transcription using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (Life Technologies). cDNA was used as a template for quantitative RT-PCR using ABsolute Blue QPCR SYBR Green Low ROX Mix (Thermo Fisher Scientific) and the Applied Biosystems 7500 Fast PCR system (Life Technologies).
MTS assay
MCF10A-MYC-ER cells were plated in 96-well plates at a density of 3,000 cells per well with vehicle or 200 nM 4OHT. The next day, media in wells were replaced with fresh media including the appropriate concentration of DMSO, P5091, or KU55933 (S1092; Selleck Chemical), along with vehicle or 200 nM 4OHT. Each condition was assayed in triplicate in each experiment. 48 h later, each well was replaced with fresh media for 1 h, followed by addition of MTS reagent included in the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (G5421, Promega). After 2 h of incubation, plates were analyzed using the uQuant plate reader (BioTek Instruments).
Cell lysis and cellular fractionation
Cells were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0), supplemented with protease and phosphatase inhibitors (Sigma-Aldrich), for 15 min on ice. Samples were sonicated (two times for 5 min each) and centrifuged for 10 min at 4°C to produce whole cell lysates. Cellular fractionation was performed as previously described (Méndez and Stillman, 2000). Briefly, cells were resuspended in buffer A (10 mM Hepes [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 5 µg of aprotinin per milliliter, 5 µg of leupeptin per milliliter, 0.5 µg of pepstatin A per milliliter, 0.1 mM PMSF) at a density of 4 × 107 cells per milliliter. Triton X-100 was added to a 0.1% concentration, and cells were incubated on ice for 5 min. Nuclei were collected by low-speed centrifugation (4 min at 1,300 × g; 4°C) to form pellet P1. The supernatant (S1) was then centrifuged at high speed (15 min at 20,000 × g; 4°C) to clarify. Nuclei in P1 were washed in buffer A and then lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and protease inhibitors as described above). The insoluble chromatin was then collected by centrifugation (4 min at 1,700 × g; 4°C), washed in buffer B, and centrifuged again. The final chromatin pellet (P3) was resuspended in Laemmli buffer and sonicated for 15 s in aTekmar CV26 sonicator using a microtip at 25% amplitude.
iSTOP CRISPR editing
Guides for the iSTOP protocol were designed from the online iSTOP database (Billon et al., 2017). Three different guides were selected and individually cloned into the B52 single-guide RNA expression plasmid (Addgene; 100708). The B525 plasmid (a generous gift from the Ciccia laboratory, Department of Genetics & Development, Columbia University Medical Center, New York, NY) was transfected into cells to transiently express the cas9-APOBEC1 fusion protein BE3, as well as a GFP fluorescent marker and a blasticidin resistance marker.
MCF10A cells, either wild type or expressing the MYC-ER fusion protein, were cotransfected with the single-guide RNA expressing B52 and the B525 plasmid using the Neon Transfection System (1,250 V; 20 ms; two pulses; 2.5 × 105 cells per transfection). Transfection efficiency was assessed by the GFP fluorescent signal encoded by the B525 plasmid. Blasticidin was added to the media after 48 h to select for transformed cells. Following 96-h selection, cells were expanded to large uncloned culture and prepared for cloning.
Cell cloning
Cells from each uncloned population were diluted to a density of 10 cells/ml and then plated onto a 96-well plate with 100 μl per well. Wells were monitored for growth and for doublets and expanded up to 24-well plates.
Doubling-time assay
MCF10A cells were seeded at low density (4,500 cells/well in a six-well dish) to ensure logarithmic growth through 6 d. Cells were cultured with either 400 nM 4OHT or ethanol (control), added immediately upon seeding. For each condition, six wells were seeded. Cells were collected on day 3 and day 6 after induction in triplicates. Cells were counted using the Countess II automated cell counter with Trypan blue dye to obtain a viable cells per milliliter count. Doubling-time calculation was used to compare the results of multiple biological replicates. For each condition, the doubling time was calculated using the following equation:
where t is the time after seeding (in hours) at the time of collection and C is the average of the cell counts from each replicate at that time point. This yields the time in hours that it takes for each cell line in each condition to complete one doubling. These values were used to statistically compare the biological replicates.
