SUMMARY
Stress granules (SGs) are cytoplasmic assemblies of proteins and non-translating mRNAs. Whereas much has been learned about SG formation, a major gap remains in understanding the compositional changes SGs undergo during normal disassembly and under disease conditions. Here, we address this gap by proteomic dissection of the SG temporal disassembly sequence using multi-bait APEX proximity proteomics. We discover 109 novel SG proteins and characterize distinct SG substructures. We reveal dozens of disassembly-engaged proteins (DEPs), some of which play functional roles in SG disassembly, including small ubiquitin-like modifier (SUMO) conjugating enzymes. We further demonstrate that SUMOylation regulates SG disassembly and SG formation. Parallel proteomics with amyotrophic lateral sclerosis (ALS)-associated C9ORF72 dipeptides uncovered attenuated DEP recruitment during SG disassembly and impaired SUMOylation. Accordingly, SUMO activity ameliorated C9ORF72-ALS-related neurodegeneration in Drosophila. By dissecting the SG spatiotemporal proteomic landscape, we provide an in-depth resource for future work on SG function and reveal basic and disease-relevant mechanisms of SG disassembly.
Graphical Abstract
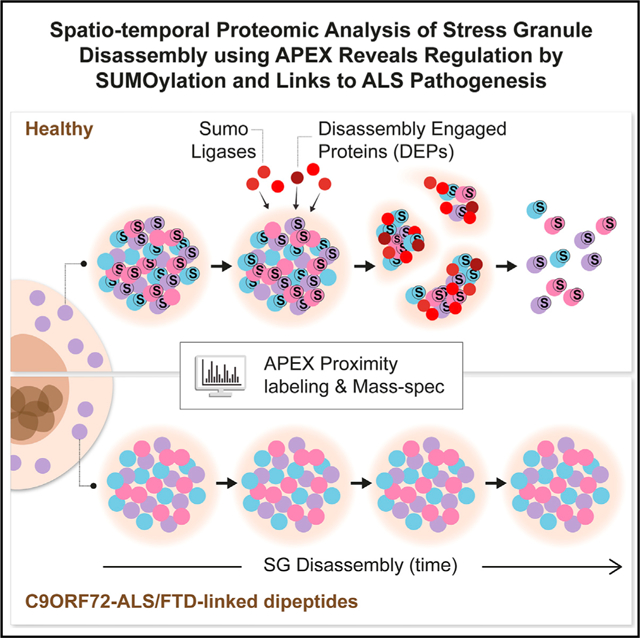
In Brief
Marmor-Kollet et al. utilize proximity proteomics to identify stress granule composition, internal organization, and mechanisms of regulated disassembly in health and disease. Disassembly-engaged proteins (DEPs), including SUMO-conjugating enzymes, are critical for normal stress granule disassembly and dysregulated in ALS-like conditions.
INTRODUCTION
Stress granules (SGs) are transient cytoplasmic membraneless condensates composed of ribonucleoproteins (RNPs) (Protter and Parker, 2016a). SGs are formed through liquid-liquid phase separation of non-translating mRNAs and RNA-binding proteins (Deniz, 2020; Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020) in response to a variety of cellular stresses and dissolve upon return to normal growth conditions (Anderson and Kedersha, 2008; Buchan and Parker, 2009; Decker and Parker, 2012). The study of SG composition, as a means of gaining insight into their function, was first explored by biochemical fractionation and functional RNAi screens, which revealed hundreds of candidate SG proteins (Jain et al., 2016; Ohn et al., 2008). More recently, proximity labeling proteomic analyses of SGs uncovered cell-type- and stress-specific SG composition (Markmiller et al., 2018; Youn et al., 2018).
Although the precise functions of SGs remain to be determined, they are thought to play a protective role during cellular stress. However, it is hypothesized that persistent and abnormal SGs may nucleate insoluble aggregates that are associated with human neurodegenerative diseases (Chew et al., 2019; Cook and Petrucelli, 2019; Li et al., 2013; Todd et al., 2020), such as amyotrophic lateral sclerosis (ALS), a neurodegenerative disease of the human motor neuron system (Alberti and Dormann, 2019; Shorter, 2019; Taylor et al., 2016; Wolozin and Ivanov, 2019).
Thus, RNA-binding proteins that are residents of SGs, such as TDP-43, HNRNPA2/B1, and FUS, are encoded by genes that are mutated in different forms of ALS and are also found in neuropathological inclusions in the brain and spinal cord of ALS patients (Li et al., 2013; Molliex et al., 2015; Ramaswami et al., 2013). In addition, the most common genetic form of ALS and frontotemporal degeneration (FTD) is C9ORF72 disease, which is characterized by repeat associated non-ATG (RAN) translation of C9ORF72-ALS/FTD-linked dipeptides, has been shown to affect SG formation (Boeynaems et al., 2017; Freibaum and Taylor, 2017).
Intrinsically disordered regions (IDRs) underlie RNA-binding proteins’ capacity to phase separate and condense within SG. Accordingly, ALS-causing mutations are prevalent in IDRs and are thought to alter phase-separation propensities and SG dynamics (Boeynaems et al., 2017; Deniz, 2020; Dormann et al., 2010; Elden et al., 2010; Guillén-Boixet et al., 2020; Hofweber et al., 2018; Kim et al., 2013; Lee et al., 2016; Molliex et al., 2015; Patel et al., 2015; Sanders et al., 2020; Wolozin and Ivanov, 2019; Yang et al., 2020). Moreover, post-translational modifications (PTMs), which show a preference toward IDRs (Bah and Forman-Kay, 2016), were shown to regulate phase separation and SG assembly (Babu et al., 2012; Banani et al., 2016; Dao et al., 2018; Han et al., 2012; Hofweber et al., 2018; Kedersha et al., 1999; Leung et al., 2011; Li et al., 2012; Sharkey et al., 2018). Accordingly, SUMOylation of eIF4A2 was shown to contribute to SG formation (Jongjitwimol et al., 2016).
Whereas much has been learned about how SGs form, many questions pertaining to basic SG biology, particularly the mechanisms underlying disassembly and how it might be linked to disease, remain unanswered. Several pathways, including autophagy, the ubiquitin-proteasome system (UPS), and SUMO-targeted ubiquitin ligase (StUbL), were implicated in SG disassembly, and ALS-associated mutations were discovered in some of the genes related to these mechanisms (Buchan et al., 2013; Ganassi et al., 2016; Hjerpe et al., 2016; Ivanov et al., 2019, Keiten-Schmitz et al., 2020; Protter and Parker, 2016a; Seguin et al., 2014; Turakhiya et al., 2018; Wang et al., 2019; Wheeler et al., 2016). However, a major gap in our understanding remains about the compositional changes SGs undergo during the process of disassembly and its link to disease conditions.
Proximity labeling proteomics approaches, such as APEX , which is based on an engineered ascorbate peroxidase (APEX2) (Lam et al., 2015; Lobingier et al., 2017), have proven valuable in uncovering new facets of membraneless organelles, particularly of RNA granules (Liao et al., 2019). This includes fine proteomic mapping of SGs with unmet resolution and the discovery of submicroscopic complexes preceding SG assembly (Markmiller et al., 2018; Padron et al., 2019).
Here, we performed an APEX proximity proteomic study with three independent SG baits to characterize the constitutive SG proteome in unprecedented resolution, and identified over 109 novel SG proteins and the composition of substructures within SGs. Furthermore, a temporal analysis of SG disassembly uncovered the existence of disassembly-engaged proteins (DEPs) that are selectively recruited upon disassembly and the impact of C9orf72 proline-arginine dipeptides that are associated with ALS and FTD (C9-ALS dipeptides) on SG disassembly. These data reveal a SUMO-dependent mechanism for the control of SG dynamics that is impaired by C9-ALS dipeptides, thus providing new insights into the pathomechanisms of ALS.
RESULTS
Comprehensive SG Proteomic Analysis by Multi-bait APEX Proximity Labeling Reveals 109 Additional SG Proteins
We characterized the protein composition of SGs in living human cells, by means of proximity labeling, utilizing engineered ascorbate peroxidase (APEX2) that is fused to a bait protein of interest. APEX facilitates free radical formation in its vicinity from hydrogen peroxide (H2O2), causing biotin-phenol (BP) radical formation that tags biomolecules with a biotin moiety. Biotinylated proteins can be purified over streptavidin beads and analyzed by mass spectrometry (MS) (Hung et al., 2016). We hypothesized that using multiple APEX baits would allow a more comprehensive, and internally validated, characterization of SGs beyond previous studies (Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2018). We engineered APEX fused to three RNA-binding proteins that have been extensively studied in SGs and displayed high SG partition coefficients in U2OS cells (Sanders et al., 2020) to increase confidence that they predominantly mark SGs, namely, Ras GTPase-activating protein-binding protein 1 (G3BP1) (Aulas et al., 2015; Candé et al., 2004; Kedersha et al., 2016; Markmiller et al., 2018; Solomon et al., 2007; Tourrière et al., 2003) (SG partition coefficient in U2OS cells ~0.5; Sanders et al., 2020), the fragile X protein FMR1 (also known as FMRP), and its autosomal homolog, FXR1 (Zhang et al., 1995) (SG partition coefficient in U2OS cells >0.55; Sanders et al., 2020), downstream of a tetracycline (tet)-inducible promoter. We also fused a nuclear export signal (NES) to APEX to serve as a cytoplasmic control bait (Hung et al., 2016), which was used previously in SG APEX studies (Markmiller et al., 2018). To avoid overexpression artifacts, including aberrations of organelle assembly dynamics or size (Anderson and Kedersha, 2008), tet-inducible FMR1-APEX and FXR1-APEX constructs were transfected to CRISPR-edited U2OS cells lacking FMR1/FXR1/FXR2 (Smith et al., 2019), whereas a tet-inducible G3BP1-APEX construct was transfected to a previously described U2OS line lacking G3BP1/G3BP2 (Kedersha et al., 2016). We titrated induction of APEX expression by tetracycline, in isolated clones, to approximate endogenous expression levels and to be comparable across all baits (Figures S1A and S1B).
All three SG-APEX baits were activated by the introduction of the APEX substrates, BP and H2O2, which appropriately demarcated SG only when cultures were stressed with sodium arsenite (NaAsO2 400 μM). Immunostaining revealed that the biotin signal co-localized with endogenous TIA1, an established SG marker (Figure 1A), and NES localized correctly to the cytoplasm (Figure S1C). We confirmed that endogenous proteins were tagged with BP by western blot analysis of pulled-down biotinylated proteins (Figures S1D and S1E). Importantly, biotinylation was specifically observed only when the APEX substrates were introduced (Figures S1D–S1F).
Figure 1. The Proteome of SGs Revealed by Multi-bait Proximity Labeling.
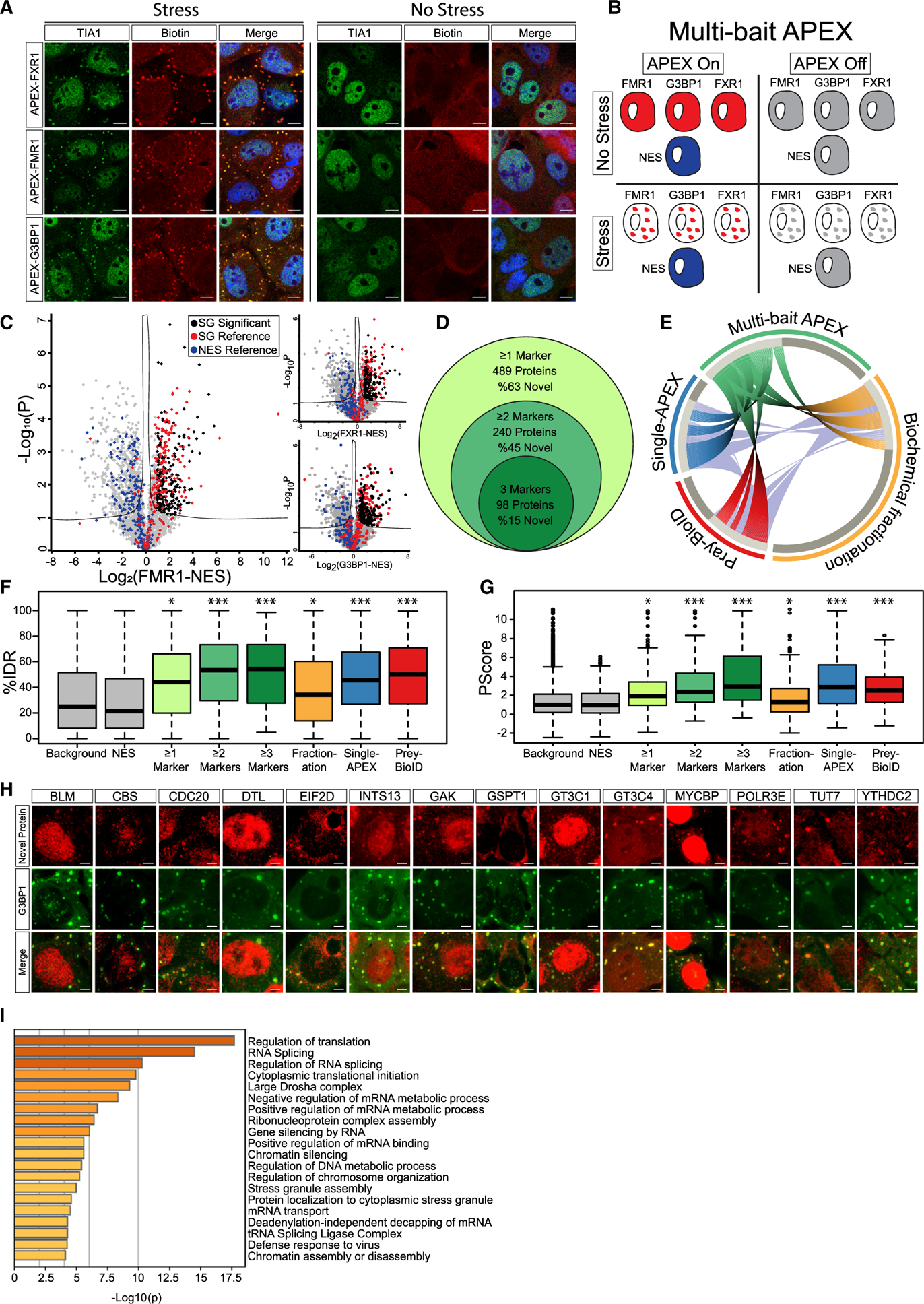
(A) Confocal micrographs depicting FXR1-APEX, FMR1-APEX, or G3BP1-APEX activity in U2OS cells, with or without sodium arsenite stress (NaAsO2 400 μM, 30 min). Immunofluorescence depiction of TIA1, neutravidin-Texas-red staining of biotinylated proteins at the proximity of the APEX bait, and merged signal demonstrating the precise localization of the APEX activity in stress granules (SGs). Lens, ×63; scale bar, 10 μm.
