Abstract
Manganese is among Earth’s most abundant elements. Its oxidation had long been theorized1, yet undemonstrated2–4, to fuel chemolithoautotrophic microbial growth. Here, an enrichment culture exhibiting Mn(II)-oxidation-dependent, exponential growth was refined to a two species co-culture. Oxidation required viable bacteria at permissive temperatures, resulting in the generation of small Mn oxide nodules to which the cells associated. The majority member of the culture, ‘Candidatus Manganitrophus noduliformans’, affiliates within phylum Nitrospirae (Nitrospirota) but is distantly related to known Nitrospira and Leptospirillum species. The minority member has been isolated, but does not oxidise Mn(II) alone. Stable isotope probing revealed Mn(II)-oxidation-dependent, 13CO2-fixation into cellular biomass. Transcriptomics reveals candidate pathways for coupling extracellular manganese oxidation to aerobic energy conservation and to autotrophic CO2-fixation. These findings expand the known diversity of inorganic metabolisms supporting life, while completing a biogeochemical energy cycle for manganese5,6, one that may interface with other major global elemental cycles.
Beijerinck and Winogradsky discovered biological redox reactions involving C, N, S, and Fe over a century ago while pioneering methods for cultivating the responsible microbiota. This led to the concept of chemolithoautotrophy7,8. The known breadth of inorganic electron accepting and donating reactions in biology has continued to expand9–13. For example, the anaerobic respiratory reduction of Mn(IV) oxides to Mn(II) by diverse microbes is now understood to be widespread and of broad biogeochemical importance5,6,14,15. Over the past century, a multitude of studies and reviews have focused on the details of Mn(II) oxidations catalysed by diverse heterotrophs2–4; however the physiological roles of those activities generally remains unclear. Despite experimental hints that the oxidation might be coupled to energy conservation in some organisms1,16–19, whether Mn(II) oxidation drives the growth of any chemolithotrophs has remained an open question [Mn(II) + ½ O2 + H2O → Mn(IV)O2(s) + 2H+ ; ΔG°′ = −68 kJ · (mol Mn)−1] (Supplementary Note 1).
Cultivation of manganese oxidisers
That unappreciated microbes from the environment might oxidise Mn(II) for energy was re-examined. A glass jar was coated with a slurry of Mn(II)CO3 and allowed to dry before being filled with municipal tap water and left to incubate at room temperature. After several months, the cream-coloured carbonate coating was oxidised to a dark Mn oxide. Serial transfer of the material into a defined medium led to the establishment of a stable, in vitro culture. Unless otherwise noted: except for trace amounts of vitamins, this medium was free of alternative organic and inorganic electron donors (e.g. nitrate was used instead of ammonia as N source to preclude the growth of nitrifiers).
To distinguish between abiological and biological oxidation, flasks of sterile, defined, Mn(II) media were either inoculated with a subculture of the promising enrichment, or left uninoculated, and incubated under oxic conditions. Because Mn oxides have been suggested to contribute to chemical autooxidation of Mn(II)20, other replicate flasks were inoculated with a steam-sterilised sub-culture with oxide products. Even after a year, oxidation did not occur in uninoculated flasks or those containing the steam-sterilised inocula, as predicted by the known chemical stability of MnCO3 under these conditions21,22. However, within 4 weeks, the flasks inoculated with “viable material” had generated dark, adherent Mn oxides (Fig. 1a). Oxidation required O2 and occurred up through 42 °C, occurring optimally between 34 °C to 40 °C (Extended Data Fig. 1a), consistent with catalysis being enzymatic. Mn(II) oxidation activity was also sensitive to exposure to antibiotics or to overnight pasteurisation at 50 °C (Extended Data Fig. 1b). Phosphate was inhibitory above 0.3 mM. When amended with MnCO3, the pH of unbuffered media ranged between 5.7 and 6.3. While pH buffer was not required, Mn(II) oxidation was faster in media buffered with 5 mM MOPS at its pKa (7.1 at 37 °C). In buffered cultures, the final pH ranged between 6.5 and 6.8. With or without buffer, increases in culture pH during or after oxidation (that might potentially lead to chemical oxidation of unreacted MnCO3) were not observed. No growth could be detected in the MOPS-buffered basal medium without addition of MnCO3 (Extended Data Fig. 2a).
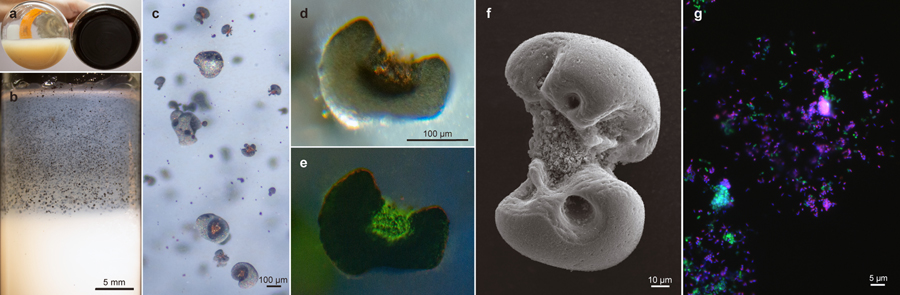
Figure 1 |. Biooxidation of MnCO3 produces Mn oxide nodules to which two species associate.
a, After incubation, comparison of an uninoculated control flask of basal medium containing bright, unreacted MnCO3 (left), with the adherent dark oxide products generated in one that had been inoculated with viable material (right). b, c, d, e, Microscopy of Mn oxide nodules generated in agarose solidified MnCO3 media. b, After incubation of tubes inoculated with viable material, the cloud of bright MnCO3 particles was clarified towards the air exposed meniscus, concomitant with the generation of larger, discrete dark oxides (enlarged in c). d, Transmitted light micrograph of an acridine orange (nucleic acid) stained Mn oxide nodule from the same agarose tube; e, epifluorescence micrograph of the same, with surface visible biomass localized to the inner clefts; material in clefts appeared orange prior to staining. f, Scanning electron micrograph of an Mn oxide nodule produced by the co-culture. g, Epifluorescence microscopy and fluorescence in situ hybridisation using species-specific rRNA-targeted probes reveal cell distributions in dissolved Mn oxide nodules: Species A (magenta), Species B (green), all DNA stained with DAPI (blue). No third species is present, via independent methods (Extended Data Fig. 3a). Each panel represents observations made from samples of multiple independent cultivation experiments (a, n >100; b,c,d,e, n=7; f, n=4; and g, n=2).
An rRNA-gene iTag community analysis of the initial enrichment culture revealed ~70 different species representing 11 bacterial phyla (Supplementary Table 1). The responsible microbes did not generate oxide-forming colonies on MnCO3 agar media, but successive rounds of serial-dilution-to-extinction in MnCO3 liquid media refined the community to a 2 species co-culture (Supplementary Table 1). Species A belongs to the phylum Nitrospirae (Nitrospirota), whereas Species B is a betaproteobacterium, occurring at cell ratio of ca. 7:1 (Supplementary Table 1; Extended Data Fig. 3a). Attempts to isolate Species A have thus far failed. Species B can be isolated from disrupted oxides as single colonies on succinate or other heterotrophic media (Supplementary Note 2), but does not oxidise MnCO3 alone. Either Species A is solely responsible for Mn(II) oxidation (Extended Data Fig. 3b), or the activity is consortial (Extended Data Fig. 3c–e). Several betaproteobacteria have proven recalcitrant to elimination from multi-species cultures: some were seemingly unimportant9,23, whereas others engaged in metabolite cross-feeding24, and at least one example central to a consortium25.
DNA harvested from both the co-culture and from a pure culture of Species B lead to near complete genome sequences for both species (Extended Data Fig. 3f), facilitating later experiments and analyses (below).
Manganese oxide nodules
Mn(II) oxidation yielded morphologically conspicuous, Mn oxide nodules ~ 20–500 µm in diameter (Fig. 1; Extended Data Fig. 4). These formed in both static and shaken liquid media, often adhering to the glass and to each other, as well as in media solidified with agarose (Fig. 1b). The surfaces were dark brown but often reflective, and typically invaginated around deeper depressions having a rough, dark orange surface (Fig. 1c,d). Attenuated total reflectance FTIR analysis revealed that Mn oxide nodules, hypochlorite-bleached to remove cellular and other organic carbon, are poorly-ordered and similar to birnessite.
Epifluorescence microscopic examination of nucleic acid-stained Mn oxide nodules localized the majority of the exposed biomass to the invaginations (Fig 1d,e), with few cells observed attached to the substrate or found planktonic. In agarose solidified media, these could be well separated from the carbonate substrate, dissolving it from a distance (Fig. 1b). The latter is partially explained by the solubility products of the MnCO3 precipitate; under the incubation conditions, these can be expected to include free Mn2+ ion, manganese bicarbonate, and soluble MnCO322. The mean concentration of dissolved Mn in uninoculated and active MnCO3 cultures was 0.214 mM (s.d. = 0.107, n=3) and 0.119 (s.d. = 0.081, n=3), respectively, before falling to 0.010 mM (s.d. = 0.009, n=3), after oxidations. Soluble Mn(II) chloride does not appear to be utilized; instead, it appears to inhibit MnCO3 oxidation when amended to active co-cultures at concentrations >2.0 mM (Extended Data Fig. 1c). No evidence for motility was observed: the oxides did not accumulate as a band across the interface between counter opposing gradients of Mn(II) and O2 in agarose-solidified media (Fig. 1b), as is commonly observed for microaerophilic iron-oxidising bacteria26. It is not yet understood whether the tight association of the cells with the oxidation product is circumstantial or is more intrinsic to the process due to some role of adsorptive, conductive, catalytic, and/or other properties of Mn oxides.
Additional biomass was revealed upon chemical dissolution of Mn oxide nodules, and was examined using fluorescence in situ hybridisation (FISH) with specific rRNA-targeted probes (Supplementary Note 3). Consistent with the iTag analysis, Species A cells were more abundant (Extended Data Fig. 3a). No stereotypic patterns of association with Species B were observed (Fig. 1g; Extended Data Fig. 4a–e). Cells of both were often pleomorphic. Species A cells were typically crescents, 1.07 µm by 0.40 µm (s.d. = 0.17 by 0.08, n=50); Species B were typically rods, 1.22 µm by 0.56 µm (s.d. = 0.20 by 0.09, n=50) in co-culture, but at high cell densities in pure cultures, cells elongate and form flocs.
Growth rates and yields
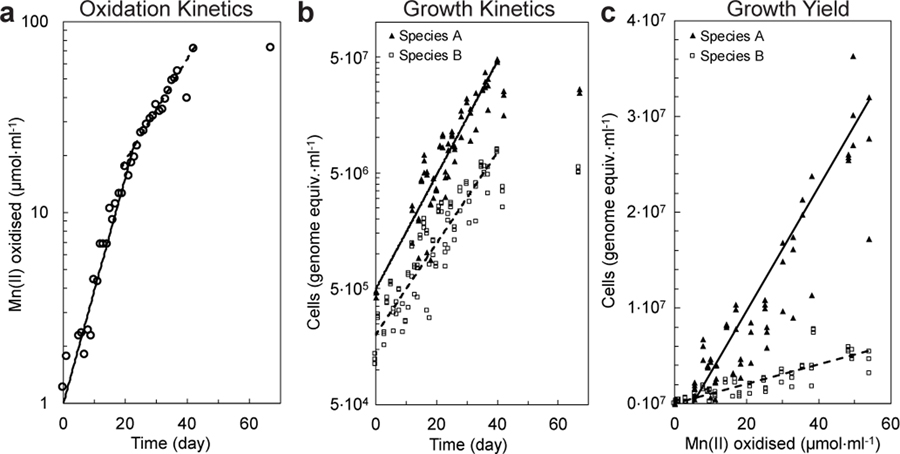
If truly chemolithotrophic, one or both species in the co-culture should exhibit a) exponential increases in the rate of Mn(II) oxidation during and in parallel to b) Mn(II)-dependent exponential growth. Indeed, after inoculation, the rates of Mn(II) oxidation in the basal media increased exponentially, initially doubling every 6.2 days (s.d. = 1.1; n = 4 replicate cultures; Fig 2a; Extended Data Fig. 2e–g; Supplementary Note 4) before decelerating to 10.8 days (s.d. = 0.1). Concurrent with Mn(II) oxidation, Species A exhibited sustained exponential growth, roughly matched by the less numerous and perhaps commensal Species B (Td = 6.1 and 7.7 day, respectively; Fig. 2b; Extended Data Fig. 2b; Supplementary Note 5). The co-culture oxidised Mn(II) at a combined rate of 3.4 to 9.0 · 10−15 mol Mn(II) · cell−1· hr−1. A linear relationship between Mn(II) oxidised by the co-culture and the cell yields was observed (Fig. 2c; Supplementary Note 6): 6.4 · 1011 cells of Species A, 1.0 × 1011 cells of Species B, for a combined yield of 7.4 · 1011 cells · (mol Mn(II) oxidised)−1. The total amount of DNA extracted from samples also increased exponentially with time and oxidation of Mn(II) (Extended Data Fig. 2c,d), yielding 3.1 · 106 ng DNA · (mol Mn(II) oxidised)−1. Based on either of two estimates (both using the known dry weight of a single cell of E. coli27), the growth yield in the co-culture is estimated to be ~100–200 mg dry biomass · (mol Mn(II) oxidised)−1. Such growth yields and normalised substrate oxidation rates are comparable with those observed for chemolithotrophic microbial nitrite oxidation (Extended Data Fig. 3g), a metabolism predicted to yield a similar free energy12.
Figure 2 |. Mn(II) oxidation coupled to co-culture growth of Species A and Species B.
a, Mn(II) oxidation rates increased exponentially over time in two distinct phases before plateauing. b, Exponential growth of Species A and Species B paralleled Mn(II) oxidation. c, Linear relationship between growth yield and the amount of Mn(II) oxidised. Symbols in b and c represent the three technical replicates for each sample. See Extended Data Fig. 2b–n for analyses on independent cultivation experiments (n=9).
