Abstract
To defend against microbial invaders but also to establish symbiotic programs, plants need to detect the presence of microbes through the perception of molecular signatures characteristic of a whole class of microbes. Among these molecular signatures, extracellular glycans represent a structurally complex and diverse group of biomolecules that has a pivotal role in the molecular dialog between plants and microbes. Secreted glycans and glycoconjugates such as symbiotic lipochitooligosaccharides or immunosuppressive cyclic β-glucans act as microbial messengers that prepare the ground for host colonization. On the other hand, microbial cell surface glycans are important indicators of microbial presence. They are conserved structures normally exposed and thus accessible for plant hydrolytic enzymes and cell surface receptor proteins. While the immunogenic potential of bacterial cell surface glycoconjugates such as lipopolysaccharides and peptidoglycan has been intensively studied in the past years, perception of cell surface glycans from filamentous microbes such as fungi or oomycetes is still largely unexplored. To date, only few studies have focused on the role of fungal-derived cell surface glycans other than chitin, highlighting a knowledge gap that needs to be addressed. The objective of this review is to give an overview on the biological functions and perception of microbial extracellular glycans, primarily focusing on their recognition and their contribution to plant–microbe interactions.
Keywords: Cell wall, chitin, extracellular polysaccharides, glucan, immunity, matrix, microbes, symbiosis
This review highlights the structural diversity of microbial glycans and glycoconjugates, and focuses on their contribution to communication processes between plants and microbes.
Introduction
Plants are immobile organisms that constantly sense and integrate information from biotic and abiotic cues in order to adapt to a dynamic environment. Monitoring of the microbial surrounding can be achieved by perception of molecular patterns that are either microbe-derived compounds [including structural microbe-associated molecular patterns (MAMPs) and microbial effectors] or host-specific molecules released or modified upon microbial activities, also referred to as damage-associated molecular patterns (DAMPs). Pattern sensing occurs via extracellular, membrane-integral pattern recognition receptors (PRRs) or intracellular receptors such as nucleotide-binding domain leucine-rich repeat-containing (NLR) proteins (Dodds and Rathjen, 2010; Cook et al., 2015). Upon recognition of these cues, subsequent receptor activation leads to signaling cascades that enable the plant to readjust its own physiological status towards defense (including pattern-triggered immunity and effector-triggered immunity) or symbiotic (i.e. mutualistic) programs (Zipfel and Oldroyd, 2017). Common features of microbe-triggered signaling are transphosphorylation cascades, ionic fluxes and membrane depolarization, apoplastic production of reactive oxygen species (ROS), transcriptional reprogramming, phytohormone signaling, production of secondary metabolites, and initiation of developmental programs (Couto and Zipfel, 2016; Zipfel and Oldroyd, 2017). Furthermore, these plant responses also hold the potential to influence and reshape the microbial composition in above- and below-ground tissues (Xin et al., 2016; Hacquard et al., 2017; Teixeira et al., 2019; Chen et al., 2020). However, the mechanisms governing the processing and integration of information obtained from the remarkable number of signaling events into an outcome that is overall favorable for the plant remain largely unclear and are under intensive investigation.
The nature of molecular patterns acting as indicators of microbial presence is highly diverse. It covers the most important classes of biopolymers, such as proteins and peptides, fatty acids, nucleic acids, and purine derivatives, as well as glycan-based substances (Gallucci and Maffei, 2017; Saijo et al., 2018; Nizam et al., 2019; Albert et al., 2020). Plants can sense an extremely wide spectrum of glycan substrates consisting of homo- and heteromeric polysaccharides and glycoconjugates. This includes endogenous sugar metabolites derived from carbon assimilation, plant cell wall components released upon damage, microbial glycoconjugate messengers dedicated to plant–microbe communication, as well as fibrous cell surface glycans involved in microbial biofilm and cell wall formation (Bolouri Moghaddam and Van den Ende, 2013; Limpens et al., 2015; Fesel and Zuccaro, 2016). Cell surface glycans in particular represent conserved and abundant targets for the plant’s surveillance system (Fesel and Zuccaro, 2016). On the other hand, microbes have evolved a plethora of tools and strategies to evade plant recognition (Rocafort et al., 2020).
The widespread potential for communication mediated by glycans lies in their structural complexity. While many biological polymers (e.g. polypeptides) are of linear nature, glycans are often built from small repeating basic units of monosaccharides that assemble into different three-dimensional flexible structures with a myriad of possible linkage types and branching patterns (Guberman and Seeberger, 2019). Chain length [also referred to as degree of polymerization (DP)], chemical modifications, and supramolecular structures have been shown to constitute relevant characteristics for specific recognition by the host (Ménard et al., 2004; Takahasi et al., 2009; Kanagawa et al., 2011; Zipfel and Oldroyd, 2017; Wanke et al., 2020).
This review provides an overview of the most important microbial glycan-derived patterns that are either directly perceived by plants or indirectly manipulate plant responses. Based on the diverse portfolio of biological and biophysical properties of microbe-derived polysaccharides, we highlight their potential to transmit information on the microbial composition (bacteria, fungi, and oomycetes) surrounding plant tissues.
Judging a microbe by its ‘cover’: cell surface glycans and glycan-binding proteins in plant–microbe interactions
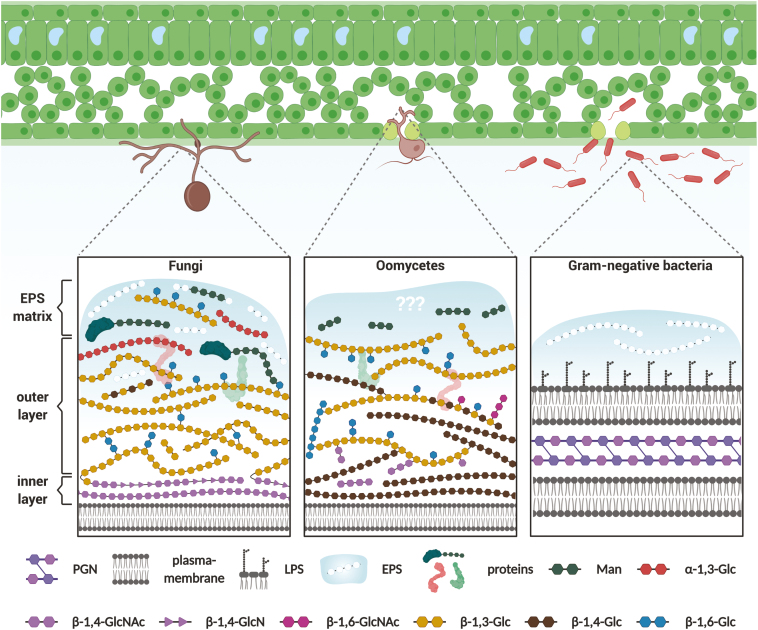
Cell walls from bacteria and filamentous microbes are multilayered structures of fibrous homo- and heteropolysaccharides (Geoghegan et al., 2017; Gow et al., 2017) (Fig. 1). They act as mechanical support maintaining the cell turgor and determining cell shape and development (Shockman and Barrett, 1983; Money, 2008; Silhavy et al., 2010; Samalova et al., 2017). Although cell walls provide mechanical protection against adverse conditions and external aggressors, they can also be considered as an ‘Achilles’ heel’ in terms of their immunological potential (Supplementary Table S1 at JXB online). Upon colonization of the plant, they are one of the very first microbial components to be in contact with plant cells, either via direct physical contact or through the release of oligomers by a fleet of plant-secreted hydrolases (Rovenich et al., 2016). The basic functional principles of cell walls are comparable between different kingdoms; however, their molecular composition can be species specific (Coronado et al., 2007; Mélida et al., 2013; Gow et al., 2017). For that reason, plants evolved a broad surveillance system to recognize structurally different carbohydrate signatures (Albert et al., 2020). In the following sections we will describe the cell surface glycan compositions of bacteria and filamentous microbes (fungi and oomycetes) and highlight those glycan structures relevant for interaction with their hosts.
Fig. 1.
Schematic overview of microbial cell surface glycans. Recognition of microbe-derived cell wall polysaccharides represents an important mechanism by which plants surveil their microbial surrounding. Since cell wall structure and function are highly interlocked, core polysaccharides are conserved within different microbial groups. This scheme illustrates these core polysaccharides and their linkage types without representing exact quantitative proportions. Cell walls of filamentous fungi and oomycetes are network-like structures consisting of highly interconnected polysaccharide fibrils. In fungi, the inner cell wall layer consists of chitin (β-1,4-GlcNAc) and chitosan polymers (β-1,4-GlcN). It is covalently linked to the outer cell wall layer, which is mainly composed of β-1,3/1,6-glucans (β-1,3/1,6-Glc) with minor amounts of β-1,4-glucose (β-1,4-Glc). The outer layer is concealed by α-1,3-glucans (α-1,3-Glc) and mixed-linkage mannose (Man) polymers. Mannose polymers often occur as heterosaccharides with minor amounts of additional sugar types (e.g. rhamnose and galactose). A highly mobile, gel-like extracellular polysaccharide (EPS) matrix is loosely attached to the outer cell wall of many fungi. In contrast to fungi, no detailed studies on the cell wall architecture of oomycetes have been performed. Cross-linked cellulose (β-1,4-Glc) and β-1,3/1,6-glucans are major components of the inner part of oomycete cell walls. Chitin (β-1,4- and β-1,6-GlcNAc) only occurs in minute amounts; most of it is assumed to be connected to β-glucan polymers. The cellulose content is reduced in the external parts of the cell wall. Instead, branched β-glucans and mannose oligomers are present in that layer. To our knowledge, no detailed information on the architecture of an EPS matrix in oomycetes has been reported. Peptidoglycan (PGN) is a conserved part of bacterial cell walls present in Gram-positive and Gram-negative bacteria. The main heteropolysaccharides consist of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid residues, which are interconnected through peptide chains. In Gram-negative bacteria, PGN is embedded between an inner (plasma membrane) and outer membrane layer. The outer membrane layer is decorated with lipid-linked polysaccharides, so-called lipopolysaccharides (LPSs). An amorphous matrix made of EPSs (e.g. xanthan and succinoglycan) encases many bacterial species. A more detailed overview on microbial cell surface glycans and their implications on plant–microbe interactions can be found in Supplementary Table S1.
