Abstract
BACKGROUND
There has been a striking generational increase in the prevalence of food allergies. We have proposed that this increase can be explained, in part, by alterations in the commensal microbiome.
METHODS
To identify bacterial signatures and metabolic pathways that may influence the expression of this disease, we collected fecal samples from a unique, well-controlled cohort of twins concordant or discordant for food allergy. Samples were analyzed by integrating 16S rRNA gene amplicon sequencing and liquid chromatography–tandem mass spectrometry metabolite profiling.
RESULTS
A bacterial signature of 64 operational taxonomic units (OTUs) distinguished healthy from allergic twins; the OTUs enriched in the healthy twins were largely taxa from the Clostridia class. We detected significant enrichment in distinct metabolite pathways in each group. The enrichment of diacylglycerol in healthy twins is of particular interest for its potential as a readily measurable fecal biomarker of health. In addition, an integrated microbial-metabolomic analysis identified a significant association between healthy twins and Phascolarctobacterium faecium and Ruminococcus bromii, suggesting new possibilities for the development of live microbiome-modulating biotherapeutics.
CONCLUSION
Twin pairs exhibited significant differences in their fecal microbiomes and metabolomes through adulthood, suggesting that the gut microbiota may play a protective role in patients with food allergies beyond the infant stage.
TRIAL REGISTRATION
Participants in this study were recruited as part of an observational study (ClinicalTrials.gov NCT01613885) at multiple sites from 2014 to 2018.
FUNDING
This work was supported by the Sunshine Charitable Foundation; the Moss Family Foundation; the National Institute of Allergy and Infectious Diseases (NIAID) (R56AI134923 and R01AI 140134); the Sean N. Parker Center for Allergy and Asthma Research; the National Heart, Lung, and Blood Institute (R01 HL 118612); the Orsak family; the Kepner family; and the Stanford Institute for Immunity, Transplant and Infection.
Keywords: Immunology
Keywords: Allergy

Introduction
Recent surveys estimate that 32 million children and adults in the United States suffer from food allergies (1, 2). This represents a marked increase in allergic responses to food in industrialized societies worldwide (3), which parallels increases in other noncommunicable diseases (NCDs), including obesity, diabetes, asthma, autism, and inflammatory bowel disease. These NCDs share an association with dysbiosis of the commensal microbiome, particularly in the gut (4). Microbes colonize the skin and all mucosal surfaces and have profound influences on basic aspects of physiology and health. Emerging evidence suggests that increased antibiotic use, low-fiber/high-fat diets, reduced exposure to infectious disease, caesarean delivery, and formula feeding have collectively depleted bacterial populations beneficial to health (4). Early childhood is a particular period of vulnerability for the maturation of the microbiota and the developing immune system, which are intimately intertwined (5, 6). The intestinal microbiome of children with food allergies may differ in important ways from genetically similar nonallergic children and age-matched controls. The aim of this study was to characterize fecal microbiomes to identify taxa that may influence the expression of food allergy in children and adults.
An association between gut microbial community changes and childhood food allergies has been reported in some epidemiological studies (7–9). The Canadian Healthy Infant Longitudinal Development study showed alterations in the gut microbial community of food-sensitized infants; in that study, Enterobacteriaceae were overrepresented in a less diverse microbial community in infants at 3 months of age, whereas Bacteroidaceae were underrepresented at 1 year (9). A Chinese infant cohort study showed that infants with food allergies had a higher abundance of Firmicutes and a lower abundance of Bacteroidetes at 6 months, but no significant difference in the total microbial diversity was found (8).
Oral immunotherapy (OIT) and epicutaneous immunotherapy, allergen-specific desensitization protocols performed by introducing small but gradually increasing doses of allergen, have been shown to safely and effectively desensitize patients with food allergies to their allergens (10–12). However, OIT requires a prolonged period of updosing (usually years), during which gastrointestinal symptoms are common and might contribute to the high withdrawal rate observed in clinical trials (13, 14). Although OIT can achieve short-term desensitization, this desensitization is not sustained without daily maintenance dosing, and long-term tolerance is not induced in the majority of the cases (11). Live microbiome-modulating biotherapeutics are already showing promise in clinical trials for a variety of diseases (15, 16). Preclinical data suggest that microbiome-modulating therapeutics have the potential for improving both the efficacy and safety of OIT. Our mouse model work shows that preventing an allergic response to food requires the induction of both a food allergen–specific immunoregulatory response and a commensal bacteria–induced intestinal barrier–protective response, which regulates epithelial permeability to food allergens (17). Thus, microbiome-modulating therapies could possibly be implemented to reduce adverse events, improve compliance, and to achieve efficacy and sustained unresponsiveness in patients with food allergies.
Toward our ultimate goal of developing novel microbiome-modulating therapeutics, in this report, we extend our work in infants (18) to a broader patient population to identify both bacterial taxa and their products associated with a healthy microbiota. We hypothesized that the intestinal microbiome of food-allergic twins would differ from genetically similar twins without food allergy (siblings raised in the same household). We examined fecal samples from a unique cohort of food-allergic and healthy twins across a broad age range and identified a distinct set of bacterial species and metabolites that distinguished the healthy and allergic groups. By integrating microbiota and metabolite abundance, we identified in healthy twins a significant enrichment in particular metabolite pathways not seen in their allergic counterparts, particularly diacylglycerol (DAG), an essential lipid second messenger, involved in numerous cell signaling cascades supporting biosynthesis of glycerolipids and regulating protein kinase C (19). We also identified 2 bacterial species more abundant in healthy twins that correlate with differentially abundant metabolites and are potential targets for future translational and clinical studies: Phascolarctobacterium faecium, an acetate/propionate-producing obligate anaerobe (20, 21) associated with increased DAG and biotin metabolism, and Ruminococcus bromii, a keystone resistant starch–degrading strict anaerobe (22, 23) associated with fatty acid, sterol, and amino acid metabolism.
Results
Healthy and allergic twins exhibit distinct fecal microbial profiles.
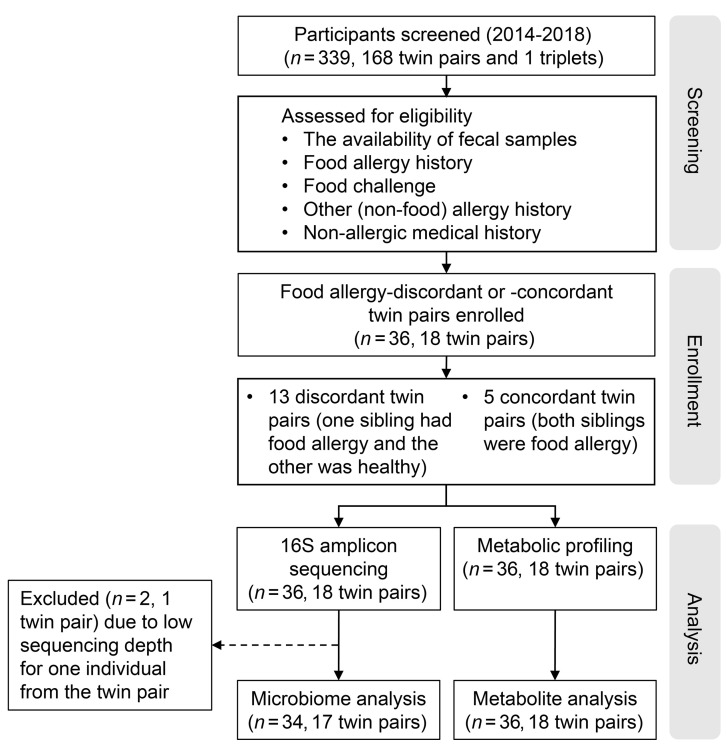
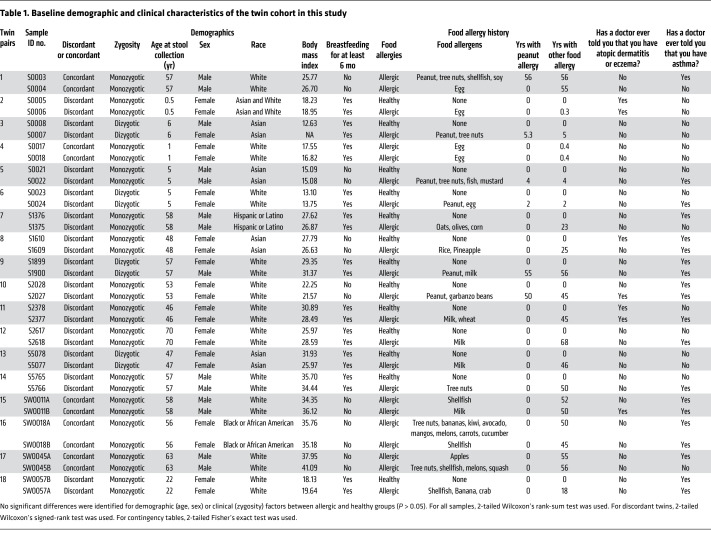
The composition of the fecal microbiota has been reported to differ in young children with food allergies compared with healthy children (whether siblings or unrelated) (24). We therefore examined both the microbial signatures and metabolomic profiles in the fecal samples from a unique collection of twin pairs that were raised in the same household, in which they equally avoided the foods to which the affected twin was found to be allergic and were either concordant or discordant for food allergy (Figure 1). Baseline demographic and clinical characteristics of the twin cohort are shown in Table 1, and a food diary prepared at the time of sample collection is shown in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI141935DS1). Interestingly, the concordant twins did not necessarily share the same food allergy (Table 1). The average age of participants at sample collection was 39.4 ± 4.1 years (mean ± SEM). All of the twins lived independently after the age of 19 years. There were no significant differences in the baseline demographic and clinical characteristics between the healthy and food-allergic twin pairs (Table 1).