Phylogenetic analyses
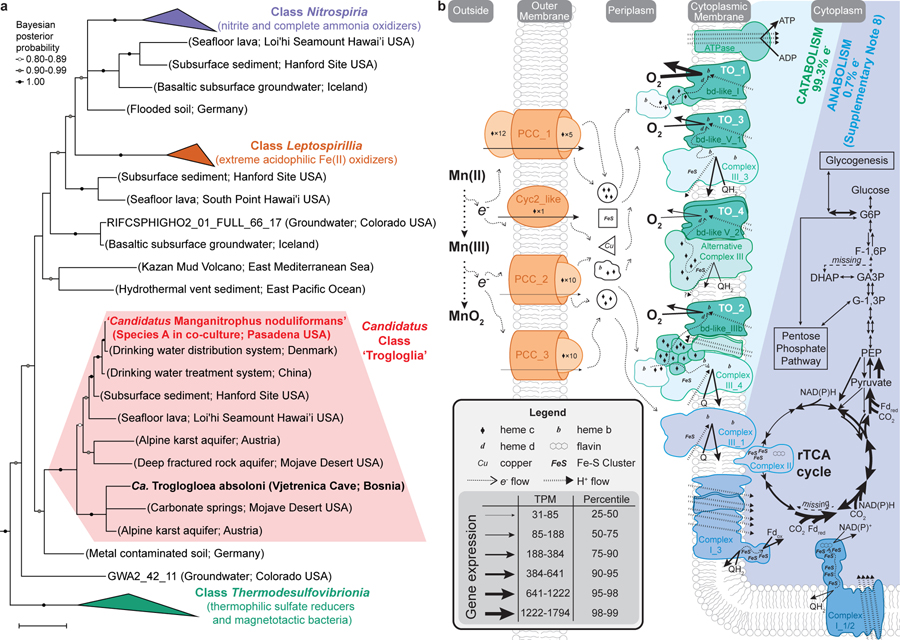
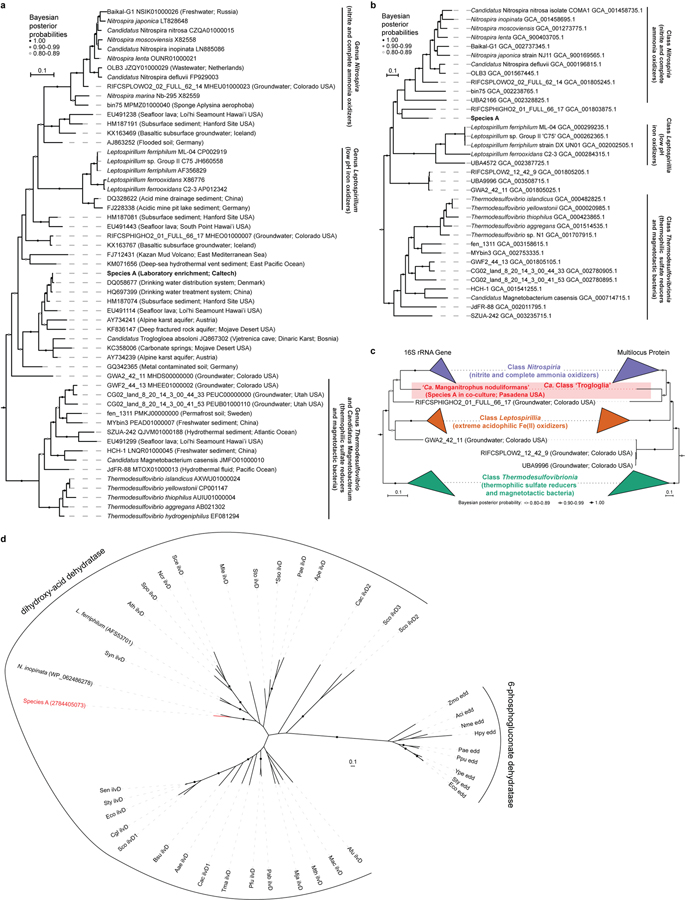
Species A affiliates remotely with the genera Nitrospira, Leptospirillum, and other members of the phylum Nitrospirae (Nitrospirota), yet shares <84% 16S rRNA identity with any cultivated organism, and <87% identity to all but ~50 16S rRNA gene sequences from not-yet-cultivated organisms (Fig. 3a; Extended Data Fig. 5a,c; Supplementary Table 2). The genome of Species A does not encode recognisable genes for the chemolithotrophic oxidation of ammonia, hydroxylamine, nitrite, or reduced sulfur compounds. Several closely related sequences have been recovered from drinking and karst-impacted groundwater, marine sites, and a subsurface oxic-anoxic transition zone (Fig. 3a; Extended Data Fig. 5a). Together, these cluster with ‘Candidatus Troglogloea absoloni’28, an uncultivated cave organism of unknown physiology. Few genome datasets are available for comparison. Species A shares <88% 16S rRNA gene identity to the best reference genomes (from metagenome-assembled genomes from groundwater29,30), and is the only genome currently available in the Candidatus Class ‘Trogloglia’ (Fig. 3a; Extended Data Fig. 5b,c).
Figure 3 |. Phylogenetic analysis and metabolic reconstruction of Species A (‘Ca. Manganitrophus noduliformans’).
a, Bayesian phylogram based on 1532 aligned 16S rRNA nucleotide positions. Sequences clustering within the three previously described classes within this phylum are collapsed into separate nodes. Species A clusters with not-yet-cultivated members of the Trogloglia, a distinct class within the bacterial phylum Nitrospirae (Nitrospirota). See Extended Data Fig. 5a–c for greater detail and identifiers. Scale bars = evolutionary distance (0.1 substitutions-per-site average). b, Hypothetical model of e− flow from extracellular Mn(II) to the energy and anabolic systems of Species A. The oxidation is hypothesized to be mediated by several expressed outer membrane complexes (orange), with subsequent e− transfers to periplasmic carriers. The bulk of e− flow is towards generating proton motive force via terminal oxidases (TO, green) during O2 respiration. The remaining e- flow would be to motive force dissipating, reverse electron transport complexes (blue), generating the low-potential e− carriers required for rTCA cycle mediated CO2-fixation. Gene expression values represent the mean of independent cultivation experiments (n=7). See Supplementary Table 4 for identifiers and transcript levels for each gene, and Supplementary Notes 7 and 8 for more detailed explanations of the diagrams.
Species B affiliates with heterotrophs from the betaproteobacterial genus Ramlibacter (Extended Data Fig. 6a,b). It exhibited H2 + O2 dependent growth; encodes genes for hydrogenases; encodes both Sox and DsrMKJOP gene clusters for the oxidation of reduced sulfur; encodes a Calvin-Benson-Bassham Cycle; and may capable of anaerobic respirations such as denitrification and dissimilatory metal reduction. The potential for facultative lithotrophy had not been previously reported for members of this genus.
Transcriptomics of Mn dependent growth
The transcriptomes of the co-culture (in particular Species A) were examined, during different stages of Mn(II) oxidation in replicate cultures (n=7; Extended Data Fig. 3h). Although both species encode genes for swimming and twitching motility, as well as for chemotaxis, they were not expressed (Supplementary Table 3), matching the observations in agarose solidified media (above). In contrast, genes for different compatibles solutes (e.g. hydroxyectoine, trehalose, and betaines) were expressed by both species and, consistent with this, co-cultures oxidised Mn when grown at a range of brackish salinities, up through nearly 40% of that of seawater.
Candidate genes that might underly Mn(II) chemolithotrophy have been identified. Species A transcribed 4 gene clusters encoding outer membrane complexes that evoke comparisons with lithotrophic iron oxidisers and respiratory metal reducers. By analogy, these might play a role in extracellular electron transfer (EET) by ferrying Mn(II)-derived electrons to periplasmic carriers (candidates for which were also expressed; Supplementary Table 4), leaving the resultant, insoluble oxide outside the cell (Fig. 3b). In iron-oxidising microbes, an outer membrane c-type cytochrome (Cyc2 or Cyt572) is often employed as the initial oxidant and carrier for the Fe(II)-derived electron31,32. Species A expressed a Cyc2 homolog with the predicted heme-binding site and outer membrane beta-barrel structure (Fig. 3b; Supplementary Table 4). In iron-oxidising anoxygenic phototrophs33,34 and in several neutrophilic iron-oxidising chemolithotrophs35, an alternative mechanism involves a porin-cytochrome c protein complex (PCC)36. Species A expresses genes for three recognisable PCCs: a porin-dodecaheme cytochrome c with no homologs in the databases (PCC_1); and two distinct porin-decaheme cytochrome c modules (PCC_2 and PCC_3; Fig. 3b). Curiously, during growth in the Mn(II) oxidising co-culture, Species B expresses an MtrABC-like PCC and other multiheme cytochromes c with greater resemblance to the complexes involved in anaerobic reduction of metals by Shewanella sp.36 (Supplementary Table 5).
After the transfer of Mn(II)-derived electrons from outside of the cell into the periplasm, their flow through respiratory complexes in the cytoplasmic membrane is central to understanding this novel mode of energy conservation. On average, the two Mn(II)-derived electrons are generally considered to be of high potential [Mn(II)/Mn(IV), E°´=+466 mV; Supplementary Note 1]. However, the energetics of each of the two sequential one-electron transfers can be impacted by inorganic and organic binding ligands22,37, leading to a degree of uncertainty here. Of the respiratory complexes, canonical respiratory Complex I is unlikely to be employed for energy conservation, leaving canonical or alternative Complex III, Complex IV, or cytochrome bd oxidases as possible candidates for generating a proton motive force during Mn(II) chemolithotrophy.
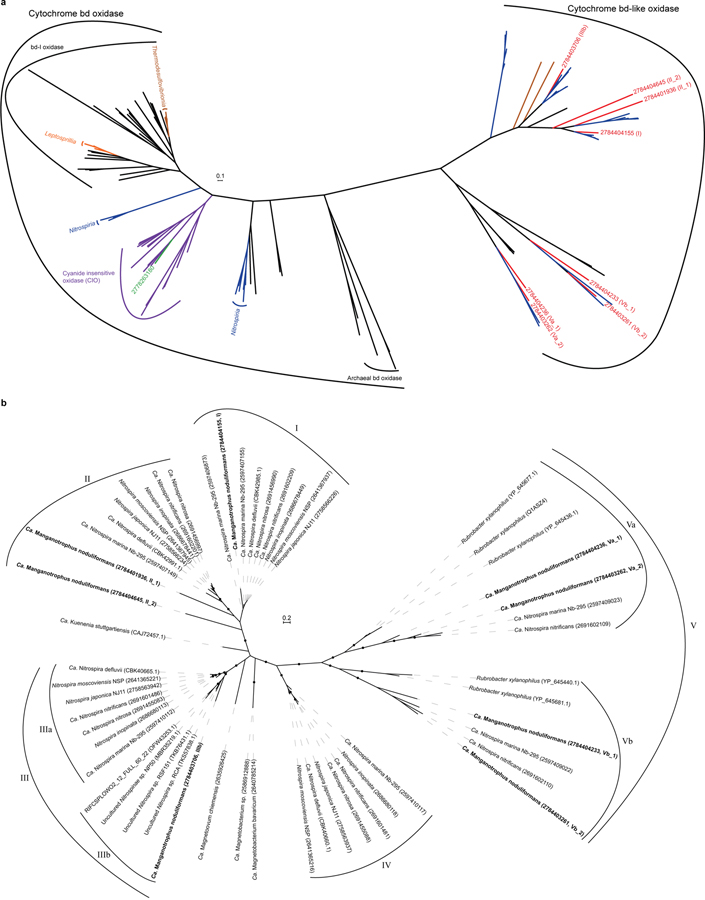
The genome of Species A contains a host gene clusters for terminal oxidases (TO) that could link the quinone pool to O2, many of which are strongly transcribed (Supplementary Table 4). Although a role in the process can’t be ruled out, the single Complex IV (cbb3-type cytochrome c oxidase) of Species A is not as well expressed (24th percentile) as four unconventional terminal oxidase complexes containing cytochrome bd-like oxidase (Fig. 3b; Supplementary Table 4; Extended Data Figs. 7 and 8; Supplementary Note 7). The most highly expressed of the latter is “TO_1” (99th percentile; Fig. 3b), a complex generally similar to those observed in ammonia- and nitrite-oxidising Nitrospirae38,39, but without the hypothesized candidate catalytic and iron-sulfur subunits, NxrAB. The second highest expressed terminal oxidase (TO_2; 83rd percentile) also misses these and is highly unusual amongst cultivated organisms, having extra heme c domains, cytochrome c, and two MrpD-like ion-pumping subunits (Fig. 3b; Supplementary Table 4) that might, hypothetically, facilitate electron transfer while generating a motive force. In both TO_1 and TO_2, a membrane attached di-heme cytochrome b may connect EET and periplasmic electron carriers to these membrane complexes (Fig. 3b; Supplementary Note 7). Whether either of these will have a high affinity for O2, as canonical bd-oxidases do, is not known. Future studies are required to examine the function of these unusual terminal oxidases, their roles in energy conservation, and how any energetic challenge of the initial oxidation of Mn(II) to Mn(III) is offset or met by the organism.
Mn(II)-oxidation-dependent CO2-fixation
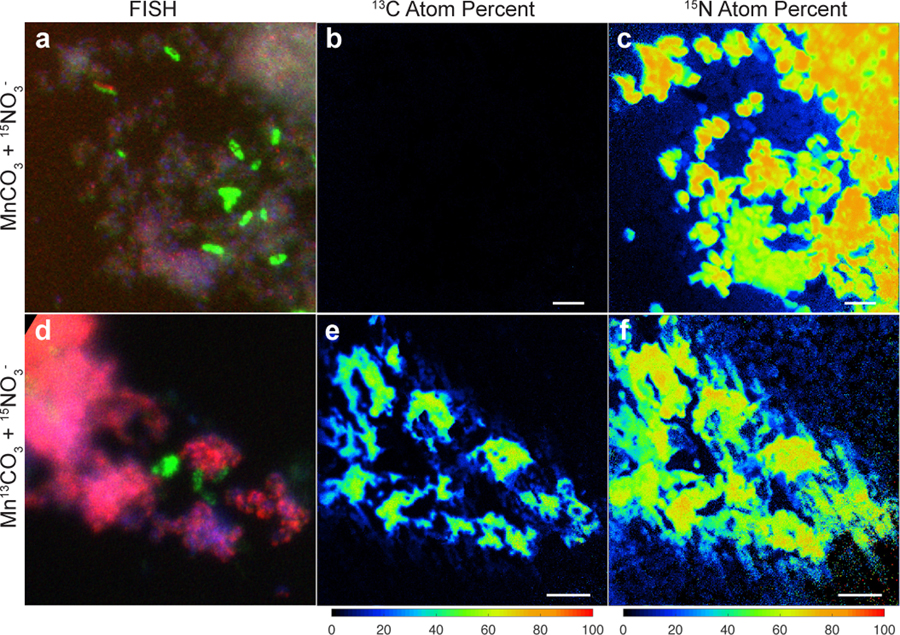
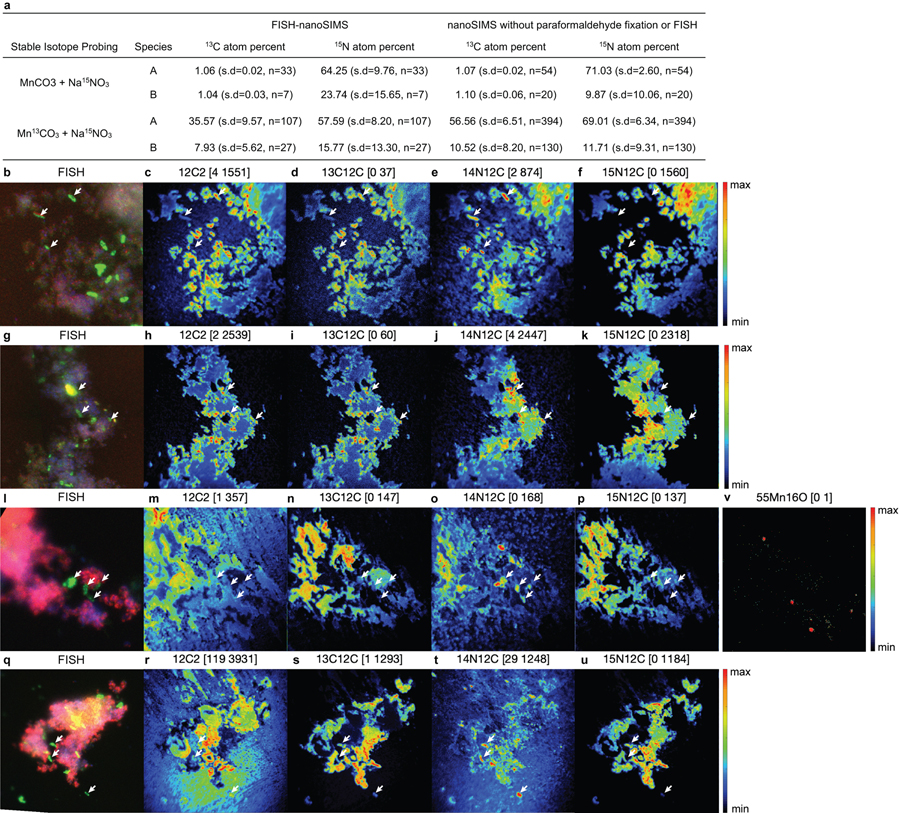
With the demonstration that Mn(II) oxidation drives lithotrophic energy metabolism and growth, that the co-culture might generate biomass via autotrophic CO2-fixation with Mn(II)-derived electrons was examined. For this, the co-culture was grown with labelled 13C-MnCO3 (and 15N-nitrate, to aid in tracking the synthesis of new biomass), with the isotopic compositions of cells visualized microscopically by species-specific FISH coupled to nanometre-scale secondary ion mass spectrometry (nanoSIMS) (Fig. 4). The results are consistent with the co-culture being autotrophic. Both Species A and Species B were confirmed to incorporate significant amounts of 13C and 15N isotopes (Fig. 4; Supplementary Note 9). Species A showed a higher enrichment for both isotopes, as compared to Species B (Extended Data Fig. 9a), suggesting that it is the main if not sole driver of Mn(II)-dependent CO2-fixation and lithoautotrophic growth in the co-culture, especially when taken together with its greater abundance (Extended Data Fig. 3a). While it can’t be ruled out that some degree of anabolic mixotrophy might occur via the uptake of trace contaminating organic carbon, it is likely that Species A has an inoperable oxidative TCA cycle: it does not encode a recognisable homodimeric 2-oxoglutarate dehydrogenase (OGDH) complex (Fig. 3b; Supplementary Note 10), a hallmark deficiency observed in many autotrophs that are unable to mineralize organic carbon (including N- and Fe-oxidising Nitrospirae species, which grow autotrophically using the rTCA pathway38,40,41).
