Kanfer et al. develop a pooled CRISPRi screening method to identify genes regulating intracellular protein localization, organelle morphology or other subcellular phenotypes. The method uses machine learning to identify genetically altered cells, a photoactivated fluorescent protein to label them, and FACS plus deep sequencing to identify the affected gene.
Abstract
Genome-wide CRISPR screens have transformed our ability to systematically interrogate human gene function, but are currently limited to a subset of cellular phenotypes. We report a novel pooled screening approach for a wider range of cellular and subtle subcellular phenotypes. Machine learning and convolutional neural network models are trained on the subcellular phenotype to be queried. Genome-wide screening then utilizes cells stably expressing dCas9-KRAB (CRISPRi), photoactivatable fluorescent protein (PA-mCherry), and a lentiviral guide RNA (gRNA) pool. Cells are screened by using microscopy and classified by artificial intelligence (AI) algorithms, which precisely identify the genetically altered phenotype. Cells with the phenotype of interest are photoactivated and isolated via flow cytometry, and the gRNAs are identified by sequencing. A proof-of-concept screen accurately identified PINK1 as essential for Parkin recruitment to mitochondria. A genome-wide screen identified factors mediating TFEB relocation from the nucleus to the cytosol upon prolonged starvation. Twenty-one of the 64 hits called by the neural network model were independently validated, revealing new effectors of TFEB subcellular localization. This approach, AI-photoswitchable screening (AI-PS), offers a novel screening platform capable of classifying a broad range of mammalian subcellular morphologies, an approach largely unattainable with current methodologies at genome-wide scale.
Introduction
Recent advances have expanded traditional genetic screens from bacteria and yeast to mammalian cells. RNAi, CRISPRi, and CRISPR screens rely on two main strategies: arrayed and pooled screens. Arrayed screens are highly specific, but require the production and, by definition, individual assortment of each RNAi or CRISPR guide separately, requiring high-throughput equipment not readily available to academic laboratories. Pooled screens are more facile, but had been restricted to phenotypes that affect cell growth rates or viability or result in a fluorescence increase that allows for isolation of hits from the population by using FACS. Single-cell RNA-based pooled screens are also useful to link genetic profiles to perturbations (Horlbeck et al., 2018; Datlinger et al., 2017; Dixit et al., 2016; Adamson et al., 2016). The use of image-based pooled genetic screens linking phenotypes to genotypes was previously reported in three independent studies in which in situ barcoded sequencing was coupled to phenotypes. First, this approach was used to identify photostable and brighter variants of a fluorescent protein by testing 60,000 mutation variants (Emanuel et al., 2017). Then, an in situ platform was integrated with CRISPR genetic screens to identify genes involved in RNA nuclear localization, while another CRISPR screen used in situ sequencing imaging to identify factors associated with nuclear factor κB translocation regulation. These later two methods screened 162 CRISPR guides in Wang et al. (2019) and 3,063 guides in Feldman et al. (2019). More recently, a semiarrayed 12,500 gRNA CRISPR screen was used to identify regulators of stress granule formation (Wheeler et al., 2020). These methods enable the investigation of protein pathways regulating subcellular organization and positioning in an unbiased manner. In addition to unbiased CRISPR screens linking microscopic phenotypes to genotypes, single-cell images linking microscopic phenotypes to genotypes were established by a new method called single-cell magneto-optical capture (Binan et al., 2019). Although these processes are elegant and will improve genetic studies, they are not well suited for high-throughput large-scale screens. Hence, we propose that a simple photoactivation of cells with desired phenotypes coupled to cell sorting will reduce image-based screen complexity. Previously, B lymphocyte isolation and characterization were conducted from photoactivatable transgenic mice by coupling photoactivation and flow cytometry (Victora et al., 2010). In addition, in a more recent study, photoactivation coupled to flow cytometry enabled the investigation of the link between the morphology response to a drug and the genetic profile at single-cell resolution (Hasle et al., 2020).
Recent advances in machine learning, and particularly in deep learning (convolutional neural networks [CNN]; Caicedo et al., 2019; Bzdok et al., 2018), offer novel strategies for identifying individual cells with altered organelle morphology or subcellular protein localization. We developed a screening method to identify genetic perturbations of subcellular morphologies that is widely applicable and high throughput. The method is divided into four steps. First, a morphology classification model is trained on single-cell images. Second, pools of CRISPRi-perturbed target cells are imaged sequentially, and the phenotypically selected cells are labeled by laser photoactivation of a fluorescent protein. Third, the photoactivated cells are sorted. Fourth, the guides within the phenotypically identified cells are amplified and sequenced. The decision to select cells is made on the fly by pretrained classification models allowing for screening of 106 cells within 12 h and the whole human genome in a week.
Results
Building the single-cell imaging screening approach
We developed a new platform that assesses images of cells and uses machine learning to distinguish their subcellular phenotypes. By using laser activation of a fluorescent probe to denote the selected cell phenotypes and FACS to separate the cells for guide sequencing, one essentially converts the individual cells exposed to pooled CRISPRi libraries into an arrayed screen (Fig. 1 a, i–iii). By using this approach, every imaged cell is referred to as an independent entity, and a predicted phenotype score is produced based on a classification machine-learning model (Fig. 1 a, ii). Making the artificial intelligence (AI) platform entails three steps: training and creation of the phenotype classification model, model deployment on pooled imaged cells, and validation of the model’s screening performance. We used Pink1-dependent Parkin translocation to mitochondria as a proof of concept (Fig. 1 b). In cells with unimpaired polarized mitochondria, Parkin is in the cytoplasm; however, upon mitochondrial depolarization, it translocates to mitochondria (Narendra et al., 2008; Fig. 1, b and c). This binary switch in the Parkin location is suitable for detection by a support vector machine (SVM) classification model. An SVM classification model was trained on images of cells with either cytosolic or mitochondrial GFP-Parkin. To build the SVM classification model, 18 features were computed from 2,500 single-cell images of cytosolic or mitochondrial GFP-Parkin (Fig. 1 d). The features were computed by using the R image processing and analysis package, EBImage (Pau et al., 2010). To prevent classifier overfitting and reduce the computational cost, five cellular features measuring the 5% intensity quantile, the SD of intensity, minimum radius, eccentricity, and area that showed distinct variation were selected (Fig. 1 e). The selected features and labeled cell images were computationally applied on a nonlinear SVM algorithm for creating the classification model (Fig. S1 a).
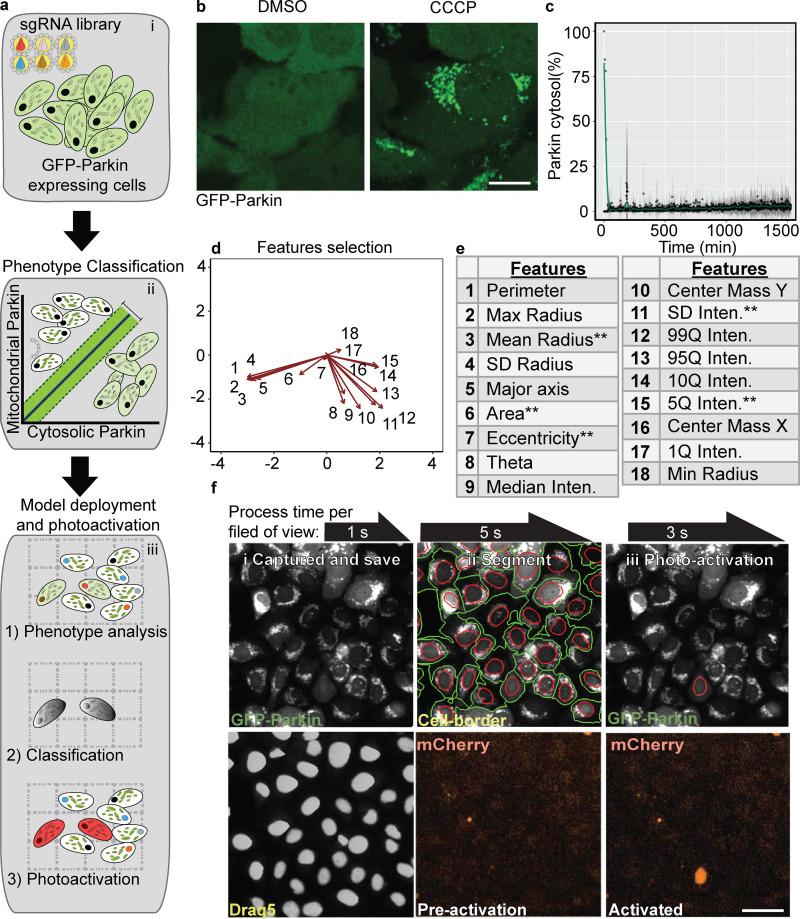
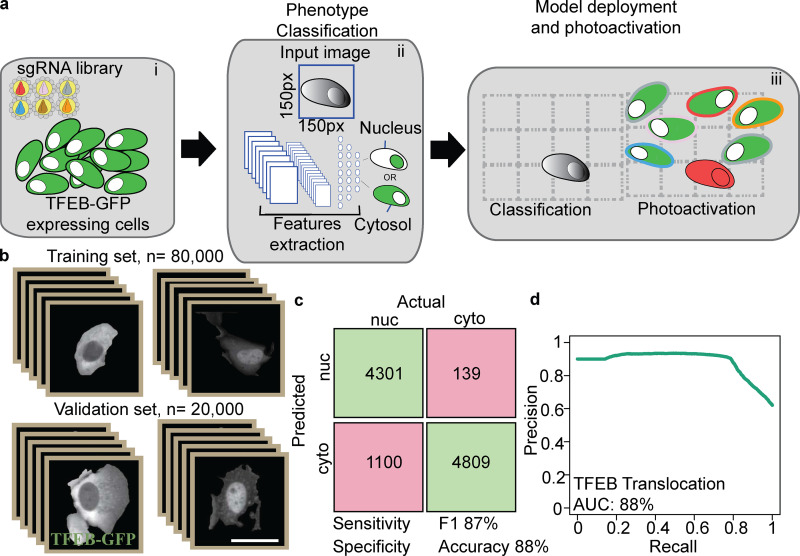
Figure 1.
Machine learning genetic screening platform for Parkin localization: proof of principle screen. (a) Screen illustration. The AI-PS platform is composed of three components i: Transduction. GFP-Parkin, pa-mCh, and dCas9-KRAB cells transduced with a subpooled library of sgRNA. ii: Machine learning modeling. Single-cell images are labeled and trained using SVM classification of mitochondrial Parkin vs. cytosolic Parkin. iii: AI-PS deployment. First, cells are imaged and segmented, and then the phenotypes of targeted cells are called and photoactivated. (b) Representative images of GFP-Parkin U2OS cells treated with DMSO or CCCP (2 h). Scale bar, 5 µm. (c) Translocation dynamic analysis of GFP-Parkin cells imaged for 25 h (1,500 min) in 1-min time lapses. Percent of cytosolic Parkin over time after 10 µM CCCP treatment supplemented with 100 nM bafilomycin A. n = 7, mean ± SD. (d) PCA analysis of 18 feature predictors calculated from 5,435 single-cell images using the R function computeFeatures from the EBImage library. The images are pooled from five different biologic repeats. (e) Table includes all the input features computed. **features selected for the model. (f) Image examples of field of view of GFP-Parkin U2OS cell screening procedure. i: Images were captured and saved on a local computer. Top: Parkin-GFP. Bottom: Draq5 dye for nucleus segmentation. ii: Cell borders were identified (green circle surrounding cell border, red circle the nucleus) following nucleus segmentation. Bottom: mCherry channel for ph-mCh protein. iii: SVM classification model was deployed and masked (red circle). Photoactivation of the SMV identified cell. Scale bar, 10 µm.
Figure S1.
SVM classification build for Parkin screen. (a) Two-dimensional representation of nonlinear hyperplane separation of mitochondrial (mito) Parkin phenotype vs. cytosolic (cyto) phenotype. Mitochondrial Parkin predicted cells are in red, cytosolic Parkin in black. The variable o represents correct classification, and x misclassification. (b) The confusion matrix is computed from a test set composed of three biological replicates pooled single-cell images (n = 4,894). Prior to the SVM classification prediction, the images were manually labeled. The green boxes are designated for the true positive single-cell counts (upper left) and the true negative (lower right). The pink boxes represent the false positive single-cell counts (upper right) and the false negative (lower left). (c) Precision recall plot (right) summarizes the same image test set. The accuracy is calculated from the precision recall curve. (d) Beeswarm plot summarizing single-cell TFEB-GFP segmentation score comparison. The image contains 79 cells from the TFEB screen, segmented with AI-PS segmentation procedure (using the R package EBImage; black) or Cellprofiler segmentation pipeline (red). Mean ± SD. (e) dCas9-KRAB–expressing U2OS cells treated with sgRNA targeting either TRANS or CDH2 and immunostained using TRANS or CDH2 antibodies. Scale bar, 20 µm. AUC, area under the curve; IOU, intersection over union.
