Abstract
Autophagy is a cellular degradation system contributing to homeostasis of tissue stem cells including haematopoietic stem cells (HSCs). It plays pleiotropic roles in HSC characteristics throughout life, but its stage-specific roles in HSC self-renewal are unclear. To investigate the effects of Atg5 deletion on stage-specific HSC functions, we compared the repopulating capacity of HSCs in Atg5f/f;Vavi-cre mice from postnatal day (P) 0–7 weeks of age. Interestingly, Atg5 deficiency led to no remarkable abnormality in the HSC self-renewal capacity at P0, but significant defects at P7, followed by severe defects. Induction of Atg5 deletion at P5 by tamoxifen administration to Atg5f/f;Rosa26-Cre-ERT2 mice resulted in normal haematopoiesis, including the HSC population, until around 1 year, suggesting that Atg5 in the early neonatal period was critical for haematopoiesis in adults. Mitochondrial oxidative stress was increased by Atg5 loss in neonatal HSC/progenitor cells. Although p62 had accumulated in immature bone marrow cells of Atg5f/f;Vavi-cre mice, p62 deletion did not restore defective HSC functions, indicating that Atg5-dependent haematopoietic regulation in the developmental period was independent of p62. This study proposes a critical role of autophagy in HSC protection against harsh environments in the early neonatal stage, which is essential for healthy long-term haematopoiesis.
Subject terms: Haematopoiesis, Haematopoietic stem cells, Autophagy
Introduction
Autophagy is a conserved catabolic system that degrades cellular components engulfed in autophagosomes within lysosomes. This system is constitutively active to maintain cellular functions, but it is also induced in response to stress condition such as oxidative stress, infection, and starvation. Induced autophagy is a prosurvival mechanism to maintain cellular homeostasis by eliminating unwanted components such as damaged organelles, protein aggregates, and intracellular bacteria, and supplying an energy source. Autophagy consists of serial steps and different autophagy-related genes (Atgs) are involved in the process. It starts with emersion and expansion of an isolation membrane/phagophore. The phagophore encloses cytoplasmic components to form double membrane vesicles called autophagosomes. The autophagosomes then fuse with lysosomes to become autolysosomes in which degradation of cargo by lysosomal enzymes occurs. In mammals, for the initial step, the Unc-51 like autophagy activating kinase (ULK) complex consisting of ULK1 or ULK2, Atg13, FIP200, and Atg101 is recruited to sites near the endoplasmic reticulum (ER) and forms an assembly. Atg9-positive vesicles derived from the trans-Golgi network and plasma membrane localize to the ULK complex assembly and contribute to the formation of autophagosome precursors1. Phosphatidylinositol 3-kinase (PI3K) complex I consisting of Beclin1, Vps34, p150, Nrbf2, and Atg14L produces phosphatidylinositol 3-phosphate (PtdIns3P) in autophagosome-biogenesis-related membranes (i.e. autophagosome precursor, isolation membrane, and omegasome) to recruit multiple PtdIns3P-binding proteins that regulate autophagy formation1. The Atg12–Atg5–Atg16L1 complex, which is formed through a ubiquitin-like conjugation system involving Atg7 and Atg10, is recruited to the autophagosome precursor/isolation membrane via PtdIns3P-binding protein WIPI2b1. The Atg12–Atg5–Atg16L1 complex promotes conjugation of phosphatidylethanolamine to LC3 (Atg8 homolog in mammals), another ubiquitin-like conjugation system involving Atg4, Atg7, and Atg31,2. The lipidation of LC3 has multiple roles in autophagy. For example, it is required for expansion of the isolation membrane and fusion of autophagosomes with lysosomes1,3. These two ubiquitin-like systems are crucial for successful autophagy.
Mammalian target of rapamycin (mTOR), a negative regulator of the autophagy pathway, which inactivates ULK1 by phosphorylation, is stimulated by insulin, growth factors, and amino acids. Lack of insulin or growth factors induces autophagy via the PI3K/Akt/mTOR axis, and a lack of amino acid induces it via regulation of Rag GTPase4, a component of mTOR complex 1 (mTORC1). mTOR also senses glucose starvation via the AMPK/mTOR axis to induce autophagy4. Generated amino acids are reused for protein and fatty acid biosynthesis, gluconeogenesis, and ATP production. Under infected conditions, attachment of bacteria to host cells5, bacterial pathogen-associated molecular patterns (PAMPs; e.g. lipopolysaccharide), and damage-associated molecular patterns (DAMPs; e.g. disturbed endosomal membrane)6, and bacterial nucleic acids (e.g. double-stranded DNA)7 trigger recruitment of autophagy machinery and rapid elimination of intracellular bacteria. Dysregulated autophagy is associated with pathogenesis in several tissues, typically neurons. In Alzheimer’s disease (AD), accumulation of amyloid beta peptide (Aβ) is thought to primarily trigger the pathogenesis. Growing evidence suggests that autophagy is a major pathway for degradation of Aβ, and dysfunction of autophagy contributes to the pathogenesis of AD8. Thus, in many situations, autophagy has important roles to protect cells from stress.
Although autophagy had been originally identified as a bulk degradation pathway, selective autophagic degradation has been identified. p62, also known as SQSTM1 (hereafter referred to as p62), is a major autophagic receptor that interacts with ubiquitinated substrates via the ubiquitin association domain and autophagosomes via the LC3 interaction region to intermediate selective engulfment of specific cargo into autophagosomes. p62 functions not only as an autophagic receptor, but also in the stress response. p62 activates nuclear factor erythroid 2-related factor (Nrf2) by competitive binding to Kelch-like ECH-associated protein (Keap) 1, promoting tumourigenesis in the liver9. Phosphorylation of S351 in p62 is observed in autophagic cargo, which increases affinity of p62 for Keap1. These findings suggest that selective autophagy and the Keap1-Nrf2 pathway are interdependent via p629. In addition to mediating selective degradation of cargo, p62 itself is degraded by autophagy. Therefore, p62 accumulates under autophagy inhibitory conditions and in autophagy-deficient tissues. p62 accumulates in Atg7-deficient hepatocytes, and p62 ablation suppresses liver injury and tumour development caused by autophagy deficiency10–12. However, accumulated p62 is not related to defective autophagy-deficient neurons10. The relationship of the phenotype caused by autophagy deficiency and p62 accumulation remains unclear.
Atg3, Atg5, Atg7, Atg12, and Atg16L1 are involved in the Atg12 conjugation system, and mice deficient for these genes (Atg3−⁄−, Atg5−⁄−, Atg7−⁄−, Atg12−⁄−, and Atg16l1−⁄−) are neonatal lethal and die within 1 day after birth13–17. Atg5-null mice (Atg5−⁄−), in which Atg5 is deficient in the whole body, fail to suckle milk and die within 1 day after birth18. Introduction of exogenous Atg5 to neurons in Atg5-null mice (Atg5−⁄−;Eno2-Atg5) recovers the suckling defect and neonatal lethality, suggesting that neuronal dysfunction triggers severe defects in Atg5-null mice. However, Atg5−⁄−;Eno2-Atg5 mice, in which Atg5 gene was restored only in neuron, start to die at around 2 months with abnormalities in multiple organs, indicating that autophagic activity is essential for development of tissues other than neuronal tissues. Additionally, because the amino acid level in Atg5-null neonates is lower and they died earlier than non-milk-fed wildtype neonates, metabolic and suckling defects trigger early neonatal lethality. Thus, it is still unclear how autophagy affects developmental processes.
