Abstract
Antibodies play a crucial role in basic research and clinical decision-making. However, there are no standardized algorithms or guidelines to ensure their accuracy and validity. There have been efforts to generate consensus, but, with the exception of clinical labs, antibody validation remains variable in the literature and sometimes in clinical practice. Here we focus on immunohistochemistry, an example of a scientific and clinical tool where validation of antibodies is critical. We describe a protocol that we use to validate antibodies specifically for immunohistochemistry, including some of the pillars of antibody validation from Uhlen et al. 2016, as an example of a rigorous approach to build antibody-based tests for both basic and translational science labs and for the clinic.
Keywords: : antibody, cancer, immunofluorescence, immunohistochemistry, protocol, validation
Antibodies are among the most commonly employed tools in scientific research, routinely used for western blot (WB), flow cytometry, immunoprecipitation, ELISA, immunohistochemistry (IHC) and immunofluorescence (IF). They also can have a critical role in clinical decision-making, as they are used broadly in anatomic pathology and laboratory medicine. IHC assays are used as diagnostic, prognostic or predictive tests and directly affect everyday management of patients in the clinical setting; for example, the assessment of PD-L1 by IHC is the definitive diagnostic test to determine whether a patient with cancer will receive an immune checkpoint inhibitor, and it costs on average $150,000 per year [1]. Thus antibody validation for IHC can be critical in both the lab and the clinic.
Data from the Human Protein Atlas indicate that at least 50% of over 2500 commercially available antibodies did not perform as expected in their intended assay [2]. Improperly validated antibodies, such as several which target estrogen receptor β, have led to nonrigorous research in promising fields [3]. Bordeaux et al. reviewed the common pitfalls of working with antibodies and common antibody validation approaches in a research lab and developed an algorithm for antibody validation for IHC and IF assays that has been downloaded by over 40,000 investigators [4]. Subsequently, Uhlen et al. convened an ad hoc International Working Group for Antibody Validation, introducing five conceptual pillars of validation to be used in an application-specific manner [5].
The FDA defines validation as ‘the collection and evaluation of data, from the process design stage through commercial production, which establishes scientific evidence that a process is capable of consistently delivering quality product’. In 2014, Fitzgibbons et al. developed laboratory practice guidelines for analytical validation and revalidation of IHC assays used in anatomic pathology clinical services [6]. Still, it is estimated that only 52% of laboratories have adopted some or all of the recommendations [7]. Sfanos et al. explained that much of the ‘reproducibility crisis’ involved with antibody use in IHC stems from end users being unaware of the need to properly validate an antibody [8].
Here we present methodology that has evolved since the work by Bordeaux et al. [4] for antibody validation in IHC and IF of formalin-fixed paraffin-embedded tumor specimens. We describe a protocol for validation that includes four distinct steps for successful antibody validation, including some of the ‘pillars of validation’ from Uhlen et al. [5] and concepts from earlier IHC methods papers [9]. Our steps include: expected localization of expression; quantitative titration; orthogonal methods of validation, genetic methods of validation, or independent epitope validation; and antibody reproducibility. Some data we present on each pillar are original and were prepared specifically for this review, and are therefore not referenced. We believe a stepwise approach to validation allows confidence in the IHC results without extreme expense.
Antibody selection considerations
For use in an IHC assay, an antibody must be highly sensitive and also highly specific to the target antigen. The best antibodies have a very high affinity and very low cross-reactivity. It is also beneficial for them to have a fast on-rate and slow off-rate. However, even with recombinant methods, it is hard and expensive to make a perfect antibody, especially for use in IHC; these assays in particular involve unique conditions for antigens, as tissue fixation can hide epitopes exposed in native or denatured forms and expose epitopes that are not exposed when the protein is in its native form in vivo [10,11].
For long-term use in a diagnostic test, and even for reproducible scientific studies, monoclonal antibodies are the best choice. Polyclonal antibodies are produced by several B-cell clones and essentially represent a pool of different antibody clones, each binding to a distinct epitope on the target antigen. While they may be good for rapid proof-of-concept studies, they represent a batch-specific mix that will ultimately be exhausted and is then impossible to reproduce exactly. Monoclonal antibodies are produced by identical B cell clones from a single parent cell and bind to a single, definable epitope on the target antigen; thus they are highly specific and consistent between experiments. Furthermore, with recombinant technologies, they can be optimized and exactly reproduced. Hence, for scientifically rigorous work, or for clinically useful diagnostic assays, a monoclonal antibody is required. As has been shown, however, monoclonal antibodies directed at a target are not all devoid of their own specificity and reproducibility issues, and care must be taken in selecting the best monoclonal antibody, which should always be validated before use [12,13].
For the average scientist or pathologist, a good monoclonal antibody directed to the ‘novel’ protein of interest can be hard to find. We routinely start by screening for all commercial antibodies available for the target of interest. For that reason, we use the web portals BenchSci and Antibodypedia [14,15]. These portals provide information about which antibodies have been tested for and used by other investigators. BenchSci is a source for the published literature in which a given commercial antibody was used, and Antibodypedia yields a ‘validation score’ for commercial antibodies in the context of particular assays. Once we have narrowed our options to a few, we purchase at least one antibody which shows the best combination of successful use in published literature and validation evidence for IHC assays. There are companies with more rigorous validation protocols that are preferred, but they may not have the ‘novel’ antibody we desire.
