Abstract
Epilepsy, one of the most common neurological diseases with spontaneous recurrent seizures, is a severe health problem globally. The present study aimed to study the role and upstream mechanism of 26S proteasome non-ATPase regulatory subunit 11 (Psmd11) in epilepsy. In the current paper, epileptic mice models were successfully established. Hematoxylin and eosin (HE) staining was performed to reveal morphology of hippocampal tissues. Nissl’s staining was performed for detection of neuron injury. Enzyme-linked immunosorbent assay (ELISA) was conducted to detect concentrations of pro-inflammatory cytokines. The expression of Psmd11 was downregulated in the hippocampal tissues of epileptic mice, and overexpression of Psmd11 improved the spatial learning and memory of epileptic mice. Further, upregulation of Psmd11 protected epileptic hippocampal neurons from injury. Moreover, Psmd11 overexpression inhibited cell apoptosis, suppressed the activities of microglia and astrocytes, as well as reduced inflammatory response in epileptic hippocampi. Psmd11 was a downstream target of miR-490-3p. Long noncoding RNA (lncRNA) Peg13 bound with miR-490-3p to upregulate Psmd11. Subsequently, rescue experiments revealed that Peg13 suppressed the progression of epilepsy via upregulating Psmd11. Furthermore, Psmd11 was verified to inactivate the Wnt/β-catenin pathway. Peg13 repressed the Wnt/β-catenin pathway via upregulation of Peg13. In conclusion, this paper illuminated the function and upstream mechanism of Psmd11 in epilepsy. Psmd11 was upregulated by Peg13 at a miR-490-3p dependent way, thus inactivating the Wnt/β-catenin pathway and alleviating epilepsy course in mice, which may be a promising approach for epilepsy treatment.
Keywords: Psmd11, miR-490-3p, Peg13, neuron injury, inflammatory response, epilepsy
Introduction
As one of the most common neurological disorders, epilepsy affects over 70 million people across the globe with elevated occurrence and mortality [1-3]. Antiepileptic drugs may inhibit seizures in epileptic patients, but cannot change long-term outcomes, and one third of the patients are resistant to medical treatment [3-5]. Some therapeutic methods, such as surgery, nerve stimulation therapy and food therapy are applied to manage chemo-resistant therapy. In addition, more and more new treatments including gene therapy, exosome therapy and molecular network targeted therapy are available for epilepsy [3,5,6]. Thus, it is crucially important to reveal the potential molecular mechanisms in the pathogenic process of epilepsy.
Based on previous papers, Pum2 [7], PTEN [8], Notch1 [9] have been indicated as putative biomarkers for epilepsy. 26S proteasome non-ATPase regulatory subunit 11 (Psmd11) has been indicated to regulate cell injury and inflammation. Psmd11 reduces pancreatic cancer cell apoptosis by activating the MEK1/ERK1/2 pathway [10]. Psmd11 is targeted by miR-451 and contributes to the inflammatory response and proliferation of glomerular mesangial cells [11]. In addition, upregulated Psmd11 is a therapeutic target for spinal cord injury [12], and is closely connected with neurodegenerative diseases [13]. However, the exact function and relevant mechanisms of Psmd11 in epilepsy need more studies.
Recently, functions of long non-coding RNAs (lncRNAs) in epilepsy have been gradually revealed. LncRNAs are longer than 200 nucleotides in length and possess limited or no protein-coding capacity. ZNF883 [14], H19 [15], ILF3-AS1 [16] are pivotal regulators in progression of epilepsy. In mechanism, lncRNAs compete with mRNAs for the same microRNAs (miRNAs) via the ceRNA pattern to regulate disease progression [17,18]. For example, lncRNA MALAT1 binds with miR-150-5p to promote inflammation and oxidative stress in pregnancy-induced hypertension via upregulation of ET-1 [19]. LncRNA HAGLROS modulates apoptosis and autophagy through binding with miR-100 and enhancing ATG10 expression in Parkinson’s disease [20]. Moreover, the ceRNA networks exert significant functions in epilepsy. An article has indicated that lncRNA FTX competitively binds with miR-21-5p to repress hippocampal neuron apoptosis by elevating SOX7 expression in rats with temporal lobe epilepsy [21]. Another article has illustrated that lncRNA UCA1 inhibits inflammation via the miR-203/MEF2C axis in epilepsy [22].
In the present study, we aimed to research the role and the underlying ceRNA mechanism of Psmd11 in epilepsy. The effects of Psmd11 on cell injury, inflammatory response of hippocampal neurons isolated from epileptic mice were probed. Moreover, the downstream signaling pathway for Psmd11 was investigated, which might shed a new insight into the epilepsy research.
Materials and methods
Animal grouping and stereotaxic injection
Healthy adult male C57/BL6 mice (25-30 g) were provided by Vital River Co. Ltd. (Beijing, China). All mice were free to food and water and were kept in a 12-h light/12-h dark cycle for one week at 22-24°C and 55% humidity. These mice were randomly divided into 9 groups (n = 6-15 per group): Normal, Epilepsy, Epilepsy+pcDNA3.1, Epilepsy+pcDNA3.1/Psmd11, Epilepsy+pcDNA3.1/Peg13, Epilepsy+pcDNA3.1/Peg13+sh-NC, Epilepsy+pcDNA3.1/Peg13+sh-Psmd11, Epilepsy+NC inhibitor and Epilepsy+miR-490-3p inhibitor groups. The study was carried out in accordance with the Animal Ethics Committee of the Affiliated Suzhou Hospital of Nanjing Medical University (Suzhou Municipal Hospital) (Jiangsu, China) and Guide for the Care and Use of Laboratory Animals.
GeneChem (Shanghai, China) constructed adeno associated viral (AAV) vectors to overexpress or knock down Psmd11, Peg13 or miR-490-3p expression. 2.5% sodium pentobarbital was intraperitoneally injected into mice in corresponding groups for anesthesia. Afterwards, the mice were fixed on stereotaxic frame and their cranial skin was sterilized with 75% alcohol. Then, a 0.5 cm incision was made to expose anterior fontanel and 2 holes of 0.8 mm in diameter were drilled. Subsequently, the vectors were administered into each side with a microsyringe at 0.5 μl/min and the needle was removed 5 min later, followed by scalp suture. When the mice wake up and were able to move freely, they were kept in their cages for two weeks until establishment of epilepsy model [23].
