Abstract
The SRY gene induces testis development even in XX individuals. However, XX/Sry testes fail to produce mature sperm, due to the absence of Y chromosome carrying genes essential for spermatogenesis. XX/Sry Sertoli cells show abnormalities in the production of lactate and cholesterol required for germ cell development. Leydig cells are essential for male functions through testosterone production. However, whether XX/Sry adult Leydig cells (XX/Sry ALCs) function normally remains unclear. In this study, the transcriptomes from XY and XX/Sry ALCs demonstrated that immediate early and cholesterogenic gene expressions differed between these cells. Interestingly, cholesterogenic genes were upregulated in XX/Sry ALCs, although downregulated in XX/Sry Sertoli cells. Among the steroidogenic enzymes, CYP17A1 mediates steroid 17α-hydroxylation and 17,20-lyase reaction, necessary for testosterone production. In XX/Sry ALCs, the latter reaction was selectively decreased. The defects in XX/Sry ALCs, together with those in the germ and Sertoli cells, might explain the infertility of XX/Sry testes.
Subject terms: Sterols, Transcriptomics
Introduction
It has been established that the SRY (sex-determining region on the Y chromosome) gene is responsible for the differentiation of the testes in mammals1,2. Indeed, injection of the Sry gene into fertilized XX mouse eggs leads to testis development in XX fetuses. However, XX mice carrying the Sry transgene (XX/Sry mice) suffer from spermatogenic failure3,4. Although the developmental defects of germ cells have been thought to be caused by the lack of Y-chromosome genes essential for spermatogenesis5, the reason for this infertility in XX/Sry mice is still under discussion. In fact, our previous study identified disfunction of XX/Sry Sertoli cells6. In general, Sertoli cells support the differentiation of germ cells by providing them with nutrients including lactate7 and cholesterol8. XX/Sry Sertoli cells were found to synthesize these substances less than XY Sertoli cells, due to lower expression levels of the genes required for their synthesis6.
In addition to Sertoli cells, testes contain Leydig cells, which are developmentally divided into two types, fetal-type (FLCs) and adult-type (ALCs). During the fetal stage, FLCs emerge within the interstitial space of the fetal testes and increase in number during embryonic development. After birth, FLCs are gradually substituted with ALCs9. Finally, in the adult stage, the testicular interstitial space is predominantly occupied by ALCs, although a small population of FLCs remains9–11. With respect to the Leydig cells in XX/Sry testes, it remains largely unclear whether the ALCs in XX/Sry mice exhibit functions equivalent to XY ALCs.
In general, ALCs are characterized by the functional capacity to produce testosterone. Four enzymes have been implicated in the synthesis of testosterone from cholesterol: cytochrome P450 family members cholesterol side-chain cleavage enzyme (CYP11A1) and 17α-hydroxylase/17,20-lyase (CYP17A1); 3β-hydroxysteroid dehydrogenase (HSD3B1 and HSD3B6); and 17β-hydroxysteroid dehydrogenase (HSD17B3)12,13. Of these enzymes, CYP17A1 uniquely mediates two distinct reactions: 17α-hydroxylation and C17,20-cleavage of steroids14. Both reactions are successively mediated by CYP17A1 in the Leydig cells of all mammalian species.
Ad4BP/SF-1 (adrenal-4 binding protein/steroidogenic factor 1/NR5A115) was initially identified as a nuclear receptor-type transcription factor that regulates the gene transcription of CYP11A1 and CYP11B1 (steroid 11β-hydroxylase)16–18. Thereafter, many studies have investigated whether other steroidogenic genes are also regulated by Ad4BP/SF-1. These studies identified HSD3B219,20, CYP17A121–23, and CYP19A124 as target genes of this factor. Thus, it has been widely accepted that Ad4BP/SF-1 plays a central role in the regulation of steroidogenic genes25,26.
All steroid hormones are synthesized from cholesterol. In addition to special usage for steroidogenesis, cholesterol is known to be an essential component of various cellular membranes27. In accordance with this broad range of requirements for cholesterol, cholesterogenic genes are expressed in a variety of cell types. Extensive investigation of cholesterogenic gene regulation in the liver has led to the identification of sterol regulatory element binding protein 2 (SREBP2, encoded by SREBF2) as the key transcription factor regulating all cholesterogenic genes28. In addition to this key molecule, Ad4BP/SF-1 has recently been shown to be involved in cholesterogenic gene regulation in steroidogenic cells29.
In this study, we investigated whether XX/Sry ALCs are functionally different from XY ALCs. Comparison of the transcriptomes obtained from these two types of cells revealed that the expression of immediate early genes and cholesterogenic genes was altered in the XX/Sry ALCs. In addition, we found that the 17,20-lyase reaction mediated by CYP17A1 was specifically affected in XX/Sry ALCs.
Results
Increase of ALCs in XX/Sry testes
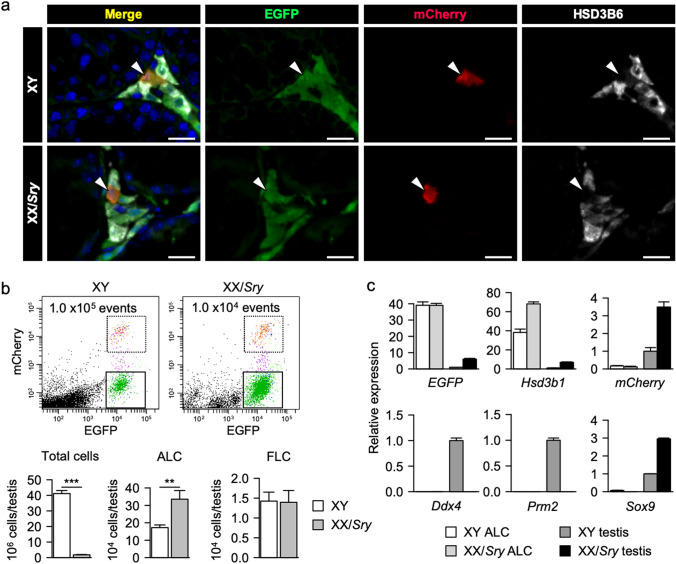
It was previously believed that FLCs are completely replaced by ALCs after birth. However, our previous studies have demonstrated that FLCs persist in adult mouse testes11,30. Therefore, to selectively investigate ALCs, we established a mouse line carrying Ad4BP-BAC-EGFP and mFLE-mCherry as transgenes. In the mouse testes, FLCs were labeled with both EGFP and mCherry, whereas ALCs were labeled with EGFP alone. This double transgenic mouse line thus enabled us to isolate ALCs and FLCs with no mutual contamination. We transferred these two transgenes into XY and XX/Sry mice to obtain XY and XX/Sry ALCs as EGFP single-positive cells. As shown in Fig. 1a, we found both EGFP single-positive and EGFP/mCherry double-positive Leydig cells in both XY and XX/Sry testes. HSD3B6, an ALC marker, was colocalized with the EGFP in the single-positive Leydig cells, indicating that these cells were ALCs.
Figure 1.
Preparation of ALCs from XY and XX/Sry testes. (a) XY and XX/Sry testes from eight-week-old Ad4BP-BAC-EGFP/mFLE-mCherry mice were immunostained with antibodies for EGFP (green), mCherry (red), and HSD3B6 (white). Nuclei were stained with DAPI (blue). Merged images are shown on the left. Arrowheads indicate FLCs (EGFP/mCherry double-positive, HSD3B6-negative). ALCs were detected as EGFP/HSD3B6 double-positive, mCherry-negative cells. Scale bars = 20 μm. (b) Whole cell preparations from the testes of 8-week-old XY and XX/Sry mice were analyzed via FACS. The cells surrounded by solid lines were recovered as EGFP single-positive ALCs, whereas those surrounded by dotted lines were recovered as EGFP/mCherry double-positive FLCs (upper panels). Total testicular cells were counted. Numbers of ALCs and FLCs per testis are calculated using the results obtained by FACS. Nine biologically independent samples (n = 9) were used for counting (lower panels). (c) The expression levels of marker genes in the sorted XY and XX/Sry ALCs, along with those of EGFP and mCherry, were examined using qRT-PCR. XY and XX/Sry testes were used as controls. Three biologically independent samples (n = 3) were used. The data were normalized to Actb and are presented as means ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, using Student’s t-test (b). R software version 3.4.3 (https://www.r-project.org) was used to draw the plots in (b,c).
