Abstract
Introduction
Updated and comprehensive data on the mechanism underlying colistin resistance is lacking in Africa.
Literature search
Herein, we aimed to review available literature on the molecular mechanisms of colistin resistance in Africa. PubMed, Google Scholar, and African Journal online databases were searched on the 15th of January 2020 for original research articles that reported mechanisms of colistin resistance in any of the 54 African countries.
Review
Of the 1473 studies identified through initial database search, 36 met the inclusion criteria. Colistin resistance was mostly observed in Escherichia coli isolated from human clinical samples. Plasmid-mediated colistin resistance mechanism (26; 72.2%) was the most frequently reported resistance mechanism. About three-quarters (27; 75.0%) of the 36 studies were done in North Africa. In this zone, the mobilized colistin resistance (mcr) genes were mostly detected in E. coli harboring three plasmid types, IncHI2, IncI2, and IncX4, from animal samples (n=9; 42.8%). Of the six studies performed in Southern Africa, four reported mcr-1 mostly detected from human samples (n=2; 50.0%) in E. coli isolates carrying IncHI2, IncI2, and IncX4 with diverse range of STs. One hitherto unknown mutation, the mutation in the I527N gene was detected in colistin resistant isolates in this region, which was absent in colistin susceptible isolates. In West and Central Africa, two and one studies, respectively, reported mcr-1 gene exclusively in Escherichia coli isolates.
Conclusions
Transferable plasmid mediated colistin resistance is rapidly emerging in Africa with mcr-1 as the predominant genetic variant in human, animals, and environmental samples.
Keywords: Colistin resistance, mechanisms, mobilized colistin resistance, Gram-negative bacteria, Africa
Introduction
The emergence of multidrug resistant bacterial infections is one of the greatest threats today to the global public health.1 It arises naturally as one of the direct consequences of antibiotic use.2 Its abuse particularly in animal production and aquaculture has further driven the emergence and spread of antibiotic resistance, leading healthcare practice towards a post-antibiotic era.3 Resistance to all important classes of antibiotics including carbapenems, has been reported particularly among the clinically important pathogens, the ESCAPE pathogens, with substantial impact on morbidity, mortality and attendant increase in healthcare cost.4 Further exacerbating the problem is the lack of new antibiotics classes in the pipeline, owing to decline in research and development of new effective anti-infective agents.5 Consequent upon this, old antibiotics such as colistin, which were initially abandoned due to patient safety concerns, were recalled as a life-saving and last resort measure against serious Gram-negative bacterial infections.6
Colistin is one of the five polymyxin antibiotics originally isolated in 1947 from the soil bacterium Paenibacillus polymyxa subsp. Colistinus. 7 It is a polycationic lipopolypeptide that acts by competitively displacing divalent cations, Mg2+ and Ca2+, from the phosphate group of lipopolysaccharides of Gram-negative cell envelope, thereby disrupting cell membrane integrity and leading to leakage of important cellular components and ultimately bacterial cell death.7 It has also been shown to act by inhibiting a key respiratory enzyme, type II NADH-quinone oxidoreductases (NDH-2).7 Colistin is highly and rapidly bactericidal against susceptible bacterial species particularly Pseudomonas aeruginosa, Acinetobacter baumannii and most members of Enterobacteriaceae family with the exception of Providencia spp., Morganella morganii, Proteus spp., Serratia marcescens, among others which are naturally resistant.8
Until recently, acquired resistance to colistin has been mostly due to chromosomal mutation in the PmrA/PmrB and PhoP/PhoQ two-component regulatory systems, or through increased production of capsular polysaccharide.8 Mutational inactivation of mgrB, a negative regulator of the PhoP/PhoQ signaling system has also been identified in several studies as a basis for colistin resistance.8-10 The transferable plasmid encoded colistin resistance (the mobilized colistin resistance or mcr-1) emerged in China in late 2015.11 Today, nine other families of mcr genes have been detected from various hosts and pathogens range.9,10,12 The mcr genes have now been globally disseminated and it continues to be increasingly reported worldwide.13
While the molecular mechanisms underlying colistin resistance have been described considerably in North America, Europe and more particularly in Asia, updated and comprehensive data on the different mechanisms of colistin resistance are lacking in Africa. We therefore aimed to systematically review available literature on the molecular mechanisms of colistin resistance in African countries, to determine the most prevalent colistin resistance mechanisms, the circulating colistin resistant Gram-negative bacteria clones and hosts in Africa.
Methods
Literature search
A systematic literature search was conducted to identify articles reporting colistin resistance in Africa. Multiple searches were conducted in PubMed, Google Scholar, and African Journal online (AJOL). Our search strategy uses different relevant keywords: “colistin resistance” OR “mobilized colistin resistance gene” OR “mcr” AND Africa, or names of the 54 African countries. The present study was designed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.14
Study selection
Publications identified were considered up to January 15, 2020. Searches undertaken were not restricted by language. Two authors independently performed the literature search. After removing duplicates, the studies identified in the initial search were first screened by title and abstract and retained if they met the predefined inclusion criteria, as follows: (i) original article published or accepted in a peer-reviewed journal, (ii) studies that described mechanism of colistin resistance in humans, animals and environment, and (iii) studies conducted in any of the 54 Africa countries. Full text of the articles that met the inclusion criteria were retrieved and further screened. Studies that reported phenotypic prevalence of colistin resistance without investigation of molecular basis underlying the resistance were excluded. Also, studies conducted outside the specified period of this systematic review were excluded. The screening process was documented in the preferred reporting items for systematic reviews and meta-analyses (PRISMA) flow chart of the study selection.
