Abstract
Nanoparticles have been widely used for preclinical cancer imaging. However, their successful clinical translation is largely hampered by potential toxicity, unsatisfactory detection of malignancy at early stages, inaccurate diagnosis of tumor biomarkers and histology for imaging-guided treatment. Herein, a targeted and in vivo copper nanocluster (CuNC) is reported with high potential to address these challenges for future translation. Its ultrasmall structure enables efficient renal/bowel clearance, minimized off-target effects in non-targeted organs, and low non-specific tumor retention. The pH-dependent in vivo dissolution of CuNCs affords minimal toxicity and potentially selective drug delivery to tumors. The intrinsic radiolabeling through the direct addition of 64Cu into CuNC (64Cu-CuNCs-FC131) synthesis offers high specific activity for sensitive and accurate detection of CXCR4 via FC131 directed targeting in novel triple negative breast cancer (TNBC) patient-derived xenograft mouse models and human TNBC tissues. In summary, this study not only reveals the potential of CXCR4 targeted 64Cu-CuNCs for TNBC imaging in clinical settings, but also provides a useful strategy to design and assess the translational potential of nanoparticles for cancer theranostics.
Keywords: copper nanocluster, CXCR4, breast cancer, positron emission tomography, translation
Graphical Abstract
INTRODUCTION
The current focus of cancer research on personalized treatment dictates the importance of early, sensitive, and specific detection of tumor-related biomarkers in primary and metastatic neoplasms. This is especially critical for highly aggressive tumors, such as triple negative breast cancer (TNBC), in which effective detection and therapy are lacking.1 TNBC patients frequently suffer from early-onset relapse and metastasis after conventional therapy, leading to an overall poor survival.2–5 Given the success of targeted therapy in other breast cancer (BC) subtypes, it is crucial to identify tumor biomarkers and develop targeted approaches for accurate TNBC detection and drug delivery to improve patient survival. Cancer theranostics that combine cancer imaging and therapy hold great potential to improve patient survival through imaging-guided treatment towards optimal dose and individualized treatment. For current TNBC research, nanoscopic agents have shown unique advantages in preclinical settings. However, the delivery of nanostructures to human tumors via the non-specific enhanced permeability and retention (EPR) effect did not replicate the data observed in mice. Their clinical translation is limited due to the inadequate tumor imaging accuracy, low drug delivery efficiency to tumors, potential nanotoxicity due to high non-specific retention in non-targeted organs, and lack of an appropriate strategy to overcome the gap between mouse cell line derived xenograft models and human tumors for translation.6–12
Of the molecular targets currently evaluated for TNBC, chemokine receptor CXCR4 is of particular significance, because the CXCR4/CXCL12 axis has a key role in not only regulating tumor growth and metastasis,2–5, 13 but also affecting tumor’s resistance to therapy.14–15 The upregulation of CXCR4 is observed in tumor biopsies from nearly 75% of TNBC patients.16–17 Patients with elevated CXCR4 expression (CXCR4high) had a significantly higher incidence of early-onset metastases than CXCR4low patients, leading to a negative correlation between tumor CXCR4 expression and patient survival.18 This suggests that CXCR4 is a compelling target for TNBC management to improve patient treatment outcomes.19–21
Ultrasmall nanoclusters have drawn substantial interest as an emerging platform for translational cancer theranostics due to rapid renal clearance, favorable in vivo pharmacokinetics and more importantly, accurate detection of tumor biomarkers.22–27 Based upon our previous work using gold nanoclusters for TNBC targeted positron emission tomography (PET) imaging,26 an copper nanocluster (CuNC) for in vivo targeted TNBC PET imaging was developed to address the potential toxicity due to high liver retention of gold nanoparticles.28 Through the intrinsic radiolabeling using 64Cu and conjugation of a clinically used CXCR4 specific binding peptide FC131,29–32 well-defined 64Cu-CuNCs-FC131 were synthesized in a more straightforward manner for feasible future translation. More importantly, we proposed our strategy assessing nanotheranostic for translational research by using three TNBC models including a mouse 4T1 cell line xenograft and two novel patient-derived xenograft models. Furthermore, de-identified human TNBC tissues were used to assess the CXCR4 binding capability of 64Cu-CuNCs-FC131 and highlight the potential for future translation.
RESULTS AND DISCUSSION
The CuNCs intrinsically radiolabeled with 64Cu (64Cu-CuNCs) were synthesized following a published procedure25–26, 33–35 with slight modifications. Bidentate thioctic acid (TA)-based ligands, i.e., TA-poly(ethylene glycol) (TA-PEG) and TA-PEG-FC131, were employed to modify the surface of the CuNCs to ensure their colloidal stability in aqueous solutions, prolong blood circulation for enhanced targeting, and enable active CXCR4 targeting. In brief, CuCl2, 64CuCl2, and TA-PEG ligands in water were reduced with NaBH4 under stirring. The incorporation of 64Cu into 64Cu-CuNCs was typically ≥ 95% and the final product were further purified to remove the unreacted reactants to reach radiochemical purity of ≥ 95% determined by instant radioactive thin layer chromatography (Radio-iTLC) for pre-clinical studies. Transmission electron microscopy (TEM) and scanning transmission electron microscopy (STEM) images of decayed 64Cu-CuNCs showed a narrow size distribution with a diameter of 2.7 ± 0.4 nm (Figure 1a,b), which led to a promoted surface area/volume ratio potentially for effective drug loading via physical loading or covalent conjugation for cancer therapy.36 The number-averaged hydrodynamic diameter measured by dynamic light scattering (DLS) was 5.5 ± 0.1 nm and ξ-potential was −7.2 ± 1.2 mV (Figure S1). There are approximately 960 Cu atoms (measured by inductively coupled plasma mass spectrometry (ICP-MS)) and 150 TA-PEG ligands per CuNC (determined by reversed-phase high performance liquid chromatography (RP-HPLC) analysis after dissolution of CuNCs) after the dissolution of CuNCs, leading to a molecular weight of ~ 175 kDa. The CXCR4-targeted 64Cu-CuNCs-FC131 were synthesized using a 1:1 molar ratio of TA-PEG-FC131:TA-PEG, leading to approximately 75 FC131 peptides per CuNC as quantified by RP-HPLC. The specific activity of 64Cu-CuNCs was well controlled (555 MBq nmol−1, Figure 1c) to enable trace amount (ca. 6.7 pmol, ca. 426 pg) administration for PET imaging in mouse models. The scaled-up radiosynthesis showed that up to 1.85 GBq (3.4 nmol of Cu, 216 ng) of uniformly sized 64Cu-CuNCs could be produced in a single reaction within 2–3 h, which demonstrated that sufficient quantities can be made for future human research. X-ray photoelectron spectroscopy (XPS) was employed to evaluate oxidation states of Cu since CuNC is prone to oxidation in aqueous solution. In addition to the characteristic 2p3/2 and 2p1/2 peaks of Cu(0) and Cu(I), 2p3/2 peak of Cu(II) and two satellites were observed (Figure S2a), suggesting the possible formation of CuO on the surface. Deconvolution of 2p3/2 peaks showing a ratio of Cu(0)/Cu(I) to Cu(II) is approximately 1:1. The formation of Cu(I) might be induced by the charge transfer between Cu and S atoms on the CuNC surface.37 The dissolution kinetics of CuNCs under varying pH values (in pH 5.5, 6.5, and 7.4 buffer systems) were assessed using ICP-MS. As shown in Figure 1d, under pH 5.5 conditions, ~55% of CuNCs were dissolved within 4 h that further reached 100% within 24 h. At pH 7.4, ~10% and 50% of CuNCs were dissolved at 4 h and 48 h, respectively. Dissolution of CuNCs was also monitored using radio-iTLC after chelation of free 64Cu2+ by addition of ethylenediaminetetraacetic acid (EDTA), which showed comparable kinetics (Figure S3, ~36% in 4 h at pH 5.5, ~14% in 4 h and ~43% in 49 h at pH 7.4) to the data shown in Figure 1, suggesting that CuNCs initially dissolved into Cu(II). In contrast to other metal nanoparticles, the mild conditions required for dissolution of the CuNCs could significantly reduce concerns for translation in vivo. Moreover, in future theranostic applications, the pH dependent dissolution kinetics of CuNCs would allow fast and targeted breakdown of the CuNCs to promote release of loaded drugs within relatively-acidic tumors or tumor microenvironment, while allowing unbound nanoclusters to remain stable at physiological pH for systemic clearance, thus reducing potential toxicity in non-targeted organs.38
Figure 1.
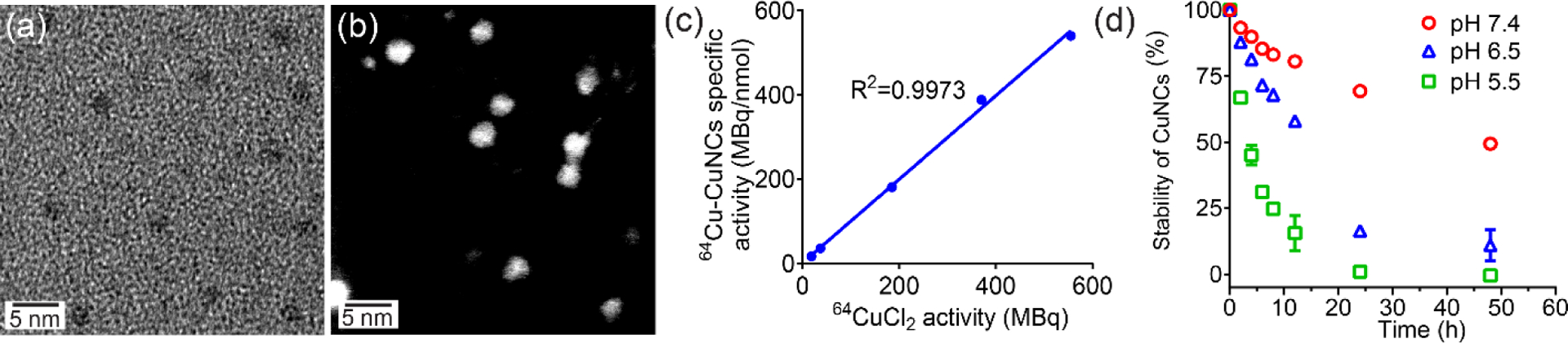
64Cu-CuNCs characterization. (a) TEM, (b) STEM (dark field), (c) Correlation between 64CuCl2 activity and specific activity of 64Cu-CuNCs, (d) Dissolution kinetics under different pH values.