Figure 4 |. Stable isotope probing of autotrophic CO2 fixation.
Cells grown in the basal medium with labelled Mn13CO3 and 15NO3− were compared to cells grown with unlabelled MnCO3 and 15NO3−. a,d, Fluorescence in situ hybridisation of cells dissolved from Mn oxide nodules using species-specific rRNA-targeted probes: Species A (magenta), Species B (green), all DNA stained with DAPI (blue). b,c,e,f, Nanometre-scale secondary ion mass spectrometry reveal incorporation of 13C and 15N into cells. Coloured scale bars indicate 13C or 15N atom percent. White scale bars = 3 µm. Areas shown in b,c and e,f correspond to that in a and d, respectively. See Extended Data Fig. 9a for analyses on independent nanoSIMS images (n=5) from the same culture incubated with inorganic 13C.
During Mn(II)-dependent growth in the co-culture, Species A expresses genes for a complete Reverse Tricarboxylic Acid Cycle (rTCA; Fig. 3b; Supplementary Table 4; Supplementary Note 10). Curiously, although its role in Mn oxidation is speculative, Species B expresses genes for the Calvin-Benson-Bassham Cycle (Supplementary Table 5).
For Species A to fix CO2 via the rTCA pathway, low potential electron carriers [e.g. NAD(P)+ and ferredoxin]42 need to be reduced with high potential, Mn(II)-derived electrons (see discussion above), possibly via a reverse electron transport chain, at the very least involving Complex I (Fig. 3b). Three distinct Complex I gene clusters can be found in Species A, two of which (Complex I_1&2; Fig. 3b; Supplementary Table 4) hypothetically might dissipate energy when operated in reverse and generate NAD(P)H (E°´=−320 mV)43 from reduced quinones with a more positive reduction potential. Similarly, a remarkably unusual Complex I (Complex I_3; Fig. 3b; Supplementary Table 4) might be used to reduce ferredoxin (E°´=−398 mV)43. This complex encodes five ion-pumping subunits (NuoN, 2 NuoM, and 2 MrpD-like subunits, but no NuoL), whereas the canonical Complex I only encodes three, NuoLMN44. Recently, rare and unusual variants of Complex I with four ion-pumping subunits (NuoLMMN) have been identified and postulated to couple the inward flow of 5 protons to drive the endergonic reduction of ferredoxin from a quinone45,46. In Species A, Complex I_3 hypothetically may use electrons from the quinone pool to drive the reduction of ferredoxin via the inward flow of 6 protons or ions (Fig. 3b). If the entry point(s) of Mn(II) derived electrons into the electron transport chain have reduction potentials more positive than the quinone pool, then additional respiratory complexes (Fig. 3b) would have to be involved to accomplish productive reverse electron flow (Supplementary Note 7).
Discussion
Whether chemolithoautotrophic manganese-oxidising microbes exist had been an open question for over a century. This study establishes their existence and provides insights into the details and dynamics of the process at cellular, physiological, genomic, transcriptomic, and isotopic levels. Manganese chemolithoautotrophy extends the known physiologies in the phylum Nitrospirae that leverage meagre differences in redox potentials between inorganic electron donors and acceptors9,12,47–49. Based on its physiology, phylogeny, genomics, and other characteristics, the epithet ‘Ca. Manganitrophus noduliformans’ is proposed for Species A (Supplementary Taxonomic Proposal).
The potential impact of Mn(II) oxidation coupled to seemingly slow exponential growth of the co-culture (Extended Data Fig. 3g) has current and past environmental implications. Starting with a single cell each of Species A and B, unrestricted chemolithotrophic growth at the observed cell doubling times and oxidation rates would be sufficient to generate Mn oxides equalling global Mn reserves within 2 years (Supplementary Note 11). Based on phylogenetic inferences, close relatives of Species A reside in many subsurface and karst environments (Fig. 3a; Extended Data Fig. 5a), including at oxic-anoxic transition zones (OATZ) such as Hanford sediments50. At such interfaces, Mn could be cycled between aerobic Mn(II)-oxidising chemolithoautotrophs and anaerobic, Mn oxide respiring (reducing) chemotrophs5,6,14,15, thereby stimulating significant electron flows through the element over even brief geological time scales. This has implications for the interconnected biogeochemical cycles of C, N, S, Fe, H, and O, with which a newfound, complete Mn energy cycle likely interacts.
Methods
No statistical methods were used to predetermine sample size, and the investigators were not blinded to allocation during experiments and outcome assessment.
Media and culture enrichment, refinement, and maintenance.
A fortuitous enrichment culture for Mn(II) lithotrophs was established over the Summer and Fall of 2015. A dense slurry of freshly precipitated MnCO3 (see below) was distributed onto the internal surface of wide mouth glass jar and the coat was allowed to dry. The jar was filled completely with unsterilised Pasadena municipal drinking water (typically a blend from surface and aquifer sources) collected and allowed to stand for approximately 10 weeks, open and without agitation in an unoccupied room maintained at ca. 21°C (GPS 34°08’12.1”N 118°07’38.3”W). Additional freshly precipitated MnCO3 was added as a dense suspension after the cream-coloured coating had blackened, and the jar was covered with a loose fitting lid and allowed to incubate for several more months, after which a small amount (ca. 5% v/v) of the dark product was used to inoculate flasks of MnCO3 suspended in municipal tap water. After this, a separate line of cultures was initiated in a defined, deionized water medium (below) incubated in the laboratory at 37 °C.
The basal medium used for routine growth and maintenance of this line and in related experiments was adapted and modified from prior formulations51,52. The medium contained, per liter deionized H2O: NaCl, 1 g; MgCl2·6H2O, 400 mg; CaCl2·2H2O, 1 g; KCl, 5 g; Na2SO4, 142 mg; FeCl3·6H2O, 2 mg; H3BO3, 30 µg; MnCl2·4H2O, 100 µg; CoCl2·6H2O, 190 µg; NiCl2·6H2O, 24 µg; CuCl2·2H2O, 2 µg; ZnCl2, 68 µg; Na2SeO3, 4 µg; Na2MoO4, 31 µg; riboflavin, 100 µg; biotin, 30 µg; thiamine HCl, 100 µg; L-ascorbic acid, 100 µg; d-Ca-pantothenate, 100 µg; folic acid, 100 µg; nicotinic acid, 100 µg; niacinamide, 100 µg; 4-aminobenzoic acid, 100 µg; pyridoxine HCl, 100 µg; lipoic acid, 100 µg; NAD, 100 µg; thiamine pyrophosphate, 100 µg; cyanocobalamin, 10 µg. As P source, a solution of potassium phosphate (pH 7.2) was added to a final concentration of 0.15 mM. As N source, NaNO3 was added to a final concentration of 1 mM; alternatively, 1 mM NH4Cl or other N sources were employed as noted. MOPS at its pKa was added as a buffer to a final concentration of 5 mM, when noted. MnCO3, or alternative, heat-stable substrate for growth was added to the basal medium prior to steam sterilisation; alternatively, heat-unstable substrate for growth was added using filter sterilised stock solutions after media had adequately cooled.
Freshly precipitated MnCO3 was employed, for the initial enrichment culture, for routine Mn(II)-dependent cultures in small volumes; or for the serial dilution-to-extinction resolution of complex cultures; or for stock and starter cultures. To prepare this, 25 g MnCl2 (Sigma-Aldrich #221279) was dissolved into 100 ml deionized water, yielding 125 ml of a 1.59 M MnCl2 solution. Over the course of several minutes, this solution was slowly poured into 3 liters of 0.33 M NaHCO3 while vigorously stirring. After the cessation of stirring after ca. 1 hour, the resultant precipitate was allowed to settle by gravity. Thereafter, the overlying reaction fluid was decanted and discarded. The precipitate was resuspended in 3 liters of deionized water, and the stirring, settling, decanting, and resuspension steps were repeated at least 10 times. After the final wash, the precipitate was resuspended in deionized water to a final volume of 100 ml, stored in a clean glass bottle, and refrigerated at 4°C in the dark. Initially, the precipitate appeared white to light pink, but aged to a light tan within ca. 24 h. Thereafter, the material remained stable for months to years. Alternatively, a hydrated MnCO3 substrate (Sigma-Aldrich #63539) was employed for larger culture volumes, and/or for reproducible mass balances, as noted.
Cells were typically cultured in 10 ml of medium in 18-mm diameter culture tubes, or in 100 ml of medium in 250-ml Erlenmeyer flasks at 37 °C, with 25 to 200 mM MnCO3, as noted. To prevent dehydration of cultures over the long periods of incubation, cut strips of Parafilm (Heathrow Scientific, Vernon Hills, IL, USA) were used to seal the bottom edge of the 18-mm plastic test tube enclosures. Cultures were incubated stationary without agitation, or with shaking at 200 rpm, as noted.
To refine the number of species in the complex manganese-oxidising enrichment, 5 successive rounds of serial tenfold dilution-to-extinction series were performed using 9 ml of MnCO3 nitrate basal media in 18-mm culture tubes, incubated at 32 °C. Culture tubes in each dilution series were scored as positive for the presence of lithotrophic manganese oxidisers when, after 2–12 weeks of incubation, the small and easily dispersed particles of the MnCO3 substrate (light cream in colour) were converted to larger Mn oxide nodules or a single continuous Mn oxide coating (dark brownish-black in colour) that typically tightly adhered to the bottom of the glass culture tubes. Mn oxides from the final dilution tube showing such oxidation were used as the inocula for the next serial-dilution-to-extinction series.
To examine whether Mn(II) oxidation in the cultures was biological in nature, an active co-culture with Mn oxides was used to inoculate 18-mm culture tubes containing the basal media with 50 mM MnCO3. Cultures were amended with either of two antibiotics, kanamycin (30 µg/ml) or vancomycin (20 µg/ml), or pasteurised overnight at 50 °C before incubation at 32 °C. To examine the impact of incubation temperature on oxidation, cultures were incubated without agitation in different incubators set at a diversity of temperatures; incubation temperatures were regularly and independently confirmed with >2 thermometers. After 2 weeks, 2 ml mixture from the cultures were sampled and stored at −80 °C for later ICP-MS analysis (below). Reported values were corrected for Mn oxides carried over in the inoculum, as ascertained by the lowest amount determined in the 50 °C pasteurisation experiments.
To examine the growth of the culture in the absence of Mn(II) substrate, a stationary co-culture (confirmed separately to be viable) was used to inoculate 4 replicates of 250-ml Erlenmeyer flasks containing 120 ml of the basal media without MnCO3. The flasks were incubated at 37 °C with shaking at 200 rpm. After inoculation and after 10 and 21 days, 20 ml mixture from the cultures were sampled and centrifuged at 5250 × g for 10 min; the pellet was stored at −80 °C. DNA was extracted from the thawed pellets using the DNeasy PowerSoil kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions, with the bead beating option using FastPrep FP120 (Thermo Electron Corporation, Milford, MA, USA) at setting 5.5 for 45 s instead of the 10 min vortex step. DNA concentration was quantified using Qubit dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA).
To examine whether MnCl2 can be oxidised by, or otherwise impacts growth: an active co-culture, which had oxidized 38% of the 35 mM MnCO3 initially provided (measured using the ICP-MS method below), was used to inoculate (10% v/v) 18-mm culture tubes containing 10 ml of the basal media with 0.5–20 mM MnCl2 instead of MnCO3. After inoculation, and after days 5 and 10, 0.5 ml of the oxides and culture fluid mixture was sampled and stored at −80 °C for later ICP-MS analysis (below). Reported values were corrected for Mn oxides carried over in the inoculum, as ascertained by the lowest amount determined in the 0.5 mM MnCl2 culture.
For attempts to observe single colony formation by Mn(II) lithotrophs on agar media, the basal medium was adjusted to contain 200 mM MnCO3 and 1.5% washed agar (BD Difco), and distributed into petri dishes after steam sterilisation.
Genomics predicts that each species in the co-culture may be able to produce compatible solutes (e.g. trehalose and hydroxyectoine, by Species A), and thus may be able to grow under a range of salinities. To examine the impact of increased salinity on the Mn(II)-oxidising lithotrophs, the basal medium was amended with NaCl to achieve final salt concentrations of 2 ppt, 2.8 ppt, 3.8 ppt, 4.6 ppt, 5.5 ppt, 9 ppt, 16 ppt, 23 ppt, 30 ppt, and 37 ppt (equivalent to 6%, 8%, 11%, 13%, 16%, 26%, 45%, 65%, 85%, and 105% of the salinity of seawater, respectively). After inoculation, oxidation in the tubes was monitored visually over time.
