Abstract
Molecular profiling of exhaled breath by electronic nose (eNose) might be suitable as a noninvasive tool that can help in monitoring of clinically unstable COPD patients. However, supporting data are still lacking. Therefore, as a first step, this study aimed to determine the accuracy of exhaled breath analysis by eNose to identify COPD patients who recently exacerbated, defined as an exacerbation in the previous 3 months.
Data for this exploratory, cross-sectional study were extracted from the multicentre BreathCloud cohort. Patients with a physician-reported diagnosis of COPD (n=364) on maintenance treatment were included in the analysis. Exacerbations were defined as a worsening of respiratory symptoms requiring treatment with oral corticosteroids, antibiotics or both. Data analysis involved eNose signal processing, ambient air correction and statistics based on principal component (PC) analysis followed by linear discriminant analysis (LDA).
Before analysis, patients were randomly divided into a training (n=254) and validation (n=110) set. In the training set, LDA based on PCs 1–4 discriminated between patients with a recent exacerbation or no exacerbation with high accuracy (receiver operating characteristic (ROC)–area under the curve (AUC)=0.98, 95% CI 0.97–1.00). This high accuracy was confirmed in the validation set (AUC=0.98, 95% CI 0.94–1.00). Smoking, health status score, use of inhaled corticosteroids or vital capacity did not influence these results.
Exhaled breath analysis by eNose can discriminate with high accuracy between COPD patients who experienced an exacerbation within 3 months prior to measurement and those who did not. This suggests that COPD patients who recently exacerbated have their own exhaled molecular fingerprint that could be valuable for monitoring purposes.
Short abstract
Exhaled breath analysis by eNose can identify COPD patients who recently exacerbated with high accuracy. This suggests that these patients have their own exhaled molecular fingerprint that could be valuable for monitoring purposes. https://bit.ly/34vTyrH
Introduction
Exacerbations have a large impact on the quality of life of COPD patients. These periods of acute worsening that require additional therapy [1] are a prominent cause for the irreversible deterioration of lung function [2, 3] and are responsible for the greatest economic burden of COPD on healthcare expenditure [1]. Several biomarkers that show an association with history and risk of exacerbations have been identified; however, problems with reproducibility ensure that to date none of these have additional value over easily obtained clinical variables [4]. Peripheral blood eosinophil levels seem promising in predicting mortality and exacerbation risk [5], yet ambiguity about cut-off levels and the use of absolute or relative values hamper their use thus far [6].
Other potential biomarkers are volatile organic compounds (VOCs) in exhaled breath. VOCs are produced via different metabolic pathways and may reflect local, systemic and exogenous processes in the airways and alveoli [7, 8]. Analysis by gas chromatography combined with mass spectrometry (GC/MS) showed specific metabolic patterns of exacerbations in COPD patients and the ability to predict asthma exacerbations in children [9, 10]. Both GC/MS and pattern analysis of so-called breathprints by an electronic nose (eNose) were able to identify ongoing exacerbations in COPD patients [11, 12]. During follow-up, both methods showed a distinct exhaled breath pattern between exacerbations and stable disease [13]. Furthermore, data from a pilot study suggest that eNose analysis could make a distinction between COPD patients with and without bacterial or viral infections [14], which are important triggers for exacerbations.
Being easy to implement and noninvasive, eNose analysis might also be an appealing monitoring tool that can help to identify patients who exacerbated. However, data to answer this question in COPD are still lacking, and as a first step, there is a need to assess whether it is possible to detect an exacerbation signal in exhaled breath by eNose analysis.
In this proof of principle study, we hypothesised that exacerbations induce changes in exhaled metabolic patterns that can be captured by eNose sensors. If so, eNose technology may be suitable as a user-friendly and noninvasive point-of-care tool to identify patients who recently exacerbated. Therefore, in this study we aimed to determine the accuracy of VOC pattern analysis in exhaled breath by eNose to identify a history of recent exacerbations in COPD patients using maintenance therapy.
Methods
Population
Data for this exploratory, cross-sectional study were extracted from the multicentre BreathCloud cohort [15] that enrolled patients with asthma, COPD and lung cancer in primary, secondary and tertiary care centres in the Netherlands during routine (outpatient) visits. Due to the noninvasive nature of the BreathCloud study, the Amsterdam UMC medical ethical review board provided a waiver for ethical approval (reference: W14_112#14.17.0147, see supplementary material). Informed consent was obtained from each patient before enrolment. Inclusion criteria for the current study were: a physician-reported diagnosis of COPD, reported use of maintenance therapy with long-acting β-agonists and/or use of long-acting muscarinic antagonists, with or without additional therapy with ICS.
