Abstract
Mammalian de novo DNA methyltransferases (DNMT) are responsible for the establishment of cell-type-specific DNA methylation in healthy and diseased tissues. Through genome-wide analysis of de novo methylation activity in murine stem cells we uncover that DNMT3A prefers to methylate CpGs followed by cytosines or thymines, while DNMT3B predominantly methylates CpGs followed by guanines or adenines. These signatures are further observed at non-CpG sites, resembling methylation context observed in specialised cell types, including neurons and oocytes. We further show that these preferences result from structural differences in the catalytic domains of the two de novo DNMTs and are not a consequence of differential recruitment to the genome. Molecular dynamics simulations suggest that, in case of human DNMT3A, the preference is due to favourable polar interactions between the flexible Arg836 side chain and the guanine that base-pairs with the cytosine following the CpG. By exchanging arginine to a lysine, the corresponding side chain in DNMT3B, the sequence preference is reversed, confirming the requirement for arginine at this position. This context-dependent enzymatic activity provides additional insights into the complex regulation of DNA methylation patterns.
INTRODUCTION
DNA methylation plays important roles during mammalian development and the perturbation of this mark is often associated with human disease. In mammals, DNA methylation is deposited by the de novo DNA methyltransferases DNMT3A and DNMT3B, while during replication, the maintenance methyltransferase DNMT1 ensures correct propagation of the methyl mark (1). Numerous genome-wide studies identified the exact position and tissue-specific dynamics of individual methyl groups on DNA. These revealed that the majority of CpGs in mammalian genomes are methylated, with the exception of active promoters and cell-type-specific enhancer elements (2,3). In addition to CpG methylation, non-CpG (or CpH) methylation has been identified in numerous tissues (2,4,5), with highest levels found in brain where it is suggested to contribute to gene regulation and neuronal function through readout by the methyl-CpG-binding protein MeCP2 (6–9).
Nevertheless, the mechanisms governing the precise deposition of DNA methylation to the genome remain to be fully understood. Although the mammalian genome is almost entirely methylated (2,3), several regions rely on active recruitment of de novo DNA methylation activity, including promoters, enhancers and CpG islands (10–12), repetitive elements (13,14), and transcribed gene bodies (15,16). These sites also show tissue-specific variability of DNA methylation and display altered methylation patterns in many diseases (3,8,17–20). These observations suggest that pathways that recruit de novo methylation vary from cell type to cell type and that the genome-wide methylation patterns are a composite picture resulting from combined activity of multiple DNMT-targeting mechanisms and mechanisms that actively or passively remove methylation (21). In previous work, we and others have shown that the de novo DNMTs associate with genomic regions that are marked by distinct chromatin modifications, resulting in enhanced deposition of methyl marks at these sites. For example, readout of H3K36me3 by DNMT3B targets DNA methylation to transcribed gene bodies (16,22), while DNMT3A shows increased preference for Polycomb-target sites and H3K36me2 (23–25).
Besides these differences in chromatin-dependent targeting of the de novo DNMT enzymes, biochemical studies suggest that DNMT3A and DNMT3B differ in their preference for CpGs based on flanking sequences. In in vitro methylation assays, DNMT3A has been found to show preferences towards CpGs flanked by pyrimidines (Y) at the +2 position, downstream of the methylated C, while studies using episomal constructs in cells indicate that DNMT3B shows higher methylation activity at sequences containing purines (R) at the same position (26–31). However, the complex interactions of DNMTs with chromatin and their site-dependent recruitment, it remains unclear if CpG-flanking preferences are observed genome-wide. Some indications come from the analysis of CpH methylation in various tissues. Non-CpG methylation is predominantly deposited by the de novo DNMTs (4,32–35) and tissues that have a predominant activity of either DNMT3A or DNMT3B show different CpH methylation flanking preferences. In tissues with high DNMT3A activity, like neurons or oocytes, the predominant CpH methylation motif occurs at CpApC, while in ES cells CpH methylation is predominantly deposited by DNMT3B and occurs at CpApG sites (2,20,35–37). Taken together, these results suggest that enzymatic activities of the de novo DNMTs could be influenced by local sequence context to shape the cell-type-specific methylomes.
Here, we have systematically interrogated the sequence preference of the de novo DNA methyltransferases DNMT3A and DNMT3B in vivo. By re-evaluating available whole genome bisulphite datasets from cells expressing de novo DNMTs in Dnmt-triple-KO mouse embryonic stem cells lacking DNA methylation, we measured how sequence context influences deposition of methylation at CpG and CpH sites, genome-wide. We identify similarities and enzyme-specific preferences of methylation activity at cytosines in different sequence contexts. We show that DNMT3A prefers to methylate cytosines at TACGYC sites while DNMT3B prefers to methylate TACGRC, resembling results obtained in vitro or using episomal constructs (26–31). The same downstream preference is retained at non-CpG sites, with pronounced preferences for TACACC and TACAGC, respectively. Through analyzing the genome-wide distribution of flanking sequence preferences, and furthermore, by investigating methylation in cells with altered targeting of DNMT3A or DNMT3B, we show that these preferences are largely independent of the genomic location or chromatin-mediated recruitment of DNMTs. The analysis of the available crystal structures and atomistic simulations of the DNMT3A catalytic domain and a DNA duplex suggest a potential mechanism underlying this specificity, which relies on a flexible arginine side chain at position 836 in human DNMT3A (832 in mouse). We confirm the requirement for this arginine in mediating DNMT3A-specific preferences and show that replacing this side chain to the lysine found at the corresponding position in DNMT3B, reverts the flanking sequence preference.
MATERIALS AND METHODS
Cell line generation and cultivation
Dnmt1,3a,3b-triple-KO cells were obtained from (38). Cells were cultured on 0.2% gelatine-coated dishes in DMEM (Invitrogen) supplemented with 15% fetal calf serum (Invitrogen), 1× non-essential amino acids (Invitrogen), 1 mM l-glutamine, leukemia inhibitory factor, and 0.001% b-mercaptoethanol. DNMT3A2- and DNMT3B-expressing Dnmt-TKO cell lines were obtained by recombinase-mediated cassette exchange (RMCE) as previously described (16).
Whole-genome bisulphite library preparation and sequencing
Whole-genome bisulphite sequencing for TKO cells expressing DNMT3A and DNMT3B was performed as described (16). In brief, 6 ug sonicated, genomic DNA were pre-mixed with SssI-methylated phage T7 and unmethylated phage Lambda DNA (both 10 ng, sonicated) and sequencing libraries were prepared using the NEB ULTRA and ULTRAII kits following manufacturer's instructions for genomic DNA library construction and using methylated adaptors (NEB E7535S). Adaptor-ligated DNA was converted by sodium bisulphite using the Qiagen Epitect bisulphite conversion kit. Converted libraries were enriched by 10 cycles of PCR with the following reaction composition: 1 μl Pfu TurboCx Hotstart DNA polymerase (Stratagene), 5 μl PfuTurbo Cx reaction buffer, 25 μM dNTPs, 1 μl universal and 1 μl index primer. PCR cycling parameters were: 95°C for 2 min, 98°C for 30 s, then 10 cycles of 98°C for 15 s, 65°C for 30 s and 72°C for 3 min, ending with one 72°C for 5 min step. The reaction products were purified twice using Ampure-XT beads. Quality of the libraries and size distribution were assessed on an Agilent High Sensitivity tape station. Libraries were sequenced on Illumina HiSeq and NovaSeq machines. All newly generated datasets (Supplementary Table S1) have been deposited to Gene Expression Omnibus (GEO), under accession number: GSE151992.
