Abstract
Extracellular vesicles (EVs) have been growingly recognized as biomarkers and mediators of alcoholic liver disease (ALD) in human and mice. Here we characterized hepatocyte-derived EVs (HC-EVs) and their cargo for their biological functions in a novel murine model that closely resembles liver pathology observed in patients with alcoholic hepatitis (AH), the most severe spectrum of ALD. The numbers of circulating EVs and HC-EVs were significantly increased by 10-fold in AH mice compared with control mice. The miRNA (miR)–seq analysis detected 20 upregulated and 4 downregulated miRNAs (P < 0.001–0.05) in AH-HC-EVs. Treatment of murine primary hepatic stellate cells (HSCs) with AH-HC-EVs induced α-SMA (P < 0.05) and Col1a1 (P < 0.001). Smad7 and Nr1d2 genes, which were downregulated in HSCs from the AH mice, were predicted targets of 20 miRs upregulated in AH-HC-EVs. Among them were miR-27a and miR-181 which upon transfection in HSCs, indeed repressed Nr1d2, the quiescent HSC marker. AH-HC-EVs were also enriched with organelle proteins and mitochondrial DNA (10-fold, P < 0.05) and upregulated IL-1β and IL-17 production by hepatic macrophages (HMs) from AH mice in a TLR9-dependent manner. These results demonstrate HC-EV release is intensified in AH and suggest that AH-HC-EVs orchestrate liver fibrogenesis by directly targeting the quiescent HSC transcripts via a unique set of miRNAs and by amplifying HSC activation via DAMP-based induction of profibrogenic IL-1β and IL-17 by HMs.
Keywords: AH, Hepatocyte-derived EVs, miRNAs, DAMPs, HSC activation, Hepatic macrophage activation
Introduction
Alcoholic liver disease (ALD) is one of the most common forms of chronic liver disease in the USA and many other countries with the global mortality estimated to exceed 0.7 million per year [1, 2]. ALD represents a wide spectrum of liver damage ranging from alcoholic steatosis, alcoholic steatohepatitis (ASH), and to alcoholic cirrhosis. ASH includes alcoholic hepatitis (AH), which is characterized by intense acute PMN infiltration and may progress to an acute on chronic liver failure with high morbidity and mortality and limited therapeutic options [3, 4]. Accumulating evidence indicates that hepatocyte (HC) stress, and eventually HC death, plays a central role in inflammation and hepatic injury during the progression of ALD [3–5]. Hepatic macrophages (HMs) and hepatic stellate cells (HSCs) are critical to the progression of ethanol-induced liver pathology [5–9]. However, the molecular signaling events involved in the crosstalk between stressed, lipid-loaded HCs and non-parenchymal cells in AH, are poorly understood. This issue is important, as identifying a factor, or factors, that communicate stress signals from HCs and initiate and perpetuate inflammatory and fibrogenic processes in the development of AH may have an obvious biomedical implication and may allow the identification of individualized therapeutic approaches in the treatment of patients with different stages of ALD. Moreover, identification of a specific signature, or “barcode”, from damaged HCs during AH may serve as a mechanism-based liver-specific and disease-specific biomarker that may revolutionize the way we diagnose and monitor AH.
Extracellular vesicles (EVs) are cell-derived membrane-bound structures containing a specific cargo including proteins, microRNAs (miRs), and DNA from the cell or origin and the stress inducing their biogenesis. EVs can act on the environment they are released via internalization into target cells and/or be released into the circulation [10]. Indeed, several groups revealed that determination of EVs and their composition in blood can be useful as potential biomarkers for ALD and for better understanding of the pathophysiology of ALD progression through HSC activation, macrophage activation, and neutrophil accumulations [11]. Furthermore, we previously demonstrated, using an intragastric mouse model of mild ASH, that specific miRs including let-7f, miR-29a, and miR-340 were enriched in HC-derived EVs (HC-EVs) and validated these miRs were also significantly upregulated in the blood EVs from patients with mild ASH [8]. Recently, a mouse model of hybrid intragastric and ad libitum feeding was developed which achieves a drastic histologic transition from chronic ASH to AH and has been utilized to disclose several pathways which are activated in this transition including pyroptosis pathway [9, 12]. We report herein that another pathway significantly activated in the chronic ASH to AH transition is EV release and that HC-EVs in AH mice carry a unique cargo composition which facilitates direct crosstalk to both HSCs and HMs culminating in profibrogenic signals as revealed by flow cytometry, transcriptomic and proteomic analyses, and ex vivo cell experiments.
Materials and methods
Animal studies
The use and care of the animals was reviewed and approved by the Institutional Animal Care and Use Committee at the University of Southern California. The AH mouse model using intragastric infusion was generated as previously described and produced by the Animal Core of the Southern California Research Center for ALPD and Cirrhosis [9]. Male C57B/6 mice were fed solid Western diet high in cholesterol and saturated fat (HCFD) for 2 weeks followed by implantation of a long-term gastrostomy catheter (iG) and iG feeding of ethanol and a high fat liquid diet (corn oil as 36% Cal) at 60% of total daily caloric intake. As a unique hybrid feeding model, mice consumed the remaining 40% Cal via ad libitum intake of HCFD. The ethanol dose was increased to 27 g/kg/day over an 8-week period. Additionally, hybrid HCFD+Alc mice were subjected to alcohol binge at a weekly interval from the 2nd week of iG feeding. For this binge, ethanol iG infusion was withdrawn for 5~6 h and a bolus dose (3.5~5 g/kg) of ethanol equivalent to what was withdrawn, was given. Following 8 weeks of intragastric alcohol feeding plus weekly binge, mice were sacrificed for isolation of liver cells and plasma EVs.
Liver and blood sample preparation
A part of liver was collected from the right lobe before liver perfusion for HC or HM isolation performed by the Integrative Liver Cell Core of the University of Southern California as described below. Liver tissue was fixed in 10% formalin for 24 h and embedded in paraffin, placed in RNAlater Solution (Thermo Fisher Scientific, Carlsbad, CA, USA) for RNA isolation, or snap-frozen in liquid nitrogen and stored in − 80 °C. Blood was collected with or without anticoagulant following centrifugation at 12,000 rpm for 15 min. The serum or plasma was stored in − 80 °C.
Real-time PCR
Total RNA was isolated from cells using Trizol and Rnase-free Dnase I (Thermo Fisher Scientific) or Quick-RNA MiniPrep (Zymo Research). The reverse transcripts (the cDNA) were synthesized from 1 μg of total RNA using the iScript cDNA Synthesis kit (Bio-Rad, Irvine, CA). Real-time PCR quantification was performed using SYBR-Green and CFX96 Thermal Cycler from BioRad. Briefly, 10 μL of reaction mix contained the following: cDNA, KAPA SYBR® FAST qPCR master mix, and primers at final concentrations of 200 nmol. The sequences of the primers used for quantitative PCR were as follows: for HSC experiments, α-SMA 5′-GTC CCA GAC ATC AGG GAG TAA-3′ and 5′-TCG GAT ACT TCA GCG TGA GGA-3′; Co1α1; 5′-GCT CCT CTT AGG GGC CAC T-3′ and 5′-CCA CGT CTC ACC ATT GGG G-3′; microglobulin 5′-CCC CAC TGA GAC TGA TAC ATA CG-3′ and 5′-CGA TCC CAG TAG ACG GTC TTG-3′; for HM experiments, pro interleukin 1-beta (IL-1β) 5′-ACT CCT TAG TCC TCG GCC A-3′ and 5′-TGG TTT CTT GTG ACC CTG AGC-3′; IL-17a 5′-TTT AAC TCC CTT GGCGCA AA-3′ and 5′-CTT TTC CCT CCG CAT TGA CAC-3′; M36B4 5′-AGA TTC GGG ATA TGC TTG GG-3′ and 5′-TCG GGT CCA AGA CCA GTG TTC-3′.