Epileptic mouse models
To establish epileptic mouse model, pilocarpine (320 mg/kg, Sigma-Aldrich, USA) was intraperitoneally injected into mice in the epilepsy groups 30 minutes after atropine (2 mg/kg, Sigma-Aldrich) injection. Epileptic seizures of the mice were assessed with the modified Racine score [24], and the models of grade IV or above were regarded successful. After status epilepticus (SE) for one hour, diazepam (4 mg/kg, Hospira, USA) was injected for SE termination. Thirty days later, the epileptic mice developed spontaneous epileptic seizures. Mice in the normal group were treated with the same amount of saline via intraperitoneal administration as controls. Videos were utilized to monitor mice.
Morris water maze test
In water maze test, a white circular pool of 120 cm in diameter and 40 cm in height was filled with water of 22-24°C. The pool was divided into four quadrants, and a transparent platform of 8 cm2 placed 1.5 cm below the water surface was put in the center of one quadrant. A tracking camera and a computer connected to it were used. Mice in Normal, Epilepsy, Epilepsy+pcDNA3.1, Epilepsy+pcDNA3.1/Psmd11 groups were trained 4 times a day for 6 consecutive days. The time to reach the underwater platform was recorded. If the time exceeded 60 seconds, the animals would be guided to the platform, and would be allowed to stay on the platform for 10 seconds. The recorded time was the escape latency, which was no longer than 60 seconds. On the 7th day, the platform was removed, and percentage of average swimming time and distance in the quadrant of platform within 60 seconds was recorded [25,26].
Tissue samples
Hippocampal tissues were obtained from the sacrificed mice in different groups, and instantly frozen in liquid nitrogen and maintained at -80°C for further assays.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from hippocampal tissues by TRIzol reagent (Invitrogen, USA). Next, complementary DNA (cDNA) was reversely transcribed from the extracted RNA with PrimeScript RT Master Mix (TaKaRa, Dalian, China). Later, LightCycler 480 instrument (Roche, Shanghai, China) with a SYBR Premix Ex Taq II kit (TaKaRa) was employed for qRT-PCR. The relative expression levels of RNAs were calculated via 2-ΔΔCt method standardized to GAPDH and U6.
Hematoxylin and eosin (HE) staining
Hippocampal tissues were fixed by 4% paraformaldehyde for one day, dehydrated and embedded in paraffin, and then cut into 5-μm sections. After being dewaxed and treated with gradient alcohol, the sections were stained by hematoxylin for 5 minutes and were washed by running water. Subsequently, alcohol solution with 1% hydrochloric acid and ammonia was employed. Next, 1% eosin was utilized for further staining, gradient ethanol for section dehydration, and neutral balsam for section mounting, followed by microscope visualization.
Nissl’s staining
Hippocampal tissue sections were treated with xylene twice for 10 min, with anhydrous ethyl alcohol, 95% alcohol, 80% alcohol and 70% alcohol for 2 min separately, and with distilled water for a moment. Then the sections were dyed with 1% toluidine blue for an hour, followed by color separation with gradient alcohol dehydration and xylene clearing before being mounted with neutral balsam. Finally, an optical microscope was applied for observation.
Western blotting
Total proteins were exacted with Radio Immunoprecipitation Assay (RIPA) lysis buffer (Beyotime, Shanghai, China) and quantified using bicinchoninic acid Protein Assay Kit (Thermo Fisher Scientific, USA). Next, the proteins were dissociated by 10% sodium dodecyl sulfate, polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Millipore) before the membranes were blocked in 5% skim milk for one hour at room temperature and cultured overnight with primary antibodies including Psmd11 (ab66346, Abcam, Shanghai, China), p53 (ab90363, Abcam), Bcl-2 (ab182858, Abcam), Bax (ab32503, Abcam), cleaved caspase-3 (ab2302, Abcam), CD68 (ab125212, Abcam), GFAP (ab7260, Abcam), IL-6 (ab208113, Abcam), IL-1β (ab9277, Abcam), TNF-α (ab1793, Abcam), β-catenin (ab32572, Abcam), Cyclin D1 (ab16663, Abcam), c-Myc (ab32072, Abcam), and GAPDH (ab181602, Abcam) at 4°C. Subsequently, the membranes were maintained with secondary antibodies (Abcam) for one hour at 37°C. Finally, ChemiDoc™ XRS+ System with Image Lab™ Software (Bio-Rad, USA) was employed for protein band analysis with GAPDH as internal control.
Enzyme-linked immunosorbent assay (ELISA)
The levels of IL-6, IL-1β and TNF-α in the supernatant of hippocampus tissues after centrifugation were evaluated with relative ELISA kits (R&D Systems, USA). A microplate reader (BioTek Instruments, USA) was employed to measure optical density (OD) value at 450 nm.
Luciferase reporter assay
HEK293 cells were co-transfected with pmirGLO-Psmd11-Wt/Mut or pmirGLO-Peg13-Wt/Mut vectors (Genechem) and NC inhibitor or miR-490-3p inhibitor. Cell transfection was performed using Lipofectamine 3000 (Invitrogen) complying with the manufacturer’s directions. Two days later, Dual-Luciferase Reporter Assay System (Promega, USA) was applied to measure the fluorescence intensity of the reporter vectors.
Statistical analysis
Statistical analysis was done via SPSS 21.0 software (SPSS, USA). Student’s t test was employed for comparison between 2 groups. Analysis of variance (ANOVA) was employed for comparisons among more than 2 groups. Spearman’s correlation analysis presented the correlations among expression of RNAs in epileptic hippocampi of mice. Data were shown as mean ± standard deviation (SD). Each in-vitro experiment was conducted at least thrice. P < 0.05 was statistically significant.
