Abstract
In plant cells, chloroplast gene expression is predominantly controlled through post-transcriptional regulation. Such fine-tuning is vital for precisely orchestrating protein complex assembly as for the photosynthesis machinery and for quickly responding to environmental changes. While regulation of chloroplast protein synthesis is of central importance, little is known about the degree and nature of the regulatory network, mainly due to challenges associated with the specific isolation of transient ribosome interactors. Here, we established a ribosome affinity purification method, which enabled us to broadly uncover putative ribosome-associated proteins in chloroplasts. Endogenously tagging of a protein of the large or small subunit revealed not only interactors of the holo complex, but also preferential interactors of the two subunits. This includes known canonical regulatory proteins as well as several new proteins belonging to the categories of protein and RNA regulation, photosystem biogenesis, redox control and metabolism. The sensitivity of the here applied screen was validated for various transiently interacting proteins. We further provided evidence for the existence of a ribosome-associated Nα-acetyltransferase in chloroplasts and its ability to acetylate substrate proteins at their N-terminus. The broad set of ribosome interactors underscores the potential to regulate chloroplast gene expression on the level of protein synthesis.
INTRODUCTION
Translation is the process by which the genetic information is decoded from linear nucleic acid strands into polypeptides, resulting in diverse three-dimensional protein structures. This process is achieved through ribosomes, the highly abundant macromolecular ribonucleoprotein machinery present in all kingdoms of life. With the rather poor correlation between mRNA levels and protein quantities, ribosomes emerged as a central regulatory hub during gene expression (1,2). Protein synthesis can be fine-tuned at any step, including translation initiation, elongation, modulation of translation-competent ribosome pools and nascent polypeptide processing (2–7). The need for rapid adjustments of protein biogenesis becomes apparent when considering that translation accounts for ∼50% of the energy consumption in bacterial cells (8) and that >10% of all proteins are supposed to impact protein synthesis at various levels (9). The composition of the translation apparatus is highly dynamic and heterogenous for the translation of the specific spatiotemporal subcellular mRNA pool (10). The regulatory network dedicated to achieving translation control is intriguingly complex, and mechanistic details are poorly understood to date.
In plants, ribosomes are found in three subcellular compartments, the cytosol, chloroplasts, and mitochondria. Due to their prokaryotic origin (11), organelles perform protein synthesis via bacterial-type 70S ribosomes. However, after the endosymbiotic event, both chloroplast and mitochondrial ribosomes increased their size and complexity to accommodate specific organellar tasks (12,13). The proteinaceous part of plastid ribosomes diversified from prokaryotic ribosomes, which led to a loss of Rpl25 and Rpl30 in most plant species and an acquisition of so-called ‘plastid-specific ribosomal proteins’ (PSRPs) (12). About one-third of all chloroplast ribosomal proteins are encoded by the chloroplast genome (plastome), whereas the remaining proteins are post-translationally imported from the cytosol. Similarly, multiple other major chloroplast protein complexes such as the photosynthetic complexes in the thylakoid membrane contain subunits of both genetic origins (e.g. 14). Thus, orchestrating plastid protein complex assembly requires substantial regulation of gene expression. Such coordination and the need to quickly respond to environmental cues is achieved by predominant post-transcriptional and translational regulatory strategies (12,15). Major players in this regulation include nuclear-encoded ’Organelle Trans-Acting Factor’ families, which frequently contain degenerated amino acid motifs of tandem repeats termed tetra-, penta- and octotricopeptide repeats (TPRs, PPRs and OPRs), respectively (16,17). Such proteins control maturation and stability of specific plastid transcripts and their translation activation. Over recent years, several of these proteins were described and mainly exhibit specific functions during the expression of one specific target transcript (18). Co-translational regulation of the protein synthesis rates might be the key step for fine-tuning gene expression in chloroplasts in order to quickly respond to external cues, such as changes in light or temperature exposure. For example, only mild changes of transcript levels, but profound changes in protein synthesis were observed upon environmental alteration (19–21), and during plant development (22). Furthermore, various ribosome profiling approaches of chloroplast translation reported severely fluctuating elongation speed over individual open reading frames interrupted by short pauses, which may reflect processing or insertion of nascent polypeptides into the thylakoid membrane (21,23–26). Certainly, the regulation of chloroplast translation is highly elaborate and dynamic, however, a deeper understanding of the regulatory principles requires a comprehensive knowledge of ribosome-associated factors.
To address this, we here established a novel technique for the fast and specific isolation of ribosomes from Chlamydomonas reinhardtii (Chlamydomonas hereafter) chloroplasts. By specifically engineering affinity tags into chloroplast-encoded ribosomal proteins of the large and the small subunit, respectively, we revealed the interaction network of chloroplast ribosomes through high resolution affinity purification-mass spectrometry (AP-MS). We uncovered a large number of proteins of known and unknown function that associate with chloroplast ribosomes, even including several transiently interacting proteins such as trans-acting factors. Through our AP-MS via tagged ribosomal proteins of the small and the large ribosomal subunit, we could attribute putative binding sites at ribosomes and further describe interactors of the assembled 70S subunit. Subsequently, we validated several of these factors, including a novel ribosome-associated enzyme which may acetylate the N-terminus of nascent chains. The data accentuate the multiple layers of control which fine-tune protein synthesis in plastids.
MATERIALS AND METHODS
Cells and culture conditions
For the construction of the Rpl5-HA (L5-HA) and Rps3-HA (S3-HA) lines, cw15 mt- strain CC4533 was used (27). For the nuclear expression of HA-tagged candidate proteins, UVM4 was used (28). Cw15 CF185 (29) was used for polysome gradients and ribosome binding assays. For all biological replicates, cultures were grown independently. If not stated elsewhere, cells were grown photomixotrophically in TAP Medium (30) on a rotary shaker at 25°C and under an illumination of 50–60 μmol of photons m−2 s−1. For polysome analyses, cells were grown under 30 μmoles of photons m−2 s−1. For experiments with FA crosslinking, cells were grown in HAP-medium containing 20 mM HEPES, 17.5 mM acetate, 1 mM K-phosphates, Beijerinck salts (7 mM NH4Cl, 0.34 mM CaCl2 and 0.71 mM MgSO4) and trace salt solution as in (30).
Plasmid construction and genomic integration
Genomic integration of the triple HA-tag coding sequence at the 3′-end of the rpl5 and rps3 coding sequences, respectively, was achieved via homologous recombination at the endogenous locus of the chloroplast genome by using the plasmid pUCatpXaadA (31). Cloning of HA-tagged cpNAT1 was achieved with the MoClo strategy (32). For heterologous expression of cpNAT1, the coding sequence of Cre14.g614750 (lacking the sequence for the putative N-terminal 57 amino-acid transit peptide) was synthesized (IDT) and cloned into NdeI/EcoRI digested pTyb21 (NEB), giving pFW214. Protein expression and purification of cpNAT1 was performed according to published protocols (33) (all details see Supplemental Methods). All primers are listed in Supplemental Table S2.
Isolation of affinity-tagged ribosomes
All replicates were conducted with independent cultures and on separate days, respectively. Cells were grown in logarithmic phase and were pretreated for 5 min with 100 μg/ml (w/v) chloramphenicol (CAP) or 100 μg/ml (w/v) puromycin, respectively. Formaldehyde was added to 0.37% (v/v) final concentration and cells were kept for an additional 10 min in light. Crosslinking was quenched by addition of 100 mM Tris–HCl pH 8.0 for 5 min and cells were harvested via rapid cooling over plastic ice cubes and agitated until the temperature dropped to 4°C. Cells were pelleted at 4000 g and 4°C for 2 min and washed in lysis buffer (50 mM HEPES pH 8.0, 25 mM KCl, 25 mM MgCl2, 25 mM EGTA, 1 mM PMSF and 100 μg/ml CAP, or 800 mM of KCl and 100 μg/ml puromycin instead of CAP for the high salt condition). Before lysis, 200 μg/ml Heparin and 100 U SUPERase·In™ RNase Inhibitor (Invitrogen, Thermo Fisher Scientific) were added to each sample, except for the RNase digest samples. Cells were lysed in the respective lysis buffer including protease inhibitors (cOmplete™ EDTA-free Protease Inhibitor Cocktail, Roche and 1 mM PMSF) by pressure homogenization at 3 bar. After lysis, 1% (w/v) n-dodecyl-β-maltoside was added and incubated for 5 min rotating at 4°C. The lysates were precleared by 15 min centrifugation at 4°C and 15 000 g. For the RNase treatment, the RNA concentration was measured with a photometer and afterwards 1 U Ambion® RNase I (Thermo Fisher) per μg of RNA and 0.02 U TURBO™ DNase (Thermo Fisher) per μl lysate were added. Affinity purification was done with Pierce™ anti-HA magnetic beads (Thermo Scientific) for 90 min at 4°C and constant gentle mixing. Beads were thoroughly washed three times with ice-cold HKM-T buffer containing 50 mM HEPES pH 8.0, 25 mM KCl, 25 mM MgCl2 and 0.05% (v/v) Tween20 and another three times with the same buffer lacking Tween20. During the first washing step, 10 μl of SUPERase·In™ were added to the RNAse treatment samples and incubated for 10 min to quench the RNase. Proteins were eluted with 2× SDS-PAGE buffer (125 mM Tris–HCl pH 6.8, 20% (v/v) glycerol, 4% (w/v) SDS, 0.005% (w/v) bromphenol blue) and incubated for 5 min at 96°C. After transfer into fresh tubes, protein crosslinks were reverted by additional incubation for 5 min at 96°C in the presence of 0.1 M DTT.
Mass-spectrometric analysis
HA-affinity purification samples were briefly separated by 10% SDS-PAGE and excised into a low molecular weight (<55 kDa) and a high molecular weight (>55 kDa) gel slice. Tryptic digest and peptide elution were described before (34). Samples were treated as described in Supplemental Methods and were directly injected into a Q Exactive HF spectrometer (Thermo Scientific). A 90 min gradient of 2–95% buffer B (80% acetonitrile, 0.5% formic acid) at a constant flow rate was used to elute peptides. Mass spectra were acquired in a data-dependent fashion using a top15 method for peptide sequencing. Raw data was processed with MaxQuant Version 1.6.3.3 using default parameters (35). MS/MS spectra were searched against a Chlamydonomas database (https://phytozome.jgi.doe.gov/pz/portal.html) concatenated with reverse copies of all sequences and a list of amino acid sequences of frequently observed contaminants (minimal peptide length = 7, minimal peptide = 1 (razor or unique), PSM FDR = 0.01). Label-free quantification (minimal ratio count = 2) and ‘match between runs’ (matching time window = 0.7 min, alignment time window = 20 min) was enabled (35). All raw files, MaxQuant results and parameter files are available at ProteomeXchange (see Data Availability).