Cell surface glycans in bacteria
Plants are able to perceive a complex array of glycan-based molecules that can be found in different layers of bacterial cell walls. Two major representatives are the glycoconjugates peptidoglycan (PGN) and lipopolysaccharide (LPS) (Fig. 1). PGN consists of a polymeric glycan-based backbone composed of alternating β-1,4-glycosidic-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues (Vollmer et al., 2008). Oligopeptide bridges cross-link MurNAc residues of separate backbone polymers resulting in a multilayered structure (Vollmer et al., 2008). LPS, the second key building block, is composed of three covalently linked parts: Lipid A, the core oligosaccharide, and the O-antigen polysaccharide (Whitfield and Trent, 2014). Lipid A anchors LPS to the outer membrane via glucosamine-linked fatty acid chains. It is linked via 2-keto-3-desoxy-octonat (Kdo) to the core oligosaccharide which is composed of various sugar moieties. Lipid A and the core oligosaccharide together are also referred to as lipooligosaccharide and can be attached to a terminal O-antigen polysaccharide that is variable in size (Whitfield and Trent, 2014). While LPS is specific to Gram-negative bacteria, PGN exists in both Gram-positive and Gram-negative bacteria. Additionally, some bacterial species have capsular polysaccharides, which are tightly associated with the bacterial membrane and contain neutral or acidic polysaccharides (D’Haeze and Holsters, 2004). The composition of capsular polysaccharides varies among bacterial species and strains, but the presence of Kdo residues is commonly observed (Lopez-Baena et al., 2016). Moreover, a highly hydrated layer of polymeric substances, often referred to as extracellular matrix, surrounds bacterial cells. This layer includes proteins, extracellular DNA, and polysaccharides (Flemming and Wingender, 2010). Bacterial biofilm formation is supported by the extracellular matrix and creates a three-dimensional microenvironment where cell–cell communication takes place and abiotic and biotic stresses are tackled (Flemming and Wingender, 2010).
The complex role of PGN and LPS in plant immunity
Bacterial cell wall glycans and glycoconjugates form conserved and unique structures in prokaryotes, which has driven the evolution of specialized surface-localized receptors in the hosts (Erbs and Newman, 2012; Ranf, 2016; Bacete et al., 2018). In Arabidopsis thaliana (hereafter Arabidopsis), PGN is perceived by a tripartite receptor complex consisting of lysin motif (LysM) domain receptor-like proteins LYM1 and LYM3 and the LysM receptor kinase CERK1 (Willmann et al., 2011). While LYM1 and LYM3 bind PGN, the transmembrane LysM receptor kinase CERK1 mediates intracellular signal transduction across the plasma membrane (Willmann et al., 2011). Similarly, PGN perception in rice is mediated through the respective orthologs LYP4, LYP6, and CERK1 (B. Liu et al., 2012; Ao et al., 2014). PGN signaling results in initiation of canonical immune responses such as medium alkalinization, cytoplasmic calcium influx, production of nitric oxide, phosphorylation of mitogen-activated protein kinases (MAPKs), camalexin accumulation, and induction of various defense-related genes (Gust et al., 2007; Erbs et al., 2008; Willmann et al., 2011; B. Liu et al., 2012; Ao et al., 2014; Liu et al., 2014). In addition to defense activation by complete PGN, there is evidence that specific PGN substructures trigger immune responses in Arabidopsis. Muropeptides derived from Xanthomonas campestris pathovar (pv.) campestris (Xcc) and Agrobacterium tumefaciens PGN preparations showed immunogenic potential in Arabidopsis (Erbs et al., 2008). Furthermore, glycan backbone structures from PGN preparations of Staphylococcus aureus also elicit immune responses in Arabidopsis (Gust et al., 2007). It has recently been shown that the Arabidopsis chitinase LYS1 can hydrolyze the O-glycosidic β-1,4-bond between MurNAc and GlcNAc residues generating soluble PGN products that mediate plant recognition (Liu et al., 2014). In line with PGN receptor mutants (Willmann et al., 2011), knockdown of LYS1 results in super-susceptibility to the phytopathogen Pseudomonas syringae pv. tomato DC300 (Willmann et al., 2011; Liu et al., 2014). However, overexpression of LYS1 also enhances susceptibility, indicating that quantitatively regulated PGN hydrolysis is important to generate immunogenic PGN fragments of a particular DP (Liu et al., 2014).
Upon recognition, LPSs can initiate classical immune responses such as cytosolic calcium influx, ROS generation, phosphorylation of MAPKs, and induction of several defense-related genes (Newman et al., 1995; Zeidler et al., 2004; Braun et al., 2005; Silipo et al., 2005; Madala et al., 2012; Desaki et al., 2018; Kutschera et al., 2019). Early studies promoted the notion that the Lipid A domain from LPSs triggers immune responses via the plant-specific bulb-type lectin kinase LORE in Arabidopsis (Ranf et al., 2015). However, later studies revealed that LPS and Lipid A preparations contained co-purified medium-chain 3'-OH-fatty acids (Kutschera et al., 2019). In fact, LORE mediates signal transduction upon recognition of medium-chain 3'-OH-fatty acids (fatty acid chain length: C8:0–C12:0). The LPS-bound fatty acid chains display the potential to be recognized by LORE, yet it has not been shown that they are hydrolytically released from LPS into the extracellular environment (Silipo et al., 2010; Ranf, 2016; Kutschera et al., 2019). Of note, two further LPS-binding proteins, LBR-1 and LBR-2, which share structural similarity with animal LPS-binding proteins (Iizasa et al., 2016), were identified in Arabidopsis (Iizasa et al., 2016, 2017). The impact on ROS generation and defense gene expression in lbr mutants upon LPS treatment suggests an active contribution of LBR1 and LBR2 to LPS recognition in Arabidopsis (Iizasa et al., 2016, 2017). A different perception mechanism was observed in rice, where the LysM receptor-like kinase CERK1 is required for LPS recognition (Desaki et al., 2018). Purified LPS induced ROS production and LPS-dependent transcriptional changes in wild-type-derived but not cerk1 mutant-derived rice suspension-cultured cells (Desaki et al., 2018). However, whether CERK1 induces defense signaling through direct binding of LPS remains unknown. Additionally, the core oligosaccharides and O-antigen polysaccharides of LPS were reported to trigger plant defense responses (Bedini et al., 2005; Silipo et al., 2005; Madala et al., 2012; Kutschera et al., 2019). For instance, purified core oligosaccharides from Xcc are immunogenic in Arabidopsis (Silipo et al., 2005), and application of truncated LPS versions derived from Xcc mutants to tobacco cells suggests that the core oligosaccharide is important for LPS recognition (Braun et al., 2005; Silipo et al., 2005). The P. aeruginosa H4 strain is devoid of the O-antigen polysaccharide, lacks further sugar residues in the core oligosaccharide, and is not immunogenic, underpinning the importance of this region for immunorecognition in Arabidopsis (Kutschera et al., 2019). Transcriptional studies with purified LPS substructures from Burkholderia cepacia demonstrated defense gene induction in Arabidopsis by the entire polysaccharide (core oligosaccharide and O-antigen polysaccharide) in addition to LPS and Lipid A (Madala et al., 2012). This is supported by a study applying synthetic O-antigen polysaccharides (oligo-rhamnans) that induce defense-related gene expression in Arabidopsis in a DP-dependent manner, suggesting that the coiled structure is recognized by the plant (Bedini et al., 2005). Besides their immunogenic activity, the core oligosaccharide from Xcc and the synthetic O-antigen polysaccharides harbor the potential to suppress the hypersensitive response induced by bacterial infection (Bedini, 2005; Silipo, 2005). Furthermore, purified LPSs from phytopathogenic Xylella fastidiosa evoke defense responses independent of the presence of the O-antigen polysaccharide, while in vivo studies demonstrated the importance of this polysaccharide chain in shielding immunogenic surface structures and, thereby, delaying immunorecognition (Rapicavoli et al., 2018). Additionally, the rhizobial LPS from Sinorhizobium fredii HH103 is unique in its structure and was recently reported to have a role in the symbiotic establishment with its host legume Macroptilium atropurpureum and Cajanus cajan (Di Lorenzo et al., 2020). Sinorhizobium fredii HH103 core oligosaccharide differs from previously described rhizobial core oligosaccharides by the presence of a high number of hexuronic acids and a β-configured pseudaminic acid derivative. LPS from the S. fredii HH103 rkpM mutant lacks the pseudaminic acid derivative residues, leading to severely impaired nodule formation and symbiosis (Di Lorenzo et al., 2020). While previous studies already reported symbiotic impairments of the S. fredii HH103 rpkM strain with different legume hosts, now the responsible structural motifs were described (Margaret et al., 2012; Acosta-Jurado et al., 2016; Di Lorenzo et al., 2020). Although these data suggest recognition of LPS sugar modules, questions regarding the components mediating their recognition and signaling remain.