Figure 1. Flow diagram of study design and participating patients.
Table 1. Baseline demographic and clinical characteristics of the twin cohort in this study.
We first performed 16S rRNA gene amplicon sequencing on fecal samples from 13 healthy and 23 food-allergic individuals consisting of 18 twin pairs. After excluding 1 sample with low sequencing depth and the corresponding twin pair, we included 34 samples for analysis, including 24 samples from 12 discordant twin pairs (1 allergic, 1 healthy) and 10 samples from 5 concordant twin pairs (both allergic). An overview of the analytical workflow is shown in Supplemental Figure 1. The composition of commensal microbiota is shown in Figure 2A, with quantitative measures provided in Supplemental Table 2. While there was sample-to-sample variation, the presence of major families, such as Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae, was consistent with that reported in previous studies on fecal samples (25). For each twin pair, we compared the relative abundance of operational taxonomic units (OTUs), which represent groups of microbes between closely related individuals. We calculated within-pair, sibling-wise OTU correlation between the 2 siblings from each twin pair. The within-pair OTU correlation did not differ significantly between discordant and concordant twin pairs or between dizygotic and monozygotic twin pairs (Figure 2, B and C). Across all samples or within discordant twin pairs only, no significant differences were detected in the α diversity (Shannon diversity, Figure 2, D and E). β Diversity (weighted UniFrac distance) metrics also did not differ between allergic and healthy groups (Supplemental Figure 2).
Figure 2. Relative abundance of microbial composition of healthy and allergic twins does not differ at the family level.
(A) Relative abundance of taxonomy at the family level. Sample IDs are shown on the x axis (n = 34). Discordant twins (12 pairs, n = 24), for which one member was healthy and the other member was allergic; concordant twins (5 pairs, n = 10), for which both members were allergic. Of 36 total samples in the twin cohort, 1 sample (S5077) failed sequencing and yielded 0 reads, hence the corresponding twin pair (no. 13) was excluded from 16S analysis. (B and C) Correlation of OTU abundance between members from each twin pair, with the comparison between concordant and discordant twin pairs shown in B and the comparison between dizygotic and monozygotic twins shown in C. Each dot denotes 1 twin pair (17 pairs shown). (D and E) Shannon α diversity index between healthy and allergic groups, with all samples are shown in D (n = 34) and only discordant twins shown in E (n = 24). Each dot denotes 1 sample. In B–E, the bounds of the boxes represent the 25th and 75th percentiles, the horizontal centers line indicate the medians, and the whiskers extend to data points within a maximum of 1.5 times the IQR. Two-tailed Wilcoxon’s rank-sum test was used in B–D, and two-tailed Wilcoxon’s signed-rank test was used in E.
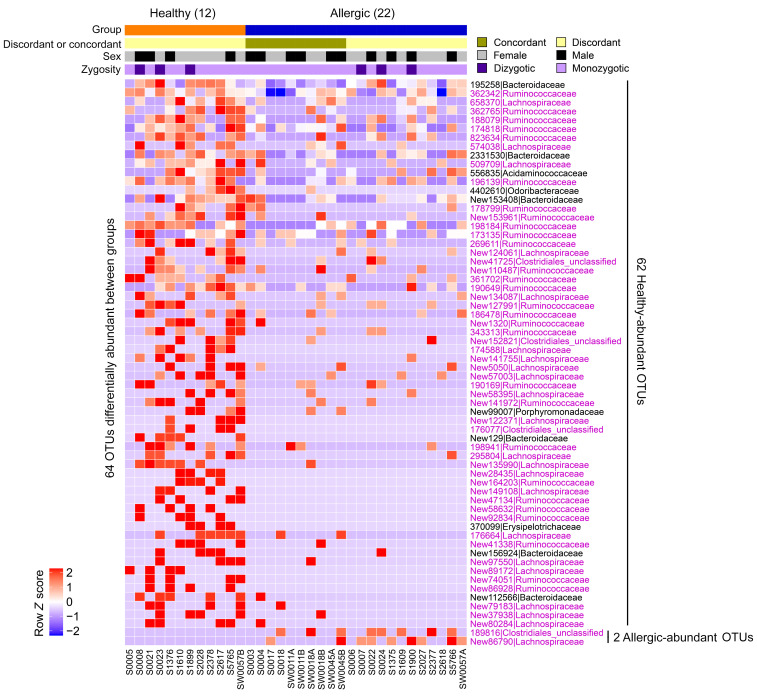
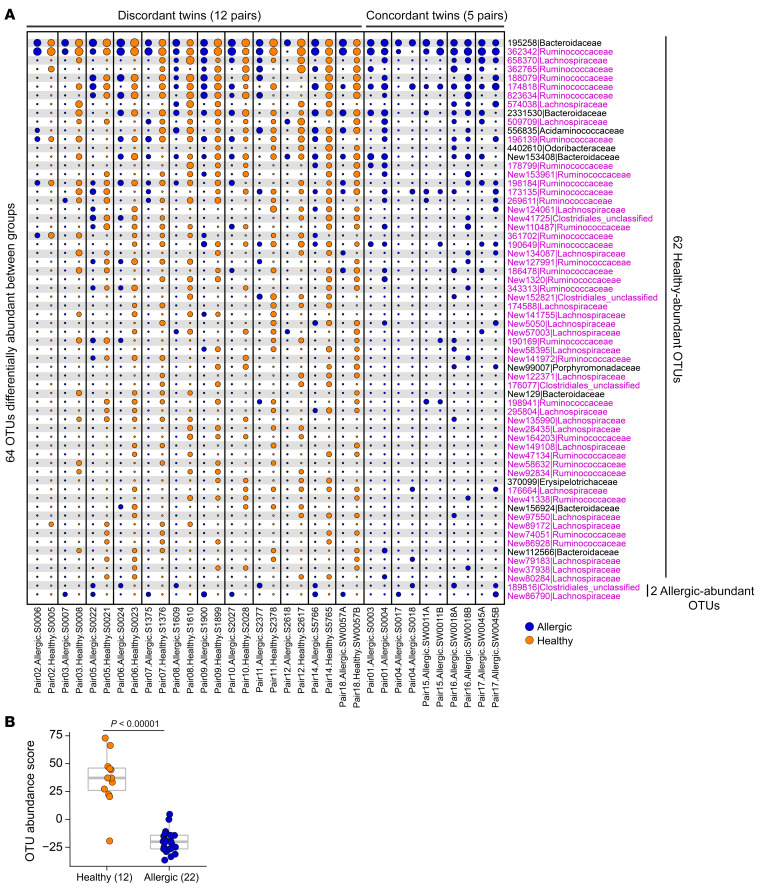
We next compared the microbial composition between allergic and healthy twin pairs and identified 64 OTUs differentially abundant between the 2 groups, with 62 OTUs higher in healthy twins (hereafter referred to as “healthy-abundant” OTUs), and 2 OTUs higher in allergic twins (hereafter referred to as “allergic-abundant” OTUs); this is shown in Figure 3, in the binary presence/absence heatmap in Supplemental Figure 3, and in Supplemental Tables 3 and 4. To better illustrate the within-pair OTU abundance differences, we show in Figure 4A that these 62 healthy-abundant OTUs were more abundant in the healthy twins compared with their allergic siblings, and the 2 allergic-abundant OTUs were more abundant in the allergic twins than their healthy siblings. Families in the Clostridia class constituted 84% of the healthy-abundant OTUs; these were annotated as Lachnospiraceae (n = 21), Ruminococcaceae (n = 28), or unclassified Clostridiales (n = 4) (Figure 4A, highlighted in pink). To develop an aggregated microbiome signature, we calculated a microbiota abundance score, taking into consideration the relative abundance of the 64 differentially abundant OTUs and their change in direction between groups (see Methods). The OTU abundance score was significantly higher in healthy relative to allergic twins across all samples (P < 0.00001) (Figure 4B) or within discordant twins only (P = 0.00049) (Supplemental Figure 4), as expected, because the score was calculated from preselected OTUs. Variance exists in the relative abundance scores for the discordant twin pairs (Supplemental Figure 4) because the majority of the differentially abundant OTUs are present in the healthy twins and absent in the allergic twins. If statistical comparisons are restricted to monozygotic twins only (14 pairs, 28 samples), the test statistics for the 62 healthy-abundant OTUs and the 2 allergic-abundant OTUs correlated with those of all twins (17 pairs, 34 samples) (Supplemental Figure 5 and Supplemental Table 4), and the OTU abundance scores remained significantly different between the healthy and allergic twin groups (Supplemental Figure 6).
