Abstract
The current pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a global health crisis and will likely continue to impact public health for years. As the effectiveness of the innate immune response is crucial to patient outcome, huge efforts have been made to understand how dysregulated immune responses may contribute to disease progression. Here we have reviewed current knowledge of cellular innate immune responses to SARS-CoV-2 infection, highlighting areas for further investigation and suggesting potential strategies for intervention. We conclude that in severe COVID-19 initial innate responses, primarily type I interferon, are suppressed or sabotaged which results in an early interleukin (IL)-6, IL-10 and IL-1β-enhanced hyperinflammation. This inflammatory environment is driven by aberrant function of innate immune cells: monocytes, macrophages and natural killer cells dispersing viral pathogen-associated molecular patterns and damage-associated molecular patterns into tissues. This results in primarily neutrophil-driven pathology including fibrosis that causes acute respiratory distress syndrome. Activated leukocytes and neutrophil extracellular traps also promote immunothrombotic clots that embed into the lungs and kidneys of severe COVID-19 patients, are worsened by immobility in the intensive care unit and are perhaps responsible for the high mortality. Therefore, treatments that target inflammation and coagulation are promising strategies for reducing mortality in COVID-19.
Keywords: Innate immunology, COVID-19, Neutrophils, Monocytes, NK cells, Macrophages
Graphical Abstract
Graphical Abstract.
Highlights
NK cells are thought to undergo mass migration to the lungs, depleting circulating populations and may contribute to tissue damage
Complement activation is linked to virus removal, but is implicated in neutrophil activation and thrombosis
A robust early macrophage IFN-response is associated with mild disease, while high levels of the cytokines IL-6, IL-10 and IL-1β in the early stages of COVID-19 are linked to poor outcome
Neutrophil activation causes tissue damage and an increased risk of potentially fatal immunothrombotic events in the lungs and kidneys of COVID-19 patients
Monocyte HLA-DR expression, neutrophil activation and the ratio of neutrophils to lymphocytes might be a potential identifier of high-risk patients
Box 1: Why does your reviewed topic matter in the pandemic?
The first responders to viral infection in coronavirus disease 2019 (COVID-19) are the cells of the innate immune system, including natural killer (NK) cells, monocytes, macrophages and neutrophils. After migrating to the site of infection and recognizing the virus from conserved features, innate cells initiate responses to eliminate the virus and coordinate the adaptive immune system. However, in some cases, innate cells have been shown to contribute to viral dissemination and tissue damage which is associated with severe COVID-19 disease and fatal outcomes. Therefore, it is critical to evaluate innate immunity in the context of COVID-19 to predict outcome and facilitate development of therapeutic interventions.
Box 2: What is the consensus?
A hallmark of life-threatening COVID-19 is a pathological hyperactive immune cell response brought about by the inability to control virus. NK cells that are lost in the circulation traffic to the lungs where they may contribute to inflammatory damage. Alveolar macrophages and monocytes are implicated in cytokine propagation. Macrophage virus infection and activation may inhibit their ability to cloak inflammation. Non-classical monocytes and neutrophils are increased in the blood of severe COVID-19 patients suggesting high bone marrow turnover. A cytokine storm has also been reported in COVID-19. Though high cytokine levels such as interleukin (IL)-6, IL-10 and IL1-β are linked to disease severity, these cytokine elevations are thought to only occur early in the disease followed by reported immunosuppression. Complement, damage and cytokines are implicated in excessive neutrophil activation, triggering fibrosis and damaging thrombotic events, thought to be the key drivers of mortality in COVID-19. As such, a high ratio of neutrophils to lymphocytes (including NK cells and T cells) and high D-dimer levels can predict COVID-19 disease outcome.
INTRODUCTION
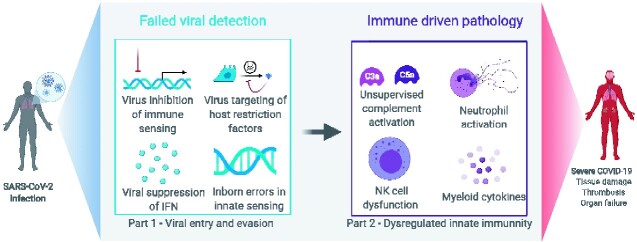
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel betacoronavirus causing coronavirus disease 2019 (COVID-19). COVID-19 has had devastating effects on human health and social and economic aspects worldwide. SARS-CoV-2 is highly infectious and while a proportion of patients present with asymptomatic disease, clinical manifestations can include fever, cough, loss of smell/taste, fatigue, difficulty breathing and organ dysfunction [acute respiratory distress syndrome (ARDS)], which for a small percentage of patients will be fatal. The risk of developing severe disease is associated with old age, gender, ethnicity and underlying health conditions including diabetes, obesity and heart disease. Severe outcomes likely involve a defective viral sensing and a dysregulated immune response. Part 1 of this review discussed the mechanisms of SARS-CoV-2 host cell entry and immune evasion. Part 2 assesses the current understanding of how innate immune responders [natural killer cells (NK), myeloid cells and neutrophils] contribute to pathogenesis.
The innate immune system provides the first line of defence against infection, rapidly respond to pathogens through recognition of conserved features, but are also important for adaptive immune responses. However, such lack of specificity can contribute to tissue damage and undermine adaptive immunity, thus a fine balance is required to protect the host.
The complement system and ‘cytokine storms’ in COVID-19
The complement system comprises several plasma proteins that opsonize or lyse pathogens and orchestrate host defence [1–3]. Dysregulated activation of complement plays a role in chronic inflammation, endothelial cell dysfunction, intravascular coagulation and thrombus formation eventually leading to multiple organ dysfunction syndromes and death [4], and has been implicated in the pathophysiology of Middle East respiratory syndrome, SARS [5–8] and COVID-19 [5]. Immunohistochemistry analysis of lung tissue from patients who died of COVID-19 revealed deposition of complement components mannose-binding lectin, C4, C3 and the terminal membrane attack complex C5b-9 in alveolar epithelial/inflammatory cells and alveolar spaces [9]. Additionally, elevated plasma C5a and sC5b-9 have been observed in patients with moderate and severe COVID-19 [10]. C5a is a potent signalling molecule that activates immune cells to release cytokines, including tumour necrosis factor (TNF), interleukin (IL)‐1β, IL‐6 and IL‐8 within hours of infection [11]. Despite some suggestive evidence, the complement system has received little attention in the search for effective anti-inflammatory treatment strategies despite multiple intuitive targets in COVID-19 [12], listed in Table 1.
