Abstract
The quantitative analysis of blood vessel networks is an important component in many animal models of disease. We describe a nondestructive technique for blood vessel imaging that visualizes in situ vasculature in harvested tissues. The method allows for further analysis of the same tissues with histology and other methods that can be performed on fixed tissue. Consequently, it can easily be incorporated upstream to analysis methods to augment these with a three-dimensional reconstruction of the vascular network in the tissues to be analyzed. The method combines iodine-enhanced micro-computed tomography with a deep learning algorithm to segment vasculature within tissues. The procedure is relatively simple and can provide insight into complex changes in the vascular structure in the tissues.
Keywords: : blood vessels, convolutional neural networks, imaging, microCT, microvasculature, nondestructive, vasculature, visualization
METHOD SUMMARY
A nondestructive technique for blood vessel imaging that visualizes in situ vasculature in harvested tissues while preserving tissue structures and epitopes for other histological analyses and outcomes.
The quantitative analysis of blood vessel networks is a key aspect of many preclinical models of disease, including those for vascular disease [1], ischemia [2], angiogenesis [3] and cancer [4,5]. Histological methods are most commonly used to assess changes in the vasculature in animal models, often coupled with techniques such as laser speckle or laser Doppler imaging that can assess changes in perfusion in the superficial layers of the tissue [6–8]. However, the complex changes in blood vessel networks are often difficult to assess from thin sections of tissue, and alterations in perfusion of deeper tissues are difficult to visualize for smaller blood vessels in many animal models. One standard method for visualizing smaller blood vessel networks is to perfuse a radiopaque polymer through the vasculature followed by the use of x-ray micro-computed tomography (microCT) [9–12]. The high contrast of the polymer allows for high-resolution imaging of the vascular networks. However, a major drawback of the method is that it generally precludes most other analyses of the tissues due to the presence of polymer in the tissue. An alternative method that has arisen in recent years is tissue clearing, followed by immunostaining and light sheet microscopy to allow the visualization of three-dimensional aspects of a tissue [13,14]. This technique can achieve impressive visualization of the tissue structure but also requires relatively long processing times and specialized equipment. Other methods, such as two-photon imaging, have been developed for visualizing live tissues; however, the comparatively short imaging depth and requirement for fluorescent labeling limit its application to specific tissue types or tissue layers [15].
In this work we have developed an optimized, nondestructive technique for tissue vasculature imaging that utilizes microCT imaging in conjunction with deep learning network segmentation. While this method does not achieve the very high resolution of other imaging techniques, it is simple and can be performed before standard histological methods. The key advantages of the method are its short processing times and use of readily available imaging systems, and that it can be performed prior to analysis of the tissues with standard histological techniques. It provides a reasonably good visualization of the vascular network in three dimensions, allowing researchers to visualize the overall network prior to analyzing specific areas histologically. The method also incorporates the use of a convolutional neural network [16–18], which provides a rapid identification of the vascular network and can provide quantitative information about structural changes in the vasculature.
Materials & methods
Tissue harvest & processing
All experimental procedures were approved and conducted according to animal care guidelines of The University of Texas at Austin Institutional Animal Care and Use Committee (Protocol ID AUP-2019-00328). Male Sprague-Dawley rats at 3 months of age were anesthetized, heparinized and perfusion-fixed with 10% phosphate-buffered formalin. Tissues were further fixed at 4°C for 1 day and then stained in 2.5% Lugol's iodine solution for the following times based on the tissue types: sciatic nerve (2 days), heart (3 days) and brain (5 days). Prior to imaging the samples were rinsed in phosphate-buffered saline (PBS) and imaged semi-dry.
MicroCT
All tissues were imaged at the University of Texas High-Resolution X-ray Computed Tomography Facility and were scanned at room temperature. No calibration phantom was used and the references for all of samples were acquired through air. Each sample varied in the degree of staining; therefore each scan was individually optimized at reconstruction to utilize the full 16-bit dynamic range of the detector. For the sciatic nerve, the tissues were scanned on an Xradia MicroXCT 400 (Zeiss) using the 4× objective and the MicroXCT controller software (Controller MicroXCT 8.2.2720.12315 ©2010). The tissue sample was cut to 6 mm in length and had a diameter of approximately 1 mm. The sample was inserted into a standard 200-μl micropipette tip and sealed with a piece of tape to minimize desiccation. The bottom of the pipette tip was secured using a stage specific pin vise. The source–object distance was 37 mm and the detector–object distance was 8 mm. This resulted in 5.54 μm resolution. The x-ray source was set to 70 kV and 10 W and no x-ray prefilter was employed. A total of 1261 views were acquired over 360 degrees of rotation, at 2 s/view. During reconstruction (Xradia XMReconstructor – Cone Beam © 2010), a beam-hardening correction of 3 was applied and the data were byte-scaled to a range of -50 to 2800.