Figure 3. Healthy twins exhibit a fecal microbial profile distinct from allergic siblings.
Relative abundance heatmap of the 64 OTUs identified to be differentially abundant between healthy (n = 12) and allergic (n = 22) twins. Of these 64 OTUs, 62 were more abundant in the healthy group (healthy-abundant OTUs), and 2 were more abundant in the allergic group (allergic-abundant OTU). OTU IDs are shown on the row in the format of “OTU_ID|Family,” and those annotated with the Clostridia class (Lachnospiraceae, Ruminococcaceae, unclassified Clostridiales) are highlighted in pink. Sample IDs are shown on the column, with annotation bars above the heatmap indicating concordant/discordant twin members, sex, and zygosity. A binary presence/absence heatmap of the 64 OTUs is shown in Supplemental Figure 3. Of 36 samples total in the twin cohort, 1 sample (S5077) failed sequencing and yielded 0 reads; therefore, the corresponding twin pair (no. 13) was excluded from 16S analysis. DS-FDR was used on all samples (P < 0.05) and 2-tailed Wilcoxon’s signed-rank test was used on discordant twin pairs (P < 0.10), respectively. Unadjusted P value thresholds were used to filter for OTUs of interest. After BH-FDR correction, no OTUs passed the FDR cutoff of 0.10 threshold, potentially due to small sample size.
Figure 4. Healthy and allergic twins exhibit within-twin pair differences in microbial composition.
(A) Bubble plot showing the per–twin pair abundance differences of the 64 OTUs shown in Figure 3 between the healthy and allergic groups. The size of each circle corresponds to the relative abundance of an OTU. Samples were arranged as discordant twins (12 pairs, n = 24), where one member is healthy and the other member is allergic; concordant twins (5 pairs, n = 10), where both members are allergic. (B) The aggregated OTU abundance score was significantly higher in healthy (n = 12) relative to allergic twins (n = 22). The score was calculated using the 64 differentially abundant OTUs from A. The score for discordant twin pairs only is shown in Supplemental Figure 4. Each dot denotes 1 sample. The bounds of the boxes represent the 25th and 75th percentiles, the horizontal center lines indicate the medians, and the whiskers extend to data points within a maximum of 1.5 times the IQR. In A, DS-FDR was used on all samples (P < 0.05) and 2-tailed Wilcoxon’s signed-rank test was used on discordant twin pairs (P < 0.10), respectively. Unadjusted P value thresholds were used to filter for OTUs of interest. After BH-FDR correction, no OTUs passed the FDR cutoff of 0.10 threshold, potentially due to small sample size. In B, 2-tailed Wilcoxon’s rank-sum test was used on all samples.
Healthy and allergic twins exhibit differential enrichment in fecal metabolic pathways.
Bacteria produce many metabolites that modulate the immune system and profoundly influence human health (26). Limited data exist on unbiased systematic profiling of fecal metabolites in patients with and without food allergy. We performed liquid chromatography–tandem mass spectrometry (LC-MS/MS) to measure the abundance of compounds in the same set of fecal samples from the twin cohort (Supplemental Figure 7). We identified 97 metabolites differentially abundant between the healthy and allergic twins, with 33 more abundant in healthy twins, and 64 more abundant in allergic twins (Figure 5 and Supplemental Tables 5 and 6). If statistical comparisons are restricted to monozygotic twins only (14 pairs, 28 samples), the test statistics for the 33 healthy-abundant metabolites and the 64 allergic-abundant metabolites correlated with those of all samples (18 pairs, 36 samples) (Supplemental Figure 8 and Supplemental Table 6). Among these 97 metabolites, 32 (16 higher in healthy, 16 higher in allergic) also reached a significance level of 0.10 within discordant twin pairs only (Supplemental Figure 9).
Figure 5. Healthy and allergic twins exhibit differential enrichment in fecal metabolic pathways.
(A) Of 36 samples, 33 metabolites were more abundant in the healthy (n = 13) group relative to the allergic (n = 23) group. Metabolites are shown on the row in the format of “COMP_ID|Biochemical_Name|Super_Pathway|Sub_Pathway.” Sample IDs are shown on the column, with annotation bars above the heatmap indicating concordant/discordant twin members, sex, and zygosity. (B) Of 36 samples, 64 metabolites were more abundant in the allergic group (n = 23) relative to the healthy (n = 13) group. Same annotations as in A. In A and B, 2-tailed Welch’s 2-sample t test was used on all samples (P < 0.10) and unadjusted P value thresholds were used to filter for individual metabolites of interest. After FDR correction, no individual metabolites passed the FDR cutoff of 0.10 threshold, potentially due to small sample sizes.
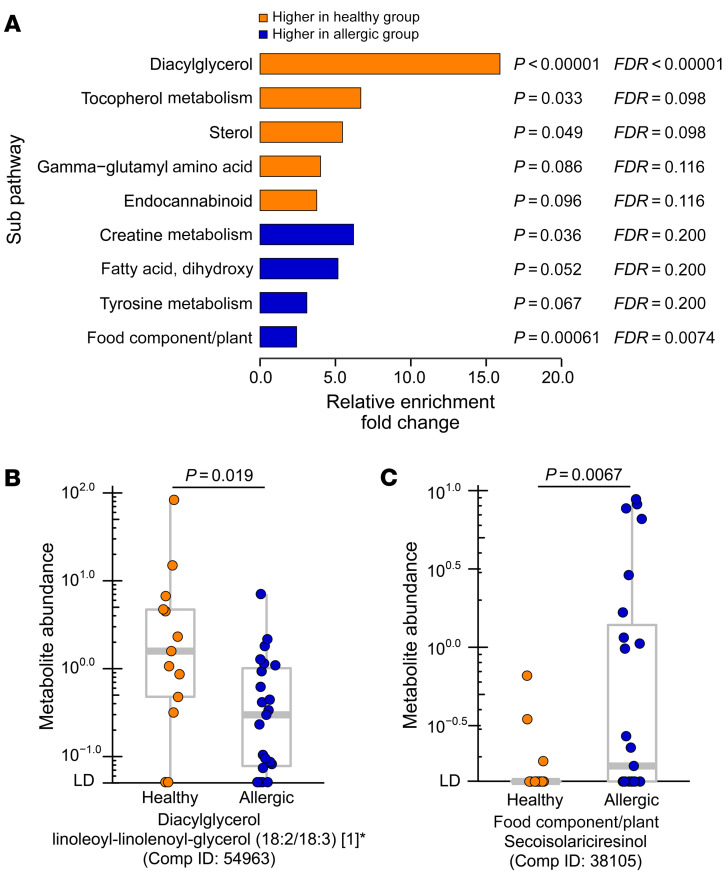
After annotating the 97 metabolites into superpathways and subpathways, healthy twins showed distinct enrichment at the pathway level compared with allergic twins (Figure 6A and Supplemental Table 7). Specifically, as shown in Figure 6A, among other pathways, the DAG subpathway was significantly enriched in the 33 metabolites more abundant in healthy twins (FDR-adjusted P < 0.00001), and the food component/plant subpathway was significantly enriched in the 64 metabolites more abundant in allergic twins (FDR-adjusted P = 0.0074). One of the DAG metabolites, linoleoyl-linolenoyl-glycerol (18:2/18:3) [1]*(Comp ID: 54963), was significantly higher (P = 0.0036) in healthy twins compared with allergic twins in discordant pairs (Supplemental Figure 10A) and was significant in the twin cohort overall (Figure 6B, P = 0.019). On the other hand, secoisolariciresinol (Comp ID: 38105) (SECO) from the food component/plant pathway was higher in allergic twins compared with healthy twins (P = 0.0067) (Figure 6C), with the same trend observed in discordant twin pairs (P = 0.094) (Supplemental Figure 10B).
Figure 6. Distinct metabolic pathways are enriched in healthy and allergic twins.