Whole-genome bisulphite data processing
Published WGBS datasets obtained from GSE57411, GSE96529 and newly-generated WGBS data including mutant DNMT3A proteins and additional independent biological replicates (Supplementary Table S1) were individually pre-processed and aligned to the mouse genome. The reason for including additional replicates (new cell lines, WGBS libraries generated by different persons and sequenced on different machines) was to exclude that the observed sequence preferences could stem from unexpected biases in library preparation or sequencing technology. For bulk analysis of CpGpN sites, the sequencing samples were processed and aligned as described in (16). In brief, reads were first trimmed to 50nt using fastx_trimmer and then aligned to the mouse genome (mm9) using BOWTIE in QuasR qAlign() with standard options and bisulfite = ‘undir’ setting (39). Methylation calls for CpGs were obtained using the qMeth() function in QuasR in ‘CpGcomb’ mode and excluding CpGs overlapping with known SNPs. Bisulphite conversion efficiency was measured by spiked-in controls of methylated T7 DNA and unmethylated lambda DNA.
For detailed WGBS analysis using 8-mer motifs, the full-length sequencing reads were pre-processed and trimmed using cutadapt v1.16 (40) and sickle v1.33 (Joshi and Fass 2011, https://github.com/najoshi/sickle) to remove adapter sequences. Read quality was measured before and after processing using FastQC v0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Processed reads were mapped against the mm9 mouse assembly using bwa-meth v0.2.0 (41) running bwa v0.7.12-r1039 (42). Mapping quality was evaluated with qualimap v2.2.1 (43). Duplicates were removed with Picard MarkDuplicates (picard-tools v1.96, http://broadinstitute.github.io/picard/) with ‘REMOVE_DUPLICATES=TRUE and REMOVE_SEQUENCING_DUPLICATES=TRUE’ flags. Alignments with MAPQ > 40 were used as input to methylation calling with methyldackel v0.3.0-3-g084d926 (https://github.com/dpryan79/MethylDackel) (using HTSlib v1.2.1) in ‘cytosine_report’ mode for all cytosines in the genome both CpG and CpH contexts (‘extract -q 40 –cytosine_report –CHH –CHG’).
Sequence-dependent DNA methylation analysis
For global analysis of methylation preferences at CpGpN sites, we first calculated the methylation scores for individual CpG instances with min coverage = 10 and max coverage = 50 in the WGBS datasets. Methylation scores were obtained as: number of methylated reads/(number of methylated +unmethylated reads) per CpG. We obtained the CpGpN sequence context by extending the CpGs by one nucleotide downstream on the + strand and querying the mm9 sequence using the Biostrings package in R (https://bioconductor.org/packages/release/bioc/html/Biostrings.html). Average methylation scores were calculated for all CpGs falling in one of the four contexts: CpGpA, CpGpT, CpGpG or CpGpC. The same analysis was performed using only CpGpNs that were only covered in both datasets according to the criteria described above (TKO-DNMT3A2_r1 and TKO-DNMT3B1_r1) and CpGpN sites that are at least 80% methylated in wild type ES cells. To evaluate the genome-wide distribution of CpGpN methylation according to H3K36me3, the mouse genome was partitioned into 1kb, nonoverlapping intervals using GenomicRanges in R (44). Intervals overlapping with satellite repeats (Repeatmasker), ENCODE black-listed and low mappability scores (<0.5) were removed in order to reduce artefacts due to annotation errors and repetitiveness. First, we calculated the signal strength of H3K36me3 at each individual interval by counting the numbers of reads from available ChIP-seq experiments (45). All genomic intervals were first ranked based on the total number of H3K36me3 ChIP-seq reads. Then, the ranked intervals were binned into groups of 1000 intervals, resulting in a total of 730 bins. Within each bin, we calculated the average methylation falling into the four possible CpGpN sequence contexts for each WGBS sample.
For the 8-mer specific analysis, first a dictionary containing a complete set of 8-mer motifs for NNNCGNNN and NNNCHNNN was created. Next, we separately counted all methylated or unmethylated individual WGBS reads aligning in the corresponding orientation to every genomic instance containing the 8-mer of interest and calculated the methylation score. The low-expression of the re-introduced DNMT3 enzymes to TKO cells and the absence of DNMT1-mediated maintenance resulted in sparse methylation, with the majority of CpG sites lacking DNA methylation (Supplemental Figure S2B). To account for the sparse methylation, we defined the methylation status of each strand-specific 8-mer as methylated when at least one methylated read was present. These genomic instances of methylated and unmethylated 8-mers were then collapsed into a list containing total methylated and unmethylated instances per motif. This was done for each WGBS dataset individually, resulting in a data frame summarising methylated (M) and unmethylated (U) reads per motif. Methylation scores per motif were obtained by calculating M/(M+U) and were used for generating motif ranks. Position weight matrices were calculated and displayed using the seqLogo package in R. Nucleotide coupling analysis upstream and downstream of the methylated target site was calculated by first counting all methylated and unmetylated instances containing NNCX or CXNN 4-mers, where X = is either G or H depending on CpG of CpH context. Finally, the methylation score at all possible combinations at –2/–1 or +2/+3 sites was calculated as described above and represented as heatmaps.
To investigate the sequence preferences at genomic sites preferentially bound by DNMT3A or DNMT3B, we made use of genomic regions we previously defined as enriched for these two enzymes (23). Next, we used these regions to separately calculate the methylation at CpGpN sites as well as for all possible 8-mer motifs in presence of DNMT3A or DNMT3B.
Molecular dynamics system preparation
The crystal structure of human DNMT3A-DNMT3L in complex with double-stranded DNA containing two CpG sites (PDB ID: 5YX2) (46) was used as the structural basis of this study. We reduced the complex to one DNMT3A methyltransferase interacting with a 10-mer DNA, containing one CpG site, and an S-adenosyl-l-homocysteine (SAH) cofactor (Supplementary Figure S10B). For the simulations, the zebularine in the crystal structure was replaced by a cytosine methylated at C5. Furthermore, four nucleotides in the DNA sequence present in the crystal structure (two nucleotides preceding and two following the methylation site) were mutated to accommodate the observed preferences of DNMT3A and DNMT3B (Supplementary Figure S10A). The mutation of the nucleotides and the reconstruction of the missing residues in the crystal structure were performed using the Maestro software (Schrödinger, release 2020-1). The methyltransferase DNMT3A, the rest of the DNA chain, the cofactor and the crystal water molecules were kept as in the crystal structure.
Molecular dynamics simulations
Overall, 34 000 node hours using 72 virtual cores per node, were used to carry out the simulations on Piz Daint at the Swiss National supercomputing Centre. All simulations were carried out using the GROMACS 2018.6 simulation package (47) and the CHARMM36m force field (48). Five independent 1-μs simulations with different initial random velocities were carried out for each system. To reproduce neutral pH conditions, the Arg and Lys side chains an N-terminus were positively charged, while the Asp and Glu side chains and the C-terminus were negatively charged. Furthermore, the 5′- and 3′-termini in the DNA sequence were capped with phosphate groups and OH groups, respectively. Each system was solvated in a cubic box (edge length of 13.9 nm) with TIP3P water molecules (49) to which 150 mM NaCl were added, including neutralizing counterions. Periodic boundary conditions were applied. Following steepest descent minimization, the systems were equilibrated under constant pressure for 2 ns, with position restraints applied on the heavy atoms of the complexes. Temperature and pressure were maintained constant at 300 K and 1 atm, respectively, by using the modified Berendsen thermostat (0.1 ps coupling) (50) and barostat (2 ps coupling) (51). For the production runs, performed in the NVT ensemble, harmonic restraints (force constant K = 1000 kJ/(mol·nm2)) were used for the heavy atoms of the end nucleotides and snapshots were saved every 50 ps. The short-range interactions were cut-off beyond a distance of 1.2 nm and the potential smoothly decays to zero using the Verlet cutoff scheme. The Particle Mesh Ewald (PME) technique (52) with a cubic interpolation order, a real space cut-off of 1.2 nm and a grid spacing of 0.16 nm was employed to compute the long-range interactions. Bond lengths were constrained using a fourth order LINCS algorithm with two iterations (53). In all simulations the time step was fixed to 2 fs.
