Abstract
Introduction
There is limited data on the prevalence and antibiotic susceptibility profile of Gram-negative bacteria in northwest Nigeria. This study thus aimed to investigate the prevalence of multidrug resistant Gram-negative bacterial infections among patients in two healthcare facilities in Sokoto, northwest Nigeria.
Methods
A total of 735 non-duplicate clinical bacterial isolates were collected between January and July 2019, from among specimens processed by the diagnostic microbiological laboratory of the two hospitals. The isolates were identified using MALDI-TOF mass spectrometry and tested against a panel of sixteen (16) antibiotics using the current EUCAST guidelines.
Results
Of the 735 randomly selected bacterial isolates, 397 (54.0%) yielded Gram-negative bacteria. In the two hospitals, E. coli 104 (26.2%) and Klebsiella spp. 58 (14.6%) were the most common Gram-negative pathogens implicated in all infections. Overall, the isolates exhibited moderate to high resistance to all tested antibiotics, the lowest was observed against amikacin (7.1%). The phenotypic test for ESBL and carbapenemase enzymes showed that 48 (24.6%) and 15 (32.6%) of the isolates were positive, with 88.9% of the isolates being multidrug resistant.
Conclusions
The study documents prevalent high multidrug resistant Gram-negative bacterial infections, predominantly caused by E. coli and K. pneumoniae in Sokoto, northwest Nigeria. The isolates were mostly MDR and exhibited ESBL and carbapenemase activities. The findings of this study call for urgent implementation of infection control measures and antibiotic stewardship in our hospitals so as to limit the spread of antibiotic-resistant bacteria in our healthcare facilities.
Keywords: Multidrug resistant, ESBL, carbapenemases, Gram-negative, Nigeria
Introduction
After decades of saving millions of lives and transforming healthcare practices, resistance of bacterial pathogens to nearly all antibiotics has emerged worldwide.1 The successes recorded in various aspects of modern medicine including surgeries, cancer chemotherapy and organ transplantation is being threatened by the emergence and spread of antibiotic resistant bacteria.2 These antibiotic resistant bacteria constitute a serious threat to the public health in developed countries as well as in resource limited setting in developing countries.1 Its prevalence is dangerously escalating worldwide leading healthcare practice towards a perilous era of post-antibiotics.2 Currently, deaths attributable to antibiotic resistance have been estimated to be more than 700,000 annually and are projected to exceed 10 million by the year 2050, if serious action is not taken to stem the tide.3 The mortality associated with infections by these difficult to treat bacteria has been shown to be more than twice that of infections in patients with susceptible bacteria.4
Although the emergence of antibiotic resistance is a natural phenomenon in most bacterial species, their spread is however being largely driven by negligent antibiotic use characterised by overuse and misuse in the healthcare systems, environment, and in agriculture/livestock practices.5 These factors alongside lack/inadequacy of infection control practices, overpopulation, poor sanitation and hygiene particularly in developing countries have exacerbated the problem.6 The poor regulatory system which enables easy accessibility to all antimicrobial agents over the counter in most developing countries including Africa and the distribution of sub-standard antimicrobials are some of the other factors driving the spread of drug resistant bacteria in the developing countries.7,8
Studies conducted in many African countries have revealed a growing prevalence of Gram-negative bacteria resistant to commonly prescribed antibiotics.9,10 In Nigeria, the Nigeria Centres for Disease Control and Prevention (NCDC) has equally documented a high rate of resistance to the commonly used antibiotics.11
Despite the untenable rate of antibiotic resistant bacterial infections reported in most Nigerian cities, there is substantial gap in the surveillance of these infections in several Nigerian cities especially in northwest Nigeria where limited research has been done on the prevalence of difficult to treat infections.12,13 In the few studies conducted, a limited number of antimicrobial classes have been tested. This study therefore aimed to comprehensively investigate the prevalence of multidrug resistant (MDR) Gram-negative bacterial infections among patients attending two major hospitals in northwest Nigeria.
Methods
Settings and study centres
This was a descriptive epidemiological study conducted between January and July 2019 among patients attending the two main tertiary healthcare facilities in the capital of Sokoto state, Sokoto, Northwest Nigeria. A total of 86 health facilities (2 tertiary, 5 secondary, and 79 primary health facilities) situated within the state are serving an estimated population of 4,998,090 people living within the state.14 The two tertiary health facilities owned respectively by the state and federal governments, Sokoto State Specialist Hospital (SHS) and Usmanu Danfodiyo University Teaching Hospital (UDUTH), are selected for this study. The hospitals are the largest hospitals located within the state. The hospitals are respectively 850 and 300-beds hospitals and rendering essential, specialized and referral medical and surgical services to residents of Sokoto state and patients from adjoining states of Zamfara, Kebbi and Niger within Nigeria and also to referral cases from the neighbouring Niger Republic. Usmanu Danfodiyo University Teaching Hospital in particular is a regional centre for many specialist cares including neurosurgery.
Bacterial collection and identification
A total of 735 clinical, non-duplicate bacterial isolates were collected from among the urine, wound swab, ear swab, high vaginal swab, and stool specimens processed by the diagnostic microbiological laboratories of the two hospitals. The isolates were randomly collected irrespective of the diagnosis and antibiotic susceptibility of the isolates or any pre-selection criteria, from both in-patients and out-patients. Socio-demographic and clinical data on gender, age and specimen types were obtained from patients’ records. The isolates were preserved on nutrient agar slants and shipped to Institute-Hospital University (IHU), Mediterranee infections, Marseille France, for further characterisation.
