Abstract
Transfer RNA (tRNA) is the most diversely modified RNA. Although the strictly conserved purine position 37 in the anticodon stem–loop undergoes modifications that are phylogenetically distributed, we do not yet fully understand the roles of these modifications. Therefore, molecular dynamics simulations are used to provide molecular-level details for how such modifications impact the structure and function of tRNA. A focus is placed on three hypermodified base families that include the parent i6A, t6A, and yW modifications, as well as derivatives. Our data reveal that the hypermodifications exhibit significant conformational flexibility in tRNA, which can be modulated by additional chemical functionalization. Although the overall structure of the tRNA anticodon stem remains intact regardless of the modification considered, the anticodon loop must rearrange to accommodate the bulky, dynamic hypermodifications, which includes changes in the nucleotide glycosidic and backbone conformations, and enhanced or completely new nucleobase–nucleobase interactions compared to unmodified tRNA or tRNA containing smaller (m1G) modifications at the 37th position. Importantly, the extent of the changes in the anticodon loop is influenced by the addition of small functional groups to parent modifications, implying each substituent can further fine-tune tRNA structure. Although the dominant conformation of the ASL is achieved in different ways for each modification, the molecular features of all modified tRNA drive the ASL domain to adopt the functional open-loop conformation. Importantly, the impact of the hypermodifications is preserved in different sequence contexts. These findings highlight the likely role of regulating mRNA structure and translation.
Keywords: MD simulations, N6-isopentenyladenosine (i6A), N6-threonylcarbamoyladenosine (t6A), Wybutosine (yW), tRNA anticodon loop
INTRODUCTION
RNA carries out many critical cellular functions, ranging from translating genetic information into molecular machines to regulating gene activity during cell development and differentiation (Murphy et al. 2004; Park et al. 2019). Although RNA and proteins both exhibit structure–function relationships, twenty canonical amino acids contribute to the structural diversity of proteins, while there are only four canonical RNA nucleobases (A, G, C, and U). Therefore, the RNA nucleobases are frequently chemically modified to impart structural, and therefore functional, diversity. Indeed, more than 100 posttranscriptional nucleobase modifications have been identified to date, in a range of RNA types and organisms (Carell et al. 2012; Duechler et al. 2016). Posttranscriptional modifications have been shown to be involved in gene regulation (Gustilo et al. 2008; Helm and Alfonzo 2014; Zhao et al. 2017; Barbieri and Kouzarides 2020), and therefore uniquely designed RNA nucleobase modifications have the potential to be used to control cellular functions with safety and precision for applications in medicine and biotechnology (Jonkhout et al. 2017; Kawasaki et al. 2020).
Several databases have been developed to assemble information about posttranscriptional modifications including chemical composition, nucleobase modification site, type of RNA and phylogenetic distribution (Sprinzl and Vassilenko 2005; Cantara et al. 2010; Sun et al. 2015; Boccaletto et al. 2017). This data reveals that posttranscriptional modifications exhibit great chemical diversity, ranging from simple isomers to nucleotide methylation to the addition of bulky chemical groups (known as hypermodifications) (Sundaram et al. 2000). Modifications can be reversible or irreversible, and alter the chemical properties of the nucleobase, as well as the surrounding RNA regions (Helm 2006). Indeed, modifications can influence RNA structure, promoting or disrupting intramolecular hydrogen-bonding, stacking, and/or hydrophobic interactions that make RNA molecules rigid or more flexible (Vendeix et al. 2009; Motorin and Helm 2010; Chawla et al. 2015; Preethi et al. 2017; Seelam et al. 2017). As a result, modifications can strongly contribute to RNA functional diversity, especially when occurring within regulatory networks, where minor structural changes can significantly affect cellular metabolism and gene expression (Lewis et al. 2017).
Transfer RNA (tRNA) is the most diversely modified RNA (Sundaram et al. 2000; Konevega et al. 2004; Gustilo et al. 2008; Manickam et al. 2015; Sarachan et al. 2016). tRNA is involved in a variety of biological processes, including a unique role in protein synthesis, decoding mRNA codons and transferring specific amino acids to the peptidyl site in the ribosome (Frank et al. 2005). tRNA is typically composed of 73–80 nt and adopts a cloverleaf–shaped secondary structure or L-shaped three-dimensional structure (Fig. 1A). In general, functional tRNA has four major domains: the acceptor stem (ACS: bases 1–9 and 66–73), the D-stem–loop (DSL: bases 10–25), the anticodon stem–loop (ASL: bases 26–44) and the T-stem–loop (TSL: bases 49–65). Many posttranscriptional modifications, such as 4-thiouridine (s4U), 2-methylguanosine (m2G) and 2-dimethylguanosine (m22G), are believed to structurally refine the DSL region (Helm 2006). In addition to the major domains, tRNA has a variable arm (bases 45–48) and exhibits tertiary interaction networks between the D-loop and the T-loop when folded into a three-dimensional structure. These interactions are affected by several modified bases in mature tRNA, including 5-methylcytosine (m5C), 5-methyluridine (m5U), and pseudouridine (ψ) in the TSL region.
FIGURE 1.
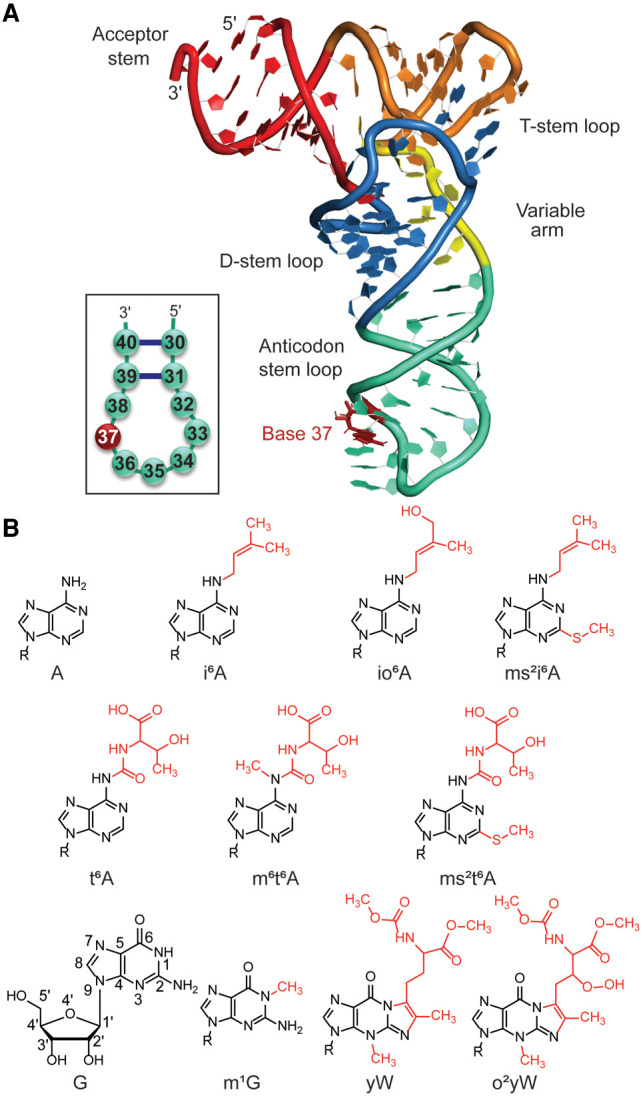
(A) Schematic representation of unmodified tRNA. (B) Chemical structures of hypermodifications found at the 37th position of tRNA considered in the present study (R = Ribose).
The ASL domain of tRNA is coaxially stacked with the DSL domain and is stabilized by five canonical base pairs. The ASL region includes seven loop nucleotides (32–38), with bases 34, 35, and 36 being the anticodon nucleotides that interact with the mRNA codon in the ribosome during protein synthesis. To ensure the efficiency and accuracy of translation, tRNA adopts a canonical U-turn structure in the ASL region, which promotes stable codon–anticodon interactions in the ribosomal A-site. A strictly conserved purine base at position 37, directly next to the anticodon nucleotides, also plays an important role in the ASL domain. Specifically, together with base pairs 31–39 and 32–38, base 37 has been proposed to help maintain an ideal anticodon conformation (Grosjean and Westhof 2016; Rozov et al. 2016). Furthermore, position 37 is key to facilitating tRNA accommodation at the ribosomal binding site, with the conserved helix 69 forming contacts with ribose in the 37th nucleotide (Rozov et al. 2016).
The largest variety and the highest density of nucleobase modifications occur in the ASL domain, which have been suggested to play important roles in maintaining the structural integrity of the anticodon region (Machnicka et al. 2014; Schweizer et al. 2017). More specifically, some modifications have been proposed to help preorder the structure of the ASL loop to facilitate tRNA accommodation in the ribosome and optimize codon–anticodon interactions (Auffinger and Westhof 1999; Sundaram et al. 2000; Stuart et al. 2003; Agris 2008; Vendeix et al. 2008; Denmon et al. 2011; Vangaveti et al. 2020). The conserved purine at position 37 undergoes a wide assortment of modifications that are phylogenetically distributed among the three domains of life (bacteria, archaea, and eukarya) (Machnicka et al. 2014). Modifications at A37 include 2-methyladenosine (m2A), N6-methyladenosine (m6A), 1-methylinosine (m1I), N6-isopentenyladenosine (i6A), N6-threonylcarbamoyladenosine (t6A), N6-glycinylcarbamoyladenosine (g6A), N6-hydroxynorvalylcarbamoyladenosine (hn6A), and associated derivatives, while G37 modified bases include 1-methylguanosine (m1G), wybutosine (yW), and associated derivatives (Boccaletto et al. 2017) (see, e.g., Fig. 1B). Many of these modifications have been proposed to enhance tRNA function by reinforcing the conserved loop structure, balancing loop flexibility and rigidity, and enhancing the stability of anticodon–codon complexes. For example, NMR data suggests that t6A at position 37 maintains the anticodon loop conformation by disrupting intraloop nucleotide interactions between U33 and A37 (Stuart et al. 2000), while crystallographic data also suggests t6A facilitates codon–anticodon interactions by stacking with A38 of tRNA and the first base of the codon (Murphy et al. 2004). In general, modifications to A37 have been proposed to be frequently used to compensate for weaker AU codon–anticodon base pairs and maintain the overall stability of the codon–anticodon duplex (Grosjean and Westhof 2016).
High-resolution crystal structures of tRNA and tRNA–protein complexes are available for select modifications at position 37 in the anticodon loop, including i6A (Fischer et al. 2016), ms2i6A (Jenner et al. 2010), t6A (Murphy et al. 2004), ms2t6A (Benas et al. 2000), and yW (Jovine et al. 2000; Mikkelsen et al. 2001; Schmeing et al. 2009). However, most tRNA functions depend on structural changes and static crystal structures do not provide a complete picture of the conformational space of the modified nucleobase, or the impact of the modification on the ASL region or codon–anticodon interactions. This leads to questions regarding the role of the modifications. For example, planarity of the added bulky chemical group with respect to the nucleobase has been proposed to be a crucial factor for maintaining the strength of codon–anticodon interactions (Stuart et al. 2000; Sundaram et al. 2000; Murphy et al. 2004; Durant et al. 2005). Indeed, a crystal structure of tRNALys containing t6A at position 37 reveals that the uriedo moiety is planar with respect to the adenosine ring and therefore might enhance stacking by acting as a third “ring” (Parthasarathy et al. 1977; Murphy et al. 2004). However, a different crystal structure of E. coli tRNA reveals a cyclic isoform of t6A that inherently contains a third ring adopts a nonplanar conformation (Matuszewski et al. 2017). Due to such discrepancies, a broad picture of the impact of different modifications to the 37th position on tRNA structure and function currently remains unclear.
Computational methods are particularly useful for uncovering molecular-level structural details about nucleobase modifications and their impact on the local tRNA environment that are difficult to access solely through experimental techniques. As a result, several computational studies have investigated the structural features of unmodified tRNA (Tewari 1988; Vermeulen et al. 2005; McCrate et al. 2006; Li et al. 2013; Zhang et al. 2014; Grzybkowska et al. 2016; Fandilolu et al. 2019; Vangaveti et al. 2020) and the conformational preferences and structural significance of hypermodifications (Morris et al. 1999; McCrate et al. 2006; Alexander et al. 2010; Allnér and Nilsson 2011; Kumbhar and Sonawane 2011; Kumbhar et al. 2013; Sambhare et al. 2014; Zhang et al. 2014; Kamble et al. 2015; Sonawane and Sambhare 2015; Fandilolu et al. 2018). However, to the best of our knowledge, these studies typically implement models that contain only a small fragment of the ASL region and consider a single trajectory of short time scale (<50 nsec), leaving a gap in our understanding of the long-range impact of modifications on the overall tRNA dynamics. In addition, most computational studies have considered combinations of modifications that exist within mature tRNA (McCrate et al. 2006; Allnér and Nilsson 2011; Sambhare et al. 2014; Zhang et al. 2014). However, evidence suggests that the impact of multiple modifications can effectively cancel each other. For example, differential scanning calorimetry showed that the ability of the i6A modification at position 37 to stabilize the stem is impacted by the presence of the pseudouridine modification at position 39 (Denmon et al. 2011). Thus, studying highly modified tRNAs prevents a thorough appreciation of the individual impact of each modification. Additionally, each previous study typically focuses on a specific modification, making comparisons within and across a broad range of families difficult.
