Abstract
Purpose
To identify tissue metabolomic profiles in biopsy specimens with IgG4-related ophthalmic disease (IgG4-ROD) and mucosa-associated lymphoid tissue (MALT) lymphoma and investigate their potential implication in the disease pathogenesis and biomarkers.
Methods
We conducted a comprehensive analysis of the metabolomes and lipidomes of biopsy-proven IgG4-ROD (n = 22) and orbital MALT lymphoma (n = 21) specimens and matched adjacent microscopically normal adipose tissues using liquid chromatography time-of-flight mass spectrometry. The altered metabolomic profiles were visualized by heat map and principal component analysis. Metabolic pathway analysis was performed by Metabo Analyst 4.0 using differentially expressed metabolites. The diagnostic performance of the metabolic markers was evaluated using receiver operating characteristic curves. Machine learning algorithms were implemented by random forest using the R environment. Finally, an independent set of 18 IgG4-ROD and 17 orbital MALT lymphoma specimens were used to validate the identified biomarkers.
Results
The principal component analysis showed a significant difference of both IgG4-ROD and orbital MALT lymphoma for biopsy specimens and controls. Interestingly, lesions in IgG4-ROD were uniquely enriched in arachidonic metabolism, whereas those in orbital MALT lymphoma were enriched in tricarboxylic acid cycle metabolism. We identified spermine as the best discriminator between IgG4-ROD and orbital MALT lymphoma, and the area under the receiver operating characteristic curve of the spermine to discriminate between the two diseases was 0.89 (95% confidence interval, 0.803–0.984). A random forest model incorporating a panel of five metabolites showed a high area under the receiver operating characteristic curve value of 0.983 (95% confidence interval, 0.981–0.984). The results of validation revealed that four tissue metabolites: N1,N12-diacetylspermine, spermine, malate, and glycolate, had statistically significant differences between IgG4-ROD and orbital MALT lymphoma with receiver operating characteristic values from 0.708 to 0.863.
Conclusions
These data revealed the characteristic differences in metabolomic profiles between IgG4-ROD and orbital MALT lymphoma, which may be useful for developing new diagnostic biomarkers and elucidating the pathogenic mechanisms of these common orbital lymphoproliferative disorders.
Keywords: IgG4-related ophthalmic disease, MALT lymphoma, metabolomics, lipids, liquid chromatography-mass spectrometry, machine learning
Lymphoproliferative diseases are common causes of orbital mass lesions. Mucosa-associated lymphoid tissue (MALT) lymphoma is the most common type, followed by IgG4-related ophthalmic disease (IgG4-ROD) in Japan.1 Both IgG4-ROD and orbital MALT lymphoma show similar clinical features and share the histologic features of small B cells and plasma cell infiltration.2,3 IgG4-related disease (IgG4-RD), which is designated as an intractable disease in Japan, is a multiorgan inflammatory condition that includes a group of disorders previously diagnosed under different names that manifest a dominant pattern of organ involvement, such as ocular adnexa including lacrimal glands, salivary glands, pancreas, kidneys, lungs, aorta, prostate, pericardium, and skin.1,4
Autoimmunity and infectious agents are generally considered immunologic triggers in IgG4-ROD.5,6 However, orbital MALT lymphoma is a type of low-grade B-cell lymphoma caused by chronic inflammatory stimulation, eventually progressing to genetic instabilities with chromosomal abnormalities, causing the transformation of a clone of normal lymphoid cells to MALT lymphoma.7 However, the pathogenic mechanisms of IgG4-ROD and orbital MALT lymphoma are poorly characterized. Notably, IgG4-ROD occasionally involves regional and/or systemic lymph nodes simultaneously or subsequently and is often clinically and/or histopathologically suspected to be malignant lymphoma. Clinically, IgG4-ROD exhibits similar clinical and imaging (magnetic resonance imaging and computed tomography) features as orbital MALT lymphoma.8,9 Furthermore, some orbital MALT lymphomas are associated with elevated serum IgG4 levels9 and infiltration of numerous IgG4-positive plasma cells in the affected tissue.10 Orbital MALT lymphoma can arise from IgG4-ROD11 and IgG4-producing MALT lymphoma.12 Elevated serum IgG4 is not sufficiently sensitive or specific for diagnostic purpose.13,14 Therefore, discriminating IgG4-ROD from orbital MALT lymphoma is sometimes challenging. Moreover, not all patients respond well to treatment, and approximately 50% of patients with IgG4-ROD will develop recurrence or progression after conventional clinical treatment, including with systemic corticosteroids or rituximab.15,16 Therefore, further elucidation of the underlying molecular mechanisms of IgG4-ROD is required.
Metabolites reflect the integration of upstream processes of genes and proteins. Metabolites function as mediator molecules and comprehensive metabolic analyses may, therefore, provide insights into the features of disease states. Metabolomics, a method for the comprehensive analysis of metabolites, has emerged as a promising tool for the identification of biomarkers, combining advanced analytical chemistry techniques with cheminformatics to characterize thousands of metabolites found in tissues and biofluids. Thus, metabolomics is widely used to characterize disease pathophysiology and metabolic pathways frequently aberrant in inflammatory diseases and cancers.17,18 Although a recent serum metabolomic study of IgG4-RD has been reported,19 a comprehensive analysis aiming to reveal the tissue metabolomic profiles of both IgG4-ROD and orbital MALT lymphoma has not been documented, to the best of our knowledge. Among the analytical methods used in metabolite characterization, liquid chromatography-mass spectrometry (LC-MS) is a suitable approach that allows simultaneous detection and quantification of a wide range of metabolites with high analytical precision. We recently demonstrated successful LC-MS–based analysis of human specimens and the identification of biomarkers of human diseases.20–23
In this study, we applied LC-MS–based metabolomics to evaluate the metabolite and lipid profiles of IgG4-ROD and orbital MALT lymphoma tissues as well as adjacent adipose tissue obtained from biopsies. We aimed to elucidate the differences in the metabolomic profile between IgG4-ROD and orbital MALT lymphoma to improve our understanding of the underlying pathophysiologic mechanisms of these disorders and identify potential molecular biomarkers.
Methods
Patients, Sample Collection, and Diagnosis
This study was approved by the institutional review board of Tokyo Medical University (approval number: 3281). Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. We enrolled a total of 40 patients, comprising 22 with newly diagnosed IgG4-ROD (9 men and 13 women; mean age, 59.7 ± 15.1 years; range, 36–85 years) and 21 with newly diagnosed orbital MALT lymphoma (9 men and 12 women; mean age, 77.0 ± 7.0 years; range, 67–87 years) at Tokyo Medical University Hospital between January 2016 and August 2020. All patients were immunocompetent Asian adults (Supplementary Tables S1 and S2). Patients with a history of treatment for IgG4-ROD or MALT lymphoma, patients with IgG4-ROD newly diagnosed with ocular MALT lymphoma, and patients with MALT lymphoma showing a tissue IgG4/IgG ratio of greater than 40% and serum IgG4 levels of greater than 135 mg/dL were excluded. For the validation cohort, 18 patients with IgG4-ROD (9 men and 9 women; mean age, 55.0 ± 13.7 years; range, 35–81 years) and 17 patients with ocular MALT lymphoma (10 men and 7 women; mean age, 72.1 ± 14.8 years; range, 35–92 years) were analyzed as the discovery dataset (Supplementary Tables S3 and S4).
Biopsy specimens were collected during debulking surgery. During surgery, the resectable lesion was removed as much as possible with a safe margin. In eight patients with IgG4-ROD and four patients with orbital MALT lymphoma, adipose tissue adjacent to the lesion was collected as a control from the same patient at the time of biopsy. Surgical biopsy specimens were delivered immediately to the Pathology and Molecular Laboratory for flow cytometry and molecular genetic analyses and were quickly cryopreserved and stored at –80 °C for metabolomic analysis. Before the metabolomic analysis, a microscopic and histologic assessment of the cryopreserved biopsy material was performed if needed, to ensure that the specimen was representative of the lesion and that the adjacent fat was not involved.
