Significance
A successful vaccine depends on productive T follicular helper (TFH) and B-cell responses to elicit protective immunity. Vaccines are delivered as live-attenuated viruses, inactivated organisms, or protein subcomponents via different routes. In the context of protein immunization, we demonstrate that Tcf1 and Lef1 transcription factors play critical roles in suppressing excessive induction of CTLA4 and LAG3 coinhibitory receptors in TFH cells and, hence, preventing undue inhibition of B cells. This function is in contrast to the requirement for Tcf1 and Lef1 in induction of Bcl6 in TFH cells elicited by viral or bacterial infections. These findings highlight that the same protein factors could be utilized to regulate distinct aspects of TFH cell differentiation depending on vaccination routes and regimens.
Keywords: follicular helper T cells, transcriptional regulation, coinhibitory pathway
Abstract
Precise regulation of coinhibitory receptors is essential for maintaining immune tolerance without interfering with protective immunity, yet the mechanism underlying such a balanced act remains poorly understood. In response to protein immunization, T follicular helper (TFH) cells lacking Tcf1 and Lef1 transcription factors were phenotypically normal but failed to promote germinal center formation and antibody production. Transcriptomic profiling revealed that Tcf1/Lef1-deficient TFH cells aberrantly up-regulated CTLA4 and LAG3 expression, and treatment with anti-CTLA4 alone or combined with anti-LAG3 substantially rectified B-cell help defects by Tcf1/Lef1-deficient TFH cells. Mechanistically, Tcf1 and Lef1 restrain chromatin accessibility at the Ctla4 and Lag3 loci. Groucho/Tle corepressors, which are known to cooperate with Tcf/Lef factors, were essential for TFH cell expansion but dispensable for repressing coinhibitory receptors. In contrast, mutating key amino acids in histone deacetylase (HDAC) domain in Tcf1 resulted in CTLA4 derepression in TFH cells. These findings demonstrate that Tcf1-instrinsic HDAC activity is necessary for preventing excessive CTLA4 induction in protein immunization–elicited TFH cells and hence guarding their B-cell help function.
Coinhibitory receptors, such as CTLA4, LAG3 and PD1, provide essential counterbalance to stimulatory signals in T cells to maintain tolerance and prevent autoimmunity (1, 2). On the other hand, excessive activation of coinhibitory pathways in chronic microbial infections and tumor microenvironments impedes eradication of microbes and transformed cells (3, 4). Precise control of coinhibitory receptor expression is therefore pivotal to achieve protective immunity while avoiding tissue damage. B cell responses are stimulated by T follicular helper (TFH) cells but restrained by regulatory T (TREG) and T follicular regulatory (TFR) cells, and CTLA4 exerts critical regulatory roles in this process through both cell-intrinsic and cell-extrinsic mechanisms (5–8). While CTLA4 is constitutively expressed in TREG cells, it is not expressed in conventional naïve CD4+ T cells but is induced upon activation (7). Induced deletion of CTLA4 in TFH cells modestly enhances the stimulatory effect on B cells in vitro (5), while forced expression of CTLA4 in TFH cells substantially compromises generation of germinal center (GC)-B cells in vivo (9). It is clear that fine-tuning CTLA4 expression in TFH cells is critical for optimal B-cell help function, yet the underlying mechanisms are not fully understood.
Tcf1 and Lef1 transcriptional factors (TFs) have versatile functions in T cells, ranging from early T cell development in the thymus to mature T cell responses in the periphery (10, 11). In CD4+ T cells, Tcf1 promotes TH2 but antagonizes TH1 and TH17 differentiation (12, 13). Recently, we and others demonstrated that although Tcf1 and Lef1 are expressed at lower levels in TREG cells than in conventional CD4+ T cells, they are necessary for the immunosuppressive function of TREG cells (14, 15). Furthermore, Foxp3-Cre–mediated ablation of Tcf1 and Lef1 greatly diminished generation of spontaneous TFR cells under homeostatic state (15). In TFH cells elicited by acute infection with lymphocytic choriomeningitis virus (LCMV-) Armstrong strain, Tcf1 is required for induction of Bcl6, the TFH-lineage defining TF, and optimal expression of Icos costimulatory receptor and IL-6 receptor to fully activate the TFH transcriptional program (16–18). However, it is also noted that Tcf1 does not seem to be necessary for TFH responses during protein immunization with alum as an adjuvant (18). It remains unknown if the discrepancy in a requirement for TFH cells is due to a compensatory effect by Lef1 or because Tcf1 and Lef1 control different aspects of TFH cell differentiation in response to protein immunization.
Tcf/Lef TFs act as transcriptional activators or repressors, depending on the interacting partners, gene, and cell context. β-catenin is a known coactivator for Tcf/Lef TFs but appears to be dispensable for activating Bcl6 transcription in infection-elicited TFH (Inf_TFH) cells because ablation of β-catenin alone, or together with its homolog γ-catenin, or Tcf1 long isoforms, of which the N termini are responsible for β-catenin interaction, did not affect Bcl6 induction (19, 20). Our recent study reveals that Ezh2, in its Ser21-phosphorylated form, functions as a coactivator with Tcf1 to induce Bcl6 in Inf_TFH cells (21). As for target gene repression, Groucho/Tle proteins are known corepressors that interact with Tcf/Lef TFs (22, 23). Among four Tle genes that encode full-length Grouch/Tle proteins in mammals, Tle3 is most abundantly expressed in T cells, with Tle4 detected at intermediate and Tle1 and Tle2 at low levels (24). Tle genes exhibit strong functional redundancy and gene dosage dependency, as observed in late stages of thymic development, where deleting Tle3 along with at least one allele each of Tle1 and Tle4 is necessary to abrogate CD8+ T cell production (24). We previously discovered that Tcf1 and Lef1 have intrinsic histone deacetylase (HDAC) activity, which is responsible for repressing CD4+ lineage-associated genes in CD8+ lineage-committed thymocytes (25). It remained unknown if Tle corepressors have a role in TFH cell differentiation and if Tcf1/Lef1 HDAC activity contributes to immune regulation beyond their role in establishing CD8+ T cell identity during thymic development.
To address these critical knowledge gaps, we adopted a protein immunization method to elicit TFH cell responses and used mouse models developed in this work to address the functional redundancy between Tcf1 and Lef1, requirements for Tcf1 HDAC activity, and roles of Tle1 to 4 corepressors in immunization-elicited TFH (Imm_TFH) cells. This systematic approach revealed that Tcf1 and Lef1 were essential for restraining CTLA4 and LAG3 expression in Imm_TFH cells and thus protecting them from undue inhibition of GC-B cell responses. The repression of CTLA4 required Tcf1 HDAC activity but did not depend on Tle corepressors, whereas loss of Tle proteins impaired proliferation of activated CD4+ T cells and hence production of TFH cells.
Results
Tcf1/Lef1-Deficient Imm_TFH Cells Are Phenotypically Normal but Functionally Impaired.