To optimize the model, we performed iterations and calculated the performance by area under the precision-recall curve. To prevent overfitting, we shuffled the featured data and split it into two unique groups, a test set and a training image set. We then fitted an SVM model on the training set and evaluated it on the test set, and then an accuracy score was calculated. This procedure was iterated 100 times where every observation was allowed to be used in the training or test set only once. On ∼5,000 single-cell images, the classifier accuracy was 99% (Fig. S1, b and c). Next, we generated an easy-to-use graphical user interface program to facilitate image segmentation, measurement, and model building (Fig. S2). The R-based script for image segmentation and analysis, as well as the SVM classification model, were deployed on the fly to identify cells exhibiting the desired phenotype—GFP-Parkin mitochondrial localization. During live-cell image acquisition, single-cell images were captured following segmentation and stored on a local computer (Fig. S2). The accuracy of the segmentation procedure was compared with the gold standard manual segmentation by using the Nikon Imaging System (NIS) elements imaging software. The segmentation was evaluated by calculating the intersection over union. Comparing the intersection over union of the current segmentation procedure with CellProfiler showed very similar segmentation scores (Fig. S1 c). The SVM-based model classified the individual cells and generated a mask corresponding to the live image field identifying the location of cells with the phenotype of interest (Fig. 1 f and Video 1). In cells identified with this mask, photoactivatable mCherry (pa-mCh) was then laser photoactivated. Selected cells were photoswitched by illumination of 50 ms/pixel dwell time with 80% UV laser intensity. This parameter was chosen so as to reduce the photoactivation time, eliminate unwanted activation of adjacent cells, and maximize signal intensity. This 10-s process was iterated across serial images of the entire chamber slide—an average of 600,000 cells for one subgenomic CRISPRi guide pool (Gilbert et al., 2014; Horlbeck et al., 2016). Finally, the photoactivated samples were sorted by using flow cytometry and were deep sequenced to determine sgRNA abundance in the activated sample compared with untreated cells.
Figure S2.
Shiny AI-PS Application and output files. Detailed instructions and the source package for the AI-PS graphical user interface (GUI) can be found at https://github.com/hbaldwin07/GK_shiny_app. The AI-PS GUI application is also hosted online at https://hab-gk-app.shinyapps.io/gk_shiny_app/. (a) Screenshots of the nucleus (left) and cell outline (right) segmentation tools. (b) Example of image classification interface. Image file(s) are segmented using predetermined parameters set in the previous panel or uploaded by the user. The individual cells (outlined in yellow) are manually selected for positive or negative classification. (c) Schematic of the integration of AI-PS output with Nikon elements software for deployment. The three files generated by the AI-PS application are the SVM model file (“Create Model” tool), the Deployment R script (downloaded from https://github.com/gkanfer/AI-PS), and the Parameter file (table of nucleus and cell segmentation parameters generated by “Save Image Parameters” in segmentation panel). All three files should be saved in a common directory on the microscope’s local computer (i.e., C:\outproc\) and directed to the Nikon elements JOB module outproc.
Video 1.
Example of AI-PS platform Parkin screen proof of principle. Nikon NIS elements JOB module screen capture during GFP-Parkin (green) acquisition. Machine learning deployed for automatic detection of sgRNA targeting PINK1 (blue) according to cell phenotype (red circle on top of GFP-Parkin image). The detected cell is photoactivated (yellow-red).
Photoactivation accuracy and performance
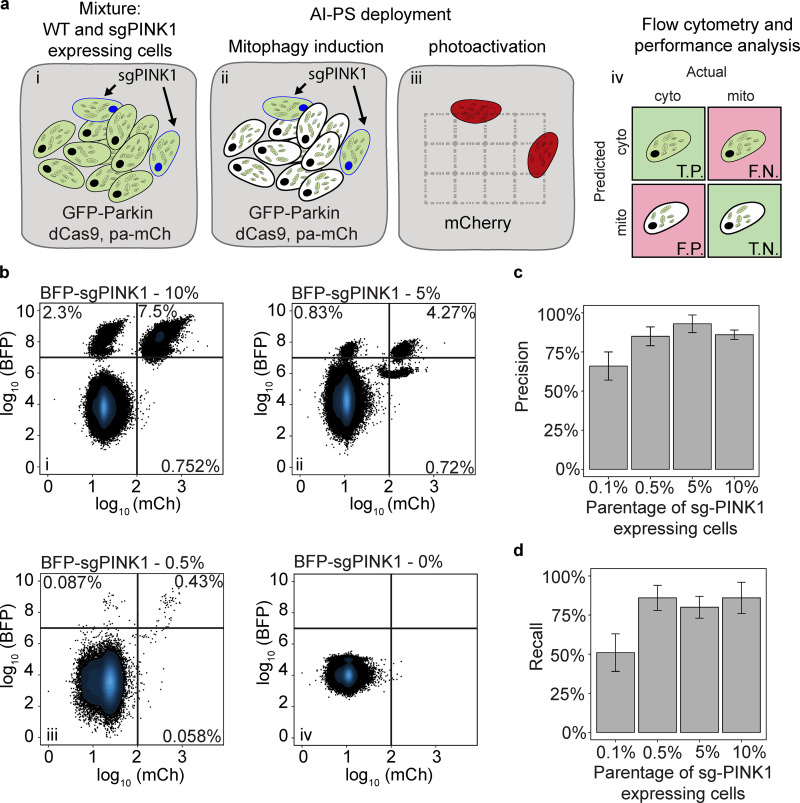
The performance of phenotype classification and sorting of photoactivated cells were evaluated separately. First, dCas9-KRAB (Horlbeck et al., 2016) was expressed and tested in U2OS cells expressing pa-mCh and GFP-Parkin (Fig. S1 d). To calculate the sorting accuracy of the detected and photoactivated cells from the entire population, we experimentally mixed cells blocked for Parkin recruitment with WT cells (Fig. 2 a). In brief, we gradated a mixture of blue fluorescent protein (BFP)–tagged gRNA-targeting PINK1 cells with WT cells expressing GFP-Parkin and dCas9-KRAB and pa-mCh in a ratio of either 0.1%, 0.5%, 5%, or 10% sgPINK1 with WT cells (Fig. 2 a, i). Then, cells were treated with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to stimulate Parkin activation (Fig. 2 a, ii). Cells with Parkin evenly spread in the cytosol that had failed to activate the PINK1-Parkin pathway were photoactivated and sorted. The sensitivity and specificity of cells were analyzed from the BFP and pa-mCh intensity ratio (Fig. 2 a, iii and iv). First, we observed that, with a phenotype penetration from 0.5% to 10%, both the precision and recall scored similarly at ∼85% (Fig. 2, b–d); however, reducing the phenotype penetration to 0.1% resulted in a reduction in both recall and precision for values to 65% and 50%, respectively (Fig. 2, b–d). Therefore, in comparison to the previous study (Hasle et al., 2020), the FACS separation performance values are slightly lower in precision (85% vs. 94%) and slightly higher in recall values (86% vs. 80%). Similar to previous work with the pa-mCh fluorescent protein that we used in the current study (Patterson and Lippincott-Schwartz, 2002), we found that UV light activation resulted in an 80-fold increase in signal intensity.
Figure 2.
Performance evaluation of AI-PS flow cytometry sorting. (a) Schematic representation of performance test. i: Mixture of GFP-Parkin cells (black nucleus) and GFP-Parkin cells expressing sgRNA targeting PINK1 (blue nucleus). ii: The color green is concentrated on the mitochondria in WT cells (black nucleus). The color green is dispersed in the cytoplasm of sgRNA-targeting PINK1 cells (blue nucleus). iii: sgPINK1 cells are detected and photoactivated. iv: The detection and flow cytometric separation of the cell populations are evaluated. T.P., true positive; T.N., true negative; F.P., false positive; F.N., false negative. (b) U2OS cells expressing dCas9-KRAB and GFP-Parkin were mixed with 10% (top left), 5% (top right), 0.5% (bottom left), and 0% (bottom left) of the sgPINK1-BFP–expressing cells. Cells with cytosolic GFP-Parkin were automatically called by the AI-PS SVM algorithm and photoactivated. The positive activated cells were gated by the BFP and RFP signals. 200,000 cells were subjected to the AI-PS procedure and the example scatter plot was set on 50,000 cells per condition. n = 3; x-axis mCh signal in log10 scale, y-axis BFP signal in log10 scale. For each scatter plot, the recovered cells are in the upper right gate, missed cells are in the upper left gate, and misclassified cells are in the bottom right. (c and d) Bar graph presenting the precision (c) and recall (d) calculated per condition. mean + SD; n = 3.
Parkin translocation screen validation
For platform validation, U2OS cells stably expressing GFP-Parkin, pa-mCh, and dCAS9-KRAB were infected with a subpool of the version 2 CRISPRi library comprising 12,775 guides targeting kinases, phosphatases, and the druggable genome (Horlbeck et al., 2016). Cells were treated with CCCP to depolarize mitochondria, and GFP-Parkin localization was assessed by using the SVM classification model (Fig. 3 a). From one batch, for example, of ∼200,000 cells, 1,132 were called, photoactivated, sorted, and sequenced (Fig. 3 b). For calculating gRNA frequency, we preformed deep sequencing and compared gRNA that was abundant between the photoactivated samples and total gRNA composition before the screen. The gRNA enrichment log2 fold change threshold was modeled based on the nontargeting negative control distribution (Fig. 3 c). The most enriched sgRNAs identified in the photoactivated samples were targeted against PINK1 (Fig. 3 d), known to be required for Parkin translocation, exhibiting a nearly 30-fold increase compared with the unsorted control sample (false discovery rate [FDR] adjusted P < 0.0001; Table S1). Thus, the single known Parkin modifier targeted in the subpool library, PINK1, was identified, validating the method. In addition, sample size estimation indicated that three biological repeats are sufficient for detecting the desired genetic link in our experimental setup (Fig. 3 e). To estimate screening performance, we evaluated AI-photoswitchable screening (AI-PS) screens by using power analysis simulation. The FDR was set to range from 5% to 15%. A power of 80% was calculated from the Parkin screen, indicating that triplicates of 200,000 cells are sufficient for screening one guide subpool comprising one seventh of the human genome; however, increasing the sample size to five repeats would increase the power and performance of this screen (Fig. 3 e).
Figure 3.
Validation of the platform with a Parkin localization screen, targeting kinases, phosphatases, and drug targets—gRNA pooled library (12,500 sgRNAs targeting 2,774 genes). (a) Schematic representation of the AI-PS platform. GFP-Parkin cells, pa-mCh, and dCas9-KRAB cells were transduced with a subgroup pooled sgRNA library. Cells with cytosolic GFP-Parkin (green color dispersed in cytosol) were photoactivated and sorted by flow cytometry and subsequently submitted to deep-sequencing analysis. (b) Flow cytometry sorted 385 cells from 77,114. Flow cytometry scatterplot representing the separation of the postscreen photoactivated from the inactivated cell population. BFP florescence signal y-axis in cyan, mCherry fluorescence signal x-axis in red. Total number of sorted cells was 2,227 (n = 3). (c) Fold-change threshold was computed from the noise model of nontargeting gRNA distribution. Mean Log2-CPM fold change in the purple line, fold-change threshold at the red vertical line represents two SDs from the mean (n = 3). Data distribution was assumed to be normal, but this was not formally tested. (d) Enrichment plot comparing sgRNA abundance in the photoactivated sample following CCCP treatment to sgRNA abundance before treatment. Vertical red line set on log2-fold change threshold; horizontal red line indicating the Benjamini-Hochberg corrected P value set on 5%. See also Table S1. The number of sgRNAs detected and filtered was 3,471 targeting 1,157 genes (n = 3). (e) Statistical power analysis by simulation on the 3,471 sgRNAs retrieved from the Parkin screen. The simulation was done using the R package PROPER (Wu et al., 2015). Effect size (log of fold-change) in the x-axis, power in the y-axis, the curved lines are color coded for the number of biologic repeats.
