Highlights
-
•
Neoadjuvant ipilimumab and IFNα were evaluated in regionally advanced melanoma.
-
•
Immune cellular profiling investigated tumor immune susceptibility and resistance.
-
•
Higher levels of peripheral Th1 cell subsets predicted favorable clinical outcomes.
-
•
Higher levels of peripheral Th2 cells was associated with poor prognosis.
-
•
Significant reductions in peripheral T-reg and MDSC were seen in responders.
Keywords: Biomarker, Ipilimumab, Neoadjuvant, Anti-CTLA4, Interferon, Melanoma, Immunotherapy, Flow cytometry
Abbreviations: : CTLA4, cytotoxic T-lymphocyte-associated protein 4; IFNα, interferon α2b; IL, Interleukin; MDSCs, myeloid-derived suppressor cell; PBMCs, peripheral blood mononuclear cells; pCR, pathological complete response; TIM-3, T-cell immunoglobulin mucin-3; Tregs, regulatory T cells
Abstract
Neoadjuvant therapy with ipilimumab in combination with high dose IFNα was evaluated in patients with locally/regionally advanced melanoma in a previously reported clinical trial [NCT01608594]. In this study, peripheral immune cell profiling was performed in order to investigate the underlying mechanisms of tumor immune susceptibility and resistance. Peripheral blood mononuclear cells (PBMCs) from treated patients (N = 28) were collected at baseline and then at 6-weeks, 3-months and 12-months. High complexity (14-color) flow cytometry, designed to detect key immunological biomarkers was used to evaluate the frequencies of immune cell subsets. Statistical significance was determined using R-package employing Kruskal's test. We found that higher levels of Th1 cells at baseline (defined as CD45RA- CCR6- CXCR3+ CCR4-) correlated with the preoperative radiological response (p = 0.007) while higher Th2 cells (defined as CD45RA- CCR6- CXCR3- CCR4+) were associated with progressive disease (p = 0.009). A multimarker score consisting of higher levels of Th1 cells and CD8+ central memory T-cells was associated with pathologic complete response (pCR) (p = 0.041) at surgical resection. On the other hand, high TIM3 expression on T-cells correlated with gross viable tumor (p = 0.047). With regard to immune related toxicity, higher levels of phenotypically naive (defined as CCR7+CD45RA+) and effector memory (defined as CCR7-CD45RO+) CD8+ T-cells (p = 0.014) or lower levels of Th2 cells were associated with lower toxicity (p = 0.024). Furthermore, a multimarker score consisting of higher CD19+ and CD8+ cells was associated with lower toxicity (p = 0.0014). In conclusion, our study yielded mechanistic insights related to the immune impact of CTLA4 blockade and IFNα and potential biomarkers of immune response and toxicity.
Introduction
Melanoma patients with clinically detected locally and/or regionally advanced melanoma have a high risk of recurrence with surgery alone that approaches 90% at 5 years [1,2]. The successes of novel targeted therapies and immune checkpoint inhibitors in the treatment of advanced metastatic melanoma leading to significant improvements in disease control and long-term survival [3], [4], [5], [6], have generated considerable interest in testing these approaches in the melanoma adjuvant and neoadjuvant settings. Adjuvant cytotoxic T-lymphocyte-associated protein 4 (CTLA4) inhibitors and Programmed cell death protein 1 (PD-1) inhibitors in patients with high-risk stage III melanoma following complete resection have been shown to improve relapse-free survival in phase III clinical trials, leading to US Food and Drug Administration approval of ipilimumab, nivolumab and pembrolizumab for resected high-risk disease [7], [8], [9]. More recently, these agents have been investigated in the neoadjuvant setting and results have been encouraging [10], [11], [12], [13], [14].
In melanoma patients, it has been observed that the quality of the host immune response differs markedly between earlier and more advanced stages of melanoma where patients with advanced disease tend to display a peripheral T cell response characterized by Th2-type polarization compared to Th1-type polarization in earlier operable stages [15,16]. Use of CTLA4 blockade and Interferon-α (IFNα) can upregulate pro-inflammatory immune responses, potentially shifting the balance towards Th1 [17,18]. IFNα affects dendritic cells at different stages of development [19], and in immature states IFNα-treated dendritic Cells can induce a ‘polarized’ Th1 cytokine microenvironment [20]. Similarly, IFNα can polarize lymphocytes toward a pro-inflammatory Th1 phenotype [21,22] and further augment antitumor CD8+ T cell-mediated cytotoxicity [23]. However, this Th1 shift in immunity induced by IFNα can be suppressed by inhibitory effects mediated by CTLA4 signaling. Hence, a potential synergistic mechanism exists whereby combining IFNα with CTLA4 blockade can potentially alter this balance by down-regulating the CTLA4 suppressive regulatory elements, thus leading to sustained antitumor responses. Based on this hypothesis, patients with locally and/or regionally advanced melanoma were treated with neoadjuvant combination immunotherapy of ipilimumab and high dose IFNα in a previously reported clinical trial [10].
Recent neoadjuvant immunotherapy studies have uncovered a number of important antitumor mechanisms utilizing peripheral blood and tumor tissue specimens collected before and during the course of treatment [14,24]. For instance, Blank and colleagues showed that neoadjuvant nivolumab and ipilimumab treatment led to peripheral expansion of CD8+ T cell clones following treatment that were identified in the primary tumor and it was associated with improved relapse free survival (RFS) [12]. Similarly, a recent study indicated that higher RFS was noted in patients with brisk lymphocyte infiltration into the tumor as compared to patients with lower lymphocyte infiltration who were undergoing treatment with neoadjuvant pembrolizumab, underpinning the hypothesis that increased T cell infiltration in tumor is associated with a more robust inflammatory response and high response rate [11].
