Abstract
Myelin water imaging is a quantitative neuroimaging technique that provides the myelin water fraction (MWF), a metric highly specific to myelin content, and the intra-/extra-cellular T2 (IET2), which is related to water and iron content. We coupled high-resolution data from 100 adults with gold-standard methodology to create an optimized anatomical brain template and accompanying MWF and IET2 atlases. We then used the MWF atlas to characterize how myelin content relates to demographic factors. In most brain regions, myelin content followed a quadratic pattern of increase during the third decade of life, plateau at a maximum around the fifth decade, then decrease during later decades. The ranking of mean myelin content between brain regions remained consistent across age groups. These openly available normative atlases can facilitate evaluation of myelin imaging results on an individual basis and elucidate the distribution of myelin content between brain regions and in the context of aging.
Subject terms: Ageing, Magnetic resonance imaging, Myelin biology and repair, Neural ageing, Brain
Myelin water imaging (MWI) is a quantitative magnetic resonance imaging (MRI) technique that uses a multi-echo T2 relaxation sequence to characterize myelin1. T2 relaxation in normal human brain generally has three main components: a short T2 component originating from myelin water (T2 < 40 ms), an intermediate T2 component attributed to intra- and extra-cellular water (40 ms < T2 < 200 ms), and a long T2 component representing cerebrospinal fluid (CSF) (T2 >> 200 ms)2. Healthy corticospinal tracts and white matter (WM) affected by pathology can also exhibit an additional component with 200 ms < T2 < 800 ms3. Myelin water fraction (MWF), the fraction of signal contribution from myelin water, has been histopathologically validated as a biomarker for myelin4, and has demonstrated significant variation in myelination between different brain structures not often apparent with other MRI techniques5. The geometric mean T2 of the intra- and extra-cellular water compartment (IET2) is sensitive to tissue water and iron, with lower values expected for lower water content and higher iron concentration6,7.
To identify changes or abnormalities in T2 relaxation metrics in the context of development, aging or disease, it is crucial to have an accurate picture of these metrics in the healthy population. One method to characterize normal brain T2 measures is to analyse MRI data using a representative reference for anatomical images (a “template”) with a corresponding representative quantitative metric map (an “atlas”). Combining datasets reduces noise from individual measurements and from biological variation between subjects, which can arise from age, sex, ethnicity, pathology, hemispheric asymmetry, or other factors. If demographic factors can sufficiently characterize biological variations, then atlases could be used to evaluate quantitative MRI metrics on an individual basis8. Voxel-wise analysis of individual MWF maps has been used to study brain and spinal cord in a variety of diseases, but with relatively small sample sizes and limited characterization of possible demographic factor influences9,10.
Atlas-based methods have been used to probe the relationship between MWF and demographic factors in healthy adult brain but with conflicting results. Opposing positive11 and negative12 linear correlations between age and MWF have been reported, while other research found relatively few brain regions where any age-MWF correlation existed13. Furthermore, some studies have suggested that an inverted U-shaped quadratic model better describes the age-MWF relationship14,15, contrary to literature where linear models performed best12.
The need for normative MWI data and the inconsistent findings of how MWF changes with age motivated the current study. We acquired high-resolution data with the reference multi-echo T2 relaxation method for MWI in a large subject group with a broad age range, and used gold-standard atlas creation methodology to:
Create an optimized anatomical template with accompanying MWF and IET2 atlases, tissue segmentations, and regions of interest (ROIs).
Investigate the strength of age and sex as explanatory variables for inter-subject MWF and IET2 variations.
Determine the extent to which ranking of MWF and IET2 between brain regions is consistent across age groups.
Results
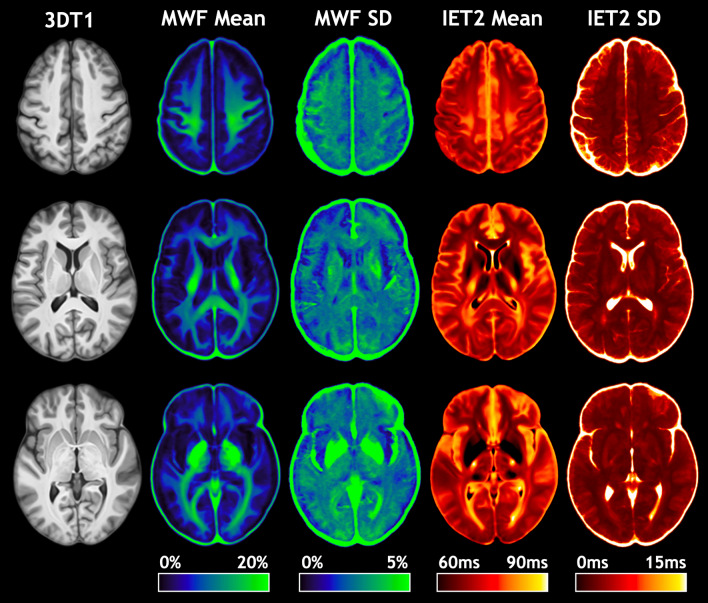
Due to incidental findings, MWF and IET2 maps from two subjects were excluded from atlas creation and subsequent analysis. Representative slices of the T1-weighted anatomical (3DT1) template and mean and standard deviation (SD) atlases for MWF and IET2 are shown in Fig. 1.
Figure 1.
Representative slices of the T1-weighted anatomical (3DT1) template, myelin water fraction (MWF) mean and standard deviation (SD) atlases, and geometric mean T2 of intra- and extra-cellular water (IET2) mean and SD atlases.
Linear regression with age
Correlation coefficients between age and mean MWF and IET2 in all ROIs are presented in Table 1. MWF correlated significantly with age in the corpus callosum (body r = 0.388 P < 0.001, splenium r = 0.337 P < 0.001), internal capsule (anterior r = 0.359 P < 0.001, posterior r = 0.369 P < 0.001, retrolenticular r = 0.306 P < 0.01), external capsules (r = 0.335 P < 0.001), fornix (r = 0.358 P < 0.001), all WM ROIs combined (r = 0.289 P < 0.01), WM mask (r = 0.326 P < 0.01), and gray matter (GM) mask (r = 0.403 P < 0.001).
Table 1.