To identify the role of autophagy in blood cells, various types of conditional Atg genes knockout mice have been used because of lethality of conventional Atg genes knockout mice13–17. It has been demonstrated that autophagy plays critical roles in the development and differentiation of various types of blood cells, including T, B, and erythroid cells19–28. Haematopoietic cell-specific Atg5 and Atg7 deletion in the foetal period using the Vav-Cre-loxP system triggers weight loss, severe anaemia, and reduced haematopoietic stem cells (HSCs) of mice and results in death27–31. In these mice, a mitochondrial status alteration has been observed30,31. In adult HSCs, for example, autophagy is induced rapidly upon starvation via the FoxO3a-driven gene expression program, which maintains the functionality of HSCs32. Furthermore, it has been suggested that autophagy degrades active healthy mitochondria and maintains low metabolic state and quiescence of HSCs33. However, knockout mice in which Atg5 and Atg12 genes re-deleted at juvenile to adult ages using the Mx1-Cre-loxP system are largely healthy, but show some phenotypes resembling aged mice, such as increased cellularity and a skewed ratio of myeloid versus lymphoid cells in peripheral blood (PB), indicating critical roles of autophagy in blood aging33.
In this study, we investigated the role of autophagy in HSC functions during developmental stages after birth using Atg5-deficient mice. Previous reports of Atg5f/f;Vav-Cre (Atg5f/f;Vav) or Atg7f/f;Vav-Cre mice have shown progressive defective haematopoiesis during developmental stages (6–9 weeks). Because the abnormalities of HSCs appeared to be more severe as the mice grew, it was assumed that autophagy is important for development of HSCs in later stages. However, the role of autophagy at early time points, such as the neonatal period, has not been clarified. In this study, by comparison of the regeneration capacity of HSCs among different timings after birth, we evaluated stage-specific effects of Atg5 deficiency on HSC functions. Additionally, because accumulation of p62 is observed in autophagy-deficient tissues10–12, we investigated the role of p62 in the response to oxidative stress induced by Atg5 deficiency in haematopoiesis by analysis of Atg5f/f;p62f/f;Vav-Cre (Atg5f/f;p62f/f;Vav) mice. As a result, we demonstrated that autophagy plays a critical role in protecting neonatal HSCs from oxidative stress in a p62-independent manner.
Results
Progressive haematopoietic failure by Atg5 deletion during developmental period
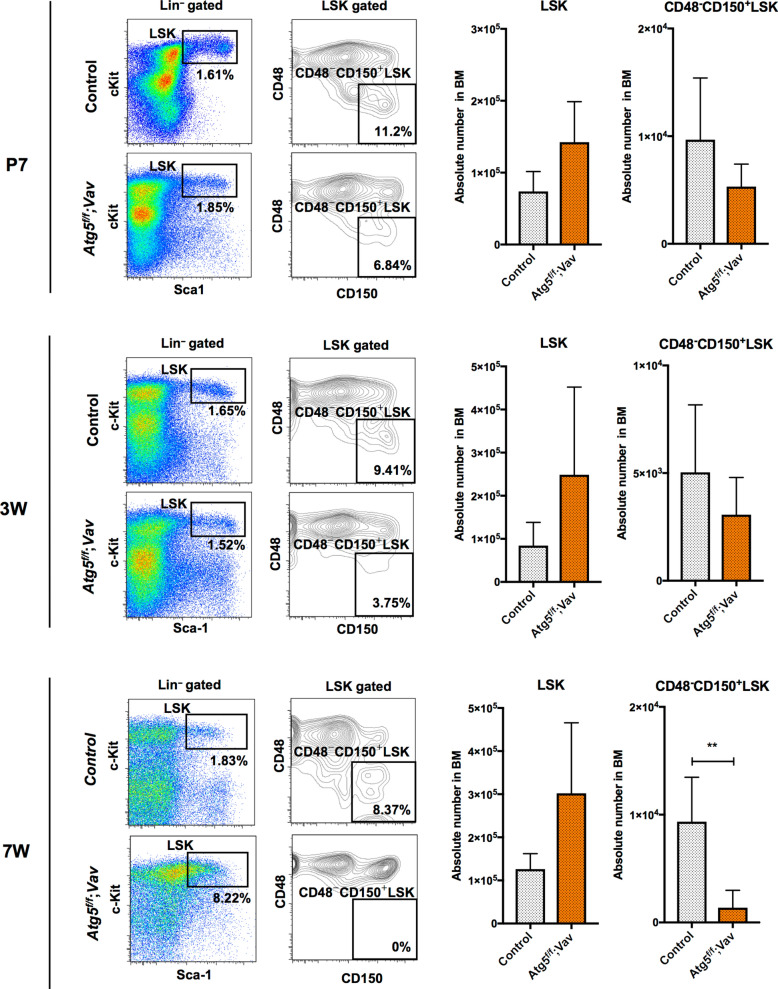
To investigate the roles of autophagy in development of haematopoiesis in mice, we analysed the effects of Atg5 deletion on haematopoiesis using the Vav-Cre-loxP system, in which Cre recombinase-mediated loxP-flanked gene deletion in haematopoietic tissue starts from around embryonic day (E) 11.534. The Atg5 deletion efficiency was sufficiently high using this system (Supplementary Figure S1). Consistent with previous reports29,30, Atg5f⁄f;Vav mice showed severe haematopoietic failure at 7 weeks of age, and most Atg5-deficient mice died presumably because of further progression of haematopoietic failure (Supplementary Fig. S2). To evaluate the effect of Atg5 on HSC development, we investigated the long-term HSC population (SLAM LSK; CD48−CD150+lineage−Sca-1+c-Kit+) in P7, 3-week-old and 7-week-old mice. In bone marrow (BM) of 7-week-old mice, SLAM LSK cells were reduced dramatically by loss of the Atg5 gene (Fig. 1) as shown previously29,30. This abnormality was not remarkable at earlier stages (Fig. 1). Therefore, as suggested in the previous reports, these data indicate that autophagy may play more critical roles in the later period rather than earlier developmental stages, such as the neonatal period.
Figure 1.
HSC population in control (Atg5f/w;Vav) and Atg5f/f;Vav mice at P7 (top), 3 weeks (middle), and 7 weeks (bottom). Left panel shows a representative flow cytometric plot of LSK (Lin−c-Kit+Sca1+) cells and CD48−CD150+LSK cells in at least three independent experiments. Right panel shows absolute numbers of LSK and CD48−CD150+LSK cells. Data are the mean ± SD (n = 3–6).
Defect in the self-renewal capacity of HSCs among BM cells of Atg5f/f;Vav mice at P7, but not P0
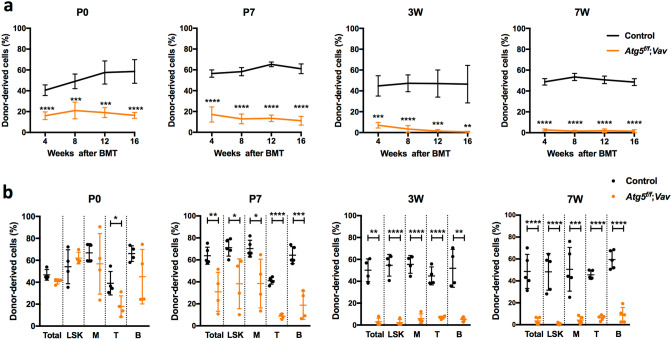
To evaluate the effect of Atg5 deficiency on HSC functions, we performed competitive reconstitution assays using BM cells of Atg5f/f;Vav mice from P0 to 7 weeks of age. When we transplanted BM from P0 to 7-week-old mice, all lineages derived from Atg5-deficient mice were reduced significantly in PB, which reduced further with age (Fig. 2a, Supplementary Fig. S3). In BM, regeneration of Atg5-deficient BM cells at 16 weeks after transplantation was reduced dramatically in 3- and 7-week-old BM cell recipients, indicating that Atg5-deficient mice lost their self-renewal capacity at 3 weeks of age (Fig. 2b). However, interestingly, its repopulation in BM was almost comparable to the control, when we transplanted Atg5-deficient BM cells at P0 (Fig. 2b). These data suggest that although production of mature cells in PB may be impaired by Atg5, development of immature cells was unaffected at P0. Hence, we analysed donor-derived HSC/progenitor cells (Lineage−Sca1+c-Kit+; LSK cells) at 16 weeks after transplantation to assess repopulation of HSCs, that is, their self-renewal capacity. We found that the frequency of donor-derived LSK cells was comparable to the control in transplantation of Atg5-deficient BM cells at P0, indicating that the self-renewal capacity of Atg5-deficient cells was normal at P0 (Fig. 2b). In transplantation of BM cells from P7 mice, we found a mild but significant reduction in repopulation of Atg5-deficient LSK cells, as well as myeloid, B, and T cells, indicating a reduction in the self-renewal capacity of HSCs (Fig. 2b). Thus, HSC dysfunction due to Atg5 loss was detected at P7, but not P0, followed by progressive dysfunction of HSCs in later stages.
