Significance
Enteropathogenic and enterhemorrhagic Escherichia coli (EPEC and EHEC) are the etiological agents of morbidity and mortality worldwide. Both pathogens use a type III secretion system (T3SS) to inject effectors into mammalian cells, taking control of cellular processes. Unlike other pathogens using T3SS, EPEC and EHEC have evolved a filamentous extension to the T3SS needle, formed by the translocator protein EspA. We determined the atomic structure of the EspA filament using cryoelectron microscopy. The EspA filament is a nearly seamless extension of the needle, although the sequences of EspA and the needle protein share little similarity. The atomic structure of the EspA filament fills a significant gap in our understanding of the architecture of the T3SS expressed by EPEC and EHEC.
Keywords: bacterial pathogenesis, intestinal epithelium, attaching/effacing pathogens
Abstract
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic Escherichia coli (EHEC) utilize a macromolecular type III secretion system (T3SS) to inject effector proteins into eukaryotic cells. This apparatus spans the inner and outer bacterial membranes and includes a helical needle protruding into the extracellular space. Thus far observed only in EPEC and EHEC and not found in other pathogenic Gram-negative bacteria that have a T3SS is an additional helical filament made by the EspA protein that forms a long extension to the needle, mediating both attachment to eukaryotic cells and transport of effector proteins through the intestinal mucus layer. Here, we present the structure of the EspA filament from EPEC at 3.4 Å resolution. The structure reveals that the EspA filament is a right-handed 1-start helical assembly with a conserved lumen architecture with respect to the needle to ensure the seamless transport of unfolded cargos en route to the target cell. This functional conservation is despite the fact that there is little apparent overall conservation at the level of sequence or structure with the needle. We also unveil the molecular details of the immunodominant EspA epitope that can now be exploited for the rational design of epitope display systems.
Enteropathogenic Escherichia coli (EPEC) is a major cause of infantile diarrhea, morbidity, and mortality in low- and middle-income countries (1), while enterohemorrhagic E. coli (EHEC) is a major cause of food poisoning in industrial countries (2). The ability of EPEC and EHEC to colonize the intestinal epithelium is encoded on a pathogenicity island named the locus of enterocyte effacement, which encodes gene regulators, the outer membrane adhesin intimin, structural components of a type III secretion system (T3SS), translocon components, chaperones, effectors, and an ATPase, which energizes protein translocation (3).
T3SS is a common virulence factor among Gram-negative pathogens of humans, animals, and plants, including Salmonella enterica serovars, Shigella, Chlamydia, and Yersinia spp., Pseudomonas aeruginosa, and Pseudomonas syringae (4). The overall architecture of the multisubunit T3SS injectisome, which spans the entire cell envelope, is highly conserved among the different pathogens. It comprises several substructures, including a cytosolic C-ring and an ATPase complex (EscN in EPEC), a basal body consisting of a series of ring structures embedded in the bacterial inner and outer membranes (including a Secretin, EscC) and a periplasmic rod (EscI) which connects the inner membrane rings with a hollow extracellular needle projection (EscF). In EPEC, the EscF needle is 8–9 nm in diameter and 23 nm in length (5–7). In most pathogenic bacteria having a T3SS, the function of the needle is to connect the basal body to a translocation pore in the plasma membrane of the eukaryotic cell (3, 4).
Electron microscopic observations of EPEC and EHEC have shown an ∼12-nm-diameter helical tube, made of the secreted translocator protein EspA (8–10), which serves as an extension to the needle, enclosing a central channel of ∼25 Å diameter (11, 12). Around 12 EspA filaments are elaborated on individual EPEC bacteria (8); when grown in vitro, EspA can vary in length and can reach 600 nm (7). Our current understanding is that the EPEC and EHEC EspA filaments evolved as an adaptation to their environment, where the needle alone would not be long enough to traverse the intestinal mucus layer. EspA filaments, like the needle, share similar helical symmetry parameters with flagellar structures and are elongated by addition of EspA subunits to the tip of the growing filament, the same mode of elongation that occurs in flagella filaments (13). Functionally, the filaments form a long flexible helical conduit which connects the tip of the needle with the translocation pore (made in EPEC by EspB and EspD), thus mediating effector translocation (3). Indeed, EspA has been shown to interact with both EspB and EspD (14, 15). In addition to their protein translocation activity, EspA filaments are important adhesins, mediating binding to both epithelial cells and edible leaves (16, 17). In the absence of EspA filaments, effectors can be secreted but not translocated; accordingly, the virulence of an espA deletion is highly attenuated in animal models (18). Moreover, EspA filaments are major antigens in vivo; antibodies against EspA were found in both human colostrum of mothers in Brazil and in serum from culture-positive patients infected with EHEC (19, 20). In animal models, IgG antibodies against EspA play a major role in clearing the pathogen (21).
EspA alone is sufficient to form filamentous structures (22). Similarly to flagellar biosynthesis (23), EspA coiled-coil interactions between N- and C-terminal α-helical segments are required for assembly of the filament (24, 25). Monomeric EspA subunits are maintained in the cytosol via interactions with the chaperone CesAB (26), which has also been called CesA (22). CesAB is essential for stability of EspA within the bacterial cell prior to secretion. A cesAB deletion cannot secrete EspA or assemble EspA filaments (26). Crystallographic analysis of the CesAB–EspA complex at 2.8 Å resolution (22) showed that the EspA α-helices are also involved in extensive coiled-coil interactions with CesAB (22). Due to disorder in the parts of EspA not directly interacting with CesAB, only 72 of the 192 EspA residues were visualized in this complex. Importantly, the ATPase EscN selectively interacts with the CesAB–EspA complex; abrogation of this interaction attenuates EspA secretion and infection (27).
Like flagella, EspA filaments show antigenic polymorphism, as EspA from different EPEC and EHEC clones show no immunological cross-reactivity (10). We have previously identified a surface-exposed hypervariable domain that contains the immunodominant EspA epitope (28). By exchanging the hypervariable domains of EspA(EPEC) and EspA(EHEC) we swapped the antigenic specificity of the EspA filaments (26). As with the Salmonella flagellin D3 domain (29), which is known to tolerate insertions of natural and artificial amino acid sequences (30), we were able to insert short peptides into the surface-exposed, hypervariable region of EspA (28).