Pearson correlation coefficients and P-values for correlation between age and mean myelin water fraction (MWF), and age and mean geometric mean T2 of intra- and extra-cellular water (IET2), for each region of interest. Statistically significant correlations are bold for P < 0.01 and bold* for P < 0.001.
| Region of Interest | MWF | IET2 | ||
|---|---|---|---|---|
| r-value | P-value | r-value | P-value | |
| Genu of corpus callosum | 0.071 | 0.488 | 0.623 | 7.15E−12* |
| Body of corpus callosum | 0.388 | 7.95E−05* | 0.437 | 6.73E−06* |
| Splenium of corpus callosum | 0.337 | 6.96E−04* | 0.290 | 3.82E−03 |
| Cerebral peduncle | 0.360 | 2.70E−04* | − 0.364 | 2.27E−04* |
| Anterior limb of internal capsule | 0.359 | 2.87E−04* | − 0.025 | 0.808 |
| Posterior limb of internal capsule | 0.369 | 1.86E−04* | 0.234 | 0.020 |
| Retrolenticular part of internal capsule | 0.306 | 2.21E−03 | 0.111 | 0.277 |
| Anterior corona radiata | 0.214 | 0.035 | 0.548 | 5.16E−09* |
| Superior corona radiata | 0.226 | 0.026 | 0.479 | 5.90E−07* |
| Posterior corona radiata | 0.221 | 0.029 | 0.331 | 8.62E−04* |
| Posterior thalamic radiation | − 0.080 | 0.435 | 0.478 | 6.54E−07* |
| Sagittal stratum | 0.115 | 0.258 | 0.328 | 9.62E−04* |
| External capsules | 0.335 | 7.36E−04* | 0.082 | 0.420 |
| Cingulum (cingulate gyrus) | 0.207 | 0.041 | 0.209 | 0.039 |
| Fornix (cres) / Stria terminalis | 0.358 | 2.92E−04* | 0.133 | 0.193 |
| Superior longitudinal fasciculus | 0.198 | 0.051 | 0.469 | 1.13E−06* |
| All white matter ROIs | 0.289 | 3.91E−03 | 0.424 | 1.33E−05* |
| White matter mask | 0.326 | 1.07E−03 | 0.200 | 0.0481 |
| Gray matter mask | 0.403 | 3.91E−05* | − 0.658 | 1.83E−13* |
IET2 correlated significantly with age in corpus callosum (genu r = 0.623 P < 0.001, body r = 0.437 P < 0.001, splenium r = 0.290 P < 0.01), corona radiata (all P < 0.001), posterior thalamic radiation (r = 0.478 P < 0.001), sagittal stratum (r = 0.328 P < 0.001), superior longitudinal fasciculus (r = 0.469 P < 0.001), all WM ROIs combined (r = 0.424 P < 0.001), and GM mask (r = − 0.658 P < 0.001).
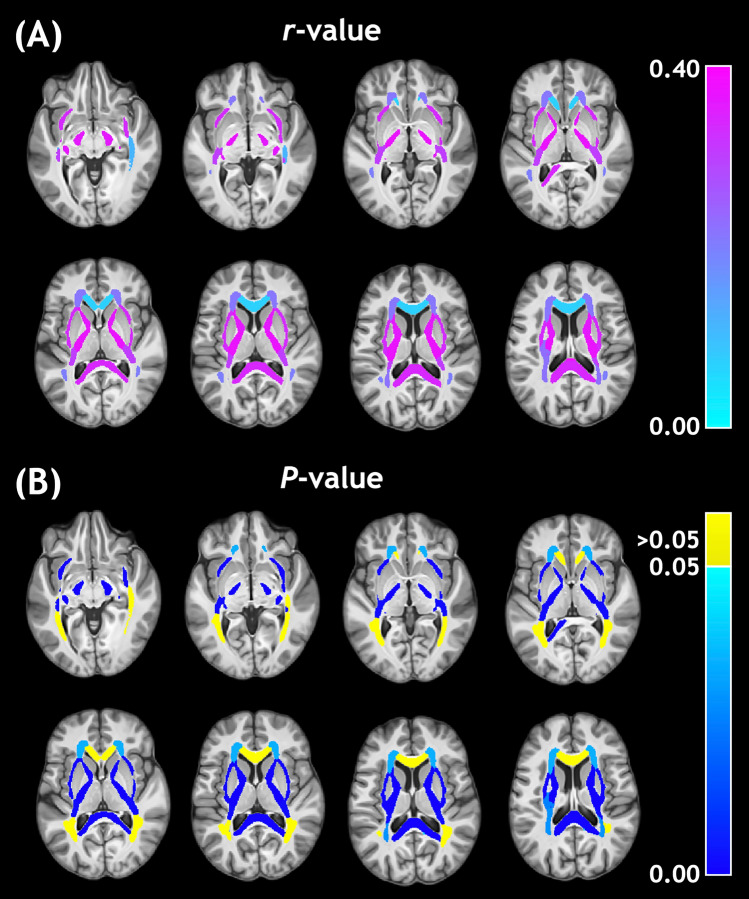
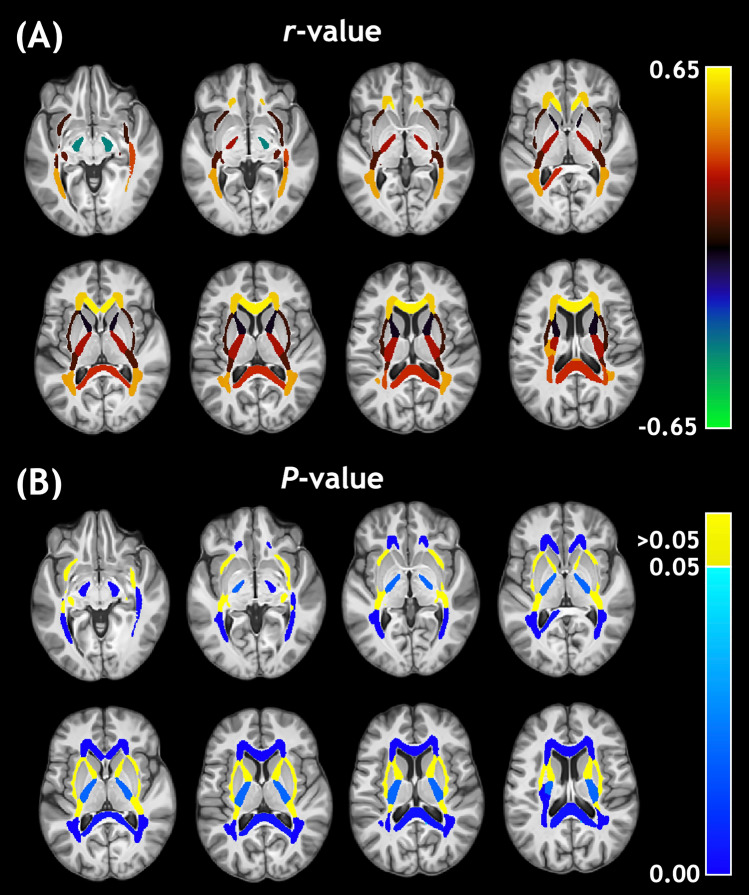
For visual assessment, these correlation values were assigned to their respective ROI masks and presented in Fig. 2 for MWF and Fig. 3 for IET2. Large ROIs composed of multiple smaller regions were omitted from Figs. 2 and 3 so that the smaller ROIs were not obscured. The strongest, highest significance age-MWF correlations in Fig. 2 were clustered towards central or central-posterior brain regions. The age-IET2 correlations in Fig. 3 showed the opposite spatial pattern, with higher significance age-IET2 correlations in peripheral brain regions and stronger correlation coefficients generally favouring anterior brain regions.
Figure 2.
Pearson (A) correlation coefficients (r-values) and (B) P-values for the relationship between age and myelin water fraction.