Results
Psmd11 presents downregulated expression in the hippocampi of epileptic mice
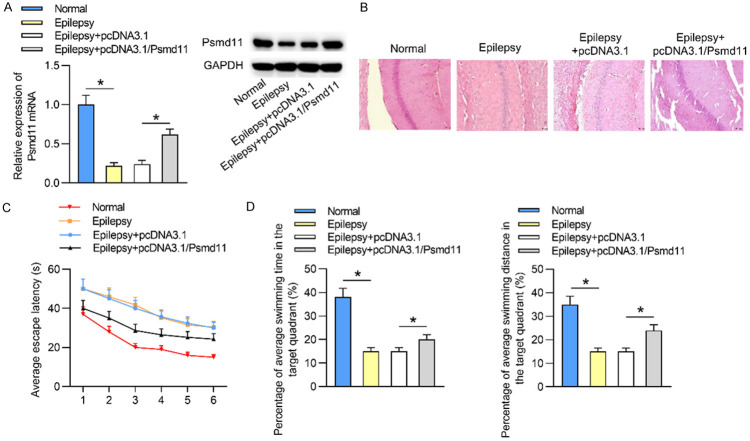
To investigate the expression status of Psmd11 in epilepsy, we established epileptic mouse models. As indicated by qRT-PCR and western blotting, Psmd11 mRNA and protein levels were underexpressed in epileptic hippocampal tissues when compared with those in control normal hippocampal tissues. Then compared to Epilepsy+pcDNA3.1 group, Psmd11 mRNA and protein levels were significantly elevated in Epilepsy+pcDNA3.1/Psmd11 group (Figure 1A). Subsequently, HE staining assay was carried out to assess the pathological features of epileptic hippocampi. As delineated in Figure 1B, the hippocampal neurons in Normal group exhibited intact structure with clearly visible nuclei, neat arrangement, clear layers and even staining. However, in Epilepsy group and Epilepsy+pcDNA3.1 group, most hippocampal neurons were damaged with pyknosis, loss and edema. Other neurons were disorganized with fuzzy layers and uneven staining. In Epilepsy+pcDNA3.1/Psmd11 group, the injury degree of neurons was alleviated than those in Epilepsy+pcDNA3.1 group: some neurons were destroyed but many nuclei were visible, and the layers were recognizable. These data indicated that Psmd11 upregulation alleviated epilepsy progression. Since hippocampi are responsible for spatial orientation and memory, we next performed Morris water maze test. Compared to the mice in Normal group, epileptic mice had longer escape latency, suggesting that the spatial positioning of the mice in Epilepsy group was worse than that of control mice. Moreover, shorter escape latency of the mice in Epilepsy+pcDNA3.1/Psmd11 group compared to those in Epilepsy+pcDNA3.1 group revealed that overexpression of Psmd11 improved the spatial orientation of epileptic mice (Figure 1C). Additionally, the percentage of average swimming time and distance in the target quadrant in Normal group was longer than those in Epilepsy group, while in pcDNA3.1/Psmd11 group, the percentage of swimming time and distance of epileptic mice in the quadrant was increased than the Epileptic mice. These data disclosed that the spatial memory of epileptic mice was improved by upregulation of Psmd11 expression (Figure 1D). In conclusion, Psmd11 was highly expressed in the hippocampal tissues of epileptic mice, and overexpression of Psmd11 improved the spatial learning and memory of epileptic mice.
Figure 1.
Psmd11 presents downregulated expression in the hippocampi of epileptic mice. (A) The mRNA and protein levels of Psmd11 in hippocampal tissues in Normal, Epilepsy, Epilepsy+pcDNA3.1 and Epilepsy+pcDNA3.1/Psmd11 groups were measured by qRT-PCR analysis and western blotting. *P < 0.05. One way ANOVA is conducted. (B) The morphological characteristics of hippocampal tissues in different conditions were evaluated via HE staining assay. (C, D) The abilities of spatial localization and memory of mice in the above groups were examined using Morris water maze test. *P < 0.05. Two way ANOVA is conducted in (C) and one way ANOVA is conducted in (D).
Upregulated Psmd11 reduces neuron injury in epileptic hippocampal tissues
We further explored the influence of Psmd11 on hippocampal neuron injury in epilepsy. According to Nissl’s staining, neurons were damaged with decreased number of Nissl bodies in epileptic hippocampal tissues, compared to those in normal hippocampal tissues. Afterwards, it was found that pcDNA3.1/Psmd11 attenuated the epileptic neuron damage (Figure 2A). Then the levels of cell apoptosis-relevant proteins were investigated. The protein levels of p53, Bax and cleaved caspase-3 were increased, and Bcl-2 level was decreased in the hippocampal tissues of epileptic mice, compared to those in normal mice. Furthermore, overexpression of Psmd11 reversed the protein levels of p53, Bcl-2, Bax and cleaved caspase-3 in epileptic hippocampal tissues (Figure 2B). These data suggested that upregulation of Psmd11 attenuated epilepsy-induced neuron injury and cell apoptosis in hippocampal tissues.
Figure 2.
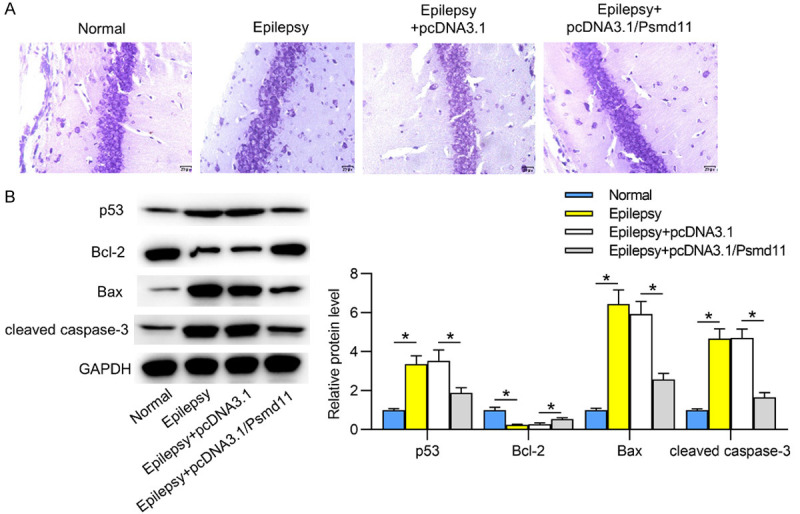
Upregulated Psmd11 reduces neuron injury in epileptic hippocampal tissues. A. Nissl’s staining was conducted in hippocampal tissues to assess neuron damage in Normal, Epilepsy, Epilepsy+pcDNA3.1 and Epilepsy+pcDNA3.1/Psmd11 groups. B. Western blotting was performed for examining the levels of cell apoptosis-relevant proteins (p53, Bcl-2, Bax, cleaved caspase-3). *P < 0.05. One way ANOVA is conducted.
