Abstract
In response to nutrient deprivation, the ubiquitous Gram-negative soil bacterium Myxococcus xanthus undergoes a well-characterized developmental response, resulting in the formation of a multicellular fruiting body. The center of the fruiting body consists of myxospores; surrounding this structure are rod-shaped peripheral cells. Unlike spores, the peripheral rods are a metabolically active cell type that inhabits nutrient-deprived environments. The survival characteristics exhibited by peripheral rods, protection from oxidative stress and heat shock, are common survival characteristics exhibited by cells in stationary phase including modifications to morphology and metabolism. Vegetative M. xanthus cells undergo a number of physiological changes during the transition into stationary phase similar to other proteobacteria. In M. xanthus, stationary-phase cells are not considered a component of the developmental response and occur when cells are grown on nutrient-rich plates or in dispersed aqueous media. However, this cell type is not routinely studied and little of its physiology is known. Similarities between these two stress-induced cell types led to the question of whether peripheral rods are actually a distinct developmental cell type or simply cells in stationary phase. In this study, we examine the transcriptome of peripheral rods and its relationship to development. This work demonstrates that peripheral rods are in fact a distinct developmentally differentiated cell type. Although peripheral rods and stationary phase cells display similar characteristics, each transcriptomic pattern is unique and quite different from that of any other M. xanthus cell type.
Keywords: Development, Transcriptomics, Sporulation, Next generation sequencing, Regulation, Metabolism
1. Introduction
Multicellular development is not limited to eukaryotes as demonstrated by the ability of Myxococcus xanthus, a Gram-negative soil bacterium, to undergo dramatic cellular differentiation forming a three-dimensional, macroscopic structure known as a fruiting body [1,2]. These ubiquitous rod-shaped cells are commonly found in complex soils [3]. If nutrients are present, cells organize into a dynamic multicellular swarm [4]. These cells are in constant search of additional macromolecules or prey during the vegetative state [4]. Nutrient deprivation induces a stringent response in M. xanthus triggering a complex developmental program [3]. Cells aggregate into discrete foci eventually forming a flat mound. The mounds give rise to the macroscopic fruiting bodies composed of at least two cell types, myxospores and peripheral rods [2]. Within the fruiting bodies, rod-shaped cells differentiate into spherical myxospores that are metabolically inactive and resistant to harsh environments [2].
Peripheral rods are ostensibly less drastic as the cells do not enter dormancy or appear to change strikingly morphologically [2,5]. Peripheral rods remain metabolically active outside of the fruiting bodies [5–7]. When nutrients become readily available, both cell types respond to the stimuli by returning to a vegetative state, albeit, peripheral rods respond more quickly than myxospores, which must undergo germination [7]. In the multicellular development of M. xanthus, cells differentiate to perform specialized functions. Peripheral rods may utilize substrates at lower concentrations than myxospores [5]. It has been proposed that this cell type is specialized to uptake minuscule nutrient influxes resulting in a short-term solution, while spores are specialized to protect the genome until the environment is more suitable for proliferation [5,7]. Bacterial cell differentiation is not limited to sporulation. Differentiation is defined as a process in which cells of an organism become different from one another or different from their previous state [5,8]. Bacterial growth phases are also examples of differentiated cell types as expression patterns among vegetative and stationary phases vary significantly and cells function differently as well [8–10]. It has been previously suggested that peripheral rod cells are a differentiated cell type that arises from the vegetative population in response to nutrient deprivation [2]. Indeed, in comparison to vegetative cells, peripheral rods meet the criteria of a distinct cell type as they (i) occur in all strains of M. xanthus, (ii) have unique patterns of expression of proteins, (iii) are structurally dissimilar from other cell types, (iv) exhibit unique responses to environmental stimuli, and (v) serve a unique function in the life cycle of M. xanthus [2].
However, stationary cells exhibit similar characteristics to peripheral rods. During the transition from exponential growth to the stationary phase, a number of morphological and physiological changes take place. The composition of the cellular envelope is altered and a series of stress-related genes is upregulated prior to or upon entering stasis [8,11,12]. As with stationary phase cells, there have been limited analyses of peripheral rods. However, there are perceivable similarities between the two cell types. Peripheral rod cells have been shown to alter their cell wall, and sigma factors (e.g. SigD) are upregulated in a manner vital to development [11–14]. Peripheral rods also possess a single chromosome and maintain a rod-shaped morphology, characteristics found in stationary cells. Due to the similarities, we address the distinction of peripheral rods as a differentiated cell type through a comparative analysis [15]. The study focuses on cell structure and response signaling induced by environmental stresses. Moreover, the use of Next Generation Sequencing (NGS) provides an in-depth look at the transcriptomic profile of M. xanthus cell types. We demonstrate that the expression patterns of the peripheral rods are different from any other cell type observed. This study also gives insight into the possible origin and developmental pathway of peripheral rods.
2. Materials and methods
2.1. Bacterial strains, growth, and media
All strains used are derivatives of the wild-type M. xanthus strain DK1622. M. xanthus strains were grown in CTTYE 1% casitone (Difco, Franklin Lakes, NJ), 10 mM Tris-HCl (pH 7.6), 1 mM KH2PO4, 8 mM MgSO4) broth or on CTTYE plates containing 1% agar. Stationary cells were passaged three times before being collected at a Klett value of 230. Low nutrient cells were grown in 0.08% CTTYE following an established protocol [16].
2.2. Microscopy
Phase contrast microscopy was used to visualize and photograph cells. Nikon Eclipse 80i light microscope with 100× oil immersion objective and 10X ocular along with a Q-Imaging MicroPublisher 3.3 RTV camera were used to image cells.
2.3. Development
Development was induced either with a submerged liquid culture buffer system [1,16] or on TPM agar plates (10 mM Tris [pH 7.6], 8 mM MgSO4, and 1 mM KH2PO4 containing 1.5% agar). Cells developed in a humidity chamber at 33°C. Cells were harvested and quick-frozen in liquid nitrogen [16].
2.4. Purification of peripheral rods
Peripheral rods were purified from myxospores in the fruiting body by using an adaptation of previous protocols [5,15]. Fruiting bodies were removed from developmental plates after four days. Cells were scraped from TPM agar with a spatula and suspended in 1 ml of 10 mM sodium phosphate, pH 7.2. This resuspension was then applied to a sucrose step gradient with levels of 60%, 30%, 15%, and 5% sucrose in 10 mM sodium phosphate, pH 7.2. Samples were subjected to centrifugation at 400 ×g for 15 min in an HB-4 rotor. The 5% sucrose fraction contains rods, and the 30–60% sucrose fractions contain myxospores. The purity of the peripheral rod samples was verified using microscopy.
2.5. RNA isolation, integrity, and quality assessment
Total RNA was extracted from N2 snap-frozen M. xanthus cells using the RNeasy mini kit (Qiagen, Valencia, CA). RNA concentrations were determined from measurements on a Nanodrop 1000 spectrophotometer.
2.6. RNA enrichment/rRNA depletion
rRNA depletion (Smaldone et al., unpublished) [17] was performed using non-overlapping synthetic DNA probes representing the entire complementary sequences of M. xanthus 16S rRNA and 23S rRNA at concentrations of 0.5 μM for each probe. One microliter of the selective depletion RNA was mixed in a volume of 5 μL 1× Hybridization Buffer (100 mM Tris-HCl, 200 mM NaCl). The mixture was heated to 95°C for 2 min, then slow-cooled to 22 °C (0.1°C/s), incubated an additional 5 min at 22 °C, and placed on ice. Ten units of Hybridase™, a thermo-stable RNaseH (Epicentre, Madison, WI), was added along with 1 μL of 10× RNaseH digestion buffer (500 mM Tris-HCl, 1 M NaCl, 200 mM MgCl2) in a final reaction volume of 10 μL, incubated at 37 °C for thirty minutes and placed on ice. DNA probes were removed by DNase treatment using an RNase-free DNase Kit (QIAGEN, Valencia, CA).
2.7. RNA-Seq library construction
The residual RNA following depletion of total RNA was used as input to prepare sequencing libraries. Libraries were prepared according to the mRNA Sequencing Sample Preparation Guide (Illumina®, San Diego, CA). Approximately 400 ng of total cellular RNA of each sample was used to generate RNA-Seq libraries using the Encore complete prokaryotic RNA-Seq DR multiplex system (NuGEN, San Carlos, CA) exactly as described in the manufacturer’s protocol. Individual indexes for multiplexing were added to each library.