Cell isolation, culture, and miRNA transfection
Liver cells were isolated as previously described [8, 13, 14]. Briefly, AH or pair-fed control mouse liver was digested with collagenase perfusion through superior vena cava and HCs were isolated by centrifugation at 50×g for 1 min. HCs were cultured for 30 min in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Camarillo, CA) containing 3% EV-free fetal bovine serum (FBS) (System Biosciences), gently washed, and cultured with fresh DMEM with 2% EV-free serum for 3 h for collection of EV released by HCs. HMs were isolated by discontinuous gradient centrifugation of a non-parenchymal cell fraction with OptiPrep (Axis-Shield PoC AS, Oslo, Norway) and purified by adherence method in DMEM with 3% FBS as described before [8]. HMs were then cultured (0.45 × 106 cells per well) in DMEM with 3% EV-free FBS and immediately tested for the effects of HC-EV. The purity exceeded 97% HCs and 96% for HMs for both groups after the initial washing in primary cultures. For HSC experiments, HSCs were isolated from B6 mice with Nicodenz and cultured in DMEM supplemented with 10% FBS (Cellgro, Manassas, VA), and penicillin and streptomycin (Gibco) at 37 °C in a 10% CO2 incubator. HSCs were serum starved for 24 h, were transfected with 1 and/or 10 nM of miRNA-lipofectamine RNAiMAX (Thermo Fisher Scientific) for 48 h, and collected for gene expression (1 or 10 nM miRNA) and immunoblot analysis (1 nM miRNA). miR-27a-3p, miR-181a-5p, and negative miRNA (control) (Thermo Fisher Scientific) were used for miRNA transfection. For HM experiments, HEK-Blue-mTLR9 cells were cultured in DMEM with 3% EV-free FBS (Cellgro, Manassas, VA), penicillin and streptomycin (Gibco), and sodium pyruvate (Gibco) at 37 °C in a 10% CO2 incubator.
For isolation of HSCs from AH vs. control mice, Col1a1-GFP mice subjected to the AH or control regimen were used. This is because isolation of pure HSCs from C57/B6 AH mice by the standard gradient centrifugation technique is impossible due to lost buoyancy of AH HSCs and contamination with fat-laden HCs and HMs. Following the two-step pronase-collagense in situ digestion and gradient ultracentrifugation using Opti-Prep™ (Millipore Sigma), the cells between 1.034 and 1.054 density were collected and subjected to FACS. GFP fluorescence was excited at 488 nm by an argon laser and measured at 530 nm while the vitamin A in HSC was excited at 350 nm by a krypton laser and measured at 450 nm. A dominant vitamin A+;GFP− population from the control Col1a1-GFP mice was collected as quiescent control HSCs while a vitamin A+;GFP+ population from the AH mice was collected as activated AH HSCs.
Immunoblot analysis
For immunoblot analysis HSC lysate was resolved by a 4–20% gradient gel, transferred to a PVDF membrane (Bio-rad), and blotted with the appropriate primary antibody, anti-Nr1d2 (Abnova, Taiwan) followed by peroxidase-conjugated secondary antibody (GE Healthcare Life Sciences, Pittsburgh, PA, USA). Protein bands were visualized using enhanced chemiluminescence reagents (Thermo Fisher Scientific) and digitized using a CCD camera (LAS4000 mini; Fuji film, JAPAN). Expression intensity was quantified by Multi Gauge (Fuji). Protein load was verified using β-actin (GeneTex) antibody.
Cell-derived EV isolation
Isolated HCs or HMs from AH or pair-fed control mice were cultured for 3 h to collect conditioned media which were subsequently centrifuged at 2000×g (4800 rpm) for 8 min at 10 °C to clear cell debris and at 16,000×g (11,400 rpm) for 30 min at 10 °C to remove mitochondria. EVs were isolated from the supernatant by centrifugation at 110,000×g (30,000 rpm) for 60 min at 10 °C (Beckman L7–65, Beckman-Coulter, Palo Alto, CA) and washed with PBS by centrifugation at 110,000×g for 45 min. Pelleted EVs were resuspended in PBS and stored at − 80 °C.
Measurement of EVs
Conditioned medium or plasma was incubated with calcein-AM (Thermo Fisher Scientifics) for 30 min at room temperature. The number of EVs was determined using 2.5-μm Alignflow alignment beads (Thermo Fisher Scientifics) as the size standards for flow cytometry, BD LSRII Flow Cytometer System (BD Biosciences, San Jose, CA). The data were analyzed using FlowJo software (TreeStar Inc., Ashland, OR).
EV size determination
For dynamic light scattering analysis, entire size was measured by Zetasizer nano ZS90 (Malvern). For transmission electron microscopy, EVs were adhered to 100 mesh Formvar and carbon-coated grids for 5 min at room temperature. Grids were washed once with water, stained with 1% uranyl acetate (Ladd Research Industries, Williston, VT) for 1 min, dried, and viewed using a JEOL 1200 EXII transmission electron microscope. Images were captured using a Gatan Orius 600 digital camera (Gatan, Pleasanton, CA).
miRNA sequencing in purified HC-EVs and data analysis
The miRNA sequencing work was carried out in the UCSD IGM Genomics Center. Quality and quantity of purified total RNA from purified HC-EVs using the method described above was assessed using an Agilent Tapestation and a NanoDrop ND-1000, respectively. Libraries were generated from 100 ng of total RNA, using the TruSeq SmallRNA Sample Prep Kit (Illumina, San Diego, CA) following manufacturer’s instructions. Library quality was assessed using a High Sensitivity DNA kit (Agilent, Santa Clara, CA). Libraries were multiplexed and sequenced at 13 pM to an average depth of 7.5 million reads with 50 basepair (bp) single end (SE) reads on an Illumina HiSeq2500 using V4 chemistry. One sample from AH-HC-EVs was not able to read miR sequencing due to technical reason.
The raw sequencing reads in FASTQ format went through preprocessing first to trim 3′ adapter and next to keep only reads whose length is between 17 and 27 bases. Using bowtie, the survived reads were aligned to reference resources such as mouse genome, miRNA precursor sequence, and non-coding RNAs, allowing only up to two mismatches. Only the reads mapped to both genome and precursor, but not to small RNAs other than miRNA, were selected for expression level quantification. Then, miRNA expression counts were quantified, normalized, calculated for fold-change between the treated and the control groups, and tested for significance of differential expression using negative-binomial model [15]. Benjamini-Hochberg method [16] was applied to adjust for multiple testing on the raw P values of differential expression test. MAGI, a web service for fast miRNA-seq analysis in a graphics processing unit (GPU) infrastructure, was used for a pipelined analysis of sequencing data [17].
The 20 significantly (raw P value less than 0.05) upregulated miRs were chosen for further analysis to predict target genes. For each miRNA, a set of target genes was created with a threshold of prediction score 0.7 against micro CDS target prediction database using DIANA TOOLS [18]. The union set comprising all 20 sets of target gene was mapped to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to derive significantly enriched pathways using Fisher’s exact test. Two heatmaps, miRNA-by-sample and miRNA-by-pathway, were drawn with Heatmapper and DIANA-miRPath, respectively.
Proteomic and mtDNA analysis in purified HC-EVs
Samples, EVs isolated from AH vs. control HC using the method described above, for proteomics analysis were prepared using the trifluoroethanol (TFE)–based protocol previously described [19]. Digested peptides were dried in a vacuum centrifuge, and the concentration was adjusted to 0.1 μg/μL before liquid chromatography–tandem mass spectrometry analysis (LC-MS/MS). Each experimental condition was represented by biological quadruplicates. All samples were block randomized before analysis by LC-MS/MS. A total of 500 ng was loaded into a trap column (5 cm by 360-μm outer diameter [OD] by 150-μm inner diameter [ID] fused silica capillary tubing [Polymicro, Phoenix, AZ]) packed with 3.6-μm Aeries C18 particles (Phenomenex, Torrance, CA). LC separation was performed in a capillary column (70 cm by 360-μm OD by 75-μm ID) packed with 3-μm Jupiter C18 stationary phase (Phenomenex) with a 100-min gradient of acetonitrile (ACN) in water containing 0.1% formic acid. Eluted peptides were analyzed online in a quadrupole-Orbitrap mass spectrometer (Q-Exactive Plus; Thermo Fisher Scientific, San Jose, CA) with settings previously described [20]. Mass spectrometry files were searched against Mus musculus Uniprot database (UniprotKB, downloaded in 2015) using MS-GF+. Identified peptides were filtered to 1% FDR and protein identifications required two peptides. Semi-quantitative analysis of protein abundance was performed using the spectral counting approach [21, 22].