Figure 2.
Competitive reconstitution analysis of BM cells from control (Atg5f/w;Vav) and Atg5f/f;Vav mice at P0, P7, 3 weeks, and 7 weeks. (a) Frequency of donor-derived cells among total PB cells were analysed every 4–16 weeks after transplantation. Data are the mean ± SD (n = 4–5). (b) Frequency of donor-derived cells among total, LSK, myeloid (M), T (T), and B (B) cells in BM were analysed at 16 weeks after transplantation. Data are the mean ± SD (n = 4–5). Horizontal line indicates the mean of values.
No remarkable abnormality in long-term haematopoiesis, including HSC functions, by Atg5 deletion after P5
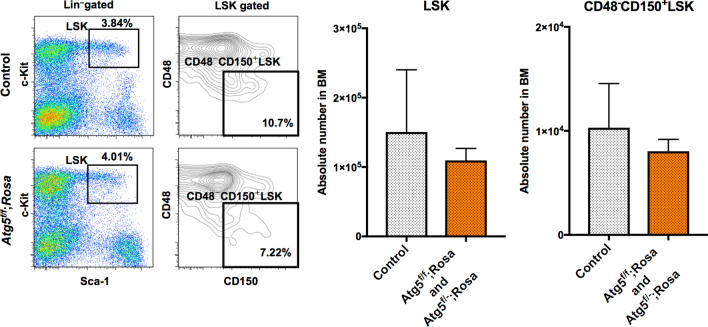
Next, we investigated the effect of Atg5 loss on haematopoiesis, when it was induced at an earlier time after birth. For this purpose, we used two systems, Atg5f/f;Mx1-Cre (Atg5f/f;Mx1), and Atg5f/f;Rosa26-Cre-ERT2 (Atg5f/f;Rosa) or Atg5f/−;Rosa26-Cre-ERT2 (Atg5f/−;Rosa), in which Atg5 gene deletion is induced by poly(I:C) and tamoxifen treatment, respectively. As the standard method, we started administration of poly(I:C) at 4 weeks of age and analysed haematopoiesis in adults. In our experiments, the Atg5 deficiency caused a mild but insignificant increase of LSK cells and LT-HSCs associated with increased myeloid cells and decreased B cells in PB, which is consistent with a previous report (Supplementary Fig. S4)33. These data indicated that deletion of Atg5 gene in later developmental periods did not remarkably affect the HSC population. To delete the Atg5 gene at the earliest timing after birth, we attempted to administrate tamoxifen at P5, followed by long-term observation of haematopoiesis. Although some mouse neonates died because of injury due to the intraperitoneal injection, most survived and appeared healthy. Analysis of PB showed normal haematopoiesis of Atg5-deficient mice. In some mice, we analysed HSCs and progenitors in BM after long-term observation (e.g., 8–12 months of age). The HSC and progenitor populations were comparable to the control (Fig. 3). To check the efficiency of gene deletion, we performed colony formation assays, followed by genotyping of individual colonies. We found that about 70% of colonies on average showed deletion of both alleles, indicating that the majority of LT-HSCs were deficient for Atg5 (Supplementary Fig. S5). Although there was the possibility that partial deletion of the Atg5 gene may fail to result in remarkable phenotypes, these data support that Atg5 was dispensable to develop and maintain HSCs when the Atg5 gene was deleted at around 1 week after birth.
Figure 3.
HSC population in control (Atg5f/w;Rosa and Atg5w/w;Rosa), and Atg5f/f;Rosa and Atg5f/−;Rosa mice at 8–12 months after tamoxifen administration at P5. Left panel shows a representative flow cytometric plot of LSK (Lin−c-Kit+Sca1+) cells and CD48−CD150+LSK cells in three independent experiments. Right panel shows absolute numbers of LSK and CD48−CD150+LSK cells. Data are the mean ± SD (n = 3).
Increased mitochondrial oxidative stress by Atg5 deletion is associated with cell death at the neonatal stage
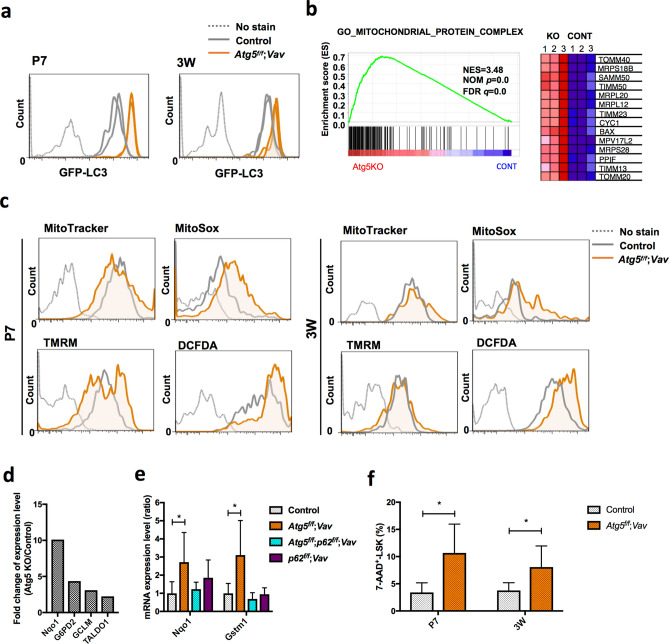
The above data prompted us to focus our analysis on HSCs at P7. First, to monitor autophagic activity in Atg5f/f;Vav and Control (Atg5f/w;Vav and Atg5f/w) mice, we used the GFP-LC3 system with FACS analysis35,36. If autophagy occurs, the GFP intensity decreases because it is digested and/or attenuated by the acidic condition in lysosomes in addition to LC3 protein, whereas GFP-LC3 accumulates upon autophagy inactivation. We compared the GFP fluorescence intensity in HSCs/progenitors (LSK) in GFP-LC3;Atg5f/f;Vav and GFP-LC3;Atg5f/w;Vav or GFP-LC3;Atg5f/w mice. We found that Atg5 deletion caused a clear increase of GFP intensity, which indicated that autophagy was impaired by Atg5 deletion (Fig. 4a). Although, haematopoietic populations of Atg5-deficient mice appeared to be normal as shown in Fig. 1, we performed gene expression profiling to investigate mechanisms underlying damage of Atg5-deficient HSCs at around P7 (P6–8). GO term analysis using microarray data showed that the expression of genes related to mitochondria, such as Tomm40 and Mrps18b, was significantly enriched in LSK cells from Atg5f⁄f;Vav mice (Fig. 4b, Supplementary Fig. S6). Additionally, we found elevated mitochondrial reactive oxygen species (ROS) detected by MitoSox staining in LSK cells of Atg5f⁄f;Vav mice at P7 (Fig. 4c). Increased mitochondrial ROS was also observed in LSK cells of 3-week-old Atg5f⁄f;Vav mice. We did not find an apparent increase of mitochondrial mass detected by MitoTracker staining or constant alteration of the mitochondrial membrane potential detected by TMRM staining caused by Atg5 deficiency at both P7 and 3 weeks of age. While p62 mediates selective degradation of autophagic cargo, it also has a role in the stress response through regulation of the Keap1-Nrf2 system that has been recognised as one of the major cellular defence mechanisms against oxidative and electrophilic stresses. It has been reported that p62 accumulation leads to activation of Nrf2, which induces many anti-oxidative genes37. It has also been reported that Nrf2 induces gene expression involved in anabolic pathways such as the pentose phosphate pathway12,38. As expected, we confirmed that p62 protein had accumulated in immature BM cells (lineage−c-Kit+) of Atg5f⁄f;Vav mice (Supplementary Fig. S7). Consistently, we found that Nrf2 target molecules, including G6pd2 and Taldo1 at around P7 (Fig. 4d) and NAD(P)H dehydrogenase quinone 1 (Nqo1) and glutathione S-transferase mu1 (Gstm1) at 7 weeks of age (Fig. 4e), were upregulated in LSK cells of Atg5-deficient mice. Furthermore, dead cells (7-AAD+) were increased by Atg5 (Fig. 4f). These data suggested that mitochondrial oxidative stress caused HSC damage in the neonatal stage, leading to a severe abnormality of haematopoiesis in adults as shown in Supplementary Fig. S2.
