Abstract

Bacteria depend on a well-regulated iron homeostasis to survive adverse environments. A key component of the iron homeostasis machinery is the compartmentalization of Fe3+ in bacterioferritin and its subsequent mobilization as Fe2+ to satisfy metabolic requirements. In Pseudomonas aeruginosa Fe3+ is compartmentalized in bacterioferritin (BfrB), and its mobilization to the cytosol requires binding of a ferredoxin (Bfd) to reduce the stored Fe3+ and release the soluble Fe2+. Blocking the BfrB-Bfd complex in P. aeruginosa by deletion of the bfd gene triggers an irreversible accumulation of Fe3+ in BfrB, concomitant cytosolic iron deficiency and significant impairment of biofilm development. Herein we report that small molecules developed to bind BfrB at the Bfd binding site block the BfrB-Bfd complex, inhibit the mobilization of iron from BfrB in P. aeruginosa cells, elicit a bacteriostatic effect on planktonic cells, and are bactericidal to cells embedded in mature biofilms.
Keywords: biofilm, Pseudomonas aeruginosa, bacterioferritin, iron homeostasis, iron metabolism
Antimicrobial resistant bacteria are a major threat to public health. The World Health Organization (WHO) has released a priority list of pathogens for which antibiotics are urgently needed.1 Carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobactereceae are at the top of the WHO priority list because of their resistance to multiple drug classes and associated mortality. P. aeruginosa, one of the leading Gram-negative pathogens associated with nosocomial infections has a propensity to form biofilms, which are populations of bacteria living in organized structures embedded in a self-produced matrix of DNA, proteins, and polysaccharides.2−4 Although the preference of bacteria for a biofilm lifestyle was reported almost 80 years ago, the ubiquitous involvement of biofilms in chronic infection has been recognized more recently.5,6 Biofilm-related diseases are typically persistent infections that develop relatively slowly, are rarely resolved by the immune system and respond poorly or transiently to antibiotics.7 Some examples are chronic wound infection, chronic otitis media, osteomyelitis, recurrent urinary tract infection, endocarditis, and cystic fibrosis-associated infections which can accelerate lung function decay in cystic fibrosis patients.8,9 Biofilms can also colonize indwelling catheters, stents, orthopedic implants, endotracheal tubes and urinary catheters,10−12 and are ubiquitous in burn wounds and in chronic nonhealing wounds, including diabetic foot ulcers, pressure ulcers and venous leg ulcers.13,14 Bacteria in biofilms are thought to be phenotypically and physiologically different from nonadhered, free living (planktonic) cells.15 Susceptibility tests have shown that the minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) of antibiotics that are required to challenge bacteria in mature biofilms can be 10- to 100-fold higher than those required for planktonic bacteria.16−18 This intrinsic tolerance to antibiotics, which is thought to be an important reason for the persistence of biofilm infections,6,10 generates an urgent need to discover novel antibiotics and to validate new targets for combatting the threat posed by multidrug resistant organisms and overcoming the limitations of conventional antibiotics to treat chronic (biofilm) infections.19−21
Iron is an essential element in biology, and a required cofactor for many enzymes that participate in important physiological processes, such as respiration, DNA synthesis, and amino acid synthesis.22 Iron homeostasis in bacteria offers a significant vulnerability because invading pathogens must obtain essential iron from the host, but host-defenses maintain the concentration of free iron at a vanishingly low level (∼10–18 M).22 Pathogens have evolved mechanisms to “steal” iron from their host, but these depend strongly on well-regulated iron homeostasis.23,24 Consequently, we have proposed iron storage proteins as a new and specific target for dysregulating bacterial iron homeostasis.23,25,26 Our work has showed that although two ferritin-like molecules coexist in P. aeruginosa, bacterioferritin B (BfrB) is the main iron storage protein.23,27 Bacterioferritin, which only exists in bacteria, is a spherical protein that can store up to ∼3000 Fe3+ ions in its interior cavity (∼80 Å diameter). Bacterioferritin is unique in that it binds 12 heme groups buried under the external protein surface, which allows the heme propionates to protrude into the interior cavity (Figure 1A).28,29 We demonstrated that mobilization of iron from BfrB requires specific interactions with the bacterioferritin-associated ferredoxin (Bfd).30,31 The structure of Bfd revealed a new class of [2Fe-2S] protein32 and the crystal structure of the BfrB-Bfd complex showed that up to 12 Bfd molecules can bind at identical sites on the BfrB surface, at the interface of subunit dimers, above a heme molecule (Figure 1B).30 The heme mediates electron transfer between the [2Fe-2S] cluster of Bfd and the Fe3+ mineral stored in the BfrB cavity, enabling mobilization of Fe2+ to the cytosol.28,30 The Bfd binding sites on BfrB are equivalent and independent, where Bfd binds with a Kd of 3 μM.33
Figure 1.

(A) BfrB from Pseudomonas aeruginosa (PDB ID 3is7) is a spherical and hollow protein assembly which can store up to 3000 Fe3+ in the interior cavity. A heme molecule (cyan) binds between two subunits, where it is buried under the exterior surface with the heme propionates exposed to the interior cavity. (B) Structure of the BfrB-Bfd complex (PDB ID 4E6K) where a Bfd molecule (blue) binds at the interface of two BfrB subunits (green), above a heme molecule (cyan). Electron transfer from the [2Fe-2S] cluster in Bfd to the Fe3+ in the interior of BfrB is mediated by the heme. The resultant Fe2+ exits the BfrB molecule for incorporation in bacterial metabolism.
Studies conducted with P. aeruginosa PAO1 and a mutant where the bfd gene has been deleted (Δbfd) showed that in the absence of Bfd iron is irreversibly accumulated in BfrB, thus demonstrating that Bfd is required to mobilize iron stored in bacterioferritin. These studies also showed that the irreversible accumulation of iron in BfrB is accompanied by depletion of free iron in the cytosol.23 Given that P. aeruginosa requires sufficient environmental and intracellular iron reserves to establish mature biofilms,34−37 we reasoned that the iron deficiency that ensues in the cytosol of the Δbfd cells might adversely affect biofilm formation. Studies conducted to pursue this idea showed that the Δbfd cells form poorly developed biofilms and that the biofilm-embedded cells experience cytosolic iron deficiency, even in iron-sufficient culture media.26 These findings, which underscored the inhibition of the BfrB-Bfd complex as a viable target to dysregulate iron homeostasis and possibly develop novel antimicrobial tools, encouraged us to conduct a fragment-based structure-guided campaign to discover inhibitors of the BfrB-Bfd complex. These efforts allowed us to identify derivatives of 4-aminoisoindoline-1,3-dione which bind BfrB selectively at the Bfd binding site,25 block the BfrB-Bfd complex in the P. aeruginosa cytosol, and inhibit the mobilization of bacterioferritin-stored iron.25 Challenging planktonic cultures of P. aeruginosa with the 4-aminoisoindoline-1,3-dione analogues elicited a dose-dependent growth inhibition phenotype, thus providing proof of concept for the usefulness of small molecules designed to inhibit the BfrB-Bfd complex as probes to dysregulate iron homeostasis and weaken bacterial cells.25 Herein we report on improvements made to the 4-aminoisoindoline-1,3-dione derivatives which increase the binding affinity for BfrB and the bacteriostatic activity against planktonic P. aeruginosa cells. Surprisingly, the 4-aminoisoindoline-1,3-dione derivatives are bactericidal to P. aeruginosa cells embedded in mature biofilms. The killing effect of the BfrB-Bfd complex inhibitors on biofilm cells, which are normally recalcitrant to several classes of commercial antibiotics, has uncovered a rare weakness that may be exploited to control biofilms.
Results and Discussion
Synthesis of 4-Aminoisoindoline-1,3-dione Derivatives
Solving the X-ray structure of the BfrB-Bfd complex30 and identifying the hot-spot residues participating at the protein–protein interface33 provided a unique molecular platform to study the mechanisms that enable mobilization of bacterioferritin-stored iron.28 The structural information also enabled a fragment based structure-guided strategy to discover inhibitors of the BfrB-Bfd complex. Fragment screening followed by efforts to cocrystallize the fragment hits with BfrB produced the X-ray crystal structure of BfrB in complex with 4-aminoisoindoline-1,3-dione (8) binding at the Bfd-binding site.25 The cocrystal structure informed a strategy for fragment growth that produced a series of 4-(benzylamino)- and 4-((3-phenylpropyl)amino)isoindoline-1,3-dione analogues represented by 11 and 16 (Table 1), which are derivatives of 8 with −(CH2)– and −(CH2)3– linkers, respectively.25,38 These analogues were shown to selectively bind BfrB at the Bfd binding site and to elicit a growth defect in P. aeruginosa planktonic cell cultures.25 To increase the binding affinity of 11 and 16 for BfrB we looked for information in the available structural data. For example, the crystal structures of 11 and 16 bound to BfrB show clear electron density defining the aminoisoindoline-1,3-dione (phthalimide) moieties. The hydroxy-substituted phenyl rings, however, are less well-defined by electron density, suggesting that conformational disorder of the phenyl ring in the cleft formed by L68 and E81 may influence the stability of the complex.25 In an attempt to lower disorder of the phenyl ring, we prepared a set of 4-aminoisoindoline-1,3-dione derivatives bearing a relatively bulky halogen atom in the phenyl ring (Table 1). A schematic summary of the representative syntheses for compounds with −(CH2)– or −(CH2)3– linkers is shown in Figure 2. For example, compound KM-5-25 was prepared from 5-chlorosalicylaldehyde by reductive amination with 8 in N,N-dimethylformamide (DMF) solution using sodium triacetoxyborohydride as the reducing agent. Compound KM-5-66 was prepared from 3-chloro-5-hydroxybenzaldehyde; the phenol was protected with tert-butyldimethylsilyl chloride (TBSCl) and the side chain elongated by Horner–Wadsworth–Emmons reaction to the α, β-unsaturated ester. Subsequent catalytic reduction of the side chain double bond followed by diisobutylaluminum hydride (DIBAL-H) reduction of the ester under carefully controlled temperature conditions furnished the requisite 3-arylpropionaldehyde. This aldehyde was then linked by reductive amination to 8 as above and deprotected with tetrabutylammonium fluoride (TBAF) in THF. Additional details are presented in the Supporting Information.
Table 1. Structure, Binding Affinity, and IC50 of 4-Aminoisoindoline-1,3-dione Derivatives.
Not determined because of low solubility (<30 μM) in PBS buffer.
Figure 2.
Schematic summary of the synthetic process to prepare 4-(benzylamino)isoindoline (i.e., KM-5-25) and 4-((3-phenylpropyl)amino)- (i.e., KM-5-66) derivatives of 4-aminoisoindoline-1,3-dione.
4-Aminoisoindoline-1,3-dione Derivatives Elicit a Bacteriostatic Effect in Planktonic P. aeruginosa Cultures
The relative strength of the association between the new analogues and BfrB was evaluated measuring the dissociation constant Kd (Table 1, Figure S1). The results show that installing a halogen atom in the phenyl ring improves the binding affinity of the derivatives relative to the previously reported analogues 11 and 16. The Kd values of halogen containing compounds with a −(CH2)– linker are on average ∼2-fold lower when compared to the Kd exhibited by 11, and the Kd values of halogen-bearing analogues with a −(CH2)3– linker are on average ∼5-fold lower than the Kd measured for analogue 16. The relative efficacy of the analogues to inhibit P. aeruginosa planktonic growth was evaluated by measuring the half maximal inhibitory concentration (IC50). Inspection of the data in Table 1 shows that all of the halogenated compounds are more active than analogues 11 and 16; the activity of KM-5-29 and KM-5-54 could not be evaluated because the relatively low aqueous solubility of these compounds prevented measurement of their IC50 values.
