Abstract
Objective
Hormone secretion from metabolically active tissues, such as pancreatic islets, is governed by specific and highly regulated signaling pathways. Defects in insulin secretion are among the major causes of diabetes. The molecular mechanisms underlying regulated insulin secretion are, however, not yet completely understood. In this work, we studied the role of the GTPase ARFRP1 on insulin secretion from pancreatic β-cells.
Methods
A β-cell-specific Arfrp1 knockout mouse was phenotypically characterized. Pulldown experiments and mass spectrometry analysis were employed to screen for new ARFRP1-interacting proteins. Co-immunoprecipitation assays as well as super-resolution microscopy were applied for validation.
Results
The GTPase ARFRP1 interacts with the Golgi-associated PDZ and coiled-coil motif-containing protein (GOPC). Both proteins are co-localized at the trans-Golgi network and regulate the first and second phase of insulin secretion by controlling the plasma membrane localization of the SNARE protein SNAP25. Downregulation of both GOPC and ARFRP1 in Min6 cells interferes with the plasma membrane localization of SNAP25 and enhances its degradation, thereby impairing glucose-stimulated insulin release from β-cells. In turn, overexpression of SNAP25 as well as GOPC restores insulin secretion in islets from β-cell-specific Arfrp1 knockout mice.
Conclusion
Our results identify a hitherto unrecognized pathway required for insulin secretion at the level of trans-Golgi sorting.
Keywords: Insulin secretion, Endosomal sorting, SNARE proteins, trans-Golgi network
Abbreviations: ARFRP1, ADP ribosylation factor-related protein 1; GOPC, Golgi-associated PDZ and coiled-coil motif-containing protein; GSIS, glucose-stimulated insulin secretion; ICG, insulin-containing granule; VPS35, vacuolar protein sorting-associated protein 35; T2D, type 2 diabetes
Highlights
-
•
β-cell specific deletion of the trans-Golgi residing small GTPase ARFRP1 leads to elevated blood glucose levels in mice.
-
•
GOPC is a newly identified ARFRP1 dependent scaffolding protein.
-
•
ARFRP1 and GOPC are required for glucose-stimulated insulin secretion from pancreatic β-cells.
1. Introduction
Insulin secretion from pancreatic β-cells is a highly dynamic and regulated process that is essential for maintaining glucose homeostasis. Insufficient insulin release results in metabolic disorders that lead to type 2 diabetes (T2D). Genome-wide association studies (GWAS) often connect single-nucleotide polymorphisms (SNPs) in genes required for protein secretion in diabetes [1,2]. However, mechanistic insights that support these studies are still sparse.
In the postprandial state, dietary glucose from the circulation enters β-cells via the glucose transporter GLUT1 in humans and GLUT2 in rodents and is immediately metabolized [3,4]. ATP-sensitive potassium channels in the plasma membrane are closed due to a rising ATP:ADP ratio, leading to membrane depolarization, followed by opening of voltage-dependent Ca2+ channels [[5], [6], [7]]. Increasing Ca2+ concentrations within β-cells are sensed by various proteins like synaptotagmin-7. This eventually leads to the fusion of insulin-containing granules with the plasma membrane via SNARE-complex formation, followed by the release of insulin into the extracellular space [8]. A well-studied SNARE complex in this context consists of VAMP2, Syntaxin1A, and SNAP25 that assemble in a 1:1:1 stoichiometry [9,10]. However, other SNARE proteins, such as Syntaxin4, are also implicated in the process of biphasic insulin secretion [11,12].
Insulin is secreted in two distinct phases: the first and acute phase is mediated by a pool of pre-docked insulin-containing granules [13]. The ensuing second phase, which is longer lasting but weaker, may be mediated by a pool of storage granules [14]. This finding, however, is highly debated. More recent research suggests that the second phase is instead governed by so-called newcomer granules [15].
Like many other membrane trafficking steps within the secretory pathway, the different stages in the lifetime of insulin granules, beginning with their biogenesis along with their movement within the cell, require the activity of regulatory GTPases. However, precise information about the individual steps is still missing. ARFRP1 is a small monomeric GTPase of the ARF family [16]. It is found at the trans-Golgi network in its active GTP-bound state. ARF family members are key regulators of cellular signaling processes as nearly every trafficking step to or from the Golgi is described to be regulated by at least one GTPase of the ARF or Rab family [17,18]. Arfrp1 is ubiquitously expressed. As suggested by different conditional knockout studies, it plays a pivotal role in glucose and lipid metabolism [[19], [20], [21]]. ARFRP1 is also involved in post-Golgi trafficking to the plasma membrane. For example, the plasma membrane trafficking of vesicular stomatitis virus glycoprotein (VSVG) and of VANGL2 in polarized cells depends on ARFRP1 [22,23]. ARFRP1 is required for the recruitment of ARL1 and its effectors, the scaffolding proteins Golgin-97 and Golgin-245, to the trans-Golgi network [[24], [25], [26]]. Our recent work suggests that ARFRP1 is required for hormone release from metabolic active tissues [27]. We therefore hypothesized that ARFRP1 is involved in the process of insulin secretion from pancreatic β-cells. Phenotypic characterization of β-cell-specific Arfrp1 knockout mice supports this hypothesis as these mice have elevated blood glucose levels and a reduced capacity in glucose-stimulated insulin secretion.
Here, by screening for potential ARFRP1-interacting proteins, we identified the Golgi-associated PDZ and coiled-coil motif-containing protein (GOPC). Functional assays revealed that this interaction is necessary for correct localization of the t-SNARE protein SNAP25 at the plasma membrane and thereby participates in the process of glucose-stimulated insulin secretion from β-cells.
2. Materials and methods
2.1. Animals
To generate β-cell-specific Arfrp1 knockout mice with a C57BL/6J background, Arfrp1flox/flox mice [28] were crossed with Ins1Cre mice [29]. Animals were housed in a 22 ± 2 °C environment with a 12:12-h light:dark cycle and unlimited access to food and water. Body weight, body composition (fat + lean mass), blood glucose, and plasma insulin levels were measured weekly from week 4 until week 14. At the age of 10 weeks, oral glucose tolerance tests were performed in male mice after a 4-h fasting period. Glucose was orally applied at 2 mg/g body weight. All metabolic measurements shown in the paper were performed in male mice between the ages of 10 and 14 weeks. Female mice showed the same effects on blood glucose and degradation of SNAP25 and GOPC proteins. Use of female mice e.g., for immunostainings or quantitative reverse transcription polymerase chain reaction (qRT-PCR), is stated in the figure legends, respectively. All animal experiments were approved by the ethics committee of the State Office of Environment, Health, and Consumer Protection (Federal State of Brandenburg, Germany).
2.2. Mass spec and LUMIER
Human ARFRP1 (Uniprot-ID Q13795) was expressed as a GST-fusion protein from pGEX-6P1 in E. coli BL21(DE3). Cells were cultured at 37 °C to an OD600 of 0.5, followed by 16 h of growth at 18 °C after addition of 100 μM of isopropyl-β-D-thiogalactopyranosid (IPTG). Cells were disrupted with a fluidizer in buffer A (50 mM of HEPES, pH 7.5, 300 mM of NaCl, 2.5 mM of 2-mercaptoethanol, 2 mM of MgCl2). Cleared lysates (96,000×g, 30 min, 4 °C) were loaded on a glutathione sepharose column. Following extensive washing (buffer A with 500 mM of NaCl), protein was eluted (buffer A with 20 mM of glutathione) and subjected to size exclusion chromatography in buffer B (20 mM of HEPES, pH 7.5, 150 mM of NaCl, 2.5 mM of 2-mercaptoethanol, and 2 mM of MgCl2). GST was expressed and purified from an unmodified pGEX-6P1 in an analogous way. Six nanomoles of GST-ARFRP1 and a GST control were bound to glutathione sepharose in buffer B. Bead-bound proteins were loaded with GTPγS by 10 min of incubation with buffer B supplemented with 200 mM of ammonium sulfate, 10 mM of EDTA, and 2 mM of GTPγS, followed by a 10-min incubation in buffer B supplemented with 200 mM of ammonium sulfate, 20 mM of MgCl2, and 2 mM of GTPγS. GST-ARFRP1 could only partially be loaded with GTPγS with this protocol, as judged by high-performance liquid chromatography (HPLC) analysis, and a substantial GDP fraction remained. Pulldowns were therefore performed with GST-ARFRP1●GTPγS compared to GST alone. Brains from two 4-week-old mice were homogenized in ice-cold lysis buffer (50 mM of Tris–HCL, pH 7.5, 150 mM of NaCl, 1.2% (w/v) octyl glucoside, 1 mM of EDTA, 1 mM of EGTA, cOmplete protease inhibitor (Roche)). Insoluble material was removed by 2 × 25-min centrifugation at 16,100×g at 4 °C, and the soluble extract was incubated with bead-bound GST-ARFRP1●GTPγS and GST for 30 min at 4 °C. Beads were washed 3 times with buffer B, and retained proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Gel bands from the GST-ARFRP1 and GST control pulldowns were excised and subjected to tryptic in-gel digestion [30]. Samples were measured by liquid chromatography tandem mass spectrometry (LC-MS/MS) on a Q Exactive Plus (Thermo) and an Orbitrap Fusion mass spectrometer (Thermo) connected to an EASY-nLC system (Thermo) as technical replicates. Database search was performed with MaxQuant version 1.5.2.8 [31]. A spectral counting approach was used to compare GST-ARFRP1 and GST control samples. Protein groups were filtered for those containing at least 10 different peptide IDs with a total intensity of ≥108. The number of peptide IDs in the GST-ARFRP1 pulldown samples was summed for each protein group and divided by the total number of peptide IDs in GST-ARFRP1 and GST control. Protein groups with a ratio of >0.9 were further refined to proteins with a known Golgi-localization (GO:0005794) and/or involvement in vesicular trafficking (GO:0016192).
From the remaining 23 mass spectrometry hits, 16 were further tested in a modified LUMIER (luminescence-based mammalian interactome) assay [32]. Briefly, a N-terminal protein A (PA)-Renilla luciferase (RL)-ARFRP1 fusion protein (PA-RL-ARFRP1) was co-expressed with V5-firefly luciferase (FL)-tagged prey proteins (V5-FL-prey) in HEK293 cells. After 48 h, cells were lysed in buffer C (1% NP40, 50 mM of HEPES/NaOH pH 7.4, 150 mM of NaCl, 1.5 mM of MgCl2, 1 mM of EDTA, 1 mM of DTT) with cOmplete protease inhibitors and Benzonase (Merck). Protein complexes were captured using immunoglobulin G (IgG, Jackson Immunoresearch) immobilized to high-binding 96-well white plates (Greiner). Bound protein complexes were washed with buffer C without NP40, and firefly luciferase activity was quantified with the Dual-Glo Luciferase Assay System (Promega) using an Infinite M200 reader (Tecan). Firefly activities were normalized to co-IP controls, employing a PA-RL fusion protein as bait and firefly-V5-tagged prey proteins. Co-IPs were performed in triplicate.
2.3. Cell culture and transfection
All cell lines were cultured at 37 °C and 5% CO2. Min6 cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM, PAN) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco). For adenoviral infection, 1.6 ∗ 105 Min6 cells were seeded per 24-well plate and 3.2 ∗ 105 cells per 12-well plate. Cells were infected with indicated multiplicities of infection (MOIs) and, if not otherwise stated, harvested after 48 h. Ins1 cells were cultured in RPMI (PAN) supplemented with 10% FCS (PAN), 1% HEPES (PAN), 2 mM of glutamine, 1 mM of Na-pyruvate, and 0.05 mM of 2-mercaptoethanol (Sigma). For microscopy experiments, 1.2 ∗ 105 cells were seeded on poly-l-lysine (Sigma) coated coverslips and cultured for 48 h. Ins1 cells were preferred over Min6 for 3D-SIM (structured illumination microscopy) since antibodies used within this study worked better in this cell line. HeLa cells were cultured in Earles medium (PAN) supplemented with 10% FCS (PAN). For co-IP experiments, 1.8 ∗ 105 cells were seeded per 6-well plate. To block lysosomal degradation, Min6 cells were treated with 10 mM of chloroquine for 16 h. For determination of protein half-life, Min6 cells were incubated for indicated time points with 100 μg/ml of cycloheximide. All cell lines were regularly tested for contamination using the Venor®GeM Advance kit (Minerva).
2.4. Plasmids
ARFRP1-expressing constructs were described previously [24]. All other constructs were purchased from Geneart (Thermo Fisher, Regensburg), and adenoviral constructs were obtained from Vector Biolabs (USA).
2.5. Western blotting and co-immunoprecipitation
Co-immunoprecipitations were performed using GFP-Traps (Chromotek) according to the manufacturer's protocol. Briefly, transfected HeLa cells were lysed in ice-cold lysis buffer (10 mM of Tris/Cl pH 7.5, 150 mM of NaCl, 0.5 mM of EDTA, 0.5% NP40), and 3 wells were pooled. Protein concentrations were determined in a BCA assay. Per pulldown, 300 μg of lysate was filled up to 500 μl, and 10 μl of GFP-traps were added. Samples were rotated overnight at 4 °C, washed 3 times, and transferred to a new tube. Immunocomplexes were eluted from beads in SDS-sample buffer by boiling for 10 min. Twenty microliters of eluate were run on an SDS-PAGE and transferred to nitrocellulose membranes by tank-blotting. Membranes were incubated with primary antibodies as indicated overnight at 4 °C and detected with the LICOR Odyssey system. Quantification of bands were performed using Image Studio Lite software (Ver. 5.2).