Bacterial extracellular polysaccharides: a way to overcome immunity
During interaction with plants, both pathogenic and symbiotic bacteria secrete substantial amounts of extracellular polysaccharides (EPSs) into the apoplast. Several functions were attributed to bacterial EPSs such as sequestration of calcium ions, ROS scavenging, prevention of cellulose-mediated cell agglutination, tolerance to acidic pH, participation in biofilm formation, and host surface attachment (Laus et al., 2005; Aslam et al., 2008; Rinaudi and Gonzalez, 2009; Lehman and Long, 2013; Perez-Mendoza et al., 2015; Hawkins et al., 2017). The impact of EPSs on the plant host has been studied for several plant–bacterial systems. Xanthan, a polysaccharide consisting of repeating pentasaccharide units, is a key component of Xcc biofilms and suppresses plant immunity (Yun et al., 2006; Aslam et al., 2008). Its backbone is built by β-1,4-linked glucose residues and an additional trisaccharide side chain (mannose–glucuronic acid–mannose) at every second glucose unit (Yun et al., 2006). The internal mannose residues are substituted by an acetyl group, and half of the terminal mannose residues are substituted by either an acetyl- or a ketal-pyruvate residue (Yun et al., 2006). Due to its anionic nature, xanthan functions as a calcium chelator in vitro (Aslam et al., 2008). This capability is dramatically reduced in truncated xanthan, which is lacking the trisaccharide side chain with anionic modifications (Aslam et al., 2008). Arabidopsis pre-treatment with xanthan but not truncated xanthan prior to Xcc infection suppresses calcium influx-mediated defense responses such as ROS accumulation, expression of the defense gene PATHOGENESIS-RELATED PROTEIN 1 (PR1), and formation of callose depositions (Yun et al., 2006; Aslam et al., 2008). Moreover, different types of EPSs such as xanthan from Xcc, amylovoran from Erwinia amylovora, alginate from Pseudomonas aeruginosa, or EPS from Ralstonia solanacearum and the symbiotic rhizobium Sinorhizobium meliloti hold the potential to reduce cytosolic calcium influx and ROS generation triggered by strong bacterial peptide elicitors such as flg22 and elf18 (Aslam et al., 2008). Since most of these EPSs have been shown to exhibit strong binding to calcium ions (Aslam et al., 2008), it is assumed that the immunosuppressive effect is conferred through apoplastic calcium chelation rather than through receptor-mediated signaling. This was also supported by the fact that xanthan did not compete with flg22 receptor binding (Aslam et al., 2008). Remarkably, a study in tomato revealed that EPSs from R. solanacearum might also trigger defense responses depending on the host plant (Milling et al., 2011). Supported by the finding that purified EPS from R. solanacearum specifically elicits salicylic acid (SA)-related defense gene expression in resistant but not in the susceptible tomato cultivars, this study suggests the emergence of an as yet unknown perception mechanism for R. solanacearum EPS in resistant tomato varieties (Milling et al., 2011).
In rhizobia–legume symbiosis (RLS), modification of bacterial EPS is commonly associated with impaired establishment of symbiosis on the level of nodule formation, infection thread initiation and elongation, bacteroid differentiation, and/or bacterial survival inside the nodules (Downie, 2010; Kelly et al., 2013; Arnold et al., 2018; Maillet et al., 2020). The mechanism governing this EPS-mediated compatibility was intensively studied for succinoglycan. Succinoglycan consists of a repeating octasaccharide monomer composed of a galactose residue and seven glucose residues that can carry additional succinyl, acetyl, and pyruvyl modifications. In interaction with the host Medicago truncatula (hereafter Medicago), S. meliloti succinoglycan- (EPS-I) deficient mutants or mutants lacking the succinyl residues fail to initiate infection thread formation and to invade the plant (Jones et al., 2008; Mendis et al., 2016; Maillet et al., 2020). Further, different early transcriptional responses to wild-type S. meliloti and a succinoglycan-deficient mutant (exoY) were observed. While the presence of functional succinoglycan leads to active changes in root metabolism, the absence of succinoglycan induced a remarkably high number of plant defense genes (Jones et al., 2008). A mechanism for sensing compatible and incompatible EPSs was suggested for the Mesorhizobium loti–Lotus japonicus (hereafter Lotus) interaction. Screenings of different EPS-deficient M. loti R7A strains supported the notion that plants distinctly distinguish compatible EPSs from non-compatible EPSs in order to appropriately modulate defense responses and allow full infection thread development and bacterial release (Kelly et al., 2013). This model was supported upon identification of the Lotus LysM receptor kinase EPR3 that senses both native and truncated monomers of polymeric M. loti EPS in order to control bacterial progression through the plant’s epidermal cell layer (Kawaharada et al., 2015, 2017). The native M. loti EPS is O-acetylated and consists of an octasaccharide repeating monomer composed of glucose, galactose, glucuronic acid, and riburonic acid residues, whereas truncated EPS is a pentaglycoside lacking the terminal riburonic acid and glucuronic acid residues as well as another glucose unit (Kawaharada et al., 2015; Muszynski et al., 2016). Specificity between these two types of EPSs could be mediated by recruitment of distinct co-receptors (Kawaharada et al., 2015). Furthermore, a recent study proposes a more general role for EPR3 in surveying microbial compatibility due to its capability to bind to EPS monomers from different bacterial species (Wong et al., 2020). These findings represent a novel control mechanism for leguminous plants to differentiate between compatible and incompatible rhizobia to initiate consequent responses leading to negative or positive effects on bacterial progression (Kawaharada et al., 2017).
Cell surface glycans in fungi
Fungal cell walls are mesh-like structures consisting of repeatedly branched glycan polymers and proteins (Fig. 1). They are not static constructions and continuously adjust according to cell type, environmental conditions, and lifestyle phases (Geoghegan et al., 2017; Gow et al., 2017). Yet, some basic structural elements are conserved and can be considered as fundamental features. Based on recent studies, three distinct layers of glycans can be found associated with the cell surface of most fungi: a stiff inner cell wall layer, a hydrated outer layer, and a highly mobile, gel-like EPS matrix, an often overlooked fungal glycan structure loosely attached to the cell wall outer layer (Kang et al., 2018; Wawra et al., 2019). The inner cell wall layer adjacent to the plasma membrane consists of a densely packed, hydrophobic layer of chitin (Osumi, 1998; Geoghegan et al., 2017). The chitin microfibrils are linear β-1,4-linked GlcNAc units, highly interconnected via hydrogen bonds, resulting in a rigid skeletal layer. Additional incorporation of melanin into this chitin layer increases resistance to oxidants and mechanical resilience as observed in the penetration structures (appressoria) of Magnaporthe oryzae (Chumley and Valent, 1990) and Colletotrichum graminicola (Ludwig et al., 2014). Some chitin fibrils are covalently linked to β-glucan chains, the predominant component of the outer cell wall layer of many fungi (~65–90% of polysaccharide content) (Bowman and Free, 2006). The β-glucans in this layer are mainly connected via β-1,3-linkages with regularly occurring β-1,6-side branches every 2–25 glucose units, thought to structurally interconnect the β-1,3-linked fibrils (Zekovic et al., 2005). In some cases, additional β-1,4-linked moieties and mixed-linkage β-1,3/1,4-glucose polymers can be observed (Fontaine et al., 2000; Pettolino et al., 2009; Kang et al., 2018). In contrast to chitin, β-glucans can aggregate into helical bundles or coils and form a branched network of non-crystalline fibrils that extends throughout the entire cell wall (Bluhm and Sarko, 1977; Bluhm et al., 1982; Osumi, 1998). On top of the β-glucan layer, interlinked α-1,3-glucans and/or mixed-linkage mannose polymers can be found (Geoghegan et al., 2017; Gow et al., 2017). These mannans can be attached to proteins (mannoproteins), galactose, and/or rhamnose moieties (Pettolino et al., 2009; Geoghegan et al., 2017; Gow et al., 2017; Mélida et al., 2018). In general, the inner and outer cell wall layer represent the fungal cell wall in the narrower sense of the term. Cytological analyses (for more information, see Box 1) have revealed the presence of an extensive gel-like EPS matrix surrounding hyphae of different human pathogenic and plant-colonizing fungi (Gow et al., 2017; Wawra et al., 2019). This highly mobile EPS matrix is not tightly bound to the cell wall and the bulk can be removed by washing. Similar to bacterial EPSs, the fungal EPS matrix is thought to be a loose scaffold for other macromolecules such as proteins, lipids, and extracellular DNA (Martins et al., 2010; Lee and Sheppard, 2016; Wawra et al., 2019). Compositional analyses of EPS matrices from different fungal species have identified glucose, mannose, galactose, rhamnose, xylose, and/or fucose as monosaccharidic building blocks (Mahapatra and Banerjee, 2013). More specifically, the EPS matrix from the saprotrophic fungus Trametes versicolor (Basidiomycota) was shown to contain a linear α-1,6-galactose backbone with mannose- and fucose-containing side chains (Scarpari et al., 2017). The EPS matrix of the root endophyte Serendipita indica (Basidiomycota) is composed of β-1,3/1,6-glucans (Wawra et al., 2019). Similarly, the plant pathogen Botrytis cinerea (Ascomycota) EPS matrix was shown to contain high molecular weight β-1,3/1,6-glucans (El Oirdi et al., 2011). While studies on the EPS matrix of plant-colonizing fungi are scarce, more information is available on the EPS matrix composition of human fungal pathogens. The EPS matrices from Aspergillus fumigatus and Candida albicans consist of complex galactans and mannans (Gow et al., 2017). Additionally, in A. fumigatus, a soluble galactosaminogalactan (GAG) made of a linear heteroglycan consisting of α-1,4-linked galactose and N-acetylgalactosamine was described (Gravelat et al., 2013). In the human pathogen Cryptococcus neoformans, the capsule (highly organized EPS matrix) contains glucuronoxylomannan (GXM) and glucuronoxylomanogalactan (GalXM). GXM is an α-1,3-mannose with β-1,2-linked xylose and glucuronic acid side chains, while GalXM consists of an α-1,6-galactan chain linked to galactomannan–xylose–glucoronic acid branches (Zaragoza et al., 2008; Denham et al., 2018; Decote-Ricardo et al., 2019). The large majority of studies report data originating from the analytics of whole-cell wall preparations without detailed knowledge of whether or not a mobile EPS matrix was present and how much of this matrix was still attached to the cell wall. It is therefore often difficult to speculate on the correct compositions of the EPS matrix or outer cell wall layer. Interestingly, the cell wall layers and EPS matrix seem to be synthesized by distinct pathways in C. albicans, accounting for the observed structural and functional differences (Taff et al., 2012; Mitchell et al., 2015).
Box 1. Making the invisible visible: light microscopic approaches to study microbial cell walls.