Stability tests of 64Cu-CuNCs performed in mouse serum showed the 64Cu radiolabel was 97.1 ± 0.4%, 91.2 ± 1.0% and 72.6 ± 1.5% stable after 1, 4, and 24 h of incubation at 37 °C, respectively, consistent with the dissolution kinetics. In vivo, the 64Cu radiolabel showed 95.0 ± 2.1%, 90.1 ± 1.8%, and 69.6 ± 2.1% stability in blood analyzed by fast protein liquid chromatography (FPLC) at comparable time points, indicating the stability of 64Cu-CuNCs up to 4 h in vivo. In addition, cytotoxicity assays of non-radioactive CuNCs and CuNCs-FC131 performed in mouse 4T1 cells using MTT (formazan) showed no noticeable change in cell viability with a concentration up to 200 μg mL−1 compared to the PBS control (per 105 cells).
In vivo pharmacokinetics and clearance of 64Cu-CuNCs and 64Cu-CuNCs-FC131 were evaluated by biodistribution studies in female C57BL/6 mice. Consistent with previous reports about the in vivo pharmacokinetics of ultrasmall nanostructures,22–25 64Cu-CuNCs showed fast blood clearance (t1/2 = 1.23 h) with retention decreasing from 12.0 ± 0.55 percent injected dose per gram (%ID g−1) at 1 h post injection (p.i.) to 3.03 ± 0.26%ID g−1 at 4 h, followed by a further decrease to 0.97 ± 0.21%ID g−1 at 24 h (Figure 2a). With the conjugation of FC131 targeting peptide, although the initial blood retention of 64Cu-CuNCs-FC131 was slightly lower (7.21 ± 1.93%ID g−−1 at 1 h p.i.) than the non-targeted counterpart, its blood clearance kinetics remained comparable (t1/2=1.5 h). Due to the ultrasmall sizes, both nanoclusters showed effective kidney clearance with the excretion of 64Cu-CuNCs (61.4%ID) being 50% higher than that of 64Cu-CuNCs-FC131 (40.4%ID) at 24 h. As we previously reported, due to the expression of CXCR4 on inflammatory cells,26, 39 the accumulations of 64Cu-CuNCs-FC131 in the liver were significantly (p < 0.05, n = 4) elevated compared to the non-targeted 64Cu-CuNCs at all four time points (Figure 2b). Both nanoclusters showed gradually decreased hepatic accumulations with less than 10%ID g−1 (< 43 pg) in the liver at 48 h, which was 3× lower than larger nanoparticles40–41 demonstrating the advantages of ultrasmall nanoclusters in reducing nanoparticle liver retention and potential toxicity. Additionally, the hepatic accumulation of 64Cu-CuNCs was also significantly (p < 0.05, n = 4) lower than that acquired with 64Cu alloyed gold nanoclusters,26 suggesting that the dissolution of 64Cu-CuNCs to 64Cu2+/Cu2+ in the liver also facilitated the clearance. At 24 h, approximately 74.9%ID of 64Cu-CuNCs and 63.2%ID of 64Cu-CuNCs-FC131 were excreted with less than 160 pg of 64Cu-CuNCs/64Cu-CuNCs-FC131 remained in the body, which significantly reduced the toxicity concerns in translational settings.
Figure 2.
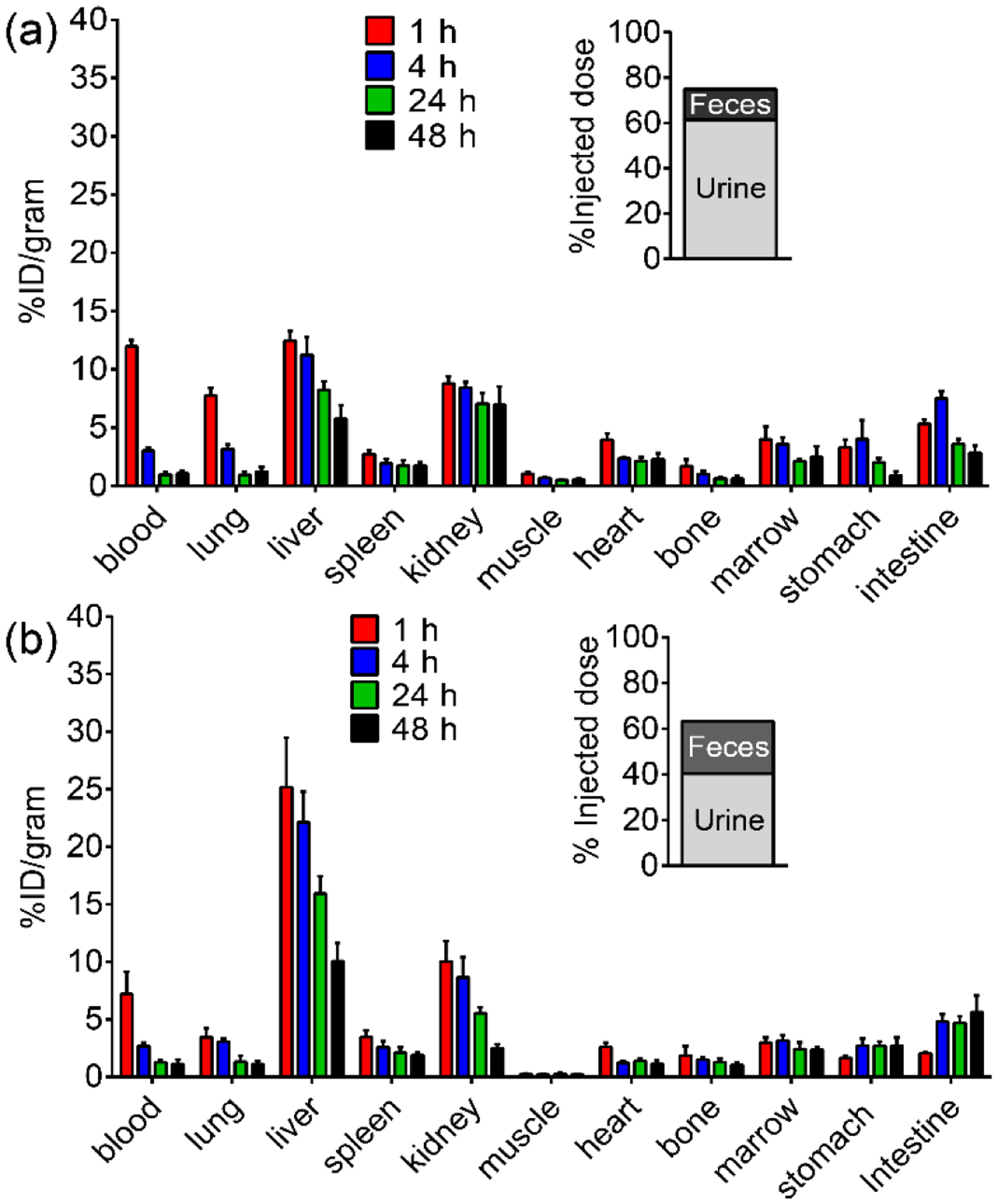
Biodistribution and clearance of (a) 64Cu-CuNCs and (b) 64Cu-CuNCs-FC131 in female C57BL/6 mice (n = 4 / group).
We next studied the intracellular fate of nanoclusters conjugated with Texas Red® (TR) in 4T1 cells since it is known that CXCR4 is subjected to rapid internalization and lysosome degradation upon the binding of its ligands.42–45 As shown in Figure 3a, non-targeted CuNCs-TR showed minimal internalization and did not colocalize with lysosomal-associated membrane protein 1 (LAMP-1, green). In contrast, the targeted CuNCs-FC131-TR internalized and significantly colocalized with LAMP-1 (yellow) at 24 h post incubation. TA-PEG-TR was also tested but did not show any internalization (data not shown). Quantification showed at least 10-fold (p < 0.001, n = 3) more TR signals in the cells incubated with CuNCs-FC131-TR compared to the non-targeted counterpart (Figure 3b), indicating CXCR4 mediated internalization of CuNCs-FC131-TR through the specific binding of FC131 peptide. This would be advantageous in future theranostic applications to facilitate the intracellular release of drugs loaded on the targeted nanoclusters to improve their therapeutic efficacy.
Figure 3.
