Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic control will require widespread access to accurate diagnostics. Salivary sampling circumvents swab supply chain bottlenecks, is amenable to self-collection, and is less likely to create an aerosol during collection compared with the nasopharyngeal swab.
Methods
We compared real-time reverse-transcription polymerase chain reaction Abbott m2000 results from matched salivary oral fluid (gingival crevicular fluid collected in an Oracol device) and nasal-oropharyngeal (OP) self-collected specimens in viral transport media from a nonhospitalized, ambulatory cohort of coronavirus disease 2019 (COVID-19) patients at multiple time points. These 2 sentences should be at the beginning of the results.
Results
There were 171 matched specimen pairs. Compared with nasal-OP swabs, 41.6% of the oral fluid samples were positive. Adding spit to the oral fluid percent collection device increased the percent positive agreement from 37.2% (16 of 43) to 44.6% (29 of 65). The positive percent agreement was highest in the first 5 days after symptoms and decreased thereafter. All of the infectious nasal-OP samples (culture positive on VeroE6 TMPRSS2 cells) had a matched SARS-CoV-2 positive oral fluid sample.
Conclusions
In this study of nonhospitalized SARS-CoV-2-infected persons, we demonstrate lower diagnostic sensitivity of self-collected oral fluid compared with nasal-OP specimens, a difference that was especially prominent more than 5 days from symptom onset. These data do not justify the routine use of oral fluid collection for diagnosis of SARS-CoV-2 despite the greater ease of collection. It also underscores the importance of considering the method of saliva specimen collection and the time from symptom onset especially in outpatient populations.
Keywords: coronavirus, COVID-19, outpatient, saliva, SARS-CoV-2
In a study of nonhospitalized COVID-19 patients, the oral fluid salivary sample type was insensitive compared to nasal-OP specimen types overall, but was improved with the addition of spit and most sensitive in the first 5 days after symptom onset.
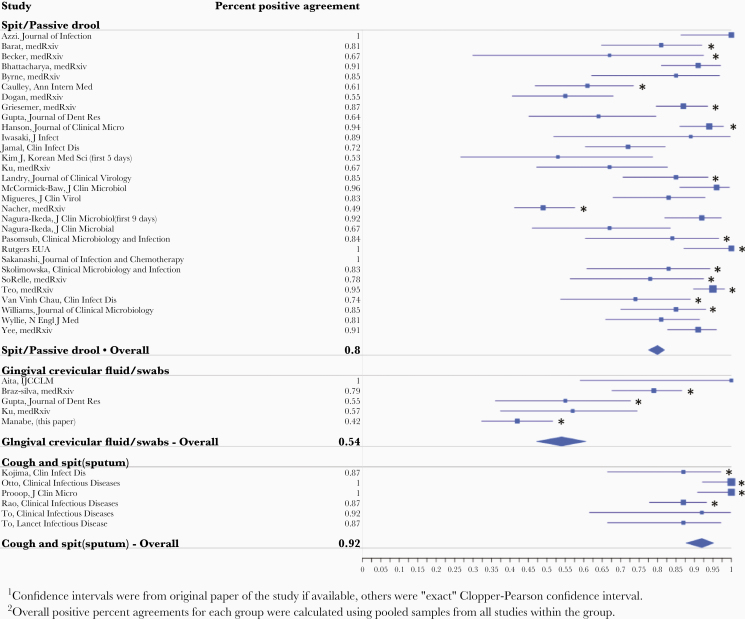
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has rapidly spread globally and resulted in significant morbidity and mortality [1]. The rapid transmission of SARS-CoV-2 has focused unprecedented attention on the importance of diagnostics that are both accurate and widely available for pandemic control [2, 3]. Although attention is often given to the diagnostic platform, the specimen collection methods are also important determinants of accuracy and availability. For molecular amplification assays, nasopharyngeal (NP) specimen collection using flocked swabs has the highest sensitivity [4]. However, there is an urgent need to replace the NP specimen type because swab supplies are limited, and the procedure is uncomfortable. Alternate sample types [5] including saliva have been used to detect SARS-CoV-2 [6–8]. More importantly, there are multiple sample types that may be called “saliva” in the literature; spit and passive drool (no material from the posterior pharynx is included), oral crevicular fluid (a sponge rubbed against the gingival crevice releasing oral fluid rich in antibodies), and expectorated spit/sputum. A review of the literature of these sample types compared against an NP molecular test qualitative gold standard is presented in Table 1 with a Forest plot of the positive percent agreement Figure 1. In many studies, saliva specimens have lower positive percent agreement, but, conversely, saliva often detected SARS-CoV-2 when the NP swab was negative in a proportion of the samples thereby decreasing negative percent agreement as well. Salivary sampling circumvents swab supply chain bottlenecks, simplifies self-collection even by children, and reduces aerosolization during collection. A few studies have also suggested salivary sampling has high diagnostic sensitivity [6]. In one hospitalized cohort of matched NP swab and saliva specimens, saliva specimens had higher mean log copies per milliliter of SARS-CoV-2 ribonucleic acid (RNA) than NP swab specimens [9]. However, the advantage of salivary collection is even greater in outpatient settings. Thus, in a cohort of ambulatory COVID-19 adults, we compared SARS-CoV-2 RNA detection and abundance in a longitudinal series of self-collected and matched oral fluid saliva and upper respiratory (nasal-oropharyngeal) specimens utilizing the Abbott Molecular RealTime SARS-CoV-2 assay followed by cell culture of RNA-positive samples. We sought to understand the sensitivity of the oral fluid saliva sample type over time in ambulatory COVID-19 patients.
Table 1.