To gain a deeper understanding of the effects of nucleobase modifications, the present study uses molecular dynamics (MD) simulations to investigate the structural dynamics of tRNA containing three hypermodified base families at the 37th position, namely i6A, t6A, and yW (Fig. 1B). The representative bases considered for each family differ in the presence of chemical substituents such as a hydroxy, methyl, thiomethyl or peroxy group. The hypermodification families were chosen based on their high occurrence frequency at position 37 in tRNA (Jühling et al. 2008). By comparing the structural dynamics of singly modified tRNAs to the corresponding unmodified variant, the molecular features imparted by individual modifications to tRNA structure can be uncovered, which may modulate tRNA function. Specifically, our detailed analysis sheds light on (i) the spatial conformations adopted by the modifications; (ii) how the modifications influence the stability of tRNA; (iii) how the modifications alter the dynamics between the four major domains of tRNA; (iv) how the modifications affect noncovalent interactions that may be involved in preordering the anticodon loop of tRNA; (v) how an increase in chemical complexity within each modification family affects tRNA structural dynamics; and (vi) how the impact of the modifications vary with sequence context. Our results reveal striking differences in the intrinsic structural dynamics and cross-strand interactions of the anticodon loop within and across different modification families. Indeed, these differences can affect the open structured anticodon loop that is required for ribosomal binding, pointing toward key roles for modifications at position 37. Overall, the atomistic details provided by our work are critical for understanding the potential biological functions of a range of tRNA hypermodifications and represent an important step toward uncovering the unique structure–function properties of different mature tRNAs. In addition to shedding light on the important biochemical roles of specific nucleobase modifications, the sometimes subtle effects of additional substituents at the modification site uncovered by examining families of modifications is valuable information for the future design of RNA-based therapeutics and biosensors (Chakraborty et al. 2016; Park et al. 2019; Kawasaki et al. 2020).
RESULTS AND DISCUSSION
Unmodified tRNAs with A or G at position 37 exhibit subtle differences in nucleotide interactions in the anticodon-stem–loop, while the m1G modification fine-tunes the functional open-loop conformation of tRNA
To provide a basis for understanding the structure and dynamics of modified tRNA, 1 μs MD simulations were initially performed in triplicate on two unmodified tRNAs that contain either A (denoted A-tRNA) or G (G-tRNA) at position 37. As described in more detail in the Materials and Methods section, this model choice is justified since the overall fold of unmodified tRNA is similar to isolated mature tRNA (Shi and Moore 2000) and tRNA bound to proteins (e.g., tRNA bound to dihydrouridine synthases [Byrne et al. 2015]) or the A-site of the ribosome (Schmeing et al. 2009), particularly the U-turn motif of the anticodon loop (Byrne et al. 2010). To permit comparison across the diverse set of modifications, models for both unmodified tRNAs were consistently generated from an X-ray crystal structure of standalone full-length unmodified Escherichia coli tRNAPhe (PDB ID: 3L0U [Byrne et al. 2010]). Nevertheless, the impact of this model choice in terms of common G modifications and sequence context are discussed in more detail below and in subsequent subsections.
The unmodified tRNAs are stable throughout the MD trajectories as indicated by the average root-mean-square deviation (RMSD) of backbone atoms with respect to the experimental starting structure (∼3 Å, Supplemental Fig. S1A; Supplemental Table S1). Sequence variation associated with A versus G at position 37 causes no significant deviation in the tRNA backbone dynamics. Both unmodified tRNAs maintain the correct global fold throughout the MD simulations, specifically the canonical L-shaped three-dimensional structure that contains all four helical domains. Furthermore, the DSL–ASL and the ACS–TSL coaxial stacks, and the interdomain tertiary contacts are preserved. Detailed analysis of the per-residue root-mean-square fluctuation (RMSF) of each nucleotide averaged over all sampled conformations reveal lower structural fluctuations for bases in the stem regions (ACS, D-stem, anticodon stem, and T-stem; average RMSF < 4 Å) than for loop nucleotides (D-loop, anticodon loop, variable arm, and T-loop; average RMSF between 4 and 11 Å, Supplemental Fig. S1B).
The domain-dependent motions were assessed by computing a residue-wise cross-correlation matrix for the C1′ atoms of all nucleotides, which highlights the positive (correlated) and negative (anticorrelated) motion of the nucleotides (Supplemental Fig. S2). In unmodified tRNA, nucleotides have significant differences in correlation, which highlights the dynamic characteristics of interdomain interactions within tRNA during the MD simulations. tRNA domains with base–base interactions exhibit positive correlation, especially helical stems with multiple base pairs, as well as long-range interdomain tertiary contacts between the DSL and the TSL or variable arm. In contrast, tRNA dynamics that result in stretching and twisting of coaxial helical domains show anticorrelated motions (e.g., between the DSL and ASL domains). While the overall pattern in the cross-correlation matrix is similar for G-tRNA and A-tRNA, there are slight differences (>0.3). For example, the positive correlation between the DSL and TSL regions is slightly enhanced for A-tRNA, while the positive correlation between the DSL and variable arm domains is slightly enhanced for G-tRNA.
In the ASL region, all five helical base pairs maintain Watson–Crick hydrogen-bonding interactions (Supplemental Tables S2–S4) and consistent glycosidic C1′–C1′ distances (10–11 Å, Supplemental Table S5) during the simulations. Even the A31–U39 base pair next to the anticodon loop is stabilized by two N–H···O/N interactions (occupancies > 90%). This suggests that the anticodon stem domain is not disrupted by the inherent dynamics of the anticodon loop or the interchange of G for A. Indeed, the backbone torsion angles are consistent throughout the loop region (base 31 to 39), with the exception of a dramatic shift between U33 and G34 (particularly in θ at U33 and η at G34, Supplemental Fig. S3). This change in the backbone conformation induces a sharp kink between bases 33 and 34. The backbone kink coupled with greater variation in the orientation of the nucleobases about the glycosidic bonds (Supplemental Fig. S4) results in the lack of stacking between bases 33 and 34 (Supplemental Fig. S5; Supplemental Table S6). The backbone kink is a well-known distinguishing feature of U-turn motifs in tRNA (Quigley and Rich 1976), and results in exposure of the anticodon triplet to the solvent.
Besides the lack of U33–G34 stacking interactions, the remaining anticodon loop bases stack with their nearest neighbors throughout the MD simulations (Supplemental Fig. S5; Supplemental Table S6). This includes the three anticodon bases, although the stacking interactions between bases 34 and 35 is reduced for A-tRNA (14%) compared to G-tRNA (84%). Base stacking within the loop region extends into the anticodon stem, as evidenced by 100% base stacking propensity between bases 31–32 and 38–39 throughout the simulations. This maintains connectivity between the anticodon loop and stem regions. In addition to stacking interactions, the first and last nucleotides of the anticodon loop, namely U32 and A38, form a base pair (Supplemental Tables S2–S4). Thus, several intrastrand interactions work together to structure the anticodon stem–loop.
The nucleobase at position 37 in the anticodon loop adopts an anti conformation throughout the trajectory (average glycosidic χ torsional angle of −172 ± 8° for A37 and −166 ± 9° for G37; Supplemental Table S7; Supplemental Fig. S1C), which is similar to the experimental starting structure of A-tRNA (Byrne et al. 2010). Furthermore, there are no significant overall deviations in the backbone torsion angles near position 37 compared to the crystal structure (Supplemental Figs. S3, S4). In the adopted conformation, base 37 is in a prime location to consistently stack between A36 and A38, regardless of the identity of the purine (100% occupancy, Fig. 2A; Supplemental Fig. S3–S5; Supplemental Table S6). Base 37 stacking interactions are slightly stronger with A38 (average interaction energy of −5.7 ± 0.9 kcal/mol for A and −6.7 ± 2.5 kcal/mol for G) than A36 (−4.6 ± 0.7 kcal/mol for A and −5.0 ± 1.1 kcal/mol for G, Supplemental Table S8). Nevertheless, the persistent stacking between bases 36 and 37 is expected to extend to the codon–anticodon helix at the tRNA–mRNA interface. Indeed, base A37 has been shown to stack with the first codon–anticodon base pair in crystal structures of tRNA bound to the ribosome (Selmer et al. 2006; Weixlbaumer et al. 2007).
FIGURE 2.
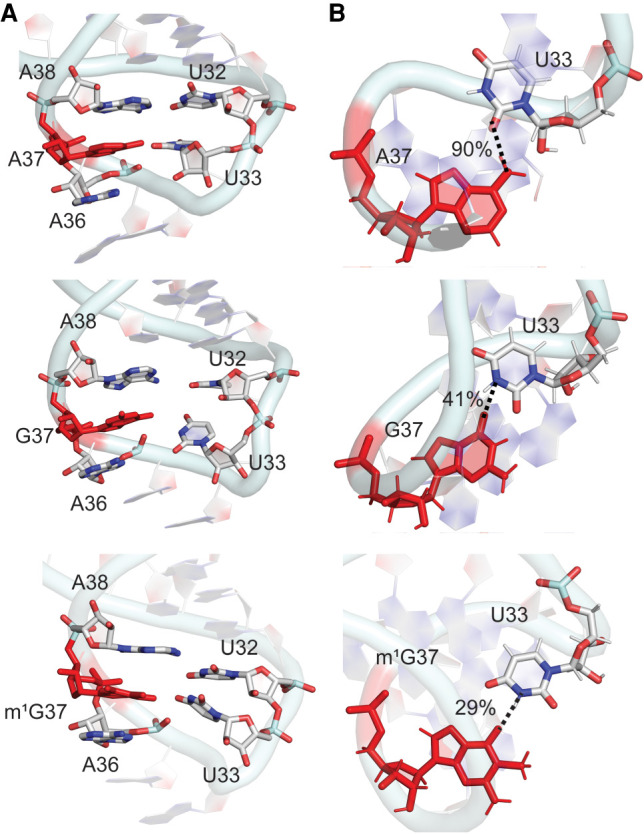
(A) Stacking interactions between base 37 and neighboring nucleobases in the anticodon loop, and (B) hydrogen bonding (with occupancies) in the 33–37 base pair for A, G, and m1G-tRNA of E. coli.
In addition to stacking, base 37 participates in cross-strand base–base hydrogen bonding in the anticodon loop. Specifically, the Hoogsteen edge of A37 (N6) forms a N–H···O hydrogen bond with the sugar edge of U33 (O2) (90% occupancy and average interaction energy of −3.1 kcal/mol; Fig. 2B; Supplemental Table S9). This A37–U33 interaction correlates with NMR chemical shifts reported for unmodified tRNA (Denmon et al. 2011) and the X-ray crystal structure of unmodified tRNA (Byrne et al. 2010). Unlike A37, base-pairing occurs between the Watson–Crick faces of G37 and U33 in G-tRNA, which results in an average interaction energy of −2.5 ± 5.7 kcal/mol. Nevertheless, this pair is more dynamic (Fig. 2B; Supplemental Table S9) and the N3(U33)–H···O6(G37) interaction is less persistent (occupancy ∼40%). Indeed, bases G37 and U33 exhibit larger deviations in the glycosidic torsion angles (standard deviations in χ of up to 16°, Supplemental Fig. S4; Supplemental Table S7) and the C1′–C1′ distance (standard deviations up to 1.0 Å, Supplemental Table S5). Overall, although both A and G at position 37 interact with U33, changes in the nature of the interaction result in a different sampling of the dominant conformation of the ASL for A versus G-tRNA (Fig. 3).
FIGURE 3.