Table 1:
Potential treatments
| Therapy | Target | Mechanism | Reference |
|---|---|---|---|
| Monalizumab | NKG2A | As upregulation of NKG2A is associated with reduced lymphocyte to neutrophil balance, inhibition of NKG2A may improve lymphocyte numbers and therefore virus control. |
van Hall et al. [13] Antonioli et al. [14] |
| Sarilumab and Tocilizumab | IL-6 | Higher blood concentrations of IL-6 were reported to be predictive of fatal outcome in COVID-19 patients therefore blocking IL-6 using antibodies may be effective. |
Ruan et al. [15] Guaraldi et al. [16] Tomasiewicz et al. [17] ClinicalTrials.gov Identifier: NCT04320615 (no statistically significant differences in ventilator-free days between the drug and placebo) |
| Adalimumab | TNF | Proven to reduce inflammation in many diseases and thus promising for severe COVID-19 | Mahase [18] |
| Anakinra | IL-1 receptor (agonist) | Competitively inhibits IL-1 binding to the IL-1 type I receptor. Increased IL-1 concentrations have been reported in COVID-19 patients. IL-1a and IL-1b have been implicated in severe COVID-19 disease. |
Giamarellos-Bourboulis et al. [19] Huang C et al. [20] Ong et al. [21] Huet et al. [22] Cavalli et al. [23] |
| IFN I and IFN II targeting drugs | IFN blocking | Late IFN responses are associated with hyperinflammation in severe COVID-19 disease. Due to potential off-target effects, blocking IFN I responses may be more effective than blocking IFN II responses. |
Hemann et al. [24] Sallard et al. [25] Prokunina-Olsson et al. [26] |
| IFN I supplementation |
IFN-β, IFN-α2b, IFN-α1b |
Severe COVID-19 patients have shown reduced IFN I responses. IFN I supplementation reduced the duration of inflammatory markers in mild disease and prevented COVID-19 infection in highly exposed individuals. |
Hung et al. [27] Zhou et al. [28] Meng et al. [29] (prevented infection in highly exposed individuals) |
| Baricitinib, Ruxolitinib | JAK 1 and JAK 2 | May prevent virus entry into cells as well as beneficial anti-inflammatory activity. Inhibits NK cell activity, DC development and function which could suppress antigen-specific T cell responses. |
Stebbing et al. [30] Elli et al. [31] |
| Gimsilumab, mavrilimumab, Sargramostim (Human recombinant GM-CSF) | GM-CSF replacement | GM-CSF promotes proinflammatory responses. GM-CSF expression increases T helper 1 cells and monocytes in COVID-19 patients, particularly those in intensive care. GM-CSF also plays a role in alveolar macrophage physiology and might protect against viral related injury in early stages. |
Zhou et al. [32] De Luca et al. [33] Clinicaltrials.gov identifier: NCT 04326920 |
| Monoclonal antibodies | Immune checkpoint blockade e.g. PD-1, NKG2A and CD39. | Immune checkpoint therapy, approved for melanoma, has been shown to upregulate effective immune responses by blocking inhibitory markers on immune cells and infected cells. In COVID-19 disease it was shown that NK cell responses could be enhanced using immune checkpoint blockade. | Demaria et al. [34] |
| Dexamethasone | Glucocorticoid receptor | Increases anti-inflammatory genes (e.g. IκB-α), reduces pro-inflammatory genes (e.g. COX2) | Johnson and Vinetz [35] |
| Hydrocortisone | Glucocorticoid receptor | Increases anti-inflammatory genes (e.g. IκB-α), reduces pro-inflammatory genes (e.g. COX2) | Mahase [36] |
| Anti-coagulants | Coagulation cascade | E.g. Warfarin, apixaban, betrixaban, dabigatran, edoxaban and rivaroxaban | Nadkarni et al. [37] |
| Ecluzimab | C5a | Inactivates C5a, removing anaphylatoxin activity |
Cugno et al. [10] Campbell and Kahwash [12] |
| High-dose IV immunoglobulin | C3b | Sequesters C3b stops both MAC activity and C5a production | Cugno et al. [10] |
In addition to complement, pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) can also induce cytokine release. Cytokines play an important role in promoting immunopathology and immune dysfunction: ‘Cytokine storms’ reported in severe SARS-CoV-2 infected patients (reviewed in detail by Ragab et al. [38] refers to an elevated and dysregulated release of pro-inflammatory cytokines and chemokines, including IL-6, TNF-α, IL-1, interferon (IFN)-γ and MCP-1. Some studies report only modest cytokine elevations [39], particularly early in disease, while other studies correlate IL-6 and TNF-α levels with disease severity [40–43] and are therefore promising targets for treatment (Table 1). However, there is no consensus on reported cytokine levels, particularly IL-6 (approximate range = 10pg/mL to >20 ng/mL), which could be attributed to analytical method or infection time-point. Indeed, it has been suggested that early IL-6 elevations are followed by cytokine immunosuppression [39]. Cytokines related to tissue repair (e.g. IL-10, IL-4 and IL-5) are also upregulated in SARS-CoV-2 infection [20]. Therefore, a ‘cytokine storm’ driving COVID-19 pathology is controversial and some studies reject this terminology [39, 44]. Yet, the inflammatory cytokine response remains important in COVID-19 as severe patients often display symptoms usually associated with sepsis [45]. Here we address the role of innate cells in this response.
NK cell dysregulation in COVID-19
NK cells are a heterogeneous subset of innate lymphocytes that play a key role in early anti-viral host defence [46, 47]. NK cells directly recognize virus-infected cells, lyse them and facilitate crosstalk between innate and adaptive immune responses [48]. Their activation is regulated by receptors that sense cell stress or viral-specific signatures and finely control the balance between activation and inhibition. In COVID-19 pathology, a functional NK response is associated with enhanced SARS-CoV-2 neutralization by antibody-dependent cellular cytotoxicity (ADCC) [49], increased numbers of the cytokine-producing Cluster of Differentiation (CD)56brightCD16- NK cell subset and upregulated IFN- and XCL2-related genes [50]. Therefore, an effective NK cell response is likely crucial to control COVID-19 disease. Without the support of healthy phagocytes ADCC may increase inflammation (e.g. through DAMP release), however, reports of this and unassisted NK cell lysis of virally infected cells are mostly absent in COVID-19 [51], including peptide-loaded MHC-1 presentation and recognition. SARS-CoV-2 ORF8-mediated downregulation of MHC-I in infected cells [52] could also either promote or inhibit NK cell-mediated lysis, though this is also not known in COVID-19.