RESULTS
De novo DNA methyltransferases display flanking sequence preferences in murine stem cells
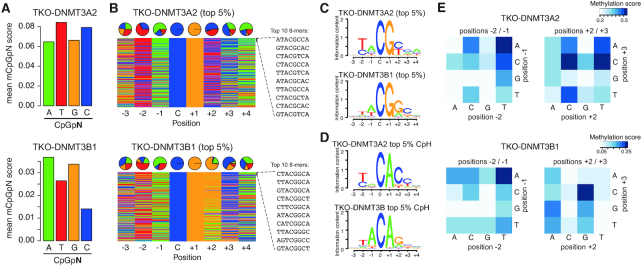
To examine DNA sequence specificities of de novo methyltransferases in vivo, we first made use of available whole-genome bisulphite sequencing (WGBS) from Dnmt-triple-KO (TKO) mouse embryonic stem cells where the de novo DNA methyltransferases DNMT3A2 or DNMT3B1 were reintroduced to the genome (16) (Supplementary Table S1). In these cells, replication-coupled maintenance of de novo methylated DNA is lacking in absence of DNMT1 and the WGBS data directly allows us to identify de novo DNA methylation activity by DNMT3A or DNMT3B. We leveraged this to investigate if the favoured and disfavoured DNA sequence contexts reported in vitro, are indeed also observed in cells. First, we calculated the average DNA methylation in the TKO-DNMT3A2 and TKO-DNMT3B1 samples at CpGs in all four CpGpN contexts. Although each CpGpN sequence context was equally represented in the TKO-DNMT3A2 and TKO-DNMT3B1 WGBS libraries (Supplementary Figure S1A-C), DNA methylation analysis indicated a sequence-preference which differed between DNMT3A and DNMT3B (Figure 1A). The DNMT3A sample showed increased methylation at cytosines in a CpGpT and CpGpC sequence-context, indicating a preference towards pyrimidines at the +2 position from the methylated cytosine (CGY), and DNMT3B at cytosines in a CpGpA and CpGpG context, containing purines at the +2 position (CGR). To further test if the observed difference could be influenced by the genomic context or different library representation of the queried sequences, we re-calculated the methylation scores only on the same CpGpNs that were covered in both, the TKO-DNMT3A2 and the TKO-DNMT3B1 libraries (Supplementary Figure S1D), and also on CpGpNs that were at least 80% methylated in wild type ES cells (Supplementary Figure S1E). The latter was necessary to exclude that active promoters and enhancers with elevated levels of DNA methylation turnover due to active DNA de-methylation (2,3,21) could have a potential influence on the calculated methylation scores. In both cases, the same sequence preference was observed when using these refined CpG sites (Supplementary Figure S1D and E). Finally, we generated new, independent Dnmt-TKO cell lines expressing DNMT3A2 and DNMT3B1 and repeated the WGBS analysis centred around CpGpN sites, resulting in identical sequence preferences and excluding potential artefacts stemming from library preparation, bisulphite conversion or sequencing technology (Supplementary Figure S1F and Table S1).
Figure 1.
De novo DNMTs display flanking sequence preferences at CpG and non-CpG sites. (A) Bar plots indicating average methylation scores at CpG sites followed by A, T, C and Gs (CpGpN). Average methylation for CpGpNs falling into one of the four categories was calculated from WGBS data obtained from Dnmt-TKO ES cells expressing either DNMT3A2 or DNMT3B1. (B) Nucleotide composition at top 5% from 8-mers ranked by CpG methylation in DNMT3A2 or DNMT3B1-expressing TKO cells. Pie charts show the distribution at the individual positions according to the methylated cytosine: C. Top 10 methylated sequences are shown. (C) Position weight matrix calculated from the top 5% 8-mers methylated by either DNMT3A2 or DNMT3B1 indicate the preferred DNA sequence motif. (D) Position weight matrix calculated from the top 5% 8-mers methylated by either DNMT3A2 or DNMT3B1 indicate the preferred DNA sequence motif at non-CpG sites. (E) Heatmap showing the effect of dinucleotide coupling at positions –2/–1 or +2/+3 on CpG methylation (at position 0/+1).
Following these results, we wanted to have a more detailed view on the sequence preferences of DNMT3A and DNMT3B. Towards this, we expanded our analysis to 8-mer strings with a fixed CpG in the centre (NNNCGNNN) and calculated the methylation scores on the forward or reverse strands separately. We first ranked the obtained sequences according to the average methylation score calculated from all 8-mer instances in the Dnmt-TKO cells expressing DNMT3A2 or DNMT3B1 (Supplementary Figure S2A-B). This ranking allowed us to identify DNA sequences that were preferentially methylated by DNMT3A and DNMT3B. Top-ranking 8-mers indicated similarities and individual preferences in nucleotides at the queried positions (Figure 1B). For example, we observed that both enzymes have a slight preference for adenines (A) at the -1 position and cytosines (C) at the +3 position. We observed an increased occurrence of thymines (T) at the –2 position at DNMT3B-preferred sites, which was even more prominent at DNMT3A targets (Figure 1B). On top of that, we again observed the +2-nucleotide preference for pyrimidines at DNMT3A targets, where Cs and Ts were equally represented, while DNMT3B displayed an increased preference for purines, especially guanines (G) (Figure 1B). These position-specific preferences flanking CpGs can be further observed when calculating the methylation difference between DNMT3A and DNMT3B for each individual nucleotide position in the 8-mer (Supplementary Figure S2C).
By grouping the ranked 8-mers into 20 bins we could furthermore calculate the sequence preferences within each bin and display the results as DNA sequence logos (Figure 1C and Supplementary Figure S3A and B). Importantly we only observed sequence preferences at the top-ranked and bottom-ranked 8-mers, but not at 8-mers ranked in the centre (Supplementary Figure S3A and B). The least methylated 8-mers allowed us to identify sequences that were disfavoured by the enzymes. These predominantly contained purines (G/A) at the –2 position for both enzymes, but we also observed differences at the –1 and +2 position, where DNMT3B showed reduced methylation preference near pyrimidines located at –1 and +2, while DNMT3A did not indicate strong occurrence of disfavoured nucleotides (Supplementary Figure S3A-B). These results are in line with previous reports obtained from incubating DNMT3A or DNMT3B with DNA substrates (29–31), indicating that the flanking preferences observed in vitro are prevalent in cells and even in presence of chromatin. Analysis of sequencing coverage distribution for all 8-mers again results in identical coverages between TKO-DNMT3A2 and TKO-DNMT3B1 (R = 0.995, Supplementary Figure S4A and B). In addition, the calculated 8-mer sequence preferences were identical between replicates of DNMT3A2 and DNMT3B1, confirming the observed preferences in independent replicates (Supplementary Figure S4C).
Finally, we extended this analysis to all sequences containing a CpH site in the centre of the 8-mer (NNNCHNNN). Ranking all sequences by their average methylation score results in a high preference for CpA methylation for both enzymes (Figure 1D and Supplementary Figure S5A), in accordance with previous results obtained from in vitro assays (54) or analysis of non-CpG methylation in mammalian genomes (55). Both enzymes show a preference for T and A at position –2 and –1, respectively (Figure 1D and Supplementary Figure S5A). In addition, we observe again a DNMT3A-specific methylation preference for CpA sequences frequently followed by a C at position +3 (CpApC), while DNMT3B prefers CpApG sites, with position +3 being less defined (Figure 1D and Supplementary Figure S5A-B). Importantly, the observed motifs resemble the reported non-CpG motifs observed in ES cells or neuronal tissues (2,5,20,55), indicating that these indeed stem from the activity of the individual de novo methyltransferases operating in these cell types. We repeated the ranking independently for sites containing CpA, CpT and CpC dinucleotides, resulting in similar DNMT3-specific sequence preferences for position +2 at CpA and CpT sites, while methylation at CpC-containing sequences was similar between both enzymes, likely due to low methylation scores in this context (Supplementary Figure S5C and D).