Statistical analysis
MS data was analyzed with Perseus version 1.6.3.2. (36). All biological replicates were grouped and Log2 Label-Free Quantification (LFQ) intensities (Supplemental Dataset) were filtered to contain valid values in at least two of the three replicates in at least one group of each comparison. To enable statistical evaluation, missing values were imputed with random numbers drawn from a normal distribution with a mean (m) and standard deviation (sd) chosen to best simulate low abundance values close to the detection limit of the instrument (m = mmeasured – 1.5, sd = sdmeasured × 0.5; for each replicate m and sd were calculated based on all measured log2 LFQ intensities). A modified t-test implemented in the Perseus software package was used to identify proteins with significantly enriched LFQ intensity in the HA pulldown reactions compared to control pulldown. Results were calculated for two significance thresholds, FDR <5%, S0 = 1 or FDR <1%, S0 = 1, respectively. All results are listed in Supplementary Datasets. Subcellular localization and domain prediction for the whole Chlamydomonas proteome was initially obtained via the functional annotator web tool (https://github.com/CSBiology/FunctionalAnnotatorWeb). Localizations were further verified by the PredAlgo results found on the most recent Chlamydomonas genome version V5.6 (https://phytozome-next.jgi.doe.gov) (37). If ambiguous localizations were predicted, further predictions were performed with ChloroP (38), and compared to Arabidopsis homologs, if available. For proteins, which had no clear functional annotation in the recent Chlamydomonas genome version, possible annotations were searched for homologous proteins (e.g. in the Arabidopsis genome or via Basic Local Alignment Search Tool, BLAST, search). In addition, several known ribosome-associated proteins of previous bacterial studies were BLAST searched against the Chlamydomonas genome. The correlation coefficients were calculated and visualized in Perseus. LFQ intensities were filtered as described above and the correlation coefficient was calculated using the coefficient of determination (R2) function, comparing all experiments and replicates against each other. Arabidopsis homologs were extracted via https://phytozome-next.jgi.doe.gov.
In vitro NAT activity assay
General N-α-acetylation and ϵ-lysine-acetylation activities were measured as described before (39). For specific N-α-acetylation of chloroplast-encoded proteins, a custom-made peptide was designed corresponding to the six N-terminal amino acids of the Arabidopsis thaliana PS II reaction center protein D2 (ATCG00270) fused to an arginine-rich sequence resembling the human adrenocorticotropic hormone (MTIALGRFRWGRPVGRRRRPVRVYP). The hydrophilic sequence facilitates peptide solubility and effective enrichment via sepharose beads, according to (40). The PS II reaction center protein D2 was selected as a target based on the previously elucidated substrate specificity of the plastid N-terminal acetyltransferase NAA70 (41). Furthermore, this MTIA N-terminus of D2 is conserved in Chlamydomonas and Arabidopsis. To determine the activity of the cpNAT1, 3–16 μg (81–324 pmol) of purified enzyme was mixed with 0.2 mM of a custom-made peptide (GeneCust), 0.2% BSA in acetylation buffer (50 mM Tris–HCl, pH 7.5, 8 mM EDTA, 1 mM DTT) and 45 μM [3H]-acetyl-CoA (7.4 GBq/mmol, Hartmann Analytics). The reaction mix was filled up to 0.1 ml with acetylation buffer and incubated at 37°C for 0.5–2 h. Subsequently, the samples were centrifuged at 1500 g for 4 min. To isolate the custom-made peptide, the supernatant was mixed with 0.1 ml SP sepharose (50% in 0.5 M acetic acid) and incubated for 5 min while shaking. After 4 min of centrifugation at 1500 g, the pellet was washed three times with 0.4 ml 0.5 M acetic acid and once with 0.4 ml 100% methanol. The amount of incorporated [3H] label was measured with a Tri-Carb 2810TR scintillation counter (PerkinElmer).
Miscellaneous
Immunofluorescence was described in (33). Primary antibodies were against HA and uL1c in 1:5000 and 1:2500 dilutions in PBS–BSA, respectively. For secondary detections, a 1:200 dilution of the tetramethylrhodamine-isothiocyanate (TRITC)-labeled goat anti-rabbit antibody or fluorescein isothiocyanate (FITC) goat anti-mouse antibody (Invitrogen, Thermo Fisher Scientific) was used, respectively. Before imaging, slides were rinsed three times with PBS followed by addition of mounting solution containing DAPI (Vectashield). Images were taken with an Olympus BX53 microscope containing the filters for TRITC and FITC and an Olympus DP26 color camera. Ribosome co-sedimentation and polysome analysis was done according to Rohr et al. (34). For SDS-PAGE loading, protein samples were adjusted based on equal protein concentrations determined by Bradford (Biorad) or BCA (Pierce) according to the manufacturer's manual. SDS-PAGE and immunoblotting was performed as published before (42). Immunodetection was done with enhanced chemiluminescence and the FUSION-FX7 Advance imaging system (PEQLAB). All antibodies used are listed in Supplemental Table S3. Chlamydomonas cpNAT1 was modelled with full-length amino acid sequences using the SWISS-MODEL and RaptorX server. The models were analyzed, and figures generated with UCSF Chimera (43).
RESULTS
Targeted isolation of chloroplast ribosomes and AP-MS
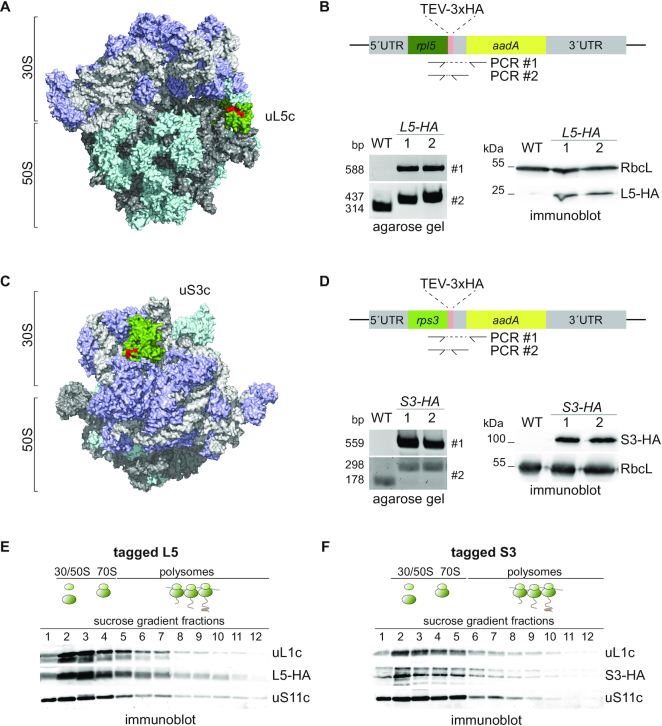
For the fast and efficient isolation of chloroplast ribosomes and their putative interactors, three repeats of a hemagglutinin (HA) affinity tag were engineered C-terminally to the chloroplast-encoded ribosomal proteins uL5c or uS3c of Chlamydomonas cells. By introducing the tag at the endogenous locus within the plastome, expression of the two ribosomal proteins remained unaltered. Expression and correct integration into all copies of the chloroplast genome was verified by immunoblotting and PCR, respectively (Figure 1A–D). Polysome profiling and growth assays confirmed that the tagged proteins assembled into fully functional ribosomes (Figure 1E and F). Even under stress conditions, such as high light or elevated temperature, both tagged strains did not show any growth defects compared to wild-type strains (Supplemental Figure S1). Thus, we were able to obtain fully functional tagged plastid ribosomes suitable for the interactome analysis.
Figure 1.
Endogenous tagging of chloroplast ribosomal proteins. (A and C) Surface-plot model of the chloroplast ribosome based on PDB file 5MMM (59). Ribosomal RNA is colored in light and dark gray, ribosomal proteins of the 30S and 50S are highlighted in purple and turquoise, respectively. uL5c (A) and uS3c (C) are highlighted in green with their surface exposed C-terminal 10 amino acids in red. (B and D) Design of the constructed DNA cassette for introduction of a 3xHA tag at the endogenous plastome locus of rpl5 (B) and rps3 (D) via homologous recombination. Correct integration was tested by PCR with oligos covering the 3′-coding sequence of the target gene and the adjacent resistance marker aadA (#1, respectively). The homoplasmic state of transformants was verified via PCR with oligos covering the 3′-coding sequence of rpl5 and the 3′ UTR of rpl5 and rps3, separating the native rpl5 or rps3 coding sequence from aadA (#2, respectively). Immunoblots with HA and RbcL antisera show expression of tagged Rpl5 or Rps3. (E and F) Polysome analysis and immunoblotting of sucrose gradient fractions with anti-HA antibody shows that L5-HA (E) and S3-HA (F) are integrated into translating ribosomes, respectively. As control, sucrose gradient fractions are detected for uL1c and uS11c. Expected positions of unassembled subunits including monosomes and polysomes in the gradient are illustrated by cartoons above the blots (n = 3).