Psmd11 inhibits inflammation in epileptic hippocampal tissues
Next, the effects of Psmd11 on activation of microglia and astrocytes in epileptic hippocampal tissues were investigated. Western blotting revealed that the protein levels of activated microglia marker CD68 and activated astrocyte marker GFAP were increased in the hippocampi of epileptic mice in contrast to those of normal mice. Moreover, CD68 and GFAP protein levels were reduced by Psmd11 overexpression in epileptic hippocampi tissues (Figure 3A). Subsequently, we probed into the change of pro-inflammatory cytokines in hippocampi. The protein levels of IL-6, IL-1β and TNF-α in epileptic hippocampi were increased compared with normal hippocampi. Besides, in the hippocampi of epileptic mice overexpressing Psmd11, the protein expression levels of these inflammatory cytokines were reduced (Figure 3B). Subsequently, ELISA reflected that the levels of IL-6, IL-1β and TNF-α were elevated in epileptic hippocampi, compared to those in normal hippocampi, and Psmd11 upregulation reduced the levels of the inflammatory cytokines in epileptic hippocampi (Figure 3C). Psmd11 overexpression inactivated microglia and astrocytes as well as inhibited the release of pro-inflammatory cytokines in epileptic hippocampi.
Figure 3.
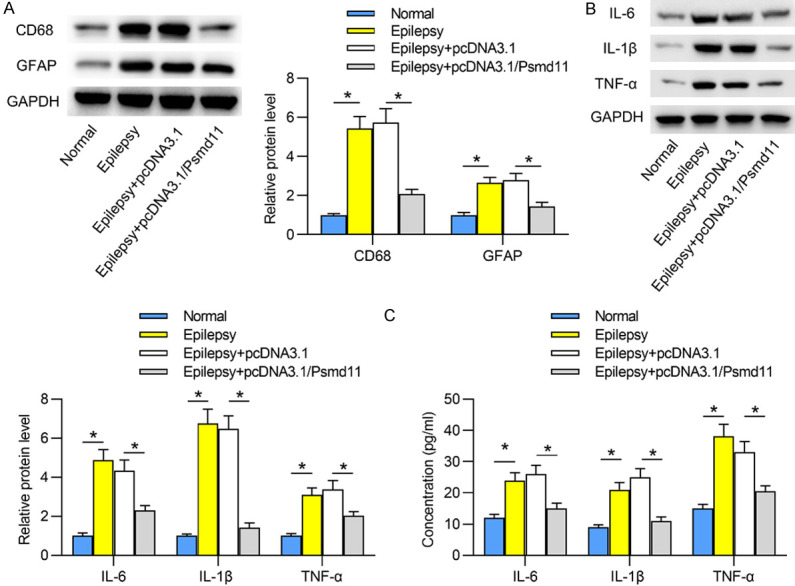
Psmd11 inhibits the activities of microglia and astrocytes in epileptic hippocampal tissues. A. Western blotting in hippocampal tissues revealed the protein levels of microglia marker CD68 and astrocyte marker GFAP in Normal, Epilepsy, Epilepsy+pcDNA3.1 and Epilepsy+pcDNA3.1/Psmd11 groups. *P < 0.05. One way ANOVA is conducted. B. Levels of inflammatory cytokines (IL-6, IL-1β and TNF-α) in hippocampal tissues were detected via western blotting. *P < 0.05. One way ANOVA is conducted. C. ELISA was employed to detect the levels of the inflammatory cytokines in hippocampal tissues. *P < 0.05. One way ANOVA is conducted.
Peg13 binds with miR-490-3p to upregulate Psmd11 in hippocampal tissues of epileptic mice
It has been recognized that lncRNAs can participate in biological processes via ceRNA crosstalk [27,28], and we hypothesized that there was a specific lncRNA acting as a ceRNA to regulate Psmd11, thus modulating epilepsy pathogenesis. Next, we sought for such a lncRNA in epilepsy. Initially, through miRmap and TargetScan databases, four miRNAs (miR-490-3p, miR-493-5p, miR-196b-5p and miR-196a-5p) were shown to have binding sites on Psmd11. Then we found that miR-490-3p showed higher expression in epileptic hippocampal tissues than the other miRNAs (Figure 4A). Subsequently, we silenced miR-490-3p expression using miR-490-3p inhibitor. A significant downregulation of miR-490-3p was induced by miR-490-3p inhibitor in HEK293 cells (Figure 4B). Afterwards, the interaction between miR-490-3p and Psmd11 was examined by luciferase reporter assay. As illustrated in Figure 4C, downregulation of miR-490-3p enhanced the relative luciferase activity of wild-type Psmd11 reporters but did not affect luciferase activity of mutant Psmd11 reporters in HEK293 cells. Further, it was revealed that the mRNA and protein levels of Psmd11 were increased when miR-490-3p was downregulated in epileptic hippocampi (Figure 4D). Then the Spearman’s correlation analysis revealed the negative correlation between miR-490-3p expression and Psmd11 expression in epileptic hippocampi (Figure 4E). Following that, we searched starBase website to explore candidate lncRNAs binding with miR-490-3p, and nineteen lncRNAs were revealed. Moreover, in epileptic hippocampal tissues, lncRNA Peg13 showed the most significant downregulation compared to the other lncRNAs (Figure 4F revealed the top 5 lncRNAs with significant changes in expression). Therefore, we focused on Peg13 in the follow-up examinations. Next, the luciferase activity of wild-type Peg13 reporters was promoted upon inhibition of miR-490-3p in HEK293 cells, which indicated that Peg13 bound with miR-490-3p at predicted sites (Figure 4G). Subsequently, we studied the effects of Peg13 on the expression of miR-490-3p and Psmd11. The results elucidated that upregulation of Peg13 decreased miR-490-3p expression and enhanced the mRNA and protein expression of Psmd11 in epileptic hippocampi (Figure 4H). Furthermore, Peg13 expression was negatively correlated with miR-490-3p expression and positively correlated with Psmd11 expression in epileptic hippocampi (Figure 4I). In conclusion, Peg13 competitively bound with miR-490-3p to elevate Psmd11 expression in hippocampi of epileptic mice.
Figure 4.