Confocal laser scanning microscopy is a common tool used to assess microbial phenotypes during axenic growth and host colonization. The two major approaches for the visualization of microbes are live cell staining and staining after tissue fixation. Live cell microscopy allows the use of living samples, enabling dynamic imaging that can be used to track developments in real time and to obtain a better understanding of biological functions. This method requires easily applicable stains which do not interfere with the observed processes. Alternatively, fluorescently tagged marker proteins that localize to the region of interest can be employed. However, many organisms are not (easily) accessible to genetic engineering, especially when exhibiting an obligate biotrophic lifestyle. In these cases, exogenously applied probes are crucial tools to study the biology of these organisms. In contrast, fixation-based approaches lead to stable but inactive tissue, thus limiting the possibilities to study dynamic processes. Fixed material can be sectioned and is suitable for high-resolution light microscopy, electron microscopy, and electron microscopy tomography (Wang et al., 2019; Franken et al., 2020; Hansel et al., 2020). Yet, sample preparation can introduce artifacts through the fixation procedure and is often labor intensive. Optimization for the respective imaging technique and specific biological sample in particular can be a bottleneck (Bell and Safiejko-Mroczka, 1997; Wisse et al., 2010).
A key prerequisite for fluorescent labeling of microbes, particularly in host tissues or complex environments, is to target specific and unique microbial structures. Most of the fluorescent probes used to study microbes target cell walls or cell wall-associated components. These probes can be divided into four distinct groups: small chemical dyes; specific antibodies; fluorescent-labeled sugar-binding lectins; and engineered neolectins. The group of neolectins was reviewed in detail elsewhere and is therefore not covered here (Arnaud et al., 2013). Small chemical fluorescent dyes have been used for decades, combining the advantage of having high photostability and easy application. Common stains used for the visualization of filamentous microbes are the β-1,3/1,4-glucan-binding fluorescent dyes Calcofluor White/Fluorescent Brightener 28 (Leigh et al., 1985) and Congo Red (Darwish and Asfour, 2013). Despite the fact that both dyes target the same type of polysaccharide, they do not always localize to the same cell wall regions (Ursache et al., 2018). Aniline blue, another widely used glycan stain, interacts with β-1,3-glucans and is also capable of detecting chitosan (Evans et al., 1984; Becker et al., 2016). Aniline blue stains cell walls and has been used for decades to screen for EPS-producing microorganisms (Nakanishi et al., 1976). Yet, the exact nature of the interaction between these dyes and the respective glycans is still unknown. In addition, most of these small molecular fluorescent probes lack a clear specificity towards a single structure and, therefore, only function as proxies for the presence of a particular type of molecule or microbe.
In contrast to small fluorescent probes, monoclonal antibodies raised against specific glycans from microbial cell walls can display a high specificity and affinity towards the respective molecule used for immunization. However, many polysaccharides are generally far less immunogenic than proteins. While proteins generally display high internal heterogeneity and therefore carry various potential antigenic structures available for the animal immune system, large polysaccharides are often constructed of regular repeating flexible units. They often need to be conjugated to protein carriers to elicit an immune response (Cunto-Amesty et al., 2001; Cobb and Kasper, 2005; Zhou et al., 2009; Rydahl et al., 2017). Currently, there are only a limited number of polysaccharide-directed monoclonal antibodies available. Thus, the choices for detection of cell walls of filamentous microbes and associated structures are limited. Furthermore, the use of monoclonal antibodies in confocal laser scanning microscopy is mostly confined to fixed samples, and is time consuming and expensive.
A promising alternative to monoclonal antibodies are lectins, which are glycan-binding proteins. In comparison with antibodies, these proteins have lower affinities towards their selective targets but have been shaped by evolution into molecules with highly specific binding. Compared with monoclonal antibodies, lectins are small and, if fluorophore tagged, easily applicable. The binding properties of lectins to their respective glycoligand are frequently defined by the geometric arrangement of their carbohydrate recognition domains. These domains as such often display rather low affinities for monosaccharides (Gabius, 1997). The biggest hurdle in using lectins as molecular probes lies in the determination of their glycan substrate specificity. The lack of commercially available and defined glycans for affinity screening largely limits the characterization of lectins binding to long and/or complex glycans. Therefore, most of the commercially available fluorescent lectins with defined affinities target small mono-, di-, or short oligosaccharides. A widely used lectin in the field of fungal biology is the GlcNAc-binding wheat germ agglutinin (WGA). However, GlcNAc is not only found in filamentous microbes, but also appears in the PGN layer of some bacteria or as building blocks in protein glycosylation (Quast and Lunemann, 2014; Lin et al., 2017). Thus, the ability of WGA to detect a short glycoligand reduces its structural specificity. To overcome this obstacle, the use of lectins that can distinctly bind long-chain glycoligands with low affinity for short-chain ligands of the same composition would be advantageous for the field. Recently, a fungal lectin exhibiting these characteristics was identified and characterized. The WSC3 protein from S. indica is a lectin composed of three WSC domains that binds to long-chain β-1,3-glucans with no measurable affinity for shorter β-1,3-linked glucose oligomers (<7) (Wawra et al., 2019). Optimization of the protein labeling procedure generated a very efficient probe, which is able to detect fungal EPS with very low background staining (Fig. 2). The only two described lectins able to detect fungal EPS are the β-1,3-glucan-binding WSC3 (Wawra et al., 2019) and the β-1,3/1,6-glucan-binding FGB1 (Wawra et al., 2016). A closer look into the microbial proteome associated with cell walls and EPSs could help to identify more lectins suitable as microscopic probes. Such probes would make a valuable contribution to the field, giving exciting insights into the structural composition of microbial cell walls and EPSs.
Cell surface glycans in oomycetes—similar but different
Despite strong morphological and lifestyle similarities to fungi, oomycetes are a phylogenetically distinct group within the Stramenopiles clade, which also includes brown algae and diatoms (Adl et al., 2012; Beakes et al., 2012). The close evolutionary relationship with algae is reflected by the presence of cellulose (linear β-1,4-glucan) as a major cell wall component (33.6–51%) (Mélida et al., 2013). Comparative analyses of the cell wall composition from different oomycete species have demonstrated the presence of β-1,3/1,4/1,6-linked homo- and heteroglucans (85.6–95%). Furthermore, chitin as well as atypical β-1,6-linked GlcNAc can be found in cell wall preparations of representative species from the Saprolegniales order (i.e. the legume pathogen Aphanomyces euteiches) (Mélida et al., 2013). Additionally, the presence of cross-linkages of β-1,3-glucan with cellulose or GlcNAc/chitooligosaccharides was reported (Mélida et al., 2013; Nars et al., 2013). This association of chitin with other cell wall polymers accounts for the soluble, non-crystalline chitin fractions observed in the mycelial cell wall. In contrast to most true fungi, crystalline and non-crystalline chitin is thought to be distributed within the cell wall of oomycetes instead of being restricted to the inner layer (Badreddine et al., 2008). While chitin is reduced or absent from cell wall preparations of species from the Peronosporales order (e.g. Phytophthora genus), these cell walls additionally contain low amounts of mannan (6.7%) and glucuronic acid (4.7%) (Mélida et al., 2013). Few studies report on the presence of an EPS matrix secreted by encysting zoospores and germinating cysts in various oomycete species (Gubler and Hardham, 1988; Estrada-Garcia et al., 1990; Durso et al., 1993). In the germ tubes and appressoria of Hyaloperonospora parasitica, the causal agent of downy mildew in many crucifers, the EPS matrix consists of β-1,3-glucans, mannan, galactose, GlcNAc, and N-acetylgalactosamine (Carzaniga et al., 2001). More work is required to further address the presence and function of the EPS matrix during later stages of plant colonization.
Hide and seek: chitin perception
In the last decades, chitin recognition in plants was intensively studied, leading to the elucidation of the main receptors and their downstream signaling pathways in several plant species (Sanchez-Vallet et al., 2015). A central player involved in chitin perception in different species is the LysM-domain receptor kinase CERK1. In Arabidopsis, recognition of chitooligomers (COs) with at least six GlcNAc units initiates the formation of ligand-induced heteromeric complexes between CERK1 and the LysM receptor kinases LYK4 and LYK5 (Miya et al., 2007; Wan et al., 2008; Petutschnig et al., 2010; Cao et al., 2014; Xue et al., 2019). Within this complex, LYK5 exhibits the highest affinity for COs, with a preference for oligomers having a DP of eight GlcNAc units (Cao et al., 2014). Binding to COs leads to the association of CERK1 with LYK5 and homodimerization of CERK1, which is needed for CERK1 autophosphorylation and further signal transduction (Petutschnig et al., 2010; T. Liu et al., 2012; Cao et al., 2014). LYK4 might act as a protein scaffold stabilizing the LYK5–CERK1 complex and thereby enhancing chitin-triggered responses (Xue et al., 2019). Furthermore, the weak, residual chitin response observed in lyk5 mutants indicates that LYK4 together with CERK1 might also function as an additional receptor pair able to compensate LYK5 loss (Cao et al., 2014). In rice, chitin perception involves the CERK1 homolog and the receptor-like protein CEBiP, which is associated with the plasma membrane through a glycosylphosphatidylinositol (GPI) anchor (Kaku et al., 2006; Shimizu et al., 2010; Hayafune et al., 2014). Upon ligand binding, two CERK1 proteins associate with a pair of CEBiP receptors cooperatively binding to a single chitin molecule (Hayafune et al., 2014). Two additional rice GPI anchor receptor-like proteins, LYP4 and LYP6, also exhibit CERK1-mediated chitin binding affinity, which might function as accessory chitin perception machineries (B. Liu et al., 2012; Kouzai et al., 2014). Interestingly, CERK1-independent chitin sensing was reported within plasmodesmal membrane microdomains of Arabidopsis (Faulkner et al., 2013). Chitin-triggered plasmodesmata closure is controlled by LYM2, LYK4, and LYK5 (Faulkner et al., 2013; Cheval et al., 2020). LYM2 is a receptor-like protein with a GPI anchor which resides in plasmodesmal membranes (Faulkner et al., 2013). In response to chitin, LYM2 is suggested to oligomerize and act as a signaling platform for LYK4 and calcium-dependent protein kinases to initiate ROS production and callose synthesis (Cheval et al., 2020). These findings emphasize how a single elicitor initiates mechanistically independent responses in different membrane microdomains.
In addition to chitin-mediated activation of defense responses, several studies have shown the potential of COs (DP 4–8) to initiate nuclear calcium oscillations, a signaling hallmark during root symbiotic interactions with nitrogen-fixing rhizobia and arbuscular mycorrhizal fungi (Genre et al., 2013; Sun et al., 2015; Feng et al., 2019; He et al., 2019).