Review of Molecular Testing of Saliva Compared With NP Specimen
| Author | Positive Percent Agreementa | Negative Percent Agreementa | Sample Size (Participants) | Setting | Symptomatic/ Asymptomatic | Comments |
|---|---|---|---|---|---|---|
| Spit/Passive Drool | ||||||
| Azzi et al [29] | 100% (25/25) (95% CI, 86.3%–100%) | - | 25 | Hospitalized, known COVID-19 | 2 positive only in saliva | |
| Barat et al [30] | 81.1% (30/37) (95% CI, 65.8%–90.5%) | 99.8% (421/422) (95% CI, 98.7%–100%) | 449 | Outpatient | Symptomatic and asymptomatic contacts | Drive-through screening (n = 380) and emergency department (n = 69) |
| Becker et al [31] | 69.2% (6/9) (95% CI, 38.6%–97.6%) | 100% (79/79) (95% CI, 95.4%–100%) | 88 | Outpatient | Symptomatic | |
| Bhattacharya et al [32] | 91.4% (53/58) (95% CI, 81%–97.1%) | 100% (16/16) (95% CI, 79.4%–100%) | 74 | Hospitalized | Symptomatic | |
| Byrne et al [33] | 85% (17/20) (95% CI, 62.1%–96.8%) | 97.6% (122/125) (95% CI, 93.2%–99.5%) | 110 (145 paired specimens) | Hospitalized (81%) and Outpatient (19%) | Symptomatic | 3 positive in saliva only |
| Caulley et al [34] | 60.7% (34/56) (95% CI, 46.8%–73.5%) | 99.3% (1869/1883) (95% CI, 98.8%–99.6%) | 1939 | Outpatient | Symptomatic and asymptomatic | 14 positive in saliva only |
| Dogan et al [35] | 54.5% (30/55) (95% CI, 40.6%–68%) | 96.6% (142/147) (95% CI, 92.2%–98.9%) | 200 | Hospitalized | Symptomatic | 5 positive in saliva only |
| Food and Drug Administration [36] | 100% (26/26) (95% CI, 87.1%–100%) | 100% (27/27) (95% CI, 87.5%–100%) | 53 | Outpatient | Symptomatic | |
| Griesemer et al [19] | 87.1% (79/91) (95% CI, 79.6%–93.6%) | 98.5% (134/136) (95% CI, 94.8%–99.8%) | 227 | Outpatient screening | Symptomatic and asymptomatic | 2 positive in saliva only |
| Gupta et al [25] | 63.6% (95% CI, 45.1%–79.6%) | 64.5% (95% CI, 45.4%–80.8%) | 33 | Symptomatic (n = 13), asymptomatic (n = 20) | ||
| Hanson et al Micro [14] | 93.8% (75/80) (95% CI, 86.0%–97.9%) | 97.8% (268/274) (95% CI, 95.3%–99.2%) | 354 | Outpatient (drive through screening) | Symptomatic | 6 positive in saliva only |
| Iwasaki et al [37] | 88.9% (8/9) (95% CI, 51.8%–99.7%) | 98.5% (66/67) (95% CI, 92%–99.9%) | 76 | Hospitalized | Known COVID-19 (n = 10), suspected (n = 66) | |
| Jamal et al [38] | 72% (52/72) (95% CI, 60.4%–82.1%) | 70.4% (19/27) (95% CI, 49.8%–86.3%) | 91 | Hospitalized | Symptomatic, known positives | 8 positive in saliva only |
| Kim J et al [28] | 53% (8/15) (95% CI, 26.6%–78.7%) | - | 15 | Hospitalized | Symptomatic | Longitudinal specimens, PPA reported corresponds to tests within the first 5 days after positive NP test |
| Ku et al [39] | 66.7% (20/30) (95% CI, 47.2%–82.7%) | 91.7% (11/12) (95% CI, 61.5%–99.8%) | 42 | Hospitalized | Known COVID-19 positive, self-collected | |
| Landry et al [20] | 84.8 % (28/33) (95% CI, 70.6%–93.7%) | 97.8% (89/91) (95% CI, 92.3%–99.7%) | 124 | Outpatient | Symptomatic | 2 positive in saliva only |
| McCormick-Baw et al [40] | 95.9% (47/49) (95% CI, 86.0%–99.5%) | 99.1% (105/106) (95% CI, 94.9%–99.9%) | 155 | ED and hospitalized (not severe) | 1 positive in saliva only | |
| Migueres et al [41] | 82.9% (34/41) (95% CI, 67.9%–92.9%) | 96.3% (79/82) (95% CI, 89.7%–99.2%) | 123 | Hospitalized and outpatient | 3 positive in saliva only | |
| Nacher et al [42] | 50% (75/152) (95% CI, 41.2%–57.6%) | 98.4% (614/624) (95% CI, 97.1%–99.2%) | 776 | Outpatient | Symptomatic and asymptomatic | 10 positive in saliva only |
| Nagura-Ikeda et al [43] | 91.8% (56/61) (95% CI, 81.9%–97.3%) (first 9 days) 66.7% (18/27) (95% CI, 46.0%–83.5%) (overall) | - | 103 | Inpatient and outpatient | Symptomatic (n = 88) and asymptomatic (n = 15) | |
| Pasomsub et al [44] | 84.2% (16/19) (95% CI, 60.4%–96.6%) | 98.9% (179/181) (95% CI, 96.1%–99.9%) | 200 | Outpatient screening | Symptomatic | 2 positive in saliva only |
| Sakanashi et al [45] | 100% (15/15) (95% CI, 87.1%–100%) | - | 12 | Hospitalized | Symptomatic | 4 positive in saliva only |
| Skolimowska et al [21] | 83.3% (15/18) (95% CI, 60.8–94.2%) | 99.1% (112/113) (95% CI, 95.2%–100%) | 131 | Outpatient | Symptomatic | 1 positive in saliva only Combined OP/NP swab |
| SoRelle et al [46] | 78% (18/23) (95% CI, 56.3%–92.5%) | 100% (43/43) (95% CI, 91.8%–100%) | 66 (paired specimens) | Outpatient | Symptomatic | |