Figure 3.
Pearson (A) correlation coefficients (r-values) and (B) P-values for the relationship between age and geometric mean T2 of intra- and extra-cellular water.
Multiple linear regression including Age2, sex
Multiple linear regression adjusted r-values as well as P-values for the age and age2 model terms are presented in Table 2, for mean MWF and IET2 values in all ROIs. Some ROIs without significant Pearson correlations reached significance for multiple regression with the quadratic age term, for MWF: superior and posterior corona radiata (both P < 0.01), sagittal stratum (P < 0.01), cingulum (P < 0.001), and superior longitudinal fasciculus (P < 0.001) and for IET2: internal capsule (anterior and posterior P < 0.001, retrolenticular P < 0.01), external capsules (P < 0.001), cingulum (P < 0.01), and WM mask (P < 0.001). Multiple regression with the additional age2 term provided a higher r-value (adjusted) than linear correlation for MWF in all ROIs and for IET2 in all ROIs except the splenium of the corpus callosum. In multiple linear regression with age and sex, the binarized sex variable was not significant for MWF (P > 0.1) or IET2 (P > 0.01) correlations in any ROI, which indicated that there were no significant differences in the age-MWF or age-IET2 relationships between sexes.
Table 2.
Coefficients and P-values for multiple linear regression between a quadratic polynomial in age and mean myelin water fraction (MWF), and between a quadratic polynomial in age and mean geometric mean T2 of intra- and extra-cellular water (IET2), for each region of interest.
| Region of Interest | MWF | IET2 | ||||
|---|---|---|---|---|---|---|
| Adjusted r-value | P-value (Age) | P-Value (Age2) | Adjusted r-value | P-value (Age) | P-Value (Age2) | |
| Genu of corpus callosum | 0.184 | 0.024 | 0.029 | 0.640 | 0.317 | 0.022 |
| Body of corpus callosum | 0.408 | 0.010 | 0.051 | 0.453 | 0.263 | 0.056 |
| Splenium of corpus callosum | 0.346 | 0.032 | 0.106 | 0.263 | 0.819 | 0.475 |
| Cerebral peduncle | 0.467 | 6.14E−05* | 5.19E−04* | 0.394 | 7.17E−03 | 0.035 |
| Anterior limb of internal capsule | 0.459 | 7.78E−05* | 6.54E−04* | 0.486 | 1.92E−07* | 1.69E−07* |
| Posterior limb of internal capsule | 0.463 | 1.08E−04* | 9.11E−04* | 0.421 | 5.16E−04* | 1.01E−04* |
| Retrolenticular part of internal capsule | 0.371 | 2.30E−03 | 9.59E−03 | 0.298 | 3.87E−03 | 1.94E−03 |
| Anterior corona radiata | 0.327 | 1.54E−03 | 0.041 | 0.628 | 3.81E−03 | 7.44E−05* |
| Superior corona radiata | 0.348 | 7.99E−04* | 2.38E−03 | 0.548 | 0.015 | 8.84E−04* |
| Posterior corona radiata | 0.323 | 2.12E−03 | 5.75E−03 | 0.402 | 0.030 | 5.81E−03 |
| Posterior thalamic radiation | 0.163 | 0.068 | 0.048 | 0.530 | 0.040 | 3.22E−03 |
| Sagittal stratum | 0.258 | 4.62E−03 | 7.20E−03 | 0.351 | 0.185 | 0.058 |
| External capsules | 0.423 | 3.69E−04* | 2.21E−03 | 0.423 | 1.75E−05* | 7.74E−06* |
| Cingulum (cingulate gyrus) | 0.369 | 2.21E−04* | 6.35E−04* | 0.318 | 0.015 | 5.08E−03 |
| Fornix (cres) / Stria terminalis | 0.441 | 3.00E−04* | 2.12E−03 | 0.178 | 0.112 | 0.068 |
| Superior longitudinal fasciculus | 0.338 | 7.31E−04* | 1.88E−03 | 0.475 | 0.438 | 0.103 |
| All white matter ROIs | 0.389 | 5.34E−04* | 2.34E−03 | 0.499 | 0.015 | 1.34E−03 |
| White matter mask | 0.464 | 2.09E−05* | 1.53E−04* | 0.401 | 5.19E−04* | 1.28E−04* |
| Gray matter mask | 0.448 | 1.49E−03 | 0.011 | 0.755 | 1.01E−10* | 1.09E−07* |
Statistically significant linear (age) and quadratic (age2) contributions to the regression model are bold for P < 0.01 and bold* for P < 0.001.
ROI value rank across age groups
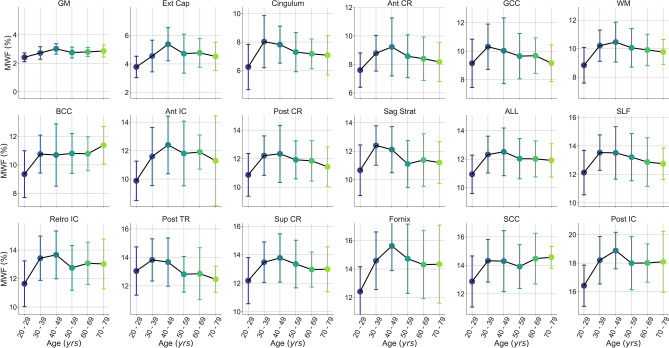
Mean metric ROI values for all subjects, grouped by decade of age, were plotted in Fig. 4A for MWF and Fig. 4B for IET2 to demonstrate the ranking of metric values between ROIs and across different age groups.
Figure 4.