Figure 4.
Gene expression and mitochondrial status in LSK cells from control and Atg5f/f;Vav mice. (a) Flow cytometric analysis of GFP-LC3 in LSK cells from control (GFP-LC3;Atg5f/w;Vav and GFP-LC3;Atg5f/w) and GFP-LC3;Atg5f/f;Vav mice at P7 and 3W. Representative data (two control and two Atg5f/f;Vav) from two independent experiments are shown (P7, n = 5–7; 3W, n = 3–5). (b) GSEA enrichment score curve for control (Atg5w/w;Vav) and Atg5f/f;Vav LSK cells analysed for the gene set associated with the mitochondrial protein complex. Heatmap of representative gene expression in the gene set is shown at the right. NES, normalised enrichment score; FDR, false discovery rate. (c) Flow cytometric analysis of MitoTracker green, MitoSox Red, TMRM, and DCFDA staining in LSK cells from control and Atg5f/f;Vav mice at P7 and 3 weeks. Representative data from at least three independent experiments are shown (n = 3–5). (d) Fold change of representative Nrf2-target gene expression levels upregulated in Atg5f/f;Vav LSK cells compared with control LSK cells from normalised microarray data. (e) mRNA expression levels of Nqo1 and Gstm1 in LSK cells from control, Atg5f/f;Vav, p62f/f;Vav, and Atg5f/f;p62f/f;Vav mice at 7 weeks. Data are the mean ± SD. n = 3–6. (f) Frequency of dead cells (7-AAD-positive cells) in LSK population from control (Atg5f/w;Vav) and Atg5f/f;Vav mice at P7 and 3W. Data are the mean ± SD (n = 4–5).
No restoration of haematopoietic abnormality in Atg5-deficient mice by p62 deletion
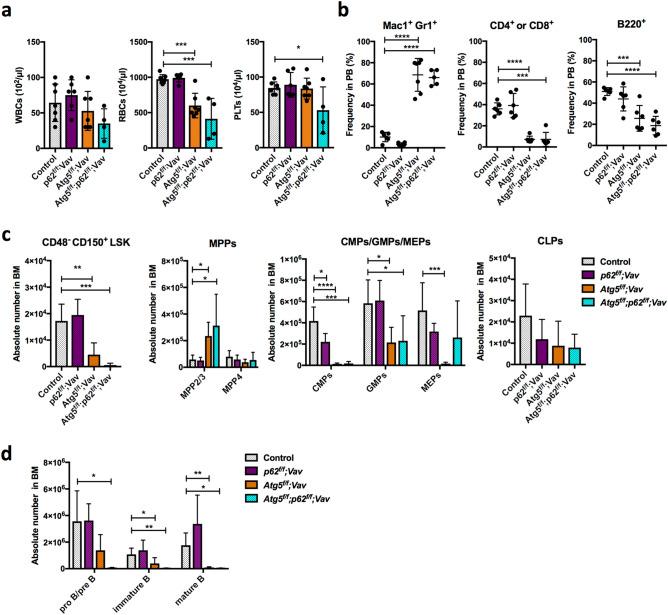
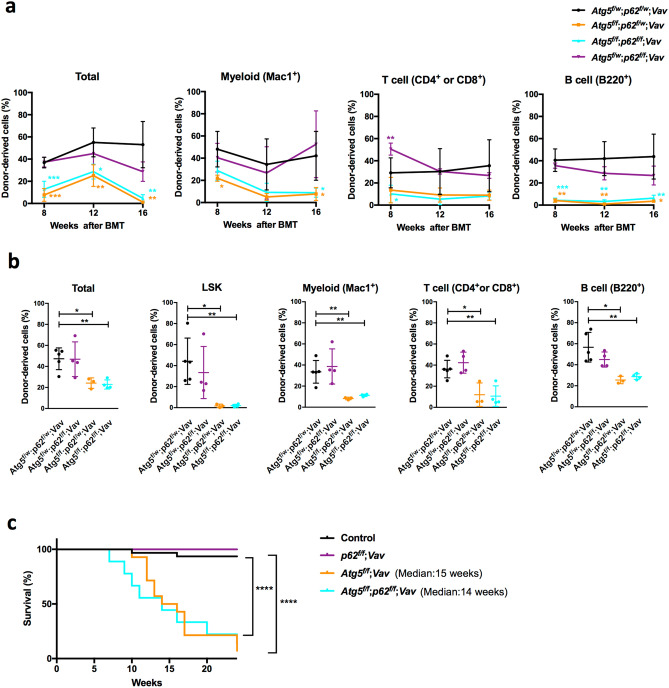
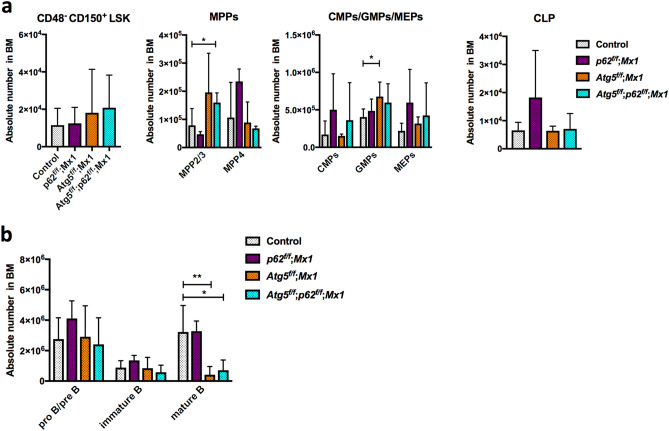
Finally, we investigated whether p62 was involved in the phenotype of Atg5 deletion by the Vav-Cre-loxP system. We confirmed sufficiently high efficiency of p62 deletion using this system (Supplementary Figure S1). p62f/f;Vav-Cre (p62f/f;Vav) mice appeared healthy and did not show any haematopoietic abnormalities such as anaemia or an altered distribution of lineage cells in PB (Fig. 5a,b). In BM, HSC/progenitor populations were almost comparable to the control, except for mildly reduced CMP in p62f/f;Vav mice at 7 weeks of age (Fig. 5c,d). Myeloid cells and lymphocytes in BM and spleen were normal (Supplementary Fig. S8). Reconstitution assays showed that the functions of p62-deficient HSCs, including self-renewal, were not impaired (Supplementary Fig. S8). Thus, p62 loss alone barely affected haematopoiesis. Hence, we assessed the effects of loss of p62 on haematopoiesis in Atg5f⁄f;Vav mice at 7 weeks of age. As shown in Supplementary Fig. S2, Atg5f⁄f;Vav mice at 7 weeks of age showed severe haematopoietic dysfunctions such as severe anaemia and leukocytopenia associated with an abnormality in balance of peripheral cell components, such as increased ratios of myeloid/B cells, compared with control (Atg5w/w;Vav) mice (Fig. 5a,b). In Atg5f⁄f;Vav mouse BM, in addition to reduced LT-HSCs29,30, significant increases of myeloid-biased multipotent progenitor (MPP) 2 and MPP3 were observed (Fig. 5c). Consistent with previous reports, granulocyte–macrophage progenitors (GMPs) were reduced significantly30, and common myeloid progenitors (CMPs)/megakaryocyte-erythrocyte progenitors (MEPs) were reduced similarly. Additionally, we found that mature B cells in BM were reduced drastically in Atg5f⁄f;Vav mice (Fig. 5d). In Atg5f/f;p62f/f;Vav mice, most haematopoietic parameters in PB, BM, and spleen appeared to be comparable with Atg5f/f;Vav mice (Fig. 5a–c, Supplementary Fig. S8), indicating that loss of p62 did not restore the defective haematopoiesis due to Atg5 deficiency. Interestingly, we found that Atg5f/f;p62f/f;Vav mice consistently exhibited more severe phenotypes, particularly in the LT-HSC population as well as proB/preB and immature B cells, suggesting that Atg5 and p62 partly support HSC functions in a collaborative manner. To investigate the roles of p62 in defective phenotypes of Atg5 deficiency, we performed reconstitution assays using Atg5f⁄f;p62f⁄f;Vav mice. While the repopulating capacity in all lineage was reduced significantly by Atg5 loss both in PB and BM of recipients, p62 loss did not restore it (Fig. 6a,b). Because of severe haematopoietic defects, Atg5f⁄f;Vav mice survived for 15 weeks on average, and lethality of Atg5/p62-deficient mice was also comparable to that of Atg5-deficient mice (Fig. 6c). Thus, we concluded that Atg5-dependent haematopoietic regulation during the developmental period was independent of p62. Additionally, we evaluated the effects of p62 deletion in Atg5-deficient mice that lost Atg5 after birth by poly (I:C) administration using the Mx1-Cre-loxP system. Consistent with a previous report33, Atg5f/f;Mx1 mice showed a slight increase of GMPs, but it was largely comparable with the control for the HSC/progenitor population (Fig. 7a). In lineage cells, a slight increase of splenic myeloid cells and altered erythroid development were observed (Supplementary Fig. S9). Apart from Atg5f/f;Vav mice, Atg5f/f;Mx1 mice also showed a reduction of mature B cells (Fig. 7b). These alterations were not restored by p62 deletion (Fig. 7a,b, Supplementary Fig. S9 and S10). In summary, haematopoietic regulation by Atg5 during both developmental and adult periods was independent of p62.