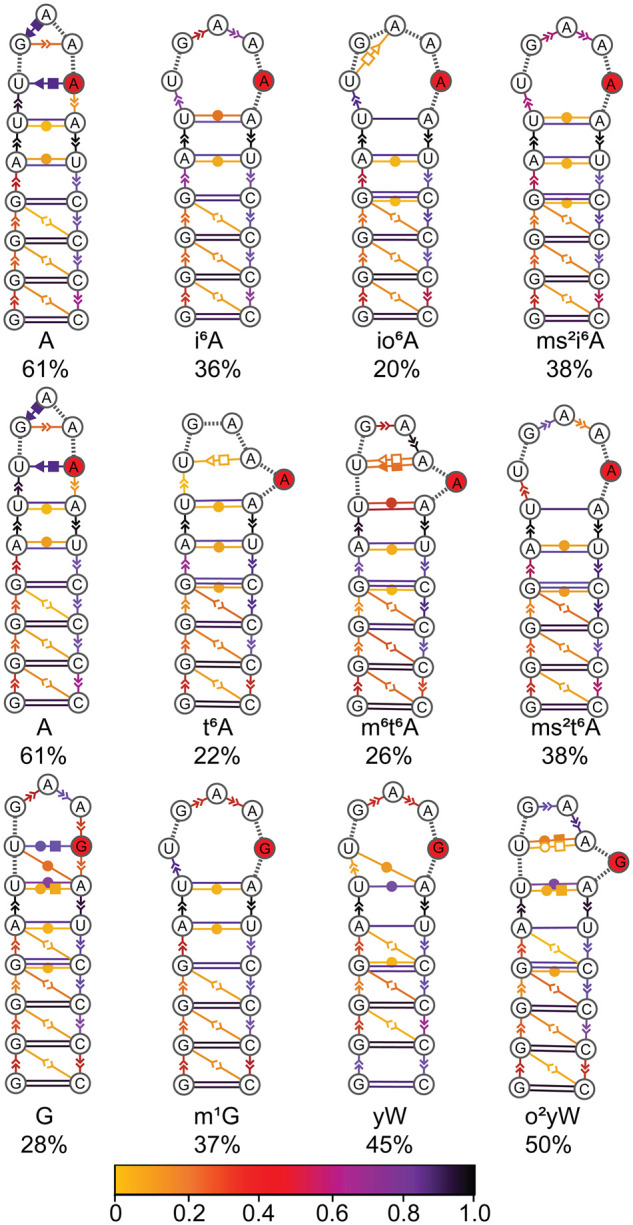
Consensus secondary structures of the anticodon stem–loop region in unmodified and modified tRNA, with the solid red circle representing the nucleotide at the 37th position of E. coli tRNA. Extended secondary structure annotation follows the Leontis–Westhof nomenclature (Leontis and Westhof 2001). The color scheme shows the probability distribution of the interactions within the cluster.
Despite interest in comparing the impact of hypermodified to unmodified tRNA with A or G at position 37, sequence analysis reveals that unmodified G is mostly absent at position 37 in the anticodon region of tRNA in all three domains of life (Jühling et al. 2008; Boccaletto et al. 2017). Instead, 1-methylguanosine (m1G) frequently occurs at the 37th position. Therefore, prior to investigating the impact of bulky hypermodifications to G, we initially compared the structural attributes of tRNA containing G and m1G at position 37. Both G and m1G-tRNA are stable throughout the MD simulations as indicated by low backbone RMSDs with respect to the experimental starting structure (∼3 Å) and have highly similar RMSFs for each nucleotide averaged over all sampled conformations (Supplemental Fig. S1). Both G and m1G-tRNA adopt the L-shaped 3D structure throughout the simulations and exhibit similar domain-dependent interactions (Supplemental Fig. S2). However, although N1-methylation of G preserves stacking interactions with bases 36 and 38, the modification reduces hydrogen-bonding interactions between the 37th nucleotide and U33 (Fig. 2). As a result, incorporation of the m1G modification enhances the stability of the open-loop structure of the ASL (Fig. 3).
Overall, sequence variation with A, G, or m1G at position 37 does not significantly alter the global tRNA structure or domain-dependent interactions. Furthermore, the predicted unmodified tRNA structures are consistent with the experimental structure for unmodified A-tRNA (Byrne et al. 2010), validating our computational approach. Nevertheless, consideration of A, G, and m1G-tRNA highlights that the nature of the base at the 37th position can affect the local architecture of tRNA in the ASL region by altering intraloop interactions (Figs. 2, 3) and further emphasizes the importance of studying the implications of nucleobase hypermodifications at this position. Regardless, the predicted structure of these tRNA will provide points of reference for understanding the impact of bulky base modifications in the anticodon loop on the structure and dynamics of tRNA as discussed in the following sections.
i6A modestly alters nucleotide conformations and nucleotide–nucleotide stacking and hydrogen-bonding interactions in the anticodon loop, with additional chemical substitutions imparting further changes
The i6A hypermodification is derived from canonical adenine with an isopentenyl substituent attached to the N6 exocyclic amino group (Fig. 1B). The two i6A-derived hypermodifications considered in the present work add a hydroxy substituent to the i6A terminal methyl group (io6A) or a thiomethyl substituent at C2 of the nucleobase (ms2i6A, Fig. 1B). Unlike i6A and ms2i6A, sequence analysis highlights that io6A is absent in E. coli (Jühling et al. 2008; Boccaletto et al. 2017). Nevertheless, since io6A is present in both bacteria (Corynebacterium fascians) and eukaryotes (Lupinus luteus and Nicotiana rustica), we consider io6A in the same sequence context as the other members of the i6A family to decipher the effect of the additional hydroxy substituent on tRNA structure. The i6A hypermodifications have the smallest chemical substituents among the families considered in the present work.
Despite the flexibility of the modification side chain (Fig. 4A,B), the modified tRNAs are stable throughout the simulation trajectories, exhibiting similar RMSDs as unmodified tRNA within the standard deviation (Supplemental Fig. S6; Supplemental Table S1). Thus, MD structures of i6A, io6A and ms2i6A-containing tRNA retain the L-shaped structure, with three-dimensional folding and interaction networks similar to the MD predicted and experimental crystal structure of unmodified A-tRNA (Supplemental Fig. S7A; Byrne et al. 2010). The dynamic cross-correlation maps indicate similar correlation patterns for the movement of modified and unmodified tRNAs (deviations <0.3, Supplemental Fig. S7B) and the time-averaged RMSFs for the individual nucleotides in modified and unmodified tRNAs exhibit similar trends (Supplemental Fig. S8). In fact, even the RMSFs for the individual nucleotides in the anticodon loop are generally similar (Fig. 4C).
FIGURE 4.
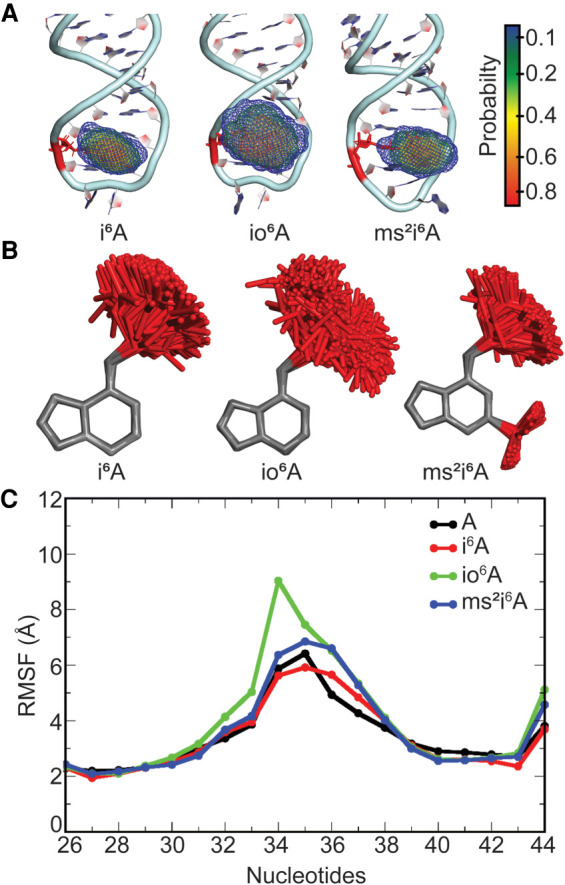
(A) Probability distribution in the location of the side chain in the i6A hypermodified nucleobase at the 37th position in E. coli tRNA. (B) Overlays of i6A, io6A, or ms2i6A modified nucleobases in tRNA with respect to the nucleobase ring for structures taken at 5 ns intervals over the MD simulations. (C) Average structural deviation of anticodon nucleotides (RMSF, Å) over the MD simulations with respect to the initial structure for each i6A modified tRNA compared to unmodified A-tRNA.
The stem base pairs in the ASL region are preserved upon tRNA modification (Supplemental Tables S2–S5), including the A31–U39 pair, which correlates with experimental evidence that the parent i6A modification does not destabilize the stem (Denmon et al. 2011). Furthermore, the modified tRNAs contain the kink in the backbone in the anticodon loop between bases 33 and 34 (Supplemental Fig. S9), indicating the presence of the U-turn in the anticodon loop. This observation is supported by NMR data highlighting the role of the parent i6A modification in promoting the U-turn conformation in the E. coli tRNAPhe anticodon loop (Cabello-Villegas et al. 2004). However, the modifications induce distinct changes in the backbone conformation in the anticodon loop depending on the level of i6A substitution. Specifically, although the backbone conformation for i6A-containing tRNA is very similar to that for unmodified tRNA throughout the loop region, deviations in both θ and η occur near U33 and G34 for ms2i6A. This correlates with previous reports that i6A does not affect the thermal stability of tRNA, while ms2i6A is destabilizing (Kierzek and Kierzek 2003). Even larger deviations from unmodified tRNA are observed for io6A throughout the three-nucleotide region spanning bases 34 to 36. Although the orientation of the nucleobase with respect to the ribose moiety at positions 31–33 and 36–39 is unimpacted by modification, larger deviations are seen in both χ and δ for G34 and A35 (Supplemental Fig. S10). This observation is supported by NMR data indicative of increased flexibility in the anticodon loop nucleotides induced by the i6A modification in E. coli tRNATyr (Denmon et al. 2011) and tRNAPhe (Cabello-Villegas et al. 2004). These deviations from unmodified tRNA are dependent on the bulky moiety, with i6A impacting G34, ms2i6A affecting A35, and io6A significantly affecting the backbone conformation of both nucleotides. Thus, the parent i6A modification alters the conformations of loop nucleotides, and small additional chemical changes (hydroxy or thiomethyl substitution) further induce flexibility in the loop region, including adjustments in the anticodon nucleotides (34–36).
Regardless of differences in the backbone conformation in the loop region, the C1′–C1′ distances between the ASL nucleotides in all i6A-modified tRNAs are close to those for unmodified tRNA (Supplemental Table S5), indicating that the overall anticodon loop conformation is maintained. Indeed, there is no stacking between bases 33 and 34 for any modified tRNA (Supplemental Fig. S11; Supplemental Table S6). Similar to solution structures of i6A-modified E. coli tRNAPhe (Cabello-Villegas et al. 2002; Denmon et al. 2011), bases 34 and 35 are coplanar, resulting in less than optimal stacking interactions as observed for unmodified A-tRNA. The remaining nearest neighbor bases stack throughout the MD simulations, which propagates into the stem as discussed for A-tRNA. The only exception is a notable reduction in the stacking occupancy between bases 35 and 36 for io6A-containing tRNA, which correlates with the previously noted highly dynamic nature of the backbone in this region of the anticodon stem–loop for this modification (Supplemental Figs. S9, S10). Regardless, hydrogen bonding is maintained between U32 and A38 for all tRNAs containing any member of the i6A family (Supplemental Tables S2–S4).
The i6A family of modifications does not affect the preferred glycosidic orientation of the nucleotide at the 37th position, with all i6A-derived nucleotides adopting the anti conformation as discussed for A-tRNA (χ > 166°, Supplemental Fig. S12; Supplemental Table S7) and observed in crystal structures of tRNA containing i6A (Jenner et al. 2010) or ms2i6A (Fischer et al. 2016). The bulky isopentenyl group in the i6A modifications is very flexible, adopting a wide range of conformations throughout the MD simulations (Fig. 4A,B; Supplemental Fig. S13). Indeed, although the sampled conformation of the isopentenyl group includes those observed in experimental structures of tRNA containing i6A (Fischer et al. 2016) or ms2i6A (Supplemental Fig. S14; Jenner et al. 2010), MD simulations emphasize the significant structural dynamics of the hypermodifications. In general, two side chain conformations are adopted with respect to the N6–C10 linkage [defined according to the torsional angle ϕ = ∠(C6N6C10C11), Supplemental Fig. S15]. For i6A, conformer-I (40° ≤ ϕ ≤ 100°) is populated for ∼40% of the simulation and conformer-II for ∼60% (160° ≤ ϕ ≤ 240°). However, the number and population of the conformations are influenced by the addition of chemical substituents to i6A. Specifically, the presence of a hydroxy substituent introduces another conformation (240° ≤ ϕ ≤ 300°), which is slightly less populated than conformers I and II, whereas a thiomethyl group at C2 reduces the N6 bulky moiety flexibility, with conformer-I being highly favored (>85% occupancy). This shows that chemical substitution directly affects the dynamical structure of the hypermodification, specifically the side chain conformation.
Despite the inherent flexibility within the hypermodified nucleobase, the backbone conformation at position 37 does not change from unmodified tRNA (Supplemental Figs. S9, S10). As a result, base 37 is stacked between 36 and 38 regardless of the i6A derivative considered (Fig. 5A). Nevertheless, all members of the i6A family considered in the present study exhibit a reduced stacking frequency with base 36 compared to canonical A (Supplemental Fig. S11; Supplemental Table S6), illustrating the impact of the flexible substituent. However, the average stacking interaction between bases 36 and 37 remains consistent with A-tRNA (Supplemental Table S8), suggesting that the bulky substituent increases the magnitude of the stacking interaction. Indeed, stacking between bases 37 and 38 is persistent regardless of the modification (Supplemental Fig. S11) and the associated interaction is statistically larger than the corresponding interaction with A (−5.7 ± 0.9 kcal/mol), with ms2i6A having the largest stacking interaction with A38 (−9.0 ± 1.6 kcal/mol) followed by io6A (−7.7 ± 2.3 kcal/mol) and i6A (−6.5 ± 1.0 kcal/mol). Thus, our simulations provide direct evidence that the dynamical i6A modification stabilizes neighboring bases A36 and A38 by enhancing stacking interactions, which supports previous proposals that modifications at the 37th position can influence the stability of the anticodon loop through stacking (Agris 2008). Furthermore, our data highlights that additional substitution of the i6A parent modification can further fine-tune intrastrand stacking interactions.