(A) Metabolites more abundant in the healthy group (from Figure 5A) or in the allergic group (from Figure 5B) were enriched in different subpathways shown. Relative enrichment fold change is shown on the x axis, and the name of subpathway is shown on the y axis. P value and FDR-adjusted P value of each subpathway enrichment are shown next to each horizontal bar. (B and C) Representative examples of metabolites in the enriched subpathways in the healthy or allergic group. (B) The linoleoyl-linolenoyl-glycerol (18:2/18:3) [1]* (subpathway: Diacylglycerol) was higher in healthy (n = 13) compared with allergic (n = 23) twin members. (C) The secoisolariciresinol (subpathway: Food Component/Plant) was higher in allergic twin pairs (n = 23) compared with healthy twin pairs (n = 13). Supplemental Figure 10 shows the result of discordant twin pairs only that correspond to metabolites shown in B and C. In B and C, units shown on the y axis represent the normalized raw area counts of UPLC-MS/MS peaks, rescaled to set the median equal to 1.00 for each biochemical (see Methods). Each dot denotes 1 sample. The bounds of the boxes represent the 25th and 75th percentiles, the horizontal center lines indicate the medians, and the whiskers extend to data points within a maximum of 1.5 times the IQR. In A, the hypergeometric test was used to compute the P values of relative enrichment of metabolite subpathways and filtered by FDR-adjusted P < 0.10. Pathways consisting of at least 2 significant metabolites were included in the statistical test. After BH-FDR multiple-testing correction DAG remained as the most significantly enriched subpathway in metabolites more abundant in healthy twins (FDR-adjusted P < 0.00001). In B and C, 2-tailed Welch’s 2-sample t test was used on all samples.
In an attempt to interpret the source of these metabolites, we compared our data with an internal microbial metabolite database from Metabolon Inc. (accessed October 24, 2019). We note that the database is under active development and does not represent a complete collection of microbiota-derived metabolites. Among the 992 metabolites with annotated pathways that we examined, 129 overlapped with the Metabolon database (Supplemental Table 8). Of the 129 overlapping metabolites, 66 were marked with discovery sites, such as colon, feces, urine, plasma, tissues, or multiple sites; 38 (58%) of the 66 metabolites were from colon or feces. In addition, of the 129 metabolites, 13 were among the 97 compounds differentially abundant between healthy and allergic twins, including 1-methylhistamine, 3-hydroxyphenylacetate, 3,4-dihydroxyphenylacetate, betaine, skatol, ethylmalonate, creatine, creatinine, putrescine, phenylacetylglycine, taurolithocholate 3-sulfate, biotin, and D-urobilin (Supplemental Table 8). Additionally, we profiled 8 short-chain fatty acids (SCFAs) using GC-MS technology: 2-methylbutyric acid, acetic acid, butyric acid, hexanoic acid, isobutyric acid, isovaleric acid, propionic acid, and valeric acid. We then compared the abundance of these SCFAs between allergic and healthy twins. Within the 13 discordant twin pairs, no SCFA reached P < 0.10, potentially due to the single point-in-time analysis (Supplemental Table 9).
Identifying microbes and metabolites associated with health.
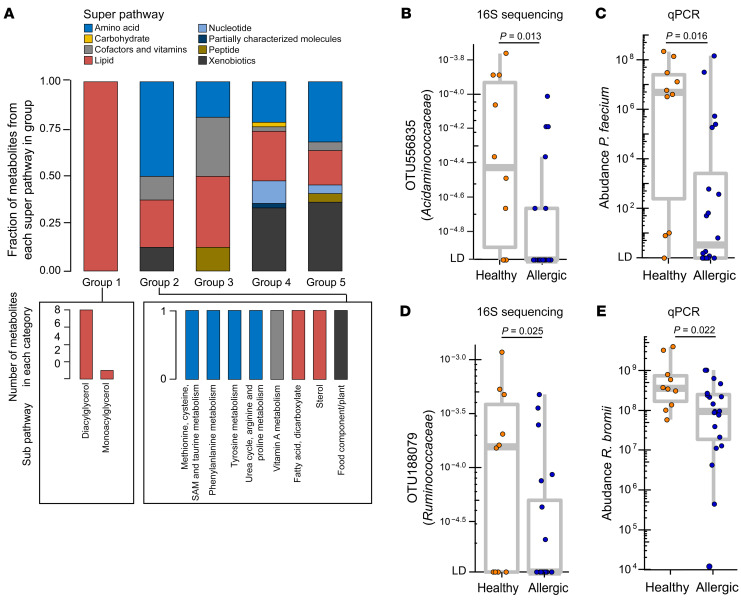
After demonstrating the differential abundance of both OTUs and metabolites in the healthy and allergic twins, we next correlated these 2 data sets to identify any bacterial species or metabolites that may be mechanistically related to health in our cohort. Overall, the OTUs differentially abundant between healthy and allergic twin groups were correlated with different sets of metabolites and pathways. We correlated the abundance of 64 differentially abundant OTUs with the 97 metabolites (Supplemental Figure 11 and Supplemental Table 10) and identified 21 healthy-abundant OTUs and 1 allergic-abundant OTU with consistent correlation across metabolites at the per-sample level (Figure 7 and Supplemental Table 11). We divided the metabolites into 5 categories based on their abundance correlation consistency among OTU clusters 1–3 (consisting of 21 healthy-abundant OTUs; cluster 4 only contains 1 OTU from an allergic-abundant taxon and was not used for metabolite group annotation). The 5 metabolite groups are as follows (Figure 7 and Supplemental Figure 12): group 1, positively correlated with the 3 OTU clusters, stronger in clusters 1 and 2 relative to cluster 3 (n = 9); group 2, positively correlated with the 3 OTU clusters, stronger in cluster 3 relative to clusters 1 and 2 (n = 8); group 3, correlated with OTU clusters with mixed patterns (n = 16); group 4, negatively correlated with the 3 OTU clusters, stronger in cluster 1 relative to clusters 2 and 3 (n = 46); and, group 5, negatively correlated with the 3 OTU clusters with similar distribution (n = 22). These 5 metabolite groups showed distinctly different distributions of metabolite superpathways and subpathways (Figure 8A). In particular, group 1 was dominated by metabolites from the lipid superpathway, including DAG and monoacylglycerol (Figure 8A), whereas amino acid metabolism, including tyrosine, phenylalanine, arginine, proline, methionine, cysteine, S-adenosylmethionine, and taurine, was enriched in group 2 (Figure 8A).
Figure 7. The OTUs differentially abundant between healthy and allergic groups are correlated with different sets of metabolites and pathways.
Of 64 OTUs from Figure 3, 4 OTUs showed a strong correlation with the 97 metabolites from Figure 5, A and B. The filtering of OTUs is illustrated in the analytical workflow (Supplemental Figure 1). Metabolites are shown on the row in the format of “COMP_ID|Biochemical_Name|Super_Pathway|Sub_Pathway,” and OTU IDs are shown on the column in the format of “OTU_ID|Family.” Three OTUs that match to bacteria species at >99% identity are bolded. On the heatmap, between each OTU and each metabolite, a positive correlation is shown in red, and a negative correlation is shown in blue. OTUs were divided into 4 clusters based on same height on the dendrogram shown on the column using R function cut.tree. Similarly, metabolites were divided into 5 groups based on same height on the dendrogram shown on the row. Annotation to metabolite groups 1–5 was added based on the distribution of Spearman’s correlation coefficient ρ among the healthy-abundant OTU clusters 1–3 consisting of 21 OTUs (Supplemental Figure 12). Cluster 4 only contains 1 OTU from allergic-abundant bacteria and, hence, was not used for metabolite group annotation. Spearman’s correlation was used.
Figure 8. Two bacterial species correlated with pathways that were differentially abundant between healthy and allergic twins.
(A) Distribution of pathways in group 1 and 2 metabolites from Figure 7. Top: Superpathways in each group. The fraction of metabolites from each superpathway on the y axis was calculated by the number of metabolites that belong to this pathway divided by the total number of metabolites in a group. Bottom: Number of metabolites that belong to each subpathway; (left) group 1, (right) group 2. SAM, S-adenosylmethionine. (B) OTU 556835 (family Acidaminococcaceae) is significantly more abundant in the healthy group compared with the allergic group by 16S sequencing. This OTU was annotated as Phascolarctobacterium faecium at the species level. (C) Quantitative PCR (qPCR) validates the abundance differences between healthy and allergic groups using P. faecium–specific primers. (D) OTU188079 (family Ruminococcaceae) is significantly more abundant in the healthy group compared with the allergic group by 16S sequencing. This OTU was annotated as Ruminococcus bromii at the species level. (E) qPCR validates the abundance differences between healthy and allergic groups using R. bromii–specific primers. Units shown on the y axis in C and E represent 2–Ct normalized to total 16S rRNA copies per gram of fecal material and multiplied by a constant (1 × 1022) to bring all values above 1 (see Methods). In B–E, n = 30 samples (15 twin pairs) with DNA available for qPCR validation are shown (10 healthy, 20 allergic). Each dot denotes 1 sample. The bounds of the boxes represent the 25th and 75th percentiles, the horizontal center lines indicate the medians, and the whiskers extend to data points within a maximum of 1.5 times the IQR. DS-FDR was used in B and D. In C and E, qPCR data were log10 transformed, and 2-tailed Wilcoxon’s rank-sum test was used.