2.8. Illumina sequencing
Ninety libraries were quantified and validated using Bioanalyzer which were further pooled in an equimolar concentration. Sequencing of pooled libraries was performed using the Illumina HiSeq2500 platform to collect single-end reads of 50 cycles following the manufacturer’s standard procedure. Real-Time Analysis was used for the base call, and quality scoring during image cycles of a sequencing run and the Illumina pipeline was used to deconvolute the library pool and generate fastq files.
2.9. RNA-Seq alignment and coverage
Adapter length varies due to the broad distribution of fragment sizes. A trimming procedure was applied to ensure maximized trimming of the adapter contents from each read using Scythe (https://github.com/vsbuffalo/scythe) and Sickle (https://github.com/najoshi/sickle). The full length of the adapter sequence was shortened one base at a time and aligned to the 3′-end of each read with varied mismatch tolerance to determine the length of the sequence to be removed
2.10. RNA-Seq analysis
Reads were aligned to the M. xanthus reference genome using DESeq2 [18]. For each sample, a count table for all genes was generated with HTSeq-count [19]. Aligned reads were filtered to remove reads with multiple alignments or ambiguous assignment. A count table for all samples of 3 biological replicates was compiled and used for differential expression analysis by the Bioconductor packages DESeq2 in a generalized linear model [18].
2.11. Differential expression analysis using DESeq2
The peripheral rods transcriptome was compared to that of other cell types. The comparison data was generated using DESeq2 contrast function; the contrast function allows us to determine how much each gene’s expression changes (log 2 fold) in juxtaposed conditions. P-values were calculated using the Wald test available in the DESeq2 internal algorithm. The Wald test compares the beta estimate divided by its estimated standard error to a standard normal distribution. For calculating Wald test p-values, the coefficients were scaled by their standard errors and then compared to a standard Normal distribution. To define differentially expressed genes, we used the criterion of adjusted P ≤ 0.05 between two samples. The tables demonstrate the differential expression under different conditions.
3. Results and discussion
During development, cells have three cell fates, cell lysis or the differentiation into either myxospores or peripheral rod cells [2,7]. Previous studies suggest that peripheral rod cells represent a unique terminal developmental cell type, based on a limited proteomic comparison to vegetative cells [2,5]. However, based on their physiology, it is possible that peripheral rod cells are related to if not, simply stationary phase cells. To distinguish between these possibilities, we took a detailed transcriptomic approach to determine the nature of peripheral rod cells and compared them to stationary phase cells and vegetative grown cells.
3.1. Comparative analysis of stationary cells and peripheral rods
One hypothesis is that stationary phase cells and peripheral rods are similar cell types based on their ecology, low nutrient environments, physiological function, and chromosome numbers [15]. Stationary phase cells are a differentiated cell type [8] not commonly studied in M. xanthus. Most bacteria have evolved intricate methods for surviving in nutrient-limited conditions; cells can enter and remain in long-term limited growth phase until eventual cell death after completely exhausting the available resources. However, this non-exponentially growing state also leads to diverse behaviors throughout the bacterial world ranging from cannibalism to sporulation [8]; all of these processes are exhibited by M. xanthus in response to limited nutrient availability. One question that remains is whether peripheral rod cells are a unique developmental cell type, as previously suggested [5] or represent a novel developmental cell type that remains in a quasi-growth state, like stationary phase cells. One way to address this question is by transcriptomic comparisons. Transcriptomic comparison of stationary phase cells and peripheral rod cells revealed two distinct cell types. The comparative analysis of stationary cells against peripheral rods demonstrated 5436 genes were differentially expressed, which is close to 75% of the genome. Of those differentially expressed genes, 5325 (73%) were upregulated and 111 (2%) were downregulated (fold-change ≥2, p ≤ 0.05).
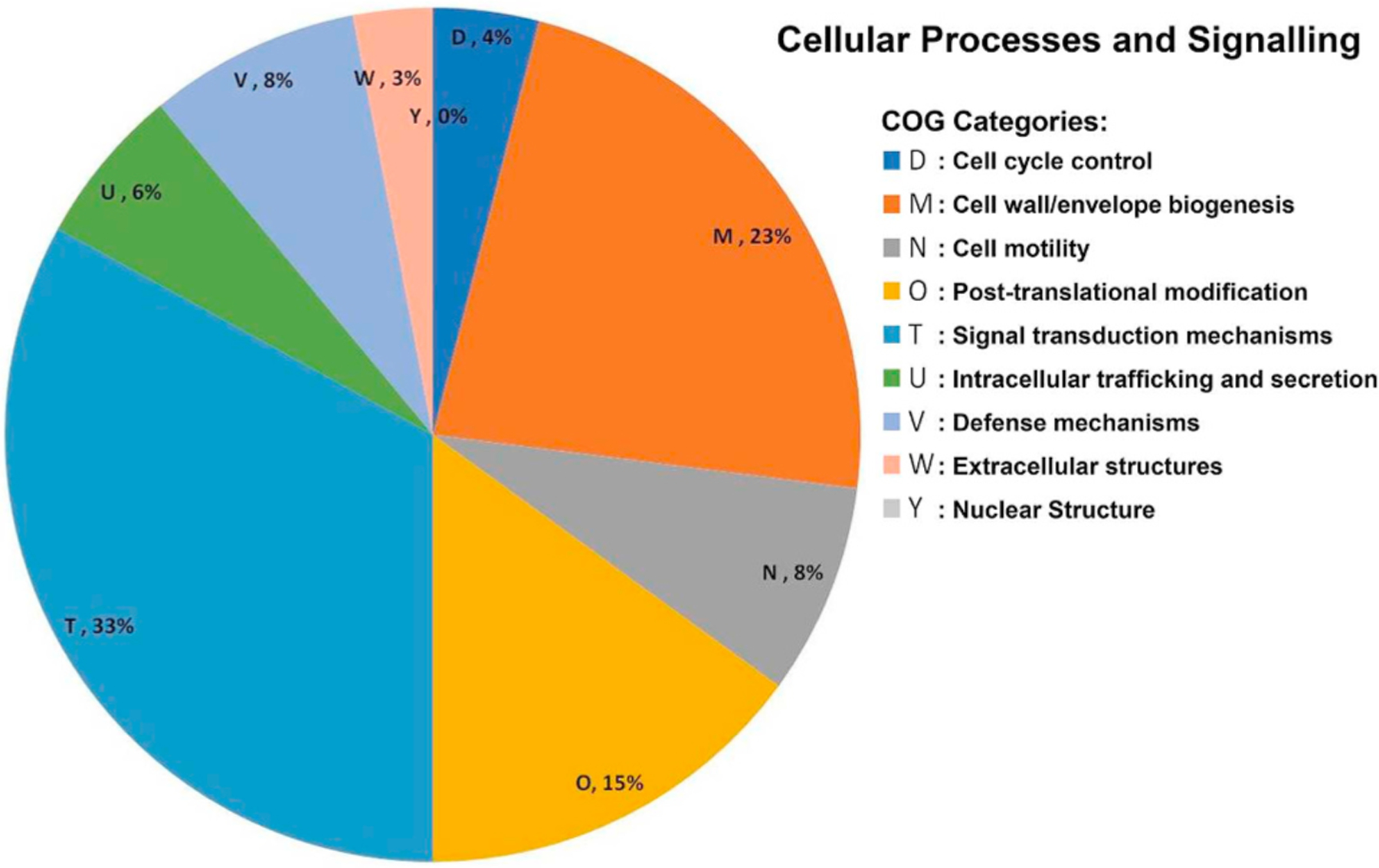
To achieve a better global analysis of the significantly different transcripts found in peripheral rod cells, we used a broad genome-wide technique that focuses on categories of genes rather than individual transcripts. The analysis displays upregulated or downregulated genes in peripheral rod cells by comparing its transcriptome to stationary phase cells. The ontology of each individual transcript was investigated and ontologies were then classified and sorted into broad categories; ontologies of individual genes were classified relating to cellular processes (Fig. 1). The results indicated that the ontologies for cell wall/envelope biogenesis and signal transduction were the strongest affected.
Fig. 1.