Identification and preservation
The isolates were re-activated and cultured primarily on prepared MacConkey agar medium (bioMérieux, France) and purified on Columbia sheep blood agar medium (bioMérieux). All isolates were identified by Matrix Assisted Laser Desorption-Ionization Time of Flight Mass Spectrometry (MALDI-TOF) following the protocol described by the manufacturer (Bruker Daltonics GmbH, Germany).
Antibiotic susceptibility testing
Antibiotic susceptibility testing was carried out using the modified Kirby-Bauer disc diffusion method as outlined in the current European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines, 2019.15 In brief, overnight culture of the test bacteria was diluted in sterile 0.85% sodium chloride solution to 0.5 McFarland standard and spread over the entire surface including the rim of a 120 mm dried Mueller Hinton Agar (MHA) medium (bioMérieux) using a sterile cotton-tipped swab bud. A panel of 16 antibiotics sourced from Biomérieux were placed on the inoculated plates using an automated disc dispenser. The antibiotics tested include the following: carbapenems: imipenem (10 µg) and meropenem (10 µg); cephalosporins: ceftriaxone (30 µg), cephalothin (30 µg) and cefepime (30 µg); fluoroquinolones: ciprofloxacin (5 µg); aminoglycosides: amikacin (30 µg) and gentamicin (10 µg); tetracyclines (doxycycline 30 µg) and polymyxin-B (colistin 50 µg). Others are trimethoprim-sulfamethoxazole (25 µg), amoxicillin-clavulanate (20 + 10 µg), fosfomycin (200 µg) and nitrofurantoin (100 µg). The inoculated plates were thereafter inverted and incubated at 37°C. The zone of inhibition formed after 16-18 h was measured and interpreted using the Interscience scan 4000 systems (Interscience, France). Isolates were classified as susceptible, intermediate or resistant according to breakpoints defined by EUCAST 2019.
The phenotypic detection of ESBL was performed using the double-disk synergy test by placing a beta-lactamase inhibitor (amoxicillin-clavulanic and piperacillin-tazobactam) discs between two third generation cephalosporin at a distance of 20 mm centre-to-centre.16 Formation of a characteristic keyhole effect or champagne-cork shaped zone of inhibition between the discs was considered as a phenotypic indication of ESBL production.
The modified Carba NP test as previously described was used to screen the carbapenem resistant isolates for phenotypic carbapenemase production.17
Operational definitions
An isolate in this study was classified as multidrug resistant (MDR) if the isolate was resistant to at least one agent in three or more different classes of antibiotics as previously defined.18 Bacterial isolates that were non-susceptible to at least one agent in all but two or fewer antimicrobial categories were regarded as extensively drug resistant (XDR) while isolates that were non-susceptible to all agents in all antimicrobial categories were regarded as pan-drug resistant (PDR).18
First-line antibiotics refer to commonly prescribed antimicrobials with good safety profile and excellent tolerability by the majority of patients. This comprises penicillins, cephalosporins, carbapenems and fluoroquinolones, resistance to which may thus necessitate the use of more toxic and expensive alternatives.19 Difficult to treat Gram-negative bacterial infections on the other hand are infections caused by bacteria resistant to the first-line antibiotics.19
Ethics statement
The Sokoto State Ministry of Health granted approval for the conduct of this study. Ethical clearance was also obtained from the Research and Ethics Committee of the two hospitals. Written informed consent is not applicable in this study as sample were not directly taken from the patients for the purpose of the study.
Statistical analysis
The data were manually checked for accuracy and completeness, analysed using IBM SPSS statistics software, version 24.0 (IBM Corporation, NY, USA) and presented as simple descriptive statistics or pictograms. Continuous variables were summarized using median and range and categorical variables were expressed using frequencies and percentages.
Results
Socio-demographic characteristics of the patients with bacterial infections
During the study period, a total of 4000 samples were received in the microbiological units of the two hospitals. Out of these, 735 were collected for this study. From among the isolates collected and shipped to the laboratory, 397 yielded Gram-negative bacteria, giving a Gram-negative bacterial infection rate of 54.0%. This comprises 267 (67.3%) and 130 (32.7%) bacterial isolates from UDUTH and SHS respectively (Table 1).