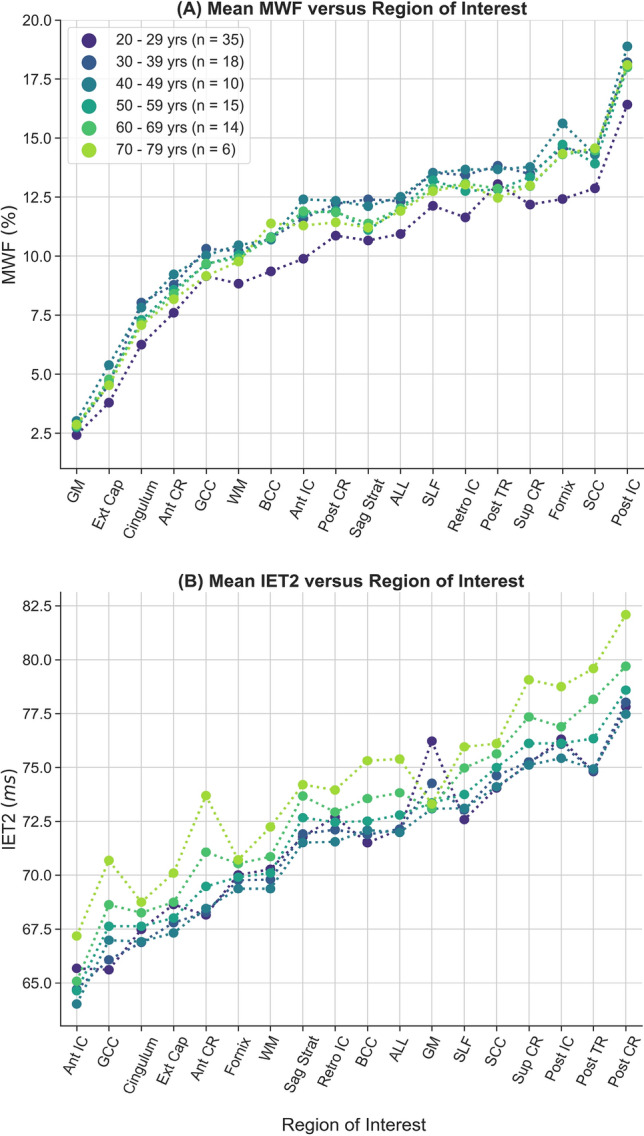
Rank of average A) myelin water fraction (MWF) and B) geometric mean T2 of intra- and extra-cellular water (IET2) for subjects grouped by decade of age within gray/white matter (GM/WM), external capsules (Ext Cap), cingulum, anterior/posterior/superior corona radiata (Ant/Post/Sup CR), anterior/posterior/retrolenticular internal capsule (Ant/Post/Retro IC), sagittal stratum (Sag Strat), all JHU labels (ALL), superior longitudinal fasciculus (SLF), posterior thalamic radiation (Post TR), fornix (Fornix), and genu/body/splenium of corpus callosum (G/B/S CC).
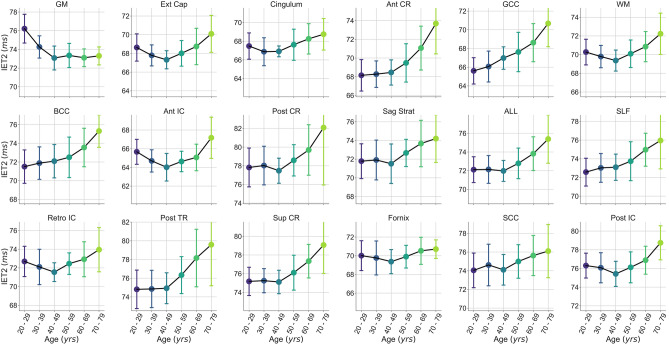
To provide a visual representation of the age-MWF and age-IET2 relationships, mean metric values were plotted versus subject age (grouped by decade) for all ROIs in Fig. 5 for MWF and Fig. 6 for IET2. In most ROIs, MWF showed an inverted U-shaped trend of increasing then decreasing MWF with age, while IET2 values generally followed the opposite pattern.
Figure 5.
Mean myelin water fraction (MWF) values versus subject age (grouped by decade) within gray/white matter (GM/WM), external capsules (Ext Cap), cingulum, anterior/posterior/superior corona radiata (Ant/Post/Sup CR), anterior/posterior/retrolenticular internal capsule (Ant/Post/Retro IC), sagittal stratum (Sag Strat), all JHU labels (ALL), superior longitudinal fasciculus (SLF), posterior thalamic radiation (Post TR), fornix (Fornix), and genu/body/splenium of corpus callosum (G/B/S CC). Uncertainty bars were calculated as the standard deviation of values across subjects within each age group.
Figure 6.
Mean geometric mean T2 of intra- and extra-cellular water (IET2) values versus subject age (grouped by decade) within gray/white matter (GM/WM), external capsules (Ext Cap), cingulum, anterior/posterior/superior corona radiata (Ant/Post/Sup CR), anterior/posterior/retrolenticular internal capsule (Ant/Post/Retro IC), sagittal stratum (Sag Strat), all JHU labels (ALL), superior longitudinal fasciculus (SLF), posterior thalamic radiation (Post TR), fornix (Fornix), and genu/body/splenium of corpus callosum (G/B/S CC). Uncertainty bars were calculated as the standard deviation of values across subjects within each age group.
Discussion
The template, atlases, tissue segmentations, and ROIs have been made openly available16 with example code for their creation17. The template was previously used for a MWI repeatability study18, and the template and atlases are being used for analyses of multiple studies at our institution.
The 3DT1 template shows excellent anatomical detail, indicative of accurate inter-subject alignment of 3DT1 images during template creation. This is demonstrated by sharp WM/GM delineation within regions with fine-scale structure, such as the cerebellum, where blurring or mixed contrast would indicate limited performance of registrations. The posterior protrusion of the left occipital pole, characteristic of the brains of right-handed individuals, is expected for a representative template of predominantly right-handed subjects19. This characteristic structure is also reflected in the mean MWF and IET2 atlas patterns, which suggests good intra-subject alignment between the 3DT1 anatomical images and gradient and spin-echo (GRASE) MWI images.
Atlas SD maps generally did not show higher-intensity borders outlining brain structures, which would indicate misalignment. With measurement noise reduced by the large number of datasets, high SD within anatomical structures is expected to originate from biological variation between subjects.
Although brain extraction generally improved template quality during our testing, inaccuracies drastically reduced registration quality. We found that intensity normalization, voxel-wise squaring to accentuate GRASE echo 1 image contrast, constraining similarity metric calculation to brain regions, and careful choice of reference template (based on image contrast, resolution, and field-of-view) facilitated consistent and accurate brain extractions.
Image registration implicitly requires that the same features are present in both inputs. High degree-of-freedom registration techniques, such as the symmetric diffeomorphic normalization transformation used here, are especially susceptible to mismatched input image features caused by brain extraction errors, pathology, or other factors. The assumptions of image registrations break down in brain regions without inter-subject correspondence, such as within cortical folds, or in those with fundamental differences in structure, such as duplication of Heschl’s gyrus20. Misregistration in regions such as these would contribute to hyperintensity in the atlas SD maps, possibly visible at the furthest anterior and posterior edges of the brain.
Diffeomorphic transformations, such as those used to create our template, intrinsically preserve the topology of input images21. Therefore, local intra-structure relationships present in quantitative maps, but not necessarily present on anatomical MRI contrast, are preserved during inter-subject alignment of large-scale MR-visible structures. Considering previous work which has suggested that the cyto-architectural layout of the brain is generally consistent22, topology-preservation is an essential characteristic for research attempting to bridge the gap between MRI macroanatomy and histological microanatomy. Although subtle quantitative metric map features could be explicitly aligned using registrations, this would introduce a circularity bias whereby the statistical significance of future comparisons to the atlas is maximized23. Furthermore, this methodology would cause naturally occurring variation between individuals to be artificially minimized, which is antithetical to the purpose of an atlas23.