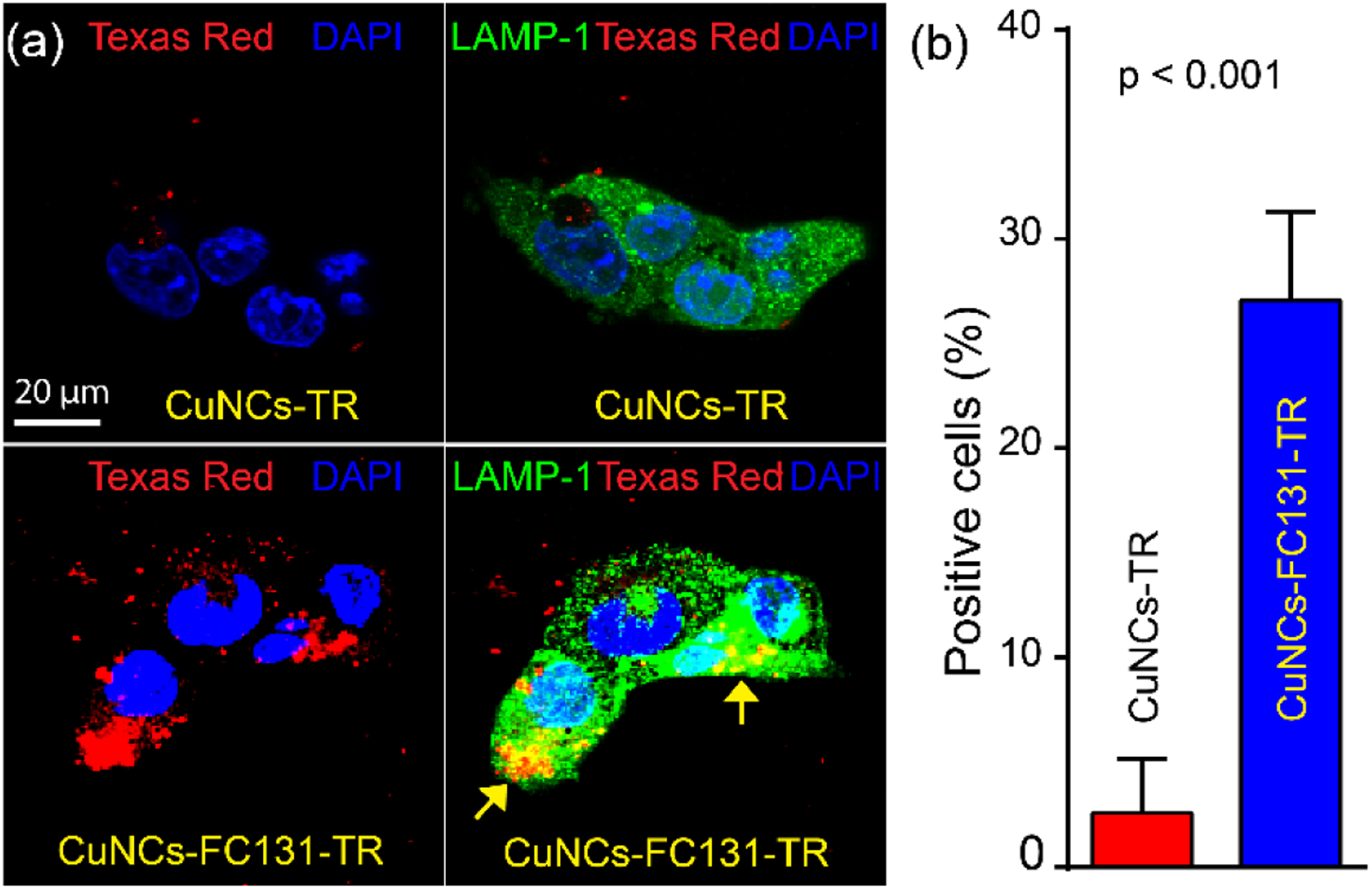
CXCR4-mediated internalization of targeted CuNCs-FC131-TR (24 h) in 4T1 cells. (a) Non-targeted CuNCs-TR showed minimal internalization while targeted CuNCs-FC131-TR showed colocalization (yellow) with lysosome marker (LAMP-1). (b) Quantification of TR positive cells. LAMP-1, green; CuNCs-TR and CuNCs-FC131-TR, red; DAPI, blue.
The CXCR4-targeted imaging of 64Cu-CuNCs-FC131 was firstly studied in a mouse 4T1 TNBC model. Consistent with our previous report,26 the non-targeted 64Cu-CuNCs showed low non-specific tumor retention (1.01 ± 0.19%ID g−1, 2.11 ± 0.22%ID g−1, and 2.44 ± 0.27%ID g−1 at 1 h, 4 h, and 24 h p.i., respectively, n = 4) while the targeted counterpart displayed significantly higher tumor accumulations at all three time points (3.65 ± 0.66%ID g−1, 5.01 ± 0.58%ID g−1, and 6.08 ± 0.87%ID g−1, p < 0.005 for all, n = 4) (Figure 4a, b). Based on the PET data, more than 80% of tumor uptake was achieved within 4 h p.i. Since 64Cu2+ has very fast clearance kinetics from the blood stream,46 and the 64Cu radiolabel was more than 90% stable up to 4 h both in vitro and in vivo, the contribution of dissolved 64Cu2+ to tumor uptake was minimal. Following our previously reported method,26 approximately 60% of tumor uptake of 64Cu-CuNCs-FC131 was mediated by CXCR4 binding at both 4 h and 24 h, indicating the CXCR4 targeting specificity of 64Cu-CuNCs-FC131. Competitive receptor blocking using 20-fold non-radioactive CuNCs-FC131 through co-injection with 64Cu-CuNCs-FC131 significantly blocked tumor uptake at 24 h p.i. to a level (2.83 ± 0.39%ID g−1, p < 0.005, n = 4) comparable to that acquired with 64Cu-CuNCs, suggesting CXCR4 targeting specificity (Figure 4c, d). Consistent with our previous report,26 histology and immunohistochemical staining showed upregulation of CXCR4 in 4T1 tumor tissues with 36% cells positive for CXCR4 (n = 4) (Figure 4e, f). The overexpression of CXCR4 in 4T1 tumor was also confirmed by western blot analysis (Figure S4), consistent with our previous report.26
Figure 4.
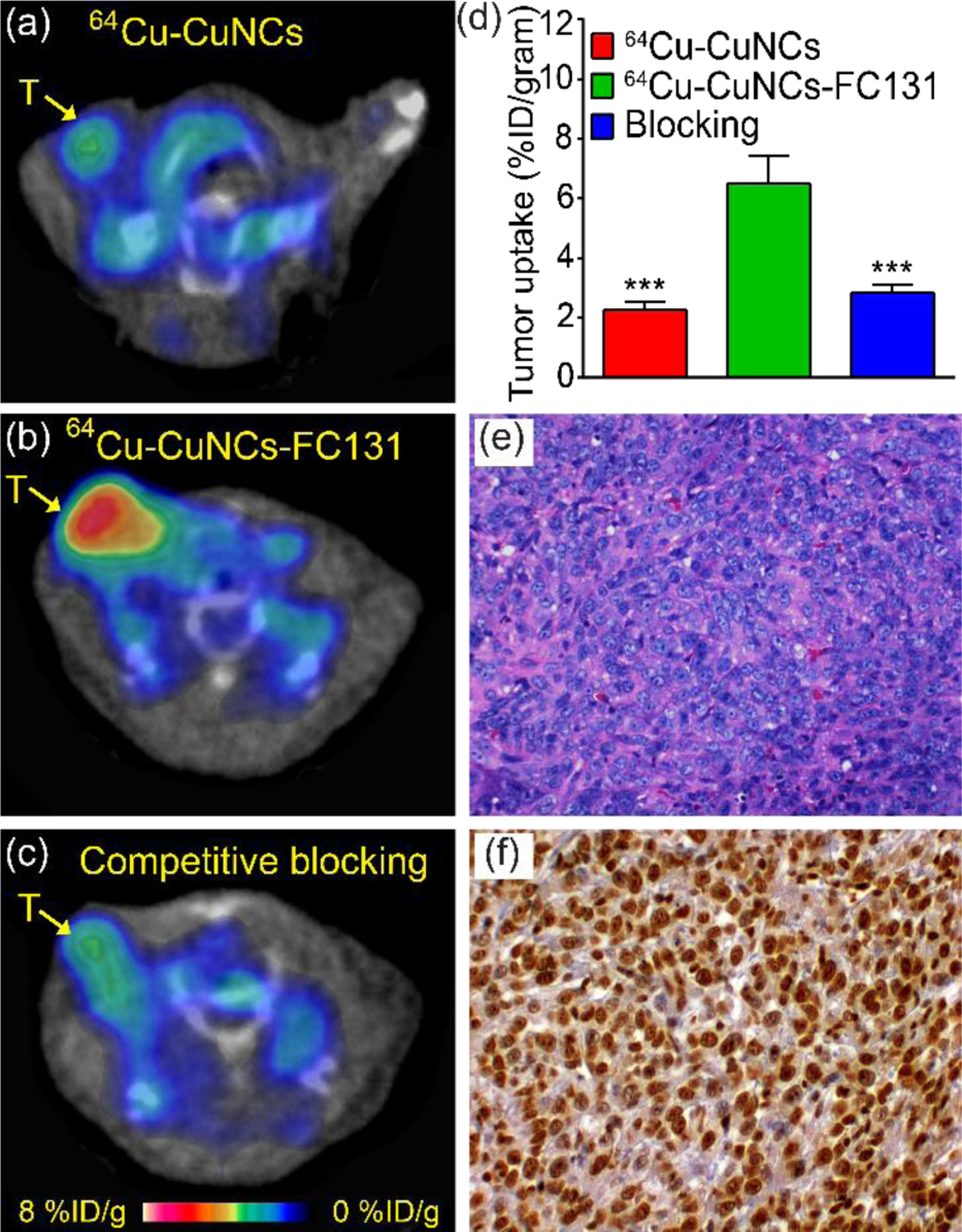
In vivo PET imaging at 24 h p.i. in a 4T1 mouse TNBC model at 2 weeks post implant and histopathological characterization of tumor tissues. (a) PET image of 64Cu-CuNCs showing weak tumor uptake. (b) CXCR4 targeted 64Cu-CuNCs-FC131 showing significant tumor uptake. (c) Competitive receptor blocking using non-radioactive CuNCs-FC131 significantly decreased tumor uptake of 64Cu-CuNCs-FC131. (d) Quantitative tumor uptake analysis, *** p < 0.005, n = 4 / group. (e) H&E and (f) CXCR4 (brown) immunohistochemical staining of 4T1 tumor (20×). T: tumor.
Given the biological difference between mice and humans, one grand challenge in translational cancer nanomedicine is whether the developed nanoprobe can achieve comparable imaging efficiency in humans as that acquired in cell line derived xenografts. The patient derived xenograft (PDX) models are ideal platforms to assess the developed nanoprobes by directly implanting human tumor cells into immunodeficient mice that preserves cancer cell morphology, vasculature, architecture, and molecular features to maximally recapitulate the original human tumor biology.47–50 The targeting efficiency of 64Cu-CuNCs-FC131 were further assessed by using novel PDX TNBC models with high or low CXCR4 expression (CXCR4high or CXCR4low) that were established based on the CXCR4 levels characterized by global gene expression array analysis in de-identified human malignant tissues. As shown in Figure 5a, non-targeted 64Cu-CuNCs showed minimal tumor accumulation (2.21 ± 0.59%ID g−1, n = 6) in the CXCR4high PDX model, which was similar to that determined in 4T1 tumors. In the CXCR4low PDX model, the tumor uptake of targeted 64Cu-CuNCs-FC131 was also low (1.79 ± 0.36%ID g−1, n = 8, Figure 5b). By comparison, in the CXCR4high PDX model, significantly higher tumor uptake was observed from 64Cu-CuNCs-FC131 (5.78 ± 0.84%ID g−1, p < 0.001 for both, n = 8) (Figure 5c, d) and approximately 62% of the tumor uptake was attributed to CXCR4 binding.
Figure 5.
