Abstract
Innate immunity serves as the rapid and first-line defense against invading pathogens, and this process can be regulated at various levels, including epigenetic mechanisms. The bromodomain and extraterminal domain (BET) family of proteins consists of four conserved mammalian members (BRD2, BRD3, BRD4, and BRDT) that regulate the expression of many immunity-associated genes and pathways. In particular, in response to infection and sterile inflammation, abnormally expressed or dysfunctional BETs are involved in the activation of pattern recognition receptor (e.g., TLR, NLR, and CGAS) pathways, thereby linking chromatin machinery to innate immunity under disease or pathological conditions. Mechanistically, the BET family controls the transcription of a wide range of proinflammatory and immunoregulatory genes by recognizing acetylated histones (mainly H3 and H4) and recruiting transcription factors (e.g., RELA) and transcription elongation complex (e.g., P-TEFb) to the chromatin, thereby promoting the phosphorylation of RNA polymerase II and subsequent transcription initiation and elongation. This review covers the accumulating data about the roles of the BET family in innate immunity, and discusses the attractive prospect of manipulating the BET family as a new treatment for disease.
Subject terms: Infection, Diseases
Introduction
The immune system is composed of special organs, cells, and chemicals that can prevent various infections (e.g., from bacteria and viruses) and injuries (e.g., wounds and trauma) by activating innate and adaptive immune responses.1 Unlike adaptive immunity, in which immune cells (e.g., B and T cells) target specific antigens through the recognition by either antibodies or cell receptors, innate immunity mediated by myeloid cells (e.g., neutrophils, monocytes, macrophages, and dendritic cells [DCs]) and natural killer (NK) cells is rapid and antigen-independent.2 Defects in or the excessive activation of the innate immune system may lead to inflammation, which is implicated in various diseases and pathological conditions, such as cancer3 diabetes,4 and sepsis.5 This process is strictly regulated at the epigenetic, transcriptional, posttranscriptional, and posttranslational levels.
Epigenetic changes in immune cells are a key component of gene activation during the inflammatory response, which causes the production of immune mediators (e.g., cytokines and chemokines) and the infiltration, polarization, or re-population of immune cells.6 The epigenetic mechanisms have many forms, such as DNA modification (e.g., methylation and oxidation), posttranslational modification of histones (e.g., acetylation, methylation, phosphorylation, ubiquitylation, and SUMOylation), nucleosome positioning, and changes in microRNA (miRNA) expression.7 Among them, histone acetylation is a reversible chromatin modification mediated by histone acetyltransferases (HATs, also called “writers”) and histone deacetylases (HDACs, also termed as “erasers”). Furthermore, acetyl-binding proteins (namely “readers”) mainly recognize acetylated histones.8 Abnormal changes in epigenetic readers modify gene expression and disrupt the cellular machinery, thereby changing the function of immune cells.
Bromodomain is an evolutionarily conserved protein-protein interaction module consisting of approximately 110 amino acids that can recognize and bind acetylated lysine residues in histones and many other proteins.9 Bromodomain-containing proteins (BRDs) serve as epigenetic readers of histone acetylation, which can recruit transcriptional regulator complexes to chromatin and bind to acetylated histones.10 In 2012, 61 bromodomain modules were identified among 46 different proteins in the human genome, and these BRDs were divided into 8 subfamilies (e.g., bromodomain and extraterminal domain [BET] subfamilies) based on the similarity of protein sequences.11 The BET family contains four related proteins (namely BRD2, BRD3, BRD4, and BRDT) that act as epigenetic readers with broad specificity on transcriptional activation (including the recruitment of positive transcription elongation factor [P-TEFb] and the control of RNA polymerase II [Pol II] transcriptional activity).12 A dysfunctional BET family member is involved in many physiological and pathological processes and has become an important therapeutic target for diseases, including immune and inflammatory diseases.13
In this review, we summarize the emerging role of the BET family in innate immunity and highlight its functions in various diseases through orchestrating pattern recognition receptor (PRR) signaling and transcriptional regulation of immune genes. We also discuss the potential application of BET inhibitors (BETis) in regulating immune homeostasis in diseases, and then look forward to future research directions in this area.
Classification and structure of BETs
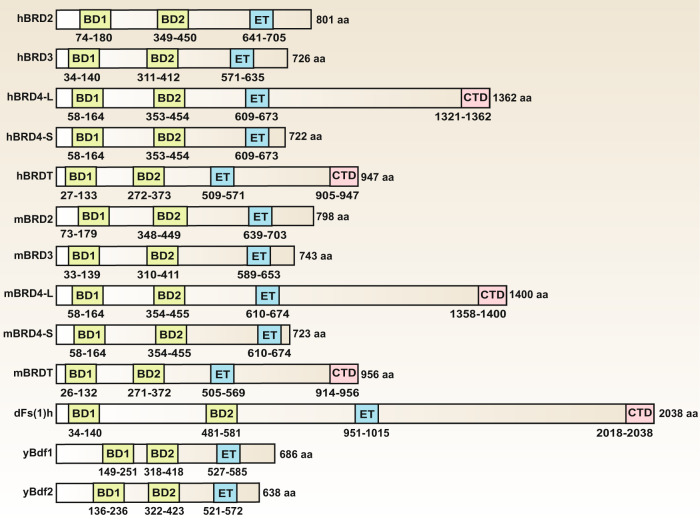
In mammalian cells, four BETs with similar gene arrangements, domain organizations, and functions have been identified.14 In humans, these genes are referred to as BRD2 (also known as FSRG1, RING3, RNF3, FSH, or D6S113E), BRD3 (also known as ORFX or RING3L), BRD4 (also known as MCAP or HUNK1), and BRDT (also known as BRD6, CT9, or SPGF21). In mice, these genes are designated as Brd2 (also known as Frg1, Fsrg1, Nat, Ring3, or Rnf3), Brd3 (also known as Fsrg2, Orfx, or Ringl3), Brd4 (also known as Fsrg4 or Mcap/Hunk1), and Brdt (also known as Fsrg3 or Brd6). Both human and mouse BRD4 have long and short isoforms, and the relative abundance of these two forms of BRD4 vary among different cell types.15,16 The long isoform of BRD4 (BRD4L) is a well-characterized coactivator of transcription (corresponding to the ordinary full-length transcript), whereas the short isoform of BRD4 (BRD4S) corresponds to an alternative splicing variant lacking exons 12–20.16 Although BRD4L and BRD4S have the same N-terminal conserved tandem bromodomains and extraterminal domain, they are not functionally redundant because the opposite roles of these two isoforms have been found in different contexts.17 Mammalian BETs are highly conserved and have homologs in other species, such as Fs(1)h in Drosophila, and BDF1 and BDF2 proteins in Saccharomyces cerevisiae, but these homologs are not fully understood (Fig. 1).
Fig. 1.
Domain architecture of BET family proteins in human (h), mouse (m), Drosophila (d), and yeast (y). Numbers indicate the amino acid boundaries of each domain in individual proteins. Alignment of amino acid sequences was based on published information using the following accession numbers retrieved from GenBank databases: hBRD2, NM_005104; hBRD3, NM_007371.4; hBRD4-L, NM_058243; hBRD4-S, NM_014299; hBRDT, NM_001242805; mBRD2, NM_010238; mBRD3, NM_001113574; mBRD4-L, NM_020508; mBRD4-S, NM_198094; mBRDT, NM_014299; dfs(1)h, NM_078523; yBdf1, NM_001182287; yBdf2, AJV11655. Abbreviations: BD1, the first bromodomain; BD2, the second bromodomain; Bdf1, chromatin-binding protein BDF1; BRD, bromodomain protein (numbers 2–4); CTD, C-terminal domain; ET, extraterminal domain; Fs(1)h, female sterile (1) homeotic
Each BET protein is characterized by the presence of two N-terminal conserved tandem bromodomains (namely the first bromodomain [BD1] and the second bromodomain [BD2]) and a unique extraterminal (ET) domain in the C-terminal moiety.14 Other protein families containing bromodomains lack this double-barrel feature. Other domains, such as motif B and Ser/Glu/Asp-rich region (SEED), are also highly conserved in BETs, whereas the C-terminal domain (CTD) and motif A are not present in each protein. The BD structure contains four alpha helices, which are separated by a variable loop region, thus forming a hydrophobic cavity. Acetylated lysine can be recognized by this central hydrophobic pocket via anchoring to a conserved asparagine residue. Of note, BETs prefer to bind to di-acetylated lysine residues closely located in the protein sequence, which is distinguished from other BRDs.11,18 Although the amino acid residues critical for binding acetyl-lysine in BD1 and BD2 are highly conserved, low homology is found between these two domains; thus they independently regulate the expression of BET-sensitive genes.19 However, BD1 and BD2 exhibit >75% identity with the homologous domains in different BETs.
In addition to acetylated lysine in histones, BETs also interact with transcription factors (TFs) and transcription elongation complexes (e.g., P-TEFb) through lysine acetylation-dependent or -independent mechanisms.20 The interaction of BDs with acetylated chromatin either at gene promoters or in long range cis regulatory elements (namely “enhancers”) allows subsequent initiation of gene transcription.21 The C-terminal ET domain is responsible for additional protein-protein interactions, thus enabling BETs as scaffolds for the recruitment of TFs and coactivators. BETs also regulate gene transcription through their intrinsic kinase activities because the regions they possess are weakly reminiscent of kinase motifs.22,23 However, these kinase motifs lack homology with other known kinase domains, and the biochemical mechanisms of the kinase activities remain elusive.
Expression of BETs in innate immunity
BRD2, BRD3, and BRD4 are significantly expressed in the nucleus, indicating that these proteins play a major role in regulating DNA events. Ordinarily, BRD2 is highly expressed in pancreatic β cells, germ cells in testis and ovaries, neurons in the cerebellum and cerebral cortex, liver, spleen, lungs, and kidney.24,25 BRD3 is commonly expressed in testis, ovaries, placenta, uterus, endocrine tissue, adipose tissue, lungs, kidney, muscle, and skin.11 BRD4 is more ubiquitously expressed and most abundant in bone marrow and lymphoid, mid-gestation embryos, testis, ovaries, adipose tissue, kidneys, and brain, whereas BRDT is selectively presented in the testis and ovaries.26 Although different BETs can be found in the same type of tissue, they are usually distributed distinctly from each other and exert different functions, suggesting that BETs are not simply redundant.27 Notably, BRD2, BRD3, and BRD4 are abnormally expressed in activated immune cells (e.g., macrophages and NK cells) as described below.
BRD4
BRD4 is the most well-studied BET protein in response to various stresses, including infection and immune stimulation (Table 1 and Fig. 2). BRD4 expression is markedly upregulated in different resident immune cells (e.g., macrophages, monocytes, T cells, and NK cells) and non-immune cells (e.g., pulmonary microvascular endothelial cells, bronchial epithelials, cardiomyocytes, and smooth muscle cells) under various stimuli (e.g., cigarette smoke extract, viruses, and listeriolysin-O).28–30 In these cells, BRD4 mainly exerts proinflammatory roles through conferring transcription activation of a variety of immune and inflammatory genes, and it may serve as a detrimental stress protein that can be used to predict disease activity. Moreover, the upregulation of BRD4 is also found in the uterus and fetal membranes induced by labor and infection,31 and severe early-onset preeclampsia placenta,32 which may lead to adverse pregnancy outcomes. In addition to normal cells, BRD4 is highly expressed in different types of tumor cells (e.g., cells of renal cell carcinoma,33 malignant pleural mesothelioma,34 pancreatic ductal adenocarcinoma,35 melanoma,36 and primary human mucoepidermoid carcinoma37), and it can protect these tumor cells against immunogenic cell death, a type of regulated cell death that enhances tumor targeting immunity. These findings indicate that BRD4 acts as a mediator of tumorigenesis and as a powerful prognostic biomarker. However, in some rare cases (e.g., in memory CD4+ T cells in HIV-1–infected individuals), BRD4 expression is downregulated, which may be a consequence of increased immune activation because of the opposing effect on expression induced by interferon (IFN) in viremic HIV-1 infection.38 It is also worthy to note that BRD4 function is not completedly determined by the change of its expression, as it also occupies essential roles in various pathophysiological processes with no detectable change in its expression, such as in pseudorabies virus infection, chronic obstructive pulmonary disease, and spinal cord injury.39–41 The expression of BRD4 changes inversely in different types of cells even in the same disease state. Hence, the immunomodulatory roles of BRD4 are highly cell-type dependent, which is regulated by unknown molecular switches.
Table 1.
