Abstract
Macropinocytosis supports the metabolic requirement of RAS-transformed pancreatic ductal adenocarcinoma cells (PDACs). However, regulators of RAS-transformation (activation) that lead to macropinocytosis have not been identified. Herein, we report that UBAP2 (ubiquitin binding associated protein 2), regulates activation of KRAS and macropinocytosis in pancreatic cancer. We demonstrate that UBAP2 is highly expressed in both pancreatic cancer cell lines and tumor tissues of PDAC patients. The expression of UBAP2 is associated with poor overall survival in several cancers, including PDAC. Silencing UBAP2 decreases levels of activated KRAS, and inhibits macropinocytosis, and tumor growth in vivo. Using a UBAP2-deletion construct, we demonstrate that the UBA-domain of UBAP2 is critical for regulation of macropinocytosis and maintaining the levels of activated KRAS. In addition, UBAP2 regulates RAS downstream signaling and helps maintain RAS in the GTP-bound form. However, the exact mechanism by which UBAP2 regulates KRAS activation is unknown and needs further investigation. Thus, UBAP2 may be exploited as a potential therapeutic target to inhibit macropinocytosis and tumor growth in activated KRAS-driven cancers.
Keywords: UBAP2, Macropinocytosis, KRAS, Pancreatic cancer, small GTPases, Therapeutic target
Introduction
PDAC is one of the most aggressive malignancies with a notoriously dismal prognosis [1]. It is well known that activating mutations of small GTPases, such as KRAS, are near universal in PDAC and critical for pathogenesis [2]. Unfortunately, even after 3 decades of intense investigation, the community has yet to develop therapeutic targets for RAS-activated proteins [3]. A new strategy for anti-RAS efforts is suggested by recent studies linking RAS with altered cellular metabolism [4–6], since aberrant metabolism is now considered as one of the hallmarks of cancer [7, 8]. To meet high metabolic demands for amino acids, PDAC cells expressing oncogenic KRAS reportedly utilize macropinocytosis to internalize these macromolecules from the extracellular environment [4, 9, 10]. Although macropinocytosis has been known for decades, the molecules underlying its mechanism and regulation remain unidentified. We recently demonstrated that gold nanoconjugates partially covered with an anti-EGFR antibody C225 (Au-C225-P), followed a dynamin-2 independent but Cdc42 dependent pinocytotic pathway of uptake by pancreatic cancer cells. In contrast, gold nanoconjugates with complete surface coverage (Au-C225-C) utilized a dynamin-2 dependent caveolar uptake pathway [11, 12]. We identified UBAP2 as one of the proteins exclusively associated with Au-C225-P and consequently investigated its role in macropinocytosis.
Bioinformatics analysis suggests that UBAP2 contains a ubiquitin-binding associated domain (UBA-domain), although the function of this domain has yet to be characterized. Ubiquitination has diverse roles in health and disease by controlling a wide range of biological processes including protein degradation, endocytosis, DNA repair, transcription, and immune responses [13, 14]. Over twenty distinct families of UBA-domain containing proteins have been identified and shown to decode ubiquitinated target signals into biochemical cascades in the cell [13, 14]. Herein, we identify UBAP2 as a molecular switch that regulates macropinocytosis and tumor growth, and activates KRAS in pancreatic cancer. Some recent reports suggested a role for UBAP2 in cancer; one report demonstrated that UBAP2 was elevated in metastatic prostate cancer and a second report showed that in hepatic cancer UBAP2 expression inversely correlated with prognosis [15, 16]. However, a role of UBAP2 in regulating macropinocytosis and small GTPase activation has not been previously described. UBAP2 may be exploited as a novel therapeutic target to inhibit activation of small GTPases, limit macropinocytosis and reduce PDAC growth.
Materials and Methods
Cell lines, patient samples and reagents.
All cell lines used in this study were purchased from ATCC (Manassas, VA, USA) and maintained under the manufacturer’s instructions. The pancreas disease spectrum (pancreatic cancer progression) tissue array (PA2081a) was purchased from US Biomax, Inc (Derwood, MD, USA).
Targeting sequences for RNA interference:
The siRNA targeting human UBAP2 and the negative control siRNA (#1027281) were purchased from Qiagen (Germanwood, MD, USA). The pLKO.1-puro carrying human UBAP2-specific shRNA constructs were purchased from Sigma-Aldrich (Burlington, MA, USA). The control pLKO.1-puro construct is from Addgene (Watertown, MA, USA). The siRNA sequences targeting human UBAP2 are 5′-CAGACUGGCUACUGUAUGUAA-3’ (siUBAP2, specifically targeting 3’-UTR) and 5’-CAGCGGUAUCCCUCCACCCAA-3’ (siUBAP2–2). The shRNA sequences targeting human UBAP2 is 5’-AGAACACACGAGCACGTATTT-3’ (shUBAP2, specifically targeting 3’-UTR); and the sequence of control shRNA is 5’-TAAGGTTAAGTCGCCCTCG-3’. The siRNA targeting human UBAP1 (sc-92628) was purchased from Santa Cruz Biotechnology, Inc (Dallas, TX, USA). Nocodazole (M1404–2MG), Chlorpromazine (C8138–5G), MG-132 (M7449–200U), were purchased from Sigma (Burlington, MA, USA). His-RAS P21 purified protein was purchased from (Cytoskeleton, Inc # RS01, Denver, CO, USA).
UBAP2 DNA was amplified from human cDNA by PCR and subcloned into pcDNA3.1, pEGFP, pET28b and pET42c. Other constructs (HA-KRAS G12V), were purchased from Addgene (Watertown, MA, USA). All plasmids were confirmed by sequencing.
Mouse Monoclonal antibodies against UBAP2 were generated using peptide DTPKTTGPPSAL (human UBAP2 656–667) as immunogen by Abmart (Berkeley Heights, NJ, USA) and used for IF. The following commercial primary antibodies were used: anti-UBAP2 (1:2000 for WB; 1:200 for IF; 1:200 for IHC), anti-β-Actin (1:5000 for WB), and anti-GAPDH (1:50000 for WB) were all from Sigma-Aldrich Co (Burlington, MA, USA); anti-myc-tag (1:2000 for WB; 1:800 for IF) was from Cell Signaling Technology, Inc.; anti-cytokeratin-19 (1:600 for IF) and anti-KRAS (1:1000 for WB) were from proteintech, (Rosemont, IL 60018, USA); and anti-RAS (1:1000) was from abcam, (Cambridge, MA)
Transfection and Infection.
For transient gene overexpression, 5×105 cells were transfected with 5 μg plasmid using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. For siRNA transfection, 2×105 cells were transfected with 90 nM siRNA using DharmaFECT™ Transfection Reagents (Dharmacon, Lafayette, CO, USA) according to the manufacturer’s instructions. To produce lentiviruses, 293T cells were co-transfected with pLKO.1-puro carrying control or UBAP2-specific shRNA, pCMV-VSVG, and pCMV-dR8.91 dvpr (Addgene, Watertown, MA, USA) at a weight ratio of 2:1:1 using Fugene 6 transfection reagent. Supernatants were collected 48–72 h after transfection and concentrated using LentX-concentrators (Clontech, Mountain View, CA, USA). Then AsPC1 cells were infected by the above lentivirus in the presence of 8 μg/ml polybrene (Sigma-Aldrich, Burlington, MA, USA). To achieve better depletion of UBAP2, stable multi-AsPC1 cell clones were selected by 2.0 μg/ml puromycin 48 h postinfection.
Cell Proliferation Assay.
Pancreatic cancer cells (2×104) were reverse transfected with siRNA and seeded in 24-well plates. Forty-eight hours after transfection, the media was replaced with 1 ml of fresh serum-free media containing 1 μCi [3H]-thymidine and cultured for 4 more hours. Then cells were washed with chilled PBS, fixed with 100% methanol, lysed with 0.1 N NaOH and collected for measurement of radioactivity in Biosafe II scintillation liquid. Experiments were repeated at least three times and each time in triplicate.
Migration and Invasion Assay.
48 hours post transfection, pancreatic cancer cells were moved to FBS free medium where they were starved for 16 hours prior to assay. For the migration assay, 105 cells in FBS free medium were plated in the upper chamber onto the noncoated membrane (24-well insert; pore size, 8 mm; Corning Costar Tewksbury, MA, USA) and allowed to migrate toward 2% serum-containing medium in the lower chamber. Parallel wells containing FBS free medium in the lower chamber were set up as the control. After 4 hours (for AsPC1 and Panc04.03) or 6 hours (for PANC1) of incubation, cells that did not pass through the pores were removed with a cotton swab. Cells on the lower surface of the membrane were fixed with methanol and stained with 0.1% crystal violet (2 mg/mL, Sigma-Aldrich, Burlington, MA, USA). Cells migrating through the membrane were counted under a light microscope. In individual experiments migration was defined as the mean number of cells that migrated toward medium containing FBS minus the mean number of cells that migrated in the parallel control. For invasion assays, Matrigel-coated Transwells (BD Bioscience, San Jose, CA, USA) were pre-incubated with FBS free medium for four hours and then 105 cells were plated in the upper chambers. The lower chambers contained 2% FBS or no FBS (as control). The inserts were incubated for 16 hours (for AsPC1 and Panc04.03) or 24 hours (for PANC1). Data were collected and analyzed as for the migration assay.
Macropinosome visualization and quantification.
Macropinosomes were visualized and quantified as described by Commisso, C. et al. [4, 17] with modifications. Fluid-phase uptake in culture cells was measured by the uptake of fluorescence-conjugated Dextran (MW 70 kDa, Life Technologies, Carlsbad, CA, USA) and BSA (Life Technologies, Carlsbad, CA, USA). Briefly, cells were starved in FBS free medium then incubated with fluorescence-conjugated Dextran or BSA for 30 min at 37 °C. Following extensive washes with ice-cold PBS and immediate fixation in 4% formaldehyde, the slides with cells were mounted using Mounting Media with DAPI (Vector).