In this present study, we conducted an in-depth immune monitoring study centered on peripheral blood of patients with locally-regionally advanced melanoma undergoing neoadjuvant treatment with ipilimumab and IFNα. Peripheral blood was collected at baseline and different time points along the course of treatment and high complexity (14-color) flow cytometry was employed to determine distinct T-cell subsets. Our aim was to identify associations of circulating immune cell populations with both radiological and pathological responses, and to better understand the underlying biologic processes driving responses, resistance and the risk of immune related toxicities.
Patients and methods
Patients
Patients with operable locally-regionally advanced melanoma were enrolled in this study and were treated with 3 or 10 mg/kg ipilimumab given concurrently with IFNα [10]. Two doses of ipilimumab were administered 3 weeks apart, followed by melanoma resection surgery at about 6–8 weeks from the first dose. Two additional doses of ipilimumab were administered after surgery, also given 3 weeks apart, followed by up to 4 additional doses given 12 weeks apart. IFNα was administered concurrently at a dose of 20 MU/m²/day intravenously for 5 consecutive days per week for 4 weeks, followed by 10 MU/m²/day subcutaneously (S.C.) every other day, 3 times each week for 2 weeks prior to definitive surgery. After surgery, IFNα was resumed at 10 MU/m²/day S.C., every other day three times a week for 46 additional weeks. The study was initiated after approval from the institutional review board (IRB) and was conducted in accordance with the Declaration of Helsinki. A University of Pittsburgh IRB approved written informed consent (IRB# PRO12020161) was obtained from all patients participating in the study [10].
Response and toxicity assessment
The tumor response assessment was performed using modified World Health Organization (mWHO) criteria on imaging studies conducted at baseline, 6–8 weeks after initiation of therapy (before surgery) and every 3 months thereafter. Radiological responses were categorized as complete response (CR), partial response (PR), stable disease (SD) or progressive disease (PD). Pathologic complete response (pCR) was assessed at the time of definitive surgery and defined as no viable malignant cells on hematoxylin and eosin staining by histological assessment. In addition, overall survival (OS) and relapse-free survival (RFS) data were tracked. The descriptions and grading scales of the NCI Common Terminology Criteria for Adverse Events (CTCAE) version 4.0 were used for adverse event grading and reporting.
Collection of peripheral blood samples
Blood was drawn into heparin (for peripheral blood mononuclear cells; PBMC) tubes and processed by the Immunologic Monitoring Lab upon receipt. PBMC from eligible and treated patients (N = 28) on this trial were collected at baseline (before initiating neoadjuvant therapy), and then at 6-weeks, 3-months and 12-months (following the initiation of neoadjuvant ipilimumab-IFNα).
Flow cytometry
Flow cytometry was performed on cryopreserved PBMCs at Navigate BioPharma Laboratory, Carsbad, CA. Briefly, cryopreserved PBMCs were thawed quickly in a 37 °C water bath and washed by centrifugation with pre-warmed RPMI medium containing 10% FBS. After washing, cells were stained with fixable viability dye eFluor™506 (Thermo Fisher Scientific, San Diego, CA) at a dilution of 1:400 in wash buffer (phosphate-buffered saline containing 0.1% sodium azide and 2% fetal bovine serum) followed by staining with fluorochrome tagged antibodies for approximately 30 min at room temperature in the dark. After incubation, cells were washed once by centrifugation in wash buffer and fixed using 0.5% formalin buffer. For characterization of different immune cell subsets including phenotypically Th1 & Th2 cells, cell surface markers based on standardized immunophenotyping recommendations as described in Maecker et.al., [25] were included. The following antibodies were used; CD3 (UCHT1), CD4 (RPA-T4), CD8 (SK1), CD11b (ICRF44), CD14 (M5E2), 63G8, CD19 (SJ25C1), CD25 (M-A251), CD33 (WM53), CD45 (HI30), CD45RA (HI100), CD45RO (UCHL1), CD183 (1C6/CXCR3), CD194 (1G1/CCR4), CD197 (150,503/CCR7)), CD366 (7D3/Tim-3), HLA-DR (G46–6), from BD Biosciences (San Jose, CA). Additional antibodies were obtained from Biolegend (San Diego, CA), including CD27 (O323), CD196 (G034E3/CCR6), or from ThermoFisher (Carlsbad, CA), CD123 (6H6), CD127 (eBioRDR5). Stained Samples were acquired on a BD LSRFortessa X-20 equipped with 5 lasers (BD Biosciences, San Jose, CA) and data were analyzed using FlowJo software (FlowJo LLC, Ashland, OR).
For data analysis, after exclusion of debris and dead cells, B cells were gated based on the expression of CD19 and T cells were gated based on the expression of CD3. T cells were further gated for CD4+ and CD8+ subsets. Both CD4+ and CD8+ cells were then subsequently analyzed for the expression of various markers to define helper T cell subsets, regulatory T cells (Tregs), memory T cell subsets and checkpoint inhibitor expression. MDSCs were gated from HLA-DR- CD123-CD16- cells based on the expression of CD33, CD11b and CD14. Gate placement for all checkpoint inhibitors was set based on isotype controls with a summary of gating strategy for different populations shown in supplementary Figure 1. Phenotypic characterization of each of the populations reported is included in supplementary Table 1. Mean frequencies of representative biomarker populations at baseline as a fraction of total WBCs (leukocytes) are shown in supplementary Table 2.
Statistical data analysis
Multimarker assessments were performed by calculating Z-score for each biomarker (z score = (xi – mean) / sd) to analyze all biomarkers on the same scale. After Z-score transformation, each biomarker has a distribution with mean zero and standard deviation of 1. Following which, P-values of the combined biomarkers (multi-marker assessments) were calculated using Kruskal-Wallis to assess the differences between groups. Briefly, the statistical significance of the associations of the tested biomarkers to patient response or toxicity was determined using R-package employing a non-parametric Kruskal's test, with a p-value below 0.05 considered to be of statistical significance.