Mechanism and function of BRDs in immunity and disease
| BET | Expression | Disease model | Cell type | Treatment | Transcription factor | Binding sites in histones | Target genes | BETi | Refs |
|---|---|---|---|---|---|---|---|---|---|
| Infectious diseases | |||||||||
| Bacterial infection | |||||||||
|
BRD2 BRD4 |
N/A | Gram-negative bacterial infection | Mouse SIM-A9 microglial cell line, mouse primary astrocytes | LPS | N/A | N/A | NOS2, COX2, IL1B, TNF, IL6, CCL2, MMP9, SERPINE1 | dBET1, JQ1 | 46,131 |
| BRD2 BRD3 BRD4 | N/A | Endotoxemia | Mouse bone marrow derived macrophages (BMDMs) | LPS | RELA, IRF4, IRF8 | H3Ac, H4K5Ac, H4K8Ac, H4K12Ac, H4Ac | TNF, IL6, IFNB1, CCL2, IL1B, IL12A, CXCL9, CCL12 | I-BET, JQ1 | 69 |
| BRD4 | N/A | Sepsis, group B Streptococcus (GBS) infection | Mouse BMDMs | GBS, LPS, cecal ligation puncture | RELA | N/A | IL6, IL1A, IL12B, CXCL9, IL23A | N/A | 97 |
|
BRD2 BRD4 |
Upregulation | Pseudomonas aeruginos infection | Human bronchial epithelial cells | IL17 | N/A | N/A | IL17A, IL22, CXCL1, CXCL5, CXCL8, CCL2, CSF3, IL6 | CPI-203 | 186 |
| BRD2 BRD3 BRD4 | N/A | Heat-killed L. monocytogenes infection | Mouse BMDMs | IFNB | RELA | N/A | NOS2, IL6, IL1RN | JQ1 | 94 |
| Virus infection | |||||||||
| BRD4 | Upregulation | RNA virus (respiratory syncytial virus infection) | Human small airway epithelial cells, nonciliated secretoglobin (Scgb1a1)-expressing bronchiolar epithelial cells | Poly(I:C) | RELA, IRF1, IRF7, IRF3 | H3K122Ac | IL6, ISG54, CIG5, RANTES, FN1, COL1A, MMP9, VIM, ACTA2, HEXIM1, KC, CSF3, CSF2, CXCL1, CXCL2, CXCL8, CCL2 | ZL0513, ZL0516, ZL0420, ZL0454, JQ1 | 55–58,110,187 |
| BRD4 | No change | Virus infection | Pig PK15 cells | Pseudorabies virus, herpes simplex virus, ectromelia virus, etc. | IRF3, RELA | H3K9, H3K27, H4K8, H4K12, H4K16 | IL1B, IFNB, ISG15 | JQ1, OTX-015, I-BET 151 | 39 |
| BRD4 | Downregulation | HIV-1 infection | Memory CD4+ T cells | N/A | N/A | N/A | N/A | N/A | 38 |
| BRD4 | N/A | HIV-1 infection | J-Lat A2 cell line | N/A | N/A | N/A | HIV | JQ1 | 107 |
| BRD3 | Downregulation | Virus infection | Mouse macrophage cell line RAW 264.7 | Sendai virus, vesicular stomatitis virus, herpes simplex virus | IRF3 | H3/H4 | IFNB1 | N/A | 50 |
| Fungi and parasitic infection | |||||||||
|
BRD2 BRD3 BRD4 |
N/A | Candida albicans and Aspergillus fumigatus infection |
Whole blood cells, peripheral blood mononuclear cells, monocytes |
N/A | N/A | H3K4Me3, H3K27Ac | TNF, IL6 | I-BET151, JQ1 | 129 |
| BRD2 | Upregulation | Plasmodium yoelii and Toxoplasma gondii-infected liver | N/A | N/A | N/A | N/A | TNF, IFNG | N/A | 140 |
|
BRD2 BRD3 BRD4 |
N/A | Schistosoma japonicum infection | Human Th17 cells | N/A | RORC | N/A | IL17, IL21, CSF2 | JQ1 | 141 |
| Non-infectious diseases | |||||||||
| Cancer | |||||||||
| BRD4 | Upregulation | Renal cancer | Human renal cell carcinoma cell lines | LPS | RELA | H3K27Ac | CXCL1, CXCL8, CXCR2, CSF2, CSF3, NLRP3 | JQ1 | 33,148 |
| BRD4 | N/A | Prostate cancer | Human prostate cancer cell lines (DU145, PC3) | N/A | N/A | N/A | CD274, HLA-A, HLA-C, IFNG | JQ1 | 152 |
| BRD4 | N/A | Pancreatic cancer | Pancreatic cancer cell lines (PANC-1, BxPC-3, MIA PaCa-2), pancreatic stellate cells | IFNG | IRF1 | N/A | CD274 | JQ1, IBET | 153,154 |
|
BRD2 BRD3 BRD4 |
N/A | Pancreatic cancer | Aspc-1, PANC-1, CAPAN-1 cells, mouse pancreatic cancer cell lines PanAsc 2159 and Panc 1343, RAW 264.7 | IFNG, LPS | STAT3 | N/A | IL6, CCL2, CSF2 | JQ1, I-BET 762 | 35,102 |
| BRD3 | N/A | Gastric adenocarcinoma | Microsatellite instability high gastric cancer cells | N/A | N/A | N/A | CD274 | N/A | 46 |
|
BRD2 BRD3 BRD4 |
N/A | A549 tumor-bearing nude mice, neuroblastoma | N/A | N/A | N/A | N/A | CD274 | JQ1 | 155 |
| BRD4 | N/A | Triple-negative breast cancer, hepatocellular carcinoma | Peripheral blood mononuclear cells, monocytes | N/A | CEBPB | H3K27Ac | CD274 | JQ1, I-BET762 | 115,156 |
| BRD4 | N/A | Triple-negative breast cancer | Human breast cancer cell line | IL6 | N/A | N/A | JAG1 | JQ1 | 147 |
|
BRD2 BRD4 |
Upregulation | Malignant pleural mesothelioma | Human primary malignant pleural mesothelioma cells | N/A | N/A | N/A | CD274, PDCD1 | JQ1, OTX015 | 34 |
| BRD2 | N/A | Melanoma | Human melanoma cell lines (Mel-RMu, SK-Mel28, Mel-RM, Mel-JD, Me1007) | N/A | RELA | N/A | IL6, IL8, VEGF, CXCL10, RANTES | I-BET151 | 87 |
|
BRD2 BRD3 BRD4 |
N/A | Human Ty-82 xenografts | Ty-82, SKOV3, A549, MDA-MB-231 cells | N/A | N/A | H3K27Ac | IDO1 | ABBV-075, JQ1, OTX015 | 126 |
| BRD4 | Upregulation |
Breast cancer with T-bet+ tumor-infiltrating T lymphocytes |
T-bet+ TILs | N/A | N/A | N/A | JAG1 | N/A | 149 |
| BRD4 | Upregulation | Mucoepidermoid carcinoma | Human mucoepidermoid carcinoma cells | N/A | RELA | N/A | N/A | I-BET762 | 37 |
|
BRD2 BRD3 BRD4 |
N/A | Neuroblastoma | NK cells | N/A | MYC, TP53 | N/A | ULBP1, ULBP3, PVR, NECTIN2 | JQ1 | 188 |
| BRD2 BRD3 BRD4 | N/A | Primary effusion lymphoma | N/A | N/A | RELA | N/A | IL6 | JQ1 | 189 |
|
BRD2 BRD3 BRD4 |
N/A | Myeloproliferative neoplasms | JAK2V617F-positive SET-2 cells | N/A | RELA | H3K27Ac | CCL2, CCL3, CCL4, CCL5, IL10, IL6, IL13, CXCL9, CSF3, II15, CXCL10, II1A, CXCL2, CXCL5 | JQ1 | 61 |
| BRD4 | N/A | Multiple myeloma | Human myeloma cell lines (SKO-007(J3), U266, ARP-1, RPMI-8226), human multiple myeloma cell line (JJN-3) | N/A | MYC | N/A | MICA | ARV-825 (PROTAC), JQ1, I-BET151 | 133 |
| Cardiovascular diseases | |||||||||
| BRD4 | Upregulation | Pathological cardiac hypertrophy | Neonatal mouse cardiomyocytes | Aortic banding, angiotensin-II | RELA | N/A | TNF, IL1B | N/A | 158 |
| BRD4 | Upregulation | Pulmonary arterial hypertension | Human microvascular endothelial cells, smooth muscle cells | N/A | FOXM1 | N/A | FOXM1, PLK1, IL8, MCP1, CCL5 | RVX208 | 159 |
|
BRD2 BRD3 BRD4 |
N/A | Pulmonary arterial hypertension | Human pulmonary microvascular endothelial cells | N/A | RELA | N/A | IL6, IL8 | JQ1 | 62 |
| BRD2 BRD3 BRD4 | N/A | Cardiovascular disease | Human monocytic cell line (THP-1), human umbilical vein vessel endothelial cells, human artery endothelial cells | TNF, LPS, IL1B | RELA | N/A | VCAM1, CCL2, IL8, SELE, IL1B | Apabetalone, MZ-1 | 132 |
| BRD2 BRD3 BRD4 | N/A |
Vascular inflammation |
Human umbilical vein vessel endothelial cells | TNF, LPS | RELA | N/A | ICAM1, VCAM1, SELE | JQ1 | 13 |
| BRD4 | Upregulation | Hypertension | Rat vascular smooth muscle cells | Angiotensin II | JUN, RELA, STAT1, CDX2, FOXL1, LIN54, ETS1 | H3K27Ac | ESM1, SPRY2, TGIF1, FST, FGF2, EGR2, FLT, FOXP1 | JQ1 | 190 |
| BRD2 BRD3 BRD4 | N/A | Heart failure (prolonged pressure overload, massive anterior myocardial infarction) | Human induced pluripotent stem cell-derived cardiomyocytes | ET1 | RELA, JUN, STAT1 | N/A | NPPB, CTGF | JQ1 | 101 |
| BRD2 BRD4 | Upregulation | Acute myocardial infarction | N/A | N/A | N/A | N/A | TLR4, TRAF6, RELA, CRP, IL6 | JQ1 | 66 |
| BRD4 | N/A | Atherogenesis | Human umbilical vein vessel endothelial cells | TNF | RELA | H3K27Ac | VCAM1, SOX18, CCL2 | JQ1 | 63 |
| Respiratory diseases | |||||||||
| BRD4 | Upregulation | Chronic obstructive pulmonary disorder | Human pulmonary microvascular endothelial cells | Cigarette smoke extract | N/A | N/A | IL6, IL8, TNF | N/A | 29,30 |
| BRD2 BRD3 BRD4 | No change | Chronic obstructive pulmonary disorder | Alveolar macrophages | LPS | N/A | N/A | IL6, CSF2 | JQ1 | 40 |
| BRD4 | N/A | Asthma | Primary human small airway epithelial cells | Cat dander extract | RELA | H3K122Ac | COL1, FN1, ZEB1, SNAI1, VIM, IL6 | ZL0454 | 163 |
| BRD2 BRD3 BRD4 | N/A | Asthma | CD4+ T cells, human bronchial epithelial cells | Cockroach allergen extract | N/A | N/A | IL4, IL17A, RORC, IL23R | JQ1 | 191 |
| BRD4 | N/A | Asthma | Asthmatic airway smooth muscle cells | FCS, TGFB | N/A | N/A | IL6, IL8 | JQ1, I-BET762 | 192 |
| BRD2 BRD3 BRD4 | N/A | Neutrophil-dominant allergic airway disease | CD4 + CD62L + naïve T cells | IL23, TGFβ, IL6, anti-IL4, anti-IFNG | N/A | N/A | IL1A, IL1B, IL2, IL6, IL10, IL12B, IL13, IL17A, CCL11, CSF3, CXCL1, CCL4, CCL5 | CPI-203 | 193 |
| BRD4 | N/A | Pulmonary fibrosis | Lung fibroblasts | Bleomycin | N/A | H4K5Ac | IL6 | JQ1 | 124 |
| Neurological diseases | |||||||||
| BRD2 | Upregulation | Parkinson’s disease | Mouse primary microglial cells | Alpha-synuclein (αSynAgg) | STAT3 | N/A | ACOD1, IFIT1, PYHIN, CDC123, SOD1, GRN | N/A | 44 |
| BRD2 BRD3 BRD4 | N/A | Systemic sclerosis | Human monocytes of systemic sclerosis patients | N/A | STAT1, STAT2, IRF | H3K27Ac | MX1, CMPK2 | JQ1 | 109 |
| BRD4 | Upregulation | Cerebral ischemia/reperfusion injury | Mouse astrocytes, microglial BV2 cells | Oxygen-glucose deprivation/reperfusion | RELA | N/A | IL6, IL1B, IL18, TNF | JQ1 | 75 |
| BRD2 BRD3 BRD4 | N/A | Spinal cord injury | BMDMs | LPS | N/A | N/A | IL6, IL1B, TNF, IL4, IL13 | JQ1 | 166 |
| BRD2 BRD3 BRD4 | No change | Spinal cord injury | Primary cerebellar granule neurons, astrocytes, oligodendrocytes, microglias | IL1B, TNF | N/A | N/A | IL6, IL1B, CCL5, CCL2, CXCL10, TNF | JQ1 | 41 |
| BRD2 | Upregulation |
Neuronal damage after deep hypothermic circulatory arrest |
Mouse microglial BV2 cells, human neuroblastoma SH-SY5Y cells | Oxygen-glucose deprivation | RELA | N/A | TNF, IL5, IL10, IL13 | JQ1 | 88 |
| BRD2 BRD3 BRD4 | N/A | Autoimmune encephalomyelitis | N/A | N/A | N/A | N/A | IL6, IL17, CCL2, CSF2, IFNG, TNF | RVX-297 | 194 |
| BRD2 BRD3 BRD4 | N/A | Alzheimer’s disease | N/A | N/A | N/A | N/A | IL1B, IL6, TNF, CCL2, NOS2, PTGS2 | JQ1 | 165 |
| Kidney diseases | |||||||||
| BRD4 | N/A | Experimental renal damage | Human renal proximal tubular epithelial cells (HK2) | Nephrotoxic serum, TNF | RELA | N/A | IL6, CCL2, CCL5 | JQ1 | 95 |
| BRD2 BRD3 BRD4 | N/A | Stage 4 or 5 chronic kidney disease | N/A | N/A | N/A | N/A | IL6, PAI1, OPN | Apabetalone | 14 |
| BRD4 | Upregulation | Lupus nephritis | N/A | N/A | N/A | N/A | IL1B, IL6, IL17, IL10, INFG | JQ1 | 168 |
| BRD4 | N/A | HIV-associated kidney disease | Human primary renal tubular epithelial cells | TNF, HIV | RELA | H3K9Ac, H3K18Ac, H3K4me3, H3K27me3 | IL1A, IL1B, LTA, LTB, CCL2, CCL3, CCL20, CXCL3, CXCL11, CCL2, CCL20, IL8 | MS417 | 167 |
| Digestive diseases | |||||||||
| BRD4 | Upregulation |
Acute liver injury |
Mouse Kupffer cells | Listeriolysin-O | RELA | TNF, IL6, IL1B, IL18, CCL2, CCL8 | JQ1 | 136 | |
| BRD4 | N/A | Colitis | Mouse intestinal epithelial cells | Dextran sulfate sodium | RELA | H3K9Ac | TNF, IL1B | N/A | 169 |
| BRD4 | N/A | Colitis | Th17 cells | N/A | N/A | H4K5Ac, H4K8Ac | IL17, IFNG, IL21, IL22, RORC, TBX21, IL6, GATA3 | MS402 | 20 |
| BRD2 BRD3 BRD4 | N/A | Colitis | Mouse BMDCs, human monocyte-derived DCs | LPS | N/A | N/A | IL6, IL12B, IL10 | I-BET151 | 195 |
| BRD2 BRD3 BRD4 | N/A | Non-alcoholic fatty liver disease, liver fibrosis | N/A | N/A | N/A | N/A | IFNG, CCL2, TNF, RSAD2, LY6A, CD4, CD7, CXCL10, STAT1, CCL5 | I-BET151 | 100 |
| BRD2 BRD3 BRD4 | N/A | Acute pancreatitis | N/A | N/A | N/A | N/A | IL6, IL10, CCL2, CXCL1 | I-BET762 | 170 |
| Metabolic diseases | |||||||||
| BRD2 BRD4 | N/A | Cancer cachexia | N/A | N/A | FOXO3 | N/A | IL6, TNF, IL1B, PTHLH | JQ1 | 174 |
| BRD2 BRD4 | N/A | Obesity, insulin resistance, diabetes | INS-1 cells, mouse adipocytes (3T3-L1) | TNF | PPARG, RELA | N/A | INS, TNF, CXCL2, IL1B, IL6, CCL2 | JQ1 | 24,89,172 |
| BRD2 BRD3 BRD4 | N/A | Type 1 diabetes | Pancreatic macrophages, β cells | LPS | RELA | N/A | CCL2, CCL5, CXCL1, CXCL2, IFNB1, IL1A, IL1B, NFKBIA, TNF, TNFAIP3, TNFRSF9, VCAM1, CXCL9, IL12B, IL6, MX1, MX2, RSAD2 | I-BET151 | 171 |
| Osteoarthritis | |||||||||
| BRD4 | Upregulation | Intervertebral disc degeneration | Nucleus pulposus cells |
TNF, advanced glycation end products |
RELA | N/A | NLRP3, CASP1, MMP13 | JQ1 | 28,89 |
| BRD4 | N/A | Acute gouty arthritis | Human monocytic cell line (THP-1) | Monosodium urate | RELA | N/A | IL1B |
Benzo[cd]indol-2(1H)-ones, Pyrrolo[4,3,2- dequinolin-2(1H)-ones |
74 |
| BRD4 | Upregulation | Periprosthetic osteolysis | Mouse macrophage RAW264.7 cells | Titanium particles | RELA | N/A | TNF, IL1B, IL6 | JQ1 | 196 |
|
BRD3 BRD4 |
N/A | Osteoarthritis | Human chondrosarcoma cells (SW1353) | IL1B, TNF | H4K5Ac, H4K8Ac, H4K12Ac | MMP1, MMP3, MMP13, ADAMTS4 | I-BET151 | 118 | |
| BRD2 BRD3 BRD4 | N/A | Failure of bone healing | C2C12 and MC3T3-E1 cell lines, BMDMs | TNF | RUNX2, SP7 | N/A | ALP, RUNX2, SP7 | N-methylpyrrolidone and N, N-dimethylacetamide, JQ1 | 197 |
| BRD4 | Upregulation | Articular cartilage of osteoarthritis | Human chondrocyte cell line sw1353 cells | IL1B | RELA | H3K27Ac | HMGB1, IL1B | JQ1 | 176 |
|
BRD2 BRD3 BRD4 |
Upregulation | Rheumatoid arthritis | Human rheumatoid arthritis fibroblast-like synoviocytes | TNF, IL1B | JUN, RELA | N/A | TNF, IL1B, IL6, IL8, IL17, IL18, MMP1, MMP3, MMP13 | JQ1, I-BET151 | 51,98,177 |
| Others | |||||||||
| BRD2 BRD3 BRD4 | Upregulation | Spontaneous preterm birth | Myometrial cells, amnion epithelial and mesenchymal cells | LPS, IL1B | RELA | N/A | IL6, CCL2, CXCL1, CXCL8 | JQ1 | 31 |
| BRD4 | Upregulation | Preeclampsia | Primary trophoblasts, human umbilical vein vessel endothelial cells | TNF | N/A | N/A | IL6, CXCL8, CCL2, CXCL1, FLT1 | N/A | 32 |
| BRD4 | N/A | Autoimmune uveitis (EAU) | Human CD4 + T cells, human Th17-polarized cells | Anti-CD3/CD28 | RORC | N/A | IL17A, IL22, RORC, IL10 | GSK151, JQ1 | 178 |
| BRD2 | Upregulation | Inherited retinal degeneration | Mouse primary microglias | LPS | N/A | N/A | TNF, CCL2, IL1B, IL6, RANTES | JQ1, RVX-208 | 45 |
| BRD2 BRD3 BRD4 | N/A | Age-related macular degeneration | Human retinal pigment epithelial cell line (ARPE-19) | Etoposide | RELA | N/A | IL6, IL8 | JQ1, PFI-1, I-BET151 | 179 |
| BRD2 | N/A | Psoriasis | Primary human keratinocytes, HS27 dermal fibroblasts | IL17A, TNF, IL22 | RELA | IL8, CXCL1, CXCL2, CXCL6, CXCL8, GCSF, CCL20 | JQ1, I-BET151 | 198 | |
| BRD2 BRD3 BRD4 | N/A | Psoriasis-like inflammation | N/A | Imiquimod | RORC | N/A | IL17A, IL22 | JQ1 | 180 |
| BRD4 | N/A | Periodontitis | RAW264.7 cells | LPS | RELA | N/A | IL1B, IL6, TNF, TLR2, TLR4 | JQ1 | 51 |
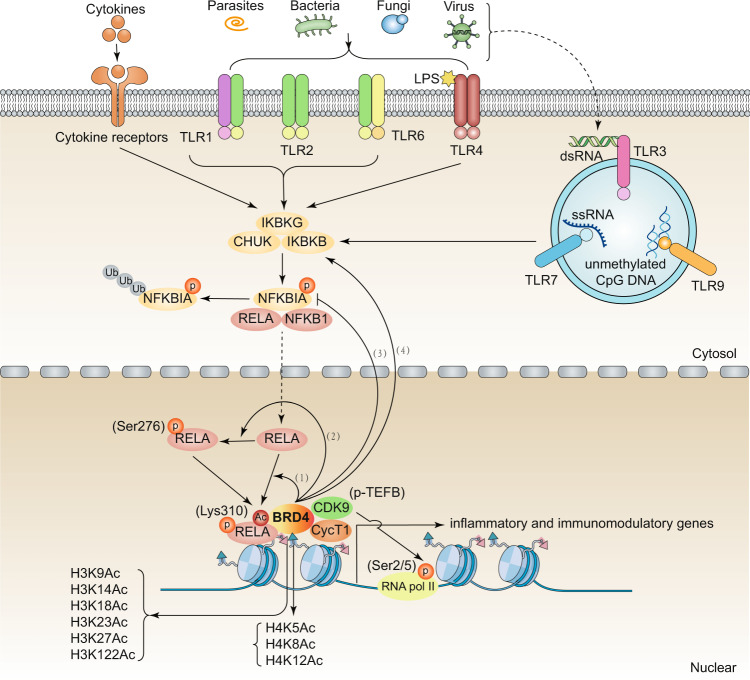
Fig. 2.