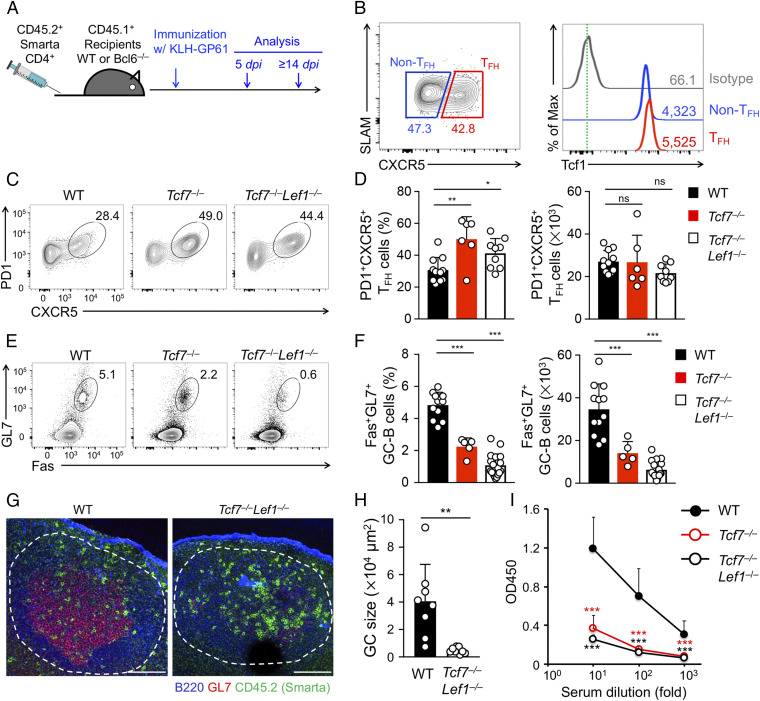
Tcf1 and Lef1 are known to have redundant functions, with Tcf1 showing a more predominant regulatory effect. As shown previously, in the process of establishing thymic CD8+ T cell identity (25), generation of CD8+ memory precursor cells (26), immunosuppressive function of regulatory T cells (14), and differentiation of Inf_TFH (17), ablation of Lef1 alone shows little to modest impact, but loss of Lef1 exacerbates the more evident defects caused by Tcf1 deficiency. To evaluate if the lack of impact of Tcf1 ablation on Imm_TFH cells (18) was due to compensation by Lef1, we used hCD2-Cre to ablate Tcf7 and/or Lef1 genes (encoding Tcf1 and Lef1, respectively) in mature T cells so as not to affect thymic development (10, 17). Preliminary studies showed that adoptively transferred hCD2-Cre+ Tcf7FL/FL Lef1FL/FL (Tcf7−/−Lef1−/−) CD4+ T cells homed to secondary lymphoid organs at ∼50% efficiency of wild-type (WT) cells, with similar survival capability (SI Appendix, Fig. S1). We therefore transferred Tcf7−/− or Tcf7−/−Lef1−/− Smarta CD4+ T cells at 2-fold as many as WT Smarta cells in immunization experiments with keyhole limpet hemocyacin (KLH) conjugated with LCMV glycoprotein 61–80 peptides (KLH-GP61) (Fig. 1A) so that they were activated at similar precursor frequencies in the draining popliteal lymph nodes (dLNs). Tcf1 expression was similar between WT CXCR5+ TFH and CXCR5– non-TFH cells on day 5 postimmunization (Fig. 1B) but was effectively ablated in Tcf7−/− or Tcf7−/−Lef1−/− cells, as validated by intranuclear staining (SI Appendix, Fig. S2A). Unlike Inf_TFH cells that are greatly diminished in the absence of Tcf1 and Lef1 (16–18, 27), similar numbers of CXCR5+PD1+ TFH cells were detected in response to protein immunization in recipients of WT, Tcf7−/−, or Tcf7−/−Lef1−/− Smarta cells, albeit Tcf1- or Tcf1/Lef1-deficient TFH cells were detected at modestly elevated frequency than WT TFH cells (Fig. 1 C and D). Icos expression also showed a modest increase in Tcf7−/−Lef1−/− TFH cells than WT cells (SI Appendix, Fig. S2B). In addition, similar numbers of CXCR5+Bcl6+ germinal center TFH (GC-TFH) cells were observed among all genotypes, and Bcl6 expression levels were similar among WT, Tcf7−/−, and Tcf7−/−Lef1−/− TFH cells (SI Appendix, Fig. S2 C and D). WT and Tcf7−/−Lef1−/− Smarta CD4+ T cells were detected at similar frequency in B cell follicles in dLNs, as determined by immunofluorescence staining (SI Appendix, Fig. S2E). These data collectively indicate that Tcf1 and Lef1 are not required for generation of phenotypic Imm_TFH cells, their migration into B cell follicles, and the TFH lineage–defining Bcl6 expression. This is in key contrast to an indispensable role of Tcf1 and Lef1 in regulating these aspects in Inf_TFH cells.
Fig. 1.
Tcf1 and Lef1 are required for B-cell help function by TFH cells elicited by protein immunization. (A) Experimental design. WT (1 × 105) or Tcf7−/− or Tcf7−/−Lef1−/− (2 × 105) Smarta CD4+ T cells were adoptively transferred into CD45.1+ WT or Bcl6−/− B6.SJL recipients, which were immunized with KLH-GP61 24 h later. TFH and GC-B cell responses were examined on day 5 and ≥14 postimmunization, respectively. (B) The detection of Tcf1 expression in TFH and non-TFH cells was derived from WT Smarta CD4+ T cells in dLNs on day 5 postimmunization. The values in the contour plot denote percentage, and those in histogram denote geometric mean fluorescence intensity. (C and D) The detection of CXCR5+PD1+ TFH cells in CD45.2+GFP+ Smarta cells in dLNs of WT recipients on day 5 postimmunization. Representative contour plots are from two to three independent experiments (C). Cumulative data on TFH cell frequency (Left) and numbers (Right) are means ± SD (D). (E and F) The detection of Fas+GL7+ GC-B cells in B220+CD19+ B cells in dLNs of Bcl6−/− recipients on day 14 postimmunization. Representative contour plots are from two to three independent experiments (E), and cumulative data on GC-B cell frequency (Left) and numbers (Right) are means ± SD (F). (G) The detection of B cell follicle (blue), GC (red), and Smarta CD4+ T cells (green) by immunofluorescence staining of dLNs from Bcl6−/− recipients on day 14 postimmunization. (Scale bars, 100 µm.) Data are representative from three experiments. (H) The cumulative data on GC sizes from at least two independent experiments, with ≥2 LN sections measured for each genotype. Note that in recipients of Tcf7−/−Lef1−/− Smarta cells, well-structured GCs are rarely observed, and instead, areas of GL7+ cell aggregates were measured. (I) The detection of KLH-specific IgG in sera of Bcl6−/− recipients on day 14 postimmunization. The data are from two experiments (n = 4 to 6). OD450, optical density at 450 nm. Statistical significance was first determined with one-way ANOVA for multigroup comparison, and as post hoc correction, Tukey’s test was used for indicated pair-wise comparison. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not statistically significant.
We next investigated if the phenotypically “normal” Tcf7−/− and Tcf7−/−Lef1−/− Imm_TFH cells were adequate in conferring B-cell help. To this end, we used CD45.1+ Cd4Cre+ Bcl6fl/fl (called Bcl6−/− hereafter) mice as adoptive transfer recipients, where the endogenous TFH and TFR responses are abrogated but B cells remain functional (21). Following transfer of Smarta CD4+ T cells and KLH-GP61 immunization, we analyzed humoral responses in the recipients on day 14 postimmunization. The formation of Fas+GL7+ GC-B cells was reduced in frequency and numbers in recipients of Tcf7−/− cells and were further diminished in those of Tcf7−/−Lef1−/− cells (Fig. 1 E and F). In line with this observation, whereas highly structured GCs were detected in recipients of WT Smarta CD4+ T cells, only scattered GC-B cells were in follicles without clearly definable structures in recipients of Tcf7−/−Lef1−/− cells (Fig. 1 G and H). In addition, KLH-specific antibodies were greatly diminished in recipients of Tcf7−/− or Tcf7−/−Lef1−/− cells (Fig. 1I). These analyses suggest that Tcf1 and Lef1 are necessary for TFH function as B-cell helpers but dispensable for TFH differentiation in response to protein immunization.