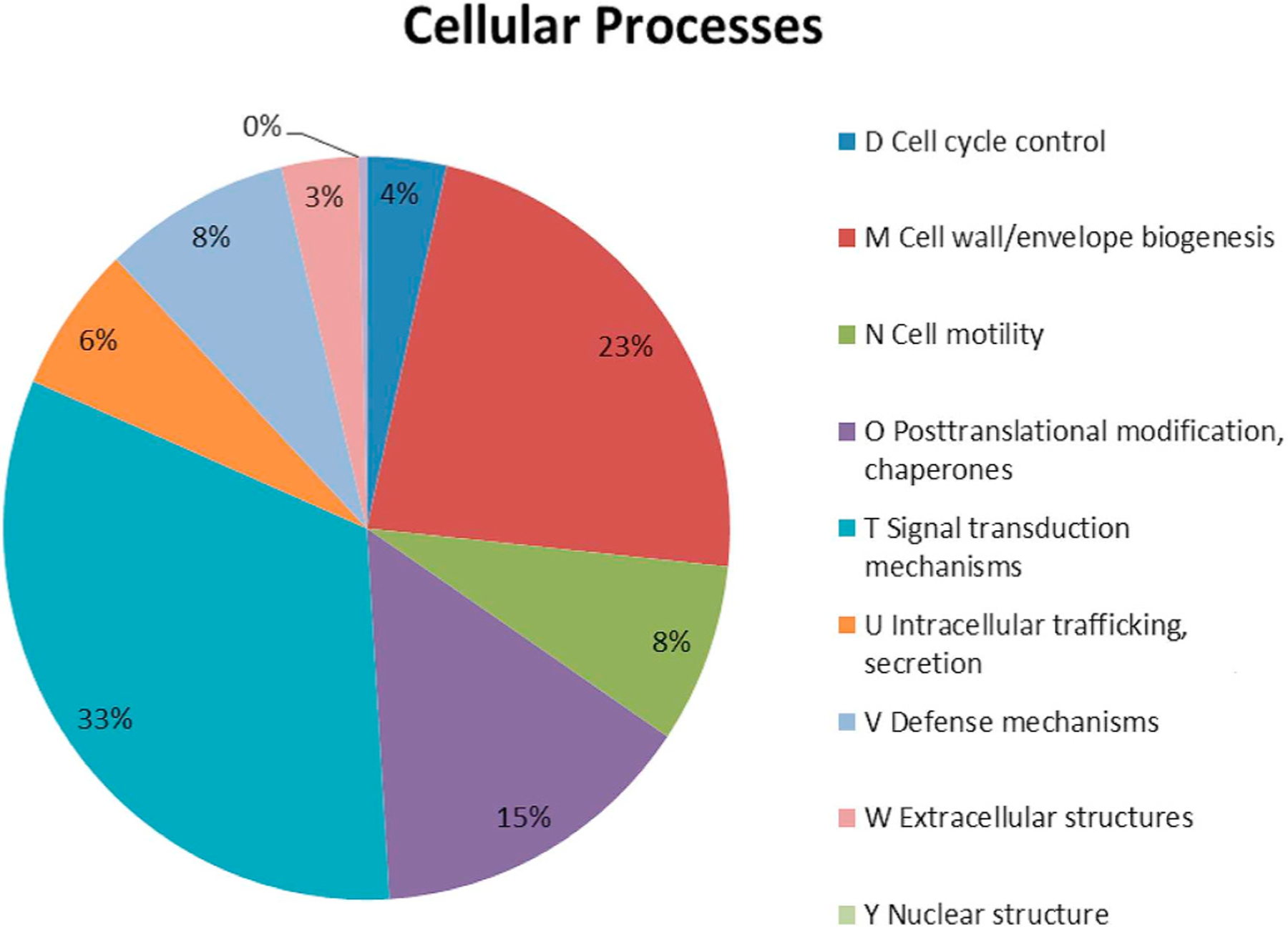
Significantly differentially expressed cellular process transcripts in peripheral rods as classified using Clusters of Orthologous Genes (COGs): Significant upregulation or downregulation of transcripts in peripheral rods mean a fold-change ≥2 with p-value ≤0.05.
Differential expression was predominantly seen among cellular envelope and signal transductions categories according to Clusters of Orthologous genes (COG) database. Although both stationary cells and peripheral rods alter their cellular envelope and induce starvation response signals to adapt to nutrient-deprived environments, there are key differences in the transcriptomes of these cell types. The differences between the two cell types were further examined by investigating specific genes and gene sets involved in various adaptive responses and cell envelope biogenesis on an individual level.
3.2. Analyzing stress response: global stress, heat stress, nutrient stress, oxidative stress, and DNA damage response pathways
To ensure survival in nutrient-poor conditions, bacteria have evolved diverse signaling cascades to regulate gene expression. In particular, the myxobacteria have some of the largest numbers of response regulators and sensory transduction pathways involved in a variety of functions [20,21]. Stress-related genes are critical to cells transitioning into stationary phase and development. One of the most common types of master regulators is sigma factors, which act to control a variety of stress responses such as oxidative stress, heat shock, and endospore formation. Sigma factors are classified into two structurally unrelated families: σ54 and σ70 families [8]. The σ54 family contributes to a diverse array of metabolic processes; the σ70 family proteins are primarily responsible for physiological changes of the cell [8]. Both, stationary phase and development are regulated by sigma factors in M. xanthus, which have been identified by genome inspection. We observed that the expression patterns of these sigma factors differ between the two cell types. Among the full repertoire, sigD is expressed in a similar manner; other sigma factors including rpoE, rpoN, sigB, and sigC, which were all upregulated in peripheral rods in comparison to stationary cells (Table 1). RpoE (σE) is necessary for the exocytoplasmic stress response, specifically heat resistance in E. coli [22,23]. In M. xanthus, the extracytoplasmic function (ECF) sigma factor RpoE is believed to play a role in the transcriptional regulation of genes involved in motility and aggregation behavior during development [13] and RpoN is essential for vegetative growth of the bacterium [24]. Under nutrient-deprived conditions, expression of rpoN is increased in the peripheral rod cells, but not in stationary phase cells. Although SigB and SigC exhibit similarities to heat shock sigma factors, the expression of sigB and sigC are essential for sporulation and early fruiting body formation, respectively [13]. Expression of these sigma factors encoding genes is the prominent difference between these two cell types, exhibiting a change of 55-fold (log2 fold difference > 5) (Table 1).
Table 1.
Relative expression studies of well-known sigma factors in stationary cells as compared with peripheral rods in M. xanthus.
| Protein function or tag | MxDK1622 encoded protein | Stationary/Peripheral |
|---|---|---|
| RpoN | MXAN_1061 | −2.03 |
| SigD | MXAN_2437 | 0.00 |
| SigB | MXAN_3357 | −5.84 |
| RpoE1 | MXAN_4147 | −1.97 |
| RpoD | MXAN_5204 | 0.00 |
| SigC | MXAN_6209 | −5.54 |
The negative sign and value in the last column represent downregulation during stationary phase and log2-fold change (unit).
Adaptation to nutrient-depleted conditions and oxidative stress is important to the survival of stationary-phase cells and presumably to peripheral rods. Atypical of most δ-proteobacteria, myxobacteria perform aerobic respiration and consequently produce reactive oxygen species (ROS). ROS accumulate in nutrient-depleted environments and become concentrated by the lack of replication in the biofilm. In E. coli, soxRS and oxyR comprise the major oxidative stress regulons responsible for removing ROS [25]. The SoxRS response is activated by superoxide agents and later on, it protects the cell against the same [25]. The sensor molecule, SoxR, induces soxS expression, which activates transcription of the regulon [25]. The SoxR protein belongs to the MerR family of proteins, which is extensively distributed across several bacterial phyla, including the Proteobacteria [26,27]. The M. xanthus genome is predicted to encode six homologs of MerR-family regulators, which are significantly overexpressed (ranging from 10–40 fold) in peripheral rod cells (Table 2). The SoxS protein belongs to the AraC family of one-component regulators also involved in the control of stress responses and found in M. xanthus [28,29]. Genome analysis revealed the presence of 18 AraC-family homologs, where all except MXAN_6206 are significantly overexpressed in peripheral rod cells as compared to stationary phase cells (Table 2). The final oxidative stress regulator examined was OxyR, a LysR-type transcriptional regulator (LTTR) found throughout Gram-negative bacteria. LTTRs are key regulators in the oxidative stress response [30,31]. M. xanthus is predicted to have 26 LysR-type family regulators, 24 out of which (except MXAN_6468 and MXAN_6715) are overexpressed in peripheral rod cells ranging from 4-fold to almost 100-fold compared to stationary phase cells (Fig. 2). Superoxide dismutase (SOD) and catalase are antioxidant enzymes central to defending the cell against oxidative stress. SOD and catalase enzymes degrade superoxide and hydrogen peroxide, respectively. However only the catalases are differentially expressed in this analysis; both MXAN_4389 and MXAN_6188 are upregulated in peripheral rods, exhibiting fold changes greater than 128 (Table S1).