Peg13 binds with miR-490-3p to upregulate Psmd11 in hippocampal tissues of epileptic mice. A. By referring to miRmap and TargetScan databases, four miRNAs (miR-490-3p, miR-493-5p, miR-196b-5p and miR-196a-5p) can bind with Psmd11, and qRT-PCR analysis was utilized for detecting expression of these miRNAs in Normal and Epilepsy groups. *P < 0.05 vs. Normal group. Student’s t test was conducted. B. The knockdown efficiency of miR-490-3p via miR-490-3p inhibitor in HEK293 cells was tested through qRT-PCR analysis. *P < 0.05 vs. NC inhibitor group. Student’s t test was conducted. C. The interaction between miR-490-3p and Psmd11 was identified by luciferase reporter assay. *P < 0.05 vs. NC inhibitor group. Student’s t test was conducted. D. The influences of miR-490-3p on the mRNA and protein expression of Psmd11 in epileptic hippocampal tissues were revealed via qRT-PCR analysis and western blotting. *P < 0.05 vs. NC inhibitor group. Student’s t test was conducted. E. The correlation between expression of miR-490-3p and Psmd11 in epileptic hippocampal tissues was investigated by Spearman’s correlation analysis. F. There were nineteen lncRNAs which can bind with miR-490-3p, as predicted from starBase website. The expression levels of these potential lncRNAs in Normal and Epilepsy groups were presented by qRT-PCR analysis. *P < 0.05 vs. Normal group. Student’s t test was conducted. G. Luciferase reporter assay was employed to detect the influence of miR-490-3p inhibitor on luciferase activity of pmirGLO-Peg13-Wt and pmirGLO-Peg13-Mut plasmids. *P < 0.05 vs. NC inhibitor group. Student’s t test was conducted. H. The effects of Peg13 on expression of miR-490-3p and Psmd11. *P < 0.05 vs. pcDNA3.1 group. I. Spearman’s correlation analysis revealed the correlation between expression of Peg13 and miR-490-3p as well as the correlation between expression of Peg13 and Psmd11 in epileptic hippocampal tissues.
Peg13 alleviates epilepsy progression via positive regulation on Psmd11
Based on the above findings, whether Peg13 affects the progression of epilepsy via Psmd11 was examined. First, we detected the expression of Psmd11 in epileptic hippocampal tissues in different groups via qRT-PCR. Overexpression of Peg13 increased Psmd11 mRNA and protein expression levels, while cotreatment of pcDNA3.1/Peg13+sh-Psmd11 decreased levels of Psmd11 in epileptic hippocampi (Figure 5A). Subsequently, results from Nissl’s staining assay disclosed that neuron injury was suppressed by upregulated Peg13, but such effect was rescued by downregulation of Psmd11 in epileptic hippocampal tissues (Figure 5B). Inhibition of Psmd11 countervailed the inhibitory effects of Peg13 on apoptosis in the hippocampi of epileptic mice (Figure 5C). In addition, Peg13-induced inactivation of microglia and astrocytes as well as the release of pro-inflammatory cytokines were reversed by silencing of Psmd11 in epileptic hippocampi (Figure 5D, 5E). These findings demonstrated that Peg13 upregulated Psmd11 expression to inhibit the progression of epilepsy.
Figure 5.
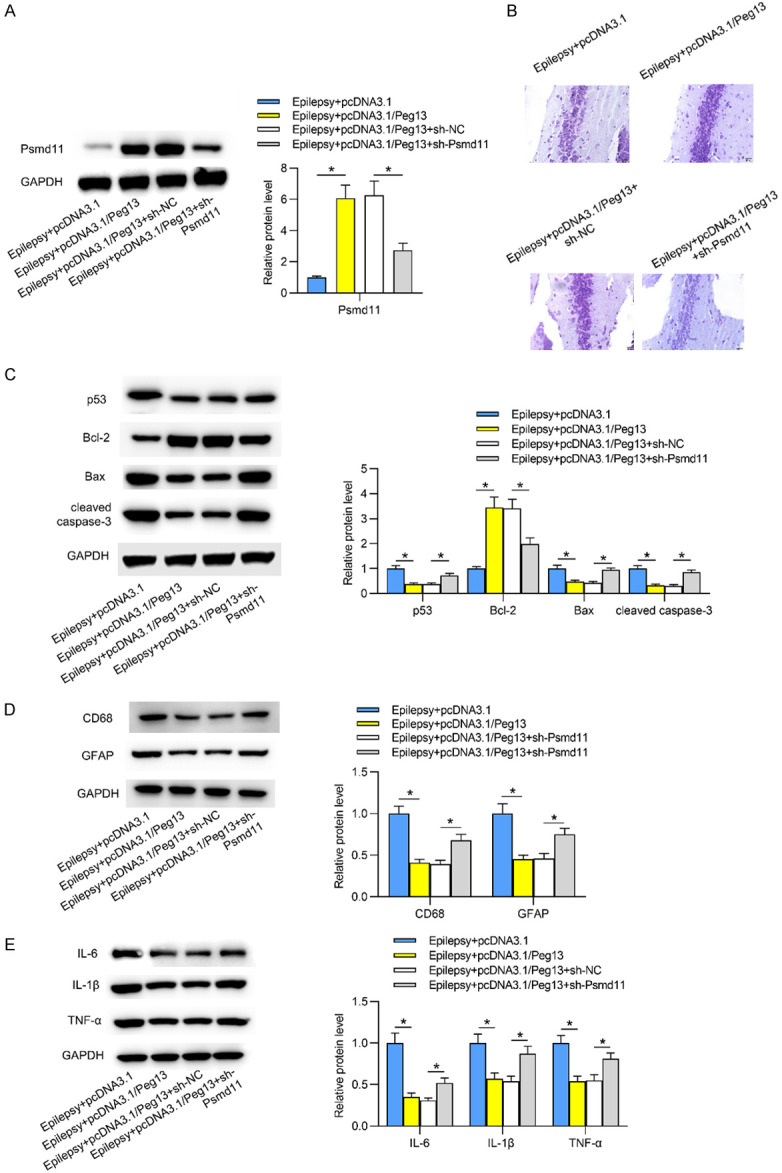
Peg13 inhibits epilepsy progression via upregulation of Psmd11. A. The expression of Psmd11 in Epilepsy+pcDNA3.1, Epilepsy+pcDNA3.1/Peg13, Epilepsy+pcDNA3.1/Peg13+sh-NC and Epilepsy+pcDNA3.1/Peg13+sh-Psmd11 was detected by qRT-PCR analysis in hippocampal tissues. *P < 0.05. One way ANOVA is conducted. B. Neuron injury in hippocampal tissues was evaluated by Nissl’s staining assay in abovementioned groups. C. Cell apoptosis-associated protein levels were measured via western blotting in the four groups. One way ANOVA is conducted. D, E. Western blotting in different groups displayed the protein levels of microglia and astrocyte markers as well as inflammatory cytokines in hippocampal tissues. *P < 0.05. One way ANOVA is conducted.