Positive correlations between age and mean MWF (Table 1, Fig. 2) are consistent with the results of Flynn et al., who found positive linear correlations between MWF and age with 27 subjects (mean age 36.5 years) acquiring a single 10 mm thick slice of 2D MWI data11. Billiet et al. (n = 59, age range 17–70 years) used a 3D gradient and spin-echo (GRASE) sequence to acquire high-resolution full-brain T2 relaxation MWI data in under 12 min, but found relatively few regions with positive correlations between MWF and age13. Our results disagree with those of Faizy et al. (n = 45, median age 36 years), which showed strong negative correlations between MWF and age using data from two different 3D GRASE sequences, acquired with lower 3 and 5 mm isotropic resolution12. However, relatively low-resolution data (27 mm3 and 125 mm3 versus our 10 mm3 MWI voxel volume) could have influenced their correlations by decreasing MWF values for older subjects with brain atrophy, due to partial-volume effects with CSF.
Our significant multiple linear regressions with a quadratic age term, especially strong for some ROIs (Table 2), are supported by the inverted U-shape relationship between age and mean MWF shown in Fig. 5. These results agree with those reported by Arshad et al., who investigated six brain regions (n = 61, 18–84 years) and found a quadratic increase then decrease in MWF with age14. Papadaki et al. found a similar quadratic relationship in a larger subject group (n = 90, 22–81 years) but were limited to coarse spatial analysis of MWF in each brain lobe, due to acquiring five 2D MWI slices with 8 mm thickness and an 8 mm inter-slice gap15. Various limitations, such as limited brain coverage, low resolution, limited age range, and modest sample size, make it difficult to draw clear conclusions from the conflicting results previously reported for the age-MWF relationship. However, our results suggest that MWF does in fact follow a rapid increase during the third decade of life, followed by a non-linear trend where myelin content plateaus then decreases in later decades. Our observed trend in MWF agrees with literature suggesting that myelination continues until the sixth decade of life24 and is corroborated by the parabolic relationship found with age for previous studies using alternative quantitative MRI measures sensitive to myelin25,26. The location of maximum MWF appears to differ somewhat between brain regions in Fig. 5. However, we avoided assigning an age of peak MWF in each ROI because the exact value of the quadratic peak would be “substantially affected by seemingly irrelevant factors, such as the age-range sampled”27 and therefore biased by the specific characteristics of our subject group.
The myelin-age relationship has also been studied using an alternative approach to MWI known as multicomponent-driven equilibrium single-component observation of T1 and T2 with Bayesian Monte Carlo analysis (BMC-mcDESPOT)28, which uses a more efficient steady state acquisition to provide a surrogate measure for the MWF29. Using a large sample and wide age range, Bouhrara et al. (n = 106, range 22–94 years) found a quadratic association between myelin values and age28, similar to that found by Arshad et al. and Papadaki et al.14,15. Although Bayesian Monte Carlo analysis improves the stability of mcDESPOT, which has been shown to provide biased myelin estimates with unpredictable fluctuations30, BMC-mcDESPOT values remain overestimated compared to the reference multi-echo T2 relaxation method31 and have not been validated with comparison to histological staining. Furthermore, BMC-mcDESPOT estimates of MWF show little contrast between white matter regions, for example the genu and splenium of the corpus callosum28,29, which is inconsistent with T2 relaxation MWF results14 and is not reflective of myelin content variations reported by post mortem studies32.
Compared to the BMC-mcDESPOT results of Bouhrara et al., our MWF in WM ROIs shows much more variation between individual subject values28. This could reflect the increased myelin-specificity of our 3D multi-echo T2 relaxation MWI method, which is less sensitive to magnetisation transfer or other effects that could explain the unusually high, homogeneous MWF provided by mcDESPOT and its derivative techniques28. Our results also show increased variation in the trajectory of MWF with age between regions, which may better reflect the heterochronous brain development trajectories expected from literature33.
Mean IET2 increased with age in most WM ROIs (Table 1, Fig. 3), possibly due to increasing water content with age, and decreased in GM, likely related to increasing iron concentration with age34. Differences in axon diameter, which would affect the rate of exchange between the myelin and intra- and extra-cellular water compartments, likely also play a significant role in driving IET2 regional differences35. Our findings indicate that controlling for age can account for a significant amount of inter-subject MWF and IET2 variations.
Excluding subjects aged < 25 years (26 subjects) eliminated significance of both Pearson correlations and multiple linear regression for MWF (P > 0.05, except for posterior thalamic radiation P < 0.01) but did not drastically change IET2 results. Although this suggests that the age-MWF relationship is driven by subjects < 25 years of age, it could also be related to having relatively few subjects > 70 years of age and none > 78 years of age. Although some ROIs show a roughly inverse pattern between MWF in Fig. 5 and IET2 in Fig. 6, different dependence of the age-MWF and age-IET2 relationships on subjects < 25 years of age confirms that MWF and IET2 provide unique, complementary information.
The addition of a binarized sex variable did not significantly affect the age-MWF (P > 0.1) or age-IET2 (P > 0.01) relationship. Faizy et al. also reported the absence of sex-related differences for MWF in any WM ROIs12. Although distinct developmental patterns have been reported between sexes during early childhood36 and for young adults with median age < 25 years37, our results suggest that male and female distributions of myelin content converge at adulthood. Our results emphasize the importance of controlling for age, but not necessarily sex, when comparing MWF and IET2 values between adults.
Mean MWF and IET2 both demonstrate clear ranking between brain structures for different age groups (Fig. 4), indicating that relative metric values are generally consistent for adults. Offsets between ranking of subjects grouped by age further support the strength of the age-MWF and age-IET2 relationships. However, this relationship may not hold in children or adolescent brains, which develop with a spatially and temporally non-uniform pattern36,38,39.
A number of limitations should be considered with respect to our study. The presence of iron can artificially increase MWF values by decreasing T2 relaxation times7. Although most iron in WM is found in oligodendrocytes, co-localized with myelin, iron content is an important consideration in processes where iron and myelin changes occur independently, such as aging34. This highlights the value in characterizing healthy brain MWF and IET2 values across a wide range of ages, as provided by our atlas. Future study could combine MWI data with diffusion models to correct some of the effect of orientation on T2 relaxation values, and subsequently MWF and IET2 values40. Kumar et al. have developed an analysis approach using 3D spatial correlations to improve robustness to the spatial and temporal noise in MWI data41. Although the resulting maps are both aesthetically and quantitatively improved, the prohibitive analysis time was not practical for use in this study, at least until computational time can be reduced using GPU acceleration or other approaches.
Our atlases can be supplemented with additional datasets by processing images, registering to the template, and re-calculating the atlases using additional quantitative maps. Age-specific atlases can be generated by simply re-calculating atlases using a subset of the available data, from subjects within an appropriate age range. Although the subject group presented here has a relatively even spread of subjects between the third to eighth decade of life, future studies could benefit from additional subjects over 70 years of age and the inclusion of datasets from children and adolescents. Standardized collection of more demographic information, such as years of education, could prove similarly beneficial to developing expectations for myelin in the brain.
With enough data, a spatiotemporal “4D atlas” could be used to generate a continuous spectrum of age-specific atlases. Similarly, longitudinal data could better characterize MWF evolution across time by removing noise from inter-subject biological variability.
