Abstract
Pancreatic ductal adenocarcinoma is the third leading cause of cancer-related death in the United States. As one of the most lethal cancer types, the prognosis for patients diagnosed with pancreatic cancer remains dismal and novel investigations are urgently needed. Evidence for an association of microbes with pancreatic cancer risk, development, treatment response, and post-treatment survivorship is rapidly developing. Herein, we provide an overview on the role of the microbiome as it relates to the natural history of pancreatic cancer, including host immune interactions, alterations in metabolism, direct carcinogenic effect, and its role in treatment response.
Keywords: Pancreatic ductal adenocarcinoma, Microbiota, Oncobiome, Microbiome, Carcinogenesis
Abbreviations: GF, germ free; PDAC, Pancreatic ductal adenocarcinoma; POPF, postoperative pancreatic fistula; qPCR, quantitative polymerase chain reaction; SPF, specific pathogen free; WT, wild-type
Introduction
The current clinical impact and treatment of pancreatic ductal adenocarcinoma
Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer-related death in the United States, with an estimated 57,600 people diagnosed in 2020 [1]. While the overall cancer survival rate has shown moderate improvement with multimodality treatment [2,3], the overall 5-year survival rate for patients diagnosed with PDAC remains stagnant at 7% to 9%. While surgical resection offers the only hope for cure, this should be applied in the context of multimodal therapy to include chemotherapy, chemoradiation, and potential targeted therapies. Even in this best-case scenario in which a patient completes all aspects of multimodality care, the median survival is 30 to 40 months [4], [5], [6], compared to 10 to 13 months for patients with metastatic or unresectable disease who receive chemotherapy alone [5,7]. While targeting the immune system and tumor stroma have been met with excitement to improve these survival statistics, their real-world results have given pause to these treatment modalities [8,9]. Thus, as one of only 2 gastrointestinal malignancies to witness a rising incidence and death rate [10], novel diagnostic and treatment modalities are urgently needed and require new experimental approaches.
Historically, only ∼20% of patients are candidates for potentially curative upfront surgical resection of their pancreatic cancer [11]. With the advent of multimodal therapy centered on systemic chemotherapy and surgery for patients even with vascular involvement, resection rates approach 70% [12]. While some studies have proven controversial and have their own limitations, the use of a multidisciplinary approach to PDAC treatment has improved survival compared to an initial surgical approach. Additionally, even patients with major mesenteric vessel involvement (superior mesenteric artery, superior mesenteric vein, portal vein) were once considered unresectable but are now able to achieve similar oncologic outcomes in highly selected cases as patients without vessel involvement [13,14]. Given that our current technical prowess for pancreatectomy has likely reached a limit, there is still areas for improvement in terms of optimizing patients before surgery, minimizing perioperative morbidity, and postoperative surveillance. Additionally, improvement in the multimodal care using systemic chemotherapeutic/biologic agents with/without radiation deserves continued attention. However, there are several areas during the treatment timeline of patients with PDAC that are candidates for paradigm-shifting investigations to improve outcomes for these patients.
The microbiome and its relationship with cancer
The microbiota, the collection of trillions of microorganisms that include bacteria, virus, fungi, and archaea which live on and within every human, has been shown to play an essential role in host physiologic homeostasis. A homeostatic imbalance of the microbiota, often referred as “dysbiosis,” contributes to the pathogenesis of many diseases [15], including cancer [16]. The majority of the human microbiota resides in the gastrointestinal tract [17] while microbial communities have also been shown to occupy nonintestinal organs [18,19]. The collection of microbial genes within each of these locations is defined as the microbiome, and the advent of next-generation sequencing has tremendously enhanced our understanding of the collective human microbiome [17].
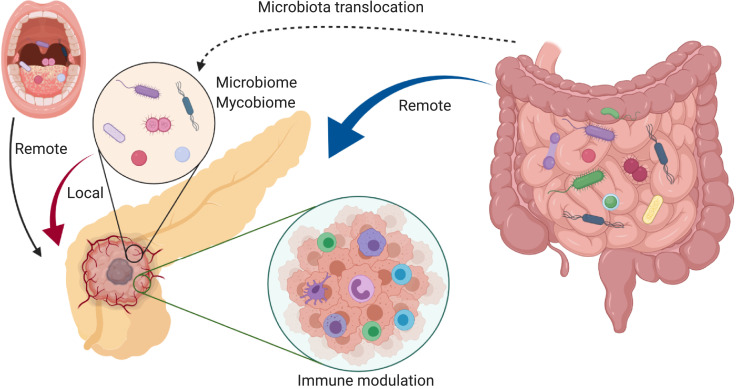
The oncobiome, the term coined to describe the field of research investigating the role of the microbiome on human cancer development, has seen rapid expansion in the past decade [20]. The field of oncobiome research, initially focused on colorectal cancer, has rapidly expanded into other malignancies as well [21]. Mechanisms hypothesized to address how the oncobiome influences cancer development include (1) direct impact of bacterial toxins/metabolites on cancer initiation and growth [22], [23], [24], (2) modulation of the host local and systemic immune response [25], [26], [27], and (3) alteration of microbial and host metabolism [28], [29], [30]. These host-microbe interactions in cancer development could occur in a local (organ dependent) and/or remote (organ independent) fashion [16] (Figure 1). Taking advantage of the microbiota through its effect on its host has emerged as a potential strategy to augment cancer therapeutics [21]. Given that PDAC remains one of the deadliest cancers, ongoing research into its mechanisms of progression and poor response to traditional therapies is desperately needed and investigations into the pancreatic oncobiome, while in their infancy, may provide novel insights into pancreatic carcinogenesis.
Fig. 1.
Microorganisms have been hypothesized to influence pancreatic cancer progression in both a remote (nonpancreatic) and local (intrapancreatic) manner. Distinct oral and gut microbiome compositions have been associated with pancreatic cancer risk, which potentially provides a noninvasive modality for pancreatic cancer early detection [32,41,43,55]. Mechanistically, gut microbiota can suppress the host anti-tumor immunity, facilitating PDAC progression. Local pancreatic microorganisms may also play an important role in pancreatic carcinogenesis in a similar manner.
Herein the current state of the literature regarding the microbiome and pancreatic tumorigenesis will be discussed and how the microbiome may impact the management of this disease along the surgical continuum from screening, to neoadjuvant/adjuvant treatment, surgical outcomes, and surveillance. Finally, future areas of needed research will be discussed to provide insight into the management of this deadly disease.
Pancreatic microbiota and pancreatic cancer
Once considered a sterile organ, recent studies have demonstrated that the pancreas harbors a microbiota, but its role is uncertain. Geller and colleagues examined 113 human PDAC samples obtained during surgery and 20 normal pancreas samples from organ donors by performing bacterial 16S ribosomal DNA quantitative polymerase chain reaction (qPCR) [31]. They found that 76% of the pancreatic tumor samples and 15% of the normal pancreas harbored bacteria. Further 16S rRNA gene sequencing of 65 PDAC samples showed high abundance of the bacterial class Gammaproteobacteria (51.7% of all reads) and primarily the Enterobacteriaceae and Pseudomonadaceae families in these specimens. An independent 16S rRNA gene sequencing study on 12 PDAC samples by Pushalkar et al. also demonstrated the presence of microbes but identified the phyla Proteobacteria, Bacteroidetes, and Firmicutes as prevalent in all samples [32]. By comparing the bacterial composition of the 12 pancreatic tumor samples to 5 healthy controls, a distinct difference in microbial composition was revealed between the human pancreatic tumor specimens and normal human pancreas [32]. The presence of pancreatic microbes has been further confirmed by Thomas and colleagues. However, by comparing 16 PDAC samples to 7 normal nonmalignant pancreatic tissue controls, Thomas and colleagues did not find microbial discrimination at the genus level after statistical scrutiny with False Discovery Rate correction [33]. The data regarding the compositional difference in intrapancreatic bacteria between pancreatic cancer and normal pancreas is still a continued area of debate both from a perspective of bacterial source and function. Study variances include patient sample source, limited quantity of tissue, and the inconsistent definition of “normal control” (healthy organ donor pancreas or normal pancreatic tissue from patients which may be derived from the nonmalignant portion of pancreas as part of a resected cancer specimen).