For isolation and maintenance of strains of “Species B” (Ramlibacter lithotrophicus) from the co-culture (Supplementary Note 2), plates of agar-solidified (1.5% agar w/v, BD Life Sciences) basal media were employed except that sodium succinate (10 mM, final conc.) or tryptone (0.5% w/v, BD Life Sciences) were used in place of MnCO3. Viable cells of this bacterium could rarely be retrieved from the co-culture as planktonic cells overlying Mn oxide nodules. For clonal isolation, 200 μl of a dense suspension of the dark Mn oxides produced by the co-culture were spotted onto the surface of succinate agar medium and allowed to dry, after which the Mn oxides were vigorously and heavily streaked over the agar surface and monitored for the development of colonies thereafter. After 3–5 days of incubation, colonies of Species B appeared small, leathery, and adherent to the agar surface; transfer of cells to new plates or liquid media was facilitated by the use of a sterile syringe needle for the removal of an entire single colony from the agar surface. In liquid, newly isolated strains of Species B can be grown with tryptone (0.5% w/v) or acetate (10 mM, final conc.) (Extended Data Fig. 6c,d), and can form fabric-like biofilms blanketing at the bottom of culture tubes. Transfer of such material proved challenging, as the fabric like biomass typically adhered to the insides of both plastic and glass pipette surfaces, leading to the rapid selection for strain variants that do not form flocs.
For the examination of anaerobic growth of Species B, 10 ml of basal medium is dispensed into 18-mm glass “Balch” tubes (Bellco, Vineland, NJ, USA) and stoppered with 1 cm butyl rubber stoppers under an N2 headspace. Autotrophic growth of the isolate using H2 + O2 + CO2 was examined similarly, except that an air headspace, periodically spiked with 1 ml of H2 + CO2 (80%/20%, v/v), was used. Colonies of Species B did not develop on standard lysogen broth (LB) agar, or on plates of the basal medium amended with 5 g/L yeast extract (BD Biosciences), traits that were used to monitor for culture purity or contamination.
For growth of manganese-oxidising lithotrophs in agarose tubes, the basal medium was amended to contain 150 mM freshly prepared MnCO3 and 0.38% w/v agarose (Aquapor LE Ultrapure; National Diagnostics, Atlanta, GA, USA). After steam sterilisation, 15 ml aliquots of the molten agar medium was dispensed into sterile 18-mm glass culture tubes fitted with plastic caps. After cooling to 45°C in a water bath, the molten agar was inoculated with 0.5 ml of a dense suspension of Mn oxides from an actively growing lithotrophic culture, gently vortexed, and allowed to harden on ice before incubation at 37°C. Tubes were sealed with parafilm to avoid desiccation over the long periods of incubation, and monitored for both clearing of MnCO3 as well as formation (or changes in the size, shape, and distribution) of Mn oxide nodules.
Examination of Mn oxide nodules
In all cases examined, the generation of the dark, granular product was coincident with the generation of Mn oxides from Mn(II)CO3, as determined via colourimetry53, FTIR-ATR spectroscopy (below), reactivity with H2O2, and/or ICP-MS (below).
To visualize, with minimal disturbance, cells on Mn oxide nodules generated in agarose tubes, cores of regions where the carbonate cleared and nodules developed were sampled with sterile glass Pasteur pipettes. Agar cores containing undisrupted nodules were extruded carefully into a plastic weight boat and soaked in the basal medium (without MnCO3) amended to contain 10 μg/μl acridine orange54 for 30 minutes. The dye solution was decanted, and the core was soaked for 30 minutes in buffered medium without dye. The core was carefully transferred to glass microscope slides and gently covered with coverslips before examination via light microscopy. Light microscopy of Mn oxide nodules in agarose cultures was performed using both a Zeiss Stemmi 2000-C stereomicroscope and a Zeiss Axioplan 2 Imaging Microscope fitted with a HBO 100 mercury arc lamp housing and an ebq 100 lamp power supply (Lighting & Electronics Jena, Jena, Germany). For epifluorescence microscopy of acridine orange stained nodules, a blue (FITC) long pass filter set was employed [excitation, filter D470/40 Lot 33820 (Chroma Technology, Bellows Falls, VT, USA); emission, long pass filter OG515 (515 nm cut-on; Schott, Elmsford, NY, USA)]. Photodocumentation was with Panasonic Lumix GH3 and G85 microfourthirds-lens-mount cameras, mounted onto the research microscope via a combination of Zeiss to c-mount (1.0 X D10ZNC; Diagnostic Instruments, Sterling Heights, MI, USA) and “c-mount to microfourthirds-mount” (Fotodiox, Gurnee, IL, USA) adaptors. RAW images were imported into Lightroom 9 (Adobe, San Jose, CA, USA). Global adjustments to RAW images were made in Lightroom, e.g. to improve clarity through the opaque agarose. The global changes to RAW images in this line of experiments involved correction for white balance and adjustments via the texture-, clarity-, dehazing-, sharpening-, and “colour-noise reduction”- sliders.
For mineralogical analyses, Mn oxide nodules were harvested from cultures and allowed to settle by gravity, washed three times with distilled water, incubated in consumer-grade concentrated bleach for 2 hours at room temperature (to remove organic biomass carbon), washed three times with distilled water, and then dried at 80 °C. Spectroscopic analyses were performed on Nicolet Magna 860 Fourier transform infrared (FTIR) spectrophotometer (Thermo Nicolet, Madison, WI, USA) with Attenuated Total Reflectance (ATR) accessory (Durascope, SensIR Technologies, Danbury, CT, USA), and Nicolet iS50 FTIR spectrophotometer (Thermo Nicolet) with GladiATR ATR accessory (Pike Technologies, Madison, WI, USA).
For scanning electron microscopy (SEM) of the Mn oxide nodules, samples were chemically fixed on ice for 3–4 hrs in the basal medium amended with 2.5% glutaraldehyde and HEPES (25 mM, pH 7.5). Fixed samples were washed twice in the same buffered solution but without glutaraldehyde, and the final pellet was resuspended in the buffered solution with 50% (v/v) ethanol. Dehydration series were performed by increasing the ethanol concentration every 15 min to 70, 90, and finally 100% (v/v). Critical point drying (Quorum Technologies, Newhaven, UK) was performed as follows: ethanol was replaced by liquid CO2 at a pressure of 55 bar at <10 °C. Fifteen minutes after reaching the critical point, the pressure was released slowly at >35 °C until ambient pressure was reached. The samples were then deposited onto double coated carbon conductive tape (Ted Pella, Redding, CA, USA), and in some cases, were sputter coated with 10 nm of Pt/Pd (80/20 by weight, Cressington, UK). SEM was performed using a field emission SEM (1550VP, Zeiss, Germany) with SE2 detector at an operating voltage of 10 kV.
Community analysis
An iTag 16S rRNA gene sequencing approach was employed to obtain microbial community profiles. For such, a 2 ml culture sample containing Mn oxide nodules was centrifuged at 6000 × g for 5 min. DNA was extracted from the pellets and quantified as above. The V4-V5 region of the 16S rRNA genes was amplified using primers 515F-Y (5’-GTG YCA GCM GCC GCG GTA A-3’) and 926R (5’-GGA CTA CNV GGG TWT CTA AT-3’)55 with Illumina adaptor overhangs56 added to the oligonucleotides. PCR amplification was performed using 7.5 µL of Q5 High Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA, USA), 0.75 µL of each forward and reverse primers (10 µM), 5 µL of PCR-grade water and 1 µL of DNA with a concentration about 1 ng/µL. PCR cycling condition was as following: initial denaturation at 98 °C for 2 min, 25 cycles of 98 °C for 10 sec, 54 °C for 20 sec, 72 °C for 20 sec, and a final extension of 72 °C for 2 min before cooling down to 6 °C. Duplicate PCR reactions for each samples were run; after confirming successful and comparable amplification using gel electrophoresis, the duplicates were combined for Illumina Nextera XT barcoding56 using PCR condition as above except in 25 µL reaction volume and 10 cycles with annealing at 66 °C. Barcoded samples were quantified using QuantIT PicoGreen dsDNA Assay (Thermo Fisher Scientific) on the C1000 Thermal Cycler with CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA), combined in equal molar amounts, and purified with QIAquick PCR Purification Kit (Qiagen). Sequencing was performed on the MiSeq platform (Illumina, San Diego, CA, USA) with paired 250 bp reads after PhiX addition of 15–20% (Laragen, Culver City, CA, USA).
Sequence data was processed using QIIME57 v1.8.0. Raw sequences were first joined and quality-trimmed using the default read pair joining and quality-trimming parameters. Processed sequences were clustered into de novo operational taxonomic units (OTUs) at 99% sequence identity using UCLUST58 v7.11.0.667, with the most abundant sequence picked as the representative sequence for each OTU. OTUs with only a single sequence (“singletons”) was removed. Taxonomic identification for the representative sequences were done using SILVA59 Ref NR 99 Database Release 119.
Cloning of near full-length 16S rRNA genes of Species A and Species B
Near full-length 16S rRNA genes were amplified and cloned from the co-culture. First, DNA was extracted as above. Second, 16S rRNA gene was amplified using primers BACT27F (5’-AGA GTT TGA TYM TGG CTC-3’) and U1492R (5’-GGY TAC CTT GTT ACG ACT T-3’) modified from published versions60. PCR was performed using the Expand High Fidelity System (Roche Molecular Systems, Pleasanton, CA, USA) with the following conditions: 2.5 µl of 10 × buffer, 0.35 µl of Taq polymerase, 0.55 µl of 10 mM dNTP, 0.50 µl of each forward and reverse primer, 18.6 µl of PCR-grade water and 2 µl of a DNA extract. The cycling condition was as following: 95 °C for 2 min, followed by 30 cycles of 94 °C for 15 sec, annealing at 54 °C for 30 sec and extension at 72 °C for 45 sec, and a final extension step at 72 °C for 4 min before cooling down to 4 °C. Third, the PCR products were immediately purified using QIAquick PCR purification kit (Qiagen), ligated into pGem-T Easy Vector (Promega, Madison, WI, USA), and the resulting plasmid was transformed into JM209 competent Escherichia coli cells (Promega) following the manufacturer’s instructions. Over 50 white colonies were observed on X-gal containing plates from 50 µl of transformed cells. Clones were grown overnight at 37 °C in LB medium with 10% glycerol and 0.1 mg/ml ampicillin following the manufacturer’s instructions (Promega). Lastly, PCR was performed using the NEB Taq Polymerase kit (New England Biolabs) with the following conditions: 3.0 μl of 10 × buffer, 0.66 μl of dNTP, 0.30 μl of Taq polymerase, 0.60 μl of each M13 forward and reverse primers (10 µM), 0.30 µl of 10 µg/µl bovine serum albumin, 23.95 μl of PCR water, and 0.60 μl of cells. The cycling conditions were the same as the cloning reaction above. The PCR products were purified using Multiscreen HTS PCR 96-well plates (Millipore, Billerica, MA, USA). Sanger sequencing was performed on the purified PCR products to confirm their sequence identities using both M13 forward and reverse primers (Laragen).
Fluorescence in situ hybridisation (FISH)
Three oligonucleotide probes were developed or tested in this study in order to visualize and differentiate cells representing Species A and Species B in Mn oxide nodules (Supplementary Note 3; Extended Data Fig. 10a–d). SILVA59 Ref NR 99 Database Release 128 and the PT Server function of ARB software61 v6.0.2 was used for the development. The Species A oligonucleotide probe (NLT499: 5’-ACA GAG TTA GCC GTG GCT-3’) was developed and designed to target the 16S rRNA genes from members of the classes Leptospirillia, Thermodesulfovibrionia, as well as other uncultivated classes within the phylum Nitrospirota62. Two oligonucleotide probes were used to target members of the order Betaproteobacteriales, including Species B. Previously, a probe (BETA359 or Beta1: 5’-CCC ATT GTC CAA AAT TCC CC-3’)63,64 had been reported, but the optimal hybridisation conditions were not. A second probe was designed for this study (BETA867: 5’-AGG CGG TCA ACT TCA CGC-3’). A previously developed probe that targets most all Bacteria (EUB338: 5’-GCT GCC TCC CGT AGG AGT-3’)65 was also employed for FISH. All probes were double labelled66: with Cy3 dye for NLT499, FITC dye for BETA359 and BETA867, and Cy5 dye for EUB338 (Integrated DNA Technologies, San Diego, CA, USA).
The Clone-FISH method67 was used to evaluate the specificity and hybridisation condition of these three oligonucleotide probes. Transformed JM209 E. coli (above) containing either 16S rRNA gene of Species A or Species B were grown overnight at 37 °C with shaking at 200 rpm in ampicillin-containing LB media. Plasmids were isolated using the QIAprep Spin Miniprep Kit (Qiagen) following manufacturer’s instructions. Isolated plasmids were then transformed into NovaBlue(DE3) E. coli competent cells (EMD Millipore, Billerica, MA, USA) following manufacturer’s instructions. 16S rRNA gene sequence of transformed NovaBlue(DE3) cells were confirmed as above by Sanger sequencing (Laragen). Cells for Clone-FISH were prepared as previously described67. Briefly, overnight cultures of OD600≈0.4 were induced with IPTG (1 mM) for 2 h, and chloramphenicol (170 mg l−1) was added to the cultures and incubated for 5 h. The cells were then harvested by centrifuging at 8000 × g for 10 min, fixed with 4% paraformaldehyde in 1 × PBS, washed twice with PBS and stored in PBS:ethanol (1:1) at −20 °C. FISH reactions were performed as previously described68 using a BX51 epifluorescence microscope (Olympus, Shinjuku, Japan) with 100× (UPlanFL N) oil immersion objective. Each point in the dissociation profile represents the mean of fluorescence intensities of at least 100 single cells in 5 microscopic fields evaluated using the software daime69 v2.1 with default automatic segmentation settings of threshold algorithm Isodata and manually size filtering resulting regions of interest for single cell analysis.
To prepare Mn oxide nodules for FISH visualisation, Superfrost Micro Slide (VWR Scientific, Radnor, PA, USA) were dipped in 0.2% UltraPure agarose (Thermo Fisher Scientific) at 45 °C. Mn oxide nodules were fixed with paraformaldehyde (above), except stored in PBS at 4 °C instead for no longer than 2 weeks. Fixed Mn oxide nodules in PBS were immediately pipetted onto the slide and cooled to room temperature. After this, the slide was dipped in freshly prepared DCBE buffer (0.05 M sodium dithionite, 0.1 M sodium citrate, 0.1 M sodium bicarbonate and 0.1 M EDTA at pH 7; a modification of DCB buffer70) for 5 min at room temperature to dissolve away the Mn, leaving nodule-associated cells constrained within the agarose. The cells were then permeabilized with protease K (15 µg/ml) for 10 min at room temperature, and lysozyme (10 mg/ml) for 30 min at 37 °C. The slide was washed once in 0.01 M HCl for 15 min and three times in water for 1 min each before drying at 37 °C. The permeabilisation and HCl washing is not necessary for FISH reaction to work, and can be omitted. FISH reactions, using NLT499-Cy3, BETA867-FITC, and EUB338-Cy5 probes, were performed in 35% formamide hybridisation buffer68 with corresponding salt concentration in the washing buffer68. DAPI-citifluor (5 μg/ml) was added to the FISH samples prior to epifluorescence microscopy using a BX51 epifluorescence microscope (Olympus) with 100× (UPlanFL N) oil immersion objective.