Some studies suggest a preferential reduction in circulating NK cells and reduced NK functionality are associated with severe COVID-19 [34, 53–55] disease but it is unclear whether this is due to cell trafficking to infected organs or NK cell death. Transcriptomic analysis suggested increased apoptosis-related pathways in circulating lymphocytes from COVID-19 patients [56]. Conversely, a recent scRNAseq analysis suggested increased NK cell representation in the lungs of COVID-19 patients [56] and genes involved in cell trafficking were enriched in bronchoalveolar lavage (BAL) NK cells from severe COVID-19 patients [57]. BAL fluids from COVID-19 patients also contained more chemokines which potentially could increase NK cell recruitment [58]. Though the mechanism has yet to be elucidated, CXCR3 ligand-producing cells are increased in COVID-19 patient’s lungs [58], which may indicate a CXCR3-dependent mechanism for NK cells recruitment to the lung as described in influenza A infection [59]. NK cells in the periphery may migrate to the lung and may contribute to immune-mediated lung damage in COVID-19 though this is not yet understood.
SARS-CoV-2 infection also modulates the NK cell phenotype causing functional changes that could affect patient outcome. Recent data from COVID-19 patients with pneumonia or ARDS suggest an increase in NK cell exhaustion, characterized by elevated NKG2A, PD-1 and CD39 expression in patients with severe infection [34]. Targeting these molecules has been suggested as therapeutic strategy for COVID-19 (Table 1). In keeping with an exhaustion phenotype, NK cells in COVID-19 patients are functionally impaired [53, 60], which could be explained by hyperinflammation, particularly the persistent elevation of IL-6 in COVID-19 patients [60, 61]. In contrast, ‘adaptive’ NK subpopulations with enhanced ADCC responses, actively proliferated and expanded in the periphery of severe COVID-19 patients seropositive for cytomegalovirus (CMV) [57]. This expansion did not correlate with CMV IgG, suggesting these cells could be repurposed towards combating SARS-CoV-2. Consistent with this, HLA-E, a major ligand for NKG2C-expressing adaptive NK cells, is upregulated in BAL fluid of COVID-19 patients [57]. Additionally, increased prevalence of the NKG2Cdel variant, associated with reduced NKG2C expression, has been found in severe COVID-19 which may highlight the importance of the NKG2C–HLA-E axis on antiviral activity [62].
In severe disease, CD56bright NK cells, (enhanced in mild cases and usually associated with cytokine release/immunoregulation) exhibited higher levels of cytotoxic mediators [57], suggesting that NK cell regulation is abandoned for cytotoxicity, perhaps further promoting tissue damage. However, overall, while NK cell dysregulation has been described in SARS-CoV-2 patients there is still lack of understanding of early NK cell-mediated responses and the balance between pathological versus protective NK cell responses at the sites of infection.
Monocytes and macrophages contribute to dysregulated inflammation
Macrophages, monocytes, and dendritic cells (DCs) are sentinels of the immune system that express a broad repertoire of PAMP, DAMP and complement receptors. They also present antigen to trigger adaptive immune responses and are equipped with antibody Fc receptors to be effector cells. Under physiological conditions, macrophages are tissue accessory cells that dampen inflammation via scavenging of dead cells and DAMPs such as ATP [63]. Tissue-resident macrophages are likely the first to encounter SARS-CoV-2 in the lung: alveolar macrophages can detect viral components via PRRs and produce Type I IFNs (IFNs I; IFN-α and IFN-β): essential cytokines in the antiviral immune response [64]. These cells will also recruit other immune cells to trigger inflammation [65], which overrides their usual anti-inflammatory function, as the ability of macrophages to cloak inflammation is ineffective when the tissue damage is considerable [63]. Reports of two fatal COVID-19 cases showed increased macrophage recruitment in alveolar cavities expressing high levels of CD68, indicating an activated state [66]. Those macrophages were shown to secrete moderate amounts of pro-inflammatory IL-6 and TNF-α but high levels of anti-inflammatory IL-10.
As macrophages express ACE-2, direct infection with SARS-CoV-2 could prevent IFN I production (a viral evasion mechanism discussed in Part 1 of this review) impairing viral control and causing hyperinflammation [67]. Mild SARS-CoV-2 infection has been associated with robust IFN I responses [68] which might account for the higher proportion of non-severe cases of COVID-19 compared to SARS-CoV. However, defective IFN I responses have been reported in severe COVID-19 cases, where patients lacking IFN-α production had poorer outcomes [69, 70]. Murine SARS-CoV studies suggest early IFNs I support clearance of virus, while delayed or absent responses cause elevated cytokine levels and impaired adaptive immune responses [67]. Consequently, blocking IFN I at late stages of the disease may be advantageous (Table 1). While alveolar macrophages can produce IFN I, they are not the primary source during viral infection. Therefore, viral downregulation of IFN I in macrophages unlikely drives the dampened immune response and macrophages will not be the primary source of cytokine production, being outnumbered by more numerous tissue and stromal cells. Despite this, macrophages produce many of the cytokines associated with COVID-19 infection [66] and the failure of these cells to fully contain cellular damage is a likely contributor to disease outcome.
Circulating monocytes are recruited to the lungs of COVID-19 patients where monocyte attracting chemokines including CCL2 and CCL7 are increased [68]. Monocytes are classified by their expression of cell surface receptors: classical monocytes (CD14+CD16−) pass through an intermediate state (CD14+CD16+) to eventually develop into non-classical monocytes (CD14−CD16+). Increased demand for monocytes in inflammation is associated with elevated intermediate and non-classical monocytes in the blood [71] and observed in COVID-19 patients [32]. Intermediate monocytes increase in proportion with disease severity and produce granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-6. In addition, the population of large, vacuolated monocytes were observed in the peripheral blood of COVID-19 patients but not healthy individuals [72]. These cells express several macrophage markers including CD163 and CD206 in addition to CD68 and CD80 and produce a mixture of cytokines including IL-6, TNF-α and IL-10. Further studies are required to validate this subset.