Previous studies have used quantitative MRI to generate maps related to human brain myelo-architecture42, usually by combining multiple imaging modalities. In vivo myelo-architecture maps could potentially be generated from MWI data by modelling the rate of exchange between the myelin water and intra- and extra-cellular water compartments, which is thought to be related to the underlying axon radius and myelin sheath thickness35. Accounting for exchange could also facilitate more robust myelin quantification in regions with especially fast or slow exchange rates.
Atlases could be aligned with atlases of spatially localized gene expression information43,44, even in tandem with temporal information. This could accelerate synergistic investigations that combine MWI metrics with genetics, which have already proven valuable by demonstrating an association between hemispheric asymmetries and genetic variation in PLP145.
In conclusion, our template, atlases, tissue segmentations, and ROIs provide an optimal framework for using MWI to study healthy myelin development or demyelinating disease. Age, but not sex, can be used to better characterize expected values for healthy adult MWF and IET2. MWF generally increases during the third decade of life, plateaus around the fifth decade, then decreases in later decades. Lastly, we showed that the ranking of MWF and IET2 values between brain structures remains consistent across different adult age groups.
Methods
MRI
Data was collected retrospectively from studies performed at the UBC MRI Research Centre on a 3.0 T Achieva (Philips Healthcare, Best, The Netherlands) using an 8-channel head coil. Each study was approved by the University of British Columbia Clinical Research Ethics Board and subjects provided written informed consent. All methods were performed in accordance with the relevant guidelines and regulations.
Inclusion criteria required that subjects had no history of brain disease or injury and that datasets included a high-resolution, sagittal T1-weighted MPRAGE sequence (3DT1) and a specific 48 echo gradient and spin-echo (GRASE) MWI sequence, with scan parameter criteria as follows.
3DT1 images had 0.8 or 1.0 mm isotropic resolution and field-of-view (FOV) = 256x256x160 mm3 or larger. Although 3DT1 MPRAGE sequence parameters varied slightly between studies, most were acquired with readout repetition time (TR) / echo time (TE) = 8.1/2.5 ms, inversion time (TI) = 1052 ms, shot interval = 3000 ms, acquisition time = 6 min 26 s. MWI data were acquired using a 3D multi-echo GRASE sequence with TR = 1073 ms, TE = 8 ms, ΔTE = 8 ms, 48 echoes, slice oversampling factor = 1.3, FOV = 230x190x100 mm3, acquired resolution = 1x2x5 mm3 (20 slices), reconstructed resolution = 1 × 1x2.5 mm3 (40 slices), sensitivity encoding acceleration factor of 2, and acquisition time = 7 min 31 s46.
Data from 100 healthy subjects were included in our study. MRI data were collected in PAR/REC format to ensure that all conversion, processing, and analysis was identical. For subjects with data available from multiple visits, only the first was included.
Age and sex demographic information was collected for all subjects (median age 38 years, range 20—78 years, 58F/42 M). Handedness information was available for 72 subjects but was not diverse enough for meaningful investigation (70 right-handed, 2 left-handed). The subject group varied in age from the third to the eighth decade of life (number of subjects: 35, 19, 11, 15, 14, and 6, respectively) and was well spread across the fourth to seventh decades.
MWI analysis
T2 distributions were produced from GRASE data in native space, before any transformations were applied, using a temporally-regularized non-negative least-squares fitting algorithm with stimulated echo correction (available upon request at https://mriresearch.med.ubc.ca/news-projects/myelin-water-fraction/)47. The analysis used 40 T2 relaxation delta functions logarithmically spaced from 15 to 2000 ms, 8 refocusing flip angles between 90° and 180°, and a χ2 regularization factor of 1.02. MWF was calculated as the fraction of the T2 distribution with 15 ms ≤ T2 < 40 ms. IET2 was calculated as the geometric mean T2 of components with 40 ms ≤ T2 < 200 ms.
Template and atlas creation
Advanced Normalization Tools software (ANTs) was used for template creation21, in addition to some FSL tools48. The ANTs symmetric diffeomorphic normalization (SyN) transformation consistently performs similarly to, or better than, alternative techniques across a wide range of applications21,49–51. No quantitative maps were involved in registration, to prevent introduction of circularity bias23.
The template creation process is visually depicted in Supplementary Material Fig. 1. To optimize template creation, empirical testing was performed using a subset of the 100 datasets (n = 6) to reduce computational demand (processing time). Testing was evaluated by computing and comparing 3 different quantitative metrics to assess how well the images were aligned. Specifically, neighborhood cross correlation, mutual information, and global correlation similarity metrics between the 3DT1 template and each subject’s 3DT1 in template space were compared for a range of template creation parameters.
3DT1 and GRASE echo 1 images underwent N4 bias field correction to mitigate low frequency intensity nonuniformities that would affect brain extraction and registration quality52. GRASE echo 1 voxel intensities were squared to accentuate tissue contrast, which improved brain extractions and intra-subject registrations. This process was motivated by previous studies that manipulated image intensities for similar purposes8.
3DT1 and GRASE echo 1 brain extractions were guided by both structure and intensity; initialized by registration to the OASIS template and priors53 then refined using Atropos n-tissue segmentations54, which also provided WM, GM, and CSF segmentations for each 3DT1. The registrations used iterative rigid, affine, and SyN transformations with optimal similarity metrics for the linear (mutual information) and SyN (neighbourhood cross-correlation) stages21. Similarity metric calculation was constrained to within a brain and skull mask, to minimize registration errors that could arise from FOV differences between our 3DT1 images and the OASIS template.
GRASE echo 1 brain extraction masks were dilated to include CSF to prevent accidental removal of brain regions during the less accurate GRASE brain extraction. Each subject’s brain extracted GRASE image was aligned with its corresponding brain-extracted 3DT1. A rigid transformation was sufficient because neither image had evidence of geometric distortion and higher degree-of-freedom registrations did not improve the alignment. CSF regions of GRASE data (from the aforementioned generous brain extraction mask) were removed using the more accurate 3DT1 brain mask.
Input images intensities were normalized to account for differences in windowing and intensity changes from N4 correction, then rigidly aligned and averaged to create an unbiased initial template starting point. The 3DT1 template was created from unbiased co-registration of all brain-extracted 3DT1 images using the same transformations and similarity metrics as for brain extraction. The resulting 0.8 mm isotropic resolution template was created using four iterations with decreasing degrees of down sampling and smoothing. This allowed registrations to align course-resolution structure before refining fine-resolution detail. Template creation was completed in 366 h running on 21 CPU cores (2.40 GHz).
GRASE-3DT1 and 3DT1-template transformations were concatenated to warp quantitative maps from GRASE space to template space with a single interpolation step. In template space, MWF and IET2 atlases were created by calculating the voxel-wise mean, as well as median and SD, across subjects.
Binary GRASE masks were also transformed to template space and were used to exclude atlas voxels not covered by all subject GRASE datasets. This mainly affected the furthest inferior edges, where coverage was affected by variations in head size.