The role BRD4 in regulating the NF-κB pathway in inflammation and immunity. BRD4 regulates the activation of the NF-κB pathway caused by TLR ligands through at least four pathways: (1) BRD4 directly acetylates RELA through its atypical histone acetyltransferase activity; (2) BRD4 directly promotes the phosphorylation of RELA; and (3–4) BRD4 promotes the phosphorylation of RELA through inhibiting the translation of NFKBIA and increasing the phosphorylation of NFKBIA and IKBKB. Abbreviations: CDK9, cyclin-dependent kinase 9; CHUK, component of inhibitor of nuclear factor kappa B kinase complex; CycT1, cyclin T1; IKBKB, inhibitor of nuclear factor kappa B kinase regulatory subunit beta; IKBKG, inhibitor of nuclear factor kappa B kinase regulatory subunit gamma; NFKBIA, NFKB inhibitor alpha; NFKB1, nuclear factor kappa B subunit 1; p-TEFb, positive transcription elongation factor b; RELA, RELA proto-oncogene; RNA pol II, RNA polymerase II; TLR, toll-like receptor (numbers 1–9)
Although the regulatory mechanism controlling BRD4 expression is not clear, different miRNAs act as posttranscriptional regulators of BRD4 expression under various conditions. For example, miR-218-5p, miR-29a, miR-29b, and miRNA-302e inhibit BRD4 expression in activated human pulmonary microvascular endothelial cells, primary hepatic stellate cells, human bronchial epithelial cells, and A549 cells, respectively.30,42,43 We still need to clarify the TFs responsible for BRD4 expression in innate immunity.
BRD2
Compared with BRD4, less attention is paid to the expression of BRD2 and BRD3, which are also implicated in the regulation of immune response. Early studies showed that BRD2 is highly expressed in pancreatic β cells, thereby inhibiting mitosis and insulin transcription.24 Recent studies indicate that α-synuclein induces BRD2 expression, thus initiating the neuroinflammation in Parkinson’s disease.44 The expression of BRD2 (but not BRD3 and BRD4) is dominantly increased during photoreceptor degeneration that occurs postnatally in lipopolysaccharide (LPS)-stimulated mouse primary astrocytes.45,46 Elevated BRD2 expression is also detectable in malignant pleural mesothelioma, melanoma, and cardiomyocytes following acute myocardial infarction, which is concordant with BRD4.45,46 Moreover, the upregulation of BRD2 may be the main driver of LPS- or spinal cord injury-induced gene expression in macrophages or neuronal cells.40,47 Although the mechanism of BRD2 upregulation is unclear, it seems that BRD2 acts as a stress-sensitive metabolism-related protein after inflammatory stimulation.
BRD3
While BRD3 and BRD2 have overlapping cellular functions, their expression patterns exhibit some differences. Frameshift mutations of BRD3 have been found in gastric cancer, which is negatively related to the expression of CD274 (also known as programmed death ligand 1 [PD-L1]), a transmembrane protein that downregulates immune responses.48 BRD3 expression is significantly downregulated during endothelial differentiation49 or virus infection in macrophages.50 In contrast, BRD3 expression is upregulated in activated lymphocytes, indicating a potential role of BRD3 in adaptive immunity.49 BRD3 is also detected in the macrophages in synovial tissues derived from rheumatoid arthritis patients and osteoarthritis patients, which is similar to BRD2 and BRD4 expression,51 suggesting that different BETs may have a synergistic effect in autoimmune diseases. Notably, BRD2 and BRD3 can exert roles with no detectable change in their expressions.41 Therefore, both the expression and structure of BETs occupy essential roles in the regulation of gene expression and immune response. More in-depth studies are needed to clarify the regulatory mechanisms of their expression in the future.
Function of BETs in innate immunity
The innate immune response is mainly triggered by the recognition of various extracellular or intracellular danger signals through PRRs expressed in immune and non-immune cells.52 The surface-expressed PRRs include toll-like receptors (TLRs), C-type lectin receptors (CLRs), and advanced glycosylation end-product specific receptors (AGER/RAGE), whereas the intracellular PRRs include nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), retinoic acid-inducible gene (RIG) I-like receptors (RLRs), AIM2-like receptors (ALRs), and cyclic GMP-AMP synthase (CGAS). These PRRs directly or indirectly recognize the evolutionary conservative structure on pathogens (namely pathogen-associated molecular patterns [PAMPs], such as microbial nucleic acids, LPS, and carbohydrates) or endogenous molecules (namely damage-associated molecular patterns [DAMPs], such as high-mobility group box 1 [HMGB1], histones, host nucleic acids, and ATP). In response to PAMPs or DAMPs, gene transcription is activated and precisely controlled by TFs and coactivators coupled to BETs.
BETs in TLR signaling
TLR3
TLR3, a sensor of viral infections, is preferentially activated by dsRNA derived from the extracellular RNA viral genome, and it triggers the production of type I IFNs.53 TLR3 is mainly expressed in hematopoietic cells, particularly in a subset of DCs, but is also expressed in some stromal cells. The specificity of TLR3 for dsRNA allows its recognition of various RNA viruses, such as respiratory syncytial virus (RSV), influenza A virus, West Nile virus, and rhinovirus.54 Thus, the long-term activation of TLR3 is implicated in various respiratory diseases.
BETs link the TLR3 signaling pathway to chromatin remodeling and specific inflammatory gene transcription. BRD4 seems to play a major role in TLR3-induced acute airway inflammation and remodeling, and specific BETis have been developed to inhibit this process. BRD4 mediates poly(I:C)-induced airway inflammation by promoting the transcription of CIG5, IL6, KC, CCL2, ORM2, CXCL2, IFNB, ISG54, and CCL5 in airway epithelial cells or lung tissues and further increases the secretion of inflammatory cytokines in bronchoalveolar lavage fluid.55 Selective BRD4 inhibitor targeting of BD1 obviously alleviates inflammatory response by reversing cytokine expression.56,57 RSV also induces neutrophilic inflammation and the production of chemokines (CSF2, CXCL2, IL8, CCL2, and CCL5) and mucosal IFN in the nonciliated SCGB1A1-expressing epithelium through the binding of RELA/p65 to BRD4.58 In addition to BRD4, BRD3 promotes the production of type I IFN in macrophages during vesicular stomatitis virus infection.50 I-BET151 (a pan-BETi) suppresses the expression of cytokines (IL6 and IL8) and MMP3 in rheumatoid arthritis synovial fibroblasts in response to poly(I:C).51 In addition, I-BET151 inhibits the association of BRD4 with interferon beta 1 (IFNB1) promoter, thereby reducing IFNB1-mediated gene transcription in macrophages following poly(I:C) or LPS stimulation.59 These findings suggest that BRD4 is essential for TLR3-stimulated activation of the IFNB1 pathway and its antitumor activity.
TLR4
TLR4 is expressed in almost all innate immune cells, and it specifically recognizes bacterial LPS, several other components of pathogens, and DAMPs derived from tissue damage.60 TLR4 binds to its ligands, which ultimately leads to the activation of the nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) signaling pathway and subsequently the production of inflammatory cytokines involved in innate immune response. RELA (also known as p65) is a member of the NF-κB family and the main subunit of NF-κB transcription factor complex. BETs are coactivators of RELA (discussed separately below), which is recruited to the promoter of target genes and enhances their transcriptional activation.61–64 In addition to mediating RELA activation, BETs positively regulate TLR4 expression through different molecular mechanisms. For example, in pancreatic cancer cell lines (PANC-1 and BxPC-3), BRD4 promotes the expression of TLR4 through the transcriptional activation of CD276 (also known as B7-H3), which supports the role of the BRD4-TLR4 pathway in the regulation of immunotherapy and chemotherapy in pancreatic cancer.65 JQ1 (a pan-BETi) significantly reduces the protein expression of TLR4, TRAF6, and NF-κB in the heart of rats with acute myocardial infarction.66 Moreover, JQ1 reduces the expression of TLR4 and the production of inflammatory cytokines (e.g., IL1B, IL6, TNF, and IL10) in LPS-induced macrophages.67 Overall, these findings indicate that BRD4 is implicated in both infectious and sterile inflammation through controlling TLR4 expression and activation.
TLR1, TLR2, and TLR6
TLR2 in association with TLR1 or TLR6 is implicated in the recognition of a wide range of components (e.g., di- and triacylated lipoproteins and lipoteichoic acid) from Gram-positive or -negative bacteria.68 Although the effect of BETs on these TLRs has not been fully explored, JQ1 may reduce the expression of TLR2, which is proposed to be implicated in the production of inflammatory cytokines in diseased gingival tissues.67 In addition, in LPS-induced bone marrow-derived macrophages, the BETi (e.g., I-BET) reduces Tlr6 mRNA expression to 2.3 folds, but the precise mechanism and pathological role of this downregulation has not been investigated.69 It is unclear whether the genetic depletion of BET protein has a similar effect to that of BETis in the regulation of TLR2 and TLR6 expression as well as their activation-mediated innate immune responses.
TLR7 and TLR9
TLR9 mainly recognizes unmethylated CpG motifs, which are abundant in bacterial or viral DNA.70 Unlike TLR9, TLR7 is an endosomal innate immune sensor used to detect single-stranded ribonucleic acid.71 Both TLR7 and TLR9 stimulation facilitate the production of type I IFNs and other inflammatory cytokines, which greatly contribute to the antiviral immune responses. While there is no direct evidence that BETs regulate the expression of TLR7 and TLR9, the roles of BETs in the production of IFNs have been demonstrated. In particular, in the human plasmacytoid DC line Gen2.2, different pan-BETis (e.g., JQ1 and I-BET151) block the production of IFNB caused by TLR7 or TLR9 agonists (e.g., CL097 and ODN1826).59 BRD3 is also found to promote the transcription of IFNB1 in macrophages during vesicular stomatitis virus, sendai virus, or herpes simplex virus infection.50 These findings provide a line of evidence indicating that BETs link TLR7 and TLR9 activation to mount inflammation and immune responses through the transcription activation of IFNs. It will be interesting to learn whether BETs also regulate TLR8-, TLR10- and TLR11-dependent signaling pathways in innate immunity.
BETs in NLR signaling
NLRs are cytoplasmic PPRs that can recognize PAMPs and DAMPs, and they also play a crucial role in initiating the innate immune response. According to the structure of the N-terminal domain, NLRs can be divided into four subfamilies, namely NLRA, NLRB, NLRC, and NLRP.72 The NLRP subfamily contains NLRP1 through NLRP14, which are involved in the formation of inflammasomes, multiprotein oligomers of the innate immune system that are responsible for the activation of inflammatory responses.73 Among the NLRPs, NLRP3 is the most studied and best characterized inflammasome, which is triggered by various inflammatory stimuli. NLRP3 inflammasome activation further promotes caspase 1 (CASP1) or caspase 11 (CASP11) activation, leading to the induction of pyroptosis and the release of proinflammatory cytokines (e.g., IL1B and IL18) and DAMPs (e.g., HMGB1).
The pharmacological or genetic inhibition of BRD4 alleviates inflammatory response by inhibiting NLRP3 signaling pathways in varous conditions, such as in TNF-primed rat nucleus pulposus cells,28 monosodium urate-induced acute gouty arthritis,74 and middle cerebral artery occlusion-mediated glial activation.75 Mechanistically, BRD4 inhibition decreases the expression of NLRP3 and CASP1 by limiting the transcriptional activity of RELA. Of note, in different situations, BETs may have an effect that is opposite to that of limiting NLRP3 inflammasome activation. For instance, in renal cell carcinoma tissues and cells, the inhibition of BRD4 through genetic knockdown or JQ1 prevents proliferation and epithelial mesenchymal transition (EMT) by increasing RELA-mediated NLRP3 expression and subsequent pyroptosis.33 Collectively, the role of BETs in NLRP3 inflammasomes is cell type-specific, which depends on different transduction signals (Fig. 3). There remain challenges to indentifying whether BETs involved in orchestrating other inflammasomes (e.g., NLRP1, NLRC4, and absent in melanoma 2 [AIM2])-mediated immune responses.
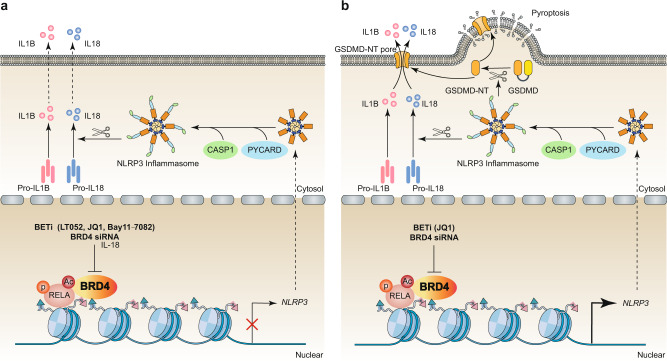
Fig. 3.
The role BRD4 in regulating the NLRP3 pathway in inflammation and pyroptosis. a In TNF-primed rat nucleus pulposus cells, in which monosodium urate induced acute gouty arthritis, and middle cerebral artery occlusion induced glial activation, BRD4 inhibits RELA-mediated NLRP3 transcription and subsequent CASP1-dependent inflammasome activation. b In renal cell carcinoma tissues and cells, BRD4 is required for RELA-mediated NLRP3 transcription and subsequent CASP1-dependent inflammasome activation and GSDMD-mediated pyroptosis. Abbreviations: BETi, bromodomain and extraterminal inhibitor; BRD4, bromodomain containing 4; CASP1, caspase 1; GSDMD, gasdermin D; GSDMD-NT, gasdermin D N-terminal domain; IL1B, interleukin 1 beta; IL18, interleukin 18; NLRP3, NLR family pyrin domain containing 3; PYCARD, PYD and CARD domain containing; RELA, RELA proto-oncogene; TNF, tumor necrosis factor
BETs in CGAS signaling
CGAS is a cytosolic DNA sensor that catalyzes the synthesis of cyclic dinucleotide cGMP-AMP (ultimately 2’3’-cGAMP) after activation.76 As a second messenger, 2’3’-cGAMP binds and activates an endoplasmic reticulum membrane adaptor protein, namely, stimulator of interferon response CGAMP interactor 1 (STING1, also known as STING or TMEM173) by inducing its conformational changes.77 The CGAS-STING1 pathway-mediated DNA-sensing signaling pathway is crucial for the production of type I IFNs and host antiviral responses.78 Interestingly, blocking BRD4 widely inhibits the attachment of various DNA and RNA viruses (e.g., pseudorabies virus) by activating nucleic acid-dependent antiviral innate immunity in vitro and in vivo. The role of BRD4 inhibition in antiviral immunity is partly mediated by the activation of the CGAS-STING1 pathway39 (Fig. 4). Excessive activation of the CGAS-STING1 pathway by bacterial cyclic dinucleotides causes cytokine storms and systemic coagulation, leading to sepsis and septic shock.79 During bacterial infections and DAMP-mediated sterile inflammation, it remains to be seen whether BRD4 and other BETs play a similar role in regulating the activation of the CGAS-STING1 pathway.