In mild COVID-19, circulating monocytes express high levels of Human Leukocyte Antigen (HLA)-DR, CD11c and CD14 and are characterized by an IFN-driven transcriptional programme. In sharp contrast, patients with severe COVID-19 monocytes showed low expression of HLA-DR while expressing anti-inflammatory associated genes [19, 73, 74]. Low monocyte HLA-DR expression has been linked to monocyte anergy and associated with increased risk of secondary infections and death after sepsis [75]. Also termed monocytic myeloid-derived suppressor cells, these monocytes proliferate rapidly in the plasma of COVID-19 patients which potentially enables identification of high-risk patients [76].
SARS-CoV-2 spike protein can induce IL-8, IL-6 and TNF-α secretion in monocyte-derived macrophages (MDMs) and specifically primes COVID-19 patient-derived MDMs for IL-1β production in vitro [77]. IL-6 is heavily involved in the transition from innate to adaptive immune response biasing the differentiation of monocytes into macrophages, in addition to driving recruitment of neutrophils and monocytes [78]. IL-6 is also important for T cell and B cell differentiation and antibody production [79]. Macrophage produced TNF-α causes apoptosis of effector T cells [80], while IL-10 produced by lung macrophages in severe COVID-19 prevents T cell responses [81]. Cytokine production is important to elicit an effective COVID-19 immune response, however, an excess or a dysregulation of production can contribute to pathology and tissue damage. Consequently, inhibiting the function of these cytokines is predicted to provide a therapeutic benefit in COVID-19 (Table 1).
Much less data have been collected on the role of DCs in COVID-19, though studies demonstrate a decrease of plasmacytoid DCs (pDCs) in the lung and blood of patients [43, 82], which usually produce high amounts of IFN I in response to viral infection [83]. DCs’ ability to mature and activate adaptive immune cells could also be impaired in COVID-19, illustrated by reduced expression of co-stimulatory CD86 [82]. Furthermore, while it has been theorized that mast cells, basophils and eosinophils are important in COVID-19 [84, 85], reports on these cells are limited, though one study proposed that eosinophils and basophils may be important for recovery [86].
Neutrophil extracellular traps drive COVID-19 pathology
Neutrophils serve as the first wave of immune cells recruited to infection and are prominent in viral defence and response to cellular damage [87]. A rat coronavirus infection model demonstrated that neutrophils were both involved in viral control and pathology including haemorrhagic lesions, epithelial barrier permeability and lung inflammation [88].
In COVID-19, neutrophil activation/degranulation is the largest gene signature in blood cells [89] predicting disease severity [90] and mortality [91]. An increasing number of reports have identified a high neutrophil to lymphocyte ratio to be associated with severe COVID-19 [92, 93]. Proteomic analysis has also revealed an IFN I signature in neutrophils in COVID-19 [94]. This is interesting because IFNs I were shown to be targeted by existing autoantibodies in some patients [95]. Anti-nuclear autoantibodies were also detected in COVID-19 presumably promoted by cell death or NETosis [96]. Neutrophil extracellular traps (NETs) are pyroptotically released strings of DNA/chromatin, toxic granules and fibres that are assisted by complement to capture and inactivate viruses [97]. As neutrophils express antibody binding Fc receptors, both autoantibody observations may indicate that neutrophils are forming a positive feedback loop of immune activation that leads to pathology. This overactivation could also be attributed to the phenotype of the neutrophils themselves, likely driven by persistent inflammation, elevated GM-CSF [20] and evidenced by the strange presence of hyperactive (spontaneous NET) low-density neutrophils [98] and immature neutrophils [94].
Along with activation, markers of systemic NETosis have been identified in COVID-19 patients’ blood and are linked to disease severity [99–101] (recently reviewed by Tomar et al. [102]. Extensive pathology may occur when large numbers of neutrophils swarm to damaged or infected tissue [63], the resulting NETosis can promote fibrosis or form a chain reaction with platelets, complement and coagulation factors leading to a thrombosis [103] that can be anti-microbial [104]. NETs are also associated with thrombotic events in COVID-19, consisting of platelets, complement (C3) and tissue factor in blood [8, 105–107]. A lower platelet count correlates with severe COVID, and may be indicative of this thrombosis [108], as are higher D-dimer levels [109, 110], which alone can predict disease outcome [111]. Thrombotic events are extremely dangerous in an intensive care unit (ICU) setting, as patients are immobile and at high risk of developing fatal clots [112–114]. Such as the NET-associated thrombotic events in the lung microvasculature, which are thought to be a key driver of lung pathology and subsequent death [99, 113, 115, 116].
Chronic kidney disease is another common outcome in COVID-19 [117]: as NETs are thought to drive kidney pathology in other diseases, they are likely to contribute to renal failure via immunothrombosis in COVID-19 [118, 119]. It is not yet clear what causes NETosis in COVID-19, though viral-induced cell death, DAMP release (e.g. Haem and ATP) and elevated cytokine levels are all implicated, especially IL-8: a potent neutrophil chemoattractant [39]. A recent study suggests that IL-6-driven complement elevation plays a role in NET formation, as activated neutrophils are known to have surfaces liable to complement binding, but also will respond to anaphylatoxins (C3a and C5a) via complement receptors [120].
CONCLUSIONS
In Part 1 of this review, we concluded that SARS-CoV-2 deliberately sabotages early innate immunity, which along with inborn errors of innate sensing and IFN signalling results in increased viral load in severe COVID-19. The heavy viral burden causes cellular damage and triggers a hyperinflammatory response involving several innate immune cell types, summarized in Fig. 1. Importantly, we focus on innate immunity because there is a bias towards these cells in COVID-19, however, these findings should be taken in context with the entire immune response, as adaptive systems likely activate innate cells through antibodies and cytokines.