To respond to the widespread capacity of plants to detect microbial cell wall components, evolution has driven the development of a multilayered potpourri of microbial strategies to evade unintended recognition by plant receptors (Rovenich et al., 2016). Commonly observed mechanisms to avoid chitin-triggered immunity include inhibition or degradation of plant chitinases (Naumann, 2011; Jashni et al., 2015; Okmen et al., 2018) and replacement or partial conversion of chitin to chitosan, a deacetylated version of chitin with lower immunogenic potential in most plants (El Gueddari et al., 2002; Cord-Landwehr et al., 2016; Gao et al., 2019; Xu et al., 2020). Since plants lack the enzymatic toolset to degrade α-1,3-glucans, decoration of exposed cell wall regions of invading hyphae with these polysaccharides acts as a protective layer to circumvent chitin hydrolysis as was shown for M. oryzae (Fujikawa et al., 2012). A further layer of immunity evasion is conveyed by fungal chitin-binding effectors that are able to mask their own cell wall molecules (van den Burg et al., 2006; Marshall et al., 2011; Kombrink et al., 2017; Romero-Contreras et al., 2019; Zeng et al., 2020) or to sequester released COs (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016; Romero-Contreras et al., 2019). Moreover, microbial effectors (Masachis et al., 2016; Fang et al., 2019; Irieda et al., 2019) or small RNAs (Jian and Liang, 2019) were shown to manipulate the host’s chitin perception and intracellular signaling machinery.
The complexity behind perception of β-glucans from filamentous microbes
Despite the fact that β-glucans are predominant components of cell walls and EPS matrices in fungi and oomycetes, comparatively little is known about their perception by plants. Early studies could show that purified β-glucans from plant-colonizing fungi and oomycete cell walls are potent inducers of plant defense (Ayers et al., 1976b; Anderson, 1978). Pioneering work was performed on Phytophthora sojae culture filtrates and cell wall hydrolysates which identified β-1,3/1,6-glucans as potent elicitors of plant immunity resulting in phytoalexin accumulation (Ayers et al., 1976a, b; Keen and Yoshikawa, 1983; Okinaka et al., 1995). The smallest fragment with immunogenic capability was shown to be a purified β-glucan heptaglucoside composed of a β-1,6-linked backbone with two β-1,3-linked glucose side branches (Cheong et al., 1991; Sharp et al., 1984a, b). Later, a high-affinity β-glucan-binding protein (GBP) isolated from soybean root membrane fractions was proposed to act as part of a multimeric β-glucan receptor complex (Umemoto et al., 1997; Cheong et al., 1993; Fliegmann et al., 2004). GBP has a glucan-binding domain and β-1,3-glucanase activity (Umemoto et al., 1997; Fliegmann et al., 2004). Despite the lack of membrane targeting and secretion signals, GBP localizes to both the plasma membrane and apoplast (Umemoto et al., 1997). GBP inhibition via a specific antibody diminished β-glucan-elicited phytoalexin synthesis in soybean, indirectly validating its role in glucan perception (Umemoto et al., 1997). Although most plant species encode GBP homologs in their genomes, only legumes mount defense responses upon elicitation with the oomycete-derived β-glucan (Fliegmann et al., 2004). Whether this specificity is due to missing components of the receptor complex in other plant species or whether GBPs from non-leguminous plants bind to distinct β-glucan structures remains open. Alternatively, GBP activity could produce tailored β-glucan structures subsequently recognized by plant species-specific receptor systems. Other glucans such as short and unbranched β-1,3-linked glucose pentamers (laminaripentaose), hexamers (laminarihexaose), and laminarin, a β-1,3-glucan (DP 20–30) with β-1,6-linked side branches, can elicit immune responses in a wide range of plant species (Fesel and Zuccaro, 2016). Laminarin is a storage carbohydrate derived from marine brown algae which bears a remarkable resemblance to β-glucan structures present in many cell walls of filamentous microbes and, therefore, has often been used to investigate β-glucan-triggered immunity in plants and animals. Studies in monocot and dicot species have demonstrated that laminaripentaose (Klarzynski et al., 2000) and laminarihexaose (Mélida et al., 2018; Wanke et al., 2020) as well as laminarin (Klarzynski et al., 2000; Aziz et al., 2003; Ménard et al., 2004; Gauthier et al., 2014; Wawra et al., 2016; Wanke et al., 2020) activate immune responses such as calcium influx, ROS production, MAPK activation, medium alkalinization, expression of pathogenesis-related genes, phytoalexin synthesis, and phytohormone signaling. Moreover, laminarin is able to prime plant immunity and induces resistance to fungal plant pathogens and viruses in Nicotiana tabacum and Vitis vinifera (Aziz et al., 2003; Ménard et al., 2004; Gauthier et al., 2014). While the perception of laminarihexaose was shown to be mediated by CERK1 and an as yet unknown co-receptor in Arabidopsis (Mélida et al., 2018), long branched and unbranched β-glucans are recognized via a CERK1-independent pathway in N. benthamiana and rice (Wanke et al., 2020). Despite the fact that most plants are able to respond to β-glucans, it is striking that there are species-specific differences regarding the length, branching pattern, and presence of chemical modification (e.g. sulfation) of the perceived structures (Yamaguchi et al., 2000; Ménard et al., 2004; Wanke et al., 2020). The degree of variation in β-glucan recognition reflects the high structural diversity of microbial cell wall architectures between species. This implies that while chitin acts as a more general elicitor of plant immunity, β-glucan cues could allow for a rather specialized discrimination between different microbial groups.
A novel, chimeric type of elicitor was identified in glucanase- and chitinase-treated cell wall preparations of A. euteiches (Nars et al., 2013). This group of chito-glucans consists of a β-1,3/1,4-mixed-linkage glucan backbone with β-1,6-linked GlcNAc branches and is able to induce defense gene expression and nuclear calcium oscillation in Medicago. In contrast to lipochitooligosaccharides, chito-glucan-mediated nuclear calcium oscillations are not dependent either on the receptor kinase NFP or on the common symbiosis signaling pathway (Nars et al., 2013).
Cell wall β-glucans are crucial for microbial development and the formation of infection structures, but at the same time they pose the risk of disclosing the microbe’s presence to the plant. To successfully manage this trade-off, cell wall synthesis is highly synchronized to microbial development in a spatial and temporal manner. Investigations of glucan synthesis in the hemibiotrophic maize pathogen C. graminicola demonstrated that the glucan synthases GLS1, KRE5, and KRE6 participate in the production of β-1,3- and β-1,6-glucans. Invading biotrophic hyphae of this fungus coordinately down-regulate GLS1, KRE5, and KRE6 expression, resulting in the formation of hyphae with little or no exposed β-1,3/1,6-glucans (Oliveira-Garcia and Deising, 2013, 2016). Overexpression of these glucan synthesis genes in biotrophic hyphae provokes callose production and defense gene expression. This highlights that dynamic remodeling of the cell wall architecture is an important strategy to subvert plant immunity (Oliveira-Garcia and Deising, 2013, 2016). Since plants have not been reported to possess β-1,6-glucanases, frequent branching patterns might spatially hinder host β-1,3-glucanases binding to their respective substrates and thus protect the microbe against cell wall hydrolysis (Nars et al., 2013). A further microbial strategy to safeguard its cell wall integrity is mediated via glucanase inhibitor proteins (Ham et al., 1997; Rose et al., 2002). These secreted proteins identified in the culture filtrate of P. sojae prevent cleavage of cell wall glucans by binding to host β-glucanases. The high-affinity complex formation is driven by a serine protease domain without a functional catalytic triad necessary for protein cleavage (Rose et al., 2002). More recently, the dual-function effector FGB1 from the beneficial root endophyte Serendipita indica was reported to mediate cell wall resistance to stress and to suppress β-glucan-triggered immunity in Arabidopsis and N. benthamiana (Wawra et al., 2016). FGB1 is a fungal lectin with a strong binding affinity for β-1,6-linked glucose residues (Wawra et al., 2016). This study draws attention to the fact that not only phytopathogenic but also beneficial fungi evolved mechanisms to circumvent recognition of their own cell wall components.
The EPS of filamentous microbes: an emerging player in plant microbe-interaction?
Although several reports document the presence of highly mobile, gel-like EPS matrices surrounding the hyphae of plant-colonizing microbes, their function is largely unknown (Carzaniga et al., 2001; Wawra et al., 2019). Different types of EPS matrices were shown to contribute to host adhesion (Deising et al., 1992; Moloshok et al., 1993; Epstein and Nicholson, 1997; Doss, 1999; Carzaniga et al., 2001), act as an enzymatic scaffold (Nicholson and Moraes, 1980; Moloshok et al., 1993; Doss, 1999), or confer resistance to environmental stresses (Nicholson and Moraes, 1980) in different fungi and oomycetes. Still, little is known about their direct implications in interactions with plants. Analyses of β-glucan-containing EPS purified from culture supernatants of an endophytic Fusarium species (Li et al., 2014) and B. cinerea (El Oirdi et al., 2011) revealed activation of defense-related responses in their respective hosts. In B. cinerea, the EPS matrix induces accumulation of SA, an important player in the complex phytohormone signaling network regulating different defense pathways (El Oirdi et al., 2011). Whereas SA signaling often leads to host cell death and thereby restricts growth of biotrophic invaders, jasmonic acid (JA)-related defense generally accounts for resistance against necrotrophic pathogens (Glazebrook, 2005). Antagonistic crosstalk between these two phytohormones presents a fine-tuning mechanism to adapt immune responses according to different pathogenic lifestyles (Koornneef and Pieterse, 2008). This counteractive relationship between SA and JA is utilized by B. cinerea to increase disease progression in tomato through EPS-triggered SA accumulation and consequent suppression of JA-mediated pathways (El Oirdi et al., 2011).