While EspA was first identified in 1996 (31), and the EspA filaments were first described in 1998 (8), low-resolution structures (at ∼15–25 Å resolution) were reported about 15 y ago (11, 12). The aim of this study was to obtain a high-resolution cryoelectron microscopy (cryo-EM) structure of the EPEC EspA filament.
Results
Cryo-EM Reconstruction of the EspA Filament.
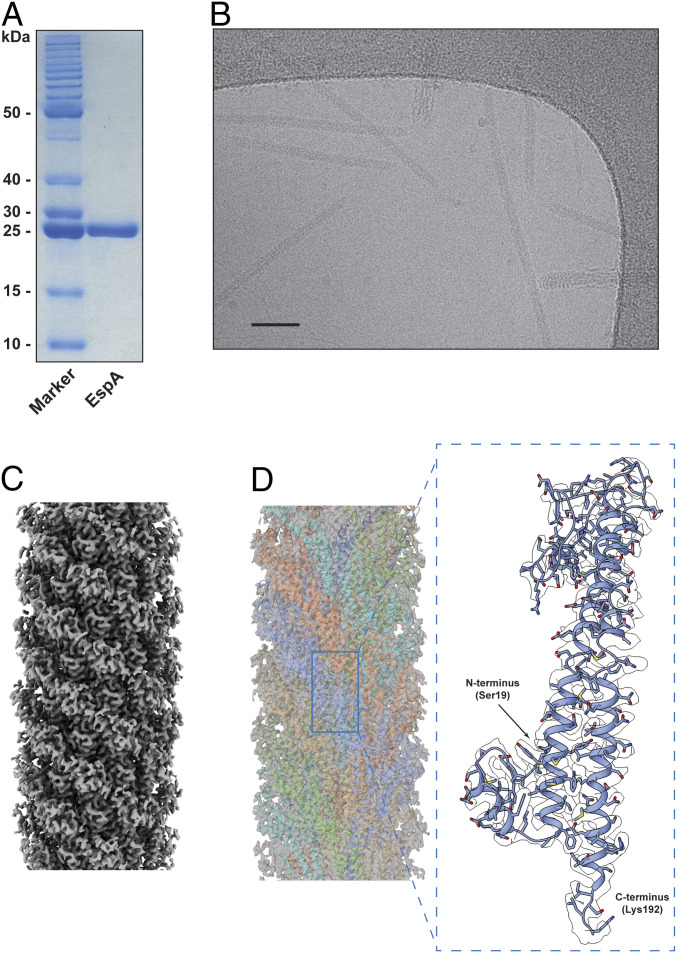
We used cryo-EM to determine the structure of the EspA filament at 3.4 Å resolution (Fig. 1, SI Appendix, Fig. S1, and Table 1). Cryo-EM images showed that the EspA filaments are straight helical rods with a narrow central channel (Fig. 1B). The three-dimensional (3D) density map clearly shows many side chains as well as the helical packing of the EspA filament (Fig. 1D), generated by a right-handed 1-start helix with a rise of 4.4 Å and twist of 64.3° per subunit. Therefore, the EspA filament has ∼5.6 subunits per turn of an ∼25 Å pitch helix, which is quite similar to the values found for a number of bacterial flagellar filaments (32).
Fig. 1.
Purification of the EspA filament and cryo-EM reconstruction at 3.4 Å. (A) SDS-PAGE of purified EspA filament. (B) Electron micrograph of EspA pili. (Scale bar, 50 nm.) (C) Side view of the EspA filament cryo-EM map. (D) The EspA filament cryo-EM map was fitted with the derived atomic model (Left), and the five strands are colored in cyan, orange, blue, brown, and green. Details of the EspA subunit density (Right) with the EspA model built into the density from Ser19 to Lys192 and shown in ribbon representation.
Table 1.
Cryo-EM data collection, processing, and model refinement statistics of the EspA filament
| Parameter value | |
| Data collection and processing | |
| Microscope | Titan Krios |
| Camera | K3 |
| Voltage (kV) | 300 |
| Electron exposure (e− Å−2) | 20 |
| Pixel size (Å) | 1.08 |
| Defocus range (μm) | −1.0 to −2.5 |
| Helical rise (Å) | 4.4 |
| Helical twist (°) | 64.3 |
| Map Resolution (Å) | 3.4 |
| Map B factor | −139 |
| Refinement and Model Validation | |
| Bond lengths rmsd (Å) | 0.007 |
| Bond angles rmsd (°) | 1.265 |
| Clashscore | 21.96 |
| Poor rotamers (%) | 0 |
| Ramachandran favored (%) | 90.70 |
| Ramachandran outlier (%) | 0 |
| MolProbity score | 2.43 |
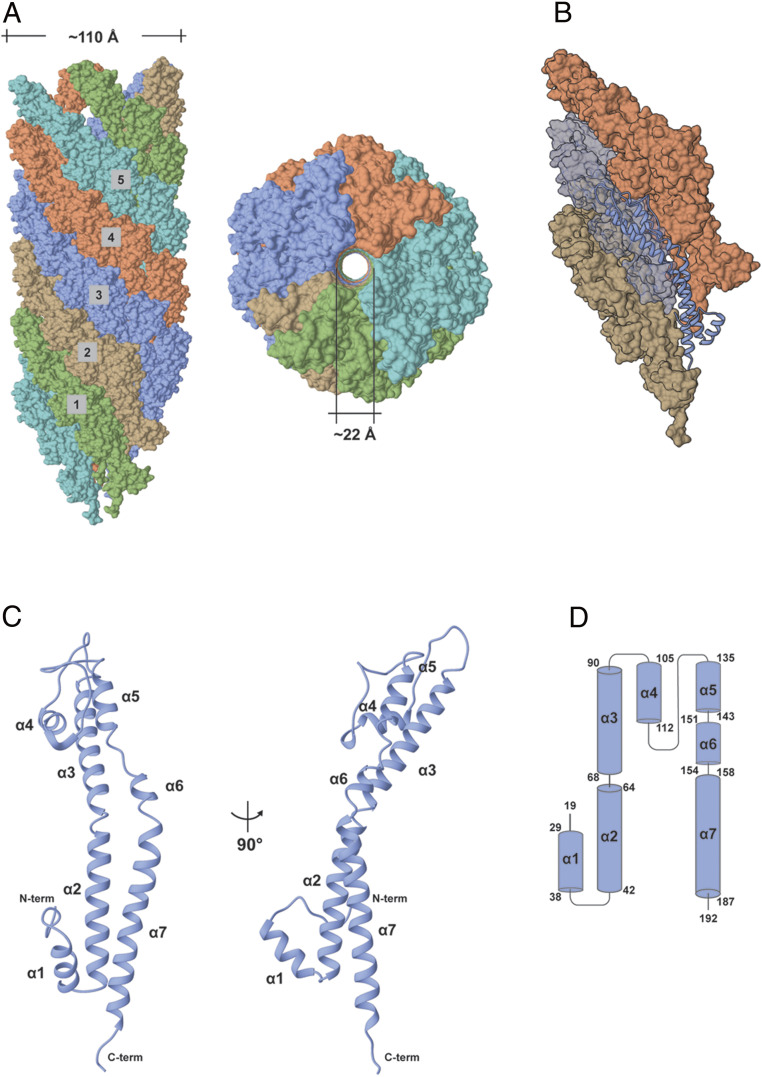
General Architecture of the EspA Filament and Subunit.