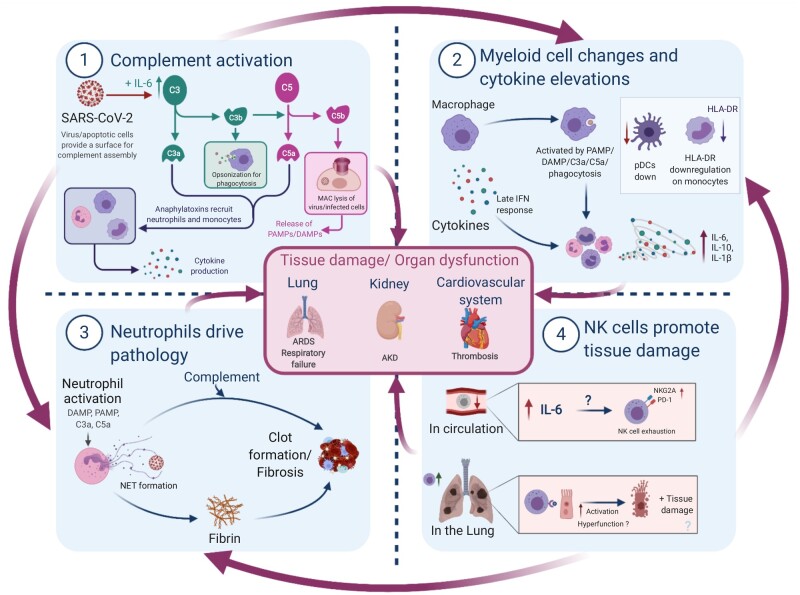
Figure 1:
Summary of innate immune-driven pathology. Graphical summary depicting the role of innate immune components in COVID-19 pathology. (1) Complement is elevated by IL-6 and likely activated by viral/apoptotic cell membranes to initiate inflammation via anaphylatoxins. (2) Macrophages can sense tissue damage (DAMPs), complement (anaphylatoxins or C3b opsonization) or virus (PAMPs) to release cytokines; direct infection delays IFN responses. Monocytes display signs of activation or anergy through HLA-DR downregulation, while anti-viral pDCs are decreased. (3) Neutrophils respond to damage, complement and cytokines by releasing NETs consisting of fibres and DNA. NETs may act as a nucleation point for complement and coagulation factors to drive thrombosis—thought to be a major driver of mortality in COVID-19. (4) NK cells are reduced in circulation and display markers of exhaustion, which is possibly driven by elevated IL-6. These cells traffic to infected lungs where they clear virus-infected cells in mild COVID-19. However, in severe disease, these cells are possibly less effective or may contribute to tissue damage and further inflammation via DAMP release from apoptotic cells, especially if phagocytes are dysfunctional. Importantly, NK cell mechanisms are still poorly understood in COVID-19. All these innate immune factors together promote the hyperinflammation, tissue damage and organ dysfunction observed in severe COVID-19, though such inflammation may be followed by immunosuppression. AKD, acute kidney disease.
NK cells are principal anti-viral innate lymphocytes and display reduced circulating numbers perhaps attributed to lung recruitment in COVID-19. In severe cases, NK cells display ‘exhausted’ phenotypes and an enrichment of specialized subsets with a bias towards cytotoxicity. Without healthy phagocyte clearance of NK-targeted apoptotic host cells, such activity may result in further inflammation, however, further research is needed to evaluate the role of NK cells in COVID-19. NK cells also likely join macrophages and monocytes in contribution to the cytokine environment identified in COVID-19. There is no consensus for a cytokine storm in COVID-19. Though certain cytokines, such as high early IL-6 levels can predict the worse outcomes. In mild cases, macrophages have an early robust IFN I response, associated with viral clearance, i.e. lacking in severe patients. Additionally, low monocyte HLA-DR expression, neutrophil activation, and the ratio of neutrophils to lymphocytes have been suggested as potential identifiers of high-risk patients.
Neutrophils express an IFN I signature and produce NETs which have been implicated in immunopathology, particularly the formation of potentially fatal thrombotic events in COVID-19 patients. It is likely that activated leukocytes and NETs provide a nucleation point for the complement and coagulation pathways, and these resulting thrombi can become clogged in the small capillaries of inflamed lungs and kidneys. The heightened systemic inflammation and poor circulation in ICUs increase the likelihood of such clotting events, and thus treatments (Table 1) which target inflammation (e.g. dexamethasone), complement and coagulation are keys to reduce mortality in COVID-19.
ACKNOWLEDGEMENTS
We would like to thank all members of the Oxford-Cardiff COVID19 Literature Consortium for their hard work and commitment during the pandemic. Figures were created using Biorender.com.
AUTHORS’ CONTRIBUTIONS
P.R.S.R., A.A., E.P. and V.M.T.B. conceptualized the review, wrote the original draft and reviewed and edited the manuscript. R.J. reviewed and edited the manuscript. C.C., F.L., M.T. and S.M.-T. were involved in conceptualization and reviewed the manuscript. F.C.R. and D.O.S. administered the project and reviewed and edited the manuscript. E.G.-M. facilitated conceptualization and reviewed and edited the manuscript. L.C.D. conceptualized the review, wrote the original draft, reviewed and edited the manuscript and administered the project.
DATA AVAILABILITY
All data are contained within the manuscript. This review was facilitated by weekly releases of the Oxford-Cardiff COVID19 Literature Consortium journal club—a database of reviewed articles and journals will be made available on request.