To visualize macropinocytosis in vivo, 6~8 week old female homozygous NCr nude mice were injected with 1 million cells in a total volume of 100 μL at the pancreas head. When tumor volume had attained approximately 500 mm3, 1 mg TMR-Dextran (70 kDa) in 100 μl HBSS was injected into the tumor [17]. Ninety minutes after injection, mice were sacrificed and the tumors were removed and frozen in O.C.T. (TissueTek, Torrance, CA, USA) rapidly. Tissues were sectioned and the slides were fixed in 4% formaldehyde. Cell membranes were visualized by staining with a human specific anti-cytokeratin-19 primary antibody followed by an Alexa Fluor 488-conjugated secondary antibody. Images were captured using an Axiovert 200 inverted fluorescent microscope (Zeiss, Pleasanton, CA, USA) or LSM710 confocal microscope (Zeiss) and the staining areas were analyzed by ImageJ (National Institutes of Health) using the ‘Analyze Particles’ feature. In vitro dextran uptake assays were repeated three times, and images from 5–10 random fields of each slide were captured. For the in vivo assay, three tumors were injected with dextran for each group and each tumor was sectioned in three directions. At least 100 cells in each slide were quantified. The area percentage of dextran signal in each cell was calculated using the formula Area Percentage of Dextran = (AREAdextran ÷ AREAcell) × 100%; results are presented as “Relative Dextran Uptake” by normalizing the area percentage of control groups as 1.0.
Immunofluorescence.
Cells growing on coverslips were fixed in 4% paraformaldehyde at room temperature for 15 min, washed, permeabilized for 15 min with 0.2% Triton X-100, and blocked with 3% bovine serum albumin (BSA) in PBS for 30 min at room temperature. The coverslips were incubated sequentially with appropriate primary and secondary antibodies and images were acquired by an Axiovert 200 inverted fluorescent microscope (Zeiss, Pleasanton, CA, USA) or LSM710 confocal microscope (Zeiss).
Immunoprecipitation and immunoblotting.
Cells were lysed in Perce IP Lysis Buffer (Thermo Fisher Scientific, Carlsbad, CA, USA) for use in immunoprecipitation studies. The immunocomplexes were separated by SDS–PAGE and analyzed. Unless otherwise stated, immunoprecipitation was performed using 1 ml of cell lysate containing 1.5mg total proteins. For immunoblotting, the blots were probed with appropriate primary antibodies and HRP-conjugated secondary antibody (Sigma-Aldrich, 1:10000, Burlington, MA, USA), and bound antibody was detected using enhanced chemiluminescence (Bio-Rad, Desplaines, IL, USA) according to the manufacturer’s protocol.
GTP-binding small GTPases pull-down assays.
GTP-binding small GTPases were evaluated using pull-down based Activation Assay Biochem Kits (Cytoskeleton, Inc., Denver, CO, USA; Briefly, 48 hours post transfection, cells were washed three times with ice cold PBS and lysate was collected as per the manufacturer’s protocol. The clarified lysates were incubated with GST-Raf-RBD (for RAS), immobilized on glutathione–agarose beads for 1 hour at 4°C. GTP-bound RAS content was determined by Western blot using the proteins from pulldown components.
Endocytosis
AsPC1 cells were transfected with either scrambled RNA (siControl) or UBAP2 siRNA (siUBAP2). 48 hours post transfection, cells were starved for 30 min at 37 ° C in pre-warmed serum free media supplemented with 25 mM HEPES pH 7.4 and 0.5% BSA. Cells were pulsed with Alexa fluor-488 conjugated transferrin (50 μg/ml) in serum free media for 40 min at 37 °C. Following incubation, samples were extensively washed with ice-cold PBS, immediately fixed in 4% formaldehyde, and mounted onto slides using Mounting Media with DAPI (Vector). Cells were treated with 40 μM chlorpromazine for 40 min, washed with PBS then chased with transferrin.
Immunohistochemistry.
Xenograft tumor samples were fixed in 10% formalin solution. Then tissues were embedded in paraffin wax and 4-μm sections were prepared. Tissue sections were deparaffinized and rehydrated in an ethanol series. Immunohistochemistry was performed according to standard protocols. Antigen retrieval was achieved by heating sections in 95 ˚C citrate buffer for 15 minutes. Sections were incubated with specific antibodies overnight at 4 ˚C. For UBAP2 (1:200) staining, the dark brown signal was revealed after incubation with the ABC kit (Vector), followed by a diaminobenzidine (DAB) and hydrogen peroxide reaction using the DAB detection kit (Vector). Counterstaining was performed by incubating the slides in hematoxylin for 5 min. Appropriate controls were used in all cases by incubating sections with all reagents except the primary antibodies; no staining was observed under these conditions.
Mouse xenograft tumor model.
Female athymic nude mice (NCr-nu; 6 to 8 wks old) were purchased from the National Cancer Institute-Frederick Cancer Research and Development Center (Frederick, MD). All mice were housed and maintained under specific pathogen-free conditions in facilities approved by the American Association for Accreditation of Laboratory Animal Care and in accordance with current regulations and standards of the U.S. Department of Agriculture, U.S. Department of Health and Human Services, and NIH. All studies were approved and supervised by the Institutional Animal Care and Use Committee at the University of Oklahoma. For the generation of orthotopic pancreatic tumor models, AsPC1 cells, stably expressing either control or UBAP2 shRNA, were injected into the pancreas heads of nude mice. Mice weights were recorded twice weekly and their health/behaviors were monitored daily. After assessing tumor growth in these animals, mice were sacrificed by CO2 inhalation and tumors and tissue were immediately harvested for further analysis. Tumor volume was calculated using the formula: Volume = Width2 × Length ÷ 2.
TCGA data analysis.
Analyses of TCGA data (the Cancer Genome Atlas Project, TCGA; http://tcga-data.nci.nih.gov/) were performed in R (version 3.0.1) (http:///www.r-project.org/) and the statistical significance was defined as a p-value less than 0.05. Patients were grouped into percentiles according to mRNA expression. The Log-rank test was employed to determine the association between mRNA expression and overall survival and the Kaplan- Meier method was used to generate survival curves.
Identification of NanoParticle (NP)-Bound Proteins by Mass Spectrometry
The identification of proteins bound to NP was performed as previously described [18]. In brief, NPs were incubated with cell lysates, pelleted and washed once with ddH2O. The resulting pellet was used for identification of bound proteins by nanoLC-MS/MS with hybrid orbitrap/linear ion trap mass spectrometry as described previously [19]. Tandem mass spectra were extracted by BioWorks version 3.2. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.2.04), Sequest (ThermoFinnigan, San Jose, CA; version 27, rev. 12), and X! Tandem (www.thegpm.org; version 2006.09.15.3). X!
Statistical analysis.
All values are expressed as means ± SD. The association of tumor type (malignant, metastatic and benign), stage (I, II, III and IV), other types (inflammation, normal adjacent tissues, and normal), and differentiation (well, moderately, and poorly differentiated) with staining intensity was assessed using the (exact) two-sided Pearson’s Chi-square test. For animal experiments, 7 mice were assigned per group, unless otherwise noted, and the significance was determined by a one-way ANOVA model using Tukey’s method for multiple comparisons. Other statistical significance was determined using Student’s t-test for comparison of two independent groups.
Results
UBAP2 functions as a regulator of macropinocytosis
Using methodology we have previously described and published, we utilized nanoconjugates as baits to catch and identify the molecular machineries involved in the caveolar or macropinocytotic processes [11, 12]. Comparative proteomic analysis of the proteins pulled down by gold nanoparticles, either completely (Au-C225-C) or partially (Au-C225-P) coated in anti-EGFR antibodies (Fig. S1A), from PANC1 cell lysates identified UBAP2 as one of the proteins exclusively present in the Au-C225-P group (Fig. S1B and Table S1).