2.6. Measurement of insulin secretion via perifusion
Isolation of pancreatic islets from 12- to14-week-old male mice was performed as described [33] and recovered overnight. Thirty to 40 islets per mouse were equilibrated in KRBH buffer with low glucose (2.8 mM) before transferring to chambers of a perifusion system (PERI-4.2, Biorep). Glucose-stimulated insulin secretion (GSIS) was recorded with a continues flow of 100 μl/min under indicated glucose concentrations over indicated time points. Islets were collected from beads with acid ethanol (0.18 M of HCl in 70% ethanol) for determination of residual insulin. Area under the curve (AUC) was calculated in GraphPad Prism 8.1.2 for the 1st phase from 20 to 34 min and for the 2nd phase from 37 to 58 min.
2.7. GSIS
GSIS from Min6 cells was performed 48 h post adenoviral infection. Cells were washed with KRBH buffer containing 2.8 mM of glucose and equilibrated for 1 h. Glucose was added to a final concentration of 20 mM, and supernatants were collected after another 60 min. Supernatants were subjected to enzyme-linked immunosorbent assay (ELISA) experiments, and cells were lysed for DNA concentration measurements using the Quant-IT PicoGreen Kit (Invitrogen). GSIS from isolated islets was performed 48 h post adenoviral infection in a 12-well plate. Two times 10 islets per mouse were pre-incubated for 1 h in 1 ml of KRBH buffer containing 2.8 mM of glucose before transfer into a fresh well containing the same buffer. After 1 h, supernatant was collected, representing basal insulin release, and islets were transferred into KRBH buffer containing 20 mM of glucose for 1 h. Afterwards, supernatant was collected, and islets were transferred into 200 μl of acidic ethanol for measuring residual insulin. Released insulin was measured in duplicates and normalized to residual insulin.
2.8. Measurement of insulin concentration via ELISA
Insulin concentrations from GSIS in Min6 cells and isolated islets were measured from supernatants with a mouse high range and ultrasensitive insulin ELISA kit (Alpco) according to the manufacturer's instruction and normalized to DNA concentration (ng/ml). Insulin concentrations from perifusion experiments were measured with a mouse ultrasensitive insulin ELISA kit (Alpco) according to the manufacturer's instructions and normalized to residual insulin from islets.
2.9. Morphometric analysis of pancreatic sections
For determination of islet size and abundance, pancreas from 3 mice per genotype were isolated, fixed, and embedded into paraffin blocks. From each pancreas, 4-μm-thick sections were taken in triplicates from the apical, mid, and distal parts of the organ. Sections were deparaffinized and stained for insulin by DAB (3,3′-diaminobenzidine) labeling. After incubation with primary antibody for 1 h at room temperature, sections were washed and incubated with peroxidase-coupled secondary antibody (Histofine Simple Stain MAX PO anti mouse) for 30 min. Sections were then incubated with substrate solution (DakoCytomation Liquid DAB + Substrate Chromogen System). Images of the whole sections were recorded with a MIRAX MIDI scanner (Zeiss) and analyzed using the MIRAX Viewer software.
2.10. Confocal microscopy and SIM
For microscopy experiments, cells were fixed for 10 min in 4% paraformaldehyde (PFA) and washed 3 times in phosphate-buffered saline (PBS). Cells were incubated 3 times in 50 mM of NH4Cl/PBS, permeabilized for 6 min in 0.2% saponin/PBS, and incubated for 5 min in 0.02% saponin/PBS. After blocking for 45 min in 2% bovine serum albumin (BSA) in 0.02% saponin/PBS, cells were incubated with primary antibodies diluted in blocking buffer overnight at 4 °C or 2 h at room temperature (RT). Cells were washed 3 times and incubated for 1 h with secondary antibodies diluted in blocking buffer supplemented with 4′,6-diamidino-2-phenylindole (DAPI). Cells were mounted in ProLong™ Glass Antifade Mountant und subjected to confocal or SIM microscopy. Confocal microscopy was performed with a Leica TCS SP8 microscope equipped with a white light laser and a 405 diode. Fluorophores were excited at respective wavelength and sequential scans for image collection performed with a 63x objective with a numerical aperture of 1.4 using oil immersion, speed of 100 Hz and a frame average of 4 at RT.
Three-dimensional (3D) 3-color SIM images were acquired using 488 nm, 568 nm, and 642 nm laser lines, standard filter sets, and 125-nm z-sectioning of the OMX V4 Blaze (GE Healthcare) system. One hundred-nanometer fluorescent beads (Tetraspeck, T7284, Thermo Fischer Scientific) were used for registration of individual channels, achieving less than 40 nm registration, respectively. Image and movie processing, including 3D-rendering was done using Arivis Vision4D. Objects were identified in ImageJ with a histogram-based threshold procedure (Otsu's method) on the maximum projection of the 3D SIM stack. Holes up to one pixel (40 × 40 nm) were closed. Only objects with a surface area above 0.008 μm2 were considered as signals. In-focus planes were automatically extracted from 3D-SIM stacks based on their signal-to-noise ratio, using a custom-written ImageJ script. Objects were identified in Arivis Vision 4D by Otsu thresholding. Only objects with a volume larger than 0.0025 μm³ were included in the analysis. Distances from surfaces of all possible combinations of objects were exported from Arivis Vision 4D and nearest neighbor (NN) distances were calculated using a custom-written python script. The same procedure was repeated after randomization by a toroidal shift of 3 μm. NN distances are displayed as mean of the “corrected NN Distance”, which was calculated as the mean of all NN distances after subtracting the distance from random object distributions.
2.11. RNA extraction and RT-qPCR
RNA extraction from freshly isolated islets was performed using the Qiagen RNeasy MinElute Kit according to the manufacturer's instructions. cDNA was generated using M-MLV (M3683) reverse transcriptase from Promega. Gene expression of Arfrp1, Snap25 and insulin was measured by RT-qPCR and normalized to housekeeping genes Ppia and Eef2 using specific probes (Taqman and IDT).
2.12. Antibodies
Antibodies used within this study for Western blotting and stainings are listed with their respective dilutions in Table 1.
Table 1.
Antibodies used within this study.
| Antibody | Company | Catalog number | Application | Dilution |
|---|---|---|---|---|
| α-Tubulin | Sigma | T 6074 | WB (loading) | 1:500 |
| ARFRP1 | Abcam | ab108199 | WB | 1:500 |
| ARFRP1-A647 | Abcam | ab108199 | IF | 1:300 |
| EXOC7 | Millipore | MABT186 | IF | 1:100 |
| GAPDH | Ambion | AM4300 | WB (loading) | 1:10,000 |
| GFP | Santa Cruz | sc-9996 | WB | 1:500 |
| Glucagon | Santa Cruz | sc-7779 | IHC | 1:50 |
| GOPC | Abcam | |||
| Insulin | Sigma | I 2018 | IF/IHC | 1:50,000 |
| Myc-Tag | Cell Signaling | #2272 | WB | 1:1,000 |
| PIST | HJK | / | IF | 1:1,000 |
| SNAP25 | BioLegend | 836304 | IF/WB | 1:100/1:1,000 |
| Syntaxin1 | Sigma | So664 | WB | 1:1000 |
| VAMP2 | Abcam | ab3347 | WB | 1:1000 |
| VPS35 | Abcam | ab57632 | IF/WB | 1:200/1:1,000 |
WB = Western blot, IF = immunofluorescence, IHC = immunohistochemistry.
3. Results
3.1. ARFRP1 function in β-cells is required for correct insulin secretion in mice
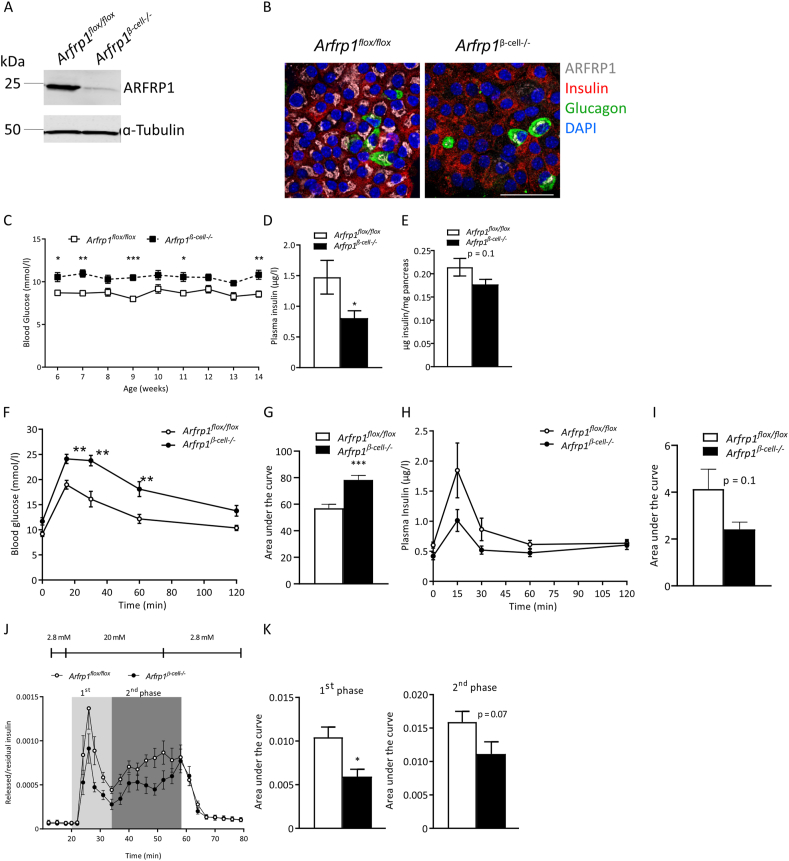
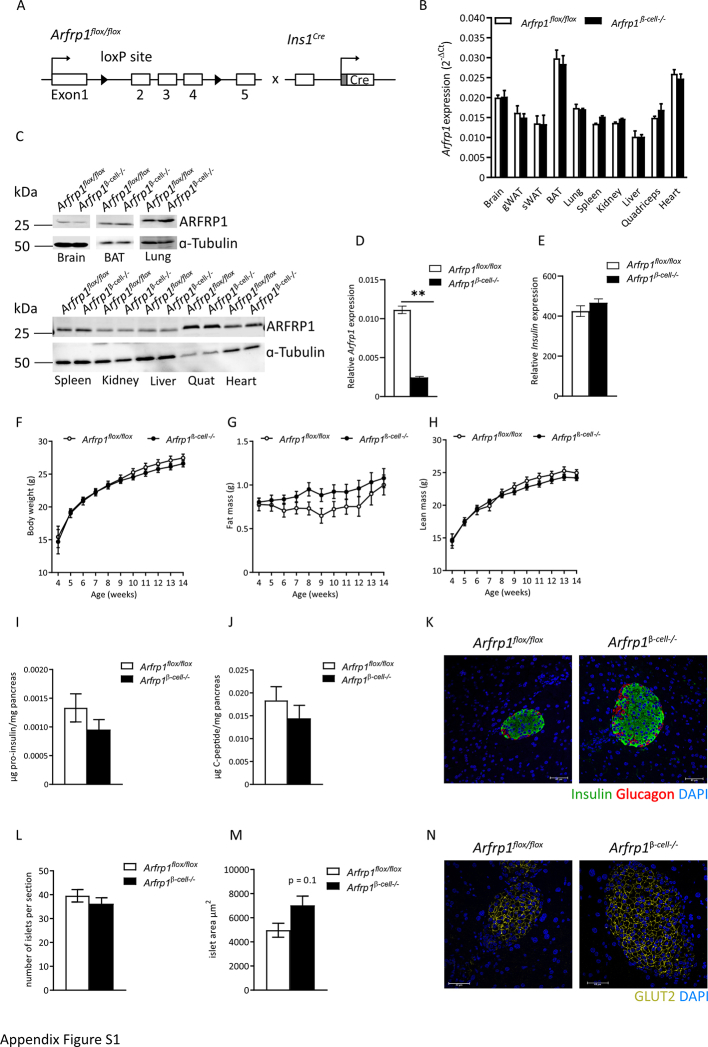
To characterize a possible role of ARFRP1 for insulin secretion, β-cell-specific knockout mice (Arfrp1β-cell−/−) were generated. A mouse strain with a floxed Arfrp1 allele (Arfrp1flox/flox) [28] was intercrossed with mice expressing the Cre recombinase specifically in β-cells (Ins1Cre mice) [29] (Appendix Fig. S1A). Knockout efficiency and specificity were proven by qRT-PCR and Western blot analysis of islets and different tissues (Figure 1A and Appendix Fig. S1B–D). Whereas ARFRP1 expression was essentially not altered in most of the tested tissues, it was greatly reduced in islets from Arfrp1β-cell−/− mice (Appendix Fig. S1C + D, Figure 1A). The residual ARFRP1 signal may stem from non-β-cells, such as α-cells that secrete the insulin-counteracting peptide hormone glucagon. These cells are also abundantly found in islets as they make up 15–20% of islet mass [34]. Supporting this assumption, immunostainings of isolated islets from Arfrp1β-cell−/− mice showed a juxtanuclear staining for ARFRP1 (white) in glucagon-positive α-cells (green) (Figure 1B).
Figure 1.