To annotate the 22 metabolite-correlated OTUs at species-level resolution, we searched the assembled 16S sequence of each OTU against NCBI’s Bacteria/Archaea 16S reference database using BLAST (27). At a sequence identity of 99% or higher, OTU556835 was matched to P. faecium (accession ID NR_026111.1) and both OTU188079 and OTU823634 were matched to R. bromii (accession ID NR_025930.1). The other OTUs (abundant in either healthy or allergic twins) did not have matches meeting the identity threshold. Quantitative PCR (qPCR) validated the significantly higher abundance of P. faecium in healthy twins compared with allergic twins (P = 0.016) (Figure 8, B and C; Supplemental Figure 13; and Supplemental Table 12). P. faecium is an obligate anaerobic non-spore-forming bacterium that consumes succinate and produces SCFAs, including acetate and propionate (20, 21). P faecium was grouped in cluster 1 and was most highly correlated with a number of DAG metabolites. P faecium was also strongly positively correlated with tocopherol and negatively correlated with a variety of metabolites, including those from the secondary bile acid metabolism pathway. R. bromii was also qPCR validated to be enriched in the healthy compared with the allergic twin group (P = 0.022) (Figure 8, D and E; Supplemental Figure 14; and Supplemental Table 12). R. bromii is a strictly anaerobic, spore-forming Clostridia important for the degradation of dietary resistant starch (22). It was associated with group 2 metabolites involved in fatty acid, amino acid, and sterol metabolism.
Discussion
From our investigation of bacterial abundance in fecal samples from well-characterized healthy and allergic twins, we identified a unique microbiota signature consisting of 64 OTUs (62 healthy-abundant and 2 allergic-abundant) that was significantly different between the 2 groups using 16S rRNA gene amplicon sequencing. In addition, we developed a microbial abundance score that is higher in healthy twins compared with that in allergic twins. The 64 OTUs showed marked enrichment of bacteria in the Clostridia class, particularly the families Lachnospiraceae and Ruminococcaceae, in the healthy twins. We first described a role for mucosa-associated intestinal bacteria in the Clostridia class in protecting mice from allergic sensitization to peanuts (17). We went on to show that the composition of the fecal microbiota is altered in infants with cow’s milk allergy (28). When we colonized germ-free mice with the feces of healthy or cow’s milk allergic (CMA) infants we discovered that mice colonized with CMA infants’ microbiota produced an anaphylactic response to the cow’s milk allergen β-lactoglobulin, while mice colonized with healthy infants’ microbiota were protected against such an allergic response (18). We developed a microbiota signature that distinguished the CMA from healthy populations in both human donors and colonized mice. Correlating ileal bacterial taxa with differentially expressed genes in the ileal epithelium from healthy-colonized mice allowed us to identify a Clostridial species, Anaerostipes caccae, that mimicked the effects of the healthy microbiota, thus providing proof of concept that bacteria or their products can protect against an allergic response to food. Our results align with findings from other groups (24, 29, 30), suggesting that fecal microbiome signatures are different between healthy children and children with food allergies.
In this report we have used positive correlation with distinct metabolic pathways to identify two potentially new allergy protective bacterial species, the Clostridia R. bromii and P. faecium, a species from a taxon not previously associated with protection against allergy. R. bromii has been described as a keystone species in fiber degradation (22), and its abundance in feces significantly increases upon dietary intervention with resistant starch (31, 32). Individuals that do not have detectable R. bromii in the feces before beginning a resistant starch–supplemented diet consume about 40% less total starch than individuals colonized with R. bromii, demonstrating that this species alone is responsible for much of the total starch digestion in the colon (31, 32). R. bromii produces acetate and propanol but does not produce butyrate (23). However, R. bromii is involved in the primary stages of resistant starch digestion, which is correlated with increased butyrate production downstream (33). Interestingly, in the Canadian Healthy Infant Longitudinal Development birth cohort, depletion of R. bromii early in life (3 and 12 months of age) was associated with the development of atopy and reduced genetic potential to produce butyrate (34).
Mouse model studies from our laboratory showed that protection against allergic sensitization to food requires a bacteria-induced barrier protective response mediated by the cytokine IL-22 (17). Interestingly, recent work has shown that P. faecium’s ability to protect against Clostridium difficile infection in mice depends on IL-22–mediated glycosylation of intestinal mucus in the absence of succinate (35). Genes involved in this glycosylation and IL22RA2 are differentially expressed in colonic tissue of patients with active or inactive ulcerative colitis (35), which correlates well with previous data showing decreased abundance of Phascolarctobacterium species in adults with inflammatory bowel diseases (36).
To the best of our knowledge, no data yet exist on unbiased systematic profiling of fecal metabolites in patients with food allergy compared with controls. Among the metabolites that were significantly differentially abundant between healthy and allergic twins, we identified those derived from microbiota, which included but are not limited to histamine metabolites (1-methylhistamine), 3,4-dihydroxyphenylacetate, betaine, skatol, creatine, creatinine, putrescine, phenylacetylglycine, and taurolithocholate 3-sulfate, all of which are higher in allergic siblings, and biotin, 3-hydroxyphenylacetate, ethylmalonate, and D-urobilin, all of which are higher in healthy siblings.
The DAG subpathway was the most significantly different between healthy and allergic twins (FDR-adjusted P < 0.00001) and was enriched in the 33 metabolites more abundant in healthy twins. In addition to P. faecium (cluster 1), metabolites in the DAG subpathway were strongly correlated with other bacteria in OTU clusters 1 and 2, many of which are from the Clostridia family. Several reports have shown that Clostridia can produce DAG (37) and may make DAG more bioavailable to the host by facilitating its conversion from dietary phospholipids (38). Whether DAG metabolism is reflective of a healthy microbiota and whether or how it contributes to protection against allergy will be the subject of future studies. However, the identification of readily measurable metabolites that distinguish healthy and allergic twins has important implications for the development of microbiome-modulating therapeutics because of their potential as biomarkers, particularly in clinical trials. Laboratory-based assays measuring, in particular, DAG may have great utility as biochemical indicators of therapeutic interventions that shift the microbiota toward health.
The food component/plant pathway was most significantly enriched in allergic twins after FDR correction (FDR-adjusted P = 0.0074), particularly the metabolite SECO. SECO is commonly observed as an intermediate product in the bacteria-mediated breakdown of plant-derived lignans into enterolignans, such as enterodiol and enterolactone, which have numerous benefits for human health (39–41). Several bacterial species and genes are involved in the multistep process of lignan metabolism (40, 42, 43). Of particular interest, the gene glm codes for an enzyme that methylates SECO into dmSECO, allowing further biotransformation into enterodiol by other bacteria. Phylogenetic analysis suggests that glm is expressed by a wide variety of bacteria, the majority of which are Lachnospiraceae (43). The abundance of SECO in feces was negatively correlated with the abundance of several OTUs in healthy individuals (many of which were Lachnospiraceae or Ruminococcaceae). We infer that the high abundance of SECO in the feces of allergic twins supports our taxonomic analysis, as the buildup of this intermediate product may be a direct effect of the lower abundance of Clostridia in these individuals.
A strength of our study is that this twin cohort allows us to examine lasting differences between genetically similar “littermate-controlled” subjects with similar lifestyle in childhood in a way that is not often possible with human samples. We have controlled for many potential demographic variables by sampling healthy and allergic twins across a broad age range. It is known that healthy and allergic children have differences in the abundance of allergy-protective Clostridia (18, 24, 28–30), and here we show that these same signatures persist into adulthood. The broad age range in this study provides a unique context to study how these early-life changes persist, despite years of separation and lifestyle changes between twins/siblings. We acknowledge, however, that the small sample size is a limitation to our report.
In summary, we performed an integrated microbial and metabolomic analysis of human fecal samples from allergic and healthy twins and identified differentially abundant bacteria and metabolic compounds as well as pathways between the 2 groups. We developed a microbiota abundance score dominated by bacteria with greater abundance in healthy twins and used positive correlation with distinct metabolite pathways to identify P. faecium and R. bromii as significantly enriched in the healthy twins. Our data demonstrate that the gut microbiota may play a protective role in patients with food allergies beyond the infant stage and through adulthood. Our findings warrant further studies in larger populations to uncover the mechanism(s) underlying microbiota-mediated modulation of systemic effects in food allergy and to provide insight into new interventions to treat and prevent food allergy.
Methods
Study design.
The design of the analytical workflow is shown in Supplemental Figure 1, which illustrates the enrollment of the twin pairs that were food allergic and those that were healthy. Collected fecal samples were analyzed for microbes and metabolites as described below. Wilcoxon’s rank-sum tests were used as statistical tests for analysis of data.
Human fecal sample collection.