TFEB nuclear localization screen: CNN-based screen
To explore a subcellular phenotype with more complex regulation, we screened for genes affecting the nuclear localization of the transcription factor EB, TFEB. Upon nutrient starvation, TFEB moves from the cytosol to the nucleus, where it activates the transcription of lysosome- and autophagy-related genes (Settembre et al., 2011). Upon prolonged starvation, mammalian target of rapamycin (mTOR) is reactivated, presumably due to replenishment of nutrients through autophagy, lysosomes repopulate the cells (Yu et al., 2010), and TFEB returns to the cytosol (Fig. S3, a and b; and Video 2). As the import of TFEB to the nucleus is well elucidated (Puertollano et al., 2018), we assessed TFEB reappearance in the cytosol following prolonged starvation-induced nuclear import. U2OS cells stably expressing GFP-tagged TFEB, pa-mCh, and dCas9-KRAB (designated as TFEB-GFP) were infected with a lentiviral library expressing sgRNAs against the entire genome divided in seven separate subpools (Horlbeck et al., 2016). The screen was split into seven subscreens, one per day for 7 d. To increase reproducibility, each subpool screen was repeated at least three times.
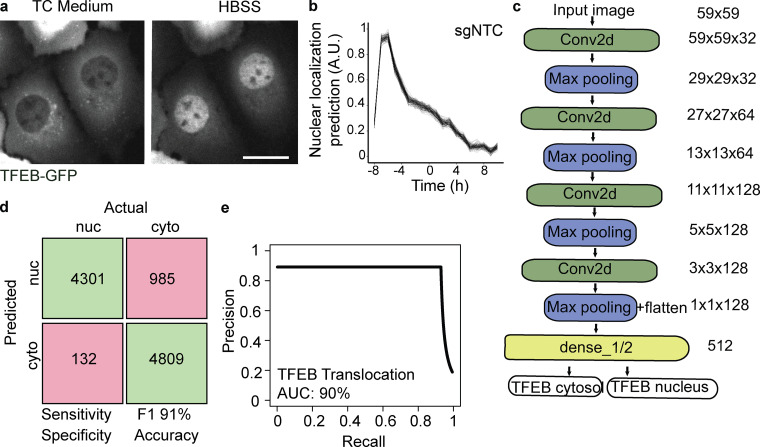
Figure S3.
GFP-TFEB translocation and parametric analysis performance. (a) Representative image of nuclear (nuc) TFEB-GFP in cells treated with HBSS (1 h) or cytosolic (cyto) TFEP-GFP in cells in complete medium. Scale bar, 5 µm. TC, tissue culture. (b) Translocation dynamic analysis of GFP-TFEB cells imaged for 18 h in 1-h time lapses. Nuclear localization prediction is computed using AI-PS segmentation and CNN prediction algorithm. n = 7, mean ± SD. (c) ImageNet-like CNN architecture is composed of four sets of convoluted processes followed by the max pooling procedure. The phenotype decision is based on probability value. (d) The confusion matrix is calculated from a test set composed of 9,110 single-cell images collected and pooled from three biologic replicates. Prior to computing the pixel intensity, the images were manually labeled. The green boxes designate the true positive single-cell count (upper left) and the true negative (lower right). The pink boxes designate the false positive single-cell count (upper right) and the false negative (lower left). (e) A precision recall plot summarizes the same image test set. The accuracy is calculated from the precision recall curve. AUC, area under the curve.
Video 2.
Live-cell images of TFEB-GFP U2OS cells under starvation conditions. TFEB-GFP–expressing U2OS cells were starved for 18 h and images were acquired every hour. Single-cell CNN prediction scores are marked in red for nuclear TFEB and in green for cytosolic TFEB. Dynamic bar chart indicates the cumulative distribution of TFEB translocation in the represented cell population.
CNN classification model for TFEB translocation prediction
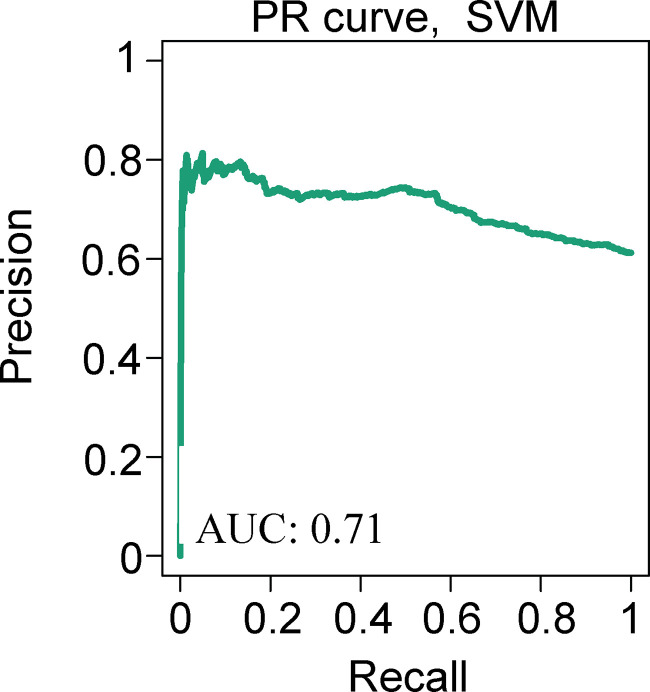
Because SVM classification failed to predict TFEB nuclear localization accurately (performance comparison between area under the precision-recall curve of 72% for the TFEB SVM classification model vs. 99% for the Parkin model; Fig. S4 and Fig. S1 b), we used deep learning via a CNN (Fig. 4 a, i-iii; and Fig. S3 c). The training set was composed of 100,000, 150-pixel × 150-pixel single-cell images using two data sets, one for each phenotypic classification (Fig. 4 b). The single-cell images were generated by using the R-based segmentation script deployed by AI-PS and manually classified. The CNN architecture was based on ImageNet (Deng et al., 2009) architecture and composed of three deconvolutions and four Max pooling processes, which were followed by a fully connected dense network (Fig. S3 c).
Figure S4.
GFP-TFEB localization SVM model prediction. GFP-TFEB phenotype classification performance by SVM, precision-recall (PR) curve from 7,848 single-cell images obtained from HBSS starved and fed cells. For the starved cell population, image collection began 8 h after starvation was initiated and continued for another 10 h. Accuracy is computed from the integral area under the precision-recall curve. AUC, area under the curve.
Figure 4.
Deep learning genetic screening platform for TFEB localization. (a) AI-PS screening platform for TFEB translocation. i: Cells were transduced with subpooled sgRNA libraries. ii: Machine learning model. Single-cell images were labeled and trained using CNN classification of nuclear (nuc) TFEB vs. cytosolic (cyto) TFEB. iii: AI-PS deployment. First, cells were imaged and segmented, and then the phenotype of target cells was called and they were photoactivated. (b) Learning set composed of 100,000 single-cell images was used for CNN classification. ImageNet-like CNN architecture was composed of four sets of convolution processes followed by the max pooling procedure. The phenotype decision is based on probability value. A low probability value was assigned to cells with cytosolic GFP-TFEB and a high probability value for cells containing nuclear GFP-TFEB. (c) The CNN classification model was applied on the test set composed of single-cell images of cytosolic or nuclear GFP-TFEB. The confusion matrix was calculated from the test sets, which were collected and pooled from five biologic replicates. Prior to the CNN classification prediction, the images were manually labeled. The green boxes are designated for the true positive single-cell counts (upper left) and the true negative (lower right). The pink boxes represent the false positive single-cell count (upper right) and the false negative (lower left). (d) Precision recall plot summarizes the same image test set. The accuracy is calculated from the precision recall curve. AUC, area under the curve.
Next, for testing our CNN classification model, single-cell images of GFP-TFEB test set were used to predict CNN classification performance compared with a parametric classification approach of the same test set. We performed this analysis and compared our CNN classification model to average pixel intensity in the nucleus vs. the cytoplasm-based prediction. Comparing CNN performance to pixel intensity computing yielded no significant difference in performance (CNN model prediction in Fig. 4, c and d; and parametric model prediction in Fig. S3, d and e). These results indicate that, in the case of the TFEB translocation classification problem, both methods preform equally and sufficiently for this task. The accuracy of pixel intensity computation of the parametric model is slightly greater than the CNN model (90% vs. 88%), whereas the classification prediction of the CNN model is better in specificity (97% vs. 83%). In the nature of the current screen, as the frequency of the desired cell phenotype is low, specificity is more of interest relative to sensitivity (Fig. 4, c and d; and Fig. S3, d and e). Overall, it is not clear why the CNN model shows higher performance than the SVM model. We speculate that the difference is most likely because of the segmentation step of our CNN model, where uneven illumination of image examples was introduced in the training set.
TFEB translocation primary screen
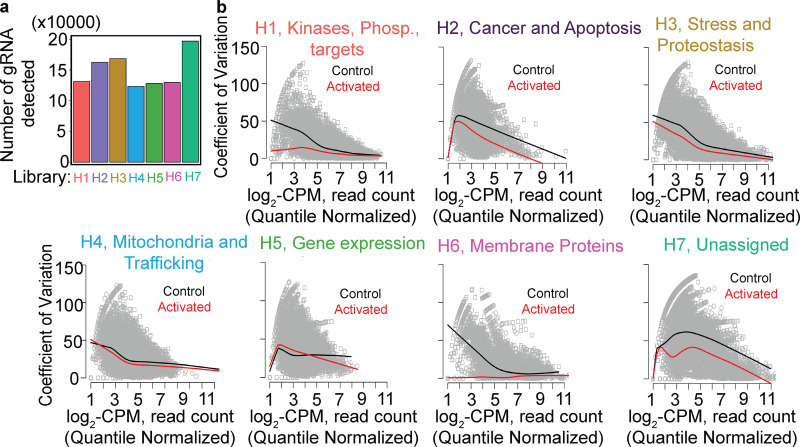
GFP-TFEB cells expressing guide libraries were grown under complete nutrient deprivation conditions for 8 h before the commencement of screening, after which those cells retaining TFEB in the nucleus were photoactivated (Fig. 5 a), isolated by FACS, and deep sequenced (Fig. 5, b and c; and Video 3). To assess the variation between the triplicate read counts of each subpool, we computed the coefficient of variation between the triplicate screens with the log2-CPM (count per million) normalized mean count per sgRNA. Every subpooled library contains 500 nontargeting gRNAs. The distribution of these gRNAs and the number of detectable gRNA per subpooled library supports a minimal variation (Fig. 6 a). From this analysis, we conclude that the overall in-group variation between the triplicate screens is minimal (Fig. 6 b, i-vii); however, there is considerable variation between the different guide subpool samples comparing photoactivated and control unactivated cells. The between-subpool sgRNA variation is reflected in the abundance analysis, since in one pool, the membrane protein-related genes were highly enriched in our gene set analysis, indicating a higher false positive rate and a higher false negative rate than, for example, in subpool H3 or H4 (Fig. 6 b, iii, iv, and vi). Thus, we cannot exclude that some hits were missed in our analysis.
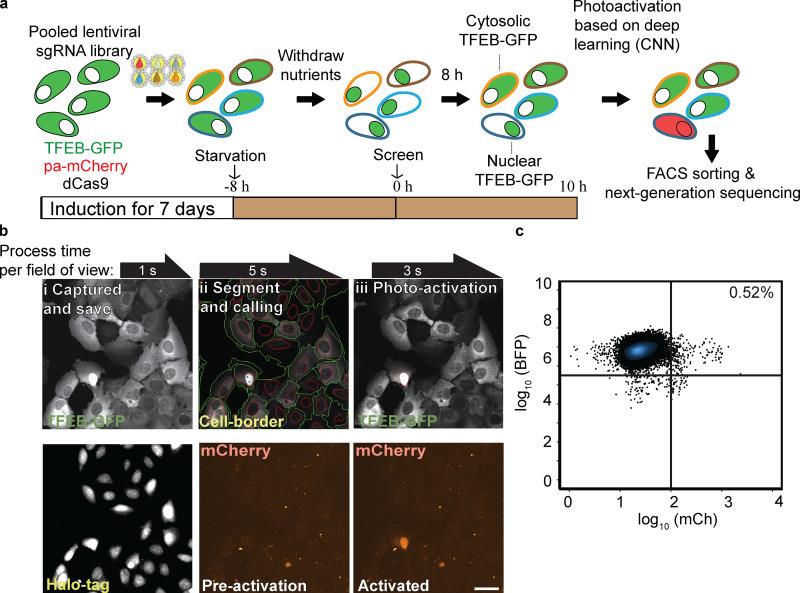
Figure 5.