Data extraction
Two reviewers independently extracted the relevant data, using a standardized collection form to extract data from the included articles. The following data were extracted from each study: name of country in which the study was conducted, the first author’s name, year of publication, sources of isolates (human/animal/environment), bacterial species, plasmid type, genes detected, number of mcr positive bacteria, sequences types (ST), number of isolates exhibiting colistin resistance, medical condition (for human), and result of antibiotic susceptibility testing.
Data analysis
The data collected, including the countries, sources of isolates, bacterial species, plasmid type, genes detected, number of mcr genes positive bacteria, and sequences types (ST), were analysed using Microsoft Excel 2013. Descriptive statistics including frequencies and percentages were used in the analysis.
Results
Literature search and study selection
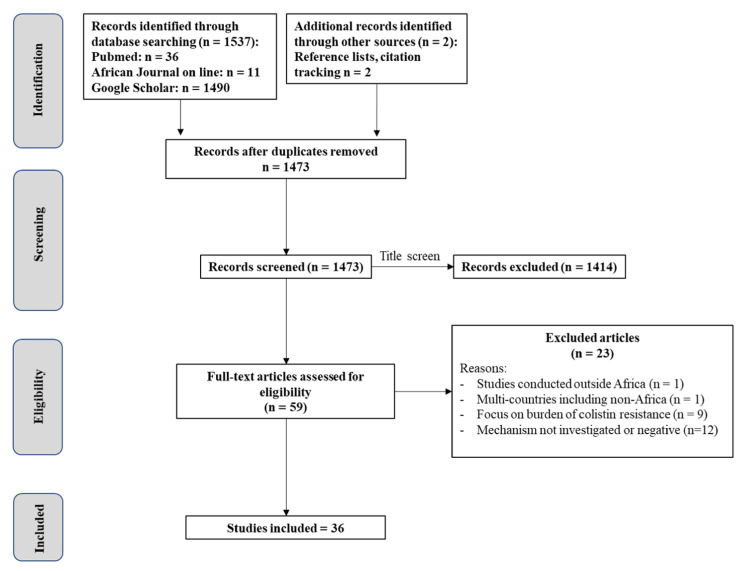
A total of 1493 non duplicate, potentially relevant studies were identified and retrieved from the databases. After screening of the titles and abstracts, 1414 articles were considered irrelevant and excluded. Of the 59 full text articles considered of interest and assessed for eligibility, only 36 articles met the inclusion criteria and were included in the final analysis (Figure 1). Based on the included studies, the first article on colistin resistance in Africa was published in 2014.
Figure 1. Literature research and study selection.
Characteristics and distribution of studies describing colistin resistance in Africa
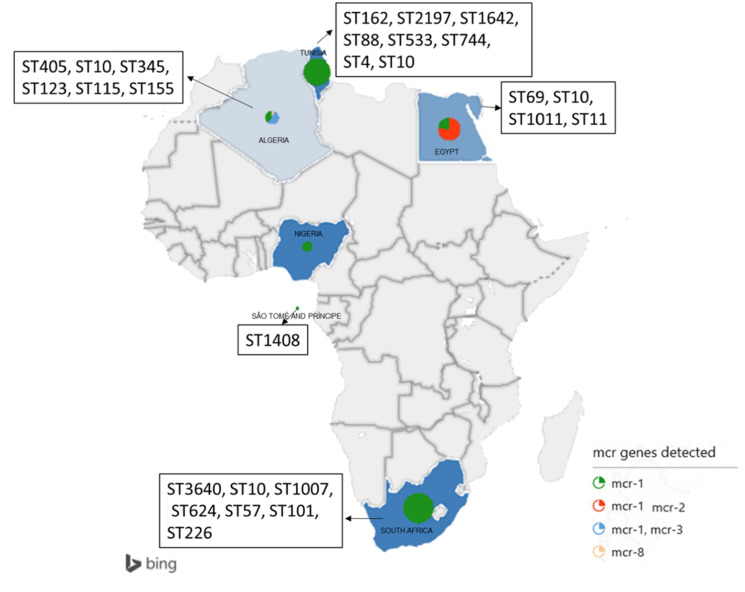
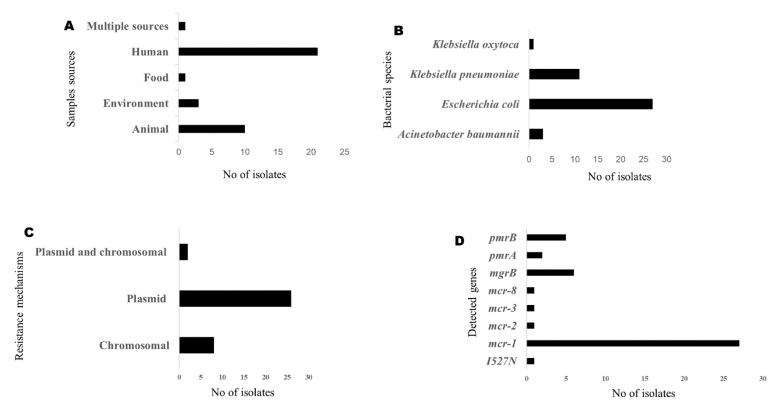
This systematic review involved a total of 36 studies conducted in seven countries namely Tunisia (n=7), South Africa (n=6), São Tomé and Príncipe (n=1), Nigeria (n=2), Libya (n=1), Egypt (n=8), and Algeria (n=11) (Figure 2). The studies were published between 2014 and 2019. The studies documented colistin resistance mechanisms in bacterial isolates obtained from humans (21; 58.3%), animals (10; 27.8%) and environmental samples (3; 8.3%). In the included studies, colistin resistance mechanism was described in mainly three bacterial species comprising A. baumannii, Escherichia coli and Klebsiella pneumoniae. Colistin resistance was most frequently observed in E. coli isolated from human clinical samples. Both chromosomal and plasmid mediated mechanisms were reported, with plasmid-mediated colistin resistance mechanism (26; 72.2%) most frequently reported (Figure 3).