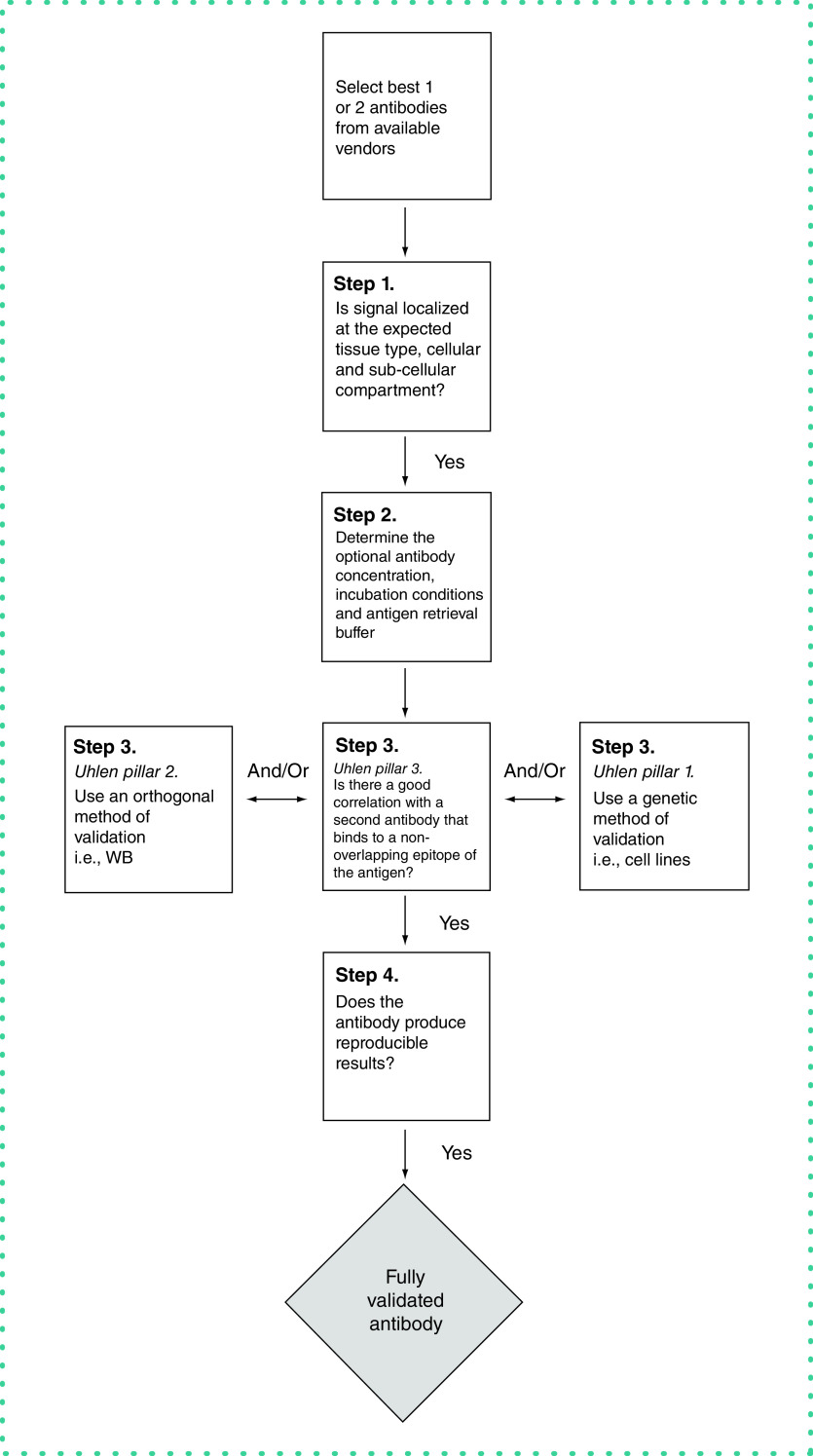
Once we have antibodies in hand, we use a series of steps (the Rimm lab antibody validation protocol) for validation (Figure 1 & Table 1). We define four particular steps that are required prior to publication of antibody-based studies of expression in human tumor cohorts.
Figure 1. . The Rimm Lab Protocol for antibody validation for immunohistochemistry or immunofluorescence.
WB: Western blotting.
Table 1. . Steps of antibody validation for immunohistochemistry, integrating some of the pillars from Uhlen et al.
| Steps of antibody validation | Summary | Common pitfalls | Significance |
|---|---|---|---|
| Step 1. Architectural or subcellular localization of expression |
Illustration of the expected biologic localization of signal for the target of interest Includes tissue type, histologic subtype, cellular and subcellular compartments |
Nonspecific staining patterns due to suboptimal antibody concentration | Provides early proof of antibody specificity Justifies further validation efforts |
| Step 2. Antibody optimization |
Optimization of assay conditions Includes optimization of antigen retrieval buffer, antibody concentration and incubation conditions |
Proper assay conditions often not indicated by the vendor | Ensures that all subsequent validation steps will be conducted under optimal assay conditions |
| Step 3. Uhlen pillar 2 Orthogonal methods of validation and/or |
Utilization of independent methods to prove antibody specificity. Includes western blot, mass spectrometry etc. |
Positive band in a western blot for nonspecific antibodies; multiple bands in a western blot for specific antibodies | Provides additional proof of antibody specificity by one or more independent methods |
| Step 3. Uhlen pillar 1 Genetic methods of validation and/or |
Genetic manipulation of the expression of the target of interest to generate positive and/or negative controls | Overexpression or underexpression of the target of interest in wild-type cell lines | Links the genetic basis of the target of interest with the corresponding protein product |
| Step 3. Uhlen pillar 3 Independent epitope validation |
Correlation of multiple antibodies with nonoverlapping epitopes for the target of interest | Identification of a second antibody for the target of interest | Provides substantial proof for the specificity of both antibodies |
| Step 4. Antibody reproducibility |
Demonstration of antibody sensitivity and specificity across different runs, operators, manual vs automated staining methods and lots | Inherent heterogeneity of the target of interest; vendor lot variability | Proves that assay is robust and ready to use |
Step 1. Architectural or subcellular localization of expression
The first step toward antibody validation consists of demonstrating the expected localization of signal (Figure 2A). While this step was not included in Uhlen et al., we have found it is a critical first step in the IHC validation process. Based on current literature, we form a hypothesis for the expected localization of expression of the target of interest. This hypothesis describes the anticipated tissue type (e.g., non-small-cell lung cancer vs breast cancer), histologic subtype (e.g., squamous cell carcinoma vs adenocarcinoma), and cellular (tumor vs stroma) and subcellular (cytoplasmic, membranous or nuclear) localizations of the signal. These hypotheses are based on published literature related to the marker, or data on mRNA from the Cancer Genome Atlas or similar online resources [16]. Generally, more than one source is helpful because RNA does not always correspond to protein. For example, if we were interested in EGFR antibodies, we would typically expect the candidate molecules to show a membranous staining pattern of tumor cells or cell lines known to overexpress EGFR based on mRNA data [17,18]. Because the candidate antibody has not yet been titrated, we often just select the vendor's recommended concentration for a first screen. Investigators should be extra cautious at this step, as application of the candidate antibody in substantially increased concentration may result in nonspecific staining patterns (see Step 2).
