Significance
Electron bifurcation is a recently recognized mechanism of biological energy conservation that is widespread in anaerobic microorganisms and provides low-potential, high-energy electrons to drive otherwise nonspontaneous metabolic reactions. Here we describe the structure of a membrane-associated bifurcating enzyme from an aerobic microorganism. Termed Fix/EtfABCX, this enzyme is used by some nitrogen-fixing microbes to drive the low-potential reduction of nitrogen gas to ammonia. Fix/EtfABCX is a superdimer and a catalytic mechanism is proposed involving six flavins, two of which bifurcate, four iron-sulfur clusters, and two menaquinones. Fix/EtfABCX shares structural similarity with mammalian quinone oxidoreductase involved in mitochondrial fatty acid oxidation, suggesting an unexpected evolutionary link between bifurcating and nonbifurcating systems.
Keywords: electron bifurcation, flavin, iron-sulfur cluster, menaquinone, Thermotoga
Abstract
The electron-transferring flavoprotein-menaquinone oxidoreductase ABCX (EtfABCX), also known as FixABCX for its role in nitrogen-fixing organisms, is a member of a family of electron-transferring flavoproteins that catalyze electron bifurcation. EtfABCX enables endergonic reduction of ferredoxin (E°′ ∼−450 mV) using NADH (E°′ −320 mV) as the electron donor by coupling this reaction to the exergonic reduction of menaquinone (E°′ −80 mV). Here we report the 2.9 Å structure of EtfABCX, a membrane-associated flavin-based electron bifurcation (FBEB) complex, from a thermophilic bacterium. EtfABCX forms a superdimer with two membrane-associated EtfCs at the dimer interface that contain two bound menaquinones. The structure reveals that, in contrast to previous predictions, the low-potential electrons bifurcated from EtfAB are most likely directly transferred to ferredoxin, while high-potential electrons reduce the quinone via two [4Fe-4S] clusters in EtfX. Surprisingly, EtfX shares remarkable structural similarity with mammalian [4Fe-4S] cluster-containing ETF ubiquinone oxidoreductase (ETF-QO), suggesting an unexpected evolutionary link between bifurcating and nonbifurcating systems. Based on this structure and spectroscopic studies of a closely related EtfABCX, we propose a detailed mechanism of the catalytic cycle and the accompanying structural changes in this membrane-associated FBEB system.
Electron bifurcation is a novel mechanism of biological energy coupling that leads to energy conservation (1–3). The concept of bifurcating electrons was originally proposed by Peter Mitchell in the 1970s to explain in respiratory complex III how the free-energy change of exergonic quinol (QH2) oxidation in the mitochondrial inner membrane is captured to drive endergonic quinone (Q) reduction in the lumen, thereby resulting in an overall thermodynamically favorable sequence of multielectron transfer reactions (4). It has recently become clear, however, that electron bifurcation is not limited to the quinone-containing complex III. It is found widely in anaerobic and also in some aerobic microorganisms and is mediated by flavin-containing protein complexes that catalyze more than a dozen different reactions (1). All use a central flavin (FAD or FMN) to bifurcate electrons to two different energy levels. For example, this enables bifurcating enzymes to use NADH (E°′ −320 mV) to couple the reduction of high-potential substrates (E > ∼−200 mV) in an exergonic reaction to the simultaneous reduction of the low-potential redox protein ferredoxin (E ∼−450 mV) in an endergonic reaction. Ferredoxin oxidation drives many global microbial processes, including the production of methane and hydrogen gas, acetogenesis, and nitrogen fixation. Flavin-based electron bifurcation (FBEB) therefore plays critical roles in many fundamental metabolic processes and is a general mechanism to provide low-potential, high-energy electrons to drive otherwise nonspontaneous reactions in microbial physiology.
FBEB enzymes can be divided into four independently evolved groups that are unrelated phylogenetically: 1) electron transfer flavoproteins containing EtfAB; 2) [FeFe]-hydrogenases and formate dehydrogenases containing HydABC; 3) heterodisulfide reductases containing HdrA; and 4) transhydrogenases containing NfnAB. X-ray–based structures have been reported for three of the four classes: for butyryl-CoA dehydrogenase (EtfAB-Bcd) (5) and caffeyl-CoA reductase [CarCDE; where CarDE corresponds to EtfAB (6)], for NfnAB (7), and for the MvhADG–HdrABC complex (8). No structural information is available for a bifurcating HydABC-type enzyme. Biochemical and spectroscopic studies have provided the basic mechanistic insights of this new mode of energy coupling. The FBEB enzyme Nfn is the best understood in terms of the overall bifurcation mechanism at the flavin site (7).
Herein we focus on the EtfAB class of bifurcating enzyme. These are part of a large family of electron-transferring flavoproteins, only a small subset of which carry out FBEB. The nonbifurcating EtfAB-type complexes contain one FAD and one adenosine monophosphate (AMP) and the crystal structures of several of them have been determined (5, 6, 9–15). In the bifurcating EtfAB enzymes, AMP is replaced by a second FAD and this is the site of electron bifurcation. The bifurcating EtfAB-type complexes EtfAB-Bcd (5) and CarCDE (6) oxidize NADH and simultaneously reduce ferredoxin and a high-potential CoA derivative (E°′ ∼0 mV). NADH is oxidized by the EtfB subunit, which contains the bifurcating flavin (BF-FAD), while the EtfA subunit contains a second flavin that serves an electron transfer role (ET-FAD). The structures of EtfAB-Bcd and CarCDE revealed that BF-FAD and ET-FAD are too far apart (18 to 38 Å) for efficient electron transfer and so the catalytic cycle must involve a significant conformational change within EtfAB. After bifurcation by BF-FAD, the ET-FAD transfers the high-potential electron to a third FAD in the subunit (Bcd or CarC) that reduces the CoA derivative (5, 16). It is not clear how the low-potential electron generated by BF-FAD reduces ferredoxin. In CarBCE, it was initially assumed that electrons are transferred from BF-FAD to ferredoxin via two [4Fe-4S] clusters in CarE (EtfB) but this was shown by a mutagenesis study not to be the case (6).
Another type of EtfAB-based bifurcating enzyme reduces quinones rather than acyl-CoA derivatives and, in further contrast, it is membrane-associated. In some nitrogen-fixing microbes, this enzyme is referred to as FixABCX. It enables NADH to provide electrons for the endergonic reduction of N2 via ferredoxin driven by exergonic quinone reduction (17, 18). Note that FixAB corresponds to EtfBA, where FixA is equivalent to EtfB and binds NADH (Fig. 1A) and FixCX is the quinone oxidoreductase. From sequence analysis, FixX is proposed to contain two [4Fe-4S] clusters. The “Fix” designation refers to the role of this enzyme in nitrogen fixation, but many homologs of FixABCX have been found in microorganisms that do not fix nitrogen. These include Pyrobaculum aerophilum (Pae), a microaerophilic crenarchaeote that grows at 100 °C, and Thermotoga maritima (Tma), a fermentative bacterium growing optimally at 80 °C (both reviewed in ref. 19). To better describe their general role in energy metabolism, all of these EtfAB-type complexes, including that of the Fix system, are now referred to as EtfABCX (20). EtfABCX is the only membrane-associated bifurcating system among the FBEB family (17). The reaction catalyzed by EtfABCX is shown in Eq. 1, where Fd is ferredoxin and MQ is menaquinone:
| [1] |
While it assumed that the EtfA subunit of EtfABCX complexes contains the BF-FAD and that its bifurcation mechanism is essentially the same as that of the BF-FAD in Nfn, the best studied FBEB system (7), it is now clear that the catalytic cycle of EtfABCX is distinctly different from that of Nfn (20). For example, NAD forms a stable charge transfer (CT) complex with the BF-FAD in Pae EtfABCX but not in Nfn, the resting and bifurcation-ready redox states of the two enzymes are not the same, and how electron flow is gated and the nature of the two two-electron cycles for the overall four-electron catalytic reaction must also be different in EtfABCX and Nfn, although how is not exactly clear. In addition, in EtfABCX, how EtfCX is orientated relative to EtfAB and the exact pathway of electron transport from BF-FAD to ferredoxin are not known. In a previous biochemical and biophysical study of the Pae EtfABCX (20), we suggested based on redox considerations that the two [4Fe-4S] clusters in EtfX, which were assumed to be of low potential, form part of the bifurcating low-potential branch to oxidized Fd, although there was no experimental evidence for this. To better understand the properties and mechanism of the unique membrane-associated and quinone-reducing bifurcating complex EtfABCX, we carried out a cryoelectron microscopy (cryo-EM) analysis of the Tma enzyme. The 2.9-Å structure of the holoenzyme reveals how a bifurcating EtfAB subcomplex is coupled to the quinone oxidoreductase (EtfCX) subcomplex.