Although the compounds synthesized so far do not include all possible substitution isomers, some insights of the governing structure activity relationships have begun to emerge (Table 1): (i) Among the compounds with a −(CH2)– linker, the data reveal that when the hydroxyl group is at position 2 relative to the linker (KM-5-29, JAG-5-7, KM-5-25, and KM-5-30) a bulkier Cl atom at position 5 (KM-5-25) imparts ∼2-fold higher binding affinity for BfrB than a smaller F atom at the same position (JAG-5-7). In line with the nearly 2-fold lower Kd, the IC50 of KM-5-25 is ∼2-fold lower than that of JAG-5-7. In comparison, a Cl atom at position 3 (KM-5-30) imparts a Kd similar that of KM-5-25, but an IC50 ∼ 2.5-fold larger, suggesting that KM-5-30 is less efficient at penetrating or accumulating in P. aeruginosa cells. Installing a Cl atom at position 6 (KM-5-29) renders the compound poorly soluble in aqueous solution. It is also interesting to compare the two analogues with a hydroxyl group at position 3. The presence of a Cl atom at position 5, KM-5-50, lowers the Kd by a factor of 2 relative to BN-XIV-53 and improves the activity vs planktonic cells significantly. (ii) Examining the compound series with a −(CH2)3– linker shows that when the hydroxyl group is at position 3, a Cl atom at position 5 (KM-5-66) decreases the Kd ∼ 4-fold and the IC50 ∼ 3-fold relative to compound 16. In comparison, the presence of a second hydroxyl group at position 5 (KM-5-57) increases the Kd ∼ 1.7-fold relative to 16 and renders the compound inactive. Given that the Kd measured for KM-5-57 is similar or lower than Kd values measured for other active compounds in Table 1, the immeasurable activity of KM-5-57 suggests that it cannot penetrate or accumulate in P. aeruginosa cells. Finally, comparison of compounds where the hydroxyl group is at position 2 (KM-5-28, JAG-5-6, and KM-5-54) also shows that a halogen at position 5 improves binding affinity. A bulkier Cl atom at position 5 increases the binding affinity of KM-5-54 2-fold relative to the compound with a F at the same position (JAG-5-6). Comparing the IC50 values corresponding to KM-5-28 and JAG-5-6 reveals that the ∼3-fold lower Kd caused by installing a F atom at position 5 is accompanied by ∼2-fold decrease in the IC50. Attempts to determine whether the lower Kd obtained when a Cl atom at position 5 would bring an additional decrease in the IC50 were stymied by the poor water solubility of KM-5-54.
The structure–activity relationships (SAR) information available thus far indicates that a halogen in the aryl ring of the 4-aminoisoindoline-1,3-dione derivatives invariably improves binding affinity for BfrB. In compounds with a −(CH2)– linker, when the hydroxyl group is at position 2, a Cl atom at position 5 imparts favorable properties, a Cl atom at position 3 renders the compound poorly active vs P. aeruginosa, despite relatively favorable Kd, and a Cl atom at position 6 imparts poor aqueous solubility. The preparation of compounds with a −(CH2)3– linker is more elaborate and accessibility to suitable starting materials at reasonable prices is also more limited. The information available thus far indicates that although a Cl at position 5 is favorable whether the hydroxyl group is at position 2 or 3, poor aqueous solubility impairs the potential biological activity when the hydroxyl is at position 2 (KM-5-54), despite having the most favorable Kd of all compounds in Table 1. Attempts to increase aqueous solubility by installing a hydroxyl group at position 5 succeeded in increasing solubility but rendered a compound (KM-5-57) with lower affinity for BfrB and inactive against P. aeruginosa cells. As new compounds become available, similar evaluation of binding affinity and activity against bacterial cells will continue to shed light on the structural requirements that simultaneously enhance target affinity in vitro and activity against P. aeruginosa cells. For the purposes of the studies described below, we chose to work with compounds KM-5-25, the most active of the analogues containing a −(CH2)– linker, and KM-5-66, the most active of the analogues harboring a −(CH2)3– linker.
Structures of Representative 4-Aminoisoindoline-1,3-dione Derivatives Bound to BfrB
To gain additional evidence for the selectivity of the 4-aminoisoindoline-1,3-dione derivatives for the Bfd binding site on BfrB, we carried out ligand soaking experiments aimed at obtaining cocrystals of compound bound to the bacterioferritin. These experiments resulted in the X-ray crystal structures of KM-5-28, KM-5-66, KM-5-50, and JAG-5-7 bound to BfrB; refinement statistics are summarized in Table S1. In all four structures the 4-aminoisoindoline-1,3-dione (phthalimide) moiety binds at the Bfd binding site with a nearly identical pose, forming hydrogen bonding interactions to the O atom of P69 and the backbone N atoms of L71 and N70 (Figures 3A,B, 4A,B, and S2). This binding mode, which is nearly identical with that observed in the previously reported structures of 11, 16, and several other analogues,25 provides strong evidence for the selectivity of the 4-aminoisoindoline-1,3-dione derivatives for the Bfd site on BfrB. The linkers and phenyl rings also bind BfrB at the Bfd binding site, where they fit in a cleft formed by the side chains of L68 and E81. As will be discussed below, the details of binding are particular to whether the compound harbors a −(CH2)– or a −(CH2)3– linker and to the substitution pattern on the ring.
Figure 3.
Binding of KM-5-66 to the Bfd binding site on BfrB. Subunits A and B of a BfrB subunit dimer are rendered as surface and colored in gray and green, respectively. (A) Fo – Fc omit map contoured at 3σ (coral mesh) from KM-5-66 bound on BfrB. (B) Hydrogen bonding interactions (dashed lines) between KM-5-66 and BfrB. (C) View along the 3-hydroxyl-5-chloro phenyl ring plane of KM-5-66 (coral) depicted as a surface rendering along with residues surrounding the phenyl ring. (D) View depicting the 3-hydroxyl-5-chloro phenyl ring position relative to residues M31 and W35; pertinent distances are indicated by solid lines. The Cl atom of KM-5-66 is depicted in green.
Figure 4.
Binding of JAG-5-7 to the Bfd binding site on BfrB. (A,B) Fo – Fc omit map contoured at 3σ (coral mesh) from JAG-5-7 bound at the Bfd binding site in one of the BfrB subunit dimers, and hydrogen bonding interactions (dashed lines). The 2-hydroxyl-5-fluoro phenyl ring is observed in only one conformation at this binding site. (C,D) View of a different binding site where the 2-hydroxyl-5-fluoro phenyl ring is observed in two conformations; in one of the conformations the hydroxyl group is hydrogen bonded to the carbonyl oxygen of G80. The F atom of JAG-5-7 is depicted in green.
The structure of KM-5-66 (Figure 3) shows the −(CH2)3– linker wedged at the “floor” of the cleft formed by the side chains of L68 and E81 on BfrB (Figure 3A,B), where it engages in hydrophobic packing with the several residues forming the cleft floor and with the hydrophobic portion of the L68 and E81 side chains. The 3-hydroxyl-5-chloro phenyl ring, which is nearly parallel with the cleft floor, enables the 3-hydroxyl group to interact with the E81 carbonyl oxygen via H-bonding, and the Cl atom to interact with the side chains of L63 and L68 (Figure 3A,B). The phenyl ring, at its closest point, is also 3.60 Å from the M31 CE atom, a favorable interaction that is also probable to hinder rotation of the ring toward a perpendicular position relative to the cleft floor (Figure 3C,D). It is also interesting to note that the 3-hydroxyl-5-chloro phenyl ring is positioned to form a pseudo edge-to-face interaction with the indole ring of W35; the phenyl ring, at its closest position, is 3.77 Å from the indole ring CZ2 atom, the distance between the centroids of the phenyl and the indole 6-member ring is 5.92 Å, and the angle between the planes of these rings is 67.4°, suggesting a weak edge-to-face interaction.
Despite significant effort, we could not obtain good diffraction data from BfrB crystals soaked in solutions containing KM-5-25. However, we were able to obtain the structure of the fluorinated analogue JAG-5-7 (Figure 4). Strong electron density consistent with JAG-5-7 was observed in 8 of the 12 subunits. The shorter −(CH2)– linker places the 2-hydroxyl-5-fluoro phenyl ring well within the cleft formed by the side chains of L68 and E81, with the ring nearly perpendicular to the cleft floor. The electron density from the 2-hydroxyl-5-fluoro phenyl ring is consistent with the ring experiencing one orientation in certain subunits (Figure 4A,B) and with the ring in two conformations in other subunits (Figure 4C,D). In one of these orientations the hydroxyl group forms a H-bond with the carbonyl oxygen of G80, whereas the fluorine atom packs against the side chain of L68. The structure of KM-5-50 (Figure S2), also places the 3-hydroxyl-5-chloro phenyl ring within the cleft formed by L68 and E81, in a similar nearly perpendicular conformation relative to the cleft floor, except that no H-bonding was observed for the 3-hydroxyl group.
Taken together, the structures of the 4-aminoisonindoline-1,3-dione derivatives reported here and those reported previously25 provide important insights. The linkers and the phenyl rings of all the compounds containing a −(CH2)3– linker adopt nearly identical binding modes, with the phenyl rings oriented parallel to the cleft floor (Figure S3) and positioned to engage the 6 member ring of the indole in W35. Superposing the structures of analogue 16 and KM-5-66 (Figure S4A) shows that the phenyl ring in 16 is notably pitched relative to the phenyl ring in KM-5-66 (the angle between the mean planes of both rings is 22.6°), and the 3-hydroxyl group is not engaged in H-bonding interactions. In comparison, the Cl atom at position 5 in KM-5-66 appears to induce a nearly parallel orientation of the ring relative to the cleft floor and a conformation that places the 3-hydroxyl within H-bonding distance of the carbonyl oxygen of E81. Together, the H-bonding engagement of the 3-hydroxyl group, the packing of the Cl atom with the side chains of L63 and L68, and the more extensive packing of the phenyl ring against the cleft floor residues are probably responsible for the higher affinity of KM-5-66 for BfrB relative to 16. It is also interesting to note that in the structures of all the compounds with a shorter −(CH2)– linker the phenyl ring is nearly perpendicular to the cleft floor. This is illustrated by superposing the structures of KM-5-66 and JAG-5-7 (Figure S4B), which shows that the phenyl rings of both compounds adopt a nearly perpendicular angle (74°) relative to one another. The structural information currently available suggests that the shorter linker and the relatively less efficient packing of the phenyl ring against the hydrophobic portions of the L68 and E81 side chains may contribute to the higher Kd values of these compounds relative to those with a −(CH2)3– linker.
Planktonic P. aeruginosa Cells Treated with 4-Aminoisoindoline-1,3-dione Derivatives Overproduce Pyoverdine
Previous studies directed at evaluating the repercussions of blocking the BfrB-Bfd complex in P. aeruginosa cells relied on deleting the bfd gene (Δbfd). These investigations showed that blockade of the BfrB-Bfd complex in planktonic Δbfd cells causes an irreversible accumulation of iron in BfrB and iron deficiency in the cytosol. The resultant phenotype is hyperproduction of pyoverdine relative to the wild type cells.23 Pyoverdine is a siderophore produced by P. aeruginosa when the cells experience iron limitation.39 A similar pyoverdine overproduction phenotype was observed when wild type P. aeruginosa cells were treated with small molecule inhibitors of the BfrB-Bfd complex (11 and 16).25 Therefore, to determine that compounds KM-5-25 and KM-5-66 inhibit iron mobilization from BfrB in the P. aeruginosa cytosol, we investigated whether cells treated with these compounds express the characteristic pyoverdine hyperproduction phenotype. To this end, planktonic cells were cultured in the presence of KM-5-25 (70 μM) or KM-5-66 (50 μM) for 27 h in M63 media and the content of the secreted pyoverdine in the cell-free spent media was analyzed by measuring the fluorescence intensity at 460 nm. Normalizing the intensity of pyoverdine fluorescence to CFU/mL shows that as expected, cells treated with KM-5-25 or KM-5-66 secrete ∼5-fold more pyoverdine than the untreated control (Figure S5), an overproduction level similar to that observed with the Δbfd mutant.23 These observations indicate that both analogues bind BfrB in the P. aeruginosa cytosol, block the BfrB-Bfd interaction and inhibit iron mobilization from BfrB, resulting in cytosolic iron limitation that is manifested in a pyoverdine hyperproduction phenotype. The cytosolic iron limitation caused by treating planktonic cultures with KM-5-25 or KM-5-66 exerts a bacteriostatic effect on the cells, as indicated by the IC50 values in Table 1. In stark contrast, when the same compounds are used to treat P. aeruginosa biofilms, a bactericidal effect is observed. The results from these experiments are discussed below.