The structure of the EspA filament shows a helical assembly with overall dimensions of ∼110 Å in diameter and an internal lumen of ∼22 Å in diameter (Fig. 2A). The filament has a 1-start helix relating every subunit, but can also be viewed as five helical strands (Fig. 2A) each with ∼10 EspA subunits per turn. The EspA protein has 192 residues, and all its amino acids could be traced in the cryo-EM electron density, with the exception of the 18 N-terminal residues that are not seen, presumably due to disorder. It has previously been shown that the first 20 amino acids in EspA contain a T3SS secretion signal (33). Replacing the 20 N-terminal amino acids in the effector Map with the corresponding 20 residues of EspA mediated protein translocation, while a random 20 residue sequence failed to support translocation. As is common in T3SS secretion signals, these 20 residues have only 10% identity between EspA and Map, quite consistent with the fact that this region is unstructured and disordered in the filament.
Fig. 2.
Overall architecture of the EspA filament and the EspA subunit. (A) Surface representations of side (Left) and top (Right) views of the EspA filament structure. Each of the five strands is shown in a different color. (B) Localization of an individual subunit within the EspA filament strand 3. (C) Ribbon representation of the EspA protein structure with the N and C terminus and all the secondary structures labeled. (D) Topology secondary structure diagram of the EspA protein with α-helices represented as cylinders and showing the first and last residues in each helix.
EspA folds into an all α-helical structure, comprising seven α helices (α1–α7) (Fig. 2 B–D). The first 10 visible residues in the EspA N terminus (residues 19–28) are followed by the α1 helix that extends to the surface of the filament. A three-residue loop between α1 and α2 folds back into the core of the protein and orients the following long α2 helix into the wall of the filament. The α2 helix forms a two-helix bundle with a large portion of α7 located at the C terminus of the protein. Interestingly, a superposition of the EspA subunit structure in the filament and when bound to its cytoplasmic CesAB chaperone (22) shows that the position of the α2 helix undergoes a translation by ∼15.6 Å (SI Appendix, Fig. S2 B and D). Another short loop made by three residues connects α2 and α3. The α3 helix extends into the filament wall before a long 13-residue loop between α3 and α4 folds the protein back onto the surface of the filament where α4 and a downstream long 21-residue loop are exposed. This region encompasses the previously identified variable and hypervariable regions of EspA that contain the immunodominant EspA epitope (28). This surface-exposed region was shown to be critical for changing antigen specificity after the hypervariable regions of EspAEPEC and EspAEHEC were swapped. The loop between α4 and α5 folds the protein back into the filament wall where α5 and α6 establish an intricate intermolecular interactions network with surrounding EspA molecules (Fig. 3). The loop between α6 and α7 orients the final helix (α7) and the final C-terminal loop into the pilus lumen suggesting a role for these residues in substrate transport through the filament lumen. Strikingly, the same short loop made by only three residues creates a 40° angle between the α6 and α7 helices that was not observed (22) when EspA was bound to its chaperone since in that complex α6 and α7 were a single continuous helix (SI Appendix, Fig. S2 A–C).
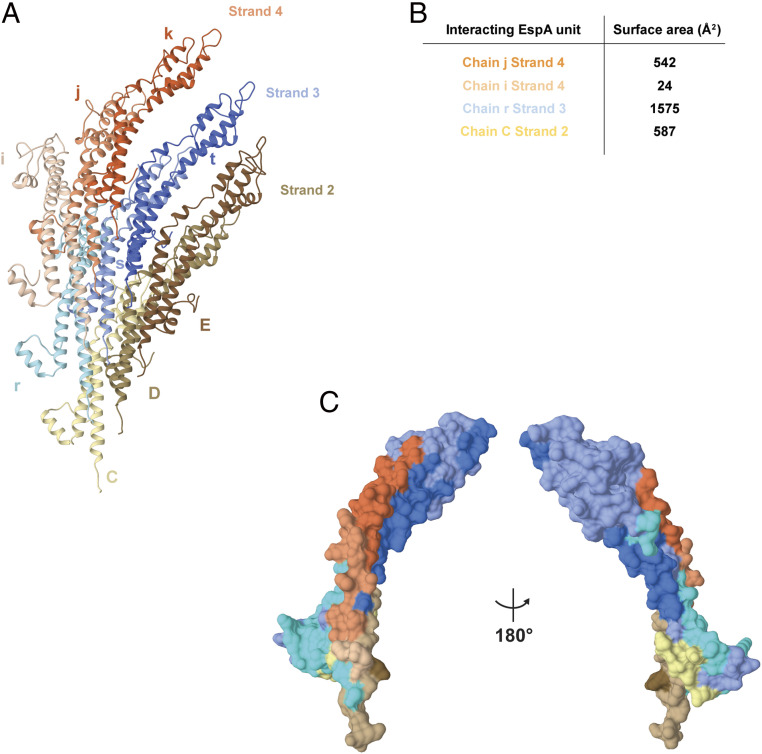
Fig. 3.