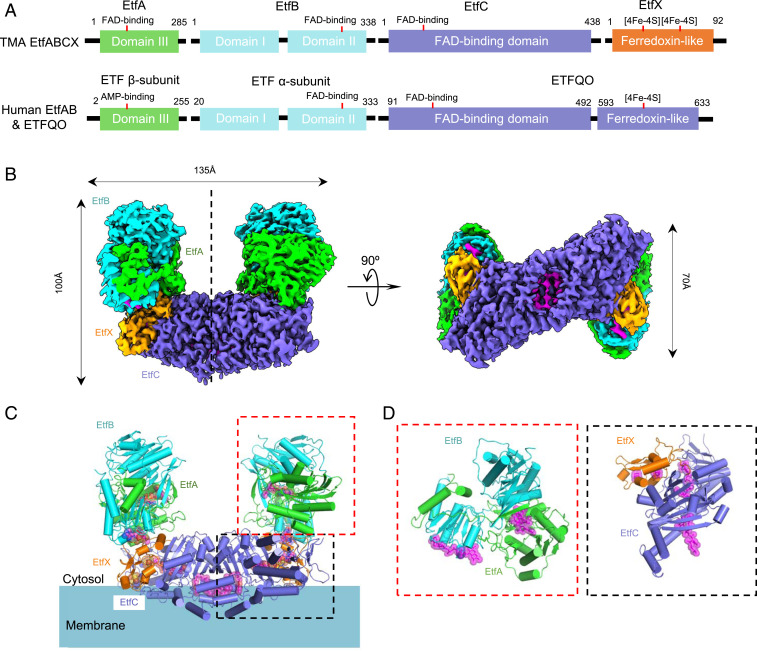
Fig. 1.
Superdimeric architecture of the Tma EtfABCX. (A) The domain architecture of EftA, B, C, and X in comparison with that of human EtfAB-QO. (B) Surface-rendered side and bottom views of the cryo-EM 3D map of the EtfABCX complex reconstituted by imposing a twofold symmetry. Subunits are individually colored with EtfA in green, EtfB in cyan, EtfC in purple, and EtfX in orange. (C) Cartoon views of the atomic model of EftABCX. The cofactors (three FAD molecules and two [4Fe-4S] clusters) are shown as magenta sticks superimposed with transparent spheres. Membrane is depicted as a cyan shade with the lipid-exposed and membrane-interacting hydrophobic side chains shown as sticks. (D) Zoomed views of EtfAB (Left) and EtfCX (Right); both are 90° rotated to better display the bound cofactors.
Results and Discussion
T. maritima EtfABCX Forms a Superdimer Complex.
An affinity-tagged form of Tma EtfABCX was recombinantly produced in the high-temperature host Pyrococcus furiosus using the same approach previously described for Pae EtfABCX (20). Interestingly, for Tma, we only purified the ABCX holoenzyme and there was no evidence for the EtfAB subcomplex (the affinity tag is on the EtfA subunit). This is in contrast to the situation with Pae ABCX, where both forms were purified separately with an AB:ABCX ratio of ∼10:1. It seems the affinity between the EtfAB and EtfCX subcomplexes is much higher in the Tma complex than it is in the Pae versions. For this reason, we carried out cryo-EM on the more stable Tma four-protein complex. In the dye-linked nonbifurcation assay in which NADH oxidation is coupled to iodonitrotetrazolium chloride reduction, the activity of the purified Tma holoenzyme complex was ∼80 U/mg at 80 °C, which is comparable to that reported for the purified Pae ABCX complex [∼15 U/mg (20)]. The sequences of the A, B, C, and X subunits of the Tma and Pae enzymes show 35, 47, 32, and 42% identities, respectively, indicating that the two complexes are likely highly similar in both structure and mechanism.
The purified Tma complex was a mixture of dimers (EtfABCX)2 (254 kDa) and tetramers (EtfABCX)2–(EtfABCX)2 (507 kDa), based on the gel-filtration profile (SI Appendix, Fig. S1). Indeed, the U-shaped dimer (EtfABCX)2 with C2 symmetry and the X-shaped tetramer (EtfABCX)2–(EtfABCX)2 with D2 symmetry accounted for the majority of raw particles in cryo-EM images (SI Appendix, Fig. S2 A and B). Additional minor species were also observed; they likely represented broken particles. Two rounds of three-dimensional (3D) classification and 3D refinement on the dimer particles resulted in a 2.9-Å average resolution 3D map of the (EtfABCX)2 complex (SI Appendix, Figs. S2C and S3 and Table S1).
The X-shaped tetramer complex (EtfABCX)2–(EtfABCX)2 is assembled end-to-end via the membrane-binding hydrophobic patch on the bottom of each of the two dimers, and therefore is most likely an in vitro artifact. Indeed, incubation of the sample in detergent (0.01% lauryldimethylamine N-oxide or 0.02% n-dodecyl-β-d-maltoside) greatly reduced or entirely eliminated the tetramer population (SI Appendix, Fig. S4). However, the dimer was stable in both detergents and, in fact, the dimer population increased due to the breakdown of the tetramers into dimers in detergents. We suggest that the dimer complex (EtfABCX)2 is the physiological unit of the enzyme. The high resolution of the 3D map, together with the availability of the structures of the homologous Acidaminococcus fermentans EtfAB and the porcine ubiquinone oxidoreductase ETF-QO, enabled building and subsequent refinement of the atomic model of the (EtfABCX)2 complex (SI Appendix, Fig. S1 A–C and Table S1). The structure reveals that two EtfC subunits mediate the dimerization of two EtfABCX complexes by forming an intermolecular β-sheet (SI Appendix, Fig. S5B). This structural feature explains the detergent stability of the dimer but not of the tetramer and lends further support to our suggestion that the dimer is the functional form of the enzyme complex while the tetramer is a preparation artifact.
Structure of Tma EtfABCX.
In the EtfABCX dimer complex, one set of EtfA, EtfB, and EtfX subunits attaches to each of the two EtfCs in the dimer, forming a characteristic U-shaped structure (Fig. 1 B and C). In each of the two EtfABCX complexes, there are three FADs, with two in EftAB and one in EtfC, two [4Fe-4S] clusters in EtfX, and one menaquinone in EtfC (Fig. 1 C and D), suggesting that each EtfABCX complex has its own independent electron transfer system. This resembles another EtfAB-containing FBEB complex, the cytoplasmic soluble CarCDE, which forms a supertetramer where each CarCDE monomer contains a complete electron transfer pathway (6). The Tma EtfAB subcomplex belongs to the electron-bifurcating subgroup of the large ETF family that is widely distributed in Archaea, Bacteria, and mitochondria (21, 22) and, as noted above, contains both bifurcating and nonbifurcating members (5, 6, 9–15). It should be noted that the Tma EtfA and EtfB subunits are named according to the gene order in the operon (Fig. 1A) but EtfA corresponds to the β-subunit of the other ETF family members and EtfB corresponds to their α-subunits (23).