Tcf1 and Lef1 Repress Coinhibitory Receptors in Imm_TFH Cells.
To determine the mechanistic requirements for Tcf1/Lef1 in Imm_TFH cells, we performed RNA sequencing (RNA-seq) on WT and Tcf7−/−Lef1−/− TFH cells isolated on day 5 postimmunization, and the replicates in each genotype formed distinct clusters based on principal component analysis (SI Appendix, Fig. S3A). Global transcriptomic analysis with GSEA showed that the gene set enriched in Inf_TFH cells (17) was negatively correlated in Tcf7−/−Lef1−/− Imm_TFH cells (SI Appendix, Fig. S3B), consistent with a known role of Tcf1 and Lef1 in inducing TFH transcriptional program. In addition, Tcf1-activated and -repressed gene sets in Inf_TFH cells (17) showed negative and positive enrichment in Tcf7−/−Lef1−/− Imm_TFH cells, respectively (SI Appendix, Fig. S3 C and D), indicating a substantial overlap of Tcf1/Lef1-dependent transcriptional programs between Inf_TFH and Imm_TFH cells. Gene set enrichment analysis (GSEA) also revealed positive enrichment of CTLA4 pathways and negative enrichment of cell cycles, E2F targets, and FOXM1 pathways in Tcf7−/−Lef1−/− Imm_TFH cells (SI Appendix, Fig. S3 E–H).
By the criteria of ≥2-fold expression changes and adjusted P < 0.05, 359 genes were down-regulated and 157 genes up-regulated in Tcf7−/−Lef1−/− over WT Imm_TFH cells. Il6st (encoding gp130) and Myb expression was diminished in Tcf7−/−Lef1−/− Imm_TFH cells (Fig. 2 A and B), exhibiting similar changes as those in Tcf1/Lef1-deficient Inf_TFH cells (SI Appendix, Fig. S3C) (17). However, Bcl6, Icos, and Pdcd1 were not among the down-regulated genes, and Cxcr5 expression was modestly elevated, if at all, in Tcf7−/−Lef1−/− Imm_TFH cells (SI Appendix, Fig. S4A), in line with immunophenotypic analysis (SI Appendix, Fig. S2A). On the other hand, Lag3, Ccl5, and Ccr6 were up-regulated in Tcf7−/−Lef1−/− Imm_TFH cells as well as Tcf1/Lef1-deficient Inf_TFH cells (Fig. 2 A and B and SI Appendix, Fig. S3D). Notably, Prdm1, which encodes the Blimp1 transcription factor and is derepressed in Tcf1/Lef1-deficient Inf_TFH cells (16–18), remained unperturbed in Tcf7−/−Lef1−/− Imm_TFH cells (SI Appendix, Fig. S4A). In contrast, Clta4, Fasl, and a few other cytokine/chemokine receptors were uniquely up-regulated in Tcf7−/−Lef1−/− Imm_TFH cells (Fig. 2 A and B). Foxp1 is reported to induce CTLA4 expression in TFH cells (28), but Foxp1 transcripts were not significantly altered between WT and Tcf7−/−Lef1−/− Imm_TFH cells (SI Appendix, Fig. S4A). The increased expression of CTLA4 and LAG3 in Tcf1/Lef1-deficient Imm_TFH cells was validated on the protein level (Fig. 2C). These observations suggest that Tcf1 and Lef1 regulate common, as well as distinct, targets in Imm_TFH and Inf_TFH cells. It is also noteworthy that Tcf1/Lef1-deficient non-TFH cells also showed elevated expression of CTLA4 and LAG3 (SI Appendix, Fig. S4B), suggesting a conserved function of Tcf1 and Lef1 in suppressing the coinhibitory pathways in immunization-activated T cells. Because of the lack of CXCR5 expression in non-TFH cells, and hence their limited access to follicle and B cells, the elevated expression of coinhibitory receptors in non-TFH cells may not contribute substantially to modulating B cell functions. We therefore focused our functional and mechanistic analyses on TFH cells in this work.
Fig. 2.
Tcf1/Lef1 deficiency leads to aberrant up-regulation of coinhibitory receptors in Imm_TFH cells. (A) A volcano plot showing differential gene expression between WT and Tcf1/Lef1-deficient Imm_TFH cells, with select genes highlighted. (B) A heatmap showing relative expression of select genes in replicates of WT and Tcf1/Lef1-deficient Imm_TFH cells. (C) The detection of CTLA4 and LAG3 proteins in WT and Tcf1/Lef1-deficient Imm_TFH cells on day 5 postimmunization. In half-stacked histographs, the dotted vertical lines mark signal strength by isotype staining, and values denote geometric mean fluorescence intensity (gMFI). The bar graphs are means ± SD of relative gMFI from two experiments. ***P < 0.001.
CTLA4 and LAG3 Blockade Rectifies B-Cell Help Defects by Tcf1/Lef1-Deficient TFH Cells.