Fluorescence-activated cell sorting (FACS) of the testicular cells enabled us to isolate two distinct Leydig cell populations, EGFP single-positive ALCs and EGFP/mCherry double-positive FLCs, from both XY and XX/Sry testes (Fig. 1b). Since the XX/Sry adult testes were hypoplastic and lacked all germ cells (Supplemental Fig. 1), the total number of cells in a single XX/Sry testis was substantially lower than that in a single XY testis. Surprisingly, however, the number of ALCs in the XX/Sry testis was close to double that in the XY testis (Fig. 1b). The purity of the ALC fraction prepared by FACS was examined using qRT-PCR for testicular cell marker genes. EGFP and Hsd3b1 were highly enriched in ALCs from both XY and XX/Sry testes, whereas mCherry was barely detectable in either group (Fig. 1c). Germ cell markers Ddx4 and Prm2 were undetectable in the ALCs, as was Sertoli cell marker Sox9. These results indicate that the ALC fraction used in this study was unlikely to have been contaminated with FLCs, germ cells, or Sertoli cells.
Differential gene expression between XY and XX/Sry ALCs
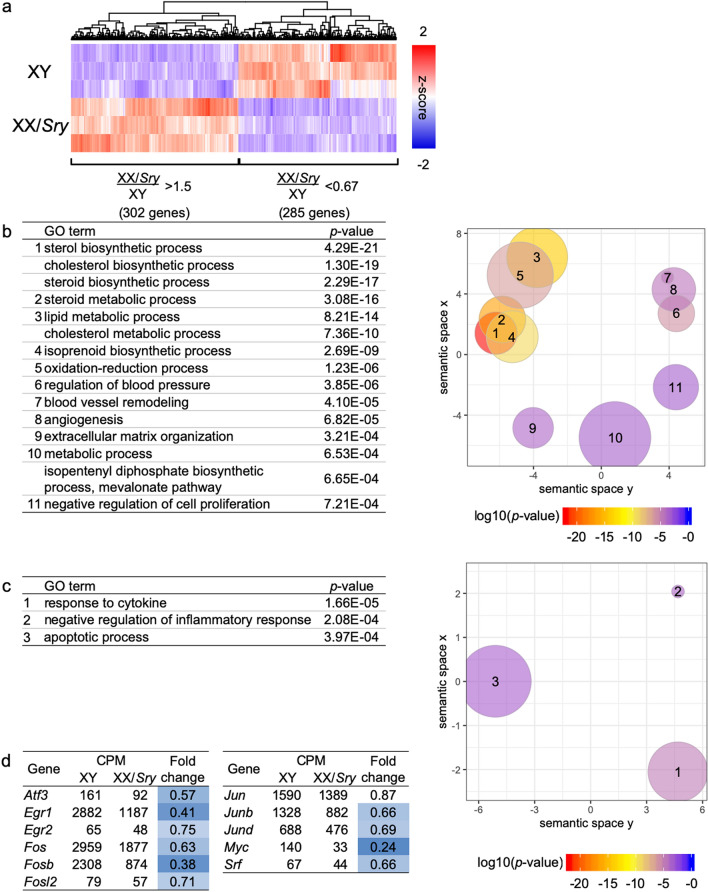
To investigate whether XX/Sry ALCs differ from XY ALCs, transcriptomes were obtained from three biologically independent sets of ALC samples each from XY and XX/Sry testes. Considering the high unique mapping rate of the sequence reads (approximately 90%) and the high reproducibility between the biological triplicates (correlation coefficient > = 0.992; Supplemental Fig. 2a and 2b), the quality of the transcriptome datasets was considered sufficient for further examination. Comparison of the transcriptomes revealed that the expression levels of 302 and 285 genes were more than 1.5-fold higher and lower, respectively, in the XX/Sry ALCs compared to the XY ALCs (Fig. 2a, Supplemental Tables 1 and 2).
Figure 2.
Genes differentially expressed in XY and XX/Sry ALCs. (a) Heatmap of differentially expressed genes in XY and XX/Sry ALCs, based on a comparison of their transcriptomes. The expression levels of 302 genes were at least 1.5 times higher in XX/Sry than XY ALCs, and those of 285 genes in XX/Sry were at most two third of those in XY ALCs. (b) GO terms identified by GO pathway analyses of the 302 upregulated genes in XX/Sry ALCs are indicated (left) and visualized in a two-dimensional plot using REVIGO (right). The top 15 GO terms (p < 0.001) were plotted by REVIGO after four redundant terms had been excluded. The numbers in the plots correspond to the numbers of the GO terms shown in the left-hand column of the table. The plot colors indicate the p-values of the GO terms as per the table, and plot sizes indicate the specificity of the terms (plots for more specific terms are smaller). (c) GO terms identified by GO pathway analyses of the 285 genes downregulated in XX/Sry ALCs are indicated and visualized as described above. The top three GO terms (p < 0.001), which were used for the analysis, are listed in the table on the left and plotted in the panel on the right. (d) Expression of immediate early genes in XY and XX/Sry ALCs. CPMs are means of biological triplicates for each cell type. Cells in the fold change column are shaded according to difference in expression: the greater the decrease, the deeper the blue shading. R software version 3.4.3 (https://www.r-project.org) was used to draw the plots in (a–c).
These differentially expressed genes were subjected to GO pathway analysis to investigate which biological processes are associated with the genes up- and downregulated in the XX/Sry ALCs. As listed in Fig. 2b (left panel), ‘sterol biosynthetic process’, ‘cholesterol biosynthetic process’, ‘steroid biosynthetic process’, and ‘steroid metabolic process’ were strongly related to the genes upregulated in the XX/Sry ALCs. Of these genes, the ones commonly associated with these processes were predominantly cholesterogenic. The next most strongly represented process was ‘lipid metabolic process’. Although the gene list for this process includes cholesterogenic genes, it also includes genes specifically required for lipid synthesis. In accordance with the sharing of cholesterogenic genes, REVIGO plot analysis suggested that these processes involving cholesterogenesis seemed to form a cluster at the top left (right panel in Fig. 2b).
Multiple terms related to blood vessels were listed, and these formed another cluster (Fig. 2b). This suggests that although we could not find any clear defect, the blood vessels of the XX/Sry testes may be affected by the differential expression of these genes. In addition to the genes included in the terms above, we noticed that the expression of extracellular matrix genes (such as those associated with several types of collagen, laminin, and biglycan) was higher in the XX/Sry ALCs, suggesting that the extracellular matrix surrounding XX/Sry ALCs is different from that surrounding XY ALCs.
A few biological processes were related to the genes downregulated in the XX/Sry ALCs, and their p-values were relatively large compared with those related to the upregulated genes (left panel in Fig. 2c). REVIGO plot analysis suggested that these biological terms were not closely related (right panel in Fig. 2c). Although any close correlations between the listed terms and Leydig cell functions were unlikely, we noticed that the term ‘response to cytokine’ includes the Fos, Junb, and Jund genes. These gene products, leucine zipper-type transcription factors, have been studied extensively and found to be activated in response to a variety of stimuli, such as serum, growth factors, and cytokines31. Since these genes have been classified into immediate early genes, we examined whether the expression levels of other genes in this group were affected in XX/Sry ALCs. Interestingly, many other immediate early genes, such as Atf3, Egr1, and Myc, were also downregulated in XX/Sry ALCs compared to XY ALCs (Fig. 2d).