(B) Diagram of experimental design. Study with U2OS cells that stably express FXR1-APEX, FMR1-APEX, G3BP1-APEX, or cytoplasmic NES-APEX. SG baits are diffusible in the cytoplasm without stress. NES-APEX remains diffusively cytoplasmic under stress conditions (NaAsO2 400 μM, 30 min.), while FXR1-APEX, FMR1-APEX, and G3BP1-APEX are recruited to SGs. APEX on: APEX peroxidase activity, induced by H2O2, causes BP radical formation that tags biomolecules in the bait vicinity with a biotin moiety. APEX off: control for nonspecific activity without BP. Experiments were performed in triplicates.
(C) Volcano plots of relative protein levels in SG APEX relative to NES-APEX samples under stress conditions (x axis log2 scale), analyzed by MS. y axis depicts the differential expression p values (−log10 scale). Black, novel SG proteins above specific marker cutoff (FMR1 = 0.96, FXR1 = 0.89, and G3BP1 = 0.88), relative to NES. Student’s t test with correction to multiple hypothesis by FDR adjusted p < 0.05; red/blue, previously known SG proteins/cytoplasmic organellar proteins.
(D) Venn diagram of multi-bait SG analysis and embedded results, revealing proteins identified by at least a single SG bait (associated with FXR1 and/or FMR1 and/or G3BP1), at least two baits, or all three baits together.
(E) Circos plot of proteome depicted by at least two baits in our multi-bait APEX study (green), single bait G3BP1-APEX (Markmiller et al., 2018), biochemical fractionation (Jain et al., 2016), or indirect (pray) analysis of data from BioID studies (Youn et al., 2018). Substantial overlap with previously known SG proteome is accompanied by the discovery of 109 novel and internally cross-validated proteins.
(F) Boxplot of intrinsically disordered region (IDR) enrichment (%IDR, by IUPred; Mészáros et al., 2018) in the data of the current study and others (Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2018). Background shows all proteins identified in our MS analyses. Upper and lower quartiles and extreme points are shown. Wilcoxon signed-rank test p < 0.005.
(G) Boxplot of SG proteome propensity to phase separate (Pscore; Vernon et al., 2018) in the data of the current study and others as in (F). Upper and lower quartiles and extreme points are shown. ANOVA with Tukey post hoc test p < 0.05.
(H) Confocal micrographs of immune-fluorescent detection of novel SG proteins in U2OS cells under stress conditions, and co-localization with the G3BP1 SG marker. Scale bar, 10 μm.
(I) Bar graph depicting the significance of enrichment in the top 20 Gene Ontology (GO) terms for the SG proteome (−log p value) by Metascape (Zhou et al., 2019).
We conducted three independent APEX experiments with the three SG baits activated in parallel, under basal and stress conditions, and controlled for both cytoplasm diffusive APEX signal (NES) and nonspecific streptavidin-bead binding (no BP; diagram in Figure 1B). We identified 5,987 proteins across all samples by label-free quantitative proteomics using a conservative analysis pipeline (Figure S2A), with very high correlation across experimental replicates (Figure S2B). We excluded proteins that were bound nonspecifically to streptavidin beads (APEX off samples, 806 proteins; Figure S2C). To define candidate SG-interacting proteins, we applied Student’s t tests (false discovery rate [FDR] cutoff p < 0.05) to proteins associated with SG-APEX samples, relative to NES-APEX samples (Figure 1C), while setting a stringent log2 fold-change cutoff for each bait (see STAR Methods). Summary proteomic data related to this study are detailed in Table S1.
Well-characterized SG proteins were clearly enriched in our data (e.g., TIA1, UBAP2L, CAPRIN1, PABPC1, FUS, and ATXN2). Overall, the MS analysis identified 215 proteins associated with G3BP1-APEX (104 novel proteins [48%] relative to the benchmark list; Youn et al., 2018), 342 proteins associated with FMR1-APEX (196 novel proteins [57%]), and 260 proteins associated with FXR1-APEX (127 novel proteins [49%]).
In total, 489 proteins were identified with at least one of the APEX baits (310 novel proteins [63%]), of which 240 were detected with at least two baits (109 novel proteins [45%]) and 98 were identified by all three SG-APEX baits (15 novel proteins [15%]) (Figure 1D). Comparison of our internally cross-validated data (i.e., 240 proteins, identified by at least two baits) with other studies that investigated SG composition using other methodologies (Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2018) demonstrated correct re-identification of ~50% of the proteins in our data and underscored the novelty of 109 previously un-characterized and internally cross-validated SG proteins (Figure 1E). Intriguingly, 73 of these unexplored proteins emerge from association with both FMR1 and FXR1 (67%), whereas only 36 of the new proteins (33%) were cross-validated by G3BP1 and either FMR1 or FXR1. These data indicate the value of using new baits (in this case, FMR1 and FXR1) that were not used in the past.
Using the IUPred and Pscore algorithms (Mészáros et al., 2018; Vernon et al., 2018), we quantified the predicted enrichment of IDRs and propensity to phase separate in our data. SG proteins displayed values above the proteins enriched in the cytoplasm control (NES) or all proteins identified by the MS (background) (p < 0.005 by Wilcoxon signed-rank test, p < 0.05 by ANOVA with Tukey post hoc test, respectively; Figures 1F and 1G). Moreover, IDRs and Pscore enrichment were significantly higher for proteins discovered by at least two or three baits, compared to the background, and resonated with IDR and Pscore values for protein lists from studies done by other methodologies. We were able to demonstrate the enrichment of 15 novel proteins in SGs using immunofluorescence microscopy (Figure 1H; Table S1).
Gene Ontology (GO) enrichment analysis (Zhou et al., 2019) showed that SG proteins in our data are associated with multiple aspects of RNA metabolism, in accordance with the known properties of SGs (Figure 1I). However, we unexpectedly observed enrichment in proteins associated with DNA or chromatin. Our data also expanded the list of m6A-binding proteins resident in SGs, in accordance with a critical role for RNA methylation in SG assembly and in phase separation (Anders et al., 2018; Ries et al., 2019), and the lists of post-translational modifiers associated with poly-ADP ribosylation, ubiquitination or phosphorylation, which were all shown to regulate SG dynamics.
Altogether, our comprehensive analysis of the SG proteome, along with the newly discovered 109 SG proteins, increase knowledge about SG composition in human cells and serve as an in-depth resource for the SG proteomic landscape.
Multi-bait APEX Analysis Allows the Characterization of Pre-stress Assemblies and Distinct SG Substructures
Macromolecular assemblies of SG proteins are known to exist under normal growth conditions, prior to induction of cellular stress (Markmiller et al., 2018; Youn et al., 2018). Therefore, we characterized the macromolecular assemblies within proximity of soluble FMR1, FXR1, or G3BP1 under basal conditions. Out of the 490 proteins that we characterized in SGs upon stress (Table S1), 153 proteins were significantly enriched with APEX-labeled FMR1, FXR1, or G3BP1 relative to APEX-NES samples in basal conditions (Figures 2A and 2B). Notably, 113 proteins were associated with G3BP1 in soluble pre-stress complexes, whereas only 55 and 41 proteins were identified in the proximity of FMR1 or FXR1, respectively (Figure 2B). 30 proteins were associated with G3BP1 and either FMR1 or FXR1 in pre-stress conditions, including several proteins that are essential for SG formation (dubbed “pre-stress seed”; Figures 2B and 2C). Moreover, bioinformatic analysis of the pre-stress seed proteins revealed a high propensity to phase separate that was significantly higher than the mature SG proteome (Pscore p < 0.0005, ANOVA with Tukey post hoc test; Figure 2D). Taken together, our data suggest the potential existence of a submicroscopic pre-stress seed composed of proteins with high-phase-separation potential.
Figure 2. APEX Study of Proteome Composition Reveals the Emergence of Distinct SG Substructures.
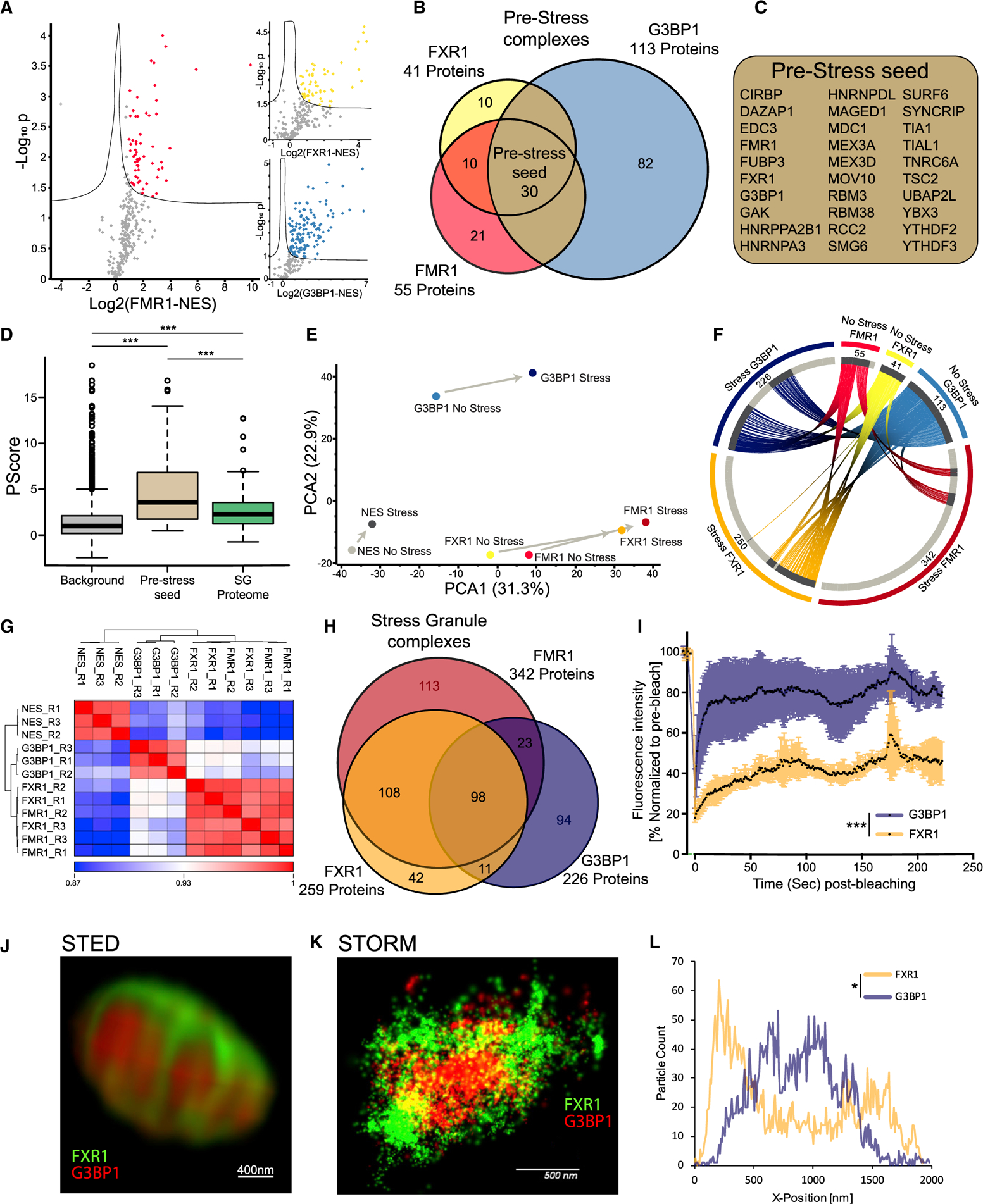
(A) Volcano plots of protein levels in FMR1, FXR1, or G3BP1 APEX samples relative to NES-APEX samples (x axis log2 scale) under non-stress conditions. y axis depicts the differential expression p values (−log10 scale). Red/yellow/blue features: proteins associated with APEX markers with at least 2-fold enrichment above values in NES. Student t test with correction to multiple hypothesis by FDR adjusted p < 0.05.
(B) Venn diagram of FMR1, FXR1, and G3BP1 proteomes under basal conditions, with the number of proteins demarcated.
(C) List of 30 pre-stress seed proteins identified by three markers in basal, pre-stress conditions.
(D) Boxplot analysis of propensity to phase separate (calculated by Pscore; Vernon et al., 2018) in the background (all proteins identified in our MS analysis, pre-stress seed [30 proteins]) and 240 proteins internally validated in mature SGs. Upper and lower quartiles and extreme points are shown. ANOVA with Tukey post hoc test p < 0.0005.