Western blotting and antibodies
Cell lysates were prepared as described, added to 5× Laemmli buffer, and heated at 95°C for 5 min. Samples were run on Tris-glycine or NuPAGE Tris-acetate gels (Invitrogen) and transferred to polyvinylidene difluoride membranes at 35 V for 2 h. Membranes were blocked in PBS–0.5% Tween–5% milk for 1 h and stained with primary antibodies (1:250–1:2,000) for 1 h or overnight, followed by staining with secondary antibodies (1:10,000) conjugated with HRP (Jackson ImmunoResearch) for 1 h. After incubation with ECL substrate (Pierce/Thermo Fisher Scientific), blots were exposed to film. ImageQuant 5.2 (Molecular Dynamics) was used for quantification. The following primary antibodies were used for immunostaining: mouse anti–β-actin (A2228; Sigma-Aldrich), mouse anti-MYC (9E10, sc-40; Santa Cruz Biotechnology), rabbit anti–histone H3 (9715; Cell Signaling Technology), rabbit anti–histone H3K4me1 (ab8895; Abcam), rabbit anti–histone H3K27Ac (ab4729; Abcam), anti–phospho-Chk2 (2661, rabbit; Cell Signaling Technology), rabbit anti–phospho-RPA32 (S4/S8, A300-245A; Bethyl Laboratories), rabbit anti–phospho-CHK1 (2341S; Cell Signaling Technology), rabbit anti-UVSSA (GTX106751; GeneTex), rabbit anti-UVSSA (NBP1-32598; Novus Biologicals), and mouse S9.6 (ENH001; KeraFAST). HRP-conjugated secondary goat antirabbit and goat antimouse antibodies were obtained from Jackson ImmunoResearch.
Clonogenicity assays
MCF10A-MYC-ER cells, parental or stably expressing shCONTROL or shUVSSA-4, were plated at a density of 500 cells per 100-mm plate either with vehicle or with 200 nM 4OHT. For UV sensitivity assays, the next day, media were replaced with 2 ml of PBS, and plates were irradiated with indicated doses of UV-C (254 nm) using the Stratalinker UV Cross-linker 2400 (Stratagene). For P5091 sensitivity assays, plates were treated with appropriate doses of P5091. After irradiation or drug treatment, plates were again replaced with media containing either vehicle or 4OHT and incubated for 10 d at 37°C and 5% CO2. At the time of harvest, cells were washed twice with PBS, fixed with 100% methanol, and stained with Crystal Violet (C3886; Sigma-Aldrich) solution, and visible colonies were counted either visually or by using ImageJ software (National Institutes of Health).
Cell cycle analysis
At the time of harvest, MCF10A-MYC-ER cells were vortexed in 0.5 ml of PBS, and 5 ml of 70% ethanol were added dropwise for fixing at −20°C overnight. The next day, cells were incubated in 1 ml of PBS for 1 h on ice and centrifuged. To the resulting pellet, RNase A was added at 1:400, and propidium iodide stock (10 mg/ml; Sigma-Aldrich) was added at 1:1,000 for flow cytometry. All analysis was performed with the BD FACSCalibur platform (BD Biosciences).
ChIP
The ChIP protocol was performed on the basis of what is described by Zhang et al., (2017). MCF10A-MYC-ER cells expressing shCONTROL or shUVSSA-4 were treated with or without 200 nM 4OHT and 1 µg/ml doxycycline for 4 wk before harvest. ChIP was performed on two independent biological replicates. Each pellet containing 50 million cells was fixed with 1% formaldehyde for 10 min at RT, following quenching with 0.125 M glycine. Pellets were washed and lysed using lysis buffer (50 mM Hepes-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8.0, 10% glycerol, 0.25% Triton X-100, 0.5% NP-40, protease inhibitors), and resuspended in shearing buffer (50 mM Tris-HCl, pH 8.1, 0.1% SDS, 10 mM EDTA, pH 8.0, protease inhibitors) before sonication using the S220 Ultrasonicator (Covaris) to chromatin fragments ranging from 200 to 500 nt in size. Fragments were then incubated overnight with 4 µg of anti-Pol II (N-20, sc-899; Santa Cruz Biotechnology) or purified rabbit IgG (011-000-002; Jackson ImmunoResearch Inc.) at 4°C. Conjugation to protein A magnetic beads, subsequent immunoprecipitation, washes, reverse cross-linking, and treatment with RNase A and proteinase K were performed as described by Zhang et al. (Zhang et al., 2017). ChIP DNA was purified using the MinElute Reaction Clean Up Kit (28204; Qiagen) and quantified using Quant-iT PicoGreen dsDNA Reagent (P7581; Life Technologies). Purified ChIP DNA was directly used as template DNA for standard SYBR Green qPCR. ChIP-qPCR was analyzed using the ChIP analysis protocol provided by Life Technologies.