Figure 2. . Validation of STING antibody.
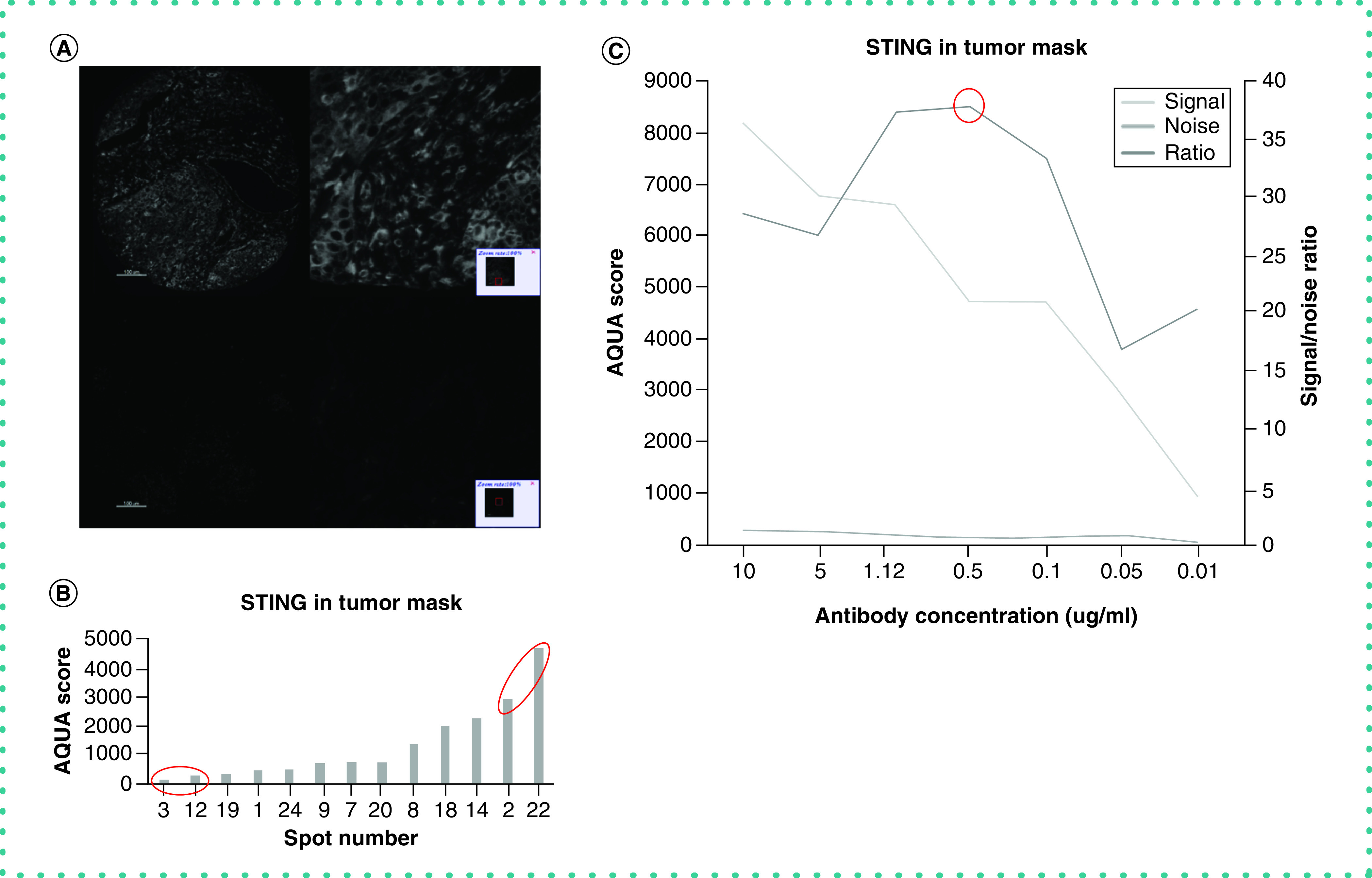
(A) Expected localization of expression. Representative images of high (top) versus low (bottom) signal intensity spots (AQUA scores = 2971.7 and 124.8, respectively). As anticipated, cytoplasmic staining pattern is observed in both tumor and immune cells in the stroma. Isotype-specific HRP-conjugated secondary antibodies were used with a tyramide-based amplification system to generate the fluorescent signal. The Cy5 channel was used for visualization of the STING antibody. (B) Dynamic range chart shows STING IF scores quantified in tumor mask on a control TMA that was created for assay validation. Signal-to-noise ratio is measured by dividing the average IF scores of the upper 10% of spots by the average IF scores of the lower 10% of spots, each indicated with a red circle. (C) Antibody optimization. Signal-to-noise ratio curve for STING antibody. We used seven different concentrations (0.01–10 μg/ml) that span over two full logs of concentration. As seen, the peak for the signal-to-noise ratio is at 0.5 μg/ml, representing the optimal concentration for this antibody.