Outcome definition
An exacerbation was defined as a worsening of respiratory symptoms requiring the use of treatment with oral corticosteroids (OCS), antibiotics or both [1] during the 3 months prior to the eNose measurement.
Exhaled breath measurement
Exhaled breath analysis was performed by the SpiroNose (Amsterdam UMC, Amsterdam, the Netherlands), an eNose that uses seven cross-reactive metal oxide semiconducting sensors and has been technically validated earlier [16]. During inclusion in the BreathCloud cohort, patients were first required to thoroughly rinse their mouth with water three times. After this, patients were instructed to perform five tidal breaths followed by full inspiration, a 5-s breath-hold and slow maximal expiration towards residual volume. The entire measurement was performed in duplicate, considering an interval of 2 min. Data was captured in real-time and directly sent to the online BreathCloud server, where it was stored. Processing of eNose sensor signals was performed as described previously [15, 16]; more details can be found in the supplementary material.
Statistical analysis
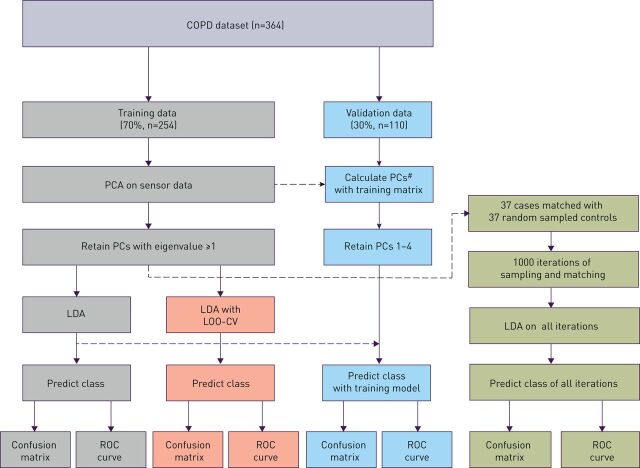
Statistical analysis was performed by R studio version 1.1.463 (Rstudio Inc., Boston, MA, USA) using R version 3.5.1 (The R Foundation for Statistical Computing, Vienna, Austria, with packages: dplyr, caret, pROC and MASS [17–19]). A schematic overview of the statistical analysis is shown in figure 1; more information on our statistical approach can be found in the supplementary material. Before the analysis, the included patients were randomly divided between a training (±70%) and validation (±30%) set. After this split, a principal component (PC) reduction of the training set eNose signal data was combined with linear discriminant analysis (LDA). Based on the Kaiser criterion [20], the training set PCs with an eigenvalue ≥1.0 were selected for further analysis. PCs from validation set sensor data were constructed by using the same parameters of standardisation and the same rotation matrix as those used to calculate the training data PCs. Discriminant scores, based on LDA, were used to construct receiver operating characteristic (ROC) curves and calculate the area under the curve (AUC). Internal validation was performed with leave-one-out cross-validation (LOO-CV); for external validation the LDA model that was constructed with the training set was applied to the validation set. Predicted classification based on the LDA was used to construct confusion matrices for the different datasets (training, training with cross-validation and validation). These confusion matrices were subsequently used to calculate overall accuracy, sensitivity, specificity, and positive- and negative predictive values.
FIGURE 1.
Flowchart of statistical analysis. PCA: principal component analysis; PCs: principal components; LDA: linear discriminant analysis; LOO-CV: leave-one-out cross-validation; ROC: receiver operating characteristic. #: Validation data principal components have been calculated by using the same parameters of standardisation and same rotation matrix as those used to calculate the training data principal components.
Sensitivity analyses
Our dataset was expected to have few cases compared to the number of controls. Therefore, to test the robustness of the model, the cases were matched with an equal amount of random sampled controls [21]. Average accuracy measures and ROC-AUC were calculated for 1000 iterations of this random sampling. The effect of current OCS and/or antibiotics use for exacerbations on the eNose signal was assessed by a secondary analysis excluding current users. Other sensitivity analyses were performed to map the effects on the discriminatory ability of the eNose of ICS use, smoking (both smoking status and pack-years), vital capacity during the measurement and clinical COPD questionnaire (CCQ) scores [22].