Whole-genome GFP-TFEB localization screen. (a) Screen workflow. GFP-TFEB cells were transduced with pooled sgRNA libraries targeting the whole genome for 7 d. Following 8 h of starvation, AI-PS screening platform was initiated. (b) Images examples of field of view of GFP-TFEB U2OS cell-screening procedure. i: Images were captured and saved on a local computer. Top: GFP-TFEB; bottom: 2 µM of JD ligand Halo-tag ligand were admitted for nucleus staining. ii: Cells borders were identified (green circle surrounding cell border, red circle the nucleus) following nucleus segmentation. Bottom: mCherry channel for ph-mCh protein. iii: CNN classification model was deployed and masked (red circle). Photoactivation of the CNN identified cell. Scale bar, 10 µm. (c) Flow cytometry sorting of 249 cells from 46,981 (flow cytometry raw data and gating instruction is stored on GitHub). Flow cytometry scatterplot representing the separation of the postscreen photoactivated from the inactivated cell population. BFP florescence signal y-axis in cyan, mCherry florescence signal x-axis in red. For three biologic repeats of seven subpooled libraries (composing the whole genome), ∼12,600,000 cells were screened and 25,579 cells were photoactivated sorted and sequenced.
Video 3.
Example of AI-PS platform for TFEB screen. Nikon NIS elements JOB module screen capture during TFEB-GFP (green) acquisition. Top: TFEB-GFP is in green and red circle is for the phenotype automatic detected cells. Bottom left corner: R-based image segmentation, machine learning prediction, and mask generation. Bottom right: Photoactivation in red (pa-mCh). Three examples are shown here.
Figure 6.
TFEB screen quality control. (a) For deep sequencing, an average of 1,218 (±305) sorted activated cells per library per biologic repeat were analyzed; number of unique sgRNAs detected per library from a whole-genome screen. sgRNA subpooled library H1 to H7 are color coded. (b) i-vii: sgRNA read counts were quantile normalized following log2-CPM treatment. The coefficient of variation was calculated as described (Robinson et al., 2010). A locally weighted linear regression (lowess) mean curve was fitted to the dispersed data using a smother spanning of 0.2. Black fitted curves designated for mean dispersion of control sample, red fitted curves designated for mean dispersion of control sample; an average number of 2,364,645 (±356,453) reads sequenced from activated sorted cells per subgroup library per biologic repeat.
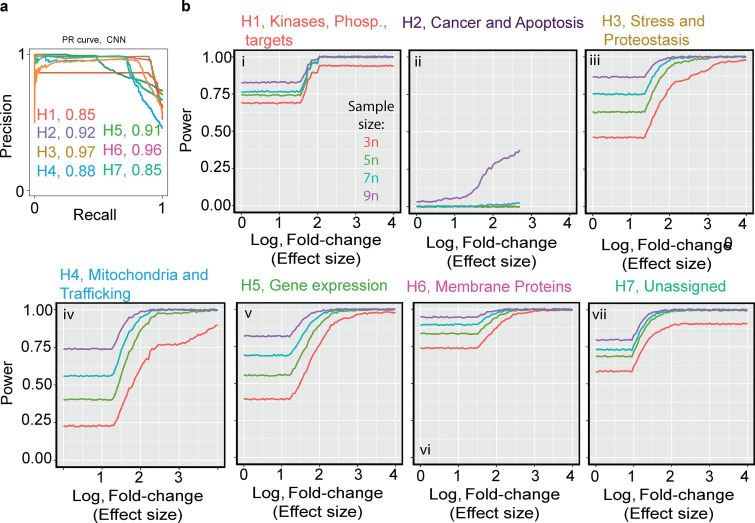
Among the seven subpooled libraries, a mean accuracy of 90% was calculated from the approximation of the area under the precision-recall curve (Fig. 7 a).
Figure 7.
TFEB screen statistical power modeling and performance. (a) GFP-TFEB phenotype classification performance by CNN. Single-cell images of GFP-TFEB induced with the designated library (color coded, H1–H7) were collected. Precision-recall curve from 5,203 single-cell images pooled from three biologic repeats. Image collection began 8 h after starvation was initiated and continued for another 10 h. The accuracy was computed from the integral area under the precision-recall curve (area under the curve [AUC]). The AUC was calculated per subpooled library (designated by color). (b) i-vii: Statistical power analysis by simulation of the sgRNA read counts retrieved from the TFEB screen. Mean unique sgRNAs detected used for the simulation was 2,680 ± 512 and average read count of 282 ± 64. The simulation was done using the R package PROPER, effect size (log of fold-change) is shown in the x-axis, power in the y-axis, the curved lines are color coded for number of biologic repeats. For each subpooled library, there was a sample size of 3 (red curve), 5 (green curve), 7 (cyan curve), and 9 (purple curve).
The power calculation simulated from the TFEB screen resulted in a power range of 50% to 80% in six of seven subpooled libraires (Fig. 7 b, i–vii); however, consistent with the power simulation of the Parkin screen, increasing biologic repeats (e.g., five biologic repeats) would improve the screening performance and reduce the FDR (Fig. 7 b and Fig. 3 e).
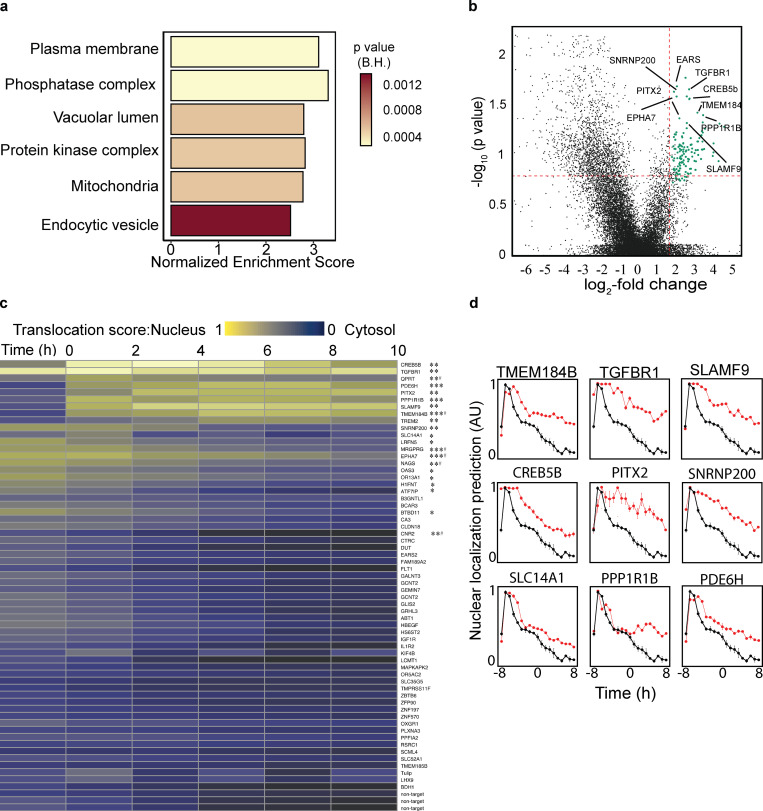
For calculating gene enrichment, we subjected the sgRNA list to the rotation gene set test provided by the R package, EdgeR (Robinson et al., 2010). The entire photoactivated and sorted gene abundance ranking list analyzed for ontology clusters revealed enrichment in mitochondrial and kinase complex gene sets (Puertollano et al., 2018; Nezich et al., 2015) that may relate to energetic consequences of mitochondrial states and TFEB post-translational regulation, respectively (Fig. 8 a and Fig. S5). Plasma membrane proteins were also enriched, perhaps related to cell division rates or nutrient import. Differential sgRNA abundance analysis between unsorted and photoactivated/sorted samples showed a significant fold-change enrichment in 64 genes (Fig. 8 b and Table S2).
Figure 8.
Whole-genome GFP-TFEB localization screen. (a) GSEA pathway analysis annotated using the gene sets derived from the GO Cellular Component database of the Molecular Signatures Database. On the x-axis is the GSEA normalized enrichment score, and the color of the bars represents the GSEA calculated FDR probabilities. (b) Volcano sgRNA plot comparing sgRNA abundance in the photoactivated samples following 8 h of starvation to sgRNA abundance in the unsorted sample. Vertical red line is set at the log2 fold-change threshold; horizontal red line is set at the Benjamini-Hochberg corrected P value of 15%. Genes selected for secondary screening are shown in green. See also Table S2. (c) Top candidates from the primary screen were selected for secondary screening. The GFP-TFEB localization CNN model probability value was used for measuring the perturbation effect on GFP-TFEB localization over time during starvation. Heatmap including all genes included in secondary screen; low probability values are shown in purple for cytosolic TFEB-GFP, high probability values (yellow) for nuclear TFEB-GFP. n = 3, *P < 0.05, **P < 0.01, or ***P < 0.0001 obtained using repeated-measures ANOVA test. P value, one sgRNA was significant. Data distribution was assumed to be normal, but this was not formally tested. (d) Triplicates of 350 images per sgRNA knockdown were imaged for 18 h during starvation. The AI-PS segmentation and CNN prediction was deployed to compute the translocation value (y-axis). GFP-TFEB translocation dynamics observed during starvation for selected gene candidates, nontargeted sgRNA in black, gRNA targeting the designated protein in red. Quantification is displayed as mean ± SEM from three independent experiments.
Figure S5.
Gene set clustering of 64 candidates enriched in TFEB-GFP translocation screen. Cytoscape analysis of enriched genes, the circle size represents fold-change enrichment and P value is color coded inside the circle. The dashed lines indicate cluster overlap.
TFEB translocation and validation
A second validation screen was conducted in the 64 enriched genes by using the top-ranked primary screened sgRNAs. As with the whole-genome screen, TFEB-GFP nuclear localization following validation guide transduction during prolonged starvation was recorded 8 h after starvation for 10 h. The perturbation effect on TFEB positioning was compared with a nontargeting control sgRNA. To validate the screen, the TFEB-GFP positioning score was computed by using a CNN-based classification algorithm. The mean prediction score over time was calculated and subtracted from the nontargeting control sgRNA. To determine if there is a significant prediction score difference between the nontargeting control sgRNA and the target sgRNA, we used repeated-measure ANOVA. We found that 21 of the 64 sgRNAs from the whole-genome analysis significantly extended nuclear TFEB retention (Benjamini-Hochberg [BH] corrected P < 0.05, repeated-measures ANOVA; Fig. 8 c).
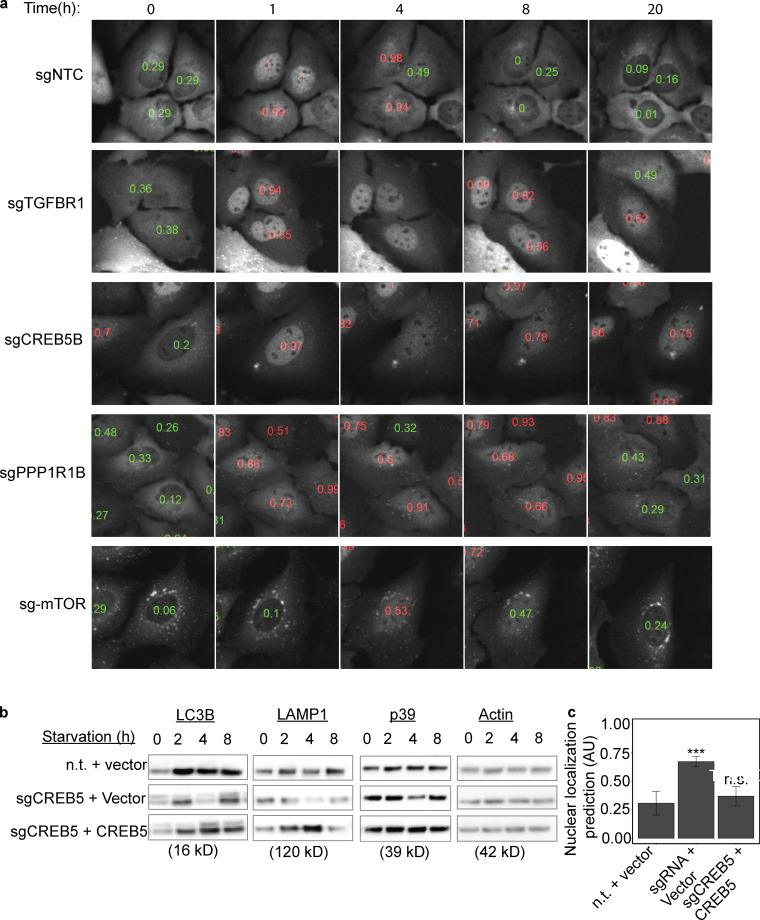
Interestingly, these 21 validated hits were among the genes with the highest-ranked P value significance in the whole-genome screen (Table S2 and Table S3). Among the validated genes, the signaling receptor, Transforming Growth Factor Beta Receptor 1 (TGFBR1), was enriched in the secondary TFEB screen (Fig. 8, c and d; Fig. 9 a; and Video 4). This may be related to a previous report of the induction of another MITF (Melanocyte-Inducing Transcription Factor) family of transcription factors member, TFE3, by the loss of TGFBR1 (Sun et al., 2016). In addition, the loss of another hit, Pitx2, in vivo causes an increase in mitophagy that has been linked to TFEB activation (Nezich et al., 2015; Chang et al., 2019). Additionally, the membrane protein, TMEM184b, has been reported previously to play a role in autophagy (Fig. 8, c and d; Bhattacharya et al., 2016; Agod et al., 2018). The loss of the phosphatase, PPP1R1B, which also scored among the top validated hits, resulted in significant retention in TFEB in the nucleus upon starvation (Fig. 8, c and d; Fig. 9 a; and Videos 5 and 6). As phosphorylation of TFEB is intimately linked to its activation and subcellular localization (Puertollano et al., 2018), this hit deserves further mechanistic study. The extensively studied TFEB regulator, mTOR, was not significantly enriched in our photoactivated samples. To explore this explicitly, live-cell imaging of starved GFP-TFEB infected with two distinct sgRNAs targeting mTOR showed an accumulation of TFEB on lysosomes, which resulted in punctate cytosolic foci, which was similar to previous reports (Martina and Puertollano, 2013; Settembre et al., 2012; Fig. 9 a and Video 7). Therefore, mTOR was not identified in the enrichment analysis owing to the lack of classification of this specific mTOR phenotype, which is distinct from the deep learning model trained for nuclear localization. A parametric pixel intensity computation model may have detected such an unanticipated phenotype.