For the heart, the tissue was scanned with a custom scanner (North Star Imaging, Inc., MN, USA). The tissue was positioned in a carved cradle of florist foam. The scan used the Fein Focus High Power source with 130 kV and 0.14 mA (no filter). X-rays were detected with a Perkin Elmer detector using 0.25 pF gain, 1 fps, 1 × 1 binning, no flip and a source-to-object distance of 143.0 mm. The overall source-to-detector distance was 1316.717 mm. A continuous computed tomography (CT) scan, with no frames averaged, 0 skip frames, 3600 projections and 5 gain calibrations was taken. A 5-mm calibration phantom was used. A data range of -40.0 to 755.0 was set and a beam-hardening correction of 0.4 was used. A post-reconstruction ring correction was applied using the following parameters: oversample = 2, radial bin width = 21, sectors = 32, minimum arc length = 8, angular bin width = 9, angular screening factor = 1. This resulted in a voxel size of 11.8 μm, leading to 1961 total slices on the transverse axis of the heart.
For the brain tissues, the sample was scanned with a custom scanner (North Star Imaging). The tissue was positioned in a carved cradle of florist foam. The scan used the Fein Focus High Power source set to 110 kV and 0.18 mA (no filter). X-rays were detected with a Perkin Elmer detector with 0.25 pF gain, 1 fps, 1 × 1 binning, no flip and a source-to-object distance of 137.5 mm. The overall source-to-detector distance was 1316.635 mm. A continuous CT scan with 2 frames averaged, 0 skip frames, 3600 projections and 5 gain calibrations was taken. A calibration phantom of 0.762 mm was used. A data range of -40.0 to 695.0 was set and a beam-hardening correction of 0.3 was used. A post-reconstruction ring correction was applied using the following parameters: oversample = 2, radial bin width = 21, sectors = 64, minimum arc length = 2, angular bin width = 9, angular screening factor = 2. The resulting voxel size was 10.5 μm, leading to 1989 slices on the coronal axis of the brain.
Convolutional neural network & visualization
Using the Deep Learning Tool in the Dragonfly 4.1 software (Object Research Systems, Inc.; QC, Canada), we developed a binary segmentation model to automatically identify the blood vessels of the three tissues following a standard U-Net architecture. The convolutional neural network model parameters are outlined in Table 1. Using five max pooling layers and five upsampling layers, we were able to develop robust models of binary segmentation in the three tissues. The max pooling layers contained two subsequent 2D convolutions followed by the 2 × 2 max pooling operation. The upsampling layers contain an up-convolution, a concatenation with a skip connection and two subsequent 2D convolutions. Training data sets were taken randomly throughout the CT slices using a random number generator to select slice numbers. The selected slices were then manually segmented to form the training dataset. To reduce the background data imbalance, the images were substantially cropped and a mask was applied that covered the regions of interest. As with previous iterations of the U-net convolutional neural network, data augmentation was used to increase the robustness of the segmentation (Table 1). After training, the model was then used to segment the respective dataset. The segmented data were exported as a binary region of interest and imported into the Avizo 9 software (ThermoFisher, NJ, USA) for visualization. In Avizo 9, volume renderings and cutaway animations were created. The blood vessel radii were also calculated using a built-in network analysis tool. The models for the brain and heart were pretrained with the nerve segmentation model before being applied to the more complex tissues. Vessel radius quantification was performed using ImageJ by measuring the pixel area of each vessel and calculating the radius of a circle with the same area.