Participants in this study were recruited as part of an observational study (ClinicalTrials.gov NCT01613885) at multiple sites (Stanford Main Hospital and Clinics, Palo Alto, California, USA and El Camino Hospital Stanford/Lucile Packard Children’s Hospital Stanford, Mountain View, California, USA) from 2014 to 2018. Food-allergic participants in this study were diagnosed with food allergy by a staged and validated food challenge (14) performed by trained center staff. Fecal samples were collected per a standard operating procedure developed by the NIH Microbiome Project (44). Fecal samples were collected from 18 pairs of twins. Among 18 twin pairs, 13 were discordant for food allergy (one sibling had food allergy and the other was healthy) and 5 were concordant for food allergy (both siblings had food allergy) (Table 1). No one was on any medications or experienced any respiratory infections (e.g., cold, flu, pneumonia) at the time of fecal sample collection. For food diary records see Supplemental Table 1. All samples that passed quality control (QC) (n = 34 for microbiota analysis; n = 36 for metabolite analysis) were used for statistical comparisons.
16S rRNA–targeted library preparation and sequencing.
Bacterial DNA was extracted from fecal samples of the twin cohort using the Power Soil DNA Isolation Kit (MoBio). 16S rRNA–targeted gene amplicon library preparation and sequencing were performed at the Environmental Sample Preparation and Sequencing Facility at Argonne National Laboratory (DuPage, Illinois, USA). The V4 region of the 16S rRNA gene was amplified by PCR with region-specific primers 515F (Parada) to 806R (Apprill) (45, 46) that include sequencer adapter sequences used in the Illumina flowcell. 151 bp paired-end reads with 12 bp barcodes were generated following previously described protocols (47) on an Illumina MiSeq instrument. On average, each sample yielded 183,952 ± 7011 (mean ± SEM; ranging from 94,917 to 268,423) read pairs. One sample (S5077, allergic sibling of a twin pair) failed sequencing and yielded 0 reads and the corresponding twin pair (no. 13) was therefore excluded from 16S data analysis. A total of 34 samples (from 22 allergic and 12 healthy twins) were kept for further analysis, including 12 discordant twin pairs (n = 24), 5 concordant twin pairs (n = 10).
16S rRNA microbial quantification and normalization.
The microbial 16S rRNA–targeted gene amplicon sequencing data were processed by Quantitative Insights into Microbial Ecology (version 1.9) (48) using a procedure similar to one we have previously described (18). In brief, low-quality bases were trimmed at 5′ end of raw paired-end reads and 3′ overlapping mates were merged by SeqPrep (v1.2) (https://github.com/jstjohn/SeqPrep; master branch, commit ID bda7a3d). The open reference OTU picking protocol was used at 97% sequence identity against the Greengenes database (08/2013 release) (49). Reads were aligned to reference sequences using PyNAST (50), and taxonomy was assigned using uclust consensus taxonomy assigner (51). Chimeric sequences were identified and removed using ChimeraSlayer (v20110519) (52). Sequences with “Unassigned” taxa at the kingdom level were also removed. Data were then rarefied to an even depth of 92,670 sequences across all samples (n = 34, the twin cohort). α Diversity (Shannon index) was compared between the allergic and healthy groups using Wilcoxon’s rank-sum test (unpaired, nonparametric) for all samples or Wilcoxon’s signed-rank test (paired, nonparametric) within the discordant twin pairs only. β Diversity metrics were calculated and compared between the 2 groups using permutational multivariate analysis of variance (PERMANOVA) with weighted UniFrac distance in R package vegan (v2.4.5) (53).
Differentially abundant microbial taxa identification.
Bacterial taxa differentially abundant between allergic and healthy groups of the twin cohort were identified using the following approach. First, OTUs present in fewer than 4 samples were removed. Second, for all samples (n = 34), relative abundance of OTUs was compared between the 2 groups using discrete FDR (DS-FDR) (54) method (hereafter referred to as test no. 1) with parameters “transform_type = normdata, method = meandiff, alpha = 0.10, numperm = 1000, fdr_method = dsfdr” (accessed 10102018) (https://github.com/biocore/dsFDR; master branch, commit ID 51521d7). The DS-FDR algorithm computes test statistics, raw P values, and estimates FDR from a permutation test (default 1000 permutations). Of 5590 OTUs total, 180 reached P < 0.05; none reached FDR cutoff of 0.10 potentially due to sample size. Within the discordant twin pairs (n = 24, from 12 pairs), for which one sibling is allergic and the other is healthy, the relative abundance of OTUs was compared between groups using Wilcoxon’s signed-rank test (paired, nonparametric) (hereafter referred to as test no. 2). Of 5590 OTUs total, 259 reached a significance level of P < 0.10. A more lenient P value cutoff (0.10) was used here considering that nonparametric rank-based method has less power than the DS-FDR method (54). After Benjamini-Hochberg FDR (BH-FDR) correction (55) for multiple testing, no OTUs passed the FDR cutoff of 0.10, potentially due to small sample sizes. Between 180 OTUs returned by test no. 1 and 259 OTUs returned by test no. 2, 64 OTUs overlapped and showed consistent direction of change in abundance (more abundant in healthy or allergic) in both tests and were kept for further analysis (Supplemental Table 4).
OTU abundance score calculation.
Of 64 OTUs differentially abundant between allergic and healthy twin members, 62 were healthy-abundant OTUs and 2 were allergic-abundant OTUs (Supplemental Table 4, Figure 3, and Figure 4A). The limited number of allergic-abundant OTUs did not warrant the calculation of an OTU ratio as we had previously (18), defined as the total number of potentially healthy-abundant OTUs divided by the total number of potentially allergic-abundant OTUs per sample. We were able to compute an OTU abundance score as an aggregated signature taking into consideration the relative abundance of 64 OTUs shown in Figure 4B. First, the rarefied absolute count matrix of OTUs was added by offset 1.0 and log10-transformed to bring data close to Gaussian distribution, and then data were scaled by dividing the value by their root mean square across samples. The abundance of allergic-abundant OTUs was multiplied by –1. Second, the sum of the transformed abundance of the 64 OTUs was calculated to generate the aggregate score.
qPCR.
The presence of P. faecium and R. bromii in fecal samples was confirmed using qPCR with species-specific primers for the 16S gene. Bacterial DNA was extracted using the Power Soil DNA Isolation Kit (MoBio), and qPCR was performed with PowerUp SYBR green master mix (Applied Biosystems) using 4 μL of each primer at 10 μM working dilution and 2 μL of bacterial DNA. Primers are listed in Supplemental Table 13. For normalization purposes, widely recognized primers 8F (56) and 338R (57) were used to quantify total bacterial abundance. For P. faecium, we used primers from a previously published study (21) and reannotated the taxonomy of the corresponding OTU556835 as family Acidaminococcaceae, genus Phascolarctobacterium. For R. bromii, we used primers from ref. 32. The cycling conditions for P. faecium–specific qPCR consisted of an activation cycle of 95°C for 2 minutes; followed by 40 cycles at 95°C for 15 seconds, 58°C for 30 seconds, and 72°C for 60 seconds; and a final extension period at 72°C for 5 minutes (21). The cycling conditions for R. bromii–specific qPCR consisted of an activation cycle of 95°C for 5 minutes; followed by 40 cycles at 95°C for 30 seconds, 52°C for 30 seconds, and 72°C for 2 minutes; and a final extension period at 72°C for 8 minutes (32). The fluorescent probe was detected in the last step of this cycle. A melt curve was performed at the end of the PCR to confirm the specificity of the PCR product. Relative abundance is expressed as 2–Ct normalized to total 16S rRNA copies per gram of fecal material and multiplied by a constant (1 × 1022) to bring all values above 1 and log10 transformed before statistical analysis.
Metabolic profiling sample preparation.
The metabolic profiling of human fecal samples was performed by Metabolon Inc. All samples were maintained at –80°C until processed. Samples were prepared using the automated MicroLab STAR system from Hamilton Company. Recovery standards were added prior to the first step in the extraction process for QC purposes. To remove protein, to dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites proteins were precipitated with methanol under vigorous shaking for 2 minutes (Glen Mills GenoGrinder 2000) followed by centrifugation. The resulting extract was divided into 5 fractions: 2 for analysis by 2 separate reverse phase/ultrahigh-performance liquid chromatography–tandem mass spectroscopy (RP/UPLC-MS/MS) methods with positive ion mode electrospray ionization (ESI), 1 for analysis by RP/UPLC-MS/MS with negative ion mode ESI, and 1 for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI; 1 sample reserved for backup. Samples were placed briefly on a TurboVap (Zymark) to remove the organic solvent. The sample extracts were stored overnight under nitrogen before preparation for analysis.
Metabolic profiling sample QA/QC.