4-Aminoisoindoline-1,3-dione Derivatives Kill P. aeruginosa Cells in Mature Biofilms
A characteristic of biofilms is their high tolerance to antimicrobial agents. Tolerance is a physiological condition which does not involve mutation and enables bacteria to survive in the presence of antibiotics.40−43 The persistent biofilm phenotype is thought to arise from several factors, including restricted penetration of antibiotic molecules due to interactions with components of the biofilm matrix, slow cell metabolism in the biofilm, differential expression of specific genes, and the presence of persister cells. In addition, biofilms are composed of distinct subpopulations that exhibit different physiological activity; cells in the biofilm interior exhibit low metabolic activity, distinct from the high metabolism of cells near the surface.40,44,45 The dissimilar metabolic activity is thought to result from a concentration gradient of O2 and nutrients, which are high at the biofilm surface and low in the deeper layers of the biofilm.45,46 Commercial antibiotics that interfere with cell replication (e.g., ciprofloxacin), or protein translation (e.g., tobramycin), preferentially kill the metabolically active bacteria in the outer biofilm layers, whereas cells in the biofilm interior survive,44,47−49 despite the ability of both antibiotics to diffuse into the inner regions of the biofilm.44,50 In contrast, some antimicrobials that affect membrane structure, such as colistin, a “last-line” therapy to treat multidrug resistant infections,51−53 can kill cells in the deeper biofilm layers.49
We tested the susceptibility of mature biofilms to treatment with analogues of 4-aminoisoindoline-1,3-dione using two platforms, biofilms cultured at the solid–liquid interface (flow cell biofilms) and biofilms cultured at the air–liquid interface (pellicles). Biofilms of P. aeruginosa cells expressing an enhanced yellow fluorescent protein (EYFP) were cultured in flow cells using AB minimal media supplemented with 15 μM Fe. 3-Day old biofilms were treated for 24 h with commercial antibiotics or with 4-aminoisoindoline-1,3-dione analogues by flowing AB media containing analogue or commercial antibiotic, 0.025% hypromellose (HPMC), 1.5% DMSO and 15 μM Fe. In most experiments the concentration of commercial antibiotics used was 25× or 50× MIC, and the concentration of 4-aminoisoindoline-1,3-dione analogues was between 0.36× and 1.2× the IC50. The treated biofilms were counterstained with the cell impermeable fluorescent nucleic acid dye Sytox Red and then imaged with the aid of confocal laser scanning microscopy (CLSM). Figure 5A depicts a representative image of the untreated control, illustrating yellow-fluorescent viable cells and red-stained dead cells and extracellular DNA. Figures 5B and 5C show representative images depicting 4-day old biofilms tolerant to 24 h treatment with ciprofloxacin or tobramycin, respectively, and Figure 5D shows 4-day old biofilms susceptible to 24 h treatment with colistin. In agreement with previously reported observations,49,54 treatment with ciprofloxacin preferentially kills cells at the biofilm surface, leaving the interior of the biofilm almost unaffected (Figure 5B). In contrast, treatment with colistin preferentially kills bacteria in the biofilm interior, leaving the biofilm surface less affected (Figure 5D). COMSTAT software was used to attempt a quantitative comparison of the biofilm biomass by estimating the biovolume, which is calculated as the overall volume/substratum area (μm3/μm2).55 Comparing the biofilm biomass as the ratio of (untreated biomass)/(treated biomass) expressed as % survival (Figure 5E) shows that ∼20% of biomass remains viable (yellow fluorescent) after treatment with colistin at 25× MIC. In comparison, ∼60% and 70% of biomass remains viable after treatment with 25× MIC ciprofloxacin or tobramycin, respectively.
Figure 5.
Biofilms cultured in flow cells are tolerant to ciprofloxacin and tobramycin and susceptible to colistin. EYFP-expressing P. aeruginosa PAO1 biofilms were cultured for 3 days by flowing AB media supplemented with 15 μM Fe and then treated for 24 h by flowing the same media containing antibiotic. Biofilms were counterstained with Sytox Red and imaged with the aid of CLSM. Top-down views (x–y plane) are depicted with side views (x–z plane) at the bottom. Viable cell mass is in yellow and dead cells and extracellular DNA in red. (A) Untreated (DMSO) control, (B) 25× MIC ciprofloxacin (19 μM), (C) 25× MIC tobramycin (27 μM), (D) 25× MIC colistin (20 μM). (E) The % survival obtained from viable biomass calculated with the aid of COMSTAT software. The scale of the bars represents 20 μm. p < 0.01 denoted by ** and p < 0.001 by *** relative to untreated.
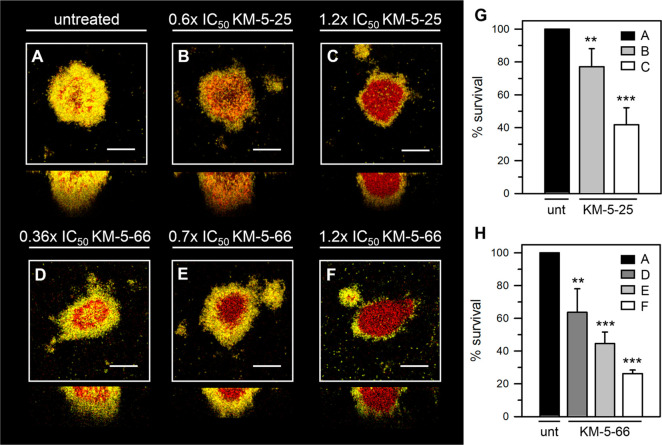
Having established that the biofilms are susceptible to colistin and significantly tolerant to ciprofloxacin and tobramycin, we treated similarly cultured 3-day old biofilms with KM-5-25 and KM-5-66 for 24 h. Compounds KM-5-25 and KM-5-66 are soluble in aqueous media to ∼110 and ∼80 μM, respectively. To ensure that the compounds remain soluble during the 24 h treatment period, KM-5-25 was used at concentrations 40 and 80 μM, equivalent to 0.6× and 1.2× the IC50, and KM-5-66 was used at concentrations 15, 30, and 50 μM, equivalent to 0.36×, 0.7×, and 1.2× the IC50 (Table 1). Representative CLSM images obtained after treating 3-day old biofilms with each of the analogues for 24 h show that both compounds kill biofilm cells in a concentration dependent manner (Figure 6). Treatment with 50 μM KM-5-66 elicits a similar level of killing as treatment with 80 μM KM-5-25. The higher efficacy exhibited by KM-5-66 agrees with its higher binding affinity for BfrB and lower IC50. Inspection of the images obtained upon treatment with the higher concentrations of KM-5-25 or KM-5-66 clearly shows that the inhibitors of the BfrB-Bfd complex kill the cells in the interior of the biofilm, leaving most of the viable cells located at the biofilm surface (Figure 6C,F). This pattern of killing is reminiscent of previously reported observations showing that treatment of biofilms with Ga3+ preferentially kills cells in the inner portion of the biofilm.56 The same authors concluded that cells in the biofilm interior are more sensitive to Ga3+ because this population experiences a more pronounced iron starvation. Therefore, we speculate that in biofilms treated with KM-5-25 or KM-5-66 the internal biofilm population is more susceptible to iron limitation caused by the nearly irreversible accumulation of iron in BfrB. It is important to underscore that although the mechanisms whereby iron starvation contribute to cell death in the biofilm interior are not yet understood, the fact remains that perturbation of iron homeostasis, either by systemic replacement of Fe3+ with Ga3+, or by selective inhibition of the BfrB-Bfd complex, leads to bacterial cell death.
Figure 6.
P. aeruginosa cells embedded in mature biofilms grown in flow cells are susceptible to 4 aminoisoindoline-1,3-dione analogues. EYFP-expressing P. aeruginosa PAO1 biofilms were cultured for 3 days by flowing AB media supplemented with 15 μM Fe and then treated for 24 h with 4-aminoisoindoline-1,3-dione analogue. Biofilms were counterstained with Sytox Red and imaged with the aid of CLSM. Top-down views (x–y plane) are depicted with side views (x–z plane) at the bottom. Viable cell mass is in yellow and dead cells and extracellular DNA in red. (A) Untreated (DMSO) control, (B) 0.6× IC50KM-5-25 (40 μM), (C) 1.2× IC50KM-5-25 (80 μM), (D) 0.36× IC50KM-5-66 (15 μM), (E) 0.7× IC50KM-5-66 (30 μM), and (F) 1.2× IC50KM-5-66 (50 μM). (G,H) The % survival obtained from viable biomass calculated with the aid of COMSTAT software for cells treated with KM-5-25 and KM-5-66, respectively. The scale of the bars represents 20 μm. p < 0.01 denoted by ** and p < 0.001 by *** relative to untreated.
To expand the observations made with flow cell biofilms into a second biofilm model we also studied the susceptibility of biofilms grown at the air–liquid interface (pellicles).57,58 Pellicle biofilms (henceforth pellicles) are an attractive alternative platform to study biofilms because pellicles are amenable to imaging by CLSM and to harvesting, which can be desirable for additional biofilm analysis.26 To determine the susceptibility of pellicles to antibiotics or inhibitors of the BfrB-Bfd complex, we cultured 2-day old pellicles of EYFP-expressing P. aeruginosa cells in PI media supplemented with 20 μM Fe, as indicated in the Methods. The pellicles were transferred onto glass coverslips by allowing the surface of the coverslip to contact a pellicle. The coverslip-adhered pellicles were subsequently exposed to treatment solution (AB media supplemented with 15 μM Fe, 0.025% HPMC, 1.5% DMSO, and antibiotic or analogue) for 24 h prior to staining with Sytox Red and imaging with the aid of CLSM. The pellicle biofilms are tolerant to ciprofloxacin and tobramycin at concentrations 25× and 50× the MIC (Figure 7B–E), as is evident by the yellow fluorescence and near complete absence of red-stained dead cells. In contrast, the pellicle biofilms are susceptible to colistin at concentrations above 10× MIC (Figure 7F–H). Analysis of the images with COMSTAT, which allowed a more quantitative comparison of cell survival upon treatment with each of the antibiotics (Figure 7I), confirms tolerance to ciprofloxacin and tobramycin, but sensitivity to colistin. Note that when the concentration of colistin is 50× MIC the fluorescence signal from viable cells expressing EYFP is undetectable. When pellicle biofilms are challenged with compound KM-5-25 of KM-5-66 bacterial cell death occurs in a concentration dependent manner (Figure 8). These results agree with the idea that the 4-aminoisoindoline-1,3-derivatives penetrate the bacterial cell and bind to their target in the P. aeruginosa cytosol. Inspection of the images and analysis with COMSTAT (Figure 8G and H) shows that compound KM-5-66 is more efficacious than KM-5-25, observations that are consistent with the lower Kd and IC50 values measured for KM-5-66. It is also important to note that when compounds KM-5-25 and KM-5-66 are used at a concentration of 80 μM and 50 μM, respectively (Figure 8G,H), which correspond approximately to 1.2× IC50, nearly 85% of the cells in the pellicle are killed. This efficacy is similar that observed with colistin when used at 20 μM, equivalent to 25× MIC (Figure 7I).
Figure 7.
Pellicle biofilms are tolerant to ciprofloxacin and tobramycin, and susceptible to colistin. Pellicles of P. aeruginosa PAO1 expressing EYFP were cultured in PI media supplemented with 20 μM Fe for 48 h, and then treated with antibiotics for 24 h. Pellicles were counterstained with Sytox Red and imaged with the aid of CLSM. Images depict top-down views (squares) and side views (rectangles) where viable cells are shown in yellow and dead cells and extracellular DNA in red. (A) Untreated (DMSO) control, (B) 25× MIC ciprofloxacin (19 μM), (C) 50× MIC ciprofloxacin (38 μM), (D) 25× MIC tobramycin (27 μM), (E) 50× MIC tobramycin (54 μM), (F) 12.5× MIC colistin (10 μM), (G) 25× MIC colistin (20 μM), (H) 50× MIC colistin (40 μM). (I) The % survival obtained from viable biomass calculated with the aid of COMSTAT software. The scale of the bars represents 20 μm. p < 0.001 denoted by *** relative to untreated.
Figure 8.
4-Aminoisoindoline-1,3-dione analogues kill P. aeruginosa cells embedded in pellicle biofilms. Pellicles of P. aeruginosa PAO1 cells expressing EYFP were cultured for 48 h in PI media supplemented with 20 μM Fe, and then treated with KM-5-25 or KM-5-66 for 24 h. Pellicles were counterstained with Sytox Red and imaged with the aid of CLSM. Images depict top-down views (squares) and side views (rectangles) where viable cells are shown in yellow and dead cells and extracellular DNA in red. (A) Untreated (DMSO) control, (B) 0.6× IC50KM-5-25 (40 μM), (C) 1.2× IC50KM-5-25 (80 μM), (D) 0.36× IC50KM-5-66 (15 μM), (E) 0.7× IC50KM-5-66 (30 μM), and (F) 1.2× IC50KM-5-66 (50 μM). (G,H) The % survival obtained from viable biomass calculated with the aid of COMSTAT software for pellicles treated with KM-5-25 and KM-5-66, respectively. The scale of the bars represents 20 μm. p < 0.01 denoted by ** and p < 0.001 by *** relative to untreated.