Figure 2. Geographical distribution of the study countries.
Figure 3. Characteristics of colistin resistance in Africa.
Overall, the studies described colistin resistance in 904 bacterial isolates, 188 (20.79%) of which harbored various mcr genes. Of the 188 mcr genes detected, mcr-1 was the most prevalent. Among the 188 isolates in which the presence of mcr genes has been reported, 88 (46.80%) reported the types of plasmid. The mcr genes were mostly harbored on an IncHI2 plasmid (n=50; 56.82%), followed by IncI2 (n=19; 21.59%).
Distribution of mcr genes in various Africa regions
North Africa
About three-quarters (27; 75.0%) of the 36 studies were done in North Africa. The prevalence of mcr genes was 17.53% among colistin resistant isolates. A total of 21 studies reported mcr genes in three countries: Tunisia (n=5), Egypt (n=8), and Algeria (n=8) (Figure 2). The mcr genes included mcr-1 (n=18; 85.9%), mcr-1 and mcr-2 (n=1; 4.7%), mcr-1 and mcr-3 (n=1; 4.7%), and mcr-8 (n=1; 4.7%). The mcr genes were equally detected from samples obtained from animals (n=9; 42.8%) and humans (n=9; 42.8%). The presence of mcr genes was reported in only Escherichia coli and Klebsiella pneumoniae. Four plasmid types were reported. All of them were reported from Escherichia coli and harbored mcr-1 with IncHI2 (n=3) as the most prevalent mcr associated plasmid encountered in North Africa (Table 1).
Table 1. Distribution of the studies describing colistin resistance in North Africa.
| Country | Year | Source | Organism | Number of isolates exhibiting colistin resistance | Mechanism | Plasmid | Genes detected | Number of isolates positives for mcr genes | ST | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Algeria | 2014 | Human | Acinetobacter baumannii | 1 | Chromosomal | NA | PmrB | NA | ND | 27 |
| Egypt | 2019 | Food | Escherichia coli | 1 | Plasmid | IncHI2A and IncHI2 | mcr-1 | 1 | ST69 | 28 |
| Algeria | 2017 | Animal | Escherichia coli | 1 | Plasmid | ND | mcr-1 | 1 | ST405 | 29 |
| Tunisia | 2019 | Animal | Escherichia coli | 5 | Plasmid | IncHI2 | mcr-1 | 5 | ST162 | 30 |
| Tunisia | 2019 | Animal | Escherichia coli | 1 | Plasmid | IncHI2 | mcr-1 | 1 | ND | 31 |
| Algeria | 2016 | Animal | Escherichia coli | 5 | Plasmid | ND | mcr-1 | 3 | ND | 32 |
| Tunisia | 2016 | Animal | Escherichia coli | 37 | Plasmid | IncHI2 | mcr-1 | 37 | ST4 | 18 |
| Tunisia | 2018 | Animal | Escherichia coli | 2 | Plasmid | IncI1 and incP | mcr-1 | 2 | ST2197 | 33 |
| Egypt | 2016 | Animal | Escherichia coli | 1 | Plasmid | ND | mcr-1 | 1 | ST10 | 34 |
| Algeria | 2019 | Environment | Escherichia coli | 103 | Plasmid | ND | mcr-1, mcr-3 | 6 | ST10, ST155, ST345 and ST405 | 35 |
| Tunisia | 2019 | Animal | Escherichia coli | 4 | Plasmid | ND | mcr-1 | 4 | ST1642 | 36 |
| Algeria | 2016 | Human | Escherichia coli | 1 | Plasmid | IncFIB | mcr-1 | 1 | ST405 | 37 |
| Algeria | 2016 | Human | Escherichia coli | 6 | plasmid | ND | mcr-1 | 1 | ST405 | 38 |
| Egypt | 2016 | Human | Escherichia coli | 1 | Plasmid | ND | mcr-1 | 1 | ST1011 | 39 |
| Algeria | 2019 | Human | Escherichia coli | 1 | Plasmid | ND | mcr-1 | 1 | ND | 40 |
| Egypt | 2019 | Human | Escherichia coli | 5 | Plasmid and chromosomal | ND | mcr-1, pmrA, pmrB | 1 | ND | 41 |
| Algeria | 2018 | Environment | Escherichia coli | 246 | Plasmid | ND | mcr-1 | 2 | ND | 42 |
| Egypt | 2019 | Human | Escherichia coli, Klebsiella pneumoniae | 40 | Plasmid and chromosomal | ND | mcr-1, mgrb | 2 | ST11 | 43 |