The diagnosis of IgG4-ROD and orbital MALT lymphoma was established by the integration of clinical, radiographic, histologic, flow cytometric, and molecular genetic analyses, including gene rearrangement. Specifically, the diagnosis of IgG4-ROD was made following the comprehensive diagnostic criteria for IgG4-RD24 and ophthalmology-specific criteria published previously.25 Briefly, the diagnosis of IgG4-ROD was based on three characteristic findings: (1) enlargement of orbit tissue and marked diffuse lymphoplasmacytic infiltrate with either fibrosis or sclerosis, (2) a tissue IgG4/IgG ratio of more than 40% and the mean number of IgG4+ plasma cells in tissue of more than 10/high-power field, and (3) an elevated serum IgG4 level (>135 mg/dL). Definite IgG4-ROD was diagnosed when (1), (2), and (3) were fulfilled, and probably IgG4-ROD when (1) and (2) were fulfilled. Thirty-nine patients were diagnosed with definitive and 1 with probable IgG4-ROD based on the criteria. Conventional histologic and immunohistochemical evaluations were conducted on biopsy tissues fixed in 10% formaldehyde to evaluate features of IgG4-ROD and orbital MALT lymphoma. Immunohistochemical staining was performed using the avidin–biotin–peroxidase technique with the following primary antibodies: monoclonal antibodies to IgG and IgG4. For orbital MALT lymphoma, B-cell monoclonal expansion was confirmed by flow cytometry analysis and immunoglobulin heavy chain (IgH) gene rearrangements using Southern blot analysis (SRL, Tokyo, Japan). Preoperative serum levels of IgG, soluble IL-2 receptor and LDH were also recorded. Patient characteristics and the presence of monoclonality are summarized in Supplementary Table S1 and Table S2. Combined computed tomography scans of the neck, chest, abdomen, and pelvis was performed in all cases, and gallium scintigraphy was performed as needed.
For further investigation of the comprehensive metabolomic analysis results, a validation cohort was studied, comprising 18 patients with IgG4-ROD and 17 patients with orbital MALT lymphoma not included in the original discovery set (Supplementary Tables S3 and S4).
Processing of Tissue Samples
Charged Metabolites
For metabolite extraction, a frozen tissue sample (approximately 4–30 mg) was immersed in methanol (tissue to methanol ratio = 1.00:14.25, w:w) containing zirconia beads (φ 5 mm × 2, φ 3 mm × 4, φ 1.5 mm × 4; Nikkato, Osaka, Japan). The samples were homogenized for six cycles (5000 rpm for 10 seconds and pause for 20 seconds) at 4 °C.
For Positive Mode
The homogenate (10 mL) was mixed with methanol (20 mL) containing 149.6 mM ammonium hydroxide (1%, v/v, ammonia solution) and 0.75 mM internal standards (d8-spermine, d8-spermidine, d6-N1-acetylspermidine, d3-N1-acetylspermine, d6-N1,N8-diacetylspermidine, d6-N1,N12-diacetylspermine, hypoxanthine-13C,15N, 1,6-diaminohexane). Following centrifugation at 15,780 × g for 10 minutes at 4 °C, the whole supernatant was transferred to another tube and vacuum dried. The sample was then reconstituted with 90% methanol (4 mL) and water (12 mL), vortexed, and centrifuged at 15,780×g for 10 minutes at 4 °C. The resulting supernatant was injected into the LC-MS system.
For Negative Mode
The homogenate (10 mL) was mixed with methanol (20 mL) containing internal standard (1.5 mM of camphor-10-sulfonic acid, and 15 mM of sulfanilic acid and methionine sulfone). Following centrifugation at 15,780×g for 10 minutes at 4 °C, the whole supernatant was transferred to another tube and vacuum dried. The sample was then reconstituted with 90% methanol (4 mL) and water (12 mL), vortexed, and centrifuged at 15,780×g for 10 minutes at 4 °C. The resulting supernatant was injected into the LC-MS system.
Lipids
For metabolite extraction, a frozen tissue sample (approximately 4–30 mg) was immersed in methanol (tissue to methanol ratio = 1.00:14.25, w:w) containing zirconia beads (φ 5 mm × 2, φ 3 mm × 4, φ 1.5 mm × 4). The samples were homogenized for six cycles (5000 rpm for 10 seconds and pause for 20 seconds) at 4 °C, and subsequently, 60 µL of the homogenate was transferred to another tube. The homogenate was mixed with 186 µL of chloroform and methanol (80:13, v:v) containing internal standards (6.54 µM of N-laurolyl-d-erythro-sphingosylphosphorylcholine and 1,2-dimyristoyl-sn-glycero-3-phosphocholine, and 65.4 µM etodolic acid) and incubated for 15 minutes at room temperature (20 ± 1 °C). The samples were then mixed with 362 µL of methanol and Milli-Q water (1.828:1.000, v:v) and incubated for 15 minutes at room temperature (20 ± 1 °C). Following centrifugation at 9,000×g for 10 minutes at 4 °C, 250 µL of the supernatant was used for LC time-of-flight (TOF) MS analysis.
Signal Selection in Lipidomics
Samples diluted 1.000, 0.500, 0.250, and 0.125 times were analyzed by LC-TOF-MS, yielding 1771 theoretical m/z values for fatty acids, phospholipids, neutral lipids, and sphingolipids with all possible combinations of fat chains from the metabolites listed in the lipid metabolism category in the Kyoto Encyclopedia of Genes and Genomes ligand database. The peaks were extracted using MassHunter (matching tolerance, 10 ppm; adduct ion, +H, +NH4 [positive] –H, +HCOO [negative], height count, 1000 or more; charge number range, 1–2). The peaks were matched among the four samples based on m/z values and retention times. The results showed that 101 peaks were detected in only 1 of the 4 samples, whereas 69, 74, and 93 peaks were detected in 2, 3, and 4, respectively, of the 4 samples. For peaks with different adduct ions at the same retention time, those with large areas were adopted. For peaks detected in three of the four samples, peaks showing high linearity (R = 0.8; n = 3, 4) among peak areas and dilution rates were selected for subsequent analyses. Peaks detected in two of the four samples were also selected. In total, 209 peaks were analyzed statistically.
LC and MS Systems
The LC system was an Agilent Technologies 1290 Infinity instrument (Agilent Technologies, Santa Clara, CA), consisting of an autosampler, quaternary pump, and column compartment. MS detection was conducted with an Agilent Technologies G6230B TOF mass spectrometer. The samples were analyzed in positive and negative ion modes. Agilent MassHunter Qualitative Analysis software (version B.08.00; Agilent Technologies) was used for data processing.
Data Analysis
Charged Metabolites
The analysis was performed using Agilent MassHunter Qualitative Analysis software. Annotation tables for both positive and negative modes were generated based on the results of the LC-MS analysis of 166 standard compounds, including amino acids, polyamines, organic acids, and several metabolites. Peaks were extracted using MassHunter software according to retention time and m/z values. Peaks out of the range for a retention time of 0.1 minute and an m/z of 50 ppm were integrated manually for area calculation. The relative peak area of each metabolite was calculated by dividing the peak area by the area of internal standard and was quantified using the relative peak areas of external standard compounds.
Lipids
The analysis was performed using an Agilent MassHunter Qualitative Analysis software. Peaks were extracted using MassHunter according to retention time and m/z. Peaks out of the range for a retention time of 0.1 minutes and an m/z of 100 ppm were manually integrated to calculate the area. Relative peak areas for each metabolite were calculated by dividing the peak area by the area of the internal standard.