HRP: Horseradish peroxidase; IF: Immunofluorescence; TMA: Tissue microarray.
Step 2. Antibody optimization (quantitative titration)
The next step for successful antibody validation is optimization of assay conditions, which must be accomplished to maximize the dynamic range of the antibody [19]. This was also not a general pillar of validation, but it is critical for IHC. Conditions for an IHC assay that must be considered include antibody concentration, antigen retrieval buffer and primary antibody incubation conditions. To find the optimal concentration for a candidate antibody, we perform a quantitative titration experiment on the AQUA platform for quantitative IF [20,21]. The AQUA platform utilizes pixel intensity quantification in compartments defined by molecular interactions, including the use of 4′,6-diamidino-2-phenylindole to define the nuclear compartment. A quantitative titration experiment can be done with any sort of quantitative software (e.g., inForm Tissue Finder, CA, USA; HALO, Indica Labs, NM, USA; VisiomorphDP, Visiopharm, Denmark), but is not easily achieved without accurate measurement [22–24]. This experiment involves using serial antibody dilutions which span at least two full logs of concentration (i.e., 1:100–1:10,000) to stain serial sections of a tissue microarray (TMA) with spots with expected varying expression of the target of interest [25]. In fact, with monoclonal antibodies, often the exact concentration of the antibody can be used (a typical range for our lab is 0.05–5 μg/ml), because expressing antibody concentrations as dilutions is a remnant from the days of polyclonal antibodies when we did not know the concentration of the antibody. Then we calculate the signal-to-noise ratio by dividing the average IF score of the top 10% of spots (which we recognize as signal) by the average IF score of the bottom 10% of spots (which we recognize as noise) for each of the serial dilutions used in the experiment (Figure 2B). Finally, we plot the results and pinpoint the maximum signal-to-noise ratio that indicates the optimal antibody concentration (Figure 2C). We have previously shown that the antibodies are most reproducible at their maximal signal-to-noise ratio [18].
Identifying the optimal antigen retrieval buffer and primary antibody incubation time and temperature can often be attained by corresponding with the antibody vendor. Vendors often have extensive information regarding their antibody products, batch concentrations and epitope information. It is often necessary to test a candidate antibody in different conditions to maximize its dynamic range and signal-to-noise ratio. Once optimal conditions have been determined, the maximum specificity and sensitivity of the candidate antibody can be utilized to proceed through further validation steps.
Step 3. Validate using the Uhlen pillars of validation
For IHC these include the following:
-
Uhlen pillar 1: genetic methods
Genetic manipulation of the expression of a target of interest in control cell lines or tissues is a powerful tool for validation of a candidate antibody in IHC. This approach directly links the genetic basis of the target with the corresponding protein product and is a compelling method to prove that an antibody binds specifically to the corresponding antigen. The expression of a target protein can be altered in various ways, including knockdown with siRNAs or knockout with CRISPR/Cas9 [26]. Abcam utilizes the CRISPR/Cas9 technology to generate custom single-gene knockout cell lines or cell lysates that can be used for antibody validation purposes [27]. These methods allow reduction or elimination of the expression of the protein of interest, creating negative controls and subsequently establishing the levels of cross-reactivity of the candidate antibody to other proteins in the cell lines. Similarly, transfection of the target gene to cell lines or tissues enables us to generate positive controls that overexpress the protein of interest (Figure 3B). Transfection can be utilized whether or not the parental cell line expresses the target protein, because in each case positive controls will be produced that have higher levels of the target relative to either the wild-type or negative parental line. This approach can be especially valuable if a range of target concentrations are expressed in different clones of the cell line that can then be used as a standard curve for future IHC assay standardization [28].
-
Uhlen pillar 2: orthogonal methods
Orthogonal methods of target assessment represent another key pillar of validation. The most common orthogonal method is the WB, but failure on a WB, where the antigen is denatured, does not necessarily invalidate an IHC antibody [29]. WB is an example of an antibody-dependent orthogonal method of validation. Detection of a single band at the expected molecular weight serves as an adequate first indication that the antibody is specific for the target of interest (Figure 3A) and is often used in product spec sheets. However, antigens recognized in the setting of a WB are denatured; thus a positive WB does not always mean that a candidate antibody will recognize the folded form of the antigen in the context of IHC. In addition, nonspecific antibodies in WB can display bands at the expected molecular weights [30,31]. Furthermore, posttranslational modifications, splice variants and breakdown products may result in multiple bands or bands at unexpected molecular weights, so similarly, the presence of multiple bands does not necessarily prove an antibody is nonspecific. As with most tests, a positive result – or in this case a single band at the correct molecular weight – is meaningful. In contrast, a negative test – or here the absence of a band or multiple bands – does not prove nonspecificity.
Targeted proteomics is another antibody-independent orthogonal method of validation that uses mass spectroscopy with labeled internal standards to measure relative protein expression across different samples [32]. In this approach, it is critical to use samples with variable expression of the target protein to ensure antibody specificity and allow for statistical analysis [33]. Results generated by this method should be correlated to protein quantification by IHC or quantitative IF. It should be noted that correlations between orthogonal methods and IHC are not explicit, but they must be within a window of confidence to show that an antibody displays specific signal with each method. Because there is variability between methods of antibody detection, and variables in selection of orthogonal method(s) that can be realistically employed, it is necessary for operators to be as rigorous as possible in selecting appropriate orthogonal methods.