| Teo et al [47] | 95.1% (117/123) (95% CI, 89.7%–98.2%) | 63% (41/65) (95% CI, 50.2%–74.7%) | 188 (paired specimens) | Outpatient | Symptomatic | 24 positive in saliva only Men only |
| Van Vinh Chau et al [22] | 74.1% (20/27) (95% CI, 53.7%–88.9%) | - | 30 | Outpatient quarantine | Symptomatic and asymptomatic | 1 positive in saliva only |
| Williams et al [48] | 84.6% (33/39) (95% CI, 70.0%–93.1%) | 98% (49/50) (95% CI, 89.4%–99.9%) | 522 | Outpatient screening | 1 positive in saliva only | |
| Wyllie et al [9] | 80.9% (34/42) (95% CI, 65.9%–91.4%) | 25% (4/16) (95% CI, 7.3%–52.4%) | 70 | Hospitalized | Symptomatic | 13 positive in saliva only Sensitivity reported for first 5 days |
| Yee et al [49] | 90.1% (79/87) (95% CI, 82.7%–96.0%) | 95.5% (213/223) (95% CI, 91.9%–97.8%) | 300 | Inpatients, outpatients, contacts | Symptomatic and asymptomatic | 10 positive in saliva only |
| Gingival Crevicular Fluid/Swabs | ||||||
| Aita et al [50] | 100% (7/7) (95% CI, 59.0%–100%) | 97.2% (35/36) (95% CI, 85.5%–99.9%) | 43 | Hospitalized | Symptomatic, known COVID-19 positive | 1 positive in saliva only |
| Braz-silva et al [51] | 78.6% (37/52) (95% CI, 67.6%–86.6%) | 87.9% (131/149) (95% CI, 81.6%–92.7%) | 70 | Outpatient | Symptomatic | 18 positive in saliva only |
| Gupta et al [25] | 54.8% (17/31) (95% CI, 36.0%–72.7%) | - | 33 | Outpatient | Symptomatic | |
| Ku et al [39] | 56.7% (17/30) (95% CI, 37.4%–74.5%) | 100% (12/12) (95% CI, 73.5%–100%) | 42 | Hospitalized | ||
| Manabe et al (this paper) | 41.6% (45/108) (95% CI, 32.3%–51.6%) | 95.2% (60/63) (95% CI, 86.7%–99.0%) | 171 (paired specimens) | Outpatient | Symptomatic | 2 positive in saliva only |
| Cough and Spit (Sputum) | ||||||
| Kojima et al [52] | 87.0% (20/23) (95% CI, 66.4%–97.2%) | 72.7% (16/22) (95% CI, 49.8%–89.3%) | 45 | Outpatient (drive-through testing) | 6 positive in saliva only | |
| Otto et al [53] | 100% (45/45) (95% CI, 92.1%–100%) | 91.4% (43/47) (95% CI, 79.6%–97.6%) | 92 | Outpatient | 4 positive in posterior OP spit only | |
| Procop et al [54] | 100% (38/38) (95% CI, 90.8%–100%) | 99.4% (177/178) (95% CI, 96.9%–99.9%) | 216 | Outpatient | Symptomatic | 1 positive in saliva only |
| Rao et al [26] | 86.9% (73/84) (95% CI, 77.8%–93.3%) | 42.9% (57/133) (95% CI, 34.3%–51.7%) | 217 | Outpatient | Asymptomatic | 76 positive in saliva only |
| To et al [27] | 91.7% (11/12) (95% CI, 61.5%–99.8%) | - | 12 | Hospitalized | ||
| To et al [55] | 87.0% (20/23) (95% CI, 66.4%–97.2%) | - | 23 | Hospitalized |
Abbreviations: CI, confidence interval; COVID-19, coronavirus disease 2019; NP, nasopharyngeal; OP, oropharyngeal.
aCompared with NP swab gold standard.
Figure 1.
Forest plot of the positive percent agreement of salivary sample types compared with nasopharyngeal swab. Asterisks denote outpatient studies.
MATERIALS AND METHODS
Patient Consent Statement
Due to the contagious nature of COVID-19 being studied under this protocol, obtaining signed informed consent form for subjects enrolled in this study was not feasible or safe initially for study staff. Instead, the study staff obtained a verbal consent using consent waiver with an alteration of the informed consent. All participants provided verbal consent after documentation of understanding as they were self-isolating at home due to COVID-19 according to a consent script that was provided in either English or Spanish. A copy of the informed consent was sent to the participants. This protocol and verbal consent were approved by the Johns Hopkins University School of Medicine Institutional Review Board (IRB). All procedures were in accordance with the ethical standards of the Helsinki Declaration of the World Medical Association.
Study Cohort
From April 21 to July 16, 2020, nonhospitalized adults who were self-isolating after receiving a positive NP SARS-CoV-2 real-time reverse-transcription polymerase chain reaction (rRT-PCR) result from the Johns Hopkins Medical Microbiology laboratory were approached for participation by telephone using a verbal consent script. Inclusion criteria were age ≥18 years, able to receive study materials while remaining in isolation, and able and willing to perform self-collection of specimens. Participants who were able to give an oral informed consent after documentation of understanding were enrolled in the study [10]. This study was approved by the IRB of the Johns Hopkins University School of Medicine.