Table 2.
Comparative distribution of transcriptional expression of soxRS regulons in M. xanthus.
| Encoded protein in M. xanthus | Stationary/Peripheral | |
|---|---|---|
| MerR transcriptional regulator | MXAN_0777 | −4.06 |
| MXAN_0903 | −4.04 | |
| MXAN_0904 | −5.69 | |
| MXAN_2912 | −5.58 | |
| MXAN_5120 | −2.99 | |
| MXAN_6983 | −4.56 | |
| AraC transcriptional regulator | MXAN_0387 | −6.21 |
| MXAN_0445 | −5.26 | |
| MXAN_0631 | −3.74 | |
| MXAN_0707 | −5.85 | |
| MXAN_1137 | −5.73 | |
| MXAN_1667 | −5.80 | |
| MXAN_1719 | −5.86 | |
| MXAN_2213 | −5.00 | |
| MXAN_2216 | −6.09 | |
| MXAN_2612 | −5.24 | |
| MXAN_3142 | −2.36 | |
| MXAN_3429 | −5.96 | |
| MXAN_4060 | −4.78 | |
| MXAN_5274 | −3.65 | |
| MXAN_5899 | −5.78 | |
| MXAN_6206 | 0.00 | |
| MXAN_6479 | −5.62 | |
| MXAN_7078 | −5.65 |
SoxR and SoxS belong to the MerR and AraC transcriptional regulator families respectively. Here stationary cells were contrasted against peripheral rods. In the second column, numbers represent log2-Fold change and the negative sign depicts the downregulation of transcripts during stationary phase.
Fig. 2.
Relative change in expression of OxyR stress regulators in M. xanthus stationary/peripheral cells: OxyR regulator proteins belong to the LysR family. In this study, transcriptional expression in stationary cells was compared to peripheral rods and represented by this plot against log2-Fold change per protein.
One survival technique associated with myxospores is resistance to DNA damage. Nutrient deprivation can also lead to stress-induced mutagenesis [32,33], and stationary phase cells often show more resistance to UV damage than vegetative cells. These transcriptional changes aid in the growth and survival of the organism under harsh conditions [25,34]. This adaptive or stationary-phase mechanism relies on the bacterial SOS response [33]. During the response, replication is arrested allowing cells to begin DNA repair, and mutagenesis occurs [33]. Such stationary-phase mutation has been well documented in E. coli [33]. The SOS response regulates several genes under the direct and indirect transcriptional control of LexA [32,33]. The LexA regulon includes the recombination and repair gene recA, and nucleotide excision repair genes uvrAB and uvrD [33]. A comparative analysis revealed the majority of SOS response genes to be upregulated in peripheral rod cells (Table 3). What is noticeable is that the transcriptional expression of lexA and recA2 is not upregulated. The recA2 gene is regulated by LexA and is essential for vegetative growth and development in M. xanthus [34]. By contrast, recA1 is not considered functional and is upregulated through a pathway independent of LexA [34]. It also appears that the uvr genes are activated through a LexA-independent pathway similar to the DNA damage-inducible genes recN (highly upregulated) and ssb (not regulated) [34].
Table 3.
Comparative Analysis of SOS Response genes.
| Protein function or tag | MxDK1622 encoded protein | Stationary/ Peripheral |
|---|---|---|
| RecA 2 | MXAN_1388 | 0.00 |
| RecA 1 | MXAN_1441 | −4.1 |
| RecA 3 | MXAN_3991 | −5.55 |
| LexA | MXAN_4464 | 0.00 |
| UvrA | MXAN_2388 | −4.55 |
| UvrA | MXAN_2609 | 3.22 |
| UvrB | MXAN_2632 | −3.59 |
| UvrC | MXAN_2633 | −5.07 |
| UvrD | MXAN_1992 | −3.36 |
| UvrD | MXAN_2617 | −5.00 |
This table deciphers the transcriptomic comparison of SOS response genes under two conditions (stationary cells as contrasted with peripheral rods) in M. xanthus in a unit of log2-Fold change (last column). Negative values demonstrate downregulation of transcripts during the stationary phase or upregulated amongst peripheral cells.
3.3. Cellular morphology and structure
Morphologically, cells growing in rich media are long flexible slender rods, between 5 and 10 μM in length. Stationary cells, cells grown under nutrient limitation and peripheral rod cells are somewhat shorter and wider as depicted in Fig. 3. Cell wall modifications are common among bacteria in stationary phase as well as in development. This study reveals that the cell wall of peripheral rods is quite different from that of stationary cells. The shape-determining structure in the cell envelope of bacteria is made up of peptidoglycan (murein), the main component of the cell wall. During the stationary phase, it is common for Gram-negative bacteria to maintain significant murein-biosynthetic activity for strengthening the cell wall by increasing the number of cross-links in the murein [35,36]. Proteins encoded by the mreB, mreC, mraY, pbp (Penicillin-binding protein) and mur genes maintain and control the bacterial exoskeleton [37]. The transcriptomic comparison of stationary phase cells to peripheral rods revealed stark differences in cell envelope activity (Table 4). The murein wall is synthesized and maintained by a cascade of proteins encoded by the mur genes (murB, murC, murD, murE, murF, murG, and murY) [37]. In peripheral rod cells, eight out of the 10 cell wall genes analyzed showed a significant upregulation in comparison to stationary cells. Only murE and murF exhibited similar transcript levels in both cell types. MreB, the bacterial actin homolog functions in spatially coordinating cell morphogenesis in conjunction with MreC, a protein that wraps around the outside of the cell within the periplasmic space, while PBP-2 is responsible for determining the rod shape of the cell [37]. Transcripts for all three genes were highly upregulated in peripheral rods as compared to stationary phase cells.
Fig. 3.
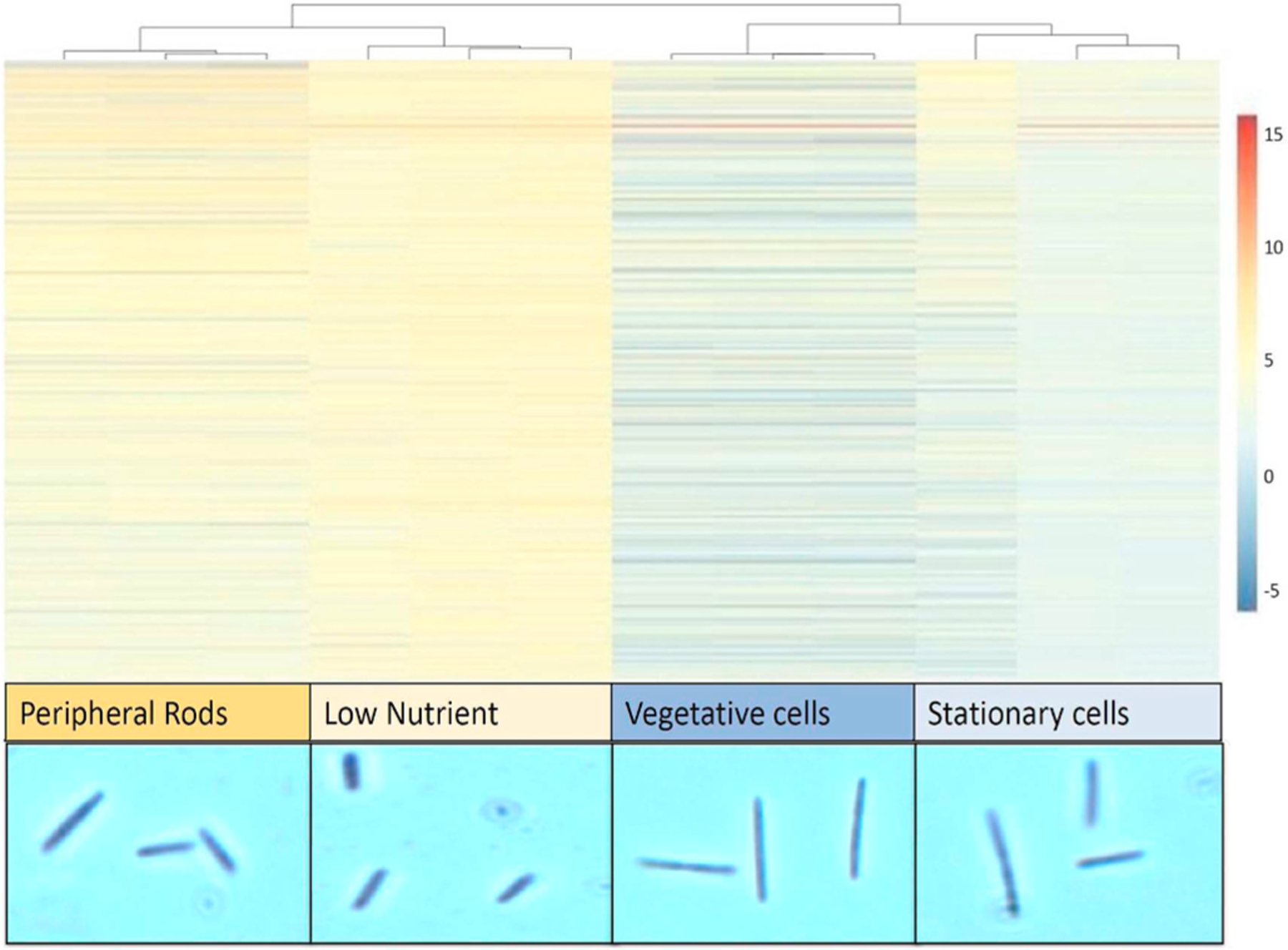
A comparative heat map depicting scaled expression for peripheral rods, low nutrient, vegetative cells and stationary cells (left to right). Low nutrient cells were maintained at the brink of vegetative growth at 0.004% casitone for 3 days (see Materials and methods). In the lowermost panel, the microscopic images of the respective cells are depicted.