(E) Principal-component analysis (PCA) of FMR1, FXR1, G3BP1, and NES proteomes with or without stress. Minimal compositional changes in the NES proteome influenced by stress (depicted as short principal component vector), in contrast to more substantial compositional changes in the FMR1, FXR1, and G3BP1 proteomes, are shown. FMR1 and FXR1 proteomes collide.
(F) Circos plot of proteins identified in basal and stress conditions, per each SG-APEX bait. Approximately 52% of the proteins associated with G3BP1 under stress are already residents of the pre-stress G3BP1 complexes, whereas most of the proteins (84%) associated with FMR1/FXR1 assemble de novo with stress.
(G) Unsupervised clustering of Pearson correlation values for proteomes captured by SG APEX baits during stress. High similarity was found between FMR1 and FXR1 proteomes, which are distinct from the G3BP1 and NES proteomes. R1–3 are the three experimental replicates for each APEX bait.
(H) Venn diagram of FMR1, FXR1, and G3BP1 proteomes in stress conditions, with the number of proteins demarcated.
(I) Fluorescence recovery of SGs after photobleaching in U2OS cells that co-express G3BP1-RFP and FXR1-YFP. Laser bleaching was defined as time 0, and snapshots were taken every 1 s. The recovery of G3BP1-RFP was monitored at ~5 s, whereas FXR1-GFP did not completely recover, even after 200 s. Mean intensity presented as the percent of the average pre-bleach signal normalized to unbleached SGs and corrected for background fluorescence. Unpaired t test with Welch’s correction *** p<0.0001. See also Video S1.
(J) Dual-color stimulated emission depletion microscopy (STED) of FXR1 (green) and G3BP1 (red) in U2OS cells. 93× lens. Scale bar, 400 nm.
(K) Dual-color stochastic optical reconstruction microscopy (STORM) of FXR1 (green) and G3BP1 (red) in U2OS cells, captured at 4,000 frames/channel. 60× lens. Scale bar, 500 nm.
(L) Representative particle count per normalized SG position (x axis) from the STORM study. Total counts are shown on the y axis, normalized between the FXR1 (green) and G3BP1 (red) channels. Distribution of particles between channels is statistically different by ANOVA of 15 SGs (Figure S3C). p < 0.05. See also Video S2.
SG are non-homogeneous and contain substructures (Cirillo et al., 2020; Jain et al., 2016; Wheeler et al., 2016). However, little is known about the properties of such substructures. While FMR1, FXR1, and G3BP1 are all SG residents, principal-component analysis (PCA) of the proteomic data revealed that G3BP1 samples are distinct from FMR1/FXR1 samples under both basal and stress conditions (Figure 2E). Notably, a substantial fraction of the proteins associated with G3BP1 under stress are already residents of pre-stress G3BP1 complexes (~52%), whereas most of the proteins associated with FMR1/FXR1 assemble only with stress (pre-stress complexes: 16% and 16%, respectively), creating de novo a differentiated protein network that is distinct from that associated with G3BP1 (Figure 2F).
In agreement with the PCA results (Figure 2E), unsupervised clustering of samples under stress, revealed highly correlated mass spectrometric signatures associated with FMR1 and FXR1, which were dissimilar to the G3BP1 samples (Figure 2G). Accordingly, the 108 proteins shared between FMR1 and FXR1 suggest a single substructure that is distinguishable from G3BP1 substructure (Figure 2H).
The distinct G3BP1 and FMR1/FXR1 proteomes may genuinely reflect distinctive SG substructures or, alternatively, may result from cytoplasmic complexes outside of SGs (see Discussion). We therefore tested whether G3BP1 and FMR1/FXR1 display distinct biophysical behaviors and spatial organization inside SGs. We analyzed SG liquidity by fluorescence recovery after photobleaching (FRAP) in cells that co-express mRFP-G3BP1 and GFP-FXR1. We observed rapid recovery of mRFP-G3BP1 after bleaching, whereas GFP-FXR1 displayed slow and incomplete recovery dynamics (Figures 2I and S3A). In addition, super-resolution microscopy revealed two distinct substructures, whereby G3BP1 was positioned centrally and FXR1 was peripheral (Figures 2J–2L and S3B; ANOVA p < 0.05). Together, proteomics, recovery kinetics, and super-resolution microscopy reveal unexpected differences between distinct G3BP1 and FMR1/FXR1 substructures, with different biomaterial properties, spatial organization, and proteomic composition. While one substructure is organized around G3BP1 under basal conditions and its composition modestly increases with stress, the other substructure assembles de novo around FMR1/FXR1 in response to stress.
Temporal Analysis of SG Disassembly Reveals DEPs
The compositional changes that SGs undergo during the process of disassembly are poorly understood. To better understand normal SG disassembly, we first monitored disassembly by mRFP-G3BP1 live imaging after washing out medium with sodium arsenite (300 μM, 30 min). We noticed that microscopically visible disassembly started ~40 min after stressor washout and was completed in ~120 min (Figure 3A). In this context, we then used APEX and MS to characterize the SG proteome at three different time points during disassembly. We chose FMR1-APEX because it exhibited the largest proximity proteome of the three SG baits and, as opposed to G3BP1, does not preferably bind to the UPS system (Kedersha et al., 2016). From the MS results, we excluded proteins that were nonspecifically bound to streptavidin beads and normalized the protein intensity values of FMR1-APEX samples to the values measured in the NES-APEX. Out of the 7,003 proteins that were identified in all MS samples, 426 were reproducibly enriched in the FMR1-APEX samples relative to levels in the NES-APEX samples by at least 2-fold at at least one of the time points (Figure S4A, pipeline description; ANOVA statistical test, FDR < 0.05). Unexpectedly, 202 of the 426 proteins were more enriched during the disassembly process than under sodium arsenite stress (Figure 3B; Table S2). We named this protein group disassembly-engaged proteins (DEPs). The recruited DEPs suggest that SG disassembly is a highly regulated process. DEPs are associated with processes that were previously linked to the turnover of SGs, including autophagy and ubiquitin pathways (see Figure 3C for examples and Table S2 for a full description). In addition, heat-shock proteins, RNA helicases, cytoskeletal proteins, and mitochondrial proteins were also observed. SG proteins were enriched with IDRs and displayed higher likelihood to phase separate than DEPs (p < 0.0005 by Student’s t test, p < 0.0005 by Wilcoxon signed-rank test, respectively; Figures 3D and 3E). We also tested the role of a manually curated list of 22 DEPs in disassembly by performing small interfering RNA (siRNA) knockdown and live-imaging studies of SG disassembly. The knockdown of nine proteins resulted in significant changes to the kinetics of SG disassembly (Figures 3F, 3G, and S4B). These hits included proteins related to the ubiquitin proteasome, autophagy, and, unexpectedly, the SUMOylation machineries. Of note, we identified SETX, an ALS-related protein (Blair et al., 2000) that has never been identified in SGs, to functionally interact during SG disassembly.
Figure 3. Temporal Resolution of SG Disassembly Reveals a Network of DEPs.
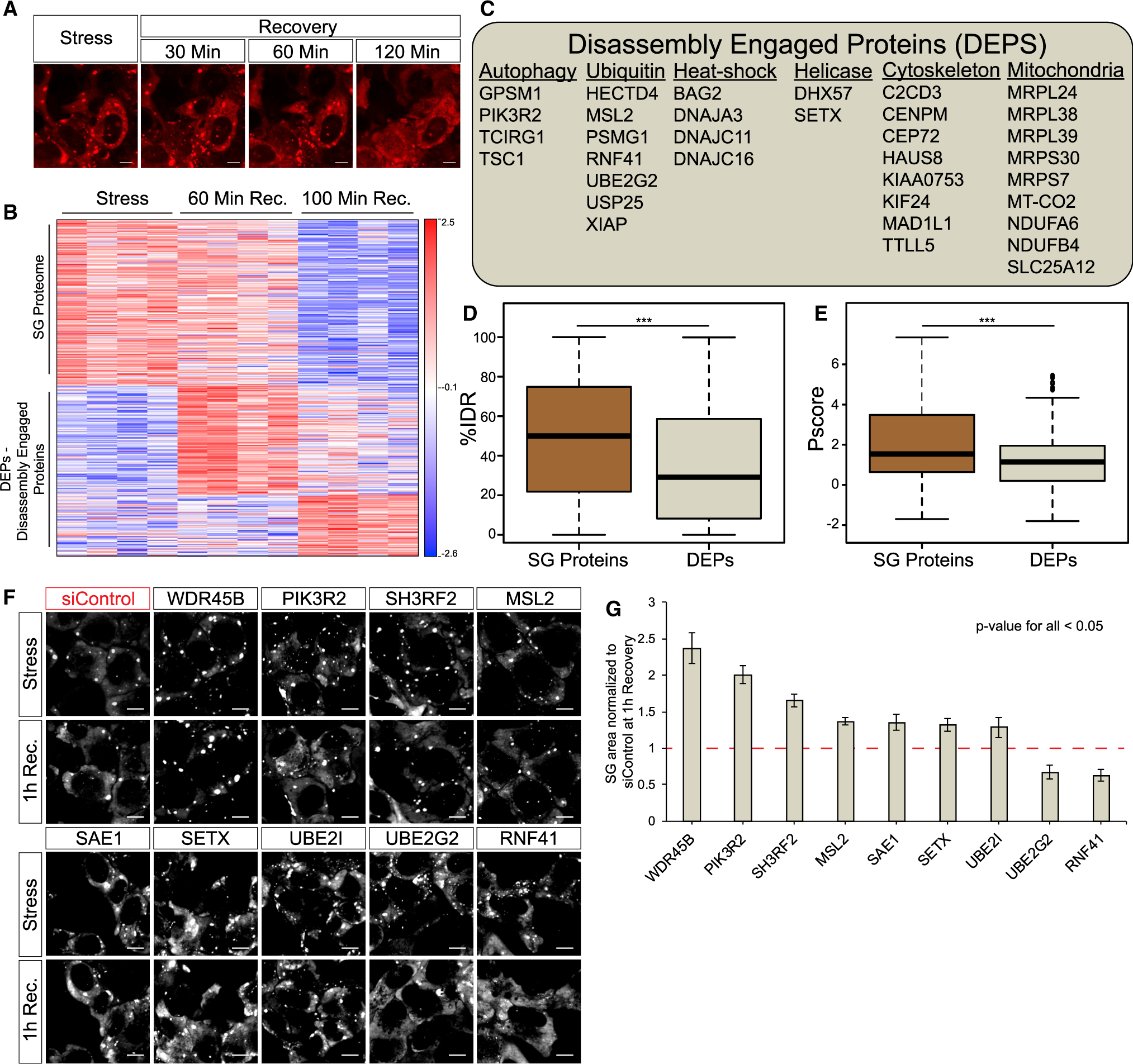
(A) Representative micrographs depicting RFP-G3BP1 in stressed U2OS cells and 30, 60, and 120 min during recovery, after the stressor was washed out; scale bar, 10 μm.
(B) Heatmap of unsupervised clustering of proteins associated with SG disassembly. Proteins specifically enriched in SG relative to the cytoplasm, if exceeding a 2-fold enrichment in FMR1-APEX/NES-APEX and p < 0.05 by Student’s t test with correction to multiple hypothesis by FDR. 224 proteins are enriched in SGs, while 202 proteins are enriched in SGs once stress is removed and disassembly dynamics ensue.
(C) A list of representative disassembly-engaged proteins (DEPs) associated with different cellular pathways.
(D) Boxplot analysis of IDR enrichment (%IDR; Mészáros et al., 2018) in 224 SG resident proteins or 202 DEPs. Upper and lower quartiles and extreme points are shown. Wilcoxon signed-rank test p < 0.0005.
(E) Propensity to phase separate (Vernon et al., 2018) in 224 SG resident proteins or 202 DEPs. Upper and lower quartiles and extreme points are shown. Two-sided Student’s t test p < 0.0005.
(F) Representative micrographs depicting GFP-G3BP1 in stressed U2OS cells and during recovery after the stressor was washed out. Cells were transfected with non-targeting siControl or specific siRNAs for the knockdown of DEPs; scale bar, 10 μm.
(G) Graph quantification of SG disassembly dynamics by live GFP-G3BP1 imaging. SG area, normalized to siControl area (y axis) at 1 h after stress washout. Three experimental repeats for measurement with four different areas per well. Representative experiment from two independent live-imaging studies tested by ANOVA repeated-measurement, p < 0.05. siRNA tested that failed statistical significance were Topors, Becn1, Bag2, Kif24, Mex3c, Ranbp2, Nbr1, Hsbp1, Dst, Cep72, Xiap, Usp25, and Traf5.
These data indicate that DEPs are extensively recruited to SGs during disassembly. While DEPs are not necessarily engaged to SGs by phase separation, the programmed engagement of at least some of the DEPs is functionally relevant to the regulation of SG disassembly. However, some DEPs may represent degradation or recycling of proteins after SGs have already been disassembled (see Discussion).
SUMOylation Controls SG Formation and Disassembly
The ubiquitin-like SUMO ligase complex consists of E1, E2, and E3 ligases (conjugating enzymes), which SUMOylate target proteins (Geiss-Friedlander and Melchior, 2007). We identified SAE1 (E1), UBE2I (SUMO-conjugating enzyme UBC9, E2), and the E3s RANBP2 and TOPORS as SG DEPs (Figure 4A), and siRNA knockdown of SAE1 (E1) and UBE2I (E2) impaired SG disassembly (Figures 3F and 3G). We therefore further investigated the involvement of SUMOylation in SG disassembly.
Figure 4. SUMOylation Controls SG Formation and Disassembly.