To assess the efficacy of antibiotics and compounds with an approach complementary to imaging with CLSM, we resorted to dispersing biofilm cells for subsequent enumeration of viable cells (CFU/mL). To this end, we cultured pellicle biofilms for 48 h in PI media containing 20 μM Fe and exposed them to AB media containing 15 μM Fe and antibiotic or compound for 24 h, as described above. The biofilms were then harvested, and the cells dispersed into sterile PBS by vortexing in the presence of zirconia beads, prior to plating the cell suspensions for subsequent enumeration of CFU/mL. The results from these experiments are summarized in the plots of Figure 9 which show the % cell survival of pellicle-embedded cells after challenges with antibiotic or compound, calculated from the fraction CFU/mL(after treatment)/CFU/mL(pretreatment). When colistin is used to treat the pellicles at concentrations equivalent to 25× and 50× MIC, the treated biofilms exhibit ∼10% and ∼1% cell survival, respectively, relative to the pretreated biofilm (Figure 9A), corroborating the sensitivity of the pellicles to colistin. In contrast, challenging the pellicles with tobramycin (25× and 50× MIC) results in ∼70% survival relative to the pretreated biofilm, and nearly identical cell survival relative to the untreated (DMSO control) pellicles (Figure 9A). These observations, which are in good agreement with those made with the aid of CLSM (Figure 7D–H) corroborate that the pellicles are tolerant to tobramycin and sensitive to colistin. Interestingly, attempts to enumerate cells after challenging the pellicles with ciprofloxacin (25× or 50× MIC) resulted in extremely low CFU/mL, findings which at first glance appear to be in conflict with the tolerance of the biofilms to ciprofloxacin observed in the CLSM images (Figure 7B,C). To reconcile these seemingly discrepant observations it is important to consider that several stressors, including ciprofloxacin, can induce a dormancy state in bacterial cells known as the viable but not culturable (VBNC) state. A characteristic of cells in the VBNC state is their inability to develop into colonies on routine culture media, even though the cells remain viable for long periods of time.59,60 Evidence that bacterial cells can enter the VBNC has been obtained by several distinct methods,60 one of which is the utilization of bacteria engineered to constitutively express bioluminescent proteins, and using the bioluminescence as a reporter of metabolic activity. Studies conducted with P. aeruginosa showed that following treatment with ciprofloxacin the bioluminescence emitted by P. aeruginosa cells decreased significantly less than the viable cell counts (CFU/mL). The perceived reduction in viable cell counts, which did not correlate with the relatively high metabolic activity reported by the small decrease in bioluminescence, indicated that challenges with ciprofloxacin induce P. aeruginosa cells to enter a VBNC state.61,62 Our observations suggest a similar situation. Imaging the pellicles with CLSM following the 24 h challenge with ciprofloxacin (Figure 7B,C) shows that most of the cells are metabolically active (yellow fluorescent), but dispersion of the cells from the pellicles for enumeration of CFU/mL shows a large reduction in culturable cells relative to the untreated control. These observations strongly suggest that treating the pellicles with ciprofloxacin induces the cells to enter the VBNC state, thus rendering them tolerant to the antibiotic. These findings, which highlight the complexities associated with biofilm embedded cells, also underscore the importance of resorting to more than one platform to study the efficacy of antibiofilm agents.
Figure 9.
Assessment of cell survival in pellicle biofilms by dispersing and counting viable cells. EYFP-expressing P. aeruginosa PAO1 cells embedded in two-day old pellicles treated for 24 h with antibiotic or 4-aminoisoindoline-1,3-dione derivatives were dispersed for enumeration of viable culturable cells (CFU/mL). The % survival is expressed as the ratio CFU/mL(after treatment)/CFU/mL(pretreatment). Pellicles were treated for 24 h with (A) tobramycin (27 or 54 μM) or colistin (20 or 40 μM), concentrations equivalent to 25× and 50× the corresponding MIC, (B) compound KM-5-25 (40 and 80 μM) concentrations equivalent to 0.6× and 1.2× IC50, and (C) compound KM-5-66 (15, 30, and 50 μM), concentrations equivalent to 0.36×, 0.7×, and 1.2× IC50. p < 0.001 denoted by *** relative to untreated.
Enumeration of CFU/mL was also carried out after challenging pellicles with 4-aminoisoindoline-1,3-dione derivatives. Treating the pellicles with KM-5-25 at concentrations equivalent to 0.6× and 1.2× IC50 results in ∼38% and ∼25% survival relative to cells in the pretreated biofilm (Figure 9B), while treating with KM-5-66 at concentrations equivalent to 0.3×, 0.6×, or 1.2× IC50 results in approximately 57%, 21%, and 15% survival relative to cells in the pellicles prior to treatment (Figure 9C). These observations, which are in good agreement with the efficacy of the compounds evaluated by COMSTAT analysis of the CLSM images, corroborate the bactericidal activity of the compounds against P. aeruginosa biofilms, and provide additional evidence indicating that KM-5-66 used at 50 μM (1.2× IC50) exhibits nearly the same efficacy as colistin used at 20 μM (25× MIC). We also used the strategy of dispersing and counting viable cells to compare the relative efficacy of analogues 11, 16, KM-5-25, and KM-5-66. To this end, pellicles formed by P. aeruginosa PAO1 cells were treated (24 h) with each of the compounds at a concentration of 50 μM. The results (Figure S6) show that the compound activity against biofilm (KM-5-66 > KM-5-25 > 16 > 11) track with the Kd and IC50 values (Table 1). Taken together, these findings suggest that additional modifications made to the structures of KM-5-25 or KM-5-66 which improve the binding affinity for BfrB have the potential to produce molecules with similar or better efficacy toward biofilms than colistin.
The results from experiments aimed at determining the efficacy of the 4-aminoisoindoline-1,3-dione derivatives presented so far have been conducted with the reference strain P. aeruginosa PAO1. To investigate whether the compounds are also active against other strains of P. aeruginosa, we cultured pellicles of several clinical isolates from JMI Laboratories. Isolates PA_1081725 and PA_1076058 were chosen for additional testing because these strains exhibit relatively high MIC values for several antibiotics (Figure 10C) and form robust pellicles under the same culture conditions used to grow pellicle biofilms of P. aeruginosa PAO1. Challenging the pellicles formed by PA_1081725 and PA_1076058 with analogues KM-5-25 or KM-5-66 at concentrations equivalent to 1.2× IC50 elicits approximately 80% reduction of viable cells (Figure 10A,B), indicating that the activity of the 4-aminoisoindoline-1,3-dione analogues is not unique to biofilms formed by the P. aeruginosa PAO1 strain. To gain a broader understanding of the potential activity spectrum of the BfrB-Bfd inhibitors against P. aeruginosa strains, BLASTp63 was used to find homologues of BfrB and Bfd sequences in the >4400 P. aeruginosa genomes in the Pseudomonas Genome Database.64 The results reveal two important facts: (i) The bfrB and bfd genes are adjacent to one another in all the P. aeruginosa strains. (ii) There is an extremely high level of conservation among the bfd and bfrB sequences. These findings allow us to predict that the structures of the BfrB and Bfd proteins, as well as the structures of the BfrB-Bfd complexes, are highly conserved across P. aeruginosa strains, thus suggesting that the 4-aminoisoindoline-1,3-dione derivatives can be expected to be broadly active against P. aeruginosa.
Figure 10.
4-Aminoisoindoline-1,3-dione analogues are likely broadly active against P. aeruginosa strains. Two-day old pellicle biofilms formed by P. aeruginosa clinical isolates (PA_1081725 and PA_1076058) were challenged for 24 h with 4-aminoisoindoline-1,3-dione derivatives prior to dispersing the cells for enumeration of viable culturable cells (CFU/mL). The % survival is expressed as the ratio of CFU/mL(after treatment)/CFU/mL(pretreatment). Pellicles of (A) PA_1081725 or (B) PA_1076058 were treated with concentrations equivalent to 1.2× IC50 of KM-5-25 (80 μM), or KM-5-66 (50 μM). (C) Antibiotic susceptibility of clinical isolates PA_1081725 and PA_1076058. p < 0.001 denoted by *** relative to untreated.
4-Aminoisoindoline-1,3-dione Derivatives Inhibit Iron Mobilization from BfrB in P. aeruginosa Cells
The results presented above demonstrate that the 4-aminoisoindoline-1,3-dione derivatives can kill cells in mature biofilms. To demonstrate that the bactericidal activity is likely a result of the compounds engaging BfrB in the P. aeruginosa cytosol, inhibiting the BfrB-Bfd complex and blocking iron mobilization from the bacterioferritin in the P. aeruginosa cytosol, we carried out experiments aimed at visualizing the iron stored in BfrB. These experiments capitalize on a strategy we reported previously to demonstrate that the Δbfd mutant of P. aeruginosa irreversibly accumulates iron in BfrB23 and to show that analogue 16 inhibits iron mobilization from BfrB in planktonic cells.25 To visualize BfrB-stored iron in biofilm-embedded cells, 2-day old pellicles of P. aeruginosa PAO1 cells were treated for 24 h with analogue KM-5-25 (40 μM) or KM-5-66 (20 μM) at a concentration predicted to kill approximately 50% of the cells in the biofilm. The treated pellicles were dispersed in sterile PBS and the cell suspension was harvested by centrifugation after a small aliquot had been sampled to enumerate viable cells. To visualize iron stored in BfrB the harvested cells were lysed, the lysate solution supernatant was clarified by centrifugation and then loaded onto native PAGE gels for separation and subsequent staining with Ferene S, which reacts with iron to develop a blue color. Since the viable cell count dispersed from the pellicle biofilms treated with KM-5-25 or KM-5-66 was 42% and 37% of the cells in the untreated pellicle (Figure 11A), the clarified lysate supernatants from the untreated control were diluted approximately 2-fold prior to loading the native gels. Results obtained with a representative gel are shown in Figure 11B, where it can be observed that lanes loaded with lysate solutions from pellicles treated with analogues KM-5-25 or KM-5-66 exhibit significantly higher Ferene S stain intensity than the lane loaded with lysate solution from the untreated pellicle. To enable quantitative comparison we measured the relative intensities of the Ferene S-stained bands with the aid of ImageJ. Comparing the resultant peak areas (Figure 11C) shows that BfrB from the cells treated with KM-5-25 or KM-5-66 has ∼3-fold more iron relative to BfrB from cells in the untreated pellicles. These findings provide strong evidence indicating that compounds KM-5-25 and KM-5-66 bind BfrB in the P. aeruginosa cytosol, inhibit the formation of the BfrB-Bfd complex required to mobilize iron from BfrB, and lead to nearly irreversible iron accumulation in BfrB. Consistent with the nearly irreversible accumulation of iron in BfrB in cells treated with KM-5-25 or KM-5-66, quantification of the total intracellular iron and normalizing the values to viable cell counts demonstrates that P. aeruginosa cells dispersed from pellicles treated with KM-5-25 or KM-5-66 harbor ∼2.5-fold more intracellular iron than cells obtained from untreated biofilms (Figure 11D). Taken together, these observations support the idea that 4-aminoisoindoline-1,3-derivatives dysregulate iron homeostasis by inhibiting the BfrB-Bfd complex, causing the accumulation of unusable iron in the bacterial cell.
Figure 11.

4-Aminoisoindoline-1,3-dione analogues penetrate the P. aeruginosa cell, bind BfrB, and inhibit mobilization of BfrB-stored iron. (A) Treating pellicles with 0.6× IC50KM-5-25 (40 μM) or 0.5× IC50KM-5-66 (20 μM) for 24 h reduces the number of viable cells to <50%. (B) The iron stored in BfrB in the viable cells was visualized with the aid of native PAGE gels stained with Ferene S, which stains the iron in the interior cavity of BfrB. Recombinant BfrB (BfrBrec) was used as a standard for the electrophoretic mobility of BfrB. The lane corresponding to untreated control was loaded with 0.5× the volume of the lanes loaded with lysates from treated pellicles to account for the ∼2-fold larger number of viable cells in the untreated pellicles. Lanes loaded with treated pellicle lysates show greater accumulation of iron in BfrB relative to untreated cells. (C) Peak areas obtained from densitometry analysis (ImageJ) of the bands in the native PAGE gel of panel B indicate that there is ∼3-fold more iron stored in BfrB in the treated cells relative to the untreated control. (D) Analysis of total intracellular iron levels normalized to CFU/mL indicates ∼2.5-fold higher iron levels in the pellicle-embedded cells treated with the 4-aminoisoindoline-1,3-dione analogues relative to untreated control. Panel B shows results from a representative experiment from 3 biological replicates. Panels A, C, and D show the average of 3 biological replicates. p < 0.001 is denoted by *** relative to untreated.