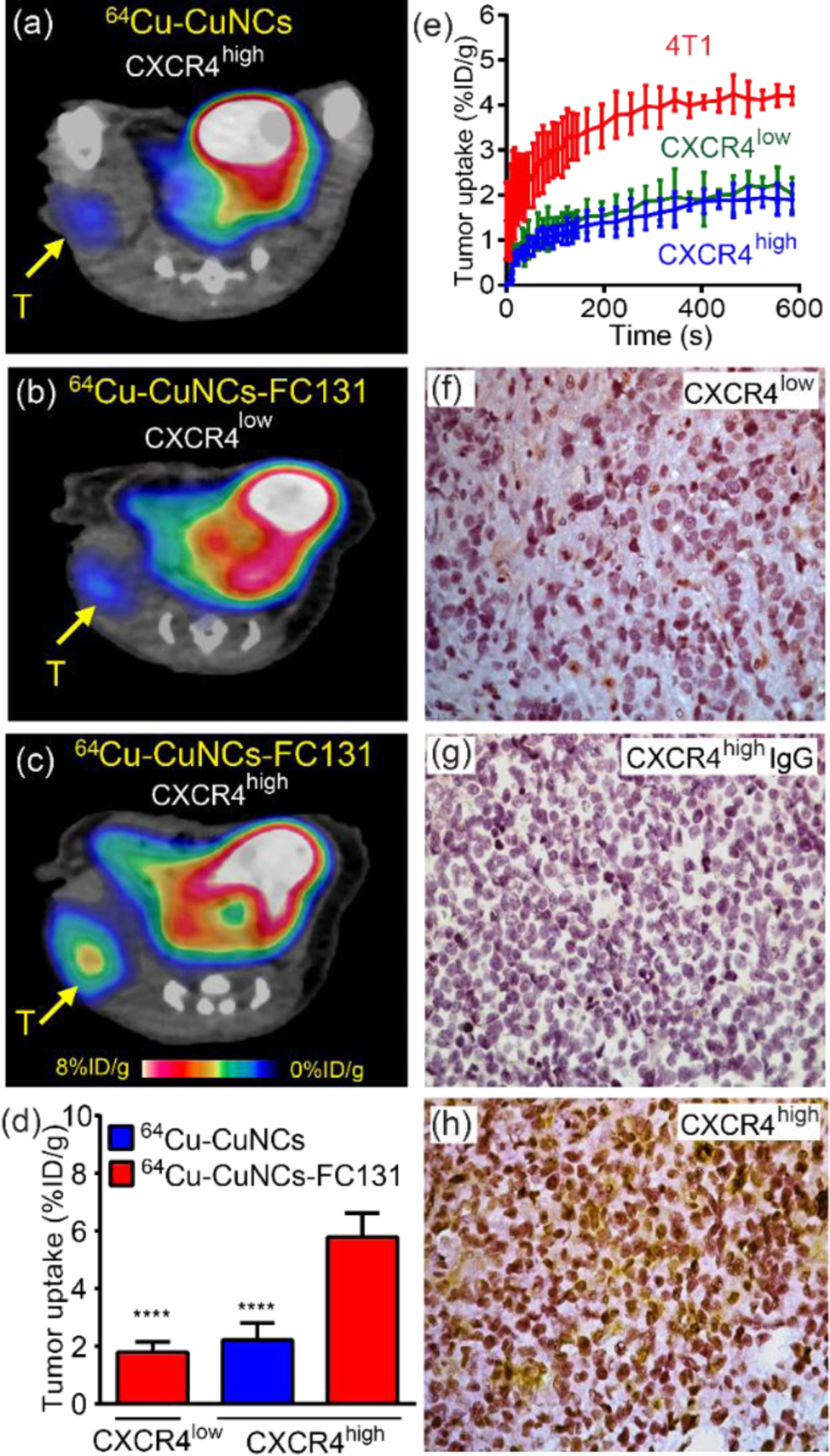
PET imaging and characterization of TNBC PDX models. (a) Non-targeted 64Cu-CuNCs showing minimal tumor uptake in the CXCR4high PDX model. Targeted 64Cu-CuNCs-FC131 showing (b) low uptake in the CXCR4low PDX model and (c) significant tumor uptake in the CXCR4high PDX model. (d) Quantitative tumor uptake analysis (n = 6–8). (e) Time-activity-curve of 15O-H2O PET in 4T1, CXCR4low, and CXCR4high PDX TNBC models showing the difference of tumor blood flows (n=4). Immunohistochemical staining showing (f) low CXCR4 (brown) expression in CXCR4low PDX tumor, (g) negative signal in CXCR4high tumor using control IgG and (h) upregulation of CXCR4 signals in CXCR4high PDX tumor. All panels are at 20×. **** p < 0.005 (n = 4)
Previously, we reported a 2-fold difference in tumor uptake between 4T1 and breast cancer PDX models using gold nanocages (~ 33 nm diameter) via non-specific EPR effects,51 reasonably due to the variance of blood flows. Herein, we quantitatively determined the difference in blood flow between 4T1 and PDX models using 15O-H2O PET imaging and assessed their effect on tumor uptake of 64Cu-CuNCs-FC131.52 As shown in Figures S5 and 5e, the blood flow in the 4T1 tumor was approximately twice as much as those determined in both PDX models which were comparable to each other. Immunohistochemical staining of CXCR4low PDX tumor tissue showed little positive signals (Figure 5f), consistent with the information identified in human malignant tissues. Quantitative analysis showed that 2% and 36 % of cells were CXCR4 positive in the CXCR4low and CXCR4high PDX tumors, respectively (Figure 5g, h), which was also verified by western blot analysis (Figure S4). Therefore, in contrast to the EPR effect dominated tumor delivery of nanoparticles which was significantly affected by blood flow, the tumor uptake of ultrasmall 64Cu-CuNCs-FC131 was largely driven by the specific CXCR4-mediated targeting effect which is independent of blood flow, a distinct advantage for translational imaging in individual settings.
To further demonstrate the potential of CXCR4 as a biomarker for human TNBC imaging, we characterized the expression of CXCR4 in human TNBC tissues. As shown in Figure 6a–c, intense CXCR4 positive staining was observed in the tumor regions, while the surrounding areas showed decreased CXCR4 staining, which were most likely due to infiltrated inflammatory cells such as macrophages. Quantitative analysis (n = 7) showed that 32% of cells were positive for CXCR4. Thus, the comparable expression pattern of CXCR4 across 4T1, CXCR4high PDX and human TNBC specimens not only confirmed the potential of CXCR4 as a biomarker for human TNBC imaging and therapy, but also highlighted the unique advantage of our approach to assess the translational potential of developed nanoclusters. Autoradiography of CXCR4 targeted 64Cu-CuNCs-FC131 on the malignant human TNBC tissue displayed strong binding to the cancerous regions in a profile similar to CXCR4 staining, indicating the CXCR4 binding sensitivity. Through competitive ex vivo receptor blocking using 100× non-radioactive CuNCs-FC131, the radioactivity bound to the tissue was largely reduced (56.0 ± 4.2%, n = 7), suggesting specific binding of 64Cu-CuNCs-FC131 to CXCR4 on human TNBC specimens (Figure 6d, e, f). Moreover, the binding of 64Cu-CuNCs to human TNBC tissues was also assessed and compared to 64Cu-CuNCs-FC131 via autoradiography. Low background retention on the tissue was determined for the non-targeted 64Cu-CuNCs while intensive signal was observed for 64Cu-CuNCs-FC131 (Figure S6), further confirming the binding specificity of the targeted nanoprobe to human cancerous specimens. Immunohistochemcial staining on additional human malignant tissues showed mostly positive signals of CXCR4 (Figure S7).
Figure 6.
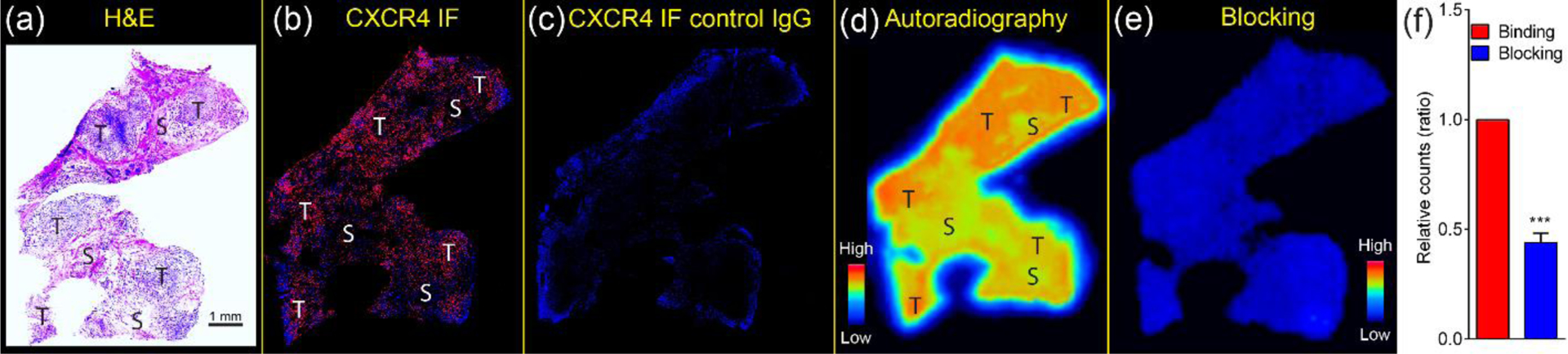
Ex vivo analysis of representative human TNBC tissue. (a) H&E staining. (b) CXCR4 immunofluorescent staining (red) showing strong expression of CXCR4. (c) Control IgG staining showing negative signal of CXCR4. Autoradiography of (d) binding and (e) blocking of 64Cu-CuNC-FC131 to the tissue was assessed and (f) quantified using ImageJ. *** p < 0.005, n = 7. T, tumor; S, surrounding area.
FC131 peptide based imaging agent has been used for targeting both mouse53 and human54 CXCR4 receptor. However, the binding capabilities of FC-131 peptide to both receptors have not been compared. Thus, computational studies were conducted to assess the binding of FC131 derivative (FC131-N-Me-D-Orn) used in this study to both human and mouse CXCR4 receptors. Since most computational studies have focused on human receptors, we developed an approach suitable for both the human and murine receptors through docking studies with the FC131 derivative to identify binding sites of CXCR4 receptors. The contacts between FC131-N-Me-D-Orn and both human and mouse CXCR4 receptors for the best scoring pose are summarized in Tables S1–2. Figure S8 is a diagram of the binding sites and best binding pose found for the FC131-N-Me-D-Orn in human and murine receptors. These results show that FC131-N-Me-D-Orn binds to both human (93.79) and murine (90.32) CXCR4 receptors in a comparable manner based on the GoldScore.