Stable isotope probing and nanoSIMS analyses
Mn-oxidising co-cultures were grown in basal medium containing Mn13CO3 and 15NO3− to visualize the assimilation of 13C and 15N into the cells of Species A and Species B. For these, 0.5 ml samples from of a stationary culture grown in basal medium with 26 mM unlabelled MnCO3 was transferred into 18-mm glass “Balch” tubes containing 9.6 ml of autoclaved basal media with 13 mM Mn13CO3 (prepared using 13C-NaHCO3, 99 atom % 13C, Cambridge Isotope Laboratories, Tewksbury, MA, USA). For N source, the media contained 1 mM Na15NO3 (98 atom % 15N, Sigma-Aldrich Isotec, #364606). The culture tubes were stoppered under ca. 20 ml of air headspace with 1 cm butyl rubber stoppers, and incubated at 37 °C without shaking until, by visual examination, no MnCO3 appeared to remain. Resulting Mn oxide nodules were fixed with paraformaldehyde, dissolved with DCBE buffer (above) by shaking at 37 °C for 1 hr, and filtered onto 0.2 µm polycarbonate filters (Isopore GTBP02500, Millipore) that have been sputter coated with 10 nm of Au. FISH reactions were performed on the filters (as describe above but without permeabilisation and HCl washing) in 25% formamide hybridisation buffer. As evaluated in Clone-FISH, no cross-reaction was observed in 25% formamide hybridisation buffer and may increase signal intensity. A culture grown in the presence of 13 mM unlabelled MnCO3 (prepared using NaHCO3, Thermo Fisher Scientific, #S233–500) and 1 mM Na15NO3 was used for comparison and as a control. DAPI-citifluor (5 μg/ml) was added to the FISH samples prior to epifluorescence microscopy using a Elyra S1 (Zeiss, Jena, Germany) fitted with a Plan-Apochromat 63×/1.4 Oil immersion DIC objective. In parallel, in order to both assess and to minimize the dilution of signal 13C and 15N in labelled biomass (e.g. by certain unlabelled reagents such as paraformaldehyde and reagents for FISH) during post-incubation sample preparations: Mn oxide nodules were dissolved as above with DCBE buffer but without paraformaldehyde fixation, with the resultant cells subsequently stained for epifluorescence microscopy with DAPI-citifluor only (i.e. no FISH reagents). However, the dissolution of nodules for analyses required the use of DCBE buffer which contains unlabelled citric acid, bicarbonate, and EDTA, residuum from which, if associated with biomass from the nodules, would artifactually lower estimates of the extent of the labelling especially for 13C. Finally, filters were washed in ethanol with increasing concentrations (50%, 80%, 100%) each for 3 minutes before mounting on nanoSIMS holder using double coated carbon conductive tape (Ted Pella).
NanoSIMS analysis was performed using a CAMECA NanoSIMS 50L at the Caltech Microanalysis Center. A focused primary Cs+ beam of 100–1000 pA was used for pre-sputtering the sample until 12C14N– counts stabilized, and 2–4 pA was used for image collection with rasters of 512 × 512 pixels. Secondary ions 12C12C−, 13C12C–, 12C14N–, 12C15N– and 32S− were measured simultaneously for at least 3 image frames. In one instance, a region was measured a second time with 55Mn16O− added to the analysis to confirm the absence of Mn in the sample. Individual ion image frames in the resulting data were aligned using the 12C14N− ion and epifluorescence microscopy images were transformed to match that of nanoSIMS ion images using Look@NanoSIMS71 v2019-05-14; regions of interest (ROIs) corresponding to cells of Species A or Species B were defined manually. Final ion counts per ROI were calculated by summation of ion counts over all image frames. Atom percent of 13C or 15N was calculated from the individual ion counts as described in Supplementary Note 9.
Kinetics of Mn(II) oxidation and cell growth
Glassware used in the kinetic experiments were acid washed with 3.7% HCl and combusted at 550 °C to eliminate residue metals and organic carbon. All cultures were incubated at 37 °C with shaking at 200 rpm.
The kinetic experiments were performed in 1 L flasks with 0.4 L of the basal media using commercial MnCO3 (Sigma-Aldrich) with a final content of 100 mM Mn(II) as determined using the ICP-MS method (below). The basal media contain either 1 mM NaNO3 (n=4) or 1 mM NH4Cl (n=5) as the N source. Each culture flask was inoculated with 5 ml of a Mn oxide slurry sampled from a flask of an active co-culture grown with 1 mM nitrate and 1 mM urea as the N source. Culture material was sampled from each flask daily for the first 36 days, followed by 3 final, well separated time points over the final 30 days. At each time point, culture flasks were removed from the shaking incubator and swirled. Immediately thereafter, a total of 3 ml of oxides and culture fluid mixture was aseptically sampled from the flask via 5 ml disposable pipette. Of this, 1 ml of the sample was saved at −80 °C for later ICP-MS analysis (below); 2 ml of the sample was centrifuged at 8000 × g for 5 min and the pellet was stored at −80 °C for later quantitative PCR analyses. DNA was extracted from the pellets and quantified as above.
Chemical analysis of Mn
Reduced Mn(II) and oxidised Mn(III)/Mn(IV) pools were measured using a previously described method6 and evaluated (Supplementary Note 4; Extended Data Fig. 10e). Briefly: 0.1 ml of oxide and culture fluid mixture was mixed with 0.9 ml of 0.5 N HCl. After reacting for at least 10 min, the mixture was centrifuged at 16100 × g for 3 min. The supernatant (“acid-soluble fraction”, representing Mn(II)) was pipetted out into a separate tube. The pellet (“acid-insoluble fraction”, representing Mn(III/IV)) was then reacted with 1 ml of 0.25 N NH2OH⋅HCl in 0.25 N HCl. The acid-soluble and acid-insoluble fractions were then centrifuged again at 16100 × g for 3 min to avoid any carryover. From each fraction, 0.1 ml was sampled and then diluted into 10 ml of 2% HNO3. The Mn contents were measured using an Agilent 8800 inductively coupled plasma mass spectrometer (ICP-MS; Agilent Technologies, Santa Clara, CA, USA) with the helium gas collision mode and quantified using a Mn standard solution (Sigma-Aldrich, Supelco 1.19789). Attempts were also made to measure total Mn content using the formaldoxime method72, and oxidised Mn(III)/Mn(IV) content using the leucoberbelin blue dye53,73. However, both methods resulted in underestimates of the manganese content when comparing to the ICP-MS method and standards. In large part, this was due to relatively large Mn oxide nodules be challenging to both dissolve and to react with those reagents to completion.
For determining dissolved Mn concentrations in particle free fluids associated with MnCO3 and spent cultures: fluids were sampled from uninoculated and inoculated cultures containing 20–100 mM MnCO3. From each sample, 0.1 ml was subsampled after centrifuging at 16000 × g for 3 min, subsequently filtered through a 0.22 µm filter, and each fraction dissolved into 10 ml of 2% HNO3. The ICP-MS measurements for both Mn(II) and Mn(III/IV) fractions were performed as above.
Quantitative PCR
To obtain standards and test quantitative PCR specificity, plasmids with 16S rRNA gene of either Species A or Species B were purified from transformed JM209 E. coli as above. Purified plasmids were then linearized using 150 units of restriction enzyme SacI in 50 µl reactions containing 1 × NEB Buffer 1 (New England Biolabs) overnight at 37 °C. The restriction digest reactions were heat inactivated at 65 °C for 20 min and purified using Multiscreen HTS PCR 96-well plates (Millipore). Concentration of the linearized and purified plasmids were quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) to make DNA template standards containing known copies of 16S rRNA genes of Species A or Species B.
To track the growth of all Bacteria in Mn-oxidising cultures, quantitative PCR assays were performed based on an assay developed and optimized previously74 (Supplementary Note 5; Extended Data Fig. 10g–j). All primers and probes were obtained from Integrated DNA Technologies (San Diego, CA, USA) and diluted in 10 mM Tris·HCl (pH 8.0). The forward and reverse primers used, BACT1369F1 (5’-CGG TGA ATA CGT TCC CGG-3’) and PROK1492R1 (5’-GGC TAC CTT GTT ACG ACT T-3’), were one version of the previous degenerate primers BACT1369F and PROK1492R. The TaqMan probe for prokaryotes, TM1389F (TM1389F-FAM-ZEN, 5’-CTT GTA CAC ACC GCC CGT C-3’), was modified on the 5’ with 6-FAM fluorophore, 3’ with Iowa Black FQ dark quencher and internally with ZEN quencher. Also, TaqMan probes that specifically target Species A and Species B were developed. The specific Species A TaqMan probe TM1484R-A (5’-ATC ACC AAT CAT ACC TTG GGT GCC TG-3’) was modified on the 5’ with HEX fluorophore, 3’ with Iowa Black FQ dark quencher and internally with ZEN quencher. The specific Species B TaqMan probe TM1484R-B (5’-GTC ACG AAC CCT GCC GTG GTA ATC-3’) was modified at the 5’ with Texas Red-X fluorophore and 3’ with Iowa Black RQ dark quencher. Optimized quantitative PCR reaction mixtures contain 10 µl of PrimeTime Gene Expression Master Mix (Integrated DNA Technologies), 1 µl of each forward and reverse primer (10 µM), 0.5 µl of each of the three TaqMan probes, 5.5 µl of PCR-grade water, and 1 µl of template DNA. The reactions were run in triplicates at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 sec and 62 °C for 30 sec, on the C1000 Thermal Cycler with CFX96 Real-Time System (Bio-Rad). The amplification efficiency was calculated [Amplification Efficiency = 10(−1/slope) – 1]. The upper and lower limits of quantification ranges were determined based on dilution of standards with amplification efficiencies in the log-linear phase between 90%−105%. To convert to cell number, the 16S rRNA gene copies quantified in quantitative PCR assays were divided by 1 or 2 for Species A or Species B, respectively, based on the number of 16S rRNA gene copy per genome.
Genomics
To obtain genomic DNA from Species B, a flask with 200 ml of the basal medium containing 5 g/L of both tryptone and yeast extract was inoculated with a single colony of isolated Species B from a succinate nitrate medium plate. The culture was grown at 37 °C with shaking at 200 rpm until early stationary phase, then harvested by centrifuging at 5250 × g for 30 min at room temperature. DNA from the cell pellets was extracted following the Bacterial genomic DNA isolation using CTAB protocol version 3 as described75. Ethanol-precipitated DNA was additionally purified using the PureLink PCR Purification Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. To obtain genomic DNA from the co-culture, Mn oxide nodules were harvested from an early stationary phase culture: 2 ml of culture containing ca. 0.15 g of Mn oxide nodules was centrifuged at 5000 × g for 10 min at room temperature. DNA was extracted from the pellet using the DNeasy PowerSoil kit (Qiagen) as above. DNA extract was purified and concentrated using Protocol A of the CleanAll DNA/RNA Clean-up and Concentration Micro Kit (Norgen Biotek, Thorold, Ontario, Canada) following the manufacturer’s instructions.
Purified genomic DNA samples (2–50 ng) were fragmented to the average size of 600 bp via use of a Qsonica Q800R sonicator (power: 20%; pulse: 15 sec on/15 sec off; sonication time: 3 min). Libraries were constructed using the NEBNext Ultra™ II DNA Library Prep Kit (New England Biolabs) following the manufacturer’s instructions. Briefly, fragmented DNA was end-repaired using a combination of T4 DNA polymerase, E. coli DNA Pol I large fragment (Klenow polymerase) and T4 polynucleotide kinase. The blunt, phosphorylated ends were treated with Klenow fragment (3’ to 5’ exo minus) and dATP to yield a protruding 3’ ‘A’ base for ligation of NEBNext Multiplex Oligos for Illumina (New England Biolabs) which have a single 3’ overhanging ‘T’ base and a hairpin structure. After ligation, adapters were converted to the ‘Y’ shape by treating with USER enzyme and DNA fragments were size selected using Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN, USA) to generate fragment sizes between 500 and 700 bp. Adaptor-ligated DNA was PCR amplified with 8 to 12 cycles depending on the input amount followed by AMPure XP bead clean up. Libraries were quantified with Qubit dsDNA HS Kit (Thermo Fisher Scientific) and the size distribution was confirmed with High Sensitivity DNA Kit for Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed on HiSeq2500 platform (Illumina) with paired 250 bp reads following manufacturer’s instructions. Base calls were performed with RTA v1.18.64 followed by conversion to FASTQ with bcl2fastq v1.8.4 (Illumina). In addition, reads that did not pass the Illumina chastity filter as identified by the Y flag in their fastq headers were discarded. The resulting reads were uploaded to the KBase platform76, trimmed using Trimmomatic77 v0.36 with default settings and adaptor clipping profile Truseq3-PE, and assembled using Spades78 v3.11.1 with default settings for standard dataset. Manual binning and scaffolding were performed using mmgenome v0.7.179, using differential coverage from isolated Species B vs the Species A + Species B co-culture, to generate genome bins for Species A and Species B. Trimmed reads were aligned to either the Species A or Species B genome bin using bowtie280 v2.3.4.1 with default settings. Finally, the resulting reads were reassembled using Spades78 v3.11.1 with --careful setting and manually binned with mmgenome79 v0.7.1 once again, excluding and contigs <500 bp. Reconstructed genomes were annotated using the IMG81 Microbial Genome Annotation and NCBI82 Prokaryotic Genome Annotation Pipelines; beta-barrel protein prediction was performed using PRED-TMBB83.
Phylogenetic analyses
For Species A phylogenies, 275 publicly available genome assemblies in NCBI Assembly Database82 (as of March 26, 2019) were analysed. These fell within the phylum Nitrospirae (Taxonomy ID 40117)84, which corresponded to the phylum under the headings Nitrospirota and Nitrospirota_A in the Genome Taxonomy Database (GTDB)62 v0.2.2. Genome assemblies with estimated completeness of <60% and contamination of >5% (based on CheckM85 v1.0.6) were excluded. For 16S rRNA gene phylogeny, 16s rRNA genes from the genome of Species A or Species B, as well as the genome assemblies were retrieved using CheckM85 v1.0.6 ssu_finder utility. Sequences less than 900 bp and contain more than 2 N were excluded. The 16S rRNA gene sequences were aligned using SINA86 v1.2.11 and imported into SILVA59 Ref NR 99 Database Release 128. 60 16S rRNA gene sequences, including 5 different outgroup sequences (Desulfovibrio vulgaris, Ramlibacter tataouinensis TTB310, Nitrospina gracilis 3/211, Acidobacterium capsulatum, Candidatus Methylomirabilis oxyfera), with 1532 nucleotide positions were exported with bacteria filter in the SILVA Database. Bayesian phylogenetic trees were constructed using MrBayes87 v3.2.6 with evolutionary model set to GTR + I + gamma, burn-in set to 25% and stop value set to 0.01, and edited in iTOL88. For concatenated multilocus protein phylogeny, marker proteins from 40 genomes including the same 5 outgroup species were identified and aligned using a set of 120 ubiquitous single-copy bacterial proteins in GTDB62 v0.2.2. The protein alignment was filtered using default parameters in GTDB62 v0.2.2 (the full alignment of 34744 columns from 120 protein markers were evenly subsampled with a maximum of 42 columns retained per protein; a column was retained only when the column was in at least 50% of the sequences, and contained at least 25% and at most 95% of one amino acid). The resulting alignment with 5036 amino acid positions was used to construct the multilocus protein phylogeny using MrBayes87 v3.2.6 as above except the evolutionary model was set to invgamma and a mixed amino acid model.
For Species B phylogenies, 60 publicly available genome assemblies were selected from the NCBI Assembly Database82 under the class Betaproteobacteria (Taxonomy ID 28216)84, which corresponded to the order Betaproteobacteriales in GTDB62 v0.2.2. These 60 genomes assemblies had >93% completeness and <3% contamination based on CheckM85 v1.0.6, and represented different genera. The same 5 outgroups were used as above, except Ramlibacter tataouinensis TTB310 was replaced with Nitrospira inopinata. Bayesian 16S rRNA gene phylogeny was performed as above with 1532 aligned nucleoic acid positions. Maximum likelihood multilocus protein phylogeny was constructed using 5035 amino acid positions using RAxML89 v8.1.7 with protein model GAMMALGF and rapid bootstrapping of 100 replicates.