Three types of controls were analyzed together with the experimental samples: (a) a pooled matrix sample generated by taking a small volume of each experimental sample as a technical replicate throughout the data set, (b) extracted water samples as process blanks, and (c) a cocktail of QC standards carefully chosen not to interfere with the measurement of endogenous compounds was spiked into every analyzed sample, allowing for monitoring of instrument performance and facilitating chromatographic alignment. Instrument variability was determined by calculating the median relative standard deviation (RSD) for the standards that were added to each sample prior to injection into the mass spectrometers (3% median RSD in this study). Overall process variability was determined by calculating the median RSD for all endogenous metabolites (i.e., noninstrument standards) present in 100% of the pooled matrix samples (7% median RSD in this study). All 36 fecal samples passed QC and were included in the metabolic data analysis.
UPLC-MS/MS.
All methods utilized a Waters ACQUITY UPLC and a Thermo Scientific Q-Exactive high-resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. The sample extract was dried and reconstituted in solvents compatible with each of the 4 methods. Each reconstitution solvent contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 μm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). Another aliquot was also analyzed using acidic positive ion conditions; however, it was chromatographically optimized for more hydrophobic compounds. In this method, the extract was gradient eluted from the same previously mentioned C18 column using methanol, acetonitrile, water, 0.05% PFPA, and 0.01% FA and was operated at an overall higher organic content. Another aliquot was analyzed using basic negative ion optimized conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using methanol and water, however, with 6.5 mM ammonium bicarbonate at pH 8. The fourth aliquot was analyzed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10 mM ammonium formate at pH 10.8. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slightly between methods but covered 70–1000 m/z.
Compound identification and curation.
UPLC-MS/MS raw data were extracted, peak-identified, and QC-processed by Metabolon Inc. Compounds were identified by comparing them to internal library entries of purified standards or recurrent unknown entities. The library was based on authenticated standards that contain the retention time/index (RI), mass-to-charge ratio (m/z), and chromatographic data (including MS/MS spectral data) on all molecules present in the library. Furthermore, biochemical identifications were based on 3 criteria: (a) retention index within a narrow RI window of the proposed identification, (b) accurate mass match to the library ± 10 ppm, and (c) the MS/MS forward and reverse scores between the experimental data and authentic standards. The MS/MS scores were based on comparing the ions in the experimental spectrum to the ions in the library spectrum. While there may be similarities among these molecules based on one of these factors, the use of all 3 data points can be used to distinguish and differentiate biochemicals. More than 3300 commercially available purified standard compounds were acquired and registered for analysis on all platforms to determine their analytical characteristics. Additional mass spectral entries were created for structurally unnamed biochemicals, which were identified by virtue of their recurrent nature (both chromatographic and mass spectral). Entries were further processed by manual curation to ensure accurate and consistent identification of true chemical entities and to remove those representing system artifacts, misassignments, and background noise.
Metabolite quantification and normalization.
UPLC-MS/MS peaks were quantified by the area under the ROC curve. Data were normalized to correct for variations resulting from instrument interday tuning differences using the median-centered method. In this method, each compound was corrected in run-day blocks by registering the median as 1.00 and normalizing each data point proportionately, hereafter referred to as block correction, and further normalized to account for differences in metabolite levels due to differences in the quantities of material in each sample.
Differentially abundant metabolite identification.
Metabolites differentially abundant across all samples from the allergic and healthy twin groups (n = 36) were identified using Welch’s 2-sample t test after log10 transformation. A total of 1308 metabolites were quantified. After removing metabolites without pathway annotation, 992 metabolites were kept for statistical comparisons. Among those, in comparing allergic with healthy groups, 97 metabolites reached a significance level of P < 0.10 and were kept for further analysis. After BH-FDR correction for multiple testing, none of the metabolites passed the FDR cutoff of 0.10, potentially due to small sample size. Additionally, the abundance of metabolites was compared between the 2 groups within discordant twins only (n = 26, from 13 twin pairs) using paired t test. A sample (S5077) and the corresponding twin pair (no. 13) excluded from microbial 16S data analysis were kept in the metabolic profiling analysis.
Correlation of bacterial taxa and metabolite abundance.
Pairwise Spearman’s rank correlation was computed among the 64 OTUs and 97 metabolites differentially abundant between allergic and healthy twins. OTUs were further prioritized using the following approach. First, OTUs were filtered for those that showed a correlation at P < 0.05 with at least 5 differentially abundant metabolites from the designated group. For analysis of potentially healthy-abundant OTUs (more abundant in the healthy group), OTUs were correlated with metabolites from group “up in healthy” or “down in healthy”; the potentially allergic-abundant OTUs (more abundant in allergic group) were correlated with metabolites from group “Up in allergic” or “Down in allergic.” Second, OTUs that passed step 1 were filtered further for those that showed a relatively consistent trend of positive correlation (Spearman’s ρ > 0.20) across at least 30% of the metabolites from the designated group. For analysis of potentially healthy-abundant OTUs, OTUs were correlated with metabolites from group “up in healthy” and “down in allergic” joined; the potentially allergic-abundant OTUs were correlated with metabolites from group “up in allergic” and “down in healthy” together. Third, OTUs that passed steps 1 and 2 were further filtered to select those at P < 0.05 from the DS-FDR method comparing allergic to healthy groups across all samples. Of the 64 OTUs, 22 passed these correlation filters and are shown in Figure 7. After a BLAST search of assembled 16S sequences against the NCBI database (16S ribosomal RNA, Bacteria and Archaea) (accessed September 12, 2020) using blastn (27), 3 of 22 OTUs were matched to bacterial species at 99% or higher sequence identity: OTU556835, matched to P. faecium (accession ID NR_026111.1, 99.21% identity); OTU188079 and OTU823634, both matched to R. bromii (accession ID NR_025930.1; 99.21% identity).
Data availability.
The 16S rRNA–targeted sequencing raw FastQ data files have been deposited into the NCBI Sequence Read Archive (SRA ID PRJNA663708) and made publicly available. Processed data files are provided as Supplemental Tables 2–12.
Code availability.
The open-source analysis software used in this study is publicly available and referenced as appropriate. Custom codes are available from the corresponding author upon request.
Statistics.
The DS-FDR method (54) was used to identify differentially abundant OTUs by comparing allergic to healthy twins. Welch’s 2-sample t test was used to identify differentially abundant metabolites comparing allergic to healthy twins. Unless stated otherwise, Wilcoxon’s rank-sum test was used for comparing groups using all samples; if only within discordant twins, Wilcoxon’s signed-rank test was used. For metabolites, paired t test was used to compare metabolite abundance between the 2 groups within discordant twins after log10 transformation. We analyzed metabolic subpathway enrichment using the hypergeometric test, requiring at least 2 metabolites annotated with each subpathway. Following Wilcoxon’s rank-sum test or Wilcoxon’s signed-rank test, and hypergeometric test, we used the BH-FDR method (55) for multiple-testing correction. For pairwise comparisons of metabolite Spearman’s correlation coefficients between OTU clusters, Tukey’s honestly significant difference test was used. Other statistical tests used included PERMANOVA, as indicated in the figure legends. Two-tailed Fisher’s exact test was used for contingency tables. For comparison of healthy and allergic groups across all samples in 16S analysis, qPCR validation, and Spearman’s correlation between OTUs and metabolites, a P value less than 0.05 was considered significant. For comparison of healthy and allergic groups across all samples in metabolite analysis, comparison of healthy and allergic groups within discordant twin pairs in 16S or metabolite analysis, and SCFA analysis, a P value less than 0.10 was considered significant. For metabolite subpathway enrichment analysis, an FDR-adjusted P value less than 0.10 was considered significant. All tests were 2 tailed.
Study approval.
All aspects of this study were approved by the ethics committee of Stanford University and Stanford University Institutional Review Board (IRB-19495). Written informed consent was obtained from the parents and/or guardians of all children involved in the research.
Author contributions
RB, KCN, and CRN designed the study. LAH performed experiments and analyses of the qPCR data. RB performed data acquisition, organization, and all bioinformatic analysis of the 16S rRNA gene amplicon sequencing and metabolite data. RB, LAH, KCN, and CRN interpreted results. KCN cared for patients and provided donor fecal samples. XZ assisted with recruitment of patients with KCN and prepared clinical tables. RB, LAH, ZH, KCN, and CRN wrote the manuscript. All authors read and commented on the manuscript.
Supplementary Material
Acknowledgments
We thank M. Bauer for preparing the bacterial DNA from the fecal samples for both the taxonomic and metabolomic analyses. We thank M. Mimee and S. Light (University of Chicago) for critical review of the manuscript. We are grateful to A. Mirmira (University of Chicago) and C. Dant (Stanford University) for their edits. We thank the children and families for their participation in this study. The bioinformatics analysis was performed on the high-performance computing (HPC) cluster at the Gardner at Center for Research Informatics (University of Chicago) and on the HPC cluster HTC at the Center for Research Computing (University of Pittsburgh). We thank M. Jarsulic (University of Chicago) and F. Mu (University of Pittsburgh) for their technical assistance in software installation and job execution on the HPCs. This work was supported by the Sunshine Charitable Foundation, the Moss Family Foundation, and National Institute of Allergy and Infectious Diseases (NIAID) (R56AI134923 to CRN) as well as the Sean N. Parker Center for Allergy and Asthma Research, the National Heart, Lung, and Blood Institute (R01 HL 118612), the Orsak Family, the Kepner Family, the Stanford Institute for Immunity, Transplant and Infection, and NIAID (R01AI 140134 to KCN).