In order to partially address this controversy and provide insight into the pancreatic microbiota and its association with PDAC, Pushalkar et al. found an increased abundance of intrapancreatic bacteria in the Ptf1aCreLSL-KrasG12D (KC) mouse model of pancreatic cancer compared to C57BL/6 (H-2Kb) wild-type (WT) mice by 16S rRNA fluorescent in situ hybridization [32]. This raises the question regarding the source of the intrapancreatic bacteria and the potential requirement for a pre-existing pancreatic abnormality/pathology, such as the presence of malignancy, in order to create a niche for microbes to colonize. Since one may hypothesize that microbes can gain access to the pancreas via the upper gastrointestinal tract, Pushalkar et al. orally gavaged WT mice with 2.5 × 108 colony-forming units GFP-labeled Escherichia coli daily and euthanized them in a time-course based manner to determine pancreatic colonization. Peak GFP signal was detected in the WT mice pancreas at 0.5 hour after the oral gavage procedure and subsequently decreased but stayed at a relatively lower level which persisted from 6 to 72 hours. However, WT mice grown in specific pathogen free (SPF) housing conditions had little to no bacterial DNA detection. While this demonstrates the ability of bacteria to translocate into the pancreatic parenchyma through artificial gavage using a murine system of PDAC, the short-term translocation in WT mice is likely due to the large amount of Escherichia coli orally administered with concomitant reflux into the pancreatic duct. This short course experiment therefore does not prove long-term colonization or that the bacteria were viable as no confirmatory culture was performed.
Because upper gastrointestinal colonization may not be the only modality for pancreatic colonization, Thomas et al. transferred germ free (GF) 129SvEv mice into SPF housing, followed by a single oral gavage of 1 × 105colony-forming units SPF microbiota and maintained them in SPF housing conditions for 1, 2, 4, and 8 weeks [33]. Through universal 16S rRNA gene PCR amplification of murine stool, Thomas et al. detected the establishment of an intestinal microbiota in the GF mice but no microbial colonization in pancreas. This demonstrated that a naturally acquired microbiota in mice with a nondiseased pancreas may not lead to microbial acquisition in the pancreas, unlike the Pushalkar study. In addition, to assess if microbe translocation could be induced with defective intestinal permeability, as has been suggested in pancreatitis [34], Thomas and colleagues orally gavaged Il10−/− mice with Campylobacter jejuni in order to induce colitis [35,36]. Even though the Il10−/− mice administrated with Campylobacter jejuni had developed severe colitis with increased intestinal permeability, as demonstrated on histology and by increased serum FITC-dextran measurement, universal 16S rRNA gene PCR amplification failed to detect bacteria translocation in the pancreas. Taken together, the acquisition of microbes in the pancreas does not appear to require defective intestinal function but may require pathologic perturbations in the pancreas itself to provide an opportunistic window for potential colonization. Nevertheless, what is congruent among different studies is that the human pancreas possesses a microbiota but the mechanisms whereby this is established is likely multifactorial [37]. Whether pancreatic cancer patients harbor a unique microbial composition compared to noncancerous patients, how the pancreas microbiota acquisition occurs, and if this plays a role in pancreatic carcinogenesis or just an associative finding remains to be defined and further research is needed to fill this knowledge gap.
The microbiome as a biomarker and screening tool for PDAC
As research involving the oncobiome matures, it is natural to hypothesize that the microbiome may provide insight into its use as a screening tool for individuals at risk for cancer, including PDAC [38]. Some of the confounding factors responsible for the poor prognosis of PDAC include its late detection, lack of screening methods, and paucity of risk factors in the majority of patients that render the potential for surgical cure futile once a diagnosis is established [39]. Utilizing the microbiota as a screening test may provide important clues for the early detection of PDAC as it would be easy to administer with low morbidity with regards to sampling the fecal or oral microbiota. Such testing has shown promise in colorectal cancer detection but its utility in PDAC remains unclear [40].
Previous studies have provided an association between the oral microbiome and the presence of PDAC [41,42]. By comparing the salivary microbiota of pancreatic cancer patients (n = 10) and healthy control subjects (n = 10), Farrell et al. identified variations of oral microbes associated with PDAC. The combination of two microbial biomarkers (Neisseria elongate and Streptococcus mitis) showed a 96.4% sensitivity and 82.1% specificity in differentiating pancreatic cancer patients from healthy controls [43]. Furthermore, Fan and colleagues characterized the oral microbiome of 361 PDAC patients and 371 matched healthy controls by 16S rRNA gene sequencing and found the oral pathogens Porphyromonas gingivalis (P= 0.0048) and Aggregatibacter actinomycetemcomitans (P= 0.016) were associated with a higher risk of pancreatic cancer while the abundance of the phyla Fusobacteria was associated with a decreased risk of PDAC (P= 0.0079) [41]. The different oral microbial signatures associated with PDAC found between various investigations may be due, in part, to differences in patient sample size, geographic locations with associated dietary and environmental variability, and specimen source (saliva vs oral wash samples). In order to examine the relationship between host immune response, oral microbes, and pancreatic cancer risk, Michaud and colleagues measured antibodies to oral bacteria in blood samples from 405 pancreatic cancer cases, prior to their diagnosis, and 416 matched healthy controls. They found that patients with high levels of Porphyromonas gingivalis antibodies had a 2-fold higher risk of pancreatic cancer (OR 2.14; 95% Cl 1.05, P= 0.05) [44]. Taken together, detection of antibodies against these oral bacteria may serve as a biomarker to identify people with high risk of pancreatic cancer but the impact of P. gingivalis on pancreatic carcinogenesis, if any, is unknown.
Whereas the aforementioned studies relied on the oral microbiome, increasing evidence has associated changes in the gut microbiota to pancreatic diseases as well [37]. It is thus natural to hypothesize that alteration of the gut microbiota may also associate with PDAC and provide a potential screening mechanism. To investigate a potential link between the gut microbiota and PDAC, Ren and colleagues characterized the fecal microbiota in 85 PDAC patients and 57 matched healthy controls through MiSeq sequencing to elucidate any gut microbiota alterations in pancreatic cancer patients [45]. They illustrated that the gut microbiota in PDAC patients harbored a decreased microbial diversity, increased abundance of potential pathogens, and a decrease of beneficial bacteria when compared to healthy controls [45]. In particular, the fecal contents of PDAC patients had an increase in lipopolysaccharide-producing bacteria which are often associated with increased risk for cancer [46], [47], [48], and a decrease of butyrate-producing bacteria which has demonstrated anticancer effects [49], [50], [51]. In addition, 40 genera of bacteria associated with PDAC patients achieved a high correlation (area under the curve = 0.842) to distinguish these PDAC patients from healthy controls. The potential utility of a stool-based screening method for PDAC detection was further strengthened with the use of a murine genetic mouse model of PDAC. It is important to note that there are inherent differences between the human intestinal microbiota in terms of abundance and diversity of microorganisms when compared to these preclinical models. Special consideration should be made when translating the knowledge from such murine models to human studies given the fact that as little as 4% of the bacterial metagenome is shared between humans and mice [52,53]. In addition, the mouse genetic background(s), housing conditions, and ages all have an impact on their gut microbiota composition [54]. Therefore, focusing on common metabolic pathways affected by the microbiota, regardless of microbiota dissimilarities, may provide more translational impact when murine models are utilized compared to focusing solely on microbiota composition. A key component of any screening test is to detect the disease of interest prior