Cholesterogenic gene expression increased in XX/Sry ALCs
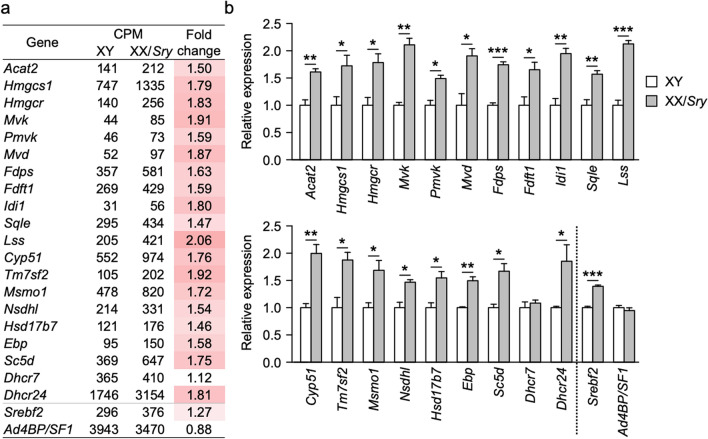
Since cholesterogenic pathway is involved in the biological functions activated in the XX/Sry ALCs, we examined the expression of cholesterogenic genes in the XY and XX/Sry ALCs. The transcriptome data indicated that almost all the cholesterogenic genes were upregulated more than 1.5-fold in the XX/Sry ALCs (Fig. 3a). This increased expression was confirmed by qRT-PCR (Fig. 3b). Numerous studies have demonstrated that SREBP2, encoded by Srebf2, plays a crucial role in cholesterogenic gene regulation28. In fact, it has been demonstrated that SREBP2 accumulates in the regions upstream of cholesterogenic genes32. In addition, we recently demonstrated that Ad4BP/SF-1 also accumulates at cholesterogenic gene loci in steroidogenic cells, including ALCs29. Therefore, we expected that at least one of these two transcription factors would also be upregulated in the XX/Sry ALCs. Although the expression of Ad4BP/SF-1 was unaltered in these cells, Srebf2 expression was slightly higher in the XX/Sry ALCs. This altered expression of Srebf2 could be responsible, at least in part, for the observed enhanced expression of cholesterogenic genes in the XX/Sry ALCs.
Figure 3.
Cholesterogenic gene expression increased in XX/Sry ALCs. (a) Expression of cholesterogenic genes as well as Srebf2 and Ad4BP/SF-1 was extracted from the transcriptome datasets for the XY and XX/Sry ALCs. CPMs are means of biological triplicates. Cells in the fold change column are shaded according to difference in expression: the greater the increase, the deeper the red shading. (b) Expression of the cholesterogenic genes, Srebf2, and Ad4BP/SF-1 was validated using qRT-PCR. The data were normalized to Actb and are presented as means ± SEM. Three biologically independent samples (n = 3) were used. *p < 0.05, **p < 0.01, ***p < 0.001, using Student’s t-test (b). R software version 3.4.3 (https://www.r-project.org) was used to draw the plots in (b).
Differential effects on gene expression between XX/Sry ALCs and Sertoli cells
We previously compared gene expression between XY and XX/Sry Sertoli cells and found that cholesterogenic genes were downregulated in the latter6. Accordingly, the present study showed that cholesterogenic gene expression was affected in opposite ways between the XX/Sry ALCs and Sertoli cells. We graphically compared whole-gene expression changes in the two types of cells. Fold changes in gene expression levels (XX/Sry vs. XY) for Sertoli cells were plotted on the vertical axis and for ALCs on the horizontal axis (Fig. 4). If genes were up- or downregulated in both types of XX/Sry cells, they would be lie on or near the red broken line in Fig. 4a. However, there was no particular pattern of distribution along this line. Instead, a considerable number of genes were aligned along the lines x = 0 or y = 0, suggesting that the alteration of gene expression was probably cell-type specific.
Figure 4.
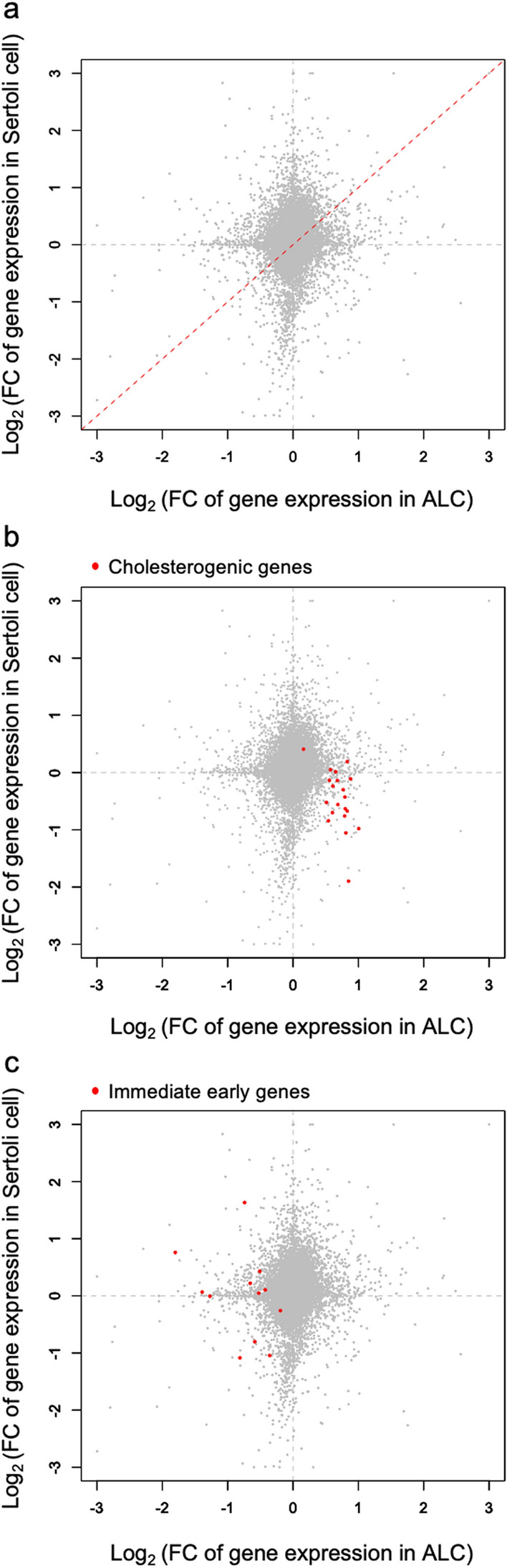
Differentially affected gene expression in XX/Sry ALCs and Sertoli cells. (a–c) Fold changes in gene expression were calculated as the ratio of the CPM values for XX/Sry Sertoli cells to those for XY Sertoli cells, and of CPM values for XX/Sry ALCs to those for XY ALCs. (a) Whole genes were plotted according to the calculated values. The dotted red line indicates where genes that are similarly expressed in the two cell types would fall. The horizontal and vertical axes represent log2 fold change (FC) in gene expression in the ALCs and Sertoli cells, respectively. A dot corresponds to a single gene. (b) Cholesterogenic genes are depicted as red dots. (c) Immediate early genes are depicted as red dots. R software version 3.4.3 (https://www.r-project.org) was used to draw the plots in (a–c).
Cholesterogenic genes are indicated with red dots in the plot shown in Fig. 4b. As expected, many of these genes are localized in the lower right quadrant, which is consistent with our finding that cholesterogenic genes were upregulated in the XX/Sry ALCs but downregulated in the XX/Sry Sertoli cells. As mentioned above, immediate early genes were downregulated in the XX/Sry ALCs. However, no biased expression of this group was detected in the XX/Sry Sertoli cells. Consistent with this, the immediate early genes are distributed within the left half of the plot area (Fig. 4c).