Specimen Collection
Participants were mailed a sample collection kit that included an international air transport association (IATA)-approved biologic sample container as well as sample collection materials and written instructions for sample collection. In addition, study coordinators provided verbal sample collection instructions and observed participants by video call when possible. The study coordinators recorded their assessment of the quality of self-collection. Participants self-collected mid-turbinate nasal and oropharyngeal (nasal-OP) swabs; both swabs were placed in 3 mLs viral transport medium ([VTM] [11]). Self-collected samples have been previously validated and published [11–13]. By placing them in the same media, this combination aimed to optimize detection and approximated the NP clinician-collected sample [14]. Participants also collected oral crevicular saliva fluid via the Oracol saliva collection system (oral fluid) (Malvern Medical Developments Ltd., Worchestershire, UK), a transport buffer-free sample collection system. All samples were immediately placed in the IATA container and stored in the participant’s freezer before shipping. Participants self-collected samples on the day they received the collection materials (day 0) and then subsequently on study days 3, 7, 14. On day 14, the participant shipped the collected samples on ice-cold packs to Johns Hopkins University for analysis using an overnight courier service. A final in-person collection occurred between day 28 and 60 when a clinician-collected NP swab and a self-collected Oracol was performed. Participants were instructed to open and remove the saliva collection sponge from the device container, rub their gums for 1–2 minutes with the sponge, then reinsert the swab back into the device container and closes the container. This collection method targets gingival crevicular fluid, which leaks from the space between the gums and teeth and is enriched with immunoglobulin G antibodies derived from blood. Based on publication of the spit saliva sample type [9], participants were instructed to spit one time into the Oracol after gum collection from June 1, 2020 onward. Clinical information was collected using a standardized Flu-PRO [15] instrument on the same days as sample collection, in addition to patient history in a predesigned database.
Specimen Testing
The nasal-OP swab VTM was aliquoted into multi-Collect tubes (Abbott Molecular, Des Plaines, IL) in 600-µL volumes before testing with the Abbott Molecular RealTime SARS-CoV-2 assay. Nucleic acid was extracted from the multi-Collect tubes utilizing the Abbott Molecular m2000sp, followed by amplification and analysis on the Abbott m2000rt; both extraction and amplification were performed per the manufacturer’s instructions. A positive reaction was defined as a reaction having a cycle number (CN ) <31.5 based on the manufacturer’s definition of a positive result. Oracol collection devices were centrifuged upon receipt at 1500 ×g for 10 minutes. The majority of the participants (60 of 71) were able to produce Oracol volumes between 500 µL and 1 mL; 200-µL undiluted volumes of oral fluid were aliquoted into Abbott multi-Collect tubes and were tested on the Abbott Moelcular m2000 platform.
Cell Culture
VeroE6-TMPRSS2 [16] cell culture model was used to assess viable virus when incubated with VTM (nasal-OP samples only). The SARS-CoV-2-specific growth was verified by indirect immunofluorescence for SARS-CoV-2 antigen (nucleocapsid and spike proteins) [17].
Statistical Analysis
Median CN value and corresponding interquartile range (IQR) for concordantly positive pairs were calculated for both nasal-OP and oral fluid samples. Difference of CN value between nasal-OP and oral fluid samples in matched pairs were tested using Wilcoxon signed-rank test in all samples, prespit samples, and postspit samples. The 0.05 significance level was used. Analyses were performed using R 3.6.2 statistical software.
RESULTS
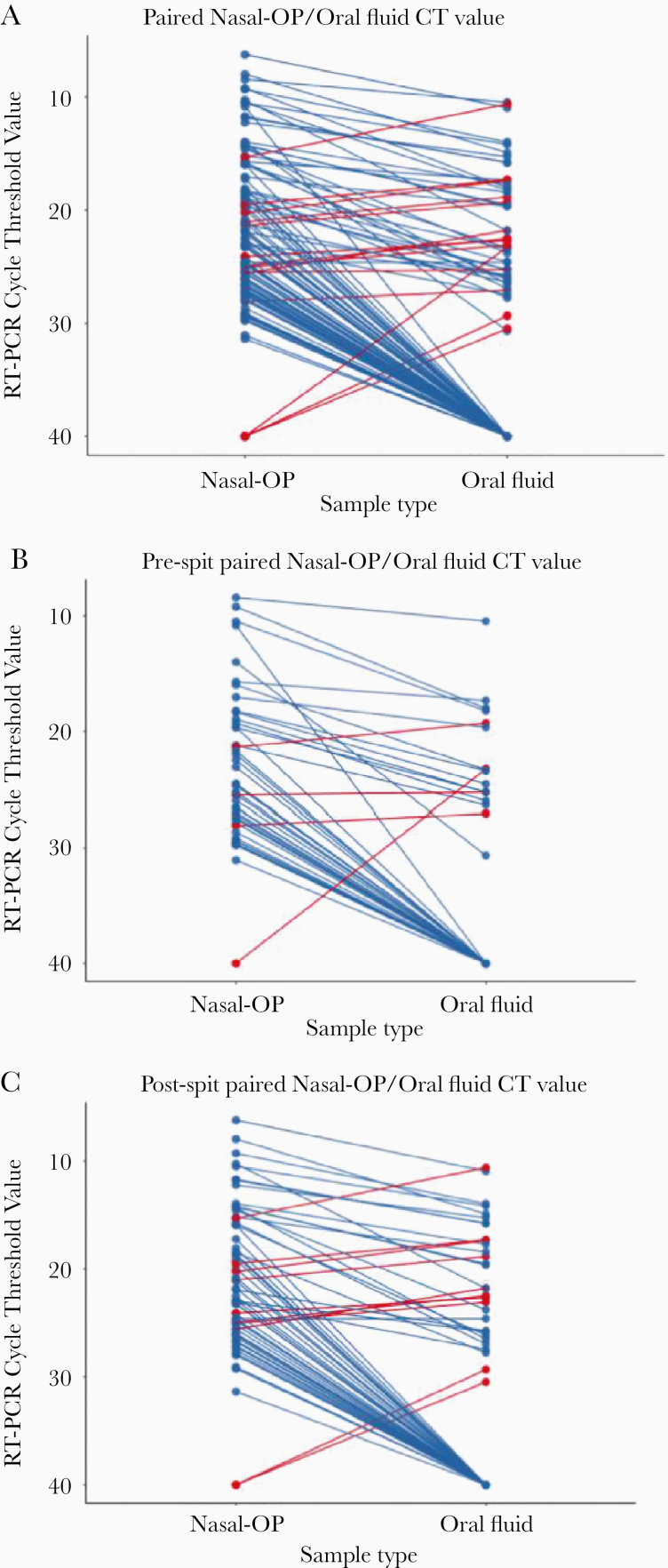
Of the 118 participants enrolled that were previously described [9], 71 participants had at least 1 sample that was rRT-PCR positive; 60 had matched saliva and nasal-OP specimens and were included in this analysis (Supplemental Figure 1). The median age was 59 (IQR, 51–66) years, 53% were women (32 of 60). From these 60 persons, there were 342 matched self-collected nasal-OP swabs and oral fluid samples (171 pairs). Of the matched samples, 60 were concordantly negative, 45 concordantly positive, and 66 discordant (63 nasal-OP pos/oral neg and 3 nasal-OP neg/oral pos). The SARS-CoV-2 RNA estimates were generally higher in nasal-OP samples. For example, among the 45 samples that were concurrently positive, the median cycle threshold (Ct) of nasal-OP swab samples was 15.98 (IQR, 13.96–21.21) versus 21.81 (IQR, 17.35–25.27) for oral fluid (P < .001). (Figure 2A) Likewise, of 111 samples when at least 1 of the tests was positive, only 14 (12.6%) had higher RNA abundance in oral fluid compared with nasal-OP swab samples. Considering nasal-OP as a reference, the sensitivity of the first oral fluid specimen from each participant was 62.1% (18 of 29), with a specificity of 80% (8 of 10) (Supplemental Figure 2).