to its development so that treatment can be instituted expeditiously. As such, Mendez and colleagues sought to determine temporal changes in the gut microbiota in the KRASG12D/+; Trp53R172H/+; Pdx1-Cre (KPC) mouse model of pancreatic cancer [55]. They analyzed the gut microbiota in KPC mice and age-matched control mice in a time course throughout the process of tumor development. First, they performed 16S rRNA gene sequencing on the fecal samples from 1 and 6 month old KPC and age-matched control (Pdx1-Cre) mice. There was significant reduction of bacterial richness in these KPC mice compared with control at 6 months of age but not at 1 month of age. When compared to 1 month old KPC mice, the 6 months old KPC mice showed a significant relative expansion of 6 genera and diminished relative richness of 19 genera. Next, to increase the resolution at the microbial genus/species level and to identify relevant bacterial species functionally related to preneoplasia time points, they applied shotgun metagenomics analysis. A whole-genome sequencing on control and KPC mice at 2, 3, and 4 months showed that the KPC mice had significant microbial composition changes as they aged (2 months vs 4 months) but not the control mice by using Bray–Curtis principal co-ordinate analysis. Finally, the predictive metabolomic analysis using HUMAnN2 pipeline [56] on the identified bacterial species revealed that the dominated metabolic pathway identified in 4 month old KPC mice is polyamine biosynthesis while the 2 month old KPC mice had a distinct dominance of energy metabolism pathways. However, the whole-genome sequencing was not applied to the mice aged at 1 or 6 months of age and therefore the full picture of longitudinal KPC murine gut microbiota development at a microbial species/metabolism level remains to be defined. To validate their finding, serum polyamine detection was performed, and they found significantly elevated serum polyamine concentrations in KPC mice aged 4 months and older when compared to 1 to 2 month old mice (1–2 month vs 4–5 month, P= 0.0025; 4–5 month vs 6+ month, P= 0.0242). Although transgenic PDAC murine models have been shown to mimic the clinical PDAC histology which serves as an important preclinical tool, the applicability of such studies to humans is still somewhat limited. Therefore, Mendez et al measured the serum polyamine levels in PDAC patients which demonstrated significantly increased polyamine concentrations when compared to normal healthy volunteers (P= 0.0011, n = 8 per group). The application of serum polyamine detection in the clinical setting as a PDAC biomarker brings promise for early detection of PDAC but requires further validation in the setting of clinical trials. Finally, a recent study by Poore et al. found the potential of utilizing microbial DNA isolated from blood and tissue to diagnose cancer. By reexamining the whole-genome and whole-transcriptome sequencing studies in The Cancer Genome Atlas of 33 different types of cancer, they found unique microbial signatures that could discriminate between cancer and healthy patients [57]. Although PDAC was classified with all evaluated gastrointestinal cancers, which was found as a group to be associated with the Fusobacterium genus. In addition, Nejman and colleagues found that intratumoral bacterial composition varies based on tumor type [58]. They detected the presence of intratumoral bacteria by using bacterial 16S ribosomal DNA qPCR in various types of cancers including pancreatic cancer, which is in line with the findings of other groups [[31], [32], [33],59]. However, the intratumoral microbial signature of pancreatic cancer was not the focus of this study as there was not normal tissue for comparison. Further studies focusing on PDAC intratumoral signature are needed, which may be utilized as biomarker for pancreatic cancer detection, albeit invasive. Taken together, these findings suggest that the development of PDAC results in alterations of the microbiome which may prove useful in the early detection of PDAC [55].
Gut microbiota and pancreatic cancer development
Whereas earlier studies had been limited to associations of gut microbial changes with the presence of PDAC, several groups have recently established a more causative role. Pushalkar et al. compared PDAC growth in mice with an intact microbiota, to mice either born in GF conditions or having antibiotic-mediated ablation of the gut microbiota. They found that the gut microbiota accelerated pancreatic cancer development and produced an overall suppression of antitumor immunity. This was demonstrated by increased myeloid derived suppressor cell infiltration which altered tumor-associated macrophages toward a pro-tumorigenic M2-like phenotype [60,61] and reduction of antitumor cytotoxic CD8+ T cells [32,[62], [63], [64]]. Furthermore, Pushalkar and colleagues identified that the fecal bacterial extract from mice which had developed PDAC had a suppressive impact on macrophage function as evidenced by reduced expression of MHC class 2, which is critical for the initiation of the antigen-specific anti-tumor immune response and increased pro-inflammatory cytokine IL10 expression through Toll-like receptor-dependent signaling. Furthermore, splenic macrophages treated with fecal cell-free microbial extract from PDAC mice mitigated CD4+ and CD8+ T cell activation which are important to exert anticancer immunity [65], [66], [67]. The fecal microbial components had a negative effect on the host antitumor immune cell activity which may be attributed to the enhanced pancreatic carcinogenesis phenotype in preclinical murine models.
In an independent study by Thomas and colleagues, they found that the ablation of the gut microbiota by a cocktail of wide spectrum antibiotics reduced the pancreatic cancer burden in both a xenograft and the KrasG12D/+;PTENlox/+;Pdx1-Cre genetic mouse model of pancreatic cancer [33]. Despite evidence for the existence of intratumoral microbes, debated as a potential instigator of pancreatic carcinogenesis [31,32], Thomas et al did not detect microbes by culture or qPCR in their subcutaneous PDAC xenografts, yet the same pro-tumor phenotype of gut microbiota was observed, suggesting that the gut microbiota plays a role in pancreatic carcinogenesis independent of the local tumor microenvironment [33]. In addition, they identified a higher immune cell infiltration in the tumor microenvironment after antibiotic-mediated microbiota depletion in NOD-SCID mice harboring human PDAC xenografts. Since NOD-SCID mice only possess innate immunity, their study highlights the potential microbial impact on host anti-PDAC innate immunity. However, a study conducted by Sethi and colleagues found that antibiotic-mediated gut microbiota depletion did not decrease the size of KPC-derived xenograft growth in Rag1−/− mice, which also possess only innate immunity [68]. This conflict may be due to differences in the regimen and delivery of the antibiotics for their microbiota depletions, housing conditions, or nutrition [69], [70], [71]. Although antibiotic-mediated microbial depletion has been utilized widely in the field to study the function of the microbiome, the criticism that the use of antibiotics may theoretically impact the host metabolism and therefore tumor growth exists [72], [73], [74]. The use of GF mouse models are therefore required to resolve this criticism. From a translational standpoint, long-term broad-spectrum antibiotics treatment in PDAC patients to alter the course of the disease is not a viable option given evidence that suggests increased risk and mortality in cancer patients [75], [76], [77]. In addition, the broad disruption of the gut microbiota through antibiotic use may cause an impaired antitumor immune response and also affect the response to immunotherapy [21,78]. Routy and colleagues found that lung and kidney cancer patients treated with antibiotics (before, during, or after anti-PD1/PDL1 treatment) had significantly lower overall survival [79]. Studies by Matson et al. and Gopalakrishnan et al. found similar results in melanoma patients receiving anti-PD-1 treatment [80,81]. Based on the above studies that components of the microbiota accelerate pancreatic carcinogenesis, it may be natural to think that the microbiota could be depleted in patients with PDAC to improve survival. However, this simplistic view does not take into account the off-target effects as described and more investigations are needed to determine a more precise way to modulate the microbiota and create a beneficial impact for PDAC patients. Nevertheless, based on these major studies, harnessing the gut microbiota for potential treatment purposes or to alter PDAC progression warrants further investigation.