Statistical Analysis
Differences between lesion tissue and matched adipose tissue were evaluated using Wilcoxon's matched-pair signed-rank test. Differences between lesion tissues from patients with IgG4-ROD and MALT lymphoma were evaluated using Mann-Whitney U tests. A principal component analysis was performed using Z score-normalized metabolite concentrations.
Biomarker Selection and Creation
Candidate metabolite biomarkers were selected from 101 charged metabolites. They were identified and quantified using external standards. The discrimination ability of each metabolite was assessed by the area under the receiver operating characteristic curve (AUC). First, among the metabolites with marked differences in concentration (P < 0.001 and fold change of <0.5 or >2.0), single metabolites with the highest AUC were selected. Second, a random forest model was used to select a combination of multiple metabolites with the best discrimination ability. R software (3.6.2) with the Random Forest package (version 4.6–14; https://cran.r-project.org/web/packages/randomForest/) was used. Boruta package of R software (https://cran.r-project.org/web/packages/Boruta/) was used for statistical selection of features based on importance in the random forest model. For robustness and pruning, we incorporated the metabolites with significant concentration differences and those selected by Boruta into the random forest model. The analysis dataset including 43 patients was randomly divided into a training set and a test set at a 7:3 ratio. The prediction was tested using the test data and repeated 1000 times, and the results of the prediction were used to generate receiver operating characteristic curves.
Using the validation cohort, we validated the five tissue metabolites selected as candidate biomarkers using the discovery set, by comparing the concentrations of those metabolites between IgG4-ROD and orbital MALT lymphoma using Mann–Whitney U tests, and quantified their ability in discriminating between IgG4-ROD and orbital MALT lymphoma using areas under the receiver operating characteristic curves.
Pathway Analysis
Pathway analysis was performed using MetaboAnalyst 4.0.26 The metabolites with a P value of less than 0.05 in the Mann–Whitney U test were used in this analysis. The charged metabolites and the lipids were analyzed independently and the respective analytical results were obtained. Statistical significance was assessed based on the hypergeometric test of overexpression analysis, and pathway topology analysis was performed based on relative betweenness centrality.
MeV (Version 4.9.0, http://mev.tm4.org/; Center for Cancer Computational Biology, Dana-Farber Cancer Institute, Boston, MA), JMP14.1.0 (SAS Institute Inc., Cary, NC), GraphPad Prism (Version 8.1.2; GraphPad Software, San Diego, CA), and R (3.6.2) were used for these analyses.
Results
General Metabolic Profiles of Lesions and Adjacent Adipose Tissue Obtained from Patients with IgG4-ROD and Orbital MALT Lymphoma
Clinical data from individual patients analyzed are summarized in Supplementary Tables S1 and S2. In total, 310 metabolites were identified in surgical biopsy specimens from patients with IgG4-ROD and orbital MALT lymphoma (Supplementary Fig. S1 and Supplementary Tables S5 and S6).
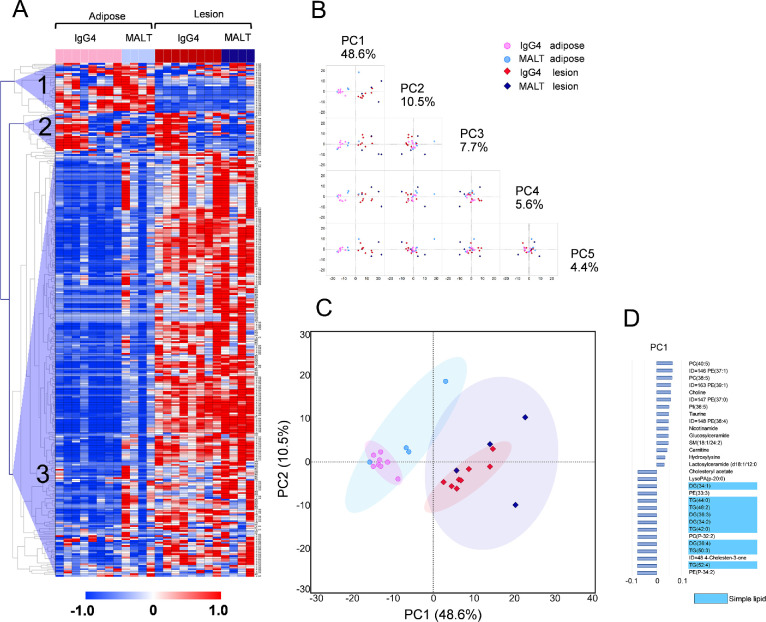
Next, we conducted hierarchical clustering of the 310 metabolites. A heatmap of the results is shown in Figure 1A. A few metabolites tended to be abundant in adipose tissue (cluster 1), whereas the majority tended to be abundant in lesion tissue (cluster 3). Some of the biopsy tissues from patients with IgG4-ROD had a metabolite profile similar to that of normal adipose tissue adjacent to the lesion (cluster 2). Principal component analysis was applied to further analysis of the different metabolic profiles between tissues. Adipose and lesion tissues were separated by principal component 1 (Figs. 1B, C). The nine simple lipids separating adipose and lesion tissues exhibited the characteristic metabolic profile of adipose tissue (Fig. 1D).
Figure 1.
Metabolome profiling of normal adipose tissue and corresponding biopsy specimens from patients with IgG4-ROD and orbital MALT lymphoma. (A) Heatmap of 310 metabolites from biopsy specimens from patients with IgG4-ROD and orbital MALT lymphoma. Column and row represent metabolite and sample, respectively. Metabolite concentrations were normalized to Z scores and vertically clustered by Spearman correlation. The red, blue, and white colors indicate higher, lower, and average values, respectively. The three distinct clusters observed are labeled as 1, 2, and 3. (B, C) The score plot of principal component(PC) analysis. PC 1 shows a natural separation between surgical biopsy tissues from IgG4-ROD and orbital MALT lymphoma, and normal adipose tissue adjacent to the lesion. Each point corresponds to a sample. Shading represents the 95% confidence region. (D) PC loading of the top 15 metabolites explaining the difference in metabolomes based on PC 1.
Targeted Metabolomics Profiles in Lesions from Patients with IgG4-ROD and Orbital MALT Lymphoma
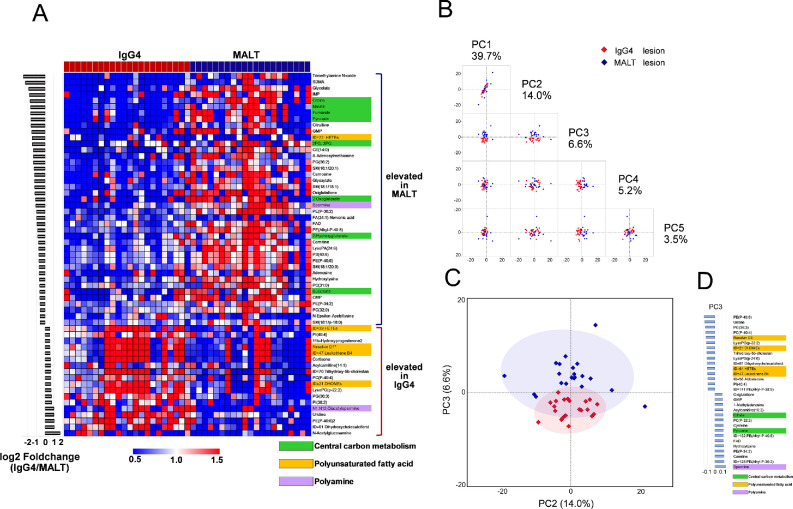
There were significant differences in 94 metabolites between IgG4-ROD and orbital MALT lymphoma, 34 of which (36.2%) were elevated in IgG4-ROD and 60 (63.8%) were elevated in orbital MALT lymphoma (Supplementary Table S7). A principal component analysis and heatmap analysis showed that the tissue metabolites in IgG4-ROD and orbital MALT lymphoma could separate patients with IgG4-ROD from those with orbital MALT lymphoma (Fig. 2), exhibiting distinct tissue metabolomic patterns with almost no overlap between IgG4-ROD and orbital MALT lymphoma. IgG4-ROD and MALT lymphoma were distinguished by principal component 3. The metabolites found in principal component 3 included polyunsaturated fatty acids (PUFAs), metabolites involved in central carbon metabolism, and polyamine (Fig. 2D), consistent with the heatmap results.
