To the Editor:
Pathway enrichment analysis is a common technique for interpreting gene lists derived from high-throughput experiments1. Its success depends on the quality of gene annotations. We analyzed the evolution of pathway knowledge and annotations over the past seven years and found that the use of outdated resources has strongly affected practical genomic analysis and recent literature: 67% of ~3,900 publications we surveyed in 2015 referenced outdated software that captured only 26% of biological processes and pathways identified using current resources.
Pathway analysis assesses the statistical enrichment of biological processes and pathways in a given gene list on the basis of information in Gene Ontology2 (GO) and pathway databases such as Reactome3 and PathwayCommons. GO is updated daily and Reactome versions are released quarterly, but many software tools interpret gene lists using functional information that has not been updated for years.
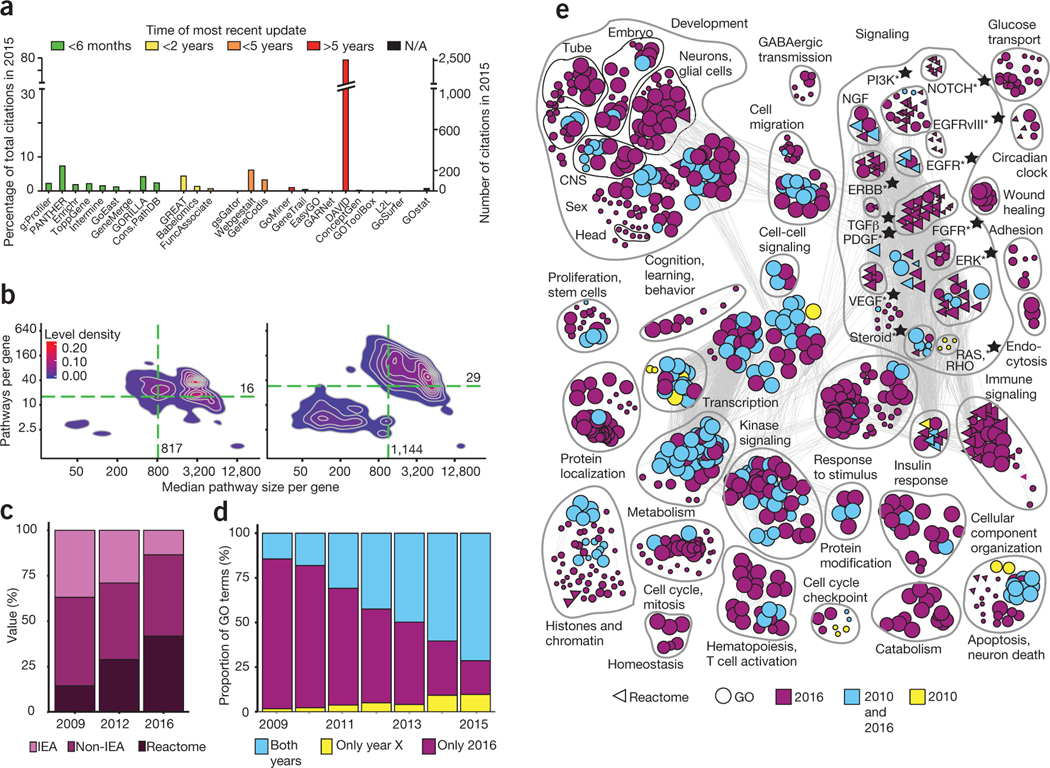
We surveyed the update times of 25 web-based pathway enrichment tools and citations of these tools in 3,879 publications (Fig. 1a and Supplementary Tables 1 and 2). Although nine tools (for example, g:Profiler4 and PANTHER5) provided gene annotations that had been revised within six months (September 2015 through February 2016), most tools were outdated by several years. Ten (42%) were outdated by five or more years, including the very popular DAVID6 tool, revised in January 2010 (DAVID was updated again recently, while this paper was under consideration). Remarkably, a total of 2,601 publications from 2015 (67%) cited severely outdated tools.
Figure 1 |.
Outdated pathway analysis resources strongly affect practical genomic analysis and literature. (a) The majority of public software tools for pathway enrichment analysis use outdated gene annotations, and the majority of surveyed papers published in 2015 used annotations that were more than five years old. (b) Density plots showing the evolution of pathway knowledge (GO + Reactome) between 2009 (left) and 2016 (right). The values for the median gene are indicated by green dashed lines. The bottom left group in the 2016 plot corresponds to Reactome pathways. (c) Gene annotation quality is improving rapidly as manually curated Reactome annotations are becoming more frequent and fewer genes in GO are IEA. (d) Pathway enrichment analysis of frequently mutated GBM genes showing the proportion of results missed in outdated GO annotations. Each bar compares annotations from a given year to 2016 annotations. (e) Enrichment map of frequently mutated GBM pathways and processes according to gene annotations from 2010 and 2016. Three-quarters of current findings are missed in out-of-date analyses (purple). Nodes represent processes and pathways, and edges connect nodes with many shared genes. Stars indicate clinically actionable pathways.