| Egypt | 2019 | Animal | Escherichia coli, Klebsiella pneumoniae | 34# | Plasmid | ND | mcr-1, mcr-2 | 34 | ND | 44 |
| Tunisia | 2017 | Human | Klebsiella pneumoniae | 7 | Chromosomal | NA | mgrb | NA | ND | 45 |
| Tunisia | 2018 | Human | Klebsiella pneumoniae | 13 | Chromosomal | NA | mgrb | NA | ND | 46 |
| Algeria | 2018 | Human | Klebsiella pneumoniae | 3 | Chromosomal | NA | mgrb, pmrA/B | NA | ST101 | 47 |
| Algeria | 2019 | Human | Klebsiella pneumoniae | 1 | Plasmid | ND | mcr-8 | 1 | ND | 15 |
| Algeria | 2018 | Human | Klebsiella pneumoniae | 2 | Chromosomal | NA | mgrb, pmrB | NA | ST2620 and ST3242 | 48 |
| Egypt | 2018 | Human | Escherichia coli, Klebsiella pneumoniae | 50 | Plasmid | ND | mcr-1 | 2 | ND | 49 |
| Egypt | 2019 | Human | Escherichia coli | 34 | Plasmid | ND | mcr-1 | 1 | ND | 50 |
| Libya | 2018 | Human | Klebsiella pneumoniae, Acinetobacter baumannii, Klebsiella oxytoca | 11 | Chromosomal | NA | mgrb | NA | ST101 | 51 |
NA - not applicable; ND - not determined; ST - sequence type.
#This figure corresponds to the number of resistance genes found because the study didn’t specify the exact number of isolates resistant to colistin.
The most commonly reported sequence type was ST10 reported in all three countries. Tunisia reported the most diverse range of STs (8 types), followed by Algeria (5 types). The presence of mgrB mutation was detected in Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Acinetobacter baumannii.
Southern Africa
In Southern African, six (16.6%) of the 36 studies were conducted in the region. The prevalence of mcr genes was 31.85% among colistin resistant isolates. Four studies reported mcr genes, exclusively mcr-1. They were mostly detected from human samples (n=2; 50.0%) followed by animal sample (n=1; 25.0%), environment sample (n=1; 50%) and food (n=1; 4.4%). Diverse range (8 types) of STs including ST1, ST10, ST14, ST57, ST101, ST226, ST624, ST1007 were reported. The presence of mcr-1 genes was reported in Escherichia coli and Klebsiella pneumoniae. Three plasmid types including IncHI2, IncI2, and IncX4 were reported. The three plasmids were contained in E. coli isolates and harbored mcr-1 (Table 2). Interestingly, one hitherto unknown mutation, the mutation in the I527N was detected in colistin resistant isolates in this region, which was absent in colistin susceptible isolates. Since then, this gene has not been detected in other regions.
Table 2. Distribution of the studies describing colistin resistance in Southern Africa.
| Country | Year | Source | Organism | Number of isolates exhibiting colistin resistance | Mechanism | Plasmid | Genes detected | Number of isolates positives for mcr genes | ST | References |
|---|---|---|---|---|---|---|---|---|---|---|
| South Africa | 2020 | Human | Acinetobacter baumannii | 26 | Chromosomal | NA | I527N | NA | ST1 | 52 |
| South Africa | 2016 | Animal | Escherichia coli | 108 | Plasmid | IncI2 | mcr-1 | 19 | ND | 53 |
| South Africa | 2016 | Human | Escherichia coli | 7 | Plasmid | IncHI2, incI2, IncX4 | mcr-1 | 7 | ST10, ST1007, ST624, ST57, ST101, ST226 | 54 |
| South Africa | 2017 | Human | Escherichia coli, Klebsiella pneumoniae | 19 | Plasmid | ND | mcr-1 | 15 | ND | 55 |
| South Africa | 2014 | Human | Klebsiella pneumoniae | 1 | Chromosomal | NA | PmrB | NA | ST14 | 56 |
| South Africa | 2018 | Environment | Escherichia coli | 65 | Plasmid | ND | mcr-1 | 31 | ND | 57 |
NA - not applicable; ND - not determined; ST - sequence type.