For functional gene phylogeny, protein sequences were aligned using ClustalO90 v1.2.4 and maximum likelihood trees were constructed using RAxML89 v8.1.7 as above.
Transcriptomics
Glassware treatment and culturing condition for the transcriptomics experiments were as described in the kinetics experiments (above). Three replicates from different, actively Mn-oxidising cultures were used as inocula for RNA experiments. A 5% v/v inoculum from two active Mn-oxidising cultures was transferred into triplicate flasks containing the basal medium amended with 100 mM freshly prepared MnCO3, for a total of 6 flasks (sample IDs: Mn03/Mn06/Mn08 and Mn09/Mn10/Mn11). A 1% v/v inoculum from another active Mn-oxidising culture was transferred into a 7th flask of the same medium (sample ID Mn12). For harvesting biomass before complete oxidation of Mn(II) (Extended Data Fig. 3h), shaking of the incubator was paused for 3 minutes to allow Mn oxide nodules to settle by gravity. The overlaying medium was then decanted off until about 10 ml of culture and oxides remained. First 35 ml and then 15 ml of LifeGuard Soil Preservation Solution (Qiagen) were added, sequentially, to maximize transfer efficiency of Mn oxide nodules from the culture into a new 100 ml centrifuge tube. The culture-LifeGuard mixture was stored at 4 °C for less than a week, a duration for which RNA was expected to remain well preserved, according to the manufacturer. Initially, a series of different trials were performed [using multiple methods and variations, including the RNeasy Mini Kit (Qiagen), the RNeasy Powersoil Kit (Qiagen), and a previous customized procedure91] to directly extract RNA from Mn oxide nodules. These all failed to extract measurable RNA from samples. When an early chemical dissolution step was added to the protocol extraction of RNA from Mn oxide nodules was successful in combination with the procedures below. Before RNA extraction: the culture-Lifeguard mixture was first centrifuging at 5250 × g for 10 min at 4 °C. 400 ml of freshly prepared 0.22 µm filtered DCBE solution (recipe above) was added to the pellet and incubated at 37 °C with shaking at 200 rpm for 10 min to dissolve the Mn oxide nodules. The dissolved samples were then centrifuged at 5250 × g for 10 min at 4 °C. The supernatant was decanted, and the remaining 5 ml material was mixed with 10 ml of 0.22 µm filtered and autoclaved CBE solution (similar to the DCBE solution, but without sodium dithionite). The mixture was then centrifuged at 5250 × g for 10 min at 4 °C. The supernatant was discarded, and 0.9 ml of RLT solution with 2-mercaptoethanol from the RNeasy Mini Kit (Qiagen) was added to the pellet. The mixture was transferred into Glass 0.1 mm PowerBead Tubes (Qiagen) and bead beat using FastPrep FP120 (Thermo Electron Corporation) at setting 5.5 for 45 s. RNA extraction then proceeded using the RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. DNA from the extracted RNA was removed using the DNase Max Kit (Qiagen). The RNA extracts were purified and concentrated using Protocol C of the CleanAll DNA/RNA Clean-up and Concentration Micro Kit (Norgen Biotek) and stored at −80 °C until sequencing library preparation.
For RNA sequencing library preparation, RNA integrity was first assessed using RNA 6000 Pico Kit for Bioanalyzer (Agilent Technologies). RNA-seq libraries were constructed using NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs) following manufacturer’s instructions. Briefly, 1–10 ng of total RNA was fragmented to the average size of 200 bp by incubating at 94 °C for 15 min in first strand buffer, cDNA was synthesized using random primers and ProtoScript II Reverse Transcriptase followed by second strand synthesis using NEB Second Strand Synthesis Enzyme Mix. Resulting DNA fragments were end-repaired, dA tailed and ligated to NEBNext hairpin adaptors (New England Biolabs). After ligation, adaptors were converted to the ‘Y’ shape by treating with USER enzyme and DNA fragments were size selected using Agencourt AMPure XP beads (Beckman Coulter) to generate fragment sizes between 250 and 350 bp. Adaptor-ligated DNA was PCR amplified followed by AMPure XP bead clean up. Libraries were quantified with Qubit dsDNA HS Kit (Thermo Fisher Scientific), and the fragments were determined to a mean of 350 bp with standard deviation of 70 bp using High Sensitivity DNA Kit for Bioanalyzer (Agilent Technologies). Sequencing was performed on the HiSeq2500 platform (Illumina) with single-end 50 and 100 bp reads following manufacturer’s instructions. Base calls were performed with RTA 1.18.64 followed by conversion to FASTQ with bcl2fastq 1.8.4 (Illumina). Low quality reads and TruSeq3-SE adaptors were removed using Trimmomatic77 v0.36 with default settings. rRNA was removed using sortmerna92 v2.0 with supplied SILVA databases and default settings. Read mapping of the non-rRNA was performed using kallisto93 v0.44.0 with 100 bootstraps, fragment mean of 130 bp and standard deviation of 70 bp. Despite that the fragment mean was determined to be 230 bp using Agilent Bioanalyzer above after adaptor removal (120 bp), this caused issue in evaluating gene expression of genes smaller than 230 bp in kallisto93 v0.44.0. The fragment mean was therefore decreased while not affecting the overall transcript expression (Extended Data Fig. 10f). Final analysis and normalisation of different RNA samples was performed using sleuth94 v0.30.0, with either the reconstructed genomes of both species or only of Species B. Supplementary Table 5 includes RNA processing statistics of the processed files where reads mapping was done using BBMap (https://sourceforge.net/projects/bbmap/) v37.93 with minid=0.97 ambiguous=toss settings.
Data availability.
All sequencing data has been deposited at National Center for Biotechnology Information (NCBI) under BioProject PRJNA562312. The cloned 16S rRNA gene sequences of “Ca. Manganitrophus noduliformans” (Species A) and Ramlibacter lithotrophicus (Species B) from the co-culture have been deposited at GenBank under accession numbers MN381734 and MN381735, respectively. The iTAG sequences from 3 different enrichments has been deposited at SRA under accession numbers SRR10031198-SRR10031200. Genome sequences of the co-culture, from which the genome of Ca. M. noduliformans was reconstructed, have been deposited under BioSample SAMN12638105 with raw sequences deposited at SRA under accession number SRR10032644; the reconstructed genome of Ca. M. noduliformans has been deposited at DDBJ/ENA/GenBank under accession number VTOW00000000. Genome sequences of R. lithotrophicus strain RBP-1 have been deposited under BioSample SAMN12638106 with raw sequences deposited at SRA under accession number SRR10031379; the reconstructed genome of R. lithotrophicus strain RBP-1 has been deposited at DDBJ/ENA/GenBank under accession number VTOX00000000. Additionally, reconstructed genomes have been deposited in Joint Genome Institute (JGI) Genomes Online Database Study ID Gs0134339, with Integrated Microbial Genome ID 2784132095 for Ca. M. noduliformans and ID 2778260901 for R. lithotrophicus strain RBP-1. Transcriptome sequence data for the 7 biological replicates have been deposited at SRA under accession numbers SRR10060009, SRR10060010, SRR10060011, SRR10060012, SRR10060013, SRR10060017, SRR10060018. Unique biological materials are available upon reasonable request.
Extended Data
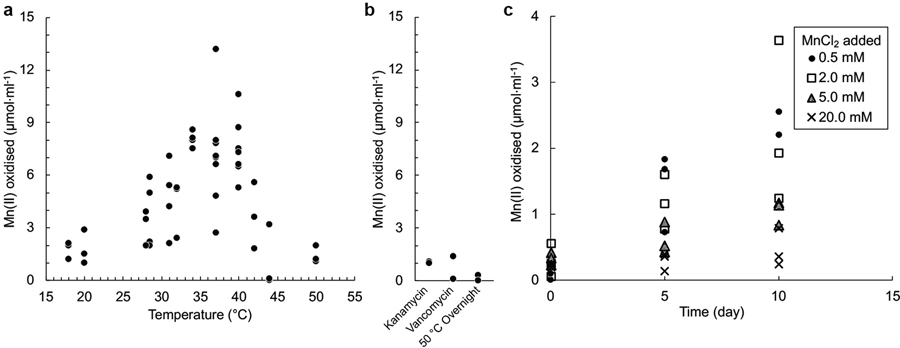
Extended Data Fig. 1 |. Effect of temperature, anti-bacterials, and Mn(II)Cl2 on biological Mn(II)CO3 oxidation.
a, Incubation temperature influences oxidation. A optimum between 34 °C and 40 °C was observed, but above these temperatures oxidation was inhibited. In contrast, non-biological reactions would generally be predicted to continue to increase in rate with increasing temperature. b, Sensitivity of Mn(II) oxidation to the presence of either of two antibiotics, or to prior pasteurisation before extended incubation at 32 °C. c, When amended to active co-cultures at concentrations >2.0 mM, MnCl2 appeared to inhibit MnCO3 oxidation when an active culture containing 2.2 mM unreacted MnCO3 was used as the inoculum. Number of points for each experimental condition represent independent cultivation experiments.
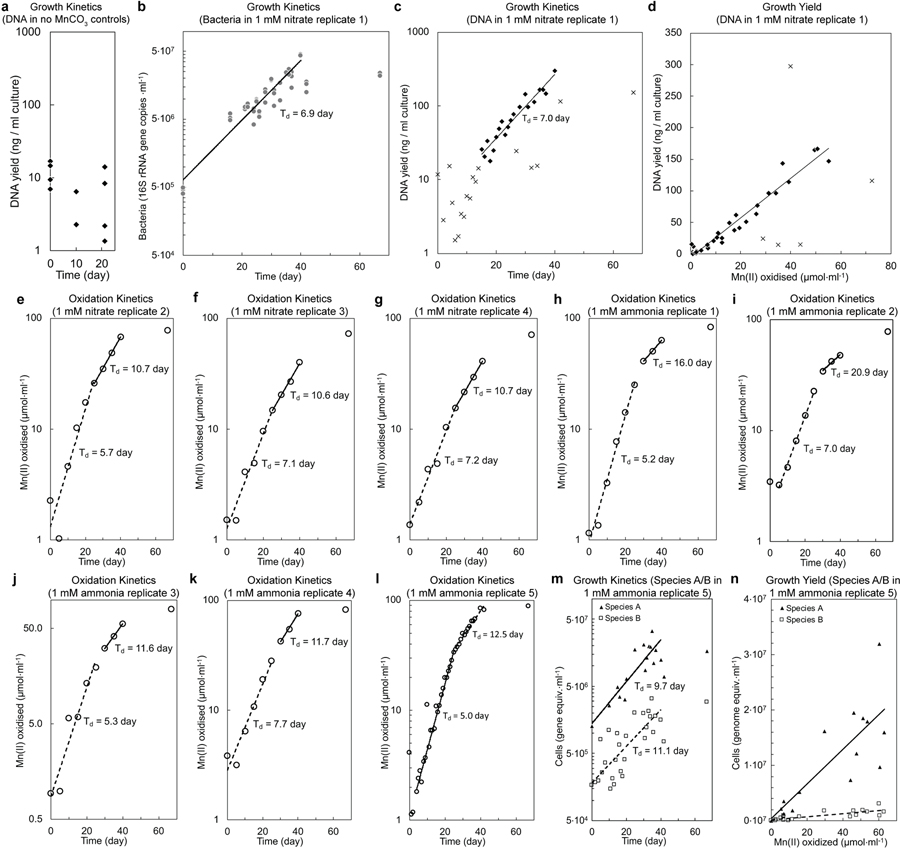
Extended Data Fig. 2 |. Mn(II) oxidation and growth by the co-culture.
a, DNA yield of the two species co-culture incubated in MOPS-buffered basal media in the absence of Mn(II) substrate. No statistically significant changes in the mean DNA yields (p=0.06, day 0 vs 10; p=0.70, day 10 vs 21; p=0.20, day 0 vs 21; two-tailed t-test with equal variance) are observed over the incubation period. b, c, Exponential increase in Bacteria and biomass yields in a Mn(II)-oxidising culture, which is coupled to exponential increases Mn(II) oxidation (same culture analysed in Fig. 2). Bacteria was measured via 16S rRNA gene copies using a general Bacteria probe in quantitative PCR; points represent 3 technical replicates. Biomass was measure via DNA yield from same culture volumes. d, Exponential increases in Mn(II) oxidation (Fig. 2a) and DNA yields by this same culture (1 mM nitrate replicate 1, c) correlate. Similar relationships were observed in samples from independent cultivation experiments (n=2). e-l, Kinetics of Mn(II) oxidation by the co-culture in basal media; two phases of exponential Mn(II) oxidation were observed. e-g, Basal media with 1 mM nitrate (n=4; for replicate 1, see b–d and Fig. 2). h-l, Basal media with 1 mM ammonia (n=5). m, Exponential growth of Species A and Species B paralleled Mn(II) oxidation in basal medium with 1 mM ammonia as N source (1 mM ammonia replicate 5, l), rather than 1 mM nitrate. n, Linear relationship between cell growth and the amount of Mn(II) oxidised (1 mM ammonia replicate 5, l and m). Values in n were normalised by subtracting the initial cell number and Mn oxide concentrations at the onset of the experiment, and negative values after normalisation were excluded from the analysis. All data points included in the line fits are used to calculate the doubling times (Td), unless otherwise noted by x symbols.