Figure 9.
Selected sgRNA targeting genes validated in the TFEB-GFP secondary screening and CREB5 expressing rescuing TFEB nucleus retention upon starvation. (a) Images selected from triplicates of 350 images per sgRNA knockdown (designated in black) made over an 18-h time course during starvation. Low probability values are shown in green for cytosolic GFP-TFEB, high probability values (red) for nuclear TFEB-GFP. TFEB-GFP–expressing U2OS cells treated with the designated sgRNA were starved in HBSS for 18 h and images were acquired every hour. Scale bar, 5 µm. (b) The mean CNN-based prediction values were calculated from 2,500 single-cell images per condition HBSS starved for 1–8 h. Mean ± SEM from three independent experiments. (c) Cell lines from b were analyzed by immunoblotting with antibodies against LC3B, LAMP1, p39, and loading control actin. n = 2. CREB5, pHAGE vector expressing full length CREB5; n.t., nontargeting sgRNA; vector, mock pHAGE vector expressing RFP only. ***P < 0.0001 (one-way ANOVA).
Video 4.
Live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting TGFBR1 under starvation conditions. sgTGFBR1-TFEB-GFP–expressing U2OS cells were starved for 18 h and images were acquired every hour. Single-cell CNN prediction scores are marked in red for nuclear TFEB and in green for cytosolic TFEB. Dynamic bar chart indicates the cumulative distribution of TFEB translocation in the represented cell population.
Video 5.
Live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting CREB5 under starvation conditions. sgCREB5-TFEB-GFP–expressing U2OS cells were starved for 18 h and images were acquired every hour. Single-cell CNN prediction scores are marked in red for nuclear TFEB and in green for cytosolic TFEB. Dynamic bar chart indicates the cumulative distribution of TFEB translocation in the represented cell population.
Video 6.
Live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting PPP1R1B under starvation conditions. sgPPP1R1B-TFEB-GFP–expressing U2OS cells were starved for 18 h and images were acquired every hour. Single-cell CNN prediction scores are marked in red for nuclear TFEB and in green for cytosolic TFEB. Dynamic bar chart indicates the cumulative distribution of TFEB translocation in the represented cell population.
Video 7.
Live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting mTOR under starvation conditions. Sg-mTOR-TFEB-GFP–expressing U2OS cells were starved for 18 h and images were acquired every hour. Single-cell CNN prediction scores are marked in red for nuclear TFEB and in green for cytosolic TFEB. Dynamic bar chart indicates the cumulative distribution of TFEB translocation in the represented cell population.
TFEB nuclear translocation is regulated by CREB5
One of the strongest hits is the little-studied transcription factor, CREB5 (Fig. 8, c and d; Fig. 9 a; and Video 5). CREB5 belongs to the transcription factor family, CAMP Responsive Element Binding Protein (CREB). CREB1 was previously reported to mediate autophagy and induce the expression of several autophagy genes, including Ulk1, Atg5, and Atg7, upstream of TFEB following starvation and TFEB itself (Seok et al., 2014). Certain autophagy genes are more predominantly activated by CREB1 and others more by TFEB. Interestingly, CREB5 knockdown in U2OS cells caused a decrease in protein expression of the autophagy protein, LC3B, and the lysosomal proteins, LAMP1 and p39, following 4 h of HBSS incubation. These protein levels were rescued by the overexpression of CREB5 (Fig. 9 b). In addition, prolonged GFP-TFEB nucleus retention by CREB5 downregulation was decreased by rescuing CREB5 expression (Fig. 9 c).
Discussion
Here, we present a platform that applies machine learning and deep learning algorithms to allow for pooled genetic screening for subcellular image phenotypes. This method, which we call AI-PS, reduces the time, cost, and complexity compared with standard screening methods that have required arrayed RNAi or CRISPR libraries. Recently, another study reported a similar concept that can be applied to detect the genetic profiles linking chemotaxis drugs and subcellular phenotypes (Hasle et al., 2020). Our study strengthens the value of photoactivation-based image screens and also shows that it can be used to investigate a large range of cell biology phenotypes.
The speed of AI-PS screening relies on the sequential execution of four steps: image capture, segmentation, generation of classification region of interest, and photoactivation of the region of interest. For a field of ∼200 cells, these four steps together take an average of 10 s, which is then iterated across an entire plate. Therefore, a screen of 600,000 cells infected with one seventh of the genome guide library, composed of 12,500 sgRNAs, takes ∼12 h. Hence, this accelerated platform, coupled with a user-friendly interface, should accelerate the utility of pooled genomic screens. The effective segmentation of live cells is critical in order to ensure efficiency in training and to avoid erroneous predictions. We found that the best way to segment mammalian cells by using the R package, EBImage, was to use two cellular markers in two different channels. Draq5 was used to mark the nuclei, which provided the seeds for segmentation. The other marker provides the cellular borders, or the cytosolic volume of the cell. The latter is important for the effective segmentation of a higher confluency of cells, which maximizes the number of cells screened. Similar two-channel approaches are commonly used in cellular segmentation (Wählby et al., 2002; Quelhas et al., 2010; Al-Kofahi et al., 2018). Three of the four most commonly differentiated channels are used for nuclei detection (far red), photoactivation (red), and CRISPR guide RNA expression (blue). Thus, AI-PS utilizes the remaining green/GFP channel to visualize both the phenotype queried and the cell borders. Deep learning models are becoming a more popular tool, but any gain in accuracy they provide is countered by the computational power and time required to deploy such models during the AI-PS segmentation step.
The method enables the detection and labeling of cells according to subcellular protein localization. We validated this by identifying PINK1 as the only known reported hit required for Parkin translocation to damaged mitochondria within the genome guide sublibrary of kinases, phosphatases, and the druggable genome, demonstrating the validity of the method.
We also used AI-PS to explore a completely different protein translocation process, one that again would be undetectable via FACS separation of whole cells based on a change in overall fluorescence intensity. The transcription factor, TFEB, is retained in the cytosol in growing cells and upon starvation relocalizes to the nucleus, where it induces transcription of lysosomal- and autophagy-related genes (Settembre et al., 2011; Sardiello et al., 2009). Upon prolonged starvation, TFEB returns to the cytosol via an undefined process. Either nuclear TFEB migrates back to the cytosol or nuclear TFEB is degraded while newly synthesized TFEB repopulates the cytosol. As we found minimal evidence for a role of cytoskeletal or nuclear transporter proteins, whether the appearance of TFEB in the cytosol is due to the physical shuttling of preexisting TFEB or to an increase in the translation of new TFEB remains an open question. Beyond protein localization screens, our method will be useful to identify genes involved in the regulation of organelle abundance, size, and shape.
Similar to the concepts presented in our study, machine learning–based image analysis has been used for the calling and sorting of cells (Ota et al., 2018; Nitta et al., 2018); however, AI-PS conveys distinct advantages. First, the microscopic resolution of AI-PS is much higher than that used during dissociated cell sorting (Ota et al., 2018; Nitta et al., 2018), allowing for the identification of more difficult to detect subcellular structures. Specifically, the detection of minor subcellular events, such as an alteration in protein distribution, positioning, and motion, requires high spatial-temporal resolution image acquisition. Previously published methods used low magnification objectives (4× and 10×) and very short exposure times (<50 ms), which resulted in low signal-to-noise ratios and are not suitable for the resolution of subcellular events.
Another advantage of the AI-PS platform is its wide accessibility—there is no need for specialized flow instrumentation, and the algorithms and code presented here can be adapted easily for a variety of microscope systems. AI-PS is compatible with adherent tissue culture cells, unlike sorting-based approaches for which cells must be in suspension, further allowing a more accurate examination of subcellular events in regular culture conditions. One current limitation of AI-PS is that the cells must be screened live to allow for trypsinization to produce single-cell suspension for FACS. Because some phenotypes would be better screened in fixed cells, we are developing methods that enable single-cell release of fixed cells to allow screening of additional cell biology processes. While machine learning methods require larger training datasets, they have a clear advantage over standard image analysis algorithms in the classification and prediction of subtle subcellular phenotypes. Classification models built with deep learning are less influenced by human bias, since they independently decide which image features are important for distinguishing between the two (or more) phenotypes.
The use of machine learning, photoconversion, and deep sequencing in separate applications is not new. Using our current method, we show improvement of the scalability of pooled optical screens in comparison to similar approaches already reported. However, we show that, compared with a previous pooled visual genetic screen (Feldman et al., 2019), only 32% of the primary screen hits were validated to directly affect TFEB translocation in a secondary assay. We cannot rule out that this lower validation rate is a result of the large scale of the current screen which increases the complexity and might increase variation. In the future, in order to increase the discovery rate of large AI-PS screens, a few considerations are recommended. First, in the current study, we observed that increasing the biological replicates from three to five resulted in a significant power increase. Second, as discussed previously, faster and larger imaging fields will allow for greater screening sample sizes. In addition, from our flow cytometry data, we learned that >0.5% frequency of the desired cell phenotype decreases false positives.
Prediction of TFEB nuclear translocation by using the deep learning approach was more accurate than the SVM classification model, possibly because of discrepancies in classification accuracy owing to the uneven fluorescence intensity of the TFEB signal. Although the cell line was carefully generated from a single clone, over several passages the TFEB expression level diverged across the population. The use of low magnification objective (20×) with a low NA value of 0.75 further amplified these variations. To address this, unevenly illuminated images were introduced into our CNN classifier builder by adding an augmentation step to our image batch generator before training. In future screening designs, there are several steps that can be used to overcome this issue. First, knocking-in GFP into the TFEB or gene of interest locus may decrease expression variability. In addition, higher magnification objectives equipped with better NA lenses would decrease the illumination heterogenicity.
Another step to improve AI-PS would be to reduce the segmentation time per image to speed up the screen. Fortunately, a huge improvement in cell segmentation, specifically the development of deep learning–based techniques, such as U-Net segmentation (Caicedo et al., 2019; Hollandi et al., 2020; Ronneberger et al., 2015), can be used in AI-PS. This new deep learning–based segmentation has the potential for at least a fivefold reduction in analysis time. Increasing the speed will make it possible to increase the sample size, thereby increasing the sgRNA coverage in the sorted samples and decreasing the FDR. Another strategy for increasing specificity would be to use single-cell DNA sequence analysis. In the future, simultaneous imaging with two CEMOS cameras will reduce capture time. Finally, large-format camera sensors with larger field-of-view capturing will greatly improve the overall screen since more cells can be screened and analyzed. Another limitation of AI-PS is that to complete a whole-genome screen, we image 600,000 cell batches in three repeats. To minimize the overall screening time, we reduced the number of fields of view to be screened by seeding cells at 90% confluency. The high confluency allows more cells to be screened, but results in a slight reduction in the accuracy of segmentation. Therefore, to allow for longer screen image acquisition times and therefore lower cell seeding density, a major improvement will be to screen fixed cells with a reversible fixation method to allow cell sorting following photoactivation.
The tool we present here is best suited for low-phenotype alteration hit rates—that is, when only 0.5% to 1% of cells are called per field of view captured to minimize photoactivation time. For example, in the current TFEB screen, a mean of three cells were detected and activated per field of view. Therefore, for the current screen, a galvo-miniscanner photoactivation unit was sufficient; however, in a scenario where the phenotype-altering hit rate is much higher, a faster photoactivation unit, such as a DMD illumination module, would be more suitable.