Results
Patient population, clinical efficacy and safety
Patient demographics and baseline disease characteristics of the 30 enrolled patients in the clinical trial are summarized in Table 1 [10]. Fifteen patients each were treated with ipilimumab at 3 mg/kg and 10 mg/kg respectively and INFα was given concurrently. Among 28 evaluable patients, 11 relapsed, of whom 5 died. The median follow-up period of 17 patients who had not relapsed was 32 months. Based on mWHO criteria, radiological response was noted in 10 patients (1 CR and 9 PR), 10 patients showed SD and 7 patients showed radiologic progression of disease. Combination neoadjuvant therapy achieved pathologic complete response in 9/28 patients (32%). With regard to patient's safety, the toxicities noted were consistent with the known profiles of the drugs and most common adverse events included maculopapular rash, elevation of the liver enzymes and gastrointestinal disturbances. Further, clinical efficacy and toxicity data were reported previously [10].
Table 1.
Patient demographics and baseline disease characteristics of the enrolled patients in the clinical trial (as previously reported in [10]).
| Variable | No. of Patients (%) | |
|---|---|---|
| Age (years); Median (range) | 61 (37–76) | |
| Cutaneous primary | 21 (70) | |
| Unknown primary | 8 (27) | |
| Mucosal | 1 (3) | |
| Gender | Female | 12 (40) |
| Male | 18 (60) | |
| Performance Status |
0 | 16 (53) |
| 1 | 14 (47) | |
| BRAF mutation (+) | 8 (27) | |
| BRAF wild type | 13 (43) | |
| Unknown | 9 (30) | |
| Recurrent disease after prior surgery | 15 (50) | |
| Presence of in-transit metastases | 16 (53) | |
| Prior adjuvant HDI | 5 (17) | |
| Estimated risk Stage |
IIIB | 3 (10) |
| IIIC | 25 (83) | |
| *IV (Not eligible) | 2 (7) | |
HDI: High-dose interferon-a; ECOG: Eastern Cooperative Oncology Group.
Correlation of immunological biomarkers with radiologic response
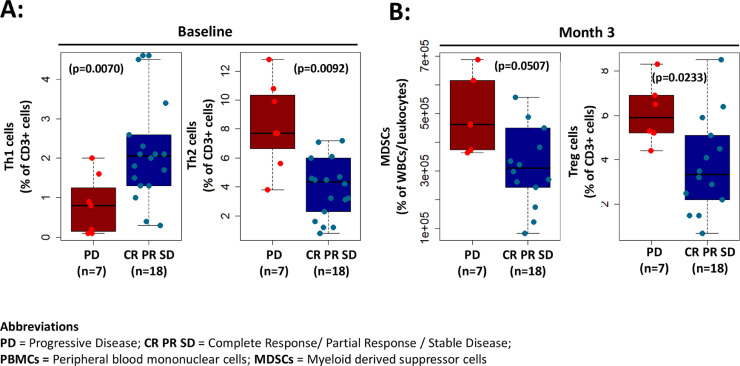
To examine whether any immunological biomarkers were associated with radiologic response, we compared peripheral immune cells between patients with pre-operative radiologic response (defined as CR, PR, SD) and patients with progression of disease (PD). We observed that patients with preoperative radiological response demonstrated higher levels of Th1 cells in baseline PBMC samples (defined as CD45RA- CCR6- CXCR3+ CCR4-) (p = 0.0070), while higher Th2 cells in baseline PBMC samples (defined as CD45RA- CCR6- CXCR3- CCR4+) were associated with progressive disease (p = 0.0092) (Fig. 1A). Interestingly, lower levels of peripheral Tregs (defined as CD4+ CD25+ CD127lo) (p = 0.0233) and MDSCs (defined as HLA-DR- CD11b+ CD33+ CD14+) (p = 0.0507) were seen at 3-months post treatment in patients exhibiting a radiologic response in comparison to radiological non-responders (Fig. 1B).
Fig. 1.
Correlation of immunological biomarkers with radiologic response. (A) Patients with higher levels of Th1 cells in baseline PBMCs samples (defined as CD45RA- CCR6- CXCR3+ CCR4-) demonstrated preoperative radiological response (RR) (p = 0.0070) while patients with higher Th2 cells in baseline PBMCs samples (defined as CD45RA- CCR6- CXCR3- CCR4+) were associated with progressive disease (p = 0.0092). (B) Patients with lower levels of MDSCs (defined as HLA-DR- CD11b+ CD33+ CD14+) (p = 0.0507) and regulatory T cells (defined as CD4+ CD25+ CD127lo) (p = 0.0233) at 3-months demonstrated radiological response.
Correlation of immunological biomarkers with pathologic complete response
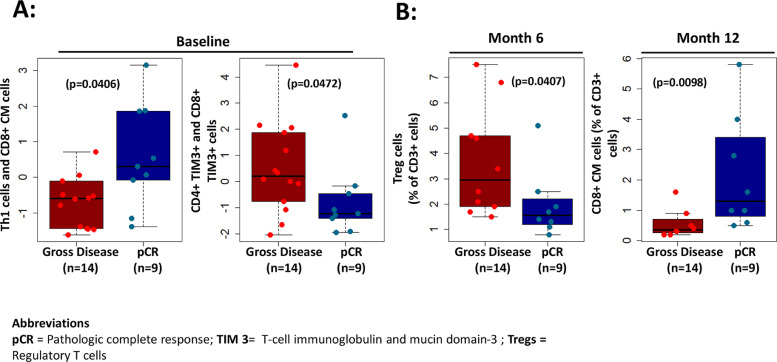
Next, we evaluated if levels of any peripheral immune phenotypes were associated with pCR. In multi-marker assessments, we found that patients with higher levels of Th1 cells and CD8+ central memory T-cells at baseline experienced superior pCR rate (p ≤ 0.05) (Fig. 2A). On the other hand, higher checkpoint inhibitor expressing T-cells including higher TIM3 expressing T-cells at baseline correlated with gross viable tumor (no pCR) (p ≤ 0.05) (Fig. 2A). Individually, lower Tregs (p = 0.0407) at 6-months as well as higher CD8 central memory cells at 12-months post treatment (p = 0.0098) correlated with pCR (Fig. 2B).