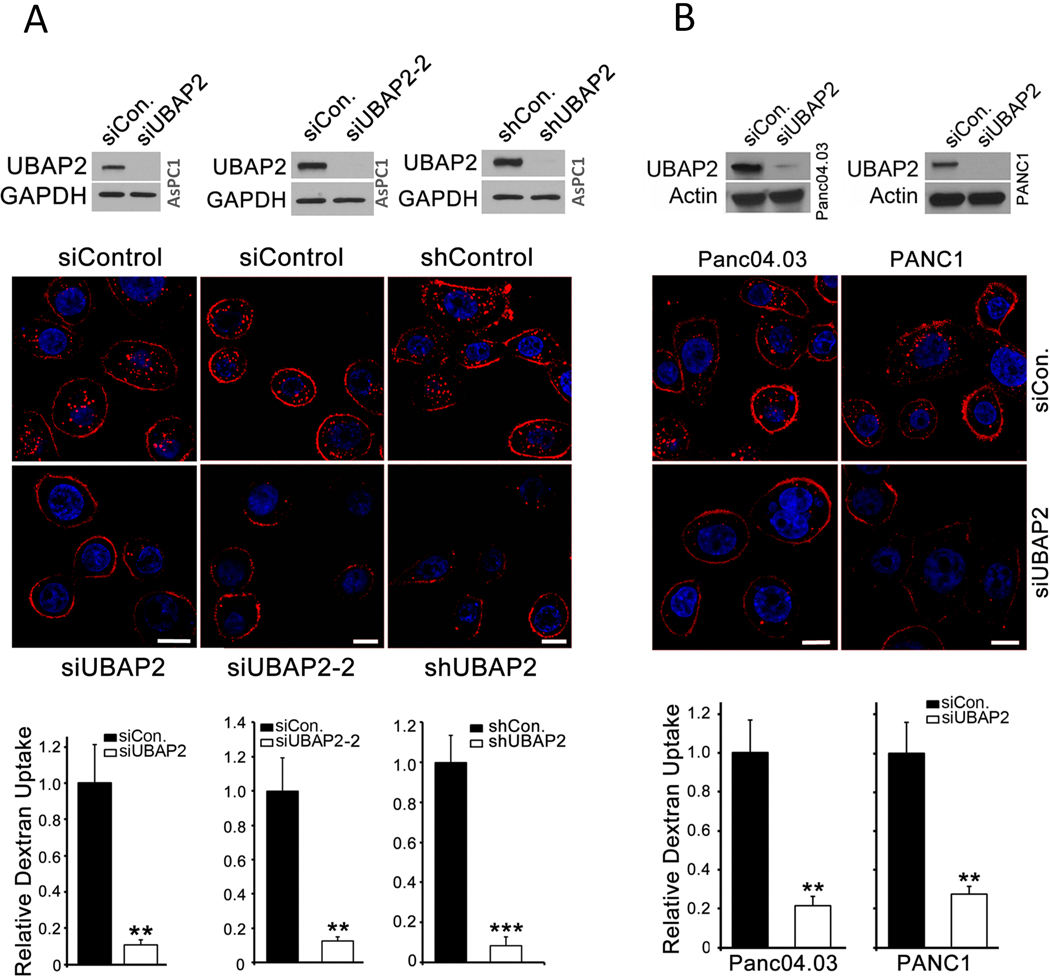
To explore any biological relevance of UBAP2, we first determined its expression in a panel of pancreatic cancer cell lines. Interestingly, higher expression of UBAP2 was observed in pancreatic cancer cells with activating KRAS mutations (AsPC1, DAN-G, HPAF-II, Mia Paca-2, Panc04.03 and PANC1) compared to the cells having wild-type KRAS (HPDE and BxPC3) (Fig. S1C), suggesting a potential link between UBAP2 and KRAS signaling. Since UBAP2 was pulled down by nanoconjugates endocytosed via the macropinocytotic pathway in pancreatic cancer cells, we wanted to investigate if there was any potential link between UBAP2 and macropinocytosis. Furthermore, since higher expression of UBAP2 was observed in pancreatic cancer cells having activating mutation of KRAS, and KRAS signaling is relevant to macropinocytosis in pancreatic cancer, we investigated whether UBAP2 played any role in macropinocytosis in pancreatic cancer cells. We silenced UBAP2 transiently by RNAi (siUBAP2) and stably using short hairpin (Sh) RNA (Sh-UBAP2) and visualized macropinosomes using the fluid-phase marker tetramethyl rhodamine (TMR)-dextran. Silencing was first confirmed by immunoblot analysis showing significant decrease in UBAP2 band both in si- and Sh-UBAP2-AsPC1 cells when compared with scrambled controls (Fig 1A, top panel). Uptake of TMR-dextran as visualized by fluorescence microscopy was significantly inhibited in AsPC1 cells upon UBAP2 silencing by different siRNAs and shRNA (Fig. 1A, middle). Quantitative analysis further demonstrated that nearly 80% of dextran uptake was inhibited upon UBAP2 silencing in AsPC1 cells (Fig. 1A, bottom panel). Similarly, UBAP2 depletion also significantly decreased dextran uptake in Panc04.03 and PANC1 cells (Fig. 1B). To understand whether UBAP2 regulated other major endocytic pathways such as clathrin mediated endocytosis (CME), we investigated the effects of UBAP2 silencing on transferrin uptake in AsPC1 cells (Fig. S2A). Transferrin uptake is facilitated through clathrin-mediated endocytosis, and can be inhibited by chlorpromazine [20]. Fluorescence imaging of Alexa fluor-488 conjugated transferrin demonstrated no inhibition of transferrin uptake in UBAP2-silenced AsPC1 (siUBAP2) cells as compared to scrambled control cells (siControl). Chlorpromazine treatment, which completely abolished transferrin uptake as expected, suggesting UBAP2 is a specific regulator of the macropinocytotic pathway in pancreatic cancer (Fig. S2A, right panel). Additionally, we also determined the kinetics of the dextran uptake as visualized from the TMR-dextran fluorescence in AsPC1 cells transfected with either Scrambled RNA (siControl) or UBAP2-siRNA (siUBAP2, Fig. S2B, C). For quantification purposes, cells with centrally-located macropinosomes were plotted against time (Fig. S2C). As expected in the control cells, dextran uptake increased with time and plateaued after 1 h, whereas in the UBAP2 silenced cells dextran uptake was inhibited over the entire period of time (Fig. S2C).
Figure 1. UBAP2 is a novel regulator of macropinocytosis.
(A and B)UBAP2 silencing decreased TMR-dextran uptake in pancreatic cancer cells. Top, silencing UBAP2 by siRNA or shRNA in pancreatic cancer cells including AsPC1 (A), Panc04.03 and PANC1 (B). Below are the corresponding, TMR-dextran uptake (red) assay indicated that depletion of UBAP2 decreased macropinocytosis. Nuclei (blue) were stained by DAPI. Scale bar, 10 μm. Bottom, Quantification of dextran uptake. Mean ± SD; **, p<0.01; *, p<0.05.
UBAP1, another ubiquitin binding-associated protein containing a UBA domain, may also play a role in macropinocytosis. To investigate this, we first analyzed the expression of UBAP1 in pancreatic cancer cells with activating KRAS mutations (AsPC1, DAN-G, HPAF-II, Mia Paca-2, Panc04.03 and PANC1) compared to the cells having wild-type KRAS (HPDE and BxPC3). Immunoblot analysis of UBAP1 (Fig. S3A) demonstrated that, unlike UBAP2, UBAP1 expression did not correlate with KRAS activation status in pancreatic cancer cells. Next, we evaluated whether UBAP1 played any role in macropinocytosis and compared its role with UBAP2. Visualization of macropinosomes by fluorescently labelled dextran (Fig. S3B), along with quantitative analysis (Fig. S3C, left panel) showed that compared with control scrambled RNA transfected cells, UBAP1 silencing by siRNA had no significant effect in AsPC1 cells whereas silencing of UBAP2 robustly inhibited dextran uptake. Silencing of both UBAP1 and UBAP2 was confirmed by immunoblot analysis in the same set of samples used for dextran uptake (Fig. S3C, right panel). In addition, UBAP1 silencing did not alter activation of KRAS in both AsPC1 and BxPC3 cells (Fig. S3D). These results further support our finding that UBAP2 is a unique UBA domain containing protein that regulates macropinocytosis in pancreatic cancer
UBAP2 promotes macropinocytosis by regulating levels of GTP-bound KRAS (Activated KRAS)
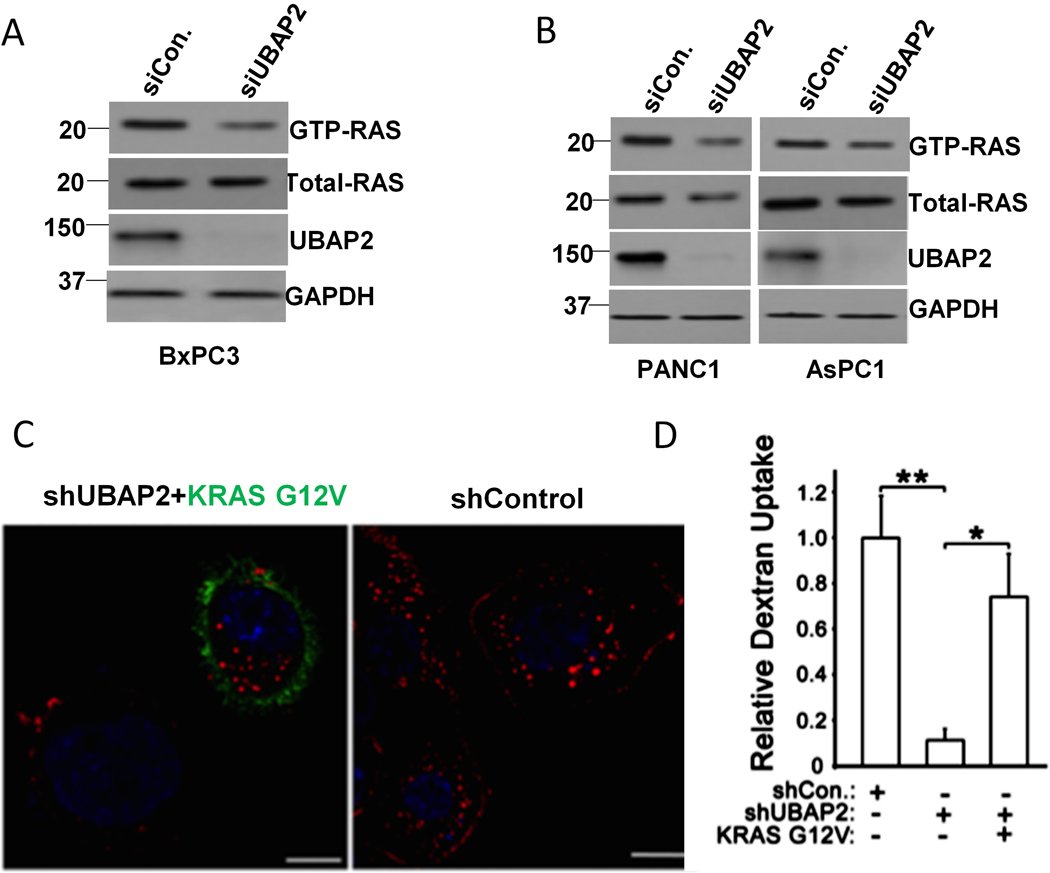
In 2013, Commisso et. al. demonstrated that high levels of macropinocytosis were dependent on oncogenic RAS signaling in pancreatic cancer [4]. As UBAP2 expression correlated with KRAS activation status in pancreatic cancer cells (Fig. S1C), we investigated whether UBAP2 regulated KRAS activation. GTP-bound RAS (activated RAS) was pulled down in UBAP2-silenced cells and its levels were determined by immunoblot analysis. In UBAP2-silenced BxPC3 cells having wild type KRAS, we observed a decrease in activated RAS (GTP-RAS) levels, while the total levels of RAS protein did not change (Fig. 2A). Quantitation of band intensity by densitometric analysis further confirmed statistically significant decreases in GTP-RAS levels (Fig. S4A), whereas total RAS protein level did not change significantly (Fig. S4B), suggesting that UBAP2 regulated RAS activation in these cells. Similar to BxPC3 cells, silencing of UBAP2 in AsPC1 and PANC1 cells harboring constitutively active KRAS mutations, decreased the levels of GTP-RAS while the total levels of RAS did not change (Fig. 2B, Fig. S4A and S4B). We observed a slight decrease in expression of total-RAS in AsPC1 and PANC1 cells which, upon densitometric analysis, was found to be statistically non-significant (Fig. S4B). We also calculated the ratio of GTP-RAS/Total-RAS in both wild type (BxPC3) and mutant (AsPC1 and PANC1) cell lines. As shown in Fig.S4C, there was a significant decrease in the ratio of GTP-RAS/Total-RAS in both wild type (BxPC3) and mutant (AsPC1 and PANC1) cell lines, confirming the role of UBAP2 in regulating RAS activation in these cell lines but not the expression of the RAS proteins.