Table 1. Age and gender distribution of the patients from whom isolates were obtained.
| UDUTH number (%) | SHS number (%) | Cumulative number (%) | |
|---|---|---|---|
| Gender | |||
| Female | 126 (47.2) | 68 (52.3) | 194 (48.9) |
| Male | 141 (52.8) | 62 (47.7) | 203 (51.1) |
| Age categories (years) | |||
| 0-9 | 33 (12.4) | 9 (6.9) | 42 (10.6) |
| 10-19 | 47 (17.6) | 19 (14.6) | 66 (16.6) |
| 20-29 | 62 (23.2) | 47 (36.2) | 109 (27.5) |
| 30-39 | 33 (12.4) | 17 (13.1) | 50 (12.6) |
| 40-49 | 36 (13.5) | 22 (16.9) | 58 (14.6) |
| 50-59 | 25 (9.4) | 6 (4.6) | 31 (7.8) |
| ≥60 | 31 (11.6) | 10 (7.7) | 41(10.3) |
| Specimen types | |||
| Ear swab | 10 (3.8) | 0 (0) | 10 (2.5) |
| Endocervical swab | 2 (0.8) | 0 (0) | 2 (0.5) |
| High vaginal swab | 9 (3.4) | 10 (7.7) | 19 (4.8) |
| Sputum | 3 (1.1) | 21 (16.2) | 24 (6.1) |
| Stool | 74 (27.7) | 29 (22.3) | 103 (26.0) |
| Urine | 126 (47.2) | 60 (46.2) | 186 (46.9) |
| Wound swab | 43 (16.1) | 10 (7.7) | 53 (13.4) |
| Total | 267 (67.3) | 130 (32.7) | 397 (100) |
SHS - Specialist Hospital Sokoto; UDUTH - Usmanu Danfodiyo University Teaching Hospital.
The isolates were obtained from 397 individuals aged between 5 weeks and 72 years old (median = 27.0 years) with a vast majority of them below the age of 30 (Table 1). The patients were approximately evenly distributed between male and female with a male to female ratio of 1.05 to 1.
The distribution of the clinical isolates recovered during routine clinical diagnostic testing in the two hospitals shows that the majority of the isolates were recovered from urine 186 (46.9%) and stool 103 (25.9%) specimens. Other members were obtained from sputum 24 (6.0%), wound swab 53 (13.4%) and ear swab 10 (2.5%) (Table 1).
Bacterial isolates
The bacterial isolates were comprised of 293 (73.8%) Enterobacteriaceae and 104 (26.2) non-fermentative bacteria. Among the members of Enterobacteriaceae family, 182 and 111 were respectively isolated from UDUTH and SHS. In the two hospitals, E. coli (26.2%) and Klebsiella spp. (14.1%) were the most common pathogens implicated in all infections (Table 2). Members of Pseudomonas species predominated among the non-fermentative bacteria. Some emerging opportunistic pathogens were also isolated (Table 2).
Table 2. Distribution of the isolated bacteria.
| Strains | UDUTH | SHS | Cumulative (%) |
|---|---|---|---|
| Enterobacteriaceae | |||
| Escherichia coli | 55 | 49 | 104 (26.2) |
| Klebsiella pneumoniae | 38 | 18 | 56 (14.1) |
| Proteus mirabilis | 26 | 11 | 37 (9.3) |
| Enterobacter cloacae | 15 | 12 | 27 (6.8) |
| Citrobacter freundii | 16 | 3 | 19 (4.8) |
| Providencia stuartii | 4 | 11 | 15 (3.8) |
| Providencia rettgeri | 13 | 0 | 13 (3.3) |
| Morganella morganii | 7 | 4 | 11 (2.8) |
| Serratia marcescens | 3 | 0 | 3 (0.8) |
| Enterobacter aerogenes | 1 | 1 | 2 (0.5) |
| Proteus vulgaris | 2 | 0 | 2 (0.5) |
| Providencia alcalifaciens | 0 | 1 | 2 (0.5) |
| Klebsiella aerogenes | 0 | 1 | 1 (0.3) |
| Klebsiella oxytoca | 1 | 0 | 1 (0.3) |
| Citrobacter braakii | 1 | 0 | 1 (0.3) |
| Non-fermentative bacteria | |||
| Pseudomonas aeruginosa | 27 | 8 | 35 (8.8) |
| Pseudomonas putida | 2 | 1 | 3 (0.8) |
| Pseudomonas otitidis | 2 | 0 | 2 (0.5) |
| Pseudomonas guariconensis | 0 | 1 | 1 (0.3) |
| Pseudomonas mendonensis | 1 | 0 | 1 (0.3) |
| Pseudomonas monteilli | 0 | 1 | 1 (0.3) |
| Stenotrophomonas maltophilia | 15 | 0 | 15 (3.8) |
| Acinetobacter baumannii | 4 | 5 | 9 (2.3) |
| Acinetobacter pittii | 1 | 0 | 1 (0.3) |
| Alcaligenes faecalis | 19 | 2 | 21 (5.3) |
| Aeromonas hydrophila | 1 | 0 | 1 (0.3) |
| Ochrobactrum intermedium | 5 | 1 | 6 (1.5) |
| Brevundimonas diminuta | 4 | 0 | 4 (1.0) |
| Peanalcaligenes suwonensis | 1 | 0 | 1 (0.3) |
| Pantoea dispersa | 1 | 0 | 1 (0.3) |
| Delftia acidovorans | 1 | 0 | 1 (0.3) |
| Wautersiella falsenii | 1 | 0 | 1 (0.3) |
| Total | 267 | 130 | 397 |
SHS - Specialist Hospital Sokoto; UDUTH - Usmanu Danfodiyo University Teaching Hospital.