DNA was extracted from EVs with a QIAmp DNA mini-kit (Qiagen) according to the manufacturer’s instructions, and qPCR was performed using SYBR Green master mix (Applied Biosystems) and mouse mt-ATP6 primers and mouse β-actin primers as previously described [23].
EV biological function assay in vitro
For demonstration of EV internalization by HSC, HC-EVs from AH were labeled with PKH26 kit (Sigma-Aldrich) according to manufacturer’s instruction and purified to remove free dyes via ultracentrifugation, 100,000×g (30,000 rpm) for 60 min at 10 °C. After 4 h labeled HC-EVs addition, HSCs were fixed with 4% PFA, stained with FITC-phallotoxins (Thermo Fisher Scientifics) according to manufacturer’s protocol, and sealed with mountain medium with DAPI (Vector). The image of HC-EV internalization was taken using confocal laser scanning biological microscope (Olympus, FV1000). For HSC activation, isolated mouse HSCs were incubated with AH-HC-EVs, Con (pair-fed control)-HC-EVs, medium as negative control, and TGF-β as a positive control for 2 days. The image of HSC morphology was taken at day 0, 1, and 2 using microscopy (Olympus). For HM activation, isolated mouse HMs were incubated with AH-HC-EVs, Con-HC-EVs, and medium as negative control for 24 h. Total mRNA was extracted from HMs for the assessment of IL-1β and IL-17a mRNA levels by qPCR described above. IL-1β and IL-17A protein levels in medium were measured using Luminex kit (R&D Systems, Minneapolis, MN) according to manufacturer’s instruction. For TLR9 reporter activity assay, HEK-Blue-mTLR9 reporter cells (InvivoGen, San Diego, CA) were incubated with LPS (500 ng/mL), ODN1826 as TLR9 agonist (InvivoGen), ODN2088 as TLR9 antagonist (InvivoGen), AH-HC-EVs, Con-HC-EVs, or medium as negative control for 24 h. TLR9 reporter activity was measured in the supernatant in a plate reader at 630 nm.
Statistical analyses
All data are expressed as mean ± SEM unless otherwise noted. Non-parametric Mann-Whitney U test was used to assess the difference between two data sets from AH mice and pair-fed control mice with different distributions. Otherwise, one-way ANOVA was used to compare the group means of the experimental variable including EV functional assay data. Graph Pad (Graph Pad Software Inc., CA, USA) for comparison of continuous variables using Bonferroni’s multiple comparison test. Differences were considered to be significant at P ≤ 0.05.
Results
Murine AH is associated with a marked increase in circulating EVs and HC-EVs
The AH mouse model develops several features of liver pathology that characterize human AH including hepatocellular damage with balloon cell degeneration, PMN inflammation, fibrosis, and ductular reaction [9]. This AH model is established by superimposing weekly alcohol binge administration to iG mice which otherwise develop chronic ASH (cASH) characterized by mononuclear cell infiltration and pericellular and perisinusoidal fibrosis, but not PMN infiltration or ductular reaction [9]. Using these two models, we searched for potential drivers which facilitate the shift from cASH to AH by performing RNA-seq analysis on liver RNA [12]. Ontology/pathway analysis for differentially regulated genes in AH vs. cASH, revealed activated pathways such as chemokines, inflammasome, and fibrosis which were expected from the histologic phenotype of AH mouse livers [12]. Another pathway activated uniquely in AH vs. cASH liver was EV-related pathway as suggested by differentially regulated genes directly or indirectly associated with EV biology per Ingenuity Ontology and Pathway Analysis (Table 1). The upregulated genes involved in EV biogenesis in the AH liver included tetraspanin-8, and annexin-3 and −4. These data suggested a potential role of EVs in the cASH to AH transition in our models.
Table 1.
Upregulated genes as EV pathway in AH liver compared with chronic ASH liver
|
We firstly assessed whether circulating EV number was changed in AH mice via flow cytometry. Indeed, the stained circulating EVs had a distinct peak in histogram with a 10-fold higher intensity of dye positivity compared with un-stained blood EVs via flow cytometry analysis (not shown). The quantification showed that the number of circulating EVs was increased more than 20-fold (P < 0.01) in AH mice compared with control mice (Fig. 1a). The characterization of circulating EVs via dynamic light scattering (Fig. 1b) identified two subpopulations of EVs: small and large EVs as described before [24].
Fig. 1.
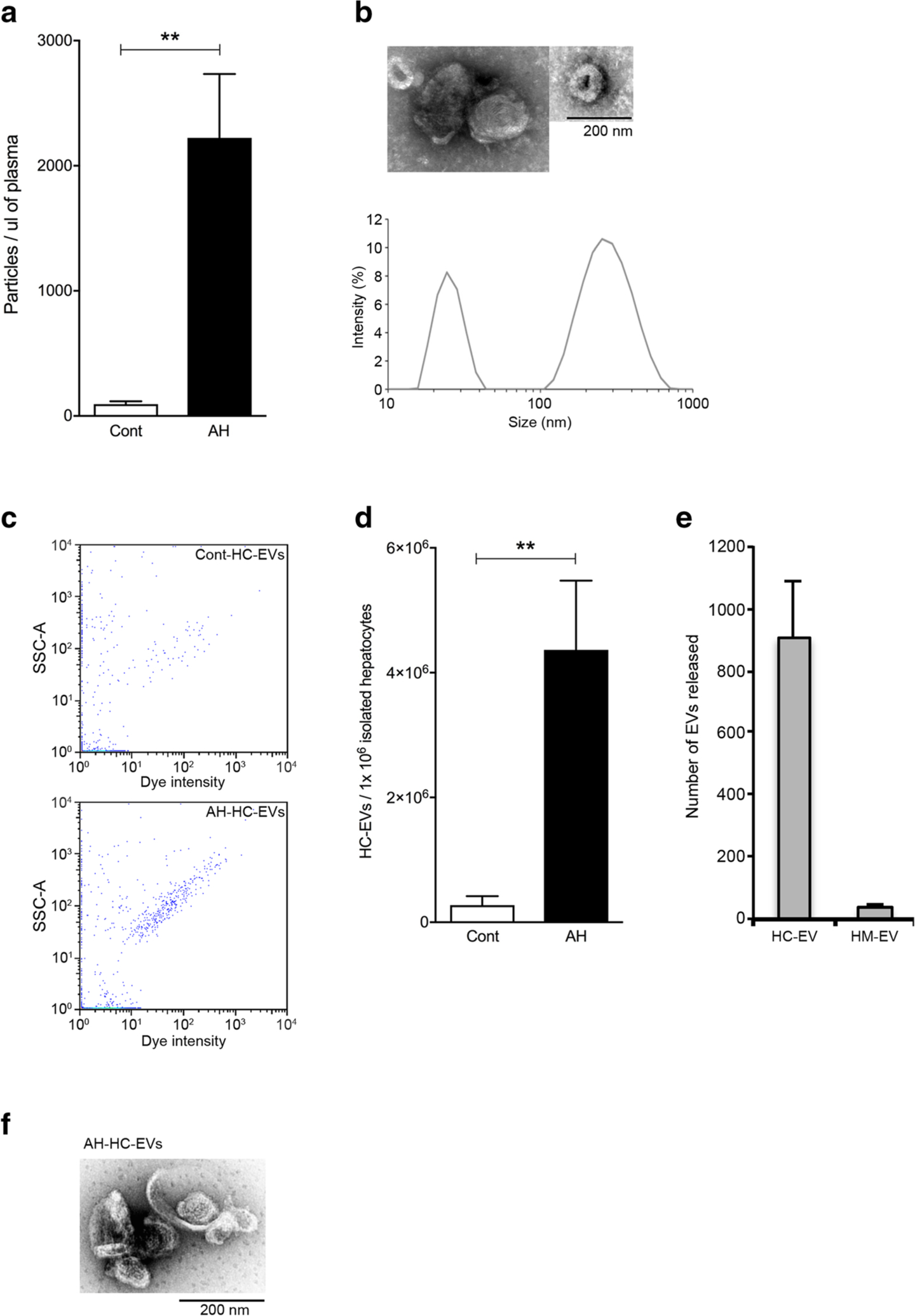