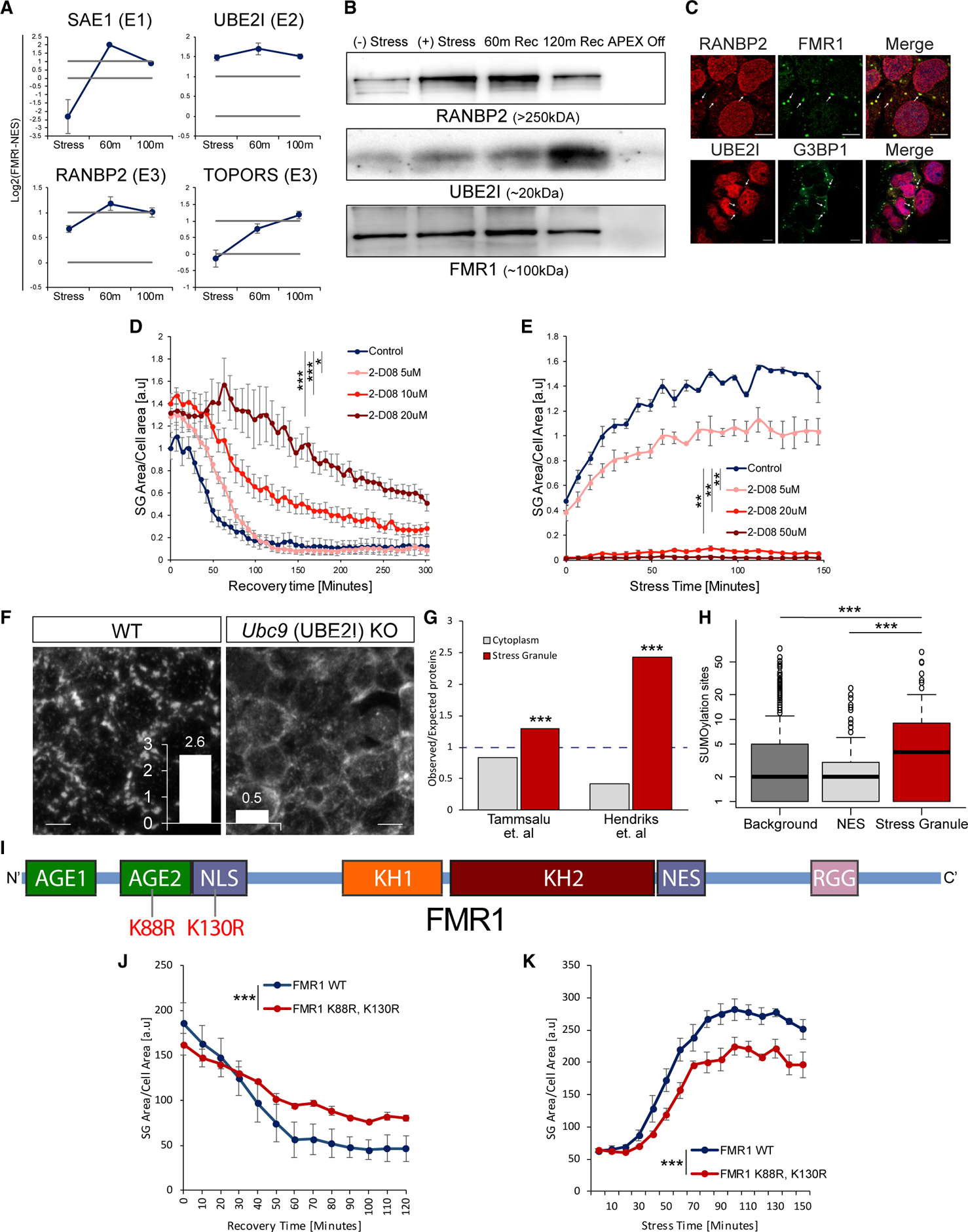
(A) Graphs of MS quantification of UBE2I, SAE1, TOPORS, and RANBP2 in U2OS SGs. Log2 fold change of label-free quantification (LFQ) intensity in FMR1 minus NES in stress conditions and two time points after washout. Lower bar shows levels in the cytoplasm; higher bar shows 2-fold enrichment. Data are presented as mean ± SEM.
(B) Western blot analysis after FMR1-APEX activity and streptavidin pull-down of biotinylated SG proteins for detection of RANBP2, UBE2I, and FMR1-APEX as loading reference. RANBP2 and UBE2I are present in SGs in response to stress and during recovery.
(C) Immunofluorescence analysis of RANBP2 and UBE2I localization in SGs. FMR1 or G3BP1 as SG markers. Merge includes demarcated nucleus (blue, DAPI). Lens ×63. Scale bar, 10 μm.
(D and E) Graph quantification of SG dynamics by live GFP-G3BP1 imaging with increasing concentrations of 2D08, a SUMOylation inhibitor. Stress induced with sodium arsenite (300 μm, for 30 min), washed out, and 2D08 was introduced (D), or 2D08 was introduced 4 h prior to induction of stress with sodium arsenite (200 μm, for 30 min) (E). SG area, normalized to cellular area (y axis), as a function of time (x axis). Repeated-measures ANOVA, * p < 0.05. ** p < 0.005, *** p<0.0005. Three experimental repeats for measurement with four different areas per well. Representative experiment from more than three independent live-imaging studies. See also Video S3.
(F) Representative images of anti-G3BP1 immunofluorescence in mouse embryonic stem cells, in which Ubc9 is conditionally nullified. Stress by sodium arsenite (300 μm, for 30 min) and quantification of SG/cell; scale bar, 10 μm.
(G) Bar graph of SUMO enrichment (expected/observed) in the SG proteome or the cytoplasm, based on SUMO moieties, characterized by Tammasalu et al. (a list of 539 proteins; Tammsalu et al., 2014) or Hendriks et al. (a list of 3,872 proteins; Hendriks et al., 2017). Hypergeometric test p < 0.0001 for both datasets.
(H) Boxplot of typical number of SUMOylated sites in total proteins identified by MS (background), cytoplasm (NES), and SG proteomes (based on Hendriks et al., 2017). p < 0.0005, Wilcoxon signed-rank test.
(I) Diagram of the FMR1 protein with main functional domains based on Prieto et al. (2020). Lysine 88 (K88) in the second N-terminal Agenet domain (AGE2) and lysine 130 (K130) of the nuclear localization signal (NLS) are known to be SUMOylated and were substituted for arginines.
(J and K) Graph quantification of SG dynamics by live GFP-FMR1 imaging with wild-type, K88R, and the K130R form of FMR1. Disassembly kinetics after washing out sodium arsenite (300 μm, for 30 min) (J), or SG formation kinetics after adding sodium arsenite (200 μm) (K). SG area, normalized to cellular area (y axis), as a function of time (x axis). Repeated-measures ANOVA, p < 0.0005. Three experimental repeats for measurement with four different areas per well. Representative experiment from more than three independent live-imaging studies.
First, we validated that UBE2I and RANBP2 are genuinely associated with SGs in a stress-dependent manner by performing western blot analysis on SG proteins that were pulled down by FMR1-APEX (Figure 4B). An immunofluorescence study further supported the presence of UBE2I and RANBP2 in SGs (Figure 4C). To test whether SUMOylation plays a functional role in SG disassembly, we performed live SG imaging in the presence of the small-molecule SUMO inhibitor 2D08, which inhibits the only E2 SUMO ligase, UBE2I (Kim et al., 2014). Introduction of 2D08 to cells reduced overall SUMOylation, as is expected (Figure S5A), while not causing cellular stress on its own. Accordingly we observed neither SG formation nor phosphorylation of eIF2 alpha (Figures S5B and S5C). When 2D08 (at 5–20 μM) was administered for 30 min along with sodium arsenite (300 μM) and included in the medium also after the stressor was washed out, disassembly was attenuated in a concentration-dependent manner (p < 0.0005, ANOVA repeated measurement; Figure 4D).
Because eIF4A2 SUMOylation contributes to SG formation (Jongjitwimol et al., 2016), we hypothesized that SUMOylation may play a role also in SG formation. We observed inhibition of SG formation when the UBE2I inhibitor 2D08 (at 5–50 μM) was introduced for 4 h prior to stress induction (sodium arsenite, 200 μM; p < 0.0005, ANOVA repeated measurement; Figure 4E). The involvement of SUMOylation in SG assembly was orthogonally demonstrated by doxycycline-inducible depletion of UBC9 in mouse embryonic stem cells, which resulted in inability to induce SGs (Figure 4F). Because of the complete blockade in SG formation, this Ubc9 genetic model could not have been used for SG disassembly analysis. Together, we conclude that SUMOylation is critical for SG assembly and disassembly.
We further demonstrated that the SUMO2/3 proteome of human cells is enriched with SG proteins (described in Figure 1D and Table S1) more than can be expected at random and that the typical number of SUMOylated sites is higher in the SG proteome than in the cytoplasm (Figures 4G and 4H). Moreover, ~20% of the SUMOylated proteins are exclusively cytoplasmic, consistent with a model in which SUMOylation takes place on the SGs (Table S2).
Finally, we mutated two reported SUMOylated lysine residues on FMR1 (Khayachi et al., 2018), so these cannot undergo SUMOylation (K88R and K130R; Figure 4I). SUMOylationdeficient FMR1 disrupted SG disassembly and formation (Figures 4J and 4K). In summary, SUMO-conjugating enzymes are recruited to SGs, and SUMOylation is critical for SG formation and disassembly.
C9ORF72-ALS Dipeptides Alter the SG Composition and Recruitment of DEPs
It was suggested that SGs serve as the origin of insoluble cytoplasmic aggregates in ALS (Buchan et al., 2013; Chew et al., 2019; Cook and Petrucelli, 2019; Li et al., 2013). Thus, better understanding of SG disassembly dysregulation may reveal mechanisms that nucleate inclusions in the diseased brain. Therefore, we stably expressed a tetracycline-inducible GFP protein fused to 50 proline-arginine dipeptide repeats (GFP-poly(PR)50; Wen et al., 2014), associated with the C9ORF72 subtype of ALS (Freibaum and Taylor, 2017; Tran et al., 2015; Wen et al., 2014). Expression of GFP-poly(PR)50 in APEX-FMR1 U2OS cells resulted in the formation of nuclear aggregates, as previously reported (Freibaum and Taylor, 2017; Tran et al., 2015; Wen et al., 2014). These aggregates neither co-localize with SGs nor cause SG formation. However, a loss of ~30% of the cultured cells was measured 3 days after induction of GFP-poly(PR)50 expression (Figure S5D). GFP-poly(PR)50 expression resulted in aberrant SG disassembly, wherein 30% of SGs failed to dissolve into the cytoplasm, even after 6 h (p < 0.0005, ANOVA repeated measurement; Figures 5A and 5B), in accordance with the behavior of other C9orf72-associated dipeptides (Zhang et al., 2018).
Figure 5. Temporal Resolution of SG Disassembly with C9-ALS-Associated Dipeptides.
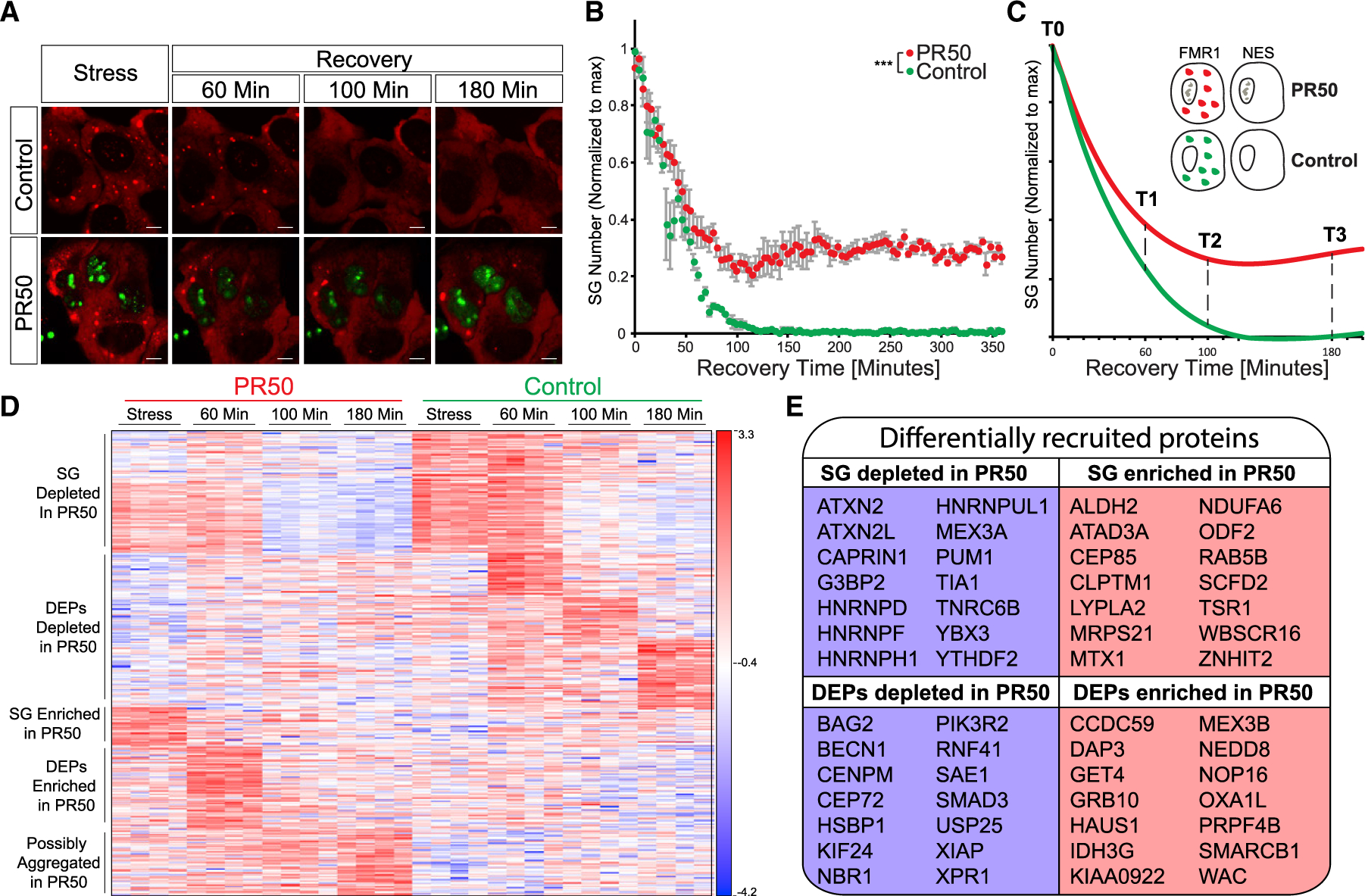
(A) Representative micrographs depicting RFP-G3BP1 in stressed U2OS cells that express GFP-poly(PR)50 and during recovery, after the stressor was washed out. Scale bar, 10 μm. See also Video S4.