Microbiome as a predictor of response to therapy
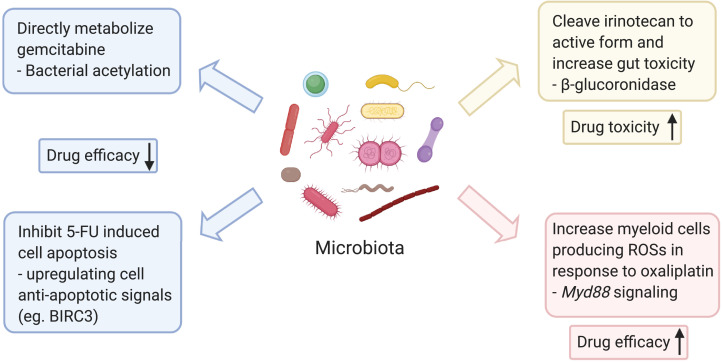
Bacteria have been previously shown to alter the efficacy of medications through their metabolic processes [82,83]. The implications for the care of cancer patients are multifaceted and include decreased/increased drug efficacy through metabolic processes or increased chemotherapy toxicity which indirectly reduces drug efficacy because of the need for dose alterations [84], [85], [86], [87]. Prior research also suggests that antibiotic exposure may have an impact on treatment efficacy in a variety of malignancies and may explain the resistance of PDAC to chemotherapy and the common issue of recurrence even after multimodality treatment that includes chemotherapy and surgery [[79], [80], [81],88]. The chemotherapy backbone for the treatment of pancreatic cancer most often involves gemcitabine or 5-fluoruracil (5FU)-based therapy [89], [90], [91], [92], [93], [94]. Recent in vitro and in vivo evidence suggests that E. coli can decrease the efficacy of gemcitabine through increased metabolism of the drug. Based on mass spectroscopic analysis and predicted gemcitabine-metabolized derivatives, it is hypothesized that bacterial acetylation is the responsible mechanism [95]. Furthermore, Geller and colleagues demonstrated that bacteria from the class Gammaproteobacteria express an isoform of cytidine deaminase which was able to metabolize gemcitabine in a colon cancer xenograft mouse model in a more rapid fashion (long form cytidine deaminase; CDDL), imparting resistance to the drug [31]. Furthermore, 16S rRNA gene sequencing was performed on 65 PDAC tissue samples derived from resected patients and demonstrated Gammaproteobacteria as the most common class (51.7%), represented primarily by the Enterobacteriaceae (which expresses CDDL) and Pseudomonadaceae families. Subsequent culture of bacteria derived from 14 of 15 fresh PDAC samples were able to render the human colorectal cancer cell lines, RKO and HCT116, resistant to gemcitabine therapy. Once the primary chemotherapy agent to treat pancreatic cancer, 5-FU was supplanted by gemcitabine after the later demonstrated a modest survival advantage compared to 5-FU in patients with advanced pancreatic cancer [96]. However, since the introduction of FOLFIRINOX (5-FU, leucovorin, irinotecan, and oxaliplatin) in the adjuvant and metastatic setting for PDAC, with both indications having superior survival outcomes to gemcitabine alone, attention has turned back to combinatorial 5-FU therapy [2,7].
Interestingly, multiple components of the FOLFIRINOX regimen have been shown to be impacted by bacteria. A thymidylate synthase inhibitor, 5-FU blocks synthesis of thymidine and results in cessation of DNA replication in exposed cells. Prior work by Yuan and colleagues demonstrated that alteration of the gut microbiota with antibiotics reduces the efficacy of 5-FU in the treatment of colorectal cancer xenografts in BALB/c mice [97]. Furthermore, evidence suggests that Fusobacterium nucleatum leads to upregulation of BIRC3, an inhibitor of apoptosis. When the human colorectal cancer cell lines, HCT116 and HT29, were infected with F. nucleatum and treated with 5-FU, the cell lines expressed greater levels of BIRC3, concomitant with decreased sensitivity to 5-FU [98]. Furthermore, high levels of F. nucleatum in resected colorectal cancer specimens subsequently correlated with 5-FU resistance as indicated by decreased recurrence-free survival in these patients. The microbiota has not only been shown to decrease drug efficacy directly, but irinotecan is a chemotherapeutic agent in which its efficacy is indirectly inhibited by reactivation of metabolic products, subsequently resulting in supratherapeutic levels that result in dose-limiting gastrointestinal toxicity. Specifically, irinotecan is inactivated in the liver by glucuronidation and its metabolites subsequently released into the gastrointestinal tract via the biliary system. These metabolites are normally excreted in feces but symbiotic bacteria in the gut that produce β-glucuronidases are able to cleave the glucuronide moiety which recreates the active form of irinotecan intraluminally, resulting in intestinal mucosal damage and dose-limiting diarrhea [99]. Despite these deleterious examples of the gut microbiota with chemotherapy efficacy, the gut microbiota could also have a positive impact, raising the possibility of using elements of the microbiome in conjunction with currently available therapeutics. Oxaliplatin efficacy has been demonstrated to be related to the microbiota by Iiad et al. [100]. Specifically, oxaliplatin treatment of mice bearing EL4 lymphoma xenografts resulted in higher reactive oxygen species (ROS) in microbiota-intact C57Bl/6 mice compared to mice with antibiotic-mediated depletion of their microbiota. These ROSs are responsible for the cytotoxic effect of oxaliplatin. Subsequently, induction of ROSs by oxaliplatin was shown to be dependent on nicotinamide adenine dinucleotide phosphate oxidase 2 which is expressed in phagocytic cells. Antibiotic treated and GF mice had decreased DNA damage and tumor infiltration of myeloid cells capable of producing ROS secondary to oxaliplatin administration, resulting in larger tumors compared to WT control. This effect was mediated through reduced microbial product sensing and signaling via Myd88, suggesting that bacteria “prime” myeloid cells capable of producing ROSs in response to oxaliplatin. These studies suggest that the microbiota may play in a role in PDAC chemotherapy resistance, efficacy, and toxicity (Figure 2). This has the potential implication for individualized chemotherapy treatment for patients with pancreatic cancer, regardless of its use in the neoadjuvant, adjuvant, or palliative setting.
Fig. 2.
Microbiota may influence the chemotherapeutic agents utilized in pancreatic cancer treatment in multiple ways. The presence of bacteria expressing cytidine deaminase can directly metabolize gemcitabine and therefore render the drug inactive [31]. Fusobacterium nucleatum can elevate cancer cell antiapoptotic signaling and induce a therapeutic resistance towards 5-FU [98]. However, not all the microbial components are deleterious toward chemotherapy efficacy. Gut microbiota could prime the action of myeloid producing reactive oxygen species in response to oxaliplatin to achieve an improved cytotoxicity [100]. In addition, the microbiota can increase the toxicity of chemotherapeutic regimens. Bacteria expressing β-glucuronidases can cleave the inactive irinotecan into its active form and induce gut toxicity and mucositis, subsequently limiting the effective dose of chemotherapy that can be delivered [85,99].
Surgical implications of the microbiome in the treatment of pancreatic cancer
Pancreatectomy for pancreatic cancer is a complex operation that, like other surgeries, has the potential for complications. The most discussed and notable complication is a postoperative pancreatic fistula (POPF) which can range in severity and outcome [101]. Prior research has demonstrated that patients who develop postpancreatectomy complications, such as POPF, are less likely to receive adjuvant therapy and subsequently have decreased survival compared to patients without postpancreatectomy complications [102]. Interestingly, patients with a POPF that is positive for bacterial growth, compared to sterile POPF collections, have a higher risk of postoperative hemorrhage (42.4% vs 21.7%, P= 0.009), sepsis (38.1% vs 6.5%, P< 0.001), and 90-day mortality (19.5% vs 4.3%, P= 0.013), respectively [103]. Whether this bacterial contamination is causative or associative is open for investigation but evidence exists for bacteria contributing to gastrointestinal anastomotic leaks [104]. Schmitt and colleagues [105] performed 16S rRNA gene sequencing on stool samples from patients prior to pancreatectomy and compared the microbiota community structure to at least two samples within the first ten days in the post-operative period as well as to additional samples when a complication was diagnosed. Three unique bacterial communities were noted with increased abundance of operational taxonomic units composed of Akkermansia, Enterobacteriaceae, and Bacteroidales as well as a decrease in Lachnospiraceae, Prevotella, and Bacteroides, in patients with increased risk of post-operative complications compared to the other two groups. Given these findings, it is imperative to mitigate risk factors that contribute to complication risk for which the microbiome may play an integral role.
The microbiome as a predictor of postoperative survival in pancreatic cancer
In addition to the potential application of gut microbiota in PDAC detection, the characteristics of intrapancreatic tumor microbiota is proposed to predict survival for patients with PDAC. Riquelme and colleagues investigated a cohort of patients who underwent surgical resection of PDAC and dichotomized them into 21 long-term survivors (LTS) and 22 short-term survivors (STS) [59]. A positive association was noted in the LTS cohort compared with STS based on increased intrapancreatic microbiota diversity. Subsequently, the finding was validated in a second independent patient cohort of 15 LTS and 10 STS from a different institution. Bacteria genus Saccharopolyspora, Pseudoxanthomona, Streptomyces, and species Baccilus clausii had excellent capacity to predict LTS after pancreatectomy by receiver operator characteristic analysis as indicated by an area under the curve of 97.5 in the discovery cohort and 99.2 in the validation cohort). Furthermore, immunohistochemistry and immunofluorescent staining with various immune cell markers revealed a significant correlation between CD3, CD8, and Granzyme B positive tissue expression and long-term PDAC survivorship after resection. Interestingly, when the antibiotic-treated mice were orally gavaged with stool derived from LTS or STS patients, the mice with orthotopic pancreatic cancer xenograft derived from syngeneic KPC mice recapitulated the patient tumor growth phenotype in vivo. Overall, the tumor and fecal microbiota of pancreatic cancer patients may serve as a potent biomarker to predict patient survivorship.