Individual and combinatorial influence of flanking nucleotides on DNMT3A and DNMT3B activity
Ranking of 8-mers suggested that the sequence context surrounding the CpG/CpH sites has a strong influence on the methylation activity. Next, we wondered if coupling of neighbouring nucleotides could have an influence on DNMT3A or DNMT3B activities and we calculated the methylation scores resulting for all possible combinations at the dinucleotides positioned immediately upstream and downstream of the CpGs (Figure 1E). This analysis indicates that for preceding positions (–2 and –1) DNMT3A and DNMT3B mainly prefer to methylate downstream of TpA dinucleotides (Figure 1E). Similar dinucleotide composition preferences can be observed upstream of CpH methylated sites (Supplemental Figure S6A). Analysis of downstream preferences indicates that DNMT3A methylation is preferentially targeted to CpGs followed by CpC or TpC > CpT or TpA > CpA (Figure 1E). In case of DNMT3B, methylation is preferentially targeted to CpG dinucleotides followed by GpC >> ApG at positions +2 and +3 (Figure 1E). These downstream di-nucleotide preferences are partially resembled at CpH sites, where DNMT3A prefers to methylate primarily non-CpGs upstream of CpC dinucleotides and DNMT3B sites followed by GpC > GpG or GpT (Supplemental Figure S6A). The results at CpH sites suggest a more defined flanking sequence preference at position +2, where only cytosines and guanines appear to be enzymatic signatures of DNMT3A and DNMT3B, respectively.
Differential localisation of de novo DNMTs influences the genomic distribution of methylated motifs
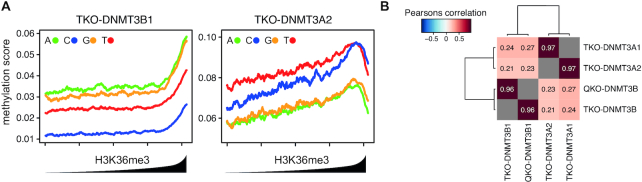
The DNMT3 proteins display distinct genome-wide localisation patterns correlating with the genomic distribution of histone modifications. In case of DNMT3B, methylation is preferentially targeted to H3K36me3 (16). To investigate how the observed sequence preference follows this genomic localisation pattern, we first analysed the genome-wide distribution of CpGpN methylation according to H3K36me3 in DNMT3A2 or DNMT3B1-reconstituted Dnmt-TKO cells. We ranked 1 kb genomic intervals according to H3K36me3 levels and calculated the mean methylation score for each CpGpN motif at each interval. In case of DNMT3B, we see a clear dependence on H3K36me3, where the total CpG methylation (independent of the nucleotide at +2 position) increases with H3K36me3, as previously reported (16) (Figure 2A). When comparing the methylation at individual CpGpN sites, we observe that although methylation at CpGpG and CpGpA sites clearly increases with H3K36me3 levels, the methylation ratio of G/A over C and T at the +2 position remains constant – independent of H3K36me3 and total methylation (Figure 2A). The methylation introduced in the DNMT3A2 reconstitution experiments shows similar constant distributions of methylated CpGpN motifs, but with reversed preferences for A/T over G/A at the +2 position (Figure 2A). The same result is observed when we investigate re-methylation at genomic regions preferentially bound by DNMT3A or DNMT3B in mouse ES cells (23). While the overall levels of CpG methylation at these sites are in agreement with the binding preference of the re-introduced DNMT3 enzyme in the Dnmt-TKO cells, the enzyme-specific sequence preferences for +2 nucleotides remain intact, independent of the binding site (Supplementary Figure S7A–C).
Figure 2.
Flanking sequence preferences scale with, but are independent of the genomic location of the DNMTs. (A) Preferential de novo methylation of purines by DNMT3B is not altered by its general preference for H3K36 tri-methylated sites. Shown are de novo DNA methylation at all four CpGpN context genome-wide in relation to H3K36me3 enrichment. 1-kb-sized genomic intervals were ranked and grouped by H3K36me3 enrichment (1000 intervals per bin) and DNA methylation was calculated per bin. Lines indicate mean DNA methylation per bin in TKO cells expressing DNMT3B1 (left) or DNMT3A2 (right). (B) Heatmap indicating strong correlation in DNA sequence preference between the DNMT3A1 and DNMT3A2 isoforms, or between DNMT3B1 in presence and absence of H3K36me3. Correlations between identical samples were removed and are shown in gray.
In general, this indicates that genomic localisation primarily determines local enzymatic activity, while differences in enzymatic flanking preference by DNMT3A or DNMT3B can result in preferred methylation of cytosines followed by pyrimidines or purines at the +2 position, respectively. This is also evident from analysing the distribution of non-CpG methylation in wild type ES cells expressing all DNMT enzymes (3), where CpApG shows slightly higher methylation at sites bound by DNMT3B (Supplementary Figure S7D-E). However, potential redundancies in presence of DNMT3A and DNMT3B isoforms as well as heterodimers between these enzymes (56,57) could result in less clear flanking preferences. To investigate how absence of either DNMT3A or DNMT3B in fully-methylated cells influences the observed flanking sequence preferences, we made use of available WGBS datasets obtained in human ES cells (hESC), hESC-derived endodermal cells (CD184) and hESC-derived motor neurons lacking either DNMT3A or DNMT3B (35,58). We observe a stronger global reduction in methylation at CpG sites followed by pyrimidines in cells lacking DNMT3A, while cells lacking DNMT3B show a stronger reduction at CpG sites followed by purines (Supplementary Figure S7F).
Preference to methylate at different flanking sequences is determined by the catalytic domain
Recruitment of de novo DNMTs to the genome is predominantly regulated through the non-catalytic N-terminal part and its interactions with chromatin. To directly test how chromatin-dependent recruitment may influence the flanking sequence preference, we first re-evaluated additional WGBS datasets where we deleted SETD2, the enzyme responsible for deposition of H3K36me3 (59), in Dnmt-TKO cells expressing DNMT3B (here termed quadruple-KO, QKO). We and others have previously shown by ChIP-seq, that in absence of SETD2, binding and methylation activity of DNMT3B to actively transcribed gene bodies is lost (16,22). This is also observed when we reanalyse the DNA methylation in the context of CpGpN, where all four sequence contexts are equally affected but retain the preference of purines over pyrimidines (Supplementary Figure S8A). By further comparing the methylation scores at 8-mers obtained from DNMT3B-QKO cells with cells containing H3K36me3, we observe the same flanking preference for CpG and non-CpG sites, despite the lack of de novo methylation targeting to H3K36me3 sites (Figure 2B and Supplementary Figure S8B–D). In addition, we have investigated if the previously-reported differential genomic localization of DNMT3A isoforms 1 and 2 (23,24) would influence flanking sequence preferences. By comparing the results obtained in TKO-DNMT3A2 with a WGBS dataset obtained from Dnmt-TKO cells expressing the longer DNMT3A1 isoform, we could not observe any differences in methylated sequence contexts (Figure 2B and Supplementary Figure S8E-G). This also holds true when we compare the datasets at genomic regions with differential de novo DNMT binding (Supplementary Figure S8H), excluding that recruitment of DNMTs based on histone modifications is responsible for the observed sequence preference at +2 position. This suggests differences in the catalytic domain of DNMT3A and DNMT3B, rather than genomic localisation, as the source of the observed difference in flanking sequence preference (Supplementary Figure S8I).