Figure 5.
Comparative analysis of PB and BM cells from control, p62f/f;Vav, Atg5f/f;Vav, and Atg5f/f;p62f/f;Vav mice. (a) Counts of white blood cells (WBCs), red blood cells (RBCs), and platelets (PLTs) in PB. Data are the mean ± SD (n = 4–7). Each dot indicates values of individual mice. (b) Frequencies of myeloid cells (Mac1+Gr-1+), T cells (CD4+ or CD8+), and B cells (B220+) in PB. Data are the mean ± SD (n = 3–7). Horizontal line indicates the mean of values. (c) Absolute numbers of CD48−CD150+LSK cells, multipotent progenitors (MPPs; MPP2/3 and MPP4), common myeloid progenitors (CMPs), granulocyte macrophage progenitors (GMPs), megakaryocyte erythrocyte progenitors (MEPs), and common lymphoid progenitors (CLPs) in BM. Data are mean ± SD (n = 4–8). (d) Absolute numbers of mature B (CD43−B220hiIgM+), immature B (CD43−B220loIgM+), and pro B/pre B (CD43−B220loIgM−) cells in BM. Data are the mean ± SD (n = 3–6).
Figure 6.
Competitive reconstitution analysis and survival assay. (a,b) Competitive reconstitution analysis of BM cells from Atg5f/w;p62f/w;Vav,Atg5f/f;p62f/w;Vav,Atg5f/f;p62f/f;Vav, and Atg5f/w;p62f/f;Vav mice at 3 weeks. (a) Frequencies of donor-derived cells among total cells and lineage cells in PB were analysed 8–16 weeks after transplantation. Data are the mean ± SD (n = 3–5). (b) Frequencies of donor-derived cells among total, LSK, myeloid, T, and B cells in BM were analysed 16 weeks after transplantation. Data are the mean ± SD (n = 3–5). Horizontal line indicates the mean of values. (c) Survival curve of control (n = 31), p62f/f; Vav (n = 6), Atg5V; Vav (n = 13), and Atg5f/f; p62f/f; Vav (n = 9) mice.
Figure 7.
Comparative analysis of HSCs, progenitors, and B cells in BM from control, p62f/f; Mx1, Atg5f/f; Mx1, and Atg5f/f; p62f/f; Mx1 mice. (a) Absolute numbers of CD48−CD150+LSK cells, multipotent progenitors (MPPs; MPP2/3 and MPP4), common myeloid progenitors (CMPs), granulocyte macrophage progenitors (GMPs), megakaryocyte erythrocyte progenitors (MEPs), and common lymphoid progenitors (CLPs) in BM. Data are the mean ± SD (n = 4–7). (b) Absolute numbers of mature B (CD43−B220hiIgM+), immature B (CD43−B220loIgM+), and pro B/pre B (CD43−B220loIgM−) cells in BM. Data are the mean ± SD (n = 5–7).
Discussion
In mice, primitive haematopoiesis begins in blood islands of the yolk sac at E7, followed by the switch from primitive to definitive haematopoiesis at E10 and 11. At the beginning of the perinatal period, the liver is still a source of extra-medullary haematopoiesis, and then the BM eventually takes over39. During the perinatal period, foetal and neonatal development and maturation are deeply affected by environmental conditions such as oxidative stress, nutritional stress, and microbial infection. Although it has been demonstrated that autophagy plays a critical role in development and aging of HSCs, the importance of autophagy in perinatal HSCs, especially during the neonatal period, has not been revealed. Based on this study, we propose a critical role of autophagy in protection of HSCs against harsh environments in the early neonatal stage, which is essential for healthy long-term haematopoiesis. The perinatal period is accompanied by dramatic environmental changes in both oxidative stress and nutritional conditions, which may affect autophagic activity. Because the transplacental nutrient supply is suddenly interrupted at birth, neonates have stress due to starving until milk feeding. Foetal life evolves under a hypoxic condition, but it provides comfortable environments for development of organs. During foetal-to-neonatal transition, asphyxia is characterised by periods of severe hypoxia that may evolve to ischemic organ impairment. In another aspect of oxygen, foetal-to-neonatal transition may cause “relative hyperoxia”. In both cases, oxidative stress occurs in neonatal tissues. Although it is unclear which factors activate autophagy, it is assumed that autophagy is important for survival of HSCs against specific harsh neonatal environments. To understand the neonatal period better, we analyzed public RNA-seq data from wild type neonatal (P7) HSCs (CD48-CD150+Lineage-Sca-1+c-Kit-) and adult HSCs deposited in GEO (GSE128762)40. We found that expression of many genes involved in glucose and glutamine metabolisms, mitochondria, and redox regulation varied significantly between at P7 and adult mice. The expression levels of most of these genes are significantly lower in neonatal HSCs compared with those in adult HSCs (Supplementary Fig. S11). It has been reported that the metabolism and redox regulation are closely related to the regulation and maintenance of HSCs in adults41–43. Mitochondria, a dynamic organelle in which many metabolic processes occur, also have critical roles in maintenance of HSCs44. The lower expression of these genes in HSCs during neonatal period suggest that the protective system of HSCs against metabolic stress may be somewhat different among the developmental stages, and that survival of neonatal HSCs may be highly dependent on autophagy. Additional analysis is needed to understand the stage-specific roles of autophagy.