Figure 2. UBAP2 promotes macropinocytosis by activating KRAS.
(A) UBAP2 knockdown using siRNA (siUBAP2) decreased the activated RAS (GTP-RAS) but not the total -RAS (Total-RAS) in BxPC3 cells using pulldown-based RAS activation assay. (B) UBAP2 knockdown by siRNA (siUBAP2) decreased KRAS in pancreatic cancer cells expressing constitutively activating KRAS mutation. (C) Activated KRAS rescued the dextran uptake upon UBAP2 silencing (shUBAP2). KRAS G12V (green) and dextran (red) was visualized by IF. Scale bar, 10 μm. (D) Quantification of dextran uptake. Mean ± SD; **, p<0.01; *, p<0.05. Ladder represents molecular weight in KDa.
RAS is known to be activated by stimulation with growth factors (GF) including EGF [21]. We sought to determine whether either refeeding serum or EGF stimulation results in enhanced GTP loading of KRAS and whether UBAP2 plays any role in GTP loading. To do so we measured active RAS(GTP-RAS) in BxPC3 cells transfected with either siControl, or siUBAP1 or siUBAP2 and under normal (10% FBS), serum starved (0% FBS), and serum starved (0% FBS) plus EGF stimulation (50ng/ml for 1 min) conditions (Fig.S5). Silencing of UBAP1 and UBAP2 was first confirmed by immunoblot analysis of the same lysate used for RAS activation assay (Fig. S5A–C). Immunoblot analysis revealed that silencing UBAP2 decreased the GTP-RAS levels as compared to scrambled control and siUBAP1 in normal serum (10% FBS). Moreover, GTP-RAS levels in UBAP1 silenced BxPC3 were similar to control suggesting that silencing UBAP1 in BxPC3 cells had no effect on GTP-RAS levels (Fig. S5A). Similarly, upon serum starvation (0% FBS), silencing UBAP2 decreased the GTP-RAS levels as compared to control and siUBAP1. Furthermore, there was no change in GTP-RAS levels upon UBAP1 silencing when compared to siControl transfected BxPC3 cells (Fig. S5B). Interestingly, upon EGF stimulation (50ng/ml for 1 min) after overnight serum starvation, we observed similar results; silencing UBAP2 decreased the GTP-RAS levels as compared to control and siUBAP1, and GTP-RAS levels in UBAP1 silenced BxPC3 were similar to control (Fig. S5C). These results further support that GTP loading of RAS is regulated by UBAP2. Silencing of UBAP2 but not UBAP1 decreased GTP loading of RAS.
To test whether KRAS activation (GTP-loading) is one of the mechanisms by which UBAP2 regulated macropinocytosis, we introduced a vector with an oncogenic KRAS mutation (HA-KRAS G12V) into stable, UBAP2-silenced AsPC1 cells. In our assays, cells overexpressing KRAS were visualized by staining the vector’s HA-tag. For visual representation, we selected a field that showed both transfected (green fluorescence) and non-transfected cell (non-fluorescent) (Fig. 2C). Importantly, we observed that successfully transfected cells (visualized by the HA-stain) showed increased macropinosomes compared to cells that were un-transfected, demonstrating that forced expression of mutant KRAS could rescue dextran uptake defects in UBAP2 silenced cells (Fig. 2C–D). We also performed immunoblot analysis of endogenous KRAS, RAS and KRAS G12V (visualized by the HA-antibody) in the UBAP2 knock-down condition in AsPC1 cells. There was no change in the expression of endogenous KRAS, RAS and KRAS G12V (visualized by the HA-antibody) in UBAP2 silenced AsPC1 cells (Fig S7E), confirming that UBAP2 did not alter the expression of the RAS protein but rather it regulated GTP-loading of RAS. Taken together these data demonstrated that UBAP2 regulated macropinocytosis through activation of KRAS without altering the RAS protein level.
To further support our finding that activation of KRAS was partly responsible for UBAP2 mediated macropinocytosis in pancreatic cancer cells, we performed dextran uptake experiments with a non-Ras, small GTPase control, Rho-A (Fig. S7C, D). Two different Rho-A plasmids, Rho-19N (dominant negative mutant) and RhoA- Q63L (constitutively active mutant) were used[22]. Validation of differential expression of Rho-19N and RhoA- Q63L was performed using qRT-PCR (Fig. S7C). In order to study any role of RhoA in macropinocytosis we only used the constitutively active RhoA- Q63L mutant. Next, sh-Control and sh-UBAP2 AsPC1 cells were transiently transfected with RhoA- Q63L and subjected to dextran uptake analysis (Fig. S7D). The data demonstrated that the forced expression of constitutively active RhoA- Q63L mutant did not affect the macropinocytosis index in either the sh-Control or sh-UBAP2 AsPC1 cells, further supporting our hypothesis that KRAS activation was a mechanism through which UBAP2 regulated macropinocytosis in pancreatic cancer cells (Fig. S7C, D). Furthermore, as discussed above and shown Fig. S3D levels of activated RAS did not change in either AsPC1 or BxPC3 cells upon UBAP1 silencing (Fig. S3D) suggesting that UBAP2 served as a unique UBA domain-containing regulator of KRAS activation in both BxPC3 (wild type) and AsPC1 (mutant) cells.
Next, we wanted to investigate whether modulation of UBAP2 expression could alter KRAS mRNA levels in wild type (BxPC3) and mutant (AsPC1, Panc04.03, and PanC1) cell lines. As shown in Figures S8A and S8B, UBAP2 silencing did not affect KRAS mRNA levels in any of the four cell lines, indicating that UBAP2 was involved in posttranslational regulation of KRAS, rather than being a transcriptional regulator. Together, the results further supported the idea that UBAP2 regulated KRAS activation to promote macropinocytosis in pancreatic cancer.
UBA domain is required for UBAP2-mediated macropinocytosis and KRAS activation
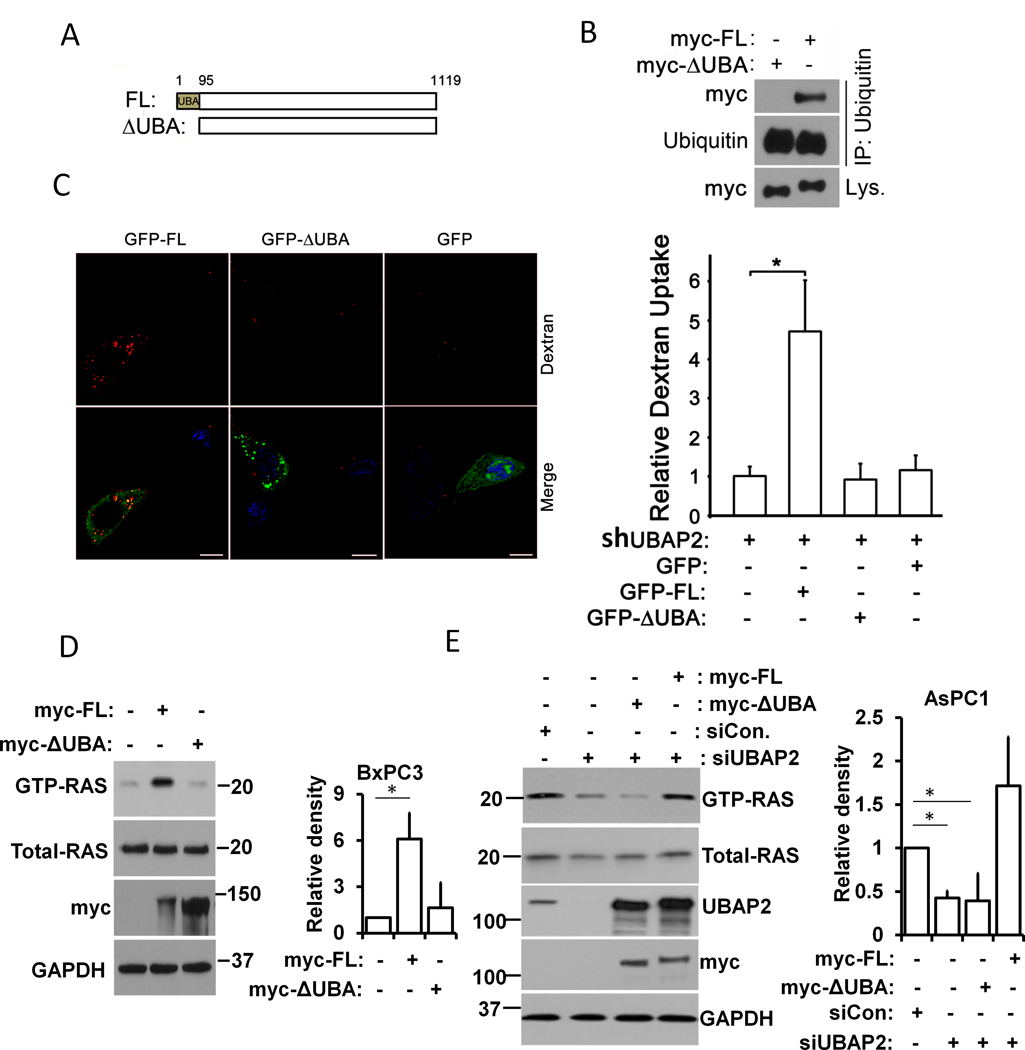
Since UBAP2 contains a UBA domain, a characteristic of proteins involved in the ubiquitination pathway, we investigated whether the UBA domain was required for UBAP2 to interact with ubiquitin leading to macropinocytosis. First, we designed myc-tag fused constructs with either the complete UBAP2 sequence (myc-FL) or UBAP2 having a UBA-domain truncation (myc-ΔUBA) (Fig. 3A); constructs were confirmed by sequencing. To determine the importance of the UBA domain for the interaction with ubiquitin, we overexpressed the myc- constructs in HEK293 cells and lysed the cells for pulldown assays. Cell lysates were immunoprecipitated with ubiquitin antibody and immunoblotted with both ubiquitin and myc antibodies. Only full length UBAP2 could be immunoprecipiated with ubiquitin antibody whereas UBA-truncation prevented this immunoprecipitation (Fig. 3B). These data showed the UBA-domain to be essential for UBAP2 to interact with ubiquitin. To demonstrate any role of the UBA-domain in the regulation of macropinocytosis, we transiently transfected GFP, GFP-fused UBAP2-FL, and UBAP2-ΔUBA vectors into our stable, shUBAP2 AsPC1 cells and evaluated their dextran uptake. Visualization of dextran uptake by florescence microscopy (Fig. 3C left panel) and its quantitation (Fig. 3C, right panel) demonstrated an enhanced dextran uptake (~ 4-fold or more) in cells expressing full length UBAP2 when compared with the GFP control (sh-UBAP2 only) (Fig. 3C). The UBA truncation vector was unable to rescue dextran uptake (Fig. 3C), confirming that the UBA-domain of UBAP2 was critical for the regulation of macropinocytosis.