Peg13 inhibits the Wnt/β-catenin pathway in epileptic mice through upregulation of Psmd11
The Wnt/β-catenin pathway has been reported to facilitate epileptogenesis [29,30]. As shown in Figure 6A, the expression levels of Wnt/β-catenin pathway-related proteins (β-catenin, Cyclin D1 and c-Myc) were reflected by western blotting. In contrast to those in normal hippocampal tissues, protein levels of β-catenin, Cyclin D1 and c-Myc were increased in epileptic hippocampal tissues, indicating that the Wnt/β-catenin pathway was activated in epileptic mice. However, protein levels of β-catenin, Cyclin D1 and c-Myc were decreased by overexpression of Psmd11 in epileptic hippocampal tissues. Figure 6B revealed that upregulation of Peg13 reduced the protein expression of β-catenin, Cyclin D1 and c-Myc. Further, depletion of Psmd11 counteracted the suppressive effects of overexpression of Peg13 on β-catenin, Cyclin D1 and c-Myc protein levels. Thus, Peg13 inhibited the Wnt/β-catenin pathway through upregulation of Psmd11 in hippocampi of epileptic mice.
Figure 6.
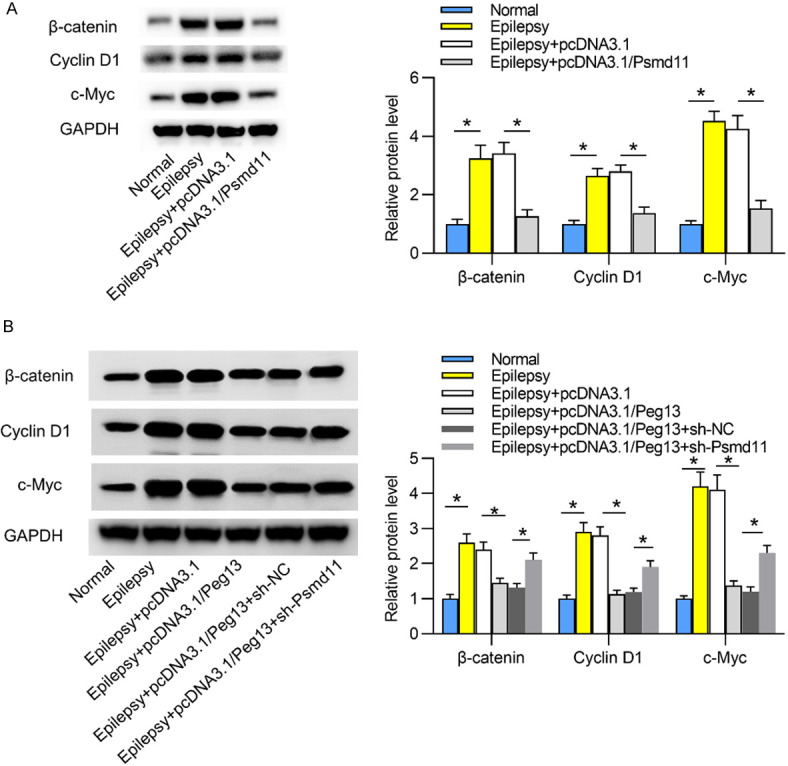
Peg13 inhibits the Wnt/β-catenin pathway in epileptic mice through upregulation of Psmd11. A. The levels of Wnt/β-catenin pathway-related proteins (β-catenin, Cyclin D1 and c-Myc) in hippocampal tissues were evaluated by western blotting in Normal, Epilepsy, Epilepsy+pcDNA3.1, Epilepsy+pcDNA3.1/Psmd11 groups. *P < 0.05. One way ANOVA is conducted. B. The levels of β-catenin, Cyclin D1 and c-Myc in hippocampal tissues were evaluated by western blotting in Normal, Epilepsy, Epilepsy+pcDNA3.1, Epilepsy+pcDNA3.1/Peg13, Epilepsy+pcDNA3.1/Peg13+sh-Psmd11 groups. *P < 0.05. One way ANOVA is conducted.
Discussion
Psmd11 plays significant roles in spinal cord injury, neurodegenerative disease and other diseases [10,12,13]. However, the influences of Psmd11 on epilepsy progression remain unclear. Therefore, the research focused on Psmd11, and initially, the expression of Psmd11 was disclosed to be downregulated in the hippocampal tissues of epileptic mice. Further, functional assays showed that Psmd11 overexpression improved the spatial localization and memory of epileptic mice, inhibited neuron damage and suppressed inflammatory response in epileptic hippocampi.
Since the ceRNA network plays a significant role in epilepsy [21], we next searched for the underlying ceRNA mechanism of Psmd11 in epilepsy. With the analysis of miRmap and TargetScan databases, miR-490-3p was hypothesized to bind with Psmd11 in epileptic mice. MiR-490-3p has been indicated to promote cell apoptosis in diverse cancers, such as hepatocellular carcinoma, colorectal cancer and ovarian cancer [31-33]. MiR-490-3p is influenced by HOTTIP to suppress the proliferation and migration of vascular smooth muscle cells in atherosclerosis [34]. MiR-490-3p inhibits the osteogenic differentiation of thoracic ligamentum flavum cells through negative regulation on FOXO1 [35]. The present study innovatively revealed that miR-490-3p targeted the 3’UTR of Psmd11 to inhibit Psmd11 expression. MiR-490-3p was upregulated in hippocampi of epileptic mice.
Next, the upstream lncRNA for miR-490-3p was investigated and lncRNA Peg13 was selected. The downregulation of Peg13 expression in hippocampal tissues of epileptic mice was shown in our study. Peg13 directly interacted with miR-490-3p and elevated Psmd11 expression by negatively modulating miR-490-3p in epileptic hippocampal tissues. Furthermore, our findings revealed that Psmd11 inhibited neuron damage, suppressed activation of glial cells and inflammation in epileptic hippocampi. Based on a previous study, Peg13 is underexpressed in brain microvascular endothelium after cerebral ischemia [36]. Paternally expressed Peg13 within the Trappc9 gene region is highly differentiated, which has been implicated in Prader-Willi nervous system disorder phenotypes in humans [37]. Peg13 is relatively high-expressed in the hippocampus, septal and hypothalamic regions, and the cerebral cortex of adult mice [38].