Antibiotic resistance profile
The antibiotic resistance profile of the fermentative Gram-negative bacteria is presented in Figure 1. The isolates exhibited divergent degree of resistance to the tested antibiotics. Overall, the highest resistance was exhibited against amoxicillin where 86.6% of the isolates were resistant. This was followed by an equally high resistance to first generation cephalosporin, cephalothin (76.0%). A moderately high resistance was observed against third generation and fourth cephalosporin antibiotics - ceftriaxone (46.2%) and cefepime (44.4%). The isolates exhibited exceptionally high resistance to amoxicillin-clavulanate (77.2% but low resistance to piperacillin-tazobactam (22.5%), another β-lactam/β-lactamase inhibitor combination. Resistance to other antibiotics such as gentamicin (32.5%), ciprofloxacin (50.5%), and trimethoprim-sulfamethoxazole (55.9%) was also high. In all isolates, amikacin remained the most active antibiotic with only 5.5% resistance. One isolate each of the Klebsiella pneumoniae and E. coli tested showed reduced susceptibility to colistin by disc diffusion test. However, the result of UMIC test showed that the MIC of the two strains (<2 µg/mL) was below the resistance cut-off as defined by EUCAST, 2019. Unsurprisingly, all strains of Proteus spp., M. morganii and Providencia species as well as S. marcescens, as expectedly, were 100% resistant to colistin.
Figure 1. Antibiotic susceptibility profile of fermentative Gram-negative bacteria. AMX- amoxicillin; AMC - amoxicillin-clavulanate; FEP - cefepime; TZP - piperacillin/tazobactam; KF - cefalotin; CRO - ceftriaxone; ERT - ertapenem; IMP - imipenem; FF - fosfomycin; F - nitrofurantoin; SXT - trimethoprim/sulfamethoxazole; AK - amikacin; CIP - ciprofloxacin; DO - doxycycline; CT - colistin; CN - gentamicin.
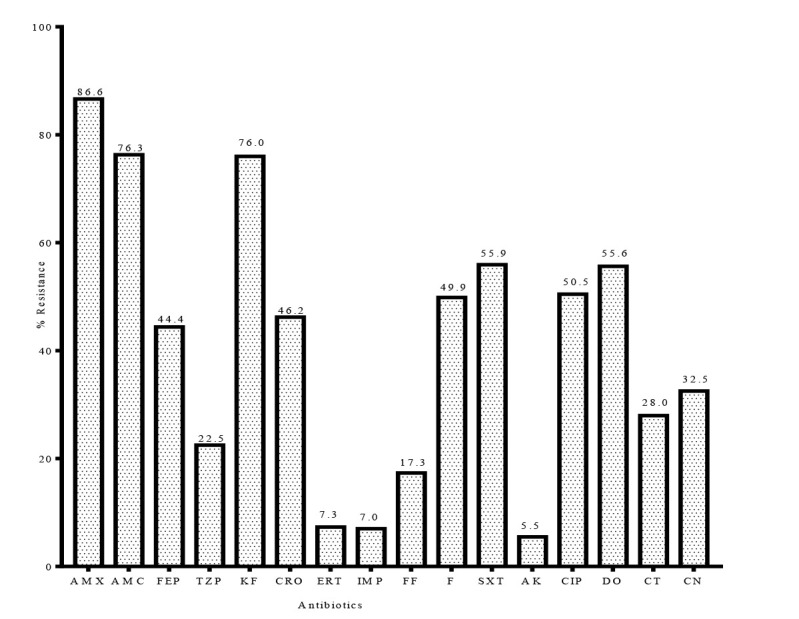
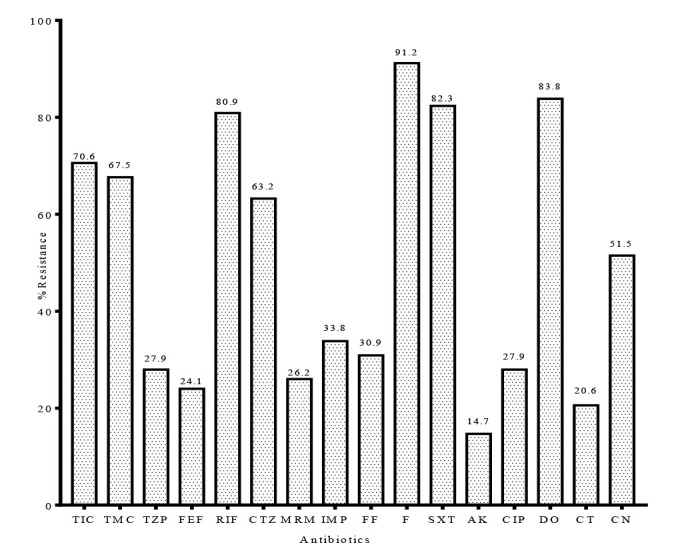
Figure 2 presents the rates of susceptibility of non-fermentative bacteria to the tested antibiotics. Similar to the Enterobacteriaceae, a high resistance rate was observed against third generation cephalosporin - ceftazidime (63.2%) - and antipseudomonal penicillin-ticarcillin (70.6%). Resistance to ticarcillin-clavulanate was equally high (67.7%).
Figure 2. Antibiotic susceptibility profile of non-fermentative bacteria. TIC- ticarcillin; TMC - ticarcillin-clavulanate; TZP - piperacillin-tazobactam; MRM - meropenem; RIF - rifampicin; CTZ - ceftazidime; FF - fosfomycin; F - nitrofurantoin; SXT - trimethoprim-sulfamethoxazole; AK - amikacin; CIP - ciprofloxacin; DO - doxycycline; CT - colistin; CN - gentamicin.