Altered expression of genes normally enriched in ALCs
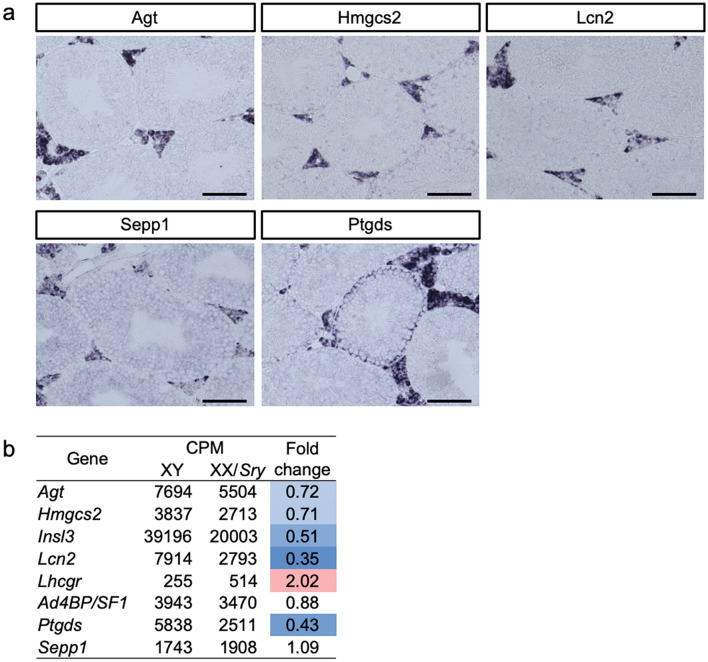
It has been established that the expression levels of Insl3, Ad4BP/SF-1, and Lhcgr (Luteinizing hormone/choriogonadotropin receptor) are enriched in ALCs33,34. In addition, we previously found several candidate genes that are probably enriched in ALCs by comparing the transcriptomes of ALCs and FLCs30. In the present study, we examined the expression of these genes via in situ hybridization. As shown in Fig. 5a, Agt (angiotensinogen) was expressed in ALCs but not in Sertoli or germ cells in adult testes. Enriched expression in ALCs has previously been observed for Hmgcs2 (3-hydroxy-3-methylglutaryl-CoA synthase 2)35,36, Lcn2 (lipocalin-2)37, and Sepp1 (selenoprotein P, plasma, 1)38. A high level of Ptgds (prostaglandin D2 synthase) expression was detected in ALCs, although the expression was also detected in Sertoli cells from some, but not all, testicular tubules39. Interestingly, the transcriptomes obtained in the present study revealed that many of these genes were downregulated in the XX/Sry ALCs (Fig. 5b).
Figure 5.
Altered expression of genes normally enriched in ALCs. (a) Expression of Agt, Hmgcs2, Lcn2, Sepp1 and Ptgds in XY adult testes was examined using in situ hybridization. Scale bars = 100 μm. (b) Expression of genes that are normally enriched in ALCs in XY and XX/Sry ALCs was obtained from the transcriptome datasets. The displayed CPMs are the means of biological triplicates. Increased and decreased gene expression in the XX/Sry ALCs is indicated in red and blue, respectively, with deeper shading for larger differences.
Steroidogenesis possibly affected by decreased steroid 17,20-lyase activity
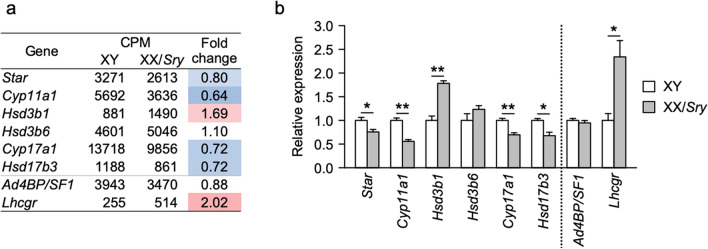
We previously demonstrated that the amount of testosterone synthesized in XX/Sry testes at postnatal day 21 was smaller than in XY testes6. We therefore extracted the expression data for steroidogenic genes from our transcriptome datasets. The expression of Star, Cyp11a1, Cyp17a1, and Hsd17b3 was decreased to approximately 70% of XY ALC levels in the XX/Sry ALCs (Fig. 6a). Similar expression profiles for these genes were obtained using qRT-PCR (Fig. 6b). The expression of Ad4BP/SF-1, a key regulator of steroidogenic gene expression, was not significantly affected in the XX/Sry ALCs, while that of Lhcgr was more than doubled.
Figure 6.
Steroidogenic gene expression is affected in XX/Sry ALCs. (a) Expression of steroidogenic genes as well as Ad4BP/SF-1 and Lhcgr in the XY and XX/Sry ALC transcriptome datasets. CPMs are the means of biological triplicates. Increased and decreased gene expression in the XX/Sry ALCs is indicated in red and blue, respectively, with deeper shading for larger differences. (b) Expression of the genes shown in (a) was validated by qRT-PCR. The data were normalized to Actb and are presented as means ± SEM. Three biologically independent samples (n = 3) were used. * p < 0.05, ** p < 0.01, *** p < 0.001, using Student’s t-test (b). R software version 3.4.3 (https://www.r-project.org) was used to draw the plots in (b).
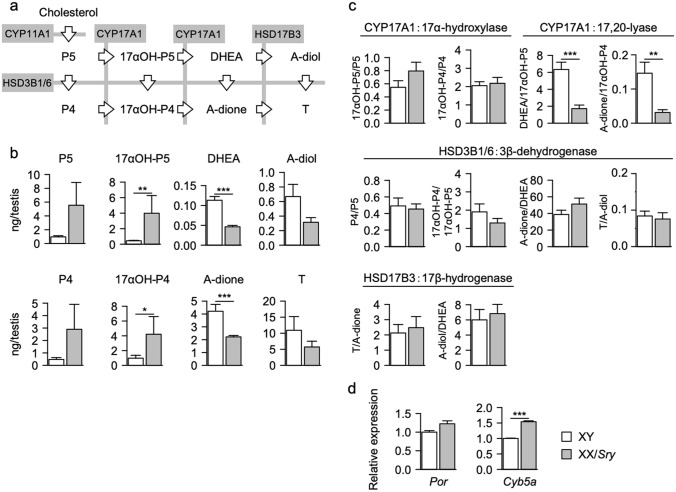
To examine whether these changes affected steroidogenesis, the quantities of steroidal molecules were determined for both XY and XX/Sry testes. Testosterone synthesis from cholesterol is mediated by multiple enzymes (Fig. 7a). As indicated in Fig. 7b, the quantities of P5 (pregnenolone), P4 (progesterone), 17αOH-P5 (17α-hydroxypregnenolone), and 17αOH-P4 (17α-hydroxyprogesterone) in the XX/Sry testes were greater than those in the XY testes. Interestingly, however, the quantities of DHEA (dehydroepiandrosterone), A-dione (androstenedione), A-diol (androstenediol), and T (testosterone) in the XX/Sry testes were smaller than those in the XY testes. Based on these steroid quantities, the enzymatic activities were evaluated by calculating metabolic ratios. While 17α-hydroxylation, 3β-dehydrogenation, and 17β-hydroxylation were not significantly altered, the 17,20-lyase reaction was substantially reduced in the XX/Sry testes (Fig. 7c). Interestingly, 17α-hydroxylation and 17,20-lyase reaction are mediated by a single enzyme, CYP17A1. Electrons from NADPH/NADH required for these reactions are transferred to CYP17A1 from POR (P450 oxidoreductase) and/or CYB5A (cytochrome b5a). The expression of Cyb5a was increased in the XX/Sry ALCs (Fig. 7d), but that of Por was not significantly altered.
Figure 7.