Although copper is an essential element for humans, copper deficiency55–58 or surplus59 may cause adverse health problems. The amount of tolerable upper intake level of copper for human is 10 mg day−1, which was equivalent to 45 μg mouse−1 (25g) based on body weight.37, 60–61 Thus, we next assessed the potential toxicity of developed nanoclusters to liver and kidneys since these two organs were the major clearance routes by performing liver and renal function tests in BALB/c mice injected with saline (n = 3) and CuNCs (375 μg, n = 3) at 24 h p.i.. The mass of Cu administered for both tests (375 μg mouse−1) was approximately 106-fold higher than the dose used for PET imaging studies (426 pg mouse−1). Serum measurements of blood urea nitrogen (BUN), creatinine, alanine aminotransferase (ALT), serum alkaline phosphatase (SAP), and total protein (TP) were analyzed using a standard clinical chemistry analyzer. Liver and kidney weights calculated as percentages of body weight were also compared as well as the histopathological examination of livers. As summarized in Figures S9 and S10, there was no significant difference between the two groups for all analyzed parameters. H&E images also showed no significant lesion in the livers examined. All these data suggested the minimal toxicity concerns of 64Cu-CuNCs-FC131 for PET imaging and potential for translation.
CONCLUSIONS
In summary, the CXCR4-targeted 64Cu-CuNCs-FC131 were developed for accurate and sensitive TNBC PET imaging and assessment using a comprehensive strategy for future clinical translation. The well-defined and straightforward preparation of 64Cu-CuNCs-FC131 enabled high radiolabeling specific activity and scale-up capability for PET imaging. The ultrasmall nanostructure afforded favorable pharmacokinetics and effective systemic clearance to minimize potential toxicity concerns. The low non-specific tumor retention, as well as sensitive and specific detection of CXCR4 in the experimental mouse TNBC models demonstrated the advantages of the targeted 64Cu-CuNCs-FC131 for accurate tumor imaging. The highly integrated assessment of CXCR4 expression from mouse models to human TNBC tissues ensured the potential of CXCR4 as a biomarker for TNBC imaging and therapy. Preliminary toxicity evaluations demonstrated the biocompatibility of 64Cu-CuNCs-FC131 for future translational research. However, our study also has several limitations. Due to repeated passages, original human stromal components such as cancer associated fibroblasts, immune and inflammatory cells might be replaced by murine cells in the CXCR4high and CXCR4low PDX models, leading to incomplete representation of human tumor microenvironment for imaging and therapy.62 Moreover, effect of complex human tumor biology on the targeting capability of as-developed nanoprobes needs further evaluation although the ex vivo human tissues serve as a valuable platform to test the binding of as-developed nanoprobes to human CXCR4.
EXPERIMENTAL SECTION
Materials.
Lipoamido-dPEG®12-TFP ester (TA-PEG12-TFP ester) and m-dPEG®12-Lipoamide (TA-PEG12-OMe) were obtained from Quanta BioDesign, Ltd. and used as received. The FC131 peptide (cyclo[2-Nal-Gly-D-Tyr-NMe-D-Orn-Arg]) was synthesized by CPC Scientific (Sunnyvale, CA). Sulforhodamine 101 (Texas Red®) Cadaverine was obtained from Biotium. All other solvents and chemicals were obtained from Fisher Scientific, Sigma-Aldrich, or TCI America and were used without further purification.
Characterization.
All the synthesized compounds were characterized with 1H and 13C NMR using 400 NMR spectrometer (Varian). Solvent resonance signals were used as reference for chemical shifts. Transmission electron microscopy (TEM) images were acquired using Tecnai G2 Spirit Transmission Electron Microscope at 120 kV (FEI, Hillsboro, OR). Additionally, higher-resolution TEM images and scanning transmission electron microscopy (STEM) images were acquired on JEOL JEM-2100F Field Emission Electron Microscope operating at an accelerating voltage of 200 kV. TEM specimens were prepared by drop-casting method on an ultrathin carbon film supported by a lacey carbon film on a copper grid. Dynamic light scattering (DLS) and ζ-potential data were acquired with Zetasizer Nano ZS (ZEN3600, Malvern instruments). ICP-MS analysis was carried out using a PerkinElmer Elan DRCII. XPS analysis was performed using a Physical Electronics 5000 VersaProbe II Scanning ESCA Microprobe. Matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) results were collected by a Bruker AutoFlex Speed in a reflector mode with positive ion detection. Radiochemical purity was measured by instant radio-thin layer chromatography (radio-iTLC) (Bioscan). Briefly, samples were spotted on a glass microfiber chromatography paper impregnated with a silica gel (Agilent Technology) and were developed in a TLC developing tank with 10% ammonium acetate and methanol mixture (1:1 volume ratio). Reverse-phase high performance liquid chromatography (RP-HPLC) analysis and purification were carried out using an Agilent 1200 Series with photodiode array detector.
Synthesis of TA-PEG12-FC131 (Scheme S1).
To a solution of FC131 (3.3 mg, 4.7 μmol) and TA-PEG12-TFP ester (4.5 mg, 4.7 μmol) in 150 μL of DMF, N,N-diisopropylethylamine (DIPEA, 4.1 μL, 24 μmol) was added at RT. After overnight stirring, the reaction mixture was precipitated into diethyl ether twice. The resulting white solid was collected by centrifugation, dissolved in water, purified by RP-HPLC with H2O/acetonitrile (with 0.1% TFA) solvent system, and lyophilized to obtain a white solid (yield = 60%). MALDI-TOF m/z: [M + H]+ Calcd for C71H112N10O20S2H 1489.8; Found 1489.7, [M + Na]+ Calcd for C71H112N10O20S2Na 1511.7; Found 1511.6.
Synthesis of TA-PEG12-Texas Red (TA-PEG12-TR) (Scheme S2).
Texas Red® cadaverine (5.0 mg, 7.2 μmol), TA-PEG12-TFP ester (6.9 mg, 7.2 μmol), and N,N-diisopropylethylamine (DIPEA, 6.3 μL, 36 μmol) were mixed in 300 μL of DMSO. After overnight stirring in the darkness, TA-PEG-TR was purified by RP-HPLC with H2O/acetonitrile (with 0.1% TFA) solvent system, and lyophilized to obtain a dark red solid (yield = 50~60%). MALDI-TOF m/z: Calcd [M + H]+ = 1478.6; Found 1478.8.
Synthesis of 64Cu-CuNCs.
The 64Cu (t1/2 = 12.7 h, β+ = 17%, β− = 40%) was obtained from the Washington University cyclotron facility. In a typical reaction, TA-PEG ligands (total 400 μL, 2.5 mM) were added dropwise at RT to a solution of CuCl2 (376 μL, 10 mM) and 64CuCl2 (in 0.1 M NH4OAc, pH 5.5 or in 20 mM NaOAc, pH 7.0, ca. 18.5 MBq μL−1) in water (2 mL). After stirring for ca. 15 min at RT, sodium borohydride (450 μL, 20 mM) was added to the mixture with rapid stirring (> 600 rpm) at RT. After ca. 25 min, the resulting nanoclusters were purified by washing 4 times with water using a centrifugal filter unit (Amicon Ultra, 10 kDa NMWL). When the radiochemical purity determined by radio-iTLC was ≥95%, the 64Cu-CuNCs were diluted in saline for tail vein injection into mice. The targeted 64Cu-CuNC-FC131 were synthesized identically with the exception of using a 1 : 2 molar ratio of TA-PEG-FC131 and TA-PEG-OMe.
Dissolution kinetics of CuNCs under different pHs.
CuNCs were incubated with 50 mM ammonium acetate buffer (pH 5.5), 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.5) and 50 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) buffer (pH 7.4) at 37 °C, respectively. At various time points, an aliquot was collected and purified using Amicon Ultra-0.5 mL centrifugal filters (3 KDa NMWL). The filtrate was analyzed by ICP-MS.
Mouse Tumor Models.
All animal procedures in this work were approved by the Washington University Animal Studies Committee. The 4T1 tumor model was developed as reported previously.63 Typically, 3 ×105 4T1 cells in 100 μL of saline were implanted into the mammary fat pad (right) of 7-week old female BALB/c mice weighing 15−20 g subcutaneously. The tumors typically grew to 200−300 mm3 within two weeks for biodistribution and PET imaging studies. The CXCR4high and CXCR4low mouse PDX models were established by the Human & Mouse Linked Evaluation of Tumors (HAMLET) Core at Washington University. To generate these mice, 1×106 CXCR4high or CXCR4low cancer cells (passages 3) re-suspended in 60 μL of Collagen: Matrigel (1:1 v/v) were injected into the right flank of 8-week old female NSG mice. The tumors were allowed to grow for 4 weeks to reach a size of 200–300 mm3 before the PET imaging studies.
Animal Biodistribution and clearance Studies.
Biodistribution and clearance studies were performed using female wild type C57BL/6 mice (Charles River Laboratory, Wilmington, MA), as previously reported.26 Typically, 64Cu-CuNCs or 64Cu-CuNCs-FC131 (approximately 740 kBq/100 μL saline (APP pharmaceuticals, Schaumburg, IL)) was injected into mice anesthetized with inhaled isoflurane through tail vein. At 1, 4, 24 and 48 h post injection, mice were euthanatized by cervical dislocation (n = 4 / time point) and organs of interest were collected, weighed, and counted in a Beckman 8000 gamma counter (Beckman, Fullterton, CA). To measure the clearance of the two nanoclusters, a group of mice (n=4) was housed in a metabolism study cage to collect urine and feces at 4, 24, and 48 h post injection. Standards for each nanocluster were prepared and counted with the biodistribution samples together to calculate percentage of the injected dose per gram of tissue (%ID g−1).
Immunofluorescent Staining of targeted CuNCs-FC131-TR in 4T1 cells.