Table 1. . U-net convolutional neural network parameters.
| Category | Parameter | Brain | Heart | Nerve |
|---|---|---|---|---|
| Augmentation | Flip horizontally | Yes | Yes | Yes |
| Flip vertically | Yes | Yes | Yes | |
| Rotation (degrees) | 0–171 | 0–171 | 0–171 | |
| Scale (%) | 90–110 | 85–115 | 90–110 | |
| Training data | # of slices | 12 | 10 | 7 |
| Validation data | Size | 20% of training data | 20% of training data | 20% of training data |
| Training | Input patch size | 64 | 64 | 64 |
| Stride to input | 0.6 | 0.6 | 0.6 | |
| Epochs | 1000 | 2000 | 100 | |
| Batch size | 64 | 64 | 64 | |
| Loss function | Categorical cross-entropy | Categorical cross-entropy | Categorical cross-entropy | |
| Optimization algorithm | Adadelta | Adadelta | Adadelta |
Immunostaining
Following imaging, the samples were washed with PBS with changes every 12 h for 3–5 days to remove the iodine staining. Tissues were then cryoprotected with progressive sucrose solutions increasing from 0 to 30% in PBS. The tissues were then placed in a corresponding tissue matrix and a cryotome blade was used to cut subsections of the tissues. Using the tissue matrix, the tissue position could be maintained and accurate distances could be measured into the tissue. The subsections were then embedded in optimal cutting temperature medium and frozen in isopentane chilled with liquid nitrogen. The samples were then further sliced into 4 μm thick sections using a cryotome (Leica CM1850 Cryostat) while tracking for tissue depth to register with the microCT imaging. The sections were then dried onto slides at 60°C for 30 min before being stored at -20°C until immunostaining. The slides were warmed to room temperature for 20 min before being rehydrated in PBS for 5 min. Antigen retrieval was performed using citric acid buffer (pH 6.0) for 20 min in a 95°C water bath. After the slides were cooled, the sections were permeabilized with 1% fetal bovine serum and 0.4% Triton™ X-100 in PBS for 5 min. The slides were washed with PBS and blocked in PBS with 20% fetal bovine serum at room temperature for 45 min. The slides were then incubated with the primary antibody overnight at 4°C. The primary antibodies used were PECAM-1 (1:100 dilution, R&D Systems AF3628) combined with β-III tubulin (1:100 dilution, Abcam ab18207) for the sciatic nerve sections, Troponin T (1:100 dilution, Abcam 47003) for heart sections, or NeuN (1:100 dilution, Abcam ab128886) for brain tissues. The next day the slides were washed three-times with 1% bovine serum albumin in PBS for 5 min each. Then the slides were incubated with Alexa Fluor® 488 and 594 (Abcam ab150073 and ab150108) dye-conjugated secondary antibodies in 1% bovine serum albumin in BSA at room temperature (1:500 dilutions). After a final wash, the slides were mounted with antifade mounting media and stored at 4°C until imaging. Images were taken with a Fluoview FV1000 confocal laser scanning microscope (Olympus) and z-stacks were created with ImageJ.
Results & discussion
Iodine-enhanced microCT imaging paired with neural network-based analysis allows the visualization of the peripheral nerve vascular network
The sciatic nerve was imaged using microCT and the vasculature segmented using a trained convolutional neural network that was optimized for each tissue technique (Figure 1). We first tested the method on the sciatic nerve from the rats because this tissue has a relatively simple vascular structure with high contrast to the nerve tissue. After the nerve CT images were reconstructed, the CT image stack was cropped to a relatively small pixel volume (317 × 317 × 955 pixels) with a voxel size of 5.54 μm3 (Figure 2A). The trained neural network was then used to segment the vessels in the rest of the nerve tissue (Figure 2B). After processing, the CT slices were reconstructed to visualize the three-dimensional morphology of the soft nerve tissue (Figure 2C). We then created a volume rendering of the segmented vascular network that could be superimposed on the standard microCT reconstruction (Figure 2D). The nerve was then cut using a tissue matrix support to ensure an accurate measurement of slicing depth. By positioning the tissue orthogonal to the reconstructed CT slices, we could match major blood vessel landmarks corresponding to CT slices (Figure 2E). The registered portion of the tissue was then stained for axons (β-III tubulin) and blood vessels (PECAM-1) (Figure 2F). Analysis of the vessel dimensions of all the segments of blood vessels segmented by our convolutional neural network showed a distribution of mean blood vessel radii from the voxel limit (2.77 μm) to 13.47 μm (Figure 2G). To assess the accuracy of the method, we compared the radii of the vessels from the reconstructed network with the radii measured at the same location in the tissue using immunostaining (Figure 2H). The average percentage difference between the radii measured from the reconstructed network and the immunostaining images was 12.6 ± 3.3%. The vessel measurements obtained by microCT were consistently slightly larger than those from histological analysis; this may reflect the effects of further tissue processing in the histology measurements.