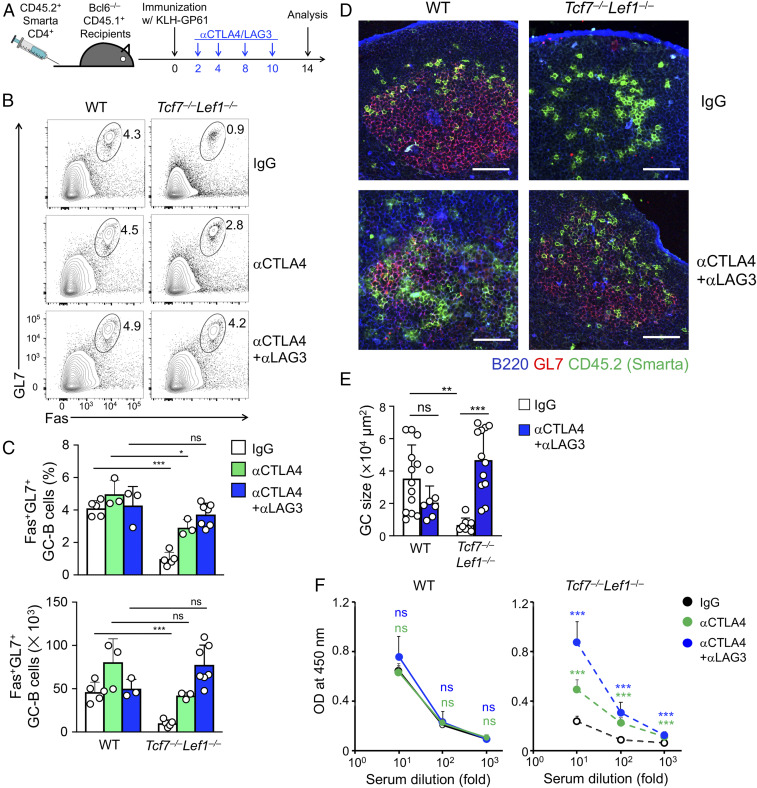
Because TFR cells are thymus derived (29), the adoptively transferred mature Smarta CD4+ T cells did not give rise to TFR cells, as confirmed by the absence of Foxp3 transcripts on RNA-seq and Foxp3 protein by intracellular staining in Smarta cells (SI Appendix, Fig. S4 A and C). In addition, the endogenous TFR response did not develop in Bcl6−/− recipients in our experimental system. We therefore hypothesized that the impaired GC-B cell responses in recipients of Tcf7−/−Lef1−/− cells were due to intrinsic effects of aberrantly induced CTLA4 and/or LAG3 on Tcf7−/−Lef1−/− TFH cells. Blocking the coinhibitory receptors is an effective approach to rectify T cell dysfunction (3), and in fact, treatment with CTLA4 antibodies enhances GC-B cell responses (6, 9). We thus tested treatment with anti-CTLA4 and/or anti-LAG3 in our adoptive transfer and KLH-GP61 immunization experimental system (Fig. 3A). When measured on day 5 postimmunization, WT and Tcf7−/−Lef1−/− TFH cells showed only a marginal increase in numbers by a single antibody treatment but were elevated approximately threefold by treatment with both antibodies (SI Appendix, Fig. S5A), with Bcl6, CTLA4, and LAG3 expression, per se, largely unaffected (SI Appendix, Fig. S5 B–D). On day 14 postimmunization, GC-B cells were not augmented in Bcl6−/− recipients of WT Smarta cells by anti-CTLA4/LAG3 treatment; in contrast, the reduced GC-B cell production in Bcl6−/− recipients of Tcf7−/−Lef1−/− Smarta cells was potently rectified by the combination therapy (Fig. 3 B and C). In addition, GC formation and KLH-specific antibody production were substantially restored in Tcf7−/−Lef1−/− recipients (Fig. 3 D–F). It is of note that treatment with anti-CTLA4 alone also partly restored GC-B cell formation and KLH antibody production (Fig. 3 B, C, and F), albeit it did not increase Tcf7−/−Lef1−/− TFH cell number (SI Appendix, Fig. S5A). These data indicate that blocking the coinhibitory pathways was sufficient to rectify B-cell help defects by Tcf1/Lef1-deficient TFH cells and indicate that a major function of Tcf1 and Lef1 TFs in Imm_TFH cells is to prevent aberrant induction of CTLA4 and LAG3.
Fig. 3.
Blocking CTLA4 and LAG3 restores B-cell help by Tcf1/Lef1-deficient TFH cells. (A) The experimental design for treatment with anti-CTLA4 and/or anti-LAG3 antibodies, where WT (1 × 105) or Tcf7−/−Lef1−/− (2 × 105) Smarta CD4+ T cells were transferred. (B and C) The detection of GC-B cells in Bcl6−/− recipients on day 14 postimmunization. The representative contour plots in B show percentages of GC-B cells in B220+CD19+ B cells in dLNs from two independent experiments, and cumulative data on percentages and numbers of GC-B cells in C are means ± SD. (D) The detection of GC formation (red) by immunofluorescence staining of dLNs from Bcl6−/− recipients on day 14 postimmunization. (Scale bars, 100 µm.) Data are representative from two to three experiments. (E) Cumulative data on GC sizes from at least two independent experiments with ≥ 2 LN sections measured for each experimental condition. (F) The detection of KLH-specific IgG in sera of Bcl6−/− recipients on day 14 postimmunization. OD450, optical density at 450 nm. Data are from two experiments (n = 3 to 5). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not statistically significant.
Tcf1 and Lef1 Restrain Chromatin Accessibility at the Coinhibitory Receptor Gene Loci.
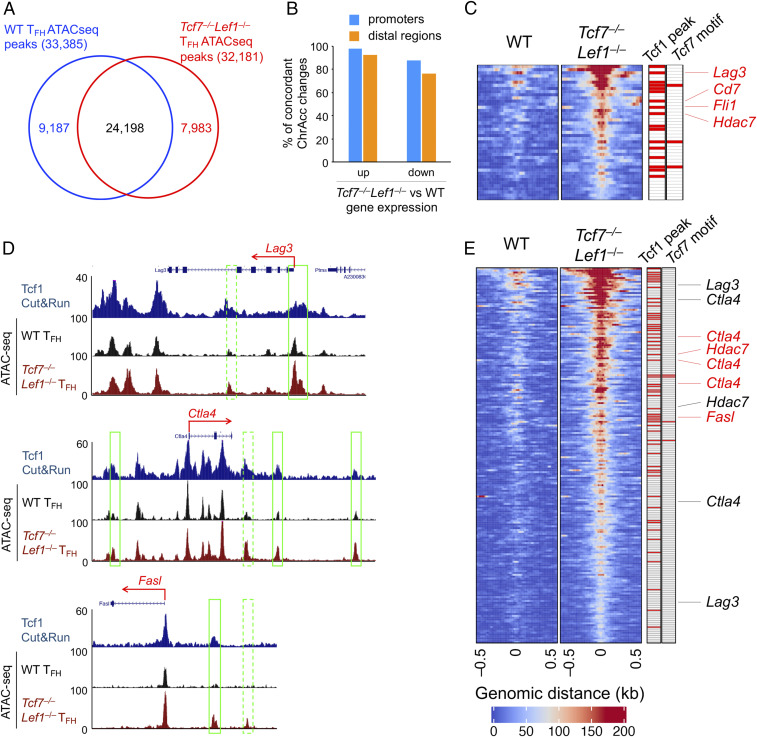
To determine the mechanisms underlying Tcf1/Lef1-mediated regulation of Imm_TFH cells, we used Cleavage Under Targets and Release Using Nuclease (CUT&RUN) to map global Tcf1-binding events in WT TFH cells and assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) to compare chromatin accessibility (ChrAcc) between WT and Tcf7−/−Lef1−/− TFH cells. Using P < 10−4 as a stringent cutoff in the Genrich algorithm, coupled with peak calling using model-based analysis of ChIP-seq version 2.0 (MACS2), a total of 21,602 Tcf1-binding peaks were identified in WT Imm_TFH cells. Taking a gene-centric approach with a focus on the differentially expressed genes (DEGs), Tcf1 peaks were found in promoter regions (defined as +/−3 kb flanking transcription start site (TSS)) of 85% down-regulated and 78% up-regulated genes in Tcf7−/−Lef1−/− TFH cells, as well as in distal regulatory regions (defined as gene body and its +/−50 kb flanking sequences) at even higher percentage of these DEGs (SI Appendix, Fig. S6A). In addition, ∼82% of Tcf1 peaks overlapped with ChrAcc sites, as determined by ATAC-seq in WT TFH cells (SI Appendix, Fig. S6B), consistent with previous reports (30, 31). A comparison between ATAC-seq peaks between WT and Tcf7−/−Lef1−/− TFH cells revealed consistent and extensive ChrAcc changes upon loss of Tcf1 and Lef1, with ∼22 and 19% of sites showing increased and decreased ChrAcc in Tcf7−/−Lef1−/− TFH cells, respectively (Fig. 4A and SI Appendix, Fig. S6C).
Fig. 4.