To determine the internalization of CuNCs-FC131-TR in 4T1 cells, immunofluorescent staining was performed. After the incubation of CuNCs-FC131-TR in 4T1 cells for indicated times, cells were washed twice in PBS, then fixed for 15 min in 4% PFA. After washing three times, cells were blocked/permeabilized in Roth Buffer for 1 h (0.25% Milk, 1% BSA, 0.3% Triton-X 100 in TBS (50 mM Tris/150 mM NaCl; pH 7.4). Lamp1 antibody (Abcam, ab24170) was diluted in Roth buffer (1:250) and incubated with cells at room temperature for 2 h. After washing, cells were incubated with Alexa Flour 488 Conjugated Donkey anti Rabbit IgG (Life Tech, A-21206) (diluted in Roth buffer, 1:250) for 1 h. With another washing and DAPI (Sigma D9542) staining, the cell-growing coverslip was put on the VECTASHIELD fluorescence mounting medium (Vector Labs, H-1000) and then fixed with nail polish. A Zeiss LSM-700 confocal microscope was used for imaging; regions of interest (ROIs) and thresholds were determined, and signal over the threshold was quantified using ZEN microscope software (Carl Zeiss Microscopy).
Micro-PET/CT Imaging.
Small animal PET imaging was carried at multiple time points (1 h, 4 h, 24 h) post the intravenous injection of 64Cu-CuNCs or 64Cu-CuNCs-FC131 (approximately 3.7 MBq/100 μL of saline) to determine the dynamic variation of tumor uptake using either a microPET Focus 220 (Siemens, Malvern, PA) or Inveon PET/CT system (Siemens, Malvern, PA), which were cross-calibrated periodically. To determine the effect of blood flow on tumor uptake, 15O-water (half-life=122.24 s) PET imaging was performed on the PDX mice (n=4) by injecting 22–37 MBq of 15O-H2O through tail vein. A 0–10 dynamic scan was acquired immediately after the injection. All the PET images were reconstructed with maximum a posteriori (MAP) algorithm to co-register with CT imaging after the correction of attenuation, scatter, normalization, and camera dead time. Tumor uptake was analyzed using Inveon Research Workplace in three-dimensional regions of interest (ROIs) and presented as percent injected dose per gram (%ID g−1) of tumor tissue. The tumor targeting specificity of 64Cu-CuNCs-FC131 was assessed by competitive receptor blocking studies through the co-injection of using 20-fold non-radioactive CuNCs-FC131 along with the radiotracer.26
Liver and renal function tests.
Liver and renal function tests were performed in BALB/c mice injected with saline (n = 3) and CuNCs (375 μg, n = 3) at 24 h post injection. Voided urine samples were collected from mice for urinalysis. The color and clarity of the urine was assessed, and sediment prepared according to standard practice. The sediment was evaluated microscopically for the number of epithelial cells, red blood cells, and white blood cells per high power field (400× total magnification), and for the presence of crystals, casts, and bacteria.
Immediately following euthanasia by CO2 inhalation, blood was collected from animals by cardiocentesis. Serum measurements of blood urea nitrogen (BUN), creatinine, alanine aminotransferase (ALT), alkaline phosphatase, and total protein were determined using the Liasys 330 liquid reagent chemistry analyzer (AMS Diagnostics, Weston, FL). The animal was weighed. Kidneys and liver were collected, weighed, and fixed in 10% neutral buffered formalin. After fixation, the tissues were trimmed, paraffin-embedded, and 5 micron sections were prepared and stained with hematoxylin and eosin for standard histopathologic evaluation.
Immunohistochemistry of tumor tissues.
Paraffin embedded tumor sections were deparaffinized in xylenes and rehydrated through a series of graded alcohols. After quenching endogenous peroxidase activity in 3% hydrogen peroxide, antigen retrieval was performed by boiling the slides in citrate buffer (pH 6.0 containing 0.1% Tween). After incubation in blocking serum for 1 h to prevent nonspecific binding (Vectastain; Vector Laboratories), the sections were further incubated overnight at 4°C with primary antibody (anti-CXCR4, Novus Biologicals 1:2000) or control IgG (anti-rabbit IgG, Novus Biologicals 1:2000). After applying secondary antibody (Vector Laboratories), the addition of diaminobenzidine develop a brown color. The sections were counterstained in hematoxylin, and digital images of the stained sections were collected using a Zeiss Light Microscope system.
Immunofluorescence of tumor tissues.
De-identified human triple negative breast cancer specimens were obtained from the Tissue Procurement Core of Siteman Cancer Center under the approval from Human Research Protection Office of Washington University. The Tissue Procurement Core consented the patients for using their biospecimens for research projects. In cased where consent was not obtainable, the Tissue Procurement Core has an Institutional Review Board issued waiver of consent, under which tumor tissues may be used anonymized biospecimens for non-genomic based studies. Paraffin embedded sections were deparaffinized in xylenes and rehydrated through a series of graded alcohols. Tissues were processed for antigen retrieval by boiling in citrate buffer (pH 6.0 containing 0.1% Tween). They were blocked in 10% donkey serum for 2 h to prevent nonspecific binding. The sections were then incubated overnight at 4 °C in primary antibody (anti-CXCR4, Novus Biologicals 1:400) or control IgG (anti-rabbit IgG, Novus Biologicals, 1:400). Anti-rabbit secondary antibody was applied (Jackson Laboratories) for 1 h at room temperature, and sections were washed in PBS, mounted in DAPI mounting medium (Vector Laboratories), and imaged using a Zeiss Confocal microscope system.
Western Blot.
4T1, CXCR4high and CXCR4low PDX tumors were collected, rinsed with saline, and frozen at −80°C. The frozen tissues were homogenized in NP40 buffer (50 mM pH 8.0 Tris buffer, 150 mM NaCl solution, 1% NP-40) supplemented with protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). All the lysates were centrifuged for 30 min at 10, 000 RPM at 4 °C. The supernatants were collected in a new microcentrifuge tube as the protein extracts. Protein content in each sample was determined using the BioRad protein assay (Biorad, Hercules, CA). For each specimen, approximately 80 μg of total protein was separated on a 12% SDS-PAGE and then electrophoretically transferred onto nitrocellulose membrane (BioRad, Hercules, CA). Nonspecific binding of the membranes was blocked with PBS-T (0.1% Tween-20) containing 5% nonfat milk powder for 1 h at room temperature and then incubated with anti-CXCR4 antibodies (dilution 1:1000, ab1670, Abcam, Cambridge, MA) at 4 °C overnight. After washing with PBS-T, the membranes were incubated with horseradish peroxidase-linked antigoat IgG (dilution 1:5000; GE Healthcare Bio-Sciences, Pittsburgh, PA) at room temperature for 45 min. The membrane was then subject to chemiluminescent detection with ECL substrate (GE Healthcare Bio-Sciences, Pittsburgh, PA) for 1 min. The images were exposed and captured immediately. In all studies β-actin was detected as internal standard in parallel blots.
Autoradiography.
Fixed human TNBC tissue sections were deparaffinized in Citrasolv and rehydrated through a series of graded alcohols and into PBS. The slides were incubated with 64Cu-CuNCs (8.3 MBq) and 64Cu-CuNCs-FC131 (8.3 MBq) for 15 min. Slides were extensively washed with water and then covered by a phosphor-imaging film plate and exposed overnight at −20 °C prior to imaging with a Molecular Dynamics Storm Imager 840.
Computational studies on the binding of FC131 to both the human and the mouse CXCR4 receptor.
The crystal structures of the chimeric human CXCR4 receptor were obtained from the PDB (3OE0, 3OE6).64 These crystal structures were used as the templates for homology modeling using the program Modeller (v 9.7).65 A total of 50 models of both the non-chimeric human and murine receptors were generated and the best scoring models were used for further study. The mouse receptor has a 10 residue loop not present in the human sequence, located at 151–161. In order to refine this region, the Modeller loop refinement protocol was used to generate 50 models of this region starting from the best original model, each model was scored with the DOPE-HR scoring method. The best models were then further refined with a multi-step molecular dynamics workflow. First, each structure was submitted individually to the H++ server at Virginia Tech where the hydrogens and protonation states were optimized at pH 6.5. In both cases a total of 7 Cl- anions were added to neutralize the charge. The receptors were then solvated in an octahedral box, and restrained molecular dynamics performed with the program NAMD using a CHARMM force field. From each production run five frames were pulled from the trajectories at RMSD minima for use in docking studies. Since most computational studies have focused on the human receptor,66 we developed an approach suitable for both the human and murine receptors.
Using the frames pulled from each trajectory, the programs mdpocket and fpocket were used to identify binding pockets which corresponded to the known peptide inhibitors CVX15 in the crystal structures 3OE0 and vMIP-II in 4RWS.67–68 A recent review of peptide docking by Hauser et al found that the best peptide docking results were obtained with the docking program GOLD and GS scoring.69 Ensemble docking studies of FC131-N-Me-D-Orn to all five snapshots of both the human and mouse CXCR4 receptor’s binding sites identified with mdpocket were then performed. One site was identified in the human receptor, while a total of three possible binding sites were identified in the murine receptor. The contacts between FC131 and the human and mouse CXCR4 receptor for the best scoring pose are summarized in Tables S1 and S2. Figure S5 is a diagram of the binding sites and best binding pose found for the peptide in the human and murine receptors. Residues in common between the human and mouse receptors are: D187, Y116. In the Thiele paper mutations to both of these residues lower the activity compared to the native human CXCR4 receptor. In summary, FC131-N-Me-D-Orn can successfully bind to both human and murine receptors while it slightly favors human over mouse (93.79 vs. 90.32). The numerical values represent the GoldScore used to identify the best pose.
Supplementary Material
ACKNOWLEDGMENTS
All animal experiments were carried out in compliance with the institutional animal care and usage committee (IACUC) guidelines of the Washington University. De-identified human triple negative breast cancer specimens were obtained from the Tissue Procurement Core of Siteman Cancer Center under the approval from Human Research Protection Office of Washington University. This work was partially performed at the Nano Research Facility (NRF), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under Grant No. ECS-0335765. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. NRF is part of the School of Engineering and Applied Science at Washington University in St. Louis. This work was performed with the support from the Siteman Cancer Center Small Animal Imaging Core at Washington University in St. Louis. We thank the staff members from Washington University cyclotron facility for 64Cu production.
The ESI-MS results were acquired by use of the NIH/NIGMS Biomedical Mass Spectrometry Resource at Washington University in St. Louis, MO, which is supported by National Institutes of Health / National Institute of General Medical Sciences Grant (8P41GM103422). Experiments were performed in part through the use of Washington University Center for Cellular Imaging (WUCCI) supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CDI-CORE-2015-505) and the National Institute for Neurological Disorders and Stroke (NS086741). The authors acknowledge financial support from Washington University in St. Louis and the Institute of Materials Science and Engineering for the use of instruments and staff assistance. The authors acknowledge the support from NCI grant (U24-CA209837) for animal models and PET imaging.