FIGURE 5.
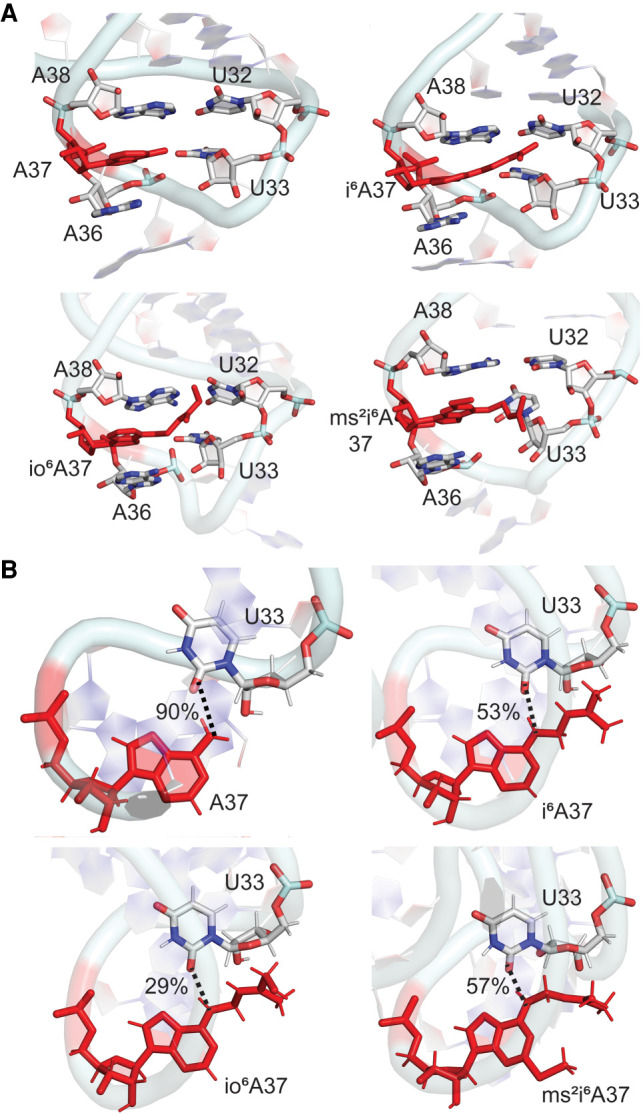
(A) Stacking interactions between base 37 and neighboring nucleobases in the anticodon loop and (B) hydrogen-bonding interactions (with occupancies) between bases 37 and 33 throughout the MD simulations for unmodified and i6A-modified E. coli tRNAs.
Beyond altering stacking interactions with the neighboring bases, the i6A modifications can impact intraloop hydrogen-bonding interactions between nucleotides A37–U33 in the anticodon region. As reported based on an NMR solution structures of i6A-modified E. coli tRNAPhe (Cabello-Villegas et al. 2002; Denmon et al. 2011), the hydrogen-bond network of the unmodified anticodon loop is disrupted by all i6A modifications. Specifically, although the hydrogen-bonding pattern of the A37–U33 base pair is the same regardless of the A37 modification, the propensity to form the N6–H···O2 hydrogen bond decreases upon introduction of a member of the i6A family (Fig. 5B). The hydrogen bond is less persistent for i6A and ms2i6A compared to canonical A, and even less persistent for io6A, likely due to the dynamic nature of the loop backbone for the latter modification. Interestingly, the sidechain of all i6A modifications can also interact with U33, either stacking above the plane of the uracil nucleobase or hydrogen bonding with O2, as well as O2′ and O4′ of ribose. Although these interactions between the modified A37 side chain and U33 will collectively provide additional stability to the loop region, each contact is relatively short-lived (<5% occupancy) due to the highly dynamical nature of the hypermodifications. As a result, the modified nucleotide at the 37th position does not participate in cross-strand interactions in the dominant conformations of the ASL, which correspond to an open-loop conformation that will enhance tRNA function (Fig. 3). Thus, the differences between modified and unmodified A-tRNA highlight the potential importance of hypermodifications for controlling noncovalent interactions in the loop region. Furthermore, our data illustrates that an increased level of complexity of the chemical modification, such as including a hydroxy group in the bulky moiety, can further refine discrete noncovalent interactions within tRNA.
Overall, although the i6A modifications do not disrupt the global fold of unmodified tRNA, domain–domain interactions, or the structural integrity of the anticodon stem–loop, the isopentenyl group along with additional chemical (hydroxy or thiomethyl) substituents can introduce local changes in the loop nucleotides that fine-tune tRNA structure and likely function (Table 1). Specifically, the i6A modifications are dynamic and adopt multiple conformations at position 37 in tRNA. In fact, accommodation of the flexible isopentenyl group requires rearrangement within the anticodon loop, with the extent of structural adjustment depending on the level of chemical substitution. Even subtle alterations in the backbone and neighboring base conformations directly impact nucleotide–nucleotide stacking and hydrogen-bonding interactions in the anticodon loop, as well as the conformation of the anticodon nucleotides. Although previous NMR studies on E. coli tRNAPhe highlight increased flexibility of the anticodon loop region upon inclusion of the parent i6A modification (Cabello-Villegas et al. 2002), our work uncovers new molecular-level details surrounding the impact of these dynamics, as well as the relative effects of different levels of i6A chemical substitution.
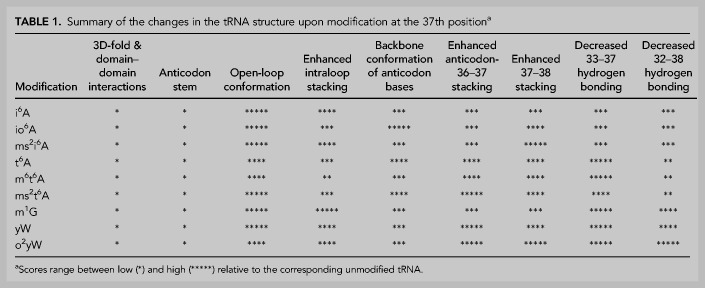
TABLE 1.
Summary of the changes in the tRNA structure upon modification at the 37th positiona
t6A modifications significantly change the conformation of anticodon loop nucleotides, as well as enhance stacking and decrease hydrogen bonding with base 37, with a dependence on the level of chemical substitution
Similar to i6A, the t6A family introduces modifications at the N6 position of adenine. Specifically, t6A contains a uriedo-threonyl moiety at N6 of A37, which is further substituted with a methyl group at N6 in m6t6A or a thiomethyl group at C2 in ms2t6A (Fig. 1B). Although sequence analysis highlights that the ms2t6A modification is absent in E. coli, it is present in bacteria (Bacillus subtilis) and eukaryotes (Drosophila melanogaster, Lupinus luteus, Triticum aestivum, and Rattus norvegicus). Nevertheless, ms2t6A will be considered in the same sequence context in this study to understand the fundamental impact of t6A thiomethyl substitution on tRNA structure. These are much larger hypermodifications than the i6A family and exhibit even greater dynamics within the bulky substituent (Fig. 6A,B). Indeed, our simulations show that the t6A modified base adopts a distal conformation throughout the simulation, where the N6-threonylcarbonyl substituent spreads away from the imidazole moiety of base A (Sambhare et al. 2014). As a result, although the t6A-modified tRNAs are stable across the simulation trajectory (Supplemental Fig. S6) and retain the global architecture of mature tRNA (Supplemental Fig. S16A), the average RMSD for the modified tRNA backbone (∼4.3–4.8 Å) is significantly larger than that for the corresponding unmodified A-tRNA (∼3 Å, Supplemental Table S1). Indeed, all t6A modifications considered in the present work increase the dynamics of most tRNA nucleotides compared to the unmodified tRNA (Supplemental Fig. S8). This is especially true for nucleotides in the anticodon loop (Fig. 6C), suggesting that the modifications (particularly ms2t6A) directly impact neighboring nucleotides. Furthermore, unlike the i6A family, all t6A modifications considered increase structural deviations of nucleotides in domains outside the tRNA anticodon stem (Supplemental Fig. S8). Nevertheless, the modifications do not significantly affect the dynamic interactions between RNA domains (Supplemental Fig. S16B).
FIGURE 6.
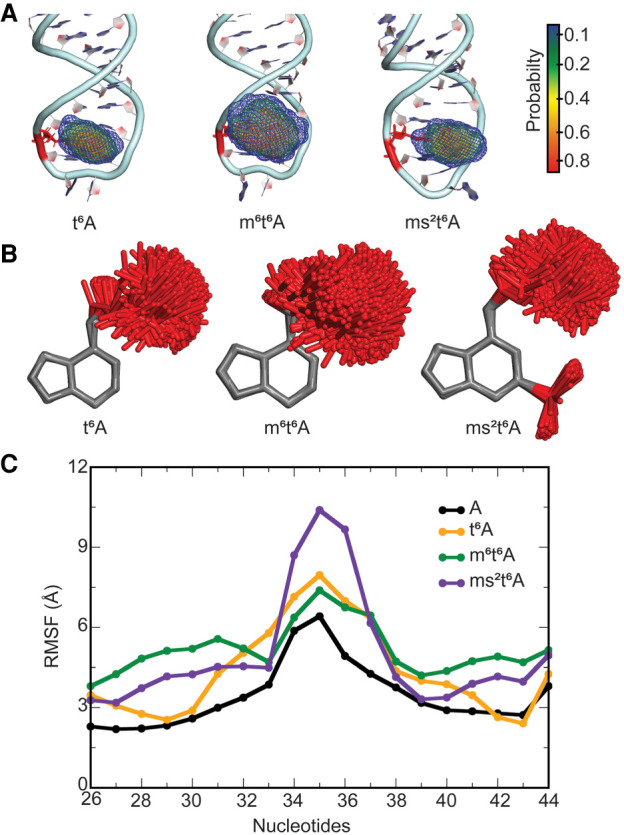
(A) Probability distribution in the location of the side chain in the t6A hypermodified nucleobase at the 37th position in E. coli tRNA. (B) Overlays of t6A, m6t6A, and ms2t6A modified nucleobases in tRNA with respect to the nucleobase ring for structures taken at 5 ns intervals over the MD simulations. (C) Average structural deviation of anticodon nucleotides (RMSF, Å) over the MD simulations with respect to the initial structure for each t6A modified tRNA compared to unmodified A-tRNA.
Despite increased overall tRNA dynamics, the base pairs in the stem region of the ASL are maintained upon introduction of t6A-derived modifications (Supplemental Tables S2–S4) and the backbone conformation induces the expected kink between nucleotides 33 and 34 (Supplemental Fig. S17), as evidenced by NMR spectroscopic data for t6A-modified human tRNALys (Stuart et al. 2000) and E. coli tRNALys (Sundaram et al. 2000). In complement to previous NMR data suggesting that the parent t6A modification leads to anticodon loop remodeling and increased dynamic flexibility of the anticodon bases (Stuart et al. 2000; Durant et al. 2005), our MD data shows that all members of the t6A family significantly affect the backbone conformation in the anticodon loop between bases 33 to 37, with smaller changes occurring at 32 and 38 (Supplemental Figs. S17, S18). Furthermore, all t6A hypermodifications impact the orientation of the base about the glycosidic bond at positions 34 and 35, while the impact of ms2t6A is further reaching, influencing χ and δ from 33 to 38, with U33 frequently adopting the syn glycosidic orientation (Supplemental Fig. S18). The shifts in the backbone in the ASL region significantly impact the stacking interactions between loop bases outside position 37 (Supplemental Fig. S19; Supplemental Table S6). Specifically, in addition to the absence of stacking between bases 33 and 34 due to the U-turn motif, stacking is greatly reduced between the 32–33 and 35–36 base neighbors, while less than optimal stacking also occurs between 34–35 as discussed for unmodified A-tRNA. Nevertheless, for all modifications, the backbone maintains the conformation of unmodified tRNA at positions 31 and 39, which suggests that the conformational change in the loop does not propagate through the stem. In fact, stacking between bases 31–32 and 38–39, and U32–A38 hydrogen bonding are maintained for all t6A modifications (Supplemental Fig. S19; Supplemental Tables S2–S4), which correlates with NMR data for t6A-modified human tRNALys (Stuart et al. 2000).