Figure 2.
Metabolic profile distinguishing between IgG4-ROD and orbital MALT lymphoma lesions. (A) Heatmap of selected metabolites in biopsy specimens from patients with IgG4-ROD and orbital MALT lymphoma. Fifty-nine metabolites showing both P < 0.05 (Mann–Whitney U tests) and a log 2-fold change of less than –0.5 or greater than 0.5 between IgG4-ROD and MALT lymphoma were used for the heatmap. One column represents one metabolite, and one row represents one sample. The red, blue, and white colors indicate higher, lower, and average values, respectively. Metabolites were sorted from top to bottom according to fold change (IgG4-RD/MALT lymphoma). (B, C) The score plot of the principal component (PC) analysis of biopsied specimens in patients with IgG4-ROD versus orbital MALT lymphoma. The plot containing PC 3 shows clear separation. Each point in the plot corresponds with a sample. Shading represents the 95% confidence region. (D) PC loadings of the top 15 metabolites explaining the differences in metabolomes based on PC 3.
Metabolic Pathway Analysis
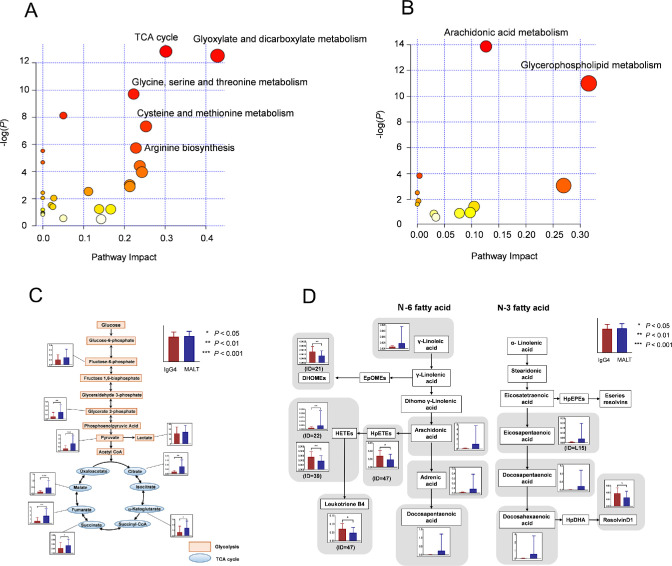
To further explore biological interpretation, we performed a functional pathway analysis using MetaboAnalyst 4.0. The most relevant pathways are shown in Figure 3A and B. A metabolic pathway analysis of 94 metabolites that differed significantly between IgG4-ROD and orbital MALT lymphoma suggested significant changes (false discovery rate of <0.05, pathway impact of >0.1) in the seven most relevant pathways comprising the tricarboxylic acid (TCA) cycle; glyoxylate and dicarboxylate metabolism; glycine, serine, and threonine metabolism; cysteine and methionine metabolism; arginine biosynthesis; arachidonic acid metabolism; and glycerophospholipid metabolism (Table 1). The quantitative data for metabolic pathways (including the TCA cycle and arachidonic acid metabolism) that altered markedly were visualized as a schematic of pathways (Figs. 3C, D). In MALT lymphoma, increases were observed in the intermediates of the glycolytic pathway and the TCA cycle (Fig. 3C). In the PUFA metabolic pathway including arachidonic acid metabolism, increases of four n-6PUFAs (12,13-dihydroxy-9-octadecenoic acid, hydroxyeicosatetraenoic acid, hydroperoxyeicosatetraenoic acid, and leukotriene B4) and one n-3PUFA (resolvin D1) were confirmed in IgG4-ROD (Fig. 3D).
Figure 3.
Differential metabolic pathways between IgG4-ROD and MALT lymphoma. (A) Metabolic pathways with significant differences between samples from IgG4-ROD and orbital MALT lymphoma using the MetaboAnalyst. (B) Lipid metabolism pathways with significant differences. The colors (varying from yellow to red) indicate metabolites with different levels of significance, with red being more significant than yellow. Differential metabolic pathways (false discovery rate [FDR] of <0.05 and a pathway impact of >0.1) are annotated. (C, D) Metabolome pathway map of the quantified metabolites, including the components of the metabolic pathways showing marked fluctuations. The TCA cycle and arachidonic acid metabolism with high statistical significance scores were visualized along with the associated metabolic pathways. Column represents average level of tissue metabolites; bars denote standard deviation. (C) TCA cycle and glycolysis. The unit for the y-axis of the figure is micromoles. (D) PUFA metabolism, including arachidonic acid metabolism. The y-axis in the figure shows relative quantitative value and has no unit.
Table 1.
Pathways Significantly Different Between IgG4-ROD and Ocular MALT Lymphoma (P < 0.05)
| Pathway Name | Match Status* | P Value | -log (p) | False Discovery Rate | Impact |
|---|---|---|---|---|---|
| Metabolic pathways with significant differences | |||||
| Citrate cycle (TCA cycle) | 6/20 | 2.63E-06 | 1.29E+01 | 1.51E-04 | 3.02E-01 |
| Glyoxylate and dicarboxylate metabolism | 7/32 | 3.60E-06 | 1.25E+01 | 1.51E-04 | 4.29E-01 |
| Glycine, serine, and threonine metabolism | 6/33 | 6.07E-05 | 9.70 | 1.70E-03 | 2.23E-01 |
| Alanine, aspartate, and glutamate metabolism | 5/28 | 2.96E-04 | 8.12 | 6.21E-03 | 5.05E-02 |
| Cysteine and methionine metabolism | 5/33 | 6.58E-04 | 7.32 | 1.11E-02 | 2.53E-01 |
| Arginine biosynthesis | 3/14 | 3.24E-03 | 5.73 | 4.54E-02 | 2.28E-01 |
| Butanoate metabolism | 3/15 | 3.99E-03 | 5.52 | 4.78E-02 | 0 |
| Arginine and proline metabolism | 4/38 | 9.33E-03 | 4.67 | 9.79E-02 | 0 |
| Pyruvate metabolism | 3/22 | 1.21E-02 | 4.41 | 1.13E-01 | 2.38E-01 |
| Glycolysis/gluconeogenesis | 3/26 | 1.92E-02 | 3.95 | 1.61E-01 | 2.43E-01 |
| Histidine metabolism | 2/16 | 4.88E-02 | 3.02 | 3.72E-01 | 2.13E-01 |
| Lipid metabolism pathways with significant differences | |||||
| Arachidonic acid metabolism | 7/36 | 9.52E-07 | 1.39E+01 | 7.99E-05 | 1.27E-01 |
| Glycerophospholipid metabolism | 6/36 | 1.68E-05 | 1.10E+01 | 7.05E-04 | 3.16E-01 |
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 2/14 | 2.18E-02 | 3.82 | 6.09E-01 | 3.99E-03 |
| Sphingolipid metabolism | 2/21 | 4.67E-02 | 3.06 | 9.82E-01 | 2.70E-01 |
The denominator represents the total number of metabolites involved in the metabolic pathway, and the numerator represents the number of metabolites with a significant difference.