(B) Graph quantification of SG disassembly dynamics by live Cherry-G3BP1 imaging after stressor washout with inducible GFP (control) or GFP-poly(PR)50 expression (PR50). SG number, normalized to maximal SG numbers per field (y axis) as a function of time after stress washout (x axis). Three experimental repeats for measurement with four different areas per well. Representative experiment from more than three independent live-imaging studies. ANOVA repeated-measurement p < 0.0005.
(C) Diagram of study design. Inducible FMR1-APEX or NES-APEX baits, with inducible GFP (control) or GFP-poly(PR)50 expression (PR50). APEX proximity labeling activity was induced at T0 (during stress) or at three time points after washout.
(D) Heatmap of unsupervised clustering of proteins that were differentially associated with SGs under normal conditions and with expression of GFP-poly(PR)50 during the course of disassembly. SG relative to cytoplasm (FMR1-APEX/NES-APEX 2-fold change) and GFP-poly(PR)50/GFP (control) by two-way ANOVA with FDR p < 0.05.
(E) A subset from the 425 proteins that were differentially associated (176 enriched/249 depleted) with SG during disassembly, with expression of GFP-poly(PR)50, relative to control conditions.
In parallel with the proteomic analysis of normal disassembly sequences that was described in Figure 3 (GFP control vector), we also performed APEX and MS in the presence of GFP-poly(PR)50 (see Figure 5C for a study design diagram and Figure S4A for pipeline analysis). Overall, we found 425 proteins that were differentially associated with normal versus GFP-poly(PR)50 SGs in at least one of the time points in the series. 176/249 proteins were relatively enriched/depleted in GFP-poly(PR)50 conditions, respectively (Figure 5D).
During stress, 59 proteins were relatively depleted from mature SG under GFP-poly(PR)50 conditions, including classical SG proteins (CAPRIN1, G3BP2, YBX3, and TIA1), while 32 proteins were relatively enriched (p < 0.05, two-way ANOVA with contrast analysis and FDR correction). When the stress was washed away to trigger SG disassembly, DEP recruitment was also impaired by GFP-poly(PR)50 expression (Figures 5D and 5E). Among the GFP-poly(PR)50-depleted DEPs, functionally validated hits include SETX and the SUMO ligases (Figure 3F). These data indicate that GFP-poly(PR)50 impacts SG proteomic composition and drives functional changes to the engagement of DEPs.
SUMOylation of SG Proteins Is Impaired by C9ORF72-ALS Proline-Arginine Dipeptides and Ameliorates an ALS Phenotype In Vivo
We found that the SUMO-conjugating enzymes SAE1, UBE2I, and RANBP2 were significantly depleted from SG upon GFP-poly(PR)50 expression by proteomics and western blot analysis of SG proteins that were pulled down by FMR1-APEX (Figures 6A and 6B). We then tested whether the SG proteome is SUMOylated in response to stress by performing FMR1-APEX pull-down and western blot analysis of SUMO modifications. We observed a stress-dependent smear of SUMO2/3 and, to a certain extent, SUMO1, primarily at protein sizes of 100–250 kDa, with reference to the APEX bait as a loading control (Figures 6C and S5E). Additionally, conjugation of SUMO2/3, but not SUMO1, to SGs was impaired by GFP-poly(PR)50 (Figures 6C and S5E). As GFP-poly(PR)50 reduces SUMOylation and both GFP-poly(PR)50 and inhibition of SUMOylation impair SG disassembly, it is plausible that reduced SG disassembly, observed with C9-associated dipeptides, is a result of inhibited SUMOylation activity.
Figure 6. SG SUMOylation Is Dysregulated by Poly(PR)50 and Ameliorates ALS Phenotype in Flies.
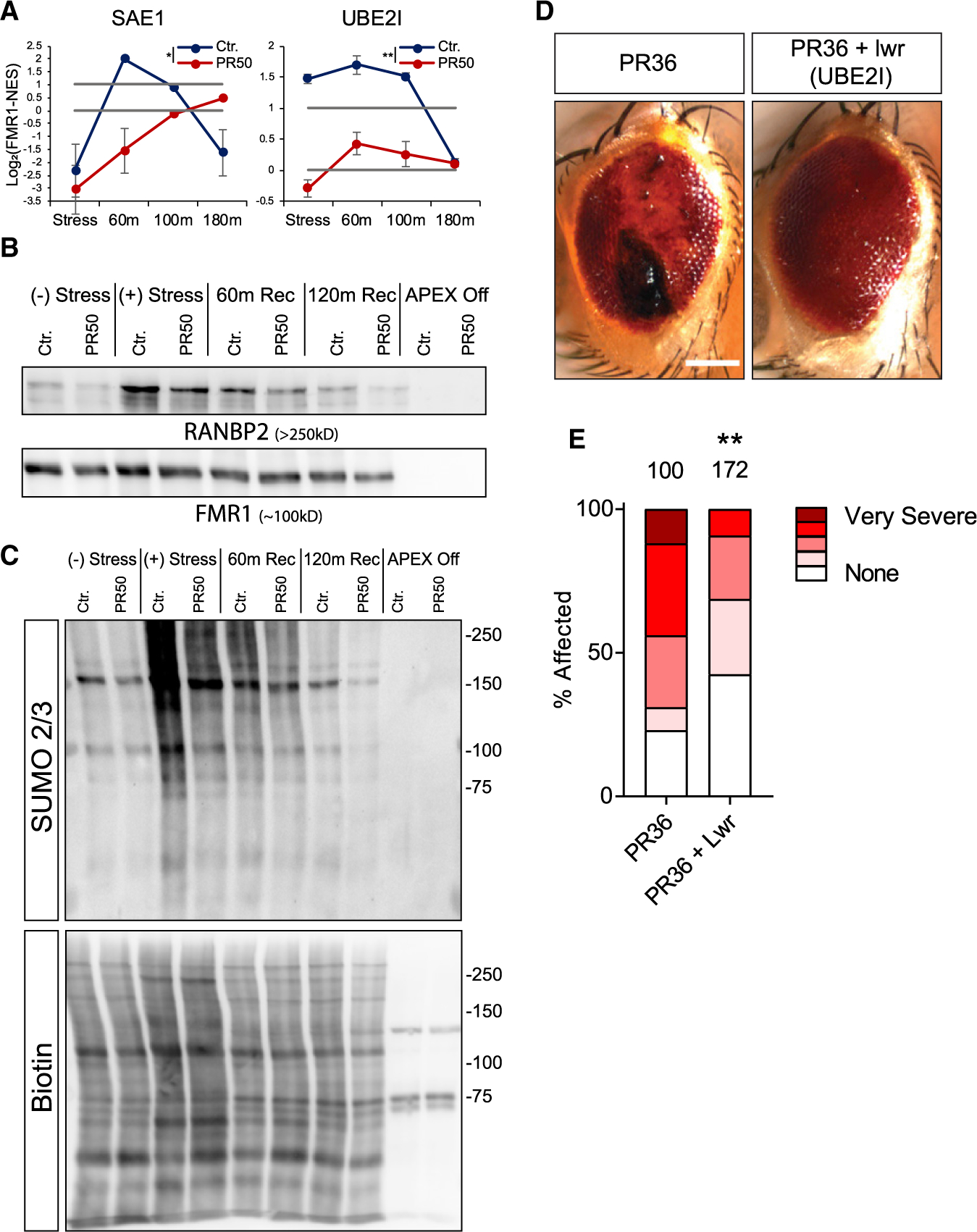
(A) Graphs of MS quantification of UBE2I, SAE1 in U2OS SGs. Log2 fold change of LFQ intensity in FMR1 minus NES in stress conditions and three time points after washout. Red, GFP-poly(PR)50. Lower bar shows levels in the cytoplasm; higher bar shows 2-fold enrichment. Data are presented as mean ± SEM. Two-way ANOVA * p <0.05; **p <0.005.
(B) Western blot analysis after FMR1-APEX activity and streptavidin pull-down of biotinylated SG proteins for detection of RANBP2 and FMR1-APEX as loading reference. RANBP2 are present in SGs in response to stress and during recovery. GFP-poly(PR)50 conditions inhibit RANBP2 recruitment.
(C) Western blot analysis after FMR1-APEX activation and streptavidin pull-down of biotinylated SG proteins for detection of SUMO2/3 -conjugated proteins (upper blot) and loading control developed with streptavidin for detection of biotinylated proteins. Extensive SUMOylation of SG proteins seen as smear at 100–250 kDa and gradual decrease associated with disassembly. GFP-poly(PR)50 expression inhibit SUMOylation. Representative blot from more than three studies.
(D) Poly(PR)36 (PR36) expression in the Drosophila melanogaster eye leads to the formation of necrotic tissue (“rough eye”). Overexpression of lesswright (Lwr) leads to a rescue of the necrosis in PR36 expressing flies (PR36 + Lwr). Scale bar, 100 μM.
(E) Quantification of percentage of flies affected with either no necrosis (none), mild, moderate, severe, or very severe necrosis. The number of flies assessed in each condition is given above the bar graph. Two-tailed Fisher’s exact test comparing the number of flies with necrosis versus no necrosis (**p = 0.0015).
Finally, to determine whether modifying SUMOylation could affect toxicity in vivo, we used an established model of C9orf72-associated ALS, expression of poly(PR)36 in the Drosophila melanogaster eye (Fumagalli et al., 2019; Mizielinska et al., 2014). When poly-(PR)36 was overexpressed using the GMR-gal4 driver, the external structure of the eye degenerated, resulting in the presence of black necrotic tissue (Figure 6D). This phenomenon was prevented by the overexpression of lesswright (Lwr), the Drosophila ortholog of UBE2I (Figures 6D and 6E). Thus, SUMOylation mitigates neurodegeneration associated with C9ORF72 poly(PR)36 in flies and suggests the in vivo relevance of control of SUMOylation as potential cellular mechanism of neurodegeneration.
DISCUSSION
Proximity proteomic mapping by multiple baits set new standards for studies of membraneless organelles. Accordingly, our study can be used as in-depth database for the spatiotemporal landscape of SGs. Particularly, we provide for the first time an analysis of SG disassembly kinetics at proteome resolution, revealing basic and disease-relevant mechanisms. We discovered more than 100 new SG proteins, establishing a resource that encompasses ~240 proteins of high confidence, gained with three different proximity-labeling baits.
We observed submicroscopic pre-stress complexes, in agreement with previous studies (Markmiller et al., 2018; Youn et al., 2018). A stringent analysis, with all three baits, revealed the identity of 30 proteins at the pre-stress seed. Many of these are classic SG proteins that collectively exhibit superior propensity to phase separate, suggesting these are relevant for SG formation. Our data are consistent with a model whereby a pre-stress seed entity emerges at the proximity of G3BP1, perhaps due to the protein’s conformational switching (Deniz, 2020; Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020).
The unique power of multi-bait APEX allowed us to characterize the mature SG proteome composition of two distinguishable substructures, at the proximity of G3BP1 and of FXR1-FMR1, consistent with substructures identified elsewhere (Cirillo et al., 2020; Wheeler et al., 2016). FRAP analysis suggested that the two substructures display different biomaterial properties and super-resolution microscopy revealed that the G3BP1-enriched substructure resides central to the FXR1-enriched one. Our data may suggest that one substructure is created by maturation of the pre-stress seed that preexists under basal conditions at the proximity of G3BP1 and only modestly changes with stress. However, the other substructure assembles de novo around FMR1/FXR1 in response to stress. The layered organization perhaps implies differences in the biophysical properties of the phases, which may facilitate sequential activity, as reported in other RNP bodies (Feric et al., 2016)
The reasoning that better understanding of SG disassembly could lead to the discovery of new pathways and therapeutic targets in neurodegeneration (Li et al., 2013) drove us to use APEX and MS to temporally study the process of SG disassembly. This analysis, which was impossible in the past, revealed a group of proteins that are recruited to SGs during disassembly, which we named DEPs. The existence of DEPs suggests that SG disassembly is a controlled process that occurs in a stepwise manner and cannot be described as passive dissolution of SG components into the cytoplasm. A substantial number of these DEPs are associated with processes that were previously linked to the turnover of SGs, including autophagy and ubiquitin. Rather unexpectedly, mitochondrial proteins make a significant group of proteins that engage with SGs during disassembly. The potential role of mitochondrial proteins in SG disassembly is unexplored and may suggest previously unappreciated links tying cellular metabolism to SG disassembly or new moonlighting functions for mitochondrial proteins. A secondary siRNA screen demonstrated that the knockdown of at least 9 of 22 tested DEPs impaired SG disassembly, providing functional evidence for the relevance of a fraction of the DEPs.