Further understanding of the roles of autophagy in neonates may uncover novel pathophysiological insights of disease that caused by autophagy-related abnormalities in the neonatal period. One such insight may be the mitochondrial behaviour of HSCs. Accumulating evidence has shown that HSCs with increased and active mitochondria have loss of haematopoietic functions such as the reconstitution ability45–47. In our study, alteration of the TMRM level was not apparent at P7 and 3 weeks of age, although an increased mitochondrial mass, TMRM level, and mitochondrial ROS were observed at 7 weeks (Supplementary Fig. S12) consistent with previous data30. Although we do not have clear evidence of the roles of oxidative stress in defective phenotypes of HSCs in Atg5f/f;Vav mice, we assume that the functional change of mitochondria may occur at the perinatal stage. Further analysis will be needed to understand the molecular mechanism.
In this study, we focused on the role of p62 in autophagy-dependent anti-oxidative effects on HSCs. p62 is a signalling hub that interacts with various partners. Previous functional analyses of p62 knockout (p62−⁄−) mice indicated that the main phenotype of the mice is obesity48. p62−⁄− mice do not show haematopoietic failure for up to 6 months after birth or transplantation49. Consistently, our analysis of p62f/f;Vav mice showed that the haematopoietic stem cells were normal and their reconstitution ability were maintained (Fig. 5c and Supplementary Fig. S8). Because it has been reported that p62 is required for amino acid sensing via the mTOR pathway50, p62 might play a critical role in the protective response to abnormalities in amino acid levels. Additionally, we analysed Atg5/p62-deficient HSCs. p62 ablation suppresses the hepatic phenotype, but not affect the neuronal defect caused by autophagy10–12. This study clarified that p62 was not the cause of the severe functional decline of VavCre-mediated Atg5-deficient HSCs. Although p62 accumulated and induced Nrf2-targeted gene expression in HSCs derived from Atg5f/f;Vav mice (Supplementary Fig. S7 and Fig. 4d,e), the gene induction was not remarkable compared with Atg5-deficient hepatic cells12. Because autophagy has pleiotropic roles in haematopoiesis, we also analysed other haematopoietic cells in addition to HSCs. For example, we found severe impairment of B cell development in Atg5f/f;Vav mice, which was consistent with the phenotype of B cell-specific Atg5-deficient mice23, but p62 loss did not rescue it. Atg5f/f;Mx1 mice also showed impairment of B cell development, but the phenotype was much more milder than that of Atg5f/f;Vav mice. p62 loss did not rescue impaired B cell development in Atg5f/f;Mx1 mice. In some cases, concomitant deletion of p62 worsens the phenotype of Atg5 loss, suggesting that p62-mediated pathways collaborate with autophagy for proper HSC development. One possibility is that p62 may have a protective role mediating mTOR signalling under autophagy-deficient conditions. It has been reported that p62 interacts with mTORC1 components Raptor and Rags, and mediates amino acid sensing50. mTORC1 has an essential role in HSC functions in vivo51,52. Because p62 deficiency did not cause apparent haematopoietic abnormalities except for a mild reduction of CMP, autophagy and p62 may act synergistically for HSC functions, at least in part. Further analysis is needed to understand how HSCs normally develop in the harsh environment during the neonatal period.
In conclusion, we demonstrated that autophagy plays a critical role in protecting neonatal HSCs from oxidative stress in a p62-independent manner. Our findings provide novel insights regarding a critical role of autophagy in protection of HSCs against harsh environments in the early neonatal stage, which is essential for healthy long-term haematopoiesis in adults.
Materials and methods
Mice
Atg5flox/flox (Atg5f/f) mice53 were bred with Vavi-Cre54 (Charles River Laboratories Japan Inc.,) or Mx1-Cre55 transgenic mice to obtain Atg5f/f;Vavi-Cre (Atg5f/f;Vav) and Atg5f/f;Mx1-Cre (Atg5f/f;Mx1) mice, respectively. p62flox/flox (p62f/f)56 mice were bred with Atg5f/f;Vav and Atg5f/f;Mx1 mice to obtain Atg5f/f;p62f/f;Vav and Atg5f/f;p62f/f;Mx1 mice. Atg5f/f mice were bred with Rosa26-CreERT257 to obtain Atg5flox/flox;Rosa26-CreERT2 (Atg5f/f;Rosa) mice. Atg5flox/wild;Rosa26-CreERT2 (Atg5f/w;Rosa) mice were administrated tamoxifen to delete the floxed Atg5 gene. Atg5−/w;Rosa mice were bred with Atg5f/w;Rosa or Atg5f/f;Rosa to obtain Atg5f/−;Rosa mice. Atg5f/f;Vavi-Cre mice were bred with CAG-GFP-LC3 mice (RBRC00806, provided by RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan) to obtain GFP-LC3;Atg5f/f;Vavi-Cre mice. Genotyping was performed on tail or PB genomic DNA. PCR primers were as follows:
Atg5 WT/floxed/deleted alleles, 5′-ATGGTGTCTCCCACATCAGTTAGC-3′, 5′-CCAATACACAGGGTACACTGAAGG-3′, 5′-AGTGAAGGAGTGGAAAGTAGTGGG-3′; p62 WT/floxed/deleted alleles, 5′-AGGCTGGCACAGTGAATCTT-3′, 5′-TCTGTGTCCTCCTAGCTTCT-3′, 5′-TCAATTCCCAGTACTGGAGG-3′; Vav-iCre, 5′-AGATGCCAGGACATCAGGAACCTG-3′, 5′-ATCAGCCACACCAGACACAGAGATC-3′; Mx1-Cre58, 5′-TCCCAACCTCAGTACCAAGC-3′, 5′-ATTCTCCCACCGTCAGTACG-3′; Rosa26-Cre-ERT2, 5′-AAAGTCGCTCTGAGTTGTTAT-3′, 5′-CCTGATCCTGGCAATTTCG-3′; GFP-LC3, 5′-ATAACTTGCTGGCCTTTCCACT-3′, 5′-CGGGCCATTTACCGTAAGTTAT-3′, 5′-GCAGCTCATTGCTGTTCCTCAA-3′.
For Mx1-Cre-mediated deletion, 4-week-old mice were injected intraperitoneally three times 2 days apart with 400 μg poly (I:C) (Sigma-Aldrich). Mx1-Cre mice were analysed at 4 weeks after poly (I:C) administration. For Rosa26-Cre-ERT2 mediated deletion, P5 neonates were injected intraperitoneally with 0.5 mg tamoxifen. The mice were analysed at 8–12 months after tamoxifen administration. To determine gene deletion efficiency, colony PCR was conducted. Sorted LSK cells were cultured in Methocult GF M3434 (STEMCELL Technologies) and single cell-derived colonies were picked up. Cells were suspended in 0.01 M Tris–HCl (pH 8.0) containing proteinase K (200 μg/ml) and incubated for 90 min at 56 °C and then for 10 min at 90 °C. DNA was obtained and analysed by PCR.
Male and female mice were used equally in all experiments. In experiments using Atg5f/f;Vav mice, Atg5f/w;Vav, Atg5w/w;Vav and Vav-negative littermates (Atg5f/f and Atg5f/w) were used equally as controls unless otherwise specified in figure legends. In experiments using Atg5f/f;p62f/f;Vav mice, Atg5f/w;p62f/w;Vav, Atg5f/w;p62w/w;Vav, Atg5w/w;p62f/w;Vav, Atg5w/w;p62w/w;Vav, and Vav-negative littermates were used equally as controls unless otherwise specified in figure legends. In experiments using Atg5f/f;Rosa and Atg5f/−;Rosa mice, Atg5f/w;Rosa and Atg5w/w;Rosa littermates were used as controls. In experiments using Atg5f/f;Mx1 mice, Atg5f/w;Mx1 and Atg5f/f littermates were used as controls unless otherwise specified in figure legends. In experiments using Atg5f/f;p62f/f;Mx1 mice, Atg5f/w;p62w/w;Mx1, Atg5w/w;p62w/w;Mx1, and Mx1-negative littermates were used equally as controls. All animal experiments were approved by the Committee on Animal Experimentation of Kanazawa University, and performed in accordance with the Guidelines for Animal and Recombinant DNA Experiments of Kanazawa University.