Results
In total, 364 patients diagnosed with COPD were enrolled in this study. The patients were randomly divided between a training (n=254) and validation set (n=110). In the training set, 37 patients (14.6%) experienced an exacerbation and in the validation set 13 patients (11.8%). Baseline characteristics of patients in the training set are shown in table 1, and characteristics of patients in the validation set are given in supplementary table S1. Mean age was 66.7±9.3 (training) and 65.9±8.7 (validation) years, percentage of females 47.6% (training) and 43.2% (validation) and median pack-years 36 (training) and 35 (validation). Pre-bronchodilator forced expiratory volume in 1 s (FEV1) (as % of predicted) was similar in the training and the validation set (61.2% and 57.1%, respectively) as was the FEV1/forced vital capacity (FVC) ratio (0.49 and 0.45, respectively). In the training set, the only difference between exacerbation/no exacerbation was found in blood eosinophil levels (0.20 cells·µL−1 versus 0.11 cells·µL−1) and the number of patients on ICS (78.4% versus 59.0%), both being higher in the exacerbation group. In the validation set, the only difference between exacerbation/no exacerbation was found in age, being higher in the exacerbation group (73.0±7.5 versus 64.9±8.5). No statistically significant differences were found in baseline clinical characteristics between the training and the validation set, although there was a borderline significant difference in pack-years (being higher in the validation set) (supplementary table S2).
TABLE 1.
Patient characteristics of the training set
| Total training | Exacerbation | No exacerbation | p-value | |
| Patients n | 254 | 37 | 217 | |
| Age years | 66.7±9.3 | 64.9±9.5 | 67.0±9.3 | 0.21 |
| Female | 121 (47.6) | 16 (43.2) | 105 (48.4) | 0.69 |
| Blood eosinophils cells·µL−1 | 0.12 (0.07–0.23) | 0.20 (0.12–0.34) | 0.11 (0.06–0.20) | 0.02 |
| Blood neutrophils cells·µL−1 | 5.83 (4.32–8.26) | 6.60 (4.50–9.88) | 5.74 (4.30–7.70) | 0.27 |
| BMI kg·m−2 | 26.4 (23.4–30.4) | 25.9 (22.7–30.5) | 26.5 (23.5–30.1) | 0.86 |
| Pre-BD FEV1 % pred | 61.2±20.9 | 57.1±21.8 | 61.9±20.7 | 0.24 |
| FEV1/FVC ratio | 0.49±0.14 | 0.45±0.14 | 0.49±0.13 | 0.15 |
| Patients with exacerbations | ||||
| Total | 37 (14.6) | |||
| OCS in the past 3 months n | 7 | |||
| AB in the past 3 months n | 9 | |||
| AB+OCS in the past 3 months n | 12 | |||
| Currently using AB n | 2 | |||
| Currently using OCS n | 6 | |||
| Currently using AB+OCS n | 1 | |||
| Patients with ICS | 157 (61.8) | 29 (78.4) | 128 (59.0) | 0.04 |
| CCQ score | 2.1±1.1 | 2.3±1.1 | 2.0±1.0 | 0.19 |
| CCQ score >3 | 55 (22.6) | 8 (21.6) | 47 (21.7) | 1.00 |
| Smoking history | 0.63 | |||
| Never | 7 (2.8) | 1 (2.7) | 6 (2.8) | |
| Ex | 168 (66.1) | 22 (59.5) | 146 (67.3) | |
| Current | 79 (31.1) | 14 (37.8) | 65 (30.0) | |
| Pack-years | 36 (20–49) | 35 (20–42) | 36 (20–50) | 0.38 |
Data are presented as mean±sd, n (%) or median (interquartile range), unless otherwise stated. BMI: body mass index; pre-BD: pre-bronchodilator; FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; OCS: oral corticosteroids; ICS: inhaled corticosteroids; AB: antibiotics; CCQ: Clinical COPD Questionnaire. p-values correspond to comparisons between exacerbation/no exacerbation.
Identification of exacerbations
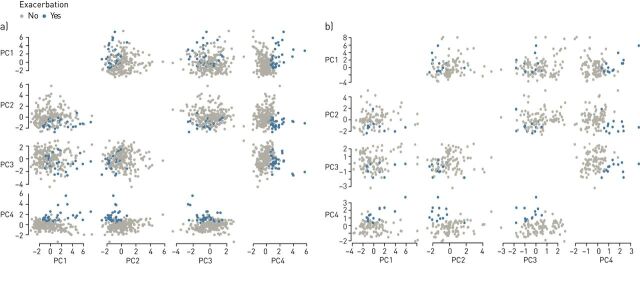
PCs 1–4 were selected to be used in the analysis. In the training set, all these principal components were significantly different between the exacerbation and non-exacerbation groups; in the validation set, this difference was only found for PCs 2–4. The distribution of patients with exacerbations and patients without exacerbations over the different principal components is shown for both sets in figure 2.