REFERENCES
- 1. Ricklin D, Reis ES, Lambris JD. Complement in disease: a defence system turning offensive. Nat Rev Nephrol 2016;12:383–401. doi:10.1038/nrneph.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kolev M, Friec GL, Kemper C. Complement—tapping into new sites and effector systems. Nat Rev Immunol 2014;14:811–20. doi:10.1038/nri3761. [DOI] [PubMed] [Google Scholar]
- 3. Kumar V. Sepsis roadmap: what we know, what we learned, and where we are going. Clin Immunol 2020;210:108264. doi:10.1016/j.clim.2019.108264. [DOI] [PubMed] [Google Scholar]
- 4. Noris M, Remuzzi G. Overview of complement activation and regulation. Semin Nephrol 2013;33:479–92. doi:10.1016/j.semnephrol.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chauhan AJ, Wiffen LJ, Brown TP. COVID-19: a collision of complement, coagulation and inflammatory pathways. J Thromb Haemost 2020;18:2110–7. doi:10.1111/jth.14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gralinski LE, Sheahan TP, Morrison TE et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio 2018;9:e01753–18.doi:10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang R, Xiao H, Guo R et al. The role of C5a in acute lung injury induced by highly pathogenic viral infections. Emerg Microb Infect 2015;4:e28. doi:10.1038/emi.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Magro C, Mulvey JJ, Berlin D et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res 2020;220:1–13. doi:10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gao T, Hu M, Zhang X et al. PREPRINT: highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv 2020. doi:10.1101/2020.03.29.20041962. [Google Scholar]
- 10. Cugno M, Meroni PL, Gualtierotti R et al. Complement activation in patients with COVID-19: a novel therapeutic target. J Allergy Clin Immunol 2020;146:215–7. doi:10.1016/j.jaci.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foreman KE, Glovsky MM, Warner RL et al. Comparative effect of C3a and C5a on adhesion molecule expression on neutrophils and endothelial cells. Inflammation 1996;20:1–9. doi:10.1007/BF01487740. [DOI] [PubMed] [Google Scholar]
- 12. Campbell CM, Kahwash R. Will complement inhibition be the new target in treating COVID-19– related systemic thrombosis. Circulation 2020;141:1739–41. doi:10.1161/CIRCULATIONAHA.120.047419. [DOI] [PubMed] [Google Scholar]
- 13. van Hall T, André P, Horowitz A et al. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J Immunother Cancer 2019;7:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Antonioli L, Fornai M, Pellegrini C et al. NKG2A and COVID-19: another brick in the wall. Cell Mol Immunol 2020;17:672–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruan Q, Yang K, Wang W et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med 2020;46:846–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guaraldi G, Meschiari M, Cozzi-Lepri A et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol 2020;2:e474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tomasiewicz K, Piekarska A, Stempkowska-Rejek J et al. Tocilizumab for patients with severe COVID-19: a retrospective, multi-center study. Expert Rev Anti-infect Therapy 2020:1–8, doi: 10.1080/14787210.2020.1800453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahase E. Covid-19: Anti-TNF drug adalimumab to be trialled for patients in the community. BMJ 2020;371:m3847. [DOI] [PubMed] [Google Scholar]
- 19. Giamarellos-Bourboulis EJ, Netea MG, Rovina N et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 2020;27:992–1000.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang C, Wang Y, Li X et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ong EZ, Chan YFZ, Leong WY et al. A Dynamic Immune Response Shapes COVID-19 Progression. Cell Host Microbe 2020;27:879–82.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huet T, Beaussier H, Voisin O et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol 2020;2:e393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cavalli G, De Luca G, Campochiaro C et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol 2020;2:e325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hemann EA, Gale M Jr, Savan R. Interferon lambda genetics and biology in regulation of viral control. Front Immunol 2017;8:1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sallard E, Lescure F-X, Yazdanpanah Y et al. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res 2020;178:104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prokunina-Olsson L, Alphonse N, Dickenson RE et al. COVID-19 and emerging viral infections: The case for interferon lambda. J Exp Med 2020;217:e20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hung IF-N, Lung K-C, Tso EY-K et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. The Lancet 2020;395:1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Q, Wei X-S, Xiang X et al. PREPRINT: Interferon-a2b treatment for COVID-19. medRxiv 2020:2020.04.06.20042580. [Google Scholar]
- 29. Meng Z, Wang T, Li C et al. PREPRINT: An experimental trial of recombinant human interferon alpha nasal drops to prevent coronavirus disease 2019 in medical staff in an epidemic area. medRxiv 2020:2020.04.11.20061473. [Google Scholar]
- 30. Stebbing J, Phelan A, Griffin I et al. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis 2020;20:400–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elli EM, Baratè C, Mendicino F et al. Mechanisms underlying the anti-inflammatory and immunosuppressive activity of ruxolitinib. Front Oncol 2019;9:1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou Y, Fu B, Zheng X et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci Rev 2020;7:998–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Luca G, Cavalli G, Campochiaro C et al. GM-CSF blockade with mavrilimumab in severe COVID-19 pneumonia and systemic hyperinflammation: a single-centre, prospective cohort study. Lancet Rheumatol 2020;2:e465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Demaria O, Carvelli J, Batista L et al. Identification of druggable inhibitory immune checkpoints on Natural Killer cells in COVID-19. Cell Mol Immunol 2020;17:995–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson RM, Vinetz JM. Dexamethasone in the management of COVID-19. BMJ 2020;370:m2648. [DOI] [PubMed] [Google Scholar]
- 36. Mahase E. Covid-19: hydrocortisone can be used as alternative to dexamethasone, review finds. BMJ 2020;370:m3472. [DOI] [PubMed] [Google Scholar]
- 37. Nadkarni GN, Lala A, Bagiella E et al. Anticoagulation, bleeding, mortality, and pathology in hospitalized patients with COVID-19. J Am Coll Cardiol 2020;76:1815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ragab D, Salah Eldin H, Taeimah M et al. The COVID-19 cytokine storm; what we know so far. Front Immunol 2020;11:1446. doi:10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Remy KE, Mazer M, Striker DA et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight 2020;5:e140329.doi:10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chi Y, Ge Y, Wu B et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis 2020;222:746–54. doi:10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han H, Ma Q, Li C et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microb Infect 2020;9:1123–30. doi:10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J, Rong L, Cui R et al. PREPRINT: dynamic changes in serum IL-6, IL-8, and IL-10 are associated with the outcome of patients with severe COVID-19 in ICU. Res Square 2020. doi:10.21203/rs.3.rs-83336/v1. [Google Scholar]