4-Aminoisoindoline-1,3-dione Derivatives Enhance the Efficacy of Colistin and Tobramycin against Biofilm-Embedded Cells
As demonstrated above and in previous reports,65,66 mature biofilms formed by P. aeruginosa cells are susceptible to colistin and tolerant to tobramycin. Since these biofilms are also susceptible to the 4-aminoisoindoline-1,3-dione inhibitors of the BfrB-Bfd complex, we asked whether these compounds would enhance the efficacy of colistin and tobramycin. To answer this question, we cultured 2-day old pellicle biofilms of EYFP-expressing P. aeruginosa PAO1 and treated them for 24 h with colistin alone, compound alone (KM-5-25 or KM-5-66), and with a combination of colistin and compound. In the combination treatment experiments, the concentration of compound was kept constant (1.2× IC50), while colistin was used at two different concentrations, equivalent to 25× and 50× MIC (Figure 12A). Treatment with each of the compounds or with colistin alone caused a reduction of viable cells similar to that reported in Figure 9A. Challenging the pellicles with a combination of colistin and compound, however, causes a significant additional reduction in the number of viable cells (Figure 12A): the combination treatment with KM-5-66 results in ∼0.3% survival when colistin is present at 25× MIC, and ∼0.02% survival when colistin is used at 50× MIC, which correspond to approximately 1.7 log and 1.9 log reduction of viable cells relative to treatment with colistin alone. In comparison, the combination treatment with KM-5-25 results in ∼1% survival when colistin is used 25× MIC and ∼0.5% survival when colistin is present at 50× MIC, which correspond to nearly 1 log and 0.7 log reduction of viable cells, respectively, compared to colistin alone. Encouraged by these results we tested whether the 4-aminoisoindoline-1,3-dione derivatives can also enhance the bactericidal activity of tobramycin. The results are shown in Figure 12B: the combination treatment with KM-5-25 results in ∼0.5% survival when tobramycin is used at 25× MIC and ∼0.003% survival when tobramycin is present at 50× MIC, which correspond to approximately 2.5 log and 4.7 log reduction of viable cells when compared to treatment with tobramycin alone. The combination treatment with KM-5-66 results in 5% survival when tobramycin is present at 25× MIC and 1% survival when tobramycin is used at 50× MIC, which correspond to approximately 1.5 log and 2 log reduction in viable cells relative to treatment with tobramycin alone. It is interesting to note that compound KM-5-66 is more effective at enhancing the efficacy of colistin (Figure 12A), whereas compound KM-5-25 is more effective at enhancing the efficacy of tobramycin (Figure 12B). Additional studies are clearly required to understand the underlying reasons.
Figure 12.
4-Aminoisoindoline-1,3-dione derivatives enhance the efficacy of colistin and tobramycin against P. aeruginosa biofilms. Two-day old pellicles of EYFP-expressing P. aeruginosa PAO1 were treated for 24 h with (A) colistin alone 25× MIC (20 μM), or 50× MIC (40 μM), KM-5-25 (80 μM) or KM-5-66 (50 μM) alone, equivalent to 1.2× IC50, or a combination of colistin and KM-5-25 or KM-5-66, and (B) tobramycin alone 25× MIC (27 μM) or 50× MIC (40 μM), KM-5-25 or KM-5-66 alone at a concentration equivalent to 1.2× IC50, or a combination of tobramycin and KM-5-25 or KM-5-66. The % survival is expressed as the ratio CFU/mL(after treatment)/CFU/mL(pretreatment). p < 0.001 is denoted by *** in the combination treatment relative to treatment with antibiotic alone.
The observations above, which indicate that the iron limitation induced by inhibitors of the BfrB-Bfd complex can increase the efficacy of colistin and tobramycin against biofilms, are in good agreement with previous studies showing that the Fe chelator HBDE is an effective colistin adjunct against P. aeruginosa,67 and the iron chelators deferoxamine and deferasirox increase the efficacy of tobramycin against P. aeruginosa biofilms.68 When taken together, the observations made in the presence of HBDE, deferoxamine, deferasirox or 4-aminoisoindoline-1,3-dione derivatives, strengthen the idea that inducing intracellular iron limitation is probably a viable strategy to enhance the efficacy of colistin or tobramycin against biofilms. Since colistin is often used as one of the very few therapeutic options available to combat multidrug resistant Gram-negative organisms such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae,69,70 it is encouraging that small molecule inhibitors of the BfrB-Bfd complex are capable of increasing the effectiveness of colistin against P. aeruginosa. In this context, it is noteworthy that the bfr and bfd genes in A. baumannii and K. pneumoniae strains are contiguous, as is the case in the >4000 P. aeruginosa genomes currently available in the Pseudomonas Genome Data Base, because it suggests that the function of the Bfr-Bfd complex in P. aeruginosa is conserved in A. baumannii and K. pneumoniae. Moreover, amino acid sequence alignment with the aid of Clustal Omega71 reveals very high conservation in the amino acid sequences of Bfr and Bfd proteins from P. aeruginosa, A. baumannii and K. pneumoniae (Figure S7–S8). Importantly, the amino acids identified as hot spot residues in the BfrB-Bfd complex of P. aeruginosa(33) are highly conserved in the Bfr and Bfd sequences of A. baumannii and K. pneumoniae, suggesting that the 4-aminoisoindoline-1,3-dione derivatives designed to inhibit the BfrB-Bfd interaction in P. aeruginosa are likely to act similarly in A. baumannii and K. pneumoniae.
Concluding Remarks
Bacterial iron metabolism is a vulnerability that may be exploited in the development of new antimicrobial therapies. The field has centered on four major avenues: (i) The development of siderophore-antibiotic conjugates aims at capitalizing on siderophore uptake receptors to enhance antibiotic cell penetration.72,73 (ii) The sequestration of iron with the aid of chelators is also directed at depleting intracellular iron, and whereas iron chelation has been shown to lead to biofilm dispersion or to prevent biofilm maturation in vitro,35,37,48,68,74 a potential concern is the probability of secondary infections fueled by the chelated iron.75,76 (iii) The perturbation of heme uptake or heme degradation aims at denying pathogens from using heme, a rich iron source during infection.77,78 (iv) The perturbation of iron homeostasis with Ga3+ aims at inducing iron limitation by replacing Fe3+ in the active site of enzymes with Ga3+ which cannot support the rich redox chemistry typical of iron.79,80 Systemic Ga3+ treatment has been shown to improve lung function in patients with chronic P. aeruginosa infection.56 Our work, which aims at adding an entirely new strategy for exploiting the bacterial iron metabolism vulnerability, is directed at inhibiting the mobilization of iron from bacterioferritin. Blocking iron mobilization from BfrB by deletion of the bfd gene has been shown to elicit a breakdown in iron homeostasis due to the irreversible accumulation of unusable iron in BfrB, which causes cytosolic iron deficiency.23 Because of the relatively high iron requirement necessary to support the biofilm lifestyle, the intracellular iron deficiency induced in the Δbfd cells has been shown to result in poorly developed biofilms.26 Encouraged by these findings our laboratories have pursued drug discovery strategies to discover small molecule inhibitors of the BfrB-Bfd complex, which led us to a series of 4-aminoisoindoline-1,3-dione derivatives capable of penetrating the P. aeruginosa cell envelope, inhibiting the BfrB-Bfd complex and eliciting a cell growth defect in planktonic cells.25 Additional structural manipulation of these 4-aminoisoindoline-1,3-dione derivatives led us to analogues with increased affinity for BfrB and enhanced bacteriostatic activity against planktonic P. aeruginosa cells (Table 1). Testing these new derivatives against mature biofilms revealed that these small molecules inhibit iron mobilization from BfrB in biofilm-embedded cells (Figure 11) and kill P. aeruginosa cells in mature biofilms cultured in flow cells (Figure 6) or in pellicles cultured at the air–liquid interface (Figure 8). These findings demonstrate the potential of small molecules developed to inhibit the BfrB-Bfd complex in P. aeruginosa as important tools to expose a rare weakness of biofilms. It is also encouraging that the small molecule inhibitors of the BfrB-Bfd complex increase the effectiveness of tobramycin and colistin because the latter is often used as one of the very few therapeutic options available to combat multidrug resistant Gram-negative organisms.69,70 The high level of conservation in the Bfr and Bfd sequences from P. aeruginosa, A. baumannii, and K. pneumoniae (Figure S7–S8) suggests that the compounds developed to block the BfrB-Bfd complex in P. aeruginosa may exert similar activity in A. baumannii and K. pneumoniae cells.
Methods
Chemicals, Bacterial Strains, and Growth Media
Chemicals were purchased from Fisher Scientific unless otherwise stated. P. aeruginosa PAO1 was obtained from the University of Washington Genome center.81 The PAO1 strain expressing enhanced yellow fluorescent protein (EYFP) was prepared previously.26 Clinical isolates of P. aeruginosa were purchased from JMI Laboratories (North Liberty, IA, USA). IC50 determinations were carried out in defined media (50 mM KH2PO4 (Sigma-Aldrich) 7.5 mM (NH4)2SO4 (Sigma, Life Sciences), 0.1% (w/v) glucose (Acros Organics, 99+%), 0.5 mM MgSO4·7H2O (Sigma-Aldrich, 99+%), 5% v/v nonessential amino acids (Gibco), 2% v/v essential amino acids (Gibco), 4 μM (NH4)2Fe(SO4)2, and 0.025% (w/v) hypromellose (HPMC, Sigma-Aldrich), pH 7.0. The media was filter-sterilized by passing through a 0.2 μm cellulose acetate membrane syringe filter (VWR). Starter cultures of P. aeruginosa PAO1 in 5 mL LB media were grown for 13 h in 50 mL conical tubes at 37 °C and 220 rpm. For biofilm experiments, a EYFP-expressing P. aeruginosa strain was routinely grown in Pseudomonas Isolation (PI) media (20 g L–1 peptone, 0.3 g L–1 MgCl2·6H2O, 10 g L–1 K2SO4, 25 mg L–1 irgasan, and 20 mL L–1 glycerol, pH 7.0). Starter cultures were grown from a single colony at 37 °C and shaking at 220 rpm for 14 h in 5 mL PI media supplemented with 10 μM Fe. Pellicle biofilms were cultured for 48 h at 30 °C in PI media supplemented with 20 μM Fe. Surface-attached biofilms were cultured in AB minimal media82 supplemented with trace metals [0.15 μM (NH4)2MoO4, 3 μM CuSO4, 2 μM Co(NO3)2, 9.4 μM Na2B4O7, and 7.6 μM ZnSO4], 3 mM glucose and 15 μM Fe. Iron supplementation was carried out by addition of a small volume of filter-sterilized 10 mM (NH4)2Fe(SO4)2 (pH ∼ 2.0) solution. The antibiotics ciprofloxacin, colistin and tobramycin were used at concentrations equivalent to 25× and 50× the reported MCI:83 ciprofloxacin MIC = 0.25 μg/mL = 0.75 μM; tobramycin MIC = 0.5 μg/mL = 1.07 μM; colistin MIC = 1 μg/mL = 0.79 μM. Compound stock solutions (100 mM or 10 mM) in DMSO (Sigma-Aldrich) were prepared weekly and stored at 4 °C. Solutions used to treat biofilms or planktonic cells include 0.025% (w/v) HPMC, and 1.5% or 2% DMSO (Sigma-Aldrich) to prevent aggregation of the analogues in aqueous solution.
Synthesis and Preparation of 4-Aminoisoindoline-1,3-dione Derivatives
Experimental details of the synthetic protocols developed to prepare the 4-aminoisoindoline-1,3-dione derivatives to be tested as inhibitors of the BfrB-Bfd complex, as well as 1H, 13C NMR and MS data, are presented in the Supporting Information.