FIGURE 2.
Scatter matrices of patients who experienced an exacerbation and those who did not in a) training set (n=254) and b) validation set (n=110). PC: principal component.
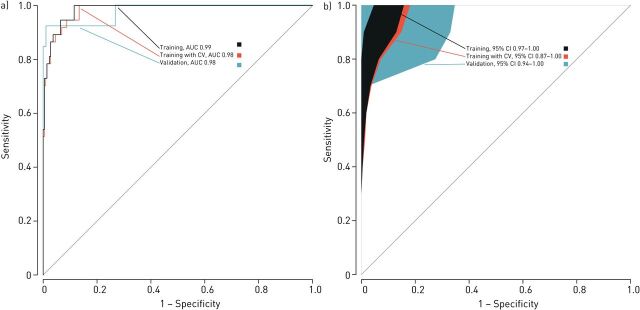
LDA was used to create a model and construct ROC curves. The ability to identify patients with a previous exacerbation showed high accuracy in the training set (ROC-AUC=0.98, 95% CI 0.97–1.00), with 95.3% of cross-validated grouped cases correctly classified after LOO-CV (95% CI 91.9–97.5%). This high accuracy was further confirmed in the validation set (AUC=0.98, 95% CI 0.94–1.00) where 95.5% of the grouped cases were correctly classified (figure 3). In comparison with other accuracy measures, sensitivity to identify patients with exacerbations was lower in both the training and validation sets (0.73 and 0.69, respectively). Specificity was 0.99 in both the training and validation sets (table 2).
FIGURE 3.
Receiver operating characteristic (ROC) analyses showing the accuracy of the linear discriminant model based on principal component reduction in the training set, internally validated training set (through leave-one-out cross-validation) and the independent validation set. a) Curves. b) 95% confidence intervals. CV: cross-validation; AUC: area under the curve; CI: confidence interval.
TABLE 2.
Accuracy parameters in the different datasets
| Data | ROC-AUC (95% CI) | Accuracy value (95% CI) | No information rate (NIR) | Sensitivity | Specificity | PPV | NPV |
| Training set | 0.99 (0.97–1.00) | 0.95 (0.92–0.98) | 0.85**** | 0.73 | 0.99 | 0.93 | 0.96 |
| Training set LOO-CV | 0.98 (0.97–1.00) | 0.95 (0.92–0.98) | 0.85**** | 0.73 | 0.99 | 0.93 | 0.96 |
| Validation set | 0.98 (0.94–1.00) | 0.96 (0.90–0.99) | 0.88** | 0.69 | 0.99 | 0.90 | 0.96 |
| 1:1 case control comparison (average of 1000 iterations on training set) (95% CI) | 0.98 | 0.91 (0.83–0.97) | 0.50**** | 0.88 (0.88–0.88) | 0.95 (0.95–0.95) | 0.95 (0.94–0.95) | 0.89 (0.89–0.89) |
ROC: receiver operating characteristic; AUC: area under the curve; CI: confidence interval; PPV: positive predictive value; NPV: negative predictive value; LOO-CV: leave-one-out cross-validation. **: AUC>NIR: p<0.01; ****: AUC>NIR: p<0.0001.
Robustness of the model
When the 37 cases in the training set were compared with 37 randomly sampled controls, AUC and accuracy value were still high. After 1000 iterations, the average sensitivity increased to 0.88 (95% CI 0.88–0.88) and average specificity somewhat decreased to 0.95 (95% CI 0.95–0.95). Both linear discriminant models (training/validation set and 1:1 comparison) showed similar loading factors for the principal components that were incorporated as predictors (PCs 1–4, see table 3), thus suggesting a similar contribution of predictors on both models. Omitting the patients that were currently using OCS/antibiotics for respiratory worsening did not impact the discriminatory ability of the eNose (accuracy measures in supplementary table S3). Sensitivity analyses did not show an effect of smoking, health status score, use of ICS or vital capacity on the discriminatory ability of the eNose sensors. The results of these sensitivity analyses can be found in the supplementary material (supplementary table S5 and figures S1–3).
TABLE 3.
Loading factors of principal components in the different models
| Training/validation model | Averaged 1:1 model | ||
| LD1 | LD1 (95% CI) | ||
| PC1 | 0.23 | PC1 | 0.11 (0.11–0.11) |
| PC2 | −0.51 | PC2 | −0.63 (−0.64–(−)0.63) |
| PC3 | −0.29 | PC3 | −0.12 (−0.13–(−)0.12) |
| PC4 | 1.17 | PC4 | 0.84 (0.84–0.84) |
LD1: linear discriminant function 1; PC: principal component; CI: confidence interval.