Distribution of resistance phenotypes
The distribution of the various resistance phenotypes is presented in the Table 3. A total of 353 (88.9%) of the 397 isolates were MDR, i.e., resistant to at least one agent in any of three or more distinct antimicrobial classes, while 1.8% and 0.3% of the isolates were extensively drug resistant and pan-drug resistant, respectively. Moreover, 30 (7.6%) of the isolates exhibited concurrent resistance to three of the four first line antibiotics while 36 (9.1%) were categorised as difficult to treat due to concurrent resistance to the four first line antibiotics.
Table 3. Resistance phenotypes of the bacterial isolates.
| Strains | MDR categories | |||
|---|---|---|---|---|
| Non-MDR | MDR | XDR | PDR | |
| Acinetobacter spp. | 0 | 9 | 1 | 0 |
| Aeromonas hydrophila | 0 | 1 | 0 | 0 |
| Alcaligenes spp. | 1 | 21 | 0 | 0 |
| Brevundimonas diminuta | 0 | 4 | 0 | 0 |
| Citrobacter spp. | 1 | 18 | 1 | 0 |
| Delftia acidovorans | 0 | 1 | 0 | 0 |
| Enterobacter spp. | 0 | 29 | 0 | 0 |
| Escherichia coli | 16 | 88 | 0 | 0 |
| Klebsiella spp. | 17 | 40 | 1 | 0 |
| Morganella morganii | 0 | 9 | 2 | 0 |
| Ochrobactrum intermedium | 0 | 5 | 1 | 0 |
| Pantoea dispersa | 0 | 1 | 0 | 0 |
| Proteus spp. | 1 | 38 | 0 | 0 |
| Providencia spp. | 0 | 29 | 0 | 0 |
| Pseudomonas spp. | 0 | 42 | 1 | 0 |
| Serratia marcescens | 0 | 3 | 0 | 0 |
| Stenotrophomonas maltophilia | 0 | 14 | 0 | 1 |
| Wautersiella falsenii | 0 | 1 | 0 | 0 |
| Total (%) | 36 (9.1) | 353 (88.9) | 7 (1.8) | 1 (0.2) |
Citrobacter spp.: C. braakii, C. freundii; Enterobacter spp.: E. aerogenes, E freundii; Klebsiella spp.: K. pneumoniae, K. oxytoca; Proteus spp.: P. mirabilis, P. vulgaris; Providencia spp.: P. stuartii, P. alkalifaciens.
Phenotypic tests for beta-lactamase enzymes
The phenotypic test for ESBL by the double disc synergy test showed that 48 (24.6%) of the 195 third generation cephalosporin-resistant isolates showed characteristic ESBL keyhole effect (Table 4). This is found to be more prevalent among the members of Enterobacteriaceae family than among the non-fermentative bacteria.
Table 4. Distribution of phenotypically ESBL and carbapenemase producing isolates.
| Strains | No. of 3GCR | ESBL positive | No. of CR-GNB | Carbapenemase producing |
|---|---|---|---|---|
| Enterobacteriaceae | ||||
| Citrobacter spp. | 10 | 3 | 4 | 2 |
| Enterobacter spp. | 18 | 8 | 5 | 3 |
| Escherichia coli | 51 | 10 | 4 | 2 |
| Klebsiella spp. | 28 | 15 | 4 | 0 |
| Morganella morganii | 4 | 0 | 1 | 0 |
| Proteus spp. | 13 | 7 | 0 | 0 |
| Providencia spp. | 3 | 0 | 1 | 0 |
| Serratia marcescens | 0 | 1 | 0 | 0 |
| Non-fermentative bacteria | ||||
| Acinetobacter spp. | 7 | 0 | 0 | 0 |
| Aeromonas hydrophila | 1 | 1 | 1 | 0 |
| Alcaligenes spp. | 16 | 0 | 2 | 3 |
| Brevundimonas diminuta | 0 | 1 | 0 | 0 |
| Delftia acidovorans | 1 | 0 | 0 | 0 |
| Ochrobactrum intermedium | 6 | 0 | 0 | 0 |
| Pantoea dispersa | 0 | 0 | 0 | 0 |
| Pseudomonas spp. | 29 | 1 | 8 | 4 |
| Stenotrophomonas maltophilia | 7 | 1 | 15 | 0 |
| Wautersiella falsenii | 1 | 0 | 1 | 1 |
| Total | 195 | 48 (24.6 %) | 46 | 15 (32.6 %) |
3GCR - third generation cephalosporin resistant; CR-GNB - carbapenem resistant Gram-negative bacteria; ESBL - extended spectrum betalactamase.
The result of Carba NP test showed that 15 (32.6%) of 46 CR-GNB with MIC above the clinical breakpoint exhibited carbapenemase activity.