ChIP-seq library preparation and Illumina sequencing
ChIP-seq libraries were prepared on the basis of the method of Zhang et al. (2017). 20 ng of ChIP or input DNA were processed for end repair, adapter ligation, and gel purification using the Illumina TruSeq ChIP Library Preparation Kit (IP-202-1012; Illumina), then quantified with the KAPA SYBR FAST Universal qPCR Kit (KAPA Biosystems). Fragment sizes were analyzed using the BioAnalyzer device (Agilent), and samples were pooled together to a final concentration of 20 nM. The samples were sequenced using the Illumina HiSeq 2000 system as single-end 100-bp reads, obtaining 29 million–42 million reads per sample.
ChIP-seq analysis
Single-end FASTQ files were aligned to the human genome assembly (hg19) using Bowtie 2 (version 2.1.0; Langmead and Salzberg, 2012). Before further analysis, the initially aligned BAM files were subjected to preprocessing that sorted and indexed using SAMtools (Li et al., 2009). To assess RNAPII binding intensity around TSSs, we used the function “pileup” from the R package “Rsamtools” to calculate ChIP-seq read depth at each nucleotide in a window of TSS plus 1 kb upstream and downstream for all human genes. Next, read counts were normalized according to the library size of the sample, followed by log2(x+1) transformation. Finally, we took the mean values for each site within the window across all genes to generate the read distribution under different conditions. The ChIP-seq data have been deposited in the Gene Expression Omnibus database under accession no. GSE121960.
SIs
SIs were calculated as the numerical ratio between the total number of reads located in two regions for all expressed genes, where “promoter” regions were defined as 300 bp upstream and downstream of the TSS and “gene body” regions were defined as 301–3,000 bp downstream of the TSS, as previously described by Sabò et al. (2014). The function “featureCounts” from the package “Subread” was adopted to count the reads (Liao et al., 2013). We then compared the empirical distributions of SIs of all expressed genes under different conditions.
siRNA
siRNA transfections were performed using jetPEI transfection reagent (101-01N; Polyplus-transfection S.A.) or Lipofectamine 2000 reagent (11668027; Thermo Fisher Scientific) with their standard protocols. Cells were seeded in six-well plates and 24 h later were transfected with 1 µg of pcDNA3 or pcDNA3-MYC-FLAG where appropriate and with 40 pmol of appropriate siRNA where denoted. siRNAs used were obtained from the Dharmacon siGENOME line (GE Healthcare). Transfected cells were then harvested at denoted times and further processed. siRNAs used in this study included: UVSSA-9 (D-024139-19), UVSSA-10 (D-024139-20), and control (D-001206-13-05).
Slot blots
After the indicated transfections or treatments, genomic DNA was isolated from cells using the QiaAMP DNA Mini Kit (51304; Qiagen). 0.5–1 µg of purified genomic DNA was then spotted onto an Amersham Hybond-N+ nylon membrane (RPN303B; GE Healthcare) using a slot blot manifold and vacuum. For RNase H treatment, genomic DNA was incubated with RNase H (M0297S; New England Biolabs, Inc.) for 20 min at 37°C before slot blot analysis. Membrane was then blocked with 5% milk–PBS–0.2% Tween for 1 h and incubated with S9.6 antibody (ENH001; KeraFAST) at a 1:1,000 dilution in 3% BSA in PBS–0.2% Tween for 1 h at RT or overnight at 4°C. Membrane was washed three times for 10 min each in PBS–0.2% Tween, then incubated with 1:10,000 goat anti mouse HRP in 5% milk–PBS–0.2% Tween for 1 h at RT. Membrane was again washed three times for 10 min each in PBS–0.2% Tween, then incubated with ECL substrate (32106; Pierce/Thermo Fisher Scientific) and exposed to film. Subsequently, membrane was stained with 1:10,000 SYBR GOLD for 10 min and imaged with the LI-COR Odyssey Fc system (LI-COR Biotechnology).
DRIP-qPCR
DRIP-qPCR was performed according to the method of Sanz et al. (2016). Briefly, MCF10A-MYC-ER cells expressing shCONTROL or shUVSSA-4 were treated with or without 200 nM 4OHT and 1 µg/ml doxycycline for 4 wk before harvest. Cells were pelleted and treated with SDS and proteinase K overnight. The next day, after phenol–chloroform extraction and EtOH precipitation, DNA was spooled carefully to preserve R-loops. Extracted DNA was digested with a cocktail of five restriction enzymes overnight at 37°C, then cleaned up with phenol–chloroform extraction followed by EtOH precipitation. For each DRIP, 4.4 µg of DNA was aliquoted with or without pretreatment by RNase H overnight at 37°C. Some DNA was saved as input, and the rest was incubated with binding buffer and S9.6 antibody for 16 h at 4°C with inversion. Then, 50 µl of protein A/G sepharose beads (20421; Pierce) were washed and added to each DRIP for 2 h at 4°C with inversion. After washing, beads were eluted with proteinase K at 55°C for 45 min with inversion. Finally, the eluate was subjected to phenol–chloroform extraction followed by EtOH precipitation. DRIP DNA was directly used as template DNA for standard SYBR Green qPCR. Data were analyzed using the percentage input method.