ARFRP1 is required for appropriate insulin secretion in mice. (A) Western blot analysis from isolated islets from ad libitum fed male Arfrp1flox/flox and Arfrp1β-cell−/− mice probed with indicated antibodies. (B) Immunostaining of dispersed islet cells from ad libitum fed Arfrp1flox/flox and Arfrp1β-cell−/− mice stained for ARFRP1 (white), insulin (red), and glucagon (green). Nuclei were visualized with DAPI (blue). Scale bar = 40 μm. (C) Blood glucose levels of 13-weeks-old ad libitum fed male Arfrp1flox/flox (n = 12) and Arfrp1β-cell−/− mice (n = 14). (D) Plasma insulin levels of 13 weeks old ad libitum male Arfrp1flox/flox (n = 11) and Arfrp1β-cell−/− mice (n = 13). (E) Total pancreatic insulin levels of ad libitum fed male Arfrp1flox/flox (n = 11) and Arfrp1β-cell−/− mice (n = 10). (F) Oral glucose tolerance test in 4h fasted male Arfrp1flox/flox (n = 6) and Arfrp1β-cell−/− mice (n = 9). Blood glucose was measured at indicated time points. Data is shown as mean ± SEM (∗ = p ≤ 0.05 2-way ANOVA). (G) Area under the curve of (F). (H) Plasma insulin levels after oGTT described in (F). (I) Area under the curve of (H). (J) Glucose stimulated insulin secretion from isolated islets of ad libitum fed 12- to 14-week-old male Arfrp1flox/flox (n = 6) and Arfrp1β-cell−/− (n = 7) mice treated with indicated glucose concentrations was measured at indicated time points in perifusion experiments. The first phase is marked in light gray, and the second phase is marked in dark grey. (K) Calculated area under the curve of first and second phase of glucose-stimulated insulin secretion measured in (J). Data are presented as mean ± SEM, ∗p ≤ 0.05 (unpaired Student's t-test, Welch's correction).
Phenotypic characterization of Arfrp1β-cell−/− and control mice revealed that deletion of Arfrp1 in β-cells led to elevated blood glucose (Figure 1C). The measured glucose levels are, however, not high enough to rank it glucotoxic [35]. Furthermore, GLUT2, which is downregulated under glucotoxic conditions, was unaltered in islets of Arfrp1β-cell−/− mice (Appendix Fig. S1N). Plasma insulin levels, as shown for 13-week-old mice, were decreased in Arfrp1β-cell−/− mice (Figure 1D). Total pancreatic insulin levels as well as Ins2 expression, however, were not significantly different between the two genotypes (Figure 1E and Appendix Fig. S1E), nor was the body weight or body composition (Appendix Fig. S1F–H). For body fat mass, higher values were observed in Arfrp1β-cell−/− mice, without reaching statistical significance. Although fat mass was slightly increased in Arfrp1β-cell−/− mice, leptin levels were not different in the plasma of the two different mice strains (2.1 ± 0.4 ng/ml in Arfrp1flox/flox mice, 2.3 ± 1.4 ng/ml in Arfrp1β-cell−/− mice). Further measurements of immature pro-insulin and cleaved C-peptide in pancreas from Arfrp1flox/flox and Arfrp1β-cell−/− mice revealed no differences, indicating that the reduced plasma insulin concentrations of Arfrp1β-cell−/− mice were not caused by an impaired maturation of insulin (Appendix Fig. S1I and J). Oral glucose tolerance tests indicated elevated fasting blood glucose levels and impaired glucose clearance capacity in Arfrp1β-cell−/− mice, presumably caused by reduced plasma insulin levels (Figure 1F–I). Morphometric analysis of pancreatic sections showed a tendency towards bigger islets in Arfrp1β-cell−/− mice, whereas the number of islets per section was unchanged (Appendix Fig. S1K–M). This might be a compensatory effect in Arfrp1β-cell−/− mice to increase the insulin content and to regulate glucose homeostasis [36].
To clarify whether the reduced plasma insulin levels in Arfrp1β-cell−/− mice were the consequence of a reduced insulin secretion, islets from both Arfrp1flox/flox and Arfrp1β-cell−/− mice were isolated and subjected to perifusion experiments to investigate their capacity for glucose-stimulated insulin release (Figure 1J). Islets from both genotypes responded to high-glucose treatment by increasing insulin release, whereas both the first and second phases of insulin release were reduced in islets from Arfrp1β-cell−/− mice (Figure 1J,K). These findings suggest that ARFRP1 influences insulin secretion from pancreatic β-cells rather than affecting its synthesis or maturation.
3.2. ARFRP1 interacts with the scaffolding protein GOPC
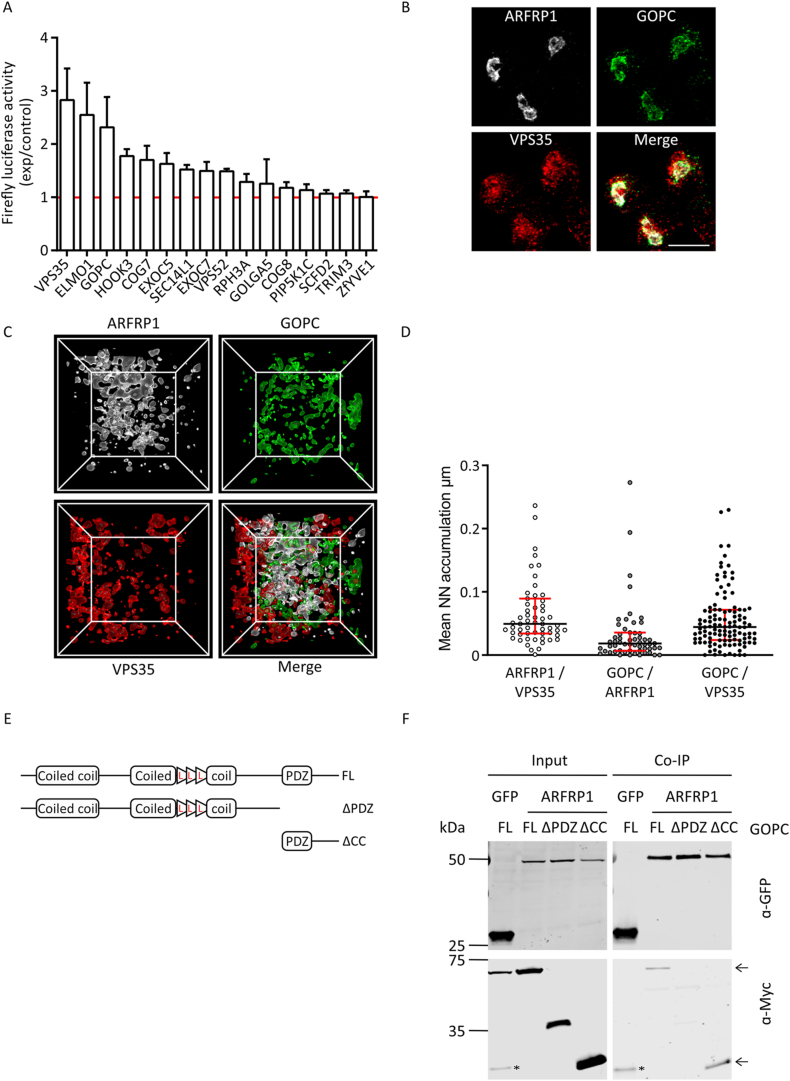
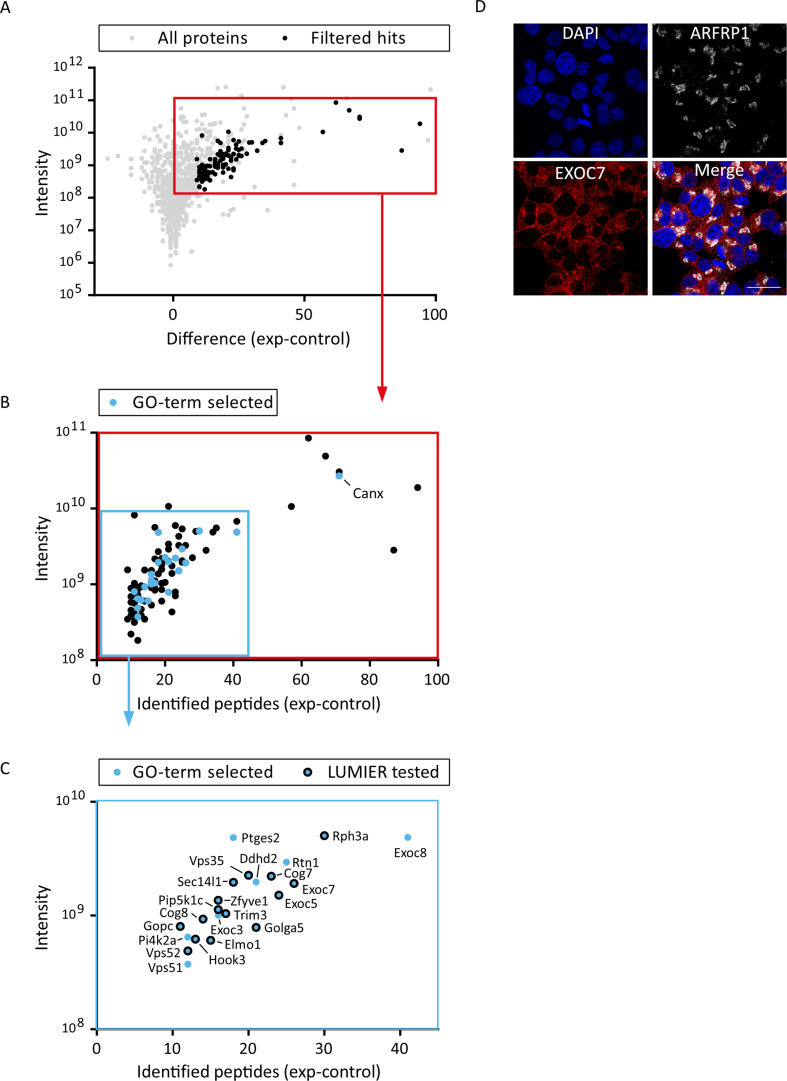
To clarify the mechanistic basis of ARFRP1 function in insulin secretion, we screened for interacting proteins. To this end, pulldown experiments were performed with purified recombinant GST-ARFRP1 and mouse brain lysates. The brain was used because of greater tissue availability and significant similarities between the exocytotic machinery in neurons and the islets of Langerhans [37]. Proteins enriched over a GST control were identified by mass spectrometry (Appendix Fig. S2A–C). We identified 98 putative interactors in the pulldown and focused on those proteins with a known Golgi-localization and/or an involvement in vesicular trafficking. From the resulting 23 gene ontology (GO)-term-selected interactors, 16 proteins were chosen as prey proteins for subsequent analysis in a luminescence-based mammalian interactome (LUMIER) assay (Appendix Fig. S2B, marked by blue dots and Appendix Fig S2C). V5-tagged candidates fused to firefly luciferase were co-expressed with protein A-tagged ARFRP1 fused to Renilla luciferase in HEK293 cells. The efficiency of co-immunoprecipitation was quantified by measuring firefly luciferase activity (Figure 2A). The top hits consisted of the retromer protein VPS35, the engulfment and cell motility protein ELMO1, the microtubule-tethering protein 3 (HOOK3), the exocyst complex components 5 and 7 (EXOC5 and EXOC7), and the Golgi-associated PDZ and coiled-coil motif-containing protein (GOPC). ELMO1 was excluded from further analysis since it is barely expressed in β-cells [38], and HOOK3 was excluded due to its described localization at the cis-Golgi [39]. EXOC7, tested representatively for the exocyst complex, showed no colocalization with ARFRP1 in Ins1 cells and was therefore excluded as well (Appendix Fig. S2D).
Figure 2.
GOPC is an ARFRP1-interacting protein. (A) Validation of ARFRP1 interacting proteins by LUMIER assay. Red line indicates baseline at 1 after normalization to illustrate fold change over control. Assay was performed in triplicates. (B) Immunofluorescent pictures of Ins1 cells stained with indicated antibodies. Scale bar represents 20 μm. (C) 3D-rendering of a ROI (region of interest) that is marked in Appendix Fig. S3B after structured illumination microscopy of Ins1 cells stained with indicated antibodies. White lines outline the 3-dimensional box containing the immunolabeled proteins. Inner box size = 3.5 ∗ 3.5 μm. (D) Nearest neighbor (NN) distances between surfaces of stained objects were calculated from SIM images. Corrected NN distances (NN distances after subtracting the random object distribution) are displayed as mean per image. Data is shown as median with interquartile range from all images (N = 60) of 3 independent experiments with 2 technical duplicates. ∗ = p < 0.05 (two-tailed Man–Whitney test). (E) Schematic of GOPC expression constructs used for immunoprecipitation assays. L = Leucine zipper. FL = full length GOPC, ΔPDZ = lacks PDZ domain, ΔCC = lacks both coiled-coil domains. Domains are not to scale. F Co-immunoprecipitation assays of Myc-tagged GOPC-constructs described in (E) and GFP-tagged ARFRP1 and GFP alone. Membranes were probed with indicated antibodies. ∗ = GFP alone in the Myc-blot results from probing the membrane first with GFP antibody, ← = co-precipitated proteins.