Current clinical trials
With the expanding knowledge of the mechanistic role of the pancreatic oncobiome in the laboratory, in order to make the translational leap to the clinical realm, a number of trials investigating the potential role of the microbiome during PDAC diagnosis and treatment have been implemented (Table 1). Studies investigating the gut microbial composition during pancreatic carcinogenesis will provide a more comprehensive insight into diagnostic microbial biomarker identification (NCT03809247, NCT03840460). An ongoing study investigating alterations of the oral and fecal microbiomes due to gastrointestinal surgery, may provide potential information on the surgical management for PDAC. In addition, an ongoing study aims to determine the role of PDAC-associated gut microbiota on anti-msln CAR-T cell functions. With these joint efforts, a fundamental understanding of the role of the microbiome during PDAC carcinogenesis and its potential role in therapy may be determined.
Table 1.
Current clinical trials involving the microbiome and pancreatic cancer.
| Study Title | Approach | Sample Collection | Location | Status | Identifier |
|---|---|---|---|---|---|
| The Mechanism of Enhancing the Anti-tumor Effects of CAR-T on Pancreatic Cancer by Gut Microbiota Regulation | 16S rRNA sequencing Stool supernatant treated anti-msln CAR-T cell function in vitro |
Stool | China | Recruiting | NCT04203459 |
| Microbial Diversity of Pancreatic Diseases | 16S rRNA and metagenomics sequencing | Stool, peripheral blood | China | Not yet recruiting | NCT03809247 |
| Microbiome analysis in esoPhageal, Pancreatic and Colorectal CaNcer Patients Undergoing Gastrointestinal Surgery | 16S rRNA sequencing | Oral, stool | Netherlands | Not yet recruiting | NCT04189393 |
| A prospective translational tissue collection study in early and advanced PDAC and pancreatic neuroendocrine tumors to enable further disease characterization and the development of potential predictive and prognostic biomarkers | 16S rRNA sequencing | Blood, urine, stool, saliva, bile, tissue | United Kingdom | Recruiting | NCT03840460 |
| Oral microbiome and pancreatic cancer | 16S rRNA sequencing | Oral | United States | Completed | NCT03302637 |
| The microbiome of pancreatic cancer: “PANDEMIC” study | Unspecified method Qualitative and quantitative measurement (unspecified) |
Oral, bile, pancreas | Italy | Recruiting | NCT04274972 |
| Colonization of bile ducts and postoperative infectious complications of pancreaticoduodenectomies | Unspecified method Bile sampling for bacterial examination |
Bile | France | Completed | NCT03525067 |
| One time injection of bacteria to treat solid tumors that have not responded to standard therapy | Intratumoral injection of clostridium novyi-NT spores | N/A | United States | Terminated | NCT00358397 |
Future areas of research
With increasing evidence demonstrating the link between microbiota and pancreatic cancer progression and therapeutic response, more comprehensive studies are needed to decipher the mechanisms of how microbiota modulation may occur and what effect could be achieved on PDAC progression and treatment. In particular, understanding which bacterial species or groups having the capacity to enhance or inhibit pancreatic cancer development and therapeutic efficacy may subsequently provide a potential microbial target or at least begin to interrogate culprit pathways. A subsequent patient-centric, microbial-based targeting strategy may offer new hope for pancreatic cancer therapeutics. Although bacteria have been the most studied component of the host microbiome, as they are the most predominant microorganism comprising the microbiota, other essential components of the human microbiome include the virome (viruses) [106] and mycobiome (fungus) [107] which have also been shown to influence host disease development. A recent study suggested that fungi may also be involved in the process of pancreatic carcinogenesis. Aykut et al. identified the presence of fungi in human pancreatic cancer tissues by using fluorescent in situ hybridization staining with 28S rRNA probe [108]. By using a KC mouse model of PDAC, ablation of the mycobiome with amphotericin B protected the mice against oncogenic progression and offered additive antitumor impact to gemcitabine. More research is needed to broaden our understanding on the function of host microorganisms on pancreatic carcinogenesis with respect to fungi and viruses.
Given that multiple studies have implicated an interaction of host microbes with the host immune system in not only PDAC, but other malignancies as well, it is of great importance to understand the impact of defined microbial compositions on host anti-tumor immunity, tumor biology, and therapeutic response. Essentially, understanding specific microbial functions through their mechanisms of action, and biochemistry of their metabolites may provide potential novel treatment algorithms for pancreatic cancer treatment. Furthermore, research on diet intervention, probiotics, and prebiotics to modulate gut microbiota and pancreatic carcinogenesis remains an under-investigated area and requires more attention, as it has the potential of being a noninvasive intervention to mitigate the risk of PDAC development or to alter treatment response.
Conclusion
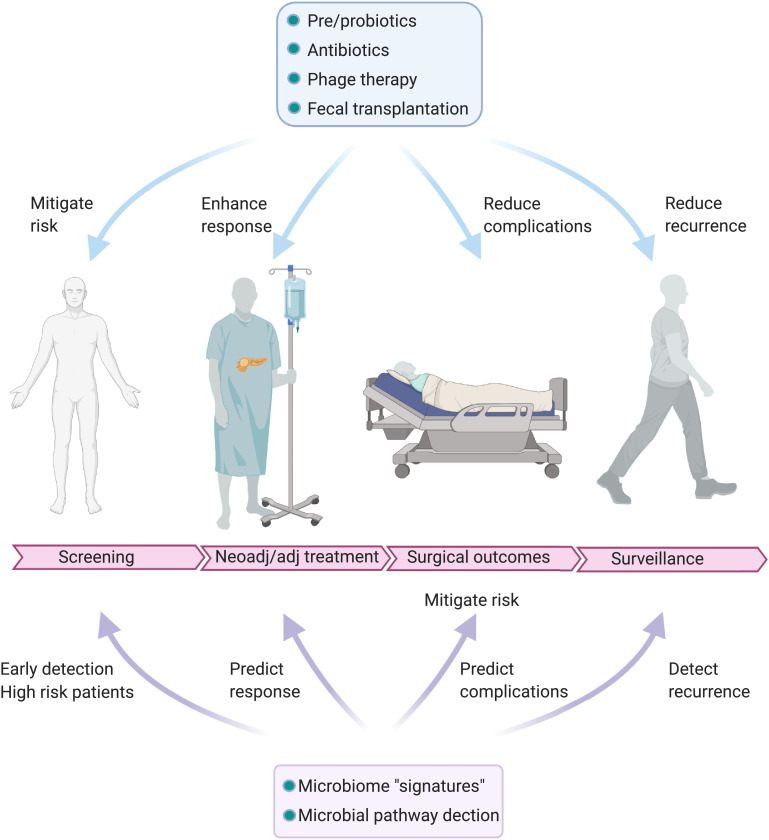
Given the deadly nature of pancreatic cancer and its limited treatment options, the microbiome may provide insight into the screening, diagnosis, and treatment of patients with this deadly disease (Figure 3). Physicians and healthcare providers who care for patients with pancreatic cancer must be aware of implications of the microbiome in pancreatic cancer and this developing field. Interventions to alter the gut microbiota may serve to modulate factors associated with pancreatic carcinogenesis and enhance treatment. Overall, microbiota modulation holds great therapeutic promise but further research regarding mechanisms and potential therapeutic approaches to microbial modification are required.
Fig. 3.
Several areas along the surgical continuum of pancreatic cancer screening, neoadjuvant/adjuvant treatment, outcomes, and surveillance lend itself to involvement of the microbiome. Evidence exists for the involvement of the microbiome in these critical areas that have the potential to impact survival for patients with PDAC.
CRediT authorship contribution statement
Qin Yu: Conceptualization, Writing - original draft. Christian Jobin: Writing - review & editing. Ryan M. Thomas: Conceptualization, Writing - review & editing, Supervision.