Immunofluorescence microscopy
10,000 cells per chamber were seeded onto eight-chamber slides with 0.5 ml of appropriate cell culture media. The next day, appropriate samples were treated with camptothecin (0.5–2 µM final) for 2 h. Cells were then washed and fixed with 4% PFA for 10 min, followed by two washes. Chamber walls were them removed, and three final washes were conducted using 0.01% Triton X-100 in PBS. The whole slide was then blocked with 5% BSA in PBS–Triton X-100 for 1 h at RT. Following three more washes, the slide was incubated with S9.6 antibody (1:1,000 in 5% BSA–PBS–Triton X-100) overnight at 4°C. The next day, following washes, the slide was incubated with Alexa Fluor 488 goat antimouse secondary antibody (ab150113; Abcam) and DAPI (both 1:1,000 in 5% BSA–PBS–Triton X-100) for 1 h in the dark. The slide was kept in the dark through three more washes and dried by aspiration, and a coverslip was applied with a few drops of VECTASHIELD mounting medium (H-1000; Vector Laboratories) and sealed with clear nail polish. Slides were then imaged using the Nikon Eclipse 80i fluorescence microscope (Nikon Instruments) equipped with the CoolSNAP HQ2 camera (Photometrics) at 60× magnification with a plan apochromatic 60×/1.40 NA oil objective. Images were subsequently analyzed using ImageJ software.
Statistics
All statistical values represented in figures were generated through appropriate t tests (paired, unpaired, one-sample, two-sample, or Student’s) using Prism 5 software (GraphPad Software). Confidence intervals were set at 95% (statistical significance, P < 0.05). Sample numbers and error bars shown in each graph are denoted in the figure legends.
Primer sequences
qRT-PCR primers: GAPDH forward, 5′-CATCTTCTTTTGCGTCGC-3′; GAPDH reverse, 5′-AAAAGCAGCCCTGGTGAC-3′; UVSSA forward, 5′-CTTGCCTTGGGCTACCACTT-3′; UVSSA reverse, 5′-TGCTTCTCCTCTTCTCTCTTCCTT-3′; ERCC8 forward, 5′-GATAATCGAATGAGGCTCTG-3′; ERCC8 reverse, 5′-CCACAGGAGACAGTGAATTT-3′; XPF forward, 5′-CTACAGCCAGTGCATCTCCA-3′. XPF reverse 5′-CACCTCGGGAAGTGAGAGAG-3′. ChIP-qPCR primers: GAPDH promoter forward, 5′-TACTAGCGGTTTTACGGGCG-3′; GAPDH promoter reverse, 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′; intergenic forward, 5′-ATGGTTGCCACTGGGGATCT-3′; intergenic reverse, 5′-TGCCAAAGCCTAGGGGAAGA-3′. DRIP-qPCR primers: RPL13A forward, 5′-AGGTGCCTTGCTCACAGAGT-3′; RPL13A reverse, 5′-GGTTGCATTGCCCTCATTAC-3′; EGR1 forward, 5′-GCCAAGTCCTCCCTCTCTACTG-3′; EGR1 reverse, 5′-GGAAGTGGGCAGAAAGGATTG-3′; MYADM- forward, 5′-TGCATCTACATCCGCAAAAG-3′; MYADM- reverse, 5′-AGAGTGGACGCTGCAGAAAT-3′. iSTOP guide RNAs: sg 1, 5′-GGATCAGAAACTTTCGAAGT-3′; sg 2, 5′-TGGATCCACGAGCACACAGC-3′; sg 3, 5′-ATCCAGGTGAGCCTCGAACC-3′. shRNA targeting sequences: UVSSA-1, 5′-TTCTCAGGATTTAGTCGGG-3′; UVSSA-3, 5′-TGAGCTTGAGTGTGTCGCG-3′; UVSSA-4, 5′-TCAGTGCGTAGTTAAACTG-3′; ERCC8-3, 5′-ATTTCTTGTCTGTGACCCT-3′; ERCC8-4, 5′-TTGACTGAAATACACAGCA-3′; ERCC8-6, 5′-ATAGTTCATAAAATCTGCT-3′; XPF-2, 5′-ACAACTTCAGGTTTGTGCT-3′.