17,20-lyase activity of CYP17A1 is reduced in XX/Sry testes. (a) The pathway of testosterone synthesis from cholesterol. The enzymes implicated in the pathway are shown in gray boxes. P5, pregnenolone; P4, progesterone; 17αOH-P5, 17α-hydroxy-pregnenolone; 17αOH-P4, 17α-hydroxyprogesterone; DHEA, dehydroepiandrosterone; A-dione, androstenedione; A-diol, androstenediol; T, testosterone. (b) The quantities of these steroids in the XY and XX/Sry testes were determined using GC–MS/MS. Eight biologically independent samples (n = 8) were used in this analysis. (c) Metabolic ratios for all reactions were calculated as the ratio of substrate to metabolite using the quantities of intratesticular steroids detected in the XY and XX/Sry testes. (d) Expression of Por and Cyb5a genes were examined using qRT-PCR. The data were normalized to Actb and are presented as means ± SEM. Three biologically independent samples (n = 3) were used for each cell type. * p < 0.05, ** p < 0.01, *** p < 0.001, using Mann–Whitney U test (b,c) and Student’s t-test (d). R software version 3.4.3 (https://www.r-project.org) was used to draw the plots in (b–d).
Discussion
In the present study, we aimed to determine whether XX/Sry ALCs are functionally equivalent to XY ALCs. To investigate it, transcriptomes obtained from XY and XX/Sry ALCs were compared. As the consequence, the expression of 302 and 285 genes was found to be up- and downregulated, respectively, in the XX/Sry ALCs compared to XY ALCs. These gene sets suggested that several biological activities and processes are affected in XX/Sry ALCs.
There are potential reasons why the number of ALCs was increased in XX/Sry testes. LH has been established to be one of the key molecules for differentiation of ALCs. In fact, ALCs were decreased in the testis of Lhcgr KO mice40. Moreover, transgenic overexpression of human chorionic gonadotropin (HCG), which potentially binds and activates LHCGR, resulted in an increase of ALCs41,42. Based on these findings, the increase of ALCs in the XX/Sry testes might be due to the increased expression of Lhcgr.
In addition to the endocrine factor above, there are several paracrine factors regulating differentiation of ALCs. Desert hedgehog (DHH), secreted by Sertoli cells, stimulates proliferation of stem Leydig cells and their differentiation into ALCs43. However, our previous study showed that the expression of Dhh was not altered in the XX/Sry Sertoli cells compared to XY cells6 (Supplemental Fig. 3a). Likewise, the expression of the hedgehog signaling components such as Ptch1/2 and Smo was not affected in the XX/Sry ALCs (Supplemental Fig. 3b). PDGF is another factor to activate proliferation of stem Leydig cells43. Although it has been unclear which cells synthesize PDGFs in adult testes, the expression of Pdgfc was increased in the XX/Sry Sertoli cells. Interestingly, the expression of the receptor gene, Pdgfra, was increased in the XX/Sry ALCs. Taken together, it was suggested that the increase of ALCs in the XX/Sry testes might be attributable to the augmentation of PDGF together with LH signaling.
ALCs actively synthesize testosterone through abundant expression of steroidogenic genes. Our transcriptomic analysis revealed that the expression of all steroidogenic genes except Hsd3b1 and Hsd3b6 was lower in the XX/Sry ALCs compared to XY ALCs. Similarly, we found that the expression of genes normally enriched in ALCs was suppressed, suggesting that the characteristic features of ALCs were affected in the XX/Sry ALCs. With respect to the reason for the suppressed expression of these genes, it is interesting to note the downregulation of immediate early genes, whose expression is activated by multiple stimuli31, in the XX/Sry ALCs. Indeed, the immediate early genes such as Fos, Jun, Junb, and Jund (AP1 family members) are activated in ALCs by hCG44. It could therefore be assumed that the gene products above activate cellular functions by enhancing the transcription of certain sets of target genes. In fact, steroidogenic gene transcription is regulated by the AP1 family members45,46. In addition to the steroidogenic genes, it has been demonstrated that LCN2 displaying ALC-enriched expression is regulated by EGR147. In the present study, we demonstrated that the expression levels of immediate early genes were decreased in the XX/Sry ALCs. The decreased expression of steroidogenic and ALC-enriched genes might therefore be caused by the downregulation of immediate early genes.
Based on this scenario, the concentration of LH secreted by the pituitary and the expression of its receptor, LHCGR, in ALCs should be considered. Our previous study showed that the serum LH concentration in the XX/Sry mice was comparable to that of the XY mice6, but the present study showed that the expression of Lhcgr was higher in the XX/Sry ALCs than in the XY ALCs. Therefore, the XX/Sry ALCs might receive more effectively the LH signal than the XY ALCs. If it is the case, gene transcription downstream of LH signal such as Fos and Jun could be activated. Nevertheless, the expression of the immediate early genes was found to be downregulated. Therefore, this inconsistent outcome suggests that intracellular signal transduction might be abnormally regulated in XX/Sry ALCs, although it remains unknown which components and/or steps may be affected.
Many transcription factors have been shown to regulate steroidogenic genes. Our transcriptome datasets revealed that the expression of Cebpb (C/EBPβ) and Fos was decreased less than 0.67-fold while that of Nr3c1 was increased more than 1.5-fold in the XX/Sry ALCs (Supplemental Fig. 4). C/EBPβ and FOS were reported to regulate positively mouse Star and human CYP11A1 genes46,48,49. Therefore, the decreased expression of Cyp11a1 and Star genes might be due to the downregulated expression of Cebpb and Fos in the XX/Sry ALCs. NR3C1 (GR) was reported to act as a suppressor of mouse Star gene transcription50. Thus, the upregulated expression of Nr3c1 might be responsible for the decreased expression of Star gene in the XX/Sry ALCs.
Steroidogenesis from cholesterol takes place via multiple enzymatic reactions. Based on our analyses of the metabolic ratios, we realized that the 17,20-lyase reaction mediated by CYP17A1 was selectively affected in the XX/Sry ALCs. CYP17A1 catalyzes two reactions: 17α-hydroxylation and 17,20-lyase reaction14. In many mammalian species, cortisol (glucocorticoid) is synthesized in the zona fasciculata of the adrenal cortex, while testosterone is synthesized in ALCs. In the former process, CYP17A1 mediates only 17α-hydroxylation, whereas in the latter process it mediates both 17α-hydroxylation and 17,20-lyase reaction.
Many studies have been performed to improve our understanding of the mechanism for selective regulation of these two reactions by CYP17A151. Some of them have focused on the two components, POR and CYB5A, which transport electrons to CYP17A1. One study reported that POR preferentially activates the 17,20-lyase reaction52, while another reported that CYB5A is responsible for this activation53. Concordantly, a KO study has shown that Cyb5a is necessary for 17,20-lyase activity in ALCs54. Unexpectedly, however, the expression of Por and Cyb5a was not decreased in the XX/Sry ALCs. Another possible regulatory mechanism of the two reactions, phosphorylation of CYP17A1 by cAMP-dependent protein kinase, p38α, and an unknown kinase activated under serum-free condition has been shown to selectively increase 17,20-lyase activity55–57. Unfortunately, however, our preliminary study failed to detect the phosphorylated CYP17A1 in the XY and XX/Sry testes. Although we could not unveil the mechanism for the selective regulation of the CYP17A1-mediated reactions, our study revealed that XX/Sry ALCs could be an excellent cellular tool for future investigation of it.
In our previous study, we examined histone modifications and showed that accumulation of H3K4me3 around the upstream regions of cholesterogenic genes was reduced in XX/Sry Sertoli cells6. Considering that H3K4me3 is a mark for an active promoter, we concluded that this reduction may have led to the decreased expression of cholesterogenic genes in the XX/Sry Sertoli cells. Interestingly, our present study demonstrated that immediate early genes and cholesterogenic genes were differentially altered in XX/Sry ALCs and XX/Sry Sertoli cells. Comparison of whole-genome histone modifications could contribute to a deeper understanding of the mechanisms underlying cell-type-specific alteration of gene expression in XX/Sry mice.