Footnotes
Funding: This work was supported by the American Cancer Society Norma and Rich DiMarco Mentored Research Scholar Grant [MRSG-17-228-01-TBG] and the UF Health Cancer Center Pilot Project Grant.
Conflicts of interest: The authors (Qin Yu, Christian Jobin, and Ryan M. Thomas) declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.American Cancer Society | Cancer Facts & Statistics.
- 2.Conroy T., Hammel P., Hebbar M., Ben Abdelghani M., Wei A.C., Raoul J.-L., Choné L., Francois E., Artru P., Biagi J.J. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 2018;379:2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 3.Dhir M., Zenati M.S., Hamad A., Singhi A.D., Bahary N., Hogg M.E., Zeh H.J., Zureikat A.H. FOLFIRINOX Versus Gemcitabine/Nab-Paclitaxel for Neoadjuvant Treatment of Resectable and Borderline Resectable Pancreatic Head Adenocarcinoma. Ann. Surg. Oncol. 2018;25:1896–1903. doi: 10.1245/s10434-018-6512-8. [DOI] [PubMed] [Google Scholar]
- 4.Ielpo B., Caruso R., Duran H., Diaz E., Fabra I., Malavé L., Ferri V., Alvarez R., Cubillo A., Plaza C. A comparative study of neoadjuvant treatment with gemcitabine plus nab-paclitaxel versus surgery first for pancreatic adenocarcinoma. Surg Oncol. 2017;26:402–410. doi: 10.1016/j.suronc.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Katz M.H.G., Pisters P.W.T., Evans D.B., Sun C.C., Lee J.E., Fleming J.B., Vauthey J.N., Abdalla E.K., Crane C.H., Wolff R.A. Borderline resectable pancreatic cancer: the importance of this emerging stage of disease. J. Am. Coll. Surg. 2008;206:833–846. doi: 10.1016/j.jamcollsurg.2007.12.020. discussion 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloyd J.M., Chen H.-C., Wang X., Tzeng C.-W.D., Kim M.P., Aloia T.A., Vauthey J.-N., Lee J.E., Katz M.H.G. Chemotherapy versus chemoradiation as preoperative therapy for resectable pancreatic ductal adenocarcinoma: A propensity score adjusted analysis. Pancreas. 2019;48:216–222. doi: 10.1097/MPA.0000000000001231. [DOI] [PubMed] [Google Scholar]
- 7.Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., Adenis A., Raoul J.-L., Gourgou-Bourgade S., de la Fouchardière C. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 8.Catenacci D.V.T., Junttila M.R., Karrison T., Bahary N., Horiba M.N., Nattam S.R., Marsh R., Wallace J., Kozloff M., Rajdev L. Randomized phase ib/ii study of gemcitabine plus placebo or vismodegib, a hedgehog pathway inhibitor, in patients with metastatic pancreatic cancer. J. Clin. Oncol. 2015;33:4284–4292. doi: 10.1200/JCO.2015.62.8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le D.T., Ko A.H., Wainberg Z.A., Picozzi V.J., Kindler H.L., Wang-Gillam A., Oberstein P.E., Morse M., Zeh H., Weekes C.D. Results from a phase 2b, randomized, multicenter study of GVAX pancreas and CRS-207 compared to chemotherapy in adults with previously-treated metastatic pancreatic adenocarcinoma (ECLIPSE Study) J. Clin. Oncol. 2017;35:345. doi: 10.1158/1078-0432.CCR-18-2992. –345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Facts & Figures 2020 | American Cancer Society.
- 11.Riall T.S., Lillemoe K.D. Underutilization of surgical resection in patients with localized pancreatic cancer. Ann. Surg. 2007;246:181–182. doi: 10.1097/SLA.0b013e31811eaa2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen Q.P., Buettner S., Suker M., Beumer B.R., Addeo P., Bachellier P., Bahary N., Bekaii-Saab T., Bali M.A., Besselink M.G. Neoadjuvant FOLFIRINOX in Patients With Borderline Resectable Pancreatic Cancer: A Systematic Review and Patient-Level Meta-Analysis. J. Natl. Cancer Inst. 2019;111:782–794. doi: 10.1093/jnci/djz073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloyd J.M., Katz M.H.G., Prakash L., Varadhachary G.R., Wolff R.A., Shroff R.T., Javle M., Fogelman D., Overman M., Crane C.H. Preoperative Therapy and Pancreatoduodenectomy for Pancreatic Ductal Adenocarcinoma: a 25-Year Single-Institution Experience. J. Gastrointest. Surg. 2017;21:164–174. doi: 10.1007/s11605-016-3265-1. [DOI] [PubMed] [Google Scholar]
- 14.Truty M.J., Kendrick M.L., Nagorney D.M., Smoot R.L., Cleary S.P., Graham R.P., Goenka A.H., Hallemeier C.L., Haddock M.G., Harmsen W.S. Factors predicting response, perioperative outcomes, and survival following total neoadjuvant therapy for borderline/locally advanced pancreatic cancer. Ann. Surg. 2019 doi: 10.1097/SLA.0000000000003284. [DOI] [PubMed] [Google Scholar]
- 15.Liang D., Leung R.K.-K., Guan W., Au W.W. Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathog. 2018;10:3. doi: 10.1186/s13099-018-0230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwabe R.F., Jobin C. The microbiome and cancer. Nat. Rev. Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ursell L.K., Metcalf J.L., Parfrey L.W., Knight R. Defining the human microbiome. Nutr. Rev. 70 Suppl 1. 2012:S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck J.M., Young V.B., Huffnagle G.B. The microbiome of the lung. Transl Res. 2012;160:258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urbaniak C., Cummins J., Brackstone M., Macklaim J.M., Gloor G.B., Baban C.K., Scott L., O'Hanlon D.M., Burton J.P., Francis K.P. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014;80:3007–3014. doi: 10.1128/AEM.00242-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas R.M., Jobin C. The microbiome and cancer: is the “oncobiome” mirage real? Trends Cancer. 2015;1:24–35. doi: 10.1016/j.trecan.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmink B.A., Khan M.A.W., Hermann A., Gopalakrishnan V., Wargo J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019;25:377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 22.Arthur J.C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J.M., Fan T.-J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graillot V., Dormoy I., Dupuy J., Shay J.W., Huc L., Mirey G., Vignard J. Genotoxicity of cytolethal distending toxin (CDT) on isogenic human colorectal cell lines: potential promoting effects for colorectal carcinogenesis. Front. Cell Infect. Microbiol. 2016;6:34. doi: 10.3389/fcimb.2016.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yong X., Tang B., Li B.-S., Xie R., Hu C.-J., Luo G., Qin Y., Dong H., Yang S.-M. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun. Signal. 2015;13:30. doi: 10.1186/s12964-015-0111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R., Zhou R., Wang H., Li W., Pan M., Yao X., Zhan W., Yang S., Xu L., Ding Y. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019;26:2447–2463. doi: 10.1038/s41418-019-0312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calcinotto A., Brevi A., Chesi M., Ferrarese R., Garcia Perez L., Grioni M., Kumar S., Garbitt V.M., Sharik M.E., Henderson K.J. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018;9:4832. doi: 10.1038/s41467-018-07305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu A.I., Zhao L., Eaton K.A., Ho S., Chen J., Poe S., Becker J., Gonzalez A., McKinstry D., Hasso M. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C., Han M., Heinrich B., Fu Q., Zhang Q., Sandhu M., Agdashian D., Terabe M., Berzofsky J.A., Fako V. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018:360. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishehsari F., Engen P.A., Preite N.Z., Tuncil Y.E., Naqib A., Shaikh M., Rossi M., Wilber S., Green S.J., Hamaker B.R. Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes (Basel) 2018:9. doi: 10.3390/genes9020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen L.J., Esterhazy D., Kim S.-H., Lemetre C., Aguilar R.R., Gordon E.A., Pickard A.J., Cross J.R., Emiliano A.B., Han S.M. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017;549:48–53. doi: 10.1038/nature23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geller L.T., Barzily-Rokni M., Danino T., Jonas O.H., Shental N., Nejman D., Gavert N., Zwang Y., Cooper Z.A., Shee K. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pushalkar S., Hundeyin M., Daley D., Zambirinis C.P., Kurz E., Mishra A., Mohan N., Aykut B., Usyk M., Torres L.E. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas R.M., Gharaibeh R.Z., Gauthier J., Beveridge M., Pope J.L., Guijarro M.V., Yu Q., He Z., Ohland C., Newsome R. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis. 2018;39:1068–1078. doi: 10.1093/carcin/bgy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koh Y.Y., Jeon W.K., Cho Y.K., Kim H.J., Chung W.G., Chon C.U., Oh T.Y., Shin J.H. The effect of intestinal permeability and endotoxemia on the prognosis of acute pancreatitis. Gut Liver. 2012;6:505–511. doi: 10.5009/gnl.2012.6.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X., Winglee K., Gharaibeh R.Z., Gauthier J., He Z., Tripathi P., Avram D., Bruner S., Fodor A., Jobin C. Microbiota-Derived Metabolic Factors Reduce Campylobacteriosis in Mice. Gastroenterology. 2018;154:1751–1763. doi: 10.1053/j.gastro.2018.01.042. .e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaser M.J., Parsons R.B., Wang W.L. Acute colitis caused by Campylobacter fetus ss. jejuni. Gastroenterology. 1980;78:448–453. [PubMed] [Google Scholar]