A look into the body of literature on EPS matrices from A. fumigatus and C. albicans reveals that their EPSs can contribute to evasion of host immunity. In A. fumigatus, GAG polymers in the EPS matrix conceal immunogenic cell wall structures from recognition by host immune receptors (Gravelat et al., 2013). Furthermore, charges within the GAG polymers can protect the hyphae from antifungal peptides via electrostatic repulsion (Lee et al., 2015). Comparative genomic analyses identified the presence of a GAG biosynthetic gene cluster in plant-pathogenic fungi, indicating that GAG-like structures could play a role in plant pathogenesis (Lee et al., 2016). In C. neoformans, the GXM of its capsule was shown to confer resistance due to its function as a ROS scavenger (Zaragoza et al., 2008; Denham et al., 2018). Furthermore, capsular GXM and GalXM harbor immunomodulatory properties and contribute to host immune evasion (Decote-Ricardo et al., 2019). Similarly, the heterogalactan EPS from Trametes versicolor functions as a pro-antioxidant agent in mammalian cell lines, plants, and fungi (Scarpari et al., 2017). To what extent EPS matrices from plant-colonizing fungi or oomycetes can be considered fundamental virulence and symbiosis factors needs to be further addressed.
Secreted microbial glycans: messengers of peace?
In addition to cell surface-attached glycans, microbes can secrete small and diffusible sugar oligomers and glycoconjugates that directly impact the outcome of plant–microbe interaction. While some function as signaling molecules to activate signaling cascades needed for the establishment of symbioses, others hold the potential to suppress immune responses in order to support successful plant colonization (Supplementary Table S1).
The symbiont’s sweet words: small secreted glycoconjugates involved in the establishment of symbiotic interactions
Many plants form intimate associations with beneficial microbes. Among different forms of symbiotic interactions, arbuscular mycorrhiza (AM) and the RLS are two widespread representatives whose signaling pathways have been intensively studied for decades. While AM fungi improve the uptake of water and soil minerals in exchange for photoassimilates and lipids, rhizobia fix inert atmospheric nitrogen in specialized organs (nodules) functioning as an environmental niche for these bacteria. AM is genetically controlled by an ancient and highly conserved signaling pathway—the so-called common symbiosis signaling pathway—for recognition and initiation of symbiotic interactions (Oldroyd, 2013). In legumes, this pathway is required for the establishment of both AM and RLS. Several plant genes required for symbiosis have therefore been conserved throughout evolution in monocot and dicot plants (Genre and Russo, 2016). Since these bacterial and fungal symbionts share many structural patterns with pathogens, successful establishment of mutual interactions is dependent on the integration of stimuli obtained from both immunogenic and symbiotic signals. The main glycoconjugate messengers involved in the establishment of symbiosis are chitooligosacharides and lipochitooligosaccharides (LCOs), the latter also referred to as myc-LCOs (in AM) or nod-LCOs (in RLS) (Liang et al., 2014). Their perception activates pre-symbiotic signaling, gene expression changes. and developmental programs facilitating infection and the formation of symbiotic structures such as arbuscules and root nodules (Liang et al., 2014; Schmitz and Harrison, 2014). In general, LCOs are short, acylated COs (DP 3–5) with a variety of chemical modifications at their reducing and non-reducing termini. They are associated with a fatty acid at their non-reducing end and often carry further chemical modifications. The high diversity in chemical structure (backbone chain length, acetylation, sulfation, etc.) is assumed to mediate compatibility between host and microbe for specific establishment of symbiosis. Analogous to the perception of microbial cell wall components, the class of LysM motif receptor kinases is also involved in LCO signal recognition. In Medicago, nod-LCO perception is mediated via a heteromeric complex of the receptor-like kinases LYK3 and NFP (in Lotus NFR1 and NFR5) (Amor et al., 2003; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006). A previous study in Lotus suggested a two-step mechanism for establishment of nodulation in which nod-LCOs initiate symbiotic signaling and EPR3-driven EPS sensing acts as a subsequent quality control mechanism. Nod-LCO perception triggers the first step of symbiotic signal transduction, including intranuclear oscillatory calcium spiking at the root hair tip (Ehrhardt et al., 1996; Sieberer et al., 2009). Further, this induces EPR3 expression and initiates nodule organogenesis within the root cortex and bacterial infection through infection threads towards the nodule primordia (Gage, 2004). During these stages, iterative EPS recognition ensures colonization with compatible bacteria and promotes the progressing infection (Kawaharada et al., 2017). This collaborative recognition of nod-LCOs and rhizobial EPS presents a sophisticated way to repetitively control the advancing endosymbiosis without risking contamination with non-compatible bacteria. Of note, endophytic colonization of non-symbiotic bacteria (e.g. in mixed-colonized nodules) is also based on host-determined compatibility of their EPS (Zgadzaj et al., 2015). In addition to this well-established role of nod-LCOs, it was shown that nod-LCOs actively suppress defense signaling in different nodulating and non-nodulating species (Liang et al., 2013).
Initiation of AM is based on perception of myc-LCOs and COs, the two major classes of diffusible symbiotic signals found within fungal exudates. In Medicago, fungal myc-LCOs have been shown to trigger nuclear calcium spiking, alter gene expression patterns, and promote lateral root formation in an NFP-dependent manner (Maillet et al., 2011; Czaja et al., 2012; Sun et al., 2015). Unexpectedly, studies in nfp mutants in Medicago revealed that overall mycorrhization was not dependent on this receptor (Amor et al., 2003; Maillet et al., 2011). Notably, a recent study revealed that the LysM receptor-like kinase LYK10 confers binding to myc-LCO in non-leguminous plants. In contrast to the lacking phenotype in the Medicago nfp mutant, lyk10 mutation impairs AM formation in tomato and petunia (Girardin et al., 2019). Since the family of LysM domain proteins is massively expanded in many plant species, it cannot be excluded that redundancy in the perception of myc-LCOs may cause the observed discrepancies.
Since perception of fungal COs also leads to oscillatory calcium fluxes, this glycoconjugate is assumed to play an important role in establishment of AM symbiosis (Genre et al., 2013). While short COs (DP 3–4) are often considered as symbiotic signals and long COs (DP 6–8) as defense patterns, recent work from Feng et al. (2019) demonstrates that COs independent of their length activate both defense and symbiotic signaling at the same time in Medicago. CO sensing involves the receptor kinases LYK9 (also CERK1) and LYR4 (Bozsoki et al., 2017). Combinatory application of both COs and LCOs was able to suppress immunity signaling in Medicago, leading to an overall symbiotic outcome (Feng et al., 2019). The integration of signaling cascades from different simultaneously perceived molecules forms an elegant mechanism supporting the plant to discriminate between detrimental or symbiotic microbes in order to respond with an adequate molecular program. Moreover, CO perception demonstrates that signaling mechanisms are entangled, often making it hard to classify signaling molecules within a dichotomous conception of immunity and symbiosis.
Cellooligomers and cyclic β-glucans: secreted glycans in the extracellular space
Cellooligomers, β-1,4-linked glucose oligomers, can induce mild defense-like responses in plants. The strength of these responses in Arabidopsis and grapevine is strictly coupled to the DP (Aziz et al., 2007; Johnson et al., 2018). So far, these molecules were mainly considered as plant cell wall-derived molecules that are perceived by membrane-bound PRRs upon activity of pathogenic cell wall-degrading enzymes or during cell wall remodeling (Aziz et al., 2007; Souza et al., 2017). Only recently, cellotriose was discovered to be produced by the root endophyte S. indica. This trisaccharide triggers cytosolic calcium elevation, weak ROS induction, and membrane depolarization in Arabidopsis and induces the up-regulation of genes involved in plant defense (Johnson et al., 2018). Interestingly, a previous report on cellobiose-induced signaling in Arabidopsis demonstrated a ROS-independent host response including the activation of MAPKs and up-regulation of defense-related WRKY transcription factors (Souza et al., 2017). Cellotriose and cellobiose harbor the potential to synergistically increase host responses to chitin (Johnson et al., 2018) and flg22 when co-applied in Arabidopsis (Souza et al., 2017; Johnson et al., 2018). Additionally, pre-treatment of grapevine with cellooligomers of different DPs (DP 7–9) was shown to promote resistance against B. cinerea (Aziz et al., 2007), and cellobiose pre-treatment of Arabidopsis conferred enhanced resistance against the hemibiotrophic pathogen P. syringae pv. tomato DC3000 (Aziz et al., 2007; Souza et al., 2017). Yet, the components that mediate cellooligomer signaling remain unknown. In Arabidopsis, CERK1 and BAK1 receptor mutants remain sensitive to cellooligomers (Souza et al., 2017). Furthermore, cellooligomer-pre-treated grapevine plants do not present refractory behavior in their defense response to plant cell wall-derived oligogalacturonides, indicating a separate recognition mechanism (Aziz et al., 2007).
The fact that S. indica colonization of Arabidopsis (Vadassery et al., 2009) and cellotriose application (Johnson et al., 2018) both result in growth benefit for the plant suggests that cellotriose signaling might be involved in growth-promoting effects. Indeed, cellotriose treatment induced Arabidopsis genes involved in cell growth and root development, suggesting a genetically regulated mechanism for growth promotion (Vadassery et al., 2009; Johnson et al., 2018).
Cyclic β-glucans (CbGs) are produced by both pathogenic and symbiotic plant-colonizing bacteria and accumulate in the periplasmic and extracellular space (Breedveld and Miller, 1994). CbGs are involved in hypoosmotic adaption (Crespo-Rivas et al., 2009), motility (Breedveld and Miller, 1994; Bhagwat et al., 1996; Gay-Fraret et al., 2012), and stress protection (Javvadi et al., 2018). Bacterial mutants affected in CbG synthesis or periplasmic secretion form no or inefficient symbiosis or have reduced virulence, which emphasizes their relevance for plant–microbe interactions (Breedveld and Miller, 1994). CbGs can be separated into two groups based on the type of glycosidic linkage and DP. Bradyrhizobial β-1,3/1,6-linked cyclic glucans are built of 10–13 glucose molecules where the majority form a β-1,3-linked ring carrying branches of β-1,6-linked glucoses (Breedveld and Miller, 1994). In contrast, rhizobial cyclic glucans consist of 17–40 β-1,2-linked glucose moieties (Breedveld and Miller, 1994). Early evidence that CbGs can act as signaling molecules was derived from studies where the β-1,3/1,6-linked cyclic glucans from Bradyrhizobium japonicum were able to suppress the immuno-eliciting activity of β-1,3/1,6-glucans from the soybean pathogen P. sojae in a concentration-dependent manner (Mithöfer et al., 1996). The observation that both β-glucans bind to the same β-glucan-binding site in soybean roots suggests that the suppressive activity is mediated by competition. Bradyrhizobial CbGs were also shown to weakly induce production of the isoflavone daidzein, a known Nod gene inducer in B. japonicum which stimulates the production of bacterial LCOs (Mithöfer et al., 1996). Their impact on symbiotic interaction with soybean is dependent on the β-1,6-linked glucoses (Bhagwat et al., 1999). In contrast to bradyrhizobial CbGs, rhizobial CbGs (β-1,2-linked) from symbiotic Rhizobium leguminosarium and the phytopathogen Agrobacterium tumefaciens did not lead to the production of isoflavonoids (Miller et al., 1994). Furthermore, S. meliloti CbGs did not compete for the same receptor-binding site with the oomycete-derived β-1,3/1,6-glucans (Mithöfer et al., 1996).