Direct comparison between DNMT3A and DNMT3B catalytic domains reveals structural constraints on methylation preference
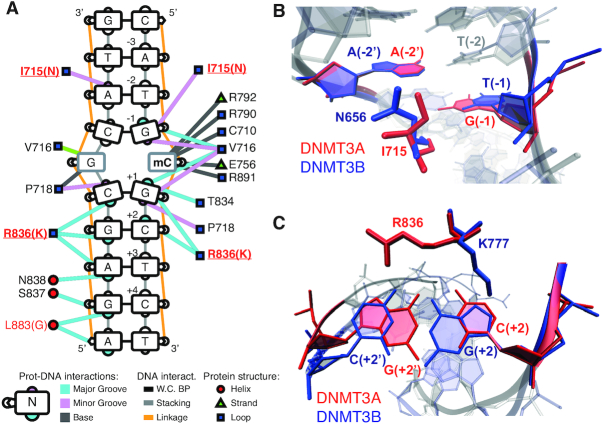
The observed sequence preferences could stem from structural differences in the catalytic domains of DNMT3A and DNMT3B. Towards this we first focused our attention on divergent amino acids in the catalytic domains of DNMT3A and DNMT3B. Of the 279 residues in the catalytic domain, 44 vary between DNMT3A and DNMT3B (Supplementary Figure S9A). We made use of available crystal structures of human DNMT3A in complex with DNA (46) (PDB ID:6F57) and used DNAproDB (60) with standard interaction criteria to identify which of the variant amino acids interact with the DNA upstream and downstream of the methylated CpG (Figure 3A). We observe that only two variant amino acids are in close contact with the nucleotide bases around the methylated CpG (Figure 3A and Supplementary Figure S9A). The catalytic loop residue I715 (N656 in human DNMT3B) sits in the minor groove of the DNA and contacts the G at –1 position on the same strand, and the A at the −2′ position on the opposite strand of the modified CpG (Figure 3A, B and Supplementary Figure S9B). The corresponding N656 in DNMT3B crystal structures in complex with DNA (61) shows a similar interaction with the minor groove and contacts the A at the −2′ position on the opposite strand (Figure 3A, B and Supplementary Figure S9C). The interactions of I715 and N656 with the adenine at position −2′ could contribute to the elevated preference for Ts at position –2 on the methylated strand, which is observed for both enzymes (Figure 1C).
Figure 3.
Structural differences between human DNMT3A and DNMT3B provide potential cues for observed sequence preferences. (A) Schematic overview of DNAproDB-reported intermolecular interactions between human DNMT3A and DNA, based on PDB ID: 6F57. Edges represent interactions between DNMT3A amino acid residues and DNA, while colours represent interactions with the major groove in blue, minor groove in pink and bases in grey. Position of interacting amino acids within DNMT3A protein secondary structures are shown as red circles for alpha helices, green triangles for beta strands and blue squares for loops. (B) Residues I715 of human DNMT3A (PDB ID: 6F57) and N656 of human DNMT3B (PDB ID: 6KDA) interact with the minor groove of the DNA by contacting the A at the –2 position on the opposite strand of the modified CpG. (C) Residues R836 of DNMT3A and K777 of DNMT3B interact with the DNA downstream of the methylated CpG through interactions with the major groove at position +2. The side chains show opposing orientation, with K777 (PDB ID 6KDA) contacting the guanine on the methylated strand and R836 (PDB ID: 6F57) contacting the guanine on the opposite strand.
The variant residue R836 in the target recognition domain (TRD) of human DNMT3A (K777 in human DNMT3B) contacts the +1 to +3 nucleotide pairs in the major groove downstream of the methylated CpG (Figure 3A). This residue was previously suggested to be involved in mediating the preference for G at the +1 position in the CpG site (46). In DNMT3B, it is rather N779 (N838 in DNMT3A) that mediates the specificity for the G nucleotide at position +1 of the methylated CpG, while K777 (corresponding to R836 in DNMT3A) contributes towards the DNMT3B-specific flanking preference for Gs at position +2 (30,61). Interestingly, a recent crystal structure of human DNMT3A together with a DNA molecule containing CpGpA shows that the R836 side chain flips away from the G at +1 to interact with the flanking nucleotides downstream of the CpG indicating increased flexibility in the TRD loop (62). Given these observations, we wanted to investigate how the potential flexibility of the R836 side chain contributes to the observed flanking preference of DNMT3A at position +2. Superimposition of available structures of DNMT3A and DNMT3B in complex with DNA (46,61) indicate that both residues (DNMT3A R836 and DNMT3B K777) can contact the nucleotides at position +2, but the side chains are oriented in opposite directions (Figure 3C). In the respective crystal structures, the DNMT3B K777 side chain contacts the guanine at position +2 on the methylated strand, while the DNMT3A R836 side chain contacts the guanine at position +2′ on the opposite strand (Figure 3C and Supplementary Figure S9D-E). The opposing orientations and interactions on opposite strands could point to a potential mechanism underlying the observed DNMT-specific preference at position +2.
Atomistic simulations reveal interaction dynamics between DNMT3A R836 and DNA
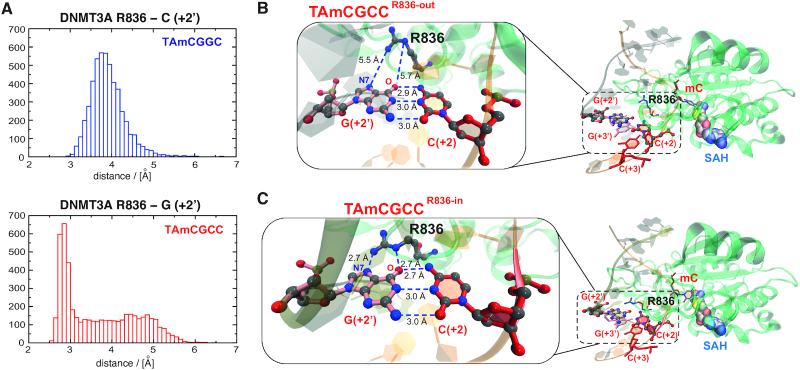
To investigate the structural plasticity of the DNMT3A-DNA interactions we performed molecular dynamics (MD) simulations starting from a DNMT3A/decameric DNA complex extracted from the crystal structure of human DNMT3A-DNMT3L-DNA (PDB ID: 5YX2) (46). Two simulation systems were prepared differing only at the position +2 of the 10-mer DNA duplex, namely CATAmCGCCCT and CATAmCGGCCT (boldface for position +2, Supplementary Figure S10A). The nucleotides preceding the methylation site were mutated to T and A to accommodate the observed preferences of DNMT3A and DNMT3B. Both systems contained also a molecule of the co-product S-adenosyl-l-homocysteine (SAH) (Supplementary Figure S10B). Five independent simulations were carried out for each system for a total sampling of 5 μs (Materials and Methods). We focused the analysis on the interaction between R836 and the DNA duplex and in particular the closest nucleotide, that is G or C at position +2′ on the unmethylated strand (Figure 3C) and calculated the shortest distance between any pairs of non-hydrogen atoms along the MD trajectories. In presence of the disfavoured sequence, the histogram of the distance between R836 and the cytosine at position +2′ shows a maximum at about 4 Å which corresponds to van der Waals contacts (Figure 4A and Supplementary Movie 1). Interestingly however, in the presence of the favoured sequence the distance between R836 and the guanine at position +2′ shows a bimodal distribution with a peak at hydrogen bond distance (∼2.7 Å) and a broad region up to 5.5 Å (Figure 4A). This indicates a potential switch in the orientation of the R836 side chain, resulting in two conformational states which we here call ‘R836-in’ and ‘R836-out’. Indeed, the simulations reveal that in presence of the preferred sequence the guanidinium group of R836 can form two hydrogen bonds, respectively, with the carbonyl oxygen and N7 nitrogen of the guanine at position +2′ (i.e. the guanine that base-pairs with C at +2) resulting in the ‘R836-in’ state (Figure 4B, C, Movies 2–3, and Supplementary Figure S10D-F). This switching of R836 is in line with the crystal structure of DNMT3A in complex with DNA containing the disfavoured CpGpA sequence, suggesting that this side chain dynamically interacts with DNA, depending on the flanking nucleotides downstream of CpG (62). In our simulations, the total interaction energy between the side chain of R836 and its surrounding (DNA, protein, and solvent) is similar for the disfavoured DNA sequence and the two states of the favoured DNA sequence (Supplementary Figure S10C). Concerning the individual contributions, the interaction energy between R836 and DNA is about 15 kcal/mol more favourable for the ‘R836-in’ state than the ‘R836-out’, and the latter is similar as in the disfavoured sequence (Supplementary Figure S10C). The difference in interaction energy originates almost only from the aforementioned hydrogen bonds. The additional hydrogen bonds with DNA (more precisely with the G at position +2′ in the favoured DNA sequence) are compensated by a loss of interactions between R836 and the solvent (Supplementary Figure S10C). Thus, the MD simulations indicate that the direct interactions between R836 and methylated DNA duplexes containing CpGpC sites are more favourable compared to CpGpG sites, while the interaction energy of R836 and the rest of the system (protein/DNA complex and solvent) is similar.