Targeted Analysis
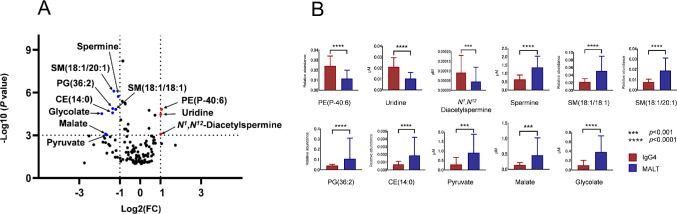
Volcano plots were generated to display differentially abundant metabolites using both fold change (<0.5 or >2.0) and P values from Mann–Whitney U tests (P < 0.001) as the criteria. We identified the top 11 metabolites expressing different concentrations in IgG4-ROD versus orbital MALT lymphoma biopsy tissues: 3 were upregulated and 8 downregulated (Fig. 4, Table 2). The diagnostic values of the potential metabolic biomarkers were assessed by AUC. Following quantification, IgG4-ROD biopsy samples had significantly increased levels of PE(P-40:6), uridine, and N1,N12-diacetylspermine, whereas orbital MALT lymphoma biopsy samples had significantly increased levels of spermine, SM(18:1/18:1), SM(18:1/20:1), PG(36:2), CE(14:0), pyruvate, malate, and glycolate. Interestingly, IgG4-ROD showed a significantly higher level of N1,N12-diacetylspermine, whereas orbital MALT lymphoma showed a significantly higher level of spermine (Fig. 4B).
Figure 4.
Eleven metabolites selected according to marked differences in concentration. (A) Volcano plot of differentially expressed metabolites in IgG4-ROD and MALT lymphoma. Only the top 11 significant metabolites are labeled and are clearly distinguished from the other metabolites. The fold change (<0.5 or >2.0) and P value from the Mann–Whitney U test (P < 0.001) were used as the cutoff. (B) A bar graph of the selected 11 metabolites. Column represents average level of tissue metabolite; bars denote standard deviation. The y-axis in the lipid figure shows relative quantitation value and has no units. Abbreviations: CE, cholesterol ester; SM, sphingomyelin; PG, glycerophospholipid.
Table 2.
Statistics for the Top 11 Metabolites
| IgG4 vs MALT | ||||
|---|---|---|---|---|
| Mann-Whitney U Test | Fold Change | AUC | ||
| Compound Name | P Value | IgG4/MALT | Area | 95% CI |
| PE(P-40:6) | 0.0000152 | 2.078 | 0.864 | 0.741–0.986 |
| Uridine | 0.0000339 | 2.018 | 0.851 | 0.734–0.967 |
| N 1, N12-Diacetylspermine | 0.0008250 | 2.016 | 0.790 | 0.640–0.940 |
| Spermine | 0.0000019 | 0.473 | 0.894 | 0.804–0.984 |
| SM(18:1/18:1) | 0.0000152 | 0.435 | 0.864 | 0.743–0.984 |
| SM(18:1/20:1) | 0.0000008 | 0.407 | 0.905 | 0.809–1.000 |
| PG(36:2) | 0.0000132 | 0.391 | 0.866 | 0.750–0.982 |
| CE(14:0) | 0.0000262 | 0.390 | 0.855 | 0.735–0.975 |
| Pyruvate | 0.0009103 | 0.319 | 0.788 | 0.651–0.924 |
| Malate | 0.0008250 | 0.307 | 0.790 | 0.643–0.937 |
| Glycolate | 0.0000299 | 0.269 | 0.853 | 0.733–0.973 |
Biomarker Signature Discriminating IgG4-ROD From MALT Lymphoma
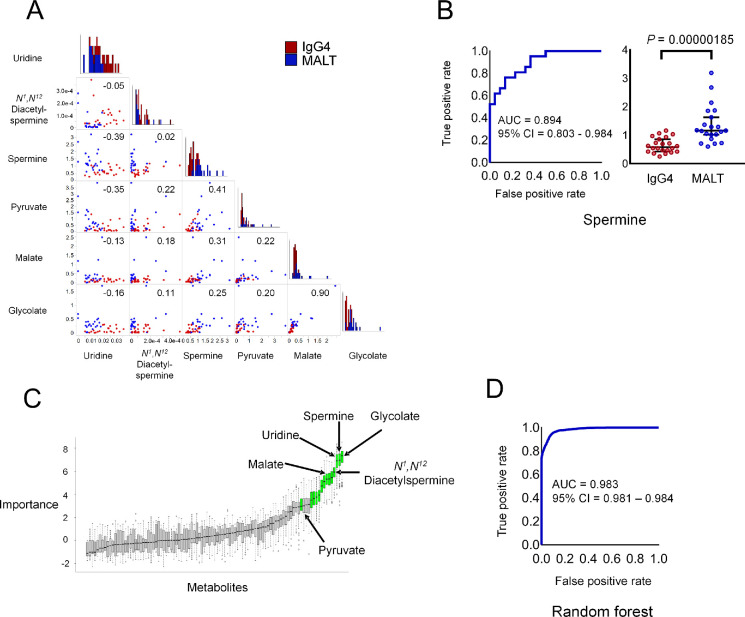
To identify the most important metabolites and test if the metabolome profile of the IgG4-ROD biopsy specimens could be differentiated from that of orbital MALT lymphoma, we attempted to identify biomarkers among the 101 metabolites detected in this study. The candidate simple biomarkers (Fig. 5A) were selected among metabolites with marked differences in abundance (Fig. 4). Among the six biomarker candidates selected, the AUC of spermine was the largest (AUC = 0.894; 95% confidence interval [CI], 0.803–0.984) (Fig. 5B), suggesting its potential as the best predictor. These results indicate that spermine is a potential biomarker for differentiating between IgG4-ROD and orbital MALT lymphoma.
Figure 5.
Discrimination ability of metabolite biomarkers and variable importance in random forest models. (A) Of the 11 metabolites with marked differences in concentration, 6 metabolites measured by external standards were used as candidate biomarkers. A scatterplot matrix is used to visualize the correlation between two metabolites. The correlation coefficient is shown in the upper right. A histogram showing the distribution of the concentration level of each metabolite is shown on the diagonal line. (B) Receiver operating characteristic (ROC) curve analysis of predictive power to distinguish patients with IgG4-ROD from those with orbital MALT lymphoma. The best metabolite that distinguishes between the two conditions is spermine. (C) Metabolites selected by Boruta from 101 metabolites measured by external standards. Box plots show the importance of each metabolite calculated during the selection process. Metabolites with confirmed significance are displayed in light green. The significance of 13 metabolites was confirmed. The six metabolites shown in (A) are indicated by arrows. Only pyruvate was not confirmed to be significant, and the other five metabolites ranked high in importance and were confirmed to be significant. We created a random forest model incorporating these five metabolites. (D) ROC curve of random forest model with five metabolites.
We identified the important combination of metabolites for predicting IgG4-ROD and orbital MALT lymphoma using a random forest model. In variable selection for the random forest model, glycolate, spermine, uridine, N1,N12-diacetylspermine, and malate ranked high with significant variable importance. Pyruvate ranked 16th and was not significant; thus, pyruvate was excluded from the model (Fig. 5C, Supplementary Table S8). The random forest model incorporating the five selected metabolites showed the highest AUC value in this study (AUC = 0.983; 95% CI, 0.981–0.984) (Fig. 5D).