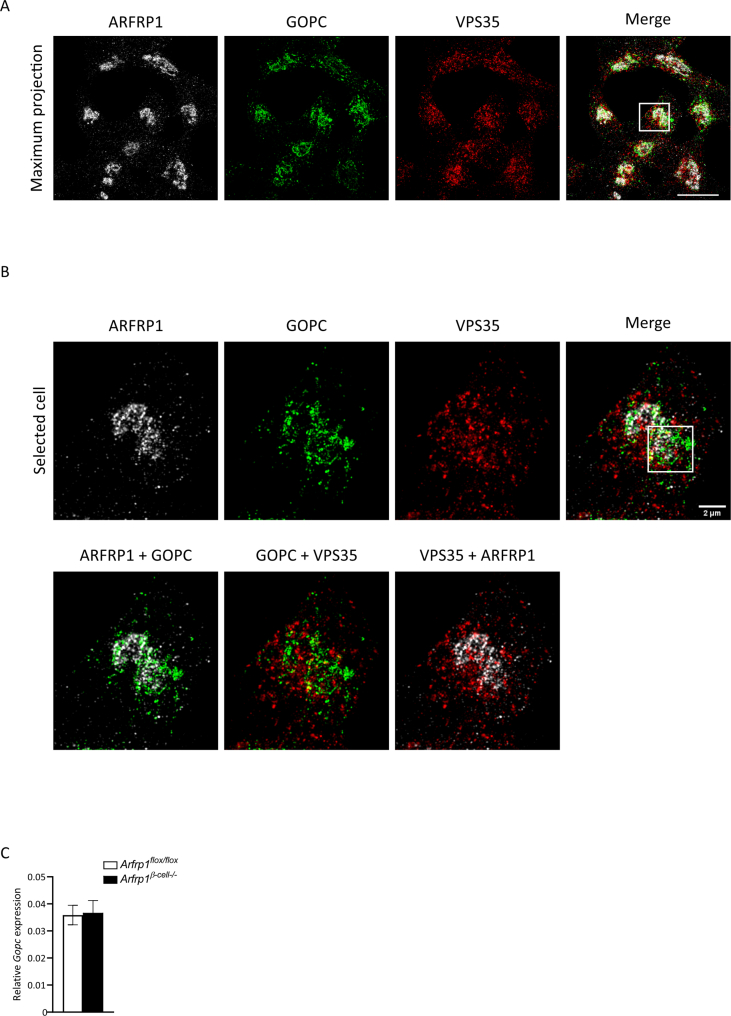
VPS35 represents the top hit with a 2.8-fold higher luciferase activity compared to control, while GOPC was enriched 2.3 times over control (Figure 2A). These two candidates were chosen for further analysis. By immunostaining of the endogenous proteins in rat insulinoma Ins1 cells, a model for pancreatic β-cells, a Golgi-like pattern for GOPC was determined, supported by colocalization with ARFRP1 (Figure 2B). VPS35 appeared in vesicular structures, distributed all over the cell, with an enrichment in proximity to ARFRP1 and GOPC (Figure 2B).
A more precise analysis of the subcellular localization of ARFRP1 and the putative interacting proteins on the nano-scale was achieved by quantitative 3D multicolor SIM (Figure 2C, Appendix Fig. 3A and B, Appendix animation 1). The quantitative analysis from the SIM images (Figure 2D) revealed a close association of the identified candidates VPS35 and GOPC towards ARFRP1, as well as of VPS35 and GOPC towards each other, which was not due to a random distribution within the cell. The analysis also indicated a closer association between ARFRP1 and GOPC than between ARFRP1 and VPS35. Based on these results, we focused on the putative ARFRP1-GOPC interaction in further experiments.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.molmet.2020.101151
The following is the supplementary data related to this article:
Initially, co-immunoprecipitation experiments were performed. With the help of the annotated domain structure of GOPC [40], truncation mutants were generated lacking either the coiled-coil domains (here ΔCC) or the PDZ domain (here ΔPDZ). GFP-tagged ARFRP1 was co-expressed with Myc-tagged GOPC variants in HeLa cells and immunoprecipitation was conducted using GFP-traps. ARFRP1 co-precipitated preferentially with full-length (FL) GOPC as well as GOPC-ΔCC (Figure 2F, marked by arrows). These results point to the PDZ domain of GOPC as interaction platform for ARFRP1.
3.3. GOPC is required for glucose-stimulated insulin secretion
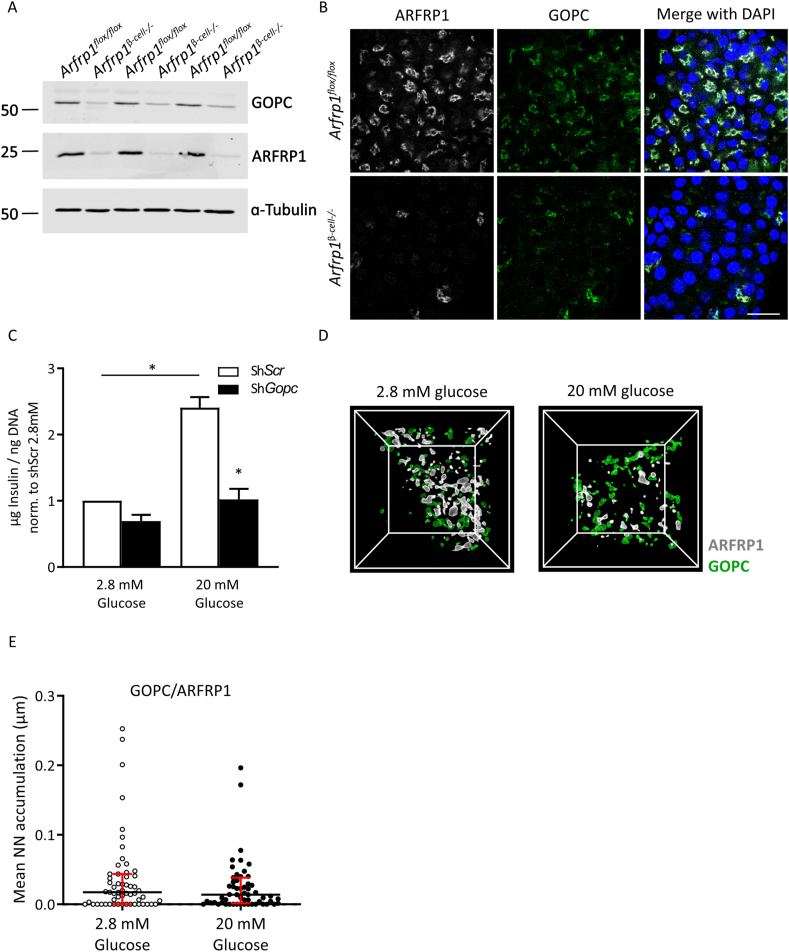
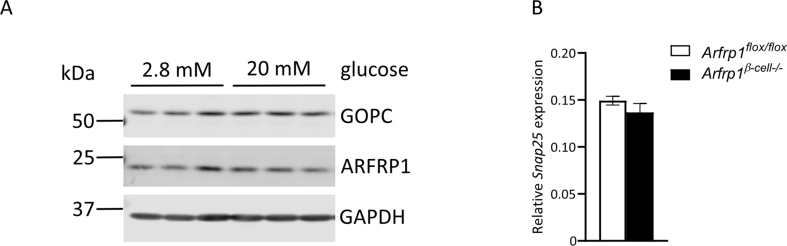
We next addressed the question of whether the ARFRP1-GOPC interaction is of functional relevance for glucose-stimulated insulin secretion. In lysates of isolated islets from Arfrp1β-cell−/− mice, GOPC levels were markedly reduced in comparison to controls, whereas no change in mRNA levels was observed (Figure 3A and Appendix Fig. S3C). Immunostainings of dispersed islets isolated from Arfrp1flox/flox and Arfrp1β-cell−/− mice underlined this observation, as signals for GOPC were only detected in cells expressing ARFRP1 (Figure 3B). These observations suggest that GOPC stability is impaired in the absence of Arfrp1. To test whether GOPC is also required for glucose-stimulated insulin secretion, its expression was suppressed in Min6 cells via adenoviral delivery of a specific shRNA. Min6 cells are an established murine model system for β-cells to measure glucose-stimulated insulin secretion. Strikingly, downregulation of Gopc led to significant reduction of glucose-stimulated insulin secretion (Figure 3C).
Figure 3.
GOPC is required for glucose-stimulated insulin secretion. (A) Western blot analysis of isolated islets from Arfrp1flox/flox and Arfrp1β-cell−/− mice probed with indicated antibodies. (B) Immunofluorescent pictures of dispersed islet cells from Arfrp1flox/flox and Arfrp1β-cell−/− mice stained with indicated antibodies. Nuclei were visualized with DAPI. Scale bar represents 20 μm. (C) Quantification of glucose-stimulated insulin secretion from Min6 cells treated with indicated adenoviruses with a MOI of 50. Data from 3 independent experiments measured in duplicates is shown as mean ± SEM. ∗p < 0.05 (2-way ANOVA). (D) 3D-rendering after structured illumination microscopy of Ins1 cells under basal (2.8 mM glucose) and stimulated (20 mM glucose) conditions stained for GOPC (green) and ARFRP1 (white). White lines outline the 3-dimensional box containing the immunolabeled proteins. Inner box size = 3.5 ∗ 3.5 μm. (E) Nearest neighbor (N/N) distances between surfaces of stained objects (GOPC and ARFRP1) were calculated from SIM images. Corrected NN distances (NN distances after subtracting the random object distribution) are displayed as mean per image. Data is shown as median with interquartile range from all images (N = 60) of 3 independent experiments with 2 technical duplicates ∗ = p < 0.05 (two-tailed Man–Whitney test).
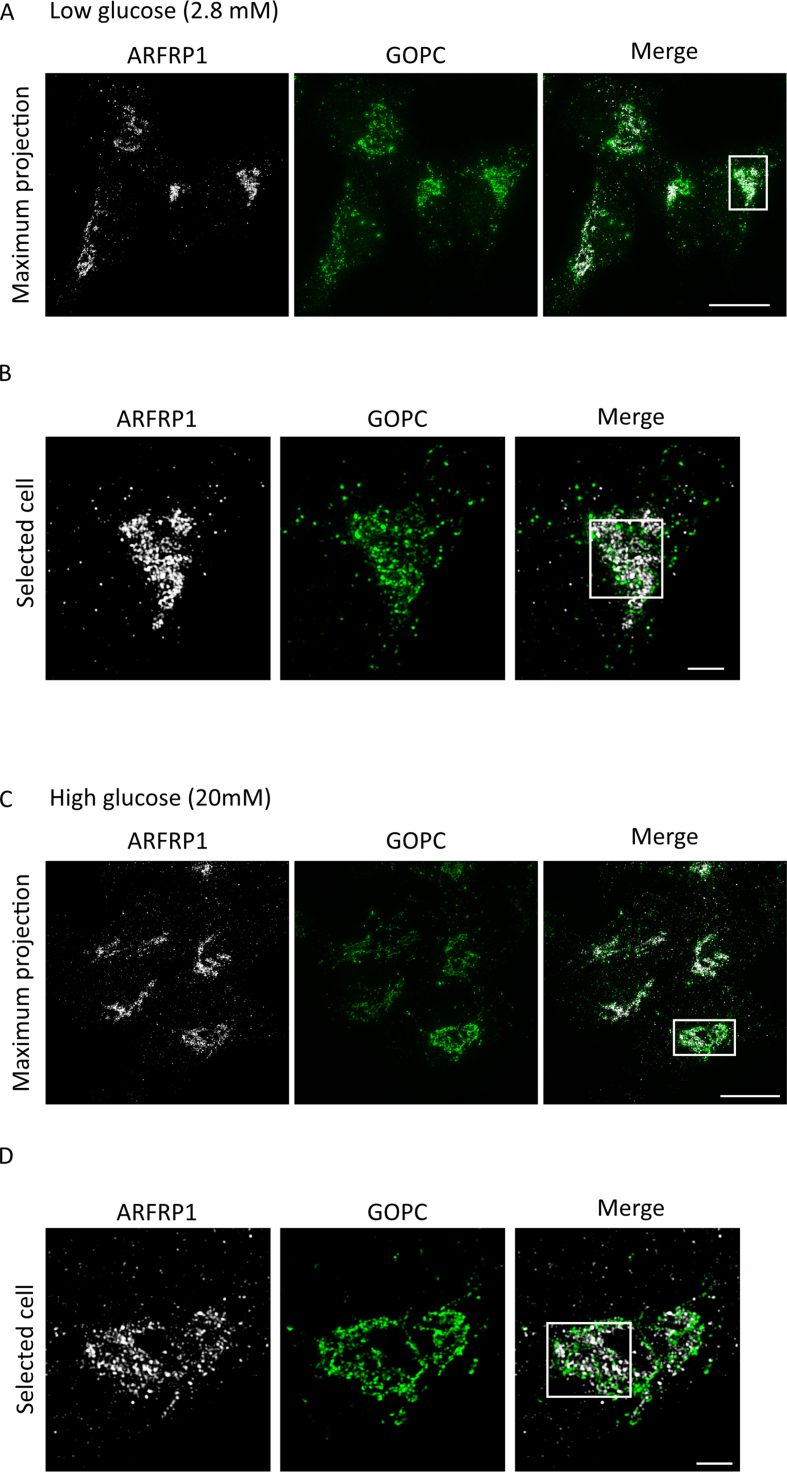
To test whether GOPC changes its localization and/or the degree of co-localization with ARFRP1 upon glucose stimulation, both proteins were stained in Ins1 cells under basal and glucose-stimulated conditions. Protein levels of ARFRP1 and GOPC were not altered in any of these conditions (Appendix Fig. S4A). 3D-SIM confirmed the close association of ARFRP1 toward GOPC (Figure 3D,E, Appendix Figure S5, Appendix animation 2 + 3) that was not altered after stimulation with high glucose levels.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.molmet.2020.101151
The following is the supplementary data related to this article:
2
Taken together, our data suggest that the Golgi-located scaffolding protein GOPC has an important role in glucose-stimulated insulin secretion.