Figure 2.
Cycle thresholds (CT) are plotted for matched nasal-oropharyngeal (OP) swab and oral fluid real-time reverse-transcription polymerase chain reaction. Viral burdens that were higher in nasal-OP or oral fluid are shown in blue and red lines, respectively, in (A) all matched specimens, (B) oral fluid only, and (C) oral fluid plus the addition of spit. Samples that were negative in both sample types are not shown.
Midway through enrollment, participants were asked to add spit into the Oracol collection tube to evaluate whether that might enhance sensitivity. Of the matched samples in which the oral fluid was supplemented with spit (n = 104), 38 were concordantly negative, 29 concordantly positive, and 38 discordant (36 nasal-OP pos/oral neg and 2 nasal-OP neg/oral pos). (Figure 2B and C) Oral fluid sample sensitivity increased from 37.2% “prespit” (n = 67) to 44.6% “postspit” (n = 104). It is interesting to note that, of the 44 prespit samples, 4 (9.1%) had lower Ct values than nasal-OP, and, in the postspit samples, this percentage increased to 14.9% (10 of the 67 samples).
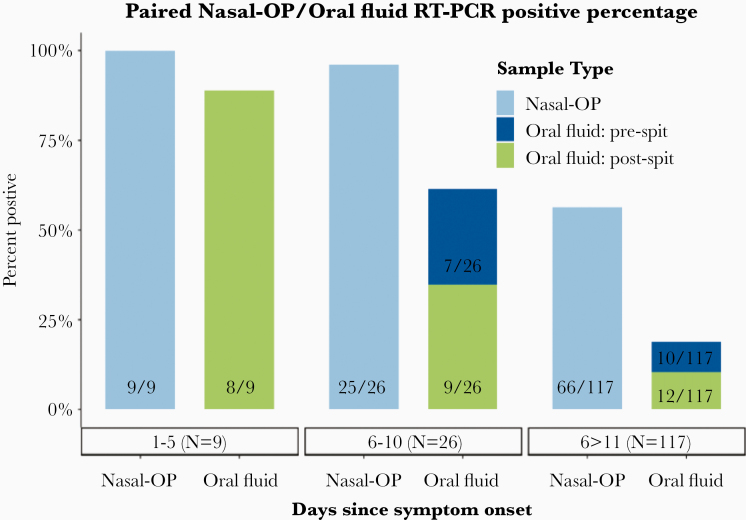
As expected, SARS-CoV-2 detection declined during follow-up (Figure 3). For samples collected more than 6 days after symptom onset, the greater SARS-CoV-2 recovery from nasal-OP compared with oral fluid was especially evident. Among 26 specimens collected 6–10 days from symptom onset, SARS-CoV-2 was detected in 25 (96.1%) by nasal-OP but in only 16 (58.3%) by oral fluid. Likewise, the SARS-CoV-2 RNA abundance in those specimens was higher in nasal-OP than oral fluid (Figure 4, Supplemental Table 1).
Figure 3.
The proportion of nasal-oropharyngeal (OP) and oral fluid matched specimens that were positive in participants who are 1–5 days, 6–10, and more than 11 days after symptom onset in samples from participants where the date of symptom onset could be determined. Eight participants without a date of symptom onset (19 specimen pairs) are not included in this figure. RT-PCR, reverse-transcription polymerase chain reaction.