West Africa
In the West Africa region, only 2 (5.5%) of the 36 studies reported the mechanism of colistin resistance during the study period. The two studies were conducted in Nigeria and exclusively reported mcr-1 genes in Escherichia coli isolates (Table 3). The prevalence of mcr genes was 26.92% among colistin resistant isolates. The genes were detected from human sample (n= 1; 50.0%) and multiple sources sample (n=1; 50.0%). The plasmid and sequence type were not determined.
Table 3. Distribution of the studies describing colistin resistance in West Africa.
| Country | Year | Source | Organism | Number of isolates exhibiting colistin resistance | Mechanism | Plasmid | Genes detected | Number of isolates positives for mcr genes | ST | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Nigeria | 2019 | Human | Escherichia coli | 21 | Plasmid | ND | mcr-1 | 2 | ND | 58 |
| Nigeria | 2018 | Multiple sources | Escherichia coli | 5 | Plasmid | ND | mcr-1 | 5 | ND | 59 |
ND - not determined; ST - sequence type.
Central Africa
In this region, only a study conducted in São Tomé and Príncipe reported the mechanism of colistin resistance. This study reported the isolation of an Escherichia coli ST1408 isolate from a human sample which harbored mcr-1 on an IncX4 plasmid (Table 4).
Table 4. Distribution of the studies describing colistin resistance in Central Africa.
| Country | Year | Source | Organism | Number of isolates exhibiting colistin resistance | Mechanism | Plasmid | Genes detected | Number of isolates positives for mcr genes | ST | References |
|---|---|---|---|---|---|---|---|---|---|---|
| São Tomé and Príncipe | 2018 | Human | Escherichia coli | 36 | Plasmid | IncX4 | mcr-1 | 1 | ST1408 | 60 |
ST - sequence type.
Discussion
Until recently, acquired resistance to colistin has been mostly due to chromosomal mutation. The transferable plasmid encoded colistin resistance emerged in China in late 2015.11 Since then, eight other families of mcr genes have been detected from various hosts and pathogens range across the world.9,15 As illustrated in this study, the diversity of colistin resistance mechanisms in the various African countries is also changing.
Our finding showed that mcr genes have been reported in six African countries. The lack of data from several African countries may not be unconnected with lack of/inadequate laboratory capacity for their detection. The emergence of these transferable colistin resistance mechanisms may be attributed firstly to the high use of colistin as growth promoters, prophylactic and other agricultural use in many African countries.16 In a survey conducted by World Organization for Animal Health, 15% of African countries authorized the use of antibiotics including colistin in food animals.16 Secondly, the importation of food from countries including France and China where colistin resistance is endemic may have also contributed to the emergence of colistin resistance.13,17 Importation of antibiotic resistant bacteria across geographical regions has been well documented in the literature.18 China for example is Africa's leading commercial partner. Thus, there are large travel volumes through which the transferable plasmid encoded colistin resistance could reach the continent. Algeria, Egypt, Nigeria and South Africa were China’s most important trading partners in Africa and consequently the countries at highest importation risk.19
In this study, the mcr genes bearing bacteria were prevalently reported in Escherichia coli and Klebsiella pneumoniae. The predominance of Escherichia coli among the isolates bearing mcr genes is consistent with a result of a previously published systematic review of literature on the global burden of mcr genes.13 Similarly, another study systematic review in Latin America and Caribbean showed that mcr genes were more predominantly harbored by Escherichia coli. 20 The prevalence of Escherichia coli and Klebsiella pneumoniae among mcr bearing isolates in this study may be related to the source of exposure of the isolates to colistin. Colistin contained in food animal’s feeds is usually the source of exposure of gut bacteria particularly Escherichia coli and Klebsiella pneumoniae to colistin.17
Similar to reports of studies in Europe,21 IncHI2 is the most frequently reported mcr bearing plasmid type in African countries. Moreover, IncHI2 plasmids are especially known for co-localization of various antibiotic resistance determinants.22 This plasmid has been isolated from contaminated food, animals and water, thereby supporting the fact that the global trade of food and animals is a major vehicle for dissemination of mcr genes and other antibiotic resistance genes in Africa.23 Thirdly, since antibiotic use, no matter how appropriate, contributes to antibiotic resistance, over prescription of polymyxins by health workers as alternative to carbapenem as a last resort agent against MDR Gram-negative bacteria infections may have contributed also to the rapidly emerging transferable colistin resistance in Africa.13
Findings from this study show that most of the studies that reported mcr genes were performed in North Africa. This may be due beside the aforementioned factors, to the North African countries’ collaboration and proximity with Europe, particularly with France. As we all know, these countries are low- or middle-income countries with poor medical and healthcare infrastructure. The proximity to Europe may account for the high prevalence of mcr genes in North Africa as the mcr bearing bacterial species could be easily imported due to high human and materials traffic between the countries. Also, the cordial collaboration between North Africa and Europe ensures access to state-of-the-art healthcare facility and improved human capacity for detection of this emerging resistance mechanism.