EspA–EspA interaction network in the filament. (A) Each EspA subunit interacts with eight other subunit chains within the filament. The reference chain s strand 3 (blue) interacts with the neighboring chains r (light blue) and t (dark blue) from the same strand 3, with chains i (light orange), j (orange), and k (dark orange) from strand 4 and with chains C (light brown), D (brown), and E (dark brown) from strand 2. (B) Buried surface area of the EspA–EspA interfaces using the same chain reference and the color coding as in A. The other four interfaces made with the reference chain s (with chains k, t, D, and E) are symmetry-related to these four. (C) Mapping of the EspA–EspA interactions in the reference subunit (chain s strand 3, blue color). The interactions made by the eight neighboring subunits are mapped in the reference subunit using the same color code as in A.
EspA–EspA Interaction Network in the Filament.
Each EspA protein molecule makes contact with eight subunits located in three different 5-start strands (labeled strands 2, 3, and 4, Fig. 3A). The subunit–subunit interaction network involves the contact of the reference subunit s with adjacent subunits from the same helical strand 3 (previous chain r and subsequent chain t), with three subunits from the helical strand 4 above (chains i, j, and k) and three subunits from the helical strand 2 below (chains C, D, and E) (Fig. 3A). This arrangement establishes that most contacts for any given subunit are with the adjacent subunits from the same helical strand (chains r and t). Quantifying the surface area buried in subunit–subunit interactions shows the large contacts with both adjacent r and t chains (strand 3) followed by chains k and j (strand 4) and chains C and D (strand 2) (Fig. 3B). The larger portion of solvent-accessible surface of the protein (i.e., the loop between α3 and α4, the α4 helix, and the loop between α4 and α5) encompasses the immunodominant epitope of EspA. The outer residues from the α1 helix are also exposed at the filament surface. The details of subunit–subunit interactions are shown in SI Appendix, Fig. S3 and Table S1.
The EspA Filament Lumen Architecture and Electrostatic Properties Resemble the T3SS Needle.
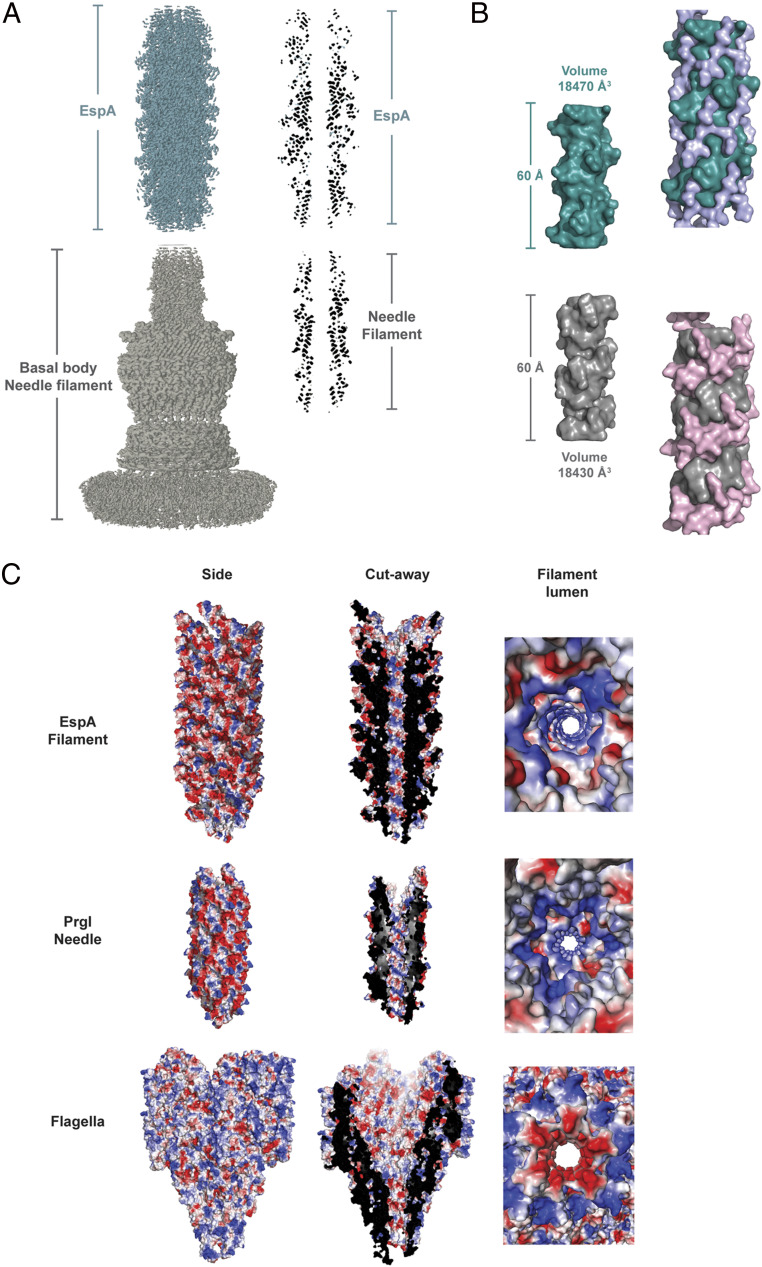
A composite high-resolution cryo-EM map was built up by aligning the T3SS apparatus (Electron Microscopy Database [EMD]-8913 and EMD-8914), the T3SS needle (EMD-8924), and the EspA filament maps; and a central longitudinal section was obtained from the final map (Fig. 4A). Ideally, we would like to build a composite structure using EPEC proteins, but the only high-resolution structures that exist for the basal body and the needle are from Salmonella. The section shown suggests that the lumen architecture of the T3SS needle and the EspA filament is conserved throughout both extracellular appendages. It was initially believed that the EspA filament was a sheath that encircled the needle and extended it (7). It can be seen that the lumen of EspA simply would not accommodate a needle within it (Fig. 4A). It is not possible at this point to model the interface between the needle and EspA, in part because we do not know if an additional protein or proteins, similar to the flagellum hook–filament junction (FlgK and FlgL) (34), are present at a very low stoichiometry at this interface. It is also not possible to look at other proteins that bind to the tip of the needle for structural insights. For example, the LcrV protein from Yersinia binds to the tip of the needle (35). However, there is vanishingly little sequence similarity between LcrV and EspA; even though both proteins contain coiled-coils these cannot be aligned, and the quaternary structure of LcrV is not known.