Materials and methods
Animals
Wild-type XY C57BL/6 and XX sex-reversed transgenic mice carrying the Hsp-Sry transgene were used58. The presence of the transgene and genetic sex were confirmed via PCR with primers for Hsp-Sry and SX59 (Supplemental Table 3). SX is a single set of primers to amplify Xlr and Sly on the X- and Y-chromosome, respectively, giving distinct banding patterns after electrophoresis. We also used Ad4BP-BAC-EGFP mice and mFLE-mCherry mice60, in which Leydig cells and FLCs are labeled with EGFP and mCherry, respectively. Sry transgenic mice were crossed with Ad4BP-BAC-EGFP;mFLE-mCherry mice to obtain EGFP single-positive ALCs from the testes of XX/Sry mice. All protocols for the animal experiments were approved by the Animal Care and Use Committee of Kyushu University. All experiments were performed in accordance with the institutional guidelines.
Cell counting and sorting
Testes were collected from eight-week-old Ad4BP-BAC-EGFP;mFLE-mCherry double-transgenic mice and dispersed with collagenase30. Total numbers of cells from XY and XX/Sry testes were counted using a Countess II FL (Thermo Fisher Scientific, Waltham, MA, USA). The dispersed cells were subjected to FACS using a BD FACS Aria SORP (BD Biosciences, San Jose, CA, USA) and FACS Diva software (BD Biosciences) to sort the cells into two populations based on EGFP and mCherry fluorescence (ALCs: EGFP single-positive; FLCs: EGFP/mCherry double-positive). 1,000,000 cells were analyzed to obtain the percentages of ALCs and FLCs, which were converted to the absolute numbers per testis by multiplying the total numbers of testicular cells. The EGFP single-positive ALCs were purified by performing two FACS cycles.
Immunofluorescence analyses
Eight-week-old mice were perfused with 4% paraformaldehyde (PFA) and their testes were collected and then immersed in 4% PFA at 4 °C for 48 h. The samples were subsequently cryoprotected in 20% sucrose at 4 °C and embedded in OCT Compound (Sakura Finetek, Torrance, CA, USA). Immunofluorescence analyses were performed as described previously11. A rabbit antibody against HSD3B661 (1:500), a chicken antibody against EGFP (ab13970, 1:1000; Abcam, Cambridge, UK), and a mouse antibody against mCherry (ab125096, 1:200; Abcam) were used as the primary antibodies. Alexa Fluor 488-labeled goat anti-chicken IgY antibody (ab150169, 1:500; Abcam), Alexa Fluor 555-labeled goat anti-mouse IgG antibody (A28180, 1:500; Life Technologies, Carlsbad, CA, USA), and Alexa Fluor 647-labeled goat anti-rabbit IgG antibody (A27040, 1:500; Life Technologies) were used as the secondary antibodies. Nuclei were stained with DAPI (4′6-diamidino-2-phenylindole; Sigma–Aldrich, St. Louis, MO, USA). Immunofluorescence was observed under a BZ-X700 microscope (Keyence, Osaka, Japan).
In situ hybridization and immunohistochemistry
In situ hybridization was performed as previously described62. RIKEN FANTOM cDNA clones for Agt (angiotensinogen, A730059G17), Hmgcs2 (3-hydroxy-3-methylglutaryl-coenzyme A synthase 2, 1300002P16), Ptgds (prostaglandin D2 synthase, 2010004I02), Lcn2 (lipocalin-2, 2G530015N18), and Sepp1 (selenoprotein P, plasma, 1; I920052L16) were purchased (DNAFORM, Yokohama, Japan). Digoxigenin-labeled riboprobes for these genes were used (Roche, Basel, Switzerland).
qRT-PCR
qRT-PCR was performed as previously described63 and conducted following the MIQE guidelines64. In brief, total RNA was isolated from the sorted cells or tissues using RNeasy Micro Kit or RNeasy Mini Kit (Qiagen, Hilden, Germany) and 50 ng of total RNA was reverse-transcribed to cDNA using random hexamers and M-MLV Reverse Transcriptase (Thermo Fisher Scientific). RNA integrity numbers (RINs) of all samples were confirmed to be higher than 7.5 using a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). qRT-PCR was performed using a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the SYBR Select Master Mix (Applied Biosystems, Foster City, CA, USA). Gene expression was determined using the standard curve method. The correlation coefficients (R2) for the standard curves were higher than 0.99. Gene expression levels were normalized to those of Actb (β-actin). The primers used for the PCR are listed in Supplemental Table 3.
mRNA sequencing
mRNA sequencing was performed as described previously30. Briefly, poly(A) RNA content was isolated from total RNA (10 ng per sample) prepared from sorted XY and XX/Sry ALCs using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Ipswich, MA, USA). Sequence libraries were constructed using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs) and NEBNext Multiplex Oligo for Illumina (Dual Index Primers Set 1; New England Biolabs). cDNA libraries were sequenced using a NovaSeq 6000 (51-bp pair-end; Illumina, San Diego, CA, USA).
Data processing
The FastQ files were mapped using STAR software65 (version 2.7.0a; standard option) to the mouse reference genome (UCSC mm10) and the genome annotation (modified to integrate the EGFP and mCherry transgenes) was downloaded from the UCSC Genome Browser. Bam files were generated using SamTools66 (version 0.3.3). Quality control, mapping, read count, and CPM (counts per million mapped reads) were computed using featureCounts67 (version 1.6.4; option ‘-O -p’), edgeR68 (version 3.20.9), and an in-house pipeline. MicroRNA and small nucleolar RNA genes were excluded from the analyses. Gene expression data are presented as CPM. Mean values for biological replicates (n = 3) were calculated, and genes with CPM values < 20 in both XY and XX/Sry ALCs were removed. Differentially expressed genes were identified based on fold change and subjected to Gene Ontology (GO) analyses using DAVID. The significantly enriched biological process GO terms with p-values < 0.001 were visualized in two-dimensional plots using REVIGO69. Fold changes in gene expression levels (XX/Sry vs. XY) for Sertoli cells were also calculated using the transcriptome data in our previous study6 (accession number: DRA004090). When comparing whole gene expression changes in the two types of cells, a pseudo-count of 10 was added to the CPM values before the fold changes were calculated.
Measurement of intratesticular sex steroids
Testes obtained from eight-week-old XY and XX/Sry mice were lyophilized using a Vacuum Centrifugal Evaporator (CVE-2000; EYELA, Tokyo, Japan) and stored at − 80 °C until later use. Gas chromatography–mass spectrometry steroid profiling was performed using a Shimadzu GC 2010 Plus gas chromatograph coupled to a triple-quadrupole GCMS-TQ8050 (Shimadzu Corporation, Kyoto, Japan) as previously described70. Quantitative results were based on absolute quantities of steroid molecules per testis, and their metabolic ratios were also calculated to express their corresponding enzymatic activities.
Statistical analysis
At least three biologically independent samples were used in all experiments. Data are presented as mean ± SEM. Differences between XY and XX/Sry cells or testes were examined using two-tailed Student’s t-tests or Mann–Whitney U tests, and statistical significance was inferred at p < 0.05. All statistical analyses were performed using R software version 3.4.3 (https://www.r-project.org).
Supplementary Information
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers JP17H06427 (K.-i.M.), JP16H05142 (K.-i.M.), JP16K08593 (T.B.), and JP19J12133 (S.Y.), Takeda Science Foundation (T.B.), and The Shin-Nihon Foundation of Advanced Medical Research (T.B.). This work was technically supported by the Research Support Center, Graduate School of Medical Sciences, Kyushu University. We are profoundly thankful to Prof. Mikita Suyama (Kyushu University, Japan) for technical support in the NGS data processing, and Profs Hitoshi Okamura and Masao Doi (Kyoto University, Japan) for kindly providing the HSD3B6 antibody.