Tma EtfAB contains three domains, of which domains I (1 to 199) and II (200 to 331) belong to EtfB and domain III (5 to 265) belongs to EtfA (Fig. 2A). Domains I and III are tightly associated via an extensive interface (∼7,900 Å2) and form the stable core of EtfAB. Domain II is flexibly linked to the core by two loops, one being the linker between domains I and II of EtfB, the other the C-terminal peptide of EtfA, which is also referred to as the EtfA arm, that partially wraps domain II. However, there is little interaction between domain II and the stable core of domains I and III; in fact, domain II is largely stabilized by the underlying EtfCX in the Tma complex structure.
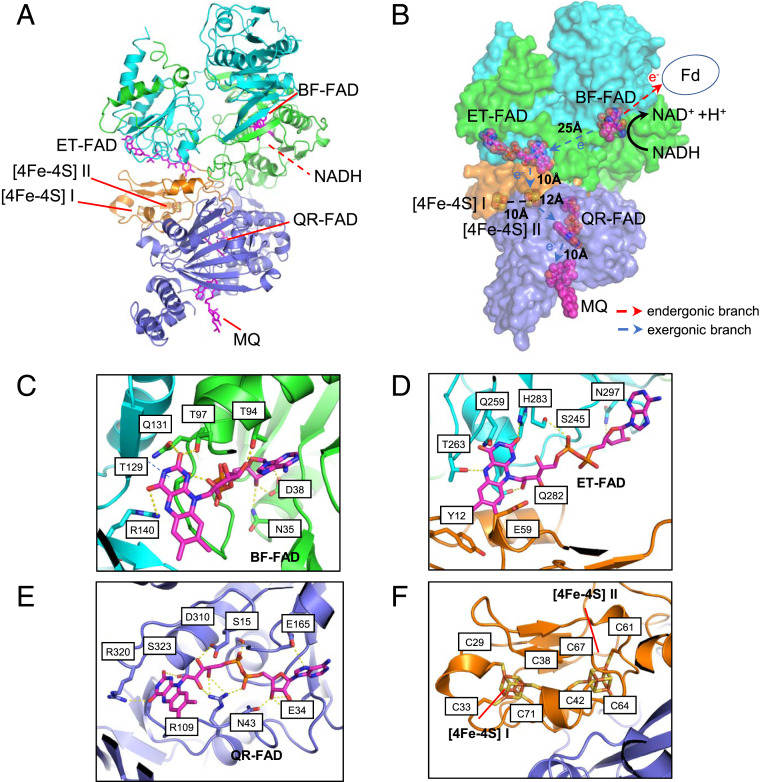
Fig. 2.
Electron transfer pathway and the cofactor coordination in the Tma EtfABCX complex. (A) Cartoon representation of one Tma EtfABCX tetramer structure, with each subunit colored differently as labeled. C-FAD, flavin of the quinone reducing subunit C. (B) Orientation and distance of the six bound cofactors. (C–F) Enlarged views of the binding pocket and coordinating residues of BF-FAD (C), ET-FAD (D), QR-FAD (E), and the [4Fe-4S] clusters (F).
EtfC shares a similar fold with the nonbifurcating porcine enzyme ETF-QO (24), but EtfC dimerizes while ETF-QO does not (Fig. 1 B and C and SI Appendix, Fig. S5). The EtfC–EtfC dimer has an extensive interface of ∼6,000 Å2, composed of three separate patches: an interprotomer β-strand interaction; two salt bridges (E97′ to K396 and E321′ to K406) and five hydrogen bonds (W98′ to Q400, N103′ to Q400, backbone of L399 and backbone of V401, and V437′ to Q410) of the C-terminal helix-turn-helix motif (residues 386 to 418); and two salt bridges (K99′ to D272 and E209′ to R204) in a short α-helix (residues 200 to 207). EtfX has a ferredoxin fold and is packed tightly with EtfC occupying ∼60% of its total surface area (Fig. 1D).
In the Tma enzyme, the EtfAB subcomplex interacts with the EtfCX subcomplex via three contacts totaling 3,200 Å2 (SI Appendix, Fig. S6). The first contact is between the EtfAB core and the N terminus of EtfC. An extended interface is formed by three loops (residues 1 to 4, 126 to 132, and 148 to 155) in the EtfC N-terminal region interacting with the EtfA helix formed by residues 195 to 204. The second contact lies between the EtfA hairpin extension and the EtfCX complex. The third contact is between domain II and EtfX and is mediated by ET-FAD. This interface is mediated by EtfB Ile-281 and Val-285 packing against EtfX Tyr-8 and several other polar/charged interactions: EtfB Arg-246 and the main chain of EtfX E16, EtfB Lys-250 and EtfX E59, and EtfB Lys-220 and EtfX E55.
Cofactor Coordination and the Electron Transfer Pathways.
The three FAD molecules found in each EtfABCX complex had well-resolved density in the 3D map and all were in extended conformations (SI Appendix, Fig. S7 A–C). Domains II and III of the EtfAB complex each contains one FAD molecule (Fig. 2 A and B). Each of these FADs is coordinated by a network of hydrogen bonds (Fig. 2 C and D) and the coordinating residues are conserved in the bifurcating EtfAB family (1, 22). Based on the published homologous EtfAB protein structures and previous biochemical and biophysical studies (16), FAD in domain III is responsible for bifurcating electrons and is called BF-FAD. FAD bound to domain II is the ET-FAD because this accepts electrons from the BF-FAD and is the first step in the transfer pathway to the final electron acceptor, MQ.
Our structure reveals how a bifurcating EtfAB family protein complex (EtfAB) is coupled to the quinone oxidoreductase (EtfCX). The EtfX subunit, which contains two [4Fe-4S] clusters, directly interacts with around half of the ET-FAD molecule as its Tyr-12 and Glu-59 residues pack against the isoalloxazine ring. This is different from the equivalent interaction in the CoA-hydrogenase subunits of the EtfAB-Bcd complex where CarC has an extended structure and covers the entire ET-FAD (SI Appendix, Fig. S8). Nevertheless, the ET-FADs in both enzyme complexes are in a similar orientation and distance (∼10 Å) from the nearest electron acceptor, which is the third or γ-FAD in CarC but is a [4Fe-4S] cluster in EtfX. The two [4Fe-4S] clusters in EtfX are 10 Å apart and will be referred to as [4Fe-4S] clusters I and II, respectively. Cluster I is covalently coordinated by Cys-29, Cys-33, Cys-38, and Cys-71, and cluster II by Cys-42, Cys-61, Cys-64, and Cys-67 (Fig. 2E). EtfC coordinates the third or quinone reductase FAD (QR-FAD) in ABCX via 12 hydrogen bonds and this is situated 10 Å from the bound menaquinone (Fig. 2 B and F). Among the two clusters in EtfX, [4Fe-4S] cluster I is located outside the electron transfer range from either ET-FAD or QR-FAD, but [4Fe-4S] cluster II is within 10 Å of ET-FAD and 13 Å of QR-FAD, allowing efficient electron coupling. For this reason, we suggest that it is [4Fe-4S] cluster II but not [4Fe-4S] cluster I that relays electrons from ET-FAD to QR-FAD (Fig. 2 B and G). Hence, based on their spatial relationship, it is clear that the two electrons from NADH are bifurcated at BF-FAD and the high-potential electron is sequentially relayed to MQ via ET-FAD, [4Fe-4S] cluster II, and QR-FAD (Fig. 2 B and G).
The structure of Tma ABCX shows that in the high-potential pathway, [4Fe-4S] cluster I is also close enough to ET-FAD to directly accept electrons although the shortest distance to QR-FAD is via cluster II. The question is, how do electrons from NADH oxidation by BF-FAD reach Fd? Clearly, the structure shows that the two [4Fe-4S] clusters of EtfX cannot be part of the low-potential pathway (Fig. 2B), as we previously proposed (20). If the first step in the high-potential pathway involves electron transfer from BF-FAD to ET-FAD, which is the closest electron-accepting cofactor, then there is no alternative but for the low-potential pathway to involve direct electron transfer from BF-FAD to Fd without any intermediate cofactor (Fig. 2B). The same situation must also exist in the EtfAB-Bcd complex (5) as it lacks an iron-sulfur cluster. Note, however, that this can only be accomplished by the BF-FAD as the reduction potentials of the standard nonbifurcating flavin system would not generate a state that was sufficiently reducing to enable reduction of a low-potential Fd. In any event, it is remarkable that an enzyme that contains [4Fe-4S] clusters does not utilize them to transfer electrons to Fd; instead, they mediate electron transfer between two flavins (ET-FAD and QR-FAD).