While the modification does not impact the anti glycosidic orientation of the 37th nucleotide for t6A and m6t6A, the conformation adopted by ms2t6A ranges from syn to near-anti (χ = 210° to 330°, Supplemental Figs. S12, S18). Although the shift in the glycosidic orientation further highlights the significant impact of the thiomethyl group, the anti conformation of ms2t6A observed in a crystal structure of HIV reverse-transcription primer tRNA (χ = 182°) (Benas et al. 2000) is sampled during the MD simulations (Supplemental Fig. S20). The uriedo-threonyl group is very flexible and covers a significant portion of conformational space throughout the simulations for all t6A modifications (Fig. 6A,B; Supplemental Fig. S13), including the orientation adopted in crystal structures of tRNA containing t6A (Murphy et al. 2004) or ms2t6A (Supplemental Fig. S20; Benas et al. 2000). Closer analysis of the side chain conformations reveals that the uriedo group primarily adopts one orientation throughout the simulations across the t6A-modified tRNAs [ω1 = ∠(N1C6N6C10) ∼ 0°, ω2 = ∠(C6N6C10N12) ∼ 0° and ω3 = ∠(N6C10N12C13) ∼ 180°, Supplemental Fig. S21], with a slight shift about the C6–N6 bond upon introduction of the N6 methyl group of m6t6A (ω1 ∼ ±30°). This conformation is consistent with crystal structures (Parthasarathy et al. 1977; Murphy et al. 2004) and NMR data (Stuart et al. 2000) suggesting that t6A maintains planarity between the side chain and the nucleobase. In contrast to the uriedo group, the threonyl group primarily adopts two conformations with respect to the 1-hydroxylethyl substituent, namely gauche +60 and gauche −60 [ω5 = ∠(N12C13C15O)], with the population of the gauche −60 conformation increasing upon further substitution of the parent t6A hypermodification (Supplemental Fig. S21). Thus, additional substitution of t6A can affect the accessible side chain orientations of the hypermodified base.
All members of the t6A family at position 37 stack between the neighboring 36 and 38 nucleobases in the anticodon loop of tRNA (Fig. 7A). NMR and crystallographic structural data showed that the t6A (Sundaram et al. 2000; Rozov et al. 2016) and ms2t6A (Benas et al. 2000; Durant et al. 2005) modifications have increased overlap with bases 36 and 38 compared to A, and therefore promote a stable anticodon loop conformation. However, our data suggests the increased structural dynamics upon base modification reduces the stacking overlap in this region (Supplemental Fig. S19). Furthermore, the standard deviations in the average 36–37 and 37–38 stacking interactions are much higher for the hypermodifications compared to A (Supplemental Table S8). Regardless, the presence of the hypermodification ensures that the average stacking interaction energies at the same position are larger in modified tRNA compared to A-tRNA (by up to ∼4.5 kcal/mol for 36 and ∼3 kcal/mol for 38, Supplemental Table S8). This suggests that the modifications contribute to the stability of the anticodon loop by enhancing stacking despite inducing structural changes in the loop nucleotides, including the syn glycosidic orientation associated with ms2t6A.
FIGURE 7.
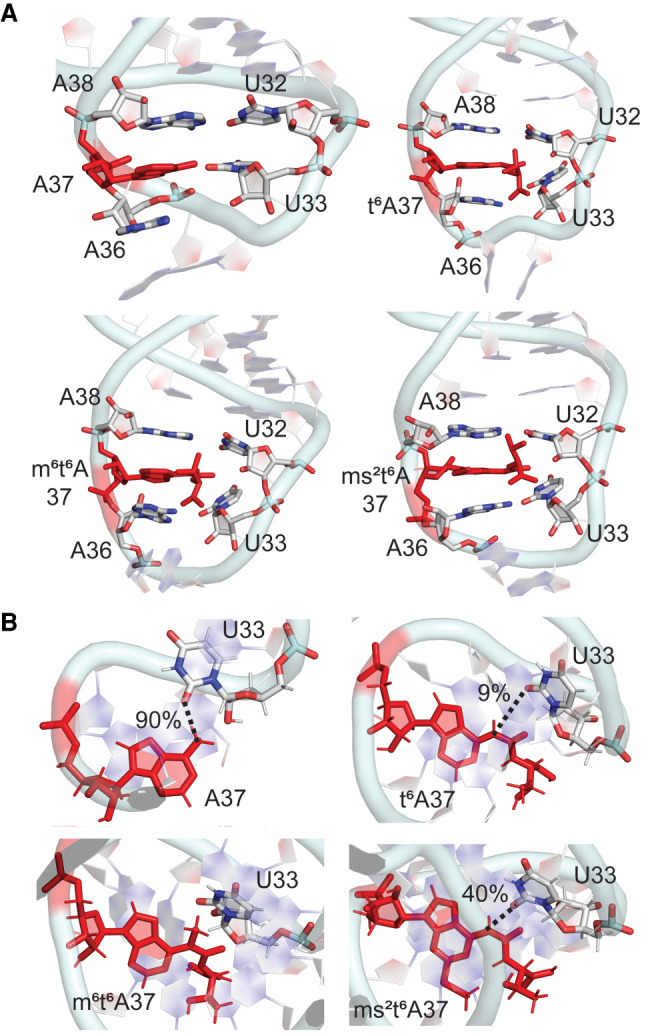
(A) Stacking interactions between base 37 and neighboring nucleobases in the anticodon loop and (B) hydrogen-bonding interactions (with occupancies) between bases 37 and 33 throughout the MD simulations for unmodified and t6A-modified E. coli tRNAs.
As discussed for the i6A family, the t6A modifications influence hydrogen-bonding interactions with base 37. Specifically, although the N6(A37) … O2(U33) hydrogen bond is maintained throughout the MD simulations for unmodified A-tRNA (90% occupancy), this interaction is significantly disrupted in the presence of t6A and ms2t6A (maximum 40% occupancy), and completely abolished in the presence of the N6 methyl group of m6t6A (Fig. 7B). The lack of the U33–t6A37 interaction in our simulations correlates with proposals that the lack of this interaction in a crystal structure of tRNALys containing t6A improves the anticodon loop conformation when bound to the ribosomal site for codon–anticodon duplex formation (Murphy et al. 2004) and suggestions that t6A prohibits canonical base-pairing based on short-time scale MD simulations on only the ASL region containing multiple modifications (Sambhare et al. 2014). Nevertheless, the magnitude of the average interaction energies between A37 and U33 in unmodified A-tRNA is maintained upon t6A substitution (∼−3 to −4 kcal/mol, Supplemental Table S9). Furthermore, the largest A37–U33 interaction is greater for the t6A modifications (up to ∼−10 ± 2.4 kcal/mol) compared to A (∼−4 ± 1.1 kcal/mol). These trends in the interaction energy occur because the side chains of the t6A modifications can also form discrete interactions with the U33 nucleotide, either stacking with the nucleobase or hydrogen bonding with O2, O2′, and O4′, as discussed for the i6A family. Nevertheless, the modified bases at position 37 do not interact with cross-strand bases in the dominant conformation of the ASL (Fig. 3), which agrees with previous reports for t6A-tRNA based on NMR spectroscopy (Sundaram et al. 2000). Interestingly, the rearrangement induced by the addition of the thiomethyl group at C2 increases sampling of the dominant open-loop conformation. Thus, the overall functional conformation of the anticodon loop is achieved in different ways for different modifications (Fig. 3).
In summary, the t6A family underscores the ability of hypermodifications to fine-tune the configuration of the anticodon bases in unbound tRNA without abolishing the structure required for function (Table 1). Specifically, the bulkier t6A modifications have more drastic and further reaching impacts on the anticodon stem–loop than members of the i6A family. Indeed, the t6A family emphasizes that loop nucleotides can adjust to accommodate a very bulky side chain at N6 of A37. Furthermore, the t6A family illustrates how tRNA hypermodifications can significantly impact many structural features in the anticodon stem–loop including increasing dynamics, changing the backbone conformation, disrupting intrastrand hydrogen bonding, and strengthening base–base stacking. These differences alter the conformation of the critical anticodon nucleotides, yet stabilize the ASL in a dominant open-loop conformation (Fig. 3). These observations for the broader t6A family complement the previously reported remodeling of the anticodon loop upon incorporation of the parent t6A modification based on NMR spectroscopic studies of tRNALys (Stuart et al. 2000; Yarian et al. 2000). Our data also further highlight that the structural changes in the loop region are not propagated into the anticodon stem, and the overall integrity of the global tRNA structure and domain–domain interactions are preserved.
Despite substantial bulk, yW modifications impart minor changes to the conformation of the anticodon stem–loop of unmodified G-tRNA in a manner similar to the smaller m1G modification, yet enhance the stability of the anticodon loop through discrete noncovalent interactions
Unlike the other families discussed thus far, the yW modifications are guanine derivatives. These modifications are among the bulkiest nucleobase derivatives occurring at the 37th position, involving the addition of a ring to the Watson–Crick binding face, as well as a flexible side chain (Fig. 1B). Among the many yW modifications that have been identified to date, yW and o2yW are considered in the present work, which differ in the additional peroxy group on the side chain of o2yW. The distribution of G modifications at the 37th position showed that m1G is the most populated G-modification and is present in bacteria (Escherichia coli, Bacillus subtilis), archaea (Halobacterium salinarum, Haloferax volcanii), and eukaryotes (Saccharomyces cerevisiae, Homo sapiens), followed by yW and o2yW in eukaryotes (Saccharomyces cerevisiae, Xenopus laevis, Homo sapiens) (Jühling et al. 2008; Boccaletto et al. 2017). Despite m1G being common at the 37th position of E. coli tRNAPhe, we draw comparisons in the present work between the yW family of modifications and unmodified G-tRNA to separate the impact of the parent nucleobase versus modification substituent. Subsequent comparison to tRNA containing m1G at the 37th position permits an understanding of the relative impact of (nonbulky) methylation versus (bulky) hypermodifications.
Both yW derivatives adopt a range of conformations at position 37 of tRNA (Fig. 8A,B). Despite the bulk and inherent flexibility of the yW modification family, the associated modified tRNAs retain the global L-shaped architecture (Supplemental Fig. S22A), average backbone RMSD (∼3 Å, Supplemental Fig. S6; Supplemental Table S1) and domain–domain interactions (Supplemental Fig. S22B) of unmodified G-tRNA. Nevertheless, these bulky modifications reduce the nucleotide dynamics, especially in the anticodon region (Fig. 8C; Supplemental Fig. S8).
FIGURE 8.
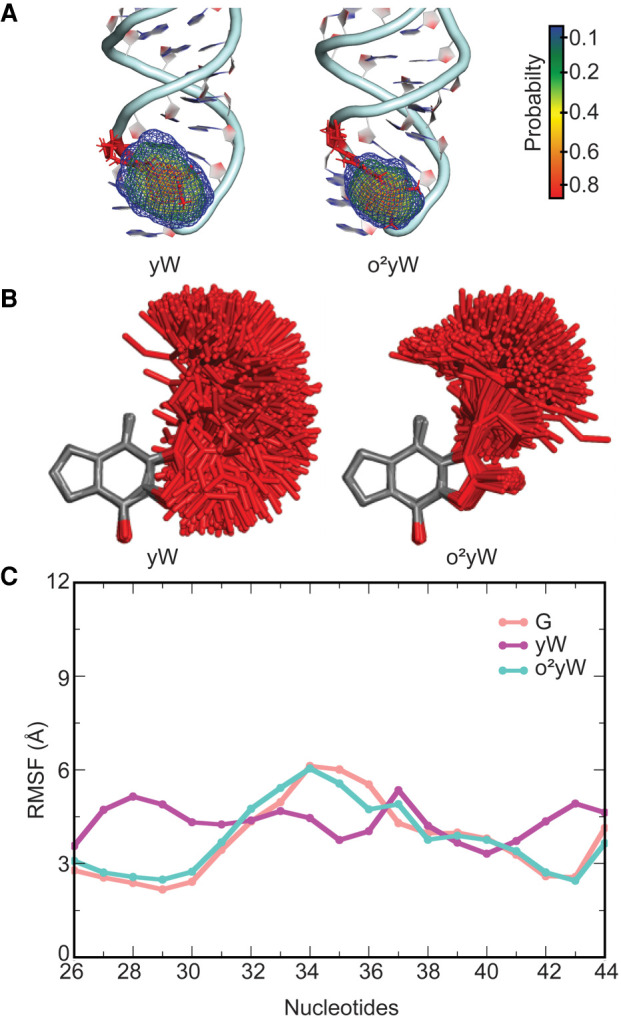
(A) Probability distribution in the location of the side chain in the yW hypermodified nucleobase at the 37th position in E. coli tRNA. (B) Overlays of yW and o2yW modified nucleobases in tRNA with respect to the nucleobase ring for structures taken at 5 ns intervals over the MD simulations. (C) Average structural deviation of anticodon nucleotides (RMSF, Å) over the MD simulations with respect to the initial structure for each yW modified tRNA compared to unmodified G-tRNA.
The structure of the stem in the ASL domain is maintained upon the introduction of a yW family modification, which includes critical nucleoside glycosidic orientations required to form Watson–Crick hydrogen bonding (Supplemental Tables S2–S4, S7). Unlike the A modifications discussed previously, the yW modifications result in similar loop backbone conformations and dynamics as unmodified G–tRNA at position 37 (Supplemental Fig. S23). The only exceptions are a slight change in δ at U33 for both modifications, as well as the glycosidic orientation of A35 for yW and G34 for o2yW (Supplemental Fig. S24). Nevertheless, these alterations in the loop conformation of G-tRNA upon modification are very modest. Importantly, the kink is preserved between bases 33 and 34 (Supplemental Fig. S23), and there are no changes in helical bases A31 or U39, which suggests the anticodon stem is preserved as seen for the other modifications. Indeed, C1′–C1′ distances and base–base stacking between the loop nucleotides more closely mimic those occurring in unmodified tRNA compared to the other modification families discussed thus far (Supplemental Tables S5–S6; Supplemental Fig. S25). Hydrogen bonding between U32 and A38 in G-tRNA is also maintained for the yW modifications (Supplemental Tables S2–S4).