3.4. Deletion of Arfrp1 leads to enhanced degradation of SNAP25
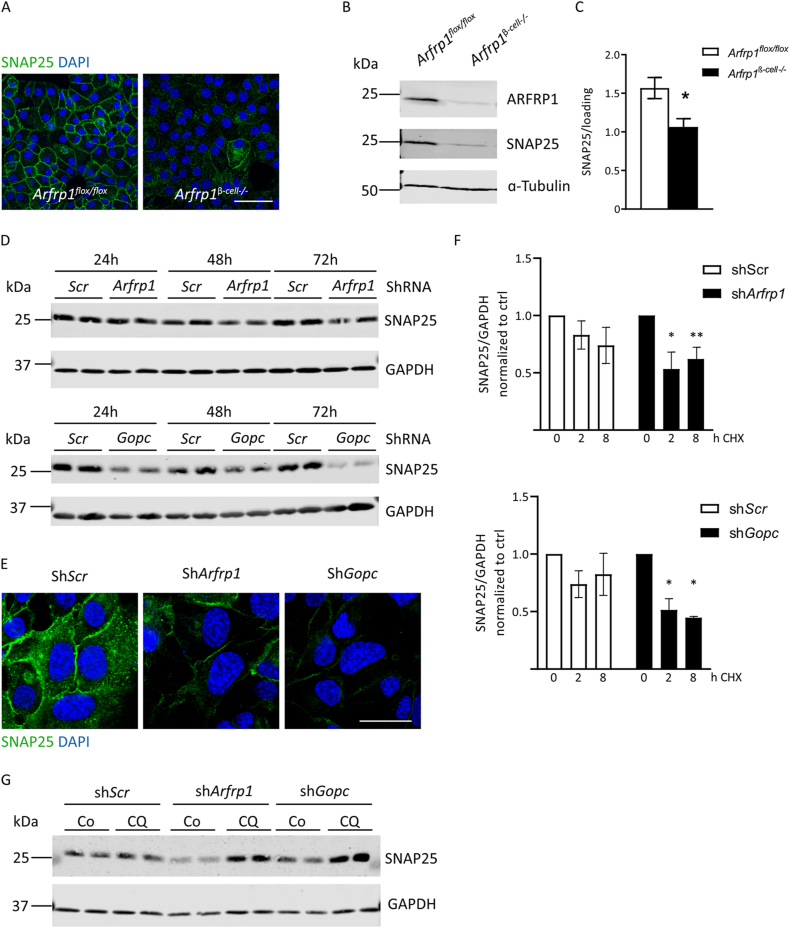
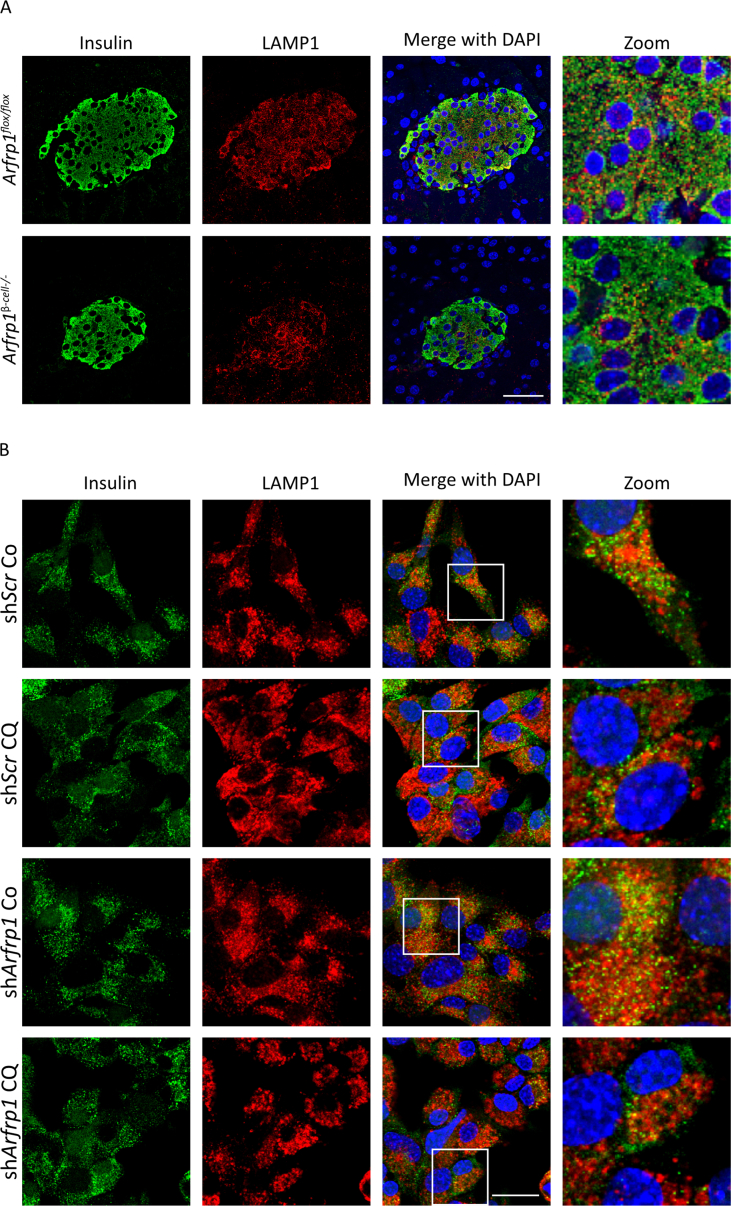
To assign ARFRP1 and GOPC to a specific step in the secretory process, the exocytotic machinery was further studied. First, we tested whether enhanced lysosomal degradation of freshly generated insulin granules might explain the reduced insulin secretion of Arfrp1β-cell−/− mice. Pancreatic sections of Arfrp1flox/flox and Arfrp1β-cell−/− mice were co-stained for insulin and the lysosomal marker LAMP1, without showing an elevated overlap of insulin with LAMP1 in islets of Arfrp1β-cell−/− mice (Appendix Fig. S6A). Similarly, no differences in the co-localization of insulin and LAMP1 were detected in Min6 cells transfected with an Arfrp1-specific shRNA and treated with chloroquine or a solvent control (Appendix Fig. S6B). These results indicate that insulin granules of Arfrp1β-cell−/− cells are not degraded to a higher degree than in controls.
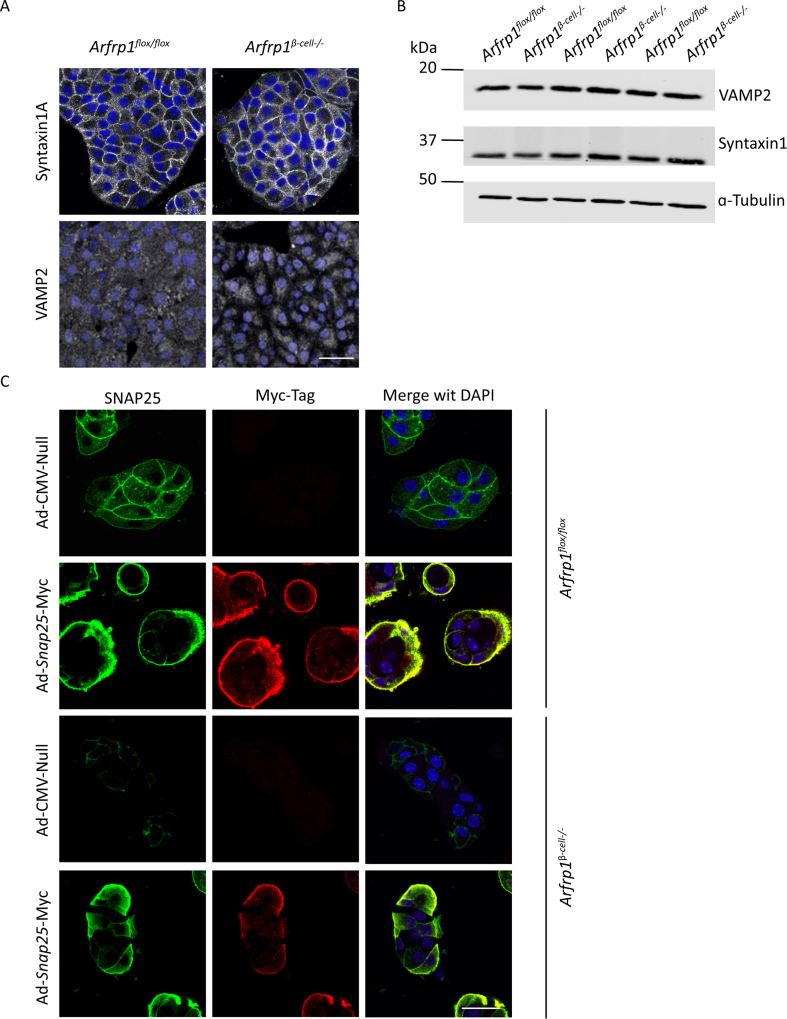
Vesicle-mediated exocytosis is comparable between β-cells and the neuronal system [37]. For calcium-dependent insulin and neurotransmitter release, the SNARE core proteins Syntaxin1A, Vamp2 and SNAP25 are required [41,42]. The important role of SNARE proteins and corresponding factors is underlined by the fact that levels of SNAP25, VAMP2, and Syntaxin1, for instance, are reduced in islets of T2D patients [43,44]. VAMP2 localizes to secretory vesicles, while Syntaxin1A is anchored to the plasma membrane. This localization was confirmed by immunostaining of both proteins in dispersed islet cells derived from Arfrp1flox/flox and Arfrp1β-cell−/− mice. No difference was noted between the two genotypes (Appendix Fig. S7A). Total protein levels of VAMP2 and Syntaxin1A were also not affected (Appendix Fig. S7B). Analysis of the subcellular localization of SNAP25, however, revealed reduced protein levels at the plasma membrane in β-cells lacking Arfrp1 (Figure 4A). Furthermore, SNAP25 protein levels were also significantly reduced in islets from Arfrp1β-cell−/− mice (Figure 4B,C), whereas mRNA expression levels were unaltered (Appendix Fig. S4B). This suggests that the changes occur at the protein level rather than at the gene expression level. A comparable effect on SNAP25 levels was observed upon downregulation of Gopc in Min6 cells, an effect which increased over time (Figure 4D). Of note is that the effect on SNAP25 levels is stronger in Gopc knockdown conditions compared to Arfrp1 knockdown conditions. This finding was recapitulated by immunostainings of SNAP25 in Min6 cells after knockdown (Figure 4E). The protein half-life of SNAP25 is 8–10 h in PC12 cells [45]. Treatment of Min6 cells with the protein translation inhibitor cycloheximide revealed a significantly reduced half-life time of SNAP25 upon downregulation of either Arfrp1 or Gopc when compared to control cells (Figure 4F). This supports the idea that SNAP25 is more rapidly degraded in the absence of ARFRP1 and GOPC. The enhanced degradation of SNAP25 upon knockdown of either Arfrp1 or Gopc could be blocked by treatment with chloroquine, an inhibitor of lysosomal maturation (Figure 4G). These data suggest that the ARFRP1-dependent Golgi-scaffold GOPC is required for the correct localization of SNAP25 at the plasma membrane and that knockdown of Arfrp1 or Gopc results in enhanced lysosomal degradation of SNAP25.
Figure 4.
Disruption of ARFRP1 dependent scaffold leads to degradation of SNAP25. (A) Immunofluorescent pictures of dispersed islet cells from Arfrp1flox/flox and Arfrp1β-cell−/− mice stained for SNAP25. Nuclei are visualized by DAPI, scale bar represents 40 μm. (B) Western blot analysis of islets isolated from Arfrp1flox/flox and Arfrp1β-cell−/− mice probed with indicated antibodies. (C) Quantification of Western blots shown in (B) of 8 mice per genotype. Relative SNAP25 levels normalized to loading control is shown as mean ± SEM. ∗ = p < 0.05 (unpaired students t-test, Welch's correction). (D) Western blot analysis of whole cell lysates from Min6 cells treated with indicated adenoviruses at a MOI of 100 and harvested at indicated time points post-infection. Knockdown efficiency for Arfrp1 is 50% after 48 h, and 60–70% for Gopc after 48 h. Membranes were probed with indicated antibodies. (E) Immunofluorescent pictures of Min6 cells infected with indicated adenoviruses. Nuclei were visualized with DAPI. Scale bar represents 20 μm. (F) Quantification of Western blots from whole cell lysates of Min6 cells treated with indicated adenoviruses and cycloheximide (CHX) for indicated time points in 3 independent experiments. Membranes were probed with indicated antibodies. (G) Western blot analysis of whole cell lysates from Min6 cells treated with indicated adenoviruses and chloroquine (CQ) or solvent control (Co = MiliQ) and harvested 36 h post-infection. Membranes were probed with indicated antibodies.