Discussion
This study shows that pattern recognition analysis of exhaled breath by eNose can discriminate with high accuracy between COPD patients on maintenance therapy who experienced an exacerbation within 3 months prior to measurement and those who did not. These results were internally validated and were confirmed in an independent validation set. Furthermore, smoking history, vital capacity during measurement, health status (CCQ score) and the use of ICS did not seem to affect this discriminatory ability. By discriminating between COPD patients who may or may not have experienced a recent exacerbation, point-of-care eNose measurements might be suitable to identify exacerbation-prone COPD patients. The next step is to prospectively collect evidence to confirm this ability.
This is the first study to use exhaled breath analysis by a point-of-care eNose to identify COPD patients who recently exacerbated. These results are in line with studies that showed an ability of eNose and GC-MS to discriminate between periods of stable COPD and acute exacerbations [12, 13]. Furthermore, the current study supports earlier findings showing differences in exhaled metabolic patterns between frequent exacerbators and stable COPD patients [9], infectious/non-infectious COPD exacerbations [11] and data from a pilot study that showed (moderate) ability to discriminate between bacterial/viral infections and stable COPD patients [14]. The ability to detect recent exacerbations shows promise as a means of monitoring COPD patients, as VOCs in exhaled breath have already proved useful in predicting asthma exacerbations in children [10] and predicting response to treatment with OCS in asthma patients [23].
A recent study by Brinkman et al. [24] further strengthens this promise, as it showed that in asthma, both GC-MS and eNose are able to correctly classify loss of control in asthma versus recovery and versus baseline measurements. The eNose used in the current study, the SpiroNose, allows real-time measurements and is easy to use in a clinical setting. In contrast, previous studies often used techniques based on GC-MS, and although real-time analysis is possible [9], a major limitation for clinical application is the requirement for highly trained personnel [25].
Strengths of this study are the relatively large sample size, the internal and external validation steps that have been taken to further support the obtained results and the quick and easy-to-use point-of-care device that was employed. The patients who have been enrolled in the BreathCloud cohort were recruited in centres with different levels of care, and there was no need to cease medication use. This resulted in a mixed population that contains patients from primary care to specialised (academic) hospitals, reflecting the general COPD population. The use of physician-reported COPD diagnosis, however, does not rule out the possibility of misdiagnosed patients. Due to the skewed ratio of patients who had experienced a (recent) exacerbation versus those who did not, “no exacerbations” was expected to have a disproportionate effect on the outcome. The no information rate indeed showed a high probability to correctly classify the majority class (no exacerbations in our case), and therefore getting similar results with equally sized groups further strengthens our findings.
This was a retrospective study and the current results do not necessarily translate into a prospective scenario. A further limitation of our study is the lack of a new and completely independent validation cohort with similar patients measured with the same type of eNose. Because to our knowledge such a cohort did not exist, we consider the second-best option of dividing our cohort into a training and a validation set justified. Another important limitation of this study is the exacerbation definition. Exacerbations are complex and heterogeneous [26, 27], and thus the use of OCS or antibiotics to treat worsening respiratory complaints may be lacking in sensitivity and/or specificity. It may also be that the detected difference in eNose signals was caused by the administered OCS and antibiotics, as these signals may be influenced by metabolites of the medication. However, we have shown that omitting current OCS/antibiotic users from the analysis did not change the outcomes. In this study, the timing of the exacerbations was not taken into account, i.e. some may have happened 3 months before measurement, some only a week, although the breath signal may be expected to change over time. Furthermore, due to the low number of subjects with >1 recent exacerbation (only 5 out of 36 subjects), we did not investigate the influence of number of exacerbations on exhaled breath profiles.
Earlier research suggested that eNose sensors are able to capture inflammatory profiles in patients [28, 29] and showed signs of a dose–response relationship with blood eosinophil and neutrophil levels [15]. This could suggest that flaring up of inflammation during an exacerbation provides a driving force in the changes in eNose fingerprints. Recently, a large study showed that VOCs identified by GC-MS in exhaled breath are able to discriminate between two important inflammatory phenotypes in asthma: eosinophilic asthma and neutrophilic asthma [30]. This suggests that inflammatory processes might show an effect on exhaled molecular breath patterns (or “breathprints”). Exacerbations have different underlying mechanisms and thus there are differences in metabolic (inflammatory) processes that are altered between different exacerbations [2, 31]. This seems contradictory to the accuracy found in the current study as these results point to a common denominator in all exacerbations. More detailed information on cause and nature of exacerbations could therefore be valuable in future studies.