Table 4.
Comparative transcriptome analysis of cell wall biosynthesis and morphology determining genes during stationary phase as contrasted with peripheral rods.
| Protein function or tag | MxDK1622 tag | Stationary/ Peripheral |
|---|---|---|
| MreC | MXAN_2645 | −4.53 |
| Penicillin-binding protein | MXAN_2647 | −3.28 |
| MreB | MXAN_2648 | −4.29 |
| MurB | MXAN_5602 | −2.29 |
| MurC | MXAN_5603 | −2.92 |
| MurG | MXAN_5604 | −3.10 |
| MurD | MXAN_5606 | −3.76 |
| MraY | MXAN_5607 | −2.04 |
| MurF | MXAN_5608 | 0.00 |
| MurE | MXAN_5609 | 0.00 |
Final column represent log2-Fold change where negative values demonstrate downregulation of transcripts during stationary phase.
3.4. Developmental pathway analysis
The developmental mechanism has been extensively investigated in M. xanthus from last three decades and several proteins involved in this complex behavior has been reported [15,38–49]. Considering the comparative transcriptomic studies of M. xanthus [49], and relevant previous literature, a set of 132 proteins has been selected for this study, known to be involved in development specific functions. Most of these proteins have homologs in other myxobacterial genomes too (unpublished data) such as M. fulvus [50], M. hansupus [51], Stigmatella aurantiaca [49], Archangium gephyra [52], Anaeromyxobacter dehalogenans [53], Haliangium ochraceum [54], Sandaracinus amylolyticus [55] and Sorangium cellulosum [56]. Here we performed the comparative transcriptional expression studies in M. xanthus stationary, vegetative and peripheral rod cells to understand the level of expression of these development genes (Tables 5 and 6). This study revealed that in contrast to the peripheral rod cells, only 17 and 22 proteins out of 132 protein showed no change in expression in stationary and vegetative cells, while rest of the proteins depicted ~4–70 fold change in expression (Table 6).
Table 5.
Transcriptome Analysis of genes encoding developmental proteins of the cellular envelope.
| Protein function or tag | MxDK1622 tag | Stationary | Vegetative |
|---|---|---|---|
| MBHA | MXAN_7061 | −3.24 | −3.65 |
| Development-specific protein S | MXAN_5432 | −7.35 | −6.43 |
| Protein C | MXAN_1956 | −4.80 | −4.73 |
In this comparison, vegetative and stationary cells were contrasted with peripheral rods. Negative values demonstrate downregulation of transcripts. The numbers in the columns represent log2-Fold change.
Table 6.
Comparative transcriptional analysis of known developmental genes among the peripheral rod cells as compared to stationary and vegetative cells.
| Protein function or tag | MxDK1622 protein tag | COG category | Stationary/Peripheral | Vegetative/Peripheral |
|---|---|---|---|---|
| Pkn13 | MXAN_0023 | T | −6.37 | −2.59 |
| PopC | MXAN_0206 | O | −1.59 | −1.76 |
| σ−54 regulator | MXAN_0353 | T | −6.47 | −4.47 |
| RedC | MXAN_0459 | T | −3.18 | −1.88 |
| RedD | MXAN_0460 | T | −2.94 | −2.5 |
| RedE | MXAN_0461 | T | −3.47 | −2.56 |
| RedF | MXAN_0462 | T | 0.00 | 0.00 |
| RR | MXAN_0710 | T | 0.00 | 0.00 |
| Hpk37 | MXAN_0712 | TK | −3.7 | −3.66 |
| SocE | MXAN_0731 | E | −4.6 | −3.41 |
| RokA | MXAN_0732 | TK | −2.61 | 0.00 |
| RodK | MXAN_0733 | TK | −2.5 | −1.29 |
| Pkn9 | MXAN_0755 | T | −2.37 | −1.35 |
| PktB6 | MXAN_0871 | T | −1.7 | −1.38 |
| PktA5 | MXAN_0930 | T | −5.87 | −3.87 |
| EspA | MXAN_0931 | T | −7.12 | −4.1 |
| EspB | MXAN_0932 | S | −4.14 | −5.67 |
| PktB8 | MXAN_0933 | T | −3.16 | −3.06 |
| AsgE | MXAN_1010 | FR | −4.3 | −4.2 |
| SdeK | MXAN_1014 | T | −3.25 | −8.36 |
| RpoN | MXAN_1061 | K | −2.03 | −2.14 |
| HK | MXAN_1077 | T | −2.86 | −3.65 |
| Nla19 | MXAN_1078 | T | −2.56 | 0.00 |
| Pab4 | MXAN_1088 | T | −3.26 | 0.00 |
| Nla28 | MXAN_1167 | T | −2.85 | −4.44 |
| Pdd3 | MXAN_1234 | T | −3.44 | −3.91 |
| SasN | MXAN_1244 | −3.14 | −2.88 | |
| SasR | MXAN_1245 | T | −3.96 | −2.28 |
| SasS | MXAN_1249 | T | −4.28 | −4.28 |
| CsgA | MXAN_1294 | IQR | 0.00 | −3.84 |
| LadA | MXAN_1402 | K | −2.9 | −2.57 |
| Pkn1 | MXAN_1467 | T | −4.86 | −8.87 |
| MazF | MXAN_1659 | V | −4.4 | −3.86 |
| Pkn8 | MXAN_1710 | R | −3.95 | −2.95 |
| Lon protease 1 | MXAN_2017 | O | −0.6 | 0.00 |
| PktE2 | MXAN_2176 | T | −5.04 | −5.56 |
| PktB4 | MXAN_2255 | T | −5.38 | −4.13 |
| MspA | MXAN_2269 | S | −3.86 | −4.23 |
| MspB | MXAN_2432 | S | −3.84 | −4.91 |
| SigD | MXAN_2437 | K | 0.00 | 0.00 |
| Prw | MXAN_2491 | R | −6 | −10.02 |
| Nla4 | MXAN_2516 | T | −3 | −4.2 |
| Pkn6 | MXAN_2550 | T | −6.38 | −4.64 |
| AsgA | MXAN_2670 | T | 0.00 | 0.00 |
| PskA12 | MXAN_2680 | T | −1.16 | 0.00 |
| PhoP2 | MXAN_2778 | TK | −4.15 | −3.09 |
| PhoR2 | MXAN_2779 | T | −6.98 | −5.28 |
| σ−54 regulator | MXAN_2902 | KT | −3.44 | −4.69 |
| AsgB | MXAN_2913 | K | −1.95 | 0.00 |
| PktA7 | MXAN_3094 | T | −2.58 | −4.82 |
| FruA | MXAN_3117 | TK | 0.00 | −7.2 |
| Monooxygenase | MXAN_3122 | HC | −3.47 | −2.35 |
| RelA | MXAN_3204 | TK | −4.09 | 0.00 |
| ActA | MXAN_3213 | TK | −6.63 | −4.97 |
| ActB | MXAN_3214 | T | −5.98 | −6.1 |
| ActC | MXAN_3215 | M | −4.27 | −3.28 |
| ActD | MXAN_3217 | R | −6.46 | −4.75 |
| FdgA | MXAN_3225 | M | −5.3 | −7.14 |
| Exo | MXAN_3227 | M | −5.71 | −6.33 |
| Hpk8 | MXAN_3290 | TK | −2.4 | −2.42 |
| SigB | MXAN_3357 | K | −5.84 | −5.93 |
| NfsA | MXAN_3371 | S | −6.02 | −4.81 |
| NfsB | MXAN_3372 | S | −5.88 | −5.99 |
| NfsC | MXAN_3373 | R | −5.21 | −5.29 |
| NfsD | MXAN_3374 | R | −7.77 | −6.96 |
| NfsE | MXAN_3375 | R | −4.94 | −5.55 |
| NfsF | MXAN_3376 | S | −5.79 | −4.92 |
| NfsG | MXAN_3377 | M | −6.36 | −5.57 |
| NfsH | MXAN_3378 | S | −3.32 | −8.18 |
| Nla18 | MXAN_3692 | T | −5.64 | −4.21 |
| RR | MXAN_3711 | TK | 0.00 | 0.00 |