Tcf1 and Lef1 negatively regulate Ctla4 and Lag3 by limiting chromatin accessibility. (A) Venn diagram showing differential chromatin accessibility between WT and Tcf1/Lef1-deficient TFH cells, as determined by ATAC-seq. (B) Bar graphs summarizing the percentage of ChrAcc sites that show an increase in up-regulated or decrease in down-regulated genes in Tcf1/Lef1-deficient TFH cells, at either promoters or distal regulatory regions. (C) Heatmaps showing increased ChrAcc at the promoters of up-regulated genes in Tcf1/Lef1-deficient TFH cells. The two columns on the right denote the presence of Tcf1-binding peaks and Tcf7 motif with red lines. Select genes associated with increased ChrAcc are marked, with those in red denoting overlap with Tcf1 peak(s). (D) Tcf1 CUT&RUN track in WT TFH cells and ATAC-seq tracks of WT and Tcf1/Lef1-deficient TFH cells at select up-regulated gene loci. The structure and transcription direction are marked on top. The green rectangles mark increased ChrAcc sites Tcf1/Lef1-deficient TFH cells, with solid ones denoting overlap with Tcf1 peak(s). (E) Heatmaps showing increased ChrAcc at the distal regions of up-regulated genes in Tcf1/Lef1-deficient TFH cells as in C.
A focused analysis of DEG-associated ChrAcc sites showed that most ChrAcc changes, at either promoters or distal regulatory regions, were concordant with gene expression changes (Fig. 4B). Among the 157 up-regulated genes in Tcf7−/−Lef1−/− TFH cells, 37 gene promoters showed increased ChrAcc (Fig. 4C and SI Appendix, Fig. S6D), as observed near the TSS of Lag3 (Fig. 4D). In addition, 92 genes had increased ChrAcc sites in distal regulatory regions in Tcf7−/−Lef1−/− TFH cells (Fig. 4E and SI Appendix, Fig. S6D), as seen in both upstream and downstream sites flanking Ctla4, and upstream sites of Fasl (Fig. 4D). By stratifying with Tcf1 peaks, a substantial portion of the increased ChrAcc sites were bound by Tcf1, such as the Lag3 promoter and some distal sites to Ctla4 and Fasl (Fig. 4 C–E and SI Appendix, Fig. S6D). These observations suggest that a major function of Tcf1 and Lef1 is to restrain the ChrAcc open state to suppress coinhibitory receptor expression in Imm_TFH cells.
Among the 359 down-regulated genes in Tcf7−/−Lef1−/− TFH cells, 46 gene promoters showed decreased ChrAcc (Fig. 5A and SI Appendix, Fig. S7A), as observed at the TSS of Cxcr6 (Fig. 5B). In addition, 125 genes had decreased ChrAcc sites in distal regulatory regions in Tcf7−/−Lef1−/− TFH cells (Fig. 5C and SI Appendix, Fig. S7B), as seen in Stim1 intron and the downstream region of Il6st (Fig. 5B). We also noticed that 45 down-regulated genes in Tcf7−/−Lef1−/− TFH cells were associated with increased ChrAcc sites (SI Appendix, Fig. S7 B and C), as seen in Sell intron (SI Appendix, Fig. S7D), where the increased ChrAcc sites may function as transcriptional suppressors as our recent study suggested (30). By stratifying with Tcf1 peaks, most decreased ChrAcc sites were bound by Tcf1, such as the Cxcr6 promoter and some distal sites to Stim1 and Il6st (Fig. 5B). When observed globally, the decreased ChrAcc sites showed more frequent overlap with Tcf1 peaks than the increased ChrAcc sites did, and this was true for both promoters and distal regions (Fig. 5D). Tcf1 peaks associated with the decreased ChrAcc sites were greater in peak height and narrower in peak width than those associated with the increased ChrAcc sites (Fig. 5E; also compare Fig. 5B with Fig. 4D). Motif analyses further showed that the Tcf/Lef motif was the most enriched in Tcf1 peaks associated with the decreased ChrAcc sites (Fig. 5F); in contrast, Tcf1 peaks associated with the increased ChrAcc sites harbored Stat, Ets, Runx, Irf, and Egr motifs (Fig. 5G). In addition, the canonical Tcf/Lef motif (TCAAAG) was found in 16.3% of Tcf1 peaks associated with decreased ChrAcc (Fig. 5 A and C) but in only 8.7% of Tcf1 peaks associated with increased ChrAcc (Fig. 4 C and E and SI Appendix, Fig. S7C). These observations suggest that for Tcf1-activated genes (i.e., down-regulated in Tcf7−/−-Lef1−/− TFH cells), Tcf1 directly binds to these loci to maintain chromatin accessibility, while for Tcf1-repressed genes (i.e., up-regulated in Tcf7−/−Lef1−/− TFH cells), Tcf1 is likely recruited as a component in protein complexes to restrain chromatin accessibility.
Fig. 5.
Tcf1 and Lef1 positively regulate TFH genes by maintaining a chromatin-accessible state. (A) Heatmaps showing decreased ChrAcc at the promoters of down-regulated genes in Tcf1/Lef1-deficient TFH cells. The two columns on the right denote the presence of Tcf1-binding peaks and Tcf7 motif with red lines. Select genes associated with decreased ChrAcc are marked, with those in red denoting an overlap with Tcf1 peak(s). (B) The Tcf1 CUT&RUN track in WT TFH cells and ATAC-seq tracks of WT and Tcf1/Lef1-deficient TFH cells at select down-regulated gene loci. The structure and transcription direction are marked on top. The green rectangles mark decreased ChrAcc sites, with solid ones denoting overlap with Tcf1 peak(s). (C) Heatmaps showing decreased ChrAcc at the distal regions of down-regulated genes in Tcf1/Lef1-deficient TFH cells as in A. (D) A bar graph summarizing the frequency of Tcf1-binding to differential ChrAcc sites at promoters and distal regulatory regions between Tcf7−/−Lef1−/− and WT TFH cells. (E) Box plots showing Tcf1 peak heights (Left) or width (Right) associated with differential ChrAcc sites between Tcf7−/−Lef1−/− and WT TFH cells. The statistical significance is determined by Student’s t test. (F and G) Motif analysis of Tcf1 peaks associated with decreased (F) or increased (G) ChrAcc sites in Tcf7−/−Lef1−/− TFH cells. The top five motifs of the transcription factor family are shown together with motif logos and statistical significance.
Groucho/Tle Corepressors Are Essential for TFH Cell Proliferation.
To investigate how Tcf1 and Lef1 achieve negative regulation of the coinhibitory pathways in Imm_TFH cells, we examined the Groucho/Tle corepressors, which are well documented to interact with Tcf/Lef TFs for transcriptional repression (22, 23). Our previous analysis of Tle1, 3, and 4 deficiency in thymic development revealed strong functional redundancy and gene dosage dependency among Tle genes, with ablating Tle3 showing stronger effect than ablating Tle1 and/or Tle4 (24). To fully address the redundancy issue, we conditionally targeted Tle2 (SI Appendix, Fig. S8 A and B), generated Smarta+Rosa26GFP hCD2-Cre+ Tle1-, 2-, 3-, and 4-floxed mice, and investigated their roles in TFH cells using adoptive transfer and the KLH-GP61 immunization approach. Deleting one allele of each Tle gene (Tle1FL/+Tle2FL/+Tle3FL/+Tle4FL/+, i.e., Tle1234+/−) or both alleles of Tle1, 2, and 4 along with one allele of Tle3 (Tle1FL/FLTle2FL/FLTle3FL/+Tle4FL/FL, i.e., Tle124−/−Tle3+/−) did not affect CD4+ T cell expansion in response to protein immunization (SI Appendix, Fig. S8C). Ablation of all four Tle proteins (Tle1FL/FLTle2FL/FLTle3FL/FLTle4FL/FL, i.e., Tle1234−/−), however, greatly diminished the expansion of Smarta CD4+ T cells (Fig. 6A). On the other hand, ablation of Tle3 together with one allele of Tle1, 2, and 4 (Tle1FL/+Tle2FL/+Tle3FL/FLTle4FL/+, i.e., Tle3−/−Tle124+/−) alleviated the expansion defects observed with Tle1234−/− cells (Fig. 6A). The defective proliferation of Tle1234-deficient CD4+ T cells was also validated when they were activated by acute viral infection and was observed at the early division stage (Fig. 6B).