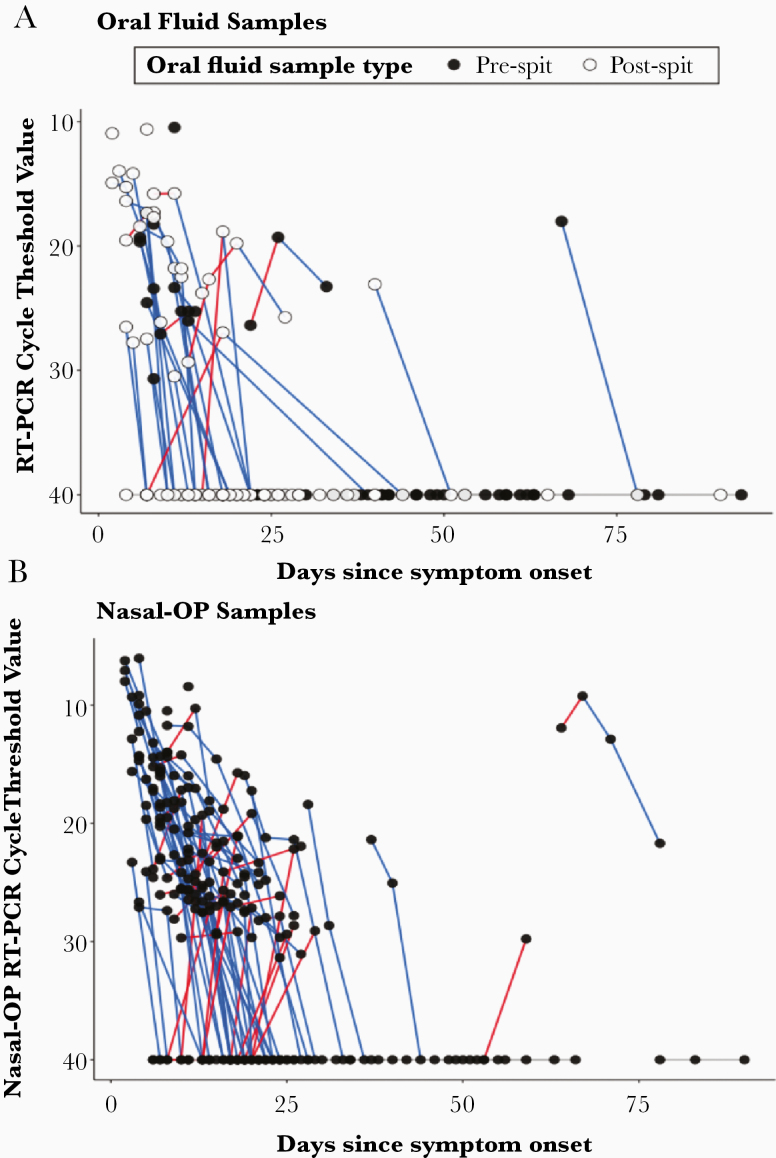
Figure 4.
(A) Cycle threshold values are shown for individual participants over time. Blue lines denote decreasing viral burden, whereas red lines represent increasing viral burden with increasing number of days after symptom onset in oral fluid. Samples where spit was added are shown in the open circles, and those with oral fluid only are in the black circles. and (B) nasal-oropharyngeal (OP) specimens.
We cultured all rRT-PCR-positive nasal-OP specimens on VeroE6 TMPRSS2 cells. All culture-positive samples (n = 16) were obtained within 11 days of symptom onset. In all matched samples in which SARS-CoV-2 was culture positive (n = 9), SARS-CoV-2 RNA was detected by rRT-PCR in both nasal-OP and oral fluid samples.
DISCUSSION
In this investigation of nonhospitalized SARS-CoV-2-infected persons, we demonstrate lower diagnostic sensitivity of self-collected oral fluid compared with nasal-OP specimens, a difference that was especially prominent more than 5 days from symptom onset. These data do not justify the routine use of oral fluid collection for diagnosis of SARS-CoV-2 despite the greater ease of collection.
Our findings are consistent with what most (but not all) other investigators have found particularly in the outpatient setting (Table 1, Figure 1) [18–23]. Studies differ in (1) whether they involve hospitalized patients, (2) the methods used to collect saliva, as well as (3) the duration after infection onset, and these factors might contribute to the discordance in results. For example, Wyllie et al [9] found even greater detection of SARS-CoV-2 in oral fluid compared with nasal-OP but collected spit in the morning in hospitalized patients. Expectorated “spit” samples collected in the morning (possible for hospitalized patients) might increase viral abundance by enrichment of deeper samples and has been used to increase yield in hospitalized patients with pneumonia [24]. That difference would be expected to be greatest when SARS-CoV-2 is replicating in lower airways such as in hospitalized patients with pneumonia. In contrast in the present study, participants self-collected oral fluid, optimized for the detection of oral crevicular fluid antibodies. Although this sample type may dilute the salivary sample and decrease its sensitivity for viral RNA detection, Gupta et al [25] showed no difference in a comparison of oral crevicular fluid with spit. In our study, we found that spit added to the oral fluid did increase sensitivity. Taken together, spit/drool sample type is better than oral fluid from the gumline. These differences are important because many in vitro devices that are currently being tested for the direct detection of SARS-CoV-2 use different salivary sample types including passive drool, spit, oral fluid collected with a sponge, and sputum from clearing the throat. Future studies for home collection should consider the added sensitivity of a first morning sputum compared with routine oral fluid collection [26].
Differences in the stage of infection may also factor into net sensitivity. Small differences in the sensitivity of tests can be inapparent in the early stages of infection when SARS-CoV-2 RNA levels are highest. Indeed, in the present study, both methods performed well in the first 5 days after symptom onset. Others have similarly suggested salivary tests have higher sensitivity in the first week of symptoms in outpatients [9, 27] and may have longer duration of positivity in inpatients [9, 28]. It remains unclear whether differences in diagnostic yield for saliva versus nasal-OP reflect shifts in SARS-CoV-2 replication from upper to lower respiratory tissues. Nonetheless, the timing of collection, time after onset of symptoms, hospitalized versus outpatient populations, and the volume of saliva/spit may all be important for optimizing diagnostic sensitivity, and device manufacturers will need to consider these factors when considering what sample types to test and when assessing assay performance. Overall, most studies of spit or passive drool reveal a lower sensitivity compared with NP but identify cases missed by the NP sample type.
Although SARS-CoV-2 testing is chiefly used for diagnostics, it has also been used to assess infectivity. In our experience using culture as a gold standard for infectivity, both collection methods were equivalent in their ability to identify patients with infectious virus; all those whose nasal-OP sample was cultured were rRT-PCR positive in both sample types.
Our paper has some important limitations. Because all participants were enrolled on the basis of a clinician-collected NP swab, this may bias against saliva sample types; if saliva is more sensitive early in infection, any participant who was initially saliva positive and NP negative would not have had the opportunity to enroll. In addition, 200 µL of neat saliva was assayed compared with 600 µL of nasal-OP swab VTM (from a total volume of 3 mL). It is possible that if a concentration capture method were applied to saliva, yields for this sample type may improve sensitivity. Based on the enrollment strategy to follow ambulatory participants, we collected specimens from when participants were enrolled. Therefore, the number of days after symptoms varied; those that were within the first 5 days of symptoms, when oral fluid with spit was most sensitive, were limited. Finally, although self-collected samples have been shown by others to be equally sensitive, Kojima et al [13] found that unsupervised self-collected oral fluid had lower sensitivity. In our study, participants were coached by phone or facetime, and the presence of human deoxyribonucleic acid in the sample was verified. Nonetheless, self-collection may have contributed to variability.