Circulating EVs and HC-EVs are increased in AH mice. a Quantification of stained circulating EVs via flow cytometry in pair-fed control or AH mice at 8 weeks plus weekly binge. **P < 0.01. b Dynamic light scattering analysis and transmission electron microscopy of isolated circulating EVs from AH mice. c Flow cytometry analysis (dot plot) of calcein+ HC-EVs from pair-fed control or AH mice. d–e Quantification of stained (d) hepatocyte-derived EVs (HC-EVs) or (e) hepatic macrophage-derived EVs (HM-EVs) from pair-fed control or AH mice. **P < 0.01. f Transmission electron microscopy of isolated HC-EVs from AH mice. Values are mean ± SEM
Hepatocyte-derived EVs contain specific miRNAs associating with fibrosis in the AH liver
Our previous finding that hepatocyte-derived EVs (HC-EVs) were released in a stress-specific manner containing a pathogenic miRNA signature [8] led us to further investigate the miRNA profile in HC-EVs in the AH mice. We isolated primary HCs and HMs from AH or pair-fed control mice and cultured them to determine the number of HC-EVs (HC-EVs) and the HM-derived EVs (HM-EVs) released into the media. We minimized the contamination of dead or dying cells, by culturing for 30 min, removing the media, gently washing the cells with PBS, and replenishing with fresh media containing 2% EV-free serum. This process improved the viability of cultured HCs to > 93% in both groups, and the conditioned medium was collected after 3 h of culture. The viability of purified HMs from the two groups after the adherence method exceeded 95%. HC-EVs or HM-EVs were analyzed using flow cytometry. The number of HC-EVs from AH was significantly increased by 15-fold compared with HC-EVs from pair-fed control animals (P < 0.01) (Fig. 1 c and d). Furthermore, the number of HC-EVs was 24 times higher than HM-EVs in AH mice (HC-EV or HM-EV number per 1 × 106 cells), suggesting that HCs are the main source of EVs (Fig. 1e). HC-EV morphology was characterized via electron microscopy (Fig. 1f).
We next performed miR-seq analysis on HC-EVs from AH (AH-HC-EVs) vs. pair-fed control mice (Cont-HC-EVs) to assess whether of differential levels of encapsulated miRs are associated with the liver pathology. The extracted RNA from HC-EVs contained predominantly small RNAs with a similar pattern in control and AH mice (Supplementary Fig. 1). A total of 633 known miRs were quantified by alignment with at least 5 reads in either group, and we found 24 miRs were significantly up- or downregulated in AH-HC-EVs compared with Cont-HC-EVs (P < 0.05) (Fig. 2a, b, and supplementary Table 1). All except Let-7f were uniquely upregulated in AH-HC-EVs as compared with HC-EVs from mild ASH mice previously studied [8], suggesting they may serve as potential novel biomarkers for AH, as well as possible drivers for the cASH-AH transition. The heatmap of miRNA expression showed high within-group similarity and clear differences between groups among 24 differentially expressed miRs (Fig. 2c and supplementary Table 2). A heatmap of miRs by pathways is shown in Fig. 2d. The row enlists top 24 differentially expressed miRs, and the column contains 18 enriched KEGG pathways. ECM receptor interaction and mucin-type O-glycan biosynthesis appeared activated, supporting the notion of pro-fibrogenic effects (Fig. 2d).
Fig. 2.
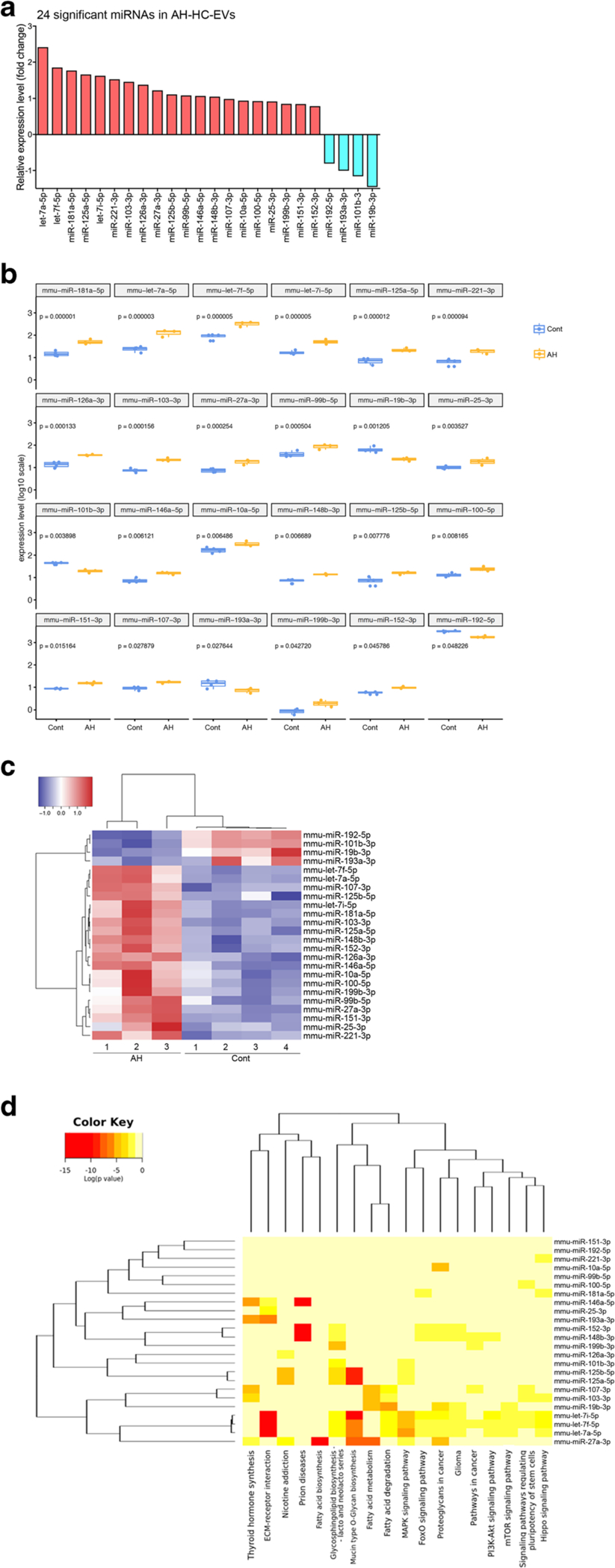
miRNA profile in HC-EVs. a A fold change of 24 significantly upor downregulated miRNAs in AH-HC-EVs compared with miRNAs in pair-fed control HC-EVs. b Relative miRNA expression level in 24 significantly up- or down regulated miRNA. ***P < 0.001, **P < 0.01, and *P < 0.05. c Heatmap clusters of 24 significantly up- or downregulated miRNAs by both rows and columns. d Target genes in KEGG cancer pathway in each 24 miRNAs
HC-EVs from AH mice activate hepatic stellate cells
The upregulation of profibrogenic miRs in AH-HC-EVs via miR-sequencing let us to further investigate HSCs as a potential target cell type of HC-EVs. HC-EVs from AH mice were labeled with red fluorescence and added to primary culture of mouse HSCs which were stained for F-actin (green) and nucleus (blue) (Fig. 3a). Labeled HC-EVs with red fluorescence were internalized into HSCs, while there was no red fluorescence in HSCs without addition of HC-EVs (control) (Fig. 3a). HSC morphology was unchanged in four groups incubated with AH-HC-EVs, Cont-HC-EVs, the medium as a negative control, or TGF-β as a positive control, at day 0, but showed an activated phenotype with AH-HC-EVs or TGF-β at day 1 and day 2 (Fig. 3b). The levels of fibrogenic genes, Acta2 (α-SMA) (P < 0.001), Col1a1 (P < 0.05), and Timp1 (P < 0.001), were significantly induced in HSCs incubated with AH-HC-EVs compared with Cont-HC-EVs or the medium (Fig. 3c).