-
Uhlen pillar 3: independent epitope validation
The most rigorous pillar of antibody validation involves the use of a second antibody which binds to an independent epitope of the target antigen. Most antibody-binding epitopes are between five and seven amino acids in sequence, and the probability that two nonspecific or cross-reacting antibodies binding to different domains of the target show a correlated signal on the same random tumor TMA is extremely low. The presence of two or more correlated antibodies binding to nonoverlapping epitopes of the target antigen shows explicitly that the antibodies are specific and provides substantial proof that the candidate antibody recognizes the target of interest (Figure 3C) [34]. Successful use of this pillar can reduce the number of pillars required to declare antibody validation.
Figure 3. . Validation of CD200R antibody.
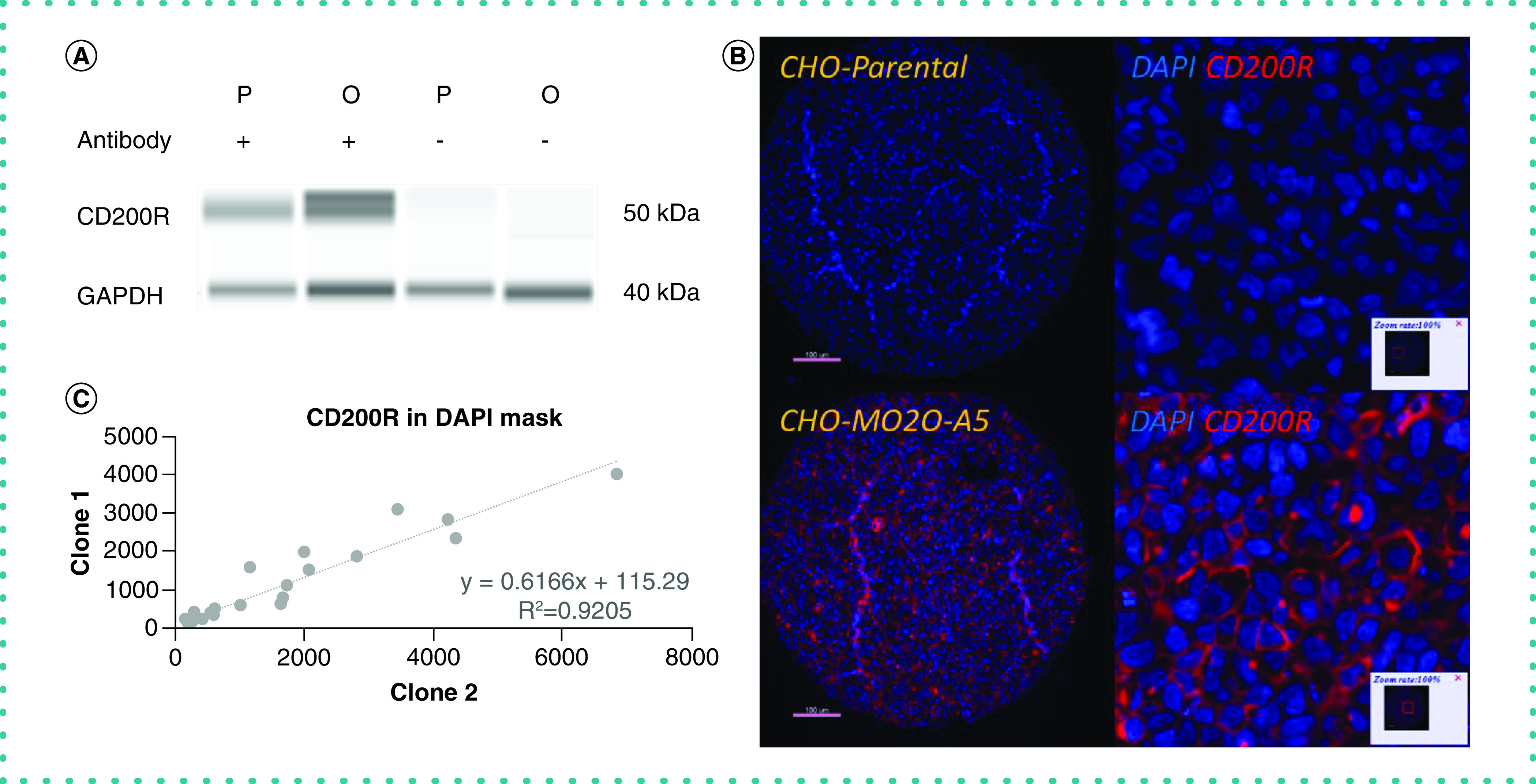
(A) Orthogonal methods of validation. Western blot exhibiting that the candidate antibody recognizes CD200R. Parental CHO cell line with anti-CD200R antibody shows weak signal (lane 1); MO2O-A5 CHO cell line with anti-CD200R antibody shows strong signal (lane 2); parental CHO cell line without antibody (lane 3) and MO2O-A5 CHO cell line without antibody (lane 4) show no signal. (B) Genetic methods of validation. CHO parental cell line has basal levels of CD200R expression and stains negative for CD200R (top); CHO MO2O-A5 cell line overexpresses CD200R and shows clear membranous staining pattern for CD200R (bottom). Isotype-specific HRP-conjugated secondary antibodies were used with a tyramide-based amplification system to generate the fluorescent signal. The Cy5 channel was used for visualization of the CD200 antibody. (C) Independent epitope validation. Scatter plot showing good correlation between two antibodies that bind to nonoverlapping epitopes of human CD200R.
CHO: Chinese hamster ovary; DAPI: 4′,6-diamidino-2-phenylindole; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; HRP: Horseradish peroxidase.