Measurement of Dissociation Constant (Kd)
Dissociation constants for the interaction between BfrB and 3-aminoisoindoline-1,3-dione derivatives (Table 1, Figure S1) were measured in vitro with a fluorescence polarization method based on the intrinsic fluorescence of the isoindoline-1,3-dione moiety, as described previously.25
Measurement of Half Maximal Inhibitory Concentration (IC50)
IC50 values (Table 1, Figure S1) were determined as reported previously25 with small modifications. In brief: precultures of P. aeruginosa PAO1 (5 mL) were grown in LB media for 13 h at 37 °C and 220 rpm in 50 mL conical tubes (VWR International, PA). The cells were centrifuged for 5 min at 4000 rpm and 4 °C, washed two times and then diluted in buffer (100 mM KH2PO4 and 15 mM (NH4)2SO4) to an optical density at 600 nm (OD600) of 0.1. A small volume of compound stock solution (10 mM) was transferred to a microcentrifuge tube, initially diluted with DMSO to 20 μL, and then diluted to 1 mL with preculture cell suspension in defined media with OD600 = 0.0001, so the final DMSO concentration is 2%. The resultant cell suspension (200 μL) was transferred to a clear-bottom polystyrene 96-well plate (VWR) covered with a lid and incubated at 35 °C and 205 cpm for 24 h in a Synergy H1 microplate reader (Biotek Instruments Inc., Vermont). The cell cultures were serially diluted and then plated on PI Agar (PIA; BD biosciences) plates for enumeration of viable cells (CFU/mL). The % growth was calculated from the ratio CFU/mL(treated)/CFU/mL(untreated control). To calculate the IC50 values, the % growth was plotted as a function of compound concentration, expressed as log[compound] (μM), and fitted to the 4-parameter logistic model describing the sigmoid-shaped response pattern (eq 1),84 where b is the slope factor, max is the upper asymptote (plateau), and min is the lower asymptote. Values are the average and standard deviation from three independent experiments.
| 1 |
Analysis of Secreted Pyoverdine in Planktonic Cultures
These experiments were conducted in 96 well plates as described above for the determination of IC50, except that the cells were cultured in M63 media (2 g/L (NH4)2SO4, 13.6 g/L KH2PO4 (Sigma-Aldrich), 2 g/L glucose, 4 g/L citric acid, 5 g/L technical grade casamino acids (BD Scientific), 0.24 g/L MgSO4 (Alfa Aesar), and 0.05% (w/v) HPMC, pH 7.0 adjusted with KOH). Cultures of P. aeruginosa PAO1 treated with KM-5-25 (70 μM) or KM-5-66 (50 μM) were grown for 27 h prior to diluting the contents of each well in PBS (pH 7.4) and plating the cells on PIA plates for enumeration of CFU/mL. The 500-fold diluted solution was clarified by centrifugation and the pyoverdine in the cell-free supernatant was analyzed by acquiring fluorescence emission spectra (430–550 nm) with excitation at 400 nm (10 nm slit width) and emission at λmax = 460 nm (10 nm slit width) using a PerkinElmer LS50B spectrophotometer.
Flow Cell Biofilm Assays
Surface-attached biofilms of P. aeruginosa PAO1 cells expressing EYFP were grown on flow cells with an 800 μm channel depth (μ-slide I0.8 Luer, Ibidi) using an automated perfusion system (Ibidi, Munich, Germany), as described previously.26 Briefly, the flow cell was inoculated with 200 μL of an overnight culture diluted to OD600 = 0.5, followed by 1 h incubation at 30 °C to allow bacterial cell attachment. The μ-slide was connected to the Ibidi Pump System and the biofilms were cultured for 3 days at 30 °C while flowing AB minimal media containing 15 μM Fe. The experimental shear stress was 0.14 dyn/cm2 (shear rate = 14 s–1, pressure = 7.1 mbar, flow rate = 0.4 mL/min) and the switch time was set to 540 s. The biofilms were treated for 24 h by flowing AB minimal media supplemented with 15 μM Fe, 0.025% HPMC, 1.5% DMSO and commercial antibiotics or 4-aminoisoindoline-1,3-dione derivatives in the concentrations indicated in the corresponding figure captions. During biofilm growth and challenge with antibacterial, the culture medium in the reservoirs was removed every 12 h and replaced with fresh prewarmed medium. Prior to imaging with the aid of CLSM the biofilms were stained with 4 mL of 2.5 nM Sytox Red (Invitrogen Life Technologies), a cell impermeable fluorescent nucleic acid dye that stains dead cells and extracellular DNA,85 for 20 min (switch time = 200 s) and then washed with AB media for 20 min to remove excess fluorescent dye. The biofilms were imaged with the aid of a Leica TCS SP8 confocal microscope (Leica Microsystems, Germany) using a HC PL apo CS2 63 × /1.4 oil objective. For detecting the EYFP fluorescence the laser line was set at 506 nm and the emission range to 520–610 nm. Sytox Red fluorescence was detected with excitation at 631 nm and emission range 637–779 nm. Image stacks were acquired with a z-step size of 0.3 μm at randomly chosen positions. The Leica Application Suite X (LAS-X) software was used for image stack processing.55 Quantitative analysis of biofilm biomass was performed using the COMSTAT computer program55 and the Otsu method of automatic thresholding.86
Pellicle Biofilm Assays
Pellicle biofilms of EYFP-expressing P. aeruginosa PAO1 or clinical isolates were grown in PI media supplemented with 20 μM Fe. Starter cultures were diluted to OD600 = 0.001 in 4 mL media, placed in 35 × 10 mm Petri dishes and incubated statically at 30 °C for 48 h. The pellicles were transferred onto circular (1.5 cm diameter) glass coverslips by gently allowing the surface of a coverslip to contact a pellicle. The pellicle-adhered coverslip was washed in PBS and then deposited on top of 1.5 mL AB challenge media contained in a well of a 12-well microplate, with the pellicle exposed to the media. Challenge media consists of AB minimal media supplemented with 15 μM Fe, 0.025% HPMC, 1.5% DMSO, and commercial antibiotic or 4-aminoisoindoline-1,3-dione derivative, used in the concentrations specified in the figure captions. The 4-aminoisoindoline-1,3-dione derivatives were prepared as 10 mM stock solutions in DMSO and then diluted in culture media to the appropriate concentrations. The coverslip-adhered pellicles were exposed to challenge media at 30 °C for 24 h, changing the challenge media every 12 h by transferring the pellicle-adhered coverslip to a new plate containing prewarmed challenge media.
Prior to imaging with the aid of CLSM, pellicles formed by EYFP-expressing PAO1 cells were washed with PBS and then stained by placing the coverslip-adhered pellicles in 1 mL of PBS containing 2.5 nM Sytox Red (20 min). Excess fluorescent dye was washed with PBS, the coverslip was mounted on a glass slide using 5 μL SlowFade (Invitrogen Life Technologies) and the edges sealed with fingernail polish. CLSM image stacks (z-step size of 0.3 μm) were acquired with the aid of a Leica TCS SP8 microscope, as described above. Quantitative analysis was performed by determination of pellicle biomass using COMSTAT55 and the Otsu method of automatic thresholding.86
Determination of Viable Cells in Pellicle Biofilms
Pellicle biofilms were grown as described above. Planktonic and loosely attached cells were washed (3 times) by immersing the coverslip-adhered pellicles (biofilm facing up) into a well of a 12-well plate containing 3 mL PBS, and incubating (5 min) with gentle rocking. To remove the pellicle from the coverslip, break the extracellular matrix and release cells from the biofilm, the coverslip-adhered pellicle was placed in a 50 mL conical tube containing a 2 mL suspension of zirconia beads (0.1 mm diameter, BioSpec Products), 10 mL PBS, 0.2 μg/mL alginate lyase and 0.2 μg/mL DNase. The resultant mixture was incubated at room temperature for 3 min, followed by vigorous vortexing for 4 min. After sedimentation of the zirconia beads, a 100 μL aliquot was used for serial dilution and plating on PIA plates for subsequent enumeration of viable cells (CFU/mL).
Imaging of Iron Stored in BfrB and Analysis of Total Intracellular Iron in Biofilm-Embedded Cells
Pellicle biofilms were grown for 48 h as described above. The pellicles were transferred onto square (2 × 2 cm) glass coverslips by gently contacting the pellicle with the coverslip. The pellicle-adhered coverslip was washed in PBS and then placed in a 50 mL conical tube containing 2 mL suspension of zirconia beads, 15 mL PBS, 0.2 μg/mL alginate lyase and 0.2 μg/mL DNase; the resultant mixture was incubated (3 min) at room temperature, and then vortexed vigorously for 4 min. After the zirconia beads had sedimented, a 100 μL aliquot was sampled from the cell suspension for plating and enumeration of viable cells and a 14 mL sample was used to harvest the cells by centrifugation (20 min, 400 rpm). The cell pellet was resuspended in 1 mL PBS, transferred to a 1.5 mL microcentrifuge tube, centrifuged for 10 min at 12 500 rpm at 4 °C and the cell pellet frozen at −80 °C. The frozen cells were subjected to three freeze–thaw cycles and then lysed by addition of 200 μL of lysis buffer (50 mM Tris-HCl buffer (pH 8.0) containing 10% (v/v) glycerol, 20 mg/mL lysozyme, 0.2 mg/mL DNase, 0.1 M NaCl, 1 mM MgSO4 and 1% (v/v) Triton-X100) and incubated at ambient temperature (30 min) and at 37 °C (30 min). Imaging of iron in BfrB was carried out as previously reported:23 lysate suspensions were clarified by centrifugation (10 min at 12 500 rpm), mixed with 10 μL loading dye (5.9 mL deionized water, 0.5 mL glycerol, 0.4 mL β-mercaptoethanol, 0.4 mL 1% (w/v) bromophenol blue, and 0.5 mL 1 M Tris–HCl (pH 6.8), and loaded onto 1.5 mm-thick native PAGE gels (4% stacking gel, 8% resolving gel). Electrophoresis was carried out at 60 V and 4 °C for 9 h, and the gels were stained in the dark by immersion (10 min) in a solution containing 0.049 g Ferene S, 250 μL thioglycolic acid, 2.4 mL acetic acid and 100 mL deionized water. Levels of total intracellular iron were determined as reported previously:23,87 The cell pellets were treated with 500 μL of freshly prepared digestion reagent (0.6 N HCl, 2.25% (w/v) KMnO4 in water), thoroughly mixed by vortexing, and then incubated at 65 °C for 3.5 h. The resultant solutions were cooled to ambient temperature, treated with 100 μL of iron detection reagent (6.5 mM Ferene S, 15.4 mM neocuproine, 2 M ascorbic acid, and 5 M ammonium acetate), incubated for 30 min at ambient temperature and centrifuged for 5 min at 12500 rpm. The iron concentration was measured from the absorbance of the Fe2+–Ferene S complex (ε593 = 34.5 mM–1 cm–1),88 normalized by the viable cell counts and reported as Fe atoms per cell.
Statistical Analysis
Statistical significance between the means and standard deviation of values obtained in experiments comparing results from untreated vs treated with antibiotic or analogue conditions was determined using one-way ANOVA followed by Tukey’s multiple post hoc test, with the aid of SigmaPlot (Systat Software, Inc. CA).
X-ray Crystallography
Crystallization screening of wild type BfrB was conducted in Compact 300 (Rigaku Reagents) sitting drop vapor diffusion plates at 20 °C using equal volumes of protein and crystallization solution equilibrated against 75 μL of the latter. Crystals displaying a prismatic morphology were obtained in 1–2 days from the Cryo 1–2 HT screen (Rigaku Reagents) condition H6 (30% (v/v) PEG 200, 100 mM Na acetate pH 4.5, 100 mM NaCl). To prepare the ligand complexes, stock solutions (100 mM) of ligand in DMSO were mixed with the crystallization solution to obtain a 10 mM ligand soaking solution. Crystals were transferred to the soaking solution and incubated for 3 h before harvesting directly from the drop and storing in liquid nitrogen. Due to the low solubility of KM-5-66, a multicomponent mixture89 composed of 25% (v/v) dioxane, 25% (v/v) glycerol, 25% (v/v) diethylene glycol and 25% (v/v) ethylene glycol (SM3) was prepared to improve the ligand solubility. The ligand stock solution (100 mM in DMSO) was mixed in a 1:1 (v/v) ratio with SM3; 4 μL of the resultant solution was mixed with 5uL of 2× crystallant solution and 1 μL of 10× buffer (1 M HEPES pH 7.5) to give a 20 mM ligand solution. Crystals were transferred to the soaking solution and incubated for 3 h before harvesting directly from the drop and storing in liquid nitrogen. Data were collected at the Advanced Photon Source beamline IMCA-CAT beamline 17-ID. Intensities were integrated using XDS90 via Autoproc91 and the Laue class analysis and data scaling were performed with Aimless.92 Structure solution was conducted by molecular replacement using the previously determined structure of wt holo-BfrB (PDB: 5D8O)33 as the search model with Phaser.93 Structure refinement and manual model building were conducted with Phenix94 and Coot,95 respectively. Structure validation was conducted with Molprobity.96
Acknowledgments
This study was supported by a grant from the National Science Foundation (MCB1837877) and a grant from the National Institutes of Health (AI125529) to MR. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-06CH11357.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00669.