Footnotes
The authors declare no conflict of interest.
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website at DOI:10.1021/acsami.8b22752.
Synthetic schemes of TA-PEG ligands (Scheme S1 and S2); DLS and ξ-potential analysis of CuNCs (Figure S1); XPS spectra of CuNCs (Figure S2); Radio-iTLC analysis of 64Cu-CuNCs (Figure S3); Western blot of 4T1, CXCR4high and CXCR4low tumors (Figure S4); 15O-H2O PET images in animal models (Figure S5); Autoradiography of non-targeted 64Cu-CuNCs vs. targeted 64Cu-CuNCs-FC131 in human TNBC tissues (Figure S6); Histology and immunohistochemistry of CXCR4 in human TNBC tissues (Figure S7); Binding site plots of FC131 (Figure S8); Liver (Figure S9) and renal (Figure S10) function tests; contacts between FC131 and CXCR4 receptor (Table S1 and S2).
REFERENCES
- (1).Bosch A; Eroles P; Zaragoza R; Vina JR; Lluch A Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat. Rev 2010, 36, 206–215. [DOI] [PubMed] [Google Scholar]
- (2).Balkwill F The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol 2004, 14, 171–179. [DOI] [PubMed] [Google Scholar]
- (3).Chatterjee SA, Babak Behnam; Nimmagadda, Sridhar. The intricate role of CXCR4 in cancer. Adv. Cancer Res 2014, 124, 31–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Salvucci O; Bouchard A; Baccarelli A; Deschenes J; Sauter G; Simon R; Bianchi R; Basik M The role of CXCR4 receptor expression in breast cancer: a large tissuemicroarray study. Breast Cancer Res. Treat 2006, 97, 275–283. [DOI] [PubMed] [Google Scholar]
- (5).Sun X; Cheng G; Hao M; Zheng J; Zhou X; Zhang J; Taichman RS; Pienta KJ; Wang J CXCL12 / CXCR4 / CXCR7 chemokine axis and cancer progression. Cancer and Metastasis Rev. 2010, 29, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chow EK-H; Ho D Cancer Nanomedicine: From Drug Delivery to Imaging. Sci. Trans. Med 2013, 5, 216rv4–216rv4. [DOI] [PubMed] [Google Scholar]
- (7).Ferrari M Cancer nanotechnology: opportunities and challenges. Nat. Rev. Cancer 2005, 5, 161–171. [DOI] [PubMed] [Google Scholar]
- (8).Grobmyer SR; Zhou G; Gutwein LG; Iwakuma N; Sharma P; Hochwald SN Nanoparticle delivery for metastatic breast cancer. Maturitas 2012, 73, 19–26. [DOI] [PubMed] [Google Scholar]
- (9).Tanaka T; Decuzzi P; Cristofanilli M; Sakamoto JH; Tasciotti E; Robertson FM; Ferrari M Nanotechnology for breast cancer therapy. Biomed. Microdevices 2009, 11, 49–63. [DOI] [PubMed] [Google Scholar]
- (10).Giljohann DA; Seferos DS; Daniel WL; Massich MD; Patel PC; Mirkin CA Gold Nanoparticles for Biology and Medicine. ngew. Chem. Int. Ed 2010, 49, 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kumar D; Saini N; Jain N; Sareen R; Pandit V Gold nanoparticles: an era in bionanotechnology. Expert Opin. Drug Deliv 2013, 10, 397–409. [DOI] [PubMed] [Google Scholar]
- (12).Miller-Kleinhenz JM; Bozeman EN; Yang L Targeted nanoparticles for image-guided treatment of triple-negative breast cancer: clinical significance and technological advances. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol 2015, 7, 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Müller A; Homey B; Soto H; Ge N; Catron D; Buchanan ME; McClanahan T; Murphy E; Yuan W; Wagner SN; Barrera JL; Mohar A; Verástegui E; Zlotnik A Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [DOI] [PubMed] [Google Scholar]
- (14).Duda DG; Kozin SV; Kirkpatrick ND; Xu L; Fukumura D; Jain RK CXCL12 (SDF1α)-CXCR4/CXCR7 Pathway Inhibition: An Emerging Sensitizer for Anticancer Therapies? Clin. Cancer Res 2011, 17, 2074–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Teicher BA Antiangiogenic agents and targets: A perspective. Biochem. Pharmacol 2011, 81, 6–12. [DOI] [PubMed] [Google Scholar]
- (16).Banin Hirata BK; Oda JMM; Losi Guembarovski R; Ariza CB; Oliveira C. E. C. d.; Watanabe MAE Molecular Markers for Breast Cancer: Prediction on Tumor Behavior. Dis. Markers 2014, 2014, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Schmadeka R; Harmon BE; Singh M Triple-Negative Breast CarcinomaCurrent and Emerging Concepts. Am. J. Clin. Pathol 2014, 141, 462–477. [DOI] [PubMed] [Google Scholar]
- (18).Xiang J; Hurchla MA; Fontana F; Su X; Amend SR; Esser AK; Douglas GJ; Mudalagiriyappa C; Luker KE; Pluard T; Ademuyiwa FO; Romagnoli B; Tuffin G; Chevalier E; Luker GD; Bauer M; Zimmermann J; Aft RL; Dembowsky K; Weilbaecher KN CXCR4 Protein Epitope Mimetic Antagonist POL5551 Disrupts Metastasis and Enhances Chemotherapy Effect in Triple-Negative Breast Cancer. Mol. Cancer Ther 2015, 14, 2473–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chen HW; Du CW; Wei XL; Khoo US; Zhang GJ Cytoplasmic CXCR4 high-expression exhibits distinct poor clinicopathological characteristics and predicts poor prognosis in triple-negative breast cancer. Curr. Mol. Med 2013, 13, 410–416. [PubMed] [Google Scholar]
- (20).Yu S; Wang X; Liu G; Zhu X; Chen Y High level of CXCR4 in triple-negative breast cancer specimens associated with a poor clinical outcome. Acta Med. Okayama 2013, 67, 369–375. [DOI] [PubMed] [Google Scholar]
- (21).Chu QD; Panu L; Holm NT; Li BD; Johnson LW; Zhang S High chemokine receptor CXCR4 level in triple negative breast cancer specimens predicts poor clinical outcome. J. Surg. Res 2010, 159, 689–695. [DOI] [PubMed] [Google Scholar]
- (22).Licciardello N; Hunoldt S; Bergmann R; Singh G; Mamat C; Faramus A; Ddungu JLZ; Silvestrini S; Maggini M; De Cola L; Stephan H Biodistribution studies of ultrasmall silicon nanoparticles and carbon dots in experimental rats and tumor mice. Nanoscale 2018, 10, 9880–9891. [DOI] [PubMed] [Google Scholar]
- (23).Shen S; Jiang D; Cheng L; Chao Y; Nie K; Dong Z; Kutyreff CJ; Engle JW; Huang P; Cai W; Liu Z Renal-Clearable Ultrasmall Coordination Polymer Nanodots for Chelator-Free (64)Cu-Labeling and Imaging-Guided Enhanced Radiotherapy of Cancer. ACS Nano 2017, 11, 9103–9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jiang X; Du B; Huang Y; Zheng J Ultrasmall noble metal nanoparticles: Breakthroughs and biomedical implications. Nano Today 2018, 21, 106–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhao Y; Sultan D; Detering L; Luehmann H; Liu Y Facile synthesis, pharmacokinetic and systemic clearance evaluation, and positron emission tomography cancer imaging of 64Cu-Au alloy nanoclusters. Nanoscale 2014, 6, 13501–13509. [DOI] [PubMed] [Google Scholar]
- (26).Zhao Y; Detering L; Sultan D; Cooper ML; You M; Cho S; Meier SL; Luehmann H; Sun G; Rettig M; Dehdashti F; Wooley KL; DiPersio JF; Liu Y Gold Nanoclusters Doped with 64Cu for CXCR4 Positron Emission Tomography Imaging of Breast Cancer and Metastasis. ACS Nano 2016, 10, 5959–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gao F; Cai P; Yang W; Xue J; Gao L; Liu R; Wang Y; Zhao Y; He X; Zhao L; Huang G; Wu F; Zhao Y; Chai Z; Gao X Ultrasmall [64Cu]Cu Nanoclusters for Targeting Orthotopic Lung Tumors Using Accurate Positron Emission Tomography Imaging. ACS Nano 2015, 9, 4976–4986. [DOI] [PubMed] [Google Scholar]
- (28).Kreyling WG; Abdelmonem AM; Ali Z; Alves F; Geiser M; Haberl N; Hartmann R; Hirn S; de Aberasturi DJ; Kantner K; Khadem-Saba G; Montenegro JM; Rejman J; Rojo T; de Larramendi IR; Ufartes R; Wenk A; Parak WJ In vivo integrity of polymer-coated gold nanoparticles. Nat. Nanotechnol 2015, 10, 619–623. [DOI] [PubMed] [Google Scholar]
- (29).Rischpler C; Nekolla SG; Kossmann H; Dirschinger RJ; Schottelius M; Hyafil F; Wester HJ; Laugwitz KL; Schwaiger M Upregulated myocardial CXCR4-expression after myocardial infarction assessed by simultaneous GA-68 pentixafor PET/MRI. J. Nucl. Cardiol 2016, 23, 131–133. [DOI] [PubMed] [Google Scholar]
- (30).Derlin T; Jonigk D; Bauersachs J; Bengel FM Molecular Imaging of Chemokine Receptor CXCR4 in Non-Small Cell Lung Cancer Using 68Ga-Pentixafor PET/CT: Comparison With 18F-FDG. Clin. Nucl. Med 2016, 41, e204–e205. [DOI] [PubMed] [Google Scholar]
- (31).Lapa C; Lückerath K; Rudelius M; Schmid J-S; Schoene A; Schirbel A; Samnick S; Pelzer T; Buck AK; Kropf S [68Ga] Pentixafor-PET/CT for imaging of chemokine receptor 4 expression in small cell lung cancer-initial experience. Oncotarget 2016, 7, 9288–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Vag T; Gerngross C; Herhaus P; Eiber M; Philipp-Abbrederis K; Graner F-P; Ettl J; Keller U; Wester H-J; Schwaiger M First Experience with Chemokine Receptor CXCR4–Targeted PET Imaging of Patients with Solid Cancers. J. Nucl. Med 2016, 57, 741–746. [DOI] [PubMed] [Google Scholar]
- (33).Aldeek F; Muhammed MAH; Palui G; Zhan N; Mattoussi H Growth of Highly Fluorescent Polyethylene Glycol- and Zwitterion-Functionalized Gold Nanoclusters. ACS Nano 2013, 7, 2509–2521. [DOI] [PubMed] [Google Scholar]