Fig. 6.
Tle corepressors are highly redundant in sustaining TFH cell expansion and B-cell help. (A) Detection of CD45.2+ Smarta CD4+ T cell expansion. WT, Tle3−/−Tle124+/−, and Tle1234−/− (1 × 105 each) Smarta CD4+ T cells were adoptively transferred and recipients immunized as in Fig. 1A. On day 5 postimmunization, CD45.2+GFP+ Smarta cells were detected in TCRβ+CD4+ cells from dLNs. Representative contour plots (Left) are from two to three independent experiments, and cumulative data on Smarta cell frequency and numbers (Right) are means ± SD. (B) The detection of CD4+ T cell activation and early division. WT or Tle1234−/− Smarta CD4+ T cells were labeled with cell trace violet (CTV) and adoptively transferred at 2 × 106 cells/recipient, followed by intravenous infection with LCMV-Armstrong. CD45.2+Smarta CD4+ T cells were identified in the recipient spleens and assessed for CD44 expression (as an activation marker) and CTV dilution 60 h later. Representative dot plots (Left) show the frequency of cells in third to fifth divisions, and cumulative data from two experiments are shown in stacked bar graphs (Right). (C) The detection of CXCR5+PD1+ TFH cells in CD45.2+GFP+ Smarta CD4+ T cells on day 5 postimmunization. Representative contour plots (Left) are from two to three independent experiments, and cumulative data on TFH cell frequency and numbers (Right) are means ± SD. (D) The detection of GC-B cells in Bcl6−/− recipients on day 14 postimmunization. Representative contour plots (Left) show percentages of GC-B cells in B220+CD19+ B cells in dLNs from two to three independent experiments, and cumulative data on GC-B cells (Right) are means ± SD. (E) The detection of KLH-specific IgG in sera of Bcl6−/− recipients on day 14 postimmunization. Data are from two experiments (n = 5 to 6). OD450, optical density at 450 nm. (F) The detection of CTLA4 and LAG3 proteins in Tle-targeted TFH cells on day 5 postimmunization. In half-stacked histographs, the dotted vertical lines mark signal strength by isotype staining, and values denote geometric mean fluorescence intensity (gMFI). The bar graphs are means ± SD of relative gMFI from two experiments. **P < 0.01; ***P < 0.001; ns, not statistically significant.
Despite the limited expansion, the activated Tle3−/−Tle124+/− or Tle1234−/− CD4+ T cells exhibited similar frequency of CXCR5+PD1+ TFH cells as WT cells (Fig. 6C), or even higher frequency of CXCR5+Bcl6+ GC-TFH cells on day 5 after immunization (SI Appendix, Fig. S8D), suggesting that Tle proteins are not necessary for activated CD4+ T cells to differentiate to the TFH lineage. Whereas the numbers of Tle3−/−Tle124+/− and WT CXCR5+PD1+ TFH or GC-TFH cells were not significantly different, the counts of Tle1234−/− TFH or GC-TFH cells were greatly diminished (Fig. 6C and SI Appendix, Fig. S8D). Consistent with numerical changes in Imm_TFH cells, Bcl6−/− recipients of Tle1234−/− cells but not those of Tle3−/−Tle124+/− cells showed diminished GC-B cells and failed to produce KLH-specific antibodies (Fig. 6 D and E). However, neither Tle3−/−-Tle124+/− nor Tle1234−/− TFH cells exhibited elevated expression of CTLA4 or LAG3 (Fig. 6F). Taken together, these observations suggest that Tle genes are highly redundant in regulating proliferative capacity of activated CD4+ T cells, likely due to gene duplication during evolution to guard this important function. As a result, Tle corepressors are required for TFH cell expansion, albeit such a requirement does not involve negative regulation of coinhibitory receptors in cooperation with Tcf/Lef TFs.
Tcf1/Lef1-Mediated Ctla4 Repression Requires Their Intrinsic HDAC Activity.
Tcf1 and Lef1 TFs have the unusual capacity to modify histones by deacetylation, and this activity is critical for suppressing CD4+ lineage-associated genes in CD8+ single-positive thymocytes (25). To investigate if Tcf1 HDAC activity contributes to broader T cell activities, we used the CRISPR/Cas9 technique to mutate five amino acids in germline (called Tcf7M allele herein, Fig. 7A). These five amino acids show a high degree of conservation between Tcf1/Lef1 HDAC domains and conventional HDAC domains, and mutating these amino acids compromised Tcf1/Lef1 HDAC activity (25) but did not detectably affect Tcf1 protein expression levels (Fig. 7B). To exclude compensatory effects by Lef1-derived HDAC activity, we generated Smarta+hCD2-Cre+Tcf7M/MLef1FL/FL (Tcf7M/MLef1−/−) mice. Because the Tcf7M allele was generated in germline, to avoid a potential impact of Tcf7M/M mutation on thymic development (to be reported elsewhere) and achieve conditional expression of Tcf1 HDAC mutant protein, we generated Smarta+hCD2-Cre+Tcf7M/FLLef1FL/FL (Tcf7M/–Lef1−/−) mice. Smarta+CD4+ T cells from both mutant strains or WT controls were adoptively transferred into congenic mice, followed by KLH-GP61 immunization, as in Fig. 1A. Both Tcf7M/MLef1−/− and Tcf7M/–Lef1−/− Smarta cells gave rise to similar numbers of CXCR5+PD1+ TFH and CXCR5+Bcl6+ GC-TFH cells on day 5 postimmunization as in WT Smarta cells, albeit both populations were detected at a modestly higher frequency in Tcf7M/MLef1−/− or Tcf7M/–Lef1−/− cells (Fig. 7 C and D). Whereas Bcl6 and LAG3 expression was similar among WT (Tcf7M/MLef1−/− and Tcf7M/–Lef1−/− TFH cells), CTLA4 expression was evidently elevated in Tcf7M/MLef1−/− and Tcf7M/–Lef1−/− TFH cells (Fig. 7 D and E), albeit the elevated CTLA4 expression was not as profound as that in Tcf7−/−Lef1−/− TFH cells. Given the similar impact on TFH cells between Tcf7M/MLef1−/− and Tcf7M/–Lef1−/− mutations, we transferred the latter into Bcl6−/− recipients. On day 14 postimmunization, generations of GC-B cells and production of KLH-specific antibodies were compromised in Bcl6−/− recipients of Tcf7M/–Lef1−/− cells (Fig. 7 F and G). Collectively, these findings support a direct role of Tcf1 HDAC activity in restraining CTLA4 expression in TFH cells and thus guarding critical TFH functions in helping B cells.
Fig. 7.