Of the ten families of mcr genes detected from various hosts and pathogens range in the world, four including mcr-1, mcr-2, mcr-3 and mcr-8 were detected in Africa. Similar to a report from other regions around the world,24 ST10 is the dominant ST in both animals and environmental samples. The ST10 bearing bacteria are known to co-produce other antibiotic inactivating enzymes such as ESBLs and carbapenemase.25,26 This may have wide range implication on the management of infections caused by these bacteria.
This systematic review is limited by a number of factors. First, this review reports data from studies published in electronic databases and indexed in Google Scholar, PubMed and AJOL. Several unpublished theses and dissertations and articles published in traditional local print journals could not be assessed and so they were not included. Secondly, studies reporting the burden of colistin resistance without the corresponding report of the molecular basis of the resistance were excluded. As such, the burden of mcr genes in Africa may have been underestimated. Nevertheless, this systematic review is the first to determine the Africa wide burden and distribution of mcr genes. It thus provides a baseline data on this rapidly emerging resistance mechanism.
Conclusions
This study shows that the transferable plasmid mediated colistin resistance is rapidly emerging in Africa with mcr-1 as the predominant genetic variant in human, animals as well as in the environmental samples. This is worrisome in a continent where alternative antibiotics are rarely available. Therefore, there is urgent need to establish African-wide antibiotic stewardship and intensify efforts to preserve the efficacy of colistin as a last resort antibiotic.
Footnotes
Authors’ contributions statement: AY and AO conceived and designed the study. AO collected the data. AY analyzed the data; AY and AO drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Conflicts of interest: All authors - none to declare.
Funding: None to declare.
References
- 1.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40:277–83. [PMC free article] [PubMed] [Google Scholar]
- 2.Roca I, Akova M, Baquero F, et al. Corrigendum to "The global threat of antimicrobial resistance: science for intervention" [New Microbes New Infect 6 (2015): 22-29] New Microbes New Infect. 2015;8:175. doi: 10.1016/j.nmni.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO) Antimicrobial resistance: global report on surveillance 2014. [Accessed on: 05 May 2020]. Available at: http://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/
- 4.Matsunaga N, Hayakawa K. Estimating the impact of antimicrobial resistance. Lancet Glob Health. 2018;6:e934–5. doi: 10.1016/S2214-109X(18)30325-5. [DOI] [PubMed] [Google Scholar]
- 5.Högberg LD, Heddini A, Cars O. The global need for effective antibiotics: challenges and recent advances. Trends Pharmacol Sci. 2010;31:509–15. doi: 10.1016/j.tips.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Karaiskos I, Lagou S, Pontikis K, Rapti V, Poulakou G. The "old" and the "new" antibiotics for MDR Gram-negative pathogens: for whom, when, and how. Front Public Health. 2019;7:151. doi: 10.3389/fpubh.2019.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–96. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aghapour Z, Gholizadeh P, Ganbarov K, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist. 2019;12:965–75. doi: 10.2147/IDR.S199844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olaitan AO, Diene SM, Kempf M, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44:500–7. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect. 2020;9:508–16. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbediwi M, Li Y, Paudyal N, et al. Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018) Microorganisms. 2019;7:461. doi: 10.3390/microorganisms7100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nabti LZ, Sahli F, Ngaiganam EP, et al. Development of real-time PCR assay allowed describing the first clinical Klebsiella pneumoniae isolate harboring plasmid-mediated colistin resistance mcr-8 gene in Algeria. J Glob Antimicrob Resist. 2020;20:266–71. doi: 10.1016/j.jgar.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Van TTH, Yidana Z, Smooker PM, Coloe PJ. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J Glob Antimicrob Resist. 2020;20:170–7. doi: 10.1016/j.jgar.2019.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Kempf I, Jouy E, Chauvin C. Colistin use and colistin resistance in bacteria from animals. Int J Antimicrob Agents. 2016;48:598–606. doi: 10.1016/j.ijantimicag.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Grami R, Mansour W, Mehri W, et al. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Euro Surveill. 2016;21:30144. doi: 10.2807/1560-7917.ES.2016.21.8.30144. [DOI] [PubMed] [Google Scholar]
- 19.Nowak W. China-Africa and India-Africa trade in the years 2000-2014. Procedia Econ Finance. 2016;39:140–6. doi: 10.1016/S2212-5671(16)30261-1. [DOI] [Google Scholar]