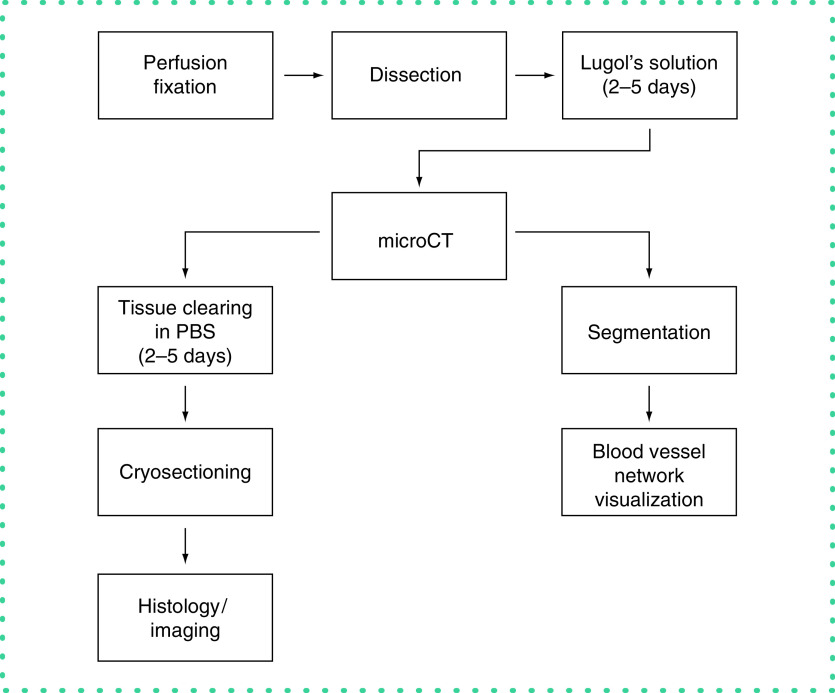
Figure 1. . Methodology overview.
Following perfusion fixation and staining in Lugol's solution, the tissue was scanned with microCT. The scans were then reconstructed and segmented with the convolutional neural network. Following imaging, the tissue was cleared of the contrast agent and sectioned for histological preparations.
Figure 2. . Application of the staining and segmentation applied to rat sciatic nerves.
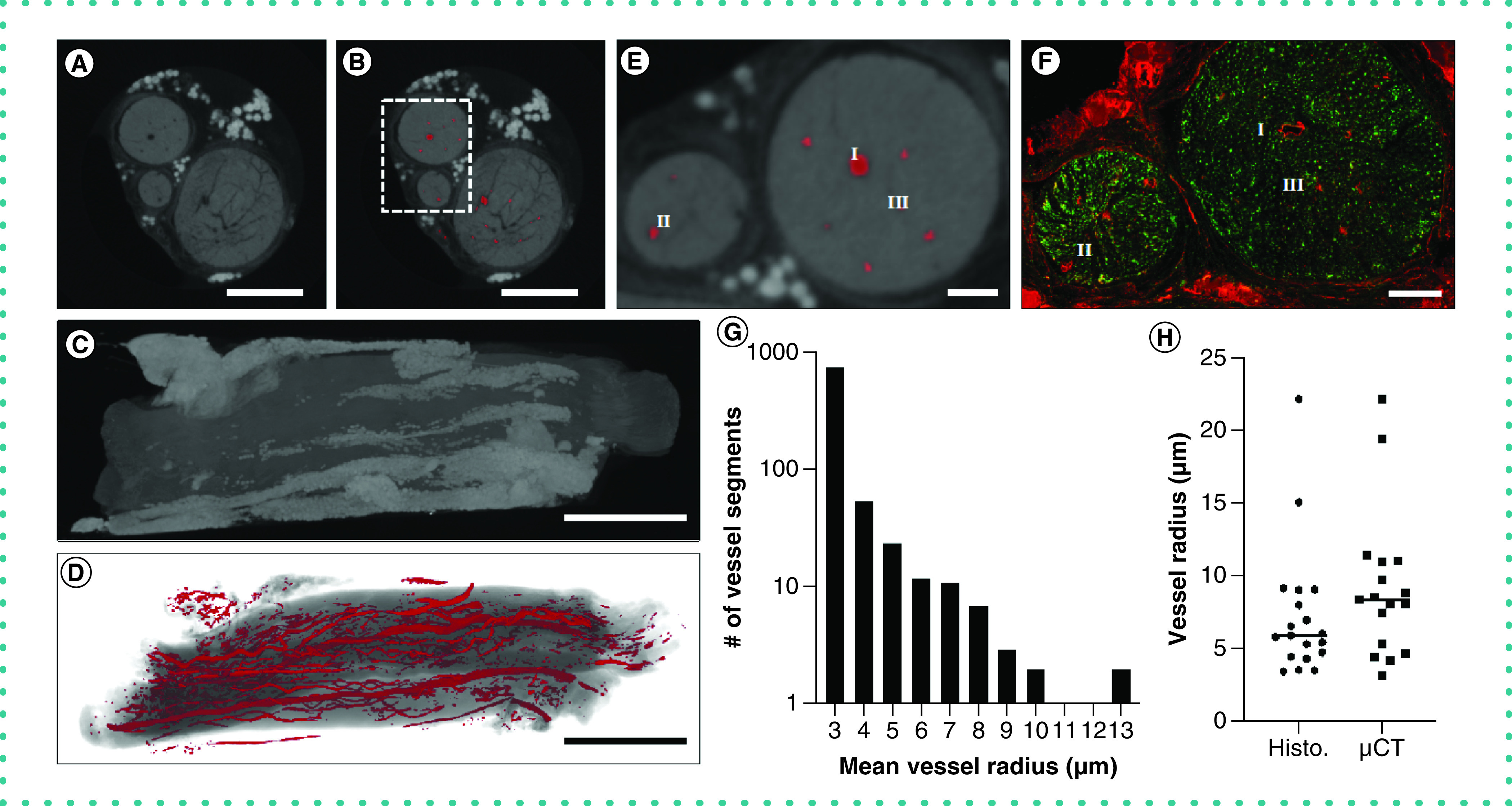
(A) The reconstructed microCT images were cropped to a reduced volume. Scale bar = 500 μm. (B) The nerve was then segmented using our convolutional neural network. Scale bar = 500 μm. (C) The maximum intensity projection of the unsegmented microCT stack. Scale bar = 1 mm. (D) The volume rendering of the segmented blood vessels. Scale bar = 1 mm. (E) A close-up of the inset in (B), showing the segmented blood vessels in greater detail. Scale bar = 100 μm. (F) The nerve was subsequently processed for histological analysis and stained for blood vessels (PECAM-1, red) or axons (β-III tubulin, green). I, II and III indicate corresponding regions in (E & F). Scale bar = 100 μm. (G) The frequency of measured radii of microCT-segmented blood vessels shows our technique can segment both vessels at the voxel size limit and larger blood vessels. (H) A measurement of vessel radii on confocal and microCT images revealed no significant difference in vessel radius.
Iodine-enhanced microCT imaging paired with neural network-based analysis allows visualization of larger vessels in the heart but is limited in resolving smaller vessels
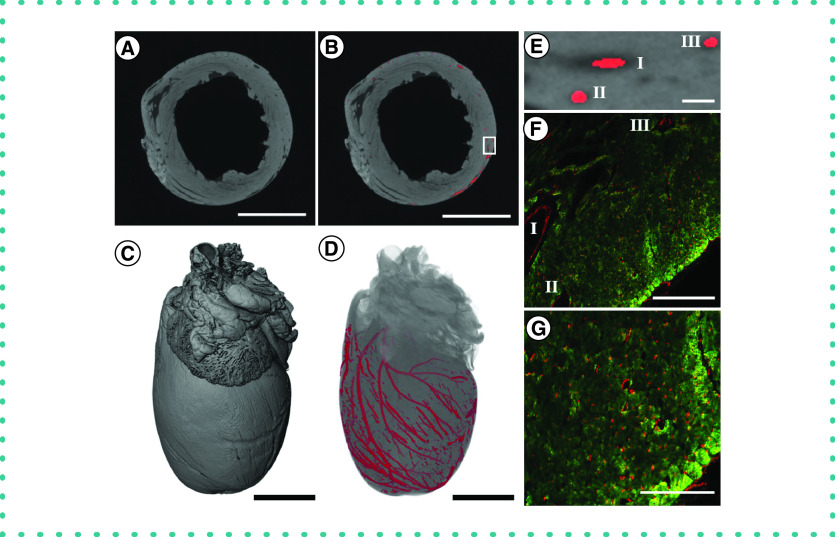
We next used similar methods to process the tissue of the heart. Visualization of the vessels of the coronary vasculature, in particular, would add valuable information to the models of vascular disease and cardiomyopathy [19,20]. We repeated the methods on the heart tissues and created a reduced volume microCT image stack (1208 × 1246 × 