CbGs from the leaf-colonizing phytopathogenic bacterium Xcc were shown to actively suppress local and systemic host defense responses. This type of CbG is structurally unique due to the presence of a single α-1,6-linked glycosyl residue among 15 β-1,2-linked glycosyl-residues (York, 1995). In N. benthamiana, CbGs from Xcc suppress PR1 induction and, locally as well as systemically, callose deposition. Accordingly, pre-treatment of Arabidopsis and N. benthamiana with physiologically relevant concentrations of bacterial CbGs increased the susceptibility to Xcc in a systemic manner (Rigano et al., 2007). The Xcc ndvB mutant strain deficient in CbG production shows earlier and longer lasting PR1 gene expression and enhanced callose deposition, resulting in lower virulence in Arabidopsis and N. benthamiana that could be restored by external application of CbGs (Rigano et al., 2007). Intriguingly, restoration of the symbiotic relationship between a CbG-deficient rhizobia strain and the legume host was not observed after external application of the respective CbGs, indicating a more complex role for rhizobial CbGs during plant–bacteria interaction (Dylan et al., 1990).
Learning from the inconspicuous—a case study: the role of extracellular polysaccharides in the lichen symbiosis
An excellent example of the important role played by extracellular polysaccharides is given by the multipartite interactions occurring in lichens. Lichens are stable, long-term interactions between one or more multicellular fungi (mycobionts) and photosynthesizing single-celled algae or cyanobacteria (photobionts). These symbioses have repeatedly evolved complex three-dimensional architectures held together by hyphal cells (which represents up to 90% of the total biomass) and hydrophilic extracellular polymeric substances in the outermost layer. Below this hydrophilic layer, typically a water-repellent, gas-filled internal space is present where the photobiont is located, a prerequisite for efficient photosynthesis. Nutrients and water are diffusing from the EPS matrix into the fungal cell wall, and through the apoplast they reach the photobiont partners. Lichens and their photobionts are tolerant to desiccation and exhibit complete physiological recovery upon rehydration. Their water content is determined by environmental water availability and they are often subjected to alternating desiccation–rehydration cycles. When hydrated, the lichen thallus expands to many times its desiccated volume. The hydrophilic properties of the EPS matrix in the outmost layer account for this effect and determine the point at which surface tension is broken and water is taken up from the environment and retained (Spribille et al., 2020). Biochemical remodeling of the EPS matrix in desiccation-tolerant lichens is species-specific and seems to play a role in the response to changes in environmental water availability. Additionally, the EPS matrix serves as a surface-maximized depot for mineral nutrients and microbial-derived secondary metabolites. Because of its affinity for positively charged molecules, the EPS matrix plays an important role in heavy metal sequestration, preventing the uptake of toxic compounds into cells. This may explain the ability of certain pioneer lichens to colonize extremely polluted sites (Backor and Loppi, 2009). The EPS matrix also acts as substrate for colonization by bacteria and basidiomycetous yeasts commonly found in lichens. As in biofilms, the lichen EPS matrix functions as the main medium by which cell–cell communication between different microbial partners takes place (Spribille et al., 2020).
The EPS matrix in lichens can be considered as a multipartite structure. Its composition is not determined by a single organism but instead depends on the participating symbionts. Fungi are considered to be the major inhabitant contributing the bulk of secreted polysaccharides. Using histological stains, strongly acidic polysaccharides were visualized with distinguishable differences in anionic density between the fungal cell walls and the EPS matrix, suggesting that fungal sulfated polysaccharides and polyuronic acids are present in the matrix. Most of the reports have identified neutral β- and α-glucans and α-mannans with varying ratios of glucose, galactose, and mannan as the major components in whole lichens (Olafsdottir and Ingolfsdottir, 2001; Carbonero et al., 2002; Reis et al., 2002). The β-glucans are mainly represented by β-1,3- or β-1,3/1,4-glucans (lichenan), while the α-glucans are represented by the α-1,3/1,4-glucans isolichenan and nigeran, and less commonly by mixed-linkage α-1,4/1,6-glucan. Branched β-1,3/1,6-glucans and the β-1,6-glucan pustulan have also been reported in Lecanoromycetidae and Umbilicariomycetidae. These glucans are structurally similar to the core cell wall polysaccharides of fungi (Gow et al., 2017). In the few basidiomycete-based lichen symbioses, different cell wall components have been found with linear α-1,3-glucan (pseudonigeran). A comprehensive review of lichen EPSs is provided by Spribille et al. (2020). Despite the large amount of fungal biomass in lichens, other microbial partners may participate in the production of the extracellular polysaccharide matrix. Green algal symbionts are often found in the hydrophobin-lined internal chambers, generally not in direct contact with the matrix. However, recent reports have shown that algal partners can also excrete EPS, including medium to small sized uronic acid-containing polysaccharides and sulfated polysaccharides. The composition and amount of the secreted polysaccharides were shown to depend, at least in part, on abiotic stresses such as nitrogen, light and water availability, and presence of heavy metals (Gonzalez-Hourcade et al., 2020). Cyanobacterial symbionts, on the other hand, are typically embedded in a self-produced matrix. Bacterial and basidiomycetous yeast-derived EPSs in lichens are less studied but their potential functions should not be underestimated. Indeed the presence of the nifH gene involved in nitrogen fixation was demonstrated in several lichen-associated α-proteobacteria, γ-proteobacteria, actinobacteria, and firmicutes (Grube et al., 2009; Almendras et al., 2018). Prominent among the α-proteobacteria are the Rhizobiales with LAR1, a lichen-associated bacterial clade (Hodkinson and Lutzoni, 2009). It is therefore plausible to think that nitrogen-fixing bacteria represent a third important partner in the lichen symbiosis.
Understanding the structure and function of this highly organized EPS matrix in the extreme examples of lichen symbioses will help to shed light on the function of EPS matrices formed during multipartite interactions in the rhizoplane (Fig. 2) and their potential function in water, secondary metabolite, and mineral nutrient storage/utilization and immunity.
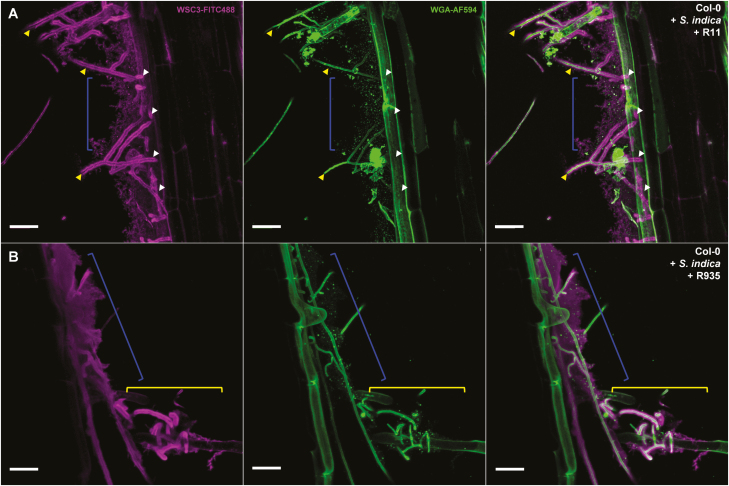
Fig. 2.
Live cell images of root-associated bacteria embedded in the fungal β-glucan matrix. Confocal laser scanning microscopy live stain images of an A. thaliana Col-0 root segment, 12 d post-colonization with the fungus Serendipita indica and root-associated bacteria R11 (Bacillus sp.) (A) or R935 (Flavobacterium sp.) (B) (Bai et al., 2015). The fungal cell wall and bacteria (dots) were stainable with WGA-AF594 (green pseudo-color). β-1,3-glucan was visualized using the FITC488-labeled lectin WSC3 (magenta pseudo-color) (Wawra et al., 2019). Fungal chitin was only detected in hyphae growing in the extracellular space (yellow arrowheads), whereas the β-glucan matrix was also detectable after the hyphae entered the root cortical cells (white arrowheads). Interestingly, the β-glucan architecture was very distinct in the presence of the individual bacterial strains (structures indicated by the blue brackets). While a fine structure in the glucan network was visible for S. indica in combination with R11, the combination with R935 resulted in an apparently extended, more dense, and amorphous looking β-glucan layer. In addition, in the presence of R935, the fungal β-glucan matrix around the S. indica hyphae appeared more diffuse and less compact compared with the matrix detected around more isolated growing hyphae (B, yellow brackets). It is currently unclear whether the additionally deposited β-glucan is produced by the bacteria themselves or whether their presence triggers enhanced production and secretion by the fungus. Scale bars=25 µm.