Although some studies suggested that blood eosinophil levels may be useful to guide corticosteroid treatment in COPD patients and to predict exacerbations [5, 32], the strong signal in our results suggests that eNoses can detect more information on metabolic (inflammatory) processes than blood eosinophils or other biomarkers that are currently used. COPD exacerbations have a distinct signature in peripheral blood that persists for months following the event [33]. This blood signature could be reflected in an exhaled metabolic signature which is in line with our finding that the eNose was able to detect an exacerbation that had occurred up to 3 months prior to measurement. A recent study by van Velzen et al. [13] could further support this, as their results suggested that exhaled breath patterns returned to baseline within 12 weeks after an exacerbation in COPD patients. This could imply that our data represent a signature of a recent previous exacerbation.
The identification of COPD patients who recently experienced an exacerbation by a noninvasive and easy-to-use tool could be of great value in monitoring of COPD patients. Early detection of patients who are unstable on treatment and prediction of risk for exacerbations have been named major clinical dilemmas in chronic diseases of airways [7] and may lead to new interventions that prevent lung deterioration and irreversible damage. A history of exacerbations is currently the best predictor for future exacerbations in COPD [34], and thus the results of the current study are promising. Our results suggest that exhaled breath analysis might be of value in predicting the risk for exacerbations. Future prospective studies should be conducted though to provide insight into the added value of exhaled breath analysis over currently used clinical symptoms. Another implication of our study is that heterogeneity in (exhaled breath composition of) COPD patients is a factor to be reckoned with in future studies. Patient selection for comparisons with other disease areas should take into account that recent exacerbations in COPD patients could influence breath profiles.
In conclusion, exhaled breath analysis by SpiroNose can accurately identify COPD patients who have experienced an exacerbation during the previous 3 months. The current study suggests that COPD patients who recently exacerbated have their own exhaled molecular breathprint that could be valuable in monitoring of COPD patients. The present data merit an independent validation of our findings by the collection of prospective longitudinal data, which will allow testing as to whether exhaled breath analysis by eNose can predict exacerbations better than we can do at the present time on the basis of clinical symptoms.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00307-2020.SUPPLEMENT (561.8KB, pdf)
Acknowledgements
The authors wish to thank and acknowledge all BreathCloud partners, in particular the staff of Diagnostiek voor U (Eindoven, the Netherlands); Medisch Centrum Den Bosch Oost ('s-Hertogenbosch, the Netherlands); Medisch Spectrum Twente (Enschede, the Netherlands); Franciscus Gasthuis and Vlietland (Rotterdam, the Netherlands); and Amsterdam UMC, locations AMC and VUmc, (Amsterdam, the Netherlands) for their help in recruitment of the COPD patients who have been enrolled in this study.
Footnotes
This article has supplementary material available from openres.ersjournals.com
Members of the Amsterdam UMC Breath Research Group: Anke H. Maitland-van der Zee, Peter J. Sterk, Paul Brinkman, Anne H. van Stuyvenberg-Neerincx, Cristina Longo, Lieuwe D. Bos, Anirban Sinha, Dominic W. Fenn, Marije Lammers, Levi B. Richards, Job J.M.H. van Bragt, Mahmoud I. Abdel-Aziz, Renate Kos, Yennece W.F. Dagelet, Marcus J. Schultz, Marry R. Smit and Laura A. Hagens (all Amsterdam UMC, University of Amsterdam, Depts of (Pediatric) Respiratory Medicine or Intensive Care Medicine).
Conflict of interest: J.J.M.H. van Bragt reports an unrestricted research grant from Boehringer Ingelheim during the conduct of the study.
Conflict of interest: P. Brinkman has nothing to disclose.
Conflict of interest: R. de Vries is COO of and has a considerable interest in the start-up company Breathomix BV.
Conflict of interest: S.J.H. Vijverberg has nothing to disclose.
Conflict of interest: E.J.M. Weersink has nothing to disclose.
Conflict of interest: E.G. Haarman has nothing to disclose.
Conflict of interest: F.H.C. de Jongh has nothing to disclose.
Conflict of interest: S. Kester has nothing to disclose.
Conflict of interest: A. Lucas has nothing to disclose.
Conflict of interest: J.C.C.M. in 't Veen reports faculty grants from Boehringer Ingelheim, Teva and Chiesi, and personal fees for an international advisory board from Sanofi, outside the submitted work.