Validation of Candidate Metabolites
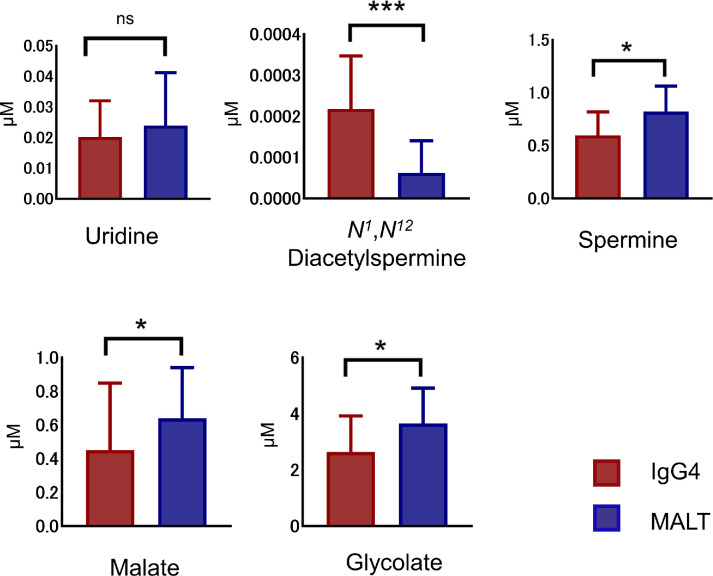
To rule out false-positive results of the 101 charged metabolites detected using the discovery dataset, we performed a validation analysis to identify the metabolites capable of distinguishing IgG4-ROD and orbital MALT lymphoma biopsy specimens by using a set of samples not included in the selection cohort. Specifically, we selected and used new biopsy specimens from 18 patients with IgG4-ROD and 17 patients with orbital MALT lymphoma. Among the five candidate tissue metabolites selected by the random forest model, N1,N12-diacetylspermine was significantly higher (P = 0.000113) in IgG4-ROD tissues, whereas spermine (P = 0.0209), malate (P = 0.0349), and glycolate (P = 0.0357) were significantly higher in orbital MALT lymphoma tissues, and only uridine was not significant different (P = 0.684) (Fig. 6). Validation of N1,N12-diacetylspermine, spermine, malate, and glycolate as biomarkers to discriminate between IgG4-ROD and orbital MALT lymphoma yielded AUCs of 0.863 (95% CI, 0.734–0.991), 0.727 (95% CI, 0.560–0.894), 0.709 (95% CI, 0.528–0.891), and 0.708 (95% CI, 0.532–0.883), respectively, indicating good performance in prediction. These results suggest that the four tissue metabolites (N1,N12-diacetylspermine, spermine, malate, and glycolate) are potential diagnostic biomarkers to discriminate between IgG4-ROD and orbital MALT lymphoma.
Figure 6.
The five most elevated tissue metabolites incorporated by random forest model in the validation cohort. The results show that four of the five metabolites; N1,N12-diacetylspermine, spermine, malate and glycolate, are consistent with untargeted groups, are significant different between IgG4-ROD and orbital MALT lymphoma. ***P < 0.001, *P < 0.05. ns, not significant.
Discussion
Because the diagnoses of IgG4-ROD and orbital MALT lymphoma are sometimes challenging and the pathophysiology of these disorders remain unclear, there is a need for novel biomarkers to improve diagnostic reliability and to better understand the metabolomic mechanisms of these orbital lymphoproliferative disorders. In this study, we performed a comprehensive measurement of charged metabolites and lipids using LC-TOF-MS and showed distinct tissue metabolomic patterns between IgG4-ROD and orbital MALT lymphoma. Despite exhibiting clinical and radiologic similarities, distinct differences in metabolomic profiles were observed between the two diseases, probably owing to the infiltration of inflammatory cells in the IgG4-ROD lesions and the proliferation of malignant B cells in the orbital MALT lymphomas. First, from the discovery set, we detected 310 metabolites from IgG4-ROD lesions and orbital MALT lymphomas and identified seven metabolic pathways that exhibited reprogramming. Based on statistical models, a panel of five metabolites, namely, N1,N12-diacetylspermine, glycolate, malate, uridine, and spermine, as potential tissue biomarkers was screened for diagnosis of IgG4-ROD and orbital MALT lymphoma and the panel seemed to have diagnostic value for both lymphoproliferative disorders with high AUCs. The AUC of the optimized model was 0.983 (95% CI, 0.981–0.984), indicating a high potential for discrimination between the two disorders. When these five candidate tissue metabolites selected by the random forest model were subjected to validation study using an independent validation set, N1,N12-diacetylspermine, spermine, malate, and glycolate were significantly different between IgG4-ROD and orbital MALT lymphoma, and only uridine was not significant different. The reason why uridine was a significant discriminator in the discovery set and not in the validation set was probably due to technical issues. (1) The uridine peak was observed at a relatively early retention time in our measurement protocol, with various peaks observed close to uridine (Supplementary Fig. S2A and B). (2) The S/N value of uridine peak was 3.8, which was slightly larger than the lower quantification limit S/N of 3 (Supplementary Fig. S2C and D). These issues may lead to an error or a variation in the measured concentrations.
Among the five candidate tissue metabolites selected by the random forest model, N1,N12-diacetylspermine was significantly higher (P = 0.000113) in IgG4-ROD tissues, and spermine (P = 0.0209), malate (P = 0.0349), and glycolate (P = 0.0357) were significantly higher in orbital MALT lymphoma tissues, and only uridine was not significant different (P = 0.684) (Fig. 6).
The IgG4-ROD lesions were associated with low levels of spermine and high levels of N1,N12-diacetylspermine. Spermine is one of the polyamine metabolites that is synthesized from ornithine by activating ornithine decarboxylase and acetylation of these metabolites are driven by spermidine/spermine N1-acetyltransferase, resulting in the production of N1,N12-diacetylspermine. The activation of these reactions was frequently observed in cancer cells27 and the latter reaction might be more activated in our IgG4-ROD samples based on the balance of these metabolites. The higher concentrations in orbital MALT of malate and glycolate, which belongs to the TCA cycle, may indicate the different activity of energy production via oxidative phosphorylation.
However, the mechanisms underlying these differences are still unclear. The observed increases in metabolite levels may be related to the infiltration of inflammatory cells in IgG4-ROD lesions and the proliferation of malignant B cells in orbital MALT lymphomas. For the first time, we identified four biopsy-based biomarkers, namely, N1,N12-diacetylspermine, spermine, glycolate, and malate, with a very high accuracy for discrimination between IgG4-ROD and orbital MALT lymphoma.
Spermine, which is produced during the biosynthesis of polyamines, is a ubiquitous small polycation with various functions, mostly linked with protein synthesis, proliferation, cell growth, and survival, which are essential for cancer growth.28,29 Spermine has been shown to be a negative modulator of B-cell antigen receptor-mediated apoptosis in lymphoma cell lines.30,31 Our findings were consistent with the roles of spermine in the pathogenesis of MALT lymphoma. A similar tendency was also observed with the TCA cycle in MALT lymphoma. We demonstrated a significant accumulation of TCA cycle intermediates, including malate, citrate, α-hydroxyglutaric acid, succinate, and fumarate, which is consistent with the observation that certain tumor cells rely heavily on the TCA cycle for energy production and macromolecule synthesis.32,33
A significant increase in PUFAs was observed in IgG4-ROD tissues. PUFAs are representative lipid mediators involved in immune regulation, and phenotypic changes in these pathways may play important roles in the termination of inflammatory processes.34 Study has shown that n-6PUFAs enhance acute inflammation and regulate the maintenance of chronic inflammation.35 In contrast, the n-3PUFA resolvin D1, which was elevated in IgG4-ROD in this study, is known to have strong anti-inflammatory effects.34,36,37 This metabolite acts on immune cells to promote differentiation into regulatory T cells,38 M2 macrophages,39 and plasma cells, resulting in the promotion of IgG secretion.40 Moreover, resolvin facilitates resolution of Th2-mediated asthmatic airway inflammation and atopic dermatitis by reducing Th2 cytokine levels.41,42 Numerous studies have reported that Th2 cells, M2 macrophages, and regulatory T cells that produce IL-4, IL-5, IL-10, IL-13, and TGF-β may contribute to the characteristic pathologic features of IgG4-RD, including IgG4-ROD, such as the proliferation and activation of B cells and plasmablasts and the upregulation of IgG4 class switch recombination; thus, resolvin may also be involved in regulating chronic inflammation, possibly via acting on cells of adaptive immunity in IgG4-ROD. Accordingly, the effects of n-3 and n-6PUFAs could explain these immunologic characteristics and may suggest that these fatty acids could act as important mediators in the pathogenesis of IgG4-ROD.