- 43. Sánchez-Cerrillo I, Landete P, Aldave B et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J Clin Invest 2020;130:6290–300. doi:10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Int Med 2020;180:1152–4. doi:10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 45. Hanidziar D, Bittner EA. Hypotension, systemic inflammatory response syndrome, and COVID-19: a clinical conundrum. Anesth Analg 2020;131:e175–6. doi:10.1213/ANE.0000000000005062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 1989;320:1731–5. doi:10.1056/nejm198906293202605. [DOI] [PubMed] [Google Scholar]
- 47. Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol 2013;132:515–25. doi:10.1016/j.jaci.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alrubayyi A, Ogbe A, Moreno Cubero E et al. Harnessing natural killer cell innate and adaptive traits in HIV infection. Front Cell Infect Microbiol 2020;10:395. doi:10.3389/fcimb.2020.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pinto D, Park YJ, Beltramello M et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020;583:290–5. doi:10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 50. Zhao XN, You Y, Wang GL et al. PREPRINT: longitudinal single-cell immune profiling revealed distinct innate immune response in asymptomatic COVID-19 patients. bioRxiv 2020. doi:10.1101/2020.09.02.276865. [Google Scholar]
- 51. Manickam C, Sugawara S, Reeves RK. Friends or foes? The knowns and unknowns of natural killer cell biology in COVID-19 and other coronaviruses in July 2020. PLoS Pathog 2020;16:e1008820. doi:10.1371/journal.ppat.1008820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Zhang J, Chen Y et al. PREPRINT: the ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv 2020. doi:10.1101/2020.05.24.111823. [Google Scholar]
- 53. Zheng M, Gao Y, Wang G et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol 2020;17:533–5. doi:10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wen W, Su W, Tang H et al. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov 2020;6:31. doi:10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilk AJ, Rustagi A, Zhao NQ et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 2020;26:1070–6. doi:10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chua RL, Lukassen S, Trump S et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat Biotechnol 2020;38:970–9. doi:10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 57. Maucourant C, Filipovic I, Ponzetta A et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol 2020;5:eabd6832. doi:10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liao M, Liu Y, Yuan J et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020;26:842–4. doi:10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 59. Carlin LE, Hemann EA, Zacharias ZR et al. ; Natural Killer Cell Recruitment to the Lung During Influenza A Virus Infection Is Dependent on CXCR3, CCR5, and Virus Exposure Dose. Front Immunol 2018;1:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Varchetta S, Mele D, Oliviero B et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol 2020;1–9. doi:10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Osman MS, van Eeden C, Cohen Tervaert JW. Fatal COVID-19 infections: is NK cell dysfunction a link with autoimmune HLH? Autoimmun Rev 2020;19:102561. doi:10.1016/j.autrev.2020.102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vietzen H, Zoufaly A, Traugott M et al. PREPRINT: NK cell receptor NKG2C deletion and HLA-E variants are risk factors for severe COVID-19. Res Square 2020. doi:10.21203/rs.3.rs-34505/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Uderhardt S, Martins AJ, Tsang JS et al. Resident macrophages cloak tissue microlesions to prevent neutrophil-driven inflammatory damage. Cell 2019;177:541–55.e17. doi:10.1016/j.cell.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kumagai Y, Takeuchi O, Kato H et al. Alveolar macrophages are the primary interferon-α producer in pulmonary infection with RNA viruses. Immunity 2007;27:240–52. doi:10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 65. McNab F, Mayer-Barber K, Sher A et al. Type I interferons in infectious disease. Nat Rev Immunol 2015;15:87–103. doi:10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang C, Xie J, Zhao L et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine 2020;57:102833. doi:10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Channappanavar R, Fehr Anthony R, Vijay R et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 2016;19:181–93. doi:10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhou Z, Ren L, Zhang L et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 2020;27:883–90.e2. doi:10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Trouillet-Assant S, Viel S, Gaymard A et al. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol 2020;146:206–8.e2. doi:10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang Q, Bastard P, Liu Z et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020:370:eabd4570. doi:10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Patel AA, Zhang Y, Fullerton JN et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 2017;214:1913–23. doi:10.1084/jem.20170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang D, Guo R, Lei L et al. PREPRINT: COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv 2020. doi:10.1101/2020.03.24.20042655. [Google Scholar]
- 73. Schulte-Schrepping J, Reusch N, Paclik D et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 2020;182:1419–40.e23. doi:10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lombardi A, Trombetta E, Cattaneo A et al. PREPRINT: early phases of COVID-19 are characterized by a reduction of lymphocyte populations and the presence of atypical monocytes. medRxiv 2020. doi:10.1101/2020.05.01.20087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Venet F, Demaret J, Gossez M et al. Myeloid cells in sepsis-acquired immunodeficiency. Ann NY Acad Sci 2020. doi:10.1111/nyas.14333. [DOI] [PubMed] [Google Scholar]
- 76. Falck-Jones S, Vangeti S, Yu M et al. PREPRINT: functional myeloid-derived suppressor cells expand in blood but not airways of COVID-19 patients and predict disease severity. medRxiv 2020. doi:10.1101/2020.09.08.20190272. [Google Scholar]
- 77. Theobald SJ, Simonis A, Kreer C et al. PREPRINT: the SARS-CoV-2 spike protein primes inflammasome-mediated interleukin-1- beta secretion in COVID-19 patient-derived macrophages. Res Square 2020. doi:10.21203/rs.3.rs-30407/v1. [Google Scholar]
- 78. Chomarat P, Banchereau J, Davoust J et al. ; IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nature Immunology 2000;1(6):510–514. doi: 10.1038/82763. [DOI] [PubMed] [Google Scholar]
- 79. Scheller J, Chalaris A, Schmidt-Arras D et al. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta 2011;1813:878–88. doi:10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 80. Mehta AK, Gracias DT, Croft M. TNF activity and T cells. Cytokine 2018;101:14–8. doi:10.1016/j.cyto.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brooks DG, Trifilo MJ, Edelmann KH et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med 2006;12:1301–9. doi:10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhou R, To KK-W, Wong YC et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity 2020;53:864–77.e5. doi:10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fitzgerald-Bocarsly P, Dai J, Singh S. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev 2008;19:3–19. doi:10.1016/j.cytogfr.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Afrin LB, Weinstock LB, Molderings GJ. Covid-19 hyperinflammation and post-Covid-19 illness may be rooted in mast cell activation syndrome. Int J Infect Dis 2020;100:327–32. doi:10.1016/j.ijid.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lindsley AW, Schwartz JT, Rothenberg ME. Eosinophil responses during COVID-19 infections and coronavirus vaccination. J Allergy Clin Immunol 2020;146:1–7. doi:10.1016/j.jaci.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rodriguez L, Pekkarinen PT, Lakshmikanth T et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep Med 2020;1:100078.doi:10.1016/j.xcrm.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Galani IE, Andreakos E. Neutrophils in viral infections: current concepts and caveats. J Leukocyte Biol 2015;98:557–64. doi:10.1189/jlb.4VMR1114-555R. [DOI] [PubMed] [Google Scholar]