Fig. 3.
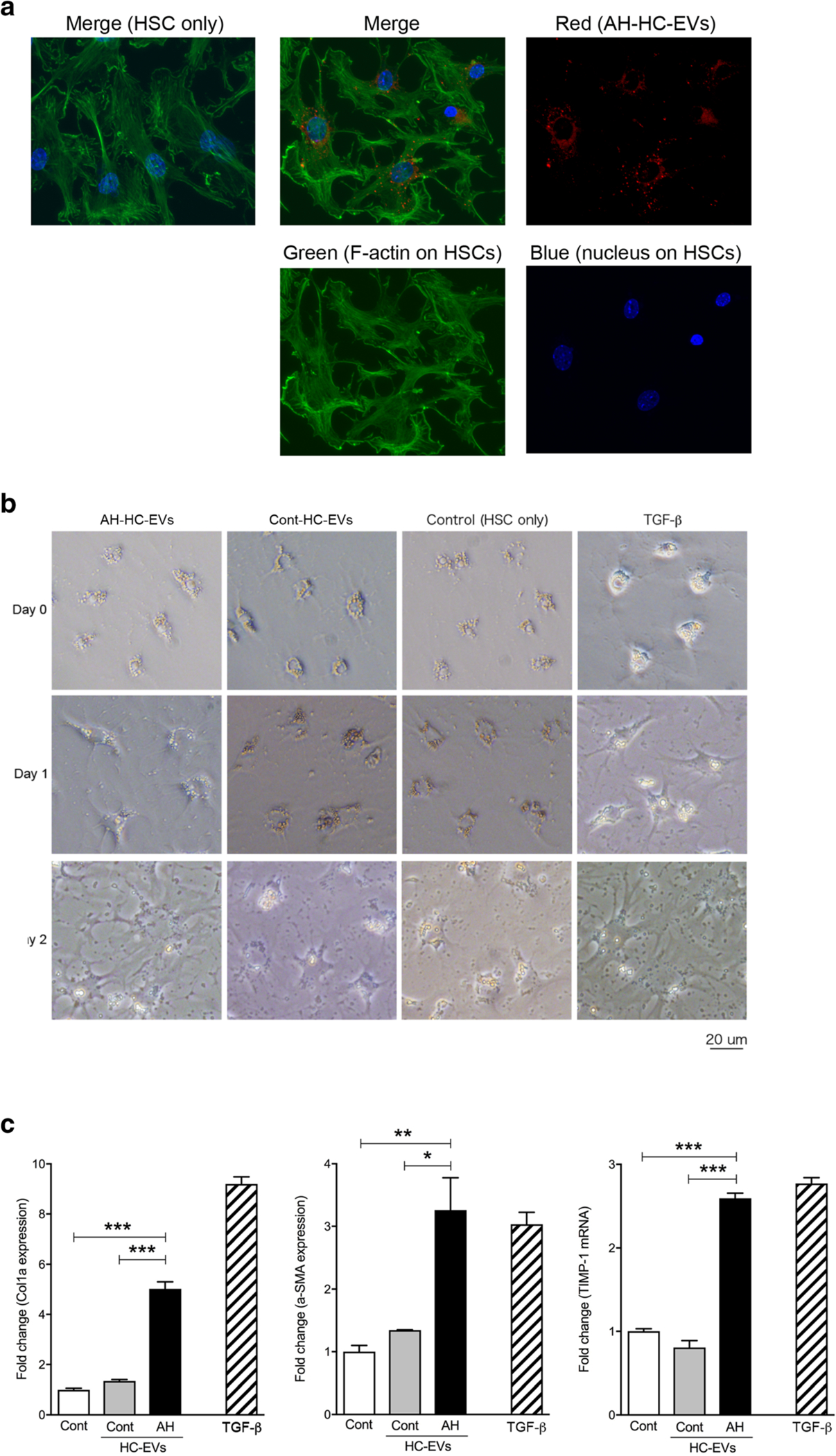
HC-EVs from AH activate primary HSC through miRNAs in HC-EVs. a Image of HSCs with labeled HC-EVs from AH mice. Green: FITC-phallotoxins and blue: DAPI in HSCs and red: HC-EVs with PKH26. b Morphology of primary HSCs with AH-HC-EVs, Cont-HC-EVs, control, and TGF-β1 at day 0, 1, and 2. c Fold change of Col1a, α-SMA, and TIMP-1 mRNA expression in primary HSCs with AH-HC-EVs, Cont-HC-EVs, control, and TGF-β at day 2. ***P < 0.001 and **P < 0.01. Values are mean ± SEM
Specific miRNA cargo in HC-EV modulates HSC activation after EV internalization
To identify genes which are targeted by the miRs carried by AH-HC-EVs and responsible for HSC activation, we screened genes that were shown to be differentially regulated by > 2-fold (P < 0.001) in either direction in HSCs from AH mice (AH-HSCs) compared with control mice (Cont-HSCs). The detailed data are reported in a separate study [25]. Using these data and the DIANA-miRPath software, we predicted target genes from 1020 down or upregulated gene in the AH-HSCs vs. Cont-HSCs DEG list which can be regulated by significantly up- or downregulated 24 miRs in AH-HC-EVs. This analysis revealed that 149 out of 360 downregulated genes in AH-HSCs were predicted targets of 20 miRNAs upregulated in AH-HC-EVs and 95 out of 660 upregulated genes in AH-HSCs as predicted targets of 4 miRNAs downregulated in AH-HC-EVs (Fig. 4a, Supplementary Table 3). The network analysis of targets in AH-HSC regulated by miRNAs in AH-HC-EVs, demonstrates a core network involving negative regulation of transcription (green color), gene expression (blue color), and cellular process (red color) (Fig. 4b). Notably, SMAD family member 7 (Smad7), which inhibits HSC activation, is a predicted target of miR-25, miR-100, and miR-181a which are all upregulated in AH-HC-EVs. Moreover, nuclear receptor subfamily 1 group D member 2 (Nr1d2), which is a quiescent HSC marker [26], is a presumed target of miR-101b, miR-27a, and miR-181a encapsulated in AH-HC-EVs (Fig. 4b). These results suggested that HSC activation in AH may be mediated by these upregulated miRs in AH-HC-EVs repressing quiescent HSC target genes. To test this notion, we examined Nr1d2 expression in primary HSCs transfected with miR-27a and miR-181a. Indeed, transfection of miR-27a and miR-181a in HSCs dose-dependently reduced Nr1d2 mRNA level and inhibited NR1D2 protein expression (Fig. 4c–e). These results suggest that AH-HC-EVs contribute to HSC activation by the ability of their cargo miRs to target and repress the quiescent HSC genes.
Fig. 4.