- 20.Mendes Oliveira VR, Paiva MC, Lima WG. Plasmid-mediated colistin resistance in Latin America and Caribbean: A systematic review. Travel Med Infect Dis. 2019;31:101459. doi: 10.1016/j.tmaid.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Hartl R, Kerschner H, Lepuschitz S, Ruppitsch W, Allerberger F, Apfalter P. Detection of the mcr-1 gene in a multidrug-resistant Escherichia coli isolate from an Austrian patient. Antimicrob Agents Chemother. 2017;61:e02623–16. doi: 10.1128/AAC.02623-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Xie M, Lv J, Wai-Chi Chan E, Chen S. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother. 2017;72:696–9. doi: 10.1093/jac/dkw509. [DOI] [PubMed] [Google Scholar]
- 23.Arcilla MS, van Hattem JM, Matamoros S, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:147–9. doi: 10.1016/S1473-3099(15)00541-1. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Liu Y, Qi X, et al. Molecular epidemiology of colistin-resistant Enterobacteriaceae in inpatient and avian isolates from China: high prevalence of mcr-negative Klebsiella pneumoniae. Int J Antimicrob Agents. 2017;50:536–41. doi: 10.1016/j.ijantimicag.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Aibinu I, Odugbemi T, Koenig W, Ghebremedhin B. Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect. 2012;18:E49–51. doi: 10.1111/j.1469-0691.2011.03730.x. [DOI] [PubMed] [Google Scholar]
- 26.Falgenhauer L, Imirzalioglu C, Oppong K, et al. Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front Microbiol. 2019;9:3358. doi: 10.3389/fmicb.2018.03358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakour S, Olaitan AO, Ammari H, et al. Emergence of colistin- and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in Algeria: first case report. Microb Drug Resist. 2015;21:279–85. doi: 10.1089/mdr.2014.0214. [DOI] [PubMed] [Google Scholar]
- 28.Hammad AM, Hoffmann M, Gonzalez-Escalona N, et al. Genomic features of colistin resistant Escherichia coli ST69 strain harboring mcr-1 on IncHI2 plasmid from raw milk cheese in Egypt. Infect Genet Evol. 2019;73:126–31. doi: 10.1016/j.meegid.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Bachiri T, Lalaoui R, Bakour S, et al. First report of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli ST405 isolated from wildlife in Bejaia, Algeria. Microb Drug Resist. 2018;24:890–5. doi: 10.1089/mdr.2017.0026. [DOI] [PubMed] [Google Scholar]
- 30.Saidani M, Messadi L, Sahmin E, et al. ESBL- and mcr-1-producing Escherichia coli in veal calves in Tunisia. J Glob Antimicrob Resist. 2019;19:104–5. doi: 10.1016/j.jgar.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Saidani M, Messadi L, Chaouechi A, et al. High genetic diversity of Enterobacteriaceae clones and plasmids disseminating resistance to extended-spectrum cephalosporins and colistin in healthy chicken in Tunisia. Microb Drug Resist. 2019;25:1507–13. doi: 10.1089/mdr.2019.0138. [DOI] [PubMed] [Google Scholar]
- 32.Chabou S, Leulmi H, Rolain JM. Emergence of mcr-1-mediated colistin resistance in Escherichia coli isolates from poultry in Algeria. J Glob Antimicrob Resist. 2019;16:115–6. doi: 10.1016/j.jgar.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Maamar E, Alonso CA, Hamzaoui Z, et al. Emergence of plasmid-mediated colistin-resistance in CMY-2-producing Escherichia coli of lineage ST2197 in a Tunisian poultry farm. Int J Food Microbiol. 2018;269:60–3. doi: 10.1016/j.ijfoodmicro.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 34.Khalifa HO, Ahmed AM, Oreiby AF, Eid AM, Shimamoto T, Shimamoto T. Characterisation of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli isolated from animals in Egypt. Int J Antimicrob Agents. 2016;47:413–4. doi: 10.1016/j.ijantimicag.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Touati M, Hadjadj L, Berrazeg M, Baron SA, Rolain JM. Emergence of Escherichia coli harbouring mcr-1 and mcr-3 genes in North West Algerian farmlands. J Glob Antimicrob Resist. 2020;21:132–7. doi: 10.1016/j.jgar.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Hassen B, Saloua B, Abbassi MS, et al. mcr-1 encoding colistin resistance in CTX-M-1/CTX-M-15- producing Escherichia coli isolates of bovine and caprine origins in Tunisia. First report of CTX-M-15-ST394/D E. coli from goats. Comp Immunol Microbiol Infect Dis. 2019;67:101366. doi: 10.1016/j.cimid.2019.101366. [DOI] [PubMed] [Google Scholar]
- 37.Yanat B, Machuca J, Yahia RD, Touati A, Pascual Á, Rodríguez-Martínez J-M. First report of the plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate in Algeria. Int J Antimicrob Agents. 2016;48:760–1. doi: 10.1016/j.ijantimicag.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Berrazeg M, Hadjadj L, Ayad A, Drissi M, Rolain JM. First detected human case in Algeria of mcr-1 plasmid-mediated colistin resistance in a 2011 Escherichia coli isolate. Antimicrob Agents Chemother. 2016;60:6996–7. doi: 10.1128/AAC.01117-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elnahriry SS, Khalifa HO, Soliman AM, et al. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother. 2016;60:3249–50. doi: 10.1128/AAC.00269-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nabti LZ, Sahli F, Hadjadj L, et al. Autochthonous case of mobile colistin resistance gene mcr-1 from a uropathogenic Escherichia coli isolate in Sétif Hospital, Algeria. J Glob Antimicrob Resist. 2019;19:356–7. doi: 10.1016/j.jgar.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Osama R, Bakeer W, Fadel S, Amin M. Association of carbapenem and colistin resistance in pathogenic Gram negative bacteria. J Pure Appl Microbiol. 2019;13:733–9. doi: 10.22207/JPAM.13.2.09. [DOI] [Google Scholar]