Domain II Moves to Form the Catalytically Competent State.
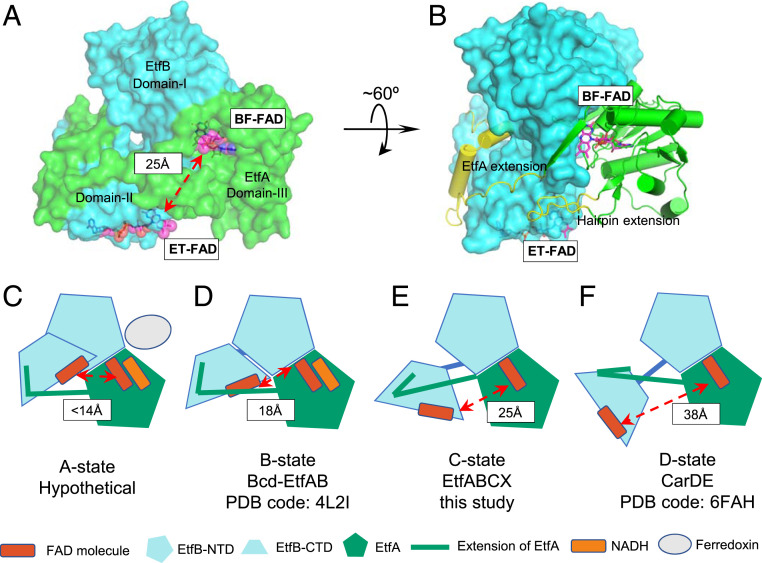
While the BF-FAD oxidizes NADH and uses one electron to directly reduce Fd, the structure also reveals that there is a major hurdle to be overcome if the other electron is to be transferred to ET-FAD and thus to MQ. The BF-FAD and ET-FAD are too far apart (25 Å) to allow direct electron transfer (Fig. 2B). Therefore, significant conformational changes, particularly of domain II, which contains the ET-FAD, are required to shorten the distance between BF-FAD and ET-FAD. The domain II density was missing in a minor cryo-EM 3D class, suggesting that this domain is mobile (SI Appendix, Fig. S9). Structural alignment of Tma EtfAB with the homologous bifurcating EtfAB-type complexes CarCDE and EtfAB-Bcd based on the shared stable core regions (domains I and III) reveals that domain II of Tma EtfAB is in a distinct conformation (5, 16) (Fig. 3 A and B). If we define the state of an EtfAB structure based on the distance between BF-FAD and ET-FAD, with the A state representing the shortest (within electron-transferring distance, <14 Å) and the D state the longest distance, then EtfAB-Bcd with a distance of 18 Å is in the B state and CarDE with a distance of 38 Å is in the D state (5) (Fig. 3 D and F). The corresponding distance in the cryo-EM structure of Tma EtfAB is 25 Å and is in the C state (Fig. 3E). Therefore, Tma EtfAB is in an intermediate state between the B and D states previously obtained by X-ray crystallography, and no experimental structure has been determined so far for the A state of a bifurcating EtfAB-based complex.
Fig. 3.
Distance between the BF-FAD and EF-FAD varies due to the movement of the EF-FAD–containing EtfB C-terminal domain (CTD). (A) Tma EtfAB is shown in surface representation with the bound FADs in magenta sticks and spheres. EtfA and EtfB are in green and cyan, respectively. (B) EtfA is shown in cartoon representation. Regions that deviate >2 Å (Cα distance) from B-state EtfAB (PDB ID code 4L2I) are highlighted in yellow, revealing a large conformational change from the B state. (C–F) Sketches showing the relative CTD of Tma EtfB in a unique C state (E), as compared with a hypothetical A state where the electron transfer can happen between two FADs (C), EftAB-Bcd (butyryl-CoA dehydrogenase; PDB ID code 4L2I) in the B state (D), and CarDE (caffeyl-CoA reductase; PDB ID code 6FAH) in the D state (E). The three structures (D–F) were aligned based on the EtfB N-terminal domain (NTD) and EtfA.
Interestingly, Tma ET-FAD is 30° and 40° rotated with respect to the ET-FAD in the D (CarCDE) and B (EtfAB-Bcd) states, respectively (Movie S1). These rotational and translational movements must be involved in reaching to the A state (<14 Å) in these enzymes. NADH is proposed to bind to domain III near the BF-FAD. There is a hairpin insertion that makes direct contact with EtfCX and the EtfA arm that partially wraps domain II (Fig. 3B). We suggest that NADH binding is the trigger for the expected conformational change that brings BF-FAD and ET-FAD into a more favorable orientation that allows electron transfer (Fig. 3C). The pulling in of domain II by the EtfA arm requires a coordinated movement of the underlying EtfCX to accommodate the domain, and the hairpin insertion is well-positioned to do so. The final position of domain II is unclear and will require the determination of the structure in the A state. This is discussed further below.
The Striking Similarity of the [4Fe-4S] Coordination in EtfX and Mammalian ETF-QO.
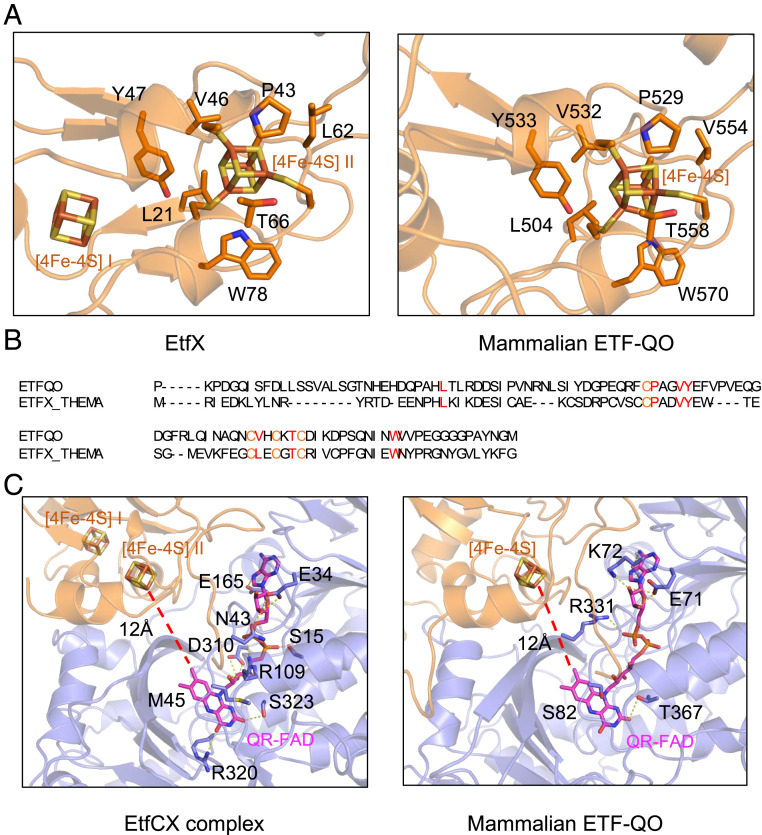
The terminal electron step in fatty acid oxidation by mitochondria is catalyzed by membrane-bound ETF-QO (24). This enzyme contains FAD and a [4Fe-4S] cluster and accepts electrons from ETF to reduce quinone. Surprisingly, we found that the structure of porcine ETF-QO is equivalent to a concatenation of Tma EtfC and EtfX (Fig. 1A). ETF-QO has apparently lost the equivalent of [4Fe-4S] cluster I in EtfX, but the structure and the relative domain orientation are remarkably conserved, with an rmsd of 1.6 Å among the 330 aligned amino acids. This conservation across evolution highlights the importance of these specific protein–iron-sulfur cluster interactions and electron transfer function. Not only are the four [4Fe-4S]–coordinating cysteines in the same positions but the second-shell residues that stabilize the four cysteines are nearly invariantly conserved (Fig. 4 A and B). These include Phe-47, Val-46, Pro-43, Leu-62, Thr-66, and Tyr-78 in the Tma EtfX with the corresponding residues Phe-533, Val-532, Pro-529, Val-554, Thr-558, and Tyr-570 in ETF-QO. This level of conservation around the [4Fe-4S] cluster is even more remarkable because the overall sequence conservation between Tma EtfCX and ETF-QO is extremely low: only 5.0% identical and 7.4% similar overall and 30.1% identical and 44.7% similar in the conserved domain. It has been proposed that in the mitochondrial ETF-QO, electrons are transferred from the FAD in ETF to the [4Fe-4S] cluster of ETF-QO and then to its FAD (QR-FAD) (25). The reduction potential of the [4Fe-4S] cluster in ETF-QO is +37 mV, which is about 400 mV more positive than that of a typical 4Fe cluster (near −400 mV) but consistent with the potentials of the QO-FAD (with potentials of +47 and −30 mV for the oxidized flavin/neutral semiquinone state [OX/NSQ] and NSQ/fully reduced hydroquinone state [HQ] couples, respectively). The virtually identical coordination of the [4Fe-4S] clusters in ETF-QO and EtfX would suggest that they are of a comparable redox potential and that the proposed high-potential electron transfer pathway in EtfABCX is the same as that in ETF-QO, with electron transfer to a high-potential [4Fe-4S] cluster prior to reduction of the quinone-reducing QR-FAD.