Quantification of Kd and IC50 for KM-5-25 and KM-5-66, superposition of structures of 4-aminoisoindoline-1,3-dione derivatives with a −(CH2)3– linker bound to BfrB, structure of KM-5-50 bound to BfrB, pyoverdine release from planktonic cells treated with KM-5-25 and KM-5-66, comparison of the efficacy of analogues 11, 16, KM-5-25, and KM-5-66 against pellicle biofilms, multiple sequence alignments of representative Bfr and Bfd proteins, X-ray diffraction data refining statistics table, synthetic procedures for the preparation of 4-aminoisoindoline-1,3-dione derivatives, 1H, 13C NMR spectra and mass spectra of synthesized compounds (PDF)
Accession Codes
Coordinate and structures factors for the following inhibitor complexes with BfrB were deposited to the Worldwide Protein Data Bank with the accession codes 7K5E (JAG-5-7), 7K5G (KM-5-28), 7K5F (KM-5-50), and 7K5H (KM-5-66).
Author Contributions
AS: Performed the pellicle and surface biofilm studies, tested the susceptibility of biofilms to antibiotics and 4-aminoisoindoline-1,3-dione derivatives, and measured the total and stored iron in bacterioferritin. HY: Measured the Kd values, the susceptibility of planktonic cells to 4-aminoisoindoline-1,3-dione derivatives, and the pyoverdine release. ANDPH: Screened 4-aminoisoindoline-1,3-dione derivatives and contributed to evaluate the susceptibility of planktonic cells. KM and JKA-G: Synthesized and purified 4-aminoisoindoline-1,3-dione derivatives. RAB: Directed the synthesis of 4-aminoisoindoline-1,3-dione-derivatives. KPB: Collected X-ray diffraction data. SL: Solved X-ray crystal structures. MR: Directed the biology and analytical biochemistry studies and wrote the manuscript. AS, HY, RAB, and SL edited the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Tacconelli E.; Carrara E.; Savoldi A.; Harbarth S.; Mendelson M.; Monnet D. L.; Pulcini C.; Kahlmeter G.; Kluytmans J.; Carmeli Y.; Ouellette M.; Outterson K.; Patel J.; Cavaleri M.; Cox E. M.; Houchens C. R.; Grayson M. L.; Hansen P.; Singh N.; Theuretzbacher U.; Magrini N. (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- Davies D. (2003) Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discovery 2, 114–122. 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- Lawrence J. R.; Korber D. R.; Hoyle B. D.; Costerton J. W.; Caldwell D. E. (1991) Optical sectioning of microbial biofilms. J. Bacteriol. 173, 6558–6567. 10.1128/JB.173.20.6558-6567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. (2001) Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3–9. 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- Lam J.; Chan R.; Lam K.; Costerton J. W. (1980) Production of Mucoid Microcolonies by Pseudomonas aeruginosa Within Infected Lungs in Cystic Fibrosis. Infect. Immun. 28, 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W.; Stewart P. S.; Greenberg E. P. (1999) Bacterial Biofilms: A Common Cause of Persistent Infection. Science 284, 1318–1322. 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Parsek M. R.; Singh P. K. (2003) Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57, 677–701. 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Konstan M. W.; Morgan W. J.; Butler S. M.; Pasta D. J.; Craib M. L.; Silva S. J.; Stokes D. C.; Wohl M. E.; Wagener J. S.; Regelmann W. E.; Johnson C. A. (2007) MBCHB for the Scientific Advisory Group and the Investigators and Coordinators of the Epidemiologic Study of Cystic Fibrosis Risk factors for rate of decline in forced expiratory volume in one second in children and adolescents with cystic fibrosis. J. Pediatr. 151, 134–139. 10.1016/j.jpeds.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Crull M. R.; Ramos K. J.; Caldwell E.; Mayer-Hamblett N.; Aitken M. L.; Goss C. H. (2016) Change in Pseudomonas aeruginosa prevalence in cystic fibrosis adults over time. BMC Pulm. Med. 16, 176. 10.1186/s12890-016-0333-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U.; Balsalobre C. (2012) Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272, 541–561. 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- Burrows L. L. (2018) The Therapeutic Pipeline for Pseudomonas aeruginosa Infections. ACS Infect. Dis. 4, 1041–1047. 10.1021/acsinfecdis.8b00112. [DOI] [PubMed] [Google Scholar]
- Boucher H. W.; Talbot G. H.; Bradley J. S.; Edwards J. E.; Gilbert D.; Rice L. B.; Scheld M.; Spellberg B.; Bartlett J. (2009) Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–11. 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- James G. A.; Swogger E.; Wolcott R.; Pulcini E.; Secor P.; Sestrich J.; Costerton J. W.; Stewart P. S. (2008) Biofilms in chronic wounds. Wound Repair Regen 16, 37–44. 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- Kadam S.; Shai S.; Shahane A.; Kaushik K. S. (2019) Recent Advances in Non-Conventional Antimicrobial Approaches for Chronic Wound Biofilms: Have We Found the ‘Chink in the Armor’?. Biomedicines 7, 35. 10.3390/biomedicines7020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe A.; Jensen P. O.; Bjarnsholt T.; Coenye T. (2019) Antimicrobial Tolerance and Metabolic Adaptations in Microbial Biofilms. Trends Microbiol. 27, 850–863. 10.1016/j.tim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Ceri H.; Olson M. E.; Stremick C.; Read R. R.; Morck D.; Buret A. (1999) The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37, 1771–1776. 10.1128/JCM.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P. S.; Costerton J. W. (2001) Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Anwar H.; Costerton J. W. (1990) Enhanced activity of combination of tobramycin and piperacillin for eradication of sessile biofilm cells of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34, 1666–1671. 10.1128/AAC.34.9.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellat M. F.; Raguz L.; Riedl R. (2016) Targeting Antibiotic Resistance. Angew. Chem., Int. Ed. 55, 6600–6626. 10.1002/anie.201506818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakemeyer M.; Zhao W.; Mandl F. A.; Hammann P.; Sieber S. A. (2018) Thinking Outside the Box-Novel Antibacterials To Tackle the Resistance Crisis. Angew. Chem., Int. Ed. 57, 14440–14475. 10.1002/anie.201804971. [DOI] [PubMed] [Google Scholar]
- Verderosa A. D.; Totsika M.; Fairfull-Smith K. E. (2019) Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 7, 824. 10.3389/fchem.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen J. J.; Rogers H. J.; Spalding P. B.; Ward C. G. (2005) Iron and Infection: The Heart of the Matter. FEMS Immunol. Med. Microbiol. 43, 325–330. 10.1016/j.femsim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Eshelman K.; Yao H.; Punchi Hewage A. N. D.; Deay J. J.; Chandler J. R.; Rivera M. (2017) Inhibiting the BfrB:Bfd Interaction in Pseudomonas aeruginosa Causes Irreversible Iron Accumulation in Bacterioferritin and Iron Deficiency in the Bacterial Cell. Metallomics 9, 646–659. 10.1039/C7MT00042A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K.; Imlay J. A. (1996) Superoxide Accelerates DNA-Damage by Elevating Free-Iron Levels. Proc. Natl. Acad. Sci. U. S. A. 93, 13635–13649. 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punchi Hewage A. N. D.; Yao H.; Nammalwar B.; Gnanasekaran K. K.; Lovell S.; Bunce R. A.; Eshelman K.; Phaniraj S. M.; Lee M. M.; Peterson B. R.; Battaile K. P.; Reitz A. B.; Rivera M. (2019) Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity. J. Am. Chem. Soc. 141, 8171–8184. 10.1021/jacs.9b00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldano A.; Yao H.; Chandler J. R.; Rivera M. (2020) Inhibiting Iron Mobilization from Bacterioferritin in Pseudomonas aeruginosa Impairs Biofilm Formation Irrespective of Environmental Iron Availability. ACS Infect. Dis. 6, 447–458. 10.1021/acsinfecdis.9b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.; Jepkorir G.; Lovell S.; Nama P. V.; Weeratunga S. K.; Battaille K. P.; Rivera M. (2011) Two Disctinct Ferritin-Like Molecules in P. aeruginosa: The Product of the bfrA Gene is a Bacterial Ferritin (FtnA) not a bacterioferritin (Bfr). Biochemistry 50, 5236–5248. 10.1021/bi2004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M. (2017) Bacterioferritin: Structure, Dynamics and Protein-Protein Interactions at Play in Iron Storage and Mobilization. Acc. Chem. Res. 50, 331–340. 10.1021/acs.accounts.6b00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeratunga S.; Lovell S.; Yao H.; Battaile K. P.; Fischer C. J.; Gee C. E.; Rivera M. (2010) Structural Studies of Bacterioferritin B (BfrB) from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center. Biochemistry 49, 1160–1175. 10.1021/bi9015204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.; Wang Y.; Lovell S.; Kumar R.; Ruvinsky A. M.; Battaile K. P.; Vakser I. A.; Rivera M. (2012) The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin. J. Am. Chem. Soc. 134, 13470–13481. 10.1021/ja305180n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeratunga S.; Gee C. E.; Lovell S.; Zeng Y.; Woodin C. L.; Rivera M. (2009) Binding of Pseudomonas aeruginosa Apobacterioferritin-Associated Ferredoxin to Bacterioferritin B Promotes Heme Mediation of Electron Delivery and Mobilization of Core Mineral Iron. Biochemistry 48, 7420–7431. 10.1021/bi900561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijerathne H.; Yao H.; Wang Y.; Lovell S.; Battaile K. P.; Rivera M. (2018) Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site. Biochemistry 57, 5533–5543. 10.1021/acs.biochem.8b00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Yao H.; Cheng Y.; Lovell S.; Battaile K. P.; Middaugh C. R.; Rivera M. (2015) Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Potein-Protein Interactions in Solution and Determination of Binding Energy Hot Spots. Biochemistry 54, 6162–6175. 10.1021/acs.biochem.5b00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E.; Vasil M. L.; Greenberg E. P. (2005) Iron and Pseuodomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 102, 11076–11081. 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.; Kirienko N. V. (2018) Interdependence between iron acquisition and biofilm formation in Pseudomonas aeruginosa. J. Microbiol. 56, 449–457. 10.1007/s12275-018-8114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. K.; Parsek M. R.; Greenberg E. P.; Welsh M. J. (2002) A Component of Innate Immunity Prevents Bacterial Biofilm Development. Nature 417, 552–555. 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Post S. J.; Shapiro J. A.; Wuest W. M. (2019) Connecting iron acquisition and biofilm formation in the ESKAPE pathogens as a strategy for combatting antibiotic resistance. MedChemComm 10, 505–512. 10.1039/C9MD00032A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekaran K. K.; Rivera M.; Bunce R. A. (2018) 4,7-Diaminoisoindoline-1,3-dione. Org. Prep. Proced. Int. 50, 372–374. 10.1080/00304948.2018.1462072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk I. J.; Guillon L. (2013) Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ. Microbiol. 15, 1661–1673. 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- Harmsen M.; Yang L.; Pamp S. J.; Tolker-Nielsen T. (2010) An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 59, 253–268. 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- Brauner A.; Fridman O.; Gefen O.; Balaban N. Q. (2016) Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330. 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- Anwar H.; Strap J. L.; Chen K.; Costerton J. W. (1992) Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob. Agents Chemother. 36, 1208–1214. 10.1128/AAC.36.6.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl J. N.; Zahller J.; Roe F.; Stewart P. S. (2003) Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 47, 1251–1256. 10.1128/AAC.47.4.1251-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M. C. 3rd; Roe F.; Bugnicourt A.; Franklin M. J.; Stewart P. S. (2003) Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47, 317–323. 