CONCLUSIONS
In summary, in what we believe may be the largest ambulatory study of its kind, we detected (1) lower SARS-CoV-2 yield in oral fluid compared with nasal-OP specimens and (2) improvements in oral fluid enriched with spit. Differences in net diagnostic sensitivity were especially notable more than 5 days after symptom onset. These results are significant because more than 80% of COVID-19 and almost all the initial diagnostics are in ambulatory persons. Thus, although our data and others demonstrate the potential pragmatic use of salivary samples to detect SARS-CoV-2 [29], they also underscore the importance of carefully considering the source of specimens and possibly time from symptom onset, especially for home detection systems.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support.This work was funded by the Johns Hopkins University School of Medicine COVID-19 Research Fund and the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases Discovery Program. Salary support for Y. C. M. was from the National Institutes of Health (Grant Numbers U54EB007958-12, U5411090366, U54HL143541-02S2, UM1AI068613).
Potential conflicts of interest. Y. C. M. has received tests from Quanterix, Becton-Dickinson, Ceres, and Hologic for research-related purposes. She also receives funding support to Johns Hopkins University from miDiagnostics. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Ambulatory COVID Study Team
Andrea Cox, Sara Keller, Jeanne Keruly, Sabra Klein, Shruti Mehta, Heba Mostafa, Nora Pisanic, Lauren Sauer, Jeffrey Tornheim, Jennifer Townsend, Derek Armstrong, Vismaya Bachu, Brittany Barnaba, Curtisha Charles, Weiwei Dai, Abhinaya Ganesan, Jeffrey Holden, Minyoung Jang, JR Johnstone, Kate Kruczynski, Oyinkansola Kusemiju, Anastasia Lambrou, Lucy Li, Kirsten Littlefield, Han-Sol Park, Amanda Tuchler, Manuela Plazas Montana, Michelle Prizzi, Rebecca Ursin.
Contributor Information
Ambulatory COVID Team:
Andrea Cox, Sara Keller, Jeanne Keruly, Sabra Klein, Shruti Mehta, Heba Mostafa, Nora Pisanic, Lauren Sauer, Jeffrey Tornheim, Jennifer Townsend, Derek Armstrong, Vismaya Bachu, Brittany Barnaba, Curtisha Charles, Weiwei Dai, Abhinaya Ganesan, Jeffrey Holden, Minyoung Jang, J R Johnstone, Kate Kruczynski, Oyinkansola Kusemiju, Anastasia Lambrou, Lucy Li, Kirsten Littlefield, Han-Sol Park, Amanda Tuchler, Manuela Plazas Montana, Michelle Prizzi, and Rebecca Ursin
References
- 1. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20:533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manabe YC, Sharfstein JS, Armstrong K. The need for more and better testing for COVID-19. JAMA 2020; 324:2153–4. doi: 10.1001/jama.2020.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daley P, Castriciano S, Chernesky M, Smieja M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J Clin Microbiol 2006; 44:2265–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perchetti GA, Nalla AK, Huang ML, et al. Validation of SARS-CoV-2 detection across multiple specimen types. J Clin Virol 2020; 128:104438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2, an observational cohort study. Lancet Infect Dis 2020. doi: 10.1016/S1473-3099(20)30196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bwire GM, Majigo MV, Njiro BJ, Mawazo A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens, a systematic review and meta-analysis. J Med Virol 2020. doi: 10.1002/jmv.26349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cevik M, Tate M, Lloyd O, et al. SARS-CoV-2, SARS-CoV-1 and MERS-CoV viral load dynamics, duration of viral shedding and infectiousness, a living systematic review and meta-analysis [preprint]. Lancet Microb 2021; 2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 2020; 383:1283–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blair PW, Brown DM, Jang M, et al. The clinical course of COVID-19 in the outpatient setting, a prospective cohort study [preprint]. medRxiv 2020. doi: 10.1101/2020.09.01.20184937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Altamirano J, Govindarajan P, Blomkalns AL, et al. Assessment of sensitivity and specificity of patient-collected lower nasal specimens for severe acute respiratory syndrome coronavirus 2 testing. JAMA Netw Open 2020; 3:e2012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCulloch DJ, Kim AE, Wilcox NC, et al. Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open 2020; 3:e2016382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kojima N, Turner F, Slepnev V, et al. Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanson KE, Barker AP, Hillyard DR, et al. Self-collected anterior nasal and saliva specimens versus healthcare worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol 2020. doi: 10.1128/JCM.01824-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Powers JH, Guerrero ML, Leidy NK, et al. Development of the Flu-PRO, a patient-reported outcome (PRO) instrument to evaluate symptoms of influenza. BMC Infect Dis 2016; 16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 2020; 117:7001–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pekosz A, Cooper C, Parvu V, et al. Antigen-based testing but not real-time PCR correlates with SARS-CoV-2 virus culture [preprint]. medRxiv 2020. doi: 10.1101/2020.10.02.20205708 [DOI] [Google Scholar]
- 18. Caulley L, Corsten M, Eapen L, et al. Salivary detection of COVID-19. Ann Intern Med 2020. doi: 10.7326/M20-4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Griesemer SB, Van Slyke G, Ehrbar D, et al. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing [preprint]. medRxiv 2020. doi: 10.1101/2020.06.16.20133041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Landry ML, Criscuolo J, Peaper DR. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol 2020; 130:104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skolimowska K, Rayment M, Jones R, et al. Non-invasive saliva specimens for the diagnosis of COVID-19, caution in mild outpatient cohorts with low prevalence. Clin Microbiol Infect 2020; 26:1711–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Vinh Chau N, Lam VT, Dung NT, et al. The natural history and transmission potential of asymptomatic severe acute respiratory syndrome coronavirus 2 infection. Clin Infectous Dis 2020. doi: 10.1093/cid/ciaa711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hung DL, Li X, Chiu KH, et al. Early-morning vs spot posterior oropharyngeal saliva for diagnosis of SARS-CoV-2 infection, implication of timing of specimen collection for community-wide screening. Open Forum Infect Dis 2020; 7:ofaa210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gupta S, Mohindra R, Chauhan PK, et al. SARS-CoV-2 detection in gingival crevicular fluid. J Dent Res 2020. doi: 10.1177/0022034520970536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rao M, Rashid FA, Sabri FSAH, et al. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS-CoV-2. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. To KK, Tsang OT, Yip CC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 2020; 71:841–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim SE, Lee JY, Lee A, et al. Viral load kinetics of SARS-CoV-2 infection in saliva in Korean patients, a prospective multi-center comparative study. J Korean Med Sci 2020; 35:e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect 2020; 81:e45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barat B, Das S, De Giorgi V, et al. Pooled saliva specimens for SARS-CoV-2 testing [preprint]. J Clin Microbiol 2020; JCM.02486–20. doi: 10.1128/JCM.02486-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becker D, Sandoval E, Amin A, et al. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting [preprint]. medRxiv 2020. doi: 10.1101/2020.05.11.20092338 [DOI] [Google Scholar]
- 32. Bhattacharya D, Parai D, Rout UK, et al. Saliva as a potential clinical specimen for diagnosis of SARS-CoV-2 [preprint]. medRxiv 2020. doi: 10.1101/2020.09.11.20192591 [DOI] [Google Scholar]
- 33. Byrne RL, Kay GA, Kontogianni K, et al. Saliva offers a sensitive, specific and non-invasive alternative to upper respiratory swabs for SARS-CoV-2 diagnosis [preprint]. Emerg Infect Dis 2020; 26:2769–70. doi: 10.3201/eid2611.203283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caulley L, Corsten M, Eapen L, et al. Salivary detection of COVID-19. Ann Intern Med 2020. doi: 10.7326/M20-4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dogan OA, Kose B, Agaoglu NB, et al. Does sampling saliva increase detection of SARS-CoV-2 by RT-PCR? Comparing saliva with oro-nasopharyngeal swabs [preprint]. J Virol Methods 2020;114049. doi: 10.1016/j.jviromet.2020.114049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Food and Drug Administration. Emergency Use Authorizations. Available at, https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#coronavirus2019. Accessed 10 November 2020.
- 37. Iwasaki S, Fujisawa S, Nakakubo S, et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect 2020; 81:e145–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jamal AJ, Mozafarihashjin M, Coomes E, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020. doi: 10.1093/cid/ciaa848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ku CW, Shivani D, Kwan JQ, et al. Validation of self-collected buccal swab and saliva as a diagnostic tool for COVID-19 [preprint]. Int J Infect Dis 2020. doi: 10.1016/j.ijid.2020.12.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCormick-Baw C, Morgan K, Gaffney D, et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol 2020; 58:e01109-20. doi: 10.1128/JCM.01109-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Migueres M, Mengelle C, Dimeglio C, et al. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. J Clin Virol 2020; 130:104580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nacher M, Mergeay-Fabre M, et al. Prospective comparison of saliva and nasopharyngeal swab sampling for mass screening for COVID-19 [preprint]. medRxiv 2020. doi: 10.1101/2020.09.23.20150961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nagura-Ikeda M, Imai K, Tabata S, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol 2020; 58:e01438–20. doi: 10.1128/JCM.01438-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pasomsub E, Watcharananan SP, Watthanachockchai T, et al. Saliva sample pooling for the detection of SARS-CoV-2. J Med Virol 2020. doi: 10.1002/jmv.26460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sakanashi D, Asai N, Nakamura A, et al. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. J Infect Chemother 2021; 27:126–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. SoRelle JA, Mahimainathan L, McCormick-Baw C, et al. Evaluation of symptomatic patient saliva as a sample type for the Abbott ID NOW COVID-19 assay [preprint]. medRxiv 2020. doi: 10.1101/2020.06.01.20119198 [DOI] [Google Scholar]
- 47. Teo AKJ, Choudhury Y, Tan IB, et al. Validation of saliva and self-administered nasal swabs for COVID-19 testing [preprint]. medRxiv 2020. doi: 10.1101/2020.08.13.20173807 [DOI] [Google Scholar]
- 48. Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00776–20. doi: 10.1128/JCM.00776-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yee R, Truong T, Pannaraj PS, et al. Saliva is a promising alternative specimen for the detection of SARS-CoV-2 in children and adults [preprint]. medRxiv 2020. doi: 10.1101/2020.10.25.20219055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aita A, Basso D, Cattelan AM, et al. SARS-CoV-2 identification and IgA antibodies in saliva, one sample two tests approach for diagnosis. Clin Chim Acta 2020; 510:717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Braz-Silva PH, Mamana AC, Romano CM, et al. Performance of at-home self-collected saliva and nasal-oropharyngeal swabs in the surveillance of COVID-19 [preprint]. J Oral Microbiol 2021; 13:1858002. doi: 10.1080/20002297.2020.1858002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kojima N, Turner F, Slepnev V, et al. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection [preprint]. Clin Infect Dis 2020: ciaa1589. doi: 10.1093/cid/ciaa1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pierre Otto MP, Darles C, Valero E, et al. Posterior oropharyngeal saliva for the detection of SARS-CoV-2 [preprint]. Clin Infect Dis 2020: ciaa1181. doi: 10.1093/cid/ciaa1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Procop GW, Shrestha NK, Vogel S, et al. A direct comparison of enhanced saliva to nasopharyngeal swab for the detection of SARS-CoV-2 in symptomatic patients. J Clin Microbiol 2020; 58:e01946-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2, an observational cohort study. Lancet Infect Dis 2020; 20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.