3.5. GOPC-based Golgi scaffold controls glucose-stimulated insulin release via SNAP25
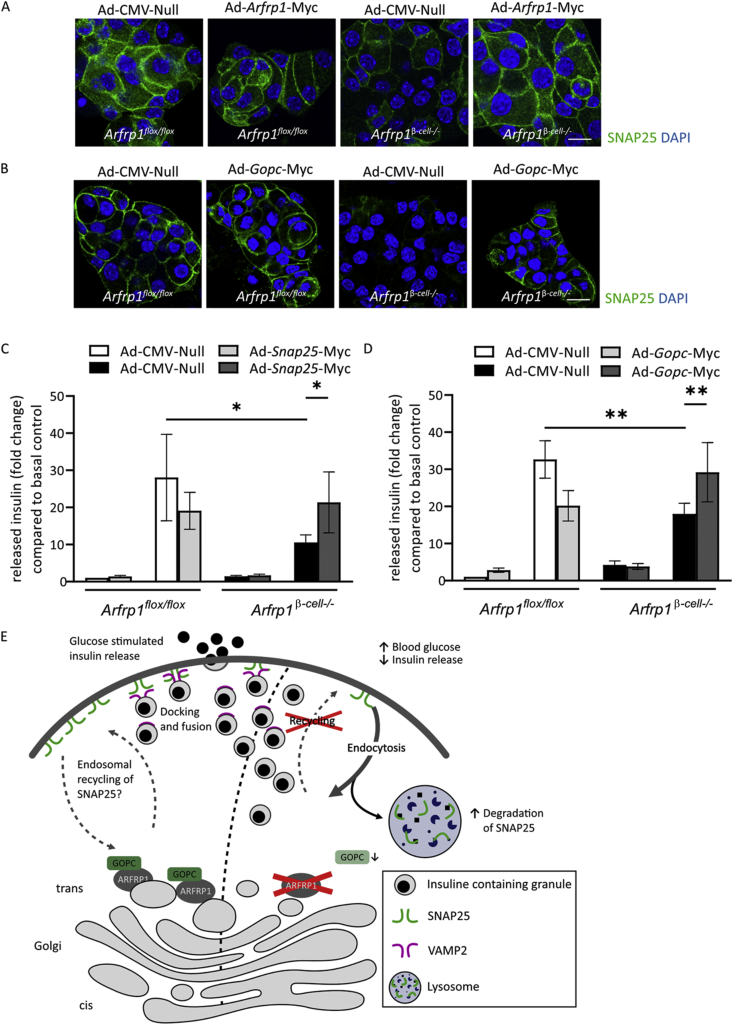
To provide evidence that ARFRP1 and GOPC are directly involved in glucose-stimulated insulin secretion by regulating plasma membrane localization and stability of SNAP25, rescue experiments were conducted. Dispersed islet cells isolated from Arfrp1flox/flox and Arfrp1β-cell−/− mice were treated with adenoviral particles encoding either an empty vector (Ad-CMV-Null) or a Myc-tagged Arfrp1 variant (Ad-Arfrp1-Myc) (Figure 5A). Immunolabeling of SNAP25 followed by confocal microscopy revealed that adenoviral re-expression of Arfrp1 rescued the loss of SNAP25: In dispersed islet cells from Arfrp1β-cell−/− mice expressing Ad-Arfrp1-Myc, an increased plasma membrane localization of SNAP25 was observed (Figure 5A, right picture). The same positive effect on SNAP25 localization was observed upon ectopic expression of Gopc in dispersed islet cells from Arfrp1β-cell−/− mice (Figure 5B).
Figure 5.
SNAP25 phenotype can be rescued by restoring ARFRP1 dependent scaffold. (A) and (B) Immunofluorescent pictures of dispersed islet cells from Arfrp1flox/flox and Arfrp1β-cell−/− mice treated with indicated adenoviruses at a MOI of 100, fixed and stained for SNAP25 48 h post-infection. Nuclei visualized with DAPI, scale bar represents 10 μm. C Quantification of glucose stimulated insulin secretion from islets isolated from ad libitum fed 11- to 12-week-old Arfrp1flox/flox (n = 4) and Arfrp1β-cell−/− (n = 5) mice infected in duplicates with indicated adenoviruses. Released insulin normalized to residual insulin is shown as fold change compared to 2.8 mM of control. Cells were stimulated with 20 mM of glucose. Data is shown as mean ± SEM. ∗ = p ≤ 0.05 (2-way ANOVA). (D) Quantification of glucose stimulated insulin secretion from islets isolated from ad libitum fed 12-week-old Arfrp1flox/flox (n = 3) and Arfrp1β-cell−/− (n = 3) mice infected in duplicates with indicated adenoviruses. Released insulin normalized to residual insulin is shown as foldchange compared to 2.8 mM of control. Cells were stimulated with 20 mM of glucose. Data is shown as mean ± SEM. ∗ = p ≤ 0.05 (2-way ANOVA). (E) Graphical summary. In the absence of ARFRP1, the GOPC scaffold cannot assemble at the TGN. This leads to enhanced degradation of SNAP25. Insulin secretion levels are reduced, and blood glucose is elevated.
Next, Snap25 was re-expressed in islets from Arfrp1flox/flox and Arfrp1β-cell−/− mice and glucose-stimulated insulin secretion was examined (Figure 5C). Quantitative measurements of released insulin recapitulated the reduced insulin secretion capacity of islets from Arfrp1β-cell−/− mice treated with the control virus. Overexpression of Snap25 in islets from Arfrp1flox/flox mice resulted in slightly reduced insulin secretion, without reaching significance, which might be explained by altered stoichiometry of the SNARE complex upon SNAP25 overexpression. Strikingly, re-expression of Snap25 was sufficient to restore glucose-stimulated insulin secretion in islets from Arfrp1β-cell−/− mice nearly to the level of controls (Figure 5C). This indicates that ARFRP1 and GOPC are not required for an appropriate delivery of SNAP25 from the Golgi to the plasma membrane. In agreement with this, overexpressed SNAP25 localized to the plasma membrane in isolated islet cells from Arfrp1flox/flox and Arfrp1β-cell−/− mice (Appendix Figure S7C).
We reasoned that lack of ARFRP1 may either enhance the internalization or impair the recycling of SNAP25, leading to its enhanced degradation. Since re-expression of Gopc in islet cells from Arfrp1β-cell−/− mice increased SNAP25 levels at the plasma membrane, glucose-stimulated insulin release was analyzed under these conditions. As seen for Snap25, overexpression of Gopc reduced glucose-stimulated insulin release in islet cells from Arfrp1flox/flox mice (Figure 5D) which again might indicate changes in stoichiometry of the SNARE complex. Insulin release from islets isolated from Arfrp1β-cell−/− mice treated with the control virus was significantly lower compared to islet cells from Arfrp1flox/flox mice. Upon re-expression of Gopc in islet cells from Arfrp1β-cell−/− mice, however, released insulin levels significantly increased (Figure 5D). Taken together, these data suggest that deletion of Arfrp1 in pancreatic β-cells disrupts a GOPC-based Golgi scaffold, leading to reduced SNAP25 levels at the plasma membrane, presumably by enhanced internalization or defective recycling and elevated lysosomal degradation of SNAP25 (summarized in Figure 5E). This might explain the higher blood glucose and lower insulin levels of Arfrp1β-cell−/− mice.
4. Discussion
This study describes the identification of a newly revealed interaction of the Golgi protein GOPC with the GTPase ARFRP1 and its impact on insulin secretion from pancreatic β-cells. ARF proteins are known regulators of cellular trafficking and signaling events [17,46]. Their impact on β-cell function is underlined by the described role of ARF6 on maintaining PI(4,5)P2 levels required for regulated exocytosis of insulin. Our data indicate that a direct or indirect interaction of the ARF-like protein ARFRP1 and the scaffolding protein GOPC is required for correct localization of SNAP25 at the plasma membrane of β-cells. This interaction involves the C-terminal PDZ-domain of GOPC. In the absence of either ARFRP1 or GOPC, SNAP25 is degraded, and insulin secretion markedly reduced.
Membrane scaffolds are spatiotemporally regulated assemblies of multidomain proteins on organellar membranes and important mediators of cellular signaling and trafficking events, such as vesicle-mediated regulated exocytosis. The scaffolding protein GOPC may represent a platform for protein–protein interactions, which is facilitated by its domain structure, consisting of two N-terminal coiled-coil domains and a C-terminal PDZ domain. GOPC is described to be a Rab6a effector protein, since the first coiled-coil domain was found to interact with Rab6a [47]. The second coiled-coil domain interacts with Syntaxin6 and the GTPase Tc10 and is required for the Golgi localization and suggested to mediate a dimerization of the protein [40,48]. The PDZ domain interacts with C-termini of different membrane proteins, such as the cystic fibrosis transmembrane regulator, glutamate receptor 2, and CD46-Cyt1 [[49], [50], [51]]. Furthermore, GOPC was shown to be involved in postendocytic trafficking as it protects the β1-adrenergic receptor from lysosomal degradation [52]. We identified the C-terminus of the protein that includes the PDZ domain of GOPC as a region to which ARFRP1 might bind. Whether this is a direct interaction needs to be further proven by biochemical assays. PDZ domains are described to bind to the C-terminal residues of target proteins [53]. ARFRP1, however, lacks a classical PDZ binding motif in its C-terminal region, which may indicate an indirect interaction between the two proteins. A sequence upstream of the PDZ domain of GOPC was shown to be required for the interaction with Stargazin, a regulator of AMPA-selective glutamate receptors [54]. This particular sequence is however not included in our ΔCC construct that contains only the PDZ domain.
Rising blood glucose levels stimulate insulin release from pancreatic β-cells, a well-known example for regulated membrane trafficking and exocytosis. Intracellular trafficking routes are often shared by different transporters or cargos. Therefore, regulatory mechanisms are required for every target to reach its correct destination, and the secretory machinery needs to quickly adapt to various extracellular stimuli. The here-identified new ARFRP1-interacting protein GOPC is required for glucose-stimulated insulin release. Disruption of this interaction leads to reduced GOPC protein levels and affects glucose homeostasis in mice, as shown by elevated blood glucose levels, impaired glucose tolerance, and reduced insulin secretion capacity in Arfrp1β-cell−/− mice. Despite this impaired insulin secretion, Arfrp1β-cell−/− mice did not become diabetic. However, we observed slightly enlarged islets in Arfrp1β-cell−/− mice, which might be a compensatory effect to guarantee an overall lower but still sufficient insulin release in order to prevent severe hyperglycemia and the development of diabetes. This compensatory capacity can be linked to the genetic background of the knockout animals (C57BL/6), that do not become diabetic because it induces β-cell proliferation and thereby increases insulin secretion when blood glucose levels rise [33,36]. Insulin is well known to control food intake and satiety [55,56]. Since plasma insulin levels are reduced in Arfrp1β-cell−/− mice, they might exhibit an elevated food intake leading to the observed elevated fat mass.
The fact that deletion of Arfrp1 reduced glucose-stimulated insulin secretion but not total pancreatic insulin levels or the maturation of insulin indicates a secretory defect. Since both the first and second phase of insulin secretion were reduced in islets from Arfrp1β-cell−/− mice, it can be concluded that the ARFRP1-GOPC interaction is required for the fusion process of insulin-containing granules with the plasma membrane. In fact, suppression of both Arfrp1 or Gopc in Min6 cells resulted in lower levels of SNAP25 (Figure 4D,E), a protein which is essential for fusion of vesicles with the plasma membrane in order to release their cargo [57]. Our data indicate that both ARFRP1 and GOPC are needed for the correct localization of SNAP25 at the plasma membrane. Since overexpressed SNAP25 in Arfrp1β-cell−/− islets localizes to the plasma membrane, we can exclude that ARFRP1 and GOPC regulate targeting of newly synthesized SNAP25 to the plasma membrane. In their absence, SNAP25 may be internalized faster or not sorted to the appropriate recycling vesicles and thereby degraded, resulting in reduced protein levels and a significantly reduced protein half-life time. In fact, the degradation of SNAP25 was blocked by treating cells with chloroquine, suggesting that SNAP25 is degraded via the lysosome in the absence of ARFRP1 or GOPC. Accordingly, ARFRP1 was described to be involved in retrograde trafficking from endosomal structures to the TGN [22,58]. SNAP25 located at the plasma membrane was described to be internalized via dynamin-independent endocytosis and to be recycled in an ARF6-dependent manner. In the same study, the SNAP25-interacting SNARE protein Syntaxin1A, which is also relevant for insulin release from pancreatic β-cells, was shown not to be co-internalized with SNAP25 [59]. This agrees with our observations that SNAP25 is the only SNARE protein of the VAMP2-STX1A-SNAP25 complex affected by Arfrp1 and Gopc knockdown (Appendix Fig. S7A + B). The fact that only SNAP25 but not VAMP2 and STX1A is degraded after suppression of Arfrp1 and Gopc1 might indicate that SNAP25 can act independently of this complex. A role for SNAP25 in endosome fusion was suggested but not further investigated [60]. We observed earlier that a related SNARE protein, SNAP23, was missorted in adipocytes from adipose tissue-specific Arfrp1 knockout mice without affecting total protein levels [19]. We did not detect an interaction of ARFRP1 or GOPC with SNAP25 by co-immunoprecipitation assays (Appendix Fig. S8). Thus, it remains to be clarified whether and how ARFRP1 and GOPC mediate SNAP25 sorting. A β-cell-specific Snap25 knockout mouse has not been described so far. However, a mouse carrying a mutation in the Syntaxin1 binding site of SNAP25 shows impaired granule exocytosis in pancreatic β-cells along with elevated blood glucose levels [61], supporting the important role of SNAP25-dependent steps for insulin secretion. Further support is given by our rescue experiments showing that re-expression of Snap25 in islets of Arfrp1β-cell−/− mice is sufficient to restore the capacity of glucose-stimulated insulin release. Comparable effects on restoring GSIS levels were observed when re-expressing either Syntaxin1a or Snap25 in islets of GK rats, a lean model for T2D that possess reduced protein levels of Syntaxin1A and SNAP25 [62].
In summary, ARFRP1 acts at the TGN by recruiting effector proteins to regulate endosomal recycling processes, which affect hormone release from metabolic active tissues, as shown earlier for adiponectin from adipocytes and IGF1 release from hepatocytes [20,27]. From the data presented in this study, we conclude that both ARFRP1 and GOPC are involved in the correct localization of SNAP25 at the plasma membrane and thereby in glucose-stimulated secretion of insulin from pancreatic β-cells. This describes a hitherto unrecognized pathway required for insulin secretion at the level of trans-Golgi sorting.