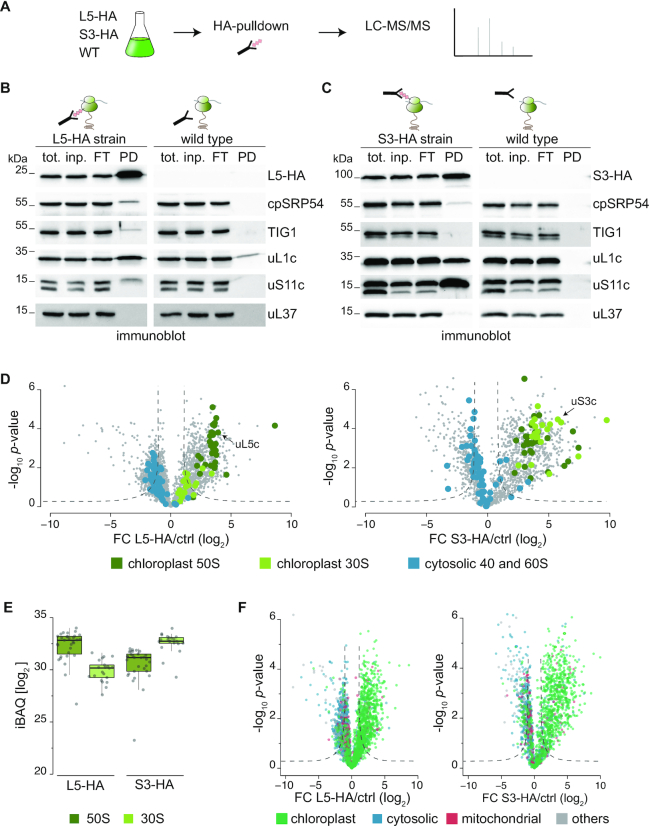
Next, we tested, whether affinity pulldown assays could yield pure and functional ribosome-nascent chain complexes (RNCs). All experiments were conducted in parallel with pulldowns from cell lysates of the L5-HA, or the S3-HA strains and the untagged parent wild type as control (Figure 2A). Chloroplast translation was arrested by treatment with chloramphenicol and brief in vivo crosslinking with 0.37% formaldehyde to maintain weak and transient interactions. Proteins were extracted in the presence of the detergent n-dodecyl-β-d-maltoside in order to yield ribosomes located in the soluble stroma and the thylakoid fraction. Immunoblots of pulldown eluates from L5-HA, S3-HA and the wild-type lysates showed that proteins of the 50S (uL1c) and 30S (uS11c) chloroplast ribosomal subunits co-eluted during affinity purification while little or no cytosolic ribosome co-purified (Figure 2B and C, see panels for the cytosolic ribosomal protein). Importantly, the two known chloroplast ribosome-associated nascent chain processing factors trigger factor (TIG1) and cpSRP54 (signal recognition particle 54) did specifically bind to purified ribosomes, indicating that the approach yielded intact RNCs (Figure 2B and C). By contrast, no signal was detected within pulldown eluates from untagged cells for all tested proteins, except for a weak background in the case of uL1c.
Figure 2.
AP-MS for the identification of the chloroplast ribosome interactors. (A) Schematic workflow of the affinity purification-mass spectrometry (AP-MS) approach. Experiments with HA-tagged strains and the respective untagged WT were done in parallel. Before harvest, translation was arrested by addition of chloramphenicol and formaldehyde crosslinking in vivo. Anti-HA affinity purification was performed from detergent-solubilized whole-cell lysates, depleted of cell debris. All experiments were performed in three biological replicates (for correlations see Supplemental Figure S2). (B and C) Test for the specific co-precipitation of functional ribosomes (uL1c and uS11c) and associated factors (TIG1 and cpSRP54) in L5-HA (B) or S3-HA (C) eluates by immunoblotting. Pulldown experiments from wild-type cells show minor background and eluates show no detectable co-elution of cytosolic ribosomes (uL37) (n = 3). (D) Volcano plots of the P-values versus enrichment (left panel: L5-HA pulldowns over pulldowns with untagged control, right panel: S3-HA pulldowns over pulldowns with untagged control). The P-values were determined by two-sided t-test, a minimal fold change S0 = 1, and a permutation-based FDR <0.05, with two valid values in first group. (E) Dot plot representing the distribution of iBAQ values of ribosomal proteins in pulldown eluates from L5-HA and S3-HA cells and untagged wild-type control. All iBAQs are averages of three independent biological replicates. (F) Volcano plots representing the predicted subcellular localization (based on the genome annotation) of proteins enriched in the L5-HA (left) and the S3-HA (right) datasets compared to proteins that were unspecifically purified with respective untagged wild-type samples. In all volcano plots, significant values are separated by dashed lines, FC = fold change.
To explore the interaction network of the chloroplast ribosomes, we employed state-of-the-art quantitative affinity-purification mass spectrometry (AP-MS), which allows comparison of protein assemblies under very mild pulldown conditions (44). In this approach, protein complexes are enriched, and experiments are normalized based on proteins that are bound non-specifically to the antibody-coupled beads in both the pull-down and control experiment (see Materials and Methods). With the high sensitivity of the Orbitrap mass spectrometer, in total more than 3,200 proteins were quantified using default MaxQuant parameters (Supplemental Dataset 1, see Methods for details). Protein identification in independent biological replicates was highly reproducible with R2 values of 0.89 for L5-HA and 0.97 for S3-HA (Supplemental Figure S2). All known 52 core proteins of the chloroplast ribosome were detected with multiple identified peptides in the HA purification experiments providing a proof of concept for the here applied affinity-based strategy for ribosome enrichment. With the S3-HA pulldown, several proteins of the large subunits were equally enriched, whereas in the L5-HA pulldown, fewer 30S proteins were enriched (Figure 2D, right versus left panel). Presumable the HA-tag of uL5c, which is located at the interface between the 50 and 30S subunits, might be less accessible in 70S ribosomes, compared to unassembled 50S. Importantly, of the 65 detectable cytosolic ribosomal proteins, only five were enriched in the S3-HA pulldown and none in the L5-HA pulldown, which demonstrates the selectivity of the applied AP-MS approach (Figure 2D). On average, ribosomal proteins of the large subunit had higher iBAQ values relative to proteins of the small subunit in the L5-HA pulldown, while S3-HA experiments showed the opposite trend (Figure 2E).
To define proteins significantly enriched in each pulldown compared to control experiments, we applied a modified t-test with a permutation-based false discovery rate cut-off (FDR < 0.05, S0 = 1). In this way, we identified 656 and 755 proteins significantly enriched in the L5-HA and the S3-HA pulldowns, respectively. The vast majority of these proteins were annotated to be localized in the chloroplast (∼82%, Figure 2F, Supplemental Figure S3A, Supplemental Dataset 1) and were selectively enriched by both strategies (see below). Application of more stringent cut-off values (FDR<0.01, S0 = 0.1) did not significantly increase the fraction of proteins with chloroplast annotation (Supplemental Figure S3B) suggesting that some non-plastid proteins were enriched due to ribosome binding after lysis (see below). For better classification, functional annotation was added to several chloroplast proteins, which were so far not described in the most recent genome version (see Materials and Methods).
Specific enrichment of proteins binding to 30S and 50S subunits
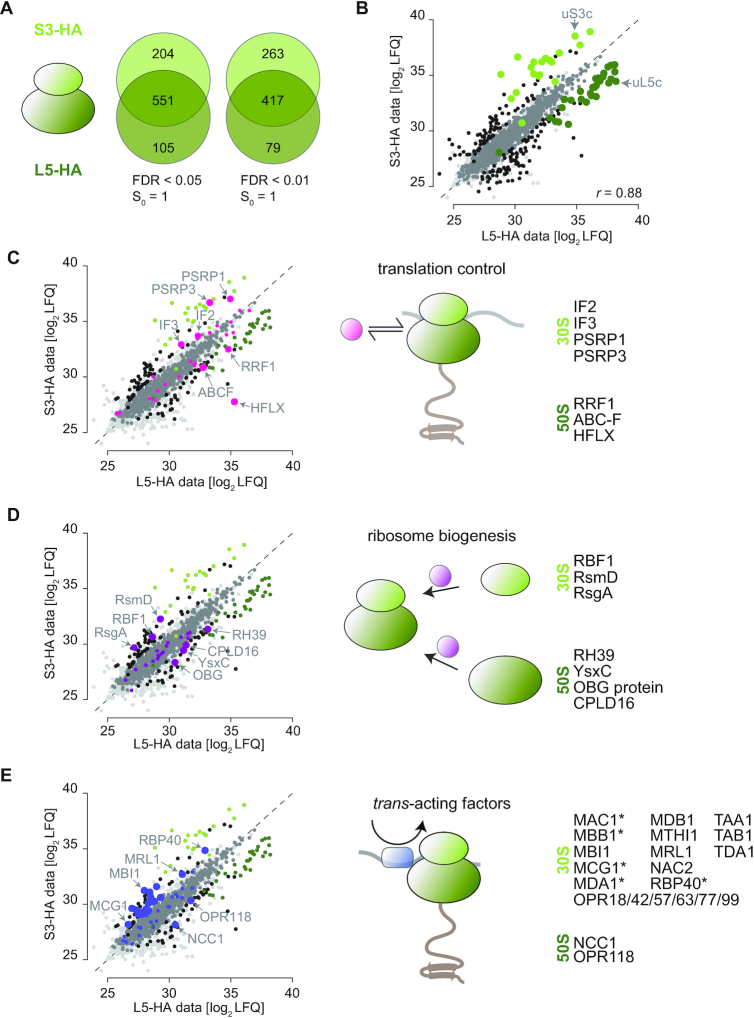
Direct comparison of enriched proteins in the L5-HA and S3-HA pulldowns showed an overlap of 551 proteins between the two datasets (84% and 73% of the proteins in the respective datasets; with a stringency of FDR < 0.05, S0 = 1; Figure 3A). Proteins that are only present in one of the datasets might be specific to unassembled ribosomal 30S or 50S or be randomly missed in one dataset due to low abundance or higher background values in the respective control. Therefore, we sought to delineate binding to either the 30S or the 50S subunit by only comparing those proteins, which had LFQ values in both pulldowns. By this, an interactor of 50S particles should be proportionately enriched in the L5-HA data when compared to S3-HA, while interactors of the assembled 70S should be similar in both datasets. According to (44), we compared all detected proteins of the L5-HA and S3-HA pulldowns, including unspecifically bound proteins, to demonstrate that overall LFQ intensities are highly comparable between the experiments (correlations of r = 0.88, Figure 3B). The bona fide ribosomal proteins of the 50S have significantly higher LFQ values in the L5-HA set, while ribosomal proteins of the 30S are over-represented in the S3-HA set (Figure 3B, Supplemental Figure S4, green dots). Importantly, several proteins with known interaction sites on the 30S or 50S, respectively, show the expected behavior in the direct comparison. For example, the canonical initiation factors IF2 and IF3 as well as the hibernation factor PSRP1 (plastid specific ribosomal protein 1) are significantly enriched in the S3-HA set when compared to the L5-HA set, which is due to their direct physical interaction with the 30S particle (12,45). In agreement with current knowledge, the ribosome recycling factor RRF1 and the 70S splitting factor HFLX are preferentially found after pulling down the L5-HA protein (46,47) (Figure 3C). Thus, stronger enrichment of newly identified proteins in either the L5-HA or the S3-HA pulldown suggests their respective place of interaction on chloroplast ribosomes. Interestingly, we enriched a protein of the ABC-F (ATP-binding cassette containing proteins) class on 50S particles. So far, the role of ABC-F proteins in chloroplasts are not known. However, this ABC-F protein shows homology to the energy-dependent translation throttle A protein EttA, which plays an important role in adjusting elongation kinetics in response to cellular energy levels (48). In total, we found four not-yet described plastid ABC-F proteins enriched in our ribosomal pulldown. A phylogenetic comparison revealed their homology to soluble bacterial-type ABC-F protein. Thus, it is possible that there is a whole class of ABC-F proteins exhibiting important regulatory functions in chloroplasts, orthologous to their putative roles in bacteria (49) (Supplemental Figure S5). We could also reveal several putative ribosome biogenesis factors as judged from homology to their bacterial counterparts, opening new avenues to study the poorly understood process of ribosome biogenesis in plastids. The chloroplast Ribosome-Binding Factor RBF1, processing the plastid 16S rRNA (50) as well as the putative ribosomal RNA small subunit methyltransferase (RsmD) and a homolog of the biogenesis factor RsgA (Ribosome small subunit-dependent GTPase A) were enriched on 30S particles but not on 50S particles. On the 50S subunit, the DEAD-box protein RH39 (RNA helicase 39), the GTPase YsxC, and the putative RNA methyltransferase CPLD16 were enriched (Figure 3D).