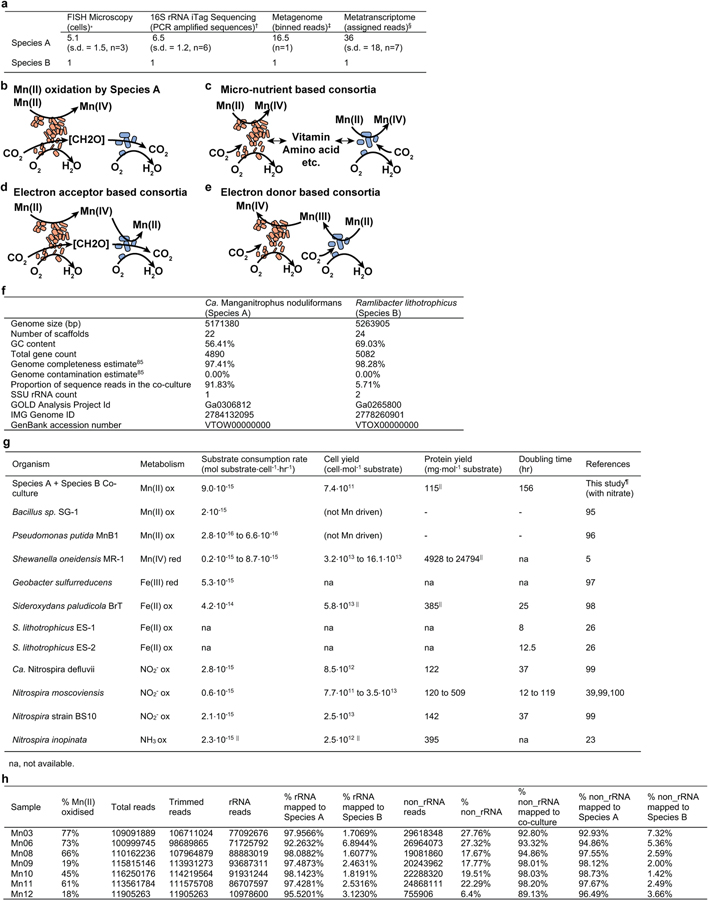
Extended Data Fig. 3 |. Properties of the refined co-culture.
a, Estimations of the relative ratio between Species A and Species B. *Slow-growing microbes, in particular Species A (which also has a smaller cell volume than Species B or Escherichia coli) could have lower number of ribosomes, resulting in lower signal intensity from rRNA-targeted fluorescent probes, relative to the fluorescent signal from DNA stain DAPI. †The two species together account for 99.7% of assigned sequence reads (Supplementary Table 1). ‡The two species together account for 97.54% of the sequence reads in the metagenome (panel f). §The two species together account for 99.576% (s.d. = 0.005%, n=7) of the rRNA sequence reads and 100.1700% (s.d. = 0.0005%, n=7) of the non-rRNA sequence reads in the co-culture metatranscriptomes (panel h). b-e, Possible metabolic interactions that may be occurring between Species A (orange) and Species B (blue). f, Genome statistics for Species A and Species B. g, Observed rates and yields of Mn(II) oxidation by the co-culture, in comparison to the literature values reported for other physiologically or phylogenetically related lithotrophs or metal-active heterotrophs. ||Conversion estimate based on Escherichia coli biomass of 2.8⋅10−13 g dry cell weight per cell, of which 55% is protein27. ¶Co-culture values correspond to results from the single independent culture with nitrate as N source for which extensive data on both oxidation kinetics and growth (genome copies) were collected. h, Transcriptome statistics for 7 co-cultures sampled at different degrees of Mn(II) oxidation.
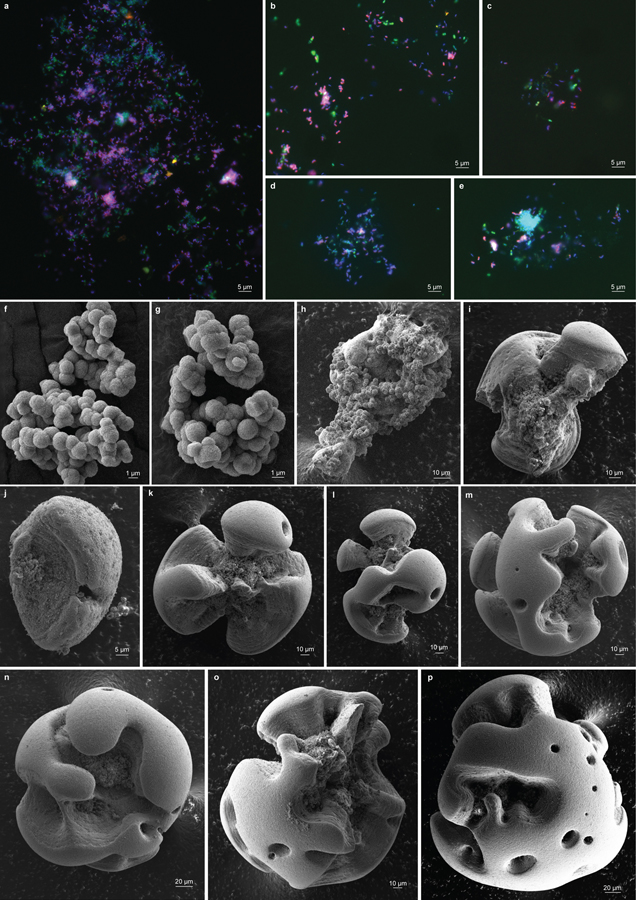
Extended Data Fig. 4 |. Microscopy of Mn oxide nodules formed by the co-culture.
a–e, Epifluorescence microscopy reveals distribution of cells of Species A and Species B associated with dissolved Mn oxide nodules. DAPI (blue) was used to stain DNA, in addition to applying species-specific FISH probes targeting the 16S rRNA of Species A (magenta) and Species B (green). Probe fluorescence for Species A was both dim and faded rapidly, but was associated with the cells that otherwise appear in photomicrographs to only be DAPI stained. No third species is present, as observed in independent cultivation experiments (n=2), and confirmed via independent methods (Extended Data Fig. 3a). Scanning electron micrographs of, f, g, Mn(II)CO3 substrate and, h-p, Mn oxide nodules harvested from liquid cultures. Representative nodules are from independent cultivation experiments (n=4).
Extended Data Fig. 5 |. Phylogenetic analyses on Species A.
a, 16S rRNA gene phylogram, based on a Bayesian analysis of 1532 aligned nucleotide positions. NCBI82 taxonomic classifications are used, and sequences shown are all from the phylum Nitrospirae. The names and known physiologies for the previously described genera in this phylum are shown on the right. NCBI accession numbers for 16S rRNA sequences are included in the node names. Source environment for the sequences are shown in brackets. b, Multilocus phylogram, based on a Bayesian analysis of 5036 aligned amino acid positions concatenated from 120 bacterial protein markers62. GTDB62 taxonomic classifications are used, and sequences shown are from the phylum Nitrospirota/Nitrospirota_A. The names and known physiologies for the previously described classes in this phylum are shown on the right. NCBI accession numbers for genome assemblies are included in the node names. For both a and b, the dots on the branches indicate posterior probabilities greater than 0.80. c, Phylogenetic analyses of the phylum Nitrospirae (Nitrospirota) limited to only those species having reconstructed genomes yield a different topology from that observed in panel a and Fig. 3a. Bayesian phylogram based on 1532 aligned 16S rRNA nucleotide positions (left); multilocus Bayesian phylogram, based on 5036 aligned amino acid positions of 120 concatenated bacterial protein markers (right). Sequences clustering within the three previously described classes within this phylum are collapsed into separate nodes. d, Protein sequence phylogeny of dihydroxy-acid and 6-phosphogluconate dehydratases. Sequences were selected based on a previous study101, with the addition of homologs found in Nitrospira inopinata, Leptospirillum ferriphilum and Species A (red). All 770 aligned amino acid positions were used in the maximum likelihood analysis. Protein accession numbers from the NCBI database or gene IDs from the IMG database of the 3 new sequences are shown in parentheses. Black dots on the branches represent bootstrap values equal to 100%. While dihydroxy-acid dehydratase and 6-phosphogluconate dehydratase are homologous, they form separate clusters phylogenetically as reported101. The homologs in Nitrospirae all belong to the dihydroxy-acid dehydratase clade, therefore are unlikely candidates for 6-phosphogluconate dehydratase activity and function in the ED pathway. All scale bars = evolutionary distance (0.1 substitutions-per-site average).
Extended Data Fig. 6 |. Phylogenetic analyses and aerobic heterotrophic growth of isolated Species B.
a, 16S rRNA gene phylogram, based on a Bayesian analysis of 1532 aligned nucleotide positions. NCBI82 taxonomic classifications are used, with sequences selected from the class Betaproteobacteria. The genus Ramlibacter, consistently identified in two phylogenetic approaches, is shaded in grey, with Species B in bold. Source environments for the species in Ramlibacter are shown in brackets. The order and family classifications are included to the right separated by a semicolon. The black dots on the branches indicate posterior probabilities great than 0.90. b, Multilocus phylogram, based on a maximum-likelihood analysis of 5035 aligned amino acid positions concatenated from 120 bacterial protein markers62. GTDB62 taxonomic classifications are used, and sequences shown are from the order Betaproteobacteriales. The GTDB family classifications are included to the right of species names. NCBI accession numbers for 16S rRNA sequences or the genome assemblies are included after the species names. The black dots on the branches indicate bootstrap values greater than 90%. Scale bars = evolutionary distance (0.1 substitutions-per-site average). c, d, Kinetics of Species B growth basal media with either c, 5 g/L of tryptone (n=3 biological replicates), or d, 10 mM acetate (n=2 biological replicates).
Extended Data Fig. 7 |. Phylogenetic analyses of cytochrome bd oxidase subunit I and cytochrome bd-like oxidases.
Only cytochrome bd-like oxidases were identified in Species A, in contrast to other classes in the phylum Nitrospirae (Nitrospirota). a, Unrooted maximum-likelihood tree, constructed using 242 amino acid positions shared between cytochrome bd and bd-like oxidases, using RAxML89 (model LGF). Deduced proteins from the genome of Species A are in red, with their IMG gene IDs and clade numbering (as shown in Fig. 3b) included in brackets. Other proteins from the phylum Nitrospirae (Nitrospirota) are coloured blue, orange or brown for classes Nitrospiria, Leptospirillia, or Thermodesulfovibrionia, respectively. Cytochrome bd oxidase of Species B, with its IMG ID, is in green; it belongs to the cyanide insensitive oxidase clade in purple. b, Phylogenetic analysis of cytochrome bd-like oxidases from Species A. Unrooted maximum-likelihood tree was constructed using 242 amino acid positions shared between different clades of cytochrome bd-like oxidases. Cytochrome bd-like oxidases are assigned to different clades, based on the phylogeny and their gene cluster structures. Species A encodes 8 cytochrome bd-like oxidases (bold), representing clades I, II, IIIb, Va and Vb; clade numbering as shown in Fig. 3b are included in brackets after the IMG IDs. Black dots on branches represent bootstrap values greater than 90%. Scale bars = evolutionary distance (substitutions-per-site average).
Extended Data Fig. 8 |. Sequence alignment of cytochrome bd and bd-like oxidases.
Cytochrome bd-like oxidase in Species A (sequence names starting with A, followed by their IMG gene ID and clade numbering as shown in brackets in Fig. 3b) and cytochrome bd oxidase subunit I in Species B (sequence name starting with B, followed by its IMG gene ID) are aligned to characterized cytochrome bd oxidases in Escherichia coli (sequence name starting with Eco, followed by its NCBI ID) and Geobacillus thermodenitrificans (sequence name starting with Geo, followed by its NCBI ID). Key features as revealed by structure102 are indicated at the top of the alignment, using E. coli protein residue numbering. The cytochrome bd oxidase subunit I sequence from Species B shows conservation of all key residues. In contrast, cytochrome bd-like oxidases in Species A do not show conservation of many key residues; instead, they are predicted to have 14 transmembrane helixes (compared to 9 in E. coli). One cytochrome bd-like oxidase in Species A has a C-terminus extension with a heme c binding motif (CXXCH).
Extended Data Fig. 9 |. Stable isotope probing of Mn(II)-oxidising co-culture measured using nanoSIMS.
a, Summary of stable isotope probing analysis of cells dissolved from Mn oxide nodules, either with paraformaldehyde fixation and FISH, or without (to avoid dilution with natural abundance isotopes). Cells of Species A and Species B were either identified by FISH or by elemental composition (Species B cells were observed to have higher 14N/15N ratios), and their isotopic compositions were obtained via nanoSIMS (n=the total number of cell regions of interest analysed in the nanoSIMS images). For FISH-nanoSIMS analyses, a total of 2 and 5 nanoSIMS images from single cultures incubated with either MnCO3 or Mn13CO3, respectively, were examined. For nanoSIMS analyses without paraformaldehyde fixation and FISH, a total of 3 and 17 nanoSIMS images from single cultures incubated with either MnCO3 or Mn13CO3, respectively, were examined. b–u, Individual secondary ion images from nanoSIMS showing incorporation of inorganic 13C and 15N into the cells of both species (dissolved from Mn oxide nodules grown in the presence of b–k, MnCO3 and 15NO3−, or l–u, Mn13CO3 and 15NO3−.), and Species B cells could have higher 14N content than Species A. Secondary ions 12C2 (mass 24 for 12C), 13C12C (mass 25 for 13C), 14N12C (mass 26 for 14N), 15N12C (mass 27 for 15N), 32S (mass 32 for 32S) were simultaneously measured. The counts of the secondary ions are shown in brackets [minimum maximum] and displayed using the colour scale shown on the right of the images. b–f and l–p correspond to the top and bottom panels in Fig. 4, respectively. White arrows indicate Species B cells identified in FISH showing high 14N in nanoSIMS. v, NanoSIMS measurement of residual Mn associated with cells grown with Mn13CO3 and 15NO3−, after dissolving from Mn oxide nodules. Same nanoSIMS image area was analysed as l–p, except 55Mn16O (mass 71 for 55Mn) was measured (n=1 nanoSIMS image) in addition to other secondary ions. Negligible amount of Mn was found in the biomass, indicating that any remaining Mn13CO3 substrate had been completely dissolved away during sample preparation, and thus did not interfere with the 13C analyses.
Extended Data Fig. 10 |. Evaluations of experimental methods.
a, b, c, d, Evaluation of FISH oligonucleotide probes. Three probes (NLT499=●, BET359=○, and BET867=□) were tested in different probe combinations and formamide concentrations, using 16S rRNA gene clones of Species A (a, b) or Species B (c, d). Each point in the dissociation profile represents the mean of fluorescence intensities of at least 100 different single cells in 5 distinct microscopic fields of 1 biological replicate. Lines connect the 95% confidence intervals of the points. No interference was found when targeting either Species A or Species B with different probe combinations and formamide concentrations. RU = relative units of fluorescence intensity. e, Evaluation of ICP-MS method to measure Mn compounds with different oxidation states. Mn(II) in its various forms can be almost entirely measured in the acid-soluble fraction with little in the acid-insoluble fraction, and any increase in the acid-insoluble fraction is an indication of oxidised Mn(II). In this study, we refer to the “acid-soluble fraction” as Mn(II), and the “acid-insoluble fraction” as Mn(II) oxidised representing Mn(III/IV). See Supplementary Note 4 for more details. f, Evaluation of transcriptome analysis software kallisto93. Average fragment length for RNA libraries was measured to be 230 bp. However, using 230 bp as the input parameter for fragment length caused a kallisto93 expression evaluation issue for genes <230 bp in length; thus, the fragment length was adjusted downward to 100 bp in order to evaluate the expression of genes <230 bp. This parameter change does not affect the overall transcript expression for genes >230 bp as seen in the correlation analysis, performed using transcriptome sample Mn03. g, h, Evaluation of quantification range and efficiency of quantitative PCR oligonucleotide probes. Three quantitative PCR oligonucleotide probes (g, Bacteria, or h, Species A or Species B specific) were tested using cloned 16S rRNA gene of either Species A (open squares, solid lines) or Species B (open triangles, dashed lines) as DNA templates. Threshold cycle (CT) versus gene copies show that all three probes had working efficiencies between 90–105% in the quantification ranges plotted. Points represent 3 technical replicates. i, j, Evaluation of specificity of quantitative PCR oligonucleotide probes. The percentage of i, Species A, and j, Species B, was estimated in reactions containing a mixture of cloned 16S rRNA genes from both Species A and Species B as DNA templates. Dashed lines represent theoretical 100% match in the expected versus measured values. The results indicate that the species-specific probes quantified their targeted species with minimal interference. Points represent 4 technical replicates.