In conclusion, our platform demonstrates the novel implementation of machine learning to improve cell biology research and discovery, and enables phenotypic-based screening at the subcellular level, an approach largely unavailable previously. Additionally, AI-PS can be implemented for drug target exploration and may prove to be valuable in methods targeting single cells within complex human samples.
Materials and methods
Cell lines, constructs, and reagents
U2OS and HEK293T cells were cultured in a humidified incubator at 37°C and 5% CO2 and maintained in DMEM (Life Technologies) supplemented with FBS (10% vol/vol; Gemini Bio Products), 10 mM Hepes (Life Technologies), 1 mM sodium pyruvate (Life Technologies), 1 mM nonessential amino acids (Life Technologies), and 2 mM glutamine (Life Technologies). Testing for mycoplasma contamination was performed bimonthly by using the PlasmoTest kit (InvivoGen).
For constituting a stably expressing dCas9-KRAB U2OS cell line, we took a similar approach to that described previously (Tian et al., 2019). In brief, pC13N-dCas9-BFP-KRAB (127968; Addgene) was integrated into the U2OS genome by using F-Talen and R-Talen (pZT-C13-R1 and pZT-C13-L1; Addgene: 62196, 62197), targeting the human CLYBL intragenic safe harbor locus between exons 2 and 3 [as described previously by Tian et al. (2019)]. The U2OS-dCas9-KRAB cell line was then subcloned and the dCas9-KRAB activity assessed to select the most potent clones for further use by live plasma membrane immunostaining (Fig. S1 d). In brief, dCas9-KRAB U2OS clones were induced with lentivirus-expressing gRNA-targeting Transferrin receptor or N-Cadherin. Following 4 d of induction, cells were seeded on an imaging chamber and immunostained with antibody against Transferrin receptor (BioLegend; #A015) diluted 1:100 or N-Cadherin (BioLegend; #8c11) diluted 1:500. Cells were single cloned and selected for dim dCas9 BFP signal that yielded the largest knockdown effect.
To generate the parental U2OS-dCas9-PA-mCh, photoactivatable-mCherry was PCR-amplified from the plasmid N-pa-mCh and assembled into the retroviral vector pBABE-puro by using HiFi DNA Assembly (E5520S; New England Biolabs). To create the stable U2OS-dCas9-PA-mCh/GFP-Parkin and U2OS-dCas9-PA-mCh/TFEB-GFP cell lines, Parkin or TFEB was inserted into the lentiviral pHAGE vector by HiFi DNA Assembly (E5520S; New England Biolabs). The cell lines were subcloned and cells expressing low levels of the GFP-tagged proteins were selected to prevent overexpression artifacts. For nucleus segmentation, we used a lentiviral plasmid expressing nuclear-localized Halo-tag, hU6-bsd-NLS-Halo. Prior to the screen, HBSS was supplemented with 2 µM of pa Janelia Dye 646, SE (Tocris). For the Parkin screen, the nucleus was detected by using 1,000× dilution of Draq5 (62251; Thermo Fisher Scientific).
For Parkin-induced mitophagy, GFP-Parkin cells were treated with 10 µM CCCP (Sigma-Aldrich) and 0.1 µM Bafilomycin A (Sigma-Aldrich). For TFEB screening, cells were starved in HBSS without calcium and magnesium (14170112; Thermo Fisher Scientific).
Parkin-GFP and TFEB-GFP positioning classification by SVM
To create the classification model, we initially trained 2,234 images of each of the binary phenotypes, Parkin or TFEB translocation. GFP-Parkin signal was mitochondrial vs. cytosolic, while TFEB-GFP was nuclear vs. cytosolic. The model was created by using the R library e1071. In brief, we used a radial basis Kernel with a cost violation of 10 computed for an example set of phenotypes using the radial Kernel formula: e(−γ|u−v|^2).
To optimize the model, we performed iterations and calculated performance by area under the receiver operating characteristic curve or precision-recall curve (in the case of asymmetric phenotype representation). The performance values were plotted against iteration to prevent data overfitting.
GFP-TFEB positioning classification by Convolutional neural network
For TFEB localization classification, an ImageNet (Deng et al., 2009) architecture CNN model was created by using TensorFlow and the software library Keras. A training set composed of 107,226 single-cell example images of GFP-TFEB in the nucleus or cytosol was produced. Of the data, 80% were used for training and 15% for validation. The remaining 5% of data were used for testing the model performance. Image input size was 150 pixels × 150 pixels, and three steps of convolution and max pooling were conducted at a learning rate of 1e−4.
Training was performed with 50 epochs and a batch size of 200. Overfitting was prevented by using the built-in Keras callbacks Application Programming Interface (API) feature to save the model weights after each epoch. The selected model was chosen from the epoch at which the validation and training loss curves were no longer decreasing. The variation in fluorescence signal intensity was accounted for by randomly applying brightness augmentation (10% to 90%) to the images in the training data set.
Model performance
To assess classification model performance, we performed a precision-recall curve in which the curve integral was a measurement of accuracy (Fu et al., 2019). In brief, 5% to 10% of images in the data set from our experiment were arbitrarily selected for performance testing. Images were collapsed into single cells. The parameters extracted for constructing the precision-recall curve were the corresponding CNN prediction value against the ground truth class. The curve and area under the curve were plotted and calculated by using the R package, PRROC (Grau et al., 2015). To train the CNN model, the files of each data set were split into three groups: training (80%), validation (15%), and testing (5%). The validation set was used during model development to evaluate the model’s performance during training and tuning classification hyperparameters. Validation accuracy was important for detecting model overfitting. After training, the model was then evaluated with the testing set. The validation and testing designated images were never used during training, allowing for the assessment of a model’s generalizability. Both the SVM and CNN models were evaluated for their performance on the testing data set—their ability to produce prediction values matching the cell image’s true class label.
After the mask was generated, the images were collapsed into single-cell images by using the EBimage function stackObjects according to the mask. The function generates 150 × 150–pixel boxes and assigned zero for all the pixels outside of the region of interest mask.
Image acquisition and model deployment
SVM deployment live-image acquisition was done on a Nikon Ti-2 CSU-W1 spinning disk confocal system equipped with a high-speed electron-multiplying charge-coupled device camera (Evolve 512; Photometrics) using a 20× air objective (NA 0.75) with an environmental control chamber (temperature controlled at 37°C and CO2 at 5%) operated by Nikon elements AR microscope imaging software.
Cells were seeded for screening at 105 cells per well on a two-well Lab-Tek chamber slide (155360; Thermo Fisher Scientific). The on-the-fly real-time capture was done by using the 488-nm laser channel for excitation and the 520-nm emission detector to collect the GFP signal, and the 647-nm excitation laser and 667-nm emission detector for the segmentation channel. Saved images were segmented live by using a bash file script (https://github.com/gkanfer/AI-PS), and the classifications were deployed by the SVM model. A mask file containing the selected cells was generated and stored on the local computer. The mask image was used to photoactivate the called regions by exciting with a 405-nm wavelength using a Bruker minscanner XY galvo photostimulation scanner. The process was iterated across more than 1,000 fields of view (512 × 512 pixels for the Parkin screen and 2048 × 2044 pixels for the TFEB screen). The NIS elements AR microscope software was used in JOB mode to allow for the integration of the deployment code on the fly (the JOB file can be found on our https://github.com/gkanfer/AI-PS/). In brief, following capture and saving of the 488-nm and 647-nm channel images on the local computer, the NIS JOB module OUTPROC was activated and directed to run the segmentation and deployment R script. Next, the region of interest mask was generated, uploaded back to the local microscope computer hard drive on the OUTPROC folder path, after which NIS-JOB continued by saving the mask coordinates and preforming the photoactivation of the selected regions of interest with a 405-nm laser. The microscope stage then moved to the next field of view to repeat the process.
Live-cell image acquisition and deployment of the CNN-based screen were performed on the Eclipse Ti2-E (Nikon) with the CSU-W1 spinning disk system equipped with an ORCA-FLASH 4.0 v3 sCMOS (Hamamatsu), an Opti-Microscan XY Galvo Scanning Unit, and a Nikon LUN-F laser unit with 90 mW 405 nm, rated 90-mW output at fiber tip, using a 20× objective (NA 0.75) and environmental control chamber (temperature controlled at 37°C and CO2 at 5%). The microscope was controlled by the NIS elements AR microscope imaging software. The on-the-fly real-time acquisition and deployment of the CNN-based screen were performed as described above with one major modification: the TensorFlow deployment script was running the backend “while-loop” throughout the acquisition (https://github.com/gkanfer/AI-PS/tree/master/TFEB_screen).
Cell segmentation analysis and processing
For image manipulation, the R package EBimage (Pau et al., 2010) was used similarly to a previous report (Laufer et al., 2013). In brief, the two-channel images were min/max-normalized and nuclear staining was used as a seed to identify individual cells. For nucleus segmentation, thresholding with a 5 × 5 filter map and Watershed transformation were applied. Then, the target channel—designated GFP—was used to identify cell borders and edges for segmentation, after which it was used for classification. High-pass filtering and local thresholding, followed by global thresholding, were used to create global and local masks. Together with the nucleus mask generated in the first step, this mask was used for the Cellprofiler (Carpenter et al., 2006)-based EBimage propagation function. To handle outlier cells, several features were computed and the outlier features were removed. To handle outlier cells, the mean intensity and area of the segmented cell outline were calculated. By using the R package SCORE, significant outlier values were calculated and removed. For SVM classification, preselected features were computed and used for classification. For the CNN classification, single cells were extracted and stacked into tensor array configuration, which is compatible with CNN-based prediction analysis.
sgRNA lentiviral production
To generate lentivirus-expressing sgRNA libraries, CRISPRi subpooled libraries were used (Horlbeck et al., 2016). On day 0, 7.5 × 107 Hek293-lentiX cells (Clontech) were seeded on 15-cm tissue culture plates. The next day (day 1), 20 µg/ml subpooled sgRNA plasmid, 14.1 µg/ml PAX2, 4.2 mg/ml MDG2, and 1.2 µg/ml pAdvantage (third-generation lentiviral vector packaging systems) were transfected by using 75 µl Lipofectamine 2000 (11668019; Thermo Fisher Scientific) in Opti-MEM (Thermo Fisher Scientific). On day 2, the medium was changed, and on day 3, the virus was harvested. A lentivirus precipitation kit (VC100; Alstem Cell Advancements) was used according to the manufacturer’s suggestions to concentrate the virus.
To determine MOI, 0.106 cells were seeded in 24-well plates and infected with four titrations of the concentrated virus. Genomic DNA was isolated by using QIAamp DNA Micro Kit (56304; Qiagen). The number of genomic viral integration sites was compared with the number of housekeeping genes by using a Bio-Rad QX200 AutoDG Droplet Digital PCR (ddPCR) System (Bio-Rad). The volume to MOI ratio was calculated by using the following formula: insertion number (from ddPCR) × dilution factor = transducing units; (desired MOI × cell number)/transducing units = virus volume.
The ddPCR primer mix for amplifying upstream of the sgRNA integration region was purchased from Bio-Rad: GAAGAAGAAGGTGGAGAGAGAGACAGAGACAGATCCATTCGATTAGTGAACGGATCGGCACTGCGTGCGCCAATTCTGCAGACAAATGGCAGTATTCATCCACAATTTTAAAAGAAAAGGGGGG (FAM). The housekeeping probe used for comparison was EiF2C1 (Assay ID: dHsaCP2500349; Cat: 10031243; Bio-Rad).
To conduct the screen, library expression, 5 × 106 dCas9-pa-mCh–expressing cells were seeded on day 0. The next day, the appropriate virus volume was added to cells to achieve an MOI less than five. Two days after infection, sgRNA-expressing cells were sorted by using a 407-nm Laser and 450/50-nm filters. Following 4 d of growth, cells were reseeded in two-well screening chambers. To maintain sufficient sgRNA representation, cells were maintained at numbers corresponding to a coverage of at least 100 cells per sgRNA.
Activated sample isolation
After screening, cells were detached by using Trypsin (Sigma-Aldrich), washed once with PBS, and filtered by using a 50-μm sieve (Corning) to obtain a single-cell suspension. The volume was adjusted to obtain up to 10 million cells per ml using PBS. Cells were kept in the dark on ice until sorting, which was done by using a BD FACS Aria cell sorter equipped with 355-nm, 407-nm, 532-nm, and 640-nm laser lines, and BD FACSDIVA software to perform aseptic cell sorting. Physical properties (forward-scatter and side-scatter parameters) of cells were used to identify and exclude debris, dead cells, and doublets. All single cells were then selected for GFP expression by using the signal from the 488-nm laser line 515/30-nm filters. mCherry signal was identified by using the 532-nm laser line and 610/25-nm filter, and BFP signal was identified by using signal from the 407-nm laser and 450/50-nm filters. Cells were purified into two populations: GFP+/BFP+/RFP+ or GFP+/BFP+/RFP− for downstream analysis.