Figure 3.
Specific enrichment of proteins binding to 30S and 50S subunits. (A) Venn diagrams, presenting overlap and specific enrichment of the L5-HA and S3-HA dataset with two different filter stringencies. (B–E) Scatterplot of mean LFQ log2 values in the L5-HA and S3-HA dataset. Black: proteins with significant difference between the two datasets based on two-sided t-test with a permutation-based false discovery rate cut-off (FDR < 0.05, S0 = 1). Medium grey: proteins enriched in the L5-HA/S3-HA datasets versus untagged control. Light grey: background proteins with no enrichment. Dark green: ribosomal proteins of the 50S, which are enriched in the L5-HA dataset based on two-sided t-test. Light green: ribosomal proteins of the 30S, which are enriched in the S3-HA dataset. r-value is PEARSON correlation (C) Pink: proteins involved in plastid translation regulation enriched (large dots) or not enriched (small dots) between the two datasets. (D) Purple: proteins involved in plastid ribosome biogenesis enriched (large dots) or not enriched (small dots) between the two datasets. The non-characterized ABC-F protein is Cre06.g265100. (E) Blue: trans-acting factors enriched (large dots) or not enriched (small dots) between the two datasets. Proteins indicated with an asterisk are not in the ribosome interactome.
Furthermore, our ribosome isolation with the two respective bait proteins co-purified a total of 21 characterized and 21 uncharacterized plastid trans-acting factors (see Supplemental Table S1 for the known factors). Uncharacterized proteins were classified as trans-acting factors, based on the presence of canonical penta- or octotricopeptide repeat containing domains. Currently, most of these factors are thought to exhibit a transcript-specific regulatory function during translation initiation (18). Indeed, 20 of these factors were significantly enriched in the S3-HA dataset including some known and several yet undescribed OPR proteins (Figure 3E). Trans-acting proteins that are only enriched in the S3-HA dataset are highlighted (Figure 3E, asterisk), which may present factors that dissociate from the small subunit once initiation is completed. We also detected NCC1 (Nuclear Control of Chloroplast gene expression) and the uncharacterized OPR118 enriched on 50S particles, which might rather have a completely different function during translation. In fact, NCC1 seems to differ from other OPRs by lower specificity to a certain chloroplast transcript and a possible alternative function during translation (51).
Features of the chloroplast ribosome interactome
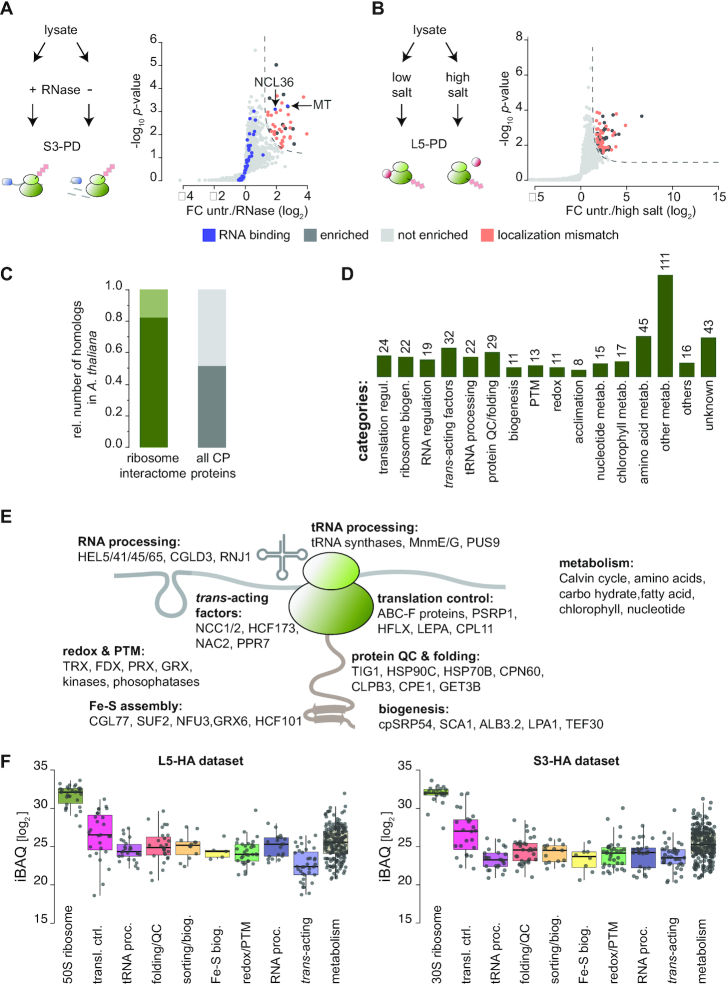
The nature of the plastid ribosome interactome was further investigated. To dissect if proteins directly bind ribosomes or rather co-purify via mRNA-binding, S3-HA pulldown samples were directly compared to parallel S3-HA pulldowns that were treated with RNaseI. All proteins that solely co-purify via the mRNA in the untreated sample should be depleted upon mRNA digestion. In fact, we observed that only 44 were clearly depleted upon RNAseI treatment (Figure 4A). Of these, six are annotated as RNA-binding proteins but not predicted to localize in plastids, and thus may have bound chloroplast ribosomes after lysis. Only two plastid mRNA binding proteins, NCL36 and a methyltransferase (MT) were significantly reduced by RNaseI treatment and thus apparently bind to translated mRNA. In addition, we performed a L5-HA pulldown with high ionic strength and puromycin to assay the stringency of protein interactions in the ribosome pulldown. Overall, enrichment scores were reduced in the ‘high salt’ pulldown (Supplemental Dataset 1), indicating that not all interactions were crosslinked to full saturation. Furthermore, several non-chloroplast localized proteins were significantly depleted, again indicating that those proteins attached to plastid ribosomes after cell lysis (Figure 4B). Upon salt wash, we observed that interactors of some categories were more depleted than others. For example, some of the trans-acting proteins or enzymes catalyzing post-translational modifications were low or even undetectable after high salt treatment. In contrast, the abundance of translation factors, protein targeting factors or many metabolic enzymes showed a similar decrease as the ribosomal proteins when the pulldown was carried out under high salt conditions (Supplemental Figure S6).
Figure 4.
Characteristics of chloroplast ribosome interactors. (A, B) Cartoons show schemes of the respective experimental procedures. Right-sided volcano plots display the enrichment at S0 = 1 and FDR <0.05 by comparison of untreated and RNaseI-treated S3-pulldowns (A) and low salt versus high salt-treated L5 pulldowns (B). Significant values are separated by dashed lines. FC = fold change. (C) Relative number of proteins with homologs in Arabidopsis found in the chloroplast ribosome interactome (dark green). For comparison, the relative number of all Chlamydomonas chloroplast proteins with homologs in Arabidopsis is shown (dark grey). All data of (C) and (D) are found in Supplemental Dataset 2. (D) Functional categories of the chloroplast ribosome interactome. Absolute numbers are given. (E) Cartoon of the chloroplast ribosome summarizing the functional categories of interactors and exemplary candidates. (F) Boxplots showing the abundance of putative ribosome-interactors in the L5-HA (left) and S3-HA (right) datasets as estimated by iBAQ distribution.
By overlapping the chloroplast-localized L5-HA and S3-HA dataset, while excluding ribosomal core proteins and RNase-sensitive proteins, we obtained a ‘chloroplast ribosome interactome’ of 438 proteins. Of this interactome, ∼82% of the identified proteins have orthologous forms in the land plant Arabidopsis thaliana (Arabidopsis), which is higher than the approximately 52% with homologs of the entire predicted Chlamydomonas chloroplast proteome (Figure 4C, Supplemental Dataset 2). This result suggests that the regulation of chloroplast translation is highly conserved within the green lineage. We classified these interactions in different categories. The functional groups go far beyond the expected categories of canonical translation regulation and ribosome biogenesis, including many factors that were previously not known to act in the context of chloroplast translation (Figure 4D) (see Discussion). By applying more stringent statistical parameters (FDR < 0.01, S0 = 0.1), none of the categories were disproportionally depleted from the ribosome interactome (Supplemental Figure S7). Comparison of iBAQ intensities of putative ribosome interactors from several functional categories shows that some groups are more abundantly associated with ribosomes compared to others. For example, translation regulators (e.g. initiation factors, elongations factors, and ABC-F proteins) or molecular chaperones are more abundant compared with trans-acting factors (Figure 4F). This could be explained by a higher specificity of the trans-acting factors for a ribosomal sub-pool, compared to a more general interaction of metabolic proteins or molecular chaperones.