Conflict of interest: P.J. Sterk reports being scientific advisor to and having a formally inconsiderable interest in the start-up company Breathomix BV.
Conflict of interest: E.H.D. Bel reports grants and personal fees from AstraZeneca, GSK, Novartis, and Teva, and personal fees from Sanofi/Regeneron, Boehringer Ingelheim, Vectura, and Sterna, outside the submitted work.
Conflict of interest: A.H. Maitland-van der Zee reports personal fees for advisory boards from AstraZeneca and Boehringer Ingelheim, and unrestricted research grants from Boehringer Ingelheim and GSK, during the conduct of the study.
Support statement: This study was supported by an unrestricted grant by Boehringer Ingelheim. The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1. Global Initiative for Chronic Obstructive Lung Disease. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease (2019 Report).
- 2.Pavord ID, Jones PW, Burgel PR, et al. Exacerbations of COPD. Int J COPD 2016; 11: 21–30. doi: 10.1183/16000617.0103-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donaldson GC, Law M, Kowlessar B, et al. Impact of prolonged exacerbation recovery in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2015; 192: 943–950. doi: 10.1164/rccm.201412-2269OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene JD, Jacobson S, Kechris K, et al. Biomarkers predictive of exacerbations in the SPIROMICS and COPDGene cohorts. Am J Respir Crit Care Med 2017; 195: 473–481. doi: 10.1164/rccm.201607-1330OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vedel-Krogh S, Nielsen SF, Lange P, et al. Blood eosinophils and exacerbations in chronic obstructive pulmonary disease. The Copenhagen General Population Study. Am J Respir Crit Care Med 2016; 193: 965–974 [DOI] [PubMed] [Google Scholar]
- 6.Bafadhel M. Eosinophils in COPD: are we nearly there yet? Lancet Respir Med 2017; 5: 913–914. doi: 10.1016/S2213-2600(17)30445-9 [DOI] [PubMed] [Google Scholar]
- 7.Van Der Schee MP, Paff T, Brinkman P, et al. Breathomics in lung disease. Chest 2015; 147: 224–231. doi: 10.1378/chest.14-0781 [DOI] [PubMed] [Google Scholar]
- 8.Bos LD, Sterk PJ, Fowler SJ. Breathomics in the setting of asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol 2016; 138: 970–976. doi: 10.1016/j.jaci.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 9.Gaugg MT, Nussbaumer-Ochsner Y, Bregy L, et al. Real-time breath analysis reveals specific metabolic signatures of COPD exacerbations. Chest 2019; 156: 269–276. doi: 10.1016/j.chest.2018.12.023 [DOI] [PubMed] [Google Scholar]
- 10.Robroeks CM, Van Berkel JJ, Jöbsis Q, et al. Exhaled volatile organic compounds predict exacerbations of childhood asthma in a 1-year prospective study. Eur Respir J 2013; 42: 98–106. doi: 10.1183/09031936.00010712 [DOI] [PubMed] [Google Scholar]
- 11.Shafiek H, Fiorentino F, Merino JL, et al. Using the electronic nose to identify airway infection during COPD exacerbations. PLoS One 2015; 10: e0135199. doi: 10.1371/journal.pone.0135199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pizzini A, Filipiak W, Wille J, et al. Analysis of volatile organic compounds in the breath of patients with stable or acute exacerbation of chronic obstructive pulmonary disease. J Breath Res 2018; 12: 036002. doi: 10.1088/1752-7163/aaa4c5 [DOI] [PubMed] [Google Scholar]
- 13.van Velzen P, Brinkman P, Knobel HH, et al. Exhaled breath profiles before, during and after exacerbation of COPD: a prospective follow-up study. COPD 2019; 16: 330–337. doi: 10.1080/15412555.2019.1669550 [DOI] [PubMed] [Google Scholar]
- 14.van Geffen WH, Bruins M, Kerstjens HAM. Diagnosing viral and bacterial respiratory infections in acute COPD exacerbations by an electronic nose: a pilot study. J Breath Res 2016; 10: 036001. doi: 10.1088/1752-7155/10/3/036001 [DOI] [PubMed] [Google Scholar]
- 15.de Vries R, Dagelet YWF, Spoor P, et al. Clinical and inflammatory phenotyping by breathomics in chronic airway diseases irrespective of the diagnostic label. Eur Respir J 2018; 51: 1701817. doi: 10.1183/13993003.01817-2017 [DOI] [PubMed] [Google Scholar]