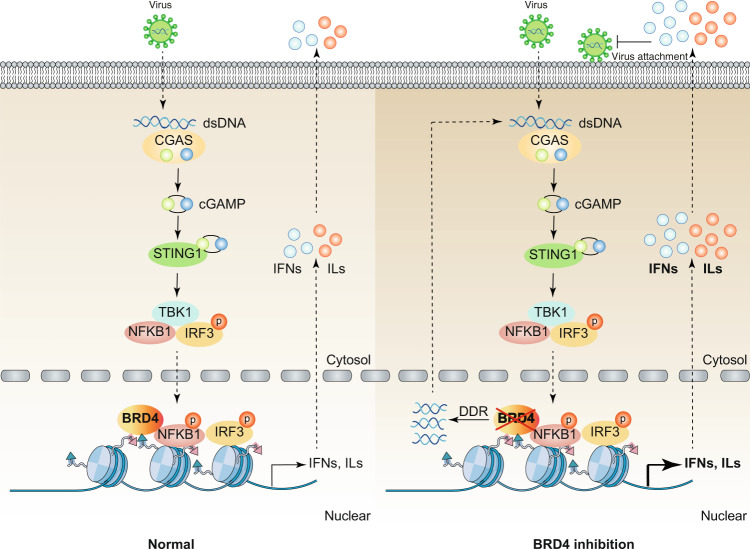
Fig. 4.
The role of BRD4 in regulating the STING1 pathway in antiviral immunity. Cytoplasmic DNA derived from various viruses activates CGAS and produces endogenous cyclic dinucleotide cGAMP, which binds to STING1 located in the endoplasmic reticulum, and then promotes the dimerization and translocation of STING1 from the ER to the perinuclear region. During trafficking, STING1 recruits and activates TBK1, stimulates the phosphorylation and nuclear translocation of IRF3, and to a lesser extent NFKB1, which leads to the production of type 1 IFN and other inflammatory cytokines (e.g., IL). The nuclear activity of IRF3 and NFKB1 is inhibited by BRD4. In addition, after BRD4 inhibition, the activation of DDR can induce the release of host dsDNA from the nucleus to the cytoplasm, leading to further activation of the CGAS-STING1 pathway to limit viral infection. Abbreviations: BRD4, bromodomain containing 4; cGAMP, cyclic GMP-AMP; CGAS, cyclic GMP-AMP synthase; DDR, DNA damage response; dsDNA, double-stranded DNA; IFN, interferon; IL, interleukin; IRF3, interferon regulatory factor 3; NFKB1, nuclear factor kappa B subunit 1; STING1, stimulator of interferon response cGAMP interactor 1; TBK1, TANK binding kinase 1
Mechanism of BETs in innate immunity
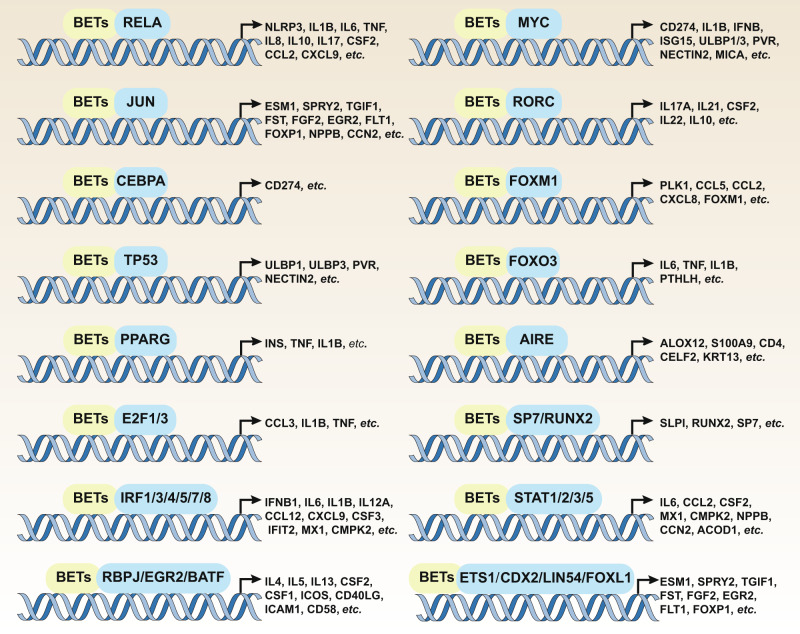
As epigenetic reader proteins, BETs recognize and bind to acetylated lysine residues on histone tails, thereby facilitating the assembly of transcription complexes, including specific TFs, mediators, and RNA Pol II-mediated transcriptional initiation machinery. BET-mediated innate immune control mainly relies on the binding of BRD4 to RELA and the P-TEFb transcription extension complex to activate RNA Pol II. In addition to RELA, the BET-mediated immune response is also involved in the regulation of other TF activators, such as the signal transduction and transcription activation (STAT) protein family, the E2F transcription factor (E2F) family, and the interferon regulatory factor (IRF) family (Fig. 5). Below, we discuss how BETs regulate gene expression and innate immunity by affecting the activity and function of TFs, transcription coactivators, and histones.
Fig. 5.
The role of BETs in regulating the expression of immune genes mediated by transcription factors. Abbreviations: ACOD1, aconitate decarboxylase 1; AIRE, autoimmune regulator; ALOX12, arachidonate 12-lipoxygenase, 12 S type; BATF, basic leucine zipper ATF-like transcription factor; CCL, C-C motif chemokine ligand (numbers 2–12); CCN2, cellular communication network factor 2; CD, CD molecule (numbers 4–274); CD40LG, CD40 ligand; CDX2, caudal-type homeobox 2; CEBPA, CCAAT enhancer binding protein alpha; CELF2, CUGBP Elav-like family member 2; CMPK2, cytidine/uridine monophosphate kinase 2; CSF1, colony stimulating factor (numbers 1–3); CXCL9, C-X-C motif chemokine ligand 9; E2F1, E2F transcription factor 1; EGR2, early growth response 2; ESM1, endothelial cell-specific molecule 1; ETS1, ETS proto-oncogene 1; FGF2, fibroblast growth factor 2; FLT1, fms-related receptor tyrosine kinase 1; FOXL1, forkhead box L1; FOXM1, forkhead box M1; FOXO3, forkhead box O3; FOXP1, forkhead box P1; FST, follistatin; ICAM1, intercellular adhesion molecule 1; ICOS, inducible T-cell costimulatory; IFIT2, interferon-induced protein with tetratricopeptide repeats 2; IFNB1, interferon beta 1; IL1B, interleukin 1 beta; IL, interleukin (numbers 4–22, including “A” variants); INS, insulin; IRF, interferon regulatory factor; JUN, Jun proto-oncogene, AP-1 transcription factor subunit; KRT13, keratin 13; LIN54, lin-54 DREAM MuvB core complex component; MX1, MX dynamin-like GTPase 1; MYC, MYC proto-oncogene, bHLH transcription factor; NECTIN2, nectin cell adhesion molecule 2; NLRP3, NLR family pyrin domain containing 3; NPPB, natriuretic peptide B; PLK1, polo-like kinase 1; PPARG, peroxisome proliferator activated receptor gamma; PTHLH, parathyroid hormone-like hormone; PVR, PVR cell adhesion molecule; RBPJ, recombination signal binding protein for immunoglobulin kappa J region; RELA, RELA proto-oncogene, NF-κB subunit; RORC, RAR-related orphan receptor C; RUNX2, RUNX family transcription factor 2; S100A9, S100 calcium-binding protein A9; SLPI, secretory leukocyte peptidase inhibitor; SP7, Sp7 transcription factor; SPRY2, sprouty RTK signaling antagonist 2; STAT, signal transducer and activator of transcription; TGIF1, TGFB-induced factor homeobox 1; TNF, tumor necrosis factor; TP53, tumor protein p53; ULBP1, UL16 binding protein (numbers 1–3)
TFs
RELA
NF-κB pathways, including the RELA-dependent canonical pathway and RELB-dependent noncanonical pathway, is critical for the modulation of immune response.80 Under normal conditions, RELA is usually sequestered in the cytoplasm in an inactive form due to its binding to IκBα. Under inflammatory conditions, various PAMPs and DAMPs activate IκB kinases (IKKs), which leads to the phosphorylation, ubiquitination, and subsequent proteasomal degradation of IκBα. This dynamic modification and expression change of IκBα further promotes the transport of RELA from the cytoplasm to the nucleus, where it can bind to the promoters of various related genes and regulate their transcription.81 Both acetylation and phosphorylation are essential for the transcriptional activity of RELA.82 RELA acetylation is regulated by the acetyltransferase activity of CREB binding protein (CREBP)/E1A binding protein P300 (EP300) and lysine acetyltransferase 2B (KAT2B), and it occurs at multiple sites, including lysine (Lys)122, −123, −218, −221, and −310.83,84 Among these, the acetylation of Lys310 is required for the full transcriptional activity of RELA. BRD4 triggers the transcription activation of RELA by specifically binding to acetylated Lys31085. BRD4 deficiency or BETi also promotes the ubiquitination and degradation of the active nuclear form of RELA,64,85 indicating a dual role of BRD4 in the regulation of RELA. However, the regulation and mechanisms of BRD4 on the ubiquitination and degradation of RELA in immune cells remains mysterious. BRD2 also plays a role in promoting RELA activation in marcophages,86 melanoma,87 microglia,88 and adipose tissue,89 and the subsequent production of inflammatory mediators. Compared with BRD2, the mechanism of BRD4 in regulating the transcriptional activity of RELA has been extensively studies (Fig. 2), as described below.
BRD4 promotes RELA acetylation
Acetylated RELA is essential for recruiting BRD4 proteins to the promoters of target genes, which initiates inflammation, leading to cardiac fibrosis, myeloproliferative neoplasms, and airway inflammation.61 Increasing the binding between RELA and BRD4 can modify the chromatin environment and further promote the acetylation of RELA lysine 310 through BRD4’s atypical histone acetyltransferase activity, resulting in the transcriptional activation of inflammation and fibrosis genes (e.g., ACTA2, COL1A, FN1, MMP, IL6, KC, and neutrophilic chemokines).55 In addition, in human airway epithelial cells, hydrogen peroxide enhances the expression of IL6 and CXCL8 induced by IL1B through promoting the acetylation of RELA and the binding of BRD4 to the promoters of IL6 and CXCL8, while BRD2 has no effect on this process.90 Together, these findings indicate that there is a positive feedback between the formation of the RELA-BRD4 complex and the production of RELA acetylation, which is important for the transcriptional activity of RELA.
BRD4 promotes RELA phosphorylation
In addition to RELA acetylation, RELA phosphorylation contributes to the transcription activation of various inflammatory and catabolic genes. BRD4 promotes gene transcription by increasing the phosphorylation of RELA under various stimuli, such as IL1B, IL6, NOS2, and COX2 in highly aggressive proliferating immortalized (HAPI) microglia cells91 induced by LPS in rats. In mammalian cells, the cyclin-dependent kinase (CDK) family is the main regulator of the cell cycle and the initiator of DNA replication. The formation of the CDK9-BRD4 complex is required for RELA phosphorylation at Ser276-mediated RELA acetylation at Lys310, which leads to downstream inflammatory gene expression during RSV infection.92 Phospho-ser276 RELA is also required for the recruitment of the CDK9-BRD4 complex to core EMT transcriptional regulators (e.g., SNAI1, TWIST1, and ZEB1) to promote transforming growth factor beta 1 (TGFB1)-induced EMT.93 In macrophages infected with monocytogenes of the intracellular bacterial pathogen Listeria, two inhibitor of NF-κB kinase subunit beta, namely BI605906 and BETi JQ1, exhibit the same effect on the recruitment of BRD4 to NOS2 promoter and its expression, indicating that RELA activation is required for BRD4 binding to the target genes.94
Apart from recruiting phosphorylated RELA to the promoters of inflammatory genes, RELA can also form inflammatory superenhancers (SEs) and modulate the activities of global enhancer by altering the occupancy of BRD4. For example, canonical proinflammatory stimuli, such as TNF-α, enhance the binding of BRD4 to the SEs of proinflammatory genes in endothelial cells, thereby promoting the development of atherogenesis.63 JQ1-mediated inhibition of RELA phosphorylation-dependent cytokine production not only abrogates experimental renal inflammation in murine models,95 but also exhibits anti-inflammatory and anti-remodeling effects on human pulmonary microvascular endothelial cells.62 BRD4 and other BET members may play overlapping or different roles in regulating the expression of NF-κB-dependent inflammatory genes. For example, the knockdown of BRD2, BRD3, and BRD4 all reduce Helicobacter pylori-induced IL1B expression, but only the depletion of BRD4 significantly impairs Helicobacter pylori-induced IL1A expression.96
BRD4 regulates NFKBIA level
Depending on the context, BETs indirectly affect the activation of the RELA pathway by increasing or decreasing the expression of its upstream kinase IKBKB. For example, the deletion of BRD4 in macrophages results in the sustained expression of MAPK interacting serine/threonine kinase 2 (MKNK2) and activation of eukaryotic translation initiation factor 4E (EIF4E), which promotes the translation of NFKBIA mRNA, thereby reducing RELA-dependent inflammatory gene expression. Consequently, mice with myeloid lineage-specific deletion of the Brd4 gene are resistant to LPS-induced septic shock and tissue injury.97 In contrast, in rheumatoid fibroblast-like synoviocytes stimulated by TNF, the knockdown of BRD4 or BRD2 reduces the phosphorylation and degradation of NFKBIA, resulting in the inactivation of RELA.98 It is still unclear which signal or checkpoint is required for BRD4-mediated up- or downregulation of NFKBIA.
STATs
The STAT family consists of seven members (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6), which act as intracellular TFs in response to selective extracellular stimuli, such as cytokines (mainly IFNs and interleukins), growth factors, and PAMPs, by the membrane receptor-associated Janus kinase (JAK).99 The activation of the JAK-STAT pathway by phosphorylation leads to the production of cytokines and chemokines, thus affecting the immune response.
BETi shows strong activity in inhibiting the phosphorylation of JAK and STAT and subsequent transcriptional activity of STAT. For example, in a non-alcoholic steatohepatitis mouse model, I-BET151 treatment significantly reduced the expression of STAT1-dependent interferon gamma (IFNG) in liver tissue.100 Similarly, JQ1 suppresses genes enriched for a network of innate immune signaling nodes with a strong convergence on RELA-, JUN-, and STAT1-mediated transcriptional responses.101 I-BET 762 blocks LPS- and caerulein-induced phosphorylation of STAT3 in oncogenetic KrasG12D-driven mice, thereby blocking the production of nitric oxide and inflammatory cytokines (e.g., IL6, CCL2, and CSF2) in both immune and pancreatic cancer cells.102 I-BET 762 also decreases the phosphorylation of STAT3 in the mammary gland of MMTV-PyMT breast cancer mice, leading to the production of antitumor T-cell populations in mammary glands and spleen.103 These findings indicate the role of BETs in regulating the STAT3-dependent inflammatory tumor microenvironment and antitumor immunity.
More importantly, the BD2 domain of BRD2 recruits STAT3 to the chromatin through interaction with STAT3-K87Ac, thereby facilitating the recruitment of STAT3 to active enhancers occupied with interferon regulatory factor 4 (IRF4) and basic leucine zipper ATF-like transcription factor (BATF) and subsequent T helper (Th) 17 cell differentiation.104,105 In mouse embryonic fibroblasts, BRD4 is recruited to the STAT3-dependent suppressor of cytokine signaling 3 gene (SOCS3) and promotes its transcription.106 Like JQ1, the knockdown of BRD4 inhibits IL6-induced STAT3 activation and subsequent inflammatory gene production, thereby inhibiting the growth of pancreatic cancer.35 These findings further support the idea that BRD4-mediated STAT3 activation promotes tumor formation.
STAT5 mainly acts as a regulator of DC activation and is a crucial survival factor for NK cells. JQ1 can inhibit LPS-stimulated phosphorylation and nuclear accumulation of STAT5 in human monocyte-derived DCs, thereby decreasing STAT5’s transcriptional activity and impairing the maturation of monocyte-derived DCs.107 BRD2 is present at the transcriptionally active Cis locus and is required for the proper recruitment of STAT5-dependent transcriptional machinery for Cis.108 In IFNA1-induced monocytes, STAT1 and STAT2 are enriched on the promoter of IFN-responsive genes MX1 and CMPK2, and this process is inhibited by JQ1.109 Overall, these findings demonstrate that BETs mediate the activation of STAT pathways in various immune cells, which is highly related to infection, immunity, and tumorigenesis. Different STATs may exert overlapping and unique roles in BET-orchestrated immunity.