Figure 3. UBA domain is required for UBAP2-mediated macropinocytosis and KRAS activation.
(A) Schematic of UBAP2 (FL) and UBA domain truncation (ΔUBA). (B) Full length UBAP2 (myc-FL) and UBA domain truncation (myc-ΔUBA) were overexpressed in HEK293 cells, respectively, and the cell lysates subjected to co-IP assay using ubiquitin antibody. The pulled down proteins were immunoblotted with myc-tag or ubiquitin antibody. (C) Full length UBAP2 (GFP-FL, green) but not UBA truncation (GFP-ΔUBA, green) promoted dextran (red) uptake in UBAP2 silencing AsPC1 cells. Scale bar, 10 μm. Left, Quantification of dextran uptake. Mean ± SD; **, p<0.01; *, p<0.05. (D) Lysates from BxPC3 cells overexpression full length UBAP2 (myc-FL) and the UBA domain truncation (myc-ΔUBA) are subjected to pulldown-based RAS activation assay. Total (Total-RAS) and activated RAS (GTP-KRAS) were detected by immunoblotting using RAS antibody. Quantification of GTP-RAS (right panel) (E) Lysates from AsPC1 cells overexpression full length UBAP2 (myc-FL) and the UBA domain truncation (myc-ΔUBA) are subjected to pulldown-based RAS activation assay. Total (Total-RAS) and activated RAS (GTP-KRAS) were detected by immunoblotting using RAS antibody. GAPDH was used as loading control. Quantification of GTP-RAS (right panel). Mean ± SD; *, p<0.05. Ladder represents molecular weight in KDa.
To further understand whether the UBA domain of UBAP2 was critical for its regulation of KRAS activation, we overexpressed full length UBAP2 and UBA domain truncation constructs in BxPC3 cells (containing wild-type KRAS), and collected lysates for pulldown based, GTP-binding RAS assays. Immunoblot analysis demonstrated that overexpression of either full length UBAP2 or UBA-domain truncation vectors had no effect on total KRAS levels (Fig. 3D). However, the full length UBAP2, despite being introduced less efficiently than the UBA-domain truncated construct (Fig. 3D, third row), had more GTP-bound KRAS (Fig. 3D, top row), suggesting that the UBA domain was necessary for UBAP2 to promote KRAS activation. To further confirm the role of the UBA domain, we co-transfected UBAP2 siRNA with myc-UBAP2-FL or myc-UBAP2-ΔUBA into AsPC1 cells. Immunoblot analysis revealed that GTP-RAS was rescued by the full length UBAP2, but not by the UBA domain truncation (Fig. 3E, top row), suggesting that the UBA domain of UBAP2 was critical for regulation of KRAS activation.
To further confirm a role of UBAP2 in regulation of macropinocytosis and KRAS activation, we overexpressed myc-tagged full length UBAP2 (myc-FL-UBAP2) in normal NIH3T3 having wild-type KRAS, low UBAP2 expression and low macropinocytotic index. Overexpression of UBAP2 enhanced the uptake of dextran and increased GTP-RAS level in NIH3T3 cells (Fig. S6 A and B) further supporting induction of macropinocytosis and RAS activation by UBAP2. Overexpression of myc-FL-UBAP2 in NIH3T3 was confirmed by immunoblot analysis showing robust expression of UBAP2 and myc in transfected cells (Fig. S6B). Taken together, these results suggested that the UBA domain was critical for UBAP2 to regulate macropinocytosis and KRAS activation. However, the exact mechanisms by which the UBA domain in UBAP2 regulates KRAS activation remain unknown.
UBAP2 promotes mouse pancreatic tumor growth
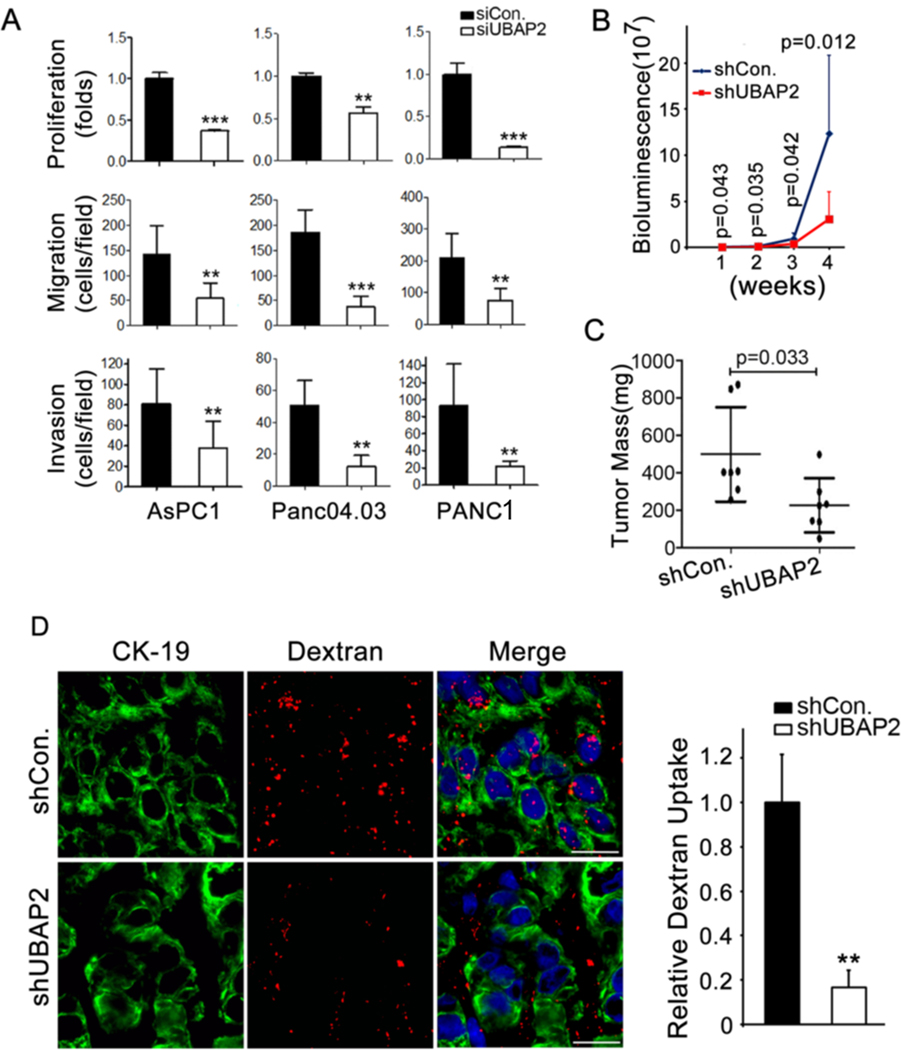
Due to their high metabolic demand, cancer cells utilize macropinocytosis as a preferred mode of nutrient uptake to support growth and survival [7, 23, 24]. Therefore, we investigated whether UBAP2 plays a role in the key steps in pancreatic cancer pathogenesis, specifically cancer cell proliferation, migration, and invasion. Silencing UBAP2 significantly inhibited the proliferation, migration, and invasion of pancreatic cancer cells (Fig. 4A), indicating a critical role of UBAP2 in pancreatic cancer progression. To verify that inhibition of proliferation, invasion and migration were UBAP2 specific, we also silenced UBAP1 in AsPC1 and Pan0403. Silencing of UBAP1 was confirmed by immunoblot analysis (Fig. S6C). Importantly, silencing of UBAP1 in AsPC1 and Pan0403 showed no effect on proliferation (Fig. S6D), invasion (Fig. S6E) or migration (Fig. S6 F) in AsPC1 and Pan0403 cells as compared to siControl. These results confirmed that inhibition of pancreatic cancer pathogenesis was UBAP2 specific.