Fig. 4.
The EspA filament and the T3SS needle form a seamless conduit. (A) Cryo-EM maps of the T3SS apparatus (EMD-8913 and EMD-8914) and T3SS needle (EMD-8924), with EspA filament map located on top of the PgrI T3SS needle (Left). Central cross-section of the same composite map (Right). (B) Surface representation of lumen segments from the EspA filament (dark green) and the T3SS needle (gray). The volumes were determined using a fixed surface height of 60 Å in both calculations (Left). Surface representation of the residues exposed in the EspA filament lumen (Arg183, Ser189, and Lys192) and PrgI needle lumen (Lys66, Gln77, Asp70, Asn78, and Arg80) highlighting the right-handed spiral interior in both filaments. (C) Distribution of residues charges within the EspA filament, PrgI T3SS needle (PDB:6DWB), and C. jejuni flagellar filament (PDB: 6X80).
To understand if the lumen cavities of the needle and EspA are indeed comparable, we calculated the luminal volume of a 60-Å-length segment from both structures. Confirming our initial observations, both luminal volumes yielded nearly identical values (Fig. 4B). Moreover, specific charged and polar residues extending along the T3SS needle lumen (Lys66, Gln77, Asp70, Asn78, and Arg80) and EspA filament lumen (Arg183, Ser189, and Lys192) define similar right-handed helical grooves (Fig. 4B). Next, we investigated the electrostatic surfaces from the PrgI T3SS needle (Protein Data Bank [PDB]: 6DWB) and the EspA filament model to determine if these were conserved in the lumens. Strikingly, the two structures show that predominantly positively charged residues are exposed in both lumens, contrasting with the predominant negatively charged environment observed in the Campylobacter jejuni flagellar lumen filament that we show for comparison, given the homology between the T3SS and the bacterial flagellar system (Fig. 4C). Protein tubes that transport DNA have been shown to have a negative electrostatic luminal surface (36, 37), which would repel the negatively charged phosphate backbone of the DNA from the walls of the tubes, “lubricating” the passage of the DNA through these structures. Based upon this, a reasonable hypothesis would be that that the substrates being transported through the needle and EspA extension are positively charged, while the molecules being transported through the flagellar filament lumen (primarily flagellin) would be negatively charged. However, the pI values for these substrates, including EspA itself, show that this is not the case (Table 2). One is therefore left with alternative possibilities. For example, mutations in Arg80 in the Salmonella needle (corresponding to Arg183 in EspA) have been shown to lead to either a constitutive phenotype (R80K) or a defect in the secretion of only a single effector (R80E) (38). The constitutive phenotype, where secretion is turned on in the absence of contact with a mammalian cell, is expected to be due to a defect in signal transduction. The interactions that occur at the tip of the needle are expected to be communicated to the basal structure by an unknown mechanism. For the long EspA extension, the same signals must be communicated over even greater distances.
Table 2.
Isoelectric point of the EPEC E2348/69 effectors and translocators
| Isoelectric point | |
| Effector | |
| Tir/EspE | 4.9 |
| EspF | 10.61 |
| EspG | 5.21 |
| EspH | 9.48 |
| EspI/NleA | 5.27 |
| EspJ | 9.67 |
| EspL | 6.33 |
| EspZ/SepZ | 4.99 |
| Map | 9.08 |
| NleB | 5.96 |
| NleC | 4.60 |
| NleD | 5.78 |
| NleE | 5.60 |
| NleF | 4.94 |
| NleG | 5.34 |
| NleH/OspG | 5.34/5.54 |
| Translocators | |
| EspA | 4.52 |
| EspB | 5.66 |
| EspD | 5.13 |
Homologous Proteins.
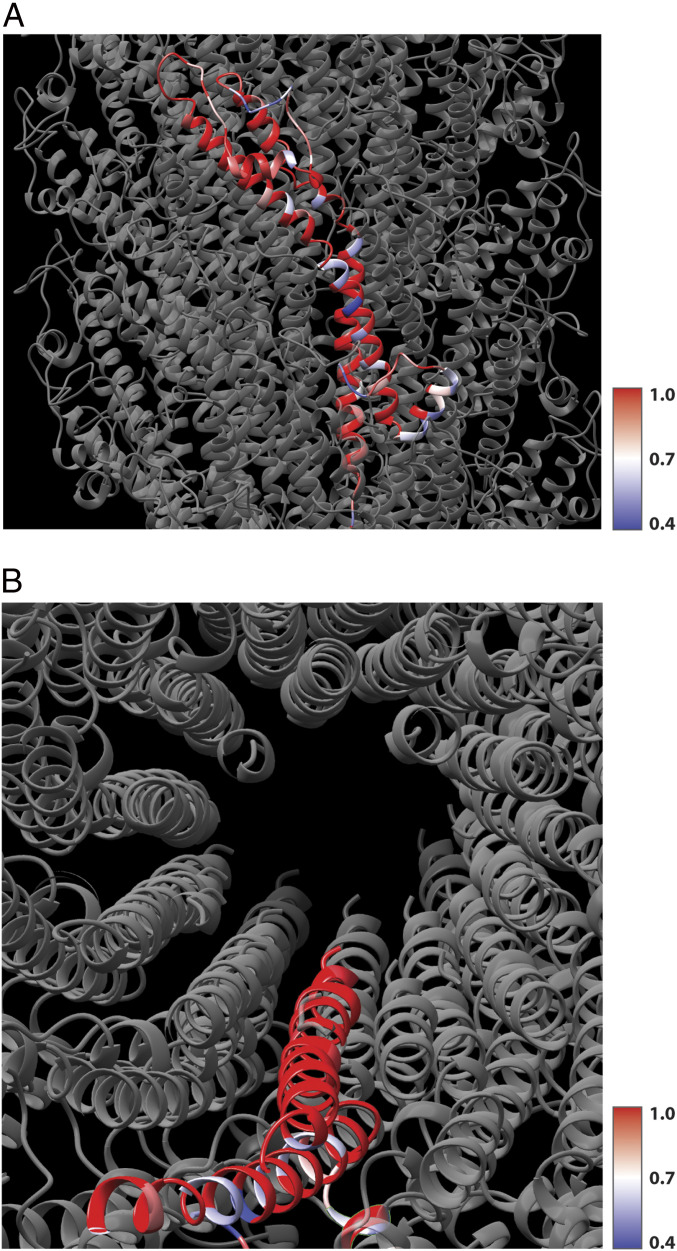
The long EspA needle extension has only been directly observed in EPEC (e.g., O127:H7, O55:H6, O55:H7) (10) and EHEC (O157:H7 and O26:H-) (28); EspA is also found in other members of the attaching and effacing pathogens: rabbit EPEC (39), the mouse pathogen Citrobacter rodentium (21), and Escherichia albertii (40). The conservation of residues among these five EspA proteins can be mapped onto a single subunit in our filament model (Fig. 5). Not surprisingly, the greatest variability in sequence is seen in exposed residues that face the outside of the filament, comprising the immunodominant region that confers antigenic polymorphism (28). In contrast, the lumen in the filament is highly conserved.