1961 pixels; Figure 3A). The pretrained convolutional neural network was further trained on ten microCT slices from the heart that were taken randomly through the CT image stack. This algorithm was then used to segment the vasculature in the heart tissue (Figure 3B). The reconstructed vascular network was then superimposed on a volume rendering of the segmented vasculature (Figure 3C & D). To assess the accuracy of the vascular segmentation, we identified a small region in the left ventricle (Figure 3E), sectioned this part of the tissue and then stained for cardiac muscle (troponin T) and blood vessels (PECAM-1) (Figure 3F & G). For vessels larger than ∼10 μm in diameter, the neural network was able to register and segment the vessels with a high degree of accuracy. However, some vessels below 10 μm in diameter were not recognized using the method (Supplementary Figures 1 & 2). There are several reasons for the reduced resolution of the vessels for the heart, including voxel size limitations due to the size of the tissue and the lower contrast of the vessels to the myocardial tissues with the iodine staining. A higher-resolution scan over a smaller volume may improve these results but this capability would depend on the microCT instrumentation used for imaging and the volume of interest.
Figure 3. . Application of the staining and segmentation method applied to a rat heart.
(A) The reconstructed microCT images were cropped to a reduced volume. Scale bar = 5 mm. (B) The heart was then segmented using the convolutional neural network. Scale bar = 5 mm. (C) The maximum intensity projection of the unsegmented microCT stack. Scale bar = 2.5 mm. (D) The volume rendering of the segmented blood vessels. Scale bar = 2.5 mm. (E) Higher magnification for inset shown in part (B). Scale bar = 200 μm. (F) Histological sections from the left ventricle of the heart. Immunostaining was performed for cardiac muscle (troponin T, green) and blood vessels (PECAM-1, red). I, II and III indicate corresponding regions in (E & F). Scale bar = 200 μm. (G) A higher-magnification image of the heart tissues, showing the microvasculature. Immunostaining was performed for cardiac muscle (troponin T, green) and blood vessels (PECAM-1, red). Scale bar = 100 μm.