Primary HSC activates by miRNAs in HC-EVs. a Pie chart of specific 344 genes (fold change > 2) in HSCs from AH mice and 67 down- or 21 upregulated with miRNAs in HC-EVs as prediction. b Protein interaction of 67 downregulated genes in HSCs from AH mice regulated by 17 upregulated miRNA in AH-HC-EVs as prediction using STRINGS. c Fold change of Nr1d2 mRNA expression in primary HSCs with negative miRNAs (Cont), and miR-27 plus miR-181. d Immunoblotting of Nr1d2 protein expression in primary HSCs with negative miRNAs (Cont), and miR-27 plus miR-181. e Quantification of Nr1d2 protein expression from immunoblotting in primary HSCs with negative miRNAs (Cont), and miR-27 plus miR-181. *P < 0.05. Values are mean ± SEM
DAMPs in HC-EVs from AH mice mediate hepatic macrophage activation through TLR9 activation
EVs have multiple mechanisms of action to mediate biologic effects on target cells based on their specific cargo content that includes not only miRs but also proteins, DNA, and DAMPs such as mitochondrial DNA (mtDNA) [27, 28]. We analyzed the protein content in AH-HC-EVs vs. Cont-HC-EVs by proteomic analysis. This analysis revealed that 142 proteins were significantly changed in AH-HC-EVs compared with Cont-HC-EVs (P < 0.05 and fold change>|2|) and 60 proteins out of 142 were significantly upregulated in AH-HC-EVs (Supplementary Table 4). The sub-cellular origins of 60 upregulated proteins were mitochondrial (46.7%), endoplasmic reticulum (ER) (25.0%), peroxisomes (8.3%), lipid droplets (3.3%), nucleus (1.7%), extracellular vesicles (3.3%), and cytosol/secretary compartment (11.7%) (Fig. 5a). These proteomic results suggested damaged organelles such as mitochondria and ER may constitute the major EV cargo content. Next we tested the presence of mtDNA by analyzing by qPCR the mitochondrial gene Atp6. As predicted, Atp6 was increased 60-fold (P < 0.001) in AH-HC-EVs compared with Cont-HC-EVs (Fig. 5b). mtDNA has been recognized as an EV-derived DAMP for macrophage activation via TLR9 activation. Thus, we tested the effects of AH-HC-EVs on proinflammatory cytokine expression by HMs isolated from AH (AH-HM) or pair-fed control mice (C-HM). AH-HC-EVs, but not Cont-HC-EVs, increased Il1b mRNA in both AH-HMs and C-HMs albeit the magnitude of the induction by the former was substantially greater (both P < 0.05) (Fig. 5c). Il17a mRNA levels were already elevated in AH-HMs (P < 0.05) vs. C-HMs without EV treatment but further induced by the treatment with AH-HC-EVs but not with Cont-HC-EVs (P < 0.05) (Fig. 5c). Il17a mRNA in C-HMs was not significantly increased by Cont-HC-EVs or AH-HC-EVs (Fig. 5c), suggesting the differential sensitivity of AH-HMs vs. C-HMs to AH-HC-EVs. Furthermore, the production of IL-1β and IL-17A proteins by AH-HCs but not C-HMs was significantly increased by AH-HC-EVs but not Cont-HC-EVs (both P < 0.05) (Fig. 5c). To test the role of TLR9 in the observed stimulatory effects of AH-HC-EVs, we used HEK-Blue-mTLR9 reporter cells and found that the reporter activity was significantly elevated by AH-HC-EVs but not by Cont-HC-EVs (P < 0.05) to the similar level seen with treatment with LPS plus the TLR9 agonist OND1826 (P < 0.01 vs. PBS vehicle treatment). Further, the TLR9 activity stimulated with AH-HC-EVs was completely abrogated with the TLR9 antagonist OND2088 (P < 0.05) (Fig. 5d). In addition, IL-17A production stimulated by AH-HC-EVs was significantly attenuated with the TLR9 antagonist (P < 0.05 vs. AH-HC-EVs) (Fig. 5e). IL-1β protein upregulation by AH-HC-EVs was only modestly (~ 20%) and insignificantly (P = 0.15) reduced with the TLR9 antagonist (date not shown). These results suggest that AH-HC-EVs enriched with DAMPs such as mitochondrial proteins and DNA, stimulate AH-HM expression of IL-17A in a manner dependent on TLR9 activity.
Fig. 5.
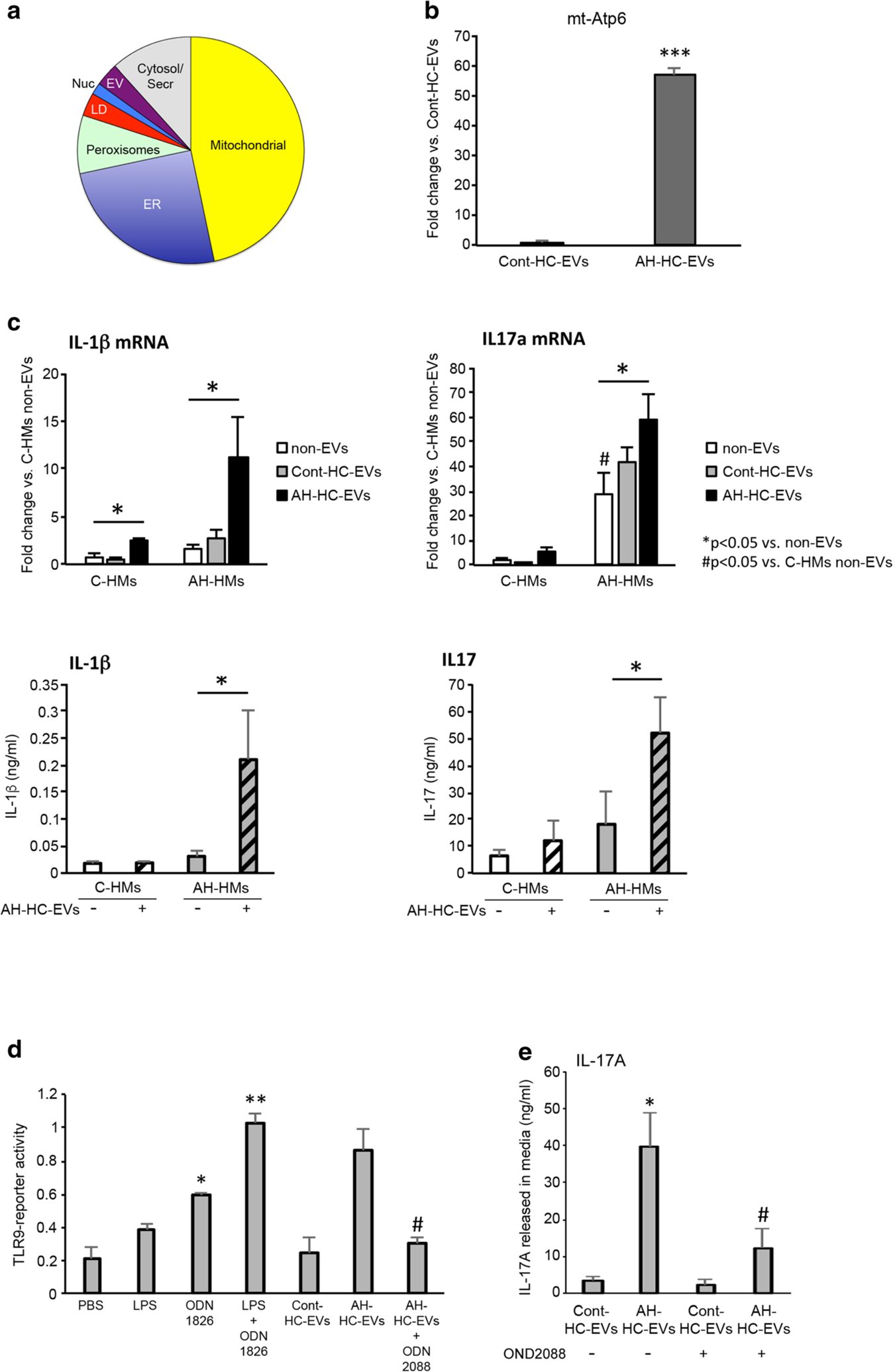
DAMPs in HC-EVs mediate hepatic macrophage activation through TL9 pathway. a Pie chart of upregulated protein composition in AH-HC-EVs (fold change >2). b Fold change of mitochondrial Atp6 (encoding the ATP synthase Fo subunit 6) DNA expression in AH-HC-EVs and Cont-HC-EVs. ***P < 0.001. c Fold change of IL-1β or IL-17a mRNA levels and protein production in hepatic macrophages (HMs) from pair-fed control or AH mice. *P < 0.05 vs. non-EVs. #P < 0.05 vs. C-HMs non-EVs. d TLR9 reporter activity in HEK-Blue-mTLR9 reporter cells incubated with LPS, ODN1826 (TLR9 agonist), LPS plus ODN1826 (TLR9 agonist), Cont-HC-EV, AH-HC-EVs, AH-HC-EVs plus ODN2088 (TLR9 antagonist). **P < 0.01 and *P < 0.05 vs. PBS. #P < 0.05 vs. AH-HC-EVs alone. e IL17A production in supernatant of AH-HMs incubated with Cont-HC-EVs and AH-HC-EVs without or with ODN2088. *P < 0.05 vs. Cont-HC-EVs. #P < 0.05 vs. AH-HC-EVs alone. Values are mean ± SEM. C-HMs, control HMs; ER, endoplasmic reticulum; LD, lipid droplets; EV, extracellular vesicle; Serc, secretary compartment
Discussion
The key finding of this study is the demonstration that in AH mice there is a conspicuously increased release of HC-EVs with a specific cargo containing a signature set of miRs and DAMPs that are involved in proinflammatory and pro-fibrogenic regulation of HSCs and HMs. We identified AH-HC-EV-derived specific miRs (e.g., miR-27a and miR-181a) targeting selective HSC mRNAs which are involved in HSC quiescence and repressed in AH-HSCs (e.g., Nr1d2), and hence most likely contributing to HSC activation in AH. AH-HC-EVs TLR-9 dependently stimulated AH-HM production of IL-17A which is implicated in HSC activation and liver fibrosis [29, 30]. Thus, these findings together support the notion that AH-HC-EV-mediated dual cross-talk with HSCs and HMs closes their loops to amplify HSC activation and liver fibrosis which are intensified in AH vs. chronic ASH mice [9]. TLR9 dependence of IL-17A expression has previously been reported in lung macrophages and blood monocytes [31, 32], but our results are the first to show it in HMs in the context of AH.