Fig. 2.
Correlation of immunological biomarkers with pathologic response. (A) Multi-marker assessments showed higher levels of Th1 cells and CD8+ central memory T-cells at baseline associated with superior pCR rate (p = 0406) in patients while higher TIM3 expression on CD4+ and CD8+ T-cells at baseline correlated with gross viable tumor (no pathologic response) (p = 0472). (B) Individually, lower levels of Tregs (p = 0.0407) at 6 months as well as higher levels of central memory CD8 cells (p = 0.0098) at 12 months post treatment correlated with pCR.
Association of peripheral immune cells with clinical toxicity
To examine any correlative findings between peripheral immune phenotypes with immune related toxicity, we stratified patients into two groups- patients with no or lower toxicity (less than Grade 2) and patients with significant toxicity (Grade 2 or higher). In multi-marker assessments, we observed that higher levels of CD8+ T cells including phenotypically naive (CD8+ CCR7+ CD45RA+) and effector memory (CD8+ CCR7- CD45RO+) CD8+ T cells (p = 0.0140) at baseline were associated with lower or no toxicity (Fig. 3). On the other hand, lower levels of Th2 cells at baseline were also associated with lower or no toxicity (p = 0.0243). In addition, a multi-marker score consisting of higher CD19+ and CD8+ cells was associated with lower toxicity (p = 0.0014) (Fig. 3).
Fig. 3.
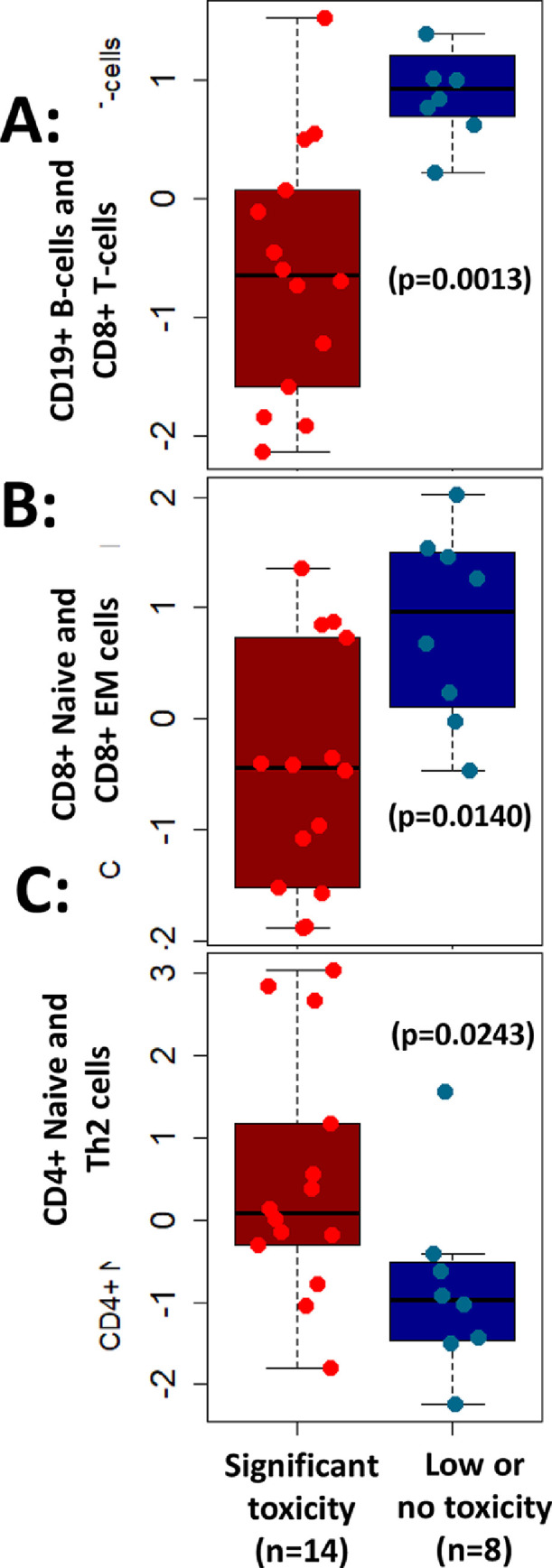
Association of peripheral immune cells with clinical toxicity. Multi-marker assessments revealed higher levels of (A) CD19+ B-cells and CD8+ T-cells (p = 0.0013) or (B) phenotypically naive CD8+ and effector memory (EM) CD8+ T-cells (p = 0.0140) associated with lower or no toxicity in patients. In addition, (C) lower levels of CD4+ naive and Th2 cells were associated with lower or no toxicity (p = 0.0243) and vice versa.
Correlation of immunological biomarkers with survival
Next, we examined if the levels of any peripheral immune cell phenotypes were associated with survival. We observed that higher levels of CD8+ central memory cells at baseline was associated with 12-month relapse free survival (P = 0.0098) (Fig. 4). On the other hand, higher expression of CD8+ TIM3+ cells was associated with worse relapse-free survival (P = 0.074) (Fig. 4). No significant association was demonstrated between peripheral immune cells and other survival parameters.