Author contributions
A.S. initiated the project. I.W. performed research and analyzed the data. T.L. performed mice phenotyping. S.G. performed GST-pulldowns and LUMIER assays with help of A.A., and supervised by O.D. and E.E.W., N.G. and J.S. devised and performed the SIM imaging and analysis. S.G., O.P., G.D. performed mass spec and analyzed the data. I.W. and A.S. wrote the manuscript. All authors critically reviewed the manuscript.
6Acknowledgments
We thank Mandy Stadion for initiating the Arfrp1flox/flox x Ins1Cre cross, Professor Hans-Jürgen Kreienkamp for sharing PIST(GOPC) antiserum, Professor Jun-ichi Miyazaki from Japan for providing Min6 cells, and Anett Helms, Andrea Teichmann, Josefine Würfel, and Eva Arlt for their excellent technical support. This study was supported by the German Research Foundation [DFG, SFB 958, A12 to O.D., Z02 to J.S., and A13 to A.S.], the German Federal Ministry of Education and Research, by the Federal State of Brandenburg [BMBF, DZD grant 82DZD00302], and the German Diabetes Society [DDG, project funding].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101151.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
(A) Breeding strategy to establish β-cell specific knockout mice (Arfrp1β-cell−/−). (B) Relative mRNA expression of Arfrp1 in different tissues from 2 mice per genotype. Data is shown as mean ± SEM. (C) Western blot analysis of indicated tissues from Arfrp1flox/flox and Arfrp1β-cell−/− mice probed with indicated antibodies. (D) Relative expression levels of Arfrp1 measured in islets of Arfrp1flox/flox (n = 3) and Arfrp1β-cell−/− (n = 6) female mice. Data is shown as mean ± SEM. ∗∗ = p < 0.01 (unpaired Student's t-test, Welch's correction). (E) Relative expression levels of Ins2 measured in islets of Arfrp1flox/flox (n = 3) and Arfrp1β-cell−/− (n = 6) female mice. Data is shown as mean ± SEM. (F) Body weight measurement of male Arfrp1flox/flox and Arfrp1β-cell−/− mice. (G) Fat mass of male Arfrp1flox/flox and Arfrp1β-cell−/− mice. (H) Lean mass of male Arfrp1flox/flox and Arfrp1β-cell−/− mice. Data in (F)–(H) are presented as mean ± SEM for Arfrp1flox/flox (n = 12) and Arfrp1β-cell−/− mice (n = 14). ∗ = p < 0.05 (2-way ANOVA). (I) Total pancreatic pro-insulin and (J) C-peptide from ad libitum fed male Arfrp1flox/flox (n = 5) and Arfrp1β-cell−/− mice (n = 5). Data is represented as mean ± SEM. (K) Immunohistochemical staining of insulin (green) and glucagon (red) in pancreatic sections of female Arfrp1flox/flox and Arfrp1β-cell−/− mice. Scale bar represents 40 μm. (L) Quantification of islet number per tissue section. (M) Quantification of islet size from pancreatic tissue sections. Data in (L) + (M) is represented as mean ± SEM from three pancreatic sections of 3 mice per genotype. ∗ = p < 0.05 (unpaired Student's t-test, Welch's correction). (N) Immunohistochemical staining of GLUT2 (yellow) in pancreatic sections of female Arfrp1flox/flox and Arfrp1β-cell−/− mice. Scale bar represents 40 μm.
figs2.
(A) Mass spectrometry analysis for GST-ARFRP1 pulldown. GTPγS-loaded GST-ARFRP1 and a GST control were used in pulldown experiments with mouse brain lysate and analyzed by mass spectrometry, using a spectral counting approach. Shown on the x-axis is the difference of the number of identified peptides for GST-ARFRP1 (exp) minus GST (control) for each protein group. On the y-axis is the total intensity of all peptides identified in exp and control. One thousand one hundred fifty-six protein groups were filtered for those containing at least 10 unique peptide IDs with a total intensity of ≥108 and having a peptide ID ratio (number of peptide IDs identified for GST-ARFRP1 over the total number of identified peptide IDs) of ≥0.9 (black dots). (B) Magnified view of the red box in A, showing the resulting 98 protein groups, which were further refined to proteins with a known Golgi-localization (GO:0005794) and/or an involvement in vesicular trafficking (GO:0016192) (blue dots). Calnexin (Canx) was not further pursued due to its known function in the endoplasmic reticulum. (C) Magnified view of the blue box in B. From the 23 GO-term selected protein groups, 16 were chosen as prey proteins for subsequent analysis in a LUMIER assay (blue dots with black outline). (D) Immunofluorescent pictures of Ins1 cells stained with indicated antibodies. Nuclei were visualized by DAPI. Scale bar represents 20 μm.
figs3.
(A) Structured illumination microscopy of Ins1 cells stained with indicated antibodies (maximum intensity projection). White rectangle marks selected cell. Scale bar = 10 μm. (B) Selected cell as shown in (A). Rectangle represents region of interest picked for 3D rendering shown in Figure 2C. Scale bar = 2 μm. (C) Relative expression levels of Gopc measured in islets of Arfrp1flox/flox (n = 3) and Arfrp1β-cell-/- (n = 6) female mice. Data is shown as mean ± SEM.
figs4.
(A) Western blot analysis of whole cell lysates from Ins1 treated with indicated glucose concentrations. Membranes were probed with indicated antibodies. (B) Relative expression levels of Snap25 measured in islets of Arfrp1flox/flox (n = 3) and Arfrp1β-cell−/− (n = 6) female mice. Data is shown as mean ± SEM.
figs5.
(A) Structured illumination microscopy of Ins1 cells under low glucose conditions stained with indicated antibodies (maximum intensity projection). White rectangle marks selected cell. Scale bar = 10 μm. (B) Selected cell as shown in (A). Rectangle represents region of interest picked for 3D rendering shown in Figure 3D. Scale bar = 2 μm. (C) Structured illumination microscopy of Ins1 cells under high glucose conditions stained with indicated antibodies (maximum intensity projection). White rectangle marks selected cell. Scale bar = 10 μm. (D) Selected cell as shown in (A). Rectangle represents region of interest picked for 3D rendering shown in Figure 3D. Scale bar = 2 μm.
figs6.
(A) Immunohistochemical staining of insulin (green) and LAMP1 (red) in pancreatic sections of female Arfrp1flox/flox and Arfrp1β-cell−/− mice. Nuclei were visualized in blue. Scale bar represents 40 μm. (B) Immunofluorescent pictures of Min6 cells treated with indicated adenoviruses and chloroquine or a solvent control and stained with indicated antibodies. Nuclei were visualized in blue. Scale bar represents 20 μm.
figs7.
(A) Immunofluorescent pictures of dispersed islet cells isolated from Arfrp1flox/flox and Arfrp1β-cell−/− mice stained with indicated antibodies, nuclei visualized in blue. Scale bar represents 20 μm. (B) Western Blots of isolated islets from Arfrp1flox/flox and Arfrp1β-cell−/− mice probed with indicated antibodies. (C) Immunofluorescent pictures of dispersed islet cells isolated from Arfrp1flox/flox and Arfrp1β-cell−/− mice, infected with control adenovirus (Ad-CMV-Null) or a Snap25-Myc encoding adenovirus (Ad-Snap25-Myc) and stained with indicated antibodies, nuclei visualized in blue. Scale bar represents 20 μm.
figs8.
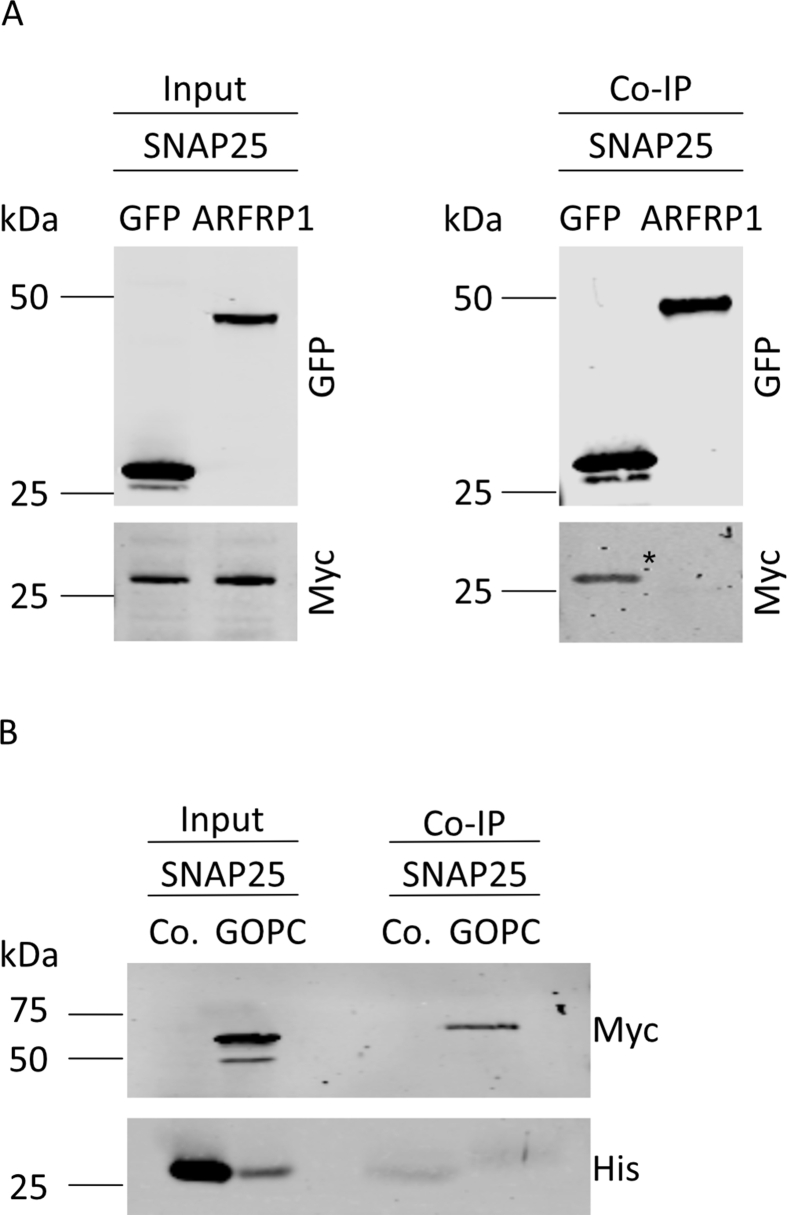
(A) Co-immunoprecipitation assays from HeLa cells co-expressing SNAP25 with GFP-tagged ARFRP1. Membranes were probed with indicated antibodies. (B) Co-immunoprecipitation assays from HeLa cells co-expressing SNAP25 and GOPC. Membranes were probed with indicated antibodies.