The Wnt/β-catenin pathway contributes to epileptogenesis [29,30]. Moreover, the lncRNA/miRNA/mRNA axes work in numerous diseases through the Wnt/β-catenin pathway. For instance, the SNHG16/miR-98/STAT3 axis promotes bladder cancer progression via the Wnt/β-catenin pathway [39]. LncRNA RP4 modulates the miR-939/Bnip3 axis and Wnt/β-catenin pathway to aggravate hypoxia injury in cardiomyocytes [40]. LncRNA MAGI1-IT1 targets the miR-302e/DKK1 axis to affect cardiac hypertrophy through the Wnt/β-catenin pathway [41]. We innovatively found that the Wnt/β-catenin pathway was activated in epilepsy and Peg13 inhibited the Wnt/β-catenin pathway through upregulation of Psmd11.
In conclusion, Peg13 attenuated epilepsy via inhibition on neuron injury and suppression on inflammation in hippocampus tissues of epileptic mice through upregulation of Psmd11 to inhibit the Wnt/β-catenin pathway, which may shed a new insight into the treatment of epilepsy.
Acknowledgements
We are grateful for support from all participants. This work was supported by CAAE Epilepsy Research fund-UCB (No. 2020004B), The Special Project for Diagnosis and Treatment of Key Clinical Diseases in Suzhou City (No. LCZX201811), and the Suzhou Science and Technology Prosperity Youth Science and Technology Project (No. KJXW2019029).
Disclosure of conflict of interest
None.
References
- 1.Tollenaere MAX, Tiedje C, Rasmussen S, Nielsen JC, Vind AC, Blasius M, Batth TS, Mailand N, Olsen JV, Gaestel M, Bekker-Jensen S. GIGYF1/2-driven cooperation between ZNF598 and TTP in posttranscriptional regulation of inflammatory signaling. Cell Rep. 2019;26:3511–3521. e3514. doi: 10.1016/j.celrep.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Coll M, Oliva A, Grassi S, Brugada R, Campuzano O. Update on the genetic basis of sudden unexpected death in epilepsy. Int J Mol Sci. 2019;20:1979. doi: 10.3390/ijms20081979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thijs RD, Surges R, O’Brien TJ, Sander JW. Epilepsy in adults. Lancet. 2019;393:689–701. doi: 10.1016/S0140-6736(18)32596-0. [DOI] [PubMed] [Google Scholar]
- 4.Kim JE, Cho KO. Functional nutrients for epilepsy. Nutrients. 2019;11:1309. doi: 10.3390/nu11061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehdizadeh A, Barzegar M, Negargar S, Yahyavi A, Raeisi S. The current and emerging therapeutic approaches in drug-resistant epilepsy management. Acta Neurol Belg. 2019;119:155–162. doi: 10.1007/s13760-019-01120-8. [DOI] [PubMed] [Google Scholar]
- 6.Johnson EL. Seizures and epilepsy. Med Clin North Am. 2019;103:309–324. doi: 10.1016/j.mcna.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Follwaczny P, Schieweck R, Riedemann T, Demleitner A, Straub T, Klemm AH, Bilban M, Sutor B, Popper B, Kiebler MA. Pumilio2-deficient mice show a predisposition for epilepsy. Dis Model Mech. 2017;10:1333–1342. doi: 10.1242/dmm.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Xu H, Ma H, Luo L, Yang L, Chen F, Qu X, Liu H, Zhang R. LncRNA CASC2 inhibits astrocytic activation and adenosine metabolism by regulating PTEN in pentylenetetrazol-induced epilepsy model. J Chem Neuroanat. 2020;105:101749. doi: 10.1016/j.jchemneu.2020.101749. [DOI] [PubMed] [Google Scholar]
- 9.Wan Y, Yang ZQ. LncRNA NEAT1 affects inflammatory response by targeting miR-129-5p and regulating Notch signaling pathway in epilepsy. Cell Cycle. 2020;19:419–431. doi: 10.1080/15384101.2020.1711578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Zhao L, Wei G, Saur D, Seidler B, Wang J, Wang C, Qi T. Homoharringtonine could induce quick protein synthesis of PSMD11 through activating MEK1/ERK1/2 signaling pathway in pancreatic cancer cells. J Cell Biochem. 2018;119:6644–6656. doi: 10.1002/jcb.26847. [DOI] [PubMed] [Google Scholar]
- 11.Wei H, Li J, Li Y, Song J. MicroRNA-451 inhibits inflammation and proliferation of glomerular mesangial cells through down-regulating PSMD11 and NF-κB p65. Biosci Rep. 2019;39:BSR20191455. doi: 10.1042/BSR20191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urso ML, Chen YW, Scrimgeour AG, Lee PC, Lee KF, Clarkson PM. Alterations in mRNA expression and protein products following spinal cord injury in humans. J Physiol. 2007;579:877–892. doi: 10.1113/jphysiol.2006.118042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Dong C, Sun L, Zhu L, Sun C, Ma R, Ning K, Lu B, Zhang J, Xu J. Quantitative proteomic analysis of age-related subventricular zone proteins associated with neurodegenerative disease. Sci Rep. 2016;6:37443. doi: 10.1038/srep37443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong L, Yang P, Hu L, Zhang C. MiR-181b suppresses the progression of epilepsy by regulation of lncRNA ZNF883. Am J Transl Res. 2020;12:2769–2780. [PMC free article] [PubMed] [Google Scholar]
- 15.Han CL, Liu YP, Guo CJ, Du TT, Jiang Y, Wang KL, Shao XQ, Meng FG, Zhang JG. The lncRNA H19 binding to let-7b promotes hippocampal glial cell activation and epileptic seizures by targeting Stat3 in a rat model of temporal lobe epilepsy. Cell Prolif. 2020;53:e12856. doi: 10.1111/cpr.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai X, Long L, Zeng C, Ni G, Meng Y, Guo Q, Chen Z, Li Z. LncRNA ILF3-AS1 mediated the occurrence of epilepsy through suppressing hippocampal miR-212 expression. Aging (Albany NY) 2020;12:8413–8422. doi: 10.18632/aging.103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorldJournal. 2014;2014:896206. doi: 10.1155/2014/896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan JY, Marques AC. miRNA-mediated crosstalk between transcripts: the missing “linc”? Bioessays. 2016;38:295–301. doi: 10.1002/bies.201500148. [DOI] [PubMed] [Google Scholar]