Fig. 4.
Comparison of the [4Fe-4S]–binding pocket in the Tma EtfX and the porcine ETF-QO. (A) The conserved coordinating residues include four [4Fe-4S]–coordinating cysteines and six largely hydrophobic supporting residues, which may tune the electric potential of the [4Fe-4S] cluster. (B) Sequence alignment. (C) Alignment of EtfCX with ETF-QO (PDB ID code 2GMH) reveals the nearly identical [4Fe-4S] coordination in EtfABCX and ETF-QO, suggesting a common electron transfer route between [4Fe-4S] and MQ or UQ. Different regions in porcine ETF-QO are colored as the corresponding regions in EtfCX.
The structural conservation between Tma EtfCX and mitochondrial ETF-QO also extends to the binding of QR-FAD, which is held in virtually the same orientation and is the same 12 Å away from the [4Fe-4S] cluster in both enzymes (Fig. 4C). However, although both QR-FADs are completely buried, the residues coordinating them differ in the proteins. In EtfC, the QR-FAD is extensively stabilized by multiple polar interactions and hydrogen bonds involving Glu-34 and Glu-165 with the adenine ring, Asn-43, Ser-15, Asp-310, and Arg-109 with the phosphate group, and Ser-323 and Arg-320 with the O atom of the isoalloxazine ring. In contrast, QR-FAD in ETF-QO has fewer polar interactions with the surrounding residues (Fig. 4C). Further, the conserved Ser-82 contacts the isoalloxazine ring likely via a hydrogen bond that could modulate the redox potential of QR-FAD (24), but Ser-82 is replaced by a nonpolar Met-45 in EtfC. All these interactions likely lead to a significant difference in the redox potentials of EtfC QR-FAD relative to ETF-QO QR-FAD. This would be reasonable because the final electron acceptor for EtfC is MQ (−80 mV), which has a more negative potential than that of ubiquinone (UQ; +100 mV), which is reduced by ETF-QO.
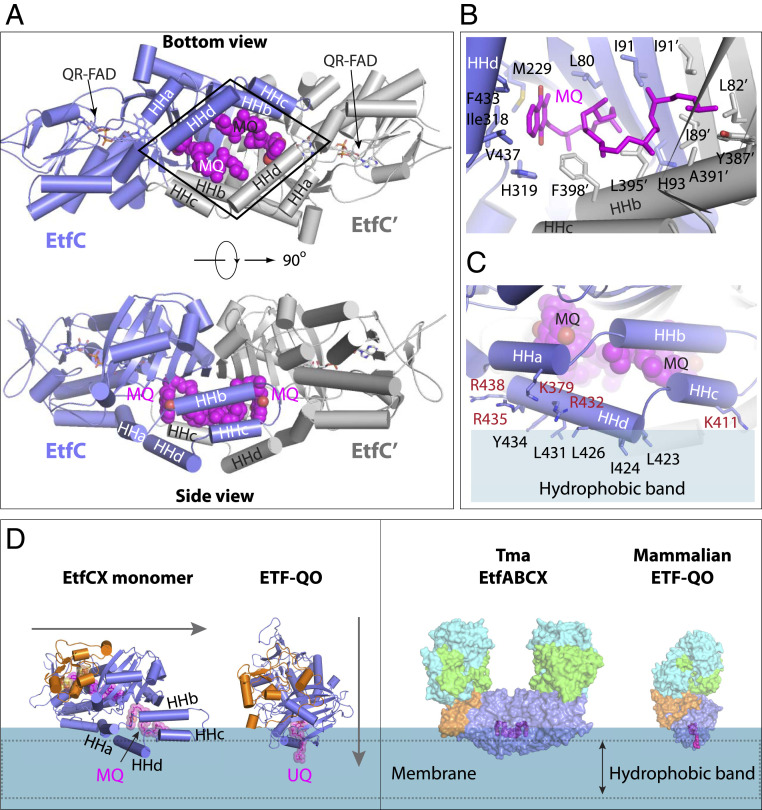
Two Menaquinones Bind in the Hydrophobic Pocket of the Membrane-Associated EtfC Dimer.
A distinguishing feature of membrane-associated Tma EtfABCX from cytoplasmic FBEB systems that have been characterized is the dimerization of the complex via EtfC–EtfC interaction. One part of the dimer interface, as mentioned above, is the intermolecular β-sheet, and another significant part is contributed by the four α-helices at the C terminus of EtfC that we term horizontal helices (HHs) a to d (Fig. 5A). These α-helices are absent in mammalian ETF-QO homologs. The four α-helices of one EtfC, together with the additional four α-helices of the other EtfC, stack on the extended cross-molecular β-sheet to form a “split rail fence,” with two HHa and two HHb α-helices forming the base rail, and two HHc and two HHd α-helices forming the outer rail. The rhombic two-rail fence encloses a large hydrophobic space to accommodate two MQ molecules. The two MQ molecules are located at the EtfC dimer interface and are related by the twofold symmetry of the superdimer complex and, therefore, interact with the EtfC dimer in an identical manner (Fig. 5B): The MQ head group is surrounded by Met-229, Ile-318, His-319, Phe-433, and Val-437 of the first EtfC and Phe-398′ of the second EtfC, and the tail is supported by Leu-80, Ile-91, His-93, Leu-82′, Ile-89′, and Ile-91′ of the underlying intermolecular β-sheet and by Tyr-387′, Ala-391′, Leu-395′, and Phe-398′ of the railing α-helix HHb from the side.
Fig. 5.
EtfC dimer interface plays a dual function in MQ binding and membrane embedding. (A) A bottom view (Top; from membrane) and side view (Bottom) of the EtfC dimer showing two MQ molecules lying flat inside the rhombic pocket formed by the C-terminal α-helices HHa to HHd. (B) Each MQ is surrounded by 15 highly hydrophobic residues. (C) A side view of one EtfC showing five positively charged residues above five hydrophobic residues of the C-terminal α-helices HHa, HHc, and HHd. Such surface property is compatible with membrane embedding. (D, Left) The presence of the four C-terminal α-helices HHa to HHd enables EtfC to insert horizontally into the membrane, as compared with the vertical insertion of the ETF-QO. (D, Right) The approximate membrane-embedding region of the Tma EtfABCX superdimer complex compared with the mammalian ETF QO.