We identified E1, E2 and E3 SUMO-conjugating enzymes as DEPs, suggesting that the broad SUMOylation of SG proteins is cytoplasmic and likely to occur directly on the SG. Recently, SUMO-primed RNF4-dependent ubiquitylation, which is part of the StUbL pathway, was shown as a new regulator of SG disassembly (Keiten-Schmitz et al., 2020), resonating our observations. Furthermore, we discovered that SUMOylation activity is functionally important not only for disassembly but also for SG formation. One compelling hypothesis is that mono-SUMOylation contributes to SG formation via enhanced valency (high network connectivity, “SUMO glue”) and phase separation, whereas poly-SUMOylation controls StUbL-dependent disassembly. The dual involvement of SUMOylation in SG on and off processes may increase reaction sensitivity in regulating the SG life cycle (Hart and Alon, 2013).
Insight into SG disassembly may be particularly informative in understanding the insoluble cytoplasmic aggregates, such as in ALS inclusions (Buchan et al., 2013; Chew et al., 2019; Cook and Petrucelli, 2019; Li et al., 2013). Thus, to better understand dysregulation of SG disassembly, we utilized APEX and MS under C9ORF7-ALS-like conditions, which impairs SG disassembly. Poly(PR)50 drove broad changes to SG proteome, which might be related to pathology. Furthermore, in contrast to normal engagement of DEPs during disassembly, the poly(PR)50 model showed aberrantly recruited DEPs that may be potentially informative about neuropathology. An initial study of a subset of aberrantly recruited DEPs demonstrated that ALS-associated RNA helicase SENATAXIN (SETX) and other DEPs are sensitive to GFP-poly(PR)50 expression and independently essential for normal SG disassembly. Therefore, DEPs characterized in our studies are primary candidates for follow-up research on pathways that are potentially relevant to the pathogenesis of ALS and FTD.
GFP-poly(PR)50 expression impaired SUMO ligase recruitment and SG SUMOylation, suggesting that SUMO plays a part in the mechanism by which C9orf72 dipeptides attenuate SG disassembly. This can further represent an unexplored molecular mechanism of neurodegeneration, acting via control of SG turnover. In addition, SUMOylation mitigates ommatidia neurodegeneration associated with the C9ORF72 poly(PR)36 model in flies. Therefore, SUMOylation is a post-translational modification with relevance to neurodegeneration in vivo. However, further studies of SUMO roles in human neuropathology are required before SUMO-based intervention may be considered in the context of experimental therapies.
Altogether, our study provides an in-depth resource for follow-up studies, dissects the SG spatiotemporal proteomic landscape and proposes basic and disease-relevant mechanisms of SG disassembly.
Limitations of Study
There are a few notable limitations to the study. Proteins labeled by SG-APEX baits under basal (stress-free) conditions resonate proposed submicroscopic pre-stress complexes (Markmiller et al., 2018; Youn et al., 2018). However, evidence is missing to directly demonstrate that these complexes indeed assemble into SGs, and the model of the pre-SG seed is open to alternative interpretations (e.g., that FMR1/FXR1 and G3BP1 form cytoplasmic complexes that are not engaged in SG). Accordingly, some of the DEPs may represent degradation or recycling of proteins after SGs have already been disassembled and not necessarily function in the control of SG disassembly. In addition, because high protein yields are required for MS, our analysis is limited to a human cell line (U2OS) and currently cannot be performed in human motor neurons differentiated from induced pluripotent stem cells (iPSCs). This limitation may be overcome by mass human neuron production (Liao et al., 2019), offering proteomic analysis in disease-relevant models.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eran Hornstein (eran.hornstein@weizmann.ac.il).
Materials Availability
Requests of plasmids and cell lines described in this study will be available upon request from the lead contact, under a standard MTA.
Data and Code Availability
The proteomic datasets generated during this study are available at the PRIDE repository: PXD017330. Original data for figures in the paper is available in Mendeley Data https://doi.org/10.17632/vvthhmwwxw.1
EXPERIMENTAL MODEL AND SUBJECT DETAILS
U2OS cells are derived from human bone osteosarcoma epithelial tissue. V6.5 mouse Embryonic Stem Cells (mESC) are derived from the inner cell mass (ICM) of a 3.5 day old mouse embryo. U2OS cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. mESCs were cultured on irradiated DR4 mouse embryonic fibroblast cells with DMEM, 20% FBS, 1% penicillin-streptomycin, 2mM glutamine, 1% Sodium Pyruvate, 1% nonessential amino acids, 0.1mM β-mercaptoethanol, 10 ng/ml recombinant human LIF. Both cells were incubated at 37°C, with 5% CO2. Drosophila flies of stocks UAS-PR36, GMR-gal4/CyO and UAS-Lwr overexpression were reared on standard Drosophila media (62.5g/L cornmeal, 25 g/L yeast, 7g/L agar, 16.9 g/L dextrose, 37.5ml/L golden syrup, 9.375 ml/L propionic acid, 1.4g/L hydroxybenzoate, 14.0 ml/L ethanol) at 25°C, 12hr/12hr light dark cycle.
METHOD DETAILS
Mammalian Cell Culture
Human Bone Osteosarcoma Epithelial Cells, U2OS (U-2 OS, ATCC HTB-96), were cultured in growth media consisting of Dulbecco’s Modified Eagle Medium (DMEM, Biological Industries, 01– 050-1A) supplemented with 10% fetal bovine serum (FBS, Biological Industries, 04-001-1A), 1% penicillin-streptomycin (Biological Industries, 03–0311B) at 37°C, with 5% CO2. Stress granules induced by NaAsO2 (300–400 μM, Sigma-Aldrich, 71287). To avoid overexpression artifacts including aberrations of organelle assembly dynamics or size (Anderson and Kedersha, 2008), tet-inducible FMR1-APEX and FXR1-APEX constructs were transfected to CRISPR-edited U2OS cells lacking FMR1/FXR1/FXR2 (Smith et al., 2019), whereas a tet-inducible G3BP1-APEX construct was transfected to a previously described U2OS line lacking G3BP1/G3BP2 (Kedersha et al., 2016). We selected single clones that displayed comparable construct expression and titrated induction by tetracycline to approximate endogenous expression levels and to be comparable across all baits (Figures S1A and S1B). Ubc9 depletion was performed in V6.5 mouse embryonic stem cells by transgenesis of an exogenous Ubc9 allele, driven by TET-OFF promoter, alongside a constitutive tTA activator. Following that, endogenous Ubc9 was specifically targeted by CRISPR/Cas9 strategy. For inducible loss of Ubc9 function, 1ug/ml doxycycline was introduced for 72 hours. Mouse embryonic stem cells (mESCs) were cultured on irradiated DR4 mouse embryonic fibroblast cells with DMEM, 20% FBS, 1% penicillin-streptomycin (Invitrogen), 2mM glutamine (Invitrogen), 1% Sodium Pyruvate (Invitrogen), 1% nonessential amino acids (Invitrogen), 0.1mM β-mercaptoethanol (Sigma), 10 ng/ml recombinant human LIF (produced in-house).
Molecular cloning
For APEX vectors, V5 epitope tag, SG bait protein (FMR1, FXR1 or G3BP1) and APEX2 were subcloned into pcDNA4.0-TetO vector downstream of Tet-On inducible promoter, using primers listed in Table S3, and transfected by using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, Cat# 11668027) to U2OS cells that express the Tet-Repressor protein. Antibiotic resistance selection against Zeocin (250μg/ml, Invivogen ZEL-41–01) or Puromycin (2ug, Invivogen, ANT-PR)) enabled the isolation of single cell clones that were taken for expansion and analysis. Mutagenesis of the K88 and K130 residues of FMR1 was performed using primers listed in Table S3.
APEX proximity labeling
APEX gene expression was induced by supplementing the medium with tetracycline for 24 hr. (50ng/ml for NES/G3BP1; 100ng/ml for FMR1/FXR1). Labeling activity was induced by supplementing Biotin-phenol (BP, 500 μM, Iris Biotech GmbH, LS-3500) for 60 minutes and H2O2 (1 mM, J.T.Baker 7722-84-1) for 1 min. APEX activity extinguish with quenching solution (QS: sodium azide (10mM, Mallinckrodt, 1953–57), sodium ascorbate (10mM, Sigma-Aldrich, A7631) and Trolox (5mM, Sigma-Aldrich, 238813) in PBS. Then, cells were scraped in PBS, centrifuged at 800 × g for 10 min at 4°C, pelleted and lysed in ice-cold RIPA lysis buffer supplemented with cOmplete Protease Inhibitor Cocktail (Roche, 4693116001) and PhosSTOP (Roche, 4906837001) and further supplemented with N-Ethylmaleimide (NEM, 2.5mg/ml, Sigma-Aldrich, E3876) in SUMO assays. Lysates centrifuged at 15,000 × g for 10 min at 4°C. Protein concentration was quantified with Bio-Rad Protein Assay Dye Reagent (Bio-Rad, 500–0006). Streptavidin-coated magnetic beads (Pierce Streptavidin Magnetic Beads, Thermo-Fisher, 88816) were incubated for pulldown experiments (SA-pulldown) with 500μl of the extract at ratio of 200μl beads per 1mg of sample with rotation overnight at 4°C.
Liquid Chromatography and Mass Spectrometry
LC-MS/MS runs were performed on the EASY-nLC1000 UHPLC (Thermo Scientific) coupled to the Q-Exactive Plus or Q-Exactive HF mass spectrometers (Thermo Scientific) (Scheltema et al., 2014). Peptides were separated with a 50 cm EASY-spray PepMap column (Thermo Scientific) using a water-acetonitrile gradient, with a flow rate of 300 nl/min at 40°c. Peptides were loaded to the column with buffer A (0.1% formic acid) and separated using a 105 min linear gradient of 7%–28% buffer B (80% acetonitrile, 0.1% formic). The resolutions of the MS and MS/MS spectra were 70,000 and 17,500 for Q-Exactive Plus, respectively. The resolutions of the MS and MS/MS spectra were 60,000 and 30,000 for the Q-Exactive HF, respectively. The m/z range was set to 300–1700 or 380– 1800 Th. MS data were acquired in a data-dependent mode, with target values of 3E+06 and 1E+05 or 5E+04 for MS and MS/MS scans, respectively, and a top-10 method.
Microscopy
SG live imaging of cultured cell images were taken by a PCO-Edge sCMOS camera controlled by VisView, installed on a VisiScope Confocal Cell Explorer system (Yokogawa spinning disk scanning unit; CSU-W1) and an inverted Olympus microscope (40 × oil objective; excitation wavelengths: GFP - 488 nm; mCherry - 560 nm). SG and cell area were analyzed using surface feature in Imaris software. Dual color (AlexaFluor647, AlexaFluor568) three-dimensional Stochastic Optical Reconstruction Microscopy (STORM) images of adherent cells on glass-bottom dishes (MatTek P35G-0.170–14-C), were recorded on Vurata SR352 system (Bruker), using 1.3 NA 60x silicon oil immersion objective (Olympus) in the presence of imaging buffer (7 μM glucose oxidase, 56 nM catalase, 2 mM cysteamine, 50 mM Tris, 10 mM NaCl, 10% glucose, pH 8). Maximal excitation of 647 nm and 561 nm lasers was 6 kW/cm2. Hamamatsu Orca Flash 4v2 camera at frame rate of 50 Hz took 4000 frames per channel. Data analyzed with Vutara SRX 6.04.19 software. Stimulated emission depletion (STED) microscopic images acquired on an inverted Leica SP8 STED3X equipped with internal Hybrid (HyD) detectors, Acusto Optical Tunable Filter (Leica microsystems CMS GmbH, Germany) and a pulsed (80 MHz) white light excitation laser (WLL). Images were acquired with a HCX PL APO 93x/1.30 GLYC STED objective (RI of 1.46 With motCORR) in sequential mode acquiring first confocal images followed by STED imaging. A 775nm pulse used for STED depletion. Excitation was at 587nm and 644nm and emission was collected at 593–631nm and 685–755nm with pinhole of 0.7 AU = 121 μm, scanning speed 400 – 600 Hz zoom of 10–11, resulting in effective field of view of 10–15 m XY 2.5 – 3 m Z and voxel size of 0.0019 m XY and 0.078 m in Z.
Fluorescence Recovery After Photobleaching (FRAP)
“Half-bleach FRAP” (Kroschwald et al., 2015), used to capture primarily inter-SG dynamics. mCherry-G3BP and tet-inducible expression of YFP-FXR1 were stably transfected to a U2OS line lacking G3BP1/G3BP2 (Kedersha et al., 2016), seeded in glass bottom dishes (CellVis cat #D35-C4-20-1.5-N) and treated with 100 ng/ml doxycycline for 15 hr to induce YFP-FXR1 expression, once at 80%–95% confluence. Stress was induced with 200 μM sodium arsenite in live cell imaging medium for 50 – 150 min. Fluorescence was bleached by 4 laser max power (wavelength 405, 488, 561, and 640 nm). Recovery images after photobleaching were collected every 1.0 s on a Zeiss LSM 800 Airyscan confocal with Zeiss Axio Observer Z1 Inverted Microscope using the 63x oil objective and Zen 2.6 software. Analysis corrected fluorescence intensity to background fluorescence and normalized to control unbleached SG with ImageJ v 2.0.0-rc-69/1.52p.
siRNA screening
G3BP1-GFP stably expressing U2OS cells were seeded one day before the transfection, 20k/well, in 96 well-plate. 48hr before imaging, 1uM of siRNAs against genes of DEP proteins and non-targeting siRNA #5 as control (Dharmacon, Cherry-peak plate, siGENOME siRNA pool) were incubated with Lipofectamine 2000 (Invitrogen) and Opti-MEME (gibco) for 20min in R.T before transfection. Meanwhile, the cells were incubated with transfection medium (DMEM, 10% FBS, without Penstrep). Then, the siRNAs were transfected and cells were incubated for 48h before experiments. For validation of knock-down, RNA was extracted from cell lysates, reverse transcribed, and was analyzed by qPCR using primers listed in Table S3. For live-imaging SG disassembly experiments, 30min before imaging, the cells were exposed to 300uM sodium-arsenite to induce SG assembly, and then were washed once with complete medium. One well per treatment was unstressed to monitor treatment side effects. Images were collected from four different sites around the well center for each well in every ~7min for at least 2hr.