- 19.Ou M, Zhao H, Ji G, Zhao X, Zhang Q. Long noncoding RNA MALAT1 contributes to pregnancy-induced hypertension development by enhancing oxidative stress and inflammation through the regulation of the miR-150-5p/ET-1 axis. FASEB J. 2020;34:6070–6085. doi: 10.1096/fj.201902280R. [DOI] [PubMed] [Google Scholar]
- 20.Peng T, Liu X, Wang J, Liu Y, Fu Z, Ma X, Li J, Sun G, Ji Y, Lu J, Wan W, Lu H. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson’s disease via regulating miR-100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif Cells Nanomed Biotechnol. 2019;47:2764–2774. doi: 10.1080/21691401.2019.1636805. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Giri V, Cui Y, Yin M, Xian Z, Li J. LncRNA FTX inhibits hippocampal neuron apoptosis by regulating miR-21-5p/SOX7 axis in a rat model of temporal lobe epilepsy. Biochem Biophys Res Commun. 2019;512:79–86. doi: 10.1016/j.bbrc.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 22.Yu Q, Zhao MW, Yang P. LncRNA UCA1 Suppresses the inflammation via modulating miR-203-mediated regulation of MEF2C/NF-κB signaling pathway in epilepsy. Neurochem Res. 2020;45:783–795. doi: 10.1007/s11064-019-02952-9. [DOI] [PubMed] [Google Scholar]
- 23.Liu AH, Chu M, Wang YP. Up-regulation of trem2 inhibits hippocampal neuronal apoptosis and alleviates oxidative stress in epilepsy via the PI3K/Akt pathway in mice. Neurosci Bull. 2019;35:471–485. doi: 10.1007/s12264-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- 25.Sakata A, Mogi M, Iwanami J, Tsukuda K, Min LJ, Fujita T, Iwai M, Ito M, Horiuchi M. Sex-different effect of angiotensin II type 2 receptor on ischemic brain injury and cognitive function. Brain Res. 2009;1300:14–23. doi: 10.1016/j.brainres.2009.08.068. [DOI] [PubMed] [Google Scholar]
- 26.Zhao T, Ding Y, Li M, Zhou C, Lin W. Silencing lncRNA PVT1 inhibits activation of astrocytes and increases BDNF expression in hippocampus tissues of rats with epilepsy by downregulating the Wnt signaling pathway. J Cell Physiol. 2019 doi: 10.1002/jcp.28264. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 28.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodges SL, Lugo JN. Wnt/β-catenin signaling as a potential target for novel epilepsy therapies. Epilepsy Res. 2018;146:9–16. doi: 10.1016/j.eplepsyres.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, Fu XH, Zhou D, Li JM. The role of Wnt/β-catenin signaling pathway in disrupted hippocampal neurogenesis of temporal lobe epilepsy: a potential therapeutic target? Neurochem Res. 2015;40:1319–1332. doi: 10.1007/s11064-015-1614-1. [DOI] [PubMed] [Google Scholar]
- 31.Ou Y, He J, Liu Y. MiR-490-3p inhibits autophagy via targeting ATG7 in hepatocellular carcinoma. IUBMB Life. 2018;70:468–478. doi: 10.1002/iub.1715. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, He B, Xu T, Pan Y, Hu X, Chen X, Wang S. MiR-490-3p functions as a tumor suppressor by inhibiting oncogene VDAC1 expression in colorectal cancer. J Cancer. 2018;9:1218–1230. doi: 10.7150/jca.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Chen X, Xiu YL, Sun KX, Zhao Y. MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 2015;362:122–130. doi: 10.1016/j.canlet.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 34.Guo X, Liu Y, Zheng X, Han Y, Cheng J. HOTTIP knockdown inhibits cell proliferation and migration via regulating miR-490-3p/HMGB1 axis and PI3K-AKT signaling pathway in ox-LDL-induced VSMCs. Life Sci. 2020;248:117445. doi: 10.1016/j.lfs.2020.117445. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Qu X, Meng X, Li M, Fan D, Fan T, Huang AY, Chen Z, Zhang C. MiR-490-3p inhibits osteogenic differentiation in thoracic ligamentum flavum cells by targeting FOXO1. Int J Biol Sci. 2018;14:1457–1465. doi: 10.7150/ijbs.26686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu T, Meng F, Li Y, Chen YE, Yin KJ. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol. 2016;277:162–170. doi: 10.1016/j.expneurol.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenc A, Linnenbrink M, Montero I, Schilhabel MB, Tautz D. Genetic differentiation of hypothalamus parentally biased transcripts in populations of the house mouse implicate the prader-willi syndrome imprinted region as a possible source of behavioral divergence. Mol Biol Evol. 2015;32:1914–1915. doi: 10.1093/molbev/msv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies W, Smith RJ, Kelsey G, Wilkinson LS. Expression patterns of the novel imprinted genes Nap1l5 and Peg13 and their non-imprinted host genes in the adult mouse brain. Gene Expr Patterns. 2004;4:741–747. doi: 10.1016/j.modgep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Feng F, Chen A, Huang J, Xia Q, Chen Y, Jin X. Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/β-catenin pathway axis. J Cell Biochem. 2018;119:9408–9418. doi: 10.1002/jcb.27257. [DOI] [PubMed] [Google Scholar]
- 40.Rong J, Xu J, Liu Q, Wu Y, Guo H, Mu T, Zhou H, Chi H. Upregulation of long noncoding RNA RP4 exacerbates hypoxia injury in cardiomyocytes through regulating miR-939/Bnip3/Wnt/β-catenin pathway. Artif Cells Nanomed Biotechnol. 2019;47:3013–3020. doi: 10.1080/21691401.2019.1640232. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, Wang F, Wang F, Wu N. Long noncoding RNA MAGI1-IT1 regulates cardiac hypertrophy by modulating miR-302e/DKK1/Wnt/beta-catenin signaling pathway. J Cell Physiol. 2020;235:245–253. doi: 10.1002/jcp.28964. [DOI] [PubMed] [Google Scholar]