Region of interest analysis
Iterative rigid, affine, and SyN transformations were used to map JHU-ICBM-DTI-81 WM labels and MNI structural regions to each subject 3DT155,56. Atropos tissue segmentations were used to mask JHU labels to WM and MNI regions to exclude CSF. Then WM, GM, and CSF tissue segmentations for each 3DT1, along with the JHU and MNI ROI, were transformed to template space. These were used as inputs to the probabilistic joint label fusion framework to estimate optimal tissue segmentations and ROIs for the template57.
Pearson correlations were calculated between age and mean metric values extracted from voxels within 18 WM ROIs, and 1 ROI encompassing deep and cortical GM, in template space. Statistical significance was defined as P < 0.01. Fits to mean ROI values were used because voxel-wise analyses generally did not improve characterization of the age-MWF relationship in previous studies12,13. Multiple linear regression was also used to model the data with both linear and quadratic age terms. The r-values were adjusted to account for the additional model parameter, and P-values for age and age2 were provided separately to show their relative strengths. Similarly, to identify potential differences in the age-MWF relationship between males and females, multiple linear regression was also performed with an additional binarized variable for sex. Left and right ROIs were combined because age-related changes are expected to occur bilaterally12,13.
Supplementary information
Acknowledgments
We thank the volunteers for their time, and the UBC MRI Research Centre and its MRI technologists for supporting this study. Adam Dvorak is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Alexander Graham Bell Canada Graduate Scholarship. Taylor Swift-LaPointe was supported by an NSERC Undergraduate Student Research Award. Shannon Kolind is supported by a Michael Smith Foundation for Health Research Scholar award and the Milan and Maureen Ilich Foundation. Funding support was also provided by an NSERC Discovery Grant [F17-05113], a Vancouver Coastal Health Research Institute Innovation and Translational Research Award [F18-00375], and the MS Society of Canada [F16-04023].
Author contributions
A.V.D. was involved in study conception, design, data acquisition, analysis and interpretation, and drafted and substantially revised the work. T.S.L. contributed to data analysis and drafting the work. I.M.V., A.L.M., and S.H.K. contributed to study conception, design, interpretation, and substantial revision. L.E.L., S.A., B.R.S., C.G., and A.W. contributed to data acquisition. H.L. contributed to study conception. C.L. contributed to study conception, interpretation, and substantial revision. D.K.B.L. contributed to study conception and interpretation. A.T. contributed to study conception, acquisition, interpretation, and substantial revision. R.T. contributed to study design and interpretation. L.A.B. contributed to data acquisition and interpretation. All authors contributed to the critical review of the manuscript.
Data availability
Ethics approval and subject consent were not granted for making individual subjects’ imaging data available. However, the anatomical template, quantitative atlases, and ROIs created and analysed in this study have been made openly available16 along with example code for their creation17.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-79540-3.
References
- 1.MacKay A, et al. In vivo visualization of myelin water in brain by magnetic resonance. Magn. Reson. Med. 1994;31:673–677. doi: 10.1002/mrm.1910310614. [DOI] [PubMed] [Google Scholar]
- 2.Whittall KP, et al. In vivo measurement of T-2 distributions and water contents in normal human brain. Magn. Reson. Med. 1997;37:34–43. doi: 10.1002/mrm.1910370107. [DOI] [PubMed] [Google Scholar]
- 3.Laule C, et al. MR evidence of long T2 water in pathological white matter. J. Magn. Reson. Imaging. 2007;26:1117–1121. doi: 10.1002/jmri.21132. [DOI] [PubMed] [Google Scholar]
- 4.Laule C, et al. Myelin water imaging of multiple sclerosis at 7 T: correlations with histopathology. Neuroimage. 2008;40:1575–1580. doi: 10.1016/j.neuroimage.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 5.O'Muircheartaigh J, et al. Quantitative neuroimaging measures of myelin in the healthy brain and in multiple sclerosis. Hum. Brain Mapp. 2019;40:2104–2116. doi: 10.1002/hbm.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanisz GJ, Webb S, Munro CA, Pun T, Midha R. MR properties of excised neural tissue following experimentally induced inflammation. Magn. Reson. Med. 2004;51:473–479. doi: 10.1002/mrm.20008. [DOI] [PubMed] [Google Scholar]
- 7.Birkl C, et al. The influence of brain iron on myelin water imaging. Neuroimage. 2019;199:545–552. doi: 10.1016/j.neuroimage.2019.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piredda GF, et al. Quantitative brain relaxation atlases for personalized detection and characterization of brain pathology. Magn. Reson. Med. 2020;83:337–351. doi: 10.1002/mrm.27927. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, et al. Myelin water atlas: a template for myelin distribution in the brain. J. Neuroimaging. 2019 doi: 10.1111/jon.12657. [DOI] [PubMed] [Google Scholar]
- 10.Dvorak AV, et al. Rapid myelin water imaging for the assessment of cervical spinal cord myelin damage. Neuroimage Clin. 2019;23:101896. doi: 10.1016/j.nicl.2019.101896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn SW, et al. Abnormalities of myelination in schizophrenia detected in vivo with MRI, and post-mortem with analysis of oligodendrocyte proteins. Mol. Psychiatry. 2003;8:811–820. doi: 10.1038/sj.mp.4001337. [DOI] [PubMed] [Google Scholar]
- 12.Faizy TD, et al. Age-related measurements of the myelin water fraction derived from 3D multi-echo GRASE reflect myelin content of the cerebral white matter. Sci. Rep.-UK. 2018;8:33. doi: 10.1038/s41598-018-33112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billiet T, et al. Age-related microstructural differences quantified using myelin water imaging and advanced diffusion MRI. Neurobiol. Aging. 2015;36:2107–2121. doi: 10.1016/j.neurobiolaging.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 14.Arshad M, Stanley JA, Raz N. Adult age differences in subcortical myelin content are consistent with protracted myelination and unrelated to diffusion tensor imaging indices. Neuroimage. 2016;143:26–39. doi: 10.1016/j.neuroimage.2016.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadaki E, et al. Age-related deep white matter changes in myelin and water content: a T2 relaxometry study. J. Magn. Reson. Imaging. 2019;50:1393–1404. doi: 10.1002/jmri.26707. [DOI] [PubMed] [Google Scholar]
- 16.Dvorak, A. V. Human Brain MRI Template and Myelin Atlas. (2020). 10.5281/zenodo.4067119.
- 17.Dvorak, A. V. avdvorak/myelin-atlas: version 1.0.1. (2020). 10.5281/zenodo.4067132.