Illumina library construction and sequencing
Following FACS sorting, samples were pelleted by centrifugation and subjected to genomic DNA isolation by using the QIAamp DNA Micro Kit (56304; Qiagen). To construct the sequencing library, genomic DNA was amplified by two-step PCR. In the first step, unique modifier identifiers (UMIs) fused with lentiviral vector integration site (step 1 Fw primer) were mixed with 7i adaptor primer fused with lentiviral vector integration 3′ integration site (step 1 Rev primer). The mixture was amplified by using 5–10 PCR cycles. The second amplification step included a forward primer complementary to the UMI primer fused to 5i (step 2 Fw primer) Illumina adaptor primers and 7i (step 2 Rev primer) and amplified by using 25 PCR cycles. DNA concentration was measured by using the NEBNext Library Quant Kit for Illumina (E7630L; New England Biolabs). Each 50-µl PCR reaction was composed of 0.5 µM primers, 0.5 µl of Phusion hot-start DNA polymerase (F549S; Thermo Fisher Scientific), and 2.5 µM dNTPs (N0447S; New England Biolabs). After 25 cycles (second PCR step), the PCR products were cleaned by using AMPure beads (A63880; Beckman Coulter) according to the manufacturer’s protocol.
Fragment size and purity were determined by using Agilent TapeStation 2200 and 4200 models, and the desired fragment size of 300 bp was extracted and eluted with a Pippin instrument (Sage Science) with HT 2% Agarose Gel, 100–600 bp (HTC2010). For the Parkin screen, we used 300 v2 Cassettes (15 million reads) on MiSeq (MS-102-2002), whereas, for the TFEB screen, Illumina paired-end sequencing was performed on a NextSeq 550 instrument with a sequencing chip of 300 Mid Output Kit v2.5 (120 million reads, cat 20024905; Illumina). The read length was 200 bp and 7 bp for the indexing primers. Custom sequencing primers were used (UMI sequence, N; Index sequence, n).
The primer set for step 1 was as follows: forward: 5′-AAGCAGTGGTATCAACGCAGAGTACNNNTNNNTNNNTNNNNNNNNGCACAAAAGGAAACTCACCCT-3′; reverse: 5′-CAAGCAGAAGACGGCATACGAGATnnnnnnnCGACTCGGTGCCACTTTTTC-3′. The primer set for step 2 was as follows: forward: 5′-AATGATACGGCGACCACCGAGATCTACACAAGCAGTGGTATCAACGCAGAGTAC-3′; reverse: 5′-CAAGCAGAAGACGGCATACGAGATnnnnnnn-3′. The sequencing primer was 5′-TTATCAACTTGAAAAAGTGGCACCGAGTCG-3′.
UMI extraction and read count generation
The sgRNA abundance analysis was split into four parts. First, the fastq file was demultiplexed according to the run sample sheet by using the FASTX Barcode Splitter. Second, by using UMI tools, the sequences were extracted and low-quality sequences were trimmed using trimmomatic. Sequences were aligned and mapped to the library data set using Bowtie and Tryhard modules as described previously (Horlbeck et al., 2016). Finally, deduplication grouping and counting were conducted by using UMI tools. The complete Unix-based bash file is available on GitHub.
Differential sgRNA abundance analysis
The differential abundance of sgRNAs between photoactivated-sorted samples and control untreated samples was assessed by using the EdgeR package. First, samples were log2- and count-per-million normalized. Sample variation was determined by covariance-based PCA analysis and read count flooring was established by modeling the noise using coverage as a function of read count. sgRNA enrichment is defined as two SDs from the mean of the distribution of nontarget sgRNA controls. For gene aggregation analysis, similar to a previous paper (Tian et al., 2019), the highest enrichment sgRNA sets were selected by bootstrapping the entire dataset. By using EdgeR (Robinson et al., 2010; Dai et al., 2014), the FDR-corrected P value was calculating by the roast function (Rotation Gene Set Test; Robinson et al., 2010) following the exactTest function of EdgeR (n = 3 or 4 replicates). Gene set analysis was performed using GSEA 4.0.3, and our whole-genome list was ranked according to FC and P value. The pathway annotation used was the MSigDB Collection (C2:C5; Reimand et al., 2019).
Experimental approach for validation
For the secondary validation, we used the best two sgRNAs with FC higher than two SDs from the nontargeting sgRNA controls and roast test FDR < 15%.
128 sgRNAs targeting 64 high-scoring hits (Table S2) identified from the primary pooled screen (two sgRNAs per gene) and two nontargeting control sgRNAs were individually cloned into the lentiviral mU6-BstXI-BlpI-BFP sgRNA vector (Horlbeck et al., 2016) and confirmed via sequencing.
Nontargeting sgRNA sequences were as follows: nontargeting control sgRNA 1, 5′-GCTGCATGGGGCGCGAATCA-3′; nontargeting control sgRNA 2, 5′-GTGCACCCGGCTAGGACCGG-3′.
All the guide sequences used in the TFEB screen can be found in Table S2. CREB5 sgRNA sequences were as follows: 1, 5′-GGAGTCTAGGAGGTACCTCT-3′; 2, 5′-GGATCTCATTTACCTGAATG-3′.
To generate virus, 2 × 106 Lenti-X 293T cells (Clontech) were seeded in six-well plates in 1.5 ml DMEM (Life Technologies) supplemented with FBS (10% vol/vol; Gemini Bio Products), 10 mM Hepes (Life Technologies), 1 mM sodium pyruvate (Life Technologies), 1 mM nonessential amino acids (Life Technologies), and 2 mM glutamine (Life Technologies). Cells were transfected the next day in the following manner by using Lipofectamine 3000 (Thermo Fisher Scientific): 1.2 µg lentiviral sgRNA plasmid, 0.8 µg psPAX2 packaging vector, 0.3 µg pMD2G packaging vector, 0.8 µg pAdvantage packaging vector, and 5 µl P3000 reagent were diluted in 150 µl Opti-MEM and incubated 5 min at RT; 3.75 µl Lipofectamine 3000 Transfection Reagent (Thermofisher) was diluted into 150 µsgRNA lentiviral productionl Opti-MEM and incubated at room temperature for 5 min, after which the diluted DNA was added, mixed via pipetting, incubated at RT for 40 min, and then added dropwise to cells. Medium was replaced the next day and harvested after 2 d and centrifuged at 4°C for 10 min at 10,000 × g to pellet cell debris. The supernatant was aliquoted and frozen at −80°C to ensure consistency throughout the validation process.
U2OS cells expressing dCas9-KRAB and PA-mCherry were seeded at 20,000 cells per well in 96-well plates on day 0, excluding all exterior wells. On day 1, cells were transduced with virus for 24 h with 8 µg/ml polybrene at two concentrations with three replicates per concentration, allowing 10 different viruses, including a control nontargeting sgRNA, to be tested per plate. Cells were checked visually on days 2 and 3 for confluency and blue nuclear signal indicating expression of the sgRNA. If crowded, cells were Trypsinized and split to one to two 96-well plates. Cells were split again on days 4 or 5 as needed into a 96-well imaging plate (PerkinElmer). A half-medium change was performed every other day if cells were not being split. On day 7, medium was removed, cells were washed three times, and then left in warm PBS without calcium and magnesium (Thermo Fisher Scientific). Cells were imaged every 60 min for 20 h using a 20× air objective (NA 0.75) on a Nikon Ti-2 CSU-W1 spinning disk system with a photometrics 95B camera operated by Nikon Elements software equipped with temperature regulation and CO2 control. For every sgRNA, nine images per well in three replicates were acquired. For TFEB translocation response compression, a fixed number of single-cell images (n = 360) per gRNA per time per biologic repeat were normalized to nontargeted control mean value. To determine if there is a significant difference between the difference values generated for the control replicates on the same plate and the difference values for a guide’s replicates on the same plate, we used repeated-measures ANOVA.
Western blotting
Cells were lysed with 1× NuPAGE LDS buffer (Thermo Fischer Scientific) containing 100 mM DTT and were boiled for 10 min. Approximately 15 µg of protein was loaded onto 4–12% Bis-Tris gels (GenScritp). Proteins were transferred to polyvinylidene difluoride membranes and were blocked with 4% skim powdered milk dissolved in Tris-buffered saline with 0.1% Tween (TBSt) buffer. Primary antibodies were incubated overnight at 4°C in 2% BSA in TBSt buffer, and secondary antibodies were incubated at RT in 4% skim milk in TBSt for 1 h. Anti-LAMP1(ab24170) and recombinant anti-ATP6V0D1/P39 (ab202897) antibodies were purchased from Abcam. Anti-LC3B (L7543-100UL) and anti-actin (MAB1501, clone C4; Millipore) were purchased from Sigma-Aldrich. Secondary HRP-linked antibodies were from GE Healthcare. Blots were developed by using peroxidase-based ECL (Pierce) and detected by using a ChemiDoc Imaging System (Bio-Rad).
Sample size power calculation
To estimate screening sample size, we conducted power calculations by using the R package, PROPER. This tool estimates the statistical power of differential guide read count data from the negative binomial distribution. The model is built on the negative binomial distribution and the per-gRNA dispersion of filtered sgRNA read counts. The sgRNA list is the same as that used for sgRNA enrichment analysis. By using the runSims function of the PROPER package, the read counts were generated based on the input data, and the number of samples were iterated 100 times. The number of repeats chosen in our study were 3, 5, 7, and 9, and the power was calculated for effect size of 0.1 to 5 with α nominal of 0.15 (similar to the FDR used in the current study).
Shiny AI-PS application
We created a graphical user interface in Shiny (by Rstudio) that performs each step—image segmentation and classification, and creation, and testing of model—required to build and test an SVM-based classification model for AI-PS. This application can be accessed directly through the website (https://hab-gk-app.shinyapps.io/gk_shiny_app/). Alternatively, the app can be run locally from the source code found at https://github.com/hbaldwin07/GK_shiny_app. Performance is better on local machines than on the network server, so this is the recommended method for those using particularly large data sets or data files (>10 MB per image). All instructions for running/using the program can be found on the GitHub website.
Data and code availability
Flow cytometry data of the TFEB screen, including gating examples and codes, can be found under https://github.com/gkanfer/AI-PS/tree/master/facs. Statistical power analysis and filtered read counts can be found under https://github.com/gkanfer/AI-PS/tree/master/Statistical%20power%20analysis. AI-PS deployment and Nikon elements module bin file for outproc are located at https://github.com/gkanfer/AI-PS/tree/master/TFEB_screen.
Online supplemental material
Fig. S1 shows the SVM classification plot and the SVM classification and segmentation performance. Fig. S2 presents a summary of the AI-PS shiny APP platform. Fig. S3 shows the CNN classification architecture and performance. Fig. S4 addresses the TFEB translocation prediction by the SVM classification model. Fig. S5 summarizes the network interaction and clustering of the hits retrieved from the whole-genome CRISPR screen. Video 1 shows an example of AI-PS platform Parkin screen proof of principle. Video 2 shows live-cell images of TFEB-GFP U2OS cells under starvation conditions. Video 3 shows an example of the AI-PS platform for TFEB screen. Video 4 shows live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting TGFBR1 under starvation conditions. Video 5 shows live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting CREB5 under starvation conditions. Video 6 shows live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting PPP1R1B under starvation conditions. Video 7 shows live-cell images of TFEB-GFP U2OS cells expressing sgRNA targeting mTOR under starvation conditions. Table S1 provides the Parkin translocation screen using an sgRNA library subpool targeting all kinases, phosphatases, and the druggable genome. Table S2 shows the TFEB translocation whole-genome screen. Table S3 lists TFEB cell numbers and next-generation sequencing read numbers.
Supplementary Material
shows the Parkin translocation screen using an sgRNA library subpool targeting all kinases, phosphatases, and the druggable genome. The rows in the Parkin screen gene enrichment spreadsheet are the proteins selected by the screen. Log2 fold-change and corresponding P values are calculated from the gene abundance analysis as described in Materials and methods and related to Fig. 3.
shows the TFEB translocation whole-genome screen. The rows in the TFEB screen gene enrichment spreadsheet are the proteins selected by the screen. Log2 fold-change and corresponding P values are calculated from the gene abundance analysis as described in Materials and methods and related to Fig. 5.
lists the TFEB cell numbers and next-generation sequencing (NGS) read numbers. In the first tab is shown the total number of cells sorted from the photoactivated samples per library per biologic repeat. In the second tab, the total number of reads detected from the NGS analysis is shown.