Figure 4.
Molecular dynamics simulations reveal potential mechanisms underlying DNMT3A preference for CpGpC sites. (A) Histograms displaying the measured distance between the R836 side chain and the base at position +2′ for the disfavored (top) and the favored (bottom) DNA duplex sequence. For the favored sequence, a broad region and a peak are observed corresponding to the state before the conformational switch (‘R836-out’ state) and after (‘R836-in’ state). (B, C) The two interactions states of DNMT3A R836 and the preferred DNA sequence obtained from the MD simulations. Magnifications show the distances between DNMT3A R836 and the guanine at position +2′ for the ‘R836-out’ (in B) and ‘R836-in’ state (in C). The hydrogen bonds between the nitrogen atoms of the guanidinium group of R836 and the carbonyl oxygen and N7 nitrogen of the guanine at position +2′ are indicated.
An arginine to lysine substitution reverts the DNMT3A sequence preference at position +2
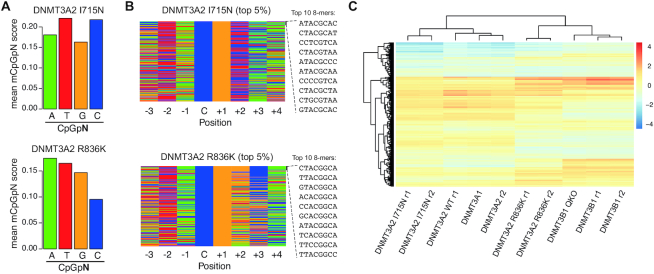
To directly test the contribution of this arginine side chain towards the observed DNMT3A flanking-sequence preference, we generated two independent Dnmt-TKO cell lines expressing mutant mouse DNMT3A2 proteins where we exchanged the arginine at position 613 (= position 836 in full length human DNMT3A) to a lysine—the corresponding side chain present in DNMT3B. We named these mutations R836K to retain consistency with the structural analysis based on human DNMT3A. In addition, we have generated two independent Dnmt-TKO cell lines expressing mouse DNMT3A2 bearing an I to N substitution at position 492, corresponding to the second variant amino acid in human DNMT3A I714, that presumably contacts the –2 and –1 nucleotides in the crystal structures (I714N, Figure 3A, B). WGBS analysis shows that, while the I715N replacement does not influence the sequence preference downstream of the CpG site, the R836 replacement reverses the preference of DNMT3A to partially resemble that of DNMT3B with decreased methylation at CpGs followed by Cs and Ts (Figure 5A, B and Supplementary Figure S11A, B). This change in sequence preference is also evident when we cluster the samples based on the methylation scores at 8-mer motifs, where DNMT3A2-R836K clusters closer to DNMT3B (Figure 5C and Supplementary Figure S11C). The clustering analysis based on the 8-mer motifs further indicates a slight change in sequence preference for the DNMT3A-I715N mutant. Direct comparison of nucleotide preferences between mutant and wild type DNMT3A at individual positions reveals a minor shift at positions –2 and –1 in the I715N mutant, where less-frequent adenines at –2 are replaced by guanines and thymines are slightly increased at –1 (Supplementary Figure S11D). However, this does not resemble the preference observed for DNMT3B, suggesting that this side chain is not relevant for the observed nucleotides at –2 and –1 (Figure 5B and Supplementary Figure S11E). In contrast, the R836K replacement results in a change in preference from pyrimidines to purines at position +2, resembling sequences observed in DNMT3B (Supplementary Figure S11E, F). Similar changes in flanking sequence preference are observed at non-CpG sequences, where R836K leads to increased preference for purines at position +2 (Supplementary Figure S12A, B), without affecting global ratios in methylation at CpG and non-CpG sites (Supplementary Figure S12C).
Figure 5.
Replacement of Arg with Lys in DNMT3A resembles DNMT3B sequence preferences. (A) Bar plots indicating average methylation scores at CpG sites followed by A, T, C and Gs (CpGpN). Average methylation for CpGpNs falling into one of the four categories was calculated from WGBS data obtained from Dnmt-TKO ES cells expressing either DNMT3A2 I714N (= residue 492 in mouse DNMT3A2) or R836K (= residue 613 in mouse DNMT3A2). (B) Nucleotide composition at top 5% from 8-mers ranked by CpG methylation in DNMT3A2 I714N or R836K -expressing TKO cells. Top 10 methylated sequences are shown. (C) Heatmap of methylation preferences at all CpG 8-mer sequences shows similarities and differences between samples analysed in this study. Shown are methylation scores scaled by sample (columns). Unsupervised clustering indicates similarities between samples (columns) and 8-mers (rows). Legend indicates values scaled per sample. QKO denotes Dnmt-TKO; Setd2-KO cells, r1 and r2 denote independent replicates.
DISCUSSION
Sequence-specific flanking preferences have been first described for the de novo DNA methyltransferases based on in vitro methylation experiments or using episomal constructs in cells (26–28,63). In addition, analysis of tissue-specific WGBS data indicates that sequence-context of non-CpG methylation strongly varies between tissues expressing DNMT3A or DNMT3B (2,20,35–37). Together, these reports suggest that DNA sequence preferences of the de novo DNMTs could provide an additional layer of methylome regulation. However, it remained to be clarified if these enzyme-specific preferences occur in vivo and genome-wide. Especially, given the reported differential localisation and cell-type-specific activities of de novo DNMTs, it remained a challenge to directly and comprehensively compare the enzymatic preferences of these enzymes in the same genomic environment. By evaluating a set of previously-published and newly-generated whole-genome bisulphite sequencing data obtained from Dnmt-TKO ES cells lacking DNA methylation with re-introduced individual de novo enzymes, we were able to characterize the genomic flanking preferences of DNMT3A and DNMT3B at unprecedented detail. Here, we show that DNMT3A prefers to methylate predominantly in a NTACGYCN context, while DNMT3B prefers NTACGRCN. We find nucleotides around the methylated site that are equally preferred by both enzymes and furthermore, highlight that the +2 nucleotide following the methylated cytosine is the strongest determinant of enzyme-specific sequence preference, confirming very recently published results using large scale in vitro measurements and WGBS datasets (30,31). The flanking sequence preference is largely consistent with the methylation patterns observed around non-CpG sites, with NTACACCN and NTACAGCN being the most-favoured sites for DNMT3A and DNMT3B, respectively. The non-CpG-specific flanking preferences are identical to non-CpG motifs reported in tissues where either DNMT3A predominantly contributes to de novo methylation, such as neurons or oocytes, or in ES cells, where DNMT3B seems to contribute to the majority of non-CpG methylation (2,5,20,55) – confirming that the methylation patterns observed in these cells are indeed a footprint of enzyme-specific de novo DNMT activities.