Iodine-enhanced microCT imaging with neural network-based analysis of the brain provides a general visualization of the larger vasculature
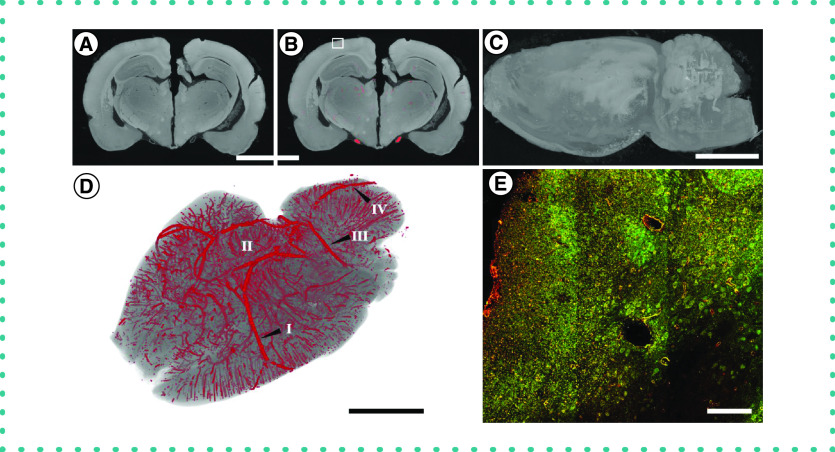
To further test the limitations of the method, we examined its ability to characterize the complex vascular network of the brain. We repeated the technique on the perfusion-fixed tissues of the brain and constructed a reduced microCT image stack (1431 × 969 × 1989; Figure 4A). The pretrained convolutional neural network was further trained on 12 CT slices from the brain and we then used this algorithm to segment the images (Figure 4B). An overall reconstruction was created for the brain (Figure 4C) and then the volume rendering of the segmented vasculature was superimposed onto this structure (Figure 4D). Anatomical landmarks within the vascular network, including the full circle of Willis, were easily visualized with this method (Supplementary Figure 3). There were similar issues as for the heart tissue, with loss of recognition for smaller vessels. However, the technique captured a substantial number of descending vessels, including in this case 10,465 individual segments (Supplemental Figure 4). The segmentation technique captured a wide range of mean blood vessel radii, from the resolution limit up to 62.7 μm. We sectioned a portion of the cortex and stained for NeuN and PECAM-1, demonstrating the maintenance of tissue epitopes in the brain after contrast-enhanced microCT processing, and qualitatively matched the major aspects of the vascular network (Figure 4E). Overall, while this method did visualize many aspects of the brain vasculature, it was far less accurate for brain tissue vasculature compared with the results for peripheral nerve or heart tissues.