Fig. 4.
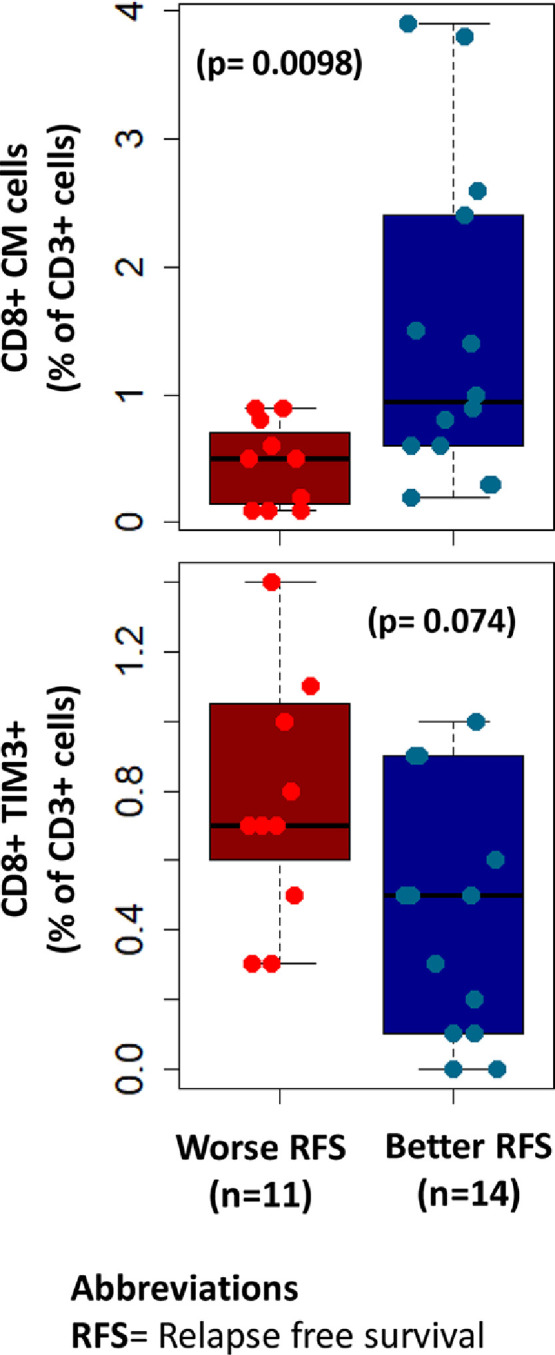
Correlation of immunological biomarkers with relapse free survival. Higher levels of CD8+ central memory cells at baseline were associated with 12-month relapse free survival (P = 0.0098). On the other hand, higher levels of CD8+ TIM3+ cells was associated with worse relapse-free survival (p = 0.074).
Discussion
In this study, we performed a detailed phenotypic analysis of the peripheral T-cell compartment in melanoma patients receiving combination ipilimumab and IFNα. A strong association with favorable clinical outcomes was the presence of higher levels of peripheral Th1 immune cell subsets at baseline while the presence of higher baseline levels of Th2 cell subsets was associated with poorer outcomes. It is known that the induction of optimal systemic antitumor immune response requires priming of both CD4+ and CD8+ T cells specific for tumor-associated antigens. Apart from cytotoxic CD8+ T cell-mediated antitumor killing, CD4+ T lymphocytes play an important role in generating effector antitumor T cell responses, in licensing dendritic cells and in maintaining immunologic memory [26], [27], [28]. CD4+ T cells mediate their effects, in part, by the production of specific cytokines. The Th1 favoring cytokines (IFN-γ, tumor necrosis factor-α, and IL-2) classically support cytotoxic T lymphocyte (CTL) responses, while the Th2 cytokines (IL-4, IL-5, IL-10) support generation of antibody responses and are antagonistic to induction of CTL responses. Further, T regulatory cells inhibit the pathogenic effect of Th1 cells by secreting anti-inflammatory cytokines [29]. Relevant to this consideration, our findings corroborated existing evidence as it was seen that the presence of higher levels of Th1 cells at baseline in the peripheral blood of melanoma patients correlated with preoperative radiological response (p = 0.0070) while higher Th2 cells at baseline were associated with progressive disease (p = 0.0092). In accordance with above, a higher multi-marker score consisting of Th1 cells and CD8+ central memory T-cells correlated with pCR (p = 0.0406). Previous studies have shown that patients with advanced measurable malignancy may have Th2-dominant responses in their peripheral blood, whereas those rendered clinically free of disease may have Th1-dominant responses [15]. In a study involving metastatic melanoma patients undergoing treatment with ipilimumab, higher baseline frequencies of CD8 effector-memory type 1 T-cells correlated with better clinical outcomes and longer OS [30]. Furthermore, we previously reported that in advanced melanoma patients treated with neoadjuvant ipilimumab monotherapy, increased expression of Th1 associated gene markers within the tumors was associated with improved RFS, although the clinical activity was very limited overall in that study with a 9% radiologic response rate and no patient achieved a pCR [31]. This suggests that IFNα can induce pro-inflammatory changes mediating antitumor immunity within the tumor microenvironment that are further enhanced with CTLA4 blockade.