- 88. Haick AK, Rzepka JP, Brandon E et al. Neutrophils are needed for an effective immune response against pulmonary rat coronavirus infection, but also contribute to pathology. J Gen Virol 2014;95:578–90. doi:10.1099/vir.0.061986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hemmat N, Derakhshani A, Bannazadeh Baghi H et al. Neutrophils, crucial, or harmful immune cells involved in coronavirus infection: a bioinformatics study. Front Genet 2020;11:41. doi:10.3389/fgene.2020.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Aschenbrenner AC, Mouktaroudi M, Kraemer B et al. PREPRINT: disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. medRxiv 2020. doi:10.1101/2020.07.07.20148395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Meizlish ML, Pine AB, Bishai JD et al. PREPRINT: a neutrophil activation signature predicts critical illness and mortality in COVID-19. medRxiv 2020. doi:10.1101/2020.09.01.20183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kong M, Zhang H, Cao X et al. Higher level of neutrophil-to-lymphocyte is associated with severe COVID-19. Epidemiol Infect 2020;148:e139. doi:10.1017/S0950268820001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Simadibrata DM, Calvin J, Wijaya AD et al. PREPRINT: neutrophil-to-lymphocyte ratio on admission to predict the severity and mortality of COVID-19 patients: a meta-analysis. medRxiv 2020. doi:10.1101/2020.09.14.20191098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Reyes L, Sanchez-Garcia MA, Morrison T et al. PREPRINT: proteomics identifies a type I IFN, prothrombotic hyperinflammatory circulating COVID-19 neutrophil signature distinct from non-COVID-19 ARDS. medRxiv 2020. doi:10.1101/2020.09.15.20195305. [Google Scholar]
- 95. Bastard P, Rosen LB, Zhang Q et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020:370:eabd4585. doi:10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhou Y, Han T, Chen J et al. Clinical and autoimmune characteristics of severe and critical cases of COVID-19. Clin Transl Sci 2020. doi:10.1111/cts.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. de Bont CM, Boelens WC, Pruijn GJM; NETosis, complement, and coagulation: a triangular relationship. Cell Mol Immunol 2019;16:19–27. doi:10.1038/s41423-018-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Morrissey SM, Geller AE, Hu X et al. PREPRINT: emergence of low-density inflammatory neutrophils correlates with hypercoagulable state and disease severity in COVID-19 patients. medRxiv 2020. doi:10.1101/2020.05.22.20106724. [Google Scholar]
- 99. Veras FP, Pontelli MC, Silva CM, et al. ; SARS-CoV-2–triggered neutrophil extracellular traps mediate COVID-19 pathology. Journal of Experimental Medicine 2020;217(12). doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang J, Li Q, Yin Y, et al. ; Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19. Frontiers in Immunology 2020;11(2063). doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zuo Y, Yalavarthi S, Shi H, et al. ; Neutrophil extracellular traps in COVID-19. JCI Insight 2020;5(11). doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tomar B, Anders H-J, Desai J, et al. ; Neutrophils and Neutrophil Extracellular Traps Drive Necroinflammation in COVID-19. Cells 2020;9(6):1383. doi: 10.3390/cells9061383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nakazawa D, Desai J, Steiger S, et al. ; Activated platelets induce MLKL-driven neutrophil necroptosis and release of neutrophil extracellular traps in venous thrombosis. Cell death discovery 2018;4:6-6. doi: 10.1038/s41420-018-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Engelmann B, Massberg S; Thrombosis as an intravascular effector of innate immunity. Nature Reviews Immunology 2013;13(1):34-45. doi: 10.1038/nri3345. [DOI] [PubMed] [Google Scholar]
- 105. Ding J, Hostallero DE, El Khili MR, et al. ; PREPRINT: A network-informed analysis of SARS-CoV-2 and hemophagocytic lymphohistiocytosis genes' interactions points to Neutrophil Extracellular Traps as mediators of thrombosis in COVID-19. medRxiv 2020:2020.07.01.20144121. doi: 10.1101/2020.07.01.20144121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Middleton EA, He X-Y, Denorme F, et al. ; Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 2020;136(10):1169-1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Skendros P, Mitsios A, Chrysanthopoulou A, et al. ; Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. The Journal of Clinical Investigation 2020. doi: 10.1172/JCI141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lippi G, Plebani M, Henry BM; Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clinica chimica acta; international journal of clinical chemistry 2020;506:145-148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yao Y, Cao J, Wang Q, et al. ; D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. Journal of Intensive Care 2020;8(1):49. doi: 10.1186/s40560-020-00466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cerda P, Ribas J, Iriarte A, et al. ; PREPRINT: D-dimer dynamics in hospitalized COVID-19 patients: potential utility for diagnosis of pulmonary embolism. medRxiv 2020:2020.09.21.20193953. doi: 10.1101/2020.09.21.20193953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Naymagon L, Zubizarreta N, Feld J, et al. ; Admission D-dimer levels, D-dimer trends, and outcomes in COVID-19. Thrombosis research 2020;196:99-105. doi: 10.1016/j.thromres.2020.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Connors JM, Levy JH; COVID-19 and its implications for thrombosis and anticoagulation. Blood 2020;135(23):2033-2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Price LC, McCabe C, Garfield B, et al. ; Thrombosis and COVID-19 pneumonia: the clot thickens! European Respiratory Journal 2020;56 (1):2001608. doi: 10.1183/13993003.01608-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wise J; Covid-19 and thrombosis: what do we know about the risks and treatment? BMJ 2020;369:m2058. doi: 10.1136/bmj.m2058. [DOI] [PubMed] [Google Scholar]
- 115. Leppkes M, Knopf J, Naschberger E, et al. ; Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine 2020;58. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Narasaraju T, Tang BM, Herrmann M et al. Neutrophilia and NETopathy as key pathologic drivers of progressive lung impairment in patients with COVID-19. Front Pharmacol 2020;11:870. doi:10.3389/fphar.2020.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Gansevoort RT, Hilbrands LB. CKD is a key risk factor for COVID-19 mortality. Nat Rev Nephrol 2020;16:705–6.doi:10.1038/s41581-020-00349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Salazar-Gonzalez H, Zepeda-Hernandez A, Melo Z et al. Neutrophil extracellular traps in the establishment and progression of renal diseases. Medicina (Kaunas, Lithuania) 2019;55:431. doi:10.3390/medicina55080431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Barnes BJ, Adrover JM, Baxter-Stoltzfus A et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med 2020;217:e20200652. doi:10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mukhopadhyay S, Sinha S, Mohapatra SK. PREPRINT: dynamic dysregulation of IL-6 and genes functional in NETosis, complement and coagulation in severe COVID-19 illness. medRxiv 2020. doi:10.1101/2020.10.13.20211425. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript. This review was facilitated by weekly releases of the Oxford-Cardiff COVID19 Literature Consortium journal club—a database of reviewed articles and journals will be made available on request.