Haematological analysis
PB was collected from the postorbital vein and suspended in a heparin solution. Complete blood count analysis was performed using a Celltac alpha (Nihon Kohden). Before antibody staining and DNA extraction, erythrocytes were lysed with ammonium chloride. BM cells were obtained from femoral and tibial bones of hind legs by flushing the BM cavity. If necessary, isolation of mononuclear cells by density gradient centrifugation using Lymphoprep (Axis-Shield) was conducted. BM cells were stained with antibodies for flow cytometric analysis or used for transplantation.
Flow cytometry
PB and BM cells were stained with monoclonal rat anti-mouse antibodies recognising each marker. Fc-block (BD Biosciences) was used to block non-specific binding of antibodies to Fcγ receptor. To stain HSCs, a mixture of biotinylated antibodies for lineage cells, streptavidin-APC780 (eBioscience), anti-c-Kit-APC (BD Biosciences), anti-Sca1-PECy7 (BD Biosciences), anti-CD48-FITC (BD Biosciences), and anti-CD150-PE (BioLegend) antibodies were used. To stain HSCs from mice at < 4 weeks of age, biotinylated anti-Gr-1 (eBioscience), anti-Ter119 (eBioscience), anti-IL7R (BD Biosciences), anti-B220 (eBioscience), anti-CD4 (eBioscience), and anti-CD8 (eBioscience) antibodies were used for lineage cell staining. To stain HSCs and progenitors (MPPs, CMP/GMP/MEPs, and CLPs) from mice at ≥ 4 weeks of age, the biotinylated anti-Mac-1 (eBioscience) antibody was added to the biotinylated antibody mixture for lineage cells. To stain MPPs, an anti-CD135-BV421 (BD Biosciences) antibody was combined with HSC-staining antibodies. For CMP/GMP/MEP staining, the mixture of biotinylated antibodies for lineage cells, streptavidin-APC780 (eBioscience), anti-c-Kit-APC (BD Biosciences), anti-Sca1-PECy7 (BD Biosciences), anti-FcγR-PE (BD Biosciences), and anti-CD34-FITC (BD Biosciences) antibodies were used. For CLP staining, the mixture of biotinylated antibodies for lineage cells except for IL7R, streptavidin-APC780 (eBioscience), anti-c-Kit-APC (BD Biosciences), anti-Sca1-PE (BD Biosciences), and anti-IL7R-PECy7 (eBioscience) antibodies were used. To analyse B cell development, biotinylated anti-CD43 (eBioscience), streptavidin-APC780 (eBioscience), anti-B220-FITC (BD Biosciences), and anti-IgM-APC (BD Biosciences) antibodies were used. For myeloid cell staining, anti-Mac1-FITC (BD Biosciences), anti-Gr-1-PE (eBioscience), or anti-Gr-1-PECy7 (eBioscience) antibodies were used. For lymphoid cell staining, anti-B220-FITC (BD Biosciences) or anti-B220-PECy7 (eBioscience), anti-CD4-APC (BD Biosciences), anti-CD8-APC (BD Biosciences), or anti-CD8-PE (BD Biosciences) antibodies were used. For erythroid cell staining, anti-Ter119-PE (BD Biosciences) and anti-CD71-FITC (eBioscience) antibodies were used. For LSK chimerism analysis, biotinylated antibodies for lineage cells, streptavidin-APC780 (eBioscience), anti-c-Kit-APC (BD Biosciences), anti-Sca-1PECy7 (BD Biosciences), anti-CD45.1-PE (BD Biosciences), and anti-CD45.2-FITC (BD Biosciences) antibodies were used. For myeloid chimerism analysis, anti-Mac-1APC (eBioscience), anti-Gr-1-PECy7 (eBioscience), anti-CD45.1-PE (BD Biosciences), anti-CD45.2-FITC (BD Biosciences), or anti-Mac1-PB (BioLegend), anti-Gr-1APC (BD Biosciences), anti-CD45.1-PE (BD Biosciences), and anti-CD45.2-FITC (BD Biosciences) antibodies were used. For lymphoid chimerism analysis, anti-B220-PECy7 (eBioscience) or anti-B220-PB (BD Biosciences), anti-CD4-APC (BD Biosciences), anti-CD8-APC (BD Biosciences), anti-CD45.2-FITC (BD Biosciences), and anti-CD45.1-PE (BD Biosciences) antibodies were used. Stained cells were washed with PBS containing 5% FCS, and dead cells were stained with 7-AAD (BD Biosciences). Samples were analysed using a FACSCanto II, FACSLyric, or FACSAria II/III (Becton Dickinson).
Transplantation
For BM cell transplantation, donor (CD45.2) whole BM cells (1 × 106/recipient) were mixed with competitor (CD45.1) whole BM cells (1 × 106/recipient) and transplanted into lethally irradiated (9 Gy) recipient (CD45.1) mice through the tail vein. Donor-derived cells were recognised by CD45.2 expression.
Western blotting
c-Kit+Lin− cells (5 × 106) were sorted by a FACSAria II and lysed with RIPA buffer (25 mM Tris–HCl, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, and 1% NP40) supplemented with Complete Mini (Roche). After mixing with SDS Sample buffer, proteins were denatured by boiling. Denatured proteins were separated on a 15% acrylamide gel and then transferred to a PVDF membrane. After blocking with 5% dry skim milk in PBST, blots were incubated with Abs against ATG5 (Novus Biologicals)59, p62 (Abnova)60, LC3 (Nanotools)61, and β-actin (Sigma-Aldrich)62. Immunocomplexes were then labelled with a HRP-conjugated anti-mouse, or with anti-rabbit antibodies, and visualised using ECL Prime Western Blotting Detection Reagent (GE Healthcare) with an ImageQuant LAS 4000 (GE Healthcare). To detect different proteins in the same membrane, conjugated antibodies were removed using stripping solution (62.5 mM Tris–HCl, pH = 6.8, 2% SDS, and 0.7% β-mercaptoethanol) by gently shaking for 50 min at 50 °C. After washing with PBST, blocking and antibody reactions were carried out.
Intracellular ROS and mitochondrial analyses
Collected BM cells were stained with antibodies (biotinylated antibodies for lineage cells, streptavidin-APC780, c-Kit-APC, and Sca1-PECy7). After the antibody staining, the cells were mixed and reacted with Mitotracker Green, tetramethylrhodamine (TMRM; Thermo Fisher Scientific), Mitosox Red (Thermo Fisher Scientific), or CM-H2CDCFDA (2′,7′-dichlorodihydrofluorescein diacetate; Thermo Fisher Scientific). Cells were then washed and analysed by the FACS Canto II or FACsLyric.
Quantitative PCR
LSK cells were sorted by fluorescence-activated cell sorting and collected by centrifugation. RNA was extracted with a RNeasy Mini Kit (Qiagen) and cDNA was synthesised with a QuantiTect Reverse Transcription Kit (Qiagen). The cDNA was used for real time PCR with TB green (Takara). PCR primers were as follows:
Nqo112, 5′-AGTACAATCAGGGCTCTTCTCG-3′, 5′-AGCGTTCGGTATTACGATCC-3′; Gstm112, 5′-CTACCTTGCCCGAAAGCAC-3′, 5′-ATGTCTGCACGGATCCTCTC-3′; Actb, 5′-CCTCTATGCCAACACAGTGC-3′, 5′-CCTGCTTGCTGATCCACATC-3′.
Microarray and gene set analyses
BM cells of Atg5f/f;Vav and Atg5w/w;Vav mice were collected and LSK cells were stained as described above. LSK cells (> 1 × 104 cells) were sorted directly into TRIzol (Invitrogen). Subsequent sample preparation and analysis were carried out by Hokkaido System Science. Details are shown in a previous report62. Normalised expression data were examined by Gene Set Enrichment Analysis (GSEA) v4.0.1 software (Broad Institute) using the GO gene set. The number of permutations was set to 1000.