We next examined the effect of in vivo crosslinking during AP-MS. Overall, crosslinking did not alter the polysome migration pattern in sucrose gradients (Supplemental Figure S8A). We furthermore performed an independent AP-MS experiment, comparing S3-HA purifications of crosslinked and non-crosslinked cell lysates. With the non-crosslinked S3-HA pulldown, still ∼70% of the chloroplast-localized factors of the crosslinked pulldown were enriched, albeit mostly with reduced enrichment scores (Supplemental Dataset 3). However, S3-HA pulldowns of non-crosslinked samples enriched more non-chloroplast localized proteins, which could be explained by increased dynamic exchanges of protein interactions upon cell lysis in these samples (Supplemental Figure S8B). The loss of putative ribosome interactors was found throughout functional groups, including canonical translation regulators and molecular chaperones as expected for transient interactors (Supplemental Figure S8C, D and Supplemental Data). There was also a reduction of abundant proteins belonging to metabolic pathways. This might suggest that some of these factors are false positives or that these proteins are secondary interactors with only transient interactions, peripheral to the ribosome. However, the here applied crosslinking AP-MS approach did not cause enrichment of abundant chloroplast-localized proteins (such as PSI, II, Cytb6f, CF0/1 ATPase), suggesting that it caused selective ribosome-attachment of proteins that were, in vivo, in close proximity to the ribosome.
Validation of selected ribosome-associated factors
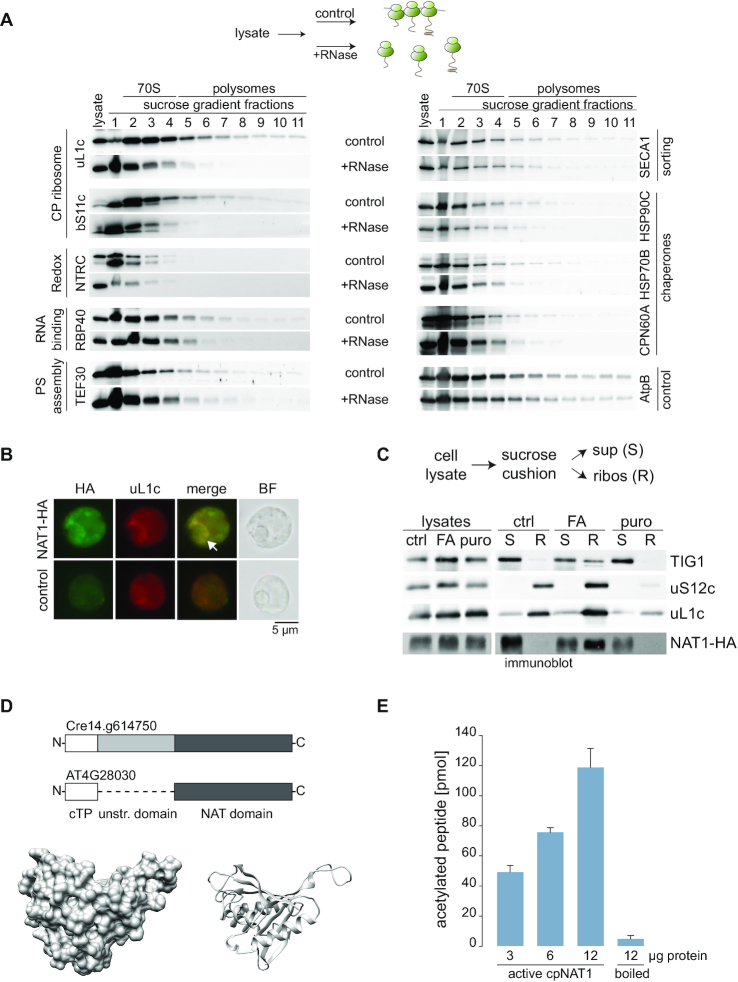
The plastid ribosome-association of 10 factors was independently confirmed to validate our AP-MS approach. In sucrose gradients, polysome co-migration was compared between untreated and RNaseI digested lysates. We applied RNaseI treatment to selectively shift the ribosome and its associated proteins from the polysomal to the monosomal fraction (scheme on top of Figure 5A). Immunological detection of ribosomal proteins and the plastid heat shock proteins HSP90C, HSP70B, the chaperonin CPN60A, the sorting factor SECA1, the PSII assembly factor TEF30 (thylakoid enriched fraction 30), and the trans-acting factor RBP40 (RNA binding protein 40) was substantially decreased in the polysomal fraction after RNaseI-treatment, demonstrating that these proteins were associated with translating ribosomes (Figure 5A). Moreover, puromycin treatment prior to polysome assays released nascent chain associated chaperones and SECA1 from polysomes as expected for chaperones assisting co-translational folding and sorting (Supplemental Figure S9) (34,52). The NADPH-Thioredoxin Reductase protein C, NTRC, was only detectable in fractions corresponding to monosomes or unassembled ribosomal subunits, both in the treated and untreated samples (Figure 5A). Thus, NTRC may act on or control the pool of non-translating ribosomes. As a control, the abundant CF1 ATPase subunit AtpB was plotted. Despite its migration into high molecular weight fractions in sucrose gradients, no profound shift was observed upon RNaseI treatment (Figure 5A), which agrees with the proteomics data that AtpB is not enriched in the ribosomal pulldowns. In addition, we confirmed ribosome association for selected newly identified interactors. We could show that HA-tagged putative phosphoribosylglycinamide formyltransferase and a ferredoxin thioredoxin reductase co-sedimented with ribosomes, at least after crosslinking and co-purified ribosomes in reverse affinity pulldowns (Supplemental Figures S10 and S11).
Figure 5.
Validation of identified proteins. Selected proteins were validated by polysome analysis of Chlamydomonas lysates. (A) Top, cartoon describing the experimental setup. Prior to harvest, translation was arrested by addition of chloramphenicol and formaldehyde crosslinking. RNaseI-treated and untreated samples were separated by sucrose density centrifugation and fractions were immunoblotted with the indicated antisera. Fractions containing monosomes or polysomes, respectively are marked above the blot (n = 4). (B) Intracellular localization of HA-tagged cpNAT1 and uL1c, as representative of chloroplast ribosomes, via immunofluorescence microscopy. Images were captured from cpNAT1-HA expressing cells (NAT-HA, top row) and UVM4 recipient strain (control, bottom row). Images from left to right: immunofluorescence using antibodies against the HA tag (FITC, green) and chloroplast-resident uL1c (TRITC, red), the merge of FITC and TRITC, and bright field (BF). The putative translation zone is marked with an arrow. Similar localization patterns were observed in 97 of 154 cells (63%). (C) Immunoblots of ribosome co-sedimentation assays and enrichment of cpNAT1 in the ribosomal fraction. Prior to harvest, translation was arrested by addition of chloramphenicol (ctrl), or addition of chloramphenicol and formaldehyde (FA). As control, release of nascent chains was mediated by incubation of lysates with 1 mM puromycin (puro). Pre-cleared cell lysates were layered on a 25% sucrose cushion (w/v) and separated into non-ribosome containing supernatant (S) and the ribosome pellet (R). Note that ‘R’ was enriched 10x compared to the sample loaded on the cushion. (D) Top: Scheme representing the domains of Cre14.g614750 and its homolog from Arabidopsis AT4G28030. White box is the chloroplast transit peptide (cTP), light grey box is the unstructured N-terminal domain, and dark grey box is the NAT domain. Bottom: Surface (left) and ribbon (right) presentation of modelled Chlamydomonas cpNAT1 based on PDB 1ghe of Pseudmonas amygdali pv.tabaci. For model parameters see Supplemental Table S4. (E) In vitro acetyltransferase activity of purified mature cpNAT1. Purified cpNAT1 was incubated for 1 h at 37°C with 45 μM [3H]acetyl-CoA and 0.2 mM of the synthetic MTIALGRFRWGRPVGRRRRPVRVYP peptide. After this incubation, the peptide was separated via SP-sepharose and the amount of incorporated [3H]acetyl in the peptide was quantified by scintillation counting. The unspecific binding of [3H]acetyl-CoA to the SP-sepharose was determined with 12 μg enzyme in the absence of peptide, and was subtracted from the measurements. As a negative control, the cpNAT1 was heat-inactivated at 95°C for 60 min (boiled). Data are presented as mean ± standard error (n = 3 for each enzyme concentration).
Identification of a ribosome-associated N-acetyltransferase in chloroplasts
We furthermore searched for a low abundant ribosome-associated protein in the dataset, to test if transient ribosome-binding can be confirmed for candidates with low enrichment scores. Cre14.g614750 is significantly enriched in the S3-HA pulldown but not in the L5-HA pulldown due to missing values (Supplemental Dataset 1). Cre14.g614750 is a putative acetyltransferase with homology to the Arabidopsis GCN5-related N-acetyltransferase (GNAT7, AT4G28030), which was recently discovered to be chloroplast-localized and active both on N-α (NTA) and ϵ-lysine (KA) amino groups (39). Since this is a promising candidate for yet-undescribed co-translational NAT function in organelles, we aimed to functionally characterize the protein (termed cpNAT1 hereafter). The full-length cpNAT1 sequence carrying a C-terminal triple HA-tag was expressed in Chlamydomonas cells (Supplemental Figure S10). Immunofluorescence microscopy confirmed chloroplast localization of cpNAT1-HA (Figure 5B). In fact, cpNAT1-HA showed a highly similar localization pattern like chloroplast ribosomes (as indicated by immunofluorescence of uL1c) with the strongest signal adjacent to the pyrenoid, displaying similar patterns like the T-zones, the spatiotemporal regions of photosystem biogenesis (53). Next, we independently confirmed the physical interaction of cpNAT1 with the ribosome by ribosome co-sedimentation assays and could demonstrate a similar behavior like the ribosome-associated chaperone TIG1 (34). Chemical crosslinking substantially increased the presence of cpNAT1-HA in ribosomal pellets suggesting an only transient interaction of cpNAT1 with the ribosome. Importantly, the puromycin-induced dissociation of RNCs entirely abolished sedimentation of cpNAT (Figure 5C), which is consistent with the action of cpNAT1-HA on the nascent chain extruding from the exit tunnel. In addition, affinity purification of cpNAT1-HA co-purified the ribosomal protein uL1c and the elongation factor TufA (Supplemental Figure S11C).