Step 4. Antibody reproducibility
Antibody reproducibility is a critical step for validation of any antibody to standardize an IHC assay, but was not mentioned as a pillar by Uhlen et al. The antibody must maintain consistent sensitivity and specificity for the desired assay on different days with different operators. Even though it is usually considered fulfilled at the time of purchase, the reproducibility of a candidate antibody for the assay of interest should be addressed before systematic use, because it can be affected by issues in shipping. We routinely establish reproducibility between different runs, different operators and manual versus automated staining methods. Furthermore, we regularly confirm reproducibility between different lots of the same antibody clone and vendor (Figure 4). Antibody reproducibility can sometimes be challenging to prove if the target is heterogeneously expressed in the target tissue. For reproducibility testing, the inherent heterogeneity of expression of a target biomarker can be reduced by using serial sections of the same TMA or cell lines in a TMA format. Beyond heterogeneity of target expression, several factors influence reproducibility in IHC stains, including tissue type and biological target localization. It is thus imperative that operators stringently decide upon an acceptable window for reproducibility.
Figure 4. . Antibody reproducibility.
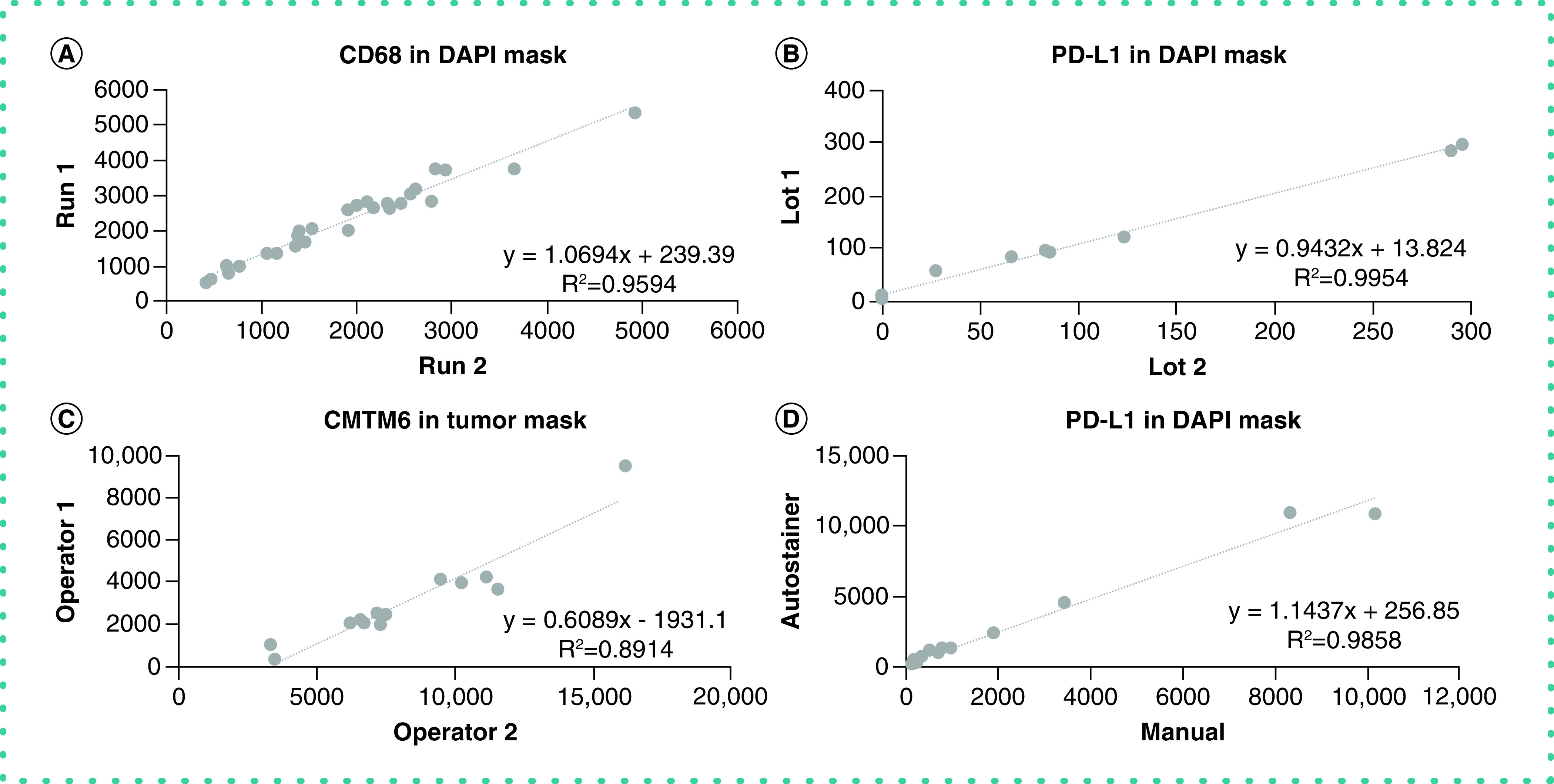
(A) Reproducibility between different days of experimentation. (B) Reproducibility between different lots of the same antibody. (C) Reproducibility between different operators. (D) Reproducibility between manual and automated staining method.
DAPI: 4′,6-diamidino-2-phenylindole.
Other considerations
There are many approaches and protocols for antibody validation. In the clinic, the works by Fitzgibbons et al. [6,35] are considered definitive and are endorsed by the College of American Pathologists for labs governed by the US Clinical Laboratory Improvement Amendments (CLIA). Research labs are generally less standardized and often less rigorous [36–38]. This may be due to extensive antibody use by junior scientists who have not yet learned the importance of antibody validation [39]. The high turnover rate of trainees provides a reason to centralize and standardize antibodies and validation [40]. Standardization processes have been proposed. For example, the Resource Identification Initiative assigns each antibody a unique identifier to allow precise identification and usage of antibodies, and these are consistent across publishers and journals [41]. Going even further, it has been proposed that because the specific sequence for each antibody can be defined and reported, disclosure of the sequence of the epitope would allow for unambiguous knowledge of the antibody being used [42]. Some authors note that a strength of recombinant antibody approaches is that sequences would be more easily reportable and would streamline validation efforts [36,43]. While reporting sequences seems like a good idea from a scientific perspective, many commercial vendors point out that the epitope is the ‘secret sauce’ that makes their antibody better than other competitors. Rather than try to patent each epitope (which is not economically feasible), they maintain confidentiality about their epitope and its sequence [44].