Statistical analysis
Unless indicated otherwise, data are shown as the mean ± SD. Dots on bar graphs indicate values of individual mice. Statistical differences between groups were determined by the Student’s unpaired t-test using Prism 5 software (GraphPad). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. In survival assays, statistical differences between groups were determined by the Mantel-Cox test.
Data availability
The accession number for the microarray data in this study is GEO: GSE153613.
Supplementary information
Acknowledgements
We are grateful to Dr. N. Mizushima (The University of Tokyo, Tokyo, Japan) and the RIKEN BRC for providing Atg5fl/fl (RBRC02975) and CAG-GFP-LC3 (RBRC00806) mice, Dr. Klaus Rajewsky (Max Delbrück Centre for Molecular Medicine) for providing Mx1-Cre mice, and Dr. Tyler Jacks (Massachusetts Institute of Technology, MA, USA) for providing Rosa26-Cre-ERT2 mice. We thank Kazue Sawa, Eri Azechi, Yukiko Takai, and Miyako Takegami for technical assistance. We also thank Mitchell Arico from Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript. C.I. was supported by a Grant-in-Aid for JSPS Fellows (JP16J07234). A.H. was supported by a Grant-in-Aid for Scientific Research (A) (JP19H01033) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT).
Author contributions
A.H. designed the study. C.I. wrote the manuscript. N.N., C.I., and T.O. performed experiments. S.K. performed gene expression analysis regarding Supplementary Fig. S11. N.N. and C.I. prepared figures. M. Komatsu. and T.Y. provided experimental animals. Y. Tadokoro., M.U., M. Kobayashi., A.K., and C.T. contributed to data interpretation and provided critical feedback. Y. Takase., K.K., and S.S. provided experimental assistance. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Naho Nomura and Chiaki Ito.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-81076-z.
References
- 1.Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020 doi: 10.1038/s41580-020-0241-0. [DOI] [PubMed] [Google Scholar]
- 2.Walczak M, Martens S. Dissecting the role of the Atg12–Atg5–Atg16 complex during autophagosome formation. Autophagy. 2013;9:424–425. doi: 10.4161/auto.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen TN, et al. Atg8 family LC3/GAB ARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 2016;215:857–874. doi: 10.1083/jcb.201607039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deuretzbacher A, et al. β 1 integrin-dependent engulfment of Yersinia enterocolitica by macrophages is coupled to the activation of autophagy and suppressed by type III protein secretion. J. Immunol. 2009;183:5847–5860. doi: 10.4049/jimmunol.0804242. [DOI] [PubMed] [Google Scholar]
- 6.Dupont N, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Watson RO, Manzanillo PS, Cox JS, Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uddin MS, et al. Autophagic dysfunction in Alzheimer’s disease: cellular and molecular mechanistic approaches to halt Alzheimer’s pathogenesis. J. Cell. Physiol. 2019;234:8094–8112. doi: 10.1002/jcp.27588. [DOI] [PubMed] [Google Scholar]
- 9.Ichimura Y, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 11.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inami Y, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sou Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.e08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malhotra R, Warne JP, Salas E, Xu AW, Debnath J. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy. 2015;11:145–154. doi: 10.1080/15548627.2014.998917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 18.Yoshii SR, et al. Systemic analysis of Atg5-null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Dev. Cell. 2016;39:116–130. doi: 10.1016/j.devcel.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Stephenson LM, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–635. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pua HH, Guo J, Komatsu M, He Y-W. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J. Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 21.Hubbard VM, et al. Macroautophagy regulates energy metabolism during effector T cell activation. J. Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei J, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 2016;17:277–285. doi: 10.1038/ni.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller BC, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 24.Pengo N, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat. Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 25.Conway KL, et al. ATG5 regulates plasma cell differentiation. Autophagy. 2013;9:528–537. doi: 10.4161/auto.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold J, et al. Autophagy is dispensable for B-cell development but essential for humoral autoimmune responses. Cell Death Differ. 2016;23:853–864. doi: 10.1038/cdd.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortensen M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc. Natl. Acad. Sci. USA. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimmey JM, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528:565–569. doi: 10.1038/nature16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson A, et al. Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Discov. 2015;1:1–10. doi: 10.1038/cddiscovery.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung HE, Shim YR, Oh JE, Oh DS, Lee HK. The autophagy protein Atg5 plays a crucial role in the maintenance and reconstitution ability of hematopoietic stem cells. Immune Netw. 2019;19:1–13. doi: 10.4110/in.2019.19.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortensen M, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warr MR, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho TT, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bustelo XR, Rubin SD, Suen KL, Carrasco D, Barbacid M. Developmental expression of the vav protooncogene. Cell Growth Differ. 1993;4:297–308. [PubMed] [Google Scholar]
- 35.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4:621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 37.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 38.Mitsuishi Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Wolber FM, et al. Roles of spleen and liver in development of the murine hematopoietic system. Exp. Hematol. 2002;30:1010–1019. doi: 10.1016/S0301-472X(02)00881-0. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, et al. Single-cell analysis of neonatal HSC ontogeny reveals gradual and uncoordinated transcriptional reprogramming that begins before birth. Cell Stem Cell. 2020 doi: 10.1016/j.stem.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oburoglu L, et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15:169–184. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Takubo K, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattes K, Vellenga E, Schepers H. Differential redox-regulation and mitochondrial dynamics in normal and leukemic hematopoietic stem cells: a potential window for leukemia therapy. Crit. Rev. Oncol. Hematol. 2019;144:102814. doi: 10.1016/j.critrevonc.2019.102814. [DOI] [PubMed] [Google Scholar]
- 44.Filippi MD, Ghaffari S. Mitochondria in the maintenance of hematopoietic stem cells: new perspectives and opportunities. Blood. 2019;133:1943–1952. doi: 10.1182/blood-2018-10-808873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vannini N, et al. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat. Commun. 2016;7:1–9. doi: 10.1038/ncomms13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sukumar M, et al. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 2016;23:63–76. doi: 10.1016/j.cmet.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang R, et al. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell. 2020;26:359–376.e7. doi: 10.1016/j.stem.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez A, et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Chang KH, et al. P62 is required for stem cell/progenitor retention through inhibition of IKK/NF-κB/Ccl4 signaling at the bone marrow macrophage-osteoblast niche. Cell Rep. 2014;9:2084–2097. doi: 10.1016/j.celrep.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duran A, et al. P62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoshii T, et al. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J. Clin. Invest. 2012;122:2114–2129. doi: 10.1172/JCI62279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng H, et al. Distinct roles of Rheb and raptor in activating mTOR complex 1 for the self-renewal of hematopoietic stem cells. Biochem. Biophys. Res. Commun. 2018;495:1129–1135. doi: 10.1016/j.bbrc.2017.11.140. [DOI] [PubMed] [Google Scholar]
- 53.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 54.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 55.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science (80-) 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 56.Harada H, et al. Deficiency of p62/sequestosome 1 causes hyperphagia due to leptin resistance in the brain. J. Neurosci. 2013;33:14767–14777. doi: 10.1523/JNEUROSCI.2954-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 58.Liu L, et al. Functional defect of peripheral neutrophils in mice with induced deletion of CXCR2. Genesis. 2013;51:587–595. doi: 10.1002/dvg.22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao J, et al. Inhibiting autophagy reduces retinal degeneration caused by protein misfolding. Autophagy. 2018;14:1226–1238. doi: 10.1080/15548627.2018.1463121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung PED, et al. Modeling germline mutations in pineoblastoma uncovers lysosome disruption-based therapy. Nat. Commun. 2020;11:1825. doi: 10.1038/s41467-020-15585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Runwal G, et al. LC3-positive structures are prominent in autophagy-deficient cells. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tadokoro Y, et al. Spred1 safeguards hematopoietic homeostasis against diet-induced systemic stress. Cell Stem Cell. 2018;22:713–725.e8. doi: 10.1016/j.stem.2018.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the microarray data in this study is GEO: GSE153613.