We furthermore show here that genomic sites preferentially bound by DNMT3A or DNMT3B show elevated methylation at cytosines with preferred nucleotides at the +2 position. However, through a series of experiments using different mutant lines, we show that flanking sequence preference does not stem from the differential localisation of the de novo DNMTs to the genome, but can be attributed to minor variations in the structure of the TRD loop in the catalytic domain. Direct comparison of protein sequences and structures of DNMT3A (46) or DNMT3B (61) together with DNA indicates a variant residue that contacts position +2 downstream of the methylated CpG in the major groove of the DNA. The arginine side chain of this residue in human DNMT3A (R836) contacts purines on the opposite strand from the methylated CpG, while a lysine in human DNMT3B at the same position (K777) contacts purines on the methylated strand. The latter interaction was recently shown to influence the flanking preference of DNMT3B for CpGpG sites in vitro (30,61), suggesting that R836 could play similar roles in determining DNMT3A preferences for purines on the opposing strand, resulting in the observed CpGpY flanking preference on the methylated strand. To shed light on the atomistic details of the DNMT3A/DNA duplex interactions we performed MD simulations with decameric DNA products containing favoured and disfavoured flanking sequences (difference at position +2). The quantification of interaction distances along the MD trajectories provides information that goes beyond interpretations of crystal structures. The favoured DNA sequence and R836 display two conformational states ‘R836-in’ and ‘R836-out’. In the first state stabilizing hydrogen bonds between the side chain of R836 and the G that base-pairs with the C at position +2 of the favoured sequence are formed, whereas in the latter only weaker van der Waals contacts are observed. This dynamic switching of R836 is fully consistent with recent crystal structures that describe flexible conformations of the R836 side chain, depending on the flanking sequence (46,62). By introducing the lysine found in DNMT3B at the corresponding residue in mouse DNMT3A2 (R836K) we show that the flanking sequence preference of DNMT3A is almost completely reverted to resemble that of DNMT3B. Together with our molecular dynamics simulations, these results support a sequence-dependent conformation of the R836 side chain, and pinpoint R836 as a critical residue in mediating the flanking sequence preferences observed for DNMT3A. Interestingly, this arginine is conserved throughout all genes annotated as Dnmt3a in jawed vertebrates, teleost species and in the jawless vertebrate Lamprey (64). The lysine in DNMT3B on the other hand, is only conserved in jawed vertebrates, while Arg, Lys or Asparagine can be found at the same position in other species (64). It will be interesting to investigate if DNA methylation is influenced by flanking-sequence preference in these species, and if this can be explained by a conserved role of the side chain at this position.
Understanding the sequence-context-dependent methylation activities of DNMT3A and DNMT3B in living cells provides novel insights into how DNA sequence could shape DNA methylation patterns. We suggest that sequence-dependent de novo methylation provides an additional regulatory layer of the methylome landscape with potential impact on gene activity through recognition of the methylated cytosine in different sequence contexts. This is for instance exemplified by MeCP2, a methyl-binding protein with in vitro and in vivo binding preference for methylated CpApC and to some extent CpApT sites (6). Methylated CpApC sites drive recruitment of this protein in neuronal cells and lack of DNMT3A has been shown to disrupt this localization, resulting in changes of gene expression relevant for neuronal function (6,65). In addition, several transcription factors (TFs) have been shown to be influenced by the presence of DNA methylation in their recognition motifs (38,66,67). One could speculate that DNMT-specific flanking sequence preferences together with their differential genomic binding and cell-type-specific activities could influence genomic binding of TFs in a context-dependent manner. Loss and gain of function mutations in the de novo DNA methyltransferases and the corresponding changes in DNA methylation patterns have been associated with numerous diseases and cancers (68,69). Future studies should help to address how the here-described differences in de novo methylation activities contribute to the observed TF binding and gene regulation differences in healthy and diseased tissues.
DATA AVAILABILITY
Published WGBS datasets were obtained from Gene Expression Omnibus (GEO) and are listed in Supplementary Table S1. Published crystal structures of DNMT3A (6F57, 5YX2) and DNMT3B (6KDA) were obtained from the Protein Data Bank (PDB) (https://www.rcsb.org). Newly generated WGBS datasets have been deposited to GEO under accession number GSE151992.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mark D. Robinson (UZH, DMLS), the Swiss National Supercomputing Centre and S3IT at the University of Zurich for providing the computational infrastructure, Martin Jinek (UZH, Biochemistry) for initial discussions on DNMT3A structures, and members of the Baubec lab for their critical input.
Contributor Information
Izaskun Mallona, Department of Molecular Mechanisms of Disease, University of Zurich, Zurich 8057, Switzerland; Department of Molecular Life Sciences, University of Zurich, Zurich 8057, Switzerland; SIB Swiss Institute of Bioinformatics, Zurich 8057, Switzerland.
Ioana Mariuca Ilie, Department of Biochemistry, University of Zurich, Zurich 8057, Switzerland.
Ino Dominiek Karemaker, Department of Molecular Mechanisms of Disease, University of Zurich, Zurich 8057, Switzerland.
Stefan Butz, Department of Molecular Mechanisms of Disease, University of Zurich, Zurich 8057, Switzerland; Life Science Zurich Graduate School, University of Zurich, Zurich 8057, Switzerland.
Massimiliano Manzo, Department of Molecular Mechanisms of Disease, University of Zurich, Zurich 8057, Switzerland; Life Science Zurich Graduate School, University of Zurich, Zurich 8057, Switzerland.
Amedeo Caflisch, Department of Biochemistry, University of Zurich, Zurich 8057, Switzerland.
Tuncay Baubec, Department of Molecular Mechanisms of Disease, University of Zurich, Zurich 8057, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [157488, 190378 to T.B.]; SNSF Sinergia [180345]; SNSF Excellence Grant [310030B-189363 to A.C.]; I.M.I. thanks the Peter und Traudl Engelhorn Foundation for a postdoctoral fellowship. Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Goll M.G., Bestor T.H.. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005; 74:481–514. [DOI] [PubMed] [Google Scholar]
- 2. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.-M. et al.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., van Nimwegen E., Wirbelauer C., Oakeley E.J., Gaidatzis D. et al.. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011; 480:490–495. [DOI] [PubMed] [Google Scholar]
- 4. Ramsahoye B.H., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R.. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schultz M.D., He Y., Whitaker J.W., Hariharan M., Mukamel E.A., Leung D., Rajagopal N., Nery J.R., Urich M.A., Chen H. et al.. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015; 523:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lagger S., Connelly J.C., Schweikert G., Webb S., Selfridge J., Ramsahoye B.H., Yu M., He C., Sanguinetti G., Sowers L.C. et al.. MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLoS Genet. 2017; 13:e1006793–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L., Chen K., Lavery L.A., Baker S.A., Shaw C.A., Li W., Zoghbi H.Y.. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. et al.. Global epigenomic reconfiguration during mammalian brain development. Science. 2013; 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kinde B., Gabel H.W., Gilbert C.S., Griffith E.C., Greenberg M.E.. Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:6800–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Antequera F., Boyes J., Bird A.. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990; 62:503–514. [DOI] [PubMed] [Google Scholar]
- 11. Mohn F., Weber M., Rebhan M., Roloff T.C., Richter J., Stadler M.B., Bibel M., Schübeler D.. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008; 30:755–766. [DOI] [PubMed] [Google Scholar]
- 12. Jeong M., Sun D., Luo M., Huang Y., Challen G.A., Rodriguez B., Zhang X., Chavez L., Wang H., Hannah R. et al.. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 2014; 46:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okano M., Bell D.W., Haber D.A., Li E.. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99:247–257. [DOI] [PubMed] [Google Scholar]
- 14. Bourc’his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H.. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001; 294:2536–2539. [DOI] [PubMed] [Google Scholar]