Fig. 5.
The conservation of residues in other EspA proteins. (A) Four other EspA proteins (having from 79 to 89% overall sequence identity with EPEC EspA) have been aligned, and the conservation at every residue is mapped onto a single EspA subunit in the filament, with red indicating 100% sequence identity at that position and blue indicating that 40% of the sequences have the same residue at this position. Almost all of the nonconserved residues are on the outer surface of the filament. (B) In contrast, a top view shows that all of the residues that line the lumen are highly conserved in these homologs.
Discussion
The atomic structure of the EspA filament can explain prior mutagenesis results. A previous study has shown that under noninducing conditions, EPEC expressing EspA mutants L39R, A45R, L47R, and M48R produced filaments indistinguishable from wild-type EspA; strains expressing EspA mutants Q43R, F49R, and Y53R produced short EspA filaments; while the F42R, A44R, and I46R mutations resulted in no visible EspA filaments, with F42R exhibiting loss of function (41). The atomic model shows that residues 42, 46, 49, and 53 are all involved in the coiled-coil interactions that exist between α2 and α7 in a single subunit, and residues 42, 43, and 44 are involved in interactions with neighboring subunits; the fact that F42 is involved in both intra- and intersubunit interactions explains its pivotal structural role (41). In contrast, residues 39, 45, 47, and 48 are near the outside of the filament and not making key interactions involved in maintaining either the overall fold of EspA or the interface with other subunits.
The common ancestry of the bacterial T3SS and the bacterial flagellar system has been discussed extensively (42–44). Since the targets of the T3SS are eukaryotic cells, which emerged long after bacterial flagellar motility, the reasonable assumption is that the T3SS evolved from the flagellar system, and not vice versa. Divergence in the flagellar system has been quite apparent, with some striking examples, such as how spirochetes have internalized the normally extracellular flagellar filaments so that motility is achieved by “reptilian” undulations of the entire cell (45). The coiled-coil core of the flagellar filaments is highly conserved in structure (with sequence identity being as low as 25%), while the outer domains are hypervariable or in some cases almost entirely absent (32, 46). The T3SS also exhibits a considerable degree of divergence, but there have been many fewer structural studies. For example, the only high-resolution structure that we have for the needle (38, 47) comes from Salmonella. But a comparison of the EPEC EcsF needle protein with the Salmonella PrgI needle protein shows only 21% sequence identity, and we do not really know the possible extent of structural divergence. We nevertheless have a reasonable expectation that the lumens will be conserved due to a conserved function.
Just as the flagellar system has undergone divergence as part of adaptations to a particular environment, the mechanical properties of Type 1 pili have been “tuned” by evolution for adaptation to different environments, such as the urinary tract versus the gut (48). The evolution of an EspA needle extension in the T3SS appears to have arisen from the need to build a specialized structure longer than the needle that can penetrate the intestinal mucus layer. Other organisms have used different T3SS adaptations. For example, the plant pathogen P. syringae elaborates a long T3SS needle, several microns in length and composed of the HrpA protein, as it has to negotiate the thick plant cell wall (49). An open question is whether the HrpA filament is actually closer to a needle or an EspA filament (50).
Understanding that the T3SS likely evolved from a flagellar system does not immediately show us the path for the evolution of the various components. While there is great similarity in helical symmetry between the EspA filament (∼5.6 subunits per turn, with an axial rise of 4.4 Å) and the flagellar filament (∼5.5 subunits per turn, with an axial rise of 4.7 Å), the structural conservation is almost nonexistent other than the presence of a coiled-coil in both. The Dali server (51) was used to find structural homologs of the EspA protein. Of the top five hits, three were flagellar proteins, and two were unrelated proteins (a phage shock protein and the p85β subunit of human phosphoinositide 3-kinase). In all cases, the structural similarity was due to a relatively small portion of the coiled-coil in EspA (SI Appendix, Fig. S5). The conclusion is simply that the two systems (flagella and T3SS) have diverged so far that the helical symmetry may be the main conserved element shared by the two.
Antibodies against EspA were found in human colostrum in regions were EPEC used to be endemic (e.g., Brazil), as well as in sera of humans with EHEC infection and of animals infected with other attaching and effacing pathogens (18–20). Considering that IgG antibodies have been shown to play a major role in clearing infection in an animal model (21), it is possible that this pressure is the driver of antigenic polymorphism in the flexible, exposed, loop of EspA, which could be exploited for presentation of foreign epitopes on the bacterial cell during infection of mucosal surfaces.
Materials and Methods
EspA Filament Production and Purification.
EspA filaments were purified from 400-mL cultures of EPEC O127:H6 ΔespA, pSA10-espA strain (27), seeded into Dulbecco's modified Eagle's medium and grown in 5% CO2 atmosphere at 37 °C for 4 h with gentle agitation every 30 min until optical density OD600 0.6. espA expression was then induced by adding 0.5 mM isopropyl β-ᴅ-1-thiogalactopyranoside for 5 h. Bacteria were harvested by centrifugation at 7,000 × g, and pellets were resuspended in 1 mL of cold phosphate-buffered saline (PBS). Filaments were sheared off from the bacteria by passage through a 25-gauge needle following a centrifugation step at 10,000 × g to remove the bacteria. At this stage, the supernatant was layered on preformed CsCl step gradients (1.0–1.3 g/cm3) and centrifuged for at least 14 h at 4 °C. The EspA filaments fraction was carefully removed and dialyzed against PBS. EspA filaments purity was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and confirmed by liquid chromatography–electrospray ionization tandem mass spectrometry.