Author contributions
S.Y., T.B. and K.-i.M. conceived and designed the experimental approach and performed experiments. S.Y. and K.-i.M. prepared the manuscript. K.I. contributed to the computational analyses for mRNA-seq. M.I. and K.Mi. performed in situ hybridization. S.H. and M.H.C. measured intratesticular sex steroids. F.T. constructed the mRNA-seq libraries. Y.K. provided the transgenic mice. Y.O. performed deep sequencing of the mRNA-seq libraries.
Data availability
The transcriptome data have been deposited in DDBJ under the accession number DRA009797 and DRA010792.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80741-z.
References
- 1.Koopman P, Münsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 2.Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Development. 2010;137:3921–3930. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- 3.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 4.Ishii M, et al. Potency of testicular somatic environment to support spermatogenesis in XX/Sry transgenic male mice. Development. 2006;134:449–454. doi: 10.1242/dev.02751. [DOI] [PubMed] [Google Scholar]
- 5.Burgoyne PS. The role of the mammalian Y chromosome in spermatogenesis. Development. 1987;101(Suppl):133–141. doi: 10.1242/dev.101.Supplement.133. [DOI] [PubMed] [Google Scholar]
- 6.Shishido Y, et al. Differential lactate and cholesterol synthetic activities in XY and XX Sertoli cells. Sci. Rep. 2017;7:41912. doi: 10.1038/srep41912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boussouar F, Benahmed M. Lactate and energy metabolism in male germ cells. Trends Endocrinol. Metab. 2004;15:345–350. doi: 10.1016/j.tem.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Keber R, Rozman D, Horvat S. Sterols in spermatogenesis and sperm maturation. J. Lipid Res. 2012;54:20–33. doi: 10.1194/jlr.r032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ariyaratne HB, Mendis-Handagama SML. Changes in the testis interstitium of sprague dawley rats from birth to sexual maturity. Biol. Reprod. 2000;62:680–690. doi: 10.1095/biolreprod62.3.680. [DOI] [PubMed] [Google Scholar]
- 10.Kerr JB, Knell CM. The fate of fetal Leydig cells during the development of the fetal and postnatal rat testis. Development. 1988;103:535–544. doi: 10.1242/dev.103.3.535. [DOI] [PubMed] [Google Scholar]
- 11.Shima Y, et al. Fetal leydig cells persist as an androgen-independent subpopulation in the postnatal testis. Mol. Endocrinol. 2015;29:1581–1593. doi: 10.1210/me.2015-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morohashi K, Baba T, Tanaka M. Steroid hormones and the development of reproductive organs. Sex. Dev. 2013;7:61–79. doi: 10.1159/000342272. [DOI] [PubMed] [Google Scholar]
- 13.Teerds KJ, Huhtaniemi IT. Morphological and functional maturation of Leydig cells: From rodent models to primates. Hum. Reprod. Update. 2015;21:310–328. doi: 10.1093/humupd/dmv008. [DOI] [PubMed] [Google Scholar]
- 14.Zuber M, Simpson E, Waterman M. Expression of bovine 17 alpha-hydroxylase cytochrome P-450 cDNA in nonsteroidogenic (COS 1) cells. Science. 1986;234:1258–1261. doi: 10.1126/science.3535074. [DOI] [PubMed] [Google Scholar]
- 15.Committee NRN. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 16.Lala DS, Rice DA, Parker KL. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol. Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 17.Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J. Biol. Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- 18.Honda S, et al. Ad4BP regulating steroidogenic P-450 gene is a member of steroid hormone receptor superfamily. J. Biol. Chem. 1993;268:7494–7502. doi: 10.1016/S0021-9258(18)53202-6. [DOI] [PubMed] [Google Scholar]
- 19.Leers-Sucheta S, Morohashi K-I, Mason IJ, Melner MH. Synergistic activation of the human type II 3β-hydroxysteroid dehydrogenase/Δ 5 -Δ 4 isomerase promoter by the transcription factor steroidogenic factor-1/adrenal 4-binding protein and phorbol ester. J. Biol. Chem. 1997;272:7960–7967. doi: 10.1074/jbc.272.12.7960. [DOI] [PubMed] [Google Scholar]
- 20.Martin LJ, et al. GATA factors and the nuclear receptors, steroidogenic factor 1/liver receptor homolog 1, are key mutual partners in the regulation of the human 3β-hydroxysteroid dehydrogenase type 2 promoter. Mol. Endocrinol. 2005;19:2358–2370. doi: 10.1210/me.2004-0257. [DOI] [PubMed] [Google Scholar]
- 21.Bakke M, Lund J. Mutually exclusive interactions of two nuclear orphan receptors determine activity of a cyclic adenosine 3',5'-monophosphate-responsive sequence in the bovine CYP17 gene. Mol. Endocrinol. 1995;9:327–339. doi: 10.1210/mend.9.3.7776979. [DOI] [PubMed] [Google Scholar]
- 22.Zhang P, Mellon SH. The orphan nuclear receptor steroidogenic factor-1 regulates the cyclic adenosine 3',5'-monophosphate-mediated transcriptional activation of rat cytochrome P450c17 (17 alpha-hydroxylase/c17-20 lyase) Mol. Endocrinol. 1996;10:147–158. doi: 10.1210/mend.10.2.8825555. [DOI] [PubMed] [Google Scholar]
- 23.Sewer MB, et al. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54 nrb /NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology. 2002;143:1280–1290. doi: 10.1210/endo.143.4.8748. [DOI] [PubMed] [Google Scholar]
- 24.Michael DM, Kilgore MW, Morohashi K-I, Simpson ER. Ad4BP/SF-1 regulates cyclic AMP-induced transcription from the proximal promoter (PII) of the human aromatase P450 ( CYP19) gene in the ovary. J. Biol. Chem. 1995;270:13561–13566. doi: 10.1074/jbc.270.22.13561. [DOI] [PubMed] [Google Scholar]
- 25.Morohashi KI, Omura T. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 1996;10:1569–1577. doi: 10.1096/fasebj.10.14.9002548. [DOI] [PubMed] [Google Scholar]
- 26.Parker KL, Schimmer BP. Steroidogenic factor 1: A key determinant of endocrine development and function. Endocr. Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 27.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell. Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 28.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/jci15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baba T, et al. Ad4BP/SF-1 regulates cholesterol synthesis to boost the production of steroids. Commun. Biol. 2018;1:18. doi: 10.1038/s42003-018-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyabayashi K, et al. Alterations in fetal leydig cell gene expression during fetal and adult development. Sex. Dev. 2017;11:53–63. doi: 10.1159/000453323. [DOI] [PubMed] [Google Scholar]
- 31.Pérez-Cadahía B, Drobic B, Davie JR. Activation and function of immediate-early genes in the nervous system. Biochem. Cell Biol. 2011;89:61–73. doi: 10.1139/o10-138. [DOI] [PubMed] [Google Scholar]
- 32.Seo Y-K, et al. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morohashi K, et al. Functional difference between Ad4BP and ELP, and their distributions in steroidogenic tissues. Mol. Endocrinol. 1994;8:643–653. doi: 10.1210/mend.8.5.8058072. [DOI] [PubMed] [Google Scholar]
- 34.O’Shaughnessy PJ, Willerton L, Baker PJ. Changes in leydig cell gene expression during development in the mouse. Biol. Reprod. 2002;66:966–975. doi: 10.1095/biolreprod66.4.966. [DOI] [PubMed] [Google Scholar]
- 35.Royo T, et al. Testis and ovary express the gene for the ketogenic mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase. J. Lipid Res. 1993;34:867–874. [PubMed] [Google Scholar]