MatureChlamydomonas cpNAT1, lacking the predicted transit peptide, shares only 15% amino acid identity and 25% amino acid similarity with its mature counterpart in Arabidopsis (Supplemental Figure S12). However, homology modeling with RaptorX and SWISS-MODEL predicted a clear GNAT conformation of the segment, which includes residues Val142 to Leu328 and the conserved acetyl-CoA binding motif RxxGxG/A (Figure 5D and Supplemental Figure S12). This modeling result is consistent with previous reports on GNATs, which are diverse in amino acid sequence although their tertiary-structure is conserved (54). The most apparent difference is an additional N-terminal extension of 135 residues, which is not found in the GNAT7 of Arabidopsis (Supplemental Figure S12).
In order to determine if cpNAT1 indeed exhibits Nα-acetylation activity, we purified the predicted mature cpNAT1 after heterologous expression in E. coli (Supplemental Figure S13). As a model substrate, we selected a peptide covering the N-terminal amino acids MTIA of PsbD/D2, which are conserved in PsbDs/D2s of Chlamydomonas and Arabidopsis. This peptide sequence closely resembles the consensus sequence of N-terminally acetylated proteins that are encoded in the plastome (41). In the absence of the ribosome, the specific activity of the purified mature cpNAT1 on this substrate was 216 ± 45 pmol min−1 mg−1 (Figure 5E). This is about the same activity range that was recently reported for other Arabidopsis GNAT enzymes (39). Furthermore, the N-terminal acetylation of the substrate was strictly dependent on incubation time and the amount of purified enzyme (Supplemental Figure S14). CpNAT1 showed no NA activity on an N-terminal alanine as well as no KA activity on the peptide substrates used by Bienvenut et al. (39) (not shown).
DISCUSSION
More than 15 years ago, pioneer studies identified the core set of the chloroplast ribosome machinery, which is now well described in terms of composition and structure (45,55–60). Due to the inherent complexity and duration of separating chloroplast ribosomes from their cytosolic counterparts, only a small group of stably binding ribosome interactors are known from ribosome isolations. This study aimed to extend the existing data about chloroplast ribosomes by targeting interactors of the protein synthesis machinery. To get around classical ribosome purification procedures via sucrose gradient fractionation, we took advantage of Chlamydomonas for the convenient engineering of the chloroplast genome to map the chloroplast ribosome interacting network via AP-MS. Furthermore, expressing affinity-tagged ribosomal proteins from their endogenous locus avoids ectopic expression-induced side effects, such as false interactions caused by orphaned affinity tagged proteins or feed-back inhibition on translation due to impaired ribosome biogenesis (61).
Comparable to the interaction network obtained by AP-MS with cytosolic ribosomes in mouse embryonic stem cells (62), we found an versatile set of ribosome interactors from diverse functional categories (Figure 4D). This network goes well beyond the list of canonical factors that govern the three major phases of protein synthesis (i.e. initiation, elongation, and termination) and the folding of emerging polypeptides (e.g. molecular chaperones). The high degree of interconnectedness is not surprising given the high abundance of ribosomes in the complex and dense environment of a cell. In logarithmically growing E. coli cells, up to 70 000 70S ribosomes exist that constitute a third of the dry mass of the whole cell and reach a concentration of 70 μM (http://book.bionumbers.org). Recently, in-cell NMR spectroscopy showed that ribosomes engage in several quinary interactions and they might directly—maybe even in a non-translating fashion—affect several biochemical processes in a cell (63). In addition, ribosomes are highly dynamic and may exhibit spatiotemporal compositions that even vary within a single ribosome population and which is dedicated for the translation of a certain pool of transcripts.
Importantly, we observed a high overlap with earlier findings describing the soluble chloroplast protein complex profile of Arabidopsis (64,65). By resolving megadalton complexes with size exclusion chromatography and subsequent mass spectrometry, translational regulators, co-translational nascent chain processing proteins, ribosome biogenesis factors and RNA processors were previously found to co-migrate with plastid ribosomes (64). All comigrating proteins with homologs in Chlamydomonas are also present in the interactome of this study, confirming the strength of previous complexome profiles and validating the here described AP-MS approach. We also found subunits of the bacterial-type RNA polymerase subunits enriched in the ribosomal pulldowns, supporting earlier findings in land plants that plastid translation and transcription might be at least partially linked, comparable to the situation in bacteria (12,66,67). However, in comparison with the proteomics study of maize nucleoids (67), none of the orthologous proteins involved in DNA stability and organization were enriched in our dataset, suggesting that ribosome affinity purification did not co-purify full particles of nucleoids.
The strict translational regulation might be best exemplified by tight coupling of chloroplast protein synthesis with the diurnal dark/light cycles, which ensures that the highly energy demanding process of protein synthesis is supplied with sufficient energy. This control was postulated to be mediated by ‘biochemical light proxies’ compounds, comprising chlorophyll or intermediates of photosynthesis such as reduced plastoquinone, reduced thioredoxin or ATP/ADP levels (reviewed in 19). The redox state directly influences transcriptional dynamics in chloroplasts, and there are also ample hints for the redox-dependent regulation of translation (reviewed in 68). Here, we found several putative BLPs that may exhibit the task of light-dependent regulation such as thioredoxins of the x-, y- and f-type, NTRC and ferredoxins (Figure 4E). NTRC was already implicated in the cascade controlling the synthesis of PsbD (69). In yeast, a thioredoxin was shown to protect ribosomes against aggregation via the peroxiredoxin Tsa1 that exhibits chaperone function during oxidative stress (70). Orthologous mechanisms could be envisioned, for example through the enriched peroxiredoxin PRX1, protecting or regulating chloroplast translation during day and night. Such control of chloroplast translation is also consistent with the ‘colocation for redox regulation hypothesis’, stating that individual organelles need to sense and adjust their components based on the redox state of their own bioenergetic membranes (71,72).
A surprisingly high number of putative ribosome-associated proteins belonging to various metabolic pathways such as carbon, amino acid, chlorophyll or nucleotide metabolism were co-purified (Supplemental Dataset). This co-isolation seems puzzling, however, a similar report describing the ribosome interaction network in mammalian cells also found several metabolic enzymes, especially of glucose metabolism, in proximity to ribosomes (62). In fact, there is accumulating evidence in literature that several metabolic enzymes exhibit RNA-binding activities and thus actively contribute to gene expression, including the subunit of the chloroplast-localized pyruvate dehydrogenase complex, DLA2, which also co-purified in our ribosomal pulldown (73,74) (Supplemental Dataset 2). In bacteria, ribosomes engage with metabolic enzymes via quinary interaction of micromolar affinity. These interactions have a direct impact on metabolic activity since ribosomes were shown to both activate and inactivate specific classes of enzymes (63). Thus, similar spatiotemporal relationships between protein synthesis and metabolic pathways can be envisioned for chloroplasts. However, further studies are required to validate the direct linkage between metabolic pathways and translation.
Expectedly, we enriched those molecular chaperones and sorting factors in our ribosomal pulldown that act early on emerging nascent polypeptides, such as plastidic trigger factor, TIG1 (33,34,75). However, we found a surprisingly diverse set of additional molecular chaperones that were not yet known to act co-translationally. This set includes two HSP70s, the co-chaperone CDJ1, the CPN60 chaperonin complex, HSP90C and the HSP100 family protein CLPB3 (Supplemental Dataset 1). Several of these chaperones were validated for ribosome interaction by independent assays (Figure 5A). In addition, a chloroplast paralog of the Guided Entry of Tail-anchored protein 3, GET3b, was present in the ribosome interactome (Figure 4E). Recently, this protein was described in Arabidopsis and may exhibit a chaperone function in plastids (76). It has been previously demonstrated that a highly oxidative environment leads to a reversible transition of the cytosolic Get3 from an ATP-dependent targeting protein to an effective ATP-independent chaperone during stress situations (77). Thus, GET3b may act as a reactive oxygen species-activated ribosome-associated chaperone in chloroplasts. Taken together, the chloroplast ribosome-associated network seems more diverse compared to the situation in bacteria (reviewed in 5). The co-translational chaperone network in chloroplasts rather resembles the cytosolic co-translational chaperone network (1), which shows a diversification of the chloroplast folding network from their ancestors and might be an essential adaptation for processing of the more complex proteome topology within plant organelles.
The potential of our ribosome interactome as source for future mechanistic studies is indicated by the characterization of the N-acetyltransferase, cpNAT1. cpNAT1 is homolog to the recently described GNAT7 of Arabidopsis (39). Based on the ribosome association and its in vitro N-acetylation activity, cpNAT1 is a promising candidate performing co-translational N-acetylation in plastids of Chlamydomonas (Figure 5B-E). However, further studies are required to confirm its direct co-translational activity. To our knowledge this is the first report of a ribosome-associated N-acetyltransferase in chloroplasts. The Arabidopsis GNAT7 appears to target a rather broad substrate range concerning the alpha-amino groups (M, A, S, T, P, V) and it exhibits an additional ϵ-lysine-acetylation activity, which was not seen for Chlamydomonas cpNAT1 (39). The importance of co-translational Nα-acetylation for the protein fate of chloroplast-encoded proteins remains elusive. In Arabidopsis, NTA of stromal proteins is frequent but the role of NTA in affecting N-degron pathways is not established yet (78–80). Also in Chlamydomonas, NTA of chloroplast proteins was detected and stromal proteins with shorter half-lives seem to have less N-terminal acetylation when compared to stable proteins (81). In Citrullus lanatus, the Nα-acetylated form of the chloroplast-encoded ATP synthase subunit AtpE is more resistant against proteolysis during drought stress when compared with the non-acetylated proteoform (82). Furthermore, the abundance of the cytosolic ribosome-associated NatA complex is tightly regulated by the drought stress-related hormone ABA (83). NTA of cytosolic proteins by the ribosome-associated complexes NatA, NatB and NatE is also essential for the responses to pathogen-attack or high salt stress (84,85). Based on these results, co-translational and post-translational NTA is supposed to affect diverse stress responses in plants (86). Thus, it will be intriguing to investigate if cpNAT1 contributes to stress adaptation in chloroplasts by imprinting of chloroplast-encoded proteins with acetylation marks.