Cryo-EM Sample Preparation and Data Collection.
Two-microliter samples of EspA filament were applied to plasma-cleaned lacey carbon grids, followed by plunge-freezing in liquid ethane using a Leica EM GP. Data collection was carried out at liquid nitrogen temperature on a Titan Krios microscope (Thermo Fisher Scientific) operated at an accelerating voltage of 300 kV. Using a K3 camera (Gatan), 40 movie frames were collected with a total dose of ∼55 electron/Å2 and sampling of 1.08 Å/pixel. The movies were collected with defocus values ranging between −1 and −2.5 μm.
Cryo-EM Image Processing and Reconstruction.
Raw movie frames with both a full dose and a dose of 20 electrons/Å2 were used in the motion correction and dose-weighting by MotionCor2 (52), respectively. Contrast transfer function (CTF) was estimated by Gctf (53), and images with poor CTF estimation were eliminated. The e2helixboxer program in EMAN2 (54) was used to box the long filaments from the full-dose images, followed by importing and extracting in RELION 3.0 software (55). A total of 232,860 256-pixel-long segments were extracted from 1,815 CTF-corrected images with a dose of 20 electrons/Å2. The 159,460 segments from Class2D were selected for Refine3D, which yield a map of recognizable secondary structure features when imposing the 1-start helical symmetry. The helical parameters converged to a twist of 64.3° and a rise of 4.4 Å per subunit. After CTF refinement and Bayesian polishing, the resolution of the final reconstruction was determined by both the Fourier shell correlation (FSC) between two independent half-maps and the FSC between model and map showing a resolution of 3.4 Å at FSC = 0.143 for map:map and a resolution of 3.5 Å at FSC = 0.38 (which is √0.143) for map:model (SI Appendix, Fig. S1).
Model Building and Refinement.
Initial model building started by building a single EspA subunit using Coot (56). As the map showed that EspA is mostly helical, polyalanine (poly-Ala) helices were placed in the density manually with Coot. The helices were then connected by manually extending the chain at the N and C termini of each helix. Once a nearly complete initial model was built, the respective side chains were assigned and further built manually to fully occupy the density of a single asymmetric unit. The model was subjected to several iterative rounds of real-space refinement in Phenix (57). Progress in refinement was tracked using Ramachandran plots and MolProbity (58). Once a single EspA subunit was successfully refined, a PDB coordinate file for the entire filament was generated by imposing the known helical parameters.
Lumen Definition and Volume Calculation.
We deployed a simple 3D grid-based procedure for the generation, visualization, and quantitative analysis of a region of the cavity. A 3D grid, at 0.5-Å spacing, was generated starting at the centroid of the molecule. As a visual inspection of the proteins showed that the approximate diameter of the channel was 21 Å, the grid size was set to 32 Å for the x and y dimensions (±16 Å either side of the centroid). These dimensions covered the area of the cavity and restricted the grid extent to the interior of the proteins. The height of the segment was set to 60 Å (±30 Å either side of the centroid along the z axis). The dimension of 60 Å was again selected by visual inspection, to restrict the generated cavity to the central region. Each grid point was treated as the position of a dummy atom, and any grid point found to be within 3.5 Å of a protein atom was excluded from any further analysis. At a distance less than 3.5 Å, the dummy atom at the grid point was deemed to be “clashing” with the protein atom. The remaining grid points were saved into a PDB format file, with the dummy atoms written out as C-alpha atoms. The program ChimeraX was then used to compute the volume of the lumen from these atoms.
Structure Analyses and Presentation.
The analysis and visualization of our cryo-EM map and the atomic model were done using University of California, San Francisco Chimera (59), ChimeraX (60), PyMol (Molecular Graphics System, Schrödinger), and PDBePISA (https://www.ebi.ac.uk/pdbe/pisa/).
Supplementary Material
Acknowledgments
Imaging was done at the Molecular Electron Microscopy Core Facility at the University of Virginia, which is supported by the School of Medicine. We thank Suhail Islam (Department of Life Sciences, Imperial College London) for his help with calculations and Paul Simpson (Electron Microscopy Facility, Centre for Structural Biology, Imperial College London) for his help with initial Cryo-EM screens. This work was supported by NIH Grant R35GM122510 (to E.H.E.) and Wellcome Trust Grants 215164/Z/18/Z (to T.R.D.C.) and 107057/Z/15/Z (to G.F.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022826118/-/DCSupplemental.
Data Availability.
The atomic coordinates have been deposited at the Protein Data Bank with accession number 7KHW. The density map has been deposited at the Electron Microscopy Data Bank with accession number EMD-22881.