Discussion
The surveillance of antibiotic resistant pathogens is immensely important. This is not only due to its use in empirical antibiotic selection but also for advocacy among the stakeholders for control of infection.20 Also, antibiotic resistance surveillance has been recognised as an important strategy for controlling antibiotic resistance.21
This study observed a high prevalence (54.0%) of Gram-negative bacterial infections among patients attending the two major hospitals in the extreme northwest, Nigeria, a significant proportion (88.9%) of which was due to MDR bacterial pathogens. This correlates with the observation of NCDC, which observed a high prevalence of resistant bacterial infections across the different states of the nation.11 Similarly, a study of antibiotic resistance in one of the leading teaching hospital in the country equally revealed a resistance rate as high as 100% to most of the commonly prescribed antibiotics.22 The high numbers of Gram-negative bacterial infections may be attributed to inadequate implementation of hospital hygiene practice and infection control.23 Furthermore, over-crowding of immunocompromised patients in the hospitals may further facilitate nosocomial transmission of resistant bacteria among the patients.24,25
Among the collected isolates, WHO classified serious life-threatening pathogens, E. coli, K. pneumoniae and Pseudomonas spp., were most prevalent pathogens implicated in all infections.26 This concurs with the report of a study at a tertiary hospital in Ibadan, south-west Nigeria where Klebsiella species (44.4%) and E. coli (13.0%) were the predominant pathogens causing clinical infections.27 A similar preponderance of E. coli, K. pneumoniae and Pseudomonas spp. has been observed in a neighbouring Republic of Ghana.28 Moreover, infections by Citrobacter spp., Enterobacter spp., Proteus spp. and Morganella morganii have also been reported among Nigerian patients.29,30 While few cases of infections by Alcaligenes faecalis (1.2%) and Aeromonas hydrophila (3.92%) have been documented in the literature, there are no reports in the literature of clinically significant infections involving Peanalcaligenes suwonensis, Brevundimonas diminuta, Ochrobactrum intermedium, Pantoea dispersa, and Delftia acidovorans in Nigeria.31,32 Elsewhere, infections by these bacteria have been described particularly among the immunocompromised and liver transplant patients and they have been described as emerging opportunistic pathogens.33,34 The isolation of these strains for the first time in Nigeria may be due to the improved and highly sensitive analytical technique (MALDITOF MS) used for the bacterial identification.35 The difficulty in identification of these emerging pathogens in microbiology laboratories is due to their taxonomic complexity and phenotypic similarity.34
The high resistance of the isolates to the commonly prescribed antibiotics calls for concern as there are limited alternatives available in the country. The prevalent high resistance to most of the antibiotics tested concurs with findings from other countries in the sub-Sahara Africa. In Ghana for example, a more than 80% resistance rate to commonly prescribed antibiotics has been reported.28 The high antibiotic resistance reported in this study may be due to easy accessibility over the counter to most antibiotics and a high selection pressure due to the extensive use of these antibiotics, mostly inappropriately for self-medications and agricultural purposes.36 The high susceptibility of the most common pathogens, E. coli and Klebsiella spp. to colistin and carbapenems may not be unconnected with their low usage in Nigeria because of their comparatively high cost compared to other agents. This may thus result in low selection and slow emergence of resistant strains to these agents. However, the occurrence of carbapenem resistance in hospitals with no history of carbapenem usage has been documented.12 The pattern of resistance observed in this study correlates with the pattern of antibiotics use in Nigeria. Generally, β-lactams, aminoglycosides and fluoroquinolones are the most prescribed antibiotics in Nigeria.37 The reported high resistance to these antibiotics in the present study gives credence to the statement that extensive antibiotic use is a major driver of emergence of resistance.38
The higher resistance of the non-fermentative Gram-negative bacteria compared to the fermentative bacteria is not surprising. These opportunistic pathogens are known to be intrinsically resistant to most important classes of antibiotics. The higher intrinsic resistance in these pathogens has been linked with their lower cellular permeability and higher efflux activities.39
The result of ESBL detection tests corresponds with a recent study where ESBL-producing Enterobacteriaceae have been shown to widely circulate in all geopolitical zones of the federation with national prevalence of 34.62%40 The circulation of the three common ESBL genes in Sokoto has also been documented.41
Globally, carbapenemase producing bacteria are increasingly reported, the prevalence of which varies from one geographical region to the other. Previous reports across the country have established varying rates. The finding of 36.32% carbapenemase-producing Gram-negative bacteria in the present study is consistent with a previous report where 38.0% of carbapenem-resistant bacteria in one of the hospitals in Sokoto were found to be carbapenemase-producing.12 While the finding of the present study corresponds to a similar prevalence rate of 25.2% reported in Yola, north-eastern Nigeria,42 it was however higher than 2% reported in Ethiopia43 and 18.9% in Ghana.44
Despite the poor drug regulatory system in Nigeria coupled with the lack of an established antibiotic stewardship, carbapenem and colistin use in both hospital and community is generally low, reserved as a last resort agent against life threatening infections by multidrug resistant bacteria.11 The observed resistance to the carbapenems in this study is troubling in a country where alternative antibiotics are rarely available.11 Furthermore, the emergence and spread of carbapenem-resistant bacteria is more worrisome because of lack of laboratory capacity for their detection.11
This study is limited, majorly, by our inability to access certain patients’ records which makes it difficult to pinpoint bacteria recovered to a particular clinical infection. It should however be noted that all samples were collected as part of patient care for diagnosis of infections and not for surveillance of bacterial colonisation. Also, the tests reported in this study are mainly phenotypic, investigation of molecular basis of the observed resistance is the focus of our future studies.