- 16.De Vries R, Brinkman P, Van Der Schee MP, et al. Integration of electronic nose technology with spirometry: validation of a new approach for exhaled breath analysis. J Breath Res 2015; 9: 46001. doi: 10.1088/1752-7155/9/4/046001 [DOI] [PubMed] [Google Scholar]
- 17.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011; 12: 12–77. doi: 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn M. Building predictive models in R using the caret package. J Stat Softw 2008; 28: 1–26. www.jstatsoft.org/v28/i05/paper. doi: 10.18637/jss.v028.i0527774042 [DOI] [Google Scholar]
- 19.Venables WN, Ripley BD. Modern Applied Statistics with S. New York City, Springer International Publishing, 2002. [Google Scholar]
- 20.Kaiser HF. The application of electronic computers to factor analysis. Educ Psychol Meas 1960; 20: 141–151. http://journals.sagepub.com/doi/10.1177/001316446002000116. doi: 10.1177/001316446002000116 [DOI] [Google Scholar]
- 21.Sanchez PM. The unequal group size problem in discriminant analysis. J Acad Mark Sci 1974; 2: 629–633. doi: 10.1007/BF02729456 [DOI] [Google Scholar]
- 22.van der Molen T, Willemse BWM, Schokker S, et al. Development, validity and responsiveness of the Clinical COPD Questionnaire. Health Qual Life Outcomes 2003; 1: 13. doi: 10.1186/1477-7525-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Schee MP, Palmay R, Cowan JO, et al. Predicting steroid responsiveness in patients with asthma using exhaled breath profiling. Clin Exp Allergy 2013; 43: 1217–1225. doi: 10.1111/cea.12147 [DOI] [PubMed] [Google Scholar]
- 24.Brinkman P, Wagener AH, Hekking PP, et al. Identification and prospective stability of eNose derived inflammatory phenotypes in severe asthma. J Allergy Clin Immunol 2019; 143: 1811–1820. doi: 10.1016/j.jaci.2018.10.058 [DOI] [PubMed] [Google Scholar]
- 25.Neerincx AH, Vijverberg SJH, Bos LDJ, et al. Breathomics from exhaled volatile organic compounds in pediatric asthma. Pediatr Pulmonol 2017; 52: 1616–1627. doi: 10.1002/ppul.23785 [DOI] [PubMed] [Google Scholar]
- 26.Hurst JR. Exacerbation phenotyping in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2011; 184: 625–626. doi: 10.1164/rccm.201106-1136ED [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Campos JL, Agustí A. Heterogeneity of chronic obstructive pulmonary disease exacerbations: a two-axes classification proposal. Lancet Respir Med 2015; 3: 729–734. doi: 10.1016/S2213-2600(15)00242-8 [DOI] [PubMed] [Google Scholar]
- 28.Plaza V, Crespo A, Giner J, et al. Inflammatory asthma phenotype discrimination using an electronic nose breath analyzer. J Investig Allergol Clin Immunol 2015; 25: 431–437. [PubMed] [Google Scholar]
- 29.Fens N, De Nijs SB, Peters S, et al. Exhaled air molecular profiling in relation to inflammatory subtype and activity in COPD. Eur Respir J 2011; 38: 1301–1309. doi: 10.1183/09031936.00032911 [DOI] [PubMed] [Google Scholar]
- 30.Schleich FN, Zanella D, Stefanuto p-H, et al. Exhaled volatile organic compounds are able to discriminate between neutrophilic and eosinophilic asthma. Am J Respir Crit Care Med 2019; 15: 444–453. doi: 10.1164/rccm.201811-2210OC [DOI] [PubMed] [Google Scholar]
- 31.Bafadhel M, McKenna S, Terry S, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med 2011; 184: 662–671. doi: 10.1164/rccm.201104-0597OC [DOI] [PubMed] [Google Scholar]
- 32.Bafadhel M, Pavord ID, Russell REK. Eosinophils in COPD: just another biomarker? Lancet Respir Med 2017; 5: 747–759. doi: 10.1016/S2213-2600(17)30217-5 [DOI] [PubMed] [Google Scholar]
- 33.Morrow JD, Qiu W, Chhabra D, et al. Identifying a gene expression signature of frequent COPD exacerbations in peripheral blood using network methods. BMC Med Genomics 2015; 8: 1–11. doi: 10.1186/s12920-014-0072-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoogendoorn M, Feenstra TL, Boland M, et al. Prediction models for exacerbations in different COPD patient populations: comparing results of five large data sources. Int J COPD 2017; 12: 3183–3194. doi: 10.2147/COPD.S142378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00307-2020.SUPPLEMENT (561.8KB, pdf)