Conclusion
The conceptual pillars of validation published by the group led by Uhlen are important and reveal significant information about the candidate antibody, but not all are essential. Our stepwise protocol is the result of a compromise between scientific rigor and lab economics. Although it was developed to assess cancer-related biomarkers on formalin-fixed paraffin-embedded tumor specimens by IHC or IF, it can be adapted to other antibody-based assays.
Here we share our routine, but fairly comprehensive, methodology for validating antibodies for IHC and IF assays that has increased confidence in our work. This review serves as an update to the methods of antibody validation detailed by Bordeaux et al. in 2010 [4] and Uhlen et al. in 2016 [5]. Because research labs in the academic sector are not subject to any certification and peer review is the only ‘regulatory body’ in that sphere, it is perhaps unrealistic to expect strict adherence to any standardization requirements. Here we provide the working ‘Rimm lab protocol’ that we believe is sufficiently rigorous and sufficiently economical that it can be used routinely in research or clinical labs. While we do not expect to see broad adoption in the way that CLIA labs have adopted the Fitzgibbons et al. guidelines, we hope that this work provides useful, stepwise protocols that can facilitate higher quality antibody validation and better basic and translational science.
Future perspective
Biomedical engineering and recombinant approaches to creating antibodies have permitted the generation of tailor-made antibodies, improving the likelihood of robust validation. Other binders can benefit from the same validation approach if they are to be used for IHC. Mini-bodies are antibody fragments that allow wider use of antigen binding in the body, but they are less common as IHC reagents [45]. Bispecific antibodies can bind to two different epitopes, creating diverse new strategies for detection of targets and for new drugs, but are not commonly used in diagnostic or research reagent applications [46].
Another key concept likely to be important in IHC in the future is antibody biochemistry. In general, the affinity and dissociation constants for most monoclonal antibodies are not known or not easily available. Optimal antibodies for IHC have a fast on-rate and slow off-rate, but the kinetics of antibody binding for IHC are essentially never published. We predict that in the future these variables will become available and will help in the competitive selection of antibodies for their optimal application. We also believe amplification systems may someday be dependent on antibody affinities or kinetics. The effort expended on antibody therapies illustrates the extent of biochemical characterization that is possible, including modification of individual amino acids to tweak affinity or modulate interactions on other regions of the molecule. These customizations will probably eventually move from therapeutic antibodies to antibodies used in the diagnostic/IHC setting.
Acknowledgments
The authors thank Lori A Charette and the staff of Yale Pathology Tissue Services for expert histology services.
Footnotes
Author contributions
D Rimm conceived the manuscript and made substantial contributions to the article. T MacNeil and I Vathiotis also reviewed the literature and made significant contributions to the manuscript and figures. All authors contributed to the content of the manuscript and reviewed the final draft.
Financial & competing interests disclosure
DL Rimm has served as an advisor for Astra Zeneca, Agendia, Amgen, BMS, Cell Signaling Technology, Cepheid, Danaher, Daiichi Sankyo, Genoptix/Novartis, GSK, Konica Minolta, Merck, NanoString, PAIGE.AI, Perkin Elmer, Roche, Sanofi, Ventana and Ultivue. Amgen, Cepheid, NavigateBP, NextCure, Nanostring, Lilly and Ultivue currently fund or have funded research in David L Rimm's lab. I Vathiotis was supported by a scholarship from the Hellenic Society of Medical Oncologists. Among the funding sources supporting D Rimm, the Yale SPORE in Lung Cancer, the Breast Cancer Research Foundation and the Eli Lilly company were particularly important for this work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Verma V, Sprave T, Haque W. et al. A systematic review of the cost and cost–effectiveness studies of immune checkpoint inhibitors. J. Immunother. Cancer 6(1), 128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglund L, Bjorling E, Oksvold P. et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell Proteomics 7(10), 2019–2027 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Andersson S, Sundberg M, Pristovsek N. et al. Insufficient antibody validation challenges oestrogen receptor beta research. Nat. Commun. 8, 15840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordeaux J, Welsh A, Agarwal S. et al. Antibody validation. BioTechniques 48(3), 197–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uhlen M, Bandrowski A, Carr S. et al. A proposal for validation of antibodies. Nat. Methods 13(10), 823–827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgibbons PL, Bradley LA, Fatheree LA. et al. Principles of analytic validation of immunohistochemical assays: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch. Pathol. Lab. Med. 138(11), 1432–1443 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Fitzgibbons PL, Goldsmith JD, Souers RJ. et al. Analytic validation of immunohistochemical assays: a comparison of laboratory practices before and after introduction of an evidence-based guideline. Arch. Pathol. Lab. Med. 141(9), 1247–1254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sfanos KS, Yegnasubramanian S, Nelson WG. et al. If this is true, what does it imply? How end-user antibody validation facilitates insights into biology and disease. Asian J. Urol. 6(1), 10–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howat WJ, Lewis A, Jones P. et al. Antibody validation of immunohistochemistry for biomarker discovery: recommendations of a consortium of academic and pharmaceutical based histopathology researchers. Methods 70(1), 34–38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks DG, Kulkarni S. HER2+ breast cancer: review of biologic relevance and optimal use of diagnostic tools. Am. J. Clin. Pathol. 129(2), 263–273 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Hsi ED. A practical approach for evaluating new antibodies in the clinical immunohistochemistry laboratory. Arch. Pathol. Lab. Med. 125(2), 289–294 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Bradbury ARM, Trinklein ND, Thie H. et al. When monoclonal antibodies are not monospecific: hybridomas frequently express additional functional variable regions. MAbs 10(4), 539–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstone L, van Wilgenburg B, James W. Several commercially available anti-CCR5 monoclonal antibodies lack specificity and should be used with caution. Hybridoma (Larchmt) 31(1), 7–19 (2012). [DOI] [PubMed] [Google Scholar]