Taken together, the present description of the large spectrum of putative ribosome interactors bears potential for advancing our understanding of how protein biogenesis is orchestrated in cells. The set of enriched proteins will be the basis for several future studies to dissect specific mechanisms and quantify ribosomal compositions on a subcellular level. Importantly, many factors of the chloroplast ribosome interactome seem homologous between the green alga Chlamydomonas and land plants, thus suggesting a high evolutionary conservation of protein synthesis and its regulation in the green linage.
DATA AVAILABILITY
All gene numbers are concise with the GenBank/EMBL data libraries given in Supplemental Dataset. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (87) partner repository with the dataset identifier PXD020377 and PXD022210.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jean-David Rochaix for antibodies against Rps12 and Rpl37, Francis-Andre Wollman for antibodies against AtpB, and Michael Schroda for antibodies against HSP90C, HSP70B, SECA1 and TEF30 and for discussion on the data. We thank Karin Gries for technical assistance with protein purification and cloning.
Author contribution: L.D.W. designed and conducted all pulldown-related experiments and wrote parts of the manuscript; V.L.G., C.H., F.R., R.T., T.K. performed validation experiments. L.A. and J.S.M. designed and executed acetyltransferase activity towards the N-terminus or internal lysine residues, respectively. M.W. and I.F. supervised L.A. and J.S.M. and contributed to manuscript writing. S.R. and J.N. helped with chloroplast transformation; M.R. and Z.S. performed mass spectrometry measurements and helped with statistical analyses; F.W. designed experiments, acquired funding and wrote the manuscript
Contributor Information
Lisa Désirée Westrich, Molecular Genetics of Eukaryotes, University of Kaiserslautern, Paul-Ehrlich-Str. 23, 67663 Kaiserslautern, Germany.
Vincent Leon Gotsmann, Molecular Genetics of Eukaryotes, University of Kaiserslautern, Paul-Ehrlich-Str. 23, 67663 Kaiserslautern, Germany.
Claudia Herkt, Molecular Genetics of Eukaryotes, University of Kaiserslautern, Paul-Ehrlich-Str. 23, 67663 Kaiserslautern, Germany.
Fabian Ries, Molecular Genetics of Eukaryotes, University of Kaiserslautern, Paul-Ehrlich-Str. 23, 67663 Kaiserslautern, Germany.
Tanja Kazek, Molecular Genetics of Eukaryotes, University of Kaiserslautern, Paul-Ehrlich-Str. 23, 67663 Kaiserslautern, Germany.
Raphael Trösch, Molecular Genetics of Eukaryotes, University of Kaiserslautern, Paul-Ehrlich-Str. 23, 67663 Kaiserslautern, Germany.
Laura Armbruster, Centre for Organismal Studies, University of Heidelberg, Im Neuenheimer Feld 360, 69120 Heidelberg, Germany.
Jens Stephan Mühlenbeck, Plant Physiology, Institute of Plant Biology and Biotechnology, University of Münster, Schlossplatz 7, 48149 Münster, Germany.
Silvia Ramundo, Department of Biochemistry and Biophysics, University of California, 600 16th St, N316, San Francisco, CA 94143, USA.
Jörg Nickelsen, Department of Molecular Plant Science, University of Munich, Grosshaderner-Str. 2-4, 82152 Planegg-Martinsried, Germany.
Iris Finkemeier, Plant Physiology, Institute of Plant Biology and Biotechnology, University of Münster, Schlossplatz 7, 48149 Münster, Germany.
Markus Wirtz, Centre for Organismal Studies, University of Heidelberg, Im Neuenheimer Feld 360, 69120 Heidelberg, Germany.
Zuzana Storchová, Molecular Genetics, University of Kaiserslautern, Paul-Ehrlich-Str. 24, 67663 Kaiserslautern, Germany.
Markus Räschle, Molecular Genetics, University of Kaiserslautern, Paul-Ehrlich-Str. 24, 67663 Kaiserslautern, Germany.
Felix Willmund, Molecular Genetics of Eukaryotes, University of Kaiserslautern, Paul-Ehrlich-Str. 23, 67663 Kaiserslautern, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Carl-Zeiss fellowship (to F.R.); Deutsche Forschungsgemeinschaft grant [TRR175 to J.N., F.W.]; Forschungsschwerpunkt BioComp (to F.W.); Work on cpNAT1 was funded by the ERA-CAPS Research Programme ‘KatNat’ [WI 3560 4/1, FI 1655/4-1, to M.W., I.F.] and project 201348542 [SFB-1036 to M.W.]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pechmann S., Willmund F., Frydman J.. The ribosome as a hub for protein quality control. Mol. Cell. 2013; 49:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stein K.C., Frydman J.. The stop-and-go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J. Biol. Chem. 2019; 294:2076–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trösch R., Willmund F.. The conserved theme of ribosome hibernation: from bacteria to chloroplasts of plants. Biol. Chem. 2019; 400:879–893. [DOI] [PubMed] [Google Scholar]
- 4. Waudby C.A., Dobson C.M., Christodoulou J.. Nature and regulation of protein folding on the ribosome. Trends Biochem. Sci. 2019; 44:914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gloge F., Becker A.H., Kramer G., Bukau B.. Co-translational mechanisms of protein maturation. Curr. Opin. Struct. Biol. 2014; 24:24–33. [DOI] [PubMed] [Google Scholar]
- 6. Preissler S., Deuerling E.. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 2012; 37:274–283. [DOI] [PubMed] [Google Scholar]
- 7. Hinnebusch A.G. Translational control 1995-2015: unveiling molecular underpinnings and roles in human biology. RNA. 2015; 21:636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russell J.B., Cook G.M.. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 1995; 59:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costanzo M.C., Hogan J.D., Cusick M.E., Davis B.P., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C. et al.. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000; 28:73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li D., Wang J.. Ribosome heterogeneity in stem cells and development. J. Cell Biol. 2020; 219:e202001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Margulis L. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth. 1970; Yale University Press. [Google Scholar]
- 12. Zoschke R., Bock R.. Chloroplast Translation: Structural and functional organization, operational control and regulation. Plant Cell. 2018; 30:745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011; 155:1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maul J.E., Lilly J.W., Cui L., dePamphilis C.W., Miller W., Harris E.H., Stern D.B.. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell. 2002; 14:2659–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eberhard S., Drapier D., Wollman F.A.. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002; 31:149–160. [DOI] [PubMed] [Google Scholar]
- 16. Hammani K., Bonnard G., Bouchoucha A., Gobert A., Pinker F., Salinas T., Giege P.. Helical repeats modular proteins are major players for organelle gene expression. Biochimie. 2014; 100:141–150. [DOI] [PubMed] [Google Scholar]
- 17. Barkan A., Small I.. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014; 65:415–442. [DOI] [PubMed] [Google Scholar]
- 18. Nickelsen J., Bohne A.-V., Westhoff P.. Chloroplast gene expression - translation. Plastid Biology: Advances in Plant Biology. 2014; Springer; 49–78. [Google Scholar]
- 19. Sun Y., Zerges W.. Translational regulation in chloroplasts for development and homeostasis. Biochim. Biophys. Acta. 2015; 1847:809–820. [DOI] [PubMed] [Google Scholar]
- 20. Schuster M., Gao Y., Schöttler M.A., Bock R., Zoschke R.. Limited responsiveness of chloroplast gene expression during acclimation to high light in Tobacco. Plant Physiol. 2019; 182:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chotewutmontri P., Barkan A.. Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet. 2018; 14:e1007555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chotewutmontri P., Barkan A.. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 2016; 12:e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zoschke R., Watkins K.P., Barkan A.. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013; 25:2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zoschke R., Barkan A.. Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gawronski P., Jensen P.E., Karpinski S., Leister D., Scharff L.B.. Plastid ribosome pausing is induced by multiple features and is linked to protein complex assembly. Plant Physiol. 2018; 176:2557–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trösch R., Barahimipour R., Gao Y., Badillo-Corona J.A., Gotsmann V.L., Zimmer D., Mühlhaus T., Zoschke R., Willmund F.. Commonalities and differences of chloroplast translation in a green alga and land plants. Nat Plants. 2018; 4:564–575. [DOI] [PubMed] [Google Scholar]
- 27. Zhang R., Patena W., Armbruster U., Gang S.S., Blum S.R., Jonikas M.C.. High-Throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell. 2014; 26:1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neupert J., Karcher D., Bock R.. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009; 57:1140–1150. [DOI] [PubMed] [Google Scholar]
- 29. Schroda M., Vallon O., Wollman F.A., Beck C.F.. A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell. 1999; 11:1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D.. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011; 66:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of chlamydomonas. Nucleic Acids Res. 1991; 19:4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crozet P., Navarro F.J., Willmund F., Mehrshahi P., Bakowski K., Lauersen K.J., Perez-Perez M.E., Auroy P., Gorchs Rovira A., Sauret-Gueto S. et al.. Birth of a photosynthetic chassis: a MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018; 7:2074–2086. [DOI] [PubMed] [Google Scholar]
- 33. Ries F., Carius Y., Rohr M., Gries K., Keller S., Lancaster C.R.D., Willmund F.. Structural and molecular comparison of bacterial and eukaryotic trigger factors. Sci. Rep. 2017; 7:10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rohr M., Ries F., Herkt C., Gotsmann V.L., Westrich L.D., Gries K., Trösch R., Christmann J., Chaux F., Jung M. et al.. The role of plastidic trigger factor serving protein biogenesis in green algae and land plants. Plant Physiol. 2019; 179:1093–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M.. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 37. Tardif M., Atteia A., Specht M., Cogne G., Rolland N., Brugiere S., Hippler M., Ferro M., Bruley C., Peltier G. et al.. PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 2012; 29:3625–3639. [DOI] [PubMed] [Google Scholar]