Figure 4. . Application of the staining and segmentation method applied to a rat brain.
(A) The reconstructed microCT images were cropped to a reduced volume. Scale bar = 5 mm. (B) The nerve was then segmented using our convolutional neural network. (C) The maximum intensity projection of the unsegmented microCT stack. Scale bar = 5 mm. (D) The volume rendering of the segmented blood vessels. The labels indicate some of the anatomical structures of the brain vasculature: (I) Middle cerebral artery, (II) Circle of Willis, (III) Superior cerebellar artery and (IV) Basilar artery. Scale bar = 5 mm. (E) Histological sections from the brain immunostained for NeuN (green) and PECAM-1 (red). Scale bar = 100 μm.
Conclusion & future perspective
Convolutional neural networks have emerged as powerful segmentation tools for biomedical imaging in situations where researchers may want to rapidly scan and identify regions of interest in huge data sets [16–18]. U-net convolutional neural network architectures require a much smaller set of training images – traditionally a bottleneck for biomedical image segmentation [21]. Furthermore, U-net-based segmentation tools are widely available and can be employed in many different software environments, including Dragonfly [22] and ImageJ [23]. Here we demonstrate the application of a convolutional neural network for segmenting vascular networks in microCT images obtained using a reversible contrast staining procedure [24–26]. With this technique, tissues can be perfusion-fixed and processed using standard methods compatible with subsequent histological analyses. The method can provide insights into vasculature in the whole tissue and can augment traditional histological vascular network analyses. The major limiters of the method used here include voxel resolution by the microCT imager for the desired field of view and the tissue-specific contrast obtainable with the reversible iodine staining technique. For a small tissue sample imaged with small voxel size and high tissue-to-vessel contrast, as in the case of the sciatic nerve, there was a high degree of correspondence with the registered histology sections. With larger voxel sizes and less contrast with the surrounding tissues, as in the case of the heart and brain, the method worked well with only the larger vessels within the vascular network. Despite these limitations, the ability to visualize the overall vascular tree in three dimensions provides important information that can be used to better understand vascular network remodeling in the entire tissue and guide targeted analyses of the tissues by histology. In addition, the overall simplicity of the technique and its nondestructive nature and relatively short processing time make the method easy to incorporate into the experimental pipelines of studies on vascular network remodeling.
Supplementary Material
Footnotes
Author contributions
A Veith performed the experiments, analyzed the data and wrote the paper. A Baker analyzed the data and wrote the paper.