Looking from the side with the twofold symmetry axis of the EtfABCX dimer complex perpendicular to the cell membrane, the two outer-rail α-helices HHd are at the lowest point of the complex structure and the lowest two-thirds of the helices are composed of hydrophobic residues (Leu-423, Ile-424, Leu-428, Leu-431, and Tyr-434) (Fig. 5 A and C). This region must be embedded in the membrane and reaches the hydrophobic core of the lipid bilayer. However, the upper one-third of the two HHd helices feature three positively charged residues: Arg-432, Arg-435, and Arg-438. At about the same height as the top of HHd, the HHa helix also contributes positively charged Lys-379, while the HHc helix contributes positively charged Lys-411. This band of positive charges must be interacting with the negatively charged phosphate groups of the lipid bilayer (Fig. 5 B and C). Therefore, the outer rail of the rhombic fence defines the membrane-binding region of the EtfABCX complex. We computationally placed phospholipid molecules surrounding the EtfC dimer structure to perform a molecular dynamics (MD) simulation. The equilibrium positions of these lipids are consistent with the proposed interactions with the fence-railing helices (HHa, HHc, and HHd), and approximately delineate the inner-leaflet position of the membrane (SI Appendix, Fig. S10).
From this structural analysis it is clear that EtfC inserts into the membrane horizontally. This is enabled by the presence of the four C-terminal extension helices (HHa to d). This EtfC arrangement makes the two MQ molecules nearly parallel to the membrane plane (Fig. 5D). The MQ binding is very different from the UQ binding in ETF-QO in which UQ is nearly perpendicular to the membrane plane and UQ is raised but partially remains in the membrane (24). Despite their different binding modes, both electron acceptors (MQ and UQ) are about the same distance (10 Å) away from their respective electron donor to QR-FAD.
A General Mechanism for the Catalytic Cycle of the EtfABCX Family.
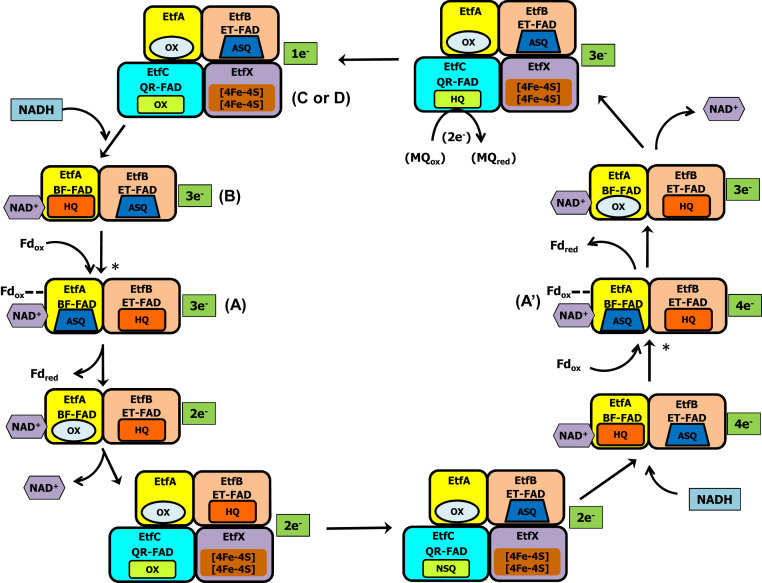
Pae EtfABCX has been previously studied extensively by transient absorption, electron paramagnetic resonance, and optical methods to monitor titrations with NADH and with inorganic reductants with and without NAD+. A reaction scheme has been proposed involving the oxidation of NADH and the transfer of one electron to Fd and one electron to MQ (20). Given the high degree of sequence identity between Tma EtfABCX and Pae EtfABCX, we assume the two complexes share the same properties including the mechanism of catalysis. In Pae ABCX, we previously showed (20) that the ET-FAD in the high-potential electron transfer branch has a reduction potential (Em) of −94 mV for the anionic semiquinone state (ASQ)/HQ couple, and the BF-FAD of the low-potential branch has an Em of −285 mV for the two-electron OX/HQ couple. This is more positive than that of NADH (Em −320 mV), indicating a favorable electron transfer with minimal energy dissipation. The lower boundary of the Em of the Pae BF-FAD OX/ASQ was estimated to be −476 mV in the Pae enzyme, which is sufficient to reduce Fd (Em ∼−450 mV). Based on detailed spectroscopic analysis (20), resting Pae EtfABCX in the one-electron reduced state was proposed to react with NADH to form a CT complex that contains two-electron reduced HQ-BF-FAD in the three-electron reduced enzyme. As shown in Fig. 6, this form of the enzyme reacts with oxidized Fd to generate the high-energy low-potential ASQ-BF-FAD. Once Fd is reduced, NAD+ is released and the two-electron reduced HQ-ET-FAD reduces QR-FAD to the NSQ state in preparation for MQ reduction and regenerates the one-electron reduced form of the EtfAB subcomplex. This completes the first half of the catalytic cycle and the enzyme is ready to bind a second molecule of NADH. We have now extended the previous mechanism (20) to include the second half of the cycle (Fig. 6). Oxidation of the second NADH generates a CT complex that contains two-electron reduced HQ-BF-FAD but, in contrast to the first half of the cycle, this time the enzyme is in the four- rather than the three-electron reduced form. After reduction of a second Fd, NAD+ is released and the two-electron reduced HQ-ET-FAD reduces QR-FAD to the HQ state. This reduces MQ in a two-electron reaction to regenerate the original one-electron reduced resting state and complete the overall four-electron oxidation reaction (Eq. 1). The key step in this mechanism is the binding of Fd to the NAD+-bound CT complex. Fd binding to the Pae EtfAB subcomplex was observed experimentally, suggesting that both NADH and Fd need to bind the resting state of the enzyme before the bifurcation reaction is initiated (20).
Fig. 6.
Catalytic cycle of EtfABCX. The complex is depicted in its resting one-electron reduced state in which the ET-FAD is in the ASQ state. Conformational changes (indicated by asterisks) are required to bring the complex into a bifurcation-ready state (ET-FAD and BF-FAD within 14 Å), designated states A and A′. We propose that this involves the binding of both NADH and Fd. The A, A′, B, C, and D states are defined in the legend to Fig. 3. For clarity, the EtfC–EtfX subcomplex is depicted only when there is a change in redox states compared with the previous step of the cycle.
Based on the spatial arrangement of the redox cofactors and the known requirement of the feasible maximal distance for electron transfer (<14 Å), our structure clearly demonstrates that it is physically impossible for the BF-FAD to transfer the low-potential electron through the EtfX [4Fe-4S] cofactors, as was previously proposed for Pae ABCX in the absence of the structure (20). Our cryo-EM structure of Tma ABCX shows a different path in which the low-potential electron from BF-FAD is most likely delivered directly to Fd rather than going through the two EtfX [4Fe-4S] clusters. We can now suggest a revised and complete catalytic mechanism for the EtfABCX family (Fig. 6) that takes into account the conformational change that accompanies catalysis, as revealed by the structures of the EtfAB-type complexes (Fig. 3). As described above, it has been shown experimentally (20) that in the first round of NADH oxidation, the BF-FAD is reduced to the HQ state and forms a charge transfer complex with NAD+. Before the first electron is transferred to the ASQ-ET-FAD to generate the HQ-ET-FAD and the ASQ-BF-FAD states, a conformational change involving domain II must occur in order to bring the ET- and BF-flavin close enough together for electron transfer, in what is now designated as the A state (Fig. 6). We now propose that this requires the binding of both NAD+ and Fdox in order to capture the low-potential electron from the HQ-BF-FAD for Fd reduction. In addition, the transfer of an electron to Fdox and/or release of Fdred could trigger another protein conformational change that increases the distance between the ET-FAD and BF-FAD back to the initial resting state (either B, C, or D). In this state, where the ET-FAD–BF-FAD distance is ≥18 Å, HQ-ET-FAD can still reduce the QR-FAD of EtfC, via the [4Fe-4S]-I clusters of EtfX, with dissociation of NAD+ to regenerate the two-electron reduced resting state at the end of the first half of the cycle (Fig. 6). Catalysis then continues with the oxidation of a second NADH and the transfer of two more electrons down the low- and high-potential branches leading to the reduction of the second Fd and the two-electron reduced QR-FAD (Fig. 6). However, in contrast to the first half, the Fdox- and NAD+-bound A state in the second half of the cycle (designated A′ in Fig. 6) is four-electron reduced as it contains NSQ-QR-FAD rather than OX-QR-FAD. To what extent the conformational states of A and A′ differ is not clear and experimentation will be required to determine this.