Supplementary Material
Acknowledgements
This work was supported by NASA Astrobiology Institute Exobiology Grant #80NSSC19K048; and by Caltech’s Center for Environmental Microbial Interactions and Division of Geological & Planetary Sciences. We thank Caltech colleagues Stephanie Connon for assistance with iTag sequencing preparations; George Rossman and Usha Lingappa for spectroscopic analyses and minerology insights; Grayson Chadwick for discussions on physiology and bioenergetics; Igor Antoshechkin and Vijaya Kumar for assistance with nucleic acid library preparation and sequencing at the Millard and Muriel Jacobs Genetics and Genomics Laboratory; Nathan Dalleska for assistance with ICP-MS analyses at the Environmental Analysis Center; Fan Gao for inputs on RNA data analysis using kallisto software at the Bioinformatics Resource Center in the Beckman Institute; Chi Ma for assistance with SEM analyses at the GPS Analytical Facility; and Yunbin Guan for assistance with nanoSIMS analyses at the GPS Microanalysis Center. We are grateful to multiple colleagues and the five reviewers for feedback prior to publication.
Footnotes
Supplementary Information is available in the online version of the paper.
Completing interests The authors declare no competing financial interests.
References
- 1.Beijerinck M Oxydation des mangancarbonates durch Bakterien und Schimmelpilze. Folia Microbiol Delft (1913).
- 2.Nealson KH, Tebo BM & Rosson RA Occurrence and mechanisms of microbial oxidation of manganese. Adv. Appl. Microbiol 33, 279–318 (1988). [Google Scholar]
- 3.Tebo BM, Johnson HA, McCarthy JK & Templeton AS Geomicrobiology of manganese(II) oxidation. Trends Microbiol 13, 421–428 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Hansel C & Learman DR Geomicrobiology of manganese in Ehrlich’s Geomicrobiology (eds. Ehrlich HL, Newman DK & Kappler A) 401–452 (CRC Press, 2015). 10.1201/b19121. [DOI] [Google Scholar]
- 5.Myers CR & Nealson KH Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Lovley DR & Phillips EJ Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol 54, 1472–1480 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winogradsky S Über schwefelbakterien. Bot Ztg 45, 489–ff (1887). [Google Scholar]
- 8.Kelly DP & Wood AP The chemolithotrophic prokaryotes in The Prokaryotes: Prokaryotic Communities and Ecophysiology (eds. Rosenberg E, DeLong EF, Lory S, Stackebrandt E & Thompson F) 275–287 (Springer Berlin Heidelberg, 2013). 10.1007/978-3-642-30123-0_63. [DOI] [Google Scholar]
- 9.Daims H et al. Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Könneke M et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Strous M et al. Missing lithotroph identified as new planctomycete. Nature 400, 446–449 (1999). [DOI] [PubMed] [Google Scholar]
- 12.van Kessel MAHJ et al. Complete nitrification by a single microorganism. Nature (2015) 10.1038/nature16459. [DOI] [PMC free article] [PubMed]
- 13.Watson SW & Waterbury JB Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch. Für Mikrobiol 77, 203–230 (1971). [Google Scholar]
- 14.Lovley DR, Holmes DE & Nevin KP Dissimilatory Fe(III) and Mn(IV) reduction in 219–286 (Elsevier, 2004). 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- 15.Henkel JV et al. A bacterial isolate from the Black Sea oxidizes sulfide with manganese(IV) oxide. Proc. Natl. Acad. Sci 116, 12153–12155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghiorse WC & Ehrlich HL Microbial biomineralization of iron and manganese. Catena Suppl 21, 75–99 (1992). [Google Scholar]
- 17.Ehrlich HL & Salerno JC Energy coupling in Mn2+ oxidation by a marine bacterium. Arch. Microbiol 154, 12–17 (1990). [Google Scholar]
- 18.Ehrlich HL Manganese as an energy source for bacteria. Environ. Biogeochem 633–644 (1976).
- 19.Dick GJ et al. Genomic insights into Mn(II) oxidation by the marine alphaproteobacterium Aurantimonas sp. Strain SI85–9A1. Appl. Environ. Microbiol 74, 2646–2658 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nealson KH The Manganese-oxidizing bacteria in The Prokaryotes (eds. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H & Stackebrandt E) 222–231 (Springer; New York, 2006). 10.1007/0-387-30745-1_11. [DOI] [Google Scholar]
- 21.van Veen WL Biological oxidation of manganese in soils. Antonie Van Leeuwenhoek 39, 657–662 (1973). [DOI] [PubMed] [Google Scholar]
- 22.Morgan JJ Kinetics of reaction between O2 and Mn(II) species in aqueous solutions. Geochim. Cosmochim. Acta 69, 35–48 (2005). [Google Scholar]
- 23.Kits KD et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 65, 3690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flagan SF & Leadbetter JR Utilization of capsaicin and vanillylamine as growth substrates by Capsicum (hot pepper)-associated bacteria. Environ. Microbiol 8, 560–565 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Kanzler BEM, Pfannes KR, Vogl K & Overmann J Molecular characterization of the nonphotosynthetic partner bacterium in the consortium “Chlorochromatium aggregatum”. Appl. Environ. Microbiol 71, 7434–7441 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emerson D & Moyer C Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol 63, 4784–4792 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidhardt FC Escherichia coli and Salmonella: Cellular and Molecular Biology vol. 1 (ASM Press, 1996). [Google Scholar]
- 28.Kostanjšek R, Pašić L, Daims H & Sket B Structure and community composition of sprout-like bacterial aggregates in a dinaric karst subterranean stream. Microb. Ecol 66, 5–18 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Wrighton KC et al. Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla. Science 337, 1661–1665 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Parks DH et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol 2, 1–1542 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Castelle C et al. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem 283, 25803–25811 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeans C et al. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J 2, 542–550 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Croal LR, Jiao Y & Newman DK The fox operon from Rhodobacter strain SW2 promotes phototrophic Fe(II) oxidation in Rhodobacter capsulatus SB1003. J. Bacteriol 189, 1774–1782 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao Y & Newman DK The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol 189, 1765–1773 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He S, Barco RA, Emerson D & Roden EE Comparative genomic analysis of neutrophilic iron(II) oxidizer genomes for candidate genes in extracellular electron transfer. Front. Microbiol 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson DJ et al. The ‘porin–cytochrome’ model for microbe-to-mineral electron transfer. Mol. Microbiol 85, 201–212 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Luther GW III Manganese(II) Oxidation and Mn(IV) Reduction in the Environment—Two One-Electron Transfer Steps Versus a Single Two-Electron Step. Geomicrobiol. J 22, 195–203 (2005). [Google Scholar]
- 38.Lücker S et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci 107, 13479–13484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mundinger AB, Lawson CE, Jetten MSM, Koch H & Lücker S Cultivation and transcriptional analysis of a canonical Nitrospira under stable growth conditions. Front. Microbiol 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch H et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345, 1052–1054 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Levicán G, Ugalde JA, Ehrenfeld N, Maass A & Parada P Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations. BMC Genomics 9, 581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg IA Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol 77, 1925–1936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thauer RK, Jungermann K & Decker K Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev 41, 100–180 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baradaran R, Berrisford JM, Minhas GS & Sazanov LA Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chadwick GL, Hemp J, Fischer WW & Orphan VJ Convergent evolution of unusual complex I homologs with increased proton pumping capacity: energetic and ecological implications. ISME J 12, 2668–2680 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lücker S, Nowka B, Rattei T, Spieck E & Daims H The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front. Microbiol 4, 27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson SW, Bock E, Valois FW, Waterbury JB & Schlosser U Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch. Microbiol 144, 1–7 (1986). [Google Scholar]
- 48.Hippe H Leptospirillum gen. nov. (ex Markosyan 1972), nom. rev., including Leptospirillum ferrooxidans sp. nov. (ex Markosyan 1972), nom. rev. and Leptospirillum thermoferrooxidans sp. nov. (Golovacheva et al. 1992). Int. J. Syst. Evol. Microbiol 50, 501–503 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Henry EA et al. Characterization of a new thermophilic sulfate-reducing bacterium Thermodesulfovibrio yellowstonii, gen. nov. and sp. nov.: its phylogenetic relationship to Thermodesulfobacterium commune and their origins deep within the bacterial domain. Arch. Microbiol 161, 62–69 (1994). [PubMed] [Google Scholar]
- 50.Lin X, Kennedy D, Fredrickson J, Bjornstad B & Konopka A Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ. Microbiol 14, 414–425 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Flagan S, Ching W-K & Leadbetter JR Arthrobacter strain VAI-A utilizes acyl-homoserine lactone inactivation products and stimulates quorum signal biodegradation by Variovorax paradoxus. Appl. Environ. Microbiol 69, 909–916 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leadbetter JR & Greenberg EP Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol 182, 6921–6926 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krumbein WE & Altmann HJ A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgoländer Wiss. Meeresunters 25, 347–356 (1973). [Google Scholar]
- 54.Emerson D & Revsbech NP Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: laboratory studies. Appl. Environ. Microbiol 60, 4032–4038 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parada AE, Needham DM & Fuhrman JA Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Illumina I 16S Metagenomic sequencing library preparation. https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html (2013).
- 57.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Quast C et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lane DJ 16S/23S rRNA sequencing. Nucleic Acid Tech. Bact. Syst 125–175 (1991).
- 61.Ludwig W et al. ARB: a software environment for sequence data. Nucleic Acids Res 32, 1363–1371 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parks DH et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Schoenmann S et al. 16S rRNA gene-based phylogenetic microarray for simultaneous identification of members of the genus Burkholderia. Environ. Microbiol 11, 779–800 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Greuter D, Loy A, Horne M & Ratteil T probeBase-an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res 44, D586–D589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amann RI et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol 56, 1919–1925 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoecker K, Dorninger C, Daims H & Wagner M Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol 76, 922–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schramm A, Fuchs BM, Nielsen JL, Tonolla M & Stahl DA Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol 4, 713–720 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Daims H, Stoecker K & Wagner M Fluorescence in situ hybridization for the detection of prokaryotes in Molecular Microbial Ecology 208–228 (Taylor & Francis, 2004). 10.4324/9780203503393-15. [DOI] [Google Scholar]
- 69.Daims H, Lücker S & Wagner M daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol 8, 200–213 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Taylor GJ & Crowder AA Use of the DCB technique for extraction of hydrous iron oxides from roots of wetland plants. Am. J. Bot 70, 1254 (1983). [Google Scholar]
- 71.Polerecky L et al. Look@NanoSIMS – a tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol 14, 1009–1023 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Brewer PG & Spencer DW Colorimetric determination of manganse in anoxic waters. Limnol. Oceanogr 16, 107–110 (1971). [Google Scholar]
- 73.Oldham VE, Miller MT, Jensen LT & Luther GW Revisiting Mn and Fe removal in humic rich estuaries. Geochim. Cosmochim. Acta 209, 267–283 (2017). [Google Scholar]
- 74.Suzuki MT, Taylor LT & DeLong EF Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5’-nuclease assays. Appl. Environ. Microbiol 66, 4605–4614 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.William S, Feil H & Copeland A Bacterial genomic DNA isolation using CTAB. Department of Energy Joint Genome Institute https://jgi.doe.gov/user-programs/pmo-overview/protocols-sample-preparation-information/ (2012).
- 76.Arkin AP et al. KBase: The United States Department of Energy systems biology knowledgebase. Nat. Biotechnol 36, 566–569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bankevich A et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karst SM, Kirkegaard RH & Albertsen M mmgenome: a toolbox for reproducible genome extraction from metagenomes. bioRxiv 059121 (2016) 10.1101/059121. [DOI]
- 80.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen I-MA et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agarwala R et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46, D8–D13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bagos PG, Liakopoulos TD, Spyropoulos IC & Hamodrakas SJ PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res 32, W400–404 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Federhen S The NCBI Taxonomy database. Nucleic Acids Res 40, D136–143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P & Tyson GW CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pruesse E, Peplies J & Glöckner FO SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinforma. Oxf. Engl 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ronquist F et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Letunic I & Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res gkw290 (2016) 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed]
- 89.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinforma. Oxf. Engl 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, (2011). [DOI] [PMC free article] [PubMed]
- 91.Lever MA et al. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front. Microbiol 6, 476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kopylova E, Noe L & Touzet H SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinforma. Oxf. Engl 28, 3211–3217 (2012). [DOI] [PubMed] [Google Scholar]
- 93.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Pimentel H, Bray NL, Puente S, Melsted P & Pachter L Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Waasbergen LG van, Hildebrand M & Tebo BM Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J. Bacteriol 178, 3517–3530 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jung WK & Schweisfurth R Manganese oxidation by an intracellular protein of a Pseudomonas species. Z. Allg. Mikrobiol 19, 107–115 (1979). [PubMed] [Google Scholar]
- 97.Esteve-Núñez A, Rothermich M, Sharma M & Lovley D Growth of Geobacter sulfurreducens under nutrient-limiting conditions in continuous culture. Environ. Microbiol 7, 641–648 (2005). [DOI] [PubMed] [Google Scholar]
- 98.Neubauer SC, Emerson D & Megonigal JP Life at the energetic edge: kinetics of circumneutral iron oxidation by lithotrophic iron-oxidizing bacteria isolated from the wetland-plant rhizosphere. Appl. Environ. Microbiol 68, 3988–3995 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nowka B, Daims H & Spieck E Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Env. Microbiol 81, 745–753 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ehrich S, Behrens D, Lebedeva E, Ludwig W & Bock E A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol 164, 16–23 (1995). [DOI] [PubMed] [Google Scholar]
- 101.Kim S & Lee SB Catalytic promiscuity in dihydroxy-acid dehydratase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Biochem. (Tokyo) 139, 591–596 (2006). [DOI] [PubMed] [Google Scholar]
- 102.Safarian S et al. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science 352, 583–586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lovley DR & Phillips EJP Manganese inhibition of microbial iron reduction in anaerobic sediments. Geomicrobiol. J 6, 145–155 (1988). [Google Scholar]
- 104.Perez-Benito JF, Arias C & Amat E A kinetic study of the reduction of colloidal manganese dioxide by oxalic acid. J. Colloid Interface Sci 177, 288–297 (1996). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data has been deposited at National Center for Biotechnology Information (NCBI) under BioProject PRJNA562312. The cloned 16S rRNA gene sequences of “Ca. Manganitrophus noduliformans” (Species A) and Ramlibacter lithotrophicus (Species B) from the co-culture have been deposited at GenBank under accession numbers MN381734 and MN381735, respectively. The iTAG sequences from 3 different enrichments has been deposited at SRA under accession numbers SRR10031198-SRR10031200. Genome sequences of the co-culture, from which the genome of Ca. M. noduliformans was reconstructed, have been deposited under BioSample SAMN12638105 with raw sequences deposited at SRA under accession number SRR10032644; the reconstructed genome of Ca. M. noduliformans has been deposited at DDBJ/ENA/GenBank under accession number VTOW00000000. Genome sequences of R. lithotrophicus strain RBP-1 have been deposited under BioSample SAMN12638106 with raw sequences deposited at SRA under accession number SRR10031379; the reconstructed genome of R. lithotrophicus strain RBP-1 has been deposited at DDBJ/ENA/GenBank under accession number VTOX00000000. Additionally, reconstructed genomes have been deposited in Joint Genome Institute (JGI) Genomes Online Database Study ID Gs0134339, with Integrated Microbial Genome ID 2784132095 for Ca. M. noduliformans and ID 2778260901 for R. lithotrophicus strain RBP-1. Transcriptome sequence data for the 7 biological replicates have been deposited at SRA under accession numbers SRR10060009, SRR10060010, SRR10060011, SRR10060012, SRR10060013, SRR10060017, SRR10060018. Unique biological materials are available upon reasonable request.