Financial & competing interests disclosure
The authors gratefully acknowledge funding through the American Heart Association (17IRG33410888), the DOD CDMRP (W81XWH-16-1-0580; W81XWH-16-1-0582) and the National Institutes of Health (1R21EB023551-01; 1R21EB024147-01A1; 1R01HL141761-01) to A Baker. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
All of the experiments performed were approved by the UT Austin Institutional Animal Care and Use Committee and in accordance with NIH guidelines.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Zamir M, Phipps S. Network analysis of an arterial tree. J. Biomech. 21(1), 25–34 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Duvall CL, Taylor WR, Weiss D, Guldberg RE. Quantitative microcomputed tomography analysis of collateral vessel development after ischemic injury. Am. J. Physiol. Heart Circ. Physiol. 287(1), H302–H310 (2004). [DOI] [PubMed] [Google Scholar]
- 3.McDonald DM, Choyke PL. Imaging of angiogenesis: from microscope to clinic. Nat. Med. 9(6), 713–725 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Toi M, Asao Y, Matsumoto Y. et al. Visualization of tumor-related blood vessels in human breast by photoacoustic imaging system with a hemispherical detector array. Sci. Rep. 7(1), 41970 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature 407(6801), 249–257 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Das S, Singh G, Baker AB. Overcoming disease-induced growth factor resistance in therapeutic angiogenesis using recombinant co-receptors delivered by a liposomal system. Biomaterials 35(1), 196–205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang E, Albadawi H, Watkins MT, Edelman ER, Baker AB. Syndecan-4 proteoliposomes enhance fibroblast growth factor-2 (FGF-2)-induced proliferation, migration, and neovascularization of ischemic muscle. Proc. Natl Acad. Sci. USA 109(5), 1679–1684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteforte AJ, Lam B, Das S. et al. Glypican-1 nanoliposomes for potentiating growth factor activity in therapeutic angiogenesis. Biomaterials 94, 45–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghanavati S, Yu LX, Lerch JP, Sled JG. A perfusion procedure for imaging of the mouse cerebral vasculature by X-ray micro-CT. J. Neurosci. Methods 221, 70–77 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Savai R, Langheinrich AC, Schermuly RT. et al. Evaluation of angiogenesis using micro-computed tomography in a xenograft mouse model of lung cancer. Neoplasia 11(1), 48–56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Wang Y, Xue C. et al. Angiogenesis in tissue-engineered nerves evaluated objectively using MICROFIL perfusion and micro-CT scanning. Neural Regen. Res. 11(1), 168–173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehling J, Theek B, Gremse F. et al. Micro-CT imaging of tumor angiogenesis: quantitative measures describing micromorphology and vascularization. Am. J. Pathol. 184(2), 431–441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat. Methods 10(6), 508–513 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Chung K, Wallace J, Kim SY. et al. Structural and molecular interrogation of intact biological systems. Nature 497(7449), 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davalos D, Lee JK, Smith WB. et al. Stable in vivo imaging of densely populated glia, axons and blood vessels in the mouse spinal cord using two-photon microscopy. J. Neurosci. Methods 169(1), 1–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Weidong C, Wang X, Zhou Y,Feng D, Mei Chen l. Medical image classification with convolutional neural network. Presented at: 13th International Conference on Control Automation Robotics & Vision (ICARCV). IEEE, Singapore; (2014). https://www.semanticscholar.org/paper/Medical-image-classification-with-convolutional-Li-Cai/701ec23ea4bde6a1c0a9817b803aacd9c8548b75 [Google Scholar]
- 17.Van Ginneken B, Setio AA, Jacobs C, Ciompi F. Off-the-shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans. Presented at: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI). IEEE; (2015) 286–289. [Google Scholar]
- 18.Milletari F, Navab N, Ahmadi S-A. V-net: fully convolutional neural networks for volumetric medical image segmentation. Presented at: 2016 Fourth International Conference on 3D Vision (3DV). IEEE; (2016) 565–571. [Google Scholar]
- 19.Weyers JJ, Carlson DD, Murry CE, Schwartz SM, Mahoney WM. Retrograde perfusion and filling of mouse coronary vasculature as preparation for micro computed tomography imaging. J. Vis. Exp. 60, e3740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sangaralingham SJ, Ritman EL, McKie PM. et al. Cardiac micro-computed tomography imaging of the aging coronary vasculature. Circ. Cardiovasc. Imaging 5(4), 518–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention, Pt Iii 9351, 234–241 (2015). [Google Scholar]
- 22.Makovetsky R, Piche N, Marsh M. Dragonfly as a platform for easy image-based deep learning applications. Microsc. Microanal. 24(S1), 532–533 (2018). [Google Scholar]
- 23.Saltukoglu, Tuan Leng T, Marco P. et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16(1), 67–70 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Heimel P, Swiadek NV, Slezak P. et al. Iodine-enhanced micro-CT imaging of soft tissue on the example of peripheral nerve regeneration. Contrast Media Mol. Imaging 2019, 7483745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gignac PM, Kley NJ. Iodine-enhanced micro-CT imaging: methodological refinements for the study of the soft-tissue anatomy of post-embryonic vertebrates. J. Exp. Zool. B Mol. Dev. Evol. 322(3), 166–176 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Degenhardt K, Wright AC, Horng D, Padmanabhan A, Epstein JA. Rapid 3D phenotyping of cardiovascular development in mouse embryos by micro-CT with iodine staining. Circ. Cardiovasc. Imaging 3(3), 314–322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.