IRFs
IFRs are the main TFs responsible for antiviral immunity by producing IFNs. In small airway epithelial cells of humans infected with RSV, BRD4 recruits CDK9 to the promoters of IRF1, IRF7, and RIG, as well as to IRF3-dependent IFN-stimulated genes (ISGs), which in turn phosphorylates RNA Pol II at Ser 2 and enhances its expression through transcriptional elongation.110 Thus, BRD4 may play a protective role in the expression of airway mucosal IFN in response to RSV infection. BRD4 inhibition also induces host DNA damage response, thereby enhancing antiviral immunity by producting IFNs and ILs mediated by the CGAS-STING1-IRF3 signaling pathway.39 In LPS-induced bone marrow macrophages, the reduced expression of IRF4 and IRF8 mediates the effect of I-BET on suppressing the initial wave of inflammatory gene expression.69 Taken together, these findings highlight the importance of BRD4 in IRF-mediated antitumor immunity.
E2Fs
E2F is a family of TFs consisting of eight family members (E2F1 though 8). It regulates the cell cycle by repression or transactivation of genes that encode cyclins, cyclin-dependent kinases, checkpoint regulators, and replication proteins.111 In addition to the cell cycle, E2F is also involved in the inflammatory response, which is regulated by BETs. For example, E2F1 is citrullinated by peptidyl arginine deiminase 4 in inflammatory cells, thereby enhancing the binding of BRD4 to the acetylation domain in E2F1, resulting in the expression of proinflammatory genes.112 Though no direct evidence for the interaction between other BETs (BRD2 and BRD3) and the E2F family as well as their transcriptional regulation of immune genes has been reported, BRD2 is found to act as an E2F1- and H2A.Z variant histone 1 (H2AZ1)-interacting protein, which promotes the transcription of cell cycle-related genes.112 In addition, after JQ1 treatment, E2F3 shows enhanced motif activity, which may be related to the transcriptional regulation of BRD2.113 The details of BET-induced E2F activation still need further study.
MYC
The MYC family of proteins, a group of basic helix-loop-helix leucine zipper TFs that mainly coordinates cellular proliferation and metabolism, consists of four members: c-Myc (MYC, also known as c-MYC), L-Myc (MYCL), N-Myc (MYCN), and S-Myc.114 MYC (a BHLH transcription factor) is frequently altered in human cancers and promotes the transcription of various cell growth, apoptosis, and metabolism-related genes. Increasing evidence shows that MYC is an essential component for BET-regulated tumor immunity. For instance, JQ1 suppresses immune checkpoint CD274 expression by inhibiting the BRD4-MYC axis, indicating that the BRD4-MYC-CD274 pathway may mediate tumor immune escape.115 In neuroblastoma, JQ1 impairs the expression of ULBP1–3 ligands for NKG2D activating receptor by inhibiting the transcriptional regulation of MYC and tumor protein P53 (TP53), thereby rendering NB cell lines more resistant to NK cell-mediated killing.116 These studies demonstrate the dual role of BETs in antitumor immunity, depending on MYC status and cancer type.
Other TFs
In addition to the TFs discussed above, other TFs (e.g., TP53, RUNX2, SP7, JUN, FOS, ETS1, ETS2, CDX2, FOXL1, LIN54, RORC, ARIE, and FOXM1) also interact with BETs to participate in the transcriptional regulation of immune-related genes (Table 1 and Fig. 5). Studies further suggest that BETs are essential for TF-mediated gene transcription, although they may function in a context-dependent manner.
Transcriptional coactivators
Transcription activation is a complex, multi-stage process, involving by hundreds of proteins (including TFs and transcription coactivators). P-TEFb generally functions as a coactivator of BET-mediated gene transcription. It is composed of CDK9 and one of several other cyclin-related partners (e.g., cyclin T1), which release the paused RNA Pol II at the proximal promoter to allow transcription.117 Mechanistically, the BETs recruit CDK9 and cyclin T1 to the RNA Pol II,20,118 then activate it through phosphorylation of the C-terminal domains Ser2 and Ser5 for transcriptional extension.119,120 BRD4 can bridge autoimmune regulator (AIRE) and P-TEFb, thus promoting AIRE-mediated gene transcription in medullary epithelial cells and inducing effective immunologic tolerance.121 However, the molecular mechanism of transcriptional co-repressors and the way the protein can be converted from a coactivator to a co-repressor are unclear.
Histones
Binding to acetylated lysine residues on histone tails is a prerequisite for BET-mediated transcription activation. The core histones (H2A, H2B, H3, and H4) form the center of nucleosomes, which are linked by histone H1.122 BETs specifically recognize acetylated lysine residues in histone H3 and H4. Acetylation of lysine positions in the histone tail is performed by histone acetyltransferase enzymes (HATs). In most species, histone H3 is usually acetylated at lysine 9, 14, 18, 23, and 27, while histone H4 is mainly acetylated at lysine 5, 8, 12, and 16.123 IL1B or TNF induce the acetylation of H4K5Ac, H4K8Ac, and H4K12Ac, then recruits BRD3 and BRD4 to the promoter of the matrix degrading enzyme genes (MMP1, MMP3, MMP13, and ADAMTS4), thereby increasing their expression in human chondrosarcoma cell lines (SW1353).118 In virus-infected macrophages, BRD3 increases the acetylation of histone H3 and H4 within the IFNB1 promoter, leading to the production of type I IFN.50 Moreover, the accumulation of H4K5Ac and BRD4 on the IL6 gene promoter is found in lung fibroblasts from idiopathic pulmonary fibrosis donors, leading to increased IL6 production and secretion.124 In airway smooth muscle cells isolated from asthmatic individuals, histone H3 acetylation (especially H3K18Ac) increases, which helps BRD3 and BRD4 bind to the promoter of CXCL8 and promotes its expression, thereby driving steroid-resistant neutrophilic airway inflammation.125
Although BRD2, BRD3, and BRD4 are preferentially recruited to H4K5Ac, H4K12Ac, and H3K14Ac, H3K27Ac has attracted increasing attention in recent years. For example, investigators have found that BRD2, BRD3, and BRD4 directly bind to H3K27Ac at the promoter of 2, 3-dioxygenase 1 (IDO1) that mediates metabolism-related immune escape in cancer.126 The combination of BRD4 and H3K27Ac also facilitates the formation of SEs, which drives the transcription of NF-κB target genes (MT-CO2 and TGFB2), and then promotes the production of the extracellular matrix, myofibroblast differentiation, and tumor-associated inflammation.127,128 In addition, BETi (e.g., I-BET151) inhibits the deposition of H3K27Ac at the promoters of proinflammatory cytokines (TNF and IL6) induced by β-glucan.129 A global analysis of lysine acetylation may help us to better understand the function of BETs in gene transcription.
In the BET family, BRD4 acts as an atypical HAT, which can acetylate histone H3 and H4 in a different mode than other HATs.128 Because BRD4 can induce the acetylation of histone H3 on Lys residue 122 (H3K122Ac), which is a posttranslational modification that destabilizes nucleosome structure, the nuclear abundance of H3K122Ac is considered to be a selective marker for the HAT activity of BRD4.15 Both BRD4 and RELA are required for various stimuli (e.g., RSV, poly(I:C), and allergen)-induced acetylation of histone H3 on Lys 122, thereby promoting airway remodeling driven by inflammation.55,58,110 The functional interaction between BRD4 and classical HATs needs further clarification.
BETi in innate immunity
I-BET was discovered in 2010 and was the first BETi that was found to mimic acetylated histones to disrupt BET binding to chromatin. I-BET exhibits an anti-inflammatory effect on LPS-induced endotoxic shock and bacteria-induced sepsis.69 Since then, a large number of BETis (e.g., JQ1, I-BET151, OTX015, and I-BET762) have been developed, which exhibit excellent anti-inflammatory and immunomodulatory activities (Table 1). Unfortunately, due to the high structural homology in the two BD domains of BETs, most BETis are nonselective. Recently, selective inhibitors targeting BD1 and BD2 of the BET proteins have been developed, and it is proposed that BD1 is primarily required for steady-state gene expression whereas both BD1 and BD2 are required for the rapid inflammatory stimuli-induced increase of gene expression. As such, selective BD1 inhibitors phenocopy the effects of pan-BETi in cancer whereas selective BD2 inhibitors are dominantly effective in inflammatory and autoimmune disease.130 However, the long-term adverse reactions of these BETis are not clear. Hence, more selective BETis that target BD1 and BD2 with fewer toxic side effects are still in great need to achieve precise treatment of different diseases.
In the pathogenesis of various diseases, the roles of individual BETs overlap but are discrete, which indicates that it is still very important to develop new isotype-selective and well-tolerated BETis. In recent years, the degradation of BETs induced by proteolytic targeting chimera (PROTAC) has shown excellent targeting ability and inhibition. PROTAC-based BETis exhibit excellent immunoregulatory activities and include dBET1,131 MZ-1,132 and ARV-825.95 Importanly, dBET1 potently reduces proinflammatory responses in LPS-activated microglia by degrading BRD2 and BRD4, while ARV-825 increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells by degrading BRD4.133 MZ-1 redesigned based on JQ1 structure prevents TNF-induced expression of adhesion molecules and inflammatory mediators in the monocytes and endothelial cells by selectively degrading BRD4. These pan- or selective BETis provide a useful tool for studying the roles of BETs in regulating inflammation and immune response in vivo.
Of note, though proteolysis targeting chimera (PROTACs) have distinct advantages over small-molecule BETis, they may be limited to proteins that contain cytosolic domains to which ligands can bind and recruit the requisite cellular components. Lysosome-targeting chimeras (LYTACs) may be a more promising strategy to treat various diseases through selectively degrading BETs. These recently investigated chimeras use conjugates that bind both a cell surface lysosome-shuttling receptor and the extracellular domain of a target protein.134 In addition, phase-separated condensates that compartmentalize and concentrate anti-neoplastic drugs may facilitate LYTACs binding to BRD4 and have selective effects on oncogenes,135 although the efficiency of this approach is still controversial. Elucidating the three-dimensional structure and complexity of BETs is expected to allow us to obtain more effective and specific BETis.
BETs in diseases
Because of their critical roles in the transcriptional regulation of genes, BETs have become promising therapeutic targets for various diseases (especially inflammation-related diseases, cancers, and metabolic diseases), as described below.
Infectious diseases
Bacteria infection
Bacteria are the most common organisms that cause local and systemic inflammation, even sepsis and septic shock. LPS, a major component of the outer membrane of Gram-negative bacteria, is widely recognized as a strong activator of innate immune response. Both the genetic and pharmacological inhibition of BETs inhibit LPS-induced systemic inflammation or organ-specific inflammatory responses. For example, pan-BETi protects against endotoxic shock, polymicrobial peritonitis, polymicrobial sepsis induced by cecal ligation and puncture,69 LPS-induced periodontitis,95 and vascular inflammation.13 The genetic inhibition of BRD4 decreases NOS2 expression and inflammation response in Listeria monocytogenes-induced macrophages in vitro.94 In vivo, mice with a myeloid lineage-specific deletion of Brd4 are more sensitive to group B Streptococcus-induced infection, but are resistant to LPS-induced endotoxic shock,97 indicating a different role of BRD4 in infection. BRD2 also drives LPS-stimulated neuroinflammation and alveolar inflammation.40,46 Apart from LPS, listeriolysin-O, a hemolysin produced by the bacterium Listeria monocytogenes, increases BRD4 expression in Kupffer cells, which may induce liver injury by promoting necroptosis, inflammation, and mitochondrial dysfunction.136 Whether the functional impairment of BETs is the main cause and prognostic determinant of common bacterial infections and subsequent multiple organ dysfunction syndrome (especially disseminated intravascular coagulation) remains to be determined.
Viral infection
Viral infections have similarities and differences with bacterial infections. Most viral infections can be prevented by the innate immune system, and when the virus replicates beyond the innate defense, the adaptive immune response can be mobilized. As mentioned earlier, BRD4 inhibition enhances innate immune response, resulting in the inhibition of the attachment of DNA and RNA viruses through the CGAS-STING1 pathway.39 RSV replication increases the expression and binding of BRD4 to RELA, thereby triggering the inflammatory response in the lower respiratory tract and promoting airway remodeling.58,110 Moreover, treatment with pan-BETi also dominantly induces the resistance to influenza A virus (H1N1 subtype; strain WSN/33) by enhancing innate immunity.94 The inhibition of BETs leads to HIV promoter activation through separate modes of action, which may be beneficial for a combined anti-retroviral therapy.137 These findings generally suggest that increased BRD4 may be detrimental to antiviral immunity.
Conversely, BETs also suppress viral infection by maintaining IFNB production. Indeed, virus infection, such as through sendai virus, vesicular stomatitis virus, and herpes simplex virus, remarkably downregulate BRD3 expression in macrophages, thereby inhibiting the production of IFNB.50 As such, BET expression level is important for the establishment of antiviral immune homeostasis, and it may be useful for disease severity assessment and prognostic prediction of viral infection.
Fungi and parasitic infection
Invasive fungal infections usually result in high morbidity and mortality among immunocompromised individuals. The fungal BET protein BDF1, a global transcriptional regulator in Saccharomyces cerevisiae, also harbors two BD domains that are essential for the viability and virulence of C. albicans. Hence, BDF1 is proposed as a drug target for antifungal therapy.138 Although the exact mechanism between fungal infection, BETs, and host immune response has not yet been established, emerging evidence suggests that BETs (e.g., BRD2 and BRD4) may be involved in the differentiation of Th17 cells that can protect mucosa from bacterial and fungal infection through producing interleukin-17A (IL17A) and IL-17F.104,139 I-BET151 also inhibits the functions of human monocytes, lymphocytes, and granulocytes as well as the production of proinflammatory cytokines after candida albicans and aspergillus fumigatus stimulation,129 indicating an integrated immune modulation after BET inhibition.
Although the exact roles of BETs in parasitic infections are still elusive, BRD2 increases in livers infected with Plasmodium yoelii and Toxoplasma gondii and participates in host immune responses related to MHC class II, indicating a role of BRD2 in linking innate and adaptive immunity.140 In the mouse model of schistosomiasis, BET inhibition displays a protective effect on liver fibrosis by attenuating EP300-mediated RAR-related orphan receptor C (RORC) acetylation, thus decreasing the expression of RORC target genes.141 These findings indicate that BET, especially BRD2, predominant orchestrates innate immunity to fungi and parasitic infection, and it may serve as a potential therapeutic target for fungal and parasitic infections.
Non-infectious diseases
In addition to mediating infection, DAMPs can also initiate sterile inflammation, which is not only essential for tissue repair and regeneration, but also results in the development of numerous inflammation-related diseases, such as cancer, cardiovascular diseases, and metabolic disorders.142 As with PAMPs, accumulating evidence suggests the potential roles of BETs in controlling DAMP-mediated innate immunity and diseases.
Cancers
The development of cancer is a multi-step process and includes internal and external causes. It has been widely accepted that BETs-mediated transcription of pro-proliferative genes and anti-apoptotic genes plays a critical role in tumorigenesis. BETs inhibition down-regulates key oncogenic transcription factor pathways, thereby providing anti-tumor activity.143–145 Furthermore, BET-mediated innate and adaptive immune responses contribute to the development of cancers through the regulation of multiple events in the tumor microenvironment (TME), especially chronic inflammation and immune surveillance.
Chronic inflammation
The TME is a key factor in tumor progression and has been increasingly regarded as an anticancer therapeutic target. BETs regulate the TME through activating the transcription of proinflammatory genes in immune cell subsets infiltrating the TME. For instance, BRD4 can bind to the promoters of ARGINASE1 and other IL4-driven macrophage genes such as IL6, RETNLB, and CHIA, which results in immunosuppression in the TME.146 BRD4, but not BRD2 or BRD3, promotes TME inflammation in triple-negative breast cancer through upregulating multiple genes involved in extracellular matrix regulation, such as COL1A2, COL3A1, COL5A2, and KRT19.147 Chronic inflammation can induce DNA damage and accelerate gene mutations, which contribute to the development and progression of cancer. Like immune cells, cancer cells have the ability to release cytokines or chemokines to trigger tumorigenesis and metastasis. This process is called cancer cell-intrinsic inflammation and can be regulated by certain BETs. For example, BRD4 inhibition decreases CXC chemokine expression in clear cell renal cell carcinoma cells, resulting in a weakened inflammatory response and a reduced metastatic cascade.148 BRD4-mediated expression of IL6, RORC, COX2, MYC, CCND1, and CD47 also promotes the development of various solid cancers, such as pancreatic ductal adenocarcinoma, breast cancer, colorectal cancer, and myeloproliferative neoplasms.35 This inflammatory process is mainly driven by a BRD4-dependent activation of the NF-κB pathway in cancer cells.61 Thus, blocking the BRD4-NF-κB pathway may limit the development of cancer cell-intrinsic inflammation.