References
- 1.Chen H. D., Frankel G., Enteropathogenic Escherichia coli: Unravelling pathogenesis. FEMS Microbiol. Rev. 29, 83–98 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Pennington H., Escherichia coli O157. Lancet 376, 1428–1435 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Garmendia J., Frankel G., Crepin V. F., Enteropathogenic and enterohemorrhagic Escherichia coli infections: Translocation, translocation, translocation. Infect. Immun. 73, 2573–2585 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng W., et al. , Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Wilson R. K., Shaw R. K., Daniell S., Knutton S., Frankel G., Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell. Microbiol. 3, 753–762 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Daniell S. J., et al. , The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3, 865–871 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Sekiya K., et al. , Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U.S.A. 98, 11638–11643 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knutton S., et al. , A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17, 2166–2176 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebel F., et al. , Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol. 30, 147–161 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Neves B. C., Shaw R. K., Frankel G., Knutton S., Polymorphisms within EspA filaments of enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 71, 2262–2265 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y. A., Yu X., Yip C., Strynadka N. C., Egelman E. H., Structural polymorphism in bacterial EspA filaments revealed by cryo-EM and an improved approach to helical reconstruction. Structure 14, 1189–1196 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Daniell S. J., et al. , 3D structure of EspA filaments from enteropathogenic Escherichia coli. Mol. Microbiol. 49, 301–308 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Stern A. S., Berg H. C., Single-file diffusion of flagellin in flagellar filaments. Biophys. J. 105, 182–184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartland E. L., et al. , The type III protein translocation system of enteropathogenic Escherichia coli involves EspA-EspB protein interactions. Mol. Microbiol. 35, 1483–1492 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Luo W., Donnenberg M. S., Interactions and predicted host membrane topology of the enteropathogenic Escherichia coli translocator protein EspB. J. Bacteriol. 193, 2972–2980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleary J., et al. , Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: Role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology (Reading) 150, 527–538 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Shaw R. K., et al. , Enterohemorrhagic Escherichia coli exploits EspA filaments for attachment to salad leaves. Appl. Environ. Microbiol. 74, 2908–2914 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abe A., Heczko U., Hegele R. G., Brett Finlay B., Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp. Med. 188, 1907–1916 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loureiro I., et al. , Human colostrum contains IgA antibodies reactive to enteropathogenic Escherichia coli virulence-associated proteins: Intimin, BfpA, EspA, and EspB. J. Pediatr. Gastroenterol. Nutr. 27, 166–171 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Jenkins C., et al. , Antibody response of patients infected with verocytotoxin-producing Escherichia coli to protein antigens encoded on the LEE locus. J. Med. Microbiol. 49, 97–101 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Caballero-Flores G., et al. , Maternal immunization confers protection to the offspring against an attaching and effacing pathogen through delivery of IgG in breast milk. Cell host Microbe 25, 313–323.e314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yip C. K., Finlay B. B., Strynadka N. C., Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 12, 75–81 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Morgan D. G., Macnab R. M., Francis N. R., DeRosier D. J., Domain organization of the subunit of the Salmonella typhimurium flagellar hook. J. Mol. Biol. 229, 79–84 (1993). [DOI] [PubMed] [Google Scholar]
- 24.Delahay R. M., et al. , The coiled-coil domain of EspA is essential for the assembly of the type III secretion translocon on the surface of enteropathogenic Escherichia coli. J. Biol. Chem. 274, 35969–35974 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Portaliou A. G., et al. , Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic Escherichia coli. EMBO J. 36, 3517–3531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creasey E. A., et al. , CesAB is an enteropathogenic Escherichia coli chaperone for the type-III translocator proteins EspA and EspB. Microbiology (Reading) 149, 3639–3647 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Chen L., et al. , Substrate-activated conformational switch on chaperones encodes a targeting signal in type III secretion. Cell Rep. 3, 709–715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crepin V. F., Shaw R., Knutton S., Frankel G., Molecular basis of antigenic polymorphism of EspA filaments: Development of a peptide display technology. J. Mol. Biol. 350, 42–52 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Newton S. M., Jacob C. O., Stocker B. A., Immune response to cholera toxin epitope inserted in Salmonella flagellin. Science 244, 70–72 (1989). [DOI] [PubMed] [Google Scholar]
- 30.Westerlund-Wikström B., Peptide display on bacterial flagella: Principles and applications. Int. J. Med. Microbiol. 290, 223–230 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Kenny B., Lai L. C., Finlay B. B., Donnenberg M. S., EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20, 313–323 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Wang F., et al. , A structural model of flagellar filament switching across multiple bacterial species. Nat. Commun. 8, 960 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munera D., Crepin V. F., Marches O., Frankel G., N-terminal type III secretion signal of enteropathogenic Escherichia coli translocator proteins. J. Bacteriol. 192, 3534–3539 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Homma M., Fujita H., Yamaguchi S., Iino T., Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J. Bacteriol. 159, 1056–1059 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derewenda U., et al. , The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure 12, 301–306 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Zheng W., et al. , Refined cryo-EM structure of the T4 tail tube: Exploring the lowest dose limit. Structure 25, 1436–1441.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa T. R., et al. , Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell 166, 1436–1444.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo E. Z., et al. , A polymorphic helix of a Salmonella needle protein relays signals defining distinct steps in type III secretion. PLoS Biol. 17, e3000351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badea L., et al. , Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34, 205–215 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Egan M., et al. , Transcriptional and posttranscriptional regulation of the locus of enterocyte effacement in Escherichia albertii. Microb. Pathog. 135, 103643 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Singh M. P., et al. , Identification of amino acid residues within the N-terminal domain of EspA that play a role in EspA filament biogenesis and function. J. Bacteriol. 190, 2221–2226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desvaux M., Hébraud M., Henderson I. R., Pallen M. J., Type III secretion: what’s in a name? Trends Microbiol. 14, 157–160 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Egelman E. H., Reducing irreducible complexity: Divergence of quaternary structure and function in macromolecular assemblies. Curr. Opin. Cell Biol. 22, 68–74 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Pallen M. J., Gophna U., Bacterial flagella and type III secretion: Case studies in the evolution of complexity. Genome Dyn. 3, 30–47 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Gibson K. H., et al. , An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete. eLife 9, e53672 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kreutzberger M. A. B., Ewing C., Poly F., Wang F., Egelman E. H., Atomic structure of the Campylobacter jejuni flagellar filament reveals how ε Proteobacteria escaped Toll-like receptor 5 surveillance. Proc. Natl. Acad. Sci. U.S.A. 117, 16985–16991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J., et al. , Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat. Commun. 9, 3840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spaulding C. N., et al. , Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions. eLife 7, e31662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown I. R., Mansfield J. W., Taira S., Roine E., Romantschuk M., Immunocytochemical localization of HrpA and HrpZ supports a role for the Hrp pilus in the transfer of effector proteins from Pseudomonas syringae pv. tomato across the host plant cell wall. Mol. Plant Microbe Interact. 14, 394–404 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Li C. M., et al. , The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J. 21, 1909–1915 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holm L., Rosenstrom P., Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang G., et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr.D.Biol.Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goddard T. D., Huang C. C., Ferrin T. E., Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates have been deposited at the Protein Data Bank with accession number 7KHW. The density map has been deposited at the Electron Microscopy Data Bank with accession number EMD-22881.