Online supplemental material
Fig. S1 shows the entire MYC-SL network community (community 9), including UVSSA and ERCC8. Fig. S2 shows shRNA knockdown levels of UVSSA and ERCC8, representative single FBCS assays for shUVSSA and shERCC8, and clonogenicity assay results using P5091. Fig. S3 shows UV sensitivity of MYC on and/or shUVSSA cells, knockdown and FBCS assay results for XPF, Western blots for CHK2 activation, clonogenicity assay results using KU55933, and representative raw results of cell cycle analyses. Fig. S4 shows quality control data from ChIP, TSS data from the perspective of either DNA strand, a schematic of the PCC analysis between replicate ChIP-seq experiments, results from PCC analysis, and supplemental results of the SI analyses. Fig. S5 shows a control for S9.6 antibody’s recognition of RNA/DNA hybrids, results from MCF10A cells, results from U2OS and 293T cells, immunofluorescence results, and signal sensitivity to RNase H1. Table S1 lists the 18 top MYC-SL network communities. Table S2 lists the 1,811 top-ranked MYC-SL shRNA clones (Log2FC less than −2 and P < 0.1) from Illumina HiSeq analysis of the MCF10A-MYC-ER SL screen.
Supplementary Material
lists the 18 top MYC-SL gene network communities.
lists the 1,811 top-ranked MYC-SL shRNA clones (Log2FC less than −2 and P < 0.1) from Illumina HiSeq analysis of MCF10A-MYC-ER SL screen.
Acknowledgments
We thank Drs. David Dominguez-Sola, Carol Ying, Bin Zheng, and Jose Silva for experimental guidance and support. We also thank Dr. Wei Gu (Institute for Cancer Genetics, Columbia Univerity Medical Center, New York, NY) for providing us with P5091. We thank Dr. Frederic Chedin for the DRIP protocol. We thank all the mentioned groups for additional assistance and support. We acknowledge support from the Genomics and High Throughput Screening Shared Resource of the HICCC.
This work was supported in part by National Cancer Institute grants CA092245 and CA167826 to J. Gautier.
The authors declare no competing financial interests.
Author contributions: M. Sato performed most of the experiments. R.C. Liebau executed CRISPR KO and the doubling time assay and helped to revise the manuscript. Z. Liu performed the bioinformatics analysis. L. Liu contributed to the molecular biology work on the RNA/DNA hybrids. R. Rabadan provided critical advice on interpreting the bioinformatic data. M. Sato, R.C. Liebau, and J. Gautier designed the study, analyzed data, and wrote the paper. All authors gave final approval for publication.
References
- Adelman, K., and Lis J.T.. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13:720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluapuri, A., Wolf E., and Eilers M.. 2020. Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 21:255–267. 10.1038/s41580-020-0215-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon, P., Bryant E.E., Joseph S.A., Nambiar T.S., Hayward S.B., Rothstein R., and Ciccia A.. 2017. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell. 67:1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski, S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., and Carrico R.J.. 1986. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 89:123–130. 10.1016/0022-1759(86)90040-2 [DOI] [PubMed] [Google Scholar]
- Burgess, R.C., and Misteli T.. 2015. Not all DDRs are created equal: non-canonical DNA damage responses. Cell. 162:944–947. 10.1016/j.cell.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari, A.J. 2016. Does transcription-associated DNA damage limit lifespan? DNA Repair (Amst.). 41:1–7. 10.1016/j.dnarep.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Campbell, K.J., and White R.J.. 2014. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb. Perspect. Med. 4:a018408. 10.1101/cshperspect.a018408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli, S., Jang I.S., Bernard B., and Grandori C.. 2014. Synthetic lethal screens as a means to understand and treat MYC-driven cancers. Cold Spring Harb. Perspect. Med. 4:a014209. 10.1101/cshperspect.a014209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compe, E., and Egly J.-M.. 2012. TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13:343–354. 10.1038/nrm3350 [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell, M., McFerrin L., and Eisenman R.N.. 2014. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4:a014357. 10.1101/cshperspect.a014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro, G., and d’Adda di Fagagna F.. 2016. Transcription and DNA damage: holding hands or crossing swords? J. Mol. Biol. 10.1016/j.jmb.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Dang, C.V. 2012. MYC on the path to cancer. Cell. 149:22–35. 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A., and Jinks-Robertson S.. 1995. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 268:1616–1619. 10.1126/science.7777859 [DOI] [PubMed] [Google Scholar]