In contrast, the predominantly Th1 immune response and increased of Th1-type cytokines, such as IFN-γ and TNF-α, have been reported in patients with Helicobacter pylori–induced gastric MALT lymphoma.43,44 Recently, it has been reported that spermine downregulated all Th1 and Th2 cytokines in a dose-dependent manner in a certain conditions.45 The association between spermine or other metabolites and Th1/Th2 balance is largely unknown, and further studies are needed to clarify the association.
In this study, we demonstrated that arachidonic acid metabolism in IgG4-ROD specimens differed markedly from that in orbital MALT lymphoma. Arachidonic acid is a free fatty acid that, when liberated from cell membranes, including membranes of infiltrated inflammatory cells, can be metabolized by cytochrome P450 mono-oxygenases, lipoxygenases, and cyclo-oxygenases to form biologically active products with the ability to regulate intracellular signaling pathways and gene transcription, but also serve as precursors for lipid mediators involved in inflammation.46–48 Thus, arachidonic acid and arachidonic acid metabolites affect the ability of dendritic cells to induce T-cell proliferation and activation and to induce Foxp3-positive regulatory T cells.49 Glycerophospholipid metabolism, which was also active in IgG4-ROD, is known to overlap with arachidonic acid metabolism, and these metabolic pathways account for many messenger metabolites essential to immune signaling.50,51 Interestingly, arachidonic acid metabolites also play key roles in lung, renal, and cardiac fibrosis.52–54 Regulatory T cells producing IL-10 and TGF-β contribute to the enhancement of IgG4 class switch recombination and the pathogenesis of IgG4-RD via B cells, as well as induction of fibrosis.55 Regulatory T cells and other inflammatory cells may also contribute to fibrosis in IgG4-ROD through arachidonic acid metabolism.
There were several limitations to this study. First, in lipidomics, we used retention times and m/z values for identification. Second, the number of samples was limited. Thus, to verify the accuracy of the metabolomic profiles, studies with larger cohorts are required. Additionally, comparisons with other benign lymphoproliferative diseases that occur in the orbit, such as reactive lymphoid hyperplasia, may contribute to identifying more disease-specific metabolic changes. Third, we only used biopsy tissues. Because blood samples can be collected in a noninvasive manner and could also reflect systemic metabolic changes, verification of biomarkers in blood samples should be performed. Nonetheless, to the best of our knowledge, this study is the first and the most comprehensive of the integrative profiles of both charged metabolites and lipids from biopsy specimens from patients with IgG4-ROD and orbital MALT lymphoma.
Collectively, our results supported the important roles of several metabolic pathways in orbital lymphoproliferative disorders. We identified validated four metabolites, namely, N1,N12-diacetylspermine, spermine, malate, and glycolate, as differentially abundant in the two disorders. These different metabolic features profiles improved our detailed understanding of the pathophysiologic processes underpinning these diseases and may facilitate the discovery of biomarkers and new therapeutic strategies for future clinical application. Further investigations using more samples from numerous institutions are required to validate these findings.
Supplementary Material
Acknowledgments
The authors thank the patients who participated in this study. We also thank Teresa Nakatani for editorial assistance.
Supported in part by a Grand-in-Aid for Scientific Research (C) 16K11330, 19K09981, and 19K09959 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Disclosure: H. Shimizu, None; Y. Usui, None; R. Wakita, None; Y. Aita, None; A. Tomita, None; K. Tsubota, None; M. Asakage, None; N. Nezu, None; H. Komatsu, None; K. Umazume, None; M. Sugimoto, None; H. Goto, None
References
- 1. Japanese study group of Ig Grod. A prevalence study of IgG4-related ophthalmic disease in Japan. Jpn J Ophthalmol. 2013; 57: 573–579. [DOI] [PubMed] [Google Scholar]
- 2. Kamisawa T, Zen Y, Pillai S, Stone JH.. IgG4-related disease. Lancet. 2015; 385: 1460–1471. [DOI] [PubMed] [Google Scholar]
- 3. Zucca E, Bertoni F.. The spectrum of MALT lymphoma at different sites: biological and therapeutic relevance. Blood. 2016; 127: 2082–2092. [DOI] [PubMed] [Google Scholar]
- 4. Wallace ZS, Deshpande V, Mattoo H, et al.. IgG4-related disease: clinical and laboratory features in one hundred twenty-five patients. Arthritis Rheumatol. 2015; 67: 2466–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Usui Y, Rao NA, Takase H, et al.. Comprehensive polymerase chain reaction assay for detection of pathogenic DNA in lymphoproliferative disorders of the ocular adnexa. Sci Rep. 2016; 6: 36621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao PX, Adzavon YM, Ma JM, et al.. IgG4 and IgE co-positive group found in idiopathic orbital inflammatory disease. Int J Ophthalmol. 2018; 11: 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stefanovic A, Lossos IS.. Extranodal marginal zone lymphoma of the ocular adnexa. Blood. 2009; 114: 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haradome K, Haradome H, Usui Y, et al.. Orbital lymphoproliferative disorders (OLPDs): value of MR imaging for differentiating orbital lymphoma from benign OPLDs. AJNR Am J Neuroradiol. 2014; 35: 1976–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verdijk RM. Lymphoproliferative tumors of the ocular adnexa. Asia Pac J Ophthalmol (Phila). 2017; 6: 132–142. [DOI] [PubMed] [Google Scholar]
- 10. Kubota T, Moritani S, Yoshino T, Nagai H, Terasaki H.. Ocular adnexal marginal zone B cell lymphoma infiltrated by IgG4-positive plasma cells. J Clin Pathol. 2010; 63: 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheuk W, Yuen HK, Chan AC, et al.. Ocular adnexal lymphoma associated with IgG4+ chronic sclerosing dacryoadenitis: a previously undescribed complication of IgG4-related sclerosing disease. Am J Surg Pathol. 2008; 32: 1159–1167. [DOI] [PubMed] [Google Scholar]
- 12. Sato Y, Ohshima K, Ichimura K, et al.. Ocular adnexal IgG4-related disease has uniform clinicopathology. Pathol Int. 2008; 58: 465–470. [DOI] [PubMed] [Google Scholar]
- 13. Culver EL, Sadler R, Bateman AC, et al.. Increases in IgE, eosinophils, and mast cells can be used in diagnosis and to predict relapse of IgG4-related disease. Clin Gastroenterol Hepatol. 2017; 15: 1444–1452. e1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Culver EL, Sadler R, Simpson D, et al.. Elevated serum IgG4 levels in diagnosis, treatment response, organ involvement, and relapse in a prospective IgG4-related disease UK cohort. Am J Gastroenterol. 2016; 111: 733–743. [DOI] [PubMed] [Google Scholar]
- 15. Hong JW, Kang S, Song MK, Ahn CJ, Sa HS.. Clinicoserological factors associated with response to steroid treatment and recurrence in patients with IgG4-related ophthalmic disease. Br J Ophthalmol. 2018; 102: 1591–1595. [DOI] [PubMed] [Google Scholar]
- 16. Wallace ZS, Deshpande V, Stone JH.. Ophthalmic manifestations of IgG4-related disease: single-center experience and literature review. Semin Arthritis Rheum. 2014; 43: 806–817. [DOI] [PubMed] [Google Scholar]