| Diguanylate cyclase | MXAN_3735 | TK | −5.48 | −4.75 |
| Abc1–2 | MXAN_3899 | HT | −3.68 | −1.61 |
| Lon protease 2 | MXAN_3993 | O | 0.00 | 1.28 |
| σ−54 regulator | MXAN_4020 | T | 0.00 | 0.00 |
| Nla6 | MXAN_4042 | T | −4.23 | −3.04 |
| Protein kinase | MXAN_4049 | T | −6.39 | −5.46 |
| CarQ | MXAN_4088 | K | −4.25 | −5.08 |
| Hypothetical | MXAN_4092 | S | −5.95 | −4.38 |
| BrgE | MXAN_4151 | H | 0.00 | 0.00 |
| Serpin | MXAN_4276 | O | −5.32 | −4.6 |
| PktB9 | MXAN_4437 | Q | 0.00 | 0.00 |
| RomA | MXAN_4462 | NT | −4.54 | −6.49 |
| GGDEF domain | MXAN_4463 | T | 0.00 | −2.55 |
| Hpk30 | MXAN_4465 | TK | −3.38 | −1.14 |
| FruE | MXAN_4486 | S | −4.48 | −2.59 |
| RfbC | MXAN_4621 | M | −5.18 | −4.01 |
| RfbB | MXAN_4622 | GM | −4.07 | −3.14 |
| RfbA | MXAN_4623 | GM | −4.59 | 0.00 |
| PhoP1 | MXAN_4777 | TK | −4.61 | −2.74 |
| PhoR1 | MXAN_4778 | T | −4.15 | −4.04 |
| PhoP | MXAN_4787 | TK | −4.21 | −2.25 |
| MXAN_4899 | MXAN_4899 | T | 0.00 | 0.00 |
| Pkn14 | MXAN_5116 | T | −3.08 | 0.00 |
| MrpA | MXAN_5123 | T | −4.94 | −1.74 |
| MrpB | MXAN_5124 | T | −4.23 | −3.5 |
| MrpC | MXAN_5125 | T | 0.00 | 0.00 |
| CheA3 | MXAN_5147 | NT | −4.85 | −4.13 |
| Mcp3B | MXAN_5148 | NT | −6.27 | −5.3 |
| MCP | MXAN_5149 | NT | −4.75 | −3.46 |
| CrdC | MXAN_5150 | S | −4.05 | −5.61 |
| CheW | MXAN_5151 | NT | 0.00 | 0.00 |
| OmpA | MXAN_5152 | M | 1.29 | 2.45 |
| CrdA | MXAN_5153 | T | −4.05 | −3.67 |
| RpoD | MXAN_5204 | K | 0.00 | 0.00 |
| RR | MXAN_5656 | TK | 0.00 | −3.43 |
| CbgA | MXAN_5828 | D | −2.84 | −3.15 |
| SigE | MXAN_5870 | K | −2.24 | 0.00 |
| PktD6 | MXAN_5886 | T | −3.25 | −3.88 |
| RR | MXAN_5889 | TK | −4.77 | −2.57 |
| PktC8 | MXAN_6009 | R | −4.47 | −5.57 |
| SigC | MXAN_6209 | K | −5.54 | −3.9 |
| PskB8 | MXAN_6312 | R | −4.96 | −3.88 |
| PhoP3 | MXAN_6413 | TK | −2.97 | −5.68 |
| PhoR3 | MXAN_6414 | T | −5.6 | −5.3 |
| Pkn2 | MXAN_6420 | T | −2.26 | −1.44 |
| PktB1 | MXAN_6428 | R | −4.01 | −6.28 |
| PktA1 | MXAN_6500 | T | −3.81 | −5.33 |
| PktC7 | MXAN_6561 | R | −7.29 | −6.83 |
| EspC | MXAN_6855 | T | −4.71 | −5.16 |
| HthA | MXAN_6889 | TK | −4.31 | −2.39 |
| HthB | MXAN_6890 | T | −2.61 | −1.38 |
| TodK | MXAN_6955 | T | −2.63 | −1.97 |
| MspC | MXAN_6969 | S | −4.06 | −4.86 |
| AsgD | MXAN_6996 | T | −3.08 | −3.6 |
| Pkn12 | MXAN_7251 | R | −5.68 | −3.4 |
| DevS | MXAN_7261 | S | −5.09 | −9.15 |
| DevR | MXAN_7262 | V | −3.8 | −7.08 |
| DevT | MXAN_7263 | S | −6.46 | −5.79 |
| PktF6 | MXAN_7269 | T | −6.78 | −3.23 |
| TCS | MXAN_7396 | TK | −2.47 | −3.89 |
| Nsd | MXAN_7402 | R | −5.17 | −3.62 |
In this study, the transcript expression in both vegetative and stationary cells is contrasted with peripheral rods. The values in the last two columns are depicted in the unit of log2-Fold change and negative sign demonstrates the upregulation of transcripts in peripheral rods. No change is expression is shown in yellow shade whereas expression factor > 4 log2-Fold change is in pink shade. Positive regulation is presented in a light blue shade. Abbreviations used- MCP: Methyl accepting chemoreceptor protein; σ: sigma; HK: Histidine Kinase; RR: Response Regulator.
Initially, we were especially interested into developmental genes associated with the cellular envelope, where we found that myxobacterial hemagglutinin (MBHA), Tps, and Protein C were highly expressed in peripheral rods in comparison to stationary cells. MBHA is a lectin-like protein that localizes on the cell surface of the cell and contributes to cell-cell recognition [2]. Protein C and Tps are major spore surface proteins induced during development [2]. Previous studies demonstrated that these spore-associated genes were found in peripheral rods and not vegetative cells. These developmental genes are also downregulated in stationary phase cells in a similar manner (Table 5). This change prompted us to extend our study into the expression of identified 132 developmental genes in both stationary-phase cells and peripheral rods. The expression pattern of the known developmental genes significantly differed between the two cell types as well (Table 6). Genes activated in Myxococcus development were downregulated or not activated at all in stationary cells. The developmental expression pattern was similar in vegetative cells as well. The developmental genes were highly upregulated in peripheral rods, indicating this differentiated cell type is the product of multicellular development and not just nutrient starvation.
3.5. Comparative distribution of metabolism genes
Only a minority of metabolic genes were downregulated in peripheral rods in comparison to stationary cells. The vast majority of these genes are either not annotated or associated with translation and ribosomal biogenesis similar to the vegetative comparisons. However, several genes associated with energy conversion and respiration were also downregulated in peripheral rods. Genes encoding all three enzymes comprising the pyruvate dehydrogenase complex (PDC) were downregulated in peripheral rods (Table 7). The PDC contains three components, pyruvate dehydrogenase, dehydrolipoate acyltransferase, and dihydrolipoate dehydrogenase that catalyze the oxidative decarboxylation of pyruvate [57]. During aerobic respiration, the PDC converts pyruvate and NAD to acetyl coenzyme A, carbon dioxide, and NADH [57]. Acetyl-CoA enters the Krebs cycle, reacts with oxaloacetate and continues oxidation to form carbon dioxide, generating ATP via electron transport [57]. We also identified a decrease in transcripts of genes encoding acetyl-CoA acetyltransferase and cytochrome C. Peripheral rods might downregulate cellular respiration to avoid additional oxidative stresses.