- de Pretis, S., Kress T.R., Morelli M.J., Sabò A., Locarno C., Verrecchia A., Doni M., Campaner S., Amati B., and Pelizzola M.. 2017. Integrative analysis of RNA polymerase II and transcriptional dynamics upon MYC activation. Genome Res. 27:1658–1664. 10.1101/gr.226035.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore, J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 146:904–917. 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola, D., Ying C.Y., Grandori C., Ruggiero L., Chen B., Li M., Galloway D.A., Gu W., Gautier J., and Dalla-Favera R.. 2007. Non-transcriptional control of DNA replication by c-Myc. Nature. 448:445–451. 10.1038/nature05953 [DOI] [PubMed] [Google Scholar]
- Egly, J.-M., and Coin F.. 2011. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst.). 10:714–721. 10.1016/j.dnarep.2011.04.021 [DOI] [PubMed] [Google Scholar]
- Fei, J., and Chen J.. 2012. KIAA1530 protein is recruited by Cockayne syndrome complementation group protein A (CSA) to participate in transcription-coupled repair (TCR). J. Biol. Chem. 287:35118–35126. 10.1074/jbc.M112.398131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, P.C., Frank S.R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., and Amati B.. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115–1129. 10.1101/gad.1067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay, M., Li Y., and Felsher D.W.. 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 4:a014241. 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates, L.A., Foulds C.E., and O’Malley B.W.. 2017. Histone marks in the “driver’s seat”: functional roles in steering the transcription cycle. Trends Biochem. Sci. 42:977–989. 10.1016/j.tibs.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman, R., Kuraoka I., Chevallier O., Gaye N., Magnaldo T., Tanaka K., Kisselev A.F., Harel-Bellan A., and Nakatani Y.. 2006. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 20:1429–1434. 10.1101/gad.378206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J., Hanawalt P.C., and Spivak G.. 2013. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 41:7700–7712. 10.1093/nar/gkt524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt, P.C., and Spivak G.. 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9:958–970. 10.1038/nrm2549 [DOI] [PubMed] [Google Scholar]
- Herold, S., Wanzel M., Beuger V., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H.-P., Haenel F., and Eilers M.. 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 10:509–521. 10.1016/S1097-2765(02)00633-0 [DOI] [PubMed] [Google Scholar]
- Higa, M., Zhang X., Tanaka K., and Saijo M.. 2016. Stabilization of ultraviolet (UV)-stimulated scaffold protein A by interaction with ubiquitin-specific peptidase 7 is essential for transcription-coupled nucleotide excision repair. J. Biol. Chem. 291:13771–13779. 10.1074/jbc.M116.724658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, T.Y., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S., Echeverria G.V., Sun T., Kurley S.J., Tyagi S., et al. 2015. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 525:384–388. 10.1038/nature14985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., Ono T., and Yamaizumi M.. 1994. A new UV-sensitive syndrome not belonging to any complementation groups of xeroderma pigmentosum or Cockayne syndrome: siblings showing biochemical characteristics of Cockayne syndrome without typical clinical manifestations. Mutat. Res. 314:233–248. 10.1016/0921-8777(94)90068-X [DOI] [PubMed] [Google Scholar]
- Itoh, T., Fujiwara Y., Ono T., and Yamaizumi M.. 1995. UVs syndrome, a new general category of photosensitive disorder with defective DNA repair, is distinct from xeroderma pigmentosum variant and rodent complementation group I. Am. J. Hum. Genet. 56:1267–1276. [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., Yamaizumi M., Ichihashi M., Hiro-Oka M., Matsui T., Matsuno M., and Ono T.. 1996. Clinical characteristics of three patients with UVs syndrome, a photosensitive disorder with defective DNA repair. Br. J. Dermatol. 134:1147–1150. 10.1046/j.1365-2133.1996.d01-922.x [DOI] [PubMed] [Google Scholar]
- Jonkers, I., and Lis J.T.. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16:167–177. 10.1038/nrm3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsantis, P., Silva L.M., Irmscher S., Jones R.M., Folkes L., Gromak N., and Petermann E.. 2016. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 7:13087. 10.1038/ncomms13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed]
- Liao, Y., Smyth G.K., and Shi W.. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.J., Nasr Z., Premsrirut P.K., Porco J.A. Jr., Hippo Y., Lowe S.W., and Pelletier J.. 2012a. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 1:325–333. 10.1016/j.celrep.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.Y., Loven J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., and Young R.A.. 2012b. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 151:56–67. 10.1016/j.cell.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn, J.A., Vermeulen W., and Tresini M.. 2017. Noncanonical ATM activation and signaling in response to transcription-blocking DNA damage. Methods Mol. Biol. 1599:347–361. 10.1007/978-1-4939-6955-5_25 [DOI] [PubMed] [Google Scholar]