Immune surveillance
Through immune surveillance, the innate immune system plays a critical role in recognizing and eliminating tumor cells. This process is coordinated by various cells and proteins in the TME. In general, BETs may facilitate tumorigenesis via evading immune surveillance. For example, BRD4 expression is substantially upregulated in lymph node-negative breast cancer with a high expression of T-box transcription factor 21 (TBX21), which finally attenuates immune surveillance by upregulating jagged canonical notch ligand 1 (JAG1) expression.149 Oncogene-induced senescence is regarded as a potent barrier to tumorigenesis due to the generation of senescence-associated secretory phenotype (SASP). BRD4 is recruited to newly activated SEs adjacent to key SASP-related genes and mediates downstream paracrine signaling, thereby contributing to tumor-suppressive immune surveillance.150 Therefore, changes in the BRD4 pathway allow tumors to evade immune surveillance and to excessively proliferate, leading to their invasion into surrounding tissue structures and eventual metastasis.
Tumor antigen-specific cytotoxic T lymphocytes (CTLs) are the main effector of antitumor immune response. BETs also alter T-cell expression and function within the TME. For example, the T-cell population in the TME is changed by pan-BETi (I-BET 762) in some cancers.103 Personalized cancer vaccine is a cancer vaccine developed by encapsulating JQ1 and indocyanine green co-loaded tumor cells with a hydrogel matrix. This vaccine suppresses tumor relapse via promoting the maturation of DCs and eliciting of tumor infiltration of CTLs.151 JQ1 also has the ability to restore an immune-active environment by increasing intratumor DCs and CD8C T lymphocytes, and decreasing myeloid-derived suppressor cells.34 In addition, BETi enhances the immunogenicity of prostate cancer cells and the susceptibility to CD8 T-cell targeting by increasing MHC I expression.152
Immune checkpoints are usually activated in cancers to hinder the nascent antitumor immune response and become promising targets for cancer therapy. The CD274 checkpoint is transcriptionally regulated by histone acetyltransferase 1 in pancreatic cancer, and BRD4 is required for this process.153 Alternatively, BRD4 directly bind to CD274 promoter and regulates its transcription in pancreatic stellate cells.154 JQ1 greatly inhibits the expression of CD274, thereby overcoming the immunosuppressive effects in prostate cancer, lung cancer, and triple-negative breast cancer during cancer therapy.115,152,155 In addition, for cancers with relatively low responsiveness to immune checkpoint blockade therapy, I-BET762 enhances the efficacy of CD274 blockade by reducing the proportion of CD14 + HLA-DR/low myeloid-derived suppressor cells.156 In addition to BRD4, the gene status of BRD3 is also related to CD274 expression in cancer cells. In microsatellite high instability gastric cancer, the number of frameshift mutations in BRD3 is negatively correlated with CD274 expression.48 Apart from CD274, BETs also directly promote the expression of IDO1, an immune checkpoint that mediates metabolic immune escape in cancer through the production of L-kynurenine.126 Collectively, the evasion of immune surveillance by BETs covers multiple immunosuppressive mechanisms. The combination of BETi and traditional chemotherapy may restore or enhance antitumor immunity.
Cardiovascular diseases
DAMPs released from myocardial necrosis initiate inflammation to repair wounds and form scars. Nevertheless, persistent inflammatory response contributes greatly to myocardial remodeling and ultimately heart failure.157 BET-mediated transcription of proinflammatory and pro-fibrogenic genes are involved in various cardiovascular diseases, such as cardiac hypertrophy,158 pulmonary arterial hypertension,62,159,160 and myocardial infarction.66 The upregulation of BRD4 in cardiomyocytes may induce cardiac hypertrophy by increasing the expression of pro-fibrotic genes and activating RELA-driven inflammation.158 BRD4-mediated IL6 expression is increased in the coronary arteries, thereby promoting coronary artery remodeling during pulmonary arterial hypertension.160 Moreover, the binding of BRD4 to the SEs and promoters of proinflammatory or adhesion molecule genes in monocytes and endothelial cells promotes atherogenesis and the incidence of major adverse cardiac events.63,132 BRD4 may also promote the senescence and lipid accumulation in LPS-induced senescent macrophages by increasing the expression of SASP in autocrine and paracrine senescence, thereby promoting the progress of atherosclerosis.161 Accordingly, JQ1 exhibits therapeutic effects on heart failure caused by prolonged pressure overload and a massive anterior myocardial infarction.66,101 Similarly, BETi apabetalone (RVX-208) not only improves the cardiovascular outcomes of patients with type 2 diabetes after acute coronary syndrome,162 but also decreases the risk of atherosclerotic plaque rupture and major adverse cardiac events.132 These findings indicate that BETi can be used to treat a wide range of cardiovascular diseases, although its long-term side effects are currently unclear. Notably, the mechanisms of BETs in these cardiovascular diseases are far from being fully understood, and current studies mainly focus on the roles of BETs in the pathogenesis of chronic sterile inflammation-related cardiovascular diseases. A better understanding of the roles and mechanisms underlying BETs in sepsis-induced acute myocardial dysfunction, cardiomyogenesis, and myocardial regeneration after myocardial infarction should also be pursued in the future.
Respiratory diseases
Although infection is the leading cause of respiratory diseases, other pathogenic factors such as cigarette smoke extract and allergens, can also induce respiratory dysfunction. BRD4-mediated activation of the NF-κB pathway promotes lung inflammation, leading to airway remodeling in allergic airway disease.163 Moreover, BRD4-driven expression and secretion of IL6, CXCL8, and IL17A contribute to the development of idiopathic pulmonary fibrosis, chronic obstructive pulmonary disease, cystic fibrosis, and neutrophilic lung diseases.26,27,101,140 After growth factor stimulation, both BRD2 and BRD4 are involved in the regulation of filamentous ACTA2 expression in lung fibroblasts and drive pulmonary fibrosis.164 It remains to be seen whether BET-mediated production of proinflammatory cytokines affects immune cell infiltration and differentiation in infection-induced acute lung injury and various chronic lung diseases.
Neurological disorders
Sterile neuroinflammation is usually present in various central nervous system diseases, such as brain ischemia reperfusion injury, neurodegenerative diseases, and traumatic brain injury. The presynaptic neuronal protein α-synuclein is a pathological marker of Parkinson’s disease. It can trigger microglial activation by increasing BRD2 expression and subsequently inhibiting SIRT1 activation,44 indicating a pathologic role of BRD2 in Parkinson’s disease. In addition to Parkinson’s disease, dysfunctional BETs are also associated with Alzheimer’s disease due to their role in maintaining chronic inflammation.165 BETs promote systemic sclerosis by decreasing the acetylation and expression of two IFN-dependent genes (MX1 and CMPK2) in monocytes,109 suggesting that BET-dependent IFN signaling is a therapeutic target for systemic sclerosis. Moreover, the activation of BET pathway, especially BRD2-mediated inflammatory response and pyroptosis, aggravates cerebral ischemia-induced brain injury and acute spinal cord injury.47,75,166 Thus it can be concluded that BET-mediated activation of inflammatory pathways is an important pathological event leading to neurological disorders, and the correlation of BETs with other neuroinflammation-associated refractory diseases, such as depression, autistic disorder, and epilepsy, can also be attempted. In addition, as the immune system is closely interconnected with the state and function of the nervous system, research on the immunomodulatory roles of BETs expressed in different types of neurons is of great importance.
Kidney diseases
It is widely accepted that inflammation plays an important role in the pathogenesis of acute kidney dysfunction and chronic kidney diseases. Persistent low-grade inflammation is a hallmark of chronic kidney diseases and accelerates the loss of nephron functionality. BET-mediated expression of RELA-dependent proinflammatory genes (e.g., IL6, CCL2, and CCL5) and the activation of the Th17 immune response have considerable roles in acute renal damage caused by unilateral ureteral obstruction, lupus nephritis, and HIV-associated kidney disease.95,167,168 In addition, BET-mediated gene expression of pro-fibrotic factors promotes renal fibrosis and exacerbates renal dysfunction. Given that the pathophysiological process of kidney disease is complicated by various forms of ion pathway change, it is necessary to investigate whether BETs play a role in the disturbance of water and ion homeostasis as well as the endocrine function of the kidneys.
Digestive diseases
Given the role of BETs in the regulation of proinflammatory and pro-fibrotic gene expression, they are also involved in a diverse range of digestive diseases, such as non-alcoholic steatohepatitis and liver fibrosis,100 inflammatory bowel disease,169 acute pancreatitis,170 and colitis.20 Gut microbiota plays a critical role in the induction, training, and function of innate immunity, but their direct correlation with BETs has not yet been established. Regarding gut microbiomes and immunity, more comprehensive and in-depth research on BETs may expand our knowledge of and techniques against digestive diseases and improve clinical outcomes in the future.
Metabolic diseases
Metabolic abnormalities, such as diabetes mellitus, are also due to chronic low-grade inflammation. BRD2 is highly expressed in pancreatic β cells and physiologically inhibits INS transcription and β-cell mitosis. The knockdown of BRD2 results in severe obesity without type 2 diabetes mellitus, because BRD2 shifts energy balance toward storage without inducing glucose intolerance.24 However, this function of BRD2 in diabetes has been challenged by recent research. For example, the overexpression of BRD2 (but not BRD3 or BRD4) initiates chronic inflammation in adipocytes by activating RELA, thereby resulting in insulin resistance.89 These findings provide evidence that the upregulation of BRD2 may increase the susceptibility to type 2 diabetes.
BETs also contribute to the pathogenesis of type 1 diabetes mellitus. BETi (I-BET151) not only mitigates the immune response against pancreatic β cells, but also enhances their proliferation and function, thereby increasing insulin secretion and inhibiting the development of type 1 diabetes.171 Although BRD2 and BRD4 have the same activity to inhibit INS transcription, only BRD2 can inhibit fatty acid oxidation in β cells.172 Intriguingly, in Drosophila melanogaster, the BET protein Fs(1)h is required in fat body cells for a normal lifespan as well as metabolic and immune homeostasis. Flies lacking fat body fs(1)h exhibits a shorter lifespan, enhanced expression of immunotarget genes, disability to metabolize triglyceride, and systemic defects in insulin signaling.173 As mentioned above, the activation of the BRD4–NF-κB pathway contributes to the development of gouty arthritis, diabetic intervertebral disc degeneration, osteoarthritis, and rheumatoid arthritis, which have abnormal metabolic properties. In addition, both BRD2 and BRD4 play key roles in the onset of cancer cachexia by increasing the transcription of catabolic genes regulated by the IL6-AMPK-FOXO3 pathway.174 However, little is known about how BETs cause the interaction between immune and metabolic dysfunction in the TME, and how they link the metabolic disturbance of immune cells to immune dysfunction.
Osteoarthritis
Although osteoarthritis is traditionally regarded as a type of non-inflammatory arthritis, emerging evidence suggests that inflammation caused by DAMPs plays a vital role in the pathogenesis of osteoarthritis. As mentioned above, it has been proved that the BRD4-NF-κB signaling pathway has a great contribution to the development of gout arthritis, diabetic intervertebral disc degeneration, osteoarthritis, and rheumatoid arthritis.51,74,98,175–177
Other diseases
BETs are also involved in other diseases due to their immunomodulatory and proinflammatory properties, such as spontaneous preterm birth, preeclampsia, retinal inflammatory disease, inherited retinal degeneration, age-related macular degeneration, and psoriasis.31,32,45,178–180 It is expected that BETs may serve as potential therapeutic targets in a variety of immune-mediated diseases.
Conclusion and perspectives
As epigenetic readers, BETs control the transcription of genes by recognizing acetylated histones and recruiting TFs and co-activators to the chromatin, thereby promoting the phosphorylation of RNA pol II and facilitating transcription initiation and elongation. BETs orchestrate various extracellular or intracellular danger signals through PRRs expressed in immune and non-immune cells in a wide range of diseases and have emerged as promising therapeutic targets. Although BRD4 is the most extensively studied member of the BET family, the exact role of other BET members (e.g., BRD2 and BRD3) in diseases and pathological conditions is still far from being fully understood. It is worth noting that a defect in BETs in mice causes embryo lethality, indicating that BETi may cause side effects. More studies are needed to develop isoform-selective and well-tolerated BETi or small-molecule PROTAC degraders and to clarify the distinctive roles of individual BETs in various cellular processes, including inflammation and immunity. High-efficiency and low-toxicity BETis may benefit the treatment of various immune-mediated inflammatory diseases in the future.
Although BETs play a crucial role in regulating gene transcription, the mechanism for this has not been fully elucidated. It is worth noting that BETs can recognize not only residues in histones, but also other acetylated nuclear proteins to control gene transcription.181 It can be supposed that other acetylation-regulated TFs, including cAMP response element binding protein 1 (CREB1), heat shock transcription factor 1 (HSF1), sterol regulatory element binding proteins (SREBPs), and carbohydrate response element binding protein (ChREBP), may also contribute to BET-orchestrated innate immunity.182 In addition to transcriptional regulatory activities, BETs also possess intrinsic kinase and lysine acetyltransferase (KAT) activities that have not been extensively studied.15,22 Hence, future research can also be extended to include the diverse transcriptional regulatory mechanisms of BETs and the roles of their kinase and KAT activities.
BETs play an emerging role in phase separation. The addition of BRD4 to acetylated chromatin can induce its liquid-liquid phase separation, so that different chromatin compartments can be established and maintained, thereby regulating gene transcription.183 The inherent disordered regions of BRD4 and MED1 can form phase-separated droplets, and then separate and concentrate the nuclear extract transcription device and control key cell-identity genes.184 In addition, LncRNA DIGIT can promote BRD3 to form a phase-separated condensate, which occupies the enhancer of endoderm transcription factor and drives the transcription of genes related to endoderm differentiation.185 These findings enhance the understanding of the pathological role of BETs in diseases and may identify new therapeutic targets.
Acknowledgements
We thank Dave Primm (Department of Surgery, UTSW) for his critical reading of the manuscript. D.T. is supported by a grant from the U.S. NIH (RO1GM127791). R.K. is supported by a grant from the U.S. NIH (RO1CA211070).
Competing interests
The authors declare no competing interests.
Contributor Information
Daolin Tang, Email: daolin.tang@utsouthwestern.edu.
Rui Kang, Email: rui.kang@utsouthwestern.edu.