- 17. Guma M, Tiziani S, Firestein GS.. Metabolomics in rheumatic diseases: desperately seeking biomarkers. Nat Rev Rheumatol. 2016; 12: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaushik AK, DeBerardinis RJ.. Applications of metabolomics to study cancer metabolism. Biochim Biophys Acta Rev Cancer. 2018; 1870: 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang P, Gong Y, Liu Z, et al.. Efficacy and safety of iguratimod plus corticosteroid as bridge therapy in treating mild IgG4-related diseases: a prospective clinical trial. Int J Rheum Dis. 2019; 22: 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fuse S, Sugimoto M, Kurosawa Y, et al.. Relationships between plasma lipidomic profiles and brown adipose tissue density in humans. Int J Obes (Lond). 2020; 44: 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishikawa S, Sugimoto M, Edamatsu K, Sugano A, Kitabatake K, Iino M.. Discrimination of oral squamous cell carcinoma from oral lichen planus by salivary metabolomics. Oral Dis. 2020; 26: 35–42. [DOI] [PubMed] [Google Scholar]
- 22. Nakajima T, Katsumata K, Kuwabara H, et al.. Urinary polyamine biomarker panels with machine-learning differentiated colorectal cancers, benign disease, and healthy controls. Int J Mol Sci. 2018; 19: 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Itoi T, Sugimoto M, Umeda J, et al.. Serum metabolomic profiles for human pancreatic cancer discrimination. Int J Mol Sci. 2017; 18: 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Umehara H, Okazaki K, Masaki Y, et al.. Comprehensive diagnostic criteria for IgG4-related disease (IgG4-RD), 2011. Mod Rheumatol. 2012; 22: 21–30. [DOI] [PubMed] [Google Scholar]
- 25. Goto H, Takahira M, Azumi A, Japanese Study Group for Ig GROD. Diagnostic criteria for IgG4-related ophthalmic disease. Jpn J Ophthalmol. 2015; 59: 1–7. [DOI] [PubMed] [Google Scholar]
- 26. Chong J, Wishart DS, Xia J.. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics. 2019; 68: e86. [DOI] [PubMed] [Google Scholar]
- 27. Casero RA Jr., Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer. 2018; 18: 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas T, Thomas TJ.. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol Life Sci. 2001; 58: 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pegg AE. Functions of polyamines in mammals. J Biol Chem. 2016; 291: 14904–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi M, Gan YJ, Davis TO, Scott RS.. Downregulation of the polyamine regulator spermidine/spermine N(1)-acetyltransferase by Epstein-Barr virus in a Burkitt's lymphoma cell line. Virus Res. 2013; 177: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nitta T, Igarashi K, Yamashita A, Yamamoto M, Yamamoto N.. Involvement of polyamines in B cell receptor-mediated apoptosis: spermine functions as a negative modulator. Exp Cell Res. 2001; 265: 174–183. [DOI] [PubMed] [Google Scholar]
- 32. Anderson NM, Mucka P, Kern JG, Feng H.. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018; 9: 216–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen JQ, Russo J.. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim Biophys Acta. 2012; 1826: 370–384. [DOI] [PubMed] [Google Scholar]
- 34. Serhan CN, Chiang N, Van Dyke TE.. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008; 8: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aoki T, Narumiya S.. Prostaglandins and chronic inflammation. Trends Pharmacol Sci. 2012; 33: 304–311. [DOI] [PubMed] [Google Scholar]
- 36. Schwab JM, Chiang N, Arita M, Serhan CN.. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 2007; 447: 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014; 510: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chiurchiu V, Leuti A, Dalli J, et al.. Proresolving lipid mediators resolvin D1, resolvin D2, and maresin 1 are critical in modulating T cell responses. Sci Transl Med. 2016; 8: 353ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Titos E, Rius B, Gonzalez-Periz A, et al.. Resolvin D1 and its precursor docosahexaenoic acid promote resolution of adipose tissue inflammation by eliciting macrophage polarization toward an M2-like phenotype. J Immunol. 2011; 187: 5408–5418. [DOI] [PubMed] [Google Scholar]
- 40. Ramon S, Gao F, Serhan CN, Phipps RP.. Specialized proresolving mediators enhance human B cell differentiation to antibody-secreting cells. J Immunol. 2012; 189: 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aoki H, Hisada T, Ishizuka T, et al.. Resolvin E1 dampens airway inflammation and hyperresponsiveness in a murine model of asthma. Biochem Biophys Res Commun. 2008; 367: 509–515. [DOI] [PubMed] [Google Scholar]
- 42. Aoki H, Hisada T, Ishizuka T, et al.. Protective effect of resolvin E1 on the development of asthmatic airway inflammation. Biochem Biophys Res Commun. 2010; 400: 128–133. [DOI] [PubMed] [Google Scholar]
- 43. Riedel S, Kraft M, Kucharzik T, et al.. CD4+ Th1-cells predominate in low-grade B-cell lymphoma of gastric mucosa-associated lymphoid tissue (MALT type). Scand J Gastroenterol. 2001; 36: 1198–1203. [DOI] [PubMed] [Google Scholar]
- 44. Jafarzadeh A, Larussa T, Nemati M, Jalapour S.. T cell subsets play an important role in the determination of the clinical outcome of Helicobacter pylori infection. Microb Pathog. 2018; 116: 227–236. [DOI] [PubMed] [Google Scholar]
- 45. Fischer M, Ruhnau J, Schulze J, Obst D, Floel A, Vogelgesang A.. Spermine and spermidine modulate T-cell function in older adults with and without cognitive decline ex vivo. Aging (Albany NY). 2020; 12: 13716–13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brash AR. Arachidonic acid as a bioactive molecule. J Clin Invest. 2001; 107: 1339–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dennis EA, Norris PC.. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015; 15: 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chilton FH, Surette ME, Winkler JD.. Arachidonate-phospholipid remodeling and cell proliferation. Adv Exp Med Biol. 1996; 416: 169–172. [PubMed] [Google Scholar]
- 49. Carlsson JA, Wold AE, Sandberg AS, Ostman SM.. The polyunsaturated fatty acids arachidonic acid and docosahexaenoic acid induce mouse dendritic cells maturation but reduce T-cell responses in vitro. PLoS One. 2015; 10: e0143741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hishikawa D, Hashidate T, Shimizu T, Shindou H.. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J Lipid Res. 2014; 55: 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robichaud PP, Boulay K, Munganyiki JE, Surette ME.. Fatty acid remodeling in cellular glycerophospholipids following the activation of human T cells. J Lipid Res. 2013; 54: 2665–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Charbeneau RP, Peters-Golden M.. Eicosanoids: mediators and therapeutic targets in fibrotic lung disease. Clin Sci (Lond). 2005; 108: 479–491. [DOI] [PubMed] [Google Scholar]
- 53. Nasrallah R, Hassouneh R, Hebert RL.. Chronic kidney disease: targeting prostaglandin E2 receptors. Am J Physiol Renal Physiol. 2014; 307: F243–250. [DOI] [PubMed] [Google Scholar]
- 54. Levick SP, Loch DC, Taylor SM, Janicki JS.. Arachidonic acid metabolism as a potential mediator of cardiac fibrosis associated with inflammation. J Immunol. 2007; 178: 641–646. [DOI] [PubMed] [Google Scholar]
- 55. Pillai S, Perugino C, Kaneko N.. Immune mechanisms of fibrosis and inflammation in IgG4-related disease. Curr Opin Rheumatol. 2020; 32: 146–151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.