Tcf1 HDAC activity is necessary for restraining CTLA4 expression in TFH cells. (A) Alignment of WT and mutant peptide sequences derived from Tcf7 exon 5. Highlighted in red are the five amino acids conserved between Tcf1/Lef1 HDAC domains and conventional HDAC domains (Top) and corresponding mutations (Bottom) by CRISPR-mediated exon 5 editing. (B) The detection of Tcf1 protein expression levels. Thymocytes were collected from WT or Tcf7M/M mice, and TCRβ+CD4+ or TCRβ+CD8+ thymocytes were intracellularly stained for Tcf1 expression. Representative half-stacked histograms are from two independent experiments, and values denote geometric mean fluorescence intensity (gMFI). (C and D) The detection of CXCR5+PD1+ (C) and CXCR5+Bcl6+ (D) TFH cells. WT, Tcf7M/MLef1−/−, and Tcf7M/–Lef1−/− Smarta CD4+ T cells were adoptively transferred into CD45.1+ WT B6.SJL recipients, which were immunized with KLH-GP61 24 h later. On day 5 postimmunization, PD1 and Bcl6 were detected in CD45.2+CXCR5+ Smarta cells in dLNs. Representative contour plots are from two to four independent experiments (Left) and cumulative data on TFH cell frequency and numbers (Right) are means ± SD. In D, the relative Bcl6 expression is shown. (E) The detection of CTLA4 and LAG3 proteins in Tcf1 HDAC-mutant TFH cells on day 5 postimmunization. In half-stacked histographs, the dotted vertical lines mark signal strength by isotype staining, and values denote gMFI. Bar graphs are means ± SD of relative gMFI from three to five experiments. (F) The detection of GC-B cells in Bcl6−/− recipients on day 14 after immunizing the recipients of WT or Tcf7M/–Lef1−/− Smarta CD4+ T cells. Representative contour plots show percentages of GC-B cells in B220+CD19+ B cells in dLNs from two experiments, and cumulative data on percentages and numbers of GC-B cells are means ± SD. (G) The detection of KLH-specific IgG in sera of Bcl6−/− recipients on day 14 after immunizing the recipients of WT or Tcf7M/–Lef1−/− Smarta CD4+ T cells. Data are from three experiments (n = 9). OD450, optical density at 450 nm. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not statistically significant.
Discussion
The coinhibitory pathways are critical for preventing overexuberant immune responses. CTLA4 is deployed by multiple cell types including TFH, TREG, and TFR cells to control the magnitude of humoral responses (5, 8). Because of the strong immunosuppressive function by TREG and TFR cells, the expression of CTLA4 in TFH cells must be carefully modulated to avoid excessive, undue inhibition of GC-B cell responses. A previous report showed that Foxp1 contributes to induction of CTLA4 in TFH cells (9). This study identified Tcf1 and Lef1 TFs as key regulators that restrain CTLA4 and LAG3 coinhibitory receptors in TFH cells elicited by protein immunization. Blocking CTLA4 alone showed marginally improved expansions of Tcf1/Lef1-deficient TFH cells but showed more pronounced enhancement of GC-B cells, suggesting both cell-extrinsic and -intrinsic effects were involved. In addition, blocking both CTLA4 and LAG3 further expanded Tcf1/Lef1-deficient TFH cells and enhanced GC-B cell responses, highlighting the requirement for Tcf1 and Lef1 to mitigate excessive cell-intrinsic inhibitory signals. On the flip side, the break that Tcf1 and Lef1 put on CTLA4 and LAG3 expression could be released in autoimmune conditions to achieve therapeutic effects (32). Our observation that Tcf1 HDAC activity contributed to Ctla4 repression makes Tcf1 a more amenable therapeutic target in pathogenic TFH cells. This is because Tcf1 HADC activity shows selective sensitivity to HDAC inhibitors, such as Vorinostat and Tubacin (25).
It has been recognized that Tcf1, especially when ectopically expressed, increases chromatin accessibility to modulate target gene expression in exhausted CD8+ T cells and even fibroblasts (30, 33). In line with these observations, a predominant function of Tcf1 and Lef1 in Imm_TFH cells was to regulate transcriptional activation of downstream genes, at least in part through maintaining chromatin accessibility at the promoter and/or distal regulatory elements. Despite prevailing Tcf1 binding to 70 to 80% of these ChrAcc sites, we did notice that only 16% of these Tcf1 peaks had the canonical Tcf/Lef motif, which is historically defined as a Wnt-responsive element. Because Wnt-stabilized β-catenin does not have prominent roles in T lineage cells (20), Tcf1 and Lef1 may engage other cofactors for transcriptional activation, which may alter their preferred DNA sequences in T cells. Additionally, nonvertebrate Tcf ortholog binds to a GC-rich element through a separate “E-tail” domain located at the C terminus of its conventional high-mobility–group (HMG) DNA-binding domain (34). Besides direct contact with DNA elements, Tcf1 and Lef1 could be recruited to these sites by interacting with other DNA-bound TFs.
In addition to transactivation, Tcf1 and Lef1 were essential to negatively control chromatin accessibility to exert transcriptional repression, to suppress counterproductive pathways in TFH cells. The Tcf1 peaks detected at the Tcf1-restrained sites were wider and lower compared with those detected at the Tcf1-opened sites; empirically, this observation suggests that Tcf1 is indirectly recruited as a component in a larger complex to the former sites. This notion is consistent with the observed requirement for Tcf1 HDAC activity, but not Tle corepressors, in suppressing CTLA4 expression in Imm_TFH cells. Collectively, we postulate that Tcf1 is recruited as an HDAC rather than in complex with Tle corepressors to limit accessibility to the Clta4 locus. We recognize that Tcf1 HDAC activity may not be the sole mechanism responsible for the transcriptional repression of the coinhibitory receptors because Tcf1 HDAC mutation did not cause induction of LAG3 and the derepression of CTLA4 was not as profound as Tcf1 null mutation. Complete elucidation of repressive means that cooperate with Tcf1 HDAC await future investigations. In TREG cells, we previously found that ablating Tcf1 and Lef1, but not either factor alone, resulted in evident elevation of CTLA4 expression (14), suggesting that Tcf1/Lef1-mediated CTLA4 repression is a conserved regulatory circuit in multiple T lineage cells.
This study also revealed stark differences in the requirements for Tcf1 and Lef1 in infection- and Imm_TFH cells, despite some shared downstream genes. In Inf_TFH cells, the prominent roles of Tcf1 and Lef1 are in two key aspects; one is transactivation of Bcl6 and Icos, and the other is repression of Prdm1 (16–18). Bcl6 and Blimp1 are mutually antagonistic, and forced expression of Bcl6 or genetic ablation of Blimp1 in Tcf1-deficient CD4+ T cells rectifies TFH differentiation defects (27). In contrast, neither Bcl6 transactivation nor Blimp1 repression was perturbed in Tcf1/Lef1-deficient Imm_TFH cells. Because Tcf1 and Lef1 are no longer needed to balance Bcl6 and Blimp1 expression in Imm_TFH cells, their functions are dedicated to transcriptional repression of Ctla4 and Lag3. This finding highlights the necessity to systematically examine molecular determinants and their functional requirements in the specific context how the TFH response is activated. This has important bearings on vaccine design because vaccines are delivered as live-attenuated viruses, inactivated organisms, or protein subcomponents via different routes (35). It remains to be elucidated why the same factors/pathways are differentially utilized in the TFH program in response to infectious agents and protein immunization, although likely mechanistic insights might be acquired by examining the differences in the strength and duration of T cell receptor (TCR) stimulation, balance and kinetics of costimulatory and coinhibitory signals, and the cytokine milieu in future investigations. Nonetheless, our studies revealed a regulatory role for Tcf1 and Lef1 TFs to restrain coinhibitory receptors in TFH cells and guard B-cell help function, and this essential function is mediated by limiting chromatin accessibility of key gene loci through their intrinsic HDAC activity.