The glycosidic orientation of base 37 remains in the anticonfiguration for the yW modifications (Supplemental Fig. S12; Supplemental Table S7). Indeed, an NMR solution structure of yW-containing yeast tRNAPhe suggests that the nucleotide conformation may be restricted due to its bulky size and intercalation between the neighboring bases (Stuart et al. 2003). However, the flexible alkyl side chains of the yW derivatives adopt a wide range of orientations relative to the base (Fig. 8A,B), resulting in a higher RMSD for the nucleotide at the 37th position (Supplemental Fig. S13). Nevertheless, the conformation observed in a crystal structure of yeast tRNAPhe (Mikkelsen et al. 2001), which contains yW at the 37th position, is covered during our MD simulations (Supplemental Fig. S26). Interestingly, the parent yW modification adopts a particularly wide spread of conformations, while the addition of the peroxy group significantly reduces the conformational flexibility of the modification side chain, which correlates with a previous short-time scale MD study on tRNA anticodon loops containing yW or o2yW at the 37th position (Fandilolu et al. 2018). Despite the many degrees of rotational freedom in the side chains of yW-derived modifications, the flexibility observed at position 37 primarily occurs in the alkyl chain closest to the base (i.e., the ψ1 [∠(N1C11C12C13)] and ψ2 [∠(C11C12C13C14)] dihedral angles, Supplemental Fig. S27). Indeed, two conformations with respect to ψ1 and ψ2 are adopted for yW, while there is one predominant orientation with respect to ψ1 and ψ2 for o2yW.
Regardless of the side chain flexibility, the stacking overlap between base 37 and the neighboring bases 36 and 38 observed in G and m1G-tRNA remains very high for yW (Fig. 9A; Supplemental Fig. S25). In contrast, although base 37 can stack between bases 36 and 38 upon the introduction of the peroxy group (Fig. 9A), the frequency of the stacking with neighboring bases is reduced for o2yW (Supplemental Fig. S25). Nevertheless, the additional ring in both yW modifications compared to G leads to a significant increase in the magnitude of the average stacking interactions at position 37 (Supplemental Table S8). Indeed, the average stacking interaction is up to ∼4.5 ± 2.4 kcal/mol more stable for yW than G, with the interaction becoming stronger with base 36 than 38, highlighting a rearrangement in the anticodon loop nucleotides. Furthermore, the average stacking interactions with o2yW are even stronger, being up to ∼−10 ± 3.6 kcal/mol, at least in part due to additional periodic interactions between the peroxy group and the stacked base. Thus, the yW family of modifications provides additional stability to the anticodon loop through increased stacking. This finding agrees with previous suggestions in the literature based on the anticipated increased nucleobase hydrophobicity and size (Dao et al. 1994) and the previously noted decreased stability when the parent yW is replaced with A (Katunin et al. 1994), as well as the trends discussed for the t6A family in the present work. Due to their large size and range of orientations in tRNA, these modified bases also possibly stack with the incoming first codon base during translation and thereby further increase the stability of the codon–anticodon duplex.
FIGURE 9.
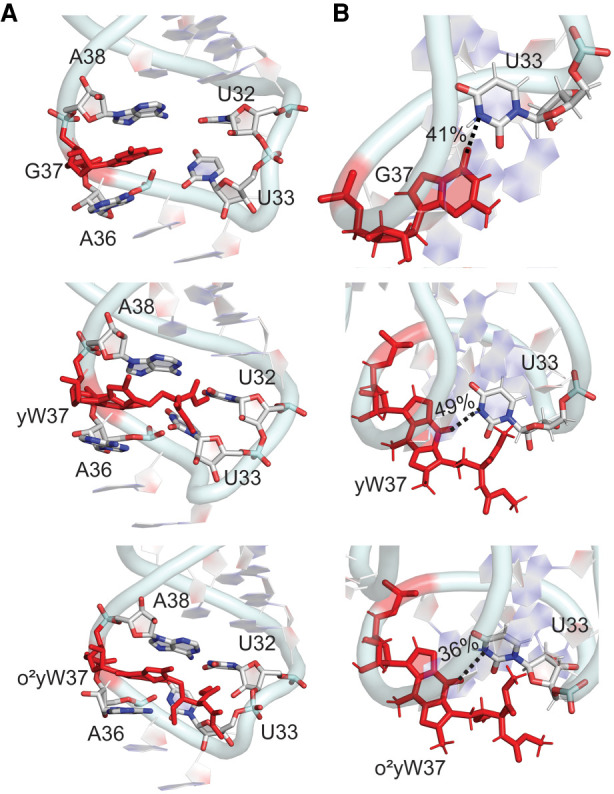
(A) Stacking interactions between base 37 and neighboring nucleobases in the anticodon loop and (B) hydrogen-bonding interactions (with occupancies) between bases 37 and 33 throughout the MD simulations for unmodified and yW-modified E. coli tRNAs.
In terms of important loop hydrogen-bonding interactions, a O6(G37) … N3(U33) hydrogen bond occasionally forms along the dynamics of G-tRNA (occupancy ∼40%, Fig. 9B). Unlike m1G-tRNA, this interaction also occurs upon incorporation of the yW or o2yW modification into the 37th position of tRNA (Fig. 9B). Furthermore, the side chains of yW and o2yW can stack or hydrogen bond with U33 as discussed for the i6A and t6A families. These noncovalent contacts can serve to stabilize the anticodon loop (Supplemental Table S9). Nevertheless, the presence of the hydrogen-bonding and stacking interactions between the modified base and U33 are highly dependent on the side chain conformation, and these interactions are present for a relatively short amount of time. Indeed, the dominant conformation of the modified-tRNAs highlight a similar open-loop conformation previously discussed for m1G (Fig. 3). Although these observations correlate with previous suggestions that changes in the G Watson–Crick binding face upon formation of yW alter interactions between base 37 and U33 (Dao et al. 1994), our simulations highlight the complicated interplay between several different noncovalent interactions to maintain the dominant anticodon loop conformation and the impact of the additional peroxy substituent.
Overall, this third family emphasizes a growing theme that tRNA hypermodifications can fine-tune the structural properties of the anticodon loop, with the addition of small functional groups (such as a peroxy group) further modulating tRNA structure and likely regulating function (Table 1). Despite inherent significant bulk and conformational flexibility, the yW family of modifications does not impact the global tRNA L-fold structure or domain–domain interactions, suggesting that the modifications have no long-range structural effects on tRNA. Although the peroxy group of o2yW introduces rigidity to the side chain of yW, both modifications enhance discrete noncovalent interactions with base 37 in the anticodon loop. More importantly, the yW modifications impart changes to the conformation of the anticodon stem–loop of unmodified G-tRNA in a manner similar to the smaller m1G modification.
Despite subtle changes in nucleotide–nucleotide interactions in the ASL, the impact of hypermodifications is preserved in different sequence contexts
There are a number of modifications that occur at the 37th position across all tRNAs and organisms, and therefore different hypermodifications occur in different sequence contexts (Jühling et al. 2008; Grosjean and Westhof 2016). As stated previously, we implemented a consistent model in the present work that was built from E. coli tRNAPhe in attempts to decipher and compare the impact of various modification families on the structure of tRNA. Nevertheless, to investigate the effects of sequence context, we have also considered unmodified G, m1G and the bulky yW modification in yeast tRNAPhe as representative comparisons since G modifications such as yW and derivatives are observed in general higher organisms (Saccharomyces cerevisiae, Brassica napus, Mus musculus, and Homo sapiens) (Jühling et al. 2008; Boccaletto et al. 2017). Importantly, yeast tRNAPhe has sequence variation in the anticodon stem–loop compared to E. coli tRNAPhe (Supplemental Fig. S28A), including G rather than A at position 37 and C rather than U at position 32.
tRNA maintains the global L-fold and similar domain–domain interactions (Supplemental Fig. S28), as well as similar average backbone RMSD and nucleotide dynamics (Supplemental Fig. S29; Supplemental Table S13), regardless of the nature of base 37 or the tRNA sequence considered. The backbones of nucleotides in the anticodon stem–loop maintain the same structural features for unmodified and modified (m1G or yW) at position 37, including the distinctive U-turn and an anti-orientation of the 37th nucleotide, and the organism has little impact (Supplemental Figs. S23, S24, S30, S31). Compared to canonical G, as well as m1G, yW is inherently highly dynamic in yeast tRNA as observed in E. coli tRNA (Supplemental Fig. S32). The orientation of yW at the 37th position observed in a crystal structure of yeast tRNAPhe (Mikkelsen et al. 2001) is sampled during the MD simulations (Supplemental Fig. S26B). Despite inherent flexibility of the hypermodification, G, m1G, and yW stack similarly and consistently with bases 36 and 38 in both tRNAs (Fig. 10; Supplemental Fig. S33; Supplemental Tables S6, S14). This equivalent relative nucleotide arrangement is anticipated to enhance stacking interactions in the case of yW in yeast tRNA due to the increased size of the 37th base as previously discussed for E. coli tRNAPhe.
FIGURE 10.
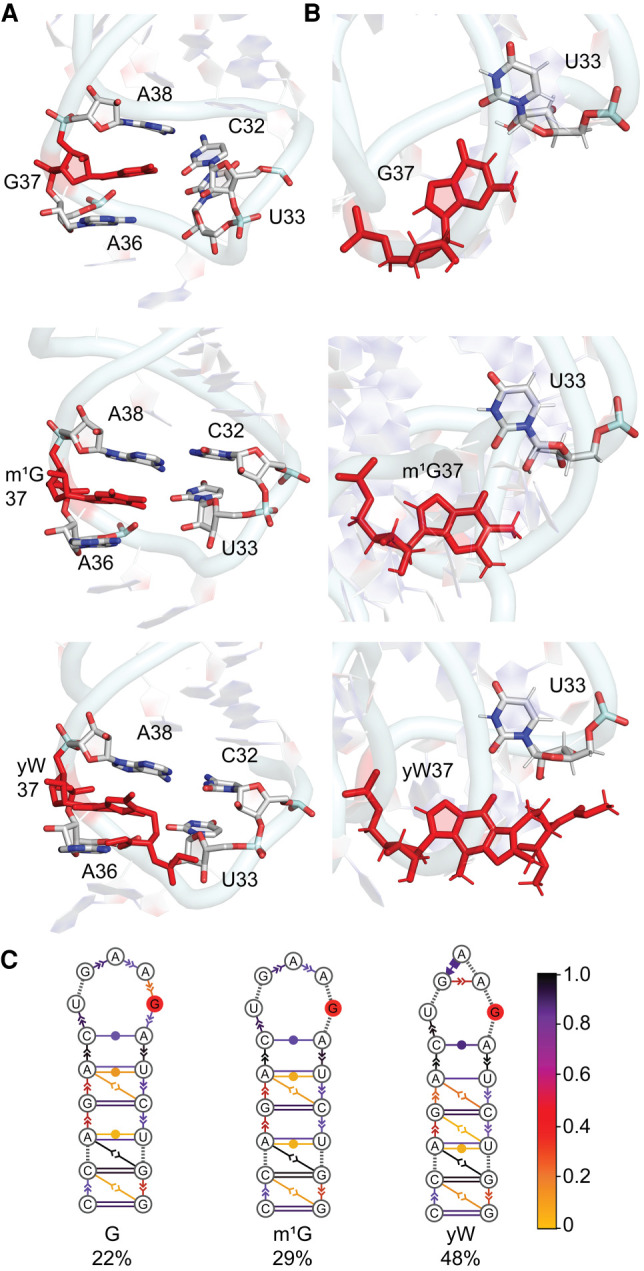
(A) Stacking interactions between base 37 and neighboring nucleobases in the anticodon loop of yeast tRNA and (B) hydrogen-bonding interactions (with occupancies) between bases 37 and 33 throughout the MD simulations for unmodified, m1G and yW-modified yeast tRNAs. (C) consensus secondary structures of the anticodon stem–loop region in unmodified and modified tRNA, with the solid red circle representing the nucleotide at the 37th position of yeast tRNA. The color scheme shows the probability distribution of the interactions within the cluster.