In summary, we have determined an atomic model of a membrane-associated electron-bifurcating complex and propose an electron transfer and catalysis mechanism for the EtfABCX family of FBEB complexes. Our study also sheds lights on the evolutionary relationship with the mammalian mitochondrial enzyme ETF-QO, mutations of which lead to multiple acyl-CoA dehydrogenation deficiency disorder (26). It has been proposed that electrons are transferred to QR-FAD by going through the [4Fe-4S] cluster in the mitochondrial enzyme (25). We have discovered that the exergonic electron transfer branch is highly conserved between the bacterial EtfCX and the mammalian mitochondrial ETF-QO, and thus our structure parallels the electron transfer mechanism in which electrons extracted from β-oxidation of fatty acids are passed to the [4Fe-4S] center in ETF-QO, before being transferred to QR-FAD and finally to the general ubiquinone pool, suggesting an intriguing evolutionary relationship between bifurcating EtfABCX in hyperthermophilic microbes and the nonbifurcating counterpart ETF-QO in mammals.
Materials and Methods
Construction of a T. maritima EtfABCX Expression Strain.
A plasmid for chromosomal expression of the Tma EtfABCX operon in P. furiosus was generated by Gibson assembly (New England Biolabs) and contained the following: the Tma EtfABCX operon under the control of the P. furiosus slp promoter (Pslp; consisting of 184 bp directly upstream of the PF1399 gene), an N-terminal 9× His tag with flanking Ala spacers encoded on the 5′ end of etfA, the T1 terminator at the 3′ end of the EtfABCX operon (5′-aatcttttttagcactttt-3′), 0.5-kb flanking regions targeting homologous recombination at P. furiosus genome region 5 (a 646-bp intergenic space between convergent genes PF1232 and PF1233), the PgdhpyrF cassette for genetic selection of uracil prototrophy in the auxotrophic background strain P. furiosus COM1, and the AprR cassette and pSC101 origin for selection of clones in Escherichia coli. The resulting plasmid, termed pGL093, was sequence verified and linearized by restriction digestion prior to transformation of P. furiosus COM1 as previously described (27). Transformants were screened and purified as previously described (20). The P. furiosus strain containing the genome-integrated Tma EtfABCX expression cassette was designated MW369. Primers used in plasmid and strain construction are listed in SI Appendix, Table S2.
Expression and Purification of Tma EtfABCX.
Fermentation on a 15-L scale of recombinant P. furiosus was performed as described previously (28) with the exception that the cells were grown at 80 °C to accommodate production of recombinant T. maritima EtfABCX. Frozen cells (30 g wet weight) were lysed in 4 volumes of 50 mM phosphate buffer (pH 7.5) with 1 mM dithiothreitol (DTT) and 50 μg/mL DNase I under strict anaerobic conditions in a Coy anaerobic chamber (95% Ar, 5% H2). Cell debris and membranes were removed by ultracentrifugation at 100,000 × g. The supernatant was loaded on a 5-mL HisTrap column (GE Healthcare) with a 50% dilution rate using 50 mM phosphate buffer (pH 7.5) with 1 mM DTT. The EtfABCX eluted around 100 mM imidazole. Based on the chromatogram, the brown/yellow fractions were pooled and concentrated to <5 mL using ultrafiltration (Millipore) under anaerobic conditions. The concentrated sample (∼200 U) was applied to a Superdex S200 column (16/60; Cytiva) and developed with 25 mM Tris⋅HCl (pH 8) containing 300 mM NaCl (with 1 mM DTT). Active fractions were diluted 10-fold with 25 mM Tris⋅HCl (pH 8) buffer (buffer A) and loaded (∼170 U) on a 5-mL QHP HiTrap column (Cytiva) and EtfABCX was eluted with a 0 to 50% gradient of 1 M NaCl in buffer A. EtfABCX eluted when 350 mM NaCl was applied. Fractions corresponding to EtfABCX were selected based on flavin content, dye-linked activity, and gel electrophoresis (Mini-PROTEAN TGX Stain-Free Gels; Bio-Rad). Thirty grams of biomass from 15 L P. furiosus culture yielded about 1 mg (80 U) of T. maritima EtfABCX.
Activity Assays.
NADH-linked activities of EtfABCX were measured anaerobically at 80 °C in 3-mL glass cuvettes sealed with rubber stoppers containing 50 mM Hepes (pH 7.5), 100 mM NaCl, 1 mM NADH, and 200 µM iodonitrotetrazolium chloride (Sigma). The formation of red formazan was followed at 500 nm (€ 19.3 mM−1⋅cm−1) in a Cary 100 ultraviolet-visible spectrophotometer with a Peltier-based temperature controller (Agilent). Activity is reported in units, where 1 U is defined as 1 µmol of iodonitrotetrazolium reduced per minute.
Electron Microscopy and Data Acquisition.
After purification, aliquots of the EtfABCX complex were further diluted with buffer (20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl, 2 mM DTT) to prepare the grids for microscopy. For negative-staining EM, 3-μL aliquots of samples after serial dilution were applied onto carbon support film copper grids (Electron Microscopy Sciences), washed, and stained with 2% uranyl acetate. The negative-staining grids were examined using a Tecnai electron microscope (TFS) operated at 120 kV, equipped with a 2k × 2k Orius 830 charge-coupled device camera (Gatan). The relative protein concentration in single-particle cryo-EM analysis is 20 times higher than that in negative staining. For single-particle cryo-EM analysis of EtfABCX, the sample at a concentration of 0.3 to 0.4 mg/mL was applied to glow-discharged holey carbon grids (Quantifoil R2/1 copper, 300 mesh), blotted for 4 s, and plunged into liquid ethane using an FEI Vitrobot Mark IV. A small dataset of 500 micrographs was collected on a 200-kV FEI Arctica electron microscope equipped with a TFS Falcon III direct detector for screening purposes. After 3D classification, 22,089 particles led to a preliminary 6.0-Å 3D map. Then a full dataset was collected during a 4-d session on a TFS Titan Krios electron microscope which is operated at 300 kV and equipped with a K2 summit camera (Gatan). The dataset was acquired using SerialEM (29) with a nominal magnification of 130,000× and a calibrated pixel size of 0.514 Å per pixel with a defocus range from −1.0 to −2.0 μm. Images were collected in the superresolution counting and movie mode with a dose rate of 2.0 electrons per Å2 per second per frame. Forty frames were recorded in each movie micrograph.
Image Processing.
A total of 9,096 movie stacks were acquired in Titan Krios and were drift corrected with electron-dose weighting and twofold binned using MotionCor2.0 (30). Contrast transfer function parameters of each aligned micrograph were calculated using CTFFIND 4.1 (31). After autopicking using RELION-3, 2,471,763 particles were obtained for 2D classification. The “good” 2D class averaged with defined structural features was selected and contained 2,143,876 particles. These particle images were imported to cryoSPARC2 for ab initio 3D model reconstruction (32). Among the five different initial 3D maps, one contained all subunits and was composed of the greatest number of particles (around 24% of the particle population). That map was used as the reference map. The corresponding particles (411,493) were converted to the RELION format using UCSF PyEM (https://github.com/asarnow/pyem). Three-dimensional classification of such particles led to 3D maps for the (EtfABCX)4 tetramer and the (EtfABCX)2 dimer. A second round of 3D classification was applied with a soft mask including only the (EtfABCX)2 dimer. The top class contained 285,377 particles and had the best structural features and the highest resolution. This class was selected for further 3D refinement with C2 symmetry. After postprocessing and Bayesian polishing, the final 3D density map reached 2.9-Å average resolution as estimated by the gold-standard Fourier shell correlation at a correlation cutoff value of 0.143 (19). The local resolution map was calculated using ResMap (33) and displayed using UCSF Chimera (34).