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K. D.; Stewart P. S.; Xia F.; Huang C. T.; McFeters G. A. (1998) Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64, 4035–4039. 10.1128/AEM.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E.; Roe F.; Bugnicourt A.; Franklin M. J.; Heydorn A.; Molin S.; Pitts B.; Stewart P. S. (2004) Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70, 6188–6196. 10.1128/AEM.70.10.6188-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M.; Wu H.; Andersen J. B.; Riedel K.; Rasmussen T. B.; Bagge N.; Kumar N.; Schembri M. A.; Song Z.; Kristoffersen P.; Manefield M.; Costerton J. W.; Molin S.; Eberl L.; Steinberg P.; Kjelleberg S.; Hoiby N.; Givskov M. (2003) Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22, 3803–3815. 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E.; Brady K. M.; Greenberg E. P. (2006) Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 72, 2064–2069. 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp S. J.; Gjermansen M.; Johansen H. K.; Tolker-Nielsen T. (2008) Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68, 223–240. 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- Vrany J. D.; Stewart P. S.; Suci P. A. (1997) Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa bofilms displaying rapid-transport characteristics. Antimicrob. Agents Chemother. 41, 1352–1358. 10.1128/AAC.41.6.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nation R. L.; Li J. (2009) Colistin in the 21st century. Curr. Opin. Infect. Dis. 22, 535–543. 10.1097/QCO.0b013e328332e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Nation R. L.; Turnidge J. D.; Milne R. W.; Coulthard K.; Rayner C. R.; Paterson D. L. (2006) Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- Ezadi F.; Ardebili A.; Mirnejad R. (2019) Antimicrobial Susceptibility Testing for Polymyxins: Challenges, Issues, and Recommendations. J. Clin. Microbiol. 57, 57. 10.1128/JCM.01390-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby N.; Bjarnsholt T.; Givskov M.; Molin S.; Ciofu O. (2010) Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Heydorn A.; Nielsen A. T.; Hentzer M.; Sternberg C.; Givskov M.; Ersboll B. K.; Molin S. (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146 (10), 2395–2407. 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Goss C. H.; Kaneko Y.; Khuu L.; Anderson G. D.; Ravishankar S.; Aitken M. L.; Lechtzin N.; Zhou G.; Czyz D. M.; McLean K.; Olakanmi O.; Shuman H. A.; Teresi M.; Wilhelm E.; Caldwell E.; Salipante S. J.; Hornick D. B.; Siehnel R. J.; Becker L.; Britigan B. E.; Singh P. K. (2018) Gallium disrupts bacterial iron metabolism and has therapeutic effects in mice and humans with lung infections. Sci. Transl. Med. 10, 10. 10.1126/scitranslmed.aat7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K.; Arai H.; Ishii M.; Igarashi Y. (2011) Trade-off between oxygen and iron acquisition in bacterial cells at the air-liquid interface. FEMS Microbiol. Ecol. 77, 83–94. 10.1111/j.1574-6941.2011.01087.x. [DOI] [PubMed] [Google Scholar]
- Friedman L.; Kolter R. (2004) Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Oliver J. D. (2010) Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 34, 415–425. 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- Ayrapetyan M.; Williams T. C.; Oliver J. D. (2015) Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 23, 7–13. 10.1016/j.tim.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Marques C. N.; Salisbury V. C.; Greenman J.; Bowker K. E.; Nelson S. M. (2005) Discrepancy between viable counts and light output as viability measurements, following ciprofloxacin challenge of self-bioluminescent Pseudomonas aeruginosa biofilms. J. Antimicrob. Chemother. 56, 665–671. 10.1093/jac/dki285. [DOI] [PubMed] [Google Scholar]
- Marques C. N. H.; Nelson S. M. (2019) Pharmacodynamics of ciprofloxacin against Pseudomonas aeruginosa planktonic and biofilm-derived cells. Lett. Appl. Microbiol. 68, 350–359. 10.1111/lam.13126. [DOI] [PubMed] [Google Scholar]
- Johnson M.; Zaretskaya I.; Raytselis Y.; Merezhuk Y.; McGinnis S.; Madden T. L. (2008) NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L.; Griffiths E. J.; Lo R.; Dhillon B. K.; Shay J. A.; Brinkman F. S. (2016) Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653. 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagensen J. A.; Klausen M.; Ernst R. K.; Miller S. I.; Folkesson A.; Tolker-Nielsen T.; Molin S. (2007) Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189, 28–37. 10.1128/JB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpen M.; Appeldorff C. F.; Brandt S.; Mousavi N.; Kragh K. N.; Aydogan S.; Uppal H. A.; Bjarnsholt T.; Ciofu O.; Hoiby N.; Jensen P. O. (2016) Increased bactericidal activity of colistin on Pseudomonas aeruginosa biofilms in anaerobic conditions. Pathog. Dis. 74, ftv086 10.1093/femspd/ftv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettrick K.; Hassan K.; Lamont I.; Reid D. (2020) The Iron-chelator, N,N’-bis (2-hydroxybenzyl) Ethylenediamine-N,N’-Diacetic acid is an Effective Colistin Adjunct against Clinical Strains of Biofilm-Dwelling Pseudomonas aeruginosa. Antibiotics (Basel, Switz.) 9, 9. 10.3390/antibiotics9040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Marquis S.; O’Toole G. A.; Stanton B. A. (2009) Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am. J. Respir. Cell Mol. Biol. 41, 305–313. 10.1165/rcmb.2008-0299OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lora-Tamayo J., Murillo O., and Ariza J. (2019) Clinical Use of Colisting in Biofilm-Associated Infections. In Polymyxin Antibiotics: From Laboratory Bench to Bedside. Advances in Experimental Medicine and Biology (Li J., Nation R. L., and Kaye K., Eds.) Vol. 1145, pp 181–195, Springer, Cham. [DOI] [PubMed] [Google Scholar]
- Li J. (2019) Reviving Polymyxins: Achievements, Lessons and the Road Ahead. In Polymyxin Antibiotics: From Laboratory Bench to Bedside. Advances in Experimental Medicine and Biology (Li J., Nation R., and Kaye K., Eds.) Vol. 1145, pp 1–8, Springer, Cham. [DOI] [PubMed] [Google Scholar]
- Madeira F.; Park Y. M.; Lee J.; Buso N.; Gur T.; Madhusoodanan N.; Basutkar P.; Tivey A. R. N.; Potter S. C.; Finn R. D.; Lopez R. (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C.; Miller P. A.; Miller M. J. (2012) Iron transport-mediated drug delivery: practical syntheses and in vitro antibacterial studies of tris-catecholate siderophore-aminopenicillin conjugates reveals selectively potent antipseudomonal activity. J. Am. Chem. Soc. 134, 9898–9901. 10.1021/ja303446w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.; Miller P. A.; Vakulenko S. B.; Stewart N. K.; Boggess W. C.; Miller M. J. (2018) A Synthetic Dual Drug Sideromycin Induces Gram-Negative Bacteria To Commit Suicide with a Gram-Positive Antibiotic. J. Med. Chem. 61, 3845–3854. 10.1021/acs.jmedchem.8b00218. [DOI] [PubMed] [Google Scholar]
- O’May C. Y.; Sanderson K.; Roddam L. F.; Kirov S. M.; Reid D. W. (2009) Iron-binding compounds impair Pseudomonas aeruginosa biofilm formation, especially under anaerobic conditions. J. Med. Microbiol. 58, 765–773. 10.1099/jmm.0.004416-0. [DOI] [PubMed] [Google Scholar]
- Windus D. W.; Stokes T. J.; Julian B. A.; Fenves A. Z. (1987) Fatal Rhizopus Infections in Hemodialysis Patients Receiving Deferoxamine. Ann. Intern. Med. 107, 678–680. 10.7326/0003-4819-107-5-678. [DOI] [PubMed] [Google Scholar]
- Visca P.; Bonchi C.; Minandri F.; Frangipani E.; Imperi F. (2013) The dual personality of iron chelators: growth inhibitors or promoters?. Antimicrob. Agents Chemother. 57, 2432–2433. 10.1128/AAC.02529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzl G. A.; Huang W.; Yu W.; Giardina B. J.; Zhou Y.; MacKerell A. D. Jr.; Wilks A.; Xue F. (2016) Iminoguanidines as Allosteric Inhibitors of the Iron-Regulated Heme Oxygenase (HemO) of Pseudomonas aeruginosa. J. Med. Chem. 59, 6929–6942. 10.1021/acs.jmedchem.6b00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centola G.; Deredge D. J.; Hom K.; Ai Y.; Dent A. T.; Xue F.; Wilks A. (2020) Gallium(III)-Salophen as a Dual Inhibitor of Pseudomonas aeruginosa Heme Sensing and Iron Acquisition. ACS Infect. Dis. 6, 2073–2085. 10.1021/acsinfecdis.0c00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y.; Thoendel M.; Olakanmi O.; Britigan B. E.; Singh P. K. (2007) The Transition Metal Gallium Disrupts Pseudomonas aeruginosa Iron Metabolism and has Antimicrobial and Antibiofilm Activity. J. Clin. Invest. 117, 877–887. 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minandri F.; Bonchi C.; Frangipani E.; Imperi F.; Visca P. (2014) Promises and failures of gallium as an antibacterial agent. Future Microbiol. 9, 379–397. 10.2217/fmb.14.3. [DOI] [PubMed] [Google Scholar]
- Stover C. K.; Pham X. Q.; Erwin A. L.; Mizoguchi S. D.; Warrener P.; Hickey M. J.; Brinkman F. S. L.; Hufnagle W. O.; Kowalik D. J.; Lagrou M.; Garber R. L.; Goltry L.; Tolentino E.; Westbrock-Wadman S.; Yuan Y.; Brody L. L.; Coulter S. N.; Folger K. R.; Kas A.; Larbig K.; Lim R.; Simith K.; Spencer D.; Wong G. K. S.; Wu Z.; Paulsen I. T.; Reizer J.; Saler M. H.; Hancock R. E. W.; Lory S.; Olson M. V. (2000) Complete Genome Sequence of Pseudomonas aeruginosa PA01, an Opportunistic Pathogen. Nature 406, 959–964. 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Clark J.; Maaløe O. (1967) DNA Replication and the Division Cycle in Escherichia coli. J. Mol. Biol. 23, 99–112. 10.1016/S0022-2836(67)80070-6. [DOI] [Google Scholar]
- Andrews J. M. (2001) Determination of Minimum Inhibitory Concentrations. J. Antimicrob. Chemother. 48 (1), 5–16. 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Sebaugh J. L. (2011) Guidelines for accurate EC50/IC50 estimation. Pharm. Stat 10, 128–134. 10.1002/pst.426. [DOI] [PubMed] [Google Scholar]
- Tawakoli P. N.; Al-Ahmad A.; Hoth-Hannig W.; Hannig M.; Hannig C. (2013) Comparison of different live/dead stainings for detection and quantification of adherent microorganisms in the initial oral biofilm. Clin Oral Investig 17, 841–850. 10.1007/s00784-012-0792-3. [DOI] [PubMed] [Google Scholar]
- Otsu N. (1979) A Threshold Selection Method for Gray-Level Histograms. IEEE Transactions on Systems, Man, and Cybernetics 9, 62–66. 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- Weinberg E. D. (2009) Iron Availability and Infection. Biochim. Biophys. Acta, Gen. Subj. 1790, 600–605. 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Hennessy D. J.; Reid G. R.; Smith F. E.; Thompson S. L. (1984) Ferene—a new spectrophotometric reagent for iron. Can. J. Chem. 62, 721–724. 10.1139/v84-121. [DOI] [Google Scholar]
- Ciccone L.; Vera L.; Tepshi L.; Rosalia L.; Rossello A.; Stura E. A. (2015) Multicomponent mixtures for cryoprotection and ligand solubilization. Biotechnol Rep. (Amst) 7, 120–127. 10.1016/j.btre.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (1988) Automatic Indexing of Rotation Diffraction Patterns. J. Appl. Crystallogr. 21, 67–72. 10.1107/S0021889887009737. [DOI] [Google Scholar]
- Vonrhein C.; Flensburg C.; Keller P.; Sharff A.; Smart O.; Paciorek W.; Womack T.; Bricogne G. (2011) Data Processing and Analysis with the AutoPROC Toolbox. Acta Crystallogr., Sect. D: Biol. Crystallogr. D67, 293–302. 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. R. (2011) An Introduction to Data Reduction: Space-Group Determination, scaling and intentisy statistics. Acta Crystallogr., Sect. D: Biol. Crystallogr. D67, 282–292. 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Brunkózci G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L.-W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. D66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowan K. (2010) Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. D66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. r.; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. (2010) MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.