Table 7.
Comparative expression of metabolism genes in Stationary cells as contrasted with Peripheral Rods.
| Protein function or tag | MxDK1622 tag | Stationary/Peripheral |
|---|---|---|
| PdhC dehydrogenase | MXAN_2666 | 2.47 |
| PdhC dihydrolipoate dehydrogenase | MXAN_2667 | 2.21 |
| PdhC dihydrolipoamide acetyltransferase | MXAN_2668 | 3.29 |
| Malate dehydrogenase | MXAN_3538 | 3.01 |
| Succinate CoA ligase | MXAN_3542 | 2.21 |
| Cytochrome c | MXAN_5560 | 2.20 |
The numbers in the final column represent log2FoldChange. Positive values demonstrate an upregulation of transcripts.
3.6. Vegetative cell analysis
The environment inhabited by peripheral rods consists of low nutrients, derived from the autolysis of nearby cells. These non-aggregate cells are physically separated by the growing and developing fruiting body encased by exopolysaccharide. It is possible that this environment contains enough nutrients to allow peripheral rod cells to survive barely, resembling cells growing at very low growth rates. We have previously shown that M. xanthus cells can grow and survive for long periods of time as biofilms with very low nutrients in our bioreactors [16]. We, therefore, compared the transcriptome of peripheral rod cells to that of vegetative M. xanthus cells growing under two distinct conditions, 1) in batch liquid culture under rich nutrient (1% casitone) conditions and 2) in our bioreactors under which an M. xanthus vegetative biofilm forms under low nutrient (0.004% casitone) conditions. The transcriptome comparisons of the nutrient-rich culture versus peripheral rod cells revealed differential expression of 75% of the genome, with 5147 (72%) of the genes being upregulated and 214 (3%) of the genes being downregulated (fold-change ≥2, p≤0.05) (Fig. 3). Surprisingly, among the genes upregulated in the nutrient-rich environment was fruA, which has previously not been detected during vegetative growth [58]. As predicted, the data show the transcriptomes of these two cell types to be highly different as established in earlier studies [2,5,59]. Similarly, when cells were grown as a vegetative biofilm under low nutrients, our analysis revealed that 64% of the genome was differentially expressed; with the majority of genes, 58%, being downregulated, while only 6% of genes were upregulated (fold-change ≥2, p≤0.05). Interestingly, while comparing the total transcriptome of the four growth states i.e. nutrient-rich, low nutrients in a biofilm, stationary phase and peripheral rod cells, the peripheral rod cells do resemble with cells growing in a low nutrient biofilm (Fig. 3), which is expected as the area outside of the fruiting body represent a low nutrient environment.
4. Conclusions
Peripheral rods are a developmentally differentiated cell type capable of responding quickly to nutrients; they exhibit upregulation of replication-associated genes (not shown). This developmental cell type appears more inclined to utilize low nutrient influxes than stationary phase cells. More importantly, they possess a global transcriptome that is unlike any other cell type found in M. xanthus (Fig. 3). Based on this work it is clear that peripheral rod cells are not a class of stationary phase cells, rather they represent a novel developmental differentiated cell type, as Zusman and O’Connor initially proposed [2,5]. Peripheral rods express a myriad of developmental genes. When examining the expression pattern of developmental genes, peripheral rods resemble low growth biofilm cells more than any other cell types. The induction of developmental genes, especially those associated with early sporulation, suggests that peripheral rods and myxospores share in part a common pathway in differentiation. We can hypothesize that peripheral rods are a cell type halted at a checkpoint in route to sporulation, a point that still allows the peripheral rod cells the ability to survive in very low nutrient environments. Sporulation is indirectly dependent upon multiple factors necessary to generate fruiting bodies. Monitoring of population density, motility, and intracellular and intercellular signaling are used to coordinate the temporal and spatial events of fruiting body development. Whether the same factors govern peripheral rod development is unknown.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2019.09.008.
Supplementary Material
Acknowledgments
The authors would like to thank Blythe Durbin-Johnson at the UC Davis bioinformatics core facility for statistical assistance, Rebecca Parales for the microscopy, and Ivy Jose for technical assistance.
Funding
This work was supported by National Science Foundation grant: IOS-1354562 to M.S.
Footnotes
Declaration of Competing Interests
Authors declare that they have no conflicts of interest.
References
- [1].Kuner JM, Kaiser D, Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus, J. Bacteriol 151 (1982) 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].O’Connor KA, Zusman DR, Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores, J. Bacteriol 173 (1991) 3318–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shimkets LJ, Control of morphogenesis in myxobacteria, Crit. Rev. Microbiol 14 (1987) 195–227. [DOI] [PubMed] [Google Scholar]
- [4].Kaiser D, Warrick H, Transmission of a signal that synchronizes cell movements in swarms of Myxococcus xanthus, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 13105–13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].O’Connor KA, Zusman DR, Behavior of peripheral rods and their role in the life cycle of Myxococcus xanthus, J. Bacteriol 173 (1991) 3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoiczyk E, Ring MW, McHugh CA, Schwar G, Bode E, Krug D, Altmeyer MO, Lu JZ, Bode HB, Lipid body formation plays a central role in cell fate determination during developmental differentiation of Myxococcus xanthus, Mol. Microbiol 74 (2009) 497–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zusman DR, Scott AE, Yang Z, Kirby JR, Chemosensory pathways, motility and development in Myxococcus xanthus, Nat. Rev. Microbiol 5 (2007) 862–872. [DOI] [PubMed] [Google Scholar]
- [8].Rittershaus ES, Baek SH, Sassetti CM, The normalcy of dormancy: common themes in microbial quiescence, Cell Host Microbe 13 (2013) 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rosenberg SM, Life, death, differentiation, and the multicellularity of bacteria, PLoS Genet. 5 (2009) e1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sung HM, Yasbin RE, Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis, J. Bacteriol 184 (2002) 5641–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kolter R, Siegele DA, Tormo A, The stationary phase of the bacterial life cycle, Annu. Rev. Microbiol 47 (1993) 855–874. [DOI] [PubMed] [Google Scholar]
- [12].Navarro Llorens JM, Tormo A, Martinez-Garcia E, Stationary phase in gram-negative bacteria, FEMS Microbiol. Rev 34 (2010) 476–495. [DOI] [PubMed] [Google Scholar]
- [13].Ueki T, Inouye S, SigB, SigC, and SigE from Myxococcus xanthus homologous to sigma32 are not required for heat shock response but for multicellular differentiation, J. Mol. Microbiol. Biotechnol 3 (2001) 287–293. [PubMed] [Google Scholar]
- [14].Ueki T, Inouye S, A new sigma factor, SigD, essential for stationary phase is also required for multicellular differentiation in Myxococcus xanthus, Genes Cells 3 (1998) 371–385. [DOI] [PubMed] [Google Scholar]
- [15].Tzeng L, Singer M, DNA replication during sporulation in Myxococcus xanthus fruiting bodies, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Smaldone GT, Jin Y, Whitfield DL, Mu AY, Wong EC, Wuertz S, Singer M, Growth of Myxococcus xanthus in continuous-flow-cell bioreactors as a method for studying development, Appl. Environ. Microbiol 80 (2014) 2461–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Morlan JD, Qu K, Sinicropi DV, Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue, PLoS One 7 (2012) e42882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15 (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Anders S, Pyl PT, Huber W, HTSeq–a Python framework to work with high-throughput sequencing data, Bioinformatics 31 (2015) 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sharma G, Khatri I, Subramanian S, Comparative genomics of myxobacterial chemosensory systems, J. Bacteriol 200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bretl DJ, Kirby JR, Molecular mechanisms of signaling in Myxococcus xanthus development, J. Mol. Biol 428 (2016) 3805–3830. [DOI] [PubMed] [Google Scholar]