- McKeown, M.R., and Bradner J.E.. 2014. Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 4:a014266. 10.1101/cshperspect.a014266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon, I., Spivak G., and Hanawalt P.C.. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 51:241–249. 10.1016/0092-8674(87)90151-6 [DOI] [PubMed] [Google Scholar]
- Méndez, J., and Stillman B.. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612. 10.1128/MCB.20.22.8602-8612.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar, J.J., Ebus M.E., Geerts D., Koster J., Lamers F., Valentijn L.J., Westerhout E.M., Versteeg R., and Caron H.N.. 2009. Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc. Natl. Acad. Sci. USA. 106:12968–12973. 10.1073/pnas.0901418106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, R., Toyoshima M., Robinson K., Gurley K.E., Howie H.L., Davison J., Morgan M., Kemp C.J., and Grandori C.. 2012. MYC-driven tumorigenesis is inhibited by WRN syndrome gene deficiency. Mol. Cancer Res. 10:535–545. 10.1158/1541-7786.MCR-11-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, Y., Sasaki K., Mitsutake N., Matsuse M., Shimada M., Nardo T., Takahashi Y., Ohyama K., Ito K., Mishima H., et al. 2012. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 44:586–592. 10.1038/ng.2229 [DOI] [PubMed] [Google Scholar]
- Nakazawa, Y., Hara Y., Oka Y., Komine O., van den Heuvel D., Guo C., Daigaku Y., Isono M., He Y., Shimada M., et al. 2020. Ubiquitination of DNA damage-stalled RNAPII promotes transcription-coupled repair. Cell. 180:1228–1244.e24. 10.1016/j.cell.2020.02.010 [DOI] [PubMed] [Google Scholar]
- Nesbit, C.E., Tersak J.M., and Prochownik E.V.. 1999. MYC oncogenes and human neoplastic disease. Oncogene. 18:3004–3016. 10.1038/sj.onc.1202746 [DOI] [PubMed] [Google Scholar]
- Nickoloff, J.A. 1992. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 12:5311–5318. 10.1128/MCB.12.12.5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai, S., Filippi S., Caputo M., Cipak L., Gregan J., Ammerer G., Frontini M., Willems D., Prantera G., Balajee A.S., et al. 2015. Identification of novel proteins co-purifying with Cockayne syndrome group B (CSB) reveals potential roles for CSB in RNA metabolism and chromatin dynamics. PLoS One. 10:e0128558. 10.1371/journal.pone.0128558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R., et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 151:68–79. 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi, T., Nakazawa Y., Sasaki K., Guo C., Yoshiura K.-I., Utani A., and Nagayama Y.. 2013. Molecular cloning and characterisation of UVSSA, the responsible gene for UV-sensitive syndrome. Seikagaku. 85:133–144 (in Japanese). [PubMed] [Google Scholar]
- Okuda, M., Nakazawa Y., Guo C., Ogi T., and Nishimura Y.. 2017. Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways. Nucleic Acids Res. 45:13043–13055. 10.1093/nar/gkx970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian, A., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C., et al. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17:1101–1114. 10.1101/gad.1066903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccez, J.D., and Zerbini L.F.. 2001. Oncogenic transcription factors: target genes. In eLS. John Wiley & Sons, Ltd., Hoboken, NJ. https://doi.org/ 10.1002/9780470015902.a0006049.pub2 [DOI] [Google Scholar]
- Phillips, D.D., Garboczi D.N., Singh K., Hu Z., Leppla S.H., and Leysath C.E.. 2013. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 26:376–381. 10.1002/jmr.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl, P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., and Young R.A.. 2010. c-Myc regulates transcriptional pause release. Cell. 141:432–445. 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman, D.S., Schulte J.H., and Eilers M.. 2018. The expanding world of N-MYC-driven tumors. Cancer Discov. 8:150–163. 10.1158/2159-8290.CD-17-0273 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Barrueco, R., Marshall N., and Silva J.. 2013. Methods and Protocols. In Methods in Molecular Biology. Vol. 980. Humana Press, Totowa, NJ. pp. 353–370. In press. [DOI] [PubMed] [Google Scholar]
- Rosvall, M., and Bergstrom C.T.. 2008. Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA. 105:1118–1123. 10.1073/pnas.0706851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann, S., Wang Y., Nasoff M., Deveraux Q.L., and Quon K.C.. 2005. A TRAIL receptor-dependent synthetic lethal relationship between MYC activation and GSK3beta/FBW7 loss of function. Proc. Natl. Acad. Sci. USA. 102:15195–15200. 10.1073/pnas.0505114102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabò, A., Kress T.R., Pelizzola M., de Pretis S., Gorski M.M., Tesi A., Morelli M.J., Bora P., Doni M., Verrecchia A., et al. 2014. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 511:488–492. 10.1038/nature13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, M. 2013. The role of Cockayne syndrome group A (CSA) protein in transcription-coupled nucleotide excision repair. Mech. Ageing Dev. 134:196–201. 10.1016/j.mad.2013.03.008 [DOI] [PubMed] [Google Scholar]