It was evident in this study that patients with higher expression of immunosuppressive markers in the peripheral blood were associated with poor outcomes. We observed that higher TIM3 expression on peripheral T-cells correlated with gross viable tumor at the time of surgery (p = 0.0472). Further, higher expression of CD8+ TIM3+ cells was associated with worse RFS (p = 0.074). TIM3 is expressed on the surface of immune cells that upon ligation, limit the duration and magnitude of Th1 and Tc1 T cell responses, thus associated with poor outcomes [32,33]. Further, TIM-3 is found to be upregulated on CD4+ T cells in cancer patients and is an exhaustion marker for Th1 cells [34]. In agreement with our results, previous work by Tallerico et al. also showed that higher expression of TIM-3 on circulating T and Natural killer cells prior to and during treatment with ipilimumab in melanoma patients was associated with poor survival outcomes [35]. These observations raise the possibility that pharmacologic inhibition of TIM-3 or its relevant downstream signals could offer potential synergistic antitumor activity by restoring T cell function and thus enhancing the clinical efficacy of the tested combination immunotherapy.
Phenotypic state of immune cells provides comprehensive information about the patient's immune status [36] Populations dominated with suppressive elements such as Tregs or MDSCs may potentially counteract the beneficial effect of ipilimumab or IFNα [37]. Hence, these are especially promising biomarker candidates because of their reasonably well-defined inhibitory mechanisms. A number of previous studies have found poor outcomes in patients with high circulating suppressive effector cells such as Tregs and MDSCs and our observations that both lower Tregs (0.0233) and lower MDSCs (p = 0.0507) at 3-months post treatment correlated with better radiological response are in alignment with previous publications. For example, in melanoma patients treated with Ipilimumab, decreased amounts of both monocytic and polymorphonuclear MDSCs correlated with superior clinical outcomes [38], [39], [40]. Similarly, lower baseline levels of circulating Tregs (CD4+CD25hi+CD39+) correlated with a better RFS (p = 0.04) in patients with locally/regionally advanced melanoma treated with neoadjuvant ipilimumab [41]. Critical mechanistic questions remain open, however, targeting the immunosuppressive cells remain a potential therapeutic strategy with the ultimate goal to boost antitumor immunity.
Finally, our analysis revealed positive correlations between levels of specific immune phenotypes at baseline with immune mediated toxicity. It was observed that patients with higher levels of CD8+ T cells in peripheral blood at baseline including phenotypically naive and effector memory CD8+ T-cells experienced lower (less than grade 2) or no toxicity (p = 0.0140). Further, patients with lower levels of Th2 cells at baseline in peripheral blood also experienced lower or no toxicity (p = 0.0243). In addition, a multi-marker score consisting of higher CD19+ and CD8+ cells was associated with lower toxicity (p = 0.0014). While a direct mechanistic explanation for these interesting findings may warrant further investigation, we speculate that a Th2 favoring immune profile state at baseline (lower CD8, CD19 cells) exacerbates the known toxicity profile associated with ipilimumab therapy. Hence, a Th1 favoring immune profile observed here at baseline not only reduced the toxicity but also resulted in better clinical response. These observations may be clinically interesting as they complement analyses in metastatic melanoma, renal cell carcinoma, and sarcoma, wherein high baseline levels of tumor-associated CD8-positive T cells and CD20-positive B cells were associated with improved survival after ICI treatment, while, patients with low B-cell tumor infiltration had a significantly increased risk of death [42] and presence of high B-cell numbers are required for optimal function of T cells [43]. Furthermore, tumors containing B-cell–rich aggregates are thought to provide a gateway for entry of naive lymphocytes and invoke sustained T-cell responses [44,45].
Several reports have corroborated possible mechanistic pathways and immune cell interactions leading to immune related toxicity in patients treated with CTLA4 inhibitors. We reported that circulating IL-17 levels at baseline significantly correlated with the incidence of high grade immune related colitis in melanoma patients treated with ipilimumab [46]. Another study found that lower baseline levels of Interleukin-6 in melanoma patients had higher risks of ipilimumab toxicity [47]. It may be hypothesized that the net rise of these proinflammatory cytokines secondary to checkpoint inhibitor therapy may be a determinant of immune toxicity. There is an unmet need of biomarkers predictive for severe autoimmunity as it may improve the individual risk/benefit assessment. Further, there have been conflicting reports of whether occurrence of autoimmune toxicity correlates with clinical benefit. A number of studies have reported a positive association between incidence of immune related adverse events (irAEs) and durable response or OS for patients treated with anti-CTLA4 [48], while others found no similar association [49,50]. Whether or not irAEs correlates with tumor response, it is becoming increasingly evident that many patients with irAEs continue to experience tumor regression/clinical benefit from immunotherapy, even after treatment is discontinued secondary to toxicity [3,51]. This is because blockade of immune pathways confers an adaptive memory immune response that recalibrates the equilibrium between the tumor and the host immune response [52,53].
Conclusion
Our analysis provides evidence that the presence of an ongoing adaptive Th1 and CD8+ central memory T cell immunity in the circulation of melanoma patients is more likely to drive benefit from anti-CTLA4-based immunotherapy. It provides additional mechanistic insights related to the immune impact of CTLA4 blockade and IFNα that may have relevance for future combination immunotherapy studies.
Declaration of Competing Interest
All authors have read the journal's policy on disclosure of potential conflicts of interest. AK has no conflicts of interest to declare. GS, JT, AP, ZA, JG, CL, CV and ND are employees of Navigate Biopharma. AAT declares contracted research support with Clinigen, OncoSec, Bristol Myers Squibb and Merck and consultancy role with Array BioPharma; BioNTech AG; Bristol-Myers Squibb; Genentech/Roche; HUYA Bioscience International; Immunocore; Incyte; Merck; Newlink Genetics; Novartis; OncoSec; Pfizer/EMD Serono; Sanofi/Regeneron.
All authors have read the journal's authorship agreement. The manuscript has been reviewed and approved by all authors.
Acknowledgments
Acknowledgments
We would like to acknowledge the patients who participated in the previously reported clinical trial [NCT01608594] and donated their biospecimens for this correlative science investigation as well as their family members.