- 37.Thomas R.M., Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat. Rev. Gastroenterol. Hepatol. 2020;17:53–64. doi: 10.1038/s41575-019-0242-7. [DOI] [PubMed] [Google Scholar]
- 38.Rajpoot M., Sharma A.K., Sharma A., Gupta G.K. Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin. Cancer Biol. 2018;52:1–8. doi: 10.1016/j.semcancer.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 39.McGuigan A., Kelly P., Turkington R.C., Jones C., Coleman H.G., McCain R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018;24:4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayanan V., Peppelenbosch M.P., Konstantinov S.R. Human fecal microbiome-based biomarkers for colorectal cancer. Cancer Prev Res (Phila Pa) 2014;7:1108–1111. doi: 10.1158/1940-6207.CAPR-14-0273. [DOI] [PubMed] [Google Scholar]
- 41.Fan X., Alekseyenko A.V., Wu J., Peters B.A., Jacobs E.J., Gapstur S.M., Purdue M.P., Abnet C.C., Stolzenberg-Solomon R., Miller G. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120–127. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaud D.S., Izard J. Microbiota, oral microbiome, and pancreatic cancer. Cancer J. 2014;20:203–206. doi: 10.1097/PPO.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farrell J.J., Zhang L., Zhou H., Chia D., Elashoff D., Akin D., Paster B.J., Joshipura K., Wong D.T.W. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michaud D.S., Izard J., Wilhelm-Benartzi C.S., You D.-H., Grote V.A., Tjønneland A., Dahm C.C., Overvad K., Jenab M., Fedirko V. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764–1770. doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Z., Jiang J., Xie H., Li A., Lu H., Xu S., Zhou L., Zhang H., Cui G., Chen X. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. 2017;8:95176–95191. doi: 10.18632/oncotarget.18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fried S., Tosun S., Troost G., Keil S., Zaenker K.S., Dittmar T. Lipopolysaccharide (LPS) promotes apoptosis in human breast epithelial × breast cancer hybrids, but not in parental cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S., Xu X., Jiang M., Bi Y., Xu J., Han M. Lipopolysaccharide induces inflammation and facilitates lung metastasis in a breast cancer model via the prostaglandin E2-EP2 pathway. Mol. Med. Rep. 2015;11:4454–4462. doi: 10.3892/mmr.2015.3258. [DOI] [PubMed] [Google Scholar]
- 48.de Waal G.M., de Villiers W.J.S., Forgan T., Roberts T., Pretorius E. Colorectal cancer is associated with increased circulating lipopolysaccharide, inflammation and hypercoagulability. Sci. Rep. 2020;10:8777. doi: 10.1038/s41598-020-65324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berni Canani R., Di Costanzo M., Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin. Epigenetics. 2012;4:4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams E.A., Coxhead J.M., Mathers J.C. Anti-cancer effects of butyrate: use of micro-array technology to investigate mechanisms. Proc Nutr Soc. 2003;62:107–115. doi: 10.1079/PNS2002230. [DOI] [PubMed] [Google Scholar]
- 51.Chen J., Zhao K.-N., Vitetta L. Effects of intestinal microbial−elaborated butyrate on oncogenic signaling pathways. Nutrients. 2019:11. doi: 10.3390/nu11051026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hugenholtz F., de Vos W.M. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol. Life Sci. 2018;75:149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen T.L.A., Vieira-Silva S., Liston A., Raes J. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagpal R., Wang S., Solberg Woods L.C., Seshie O., Chung S.T., Shively C.A., Register T.C., Craft S., McClain D.A., Yadav H. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-human Primate, and Human Feces. Front. Microbiol. 2018;9:2897. doi: 10.3389/fmicb.2018.02897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mendez R., Kesh K., Arora N., Di Martino L., McAllister F., Merchant N., Banerjee S., Banerjee S. Microbial dysbiosis and polyamine metabolism as predictive markers for early detection of pancreatic cancer. Carcinogenesis. 2019 doi: 10.1093/carcin/bgz116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abubucker S., Segata N., Goll J., Schubert A.M., Izard J., Cantarel B.L., Rodriguez-Mueller B., Zucker J., Thiagarajan M., Henrissat B. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poore G.D., Kopylova E., Zhu Q., Carpenter C., Fraraccio S., Wandro S., Kosciolek T., Janssen S., Metcalf J., Song S.J. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T., Rotter-Maskowitz A., Weiser R., Mallel G., Gigi E. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riquelme E., Zhang Y., Zhang L., Montiel M., Zoltan M., Dong W., Quesada P., Sahin I., Chandra V., San Lucas A. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795–806. doi: 10.1016/j.cell.2019.07.008. .e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao N.-B., Lü M.-H., Fan Y.-H., Cao Y.-L., Zhang Z.-R., Yang S.-M. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol. 2012;2012 doi: 10.1155/2012/948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Najafi M., Hashemi Goradel N., Farhood B., Salehi E., Nashtaei M.S., Khanlarkhani N., Khezri Z., Majidpoor J., Abouzaripour M., Habibi M. Macrophage polarity in cancer: A review. J. Cell Biochem. 2019;120:2756–2765. doi: 10.1002/jcb.27646. [DOI] [PubMed] [Google Scholar]
- 62.Farhood B., Najafi M., Mortezaee K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell Physiol. 2019;234:8509–8521. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- 63.Maimela N.R., Liu S., Zhang Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput Struct Biotechnol J. 2019;17:1–13. doi: 10.1016/j.csbj.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Leun A.M., Thommen D.S., Schumacher T.N. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer. 2020;20:218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cronin S.J.F., Penninger J.M. From T-cell activation signals to signaling control of anti-cancer immunity. Immunol. Rev. 2007;220:151–168. doi: 10.1111/j.1600-065X.2007.00570.x. [DOI] [PubMed] [Google Scholar]
- 66.Smith-Garvin J.E., Koretzky G.A., Jordan M.S. T cell activation. Annu. Rev. Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lesokhin A.M., Callahan M.K., Postow M.A., Wolchok J.D. On being less tolerant: enhanced cancer immunosurveillance enabled by targeting checkpoints and agonists of T cell activation. Sci. Transl. Med. 2015;7:280sr1. doi: 10.1126/scitranslmed.3010274. [DOI] [PubMed] [Google Scholar]
- 68.Sethi V., Kurtom S., Tarique M., Lavania S., Malchiodi Z., Hellmund L., Zhang L., Sharma U., Giri B., Garg B. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology. 2018;155:33–37. doi: 10.1053/j.gastro.2018.04.001. .e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franklin C.L., Ericsson A.C. Microbiota and reproducibility of rodent models. Lab Anim. (N.Y.) 2017;46:114–122. doi: 10.1038/laban.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friswell M.K., Gika H., Stratford I.J., Theodoridis G., Telfer B., Wilson I.D., McBain A.J. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One. 2010;5:e8584. doi: 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hufeldt M.R., Nielsen D.S., Vogensen F.K., Midtvedt T., Hansen A.K. Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp Med. 2010;60:336–347. [PMC free article] [PubMed] [Google Scholar]