- (34).Oh E; Susumu K; Goswami R; Mattoussi H One-Phase Synthesis of Water-Soluble Gold Nanoparticles with Control over Size and Surface Functionalities. Langmuir 2010, 26, 7604–7613. [DOI] [PubMed] [Google Scholar]
- (35).Wu Z; Suhan J; Jin R One-pot synthesis of atomically monodisperse, thiol-functionalized Au25 nanoclusters. J. Mater. Chem 2009, 19, 622–626. [Google Scholar]
- (36).Yang X; Yang M; Pang B; Vara M; Xia Y Gold Nanomaterials at Work in Biomedicine. Chem. Rev 2015, 115, 10410–10488. [DOI] [PubMed] [Google Scholar]
- (37).Wei W; Lu Y; Chen W; Chen S One-Pot Synthesis, Photoluminescence, and Electrocatalytic Properties of Subnanometer-Sized Copper Clusters. J. Am. Chem. Soc 2011, 133, 2060–2063. [DOI] [PubMed] [Google Scholar]
- (38).Zhang X; Lin Y; Gillies RJ Tumor pH and Its Measurement. J. Nucl. Med 2010, 51, 1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Tacke F Targeting hepatic macrophages to treat liver diseases. J. Hepatol 2017, 66, 1300–1312. [DOI] [PubMed] [Google Scholar]
- (40).Zhao Y; Sultan D; Detering L; Cho S; Sun G; Pierce R; Wooley KL; Liu Y Copper-64-Alloyed Gold Nanoparticles for Cancer Imaging: Improved Radiolabel Stability and Diagnostic Accuracy. Angew. Chem. Int. Ed 2014, 53, 156–159. [DOI] [PubMed] [Google Scholar]
- (41).Black KCL; Wang Y; Luehmann HP; Cai X; Xing W; Pang B; Zhao Y; Cutler CS; Wang LV; Liu Y; Xia Y Radioactive 198Au-Doped Nanostructures with Different Shapes for In Vivo Analyses of Their Biodistribution, Tumor Uptake, and Intratumoral Distribution. ACS Nano 2014, 8, 4385–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Scala S Molecular Pathways: Targeting the CXCR4-CXCL12 Axis--Untapped Potential in the Tumor Microenvironment. Clin. Cancer Res 2015, 21, 4278–4285. [DOI] [PubMed] [Google Scholar]
- (43).Marchese A Endocytic trafficking of chemokine receptors. Curr. Opin. Cell Biol 2014, 27, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Forster R; Kremmer E; Schubel A; Breitfeld D; Kleinschmidt A; Nerl C; Bernhardt G; Lipp M Intracellular and surface expression of the HIV-1 coreceptor CXCR4/fusin on various leukocyte subsets: rapid internalization and recycling upon activation. J. Immunol 1998, 160, 1522–1531. [PubMed] [Google Scholar]
- (45).Cheng ZJ; Zhao J; Sun Y; Hu W; Wu YL; Cen B; Wu GX; Pei G beta-arrestin differentially regulates the chemokine receptor CXCR4-mediated signaling and receptor internalization, and this implicates multiple interaction sites between beta-arrestin and CXCR4. J. Biol. Chem 2000, 275, 2479–2485. [DOI] [PubMed] [Google Scholar]
- (46).Qin C; Liu H; Chen K; Hu X; Ma X; Lan X; Zhang Y; Cheng Z Theranostics of Malignant Melanoma with 64CuCl2. J. Nucl. Med 2014, 55, 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Williams SA; Anderson WC; Santaguida MT; Dylla SJ Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab Invest. 2013, 93, 970–982. [DOI] [PubMed] [Google Scholar]
- (48).Whittle JR; Lewis MT; Lindeman GJ; Visvader JE Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 2015, 17, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Seol HS; Kang HJ; Lee SI; Kim NE; Kim TI; Chun SM; Kim TW; Yu CS; Suh YA; Singh SR; Chang S; Jang SJ Development and characterization of a colon PDX model that reproduces drug responsiveness and the mutation profiles of its original tumor. Cancer Lett. 2014, 345, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ding L; Ellis MJ; Li S; Larson DE; Chen K; Wallis JW; Harris CC; McLellan MD; Fulton RS; Fulton LL; Abbott RM; Hoog J; Dooling DJ; Koboldt DC; Schmidt H; Kalicki J; Zhang Q; Chen L; Lin L; Wendl MC; McMichael JF; Magrini VJ; Cook L; McGrath SD; Vickery TL; Appelbaum E; Deschryver K; Davies S; Guintoli T; Lin L; Crowder R; Tao Y; Snider JE; Smith SM; Dukes AF; Sanderson GE; Pohl CS; Delehaunty KD; Fronick CC; Pape KA; Reed JS; Robinson JS; Hodges JS; Schierding W; Dees ND; Shen D; Locke DP; Wiechert ME; Eldred JM; Peck JB; Oberkfell BJ; Lolofie JT; Du F; Hawkins AE; O’Laughlin MD; Bernard KE; Cunningham M; Elliott G; Mason MD; Thompson DM Jr.; Ivanovich JL; Goodfellow PJ; Perou CM; Weinstock GM; Aft R; Watson M; Ley TJ; Wilson RK; Mardis ER Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010, 464, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yang M; Huo D; D. GK; Sun X; Sultan D; Luehmann H; Detering L; Li S; Qin D; Liu Y; Xia Y Facile Synthesis of 64Cu-Doped Au Nanocages for Positron Emission Tomography Imaging. ChemNanoMat 2017, 3, 44–50. [Google Scholar]
- (52).Liu Y; Pressly ED; Abendschein DR; Hawker CJ; Woodard GE; Woodard PK; Welch MJ Targeting angiogenesis using a C-type atrial natriuretic factor-conjugated nanoprobe and PET. J. Nucl. Med 2011, 52, 1956–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Thackeray JT; Derlin T; Haghikia A; Napp LC; Wang Y; Ross TL; Schafer A; Tillmanns J; Wester HJ; Wollert KC; Bauersachs J; Bengel FM Molecular Imaging of the Chemokine Receptor CXCR4 After Acute Myocardial Infarction. JACC Cardiovasc. Imaging 2015, 8, 1417–1426. [DOI] [PubMed] [Google Scholar]
- (54).Derlin T; Sedding DG; Dutzmann J; Haghikia A; Konig T; Napp LC; Schutze C; Owsianski-Hille N; Wester HJ; Kropf S; Thackeray JT; Bankstahl JP; Geworski L; Ross TL; Bauersachs J; Bengel FM Imaging of chemokine receptor CXCR4 expression in culprit and nonculprit coronary atherosclerotic plaque using motion-corrected [(68)Ga]pentixafor PET/CT. Eur. J. Nuc.l Med. Mol. Imaging 2018, 45, 1934–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Gambling L; McArdle HJ Iron, copper and fetal development. Proc. Nutr. Soc 2007, 63, 553–562. [DOI] [PubMed] [Google Scholar]
- (56).Georgieff MK Nutrition and the developing brain: nutrient priorities and measurement. Am. J. Clin. Nutr 2007, 85, 614S–620S. [DOI] [PubMed] [Google Scholar]
- (57).Klevay LM; Inman L; Johnson LK; Lawler M; Mahalko JR; Milne DB; Lukaski HC; Bolonchuk W; Sandstead HH Increased cholesterol in plasma in a young man during experimental copper depletion. Metabolism 1984, 33, 1112–1118. [DOI] [PubMed] [Google Scholar]
- (58).Reiser S; Powell A; Yang CY; Canary JJ Effect of copper intake on blood cholesterol and its lipoprotein distribution in men. Nutr. Rep. Int 1987, 36, 641–649. [Google Scholar]
- (59).Gaetke LM; Chow-Johnson HS; Chow CK Copper: Toxicological relevance and mechanisms. Arch. Toxicol 2014, 88, 1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Institute of Medicine Panel on, M. In Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc; National Academies Press (US) Copyright 2001 by the National Academy of Sciences; All rights reserved.: Washington (DC), 2001. [PubMed] [Google Scholar]
- (61).Trumbo P; Yates AA; Schlicker S; Poos M Dietary reference intakes: vitamin A, vitamin K, arsenic, boron, chromium, copper, iodine, iron, manganese, molybdenum, nickel, silicon, vanadium, and zinc. J. Am. Diet. Assoc 2001, 101, 294–301. [DOI] [PubMed] [Google Scholar]
- (62).Jung J; Seol HS; Chang S The Generation and Application of Patient-Derived Xenograft Model for Cancer Research. Cancer Res. Treat 2018, 50, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Pang B; Zhao Y; Luehmann H; Yang X; Detering L; You M; Zhang C; Zhang L; Li Z-Y; Ren Q; Liu Y; Xia Y 64Cu-Doped PdCu@Au Tripods: A Multifunctional Nanomaterial for Positron Emission Tomography and Image-Guided Photothermal Cancer Treatment. ACS Nano 2016, 10, 3121–3131. [DOI] [PubMed] [Google Scholar]
- (64).Wu B; Chien EY; Mol CD; Fenalti G; Liu W; Katritch V; Abagyan R; Brooun A; Wells P; Bi FC; Hamel DJ; Kuhn P; Handel TM; Cherezov V; Stevens RC Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 2010, 330, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Sali A; Blundell TL Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 1993, 234, 779–815. [DOI] [PubMed] [Google Scholar]
- (66).Thiele S; Mungalpara J; Steen A; Rosenkilde MM; Vabeno J Determination of the binding mode for the cyclopentapeptide CXCR4 antagonist FC131 using a dual approach of ligand modifications and receptor mutagenesis. Br. J. Pharmacol 2014, 171, 5313–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Le Guilloux V; Schmidtke P; Tuffery P Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 2009, 10, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Schmidtke P; Bidon-Chanal A; Luque FJ; Barril X MDpocket: open-source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics 2011, 27, 3276–3285. [DOI] [PubMed] [Google Scholar]
- (69).Hauser AS; Windshugel B LEADS-PEP: A Benchmark Data Set for Assessment of Peptide Docking Performance. J. Chem. Inf. Model 2016, 56, 188–200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