To understand the impact of outdated tools, we studied how knowledge in GO and Reactome evolved during 2009–2016 (Supplementary Fig. 1, Supplementary Methods and Supplementary Table 3). We found that the number of human biological processes (BP) and molecular pathways doubled in that time (BP in GO, 6,509 to 14,735; Reactome, 880 to 1,746; Supplementary Fig. 2). The vocabulary is becoming increasingly detailed and interconnected as GO terms are connected to roots by longer paths (mean, 7.59–8.06; permutation P < 10−5) and have more parents (1.73–2.09; P < 10−5) (Supplementary Fig. 3). This affects gene list interpretation, as GO annotations are propagated to parent terms.
Knowledge of individual genes and processes has accumulated significantly in terms of annotations per gene (median 29 versus 16; P < 10−5) and sizes of annotated gene sets (1,144 versus 817; P < 10−5) (Fig. 1b and Supplementary Figs. 4 and 5). General terms previously included thousands of genes from semiautomated GO annotation pipelines, but in recent annotations a group of specific Reactome terms is also apparent that reflects complementary efforts to map details of molecular pathways (Fig. 1b and Supplementary Fig. 6). High-confidence experimental annotations are becoming more common, and fewer genes are poorly described (Fig. 1c). Between 2009 and 2016, the proportion of manually curated Reactome annotations of human genes rose from 15% to 42%, that of low-confidence ‘inferred from electronic annotations’ (IEAs) dropped from 37% to 14%, and that of protein-coding genes without annotations fell from 12.4% to 4.9% (Supplementary Fig. 7). We found that 12.2% of HGNC (HUGO Gene Nomenclature Committee) gene symbols from 2015 did not map to 2009 symbols, primarily affecting less characterized genes (P < 10−5; Supplementary Figs. 8 and 9).
We asked how outdated annotation databases influence the functional analysis of genes. We analyzed essential genes of 77 breast cancer cell lines from recent short hairpin RNA screens7 using Fisher’s exact test and annotations from 2010 (used by the DAVID software). Strikingly, 74% of enriched 2016 terms were missed on average when we tested 2010-era annotations (695 versus 191; false discovery rate P < 0.05; Supplementary Fig. 10).
To confirm our observations in a high-confidence data set, we studied 75 significantly mutated glioblastoma (GBM) genes8 using annual annotations from 2009–2016. The 2010 annotations captured only ~20% of current results (BP in GO, 172/827; Reactome, 16/128), primarily because of updated annotations of existing pathways (75%) rather than new functional vocabulary (Fig. 1d and Supplementary Figs. 11 and 12). Annotations from 2010 are often based on low-quality information, as 603/625 (96.5%) of the current results were missed when we excluded IEAs (Supplementary Fig. 13). Note that evolving gene annotations may also lead to a loss of pathway results: 12% fewer GO terms appeared in the current analysis compared with 2015, primarily owing to changes in statistical significance (Supplementary Fig. 14).
Annotations from 2010 miss biological and translational insights into GBM (Fig. 1e, Supplementary Note and Supplementary Tables 4 and 5). For example, the glucose signaling pathway enriched exclusively among current annotations helps brain-tumor-initiating cells overcome starvation9. Immune-response processes emphasize emerging opportunities in cancer immunotherapy. Further, the up-to-date analysis showed 13 potentially clinically actionable pathways, such as the Notch pathway, in which γ-secretase inhibitors are being tested in ongoing clinical trials in glioma10.
The increasing quantity and completeness of functional annotations has a crucial effect on practical data analysis. Of the 25 tools we studied, the most popular software, DAVID, used in ~2,500 publications (65%), missed the vast majority of potential results. Thus, thousands of recent studies have severely underestimated the functional significance of their gene lists because of outdated annotations, negatively impacting follow-up studies for years to come, but also providing an opportunity to generate new hypotheses and validation experiments by reanalyzing existing data.
Researchers and peer reviewers need to pay attention to the timeliness of data. Software needs to clearly indicate update times, researchers need to document these times in publications, and the bioinformatics community needs to prioritize frequent updates of gene annotations. At least semiannual updates should be required, as major databases release several versions annually. To ensure reproducibility, tools need to provide historical gene annotations. As an example of recommended practice, our g:Profiler webserver (http://biit.cs.ut.ee/gprofiler) is synchronized quarterly with the Ensembl database and maintains archived versions dating to 2011. Reliable up-to-date software allows researchers to make the best use of current knowledge of gene function and interrogate experimental data for scientific discoveries.
Supplementary Material
ACKNOWLEDGMENTS
We thank the reviewers and the bioRxiv community for insightful comments. This study was supported by the Ontario Institute for Cancer Research (Investigator Award to J.R.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Creixell P. et al. Nat. Methods 12, 615–621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M. et al. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croft D. et al. Nucleic Acids Res. 42, D472–D477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reimand J, Kull M, Peterson H, Hansen J & Vilo J Nucleic Acids Res. 35, W193–W200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mi H, Muruganujan A & Thomas PD Nucleic Acids Res. 41, D377–D386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang W, Sherman BT & Lempicki RA Nat. Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Marcotte R. et al. Cell 164, 293–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamborero D. et al. Sci. Rep 3, 2650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flavahan WA et al. Nat. Neurosci 16, 1373–1382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takebe N. et al. Nat. Rev. Clin. Oncol 12, 445–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.