- 14.BenchSci. https://www.benchsci.com
- 15.Antibodypedia. https://www.antibodypedia.com
- 16.National Cancer Institute. The Cancer Genome Atlas Program. https://www.cancer.gov/tcga
- 17.Anagnostou VK, Welsh AW, Giltnane JM. et al. Analytic variability in immunohistochemistry biomarker studies. Cancer Epidemiol. Biomarkers Prev. 19(4), 982–991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toki MI, Cecchi F, Hembrough T, Syrigos KN, Rimm DL. Proof of the quantitative potential of immunofluorescence by mass spectrometry. Lab. Invest. 97(3), 329–334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustavson MD, Rimm DL, Dolled-Filhart M. Tissue microarrays: leaping the gap between research and clinical adoption. Per. Med. 10(5), 441–451 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Yaghoobi V, Martinez-Morilla S, Liu Y, Charette L, Rimm DL, Harigopal M. Advances in quantitative immunohistochemistry and their contribution to breast cancer. Expert Rev. Mol. Diagn. 20(5), 509–522 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 8(11), 1323–1327 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Wong PF, Wei W, Smithy JW. et al. Multiplex quantitative analysis of tumor-infiltrating lymphocytes and immunotherapy outcome in metastatic melanoma. Clin. Cancer Res. 25(8), 2442–2449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nearchou IP, Lillard K, Gavriel CG, Ueno H, Harrison DJ, Caie PD. Automated analysis of lymphocytic infiltration, tumor budding, and their spatial relationship improves prognostic accuracy in colorectal cancer. Cancer Immunol. Res. 7(4), 609–620 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Labouba I, Le Page C, Communal L. et al. Potential cross-talk between alternative and classical NF-kappaB pathways in prostate cancer tissues as measured by a multi-staining immunofluorescence co-localization assay. PLoS ONE 10(7), e0131024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J. Clin. Oncol. 26(34), 5630–5637 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Kanchiswamy CN, Maffei M, Malnoy M, Velasco R, Kim JS. Fine-tuning next-generation genome editing tools. Trends Biotechnol. 34(7), 562–574 (2016). [DOI] [PubMed] [Google Scholar]
- 27.abcam. https://www.abcam.com
- 28.Martinez-Morilla S, McGuire J, Gaule P. et al. Quantitative assessment of PD-L1 as an analyte in immunohistochemistry diagnostic assays using a standardized cell line tissue microarray. Lab. Invest. 100(1), 4–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wimberly H, Han G, Pinnaduwage D. et al. ERbeta splice variant expression in four large cohorts of human breast cancer patient tumors. Breast Cancer Res. Treat. 146(3), 657–667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herkenham M, Rathore P, Brown P, Listwak SJ. Cautionary notes on the use of NF-kappaB p65 and p50 antibodies for CNS studies. J. Neuroinflammation 8, 141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Algenas C, Agaton C, Fagerberg L. et al. Antibody performance in western blot applications is context-dependent. Biotechnol. J. 9(3), 435–445 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Marcon E, Jain H, Bhattacharya A. et al. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat. Methods 12(8), 725–731 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Carr SA, Abbatiello SE, Ackermann BL. et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell Proteomics 13(3), 907–917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saper CB. An open letter to our readers on the use of antibodies. J. Comp. Neurol. 493(4), 477–478 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Fitzgibbons PL, Bradley LA, Fatheree LA. et al. Principles of analytic validation of immunohistochemical assays: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch. Pathol. Lab. Med. 1–12 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Goodman SL. The antibody horror show: an introductory guide for the perplexed. N. Biotechnol. 45, 9–13 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Hewitt SM, Baskin DG, Frevert CW, Stahl WL, Rosa-Molinar E. Controls for immunohistochemistry: the Histochemical Society's standards of practice for validation of immunohistochemical assays. J. Histochem. Cytochem. 62(10), 693–697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker M. Antibody anarchy: a call to order. Nature 527(7579), 545–551 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Freedman LP, Gibson MC, Bradbury AR. et al. The need for improved education and training in research antibody usage and validation practices. BioTechniques 61(1), 16–18 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Saper CB, Sawchenko PE. Magic peptides, magic antibodies: guidelines for appropriate controls for immunohistochemistry. J. Comp. Neurol. 465(2), 161–163 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Bandrowski A, Brush M, Grethe JS. et al. The resource identification initiative: a cultural shift in publishing. Neuroinformatics 14(2), 169–182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourbeillon J, Orchard S, Benhar I. et al. Minimum information about a protein affinity reagent (MIAPAR). Nat. Biotechnol. 28(7), 650–653 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Bradbury AR, Pluckthun A. Getting to reproducible antibodies: the rationale for sequenced recombinant characterized reagents. Protein Eng. Des. Sel. 28(10), 303–305 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Polakiewicz RD. Antibodies: the solution is validation. Nature 518(7540), 483 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Nelson AL. Antibody fragments: hope and hype. MAbs 2(1), 77–83 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Runcie K, Budman DR, John V, Seetharamu N. Bi-specific and tri-specific antibodies – the next big thing in solid tumor therapeutics. Mol. Med. 24(1), 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]