- 42.Drali R, Berrazeg M, Zidouni LL, et al. Emergence of mcr-1 plasmid-mediated colistin-resistant Escherichia coli isolates from seawater. Sci Total Environ. 2018;642:90–4. doi: 10.1016/j.scitotenv.2018.05.387. [DOI] [PubMed] [Google Scholar]
- 43.Zafer MM, El-Mahallawy HA, Abdulhak A, Amin MA, Al-Agamy MH, Radwan HH. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann Clin Microbiol Antimicrob. 2019;18:40. doi: 10.1186/s12941-019-0339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed ZS, Elshafiee EA, Khalefa HS, Kadry M, Hamza DA. Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob Resist Infect Control. 2019;8:197. doi: 10.1186/s13756-019-0657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansour W, Haenni M, Saras E, et al. Outbreak of colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J Glob Antimicrob Resist. 2017;10:88–94. doi: 10.1016/j.jgar.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 46.Jaidane N, Bonnin RA, Mansour W, et al. Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob Agents Chemother. 2018;62:e01601–17. doi: 10.1128/AAC.01601-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yousfi H, Hadjadj L, Dandachi I, et al. Colistin- and carbapenem-resistant Klebsiella pneumoniae clinical isolates: Algeria. Microb Drug Resist. 2018;25:258–63. doi: 10.1089/mdr.2018.0147. [DOI] [PubMed] [Google Scholar]
- 48.Belbel Z, Lalaoui R, Bakour S, Nedjai S, Djahmi N, Rolain JM. First report of colistin resistance in an OXA-48- and a CTX-M-15 producing Klebsiella pneumoniae clinical isolate in Algeria due to PmrB protein modification and mgrB inactivation. J Glob Antimicrob Resist. 2018;14:158–60. doi: 10.1016/j.jgar.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 49.El Sayed Zaki M, Abou ElKheir N, Mofreh M. Molecular study of colistin resistant clinical isolates of Enterobacteriaceae species. J Clin Mol Med. 2018;1:1–4. doi: 10.15761/JCMM.1000103. [DOI] [Google Scholar]
- 50.El-Sokkary RH, Gebriel MG. Colistin susceptibility and the effect of colistin-sulfadiazine combination among multidrug resistant E. coli and K. pneumoniae at Egyptian intensive care units. Egypt J Med Microbiol. 2019;28:87–93. [Google Scholar]
- 51.Kieffer N, Ahmed MO, Elramalli AK, et al. Colistin-resistant carbapenemase-producing isolates among Klebsiella spp. and Acinetobacter baumannii in Tripoli, Libya. J Glob Antimicrob Resist. 2018;13:37–9. doi: 10.1016/j.jgar.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 52.Snyman Y, Whitelaw AC, Reuter S, Dramowski A, Maloba MRB, Newton-Foot M. Clonal expansion of colistin-resistant Acinetobacter baumannii isolates in Cape Town, South Africa. Int J Infect Dis. 2020;91:94–100. doi: 10.1016/j.ijid.2019.11.021. [DOI] [PubMed] [Google Scholar]
- 53.Perreten V, Strauss C, Collaud A, Gerber D. Colistin resistance gene mcr-1 in avian pathogenic Escherichia coli in South Africa. Antimicrob Agents Chemother. 2016;60:4414–5. doi: 10.1128/AAC.00548-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 2016;60:4394–7. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newton-Foot M, Snyman Y, Maloba MRB, Whitelaw AC. Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. clinical isolates from the Western Cape region of South Africa. Antimicrob Resist Infect Control. 2017;6:78. doi: 10.1186/s13756-017-0234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother. 2014;58:4762–6. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Igwaran A, Iweriebor BC, Okoh AI. Molecular characterization and antimicrobial resistance pattern of Escherichia coli recovered from wastewater treatment plants in Eastern Cape South Africa. Int J Environ Res Public Health. 2018;15:1237. doi: 10.3390/ijerph15061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otokunefor K, Tamunokuro E, Amadi A. Molecular detection of mobilized colistin resistance (mcr-1) gene in Escherichia coli isolates from Port Harcourt, Nigeria. J Appl Sci Environ Manage. 2019;23:401–405. doi: 10.4314/jasem.v23i3.5. [DOI] [Google Scholar]
- 59.Olowe OA, Olowe RA, Oluremi AS, Adefioye OJ. A novel report of colistin-resistant Escherichia coli carrying mcr-1 gene from animal and human feacal samples in Nigeria. Pan Afr J Life Sci. 2018;1:7–10. doi: 10.36108/pajols/8102/10(0120). [DOI] [Google Scholar]
- 60.Poirel L, Aires-de-Sousa M, Kudyba P, Kieffer N, Nordmann P. Screening and characterization of multidrug-resistant Gram-negative bacteria from a remote African area, São Tomé and Príncipe. Antimicrob Agents Chemother. 2018;62:e01021–18. doi: 10.1128/AAC.01021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]