Model Building and Refinement.
The initial models were built with comparative modeling while fitting into an electron density map in the Rosetta suite (35). The template for EtfAB was the ETF protein of A. fermentans (Protein Data Bank [PDB] ID code 4L2I) (16). The template for EtfCX was the porcine ubiquinone oxidoreductase (PDB ID code 2GMH) (24). All the models were manually rebuilt in the program Coot (36) followed by real-space refinement in the PHENIX program (37). Finally, the atomic model was validated using MolProbity (38, 39). The images were generated with either UCSF Chimera (34) or PyMOL (The PyMOL Molecular Graphics System, version 1.8.x; Schrödinger). Different orientations of Tma EtfAB complex domain II were modeled using the structures of other EtfAB proteins under corresponding states as templates (see Fig. 3 for details). Domain II of the Tma EtfAB was aligned to domain II of the EtfAB model in other states, and then the connecting loops were modeled using the Rosetta suite (40). The movie was generated with UCSF ChimeraX (41).
Simulation of Lipids Surrounding the EtfC Dimer.
The lipids surrounding the EtfC dimer were equilibrated by MD simulation in NAMD (42) using a CHARMM36m force field (43). The location of the protein in the lipid bilayer was predicted with the PPM server (44). Then all the other inputs for MD simulation were prepared with the membrane builder module (45) of CHARMM-GUI (46, 47) using default parameters with 100% 1-palmitoyl-2-cis-9,10-methylene-hexadecanoic-acid-sn-glycero-3-phosphoethanolamine, an aminophospholipid often used in the computational modeling of bacterial membranes. The default six-step equilibrium simulation was carried out in NAMD (42) before checking the membrane-embedding state of the protein complex.
Abbreviations Used in This Manuscript.
SI Appendix, Table S3 provides a list of abbreviations used throughout the manuscript.
Supplementary Material
Acknowledgments
This work was funded by grants from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (DE-SC0020085 to H.L. and DE-FG02-95ER20175 to M.W.W.A.). Cryo-EM images were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite at the Van Andel Institute. We thank G. Zhao and X. Meng for help with data collection.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016978118/-/DCSupplemental.
Data Availability.
The atomic coordinates have been deposited in the PDB with accession code 7KOE and the electron microscopy map has been deposited in the Electron Microscopy Data Bank with accession code EMD-22973.
References
- 1.Buckel W., Thauer R. K., Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 118, 3862–3886 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Buckel W., Thauer R. K., Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD(+) (Rnf) as electron acceptors: A historical review. Front. Microbiol. 9, 401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters J. W., et al. , A new era for electron bifurcation. Curr. Opin. Chem. Biol. 47, 32–38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell P., Possible molecular mechanisms of the protonmotive function of cytochrome systems. J. Theor. Biol. 62, 327–367 (1976). [DOI] [PubMed] [Google Scholar]
- 5.Demmer J. K., Pal Chowdhury N., Selmer T., Ermler U., Buckel W., The semiquinone swing in the bifurcating electron transferring flavoprotein/butyryl-CoA dehydrogenase complex from Clostridium difficile. Nat. Commun. 8, 1577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demmer J. K., et al. , Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction. FEBS Lett. 592, 332–342 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Lubner C. E., et al. , Mechanistic insights into energy conservation by flavin-based electron bifurcation. Nat. Chem. Biol. 13, 655–659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner T., Koch J., Ermler U., Shima S., Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 357, 699–703 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Roberts D. L., Salazar D., Fulmer J. P., Frerman F. E., Kim J. J., Crystal structure of Paracoccus denitrificans electron transfer flavoprotein: Structural and electrostatic analysis of a conserved flavin binding domain. Biochemistry 38, 1977–1989 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Roberts D. L., Frerman F. E., Kim J. J., Three-dimensional structure of human electron transfer flavoprotein to 2.1-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 93, 14355–14360 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toogood H. S., van Thiel A., Scrutton N. S., Leys D., Stabilization of non-productive conformations underpins rapid electron transfer to electron-transferring flavoprotein. J. Biol. Chem. 280, 30361–30366 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Toogood H. S., et al. , Extensive domain motion and electron transfer in the human electron transferring flavoprotein⋅medium chain acyl-CoA dehydrogenase complex. J. Biol. Chem. 279, 32904–32912 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Leys D., Basran J., Talfournier F., Sutcliffe M. J., Scrutton N. S., Extensive conformational sampling in a ternary electron transfer complex. Nat. Struct. Biol. 10, 219–225 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Burgess S. G., et al. , Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: Role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair. Biochemistry 47, 5168–5181 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Vogt M. S., et al. , Structural and functional characterization of an electron transfer flavoprotein involved in toluene degradation in strictly anaerobic bacteria. J. Bacteriol. 201, e00326-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury N. P., et al. , Studies on the mechanism of electron bifurcation catalyzed by electron transferring flavoprotein (Etf) and butyryl-CoA dehydrogenase (Bcd) of Acidaminococcus fermentans. J. Biol. Chem. 289, 5145–5157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledbetter R. N., et al. , The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: Generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 56, 4177–4190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgren T., Nordlund S., Electron transport to nitrogenase in Rhodospirillum rubrum: Identification of a new fdxN gene encoding the primary electron donor to nitrogenase. FEMS Microbiol. Lett. 245, 345–351 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Adams M. W., Biochemical diversity among sulfur-dependent, hyperthermophilic microorganisms. FEMS Microbiol. Rev. 15, 261–277 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Schut G. J., et al. , The catalytic mechanism of electron-bifurcating electron transfer flavoproteins (ETFs) involves an intermediary complex with NAD. J. Biol. Chem. 294, 3271–3283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai M. H., Saier M. H. Jr, Phylogenetic characterization of the ubiquitous electron transfer flavoprotein families ETF-alpha and ETF-beta. Res. Microbiol. 146, 397–404 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Garcia Costas A. M., et al. , Defining electron bifurcation in the electron-transferring flavoprotein family. J. Bacteriol. 199, e00440-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sperotto R. A., Gross J., Vedoy C., Passaglia L. M., Schrank I. S., The electron transfer flavoprotein fixABCX gene products from Azospirillum brasilense show a NifA-dependent promoter regulation. Curr. Microbiol. 49, 267–273 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Frerman F. E., Kim J. J., Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc. Natl. Acad. Sci. U.S.A. 103, 16212–16217 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson M. A., Usselman R. J., Frerman F. E., Eaton G. R., Eaton S. S., The iron-sulfur cluster of electron transfer flavoprotein-ubiquinone oxidoreductase is the electron acceptor for electron transfer flavoprotein. Biochemistry 47, 8894–8901 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodman S. I., Binard R. J., Woontner M. R., Frerman F. E., Glutaric acidemia type II: Gene structure and mutations of the electron transfer flavoprotein:ubiquinone oxidoreductase (ETF:QO) gene. Mol. Genet. Metab. 77, 86–90 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Lipscomb G. L., et al. , Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: Construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77, 2232–2238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhagen M. F., Menon A. L., Schut G. J., Adams M. W., Pyrococcus furiosus: Large-scale cultivation and enzyme purification. Methods Enzymol. 330, 25–30 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Wang R. Y., et al. , Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife 5, e17219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown A., et al. , Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leaver-Fay A., et al. , ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J., et al. , CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu E. L., et al. , CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J., et al. , CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates have been deposited in the PDB with accession code 7KOE and the electron microscopy map has been deposited in the Electron Microscopy Data Bank with accession code EMD-22973.