CRediT authorship contribution statement
Arjun Khunger: Data curation, Formal Analysis, Writing - original draft, review and editing. Ghanashyam Sarikonda: Investigation, Data curation, Formal Analysis, Writing - review and editing. Jennifer Tsau: Data curation, Formal Analysis, Writing - review and editing. Anil Pahuja: Data curation, Formal Analysis, Writing - review and editing. Zeni Alfonso: Data curation, Formal Analysis, Writing - review and editing. Jane Gao: Data curation, Formal Analysis, Writing - review and editing. Christian Laing: Data curation, Formal Analysis, Writing - review and editing. Christine Vaupel: Data curation, Formal Analysis, Writing - review and editing. Naveen Dakappagari: Data curation, Formal Analysis, Writing - review and editing Ahmad A. Tarhini: Conceptualization, Methodology, Investigation, Resources, Funding acquisition, Supervision, Data Curation, Writing - Review & Editing
Funding
The original clinical trial within which this project was nested was funded by Bristol Myers Squibb and Merck. The laboratory correlates were funded by Navigate Biopharma.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101014.
Contributor Information
Ghanashyam Sarikonda, Email: shyam.sarikonda@navigatebp.com.
Jennifer Tsau, Email: jenn.tsau@navigatebp.com.
Anil Pahuja, Email: anil.pahuja@navigatebp.com.
Zeni Alfonso, Email: zeni.alfonso@navigatebp.com.
Jane Gao, Email: jane.gao@navigatebp.com.
Christian Laing, Email: christian.laing@navigatebp.com.
Christine Vaupel, Email: christine.vaupel@navigatebp.com.
Naveen Dakappagari, Email: naveen.dakappagari@navigatebp.com.
Ahmad A. Tarhini, Email: ahmad.tarhini@moffitt.org.
Appendix. Supplementary materials
References
- 1.Gershenwald J.E. Melanoma staging: evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017;67(6):472–492. doi: 10.3322/caac.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romano E. Site and timing of first relapse in stage III melanoma patients: implications for follow-up guidelines. J. Clin. Oncol. 2010;28(18):3042. doi: 10.1200/JCO.2009.26.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodi F.S. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert C. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015;372(4):320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 5.Robert C. Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 2015;372(26):2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 6.Larkin J. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eggermont A.M. Prolonged survival in stage III melanoma with ipilimumab adjuvant therapy. N. Engl. J. Med. 2016;375(19):1845–1855. doi: 10.1056/NEJMoa1611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber J. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med. 2017;377(19):1824–1835. doi: 10.1056/NEJMoa1709030. [DOI] [PubMed] [Google Scholar]
- 9.Eggermont A.M. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N. Engl. J. Med. 2018;378(19):1789–1801. doi: 10.1056/NEJMoa1802357. [DOI] [PubMed] [Google Scholar]
- 10.Tarhini A. Neoadjuvant ipilimumab (3mg/kg or 10mg/kg) and high dose IFN-α2b in locally/regionally advanced melanoma: safety, efficacy and impact on T-cell repertoire. J. Immunother. Cancer. 2018;6(1):112. doi: 10.1186/s40425-018-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang A.C. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat. Med. 2019;25(3):454. doi: 10.1038/s41591-019-0357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blank C.U. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat. Med. 2018;24(11):1655. doi: 10.1038/s41591-018-0198-0. [DOI] [PubMed] [Google Scholar]
- 13.Amaria R.N. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat. Med. 2018;24(11):1649. doi: 10.1038/s41591-018-0197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khunger A. Neoadjuvant therapy of locally/regionally advanced melanoma. Ther. Adv. Med. Oncol. 2019;11 doi: 10.1177/1758835919866959. 1758835919866959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatsumi T. Disease-associated bias in T helper type 1 (Th1)/Th2 CD4+ T cell responses against MAGE-6 in HLA-DRB10401+ patients with renal cell carcinoma or melanoma. J. Exp. Med. 2002;196(5):619–628. doi: 10.1084/jem.20012142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatsumi T. Disease stage variation in CD4+ and CD8+ T-cell reactivity to the receptor tyrosine kinase EphA2 in patients with renal cell carcinoma. Cancer Res. 2003;63(15):4481–4489. [PubMed] [Google Scholar]
- 17.Yurkovetsky Z.R. Multiplex analysis of serum cytokines in melanoma patients treated with interferon-α2b. Clin. Cancer Res. 2007;13(8):2422–2428. doi: 10.1158/1078-0432.CCR-06-1805. [DOI] [PubMed] [Google Scholar]
- 18.Hanson D.C. AACR; 2004. Preclinical in Vitro Characterization of anti-CTLA4 Therapeutic Antibody CP-675,206. [Google Scholar]
- 19.Paquette R.L. Interferon-α and granulocyte-macrophage colony-stimulating factor differentiate peripheral blood monocytes into potent antigen-presenting cells. J. Leukoc. Biol. 1998;64(3):358–367. doi: 10.1002/jlb.64.3.358. [DOI] [PubMed] [Google Scholar]
- 20.Parlato S. Expression of CCR-7, MIP-3β, and Th-1 chemokines in type I IFN-induced monocyte-derived dendritic cells: importance for the rapid acquisition of potent migratory and functional activities. Blood. 2001;98(10):3022–3029. doi: 10.1182/blood.v98.10.3022. [DOI] [PubMed] [Google Scholar]
- 21.Brinkmann V. Interferon alpha increases the frequency of interferon gamma-producing human CD4+ T cells. J. Exp. Med. 1993;178(5):1655–1663. doi: 10.1084/jem.178.5.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenner C.A. Roles of IFN-gamma and IFN-alpha in IL-12-induced T helper cell-1 development. J. Immunol. 1996;156(4):1442–1447. [PubMed] [Google Scholar]