Figure 4. UBAP2 promotes pancreatic tumor growth in vivo in mice.
(A) UBAP2 silencing decreased proliferation, migration and invasion in pancreatic cancer cells. (B) Tumor growth was monitored by non-invasive bioluminescence analysis. (C) Final tumor mass of the mice showed that knockdown of UBAP2 significantly decreased the tumor growth. Each group contains 7 mice. N=7. *p < 0.05. (D) In vivo macropinocytosis uptake assay and quantification. Section of TMR-dextran (red)-injected mouse xenograft tumors were stained with anti-CK19 (green). Quantification of dextran uptake in (right panel). Scale bar, 10 μm. **p<0.01, N=4.
Furthermore, we evaluated the role of UBAP2 in tumor growth by orthotopically implanting shControl-AsPC1 and shUBAP2-AsPC1 cells into the pancreas of 6–8-week-old nude mice (7 mice per group) and monitored the tumor growth via bioluminescence analysis. Significantly delayed and reduced tumor growth was observed in shUBAP2-AsPC1 mice as compared to the shControl-AsPC1 mice (Fig. 4B, 4C and S7A,B).
We evaluated the TMR-dextran uptake in tumor xenografts derived from either wild-type UBAP2 (shControl-AsPC1) or UBAP2 knockdown AsPC1 cells (shUBAP2-AsPC1). Uptake of dextran was significantly inhibited in shUBAP2-AsPC1 tumors when compared with the shControl-AsPC1 tumors of comparable size (Fig. 4D), suggesting that UBAP2 regulated macropinocytosis in pancreatic cancer cells in vivo.
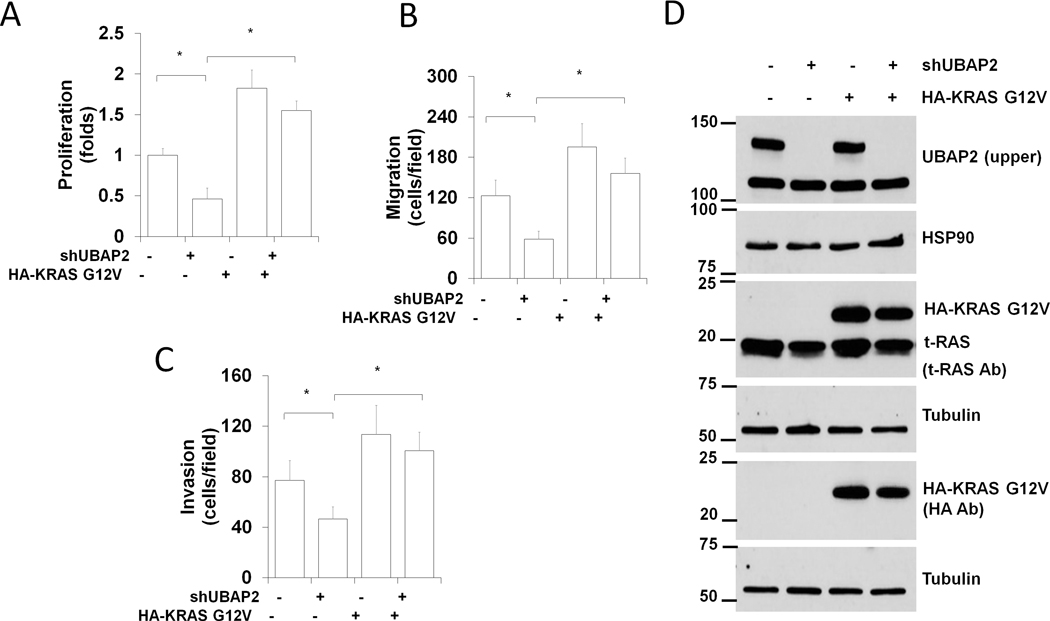
To check whether KRAS could rescue the phenotypic defects observed in UBAP2 silenced cells, we overexpressed constitutively activated KRAS (KRASG12V) in stable UBAP2 silenced AsPC1 cells. Overexpression of HA-KRASG12V was confirmed through immunoblot (Fig. 5D). Proliferation, migration and invasion of HA-KRASG12V expressed cells was higher than control cells, indicating the oncogenic potential of mutated KRASG12V. In contrast, cells transfected with shUBAP2 were low in proliferation, invasion and migration and these phenotypes were completely rescued by the forced expression of HA-KRASG12V (Fig. 5A–C). These results further confirmed activation of KRAS was one of the mechanisms through which UBAP2 regulated macropinocytosis in pancreatic cancer. Next, we measured macropinocytosis in UBAP2-depleted AsPC1 cells in which KRAS expression was also downregulated (double knocked down AsPC1 cells). Macropinocytosis was inhibited in cells with either UBAP2 or KRAS transiently silenced or silenced in tandem (double knocked down) (Fig. S7F–G). Visualization of dextran uptake by fluorescence (Fig. S7F) and its quantitation (Fig. S7G) demonstrated that overexpression of UBAP2 in the double knocked down cells partially rescued macropinocytosis. However, UBA-deleted UBAP2 could not rescue the macropinocytosis. These results further suggested that regulation of KRAS activation was a plausible mechanism by which UBAP2 regulates macropinocytosis; however, involvement of other small GTPases such as CdC42 and Rac1 could not be ruled out.
Figure 5. Activated KRAS rescues dextran uptake in UBAP2 depleted AsPC1cells.
UBAP2 silencing induced decreases in proliferation (A), migration (B) and invasion (C) in pancreatic cancer cells were rescued by HA-KRAS G12V (Activated KRAS). Quantification of proliferation, migration and invasion. Mean ± SD; *, p<0.05. (D) Corresponding Western blots of Experiments (A). Ladder represents molecular weight in KDa.
UBAP2 expression is correlated to pancreatic cancer progression and patient survival
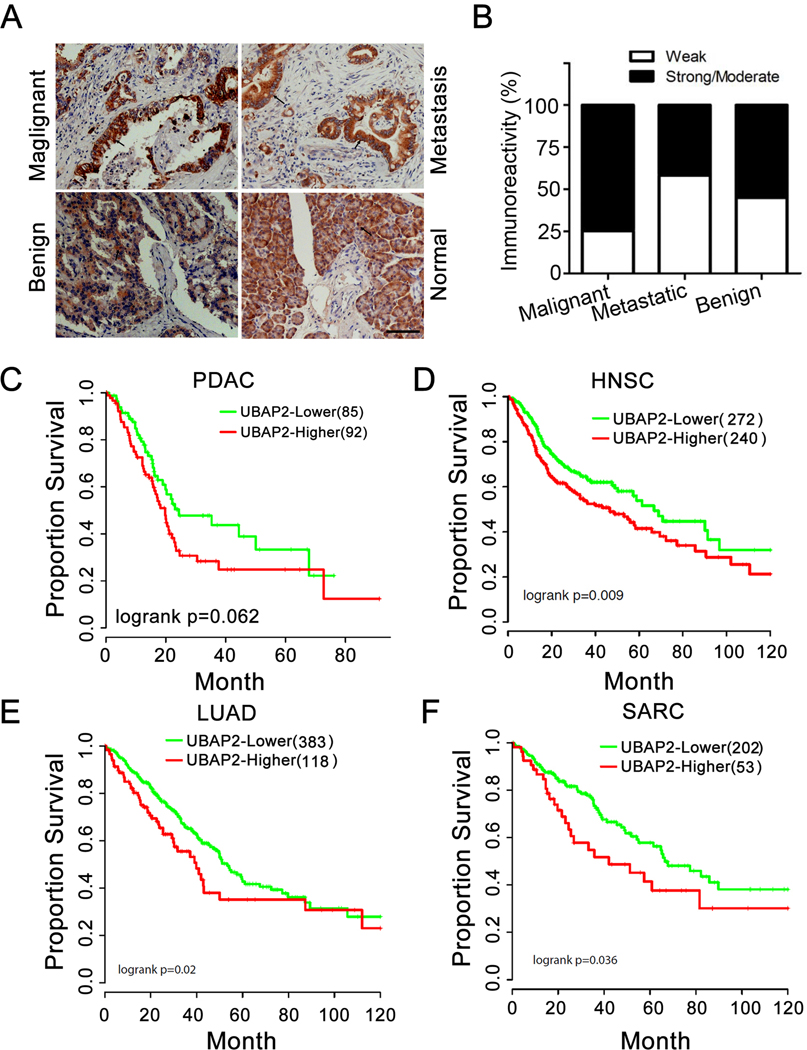
To further investigate a role of UBAP2 in pancreatic cancer prognosis, we evaluated the expression of UBAP2 in a tissue microarray consisting of 101 cases comprising normal, malignant, and metastatic pancreatic cancer tissues (Table S2). UBAP2 was expressed in PDAC, (primary and metastatic tumors, regardless of degree of tumor differentiation) and in low as well as high stage tumor (Fig. 6A). Interestingly, epithelial cells in the adjacent normal tissues did not stain for UBAP2, however, the staining was mostly localized in the acinar/islet cells (Fig. 6A). Importantly, there was no immunoreactivity in fat cells, endothelial cells, smooth muscle cells, inflammatory cells, and fibroblasts in normal, inflamed, or neoplastic pancreas. The association of tumor type (malignant, metastatic, and benign), stage (I, II, III, and IV), other types (inflammation, normal adjacent tissues, and normal), and differentiation (well, moderately and poorly differentiated) with staining intensity was assessed using the (exact) two-sided Pearson’s Chi-square test. There was a significant difference (p=0.0275) in the percentage of strong/moderate staining among the 3 tumor types with the highest percentage (74.7%) observed among malignant tumors, as compared to 41.7% among metastatic and 55% among benign tumors (Fig. 6B). No association was found between staining intensity and malignant tumor stage (p=0.98), other types (p=0.25), and differentiation (p=0.54). Furthermore, there was evidence that higher UBAP2 expression is associated with poor survival in pancreatic cancer (P=0.062) (Fig. 6C), head and neck cancer (P=0.009) (Fig. 6D), lung cancer (P=0.02) (Fig. 6E), and sarcoma (P=0.036) (Fig. 6F), although the association in pancreatic cancer did not reach statistical significance. These results implicated that UBAP2 was functionally associated with poor prognosis in numerous cancers and could be a target for cancer therapy.