- Sanz, L.A., Hartono S.R., Lim Y.W., Steyaert S., Rajpurkar A., Ginno P.A., Xu X., and Chédin F.. 2016. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 63:167–178. 10.1016/j.molcel.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin, A. 2012. UVSSA and USP7: new players regulating transcription-coupled nucleotide excision repair in human cells. Genome Med. 4:44. 10.1186/gm343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., Rodriguez-Barrueco R., Yu J., Do C., Silva J.M., and Gautier J.. 2015. MYC is a critical target of FBXW7. Oncotarget. 6:3292–3305. 10.18632/oncotarget.3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman, P., Lagarou A., Dekkers D.H.W., Raams A., van der Hoek A.C., Laffeber C., Hoeijmakers J.H.J., Demmers J.A.A., Fousteri M., Vermeulen W., et al. 2012. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 44:598–602. 10.1038/ng.2230 [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki, K., Proudfoot N.J., and Gromak N.. 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 42:794–805. 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier, J., Stork C.T., Garcia-Rubio M.L., Paulsen R.D., Aguilera A., and Cimprich K.A.. 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 56:777–785. 10.1016/j.molcel.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak, G. 2016. Transcription-coupled repair: an update. Arch. Toxicol. 90:2583–2594. 10.1007/s00204-016-1820-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini, M., Warmerdam D.O., Kolovos P., Snijder L., Vrouwe M.G., Demmers J.A.A., van IJcken W.F.J., Grosveld F.G., Medema R.H., Hoeijmakers J.H.J., et al. 2015. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 523:53–58. 10.1038/nature14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weegen, Y., Golan-Berman H., Mevissen T.E.T., Apelt K., González-Prieto R., Goedhart J., Heilbrun E.E., Vertegaal A.C.O., van den Heuvel D., Walter J.C., et al. 2020. The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II. Nat. Commun. 11:2104. 10.1038/s41467-020-15903-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vélez-Cruz, R., and Egly J.M.. 2013. Cockayne syndrome group B (CSB) protein: at the crossroads of transcriptional networks. Mech. Ageing Dev. 134:234–242. 10.1016/j.mad.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Wang, Z., Svejstrup J.Q., Feaver W.J., Wu X., Kornberg R.D., and Friedberg E.C.. 1994. Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature. 368:74–76. 10.1038/368074a0 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Engels I.H., Knee D.A., Nasoff M., Deveraux Q.L., and Quon K.C.. 2004. Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell. 5:501–512. 10.1016/S1535-6108(04)00113-8 [DOI] [PubMed] [Google Scholar]
- Yang, D., Liu H., Goga A., Kim S., Yuneva M., and Bishop J.M.. 2010. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc. Natl. Acad. Sci. USA. 107:13836–13841. 10.1073/pnas.1008366107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Putcha P., Califano A., and Silva J.M.. 2013. Pooled ShRNA screenings: computational analysis. In Pancreatic Cancer. Methods in Molecular Biology (Methods and Protocols). Vol. 980. Su G., editor. Humana Press, Totowa, NJ. 371–384., 10.1007/978-1-62703-287-2_22 [DOI] [PubMed] [Google Scholar]
- Zeller, K.I., Zhao X., Lee C.W., Chiu K.P., Yao F., Yustein J.T., Ooi H.S., Orlov Y.L., Shahab A., Yong H.C., et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. USA. 103:17834–17839. 10.1073/pnas.0604129103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Horibata K., Saijo M., Ishigami C., Ukai A., Kanno S., Tahara H., Neilan E.G., Honma M., Nohmi T., et al. 2012. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat. Genet. 44:593–597. 10.1038/ng.2228 [DOI] [PubMed] [Google Scholar]
- Zhang, J., Vlasevska S., Wells V.A., Nataraj S., Holmes A.B., Duval R., Meyer S.N., Mo T., Basso K., Brindle P.K., et al. 2017. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 7:322–337. 10.1158/2159-8290.CD-16-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Patel M., Ng N., Hsieh M.H., Orth A.P., Walker J.R., Batalov S., Harris J.L., and Liu J.. 2014. Identification of synthetic lethality of PRKDC in MYC-dependent human cancers by pooled shRNA screening. BMC Cancer. 14:944. 10.1186/1471-2407-14-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the 18 top MYC-SL gene network communities.
lists the 1,811 top-ranked MYC-SL shRNA clones (Log2FC less than −2 and P < 0.1) from Illumina HiSeq analysis of MCF10A-MYC-ER SL screen.