- 18.Dvorak AV, et al. Multi-spin echo T2 relaxation imaging with compressed sensing (METRICS) for rapid myelin water imaging. Magn. Reson. Med. 2020;84:1264–1279. doi: 10.1002/mrm.28199. [DOI] [PubMed] [Google Scholar]
- 19.Le MM, Kido DK. Asymmetries of the cerebral hemispheres on computed tomograms. J. Comput. Assist. Tomo. 1978;2:471–476. doi: 10.1097/00004728-197809000-00018. [DOI] [PubMed] [Google Scholar]
- 20.Rademacher J, Caviness V, Jr, Steinmetz H, Galaburda A. Topographical variation of the human primary cortices: implications for neuroimaging, brain mapping, and neurobiology. Cereb. Cortex. 1993;3:313–329. doi: 10.1093/cercor/3.4.313. [DOI] [PubMed] [Google Scholar]
- 21.Avants BB, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schleicher A, Morosan P, Amunts K, Zilles K. Quantitative architectural analysis: a new approach to cortical mapping. J. Autism Dev. Disord. 2009;39:1568. doi: 10.1007/s10803-009-0790-8. [DOI] [PubMed] [Google Scholar]
- 23.Tustison NJ, et al. Logical circularity in voxel-based analysis: Normalization strategy may induce statistical bias. Hum. Brain Mapp. 2014;35:745–759. doi: 10.1002/hbm.22211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yakovlev, P. The myelogenetic cycles of regional maturation of the brain. In Regional Development of the Brain in Early Life, 3–70 (1967).
- 25.Yeatman JD, Wandell BA, Mezer AA. Lifespan maturation and degeneration of human brain white matter. Nat. Commun. 2014;5:1–12. doi: 10.1038/ncomms5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carradus AJ, et al. Age-related differences in myeloarchitecture measured at 7 T. Neurobiol. Aging. 2020;96:246–254. doi: 10.1016/j.neurobiolaging.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Fjell AM, et al. When does brain aging accelerate? Dangers of quadratic fits in cross-sectional studies. Neuroimage. 2010;50:1376–1383. doi: 10.1016/j.neuroimage.2010.01.061. [DOI] [PubMed] [Google Scholar]
- 28.Bouhrara M, et al. Adult brain aging investigated using BMC-mcDESPOT-based myelin water fraction imaging. Neurobiol. Aging. 2020;85:131–139. doi: 10.1016/j.neurobiolaging.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouhrara M, Spencer RG. Rapid simultaneous high-resolution mapping of myelin water fraction and relaxation times in human brain using BMC-mcDESPOT. NeuroImage. 2017;147:800–811. doi: 10.1016/j.neuroimage.2016.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West DJ, et al. Inherent and unpredictable bias in multi-component DESPOT myelin water fraction estimation. Neuroimage. 2019;195:78–88. doi: 10.1016/j.neuroimage.2019.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Kolind SH, Laule C, MacKay AL. Comparison of myelin water fraction from multiecho T2 decay curve and steady-state methods. Magn. Reson. Med. 2015;73:223–232. doi: 10.1002/mrm.25125. [DOI] [PubMed] [Google Scholar]
- 32.Aboitiz F, Scheibel AB, Fisher RS, Zaidel E. Fiber composition of the human corpus callosum. Brain Res. 1992;598:143–153. doi: 10.1016/0006-8993(92)90178-C. [DOI] [PubMed] [Google Scholar]
- 33.Thompson PM, et al. Structural MRI and brain development. Int. Rev. Neurobiol. 2005;67:285–323. doi: 10.1016/S0074-7742(05)67009-2. [DOI] [PubMed] [Google Scholar]
- 34.Hallgren B, Sourander P. The effect of age on the non-haemin iron in the human brain. J. Neurochem. 1958;3:41–51. doi: 10.1111/j.1471-4159.1958.tb12607.x. [DOI] [PubMed] [Google Scholar]
- 35.Harkins KD, Dula AN, Does MD. Effect of intercompartmental water exchange on the apparent myelin water fraction in multiexponential T2 measurements of rat spinal cord. Magn. Reson. Med. 2012;67:793–800. doi: 10.1002/mrm.23053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean DC, et al. Characterizing longitudinal white matter development during early childhood. Brain Struct. Funct. 2015;220:1921–1933. doi: 10.1007/s00429-014-0763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F, Vidarsson L, Winter JD, Tran H, Kassner A. Sex differences in the human corpus callosum microstructure: a combined T2 myelin-water and diffusion tensor magnetic resonance imaging study. Brain Res. 2010;1343:37–45. doi: 10.1016/j.brainres.2010.04.064. [DOI] [PubMed] [Google Scholar]
- 38.Gilles F, Shankle W, Dooling E. The developing human brain. Amsterdam: Elsevier; 1983. pp. 117–183. [Google Scholar]
- 39.Morris SR, et al. Brain myelin water fraction and diffusion tensor imaging atlases for 9–10 year-old children. J. Neuroimaging. 2020;30:150–160. doi: 10.1111/jon.12689. [DOI] [PubMed] [Google Scholar]
- 40.Birkl, C., Doucette, J., Fan, M., Hernandez-Torres, E. & Rauscher, A. Myelin water imaging depends on white matter fiber orientation in the human brain. bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 41.Kumar D, et al. Using 3D spatial correlations to improve the noise robustness of multi component analysis of 3D multi echo quantitative T2 relaxometry data. Neuroimage. 2018;178:583–601. doi: 10.1016/j.neuroimage.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen-Adad J, et al. T2* mapping and B0 orientation-dependence at 7 T reveal cyto-and myeloarchitecture organization of the human cortex. Neuroimage. 2012;60:1006–1014. doi: 10.1016/j.neuroimage.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones AR, Overly CC, Sunkin SM. The Allen brain atlas: 5 years and beyond. Nat. Rev. Neurosci. 2009;10:821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- 44.Ng L, et al. An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- 45.Ocklenburg S, et al. Myelin water fraction imaging reveals hemispheric asymmetries in human white matter that are associated with genetic variation in PLP1. Mol. Neurobiol. 2019;56:3999–4012. doi: 10.1007/s12035-018-1351-y. [DOI] [PubMed] [Google Scholar]
- 46.Prasloski T, et al. Rapid whole cerebrum myelin water imaging using a 3D GRASE sequence. Neuroimage. 2012;63:533–539. doi: 10.1016/j.neuroimage.2012.06.064. [DOI] [PubMed] [Google Scholar]
- 47.Prasloski T, Madler B, Xiang QS, MacKay A, Jones C. Applications of stimulated echo correction to multicomponent T2 analysis. Magn. Reson. Med. 2012;67:1803–1814. doi: 10.1002/mrm.23157. [DOI] [PubMed] [Google Scholar]
- 48.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 49.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein A, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tustison NJ, et al. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage. 2014;99:166–179. doi: 10.1016/j.neuroimage.2014.05.044. [DOI] [PubMed] [Google Scholar]
- 52.Tustison NJ, et al. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/Tmi.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avants, B. B. & Tustison, N. J. ANTs/ANTsR Brain Templates (2018).
- 54.Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics. 2011;9:381–400. doi: 10.1007/s12021-011-9109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazziotta J, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hua K, et al. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage. 2008;39:336–347. doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Yushkevich PA. Multi-atlas segmentation with joint label fusion and corrective learning-an open source implementation. Front. Neuroinform. 2013;7:27. doi: 10.3389/fninf.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Ethics approval and subject consent were not granted for making individual subjects’ imaging data available. However, the anatomical template, quantitative atlases, and ROIs created and analysed in this study have been made openly available16 along with example code for their creation17.