References
- 1.Prasad R.B., Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 2015;6(1):87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller M.P., Rabaglia M.E., Schueler K.L., Stapleton D.S., Gatti D.M., Vincent M. Gene loci associated with insulin secretion in islets from non-diabetic mice. Journal of Clinical Investigation. 2019;130:4419–4432. doi: 10.1172/JCI129143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCulloch L.J., van de Bunt M., Braun M., Frayn K.N., Clark A., Gloyn A.L. GLUT2 (SLC2A2) is not the principal glucose transporter in human pancreatic beta cells: implications for understanding genetic association signals at this locus. Molecular Genetics and Metabolism. 2011;104(4):648–653. doi: 10.1016/j.ymgme.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 4.Thorens B. Molecular and cellular physiology of GLUT-2, a high-Km facilitated diffusion glucose transporter. International Review of Cytology. 1992;137:209–238. doi: 10.1016/s0074-7696(08)62677-7. [DOI] [PubMed] [Google Scholar]
- 5.Rorsman P., Trube G. Glucose dependent K+-channels in pancreatic beta-cells are regulated by intracellular ATP. Pflügers Archiv. 1985;405(4):305–309. doi: 10.1007/BF00595682. [DOI] [PubMed] [Google Scholar]
- 6.Arkhammar P., Nilsson T., Rorsman P., Berggren P.O. Inhibition of ATP-regulated K+ channels precedes depolarization-induced increase in cytoplasmic free Ca2+ concentration in pancreatic beta-cells. Journal of Biological Chemistry. 1987;262(12):5448–5454. [PubMed] [Google Scholar]
- 7.Drews G., Krippeit-Drews P., Düfer M. Electrophysiology of islet cells. Advances in Experimental Medicine & Biology. 2010;654:115–163. doi: 10.1007/978-90-481-3271-3_7. [DOI] [PubMed] [Google Scholar]
- 8.Dolai S., Xie L., Zhu D., Liang T., Qin T., Xie H. Synaptotagmin-7 functions to replenish insulin granules for exocytosis in human islet beta-cells. Diabetes. 2016;65(7):1962–1976. doi: 10.2337/db15-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunger A.T. Structure and function of SNARE and SNARE-interacting proteins. Quarterly Reviews of Biophysics. 2005;38(1):1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 10.Lai Y., Choi U.B., Leitz J., Rhee H.J., Lee C., Altas B. Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron. 2017;95(3):591–607. doi: 10.1016/j.neuron.2017.07.004. e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jewell J.L., Luo W., Oh E., Wang Z., Thurmond D.C. Filamentous actin regulates insulin exocytosis through direct interaction with Syntaxin 4. Journal of Biological Chemistry. 2008;283(16):10716–10726. doi: 10.1074/jbc.M709876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie L., Zhu D., Dolai S., Liang T., Qin T., Kang Y. Syntaxin-4 mediates exocytosis of pre-docked and newcomer insulin granules underlying biphasic glucose-stimulated insulin secretion in human pancreatic beta cells. Diabetologia. 2015;58(6):1250–1259. doi: 10.1007/s00125-015-3545-4. [DOI] [PubMed] [Google Scholar]
- 13.Daniel S., Noda M., Straub S.G., Sharp G.W. Identification of the docked granule pool responsible for the first phase of glucose-stimulated insulin secretion. Diabetes. 1999;48(9):1686–1690. doi: 10.2337/diabetes.48.9.1686. [DOI] [PubMed] [Google Scholar]
- 14.Bratanova-Tochkova T.K., Cheng H., Daniel S., Gunawardana S., Liu Y.J., Mulvaney-Musa J. Triggering and augmentation mechanisms, granule pools, and biphasic insulin secretion. Diabetes. 2002;51(Suppl 1):S83–S90. doi: 10.2337/diabetes.51.2007.s83. [DOI] [PubMed] [Google Scholar]
- 15.Gaisano H.Y. Here come the newcomer granules, better late than never. Trends in Endocrinology and Metabolism. 2014;25(8):381–388. doi: 10.1016/j.tem.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Schurmann A., Massmann S., Joost H.G. ARP is a plasma membrane-associated Ras-related GTPase with remote similarity to the family of ADP-ribosylation factors. Journal of Biological Chemistry. 1995;270(51):30657–30663. doi: 10.1074/jbc.270.51.30657. [DOI] [PubMed] [Google Scholar]
- 17.Sztul E., Chen P.W., Casanova J.E., Cherfils J., Dacks J.B., Lambright D.G. ARF GTPases and their GEFs and GAPs: concepts and challenges. Molecular Biology of the Cell. 2019;30(11):1249–1271. doi: 10.1091/mbc.E18-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas L.L., Fromme J.C. Extensive GTPase crosstalk regulates Golgi trafficking and maturation. Current Opinion in Cell Biology. 2020;65:1–7. doi: 10.1016/j.ceb.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hommel A., Hesse D., Volker W., Jaschke A., Moser M., Engel T. The ARF-like GTPase ARFRP1 is essential for lipid droplet growth and is involved in the regulation of lipolysis. Molecular and Cellular Biology. 2010;30(5):1231–1242. doi: 10.1128/MCB.01269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hesse D., Jaschke A., Kanzleiter T., Witte N., Augustin R., Hommel A. GTPase ARFRP1 is essential for normal hepatic glycogen storage and insulin-like growth factor 1 secretion. Molecular and Cellular Biology. 2012;32(21):4363–4374. doi: 10.1128/MCB.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaschke A., Chung B., Hesse D., Kluge R., Zahn C., Moser M. The GTPase ARFRP1 controls the lipidation of chylomicrons in the Golgi of the intestinal epithelium. Human Molecular Genetics. 2012;21(14):3128–3142. doi: 10.1093/hmg/dds140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin H.W., Kobayashi H., Kitamura M., Waguri S., Suganuma T., Uchiyama Y. Roles of ARFRP1 (ADP-ribosylation factor-related protein 1) in post-Golgi membrane trafficking. Journal of Cell Science. 2005;118(Pt 17):4039–4048. doi: 10.1242/jcs.02524. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y., Zanetti G., Schekman R. A novel GTP-binding protein-adaptor protein complex responsible for export of Vangl2 from the trans Golgi network. Elife. 2013;2 doi: 10.7554/eLife.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahn C., Hommel A., Lu L., Hong W., Walther D.J., Florian S. Knockout of Arfrp1 leads to disruption of ARF-like1 (ARL1) targeting to the trans-Golgi in mouse embryos and HeLa cells. Molecular Membrane Biology. 2006;23(6):475–485. doi: 10.1080/09687860600840100. [DOI] [PubMed] [Google Scholar]
- 25.Panic B., Whyte J.R., Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Current Biology. 2003;13(5):405–410. doi: 10.1016/s0960-9822(03)00091-5. [DOI] [PubMed] [Google Scholar]
- 26.Setty S.R., Shin M.E., Yoshino A., Marks M.S., Burd C.G. Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Current Biology. 2003;13(5):401–404. doi: 10.1016/s0960-9822(03)00089-7. [DOI] [PubMed] [Google Scholar]
- 27.Rodiger M., Werno M.W., Wilhelmi I., Baumeier C., Hesse D., Wettschureck N. Adiponectin release and insulin receptor targeting share trans-Golgi-dependent endosomal trafficking routes. Mol Metab. 2018;8:167–179. doi: 10.1016/j.molmet.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zahn C., Jaschke A., Weiske J., Hommel A., Hesse D., Augustin R. ADP-ribosylation factor-like GTPase ARFRP1 is required for trans-Golgi to plasma membrane trafficking of E-cadherin. Journal of Biological Chemistry. 2008;283(40):27179–27188. doi: 10.1074/jbc.M802108200. [DOI] [PubMed] [Google Scholar]
- 29.Thorens B., Tarussio D., Maestro M.A., Rovira M., Heikkila E., Ferrer J. Ins1(Cre) knock-in mice for beta cell-specific gene recombination. Diabetologia. 2015;58(3):558–565. doi: 10.1007/s00125-014-3468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 31.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 32.Palidwor G.A., Shcherbinin S., Huska M.R., Rasko T., Stelzl U., Arumughan A. Detection of alpha-rod protein repeats using a neural network and application to huntingtin. PLoS Computational Biology. 2009;5(3) doi: 10.1371/journal.pcbi.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kluth O., Matzke D., Kamitz A., Jahnert M., Vogel H., Scherneck S. Identification of four mouse diabetes candidate genes altering beta-cell proliferation. PLoS Genetics. 2015;11(9) doi: 10.1371/journal.pgen.1005506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brissova M., Fowler M.J., Nicholson W.E., Chu A., Hirshberg B., Harlan D.M. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. Journal of Histochemistry and Cytochemistry. 2005;53(9):1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- 35.Bensellam M., Laybutt D.R., Jonas J.C. The molecular mechanisms of pancreatic β-cell glucotoxicity: recent findings and future research directions. Molecular and Cellular Endocrinology. 2012;364(1–2):1–27. doi: 10.1016/j.mce.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Kleinert M., Clemmensen C., Hofmann S.M., Moore M.C., Renner S., Woods S.C. Animal models of obesity and diabetes mellitus. Nature Reviews Endocrinology. 2018;14(3):140–162. doi: 10.1038/nrendo.2017.161. [DOI] [PubMed] [Google Scholar]
- 37.Lang J. Molecular mechanisms and regulation of insulin exocytosis as a paradigm of endocrine secretion. European Journal of Biochemistry. 1999;259(1–2):3–17. doi: 10.1046/j.1432-1327.1999.00043.x. [DOI] [PubMed] [Google Scholar]
- 38.Schaum N. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562(7727):367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walenta J.H., Didier A.J., Liu X., Krämer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. The Journal of Cell Biology. 2001;152(5):923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neudauer C.L., Joberty G., Macara I.G. PIST: a novel PDZ/coiled-coil domain binding partner for the rho-family GTPase TC10. Biochemical and Biophysical Research Communications. 2001;280(2):541–547. doi: 10.1006/bbrc.2000.4160. [DOI] [PubMed] [Google Scholar]
- 41.Lin R.C., Scheller R.H. Structural organization of the synaptic exocytosis core complex. Neuron. 1997;19(5):1087–1094. doi: 10.1016/s0896-6273(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 42.Gerber S.H., Sudhof T.C. Molecular determinants of regulated exocytosis. Diabetes. 2002;51(Suppl 1):S3–S11. doi: 10.2337/diabetes.51.2007.s3. [DOI] [PubMed] [Google Scholar]
- 43.Ostenson C.G., Gaisano H., Sheu L., Tibell A., Bartfai T. Impaired gene and protein expression of exocytotic soluble N-ethylmaleimide attachment protein receptor complex proteins in pancreatic islets of type 2 diabetic patients. Diabetes. 2006;55(2):435–440. doi: 10.2337/diabetes.55.02.06.db04-1575. [DOI] [PubMed] [Google Scholar]
- 44.Liang T., Qin T., Kang F., Kang Y., Xie L., Zhu D. SNAP23 depletion enables more SNAP25/calcium channel excitosome formation to increase insulin exocytosis in type 2 diabetes. JCI Insight. 2020;5(3) doi: 10.1172/jci.insight.129694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lane S.R., Liu Y. Characterization of the palmitoylation domain of SNAP-25. Journal of Neurochemistry. 1997;69(5):1864–1869. doi: 10.1046/j.1471-4159.1997.69051864.x. [DOI] [PubMed] [Google Scholar]
- 46.Jackson C.L., Bouvet S. Arfs at a glance. Journal of Cell Science. 2014;127(Pt 19):4103–4109. doi: 10.1242/jcs.144899. [DOI] [PubMed] [Google Scholar]
- 47.Bergbrede T., Chuky N., Schoebel S., Blankenfeldt W., Geyer M., Fuchs E. Biophysical analysis of the interaction of Rab6a GTPase with its effector domains. Journal of Biological Chemistry. 2009;284(5):2628–2635. doi: 10.1074/jbc.M806003200. [DOI] [PubMed] [Google Scholar]
- 48.Charest A., Lane K., McMahon K., Housman D.E. Association of a novel PDZ domain-containing peripheral Golgi protein with the Q-SNARE (Q-soluble N-ethylmaleimide-sensitive fusion protein (NSF) attachment protein receptor) protein syntaxin 6. Journal of Biological Chemistry. 2001;276(31):29456–29465. doi: 10.1074/jbc.M104137200. [DOI] [PubMed] [Google Scholar]
- 49.Cheng J., Wang H., Guggino W.B. Regulation of cystic fibrosis transmembrane regulator trafficking and protein expression by a Rho family small GTPase TC10. Journal of Biological Chemistry. 2005;280(5):3731–3739. doi: 10.1074/jbc.M410026200. [DOI] [PubMed] [Google Scholar]
- 50.Joubert P.E., Meiffren G., Gregoire I.P., Pontini G., Richetta C., Flacher M. Autophagy induction by the pathogen receptor CD46. Cell Host & Microbe. 2009;6(4):354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Yue Z., Horton A., Bravin M., DeJager P.L., Selimi F., Heintz N. A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002;35(5):921–933. doi: 10.1016/s0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- 52.Koliwer J., Park M., Bauch C., von Zastrow M., Kreienkamp H.J. The golgi-associated PDZ domain protein PIST/GOPC stabilizes the beta1-adrenergic receptor in intracellular compartments after internalization. Journal of Biological Chemistry. 2015;290(10):6120–6129. doi: 10.1074/jbc.M114.605725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saras J., Heldin C.H. PDZ domains bind carboxy-terminal sequences of target proteins. Trends in Biochemical Sciences. 1996;21(12):455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 54.Cuadra A.E., Kuo S.H., Kawasaki Y., Bredt D.S., Chetkovich D.M. AMPA receptor synaptic targeting regulated by stargazin interactions with the Golgi-resident PDZ protein nPIST. Journal of Neuroscience. 2004;24(34):7491–7502. doi: 10.1523/JNEUROSCI.1255-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woods S.C., Lotter E.C., McKay L.D., Porte D., Jr. Chronic intracerebroventricular infusion of insulin reduces food intake and body weight of baboons. Nature. 1979;282(5738):503–505. doi: 10.1038/282503a0. [DOI] [PubMed] [Google Scholar]
- 56.Austin J., Marks D. Hormonal regulators of appetite. International Journal of Pediatric Endocrinology. 2009:141753. doi: 10.1155/2009/141753. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodel A. SNAP-25. The International Journal of Biochemistry & Cell Biology. 1998;30(10):1069–1073. doi: 10.1016/s1357-2725(98)00079-x. [DOI] [PubMed] [Google Scholar]
- 58.Nishimoto-Morita K., Shin H.W., Mitsuhashi H., Kitamura M., Zhang Q., Johannes L. Differential effects of depletion of ARL1 and ARFRP1 on membrane trafficking between the trans-Golgi network and endosomes. Journal of Biological Chemistry. 2009;284(16):10583–10592. doi: 10.1074/jbc.M900847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aikawa Y., Xia X., Martin T.F. SNAP25, but not syntaxin 1A, recycles via an ARF6-regulated pathway in neuroendocrine cells. Molecular Biology of the Cell. 2006;17(2):711–722. doi: 10.1091/mbc.E05-05-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aikawa Y., Lynch K.L., Boswell K.L., Martin T.F. A second SNARE role for exocytic SNAP25 in endosome fusion. Molecular Biology of the Cell. 2006;17(5):2113–2124. doi: 10.1091/mbc.E06-01-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeans A.F., Oliver P.L., Johnson R., Capogna M., Vikman J., Molnar Z. A dominant mutation in Snap25 causes impaired vesicle trafficking, sensorimotor gating, and ataxia in the blind-drunk mouse. Proceedings of the National Academy of Sciences of the U S A. 2007;104(7):2431–2436. doi: 10.1073/pnas.0610222104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagamatsu S., Nakamichi Y., Yamamura C., Matsushima S., Watanabe T., Ozawa S. Decreased expression of t-SNARE, syntaxin 1, and SNAP-25 in pancreatic beta-cells is involved in impaired insulin secretion from diabetic GK rat islets: restoration of decreased t-SNARE proteins improves impaired insulin secretion. Diabetes. 1999;48(12):2367–2373. doi: 10.2337/diabetes.48.12.2367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2