Materials and Methods
Mice.
C57BL/6J (B6), B6.SJL, Bcl6FL/FL, CD4-Cre transgenic, and Rosa26GFP mice were from the Jackson Laboratory. Tcf7FL/FL, Lef1FL/FL, Tle1FL/FL, Tle3FL/FL, and Tle4FL/FL mice were previously described (24, 36–39); Tcf1 HDAC mutant and Tle2-floxed mice were generated in this study and hCD2-Cre mice were provided by Paul E. Love (National Institute of Child Health and Human Development, NIH). All compound mouse strains used in this work were from in-house breeding at the animal care facilities of University of Iowa and Center for Discovery and Innovation, Hackensack University Medical Center. All mice analyzed were 6 to 12 wk of age, and both genders were used without randomization or blinding. All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committees of the University of Iowa and Center for Discovery and Innovation, Hackensack University Medical Center.
Supplementary Material
Acknowledgments
We thank Drs. Thomas J. Waldschmidt (University of Iowa) and Shane Crotty (La Jolla Institute) for scientific input, the University of Iowa Flow Cytometry Core facility (J. Fishbaugh, H. Vignes, and G. Rasmussen) for cell sorting, and the University of Iowa Central Microscopy Research Facility (J. Shao) for immunofluorescence imaging. This study is supported, in part, by grants from the NIH (AI112579, AI121080, and AI139874 to H.-H.X. and P30 CA16056-42 to J.W.) and the Veteran Affairs Biomedical Laboratory Research and Development Merit Review Program (BX002903 to H.-H.X.). F.L. is supported, in part, by the National Natural Science Foundation of China (31801222, 32070888, and 82022031).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014562118/-/DCSupplemental.
Data Availability.
RNA-seq, ATAC-seq, and Tcf1 CUT&RUN were performed and analyzed as detailed in SI Appendix, Supplementary Methods. All of the NextGen sequencing data are deposited at the Gene Expression Omnibus under the SuperSeries GSE146428 (40).
References
- 1.Schildberg F. A., Klein S. R., Freeman G. J., Sharpe A. H., Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 44, 955–972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Q., Vignali D. A., Co-stimulatory and co-inhibitory pathways in autoimmunity. Immunity 44, 1034–1051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callahan M. K., Postow M. A., Wolchok J. D., Targeting T., Targeting T cell co-receptors for cancer therapy. Immunity 44, 1069–1078 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Attanasio J., Wherry E. J., Costimulatory and coinhibitory receptor pathways in infectious disease. Immunity 44, 1052–1068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sage P. T., Paterson A. M., Lovitch S. B., Sharpe A. H., The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity 41, 1026–1039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C. J., et al. , CTLA-4 controls follicular helper T-cell differentiation by regulating the strength of CD28 engagement. Proc. Natl. Acad. Sci. U.S.A. 112, 524–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker L. S., et al. , Established T cell-driven germinal center B cell proliferation is independent of CD28 signaling but is tightly regulated through CTLA-4. J. Immunol. 170, 91–98 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Wing J. B., Ise W., Kurosaki T., Sakaguchi S., Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity 41, 1013–1025 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Shi B., et al. , Foxp1 negatively regulates T follicular helper cell differentiation and germinal center responses by controlling cell migration and CTLA-4. J. Immunol. 200, 586–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinke F. C., Xue H. H., From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol. Res. 59, 45–55 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Raghu D., Xue H. H., Mielke L. A., Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol. 40, 1149–1162 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., et al. , TCF-1 inhibits IL-17 gene expression to restrain Th17 immunity in a stage-specific manner. J. Immunol. 200, 3397–3406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Q., et al. , T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat. Immunol. 10, 992–999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing S., et al. , Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J. Exp. Med. 216, 847–866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang B. H., et al. , TCF1 and LEF1 control Treg competitive survival and Tfr development to prevent autoimmune diseases. Cell Rep. 27, 3629–3645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L., et al. , The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat. Immunol. 16, 991–999 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Choi Y. S., et al. , LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16, 980–990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu T., et al. , TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep. 12, 2099–2110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gullicksrud J. A., et al. , Differential requirements for Tcf1 long isoforms in CD8+ and CD4+ T cell responses to acute viral infection. J. Immunol. 199, 911–919 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X., et al. , β-catenin and γ-catenin are dispensable for T lymphocytes and AML leukemic stem cells. eLife 9, e55360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., et al. , Ezh2 programs TFH differentiation by integrating phosphorylation-dependent activation of Bcl6 and polycomb-dependent repression of p19Arf. Nat. Commun. 9, 5452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brantjes H., Roose J., van De Wetering M., Clevers H., All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29, 1410–1419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal M., Kumar P., Mathew S. J., The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 67, 472–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing S., et al. , Tle corepressors are differentially partitioned to instruct CD8+ T cell lineage choice and identity. J. Exp. Med. 215, 2211–2226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing S., et al. , Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 17, 695–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X., Xue H. H., Cutting edge: Generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J. Immunol. 189, 2722–2726 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao P., et al. , Cutting edge: Tcf1 instructs T follicular helper cell differentiation by repressing Blimp1 in response to acute viral infection. J. Immunol. 203, 801–806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., et al. , The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat. Immunol. 15, 667–675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sage P. T., Sharpe A. H., T follicular regulatory cells. Immunol. Rev. 271, 246–259 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Shan Q., et al. , Ectopic Tcf1 expression instills a stem-like program in exhausted CD8+ T cells to enhance viral and tumor immunity. Cell. Mol. Immunol. 10.1038/s41423-020-0436-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harly C., et al. , The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat. Immunol. 20, 1150–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gensous N., et al. , T follicular helper cells in autoimmune disorders. Front. Immunol. 9, 1637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson J. L., et al. , Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity 48, 243–257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atcha F. A., et al. , A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol. Cell. Biol. 27, 8352–8363 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piot P., et al. , Immunization: Vital progress, unfinished agenda. Nature 575, 119–129 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Steinke F. C., et al. , TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 15, 646–656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S., et al. , The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813–826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheat J. C., et al. , The corepressor Tle4 is a novel regulator of murine hematopoiesis and bone development. PLoS One 9, e105557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramasamy S., et al. , Tle1 tumor suppressor negatively regulates inflammation in vivo and modulates NF-κB inflammatory pathway. Proc. Natl. Acad. Sci. U.S.A. 113, 1871–1876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue H. H., Tcf1 and Lef1 regulate follicular helper T cell function elicited by vaccination. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146428. Deposited 5 March 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Xue H. H., Tcf1 and Lef1 regulate follicular helper T cell function elicited by vaccination. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146428. Deposited 5 March 2020.
Supplementary Materials
Data Availability Statement
RNA-seq, ATAC-seq, and Tcf1 CUT&RUN were performed and analyzed as detailed in SI Appendix, Supplementary Methods. All of the NextGen sequencing data are deposited at the Gene Expression Omnibus under the SuperSeries GSE146428 (40).