Fly genetics
Flies of stocks UAS-PR36, GMR-gal4/ CyO (Fumagalli et al., 2019) and UAS-Lwr overexpression (Drosophila Bloomington stock center #9324) (Mizielinska et al., 2014) were reared on standard Drosophila media (62.5g/L cornmeal, 25 g/L yeast, 7g/L agar, 16.9 g/L dextrose, 37.5ml/L golden syrup, 9.375 ml/L propionic acid, 1.4g/L hydroxybenzoate, 14.0 ml/L ethanol) at 25°C, 12hr/12hr light dark cycle. The UAS-Lwr line was crossed to w1118 for two generations to replace the X chromosome and then backcrossed for 5 further generations into w1118 before use in experiments. Flies were tipped into fresh vials and scored 7–8 days after eclosion and eye phenotyping was performed on a 5-degree scale in males based on presence of necrotic tissue, (“none,” “mild,” “moderate,” “severe” or “very severe”). Scoring was performed blinded to genotype. Images obtained using Zeiss Axio Imager M1. 5–8 independent crosses per condition.
QUANTIFICATION AND STATISTICAL ANALYSIS
Raw proteomic data processing
Raw MS data were processed using MaxQuant version 1.6.2.6 (Cox and Mann, 2008). Database search was performed with the Andromeda search engine (Cox and Mann, 2011; Cox et al., 2011) using the human Uniprot database. Forward/decoy approach was used to determine the false discovery rate and filter the data with a threshold of 1% false discovery rate (FDR) for both the peptide-spectrum matches and the protein levels. The label-free quantification (LFQ) algorithm in MaxQuant (Cox et al., 2014) was used to compare between experimental samples, except for the negative controls. Additional settings included carbamidomethyl cysteine as a fixed modification and methionine oxidation, N-terminal acetylation, and biotin-phenol as variable modifications. The “match between runs” option was enabled to transfer identification between separate LC-MS/MS runs based on their accurate mass and retention time after retention time alignment.
Proteomics statistical analysis
ProteinGroups output table was imported from MaxQuant to Perseus environment (Tyanova et al., 2016), or R (R Development Core Team, 2013). Quality control excluded reverse proteins, proteins identified only based on a modified peptide, and contaminants. Non-specific streptavidin-bead binders were excluded by the following procedure: Intensity values were log2 - transformed, and protein groups were filtered to retain only proteins with at least 2 valid values / group. Missing values were replaced by a constant low value. Student’s t test with S0 = 0.1 was performed with FDR p value ≤ 0.05 for pairs of APEX-On and corresponding APEX-Off samples. Proteins that passed all QC filters were separated for each of the SG-APEX markers and compared to the NES samples. For the stress conditions, data were filtered to retain only proteins with at least two LFQ values in at least 1 group. Missing data were imputed by creating an artificial normal distribution with a downshift of 1.8 standard deviations and a width of 0.4 of the original ratio distribution. Enriched SG proteins were called by Student’s t test (SG-APEX versus NES-APEX) with S0 = 0.1 and FDR p value ≤ 0.05 and fold-change threshold that was determined for each bait (SG-APEX - NES-APEX). Determination of thresholds was based on machine learning that used as training set positive curated SG proteins described in Table S1 (FMR1 = 0.96, FXR1 = 0.89, G3BP1 = 0.88). Stress- enriched proteins were tested for association with SG-APEX baits versus NES-APEX, under basal conditions (without NaAsO2) by Student’s t test S0 = 0.1 with FDR p value ≤ 0.05 and a minimum of 2-fold enrichment (Log2(APEX-SG - NES-APEX) > 1). Temporal SG disassembly study included proteins that were significantly enriched in FMR1-APEX over NES-APEX values at any of the time points either in GFP or GFP–poly(PR)50 conditions tested as above. Following that, FMR1-APEX values per each group were normalized by the mean of their corresponding NES-APEX values. For analysis of normal SG disassembly, we used ANOVA test with S0 = 0.1, FDR ≤ 0.05, standardized by z-score transformation and clustered by Pearson’s correlation coefficients. Two-way ANOVA was used to call proteins that were differentially recruited between GFP and GFP–poly(PR)50 conditions, with FDR ≤ 0.05. Contrast analysis was used to call proteins enriched in specific time points and between conditions, at FDR ≤ 0.05. These were standardized by z-score transformations and clustered by Pearson’s correlation coefficients.
Bioinformatics
The sequences of the proteins that were identified in the mass spectrometric experiments were downloaded from uniprot (The UniProt Consortium, 2017). Intrinsically disordered regions were predicted using iupred2A (Mészáros et al., 2018) as a stretch of ≥ 10 amino-acids with IUPred score AA ≥ 0.4, while allowing ≤ 2 consecutive structured amino-acids (IUPred score of < 0.4). Phase separation scores (Pscores) were calculated via (Vernon et al., 2018).
Statistical analysis
Statistics performed with R (R Development Core Team, 2013). Shapiro-Wilk or Levene tests were used to assess normality of the data. Pairwise comparisons passing normality test were analyzed with atudent’s t test. Wilcoxon test was used for pairwise comparison of nonparametric data. Multiple group comparisons passing normality test were analyzed using ANOVA with post hoc tests, whereas nonparametric multiple group comparisons were analyzed using the paired-Wilcoxon test when ANOVA assumptions were not met. For analysis of live-imaging disassembly, repeated-measures ANOVA test was used with contrast analysis. Statistical P values < 0.05 were considered significant. Data presented as specified in the Figure legends. Data are shown as means ± SEM or SD or graphed using boxplots, as noted in the text.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-TIA1 (C-20) | Santa Cruz | Cat#sc-1751; RRID:AB_2201433 |
| Mouse monoclonal anti-UBC9 (C-12) | Santa Cruz | Cat#sc-271057; RRID:AB_10610674 |
| Rabbit polyclonal anti-p-eif2α (Ser52) | Santa Cruz | Cat#sc-101670; RRID:AB_2096507 |
| Mouse monoclonal anti-G3BP1 (H-10) | Santa Cruz | Cat#sc-365338; RRID:AB_10846950 |
| Mouse monoclonal anti-FXR1 (B-2) | Santa Cruz | Cat#sc-374148; RRID:AB_10918113 |
| Rabbit polyclonal anti-SUMO-1 | Cell Signaling | Cat#4930; RRID:AB_10698887 |
| Rabbit polyclonal anti-SUMO-2/3 | Cell Signaling | Cat#4971; RRID:AB_2198425 |
| Rabbit polyclonal anti-RanBP2 | abcam | Cat#ab64276; RRID:AB_1142517 |
| Rabbit polyclonal anti-α-Tubulin | Sigma-Aldrich | T9026; RRID: AB_477593 |
| Mouse monoclonal anti-V5-Tag | Thermo Fisher | Cat# R960–25; RRID:AB_2556564 |
| Mouse monoclonal anti-V5-Tag | Cell Signaling | Cat# 13202; RRID:AB_2687461 |
| Rabbit polyclonal anti-ASUN | ProteinTech | Cat#19892–1-AP; RRID:AB_10638473 |
| Rabbit polyclonal anti-BLM | Bethyl labratories | Cat# A300–110A; RRID:AB_2064794 |
| Rabbit polyclonal anti-CBS | ProteinTech | Cat#14787–1-AP; RRID:AB_2070970 |
| Rabbit polyclonal anti-CBX3 | Bethyl labratories | Cat# A300–984A; RRID:AB_805792 |
| Rabbit polyclonal anti-Cdc20 | ProteinTech | Cat#10252–1-AP; RRID:AB_2229016 |
| Rabbit polyclonal anti-DTL/CDT2 | Bethyl labratories | Cat# A300–948A; RRID:AB_2093890 |
| Rabbit polyclonal anti-LGRN | ProteinTech | Cat#12840–1-AP; RRID:AB_2135302 |
| Rabbit polyclonal anti-GAK | Bethyl labratories | Cat# A304–268A; RRID:AB_2620464 |
| Rabbit polyclonal anti-GSPT1 | ProteinTech | Cat#10763–1-AP; RRID:AB_2115506 |
| Rabbit polyclonal anti-GTF3C1/TFIIIC220 | Bethyl labratories | Cat# A301–292A; RRID:AB_938040 |
| Rabbit polyclonal anti-GTF3C3/TFIIIC102 | Bethyl labratories | Cat#A301–237A; RRID:AB_890670 |
| Rabbit polyclonal anti-GTF3C4/TFIIIC90 | Bethyl labratories | Cat# A301–239A; RRID:AB_890667 |
| Rabbit polyclonal anti-MYCBP | ProteinTech | Cat#12022–1-AP; RRID:AB_2148722 |
| Rabbit polyclonal anti-OGT | ProteinTech | Cat#11576–2-AP; RRID:AB_2156943 |
| Rabbit polyclonal anti-POLR3E | Bethyl labratories | Cat# A303–708A; RRID:AB_11205449 |
| Rabbit polyclonal anti-TSC2 | Bethyl labratories | Cat# A300–526A; RRID:AB_2207795 |
| Rabbit polyclonal anti-YTHDC2 | ProteinTech | Cat#27779–1-AP |
| Rabbit polyclonal anti-ZCCHC6 | ProteinTech | Cat#25196–1-AP |
| HRP-Streptavidin | Sigma-Aldrich | Cat#RABHRP3 |
| Avidin, NeutrAvidin, Texas Red conjugate | ThermoFisher | Cat# A2665 |
| Goat-anti Mouse STAR 580 | Abberior | Cat# 52403 |
| Goat anti-rabbit ATTO647n | Sigma Aldrich | Cat# 40839 |
| Rabbit anti-Mouse Alexa Fluor 568 | Invitrogen | Cat# A-11061 |
| Goat anti-Rabbit Alexa Fluor 647 | Invitrogen | Cat# A-21245 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| (+)-Sodium L-ascorbate | Sigma-Aldrich | A7631; CAS: 134-03-2 |
| Sodium Azide OR | Mallinckrodt | 1953–57; CAS: 26628-22-8 |
| (±)-6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox) | Sigma-Aldrich | 238813; CAS: 53188-07-1 |
| N-Ethylmaleimide (NEM) | Sigma-Aldrich | E3876; CAS: 128-53-0 |
| 2-D08 | Sigma-Aldrich | SML1052; CAS: 144707-18-6 |
| Biotin | Sigma-Aldrich | B4501; CAS: 58-85-5 |
| Biotin Tyramide (Biotin-Phenol) | Iris Biotech GmbH | LS-3500; CAS: 41994-02-9 |
| Tetracycline-hydrochloride | Sigma-Aldrich | T7660; CAS: 64-75-5 |
| Sodium (meta)arsenite | Sigma-Aldrich | 71287; CAS: 7784-46-5 |
| Hydrogen peroxide 30% | J.TBaker | 2186–01; CAS: 7722-84-1 |
| Sequencing Grade Modified Trypsin | Promega | Cat#V5113 |
| LysC Trypsin mix | Promega | Cat#V5071 |
| Critical Commercial Assays | ||
| Pierce Streptavidin Magnetic Beads | Thermo Fisher | 88816 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | G7571 |
| Experimental Models: Cell Lines | ||
| U2OS-TetR-dddFMR1/FXR1/FXR2 | Paul Anderson lab | N/A |
| U2OS-TetR-ddG3BP1/G3BP2 | Paul Anderson lab | N/A |
| U2OS-TetR_TetO-APEX-NES | Paul Anderson lab | N/A |
| U2OS-TetR-dddFMR1/FXR1/FXR2_TetO-APEX-FMR1 | This paper | N/A |
| U2OS-TetR-dddFMR1/FXR1/FXR2_TetO-APEX-FXR1 | This paper | N/A |
| U2OS-TetR-ddG3BP1/G3BP2_TetO-APEX-G3BP1 | This paper | N/A |
| U2OS-TetR-dddFMR1/FXR1/FXR2_TetO-APEX-FMR1/TetO-GFP-PR50 | This paper | N/A |
| U2OS-TetR-dddFMR1/FXR1/FXR2_TetO-APEX-FMR1/TetO-GFP | This paper | N/A |
| U2OS-TetR_TetO-APEX-NES/TetO-GFP-PR50 | This paper | N/A |
| U2OS-TetR_TetO-APEX-NES/TetO-GFP | This paper | N/A |
| U2OS-TetR_TetO-GFP-PR50 | This paper | N/A |
| U2OS-TetR_TetO-GFP | This paper | N/A |
| U2OS-TetR-ddG3BP1/G3BP2_mRFP-G3BP1 | This paper | N/A |
| U2OS-TetR-ddG3BP1/G3BP2_eGFP-G3BP1 | This paper | N/A |
| U2OS-TetR-ddG3BP1/G3BP2_mRFP-G3BP1_TetO-GFP-PR50 | This paper | N/A |
| U2OS-TetR-ddG3BP1/G3BP2_mRFP-G3BP1_TetO-GFP | This paper | N/A |
| Mouse embryonic stem cells UBC9-iKO | This paper | N/A |
| U2OS-TetR-dddFMR1/FXR1/FXR2_TetO-GFP-FMR1 | This paper | N/A |
| U2OS-TetR-dddFMR1/FXR1/FXR2_TetO-GFP-FMR1-K88R,K130R | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| w; UAS-PR36, GMR-gal4/ CyO | Fumagalli et al., 2019 | N/A |
| UAS-Lwr | Drosophila Bloomington stock center | #9324 |
| Oligonucleotides | ||
| Primers used for cloning are in Table S5 | This paper | N/A |
| MSL2 siGenome siRNA | Dharmacon | M-020828–01 |
| PIK3R2 siGenome siRNA | Dharmacon | M-003021–03 |
| RNF41 siGenome siRNA | Dharmacon | M-006922–01 |
| SAE1 siGenome siRNA | Dharmacon | M-006402-00-0005 |
| SETX siGenome siRNA | Dharmacon | M-021420–01 |
| SH3RF2 siGenome siRNA | Dharmacon | M-007145–00 |
| UBE2G2 siGenome siRNA | Dharmacon | M-009095–01 |
| UBE2I siGenome siRNA | Dharmacon | M-004910-00-0005 |
| WDR45B siGenome siRNA | Dharmacon | M-017119–01 |
| siGENOME Nontargeting siRNA #5 | Dharmacon | D-001210–05 |
| Recombinant DNA | ||
| Plasmids used are in Table S5 | N/A | N/A |
| Software and Algorithms | ||
| MaxQuant | Cox and Mann, 2008 | https://maxquant.org |
| Andromeda search engine | Cox et al., 2011 | N/A |
| Perseus | Tyanova et al., 2016 | http://maxquant.org/perseus |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij |
| Other | ||
| Empore Octadecyl C18 47mm Extraction disks | Sigma-Aldrich | Cat#66883-U |
| 50 cm EASY-spray PepMap column | Thermo Scientific | Cat#ES803 |
Highlights.