Figure 6. UBAP2 expression is correlated to pancreatic cancer progression.
(A) Representative histology of UBAP2 expression (brown staining) in 101 human pancreatic tissues. Scale bar, 100 μm. (B) Statistical analysis of UBAP2 expression in human pancreatic tissues. (C-F) Analysis on TCGA data shows that UBAP2 is associated with poor survival in pancreatic cancer (C), head and neck cancer (D), lung cancer (E), and sarcoma (F).
UBAP2 silencing downregulates RAS downstream signaling
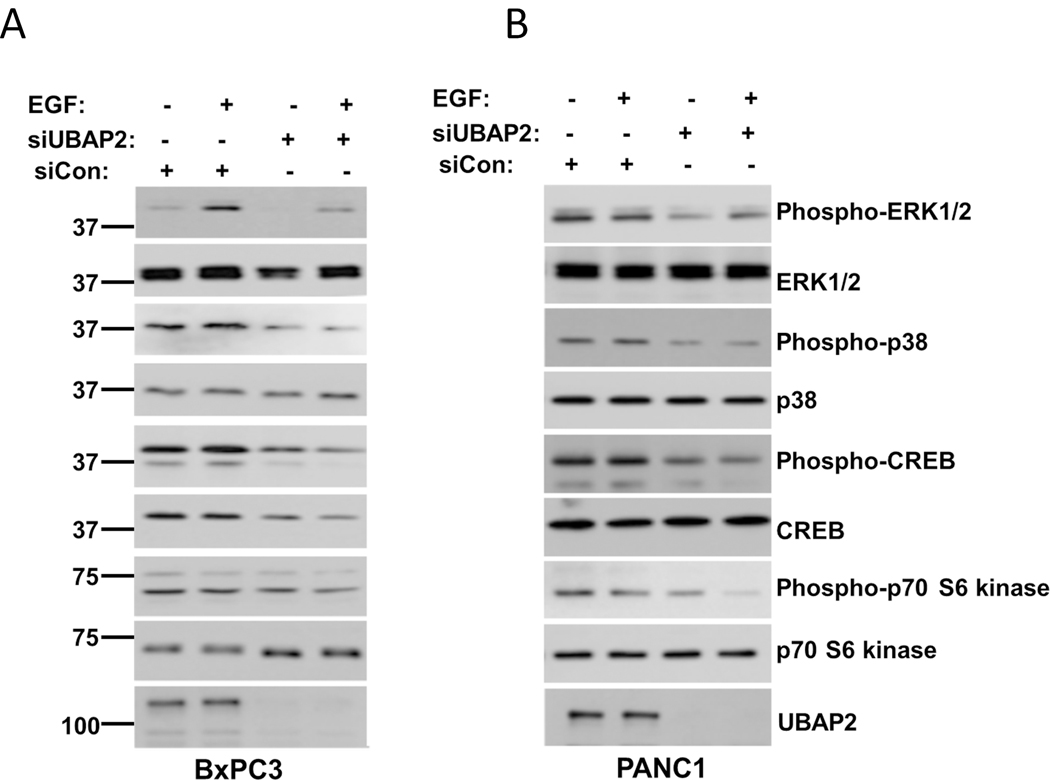
RAS activation is known to stimulate a large number of down-stream signaling cascades. In the canonical pathway, growth factors can turn on RAS by activating Guanine nucleotide Exchange Factors (GEFs), by inhibiting the GTPase Activating Proteins (GAPs), or by both mechanisms simultaneously in the process promoting its binding to downstream effectors (e.g. ErK, Raf, PI3K)[25]. The cycle is terminated by the action of RASGAPs, returning RAS to the inactive GDP-bound state[26]. Since our results demonstrated that UBAP2 regulated RAS activation and macropinocytosis in pancreatic cancer, we next investigated whether UBAP2 had any role in modulation of RAS downstream effectors. We first analyzed the expression of SOS1 (RAS-GEF) and RASGAP in both wild type (BxPC3) and mutant (AsPC1 and PANC1) cells with and without UBAP2 silencing. As shown in Fig. S8C, UBAP2 knockdown did not alter GAP and GEF expression in either wild-type or mutant cell lines. Next, we investigated RAS downstream effector molecule expression after UBAP2 silencing in both wild type and mutant cells. RAS is known to be activated by growth factors including EGF, so in order to investigate whether UBAP2 affects RAS downstream signaling, we performed EGF stimulation experiments in both control and UBAP2 silenced BxPC3 (figure 7A) and PANC1 (figure 7B) cells. As shown in figure 7, there was an increase in phospho-ERK, phospho-p38, phospho-CREB and Phospho-p70 S6 kinase upon EGF stimulation (EGF=50ng/ml, 15 mins at 37°C) in controls of both wild type (BxPC3) and mutant cells (PANC1). However, in UBAP2 silenced cells there was a significant decrease in phospho-ERK, phospho-p38, phospho-CREB and Phospho-p70 S6 kinase in both cell lines. These results collectively suggested that UBAP2 regulated RAS activation and consequently its downstream effector molecules to promote macropinocytosis in pancreatic cancer
Figure 7. UBAP2 Silencing downregulates RAS downstream signaling.
(A) BxPC3 and (B) PANC1 cells were either transfected with scrambled siRNA (siCon) or siRNA against the UBAP2 gene. 36 h post-transfection cells were starved overnight and then stimulated with EGF (EGF=50ng/ml) for 15 mins at 37°C followed by collection of cell lysate. Cell lysate were than subjected to immunoblot for phospho-ERK, ERK, phospho-p38, P38 phospho-CREB, CREB and Phospho-p70 S6 kinase and p70 S6 kinase. Efficient gene silencing was determined by immunoblotting with UBAP2 antibody. Each experiment was repeated at least 3 times. Ladder represents molecular weight in KDa.
Discussion
PDAC is one of the most aggressive malignancies, has a dismal prognosis, and is the 4th leading cause of cancer-related deaths in the United States [1]. Invasive PDAC is believed to arise from a spectrum of preneoplastic mucinous lesions with ductal morphology, namely PanIN, the most common precursor lesions observed in humans, as well as mucinous cystic neoplasia (MCN) and intraductal papillary mucinous neoplasia (IPMN) [1]. The significance of KRAS mutations for disease initiation has been demonstrated in mice, where expression of the constitutively active KRASG12D allele induces PanINs and after a significant latency period also PDA [27]. Recently, Kopp and colleagues demonstrated that acinar cells give rise to pancreatic tumors [28]. Our human pancreatic cancer TMA data revealed high UBAP2 expression across all stages of PDAC, as well as in normal acinar and islet cells, indicating a potential role of UBAP2 in pancreatic cancer origin that needs further investigation.
Although oncogenic KRAS alleles were first identified more than 20 years ago, targeting mutant KRAS remains challenging [29]. Here, for the first time, we demonstrate UBAP2 as a new regulator of macropinocytosis and KRAS activation. Only recently has a role for UBAP2 in cancer been reported; UBAP2 is elevated in metastatic prostate cancer, while in hepatic cancer, UBAP2 expression is inversely correlated with prognosis [15, 16]. Since activation of small GTPases, especially RAS, has been implicated in the oncogenesis of nearly one-third of human cancers, UBAP2 may be exploited as a unique therapeutic target to inhibit tumor growth and metastasis in human malignancies that are primarily driven by activating mutations of small GTPases.
Due to their high metabolic demand, cancer cells utilize macropinocytosis as a preferred mode of nutrient uptake to support growth and survival [7]. Hence, identifying new molecular machineries that regulate macropinocytosis in cancer cells is vital to understanding the biology of tumor progression and metastasis and provides an innovative approach to inhibit it. In that context establishing biological function for UBAP2 as a molecular switch that regulates macropinocytosis, KRAS activation and tumor growth in pancreatic cancer is significant. Recently Commisso et al. [4] demonstrated that RAS-transformed pancreatic tumor growth is dependent on the macropinocytosis mediated uptake of proteins that provide glutamine intracellularly, and suggested that targeting macropinocytosis could be efficacious. However, lack of macropinocytosis specific targets and the challenges of targeting mutant KRAS pose significant barriers to successful therapy. In this context, UBAP2 could prove to be an important target that not only regulates macropinocytosis, but also activation of small GTPases.
Activated RAS controls diverse signaling pathways including cell proliferation, survival, differentiation and motility, which in turn depend on its binding to a range of functionally diverse effector molecules such as Raf, PI3K, AF6 and RASSFs[30]. Based on the accumulating evidence, we believe there are three plausible mechanisms through which UBAP2 may regulate KRAS activation: i) activation of RAS-GEFs, ii) inhibition of Ras-GAPs, and iii) enhancing GTP loading by binding to the mono- or di-ubiquitinated KRAS [25, 31]. Since UBAP2 interacts with ubiquitin via the UBA domain and enhanced GTP-loading of RAS (Figure 3B), we speculate that UBAP2, by binding to mono- and/or di-ubiquinated KRAS via the UBA domain, enhances GTP-loading of KRAS. This speculation is supported by our data on probing GTP bound RAS under serum starvation and EGF refeeding in the presence and absence of UBAP2 (Fig. S5). Importantly, further studies are needed to demonstrate the direct interaction of UBAP2 with mono-/di-ubiquitinated KRAS or the exact mechanism by which UBAP2 regulates KRAS activation. These studies will constitute the focus of our future investigation.