- 36.Bagheri-Fam S, et al. The gene encoding the ketogenic enzyme HMGCS2 displays a unique expression during gonad development in mice. PLoS ONE. 2020 doi: 10.1371/journal.pone.0227411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang Z, Qiao N, Tan Z, Tang Z, Li Y. Expression patterns and changes of the LCN2 gene in the testes of induced cryptorchidism and busulfan-treated mice. Syst. Biol. Reprod. Med. 2017;63:364–369. doi: 10.1080/19396368.2017.1355416. [DOI] [PubMed] [Google Scholar]
- 38.Koga M, et al. Expression of selenoprotein-p messenger ribonucleic acid in the rat testis. Biol. Reprod. 1998;58:261–265. doi: 10.1095/biolreprod58.1.261. [DOI] [PubMed] [Google Scholar]
- 39.Gerena RL, Eguchi N, Urade Y, Killian GJ. Stage and region-specific localization of lipocalin-type prostaglandin D synthase in the adult murine testis and epididymis. J. Androl. 2000;21:848–854. doi: 10.1002/j.1939-4640.2000.tb03415.x. [DOI] [PubMed] [Google Scholar]
- 40.Huhtaniemi I, et al. Genetically modified mouse models in studies of luteinising hormone action. Mol. Cell. Endocrinol. 2006;252:126–135. doi: 10.1016/j.mce.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 41.Rulli SB, et al. Elevated steroidogenesis, defective reproductive organs, and infertility in transgenic male mice overexpressing human chorionic gonadotropin. Endocrinology. 2003;144:4980–4990. doi: 10.1210/en.2003-0403. [DOI] [PubMed] [Google Scholar]
- 42.Matzuk MM, DeMayo FJ, Hadsell LA, Kumar TR. Overexpression of human chorionic gonadotropin causes multiple reproductive defects in transgenic mice. Biol. Reprod. 2003;69:338–346. doi: 10.1095/biolreprod.102.013953. [DOI] [PubMed] [Google Scholar]
- 43.Li X, et al. Regulation of seminiferous tubule-associated stem Leydig cells in adult rat testes. Proc. Natl. Acad. Sci. U.S.A. 2016;113:2666–2671. doi: 10.1073/pnas.1519395113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultz R, Kononen J, Pelto-Huikko M. Induction of immediate early gene mRNAs and proteins by hCG in interstitial cells of rat testis. J. Endocrinol. 1995;144:417–424. doi: 10.1677/joe.0.1440417. [DOI] [PubMed] [Google Scholar]
- 45.Manna PR, Stocco DM. The role of JUN in the regulation of PRKCC-mediated STAR expression and steroidogenesis in mouse Leydig cells. J. Mol. Endocrinol. 2008;41:329–341. doi: 10.1677/jme-08-0077. [DOI] [PubMed] [Google Scholar]
- 46.Guo I-C, Huang C-Y, Wang C-K, Chung B-C. Activating protein-1 cooperates with steroidogenic factor-1 to regulate 3′,5′-cyclic adenosine 5′-monophosphate-dependent human CYP11A1 transcription in vitro and in vivo. Endocrinology. 2007;148:1804–1812. doi: 10.1210/en.2006-0938. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, et al. TCF7L2 and EGR1 synergistic activation of transcription of LCN2 via an ERK1/2-dependent pathway in esophageal squamous cell carcinoma cells. Cell. Signal. 2019;55:8–16. doi: 10.1016/j.cellsig.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Manna PR, Wang X-J, Stocco DM. Involvement of multiple transcription factors in the regulation of steroidogenic acute regulatory protein gene expression. Steroids. 2003;68:1125–1134. doi: 10.1016/j.steroids.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Mizutani T, et al. C/EBPβ (CCAAT/enhancer-binding protein β) mediates progesterone production through transcriptional regulation in co-operation with SF-1 (steroidogenic factor-1) Biochem. J. 2014;460:459–471. doi: 10.1042/BJ20131522. [DOI] [PubMed] [Google Scholar]
- 50.Martin LJ, Tremblay JJ. Glucocorticoids antagonize cAMP-induced Star transcription in Leydig cells through the orphan nuclear receptor NR4A1. J. Mol. Endocrinol. 2008;41:165–175. doi: 10.1677/JME-07-0145. [DOI] [PubMed] [Google Scholar]
- 51.Miller WL, Tee M. The post-translational regulation of 17,20 lyase activity. Mol. Cell. Endocrinol. 2015;408:99–106. doi: 10.1016/j.mce.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 52.Yanagibashi K, Hall PF. Role of electron transport in the regulation of the lyase activity of C21 side-chain cleavage P-450 from porcine adrenal and testicular microsomes. J. Biol. Chem. 1986;261:8429–8433. [PubMed] [Google Scholar]
- 53.Onoda M, Hall PF. Cytochrome b5 stimulates purified testicular microsomal cytochrome P-450 (C21 side-chain cleavage) Biochem. Biophys. Res. Commun. 1982;108:454–460. doi: 10.1016/0006-291x(82)90850-6. [DOI] [PubMed] [Google Scholar]
- 54.Sondhi V, et al. Impaired 17,20-Lyase activity in male mice lacking cytochrome b5 in leydig cells. Mol. Endocrinol. 2016;30:469–478. doi: 10.1210/me.2015-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang LH, Rodriguez H, Ohno S, Miller WL. Serine phosphorylation of human P450c17 increases 17,20-lyase activity: Implications for adrenarche and the polycystic ovary syndrome. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10619–10623. doi: 10.1073/pnas.92.23.10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tee MK, Miller WL. Phosphorylation of human cytochrome P450c17 by p38α selectively increases 17,20 lyase activity and androgen biosynthesis. J. Biol. Chem. 2013;288:23903–23913. doi: 10.1074/jbc.M113.460048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kempná P, Hirsch A, Hofer G, Mullis PE, Flück CE. Impact of differential P450c17 phosphorylation by cAMP stimulation and by starvation conditions on enzyme activities and androgen production in NCI-H295R cells. Endocrinology. 2010;151:3686–3696. doi: 10.1210/en.2010-0093. [DOI] [PubMed] [Google Scholar]
- 58.Kidokoro T, et al. Influence on spatiotemporal patterns of a male-specific Sox9 activation by ectopic Sry expression during early phases of testis differentiation in mice. Dev. Biol. 2005;278:511–525. doi: 10.1016/j.ydbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 59.McFarlane L, Truong V, Palmer JS, Wilhelm D. Novel PCR assay for determining the genetic sex of mice. Sex. Dev. 2013;7:207–211. doi: 10.1159/000348677. [DOI] [PubMed] [Google Scholar]
- 60.Miyabayashi K, et al. Heterogeneity of ovarian theca and interstitial gland cells in mice. PLoS ONE. 2015 doi: 10.1371/journal.pone.0128352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamura K, et al. Immunolocalization of murine type VI 3β-hydroxysteroid dehydrogenase in the adrenal gland, testis, skin, and placenta. Mol. Cell. Endocrinol. 2014;382:131–138. doi: 10.1016/j.mce.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 62.Sato Y, et al. Importance of forkhead transcription factor Fkhl18 for development of testicular vasculature. Mol. Reprod. Dev. 2008;75:1361–1371. doi: 10.1002/mrd.20888. [DOI] [PubMed] [Google Scholar]
- 63.Shima Y, et al. Contribution of Leydig and Sertoli cells to testosterone production in mouse fetal testes. Mol. Endocrinol. 2012;27:63–73. doi: 10.1210/me.2012-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bustin SA, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 65.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 68.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011 doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moon J-Y, Lee H, Kim J, Lee J, Choi M. Supported liquid extraction coupled to gas chromatography-selective mass spectrometric scan modes for serum steroid profiling. Anal. Chim. Acta. 2018;1037:281–292. doi: 10.1016/j.aca.2018.02.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data have been deposited in DDBJ under the accession number DRA009797 and DRA010792.