Resource of SG spatiotemporal proteomic landscape by multi-bait proximity labeling
Distinct substructures and >100 novel SG proteins
Disassembly-engaged proteins (DEPs) coordinate SG disassembly
SUMOylation controls SG dynamics and is dysregulated in models of C9orf72-ALS
ACKNOWLEDGMENTS
E.H. is the Mondry Family Professorial Chair and head of the Nella and Leon Benoziyo Center for Neurological Diseases at Weizmann Institute of Science. We thank Alice Ting (Stanford University) for creating and generously sharing the APEX proximity labeling technology and Davide Trotti (Jefferson University) for the 50× proline-arginine repeat vector, whose insert serves as basis of the GFP-poly(PR)50 construct. We thank Assaf Kacen, Ron Rotkof, and Avital Eisenberg (Weizmann Institute of Science), as well as Monica Gotta and Luca Cirillo (University of Geneva), for advice and protocols. We thank the Bloomington Drosophila Stock Center (National Institutes of Health grant P40OD018537). We thank Roy Beck Barkai and Eran Perlson (Tel Aviv University) for discussions, members of the Hornstein, Anderson, and Ivanov labs for helpful critiques, and life science editors for editorial assistance. Research in the Hornstein lab was supported by the Radala Foundation (project “Proteomic mapping of SG disassembly processes suggests a potential neuroprotective role of SUMOylation in C9orf72-associated ALS”); the Minerva Foundation (project “Regulation of stress granules disassembly”) with funding from the Federal German Ministry for Education and Research, ISF Legacy grant 828/17; Target ALS grant 118945; the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement 617351; the Israel Science Foundation (135/16); the ALS Therapy Alliance; AFM-Téléthon (20576); the Motor Neurone Disease Association (UK); The Thierry Latran Foundation for ALS Research; ERA-Net for Research Programmes on Rare Diseases (FP7); Yeda-Sela, Yeda-CEO; the Israel Ministry of Trade and Industry; Y. Leon Benoziyo Institute for Molecular Medicine; the Benoziyo Center Neurological Disease; the Kekst Family Institute for Medical Genetics; the David and Fela Shapell Family Center for Genetic Disorders Research; the Crown Human Genome Center; the Nathan, Shirley, Philip, and Charlene Vener New Scientist Fund; the Julius and Ray Charlestein Foundation; the Fraida Foundation; the Wolfson Family Charitable Trust; the Abney Foundation; Merck; Maria Halphen; and the estates of Fannie Sherr, Lola Asseof, Lilly Fulop, and E. and J. Moravitz. This work was further supported by EU Horizon 2020 Programme for Research and Innovation Marie Sk1odowska-Curie actions (grant 845692 to T.G.M.) and the National Institutes of Health (grant R35 GM126901 to P.A., grant RO1 GM126150 to P.I., and grant P40OD018537 to the Bloomington Drosophila Stock Center).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2020.10.032.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alberti S, and Dormann D (2019). Liquid-liquid phase separation in disease. Annu. Rev. Genet 53, 171–194. [DOI] [PubMed] [Google Scholar]
- Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian SB, and Ignatova Z (2018). Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1, e201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci 33, 141–150. [DOI] [PubMed] [Google Scholar]
- Aulas A, Caron G, Gkogkas CG, Mohamed NV, Destroismaisons L, Sonenberg N, Leclerc N, Parker JA, and Vande Velde C (2015). G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol 209, 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, Kriwacki RW, and Pappu RV (2012). Structural biology. Versatility from protein disorder. Science 337, 1460–1461. [DOI] [PubMed] [Google Scholar]
- Bah A, and Forman-Kay JD (2016). Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem 291, 6696–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional control of phase-separated cellular bodies. Cell 166, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair IP, Bennett CL, Abel A, Rabin BA, Griffin JW, Fischbeck KH, Cornblath DR, and Chance PF (2000). A gene for autosomal dominant juvenile amyotrophic lateral sclerosis (ALS4) localizes to a 500-kb interval on chromosome 9q34. Neurogenetics 3, 1–6. [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, et al. (2017). Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, and Parker R (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, and Parker R (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candé C, Vahsen N, Métivier D, Tourrière H, Chebli K, Garrido C, Tazi J, and Kroemer G (2004). Regulation of cytoplasmic stress granules by apoptosis-inducing factor. J. Cell Sci 117, 4461–4468. [DOI] [PubMed] [Google Scholar]
- Chew J, Cook C, Gendron TF, Jansen-West K, Del Rosso G, Daughrity LM, Castanedes-Casey M, Kurti A, Stankowski JN, Disney MD, et al. (2019). Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener 14, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L, Cieren A, Barbieri S, Khong A, Schwager F, Parker R, and Gotta M (2020). UBAP2L Forms distinct cores that act in nucleating stress granules upstream of G3BP1. Curr. Biol 30, 698–707.e6. [DOI] [PubMed] [Google Scholar]
- Cook C, and Petrucelli L (2019). Genetic convergence brings clarity to the enigmatic red line in ALS. Neuron 101, 1057–1069. [DOI] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, and Mann M (2011). Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem 80, 273–299. [DOI] [PubMed] [Google Scholar]
- Cox J, Michalski A, and Mann M (2011). Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom 22, 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, and Mann M (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao TP, Kolaitis RM, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, and Castañeda CA (2018). Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, and Parker R (2012). P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol 4, a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz AA (2020). Networking and dynamic switches in biological condensates. Cell 181, 228–230. [DOI] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. (2010). ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, and Brangwynne CP (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, and Taylor JP (2017). The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli L, Young FL, Boeynaems S, Decker MD, Mehta A, Swijsen A, Fazal R, Guo W, Moisse M, Beckers J, et al. (2019). C9orf72-derived arginine-containing dipeptide repeats associate with axonal transport machinery and impede microtubule-based motility. bioRxiv. 10.1101/835082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, et al. (2016). A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol. Cell 63, 796–810. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, and Melchior F (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol 8, 947–956. [DOI] [PubMed] [Google Scholar]
- Guillén-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, Kim K, Trussina IREA, Wang J, Mateju D, et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, and McKnight SL (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. [DOI] [PubMed] [Google Scholar]
- Hart Y, and Alon U (2013). The utility of paradoxical components in biological circuits. Mol. Cell 49, 213–221. [DOI] [PubMed] [Google Scholar]
- Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, and Nielsen ML (2017). Site-specific mapping of the human SUMO proteome reveals comodification with phosphorylation. Nat. Struct. Mol. Biol 24, 325–336. [DOI] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, et al. (2016). UBQLN2 mediates autophagy-independent protein aggregate clearance by the protea-some. Cell 166, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T, et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e713. [DOI] [PubMed] [Google Scholar]
- Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, and Ting AY (2016). Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc 11, 456–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, and Anderson P (2019). Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol 11, a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongjitwimol J, Baldock RA, Morley SJ, and Watts FZ (2016). Sumoylation of eIF4A2 affects stress granule formation. J. Cell Sci 129, 2407–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, and Anderson P (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, and Anderson P (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol 212, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiten-Schmitz J, Wagner K, Piller T, Kaulich M, Alberti S, and Müller S (2020). The nuclear SUMO-targeted ubiquitin quality control network regulates the dynamics of cytoplasmic stress granules. Mol. Cell 79, 54–67.e7. [DOI] [PubMed] [Google Scholar]
- Khayachi A, Gwizdek C, Poupon G, Alcor D, Chafai M, Cassé F, Maurin T, Prieto M, Folci A, De Graeve F, et al. (2018). Sumoylation regulates FMRP-mediated dendritic spine elimination and maturation. Nat. Commun 9, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Keyser SG, and Schneekloth JS Jr. (2014). Synthesis of 2′,3′,4′-trihydroxyflavone (2-D08), an inhibitor of protein sumoylation. Bioorg. Med. Chem. Lett 24, 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, and Alberti S (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, and Ting AY (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. (2016). C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organ-elles. Cell 167, 774–788.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, and Chang P (2011). Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, and Gitler AD (2013). Stress granules as crucibles of ALS pathogenesis. J. Cell Biol 201, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YC, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, Patel R, Qamar S, Nixon-Abell J, Shen Y, et al. (2019). RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell 179, 147–164.e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier BT, Huttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, and Krogan NJ (2017). An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169, 350–360.e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros B, Erdos G, and Dosztányi Z (2018). IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46 (W1), W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, et al. (2014). C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, and Anderson P (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol 10, 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padron A, Iwasaki S, and Ingolia NT (2019). Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887.e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Prieto M, Folci A, and Martin S (2020). Post-translational modifications of the fragile X mental retardation protein in neuronal function and dysfunction. Mol. Psychiatry 25, 1688–1703. [DOI] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016a). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2013). R: A language and environment for statistical computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Ramaswami M, Taylor JP, and Parker R (2013). Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, and Jaffrey SR (2019). m6A enhances the phase separation potential of mRNA. Nature 571, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltema RA, Hauschild J-P, Lange O, Hornburg D, Denisov E, Damoc E, Kuehn A, Makarov A, and Mann M (2014). The Q exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics 13, 3698–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Rasband W, and Eliceiri K (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, and Carra S (2014). Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ. 21, 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey LM, Safren N, Pithadia AS, Gerson JE, Dulchavsky M, Fischer S, Patel R, Lantis G, Ashraf N, Kim JH, et al. (2018). Mutant UBQLN2 promotes toxicity by modulating intrinsic self-assembly. Proc. Natl. Acad. Sci. USA 115, E10495–E10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J (2019). Phase separation of RNA-binding proteins in physiology and disease: An introduction to the JBC Reviews thematic series. J. Biol. Chem 294, 7113–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Curry EG, Blue RE, Roden C, Dundon SER, Rodríguez-Vargas A, Jordan DC, Chen X, Lyons SM, Crutchley J, et al. (2019). Regulation of FXR1 by alternative splicing is required for muscle development and controls liquid-like condensates in muscle cells. bioRxiv. 10.1101/818476. [DOI] [Google Scholar]
- Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, and Schrader JW (2007). Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol 27, 2324–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammsalu T, Matic I, Jaffray EG, Ibrahim AFM, Tatham MH, and Hay RT (2014). Proteome-wide identification of SUMO2 modification sites. Sci. Signal 7, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr., and Cleveland DW (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45 (D1), D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd TW, McEachin ZT, Chew J, Burch AR, Jansen-West K, Tong J, Yue M, Song Y, Castanedes-Casey M, Kurti A, et al. (2020). Hexanucleotide repeat expansions in c9FTD/ALS and SCA36 confer selective patterns of neurodegeneration in vivo. Cell Rep. 31, 107616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, and Tazi J (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol 160, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z, and Gao FB (2015). Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron 87, 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakhiya A, Meyer SR, Marincola G, Böhm S, Vanselow JT, Schlosser A, Hofmann K, and Buchberger A (2018). ZFAND1 recruits p97 and the 26S proteasome to promote the clearance of arsenite-induced stress granules. Mol. Cell 70, 906–919.e907. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, and Forman-Kay JD (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Maxwell BA, Joo JH, Gwon Y, Messing J, Mishra A, Shaw TI, Ward AL, Quan H, and Sakurada SM (2019). ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Mol. Cell 74, 742–757. e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. (2014). Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, and Parker R (2016). Distinct stages in stress granule assembly and disassembly. eLife 5, e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B, and Ivanov P (2019). Stress granules and neurodegeneration. Nat. Rev. Neurosci 20, 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e511. [DOI] [PubMed] [Google Scholar]
- Zhang Y, O’Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, and Dreyfuss G (1995). The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 14, 5358–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN, et al. (2018). Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med 24, 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomic datasets generated during this study are available at the PRIDE repository: PXD017330. Original data for figures in the paper is available in Mendeley Data https://doi.org/10.17632/vvthhmwwxw.1