The most significant difference between the behavior of base 37 in the two tRNA sequences considered herein is the presence of cross-strand hydrogen bonding with U33 in E. coli tRNA (Fig. 2) and the stark absence of hydrogen bonding with U33 in yeast tRNA (Fig. 10; Supplemental Table S15). Nevertheless, the sidechain of yW forms various noncovalent interactions with U33 in yeast that are present for short periods of time as discussed for E. coli tRNA. Furthermore, the hydrogen-bonding interactions between the 37th nucleotide and U33 are highly similar for all three bases at position 37 (Fig. 10). As a result, the dominant conformations of the ASL region in the tRNAs highlight an open-loop conformation, with an increased occupancy for m1G and especially yW-tRNA (Figs. 3, 10). Importantly, the main conclusions of the impact of the modifications hold in both sequences, namely that the bulky yW modification results in a similar conformation of the anticodon stem–loop as the smaller m1G modification. Therefore, the inherent behavior of the hypermodification and the associated impact of the modification on tRNA structure are well preserved across different sequence contexts.
Conclusions
The present work uses molecular dynamics simulations on a variety of unmodified and modified tRNA to provide an atomistic understanding of the effects of different hypermodifications at the 37th position on the overall fold and structural dynamics of tRNA, with particular emphasis on the anticodon loop. Three modification families (i6A, t6A, and yW) were considered to investigate the impact of the size of the chemical substituents added to the canonical bases. In each case, further chemical substitution of the parent modification was also investigated to uncover the role of small substituents (a hydroxy, methyl, thiomethyl or peroxy group). Regardless of the modification family or level of chemical substitution, these hypermodifications do not affect the global tRNA three-dimensional structure or the domain–domain interactions, suggesting that there are no long-range effects of modifying the 37th position. Although the overall structure of the anticodon stem remains intact regardless of the modification considered, the anticodon loop must rearrange to accommodate these hypermodifications, with the extent of the structural adjustments depending on the size of the bulky moiety and level of chemical substitution. Specifically, for adenine modifications, i6A imparts modest changes in the loop backbone and/or nucleobase interactions, while the larger t6A significantly disrupts the conformation of all anticodon loop nucleotides, including the glycosidic orientation of the anticodon nucleotides. In contrast, the yW modification derived from guanine does not significantly impact the structure of the anticodon stem–loop despite being very bulky. Regardless of the degree of loop rearrangement, the larger t6A and yW modifications provide stability to the loop through enhanced or even completely new intrastrand interactions. Furthermore, significant differences in the loop conformation and stability exist between parent modifications and associated derivatives arising from the addition of a hydroxy, methyl, thiomethyl or peroxy group. Indeed, such chemical substitutions can influence the flexibility of the hypermodification side chain and the glycosidic orientation of base 37 or anticodon nucleotides, which subsequently impact intraloop nucleobase–nucleobase hydrogen-bonding and stacking interactions. These molecular features collectively drive the ASL domain of tRNA to adopt a dominant open-loop conformation. Importantly, the impact of these hypermodifications is preserved in different sequence contexts. The changes imparted by these hypermodifications to the anticodon nucleotides are particularly important since these may regulate mRNA translation. Thus, our results provide the previously molecular-level details for how select nucleobase bulky moieties adjust tRNA structure, while maintaining the functional architecture of this important biomolecule. In addition to broadening our understanding of tRNA biochemistry, the knowledge gained from the present work regarding the ability to alter the molecular features of modified RNA through small chemical substitutions affords valuable insight that can be exploited in the future design of improved nucleic acid-based therapeutics and artificial biosensors.
MATERIALS AND METHODS
Initial RNA model
A high-resolution (3.0 Å) X-ray crystal structure corresponding to standalone full-length unmodified E. coli tRNAPhe (PDB ID: 3L0U) was used to build initial structures for MD simulations (Byrne et al. 2010). This model choice is justified based on the similarities between the structures of unbound (PDB ID: 5L4O [Monestier et al. 2017]) and bound tRNAs (PDB IDs: 4YCO [Byrne et al. 2015] and 6QNR [Rozov et al. 2019]) in a range of crystal structures (see, e.g., Supplemental Fig. S34A), as well as the similarities between our predicted tRNA structures and the crystal structures (see, e.g., Supplemental Fig. S34B). The initial structures of G-tRNA and the eight modified tRNAs were obtained by manually replacing A37 in the crystal structure with the respective base using Pymol (The PyMOL Molecular Graphics System, Version 1.8, Schrodinger LLC) and GaussView 6 (Dennington et al. 2019). The use of a single crystal structure starting point allows for an effective comparison of all modifications considered in the present work in the absence of possible structural differences arising from variations in the nucleotide sequence. Nevertheless, the impact of the sequence context was also investigated using models developed based on yeast tRNAPhe. Specifically, these models were generated from a high-resolution (1.9 Å) X-ray crystal structure corresponding S. cerevisiae tRNAPhe (PDB ID: 1EHZ) (Shi and Moore 2000), with all bases reverted to the unmodified forms, except for yW at the 37th position. Subsequently, unmodified G, and m1G were incorporated at the 37th position. In all models, the initial modified nucleotide conformation was adjusted by visual inspection to maintain the anti glycosidic conformation [χ = ∠(O4′C1′N9C4)], while minimizing steric clashes between the bulky substituent and neighboring nucleotides. Thus, 14 models were considered in total that differ in the nucleobase at position 37, including two unmodified E. coli tRNAs (containing A or G) and nine modified E. coli tRNAs (containing i6A, io6A, ms2i6A, t6A, m6t6A, ms2t6A, m1G, yW, or o2yW), as well as three yeast tRNAs (containing G, m1G, and yW).
MD simulations
All tRNA models were prepared using the tLEaP module of the AMBER 16 program package (Case et al. 2016). The ff99bsc0χOL3 force field was used for tRNA. Parameters for the modified bases were supplemented by the Generalized Amber Force Field (GAFF) (Wang et al. 2004), with partial atomic charges calculated with RED.v.III.4 using the RESP-A1 scheme (Dupradeau et al. 2010). In all simulations, the total charge was neutralized by Na+ ions and the tRNA was solvated in an explicit TIP3P octahedral water box, ensuring that the edge of the box is at least 10.0 Å from the edge of the solute in each direction. The solvent molecules and ions were initially minimized with 500 steps of steepest descent and 500 steps of conjugate gradient minimization using a nonbonded cutoff of 10 Å, while the tRNA was constrained using a 500.0 kcal mol–1 Å–2 force constant. The entire system was then minimized using 1000 steps of unrestrained steepest descent, followed by 1500 steps of unrestrained conjugate gradient minimization. Subsequently, the system was heated from 0 to 300 K during a 100 psec solute-restrained (10 kcal mol–1 Å–2) equilibration phase, using a 1 fsec time step. This was followed by a 20 nsec (unconstrained) trial MD simulation under NPT conditions (1 atm and 300 K), with a time step of 2 fsec and the SHAKE algorithm. The periodic boundary condition was used for all steps. Finally, starting from the conformation in the last frame of the corresponding trial simulation using a time step of 2 fsec and the pmemd module of AMBER 16 (Case et al. 2016), 1 µsec MD production simulations were performed in triplicate for each model. Thus, a total of 3 µsec of simulation data was collected per system for a total of 42 µsec over all tRNAs.
Trajectory analysis
Each trajectory was sampled for analysis every 20 psec over the course of the production phase. However, as the tRNA system was very flexible for the first 200 nsec, all analysis reported in the main text was performed over the last 800 nsec of the first MD simulation replicate. This data is in full agreement with analysis performed over the total 3 µsec of simulation data for each system (Supplemental Tables S10–S15; Supplemental Figs. S35–S43). Trajectory analysis was completed using the cpptraj module (Roe and Cheatham 2013) of AMBER 16. The RMSDs in the tRNA nucleotide backbone atoms were analyzed, with respect to the corresponding crystal structure coordinates for each simulation to assess system stability. Each trajectory was clustered with respect to the configuration of base 37 and the reported MD representative structures were obtained from the dominant cluster over the simulation in all cases. The density grids for the modified base side chain were obtained using a grid size of 60 Å and spacing of 0.5 Å. All average backbone RMSD, per-residue RMSFs, backbone dihedral angles, χ [∠(O4′C1′N9C4)] torsional angles, dihedral angles within the bulky substituents, hydrogen-bonding parameters and stacking propensities were calculated over the MD trajectories. To determine hydrogen-bonding occupancies, cut-offs of 120° for angles and 3.4 Å for distances were imposed. The base stacking propensity between the anticodon nucleotides was calculated based on the criteria that the distance between the center of masses of the two bases (including the side chain of the hypermodification) was ≤6 Å, and the angle between the two normal vectors of the bases was ≤40° or ≥ 140°. The magnitude of base pair hydrogen-bonding interactions and nucleobase–nucleobase stacking interactions were calculated by extracting the atomic coordinates from the simulation trajectories at 5 nsec intervals. The ribose moiety of each nucleotide was replaced with a hydrogen atom, hydrogen-only optimizations were performed using B3LYP/6-31+G(d,p), and the interaction energies (including counterpoise corrections) were calculated using B3LYP-D3(BJ)/6-311+G(2df,p). All DFT calculations were performed using Gaussian 16 (Frisch et al. 2016). Dynamic cross-correlation analysis was carried out using the cpptraj module (Roe and Cheatham 2013) of AMBER 16. The extent of correlated motion between two residues was calculated as the magnitude of the correlation coefficient between the C1′ atoms of the two nucleotides of interest. The covariance matrix, which highlights positive (correlated) and negative (anticorrelated) correlation coefficients of the nucleotides, was created using the gnuplot program. Each MD simulation was clustered to determine the dominant structure of the ASL using the Barnaba software package (Bottaro et al. 2019).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; 2016-04568), Canada Foundation of Innovation (22770), and the Board of Governors Research Chair Program of the University of Lethbridge (UofL). Computational resources from the New Upscale Cluster for Lethbridge to Enable Innovative Chemistry (NUCLEIC), and those provided by Westgrid and Compute/Calcul Canada are greatly appreciated. P.S.P. thanks RNA Innovation (NSERC-CREATE) for a postdoctoral fellowship.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078097.120.
REFERENCES
- Agris PF. 2008. Bringing order to translation: the contributions of transfer RNA anticodon–domain modifications. EMBO Rep 9: 629–635. 10.1038/embor.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RW, Eargle J, Luthey-Schulten Z. 2010. Experimental and computational determination of tRNA dynamics. FEBS Lett 584: 376–386. 10.1016/j.febslet.2009.11.061 [DOI] [PubMed] [Google Scholar]
- Allnér O, Nilsson L. 2011. Nucleotide modifications and tRNA anticodon–mRNA codon interactions on the ribosome. RNA 17: 2177–2188. 10.1261/rna.029231.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. 1999. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J Mol Biol 292: 467–483. 10.1006/jmbi.1999.3080 [DOI] [PubMed] [Google Scholar]
- Barbieri I, Kouzarides T. 2020. Role of RNA modifications in cancer. Nat Rev Cancer 920: 303–322. 10.1038/s41568-020-0253-2 [DOI] [PubMed] [Google Scholar]
- Benas P, Bec G, Keith G, Marquet R, Ehresmann C, Ehresmann B, Dumas P. 2000. The crystal structure of HIV reverse-transcription primer tRNALys,3 shows a canonical anticodon loop. RNA 6: 1347–1355. 10.1017/S1355838200000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A. 2017. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottaro S, Bussi G, Pinamonti G, Reißer S, Boomsma W, Lindorff-Larsen K. 2019. Barnaba: software for analysis of nucleic acid structures and trajectories. RNA 25: 219–231. 10.1261/rna.067678.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RT, Konevega AL, Rodnina MV, Antson AA. 2010. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res 38: 4154–4162. 10.1093/nar/gkq133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RT, Jenkins HT, Peters DT, Whelan F, Stowell J, Aziz N, Kasatsky P, Rodnina MV, Koonin EV, Konevega AL. 2015. Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases. Proc Natl Acad Sci 112: 6033–6037. 10.1073/pnas.1500161112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Villegas J, Winkler ME, Nikonowicz EP. 2002. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J Mol Biol 319: 1015–1034. 10.1016/S0022-2836(02)00382-0 [DOI] [PubMed] [Google Scholar]
- Cabello-Villegas J, Tworowska I, Nikonowicz EP. 2004. Metal ion stabilization of the U-turn of the A37N6-dimethylallyl-modified anticodon stem–loop of Escherichia coli tRNAPhe. Biochemistry 43: 55–66. 10.1021/bi0353676 [DOI] [PubMed] [Google Scholar]
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. 2010. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 39: D195–D201. 10.1093/nar/gkq1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carell T, Brandmayr C, Hienzsch A, Müller M, Pearson D, Reiter V, Thoma I, Thumbs P, Wagner M. 2012. Structure and function of noncanonical nucleobases. Angew Chem Int Ed Engl 51: 7110–7131. 10.1002/anie.201201193 [DOI] [PubMed] [Google Scholar]
- Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, T.E. Cheatham I, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, et al. 2016. AMBER 2016. University of California, San Francisco. [Google Scholar]
- Chakraborty K, Veetil AT, Jaffrey SR, Krishnan Y. 2016. Nucleic acid–based nanodevices in biological imaging. Annu Rev Biochem 85: 349–373. 10.1146/annurev-biochem-060815-014244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla M, Oliva R, Bujnicki JM, Cavallo L. 2015. An atlas of RNA base pairs involving modified nucleobases with optimal geometries and accurate energies. Nucleic Acids Res 43: 6714–6729. 10.1093/nar/gkv606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao V, Guenther R, Malkiewicz A, Nawrot B, Sochacka E, Kraszewski A, Jankowska J, Everett K, Agris PF. 1994. Ribosome binding of DNA analogs of tRNA requires base modifications and supports the “extended anticodon”. Proc Natl Acad Sci 91: 2125–2129. 10.1073/pnas.91.6.2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denmon AP, Wang J, Nikonowicz EP. 2011. Conformation effects of base modification on the anticodon stem–loop of Bacillus subtilis tRNATyr. J Mol Biol 412: 285–303. 10.1016/j.jmb.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennington R, Keith TA, Millam JM. 2019. GaussView Version 6.