- [22].Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K, The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature, J. Bacteriol 177 (1995) 2918–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Erickson JW, Gross CA, Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression, Genes Dev. 3 (1989) 1462–1471. [DOI] [PubMed] [Google Scholar]
- [24].Keseler IM, Kaiser D, et al. , Proc. Natl. Acad. Sci 94 (1997) 1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chiang SH, SM, Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria, Arch. Biochem. Biophys 525 (2012) (2012) 161–169. [DOI] [PubMed] [Google Scholar]
- [26].Zeng Q, Stalhandske C, Anderson MC, Scott RA, Summers AO, The core metal-recognition domain of MerR, Biochemistry 37 (1998) 15885–15895. [DOI] [PubMed] [Google Scholar]
- [27].O’Halloran T, Walsh C, Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization, Science 235 (1987) 211–214. [DOI] [PubMed] [Google Scholar]
- [28].Pletzer D, Schweizer G, Weingart H, AraC/XylS family stress response regulators Rob, SoxS, PliA, and OpiA in the fire blight pathogen Erwinia amylovora, J. Bacteriol 196 (2014) 3098–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ibarra JA, Perez-Rueda E, Segovia L, Puente JL, The DNA-binding domain as a functional indicator: the case of the AraC/XylS family of transcription factors, Genetica 133 (2008) 65–76. [DOI] [PubMed] [Google Scholar]
- [30].Lahiri A, Das P, Chakravortty D, The LysR-type transcriptional regulator Hrg counteracts phagocyte oxidative burst and imparts survival advantage to Salmonella enterica serovar Typhimurium, Microbiology 154 (2008) 2837–2846. [DOI] [PubMed] [Google Scholar]
- [31].Maddocks SE, Oyston PC, Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins, Microbiology 154 (2008) 3609–3623. [DOI] [PubMed] [Google Scholar]
- [32].McDougald D, Gong L, Srinivasan S, Hild E, Thompson L, Takayama K, Rice SA, Kjelleberg S, Defences against oxidative stress during starvation in bacteria, Antonie Van Leeuwenhoek 81 (2002) 3–13. [DOI] [PubMed] [Google Scholar]
- [33].Michel B, After 30 years of study, the bacterial SOS response still surprises us, PLoS Biol. 3 (2005) e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Campoy S, Fontes M, Padmanabhan S, Cortes P, Llagostera M, Barbe J, LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus, Mol. Microbiol 49 (2003) 769–781. [DOI] [PubMed] [Google Scholar]
- [35].Holtje JV, Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli, Microbiol. Mol. Biol. Rev 62 (1998) 181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cava F, de Pedro MA, Peptidoglycan plasticity in bacteria: emerging variability of the murein sacculus and their associated biological functions, Curr. Opin. Microbiol 18 (2014) 46–53. [DOI] [PubMed] [Google Scholar]
- [37].Laddomada F, Miyachiro MM, Dessen A, Structural insights into protein-protein interactions involved in bacterial cell wall biogenesis, Antibiotics (Basel) 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Higgs PI, Jagadeesan S, Mann P, Zusman DR, EspA, an orphan hybrid histidine protein kinase, regulates the timing of expression of key developmental proteins of Myxococcus xanthus, J. Bacteriol 190 (2008) 4416–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Muller FD, Treuner-Lange A, Heider J, Huntley SM, Higgs PI, Global transcriptome analysis of spore formation in Myxococcus xanthus reveals a locus necessary for cell differentiation, BMC Genomics 11 (2010) 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dahl JL, Tengra FK, Dutton D, Yan J, Andacht TM, Coyne L, Windell V, Garza AG, Identification of major sporulation proteins of Myxococcus xanthus using a proteomic approach, J. Bacteriol 189 (2007) 3187–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bellenger K, Ma X, Shi W, Yang Z, A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis, J. Bacteriol 184 (2002) 5654–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kaiser D, Coupling cell movement to multicellular development in myxobacteria, Nat. Rev. Microbiol 1 (2003) 45–54. [DOI] [PubMed] [Google Scholar]
- [43].Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB, Evolution of sensory complexity recorded in a myxobacterial genome, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 15200–15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jiang DM, Wu ZH, Zhao JY, Li YZ, Fruiting and non-fruiting myxobacteria: a phylogenetic perspective of cultured and uncultured members of this group, Mol. Phylogenet. Evol 44 (2007) 545–552. [DOI] [PubMed] [Google Scholar]
- [45].Goldman B, Bhat S, Shimkets LJ, Genome evolution and the emergence of fruiting body development in Myxococcus xanthus, PLoS One 2 (2007) e1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kaiser D, Robinson M, Kroos L, Myxobacteria, polarity, and multicellular morphogenesis, Cold Spring Harb. Perspect. Biol 2 (2010) a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Velicer GJ, Vos M, Sociobiology of the myxobacteria, Annu. Rev. Microbiol 63 (2009) 599–623. [DOI] [PubMed] [Google Scholar]
- [48].Curtis PD, Taylor RG, Welch RD, Shimkets LJ, Spatial organization of Myxococcus xanthus during fruiting body formation, J. Bacteriol 189 (2007) 9126–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Huntley S, Hamann N, Wegener-Feldbrugge S, Treuner-Lange A, Kube M, Reinhardt R, Klages S, Muller R, Ronning CM, Nierman WC, Sogaard-Andersen L, Comparative genomic analysis of fruiting body formation in Myxococcales, Mol. Biol. Evol 28 (2011) 1083–1097. [DOI] [PubMed] [Google Scholar]
- [50].Li ZF, Li X, Liu H, Liu X, Han K, Wu ZH, Hu W, Li FF, Li YZ, Genome sequence of the halotolerant marine bacterium Myxococcus fulvus HW-1, J. Bacteriol 193 (2011) 5015–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sharma G, Narwani T, Subramanian S, Complete genome sequence and comparative genomics of a novel myxobacterium Myxococcus hansupus, PLoS One 11 (2016) e0148593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sharma G, Subramanian S, Unravelling the complete genome of Archangium gephyra DSM 2261T and evolutionary insights into myxobacterial chitinases, Genome Biol. Evol 9 (2017) 1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Thomas SH, Wagner RD, Arakaki AK, Skolnick J, Kirby JR, Shimkets LJ, Sanford RA, Löffler FE, The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria, PLoS One 3 (2008) e2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ivanova N, Daum C, Lang E, Abt B, Kopitz M, Saunders E, Lapidus A, Lucas S, Glavina Del Rio T, Nolan M, Tice H, Copeland A, Cheng JF, Chen F, Bruce D, Goodwin L, Pitluck S, Mavromatis K, Pati A, Mikhailova N, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Detter JC, Brettin T, Rohde M, Goker M, Bristow J, Markowitz V, Eisen JA, Hugenholtz P, Kyrpides NC, Klenk HP, Complete genome sequence of Haliangium ochraceum type strain (SMP-2), Stand. Genomic Sci 2 (2010) 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sharma G, Khatri I, Subramanian S, Complete genome of the starch-degrading myxobacteria Sandaracinus amylolyticus DSM 53668T, Genome Biol. Evol 8 (8) (2016) 2520–2529, 10.1093/gbe/evw151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, Bode HB, Bolten CJ, Choudhuri JV, Doss S, Elnakady YA, Frank B, Gaigalat L, Goesmann A, Groeger C, Gross F, Jelsbak L, Jelsbak L, Kalinowski J, Kegler C, Knauber T, Konietzny S, Kopp M, Krause L, Krug D, Linke B, Mahmud T, Martinez-Arias R, McHardy AC, Merai M, Meyer F, Mormann S, Munoz-Dorado J, Perez J, Pradella S, Rachid S, Raddatz G, Rosenau F, Ruckert C, Sasse F, Scharfe M, Schuster SC, Suen G, Treuner-Lange A, Velicer GJ, Vorholter FJ, Weissman KJ, Welch RD, Wenzel SC, Whitworth DE, Wilhelm S, Wittmann C, Blocker H, Puhler A, Muller R, Complete genome sequence of the myxobacterium Sorangium cellulosum, Nat. Biotechnol 25 (2007) 1281–1289. [DOI] [PubMed] [Google Scholar]
- [57].Ogasawara H, Ishida Y, Yamada K, Yamamoto K, Ishihama A, PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli, J. Bacteriol 189 (2007) 5534–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ogawa M, Fujitani S, Mao X, Inouye S, Komano T, FruA, a putative transcription factor essential for the development of Myxococcus xanthus, Mol. Microbiol 22 (1996) 757–767. [DOI] [PubMed] [Google Scholar]
- [59].O’Connor KA, Zusman DR, Analysis of Myxococcus xanthus cell types by two-dimensional polyacrylamide gel electrophoresis, J. Bacteriol 173 (1991) 3334–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