References
- 1.Visvanathan K, Lewin SR. Immunopathogenesis: role of innate and adaptive immune responses. Semin Liver Dis. 2006;26:104–115. doi: 10.1055/s-2006-939755. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway C., Jr Innate immunity. N. Engl. J. Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 3.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu. Rev. Immunol. 2015;33:445–474. doi: 10.1146/annurev-immunol-032414-112043. [DOI] [PubMed] [Google Scholar]
- 4.Pino SC, Kruger AJ, Bortell R. The role of innate immune pathways in type 1 diabetes pathogenesis. Curr. Opin. Endocrinol. Diabetes Obes. 2010;17:126–130. doi: 10.1097/MED.0b013e3283372819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiersinga WJ, Leopold SJ, Cranendonk DR, van der Poll T. Host innate immune responses to sepsis. Virulence. 2014;5:36–44. doi: 10.4161/viru.25436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, Cao X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019;19:417–432. doi: 10.1038/s41577-019-0151-6. [DOI] [PubMed] [Google Scholar]
- 7.Topper MJ, Vaz M, Marrone KA, Brahmer JR, Baylin SB. The emerging role of epigenetic therapeutics in immuno-oncology. Nat. Rev. Clin. Oncol. 2020;17:75–90. doi: 10.1038/s41571-019-0266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr. Opin. Drug Discov. Dev. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 10.Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017;18:246–262. doi: 10.1038/nrm.2016.143. [DOI] [PubMed] [Google Scholar]
- 11.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi Y. The bromodomain and extra-terminal domain (BET) family: functional anatomy of BET paralogous proteins. Int. J. Mol. Sci. 2016;17:1849. doi: 10.3390/ijms17111849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M, et al. The suppression of bromodomain and extra-terminal domain inhibits vascular inflammation by blocking NF-kappaB and MAPK activation. Br. J. Pharm. 2017;174:101–115. doi: 10.1111/bph.13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 15.Devaiah BN, et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 2016;23:540–548. doi: 10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 17.Alsarraj J, et al. BRD4 short isoform interacts with RRP1B, SIPA1 and components of the LINC complex at the inner face of the nuclear membrane. PLoS ONE. 2013;8:e80746. doi: 10.1371/journal.pone.0080746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriniere J, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura Y, et al. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- 20.Cheung K, et al. BET N-terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice. Proc. Natl Acad. Sci. USA. 2017;114:2952–2957. doi: 10.1073/pnas.1615601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Disco. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 22.Devaiah BN, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc. Natl Acad. Sci. USA. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, et al. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J. 2009;425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev. Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DJ, et al. Identification of a bromodomain-containing protein 2 (BRD2) gene polymorphic variant and its effects on pork quality traits in berkshire pigs. Korean J. Food Sci. Anim. Resour. 2018;38:703–710. doi: 10.5851/kosfa.2018.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong J, et al. Bromodomain-containing protein 4 inhibition alleviates matrix degradation by enhancing autophagy and suppressing NLRP3 inflammasome activity in NP cells. J. Cell Physiol. 2020;235:5736–5749. doi: 10.1002/jcp.29508. [DOI] [PubMed] [Google Scholar]
- 29.Song J, Wang Q, Zong L. LncRNA MIR155HG contributes to smoke-related chronic obstructive pulmonary disease by targeting miR-128-5p/BRD4 axis. Biosci. Rep. 2020;40:BSR20192567. doi: 10.1042/BSR20192567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang K, Zhao J, Xie J, Wang J. Decreased miR-29b expression is associated with airway inflammation in chronic obstructive pulmonary disease. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;316:L621–L629. doi: 10.1152/ajplung.00436.2018. [DOI] [PubMed] [Google Scholar]
- 31.Lim R, Nguyen-Ngo C, Lappas M. Targeting bromodomain-containing proteins to prevent spontaneous preterm birth. Clin. Sci. 2019;133:2379–2400. doi: 10.1042/CS20190919. [DOI] [PubMed] [Google Scholar]
- 32.Liong S, Barker G, Lappas M. Bromodomain protein BRD4 is increased in human placentas from women with early-onset preeclampsia. Reproduction. 2018;155:573–582. doi: 10.1530/REP-17-0744. [DOI] [PubMed] [Google Scholar]
- 33.Tan YF, Wang M, Chen ZY, Wang L, Liu XH. Inhibition of BRD4 prevents proliferation and epithelial-mesenchymal transition in renal cell carcinoma via NLRP3 inflammasome-induced pyroptosis. Cell Death Dis. 2020;11:239. doi: 10.1038/s41419-020-2431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riganti C, et al. Bromodomain inhibition exerts its therapeutic potential in malignant pleural mesothelioma by promoting immunogenic cell death and changing the tumor immune-environment. Oncoimmunology. 2018;7:e1398874. doi: 10.1080/2162402X.2017.1398874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazur PK, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 2015;21:1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher SJ, Tiffen JC, Hersey P. Histone modifications, modifiers and readers in melanoma resistance to targeted and immune therapy. Cancers (Basel) 2015;7:1959–1982. doi: 10.3390/cancers7040870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markman RL, et al. Interfering with bromodomain epigenome readers as therapeutic option in mucoepidermoid carcinoma. Cell Oncol. 2019;42:143–155. doi: 10.1007/s13402-018-0416-2. [DOI] [PubMed] [Google Scholar]
- 38.Bachtel ND, et al. Short communication: expression of host restriction factors by memory CD4+ T cells differs between healthy donors and HIV-1-infected individuals with effective antiretroviral therapy. AIDS Res. Hum. Retroviruses. 2019;35:108–111. doi: 10.1089/aid.2018.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, et al. BRD4 inhibition exerts anti-viral activity through DNA damage-dependent innate immune responses. PLoS Pathog. 2020;16:e1008429. doi: 10.1371/journal.ppat.1008429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malhotra R, et al. Altered regulation and expression of genes by BET family of proteins in COPD patients. PLoS ONE. 2017;12:e0173115. doi: 10.1371/journal.pone.0173115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudman MD, et al. Bromodomain and extraterminal domain-containing protein inhibition attenuates acute inflammation after spinal cord injury. Exp. Neurol. 2018;309:181–192. doi: 10.1016/j.expneurol.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Lin YC, Wang FS, Yang YL, Chuang YT, Huang YH. MicroRNA-29a mitigation of toll-like receptor 2 and 4 signaling and alleviation of obstructive jaundice-induced fibrosis in mice. Biochem. Biophys. Res. Commun. 2018;496:880–886. doi: 10.1016/j.bbrc.2018.01.132. [DOI] [PubMed] [Google Scholar]
- 43.Li S, Cui W, Song Q, Zhou Y, Li J. miRNA-302e attenuates inflammation in infantile pneumonia though the RelA/BRD4/NF-kappaB signaling pathway. Int. J. Mol. Med. 2019;44:47–56. doi: 10.3892/ijmm.2019.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar S, et al. Molecular signatures of neuroinflammation induced by alphaSynuclein aggregates in microglial cells. Front. Immunol. 2020;11:33. doi: 10.3389/fimmu.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao L, et al. Photoreceptor protection via blockade of BET epigenetic readers in a murine model of inherited retinal degeneration. J. Neuroinflammation. 2017;14:14. doi: 10.1186/s12974-016-0775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi CS, et al. The epigenetic reader BRD2 as a specific modulator of PAI-1 expression in lipopolysaccharide-stimulated mouse primary astrocytes. Neurochem. Res. 2015;40:2211–2219. doi: 10.1007/s11064-015-1710-2. [DOI] [PubMed] [Google Scholar]
- 47.Jin L, et al. Identifying gene expression profile of spinal cord injury in rat by bioinformatics strategy. Mol. Biol. Rep. 2014;41:3169–3177. doi: 10.1007/s11033-014-3176-8. [DOI] [PubMed] [Google Scholar]
- 48.Cho J, et al. Four distinct immune microenvironment subtypes in gastric adenocarcinoma with special reference to microsatellite instability. ESMO Open. 2018;3:e000326. doi: 10.1136/esmoopen-2018-000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishii H, Mimori K, Mori M, Vecchione A. Differentially expressed genes in endothelial differentiation. DNA Cell Biol. 2005;24:432–437. doi: 10.1089/dna.2005.24.432. [DOI] [PubMed] [Google Scholar]
- 50.Ren W, et al. Bromodomain protein Brd3 promotes Ifnb1 transcription via enhancing IRF3/p300 complex formation and recruitment to Ifnb1 promoter in macrophages. Sci. Rep. 2017;7:39986. doi: 10.1038/srep39986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein K, et al. The bromodomain protein inhibitor I-BET151 suppresses expression of inflammatory genes and matrix degrading enzymes in rheumatoid arthritis synovial fibroblasts. Ann. Rheum. Dis. 2016;75:422–429. doi: 10.1136/annrheumdis-2014-205809. [DOI] [PubMed] [Google Scholar]
- 52.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev. Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 53.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the TLR3 pathway. Rev. Med. Virol. 2011;21:67–77. doi: 10.1002/rmv.680. [DOI] [PubMed] [Google Scholar]
- 55.Tian B, et al. Efficacy of novel highly specific bromodomain-containing protein 4 inhibitors in innate inflammation-driven airway remodeling. Am. J. Respir. Cell Mol. Biol. 2019;60:68–83. doi: 10.1165/rcmb.2017-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian B, et al. Selective antagonists of the bronchiolar epithelial NF-kappaB-bromodomain-containing protein 4 pathway in viral-induced airway inflammation. Cell Rep. 2018;23:1138–1151. doi: 10.1016/j.celrep.2018.03.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, et al. Pharmacoproteomics reveal novel protective activity of bromodomain containing 4 inhibitors on vascular homeostasis in TLR3-mediated airway remodeling. J. Proteom. 2019;205:103415. doi: 10.1016/j.jprot.2019.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian B, et al. Central role of the NF-kappaB pathway in the Scgb1a1-expressing epithelium in mediating respiratory syncytial virus-induced airway inflammation. J. Virol. 2018;92:e00441–18. doi: 10.1128/JVI.00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malik N, et al. Suppression of interferon beta gene transcription by inhibitors of bromodomain and extra-terminal (BET) family members. Biochem. J. 2015;468:363–372. doi: 10.1042/BJ20141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kleppe M, et al. Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell. 2018;33:785–787. doi: 10.1016/j.ccell.2018.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mumby S, et al. Bromodomain and extra-terminal protein mimic JQ1 decreases inflammation in human vascular endothelial cells: Implications for pulmonary arterial hypertension. Respirology. 2017;22:157–164. doi: 10.1111/resp.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown JD, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zou Z, et al. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene. 2014;33:2395–2404. doi: 10.1038/onc.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao J, et al. B7-H3 is regulated by BRD4 and promotes TLR4 expression in pancreatic ductal adenocarcinoma. Int. J. Biochem Cell Biol. 2019;108:84–91. doi: 10.1016/j.biocel.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 66.Sun Y, Huang J, Song K. BET protein inhibition mitigates acute myocardial infarction damage in rats via the TLR4/TRAF6/NF-kappaB pathway. Exp. Ther. Med. 2015;10:2319–2324. doi: 10.3892/etm.2015.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng S, et al. BET inhibitor JQ1 blocks inflammation and bone destruction. J. Dent. Res. 2014;93:657–662. doi: 10.1177/0022034514534261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takeuchi O, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 69.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abe T, et al. Involvement of the Toll-like receptor 9 signaling pathway in the induction of innate immunity by baculovirus. J. Virol. 2005;79:2847–2858. doi: 10.1128/JVI.79.5.2847-2858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petes C, Odoardi N, Gee K. The toll for trafficking: toll-Like receptor 7 delivery to the endosome. Front. Immunol. 2017;8:1075. doi: 10.3389/fimmu.2017.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saxena M, Yeretssian G. NOD-like receptors: master regulators of inflammation and cancer. Front. Immunol. 2014;5:327. doi: 10.3389/fimmu.2014.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 74.Jiang F, et al. Discovery of benzo[cd]indol-2(1H)-ones and pyrrolo[4,3,2-de]quinolin-2(1H)-ones as bromodomain and extra-terminal domain (BET) inhibitors with selectivity for the first bromodomain with potential high efficiency against acute gouty arthritis. J. Med. Chem. 2019;62:11080–11107. doi: 10.1021/acs.jmedchem.9b01010. [DOI] [PubMed] [Google Scholar]
- 75.Zhou Y, Gu Y, Liu J. BRD4 suppression alleviates cerebral ischemia-induced brain injury by blocking glial activation via the inhibition of inflammatory response and pyroptosis. Biochem. Biophys. Res. Commun. 2019;519:481–488. doi: 10.1016/j.bbrc.2019.07.097. [DOI] [PubMed] [Google Scholar]
- 76.Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Mol. Cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi H, Wu J, Chen ZJ, Chen C. Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proc. Natl Acad. Sci. USA. 2015;112:8947–8952. doi: 10.1073/pnas.1507317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, et al. TMEM173 drives lethal coagulation in sepsis. Cell Host Microbe. 2020;27:556–570. doi: 10.1016/j.chom.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen LF, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 83.Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015;115:2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giridharan S, Srinivasan M. Mechanisms of NF-kappaB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018;11:407–419. doi: 10.2147/JIR.S140188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J. Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gallagher SJ, et al. Control of NF-kB activity in human melanoma by bromodomain and extra-terminal protein inhibitor I-BET151. Pigment Cell Melanoma Res. 2014;27:1126–1137. doi: 10.1111/pcmr.12282. [DOI] [PubMed] [Google Scholar]
- 88.Liu M, et al. A novel target to reduce microglial inflammation and neuronal damage after deep hypothermic circulatory arrest. J. Thorac. Cardiovasc Surg. 2019;159:2431–2444. doi: 10.1016/j.jtcvs.2019.06.115. [DOI] [PubMed] [Google Scholar]
- 89.Sun R, et al. Bromodomain-containing protein 2 induces insulin resistance via the mTOR/Akt signaling pathway and an inflammatory response in adipose tissue. Cell Signal. 2017;30:92–103. doi: 10.1016/j.cellsig.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 90.Khan YM, Kirkham P, Barnes PJ, Adcock IM. Brd4 is essential for IL-1beta-induced inflammation in human airway epithelial cells. PLoS ONE. 2014;9:e95051. doi: 10.1371/journal.pone.0095051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, et al. BRD4 inhibition attenuates inflammatory response in microglia and facilitates recovery after spinal cord injury in rats. J. Cell Mol. Med. 2019;23:3214–3223. doi: 10.1111/jcmm.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brasier AR, et al. RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J. Virol. 2011;85:11752–11769. doi: 10.1128/JVI.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tian B, et al. BRD4 mediates NF-kappaB-dependent epithelial-mesenchymal transition and pulmonary fibrosis via transcriptional elongation. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;311:L1183–L1201. doi: 10.1152/ajplung.00224.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wienerroither S, et al. Regulation of NO synthesis, local inflammation, and innate immunity to pathogens by BET family proteins. Mol. Cell Biol. 2014;34:415–427. doi: 10.1128/MCB.01353-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suarez-Alvarez B, et al. Inhibition of bromodomain and extraterminal domain family proteins ameliorates experimental renal damage. J. Am. Soc. Nephrol. 2017;28:504–519. doi: 10.1681/ASN.2015080910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen J, et al. BET inhibition attenuates helicobacter pylori-induced inflammatory response by suppressing inflammatory gene transcription and enhancer activation. J. Immunol. 2016;196:4132–4142. doi: 10.4049/jimmunol.1502261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bao Y, et al. Brd4 modulates the innate immune response through Mnk2-eIF4E pathway-dependent translational control of IkappaBalpha. Proc. Natl Acad. Sci. USA. 2017;114:E3993–E4001. doi: 10.1073/pnas.1700109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiao Y, et al. Bromodomain and extra-terminal domain bromodomain inhibition prevents synovial inflammation via blocking IkappaB kinase-dependent NF-kappaB activation in rheumatoid fibroblast-like synoviocytes. Rheumatology. 2016;55:173–184. doi: 10.1093/rheumatology/kev312. [DOI] [PubMed] [Google Scholar]
- 99.Calo V, et al. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 100.Middleton SA, et al. BET Inhibition Improves NASH and Liver Fibrosis. Sci. Rep. 2018;8:17257. doi: 10.1038/s41598-018-35653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duan Q, et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci. Transl. Med. 2017;9:eaah5084. doi: 10.1126/scitranslmed.aah5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leal AS, et al. Bromodomain inhibitors, JQ1 and I-BET 762, as potential therapies for pancreatic cancer. Cancer Lett. 2017;394:76–87. doi: 10.1016/j.canlet.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 103.Zhang D, et al. Chemoprevention of preclinical breast and lung cancer with the bromodomain inhibitor I-BET 762. Cancer Prev. Res (Philos.) 2018;11:143–156. doi: 10.1158/1940-6207.CAPR-17-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheung KL, et al. Distinct roles of Brd2 and Brd4 in potentiating the transcriptional program for Th17 cell differentiation. Mol. Cell. 2017;65:1068–1080. doi: 10.1016/j.molcel.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fei X, Wang A, Meng X, Wang Z. Reshaping acetylated peptide selectivity between human BET Brd2 bromodomains BD-I and BD-II in glioblastoma by rationally grafting secondary anchor residues. Gen. Physiol. Biophys. 2018;37:411–419. doi: 10.4149/gpb_2017063. [DOI] [PubMed] [Google Scholar]