We have previously reported on the microRNA cargo of EVs in a murine model of mild alcoholic steatohepatitis that primarily shows severe steatosis but mild mononuclear cell (MNC) infiltration and no fibrosis [8]. In the current study we extended these findings by assessing the role of EVs and their cargo in a model that resembles human alcoholic hepatitis (AH) with features that include balloon cell degeneration, liver fibrosis, and PMN infiltration. We believe this is the consequence of heightened stress that AH-HCs experience in AH, releasing the markedly increase number of EVs. This notion is supported by the abundant protein derived from organelles such as mitochondria and ER known to undergo the pathogenic levels of stress [14, 33, 34]. Indeed, AH-HC-EVs released into the media doubled that of mASH-HC-EVs (data not shown), corroborating the RNA-seq finding on induced EV-related genes in AH livers. In our previous study, we identified 13 miRs, nine upregulated and four downregulated, in HC-EVs from mASH mice. Among them are three upregulated miRs, let-7f, miR-29a, and miR-340 which were also increased in the blood of patients with mild ambulatory ALD [8]. Current miR-seq analysis detected 20 upregulated and 4 downregulated miRs in AH-HC-EVs. All except let-7f were uniquely upregulated in AH-HC-EVs but not in mASH-HC-EVs, thus considered to be potential novel biomarkers for AH. They included profibrogenic miR-221, miR-126, and miR-27 [35, 36]. Indeed, functional studies using primary mouse HSCs demonstrated HSC morphologic and gene activation with AH-HC-EVs. We integrated this putative miR profile of AH-HC-EVs with RNA-seq data in HSCs from AH mice to identify potential targets. This effort eventually has led to the identification of novel regulation of the quiescent HSC gene Nr1d2 by miR-27a and miR-181a, although we need a future study to confirm direct NR1D2 regulation by miR-27a and miR-181a using a reporter construct.
The increased mtDNAs in AH-HC-EVs likely result from mitochondrial damage in AH livers and serve as a driver for proinflammatory activation of HMs via TLR9. Our results show the increased responsiveness of AH-HMs to this mode of activation as compared with C-HMs, suggesting the priming of AH-HMs which has taken place in vivo. Although both IL-17 and IL-1β mRNA and protein expressions were upregulated by AH-HC-EVs, the quantity of IL-17 protein released under the AH-HC-EV stimulation, was × 100 greater than IL-1β. Further, the TLR-9 antagonist clearly repressed IL-17 production stimulated with AH-HC-EVs but not IL-1β, highlighting the relative importance of TLR-9-dependent IL-17 regulation and its profibrogenic action augmented by AH-HC-EVs. Recent reports revealed that mtDNA circulates in blood inside EVs serving as potential biomarker of liver damage [37] and contributes to liver inflammation via activation of TLR-9 [23, 38]. Our results also support this paradigm. What is yet to be investigated is the roles of the organelle-derived proteins abundantly contained in AH-HC-EVs in HM and HSC activation. Proteomic analysis of oxidized and covalently modified proteins and their functional analysis are required to address this question. Growing evidence support the concept that cell to cell communication mediated by EVs contributes to the pathogenesis and progression of liver diseases [27, 39, 40]. While HC-EVs play a central role, non-parenchymal cells of the liver also release EVs that exert paracrine and autocrine effects in micro- and macro-environment [27, 39] and deserve further exploration related to AH pathogenesis.
Supplementary Material
Key messages.
Circulating EVs and HC-EVs were increased in AH mice compared with control mice
AH-HC-EVs were enriched in miRNAs, organelle proteins, and mitochondrial DNA
AH-HC-EVs increased cytokine production by AH-HMs in a TLR9-dependent manner
Acknowledgments
We thank the UCSD IGM Genomics Center, especially Kristen Jepsen for siRNA sequencing advice and assistance and Rui Yan for mtDNA and TLR9-reporter assays. The authors would like to thank the UCSD/CMM electron microscopy facility, especially Timothy Meerloo, for TEM sample preparation and imaging. This EM facility is supported by NIH equipment grant 1S10OD023527. Proteomics work was performed in W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a Department of Energy (DOE) office of Biological and Environmental Research (BER) national user facility located at Pacific Northwest National Laboratory (PNNL).
Funding information The work was supported by NIH grants P50AA011999 (Pilot Project Program), R21 AA023574, and JSPS KAKENHI Grant Number JP16H06872 to AE; JSPS KAKENHI Grant Number JP17K09419 to YT; NIH grants U54HL108460 to JK and LOM; NIH grant P41 GM103493 to RDS; NIH grants R01 AA022489 and DK082451 to AEF; NIH grants P50AA011999 (Administrative and Animal Cores), R24AA012885 (Integrative Liver Cell Core), R01AA018663, U01AA027681 and VA grants I01BX001991 (VA Merit Review) and IK6BX004205 (Senior Research Career Scientist award) to HT.
Abbreviations
- ALD
Alcoholic liver disease
- ASH
Alcoholic steatohepatitis
- HCC
Hepatocellular carcinoma
- EV
Extracellular vesicle
- HC
Hepatocyte
- HM
Hepatic macrophage
- HC-EV
Hepatocyte-derived EV
- HM-EV
Hepatic macrophages-derived EV
- miRNA
MicroRNA
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00109-020-01926-7) contains supplementary material, which is available to authorized users.