- 23.Rogge L. Selective expression of an interleukin-12 receptor component by human T helper 1 cells. J. Exp. Med. 1997;185(5):825–832. doi: 10.1084/jem.185.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amaria R.N. Neoadjuvant systemic therapy in melanoma: recommendations of the International Neoadjuvant Melanoma Consortium. Lancet Oncol. 2019;20(7):e378–e389. doi: 10.1016/S1470-2045(19)30332-8. [DOI] [PubMed] [Google Scholar]
- 25.Maecker H.T., McCoy J.P., Nussenblatt R. Standardizing immunophenotyping for the human immunology project. Nat. Rev. Immunol. 2012;12(3):191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung K. The central role of CD4(+) T cells in the antitumor immune response. J Exp. Med. 1998;188(12):2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortara L. Type 1 CD4(+) T-cell help is required for induction of antipeptide multispecific cytotoxic T lymphocytes by a lipopeptidic vaccine in rhesus macaques. J. Virol. 1999;73(5):4447–4451. doi: 10.1128/jvi.73.5.4447-4451.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoenberger S.P. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998;393(6684):480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 29.Raphael I. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine. 2015;74(1):5–17. doi: 10.1016/j.cyto.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wistuba-Hamprecht K. Peripheral CD8 effector-memory type 1 T-cells correlate with outcome in ipilimumab-treated stage IV melanoma patients. Eur. J. Cancer. 2017;73:61–70. doi: 10.1016/j.ejca.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarhini A.A. Expression profiles of immune-related genes are associated with neoadjuvant ipilimumab clinical benefit. Oncoimmunology. 2017;6(2) doi: 10.1080/2162402X.2016.1231291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson A.C. Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol. Res. 2014;2(5):393–398. doi: 10.1158/2326-6066.CIR-14-0039. [DOI] [PubMed] [Google Scholar]
- 33.Sabatos C.A. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat. Immunol. 2003;4(11):1102. doi: 10.1038/ni988. [DOI] [PubMed] [Google Scholar]
- 34.Golden-Mason L. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J. Virol. 2009;83(18):9122–9130. doi: 10.1128/JVI.00639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tallerico R. IL-15, TIM-3 and NK cells subsets predict responsiveness to anti-CTLA-4 treatment in melanoma patients. Oncoimmunology. 2017;6(2) doi: 10.1080/2162402X.2016.1261242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J. Transl. Med. 2012;10(1):146. doi: 10.1186/1479-5876-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9(3):162. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer C. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 2014;63(3):247–257. doi: 10.1007/s00262-013-1508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gebhardt C. Myeloid cells and related chronic inflammatory factors as novel predictive markers in melanoma treatment with ipilimumab. Clin. Cancer Res. 2015;21(24):5453–5459. doi: 10.1158/1078-0432.CCR-15-0676. [DOI] [PubMed] [Google Scholar]
- 40.Sade-Feldman M. Clinical significance of circulating CD33+ CD11b+ HLA-DR− myeloid cells in patients with stage IV melanoma treated with ipilimumab. Clin. Cancer Res. 2016;22(23):5661–5672. doi: 10.1158/1078-0432.CCR-15-3104. [DOI] [PubMed] [Google Scholar]
- 41.Retseck J. Long term impact of CTLA4 blockade immunotherapy on regulatory and effector immune responses in patients with melanoma. J. Transl. Med. 2018;16(1):184. doi: 10.1186/s12967-018-1563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griss J. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat. Commun. 2019;10(1):4186. doi: 10.1038/s41467-019-12160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiLillo D.J., Yanaba K., Tedder T.F. B cells are required for optimal CD4+ and CD8+ T cell tumor immunity: therapeutic B cell depletion enhances B16 melanoma growth in mice. J. Immunol. 2010;184(7):4006–4016. doi: 10.4049/jimmunol.0903009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabrita R. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 45.Xu H. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496(7446):523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- 46.Tarhini A.A. Baseline circulating IL-17 predicts toxicity while TGF-β1 and IL-10 are prognostic of relapse in ipilimumab neoadjuvant therapy of melanoma. J. Immunother. Cancer. 2015;3(1):39. doi: 10.1186/s40425-015-0081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valpione S. Sex and interleukin-6 are prognostic factors for autoimmune toxicity following treatment with anti-CTLA4 blockade. J. Transl. Med. 2018;16(1):94. doi: 10.1186/s12967-018-1467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downey S.G. Prognostic factors related to clinical response in patients with metastatic melanoma treated by CTL-associated antigen-4 blockade. Clin Cancer Res. 2007;13(22 Pt 1):6681–6688. doi: 10.1158/1078-0432.CCR-07-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horvat T.Z. Immune-related adverse events, need for systemic immunosuppression, and effects on survival and time to treatment failure in patients with melanoma treated with ipilimumab at memorial sloan kettering cancer center. J. Clin. Oncol. 2015;33(28):3193–3198. doi: 10.1200/JCO.2015.60.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber J.S. Safety profile of nivolumab monotherapy: a pooled analysis of patients with advanced melanoma. J. Clin. Oncol. 2017;35(7):785–792. doi: 10.1200/JCO.2015.66.1389. [DOI] [PubMed] [Google Scholar]
- 51.McDermott D.F. Randomized phase III trial of high-dose interleukin-2 versus subcutaneous interleukin-2 and interferon in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 2004;23(1):133–141. doi: 10.1200/JCO.2005.03.206. [DOI] [PubMed] [Google Scholar]
- 52.Kim P.S., Ahmed R. Features of responding T cells in cancer and chronic infection. Curr. Opin. Immunol. 2010;22(2):223–230. doi: 10.1016/j.coi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunn G.P., Old L.J., Schreiber R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.