- Duechler M, Leszczyńska G, Sochacka E, Nawrot B. 2016. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci 73: 3075–3095. 10.1007/s00018-016-2217-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupradeau F-Y, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. 2010. The RED Tools: advances in RESP and ESP charge derivation and force field library building. Phys Chem Chem Phys 12: 7821–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. 2005. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44: 8078–8089. 10.1021/bi050343f [DOI] [PubMed] [Google Scholar]
- Fandilolu PM, Kamble AS, Sambhare SB, Sonawane KD. 2018. Conformational preferences and structural analysis of hypermodified nucleoside, peroxywybutosine (o2yW) found at 37th position in anticodon loop of tRNAPhe and its role in modulating UUC codon-anticodon interactions. Gene 641: 310–325. 10.1016/j.gene.2017.10.072 [DOI] [PubMed] [Google Scholar]
- Fandilolu PM, Kamble AS, Dound AS, Sonawane KD. 2019. Role of wybutosine and Mg2+ ions in modulating the structure and function of tRNAPhe: a molecular dynamics study. ACS omega 4: 21327–21339. 10.1021/acsomega.9b02238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Bock LV, Maracci C, Wang Z, Paleskava A, Konevega AL, Schröder GF, Grubmüller H, Ficner R. 2016. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature 540: 80 10.1038/nature20560 [DOI] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. 2005. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett 579: 959–962. 10.1016/j.febslet.2004.10.105 [DOI] [PubMed] [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, et al. 2016. Gaussian 16 Rev. B.01. Wallingford, CT. [Google Scholar]
- Grosjean H, Westhof E. 2016. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res 44: 8020–8040. 10.1093/nar/gkw608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybkowska A, Jędrzejczyk D, Rostkowski M, Chworos A, Dybala-Defratyka A. 2016. RNA model evaluation based on MD simulation of four tRNA analogs. RSC Adv 6: 101778–101789. 10.1039/C6RA14933B [DOI] [Google Scholar]
- Gustilo EM, Vendeix FA, Agris PF. 2008. tRNA's modifications bring order to gene expression. Curr Opin Microbiol 11: 134–140. 10.1016/j.mib.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M. 2006. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34: 721–733. 10.1093/nar/gkj471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Alfonzo JD. 2014. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 21: 174–185. 10.1016/j.chembiol.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17: 555 10.1038/nsmb.1790 [DOI] [PubMed] [Google Scholar]
- Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. 2017. The RNA modification landscape in human disease. RNA 23: 1754–1769. 10.1261/rna.063503.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovine L, Djordjevic S, Rhodes D. 2000. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg2+ in 15-year old crystals. J Mol Biol 301: 401–414. 10.1006/jmbi.2000.3950 [DOI] [PubMed] [Google Scholar]
- Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. 2008. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162. 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamble AS, Kumbhar BV, Sambhare SB, Bavi RS, Sonawane KD. 2015. Conformational preferences of modified nucleoside 5-taurinomethyluridine, τm5U occur at ‘wobble’ 34th position in the anticodon loop of tRNA. Cell Biochem Biophys 71: 1589–1603. 10.1007/s12013-014-0382-x [DOI] [PubMed] [Google Scholar]
- Katunin V, Soboleva N, Mahkno V, Sedelnikova E, Zhenodarova S, Kirillov S. 1994. Effect of the nucleotide-37 on the interaction of tRNAPhe with the P site of Escherichia coli ribosomes. Biochimie 76: 51–57. 10.1016/0300-9084(94)90062-0 [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Ono H, Hirosawa M, Saito H. 2020. RNA and protein-based nanodevices for mammalian post-transcriptional circuits. Curr Opin Biotechnol 63: 99–110. 10.1016/j.copbio.2019.11.019 [DOI] [PubMed] [Google Scholar]
- Kierzek E, Kierzek R. 2003. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res 31: 4472–4480. 10.1093/nar/gkg633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI. 2004. Purine bases at position 37 of tRNA stabilize codon–anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA 10: 90–101. 10.1261/rna.5142404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhar NM, Sonawane KD. 2011. Iso-energetic multiple conformations of hypermodified nucleic acid base wybutine (yW) which occur at 37th position in anticodon loop of tRNAPhe. J Mol Graph Model 29: 935–946. 10.1016/j.jmgm.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Kumbhar BV, Kamble AD, Sonawane KD. 2013. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochem Biophys 66: 797–816. 10.1007/s12013-013-9525-8 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. 10.1017/S1355838201002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CJ, Pan T, Kalsotra A. 2017. RNA modifications and structures cooperate to guide RNA–protein interactions. Nat Rev Mol Cell Biol 18: 202 10.1038/nrm.2016.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Ge HW, Cho SS. 2013. Sequence-dependent base-stacking stabilities guide tRNA folding energy landscapes. J Phys Chem B 117: 12943–12952. 10.1021/jp402114p [DOI] [PubMed] [Google Scholar]
- Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM. 2014. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol 11: 1619–1629. 10.4161/15476286.2014.992273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickam N, Joshi K, Bhatt MJ, Farabaugh PJ. 2015. Effects of tRNA modification on translational accuracy depend on intrinsic codon–anticodon strength. Nucleic Acids Res 44: 1871–1881. 10.1093/nar/gkv1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuszewski M, Wojciechowski J, Miyauchi K, Gdaniec Z, Wolf WM, Suzuki T, Sochacka E. 2017. A hydantoin isoform of cyclic N6-threonylcarbamoyladenosine (ct6A) is present in tRNAs. Nucleic Acids Res 45: 2137–2149. 10.1093/nar/gkw1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrate NE, Varner ME, Kim KI, Nagan MC. 2006. Molecular dynamics simulations of human: the role of modified bases in mRNA recognition. Nucleic Acids Res 34: 5361–5368. 10.1093/nar/gkl580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen NE, Johansson K, Virtanen A, Kirsebom LA. 2001. Aminoglycoside binding displaces a divalent metal ion in a tRNA–neomycin B complex. Nat Struct Biol 8: 510–514. 10.1038/88569 [DOI] [PubMed] [Google Scholar]
- Monestier A, Aleksandrov A, Coureux P-D, Panvert M, Mechulam Y, Schmitt E. 2017. The structure of an E. coli tRNAfMet A1–U72 variant shows an unusual conformation of the A1–U72 base pair. RNA 23: 673–682. 10.1261/rna.057877.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RC, Brown KG, Elliott MS. 1999. The effect of queuosine on tRNA structure and function. J Biomol Struct Dyn 16: 757–774. 10.1080/07391102.1999.10508291 [DOI] [PubMed] [Google Scholar]
- Motorin Y, Helm M. 2010. tRNA stabilization by modified nucleotides. Biochemistry 49: 4934–4944. 10.1021/bi100408z [DOI] [PubMed] [Google Scholar]
- Murphy FV IV, Ramakrishnan V, Malkiewicz A, Agris PF. 2004. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11: 1186 10.1038/nsmb861 [DOI] [PubMed] [Google Scholar]
- Park SV, Yang J-S, Jo H, Kang B, Oh SS, Jung GY. 2019. Catalytic RNA, ribozyme, and its applications in synthetic biology. Biotechnol Adv 37: 107452 10.1016/j.biotechadv.2019.107452 [DOI] [PubMed] [Google Scholar]
- Parthasarathy R, Ohrt JM, Chheda GB. 1977. Modified nucleosides and conformation of anticodon loops: crystal structure of t6A and g6A. Biochemistry 16: 4999–5008. 10.1021/bi00642a010 [DOI] [PubMed] [Google Scholar]
- Preethi SP, Sharma P, Mitra A. 2017. Higher order structures involving post transcriptionally modified nucleobases in RNA. RSC Adv 7: 35694–35703. 10.1039/C6RA28478G [DOI] [Google Scholar]
- Quigley GJ, Rich A. 1976. Structural domains of transfer RNA molecules. Science 194: 796–806. 10.1126/science.790568 [DOI] [PubMed] [Google Scholar]
- Roe DR, Cheatham TE III. 2013. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9: 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G. 2016. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun 7: 10457 10.1038/ncomms10457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Khusainov I, El Omari K, Duman R, Mykhaylyk V, Yusupov M, Westhof E, Wagner A, Yusupova G. 2019. Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction. Nat Commun 10: 2519 10.1038/s41467-019-10409-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambhare SB, Kumbhar BV, Kamble AD, Bavi RS, Kumbhar NM, Sonawane KD. 2014. Structural significance of modified nucleosides k2C and t6A present in the anticodon loop of tRNAIle. RSC Adv 4: 14176–14188. 10.1039/c3ra47335j [DOI] [Google Scholar]
- Sarachan KL, Cantara WA, Vangaveti S, Spears JL, DeMirci H, Murphy FV IV, Ranganathan SV, Agris PF. 2016. Post-transcriptional modifications alter anticodon loop dynamics and codon recognition in E. coli tRNAArg1,2. FEBS J 30: 807.802. [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao Y-G, Murphy FV IV, Weir JR, Ramakrishnan V. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694. 10.1126/science.1179700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer U, Bohleber S, Fradejas-Villar N. 2017. The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol 14: 1197–1208. 10.1080/15476286.2017.1294309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelam PP, Sharma P, Mitra A. 2017. Structural landscape of base pairs containing post-transcriptional modifications in RNA. RNA 23: 847–859. 10.1261/rna.060749.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942. 10.1126/science.1131127 [DOI] [PubMed] [Google Scholar]
- Shi H, Moore PB. 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6: 1091–1105. 10.1017/S1355838200000364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane KD, Sambhare SB. 2015. The influence of hypermodified nucleosides lysidine and t6A to recognize the AUA codon instead of AUG: a molecular dynamics simulation study. Integr Biol 7: 1387–1395. 10.1039/C5IB00058K [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS. 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33: D139–D140. 10.1093/nar/gki012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF. 2000. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 39: 13396–13404. 10.1021/bi0013039 [DOI] [PubMed] [Google Scholar]
- Stuart JW, Koshlap KM, Guenther R, Agris PF. 2003. Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J Mol Biol 334: 901–918. 10.1016/j.jmb.2003.09.058 [DOI] [PubMed] [Google Scholar]
- Sun W-J, Li J-H, Liu S, Wu J, Zhou H, Qu L-H, Yang J-H. 2015. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res 44: D259–D265. 10.1093/nar/gkv1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Durant PC, Davis DR. 2000. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39: 12575–12584. 10.1021/bi0014655 [DOI] [PubMed] [Google Scholar]
- Tewari R. 1988. Conformational preferences of modified nucleic acid bases N6-(Δ2-isopentenyl) adenine and 2-methylthio-N6-(Δ2-isopentenyl) adenine by the quantum chemical PCILO calculations. Int J Quantum Chem 34: 133–142. 10.1002/qua.560340206 [DOI] [PubMed] [Google Scholar]
- Vangaveti S, Cantara WA, Spears JL, DeMirci H, Murphy FV IV, Ranganathan SV, Sarachan KL, Agris PF. 2020. A structural basis for restricted codon recognition mediated by 2-thiocytidine in tRNA containing a wobble position inosine. J Mol Biol 432: 913–929. 10.1016/j.jmb.2019.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeix FA, Dziergowska A, Gustilo EM, Graham WD, Sproat B, Malkiewicz A, Agris PF. 2008. Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry 47: 6117–6129. 10.1021/bi702356j [DOI] [PubMed] [Google Scholar]
- Vendeix FA, Munoz AM, Agris PF. 2009. Free energy calculation of modified base-pair formation in explicit solvent: a predictive model. RNA 15: 2278–2287. 10.1261/rna.1734309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, McCallum SA, Pardi A. 2005. Comparison of the global structure and dynamics of native and unmodified tRNAval. Biochemistry 44: 6024–6033. 10.1021/bi0473399 [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. 2004. Development and testing of a general amber force field. J Comput Chem 25: 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. 2007. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol 14: 733–737. 10.1038/nsmb1282 [DOI] [PubMed] [Google Scholar]
- Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, Agris PF. 2000. Modified nucleoside dependent Watson–Crick and Wobble codon binding by tRNALysUUU species. Biochemistry 39: 13390–13395. 10.1021/bi001302g [DOI] [PubMed] [Google Scholar]
- Zhang X, Walker RC, Phizicky EM, Mathews DH. 2014. Influence of sequence and covalent modifications on yeast tRNA dynamics. J Chem Theory Comput 10: 3473–3483. 10.1021/ct500107y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C. 2017. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18: 31–42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]