The use and care of the animals was reviewed and approved by the Institutional Animal Care and Use Committee at the University of Southern California.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Yin M, Wheeler MD, Kono H, Bradford BU, Gallucci RM, Luster MI, Thurman RG (1999) Essential role of tumor necrosis factor alpha in alcohol-induced liver injury in mice. Gastroenterology 117:942–952 [DOI] [PubMed] [Google Scholar]
- 2.Mathurin P (2009) Alcohol and the liver. Gastroenterologie clinique et biologique 33:840–849 [DOI] [PubMed] [Google Scholar]
- 3.Nanji AA, Su GL, Laposata M, French SW (2002) Pathogenesis of alcoholic liver disease–recent advances. Alcohol Clin Exp Res 26: 731–736 [PubMed] [Google Scholar]
- 4.Feldstein AE, Gores GJ (2005) Apoptosis in alcoholic and nonalcoholic steatohepatitis. Frontiers in bioscience : a journal and virtual library 10:3093–3099 [DOI] [PubMed] [Google Scholar]
- 5.Thurman RG, Bradford BU, Iimuro Y, Knecht KT, Connor HD, Adachi Y, Wall C, Arteel GE, Raleigh JA, Forman DT, Mason RP (1997) Role of Kupffer cells, endotoxin and free radicals in hepatotoxicity due to prolonged alcohol consumption: studies in female and male rats. J Nutr 127:903S–906S [DOI] [PubMed] [Google Scholar]
- 6.Nanji AA (2002) Role of Kupffer cells in alcoholic hepatitis. Alcohol 27:13–15 [DOI] [PubMed] [Google Scholar]
- 7.Rose ML, Rusyn I, Bojes HK, Belyea J, Cattley RC, Thurman RG (2000) Role of Kupffer cells and oxidants in signaling peroxisome proliferator-induced hepatocyte proliferation. Mutat Res 448:179–192 [DOI] [PubMed] [Google Scholar]
- 8.Eguchi A, Lazaro RG, Wang J, Kim J, Povero D, Willliams B, Ho SB, Starkel P, Schnabl B, Ohno-Machado L et al. (2017) Extracellular vesicles released by hepatocytes from gastric infusion model of alcoholic liver disease contain a MicroRNA barcode that can be detected in blood. Hepatology 65:475–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazaro R, Wu R, Lee S, Zhu NL, Chen CL, French SW, Xu J, Machida K, Tsukamoto H (2015) Osteopontin deficiency does not prevent but promotes alcoholic neutrophilic hepatitis in mice. Hepatology 61:129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J et al. (2015) Biological properties of extracellular vesicles and their physiological functions. Journal of extracellular vesicles 4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao B, Ahmad MF, Nagy LE, Tsukamoto H (2019) Inflammatory pathways in alcoholic steatohepatitis. J Hepatol 70:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanova E, Wu R, Wang W, Yan R, Chen Y, French SW, Llorente C, Pan SQ, Yang Q, Li Y, Lazaro R, Ansong C, Smith RD, Bataller R, Morgan T, Schnabl B, Tsukamoto H (2018) Pyroptosis by caspase11/4-gasdermin-D pathway in alcoholic hepatitis in mice and patients. Hepatology 67:1737–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai KKY, Kweon SM, Chi F, Hwang E, Kabe Y, Higashiyama R, Qin L, Yan R, Wu RP, Lai K, Fujii N, French S, Xu J, Wang JY, Murali R, Mishra L, Lee JS, Ntambi JM, Tsukamoto H (2017) Stearoyl-CoA desaturase promotes liver fibrosis and tumor development in mice via a Wnt positive-signaling loop by stabilization of low-density lipoprotein-receptor-related proteins 5 and 6. Gastroenterology 152:1477–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Chi F, Guo T, Punj V, Lee WN, French SW, Tsukamoto H (2015) NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J Clin Invest 125:1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300 [Google Scholar]
- 17.Kim J, Levy E, Ferbrache A, Stepanowsky P, Farcas C, Wang S, Brunner S, Bath T, Wu Y, Ohno-Machado L (2014) MAGI: a Node.js web service for fast microRNA-Seq analysis in a GPU infrastructure. Bioinformatics 30:2826–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG (2015) DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 43:W460–W466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, Camp DG 2nd, Khan AH, Sforza DM, Pallavicini M et al. (2005) Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res 4:2397–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Yin L, Lessner FH, Nakayasu ES, Payne SH, Fixen KR, Gallagher L, Harwood CS (2017) Genes essential for phototrophic growth by a purple alphaproteobacterium. Environ Microbiol 19: 3567–3578 [DOI] [PubMed] [Google Scholar]
- 21.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389:1017–1031 [DOI] [PubMed] [Google Scholar]
- 22.Schulze WX, Usadel B (2010) Quantitation in mass-spectrometry-based proteomics. Annu Rev Plant Biol 61:491–516 [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Martinez I, Santoro N, Chen Y, Hoque R, Ouyang X, Caprio S, Shlomchik MJ, Coffman RL, Candia A, Mehal WZ (2016) Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest 126:859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tkach M, Thery C (2016) Communication by extracellular vesicles: where we are and where we need to go. Cell 164:1226–1232 [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Rosenthal SB, Meshgin N, Baglieri J, Musallam SG, Diggle K, Lam K, Wu R, Pan SQ, Chen Y, Dorko K, Presnell S, Benner C, Hosseini M, Tsukamoto H, Brenner D, Kisseleva T (2020) Primary alcohol-activated human and mouse hepatic stellate cells share similarities in gene-expression profiles. Hepatology communications 4: 606–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Xu J, Brenner DA, Kisseleva T (2013) Reversibility of liver fibrosis and inactivation of fibrogenic myofibroblasts. Curr Pathobiol Rep 1:209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsova P, Ibrahim SH, Verma VK, Morton LA, Shah VH, LaRusso NF, Gores GJ, Malhi H (2016) Extracellular vesicles in liver pathobiology: small particles with big impact. Hepatology 64: 2219–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Povero D, Panera N, Eguchi A, Johnson CD, Papouchado BG, de Araujo HL, Pinatel EM, Alisi A, Nobili V, Feldstein AE (2015) Lipid-induced hepatocyte-derived extracellular vesicles regulate hepatic stellate cell via microRNAs targeting PPAR-gamma. Cellular and molecular gastroenterology and hepatology 1:646–663 e644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng F, Wang K, Aoyama T, Grivennikov SI, Paik Y, Scholten D, Cong M, Iwaisako K, Liu X, Zhang M, Österreicher CH, Stickel F, Ley K, Brenner DA, Kisseleva T (2012) Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 143:765–776 e763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabre T, Kared H, Friedman SL, Shoukry NH (2014) IL-17A enhances the expression of profibrotic genes through upregulation of the TGF-beta receptor on hepatic stellate cells in a JNK-dependent manner. J Immunol 193:3925–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhan U, Ballinger MN, Zeng X, Newstead MJ, Cornicelli MD, Standiford TJ (2010) Cooperative interactions between TLR4 and TLR9 regulate interleukin 23 and 17 production in a murine model of gram negative bacterial pneumonia. PLoS One 5:e9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salloum N, Hussein HM, Jammaz R, Jiche S, Uthman IW, Abdelnoor AM, Rahal EA (2018) Epstein-Barr virus DNA modulates regulatory T-cell programming in addition to enhancing interleukin-17A production via Toll-like receptor 9. PLoS One 13:e0200546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansouri A, Gattolliat CH, Asselah T (2018) Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology 155:629–647 [DOI] [PubMed] [Google Scholar]
- 34.Hoek JB, Cahill A, Pastorino JG (2002) Alcohol and mitochondria: a dysfunctional relationship. Gastroenterology 122:2049–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szabo G, Bala S (2013) MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 10:542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitano M, Bloomston PM (2016) Hepatic stellate cells and microRNAs in pathogenesis of liver fibrosis. J Clin Med 5 10.3390/jcm5030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MR MG, Staggs VS, Sharpe MR, Lee WM, Jaeschke H, Acute Liver Failure Study G (2014) Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology 60:1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Feng D, Li M, Gao Y, Ramirez T, Cao H, Kim SJ, Yang Y, Cai Y, Ju C, Wang H, Li J, Gao B (2017) Hepatic mitochondrial DNA/Toll-like receptor 9/MicroRNA-223 forms a negative feedback loop to limit neutrophil overactivation and acetaminophen hepatotoxicity in mice. Hepatology 66:220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eguchi A, Kostallari E, Feldstein AE, Shah VH (2019) Extracellular vesicles, the liquid biopsy of the future. J Hepatol 70:1292–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eguchi A, Feldstein AE (2018) Extracellular vesicles in nonalcoholic and alcoholic fatty liver diseases. Liver Res 2:30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