- 38. Emanuelsson O., Nielsen H., von Heijne G.. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999; 8:978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bienvenut W.V., Brunje A., Boyer J.B., Muhlenbeck J.S., Bernal G., Lassowskat I., Dian C., Linster E., Dinh T.V., Koskela M.M. et al.. Dual lysine and N-terminal acetyltransferases reveal the complexity underpinning protein acetylation. Mol. Syst. Biol. 2020; 16:e9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Evjenth R., Hole K., Karlsen O.A., Ziegler M., Arnesen T., Lillehaug J.R.. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J. Biol. Chem. 2009; 284:31122–31129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dinh T.V., Bienvenut W.V., Linster E., Feldman-Salit A., Jung V.A., Meinnel T., Hell R., Giglione C., Wirtz M.. Molecular identification and functional characterization of the first Nalpha-acetyltransferase in plastids by global acetylome profiling. Proteomics. 2015; 15:2426–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Willmund F., Schroda M.. HEAT SHOCK PROTEIN 90C is a bona fide Hsp90 that interacts with plastidic HSP70B in Chlamydomonas reinhardtii. Plant Physiol. 2005; 138:2310–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 44. Keilhauer E.C., Hein M.Y., Mann M.. Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE-MS) rather than affinity purification mass spectrometry (AP-MS). Mol. Cell. Proteomics. 2015; 14:120–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boerema A.P., Aibara S., Paul B., Tobiasson V., Kimanius D., Forsberg B.O., Wallden K., Lindahl E., Amunts A.. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat Plants. 2018; 4:212–217. [DOI] [PubMed] [Google Scholar]
- 46. Agrawal R.K., Sharma M.R., Kiel M.C., Hirokawa G., Booth T.M., Spahn C.M., Grassucci R.A., Kaji A., Frank J.. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:8900–8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blombach F., Launay H., Zorraquino V., Swarts D.C., Cabrita L.D., Benelli D., Christodoulou J., Londei P., van der Oost J.. An HflX-type GTPase from Sulfolobus solfataricus binds to the 50S ribosomal subunit in all nucleotide-bound states. J. Bacteriol. 2011; 193:2861–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boel G., Smith P.C., Ning W., Englander M.T., Chen B., Hashem Y., Testa A.J., Fischer J.J., Wieden H.J., Frank J. et al.. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat. Struct. Mol. Biol. 2014; 21:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murina V., Kasari M., Takada H., Hinnu M., Saha C.K., Grimshaw J.W., Seki T., Reith M., Putrins M., Tenson T. et al.. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic Resistance: Structural and functional diversification across the tree of life. J. Mol. Biol. 2018; 431:3568–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fristedt R., Scharff L.B., Clarke C.A., Wang Q., Lin C., Merchant S.S., Bock R.. RBF1, a plant homolog of the bacterial ribosome-binding factor RbfA, acts in processing of the chloroplast 16S ribosomal RNA. Plant Physiol. 2014; 164:201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boulouis A., Drapier D., Razafimanantsoa H., Wostrikoff K., Tourasse N.J., Pascal K., Girard-Bascou J., Vallon O., Wollman F.A., Choquet Y.. Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell. 2015; 27:984–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Teter S.A., Houry W.A., Ang D., Tradler T., Rockabrand D., Fischer G., Blum P., Georgopoulos C., Hartl F.U.. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999; 97:755–765. [DOI] [PubMed] [Google Scholar]
- 53. Sun Y., Valente-Paterno M.I., Bakhtiari S., Law C., Zhan Y., Zerges W.. Photosystem biogenesis is localized to the translation zone in the chloroplast of Chlamydomonas. Plant Cell. 2019; 31:3057–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vetting M.W., LP S.d.C., Yu M., Hegde S.S., Magnet S., Roderick S.L., Blanchard J.S.. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005; 433:212–226. [DOI] [PubMed] [Google Scholar]
- 55. Yamaguchi K. Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast Ribosome: Identification of a novel S1 Domain-Containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell Online. 2002; 14:2957–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamaguchi K., Beligni M.V., Prieto S., Haynes P.A., McDonald W.H., Yates J.R. 3rd, Mayfield S.P.. Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to th e70 S ribosome. J. Biol. Chem. 2003; 278:33774–33785. [DOI] [PubMed] [Google Scholar]
- 57. Yamaguchi K., Subramanian A.R.. The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 2000; 275:28466–28482. [DOI] [PubMed] [Google Scholar]
- 58. Yamaguchi K., Subramanian A.R.. Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. PSRP-2 (U1A-type domains), PSRP-3alpha/beta (ycf65 homologue) and PSRP-4 (Thx homologue). Eur. J. Biochem. 2003; 270:190–205. [DOI] [PubMed] [Google Scholar]
- 59. Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N.. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017; 36:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van de Waterbeemd M., Tamara S., Fort K.L., Damoc E., Franc V., Bieri P., Itten M., Makarov A., Ban N., Heck A.J.R.. Dissecting ribosomal particles throughout the kingdoms of life using advanced hybrid mass spectrometry methods. Nat. Commun. 2018; 9:2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gibson T.J., Seiler M., Veitia R.A.. The transience of transient overexpression. Nat. Methods. 2013; 10:715–721. [DOI] [PubMed] [Google Scholar]
- 62. Simsek D., Tiu G.C., Flynn R.A., Byeon G.W., Leppek K., Xu A.F., Chang H.Y., Barna M.. The mammalian Ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell. 2017; 169:1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. DeMott C.M., Majumder S., Burz D.S., Reverdatto S., Shekhtman A.. Ribosome mediated quinary interactions modulate In-Cell protein activities. Biochemistry. 2017; 56:4117–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Olinares P.D., Ponnala L., van Wijk K.J.. Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics. 2010; 9:1594–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Peltier J.B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A.J., Rutschow H., van Wijk K.J.. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics. 2006; 5:114–133. [DOI] [PubMed] [Google Scholar]
- 66. Pfalz J., Liere K., Kandlbinder A., Dietz K.J., Oelmüller R.. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006; 18:176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J.. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012; 158:156–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rochaix J.D. Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antioxid. Redox. Signal. 2013; 18:2184–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schwarz C., Bohne A.V., Wang F., Cejudo F.J., Nickelsen J.. An intermolecular disulfide-based light switch for chloroplast psbD gene expression in Chlamydomonas reinhardtii. Plant J. 2012; 72:378–389. [DOI] [PubMed] [Google Scholar]
- 70. Trotter E.W., Rand J.D., Vickerstaff J., Grant C.M.. The yeast Tsa1 peroxiredoxin is a ribosome-associated antioxidant. Biochem. J. 2008; 412:73–80. [DOI] [PubMed] [Google Scholar]
- 71. Maier U.G., Zauner S., Woehle C., Bolte K., Hempel F., Allen J.F., Martin W.F.. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol. Evol. 2013; 5:2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Allen J.F. The function of genomes in bioenergetic organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003; 358:19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bohne A.V., Schwarz C., Schottkowski M., Lidschreiber M., Piotrowski M., Zerges W., Nickelsen J.. Reciprocal regulation of protein synthesis and carbon metabolism for thylakoid membrane biogenesis. PLoS Biol. 2013; 11:e1001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Castello A., Hentze M.W., Preiss T.. Metabolic enzymes enjoying new partnerships as RNA-Binding proteins. Trends Endocrinol. Metab. 2015; 26:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ries F., Herkt C., Willmund F.. Co-Translational protein folding and sorting in Chloroplasts. Plants (Basel). 2020; 9:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xing S., Mehlhorn D.G., Wallmeroth N., Asseck L.Y., Kar R., Voss A., Denninger P., Schmidt V.A., Schwarzländer M., Stierhof Y.D. et al.. Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E1544–E1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Voth W., Schick M., Gates S., Li S., Vilardi F., Gostimskaya I., Southworth D.R., Schwappach B., Jakob U.. The protein targeting factor Get3 functions as ATP-independent chaperone under oxidative stress conditions. Mol. Cell. 2014; 56:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J.. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008; 3:e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rowland E., Kim J., Bhuiyan N.H., van Wijk K.J.. The Arabidopsis chloroplast stromal N-terminome: complexities of amino-terminal protein maturation and stability. Plant Physiol. 2015; 169:1881–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bouchnak I., van Wijk K.J.. N-Degron pathways in plastids. Trends Plant Sci. 2019; 24:917–926. [DOI] [PubMed] [Google Scholar]
- 81. Bienvenut W.V., Espagne C., Martinez A., Majeran W., Valot B., Zivy M., Vallon O., Adam Z., Meinnel T., Giglione C.. Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics. 2011; 11:1734–1750. [DOI] [PubMed] [Google Scholar]
- 82. Hoshiyasu S., Kohzuma K., Yoshida K., Fujiwara M., Fukao Y., Yokota A., Akashi K.. Potential involvement of N-terminal acetylation in the quantitative regulation of the epsilon subunit of chloroplast ATP synthase under drought stress. Biosci. Biotechnol. Biochem. 2013; 77:998–1007. [DOI] [PubMed] [Google Scholar]
- 83. Linster E., Stephan I., Bienvenut W.V., Maple-Grodem J., Myklebust L.M., Huber M., Reichelt M., Sticht C., Moller S.G., Meinnel T. et al.. Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat. Commun. 2015; 6:7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Armbruster L., Linster E., Boyer J.B., Brunje A., Eirich J., Stephan I., Bienvenut W.V., Weidenhausen J., Meinnel T., Hell R. et al.. NAA50 is an enzymatically active Nalpha-acetyltransferase that is crucial for development and regulation of stress responses. Plant Physiol. 2020; 183:1502–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huber M., Bienvenut W.V., Linster E., Stephan I., Armbruster L., Sticht C., Layer D., Lapouge K., Meinnel T., Sinning I. et al.. NatB-Mediated N-Terminal acetylation affects growth and biotic stress responses. Plant Physiol. 2020; 182:792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Linster E., Wirtz M.. N-terminal acetylation: an essential protein modification emerges as an important regulator of stress responses. J. Exp. Bot. 2018; 69:4555–4568. [DOI] [PubMed] [Google Scholar]
- 87. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All gene numbers are concise with the GenBank/EMBL data libraries given in Supplemental Dataset. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (87) partner repository with the dataset identifier PXD020377 and PXD022210.