Conclusions
The study documents prevalent high multidrug resistant Gram-negative bacterial infections, predominantly caused by E. coli, K. pneumoniae and P. aeruginosa in Sokoto, northwest Nigeria. The isolates were mostly MDR and exhibited ESBL and carbapenemase activities. The findings of this study call for urgent implementation of infection control measures and antibiotic stewardship in our hospitals so as to limit the spread of antibiotic-resistant bacteria in our healthcare facilities.
Footnotes
Authors’ contributions statement: YKEI and BOO conceived and designed the study. The research protocol as well as data acquisition and analysis were done by AO and LZN. AO drafted the first manuscript. All authors contributed to the revision of the first draft and approved the final draft for publication.
Conflicts of interest: All authors - none to declare.
Funding: None to declare.
References
- 1.Ventola CL. The antibiotic resistance crisis part 1: causes and threats. Pharm Ther. 2015;40:277–83. [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Antimicrobial Resistance: Global report on surveillance 2014. [Accessed on 12 April 2020]. Available at: https://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/
- 3.World Bank. Drug-resistant infections: a threat to our economic future. World Bank Rep. 2016 [Google Scholar]
- 4.Founou RC, Founou LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries?: A systematic review and meta-analysis. PLoS One. 2017;12:e0189621. doi: 10.1371/journal.pone.0189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roca I, Akova M, Baquero F, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015;6:22–9. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6:47. doi: 10.1186/s13756-017-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma D, Patel RP, Zaidi STR, Camerino GM, Aldo B, Moraes LA. Interplay of the quality of ciprofloxacin and antibiotic resistance in developing countries. Front Pharmacol. 2017;8:546. doi: 10.3389/fphar.2017.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osei-Safo D, Egbo HA, Nettey H, Konadu DY, Addae-Mensah I. Evaluation of the quality of some antibiotics distributed in Accra and Lagos. Int J Pharm Sci Res. 2016;7:1991–2000. [Google Scholar]
- 9.Kariuki S, Dougan G. Antibacterial resistance in sub-Saharan Africa: an underestimated emergency. Ann N Y Acad Sci. 2014;1323:43–55. doi: 10.1111/nyas.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tadesse BT, Ashley EA, Ongarello S, et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017;17:616. doi: 10.1186/s12879-017-2713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCDC. Antimicrobial use and resistance in Nigeria. 2017. [Accessed on 4 March 2020]. Available at: http://www.ncdc.gov.ng/themes/common/docs/protocols/56_1510840387.pdf.
- 12.Olowo-okere A, Abdullahi MA, Ladidi BK, et al. Emergence of metallo-b-lactamase producing Gram-negative bacteria in a hospital with no history of carbapenem usage in northwest Nigeria. Ife J Sci. 2019;21:323–31. doi: 10.4314/ijs.v21i2.6. [DOI] [Google Scholar]
- 13.Yusuf I, Arzai AH, Haruna M, Sharif AA, Getso MI. Detection of multi drug resistant bacteria in major hospitals in Kano, North-West, Nigeria. Braz J Microbiol. 2014;45:791–8. doi: 10.1590/S1517-83822014000300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nigerian Bureau of Statistics. Demographic statistics bulletin. 2017. [Accessed on: 12 April 2020]. Available at www.nigerianstat.gov.ng.
- 15.EUCAST. European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters. 2019. [Accessed on month 12 April 2020]. Available at: www.eucast.org/mic_distributions_and_ecoffs.
- 16.EUCAST. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and / or epidemiological importance. 2019. [Accessed on 12 April 2020]. Available at: https://www.eucast.org/resistance_mechanisms.
- 17.Bakour S, Garcia V, Loucif L, et al. Rapid identification of carbapenemase-producing Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii using a modified Carba NP test. New Microbes New Infect. 2015;7:89–93. doi: 10.1016/j.nmni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magiorakos A, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 19.Kadri SS, Adjemian J, Lai L, et al. Difficult-to-treat resistance in Gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin Infect Dis. 2018;67:1803–14. doi: 10.1093/cid/ciy378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tacconelli E, Sifakis F, Harbarth S, et al. Surveillance for control of antimicrobial resistance. Lancet Infect Dis. 2018;18:e99–106. doi: 10.1016/S1473-3099(17)30485-1. [DOI] [PubMed] [Google Scholar]
- 21.Ventola CL. The antibiotic resistance crisis: part 2: management strategies and new agents. P T. 2015;40:344–52. [PMC free article] [PubMed] [Google Scholar]
- 22.Olowo-Okere A, Ibrahim YKE, Sani AS, Atata RF, Olayinka BO. Prevalence of surgical site infection in a Nigerian university teaching hospital. J Pharm Allied Sci. 2017;14:2430–8. [Google Scholar]