Acknowledgments
We thank Nico Tjandra for intellectual contributions. We thank Nick Ader, Eric Bunker, Elyssa Hawk, and Sue Smith for helping with cloning, cell lines, and Lentivirus production. We thank Catherine Nezich, Hetal Shah, Jose Norbert Vargas, and Benoit Kornmann for comments on the manuscript, and all the Youle laboratory and the Lippincott-Schwartz laboratory members for critical comments. We thank Talya Chooly for supporting the project. Flow cytometry cell sorting and sample isolation was performed at the Flow Cytometry Core, National Heart, Lung, and Blood Institute. Next-generation deep sequencing was performed at the CCR Genomics Core, National Cancer Institute. We thank the National Institutes of Health (NIH)–based Nikon team for helping with imaging and integration of our external codes. This work used the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov).
This work was supported by the National Institute of Neurological Disorders and Stroke intramural program, and by National Institutes of Health, National Institute of General Medical Sciences grant DP2 GM119139 (to M. Kampmann).
The authors declare they have no competing financial interests.
Author contributions: G. Kanfer led the project and performed most of the experimental work. G. Kanfer conducted and performed all the screens and created the cell lines. Y. Maman and G. Kanfer created the SVM-based code. H. Baldwin and G. Kanfer created the Shiny APP. G. Kanfer created the deployments and CNN code. M. Kampmann and M.E. Ward built the sgRNA libraries. M. Kampmann, K.R. Johnson, M.E. Ward, and G. Kanfer conducted next-generation sequencing analysis and statistics. S.A. Sarraf and G. Kanfer planned and performed the secondary analysis validation screen. G. Kanfer, S.A. Sarraf, and R.J. Youle wrote the paper. G. Kanfer, R.J. Youle, and J. Lippincott-Schwartz designed the study. G. Kanfer and E. Dominguez Martin preformed single-hit validation. All authors discussed the results and commented on the manuscript. R.J. Youle and J. Lippincott-Schwartz supervised the project.
References
- Adamson, B., Norman T.M., Jost M., Cho M.Y., Nuñez J.K., Chen Y., Villalta J.E., Gilbert L.A., Horlbeck M.A., Hein M.Y., et al. 2016. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 167:1867–1882.e21. 10.1016/j.cell.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agod, Z., Pazmandi K., Bencze D., Vereb G., Biro T., Szabo A., Rajnavolgyi E., Bacsi A., Engel P., and Lanyi A.. 2018. Signaling lymphocyte activation molecule family 5 enhances autophagy and fine-tunes cytokine response in monocyte-derived dendritic cells via stabilization of interferon regulatory factor 8. Front. Immunol. 9:62 10.3389/fimmu.2018.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kofahi, Y., Zaltsman A., Graves R., Marshall W., and Rusu M.. 2018. A deep learning-based algorithm for 2-D cell segmentation in microscopy images. BMC Bioinformatics. 19:365 10.1186/s12859-018-2375-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, M.R.C., Geisler S., Pittman S.K., Doan R.A., Weihl C.C., Milbrandt J., and DiAntonio A.. 2016. TMEM184b promotes axon degeneration and neuromuscular junction maintenance. J. Neurosci. 36:4681–4689. 10.1523/JNEUROSCI.2893-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binan, L., Bélanger F., Uriarte M., Lemay J.F., Pelletier De Koninck J.C., Roy J., Affar E.B., Drobetsky E., Wurtele H., and Costantino S.. 2019. Opto-magnetic capture of individual cells based on visual phenotypes. eLife. 8:e45239 10.7554/eLife.45239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzdok, D., Krzywinski M., and Altman N.. 2018. Machine learning: supervised methods. Nat. Methods. 15:5–6. 10.1038/nmeth.4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, J.C., Goodman A., Karhohs K.W., Cimini B.A., Ackerman J., Haghighi M., Heng C., Becker T., Doan M., McQuin C., et al. 2019. Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nat. Methods. 16:1247–1253. 10.1038/s41592-019-0612-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.N., Singh A.J., Gross M.K., and Kioussi C.. 2019. Requirement of Pitx2 for skeletal muscle homeostasis. Dev. Biol. 445:90–102. 10.1016/j.ydbio.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Z., Sheridan J.M., Gearing L.J., Moore D.L., Su S., Wormald S., Wilcox S., O’Connor L., Dickins R.A., Blewitt M.E., and Ritchie M.E.. 2014. edgeR: a versatile tool for the analysis of shRNA-seq and CRISPR-Cas9 genetic screens. F1000 Res. 3:95 10.12688/f1000research.3928.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger, P., Rendeiro A.F., Schmidl C., Krausgruber T., Traxler P., Klughammer J., Schuster L.C., Kuchler A., Alpar D., and Bock C.. 2017. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods. 14:297–301. 10.1038/nmeth.4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J., et al. 2009. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition. 248–255. . 10.1109/CVPR.2009.5206848 [DOI] [Google Scholar]
- Dixit, A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R., et al. 2016. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 167:1853–1866.e17. 10.1016/j.cell.2016.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel, G., Moffitt J.R., and Zhuang X.. 2017. High-throughput, image-based screening of pooled genetic-variant libraries. Nat. Methods. 14:1159–1162. 10.1038/nmeth.4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, D., Singh A., Schmid-Burgk J.L., Carlson R.J., Mezger A., Garrity A.J., Zhang F., and Blainey P.C.. 2019. Pooled optical screens in human cells. Cell. 179:787–799.e17. 10.1016/j.cell.2019.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, G.H., Yi L.Z., and Pan J.. 2019. Tuning model parameters in class-imbalanced learning with precision-recall curve. Biom. J. 61:652–664. 10.1002/bimj.201800148 [DOI] [PubMed] [Google Scholar]
- Gilbert, L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. 2014. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 159:647–661. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau, J., Grosse I., and Keilwagen J.. 2015. PRROC: computing and visualizing precision-recall and receiver operating characteristic curves in R. Bioinformatics . 31:2595––2597.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle, N., Cooke A., Srivatsan S., Huang H., Stephany J.J., Krieger Z., Jackson D., Tang W., Pendyala S., Monnat R.J. Jr., et al. 2020. High-throughput, microscope-based sorting to dissect cellular heterogeneity. Mol. Syst. Biol. 16:e9442 10.15252/msb.20209442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollandi, R., Szkalisity A., Toth T., Tasnadi E., Molnar C., Mathe B., Grexa I., Molnar J., Balind A., Gorbe M., et al. 2020. nucleAIzer: A Parameter-free Deep Learning Framework for Nucleus Segmentation Using Image Style Transfer. Cell Systems. 10:453–458.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck, M.A., Gilbert L.A., Villalta J.E., Adamson B., Pak R.A., Chen Y., Fields A.P., Park C.Y., Corn J.E., Kampmann M., and Weissman J.S.. 2016. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5:e19760 10.7554/eLife.19760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck, M.A., Xu A., Wang M., Bennett N.K., Park C.Y., Bogdanoff D., Adamson B., Chow E.D., Kampmann M., Peterson T.R., et al. 2018. Mapping the Genetic Landscape of Human Cells. Cell. 174:953–967.e22. 10.1016/j.cell.2018.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer, C., Fischer B., Billmann M., Huber W., and Boutros M.. 2013. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat. Methods. 10:427–431. 10.1038/nmeth.2436 [DOI] [PubMed] [Google Scholar]
- Martina, J.A., and Puertollano R.. 2013. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200:475–491. 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra, D., Tanaka A., Suen D.F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich, C.L., Wang C., Fogel A.I., and Youle R.J.. 2015. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 210:435–450. 10.1083/jcb.201501002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta, N., Sugimura T., Isozaki A., Mikami H., Hiraki K., Sakuma S., Iino T., Arai F., Endo T., Fujiwaki Y., et al. 2018. Intelligent Image-Activated Cell Sorting. Cell. 175:266–276.e13. 10.1016/j.cell.2018.08.028 [DOI] [PubMed] [Google Scholar]
- Ota, S., Horisaki R., Kawamura Y., Ugawa M., Sato I., Hashimoto K., Kamesawa R., Setoyama K., Yamaguchi S., Fujiu K., et al. 2018. Ghost cytometry. Science. 360:1246–1251. 10.1126/science.aan0096 [DOI] [PubMed] [Google Scholar]
- Patterson, G.H., and Lippincott-Schwartz J.. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. 10.1126/science.1074952 [DOI] [PubMed] [Google Scholar]
- Pau, G., Fuchs F., Sklyar O., Boutros M., and Huber W.. 2010. EBImage - an R package for image processing with applications to cellular phenotypes. Bioinformatics. 26:979–981. 10.1093/bioinformatics/btq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano, R., Ferguson S.M., Brugarolas J., and Ballabio A.. 2018. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37:e98804 10.15252/embj.201798804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelhas, P., Marcuzzo M., Mendonça A.M., and Campilho A.. 2010. Cell nuclei and cytoplasm joint segmentation using the sliding band filter. IEEE Trans. Med. Imaging. 29:1463–1473. 10.1109/TMI.2010.2048253 [DOI] [PubMed] [Google Scholar]
- Reimand, J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C., et al. 2019. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14:482–517. 10.1038/s41596-018-0103-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger, O., Fischer P., and Brox T.. 2015. U-net: Convolutional networks for biomedical image segmentation. arXiv doi: (Preprint posted May 18, 2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- Sardiello, M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A Gene Network Regulating Lysosomal Biogenesis and Function. Science. 325:473–477. [DOI] [PubMed] [Google Scholar]
- Seok, S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S., Sun X., Yoon G., Kang Y., Zhong W., et al. 2014. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 516:108–111. 10.1038/nature13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre, C., Di Malta C., Polito V.A., Arencibia M.G., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB Links Autophagy to Lysosomal Biogenesis. Science. 332:1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre, C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z.-J., Yu G.-T., Huang C.-F., Bu L.-L., Liu J.-F., Ma S.-R., Zhang W.-F., Liu B., and Zhang L.. 2016. Hypoxia induces TFE3 expression in head and neck squamous cell carcinoma. Oncotarget. 7:11651–11663. 10.18632/oncotarget.7309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, R., Gachechiladze M.A., Ludwig C.H., Laurie M.T., Hong J.Y., Nathaniel D., Prabhu A.V., Fernandopulle M.S., Patel R., Abshari M., et al. 2019. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron. 104:239–255.e12. 10.1016/j.neuron.2019.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora, G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wählby, C., Lindblad J., Vondrus M., Bengtsson E., and Björkesten L.. 2002. Algorithms for cytoplasm segmentation of fluorescence labelled cells. Anal. Cell. Pathol. 24:101–111. 10.1155/2002/821782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Lu T., Emanuel G., Babcock H.P., and Zhuang X.. 2019. Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. Proc. Natl. Acad. Sci. USA. 116:10842–10851. 10.1073/pnas.1903808116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, E.C., Vu A.Q., Einstein J.M., DiSalvo M., Ahmed N., Van Nostrand E.L., Shishkin A.A., Jin W., Allbritton N.L., and Yeo G.W.. 2020. Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors. Nat. Methods. 17:636–642. 10.1038/s41592-020-0826-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Wang C., and Wu Z.. 2015. PROPER: comprehensive power evaluation for differential expression using RNA-seq. Bioinformatics. 31:233–241. 10.1093/bioinformatics/btu640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 465:942–946. 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the Parkin translocation screen using an sgRNA library subpool targeting all kinases, phosphatases, and the druggable genome. The rows in the Parkin screen gene enrichment spreadsheet are the proteins selected by the screen. Log2 fold-change and corresponding P values are calculated from the gene abundance analysis as described in Materials and methods and related to Fig. 3.
shows the TFEB translocation whole-genome screen. The rows in the TFEB screen gene enrichment spreadsheet are the proteins selected by the screen. Log2 fold-change and corresponding P values are calculated from the gene abundance analysis as described in Materials and methods and related to Fig. 5.
lists the TFEB cell numbers and next-generation sequencing (NGS) read numbers. In the first tab is shown the total number of cells sorted from the photoactivated samples per library per biologic repeat. In the second tab, the total number of reads detected from the NGS analysis is shown.
Data Availability Statement
Flow cytometry data of the TFEB screen, including gating examples and codes, can be found under https://github.com/gkanfer/AI-PS/tree/master/facs. Statistical power analysis and filtered read counts can be found under https://github.com/gkanfer/AI-PS/tree/master/Statistical%20power%20analysis. AI-PS deployment and Nikon elements module bin file for outproc are located at https://github.com/gkanfer/AI-PS/tree/master/TFEB_screen.