- 106.Ray S, et al. Inducible STAT3 NH2 terminal mono-ubiquitination promotes BRD4 complex formation to regulate apoptosis. Cell Signal. 2014;26:1445–1455. doi: 10.1016/j.cellsig.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Toniolo PA, et al. Inhibiting STAT5 by the BET bromodomain inhibitor JQ1 disrupts human dendritic cell maturation. J. Immunol. 2015;194:3180–3190. doi: 10.4049/jimmunol.1401635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pinz S, et al. Deacetylase inhibitors repress STAT5-mediated transcription by interfering with bromodomain and extra-terminal (BET) protein function. Nucleic Acids Res. 2015;43:3524–3545. doi: 10.1093/nar/gkv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van der Kroef M, et al. Histone modifications underlie monocyte dysregulation in patients with systemic sclerosis, underlining the treatment potential of epigenetic targeting. Ann. Rheum. Dis. 2019;78:529–538. doi: 10.1136/annrheumdis-2018-214295. [DOI] [PubMed] [Google Scholar]
- 110.Tian B, et al. BRD4 Couples NF-kappaB/RelA with Airway Inflammation and the IRF-RIG-I Amplification Loop in Respiratory Syncytial Virus Infection. J. Virol. 2017;91:e00007–e00017. doi: 10.1128/JVI.00007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 112.Ghari F, et al. Citrullination-acetylation interplay guides E2F-1 activity during the inflammatory response. Sci. Adv. 2016;2:e1501257. doi: 10.1126/sciadv.1501257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Handoko L, et al. JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states. Epigenetics. 2018;13:410–431. doi: 10.1080/15592294.2018.1469891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tian Y, et al. JQ1-loaded polydopamine nanoplatform inhibits c-MYC/programmed cell death ligand 1 to enhance photothermal therapy for triple-negative breast cancer. ACS Appl. Mater. Interfaces. 2019;11:46626–46636. doi: 10.1021/acsami.9b18730. [DOI] [PubMed] [Google Scholar]
- 116.Veneziani I, et al. The BET-bromodomain inhibitor JQ1 renders neuroblastoma cells more resistant to NK cell-mediated recognition and killing by downregulating ligands for NKG2D and DNAM-1 receptors. Oncotarget. 2019;10:2151–2160. doi: 10.18632/oncotarget.26736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Y, Liu M, Chen LF, Chen R. P-TEFb: finding its ways to release promoter-proximally paused RNA polymerase II. Transcription. 2018;9:88–94. doi: 10.1080/21541264.2017.1281864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dai J, et al. Recruitment of Brd3 and Brd4 to acetylated chromatin is essential for proinflammatory cytokine-induced matrix-degrading enzyme expression. J. Orthop. Surg. Res. 2019;14:59. doi: 10.1186/s13018-019-1091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bowman EA, Kelly WG. RNA polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation: a tail of two kinases. Nucleus. 2014;5:224–236. doi: 10.4161/nucl.29347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prelich G. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell. 2002;1:153–162. doi: 10.1128/EC.1.2.153-162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoshida H, et al. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc. Natl Acad. Sci. USA. 2015;112:E4448–E4457. doi: 10.1073/pnas.1512081112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Marino-Ramirez L, Kann MG, Shoemaker BA, Landsman D. Histone structure and nucleosome stability. Expert Rev. Proteom. 2005;2:719–729. doi: 10.1586/14789450.2.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tang X, et al. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am. J. Pathol. 2013;183:470–479. doi: 10.1016/j.ajpath.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 125.Clifford RL, et al. CXCL8 histone H3 acetylation is dysfunctional in airway smooth muscle in asthma: regulation by BET. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;308:L962–L972. doi: 10.1152/ajplung.00021.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tian CQ, et al. Inhibition of the BET family reduces its new target gene IDO1 expression and the production of L-kynurenine. Cell Death Dis. 2019;10:557. doi: 10.1038/s41419-019-1793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Felisbino MB, McKinsey TA. Epigenetics in cardiac fibrosis: emphasis on inflammation and fibroblast activation. JACC Basic Transl. Sci. 2018;3:704–715. doi: 10.1016/j.jacbts.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang C, et al. Epigenetic blockade of neoplastic transformation by bromodomain and extra-terminal (BET) domain protein inhibitor JQ-1. Biochem. Pharm. 2016;117:35–45. doi: 10.1016/j.bcp.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dominguez-Andres J, et al. Bromodomain inhibitor I-BET151 suppresses immune responses during fungal-immune interaction. Eur. J. Immunol. 2019;49:2044–2050. doi: 10.1002/eji.201848081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gilan O, et al. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science. 2020;368:387–394. doi: 10.1126/science.aaz8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.DeMars KM, Yang C, Castro-Rivera CI, Candelario-Jalil E. Selective degradation of BET proteins with dBET1, a proteolysis-targeting chimera, potently reduces pro-inflammatory responses in lipopolysaccharide-activated microglia. Biochem. Biophys. Res. Commun. 2018;497:410–415. doi: 10.1016/j.bbrc.2018.02.096. [DOI] [PubMed] [Google Scholar]
- 132.Tsujikawa LM, et al. Apabetalone (RVX-208) reduces vascular inflammation in vitro and in CVD patients by a BET-dependent epigenetic mechanism. Clin. Epigenetics. 2019;11:102. doi: 10.1186/s13148-019-0696-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Abruzzese MP, et al. Inhibition of bromodomain and extra-terminal (BET) proteins increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells: role of cMYC-IRF4-miR-125b interplay. J. Hematol. Oncol. 2016;9:134. doi: 10.1186/s13045-016-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Banik StevenM, et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature. 2020;584:291–297. doi: 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Klein IA, et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qian Z, Shuying W, Ranran D. Inhibitory effects of JQ1 on listeria monocytogenes-induced acute liver injury by blocking BRD4/RIPK1 axis. Biomed. Pharmacother. 2020;125:109818. doi: 10.1016/j.biopha.2020.109818. [DOI] [PubMed] [Google Scholar]
- 137.Archin NM, Margolis DM. Emerging strategies to deplete the HIV reservoir. Curr. Opin. Infect. Dis. 2014;27:29–35. doi: 10.1097/QCO.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mietton F, et al. Selective BET bromodomain inhibition as an antifungal therapeutic strategy. Nat. Commun. 2017;8:15482. doi: 10.1038/ncomms15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 140.Lau AO, Sacci JB, Jr, Azad AF. Host responses to Plasmodium yoelii hepatic stages: a paradigm in host-parasite interaction. J. Immunol. 2001;166:1945–1950. doi: 10.4049/jimmunol.166.3.1945. [DOI] [PubMed] [Google Scholar]
- 141.Wang X, et al. JQ1, a bromodomain inhibitor, suppresses Th17 effectors by blocking p300-mediated acetylation of RORgammat. Br. J. Pharm. 2020;177:2959–2973. doi: 10.1111/bph.15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gong T, Liu L, Jiang W, Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020;20:95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 143.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou S, et al. BET protein inhibitor JQ1 downregulates chromatin accessibility and suppresses metastasis of gastric cancer via inactivating RUNX2/NID1 signaling. Oncogenesis. 2020;9:33. doi: 10.1038/s41389-020-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Puissant A, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Joshi S, et al. SF2523: dual PI3K/BRD4 inhibitor blocks tumor immunosuppression and promotes adaptive immune responses in cancer. Mol. Cancer Ther. 2019;18:1036–1044. doi: 10.1158/1535-7163.MCT-18-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Andrieu G, Tran AH, Strissel KJ, Denis GV. BRD4 regulates breast cancer dissemination through Jagged1/Notch1 signaling. Cancer Res. 2016;76:6555–6567. doi: 10.1158/0008-5472.CAN-16-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nishida J, et al. Epigenetic remodelling shapes inflammatory renal cancer and neutrophil-dependent metastasis. Nat. Cell Biol. 2020;22:465–475. doi: 10.1038/s41556-020-0491-2. [DOI] [PubMed] [Google Scholar]
- 149.Lee M, et al. Tumoral BRD4 expression in lymph node-negative breast cancer: association with T-bet+ tumor-infiltrating lymphocytes and disease-free survival. BMC Cancer. 2018;18:750. doi: 10.1186/s12885-018-4653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tasdemir N, et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016;6:612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang T, et al. A cancer vaccine-mediated postoperative immunotherapy for recurrent and metastatic tumors. Nat. Commun. 2018;9:1532. doi: 10.1038/s41467-018-03915-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mao W, et al. Immunogenicity of prostate cancer is augmented by BET bromodomain inhibition. J. Immunother. Cancer. 2019;7:277. doi: 10.1186/s40425-019-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fan P, et al. Overexpressed histone acetyltransferase 1 regulates cancer immunity by increasing programmed death-ligand 1 expression in pancreatic cancer. J. Exp. Clin. Cancer Res. 2019;38:47. doi: 10.1186/s13046-019-1044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ebine K, et al. Interplay between interferon regulatory factor 1 and BRD4 in the regulation of PD-L1 in pancreatic stellate cells. Sci. Rep. 2018;8:13225. doi: 10.1038/s41598-018-31658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Qiao H, et al. Tumor localization of oncolytic adenovirus assisted by pH-degradable microgels with JQ1-mediated boosting replication and PD-L1 suppression for enhanced cancer therapy. Biomater. Sci. 2020;8:2472–2480. doi: 10.1039/D0BM00172D. [DOI] [PubMed] [Google Scholar]
- 156.Liu M, et al. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut. 2020;69:365–379. doi: 10.1136/gutjnl-2018-317257. [DOI] [PubMed] [Google Scholar]
- 157.Frangogiannis NG. Regulation of the inflammatory response in cardiac repair. Circ. Res. 2012;110:159–173. doi: 10.1161/CIRCRESAHA.111.243162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhu W, et al. BRD4 blockage alleviates pathological cardiac hypertrophy through the suppression of fibrosis and inflammation via reducing ROS generation. Biomed. Pharmacother. 2020;121:109368. doi: 10.1016/j.biopha.2019.109368. [DOI] [PubMed] [Google Scholar]
- 159.Van der Feen DE, et al. Multicenter preclinical validation of BET inhibition for the treatment of pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2019;200:910–920. doi: 10.1164/rccm.201812-2275OC. [DOI] [PubMed] [Google Scholar]
- 160.Meloche J, et al. Implication of inflammation and epigenetic readers in coronary artery remodeling in patients with pulmonary arterial hypertension. Arterioscler. Thromb. Vasc. Biol. 2017;37:1513–1523. doi: 10.1161/ATVBAHA.117.309156. [DOI] [PubMed] [Google Scholar]
- 161.Wang H, et al. BRD4 contributes to LPS-induced macrophage senescence and promotes progression of atherosclerosis-associated lipid uptake. Aging (Albany NY) 2020;12:9240–9259. doi: 10.18632/aging.103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ray KK, et al. Effect of selective BET protein inhibitor apabetalone on cardiovascular outcomes in patients with acute coronary syndrome and diabetes: Rationale, design, and baseline characteristics of the BETonMACE trial. Am. Heart J. 2019;217:72–83. doi: 10.1016/j.ahj.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 163.Tian B, et al. Mucosal bromodomain-containing protein 4 mediates aeroallergen-induced inflammation and remodeling. J. Allergy Clin. Immunol. 2019;143:1380–1394. doi: 10.1016/j.jaci.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tang X, et al. BET bromodomain proteins mediate downstream signaling events following growth factor stimulation in human lung fibroblasts and are involved in bleomycin-induced pulmonary fibrosis. Mol. Pharm. 2013;83:283–293. doi: 10.1124/mol.112.081661. [DOI] [PubMed] [Google Scholar]
- 165.Magistri M, et al. The BET-bromodomain inhibitor JQ1 reduces inflammation and Tau phosphorylation at Ser396 in the brain of the 3xTg model of Alzheimer’s disease. Curr. Alzheimer Res. 2016;13:985–995. doi: 10.2174/1567205013666160427101832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sanchez-Ventura J, Amo-Aparicio J, Navarro X, Penas C. BET protein inhibition regulates cytokine production and promotes neuroprotection after spinal cord injury. J. Neuroinflammation. 2019;16:124. doi: 10.1186/s12974-019-1511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang G, et al. Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J. Biol. Chem. 2012;287:28840–28851. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wei S, Sun Y, Sha H. Therapeutic targeting of BET protein BRD4 delays murine lupus. Int. Immunopharmacol. 2015;29:314–319. doi: 10.1016/j.intimp.2015.10.036. [DOI] [PubMed] [Google Scholar]
- 169.Oh SK, et al. RORalpha is crucial for attenuated inflammatory response to maintain intestinal homeostasis. Proc. Natl Acad. Sci. USA. 2019;116:21140–21149. doi: 10.1073/pnas.1907595116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Huang W, et al. Selective inhibition of BET proteins reduces pancreatic damage and systemic inflammation in bile acid- and fatty acid ethyl ester- but not caerulein-induced acute pancreatitis. Pancreatology. 2017;17:689–697. doi: 10.1016/j.pan.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 171.Fu W, et al. Epigenetic modulation of type-1 diabetes via a dual effect on pancreatic macrophages and beta cells. Elife. 2014;3:e04631. doi: 10.7554/eLife.04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Deeney JT, Belkina AC, Shirihai OS, Corkey BE, Denis GV. BET bromodomain proteins Brd2, Brd3 and Brd4 selectively regulate metabolic pathways in the pancreatic beta-cell. PLoS ONE. 2016;11:e0151329. doi: 10.1371/journal.pone.0151329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sharrock J, et al. fs(1)h controls metabolic and immune function and enhances survival via AKT and FOXO in Drosophila. Dis. Model Mech. 2019;12:dmm037259. doi: 10.1242/dmm.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Segatto M, et al. Epigenetic targeting of bromodomain protein BRD4 counteracts cancer cachexia and prolongs survival. Nat. Commun. 2017;8:1707. doi: 10.1038/s41467-017-01645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang J, et al. BRD4 inhibition regulates MAPK, NF-kappaB signals, and autophagy to suppress MMP-13 expression in diabetic intervertebral disc degeneration. FASEB J. 2019;33:11555–11566. doi: 10.1096/fj.201900703R. [DOI] [PubMed] [Google Scholar]
- 176.Jiang Y, et al. BRD4 has dual effects on the HMGB1 and NF-kappaB signalling pathways and is a potential therapeutic target for osteoarthritis. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:3001–3015. doi: 10.1016/j.bbadis.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 177.Zhang QG, Qian J, Zhu YC. Targeting bromodomain-containing protein 4 (BRD4) benefits rheumatoid arthritis. Immunol. Lett. 2015;166:103–108. doi: 10.1016/j.imlet.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 178.Eskandarpour M, Alexander R, Adamson P, Calder VL. Pharmacological inhibition of bromodomain proteins suppresses retinal inflammatory disease and downregulates retinal Th17 cells. J. Immunol. 2017;198:1093–1103. doi: 10.4049/jimmunol.1600735. [DOI] [PubMed] [Google Scholar]
- 179.Hytti M, et al. Inhibition of BET bromodomains alleviates inflammation in human RPE cells. Biochem. Pharm. 2016;110-111:71–79. doi: 10.1016/j.bcp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 180.Nadeem A, et al. Imiquimod-induced psoriasis-like skin inflammation is suppressed by BET bromodomain inhibitor in mice through RORC/IL-17A pathway modulation. Pharm. Res. 2015;99:248–257. doi: 10.1016/j.phrs.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 181.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 182.Park JM, Jo SH, Kim MY, Kim TH, Ahn YH. Role of transcription factor acetylation in the regulation of metabolic homeostasis. Protein Cell. 2015;6:804–813. doi: 10.1007/s13238-015-0204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gibson BA, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sabari BR, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Daneshvar, K., et al. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat. Cell Biol. 10.1038/s41556-020-0572-2. (2020). [DOI] [PMC free article] [PubMed]
- 186.Chen K, et al. Antiinflammatory effects of bromodomain and extraterminal domain inhibition in cystic fibrosis lung inflammation. JCI Insight. 2016;1:e87168. doi: 10.1172/jci.insight.87168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu Z, et al. Discovery of orally bioavailable chromone derivatives as potent and selective BRD4 inhibitors: scaffold hopping, optimization, and pharmacological evaluation. J. Med Chem. 2020;63:5242–5256. doi: 10.1021/acs.jmedchem.0c00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gibbons HR, et al. Bromodomain inhibitor JQ1 reversibly blocks IFN-gamma production. Sci. Rep. 2019;9:10280. doi: 10.1038/s41598-019-46516-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhou F, et al. Oncolytic reactivation of KSHV as a therapeutic approach for primary effusion lymphoma. Mol. Cancer Ther. 2017;16:2627–2638. doi: 10.1158/1535-7163.MCT-17-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Das S, et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat. Commun. 2017;8:1467. doi: 10.1038/s41467-017-01629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nadeem A, et al. Inhibition of BET bromodomains restores corticosteroid responsiveness in a mixed granulocytic mouse model of asthma. Biochem. Pharm. 2018;154:222–233. doi: 10.1016/j.bcp.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 192.Perry MM, Durham AL, Austin PJ, Adcock IM, Chung KF. BET bromodomains regulate transforming growth factor-beta-induced proliferation and cytokine release in asthmatic airway smooth muscle. J. Biol. Chem. 2015;290:9111–9121. doi: 10.1074/jbc.M114.612671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Manni ML, et al. Bromodomain and extra-terminal protein inhibition attenuates neutrophil-dominant allergic airway disease. Sci. Rep. 2017;7:43139. doi: 10.1038/srep43139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jahagirdar R, et al. RVX-297, a BET bromodomain inhibitor, jas therapeutic effects in preclinical models of acute inflammation and autoimmune disease. Mol. Pharm. 2017;92:694–706. doi: 10.1124/mol.117.110379. [DOI] [PubMed] [Google Scholar]
- 195.Schilderink R, et al. BET bromodomain inhibition reduces maturation and enhances tolerogenic properties of human and mouse dendritic cells. Mol. Immunol. 2016;79:66–76. doi: 10.1016/j.molimm.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 196.Ren Y, et al. Role of Brd4 in the production of inflammatory cytokines in mouse macrophages treated with titanium particles. Can. J. Physiol. Pharm. 2019;97:1028–1034. doi: 10.1139/cjpp-2019-0142. [DOI] [PubMed] [Google Scholar]
- 197.Chen TH, Weber FE, Malina-Altzinger J, Ghayor C. Epigenetic drugs as new therapy for tumor necrosis factor-alpha-compromised bone healing. Bone. 2019;127:49–58. doi: 10.1016/j.bone.2019.05.035. [DOI] [PubMed] [Google Scholar]
- 198.Slivka PF, et al. Small molecule and pooled CRISPR screens investigating IL17 signaling identify BRD2 as a novel contributor to keratinocyte inflammatory responses. ACS Chem. Biol. 2019;14:857–872. doi: 10.1021/acschembio.8b00260. [DOI] [PubMed] [Google Scholar]