- 23.Iliyasu G, Dayyab F, Habib ZG, et al. Knowledge and practices of infection control among healthcare workers in a Tertiary Referral Center in North-Western Nigeria. Ann Afr Med. 2015;15:34–40. doi: 10.4103/1596-3519.161724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makama JG, Iribhogbe P, Ameh EA. Overcrowding of accident & emergency units: is it a growing concern in Nigeria? Afr Health Sci. 2015;15:457–65. doi: 10.4314/ahs.v15i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abubakar HM, Musa I. Overcrowding: the need for taskforce in hospital emergency departments. Int J Med Eval Phys Rep. 2018;3:18–25. [Google Scholar]
- 26.WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017. [Accessed on 12 April 2020]. Available at: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/
- 27.Makanjuola OB, Fayemiwo SA, Okesola AO, et al. Pattern of multidrug resistant bacteria associated with intensive care unit infections in Ibadan, Nigeria. Ann Ib Postgrad Med. 2018;16:162–9. [PMC free article] [PubMed] [Google Scholar]
- 28.Agyepong N, Govinden U, Owusu-Ofori A, Essack SY. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob Resist Infect Control. 2018;7:37. doi: 10.1186/s13756-018-0324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raji MA, Jamal W, Ojemhen O, Rotimi VO. Point-surveillance of antibiotic resistance in Enterobacteriaceae isolates from patients in a Lagos Teaching Hospital, Nigeria. J Infect Public Health. 2013;6:431–7. doi: 10.1016/j.jiph.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Oli AN, Ogbuagu VI, Ejikeugwu CP, et al. Multi-antibiotic resistance and factors affecting carriage of extended spectrum β-lactamase-producing Enterobacteriaceae in pediatric population of Enugu metropolis, Nigeria. Med Sci (Basel) 2019;7:104. doi: 10.3390/medsci7110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alaka OO, Orimolade EA, Ojo OO, Onipede AO. The phenotypic detection of carbapenem resistant organisms in orthopaedic wound infections in Ile-Ife, Nigeria. Acta Sci Microbiol. 2019;2:35–42. [Google Scholar]
- 32.Mailafia S, Nafarnda W, Sugun MY. Occurrence and antimicrobial susceptibility patterns of Aeromonas hydrophila isolates among diarrhoeic patients from University of Abuja Teaching Hospital, Nigeria. J Pure Appl Microbiol. 2017;11:63–70. doi: 10.22207/JPAM.11.1.09. [DOI] [Google Scholar]
- 33.Ryan MP, Pembroke JT. Brevundimonas spp: emerging global opportunistic pathogens. Virulence. 2018;9:480–93. doi: 10.1080/21505594.2017.1419116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whistler T, Sangwichian O, Jorakate P, et al. Identification of Gram negative nonfermentative bacteria: how hard can it be? PLoS Negl Trop Dis. 2019;13:e0007729. doi: 10.1371/journal.pntd.0007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanguinetti M, Posteraro B. Mass spectrometry applications in microbiology beyond microbe identification: progress and potential. Expert Rev Proteomics. 2016;13:965–77. doi: 10.1080/14789450.2016.1231578. [DOI] [PubMed] [Google Scholar]
- 36.Ajibola O, Omisakin OA, Eze AA, Omoleke SA. Self-medication with antibiotics, attitude and knowledge of antibiotic resistance among community residents and undergraduate students in Northwest Nigeria. Diseases. 2018;6:32. doi: 10.3390/diseases6020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umar LW, Isah A, Musa S, Umar B. Prescribing pattern and antibiotic use for hospitalized children in a Northern Nigerian Teaching Hospital. Ann Afr Med. 2018;17:26–32. doi: 10.4103/aam.aam_44_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shallcross LJ, Davies DS. Antibiotic overuse: A key driver of antimicrobial resistance. Br J Gen Pract. 2014;64:604–5. doi: 10.3399/bjgp14X682561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayoub Moubareck C, Hammoudi Halat D, Akkawi C, et al. Role of outer membrane permeability, efflux mechanism, and carbapenemases in carbapenem-nonsusceptible Pseudomonas aeruginosa from Dubai hospitals: Results of the first cross-sectional survey. Int J Infect Dis. 2019;84:143–50. doi: 10.1016/j.ijid.2019.04.027. [DOI] [PubMed] [Google Scholar]
- 40.Musa BM, Imam H, Lendel A, et al. The burden of extended-spectrum β-lactamase-producing Enterobacteriaceae in Nigeria: a systematic review and meta-analysis. Trans R Soc Trop Med Hyg. 2020;114:241–8. doi: 10.1093/trstmh/trz125. [DOI] [PubMed] [Google Scholar]
- 41.Tanko N, Bolaji RO, Olayinka AT, Olayinka BO. A systematic review on the prevalence of extended spectrum beta lactamase producing Gram-negative bacteria in Nigeria. J Glob Antimicrob Resist. 2020;22:488–96. doi: 10.1016/j.jgar.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Shettima SA, Tickler IA, Dela Cruz CM, Tenover FC. Characterization of carbapenem-resistant Gram-negative organisms from clinical specimens in Yola, Nigeria. J Glob Antimicrob Resist. 2020;21:42–5. doi: 10.1016/j.jgar.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Beyene D, Bitew A, Fantew S, Mihret A, Evans M. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference. PLoS One. 2019;14:e0222911. doi: 10.1371/journal.pone.0222911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Codjoe FS, Donkor ES, Smith TJ, Miller K. Phenotypic and genotypic characterization of carbapenem-resistant Gram-negative bacilli pathogens from hospitals in Ghana. Microb Drug Resist. 2019;25:1449–57. doi: 10.1089/mdr.2018.0278. [DOI] [PubMed] [Google Scholar]