Version 1. 01/19/2021
Electronic publication
Footnotes
Conflict of interest: KCN reports grants from Allergenis and Ukko Pharma and research sponsorship by Novartis, Sanofi, Astellas, and Nestle as well as personal fees from Regeneron, Astrazeneca, Immuneworks, and Cour Pharmaceuticals. KCN is involved in clinical trials at Regeneron, Genentech, AImmune Therapeutics, DBV Technologies, AnaptysBio, Adare Pharmaceuticals, and Stallergenes-Greer. KCN is a data and safety monitoring board member at Novartis and NHLBI; is the cofounder of BeforeBrands, Alladapt, ForTra, and Iggenix; and is the director of FARE and World Health Organization Center of Excellence. CRN is the president and cofounder of ClostraBio Inc. CRN, KCN, RB, and LAH are inventors on a provisional US patent (63/122,833) filed on December 8, 2020.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(2):e141935.https://doi.org/10.1172/JCI141935.
See the related Commentary at Dysbiosis in food allergy and implications for microbial therapeutics.
Contributor Information
Riyue Bao, Email: baor@upmc.edu.
Lauren A. Hesser, Email: lahesser@uchicago.edu.
Ziyuan He, Email: ziyuanhe@stanford.edu.
Xiaoying Zhou, Email: xzhou2@stanford.edu.
References
- 1.Gupta RS, et al. The public health impact of parent-reported childhood food allergies in the United States. Pediatrics. 2018;142(6):e20181235. doi: 10.1542/peds.2018-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RS, et al. Prevalence and severity of food allergies among US adults. JAMA Netw Open. 2019;2(1):e185630. doi: 10.1001/jamanetworkopen.2018.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren CM, et al. Epidemiology and burden of food allergy. Curr Allergy Asthma Rep. 2020;20(2):6. doi: 10.1007/s11882-020-0898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iweala OI, Nagler CR. The microbiome and food allergy. Annu Rev Immunol. 2019;37:377–403. doi: 10.1146/annurev-immunol-042718-041621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wesemann DR, Nagler CR. The microbiome, timing, and barrier function in the context of allergic disease. Immunity. 2016;44(4):728–738. doi: 10.1016/j.immuni.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torow N, Hornef MW. The neonatal window of opportunity: setting the stage for life-long host-microbial interaction and immune homeostasis. J Immunol. 2017;198(2):557–563. doi: 10.4049/jimmunol.1601253. [DOI] [PubMed] [Google Scholar]
- 7.Thompson-Chagoyan OC, et al. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow’s milk protein allergy. Int Arch Allergy Immunol. 2011;156(3):325–332. doi: 10.1159/000323893. [DOI] [PubMed] [Google Scholar]
- 8.Ling Z, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80(8):2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azad MB, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. 2015;45(3):632–643. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 10.PALISADE Group Clinical Investigators. et al. AR101 oral immunotherapy for peanut allergy. N Engl J Med. 2018;379(21):1991–2001. doi: 10.1056/NEJMoa1812856. [DOI] [PubMed] [Google Scholar]
- 11.Chinthrajah RS, et al. Sustained outcomes in oral immunotherapy for peanut allergy (POISED study): a large, randomised, double-blind, placebo-controlled, phase 2 study. Lancet. 2019;394(10207):1437–1449. doi: 10.1016/S0140-6736(19)31793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeland DMH, et al. Oral immunotherapy for food allergy. Semin Immunol. 2017;30:36–44. doi: 10.1016/j.smim.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood RA. Food allergen immunotherapy: current status and prospects for the future. J Allergy Clin Immunol. 2016;137(4):973–982. doi: 10.1016/j.jaci.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Andorf S, et al. Anti-IgE treatment with oral immunotherapy in multifood allergic participants: a double-blind, randomised, controlled trial. Lancet Gastroenterol Hepatol. 2018;3(2):85–94. doi: 10.1016/S2468-1253(17)30392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez M, et al. Microbial therapeutics: new opportunities for drug delivery. J Exp Med. 2019;216(5):1005–1009. doi: 10.1084/jem.20190609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wargo JA. Modulating gut microbes. Science. 2020;369(6509):1302–1303. doi: 10.1126/science.abc3965. [DOI] [PubMed] [Google Scholar]
- 17.Stefka AT, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111(36):13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feehley T, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. 2019;25(3):448–453. doi: 10.1038/s41591-018-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishop WR, Bell RM. Functions of diacylglycerol in glycerolipid metabolism, signal transduction and cellular transformation. Oncogene Res. 1988;2(3):205–218. [PubMed] [Google Scholar]
- 20.Ogata Y, et al. Complete genome sequence of Phascolarctobacterium faecium JCM 30894, a succinate-utilizing bacterium isolated from human feces. Microbiol Resour Announc. 2019;8(3):e01487-18. doi: 10.1128/MRA.01487-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu F, Guo X, Zhang J, Zhang M, Ou Z, Peng Y. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp Ther Med. 2017;14(4):3122–3126. doi: 10.3892/etm.2017.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6(8):1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhya I, et al. Sporulation capability and amylosome conservation among diverse human colonic and rumen isolates of the keystone starch-degrader Ruminococcus bromii. Environ Microbiol. 2018;20(1):324–336. doi: 10.1111/1462-2920.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kourosh A, et al. Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatr Allergy Immunol. 2018;29(5):545–554. doi: 10.1111/pai.12904. [DOI] [PubMed] [Google Scholar]
- 25.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26(1):110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Berni Canani R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016;10(3):742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunyavanich S, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol. 2016;138(4):1122–1130. doi: 10.1016/j.jaci.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdel-Gadir A, et al. Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat Med. 2019;25(7):1164–1174. doi: 10.1038/s41591-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5(2):220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salonen A, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8(11):2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baxter NT, et al. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10(1):e02566-18. doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cait A, et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J Allergy Clin Immunol. 2019;144(6):1638–1647. doi: 10.1016/j.jaci.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 35.Nagao-Kitamoto H, et al. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat Med. 2020;26(4):608–617. doi: 10.1038/s41591-020-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vulevic J, McCartney AL, Gee JM, Johnson IT, Gibson GR. Microbial species involved in production of 1,2-sn-diacylglycerol and effects of phosphatidylcholine on human fecal microbiota. Appl Environ Microbiol. 2004;70(9):5659–5666. doi: 10.1128/AEM.70.9.5659-5666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Guryn K, et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23(4):458–469. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clavel T, Borrmann D, Braune A, Dore J, Blaut M. Occurrence and activity of human intestinal bacteria involved in the conversion of dietary lignans. Anaerobe. 2006;12(3):140–147. doi: 10.1016/j.anaerobe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Clavel T, Henderson G, Engst W, Dore J, Blaut M. Phylogeny of human intestinal bacteria that activate the dietary lignan secoisolariciresinol diglucoside. FEMS Microbiol Ecol. 2006;55(3):471–478. doi: 10.1111/j.1574-6941.2005.00057.x. [DOI] [PubMed] [Google Scholar]
- 41.Adlercreutz H. Lignans and human health. Crit Rev Clin Lab Sci. 2007;44(5–6):483–525. doi: 10.1080/10408360701612942. [DOI] [PubMed] [Google Scholar]
- 42.Woting A, Clavel T, Loh G, Blaut M. Bacterial transformation of dietary lignans in gnotobiotic rats. FEMS Microbiol Ecol. 2010;72(3):507–514. doi: 10.1111/j.1574-6941.2010.00863.x. [DOI] [PubMed] [Google Scholar]
- 43.Bess EN, et al. Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut bacteria. Nat Microbiol. 2020;5(1):56–66. doi: 10.1038/s41564-019-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha R, et al. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol Biomarkers Prev. 2016;25(2):407–416. doi: 10.1158/1055-9965.EPI-15-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18(5):1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 46.Apprill A, et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 47.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oksanen J, et al. vegan: Community Ecology Package. R package version 245. 2017. CRAN website. https://cran.r-project.org/web/packages/vegan/index.html Accessed November 10, 2020.
- 54.Jiang L, et al. Discrete false-discovery rate improves identification of differentially abundant microbes. mSystems. 2017;2(6):e00092-17. doi: 10.1128/mSystems.00092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- 56.Turner S, Pryer KM, Miao VP, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46(4):327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 57.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59(1):143–169. doi: 10.1128/MR.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA–targeted sequencing raw FastQ data files have been deposited into the NCBI Sequence Read Archive (SRA ID PRJNA663708) and made publicly available. Processed data files are provided as Supplemental Tables 2–12.