Herein, we demonstrated that UBAP2 functions as a novel regulator of macropinocytosis that sustains RAS activation. UBAP2 exhibits high expression in constitutively activated KRAS mutated pancreatic cells and in PDAC patient tissues. UBAP2 functions as an enhancer of macropinocytosis that regulates nutrient uptake to meet the high metabolic demand of cancer cells, suggesting a multifactorial role for UBAP2. In summary, UBAP2 may be exploited as a new therapeutic target to inhibit activation of small GTPases, macropinocytosis and PDAC growth. Molecular characterization of UBAP2 function will establish it as a new therapeutic target in pancreatic cancer and provide new avenues to target macropinocytosis and KRAS activation, a goal that remains elusive even after 30 years of intense investigation.
Supplementary Material
Acknowledgments
We would like to thank the Peggy and Charles Stephenson Cancer Center at the University of Oklahoma Health Sciences Center for a seed grant. An Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103639 supports the Histology and Immunohistochemistry Core, which provided immunohistochemistry and image analysis services. This research was also supported by Peggy and Charles Stephenson Endowed Chair fund to PM.
This work was supported by National Institutes of Health Grants 2CA136494, CA213278, CA220237 to P.M.
Research reported in this publication was supported in part by the Oklahoma Tobacco Settlement Endowment Trust awarded to the University of Oklahoma // Stephenson Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Oklahoma Tobacco Settlement Endowment Trust.
Research reported in this publication was supported in part by the National Cancer Institute Cancer Center Support Grant P30CA225500 awarded to the University of Oklahoma Stephenson Cancer Center and used the Biostatistics and Research Design Shared Resource and the Office of Cancer Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Nonstandard Abbreviations:
- PDACs
Pancreatic Ductal Adenocarcinoma Cells
- UBAP2
UBAP2 Ubiquitin Binding Associated Protein 2
- UBA domain
Ubiquitin Binding Associated
- GTP
Guanosine-5’-Triphosphate
- GEF
Guanine nucleotide Exchange Factor (GEF)
- GAP
GTPase-Activating Protein
- EGFR
Epidermal Growth Factor Receptor
- Au
Gold
- RNAi
RNA interference
- TMR-Dextran
TetraMethylRhodamine-Dextran
- BSA
Bovine Serum Albumin
- IP
Immunoprecipitation
- SDS–PAGE
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (PAGE)
- HRP-conjugated
Horseradish peroxidase (HRP)
- HEPES
4-(2-HydroxyEthyl)-1-PiperazineEthaneSulfonic acid)
- DAPI
4′,6-DiAmidino-2-PhenylIndole
- DAB
DiAminoBenzidine
- TCGA
The Cancer Genome Atlas
- GFP
Green Fluorescence Protein
- shRNA
Short Hairpin Ribonucleic Acid
- siRNA
Small Interfering Ribonucleic Acid
- PanIN
Pancreatic Intraepithelial Neoplasia
- MCN
Mucinous Cystic Neoplasia
- IPMN
Intraductal Papillary Mucinous Neoplasia
- IF
Immunofluorescence
- ATCC
American Type Culture Collection
- UTR
Untranslated Region
- FBS
Fetal Bovine Serum
Footnotes
Conflict of interest: No potential conflicts of interest were disclosed.
Supplementary Information
Supplementary Data contains: Two supplementary table and eight supplementary figures.
References
- 1.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK & Hruban RH (2013) Recent progress in pancreatic cancer, CA: a cancer journal for clinicians. 63, 318–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N. & Perucho M. (1988) Most Human Carcinomas of the Exocrine Pancreas Contain Mutant C-K-Ras Genes, Cell. 53, 549–554. [DOI] [PubMed] [Google Scholar]
- 3.Cox AD, Fesik SW, Kimmelman AC, Luo J. & Der CJ (2014) Drugging the undruggable RAS: Mission possible?, Nature reviews Drug discovery. 13, 828–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG & Bar-Sagi D. (2013) Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells, Nature. 497, 633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Ng SC, Kang Y, Fleming JB, Bardeesy N, Asara JM, Haigis MC, DePinho RA, Cantley LC & Kimmelman AC (2013) Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway, Nature. 496, 101-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gwinn DM, Lee AG, Briones-Martin-del-Campo M, Conn CS, Simpson DR, Scott AI, Le A, Cowan TM, Ruggero D. & Sweet-Cordero EA (2018) Oncogenic KRAS Regulates Amino Acid Homeostasis and Asparagine Biosynthesis via ATF4 and Alters Sensitivity to L-Asparaginase, Cancer cell. 33, 91-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBerardinis RJ, Lum JJ, Hatzivassiliou G. & Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation, Cell metabolism. 7, 11–20. [DOI] [PubMed] [Google Scholar]
- 8.Hensley CT, Wasti AT & DeBerardinis RJ (2013) Glutamine and cancer: cell biology, physiology, and clinical opportunities, The Journal of clinical investigation. 123, 3678–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson JA & Watts C. (1995) Macropinocytosis, Trends in cell biology. 5, 424–8. [DOI] [PubMed] [Google Scholar]
- 10.Sousa CM & Kimmelman AC (2014) The complex landscape of pancreatic cancer metabolism, Carcinogenesis. 35, 1441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya S, Bhattacharya R, Curley S, McNiven MA & Mukherjee P. (2010) Nanoconjugation modulates the trafficking and mechanism of antibody induced receptor endocytosis, Proceedings of the National Academy of Sciences of the United States of America. 107, 14541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharyya S, Singh RD, Pagano R, Robertson JD, Bhattacharya R. & Mukherjee P. (2012) Switching the targeting pathways of a therapeutic antibody by nanodesign, Angewandte Chemie. 51, 1563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husnjak K. & Dikic I. (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions, Annual review of biochemistry. 81, 291–322. [DOI] [PubMed] [Google Scholar]
- 14.Popovic D, Vucic D. & Dikic I. (2014) Ubiquitination in disease pathogenesis and treatment, Nat Med. 20, 1242–1253. [DOI] [PubMed] [Google Scholar]
- 15.Latonen L, Leinonen KA, Gronlund T, Vessella RL, Tammela TL, Saramaki OR & Visakorpi T. (2016) Amplification of the 9p13.3 chromosomal region in prostate cancer, Genes Chromosomes Cancer. 55, 617–25. [DOI] [PubMed] [Google Scholar]
- 16.Bai DS, Wu C, Yang LX, Zhang C, Zhang PF, He YZ, Cai JB, Song ZJ, Dong ZR, Huang XY, Ke AW & Shi GM (2016) UBAP2 negatively regulates the invasion of hepatocellular carcinoma cell by ubiquitinating and degradating Annexin A2, Oncotarget. 7, 32946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Commisso C, Flinn RJ & Bar-Sagi D. (2014) Determining the macropinocytic index of cells through a quantitative image-based assay, Nature protocols. 9, 182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giri K, Shameer K, Zimmermann MT, Saha S, Chakraborty PK, Sharma A, Arvizo RR, Madden BJ, McCormick DJ, Kocher JP, Bhattacharya R. & Mukherjee P. (2014) Understanding protein-nanoparticle interaction: a new gateway to disease therapeutics, Bioconjug Chem. 25, 1078–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arvizo RR, Giri K, Moyano D, Miranda OR, Madden B, McCormick DJ, Bhattacharya R, Rotello VM, Kocher JP & Mukherjee P. (2012) Identifying new therapeutic targets via modulation of protein corona formation by engineered nanoparticles, PLoS One. 7, e33650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Kim H, Chang B, Ahn N, Hwang S, Di Paolo G. & Chang S. (2006) Regulation of transferrin recycling kinetics by PtdIns[4,5]P2 availability, FASEB J. 20, 2399–401. [DOI] [PubMed] [Google Scholar]
- 21.Malik R, Luong T, Cao X, Han B, Shah N, Franco-Barraza J, Han L, Shenoy VB, Lelkes PI & Cukierman E. (2019) Rigidity controls human desmoplastic matrix anisotropy to enable pancreatic cancer cell spread via extracellular signal-regulated kinase 2, Matrix Biol. 81, 50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi X, Wang S, Huang Y, Stamnes M. & Chen JL (2013) Roles of rho GTPases in intracellular transport and cellular transformation, Int J Mol Sci. 14, 7089–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finicle BT, Jayashankar V. & Edinger AL (2018) Nutrient scavenging in cancer, Nat Rev Cancer. 18, 619–633. [DOI] [PubMed] [Google Scholar]
- 24.Torrence ME & Manning BD (2018) Nutrient Sensing in Cancer, Annu Rev Cancer Biol. 2, 251–269. [Google Scholar]
- 25.Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, Haviv S, Asara JM, Pandolfi PP & Cantley LC (2011) Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors, Sci Signal. 4, ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logsdon CD & Lu W. (2016) The Significance of Ras Activity in Pancreatic Cancer Initiation, Int J Biol Sci. 12, 338–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM & Tuveson DA (2003) Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse, Cancer cell. 4, 437–50. [DOI] [PubMed] [Google Scholar]
- 28.Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris J. P. t., Pan FC, Akiyama H, Wright CV, Jensen K, Hebrok M. & Sander M. (2012) Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma, Cancer cell. 22, 737–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson H. (2013) US National Cancer Institute’s new Ras project targets an old foe, Nat Med. 19, 949–50. [DOI] [PubMed] [Google Scholar]
- 30.Karnoub AE & Weinberg RA (2008) Ras oncogenes: split personalities, Nat Rev Mol Cell Biol. 9, 517–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, Der CJ & Cox AD (2016) The role of wild type RAS isoforms in cancer, Semin Cell Dev Biol. 58, 60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.