- 15. Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y.E.. Dnmt3a-Dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010; 329:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schübeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 17. Gu H., Bock C., Mikkelsen T.S., Jäger N., Smith Z.D., Tomazou E., Gnirke A., Lander E.S., Meissner A.. Genome-scale dna methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods. 2010; 7:133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hon G.C., Rajagopal N., Shen Y., McCleary D.F., Yue F., Dang M.D., Ren B.. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013; 45:1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Molaro A., Hodges E., Fang F., Song Q., McCombie W.R., Hannon G.J., Smith A.D.. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011; 146:1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shirane K., Toh H., Kobayashi H., Miura F., Chiba H., Ito T., Kono T., Sasaki H.. Mouse oocyte methylomes at base resolution reveal Genome-Wide accumulation of Non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013; 9:e1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ginno P.A., Gaidatzis D., Feldmann A., Hoerner L., Imanci D., Burger L., Zilbermann F., Peters A.H.F.M., Edenhofer F., Smallwood S.A. et al.. A genome-scale map of DNA methylation turnover identifies site-specific dependencies of DNMT and TET activity. Nat. Commun. 2020; 11:2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morselli M., Pastor W.A., Montanini B., Nee K., Ferrari R., Fu K., Bonora G., Rubbi L., Clark A.T., Ottonello S. et al.. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife. 2015; 4:e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manzo M., Wirz J., Ambrosi C., Villaseñor R., Roschitzki B., Baubec T.. Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J. 2017; 36:3421–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu T., Lin X., Cullen S.M., Luo M., Jeong M., Estecio M., Shen J., Hardikar S., Sun D., Su J. et al.. DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol. 2018; 19:88–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weinberg D.N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K.N., Horth C., McGuire J.T., Xu X., Nikbakht H. et al.. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019; 573:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin I.G., Han L., Taghva A., O’Brien L.E., Hsieh C.-L.. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol. 2002; 22:704–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Handa V., Jeltsch A.. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005; 348:1103–1112. [DOI] [PubMed] [Google Scholar]
- 28. Wienholz B.L., Kareta M.S., Moarefi A.H., Gordon C.A., Ginno P.A., Chédin F.. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010; 6:e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emperle M., Adam S., Kunert S., Dukatz M., Baude A., Plass C., Rathert P., Bashtrykov P., Jeltsch A.. Mutations of R882 change flanking sequence preferences of the DNA methyltransferase DNMT3A and cellular methylation patterns. Nucleic Acids Res. 2019; 47:11355–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao L., Emperle M., Guo Y., Grimm S.A., Ren W., Adam S., Uryu H., Zhang Z.-M., Chen D., Yin J. et al.. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 2020; 11:3355–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mao S.-Q., Cuesta S.M., Tannahill D., Balasubramanian S.. Genome-wide DNA methylation signatures are determined by DNMT3A/B sequence preferences. Biochemistry. 2020; 59:2541–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arand J., Spieler D., Karius T., Branco M.R., Meilinger D., Meissner A., Jenuwein T., Xu G., Leonhardt H., Wolf V. et al.. In vivo control of CpG and Non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012; 8:e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suetake I., Miyazaki J., Murakami C., Takeshima H., Tajima S.. Distinct enzymatic properties of recombinant mouse DNA methyltransferases Dnmt3a and Dnmt3b. J. Biochem. 2003; 133:737–744. [DOI] [PubMed] [Google Scholar]
- 34. Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie Bin, Zhong C., Hu S., Le T., Fan G. et al.. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2013; 17:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liao J., Karnik R., Gu H., Ziller M.J., Clement K., Tsankov A.M., Akopian V., Gifford C.A., Donaghey J., Galonska C. et al.. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 2015; 47:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He Y., Ecker J.R.. Non-CG Methylation in the human genome. Annu. Rev. Genom. Hum. Genet. 2015; 16:55–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J.-H., Park S.-J., Nakai K.. Differential landscape of non-CpG methylation in embryonic stem cells and neurons caused by DNMT3s. Sci. Rep. 2017; 7:11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Domcke S., Bardet A.F., Ginno P.A., Hartl D., Burger L., Schübeler D.. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015; 528:575–579. [DOI] [PubMed] [Google Scholar]
- 39. Gaidatzis D., Lerch A., Hahne F., Stadler M.B.. QuasR: quantification and annotation of short reads in R. Bioinformatics. 2015; 31:1130–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 41. Pedersen B.S., Eyring K., De S., Yang I.V., Schwartz D.A.. Fast and accurate alignment of long bisulfite-seq reads. 2014; arXiv doi:13 May 2014, preprint: not peer reviewedhttps://arxiv.org/abs/1401.1129.
- 42. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; arXiv doi:26 May 2013, preprint: not peer reviewedhttps://arxiv.org/abs/1303.3997.
- 43. García-Alcalde F., Okonechnikov K., Carbonell J., Cruz L.M., Götz S., Tarazona S., Dopazo J., Meyer T.F., Conesa A.. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012; 28:2678–2679. [DOI] [PubMed] [Google Scholar]
- 44. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tippmann S.C., Ivanek R., Gaidatzis D., Schöler A., Hoerner L., van Nimwegen E., Stadler P.F., Stadler M.B., Schübeler D.. Chromatin measurements reveal contributions of synthesis and decay to steady-state mRNA levels. Mol. Syst. Biol. 2012; 8:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Z.-M., Lu R., Wang P., Yu Y., Chen D., Gao L., Liu S., Ji D., Rothbart S.B., Wang Y. et al.. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018; 554:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berendsen H.J.C., van der Spoel D., van Drunen R.. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995; 91:43–56. [Google Scholar]
- 48. Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., De Groot B.L., Grubmüller H., MacKerell A.D.. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Meth. 2016; 14:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L.. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983; 79:926–935. [Google Scholar]
- 50. Bussi G., Donadio D., Parrinello M.. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007; 126:014101. [DOI] [PubMed] [Google Scholar]
- 51. Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R.. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984; 81:3684–3690. [Google Scholar]
- 52. Darden T., York D., Pedersen L.. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993; 98:10089–10092. [Google Scholar]
- 53. Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M.. LINCS: a linear constraint Solver for molecular simulations. J. Comput. Chem. 1997; 18:1463–1472. [Google Scholar]
- 54. Aoki A., Suetake I., Miyagawa J., Fujio T., Chijiwa T., Sasaki H., Tajima S.. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 2001; 29:3506–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ziller M.J., Müller F., Liao J., Zhang Y., Gu H., Bock C., Boyle P., Epstein C.B., Bernstein B.E., Lengauer T. et al.. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011; 7:e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Duymich C.E., Charlet J., Yang X., Jones P.A., Liang G.. DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat. Commun. 2016; 7:11453–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu T.-H., Liu M., Zhou X.E., Liang G., Zhao G., Xu H.E., Melcher K., Jones P.A.. Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B. Nature. 2020; 586:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ziller M.J., Ortega J.A., Quinlan K.A., Santos D.P., Gu H., Martin E.J., Galonska C., Pop R., Maidl S., Di Pardo A. et al.. Dissecting the functional consequences of de novo DNA methylation dynamics in human motor neuron differentiation and physiology. Cell Stem Cell. 2018; 22:559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Strahl B.D., Grant P.A., Briggs S.D., Sun Z.W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F. et al.. Set2 is a nucleosomal histone H3-Selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002; 22:1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sagendorf J.M., Markarian N., Berman H.M., Rohs R.. DNAproDB: an expanded database and web-based tool for structural analysis of DNA–protein complexes. Nucleic Acids Res. 2019; 79:233–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin C.-C., Chen Y.-P., Yang W.-Z., Shen J.C.K., Yuan H.S.. Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B. Nucleic Acids Res. 2020; 48:3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anteneh H., Fang J., Song J.. Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation. Nat. Commun. 2020; 11:2294–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Emperle M., Rajavelu A., Kunert S., Arimondo P.B., Reinhardt R., Jurkowska R.Z., Jeltsch A.. The DNMT3A R882H mutant displays altered flanking sequence preferences. Nucleic Acids Res. 2018; 46:3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu J., Hu H., Panserat S., Marandel L.. Evolutionary history of DNA methylation related genes in chordates: new insights from multiple whole genome duplications. Sci. Rep. 2020; 10:970–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gabel H.W., Kinde B., Stroud H., Gilbert C.S., Harmin D.A., Kastan N.R., Hemberg M., Ebert D.H., Greenberg M.E.. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015; 522:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W.T.C., Bauer C., Münzel M., Wagner M., Müller M., Khan F. et al.. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013; 152:1146–1159. [DOI] [PubMed] [Google Scholar]
- 67. Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F. et al.. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017; 356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang L., Rau R., Goodell M.A.. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E., Kandoth C., Payton J.E., Baty J., Welch J. et al.. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010; 363:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Published WGBS datasets were obtained from Gene Expression Omnibus (GEO) and are listed in Supplementary Table S1. Published crystal structures of DNMT3A (6F57, 5YX2) and DNMT3B (6KDA) were obtained from the Protein Data Bank (PDB) (https://www.rcsb.org). Newly generated WGBS datasets have been deposited to GEO under accession number GSE151992.