- 72.Morgun A., Dzutsev A., Dong X., Greer R.L., Sexton D.J., Ravel J., Schuster M., Hsiao W., Matzinger P., Shulzhenko N. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut. 2015;64:1732–1743. doi: 10.1136/gutjnl-2014-308820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zarrinpar A., Chaix A., Xu Z.Z., Chang M.W., Marotz C.A., Saghatelian A., Knight R., Panda S. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 2018;9:2872. doi: 10.1038/s41467-018-05336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujisaka S., Ussar S., Clish C., Devkota S., Dreyfuss J.M., Sakaguchi M., Soto M., Konishi M., Softic S., Altindis E. Antibiotic effects on gut microbiota and metabolism are host dependent. J. Clin. Invest. 2016 doi: 10.1172/JCI86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cao Y., Wu K., Mehta R., Drew D.A., Song M., Lochhead P., Nguyen L.H., Izard J., Fuchs C.S., Garrett W.S. Long-term use of antibiotics and risk of colorectal adenoma. Gut. 2018;67:672–678. doi: 10.1136/gutjnl-2016-313413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hasanov M., Mohindroo C., Rogers J., Prakash L., Overman M.J., Varadhachary G.R., Wolff R.A., Javle M.M., Fogelman D.R., Pant S. The effect of antibiotic use on survival of patients with resected pancreatic ductal adenocarcinoma. JCO. 2019;37:e15773. –e15773. [Google Scholar]
- 77.Petrelli F., Ghidini M., Ghidini A., Perego G., Cabiddu M., Khakoo S., Oggionni E., Abeni C., Hahne J.C., Tomasello G. Use of Antibiotics and Risk of Cancer: A Systematic Review and Meta-Analysis of Observational Studies. Cancers (Basel) 2019:11. doi: 10.3390/cancers11081174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gopalakrishnan V., Helmink B.A., Spencer C.N., Reuben A., Wargo J.A. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 80.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.-L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Enright E.F., Gahan C.G.M., Joyce S.A., Griffin B.T. The impact of the gut microbiota on drug metabolism and clinical outcome. Yale J Biol Med. 2016;89:375–382. [PMC free article] [PubMed] [Google Scholar]
- 83.Li H., He J., Jia W. The influence of gut microbiota on drug metabolism and toxicity. Expert Opin Drug Metab Toxicol. 2016;12:31–40. doi: 10.1517/17425255.2016.1121234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ciorba M.A., Hallemeier C.L., Stenson W.F., Parikh P.J. Probiotics to prevent gastrointestinal toxicity from cancer therapy: an interpretive review and call to action. Curr. Opin. Support. Palliat. Care. 2015;9:157–162. doi: 10.1097/SPC.0000000000000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wallace B.D., Roberts A.B., Pollet R.M., Ingle J.D., Biernat K.A., Pellock S.J., Venkatesh M.K., Guthrie L., O'Neal S.K., Robinson S.J. Structure and Inhibition of Microbiome β-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem. Biol. 2015;22:1238–1249. doi: 10.1016/j.chembiol.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roberts A.B., Wallace B.D., Venkatesh M.K., Mani S., Redinbo M.R. Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 2013;84:208–217. doi: 10.1124/mol.113.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Viaud S., Saccheri F., Mignot G., Yamazaki T., Daillère R., Hannani D., Enot D.P., Pfirschke C., Engblom C., Pittet M.J. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pflug N., Kluth S., Vehreschild J.J., Bahlo J., Tacke D., Biehl L., Eichhorst B., Fischer K., Cramer P., Fink A.-M. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2016.1150399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haller D.G. Chemotherapy for advanced pancreatic cancer. Int. J. Radiat. Oncol. Biol. Phys. 2003;56:16–23. doi: 10.1016/s0360-3016(03)00448-6. [DOI] [PubMed] [Google Scholar]
- 90.National Comprehensive Cancer Network. Pancreatic Adenocarcinoma (v1.2020).
- 91.Quiñonero F., Mesas C., Doello K., Cabeza L., Perazzoli G., Jimenez-Luna C., Rama A.R., Melguizo C., Prados J. The challenge of drug resistance in pancreatic ductal adenocarcinoma: a current overview. Cancer Biol. Med. 2019;16:688–699. doi: 10.20892/j.issn.2095-3941.2019.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Principe D.R., Narbutis M., Kumar S., Park A., Viswakarma N., Dorman M.J., Kamath S.D., Grippo P.J., Fishel M.L., Hwang R.F. Long-Term Gemcitabine Treatment Reshapes the Pancreatic Tumor Microenvironment and Sensitizes Murine Carcinoma to Combination Immunotherapy. Cancer Res. 2020 doi: 10.1158/0008-5472.CAN-19-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amrutkar M., Gladhaug I.P. Pancreatic cancer chemoresistance to gemcitabine. Cancers (Basel) 2017:9. doi: 10.3390/cancers9110157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grasso C., Jansen G., Giovannetti E. Drug resistance in pancreatic cancer: Impact of altered energy metabolism. Crit. Rev. Oncol. Hematol. 2017;114:139–152. doi: 10.1016/j.critrevonc.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 95.Lehouritis P., Cummins J., Stanton M., Murphy C.T., McCarthy F.O., Reid G., Urbaniak C., Byrne W.L., Tangney M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 2015;5:14554. doi: 10.1038/srep14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burris H.A., Moore M.J., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R., Cripps M.C., Portenoy R.K., Storniolo A.M., Tarassoff P. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 97.Yuan L., Zhang S., Li H., Yang F., Mushtaq N., Ullah S., Shi Y., An C., Xu J. The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed. Pharmacother. 2018;108:184–193. doi: 10.1016/j.biopha.2018.08.165. [DOI] [PubMed] [Google Scholar]
- 98.Zhang S., Yang Y., Weng W., Guo B., Cai G., Ma Y., Cai S. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res. 2019;38:14. doi: 10.1186/s13046-018-0985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wallace B.D., Wang H., Lane K.T., Scott J.E., Orans J., Koo J.S., Venkatesh M., Jobin C., Yeh L.-A., Mani S. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dusch N., Lietzmann A., Barthels F., Niedergethmann M., Rückert F., Wilhelm T.J. International Study Group of Pancreatic Surgery Definitions for Postpancreatectomy Complications: Applicability at a High-Volume Center. Scand. J. Surg. 2017;106:216–223. doi: 10.1177/1457496916680944. [DOI] [PubMed] [Google Scholar]
- 102.Hank T., Sandini M., Ferrone C.R., Rodrigues C., Weniger M., Qadan M., Warshaw A.L., Lillemoe K.D., Fernández-Del Castillo C. Association Between Pancreatic Fistula and Long-term Survival in the Era of Neoadjuvant Chemotherapy. JAMA Surg. 2019;154:943–951. doi: 10.1001/jamasurg.2019.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Loos M., Strobel O., Legominski M., Dietrich M., Hinz U., Brenner T., Heininger A., Weigand M.A., Büchler M.W., Hackert T. Postoperative pancreatic fistula: Microbial growth determines outcome. Surgery. 2018;164:1185–1190. doi: 10.1016/j.surg.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 104.Shogan B.D., Belogortseva N., Luong P.M., Zaborin A., Lax S., Bethel C., Ward M., Muldoon J.P., Singer M., An G. Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Sci. Transl. Med. 2015;7:286ra68. doi: 10.1126/scitranslmed.3010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmitt F.C.F., Brenner T., Uhle F., Loesch S., Hackert T., Ulrich A., Hofer S., Dalpke A.H., Weigand M.A., Boutin S. Gut microbiome patterns correlate with higher postoperative complication rates after pancreatic surgery. BMC Microbiol. 2019;19:42. doi: 10.1186/s12866-019-1399-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cadwell K. The virome in host health and disease. Immunity. 2015;42:805–813. doi: 10.1016/j.immuni.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cui L., Morris A., Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aykut B., Pushalkar S., Chen R., Li Q., Abengozar R., Kim J.I., Shadaloey S.A., Wu D., Preiss P., Verma N. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574:264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]