Concluding remarks and perspective
The outcome of plant–microbe interactions is strictly governed by persistent communication processes between the involved partners. Due to the inherent structural and functional complexity of glycans and glycoconjugates, they represent an important group of messengers involved in this ongoing crosstalk. The high degree of specificity within this molecular dialog is influenced by various factors. Microbial cell walls, which are diverse networks of crystalline and soluble exopolysaccharides, often represent the starting point for recognition. While many of those structural motifs are highly conserved and can be recognized by a wide range of plants, perception of other unique structural motifs is limited to a few plant species. On the other hand, microbes secrete a fleet of molecules such as enzymes, effector proteins, and soluble glycans, in order to interfere with host recognition and the activation of defense responses. Collectively, the apoplastic encounter of plant and microbes gives rise to a multitude of signaling molecules, which need to be processed by both partners. Ancient endosymbioses such as AM and RLS highlight that collaborative receptor perception, continuous scrutiny, and integration of multiple signals is required to assess microbial compatibility and guide developmental reprogramming (Kawaharada et al., 2017). While our understanding of these signaling networks and their components is constantly increasing, many questions remain unanswered. The widespread use of heterogeneous substrates instead of pure and defined glycan structures is prompting the knowledge gap regarding receptors and downstream components for several glycan-based ligands (e.g. β-glucans and LPSs). Recent technological advances made in glycoscience, glycan synthesis, and structural biology will improve the quality of glycan substrates and help to revisit our understanding of how carbohydrate signatures contribute to host–microbe interactions (Weishaupt et al., 2017; Kang et al., 2018; Guberman and Seeberger, 2019; Reichhardt et al., 2019). Furthermore, not much attention has been paid to the impact of microbial EPS matrices surrounding plant organs. Although the composition and structural organization of EPS matrices differ between different microbial classes, functional convergence can be observed. Since EPS matrices of plant-colonizing bacteria and human-pathogenic fungi have been shown to be crucial determinants of host–microbe interactions, a similarly important function can be assumed for EPS matrices of filamentous microbes interacting with plants. Interestingly, bacteria and fungi commonly form mixed, interkingdom biofilms on plant tissues and in lichens (van Overbeek and Saikkonen, 2016; Guennoc et al., 2018). As observed in the latter, the EPS architecture within such multipartite consortia is jointly shaped and highly adapted to the demands of its inhabitants. Taking into account that each partner within collaborative microbial biofilms contributes its own set of enzymes involved in glycan synthesis and hydrolysis (Bamford et al., 2019), this could fuel the generation of unique community patterns. Therefore, it would be of great interest to understand whether such community-derived glycan signatures are relevant for microbiota assembly and plant recognition. The recent technological advances made in glycoscience, glycan synthesis, and structural biology will surely help to increase our understanding of these glycan structures and shed light on their contribution to host–microbe interactions.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. Overview of microbial glycans and glycoconjugate structures and their relevance for host–microbe interactions.
Acknowledgements
We apologize to all our colleagues in this field whose work could not be cited due to word count limitation. We thank Ruben Garrido-Oter, Paul Schulze-Lefert and the DECRyPT community (SPP 2125) for providing the bacterial strains used for microscopy. AW was supported by the Max-Planck-Gesellschaft through the International Max Planck Research School (IMPRS) on ‘Understanding Complex Plant Traits using Computational and Evolutionary Approaches’ and the University of Cologne. AZ, MM, and SW acknowledge support from the Cluster of Excellence on Plant Sciences (CEPLAS) funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC 2048/1 – Project ID: 390686111 and ZU 263/11-1 (SPP DECRyPT). Graphical illustrations were designed with the BioRender online tool.
Glossary
Abbreviations
- AM
arbuscular mycorrhiza
- CbG
cyclic β-glucan
- CO
chitooligomer
- DP
degree of polymerization
- EPS
extracellular polysaccharide
- GAG
galactosaminogalactan
- GlcNAc
N-acetylglucosamine
- GXM
glucuronoxylomannan
- GalXM
glucuronoxylomanogalactan
- GPI
glycosylphosphatidylinositol
- Kdo
2-keto-3-desoxy-octonat
- LCO
lipochitooligosaccharide
- LPS
lipopolysaccharide
- LysM
lysin motif
- MAPK
mitogen-activated protein kinase
- MurNAc
N-acetylmuramic acid
- PGN
peptidoglycan
- PRR
pattern recognition receptor
- pv
pathovar
- RLS
rhizobium–legume symbiosis
- ROS
reactive oxygen species
- SA
salicylic acid
- Xcc
Xanthomonas campestris pv. campestris
Author contributions
AW, MM, and AZ developed the concept of this review. All authors wrote the manuscript, AW coordinated the writing process. AW and SW prepared the figures; SW performed the microscopy. AZ supervised the project and acquired the funding.
Conflict of interest
The authors declare no competing interests.
Data availability
All data supporting the content of this study are available within the review and within its supplementary data published online.
References
- Acosta-Jurado S, Rodriguez-Navarro DN, Kawaharada Y, et al. 2016. Sinorhizobium fredii HH103 invades Lotus burttii by crack entry in a nod factor- and surface polysaccharide-dependent manner. Molecular Plant-Microbe Interactions 29, 925–937. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AG, Lane CE, et al. 2012. The revised classification of eukaryotes. Journal of Eukaryotic Microbiology 59, 429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Hua C, Nürnberger T, Pruitt RN, Zhang L. 2020. Surface sensor systems in plant immunity. Plant Physiology 182, 1582–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendras K, Garcia J, Caru M, Orlando J. 2018. Nitrogen-fixing bacteria associated with Peltigera cyanolichens and Cladonia chlorolichens. Molecules 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Denarie J, Gough C. 2003. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. The Plant Journal 34, 495–506. [DOI] [PubMed] [Google Scholar]
- Anderson JA 1978. The isolation from three species of Colletotrichum of glucan containing polysaccharides that elicit a defense response in bean (Phaseolus vulgaris). Phytopathology 68, 189–194. [Google Scholar]
- Ao Y, Li Z, Feng D, Xiong F, Liu J, Li JF, Wang M, Wang J, Liu B, Wang HB. 2014. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. The Plant Journal 80, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Arnaud J, Audfray A, Imberty A. 2013. Binding sugars: from natural lectins to synthetic receptors and engineered neolectins. Chemical Society Reviews 42, 4798–4813. [DOI] [PubMed] [Google Scholar]
- Arnold MFF, Penterman J, Shabab M, Chen EJ, Walker GC. 2018. Important late-stage symbiotic role of the Sinorhizobium meliloti exopolysaccharide succinoglycan. Journal of Bacteriology 200, e00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, et al. 2006. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiology 142, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam SN, Newman MA, Erbs G, et al. 2008. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Current Biology 18, 1078–1083. [DOI] [PubMed] [Google Scholar]
- Ayers AR, Ebel J, Finelli F, Berger N, Albersheim P. 1976a Host–pathogen interactions: IX. Quantitative assays of elicitor activity and characterization of the elicitor present in the extracellular medium of cultures of Phytophthora megasperma var. sojae. Plant Physiology 57, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers AR, Ebel J, Valent B, Albersheim P. 1976b Host–pathogen interactions: X. Fractionation and biological activity of an elicitor isolated from the mycelial walls of Phytophthora megasperma var. sojae. Plant Physiology 57, 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A, Gauthier A, Bézier A, Poinssot B, Joubert JM, Pugin A, Heyraud A, Baillieul F. 2007. Elicitor and resistance-inducing activities of beta-1,4 cellodextrins in grapevine, comparison with beta-1,3 glucans and alpha-1,4 oligogalacturonides. Journal of Experimental Botany 58, 1463–1472. [DOI] [PubMed] [Google Scholar]
- Aziz A, Poinssot B, Daire X, Adrian M, Bézier A, Lambert B, Joubert JM, Pugin A. 2003. Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Molecular Plant-Microbe Interactions 16, 1118–1128. [DOI] [PubMed] [Google Scholar]
- Bacete L, Mélida H, Miedes E, Molina A. 2018. Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. The Plant Journal 93, 614–636. [DOI] [PubMed] [Google Scholar]
- Backor M, Loppi S. 2009. Interactions of lichens with heavy metals. Biologia Plantarum 53, 214–222. [Google Scholar]
- Badreddine I, Lafitte C, Heux L, Skandalis N, Spanou Z, Martinez Y, Esquerré-Tugayé MT, Bulone V, Dumas B, Bottin A. 2008. Cell wall chitosaccharides are essential components and exposed patterns of the phytopathogenic oomycete Aphanomyces euteiches. Eukaryotic Cell 7, 1980–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Müller DB, Srinivas G, et al. 2015. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. [DOI] [PubMed] [Google Scholar]
- Bamford NC, Le Mauff F, Subramanian AS, et al. 2019. Ega3 from the fungal pathogen Aspergillus fumigatus is an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms. Journal of Biological Chemistry 294, 13833–13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beakes GW, Glockling SL, Sekimoto S. 2012. The evolutionary phylogeny of the oomycete ‘fungi’. Protoplasma 249, 3–19. [DOI] [PubMed] [Google Scholar]
- Becker M, Becker Y, Green K, Scott B. 2016. The endophytic symbiont Epichloë festucae establishes an epiphyllous net on the surface of Lolium perenne leaves by development of an expressorium, an appressorium-like leaf exit structure. New Phytologist 211, 240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedini E, De Castro C, Erbs G, Mangoni L, Dow JM, Newman MA, Parrilli M, Unverzagt C. 2005. Structure-dependent modulation of a pathogen response in plants by synthetic O-antigen polysaccharides. Journal of the American Chemical Society 127, 2414–2416. [DOI] [PubMed] [Google Scholar]
- Bell PBJ, Safiejko-Mroczka B. 1997. Preparing whole mounts of biological specimens for imaging macromolecular structures by light and electron microscopy. International Journal of Imaging Systems and Technology 8, 225–239. [Google Scholar]
- Bhagwat AA, Gross KC, Tully RE, Keister DL. 1996. Beta-glucan synthesis in Bradyrhizobium japonicum: characterization of a new locus (ndvC) influencing beta-(1→6) linkages. Journal of Bacteriology 178, 4635–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AA, Mithöfer A, Pfeffer PE, Kraus C, Spickers N, Hotchkiss A, Ebel J, Keister DL. 1999. Further studies of the role of cyclic beta-glucans in symbiosis. An NdvC mutant of Bradyrhizobium japonicum synthesizes cyclodecakis-(1→3)-beta-glucosyl. Plant Physiology 119, 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm TL, Deslandes Y, Marchessault RH, Perez S, Rinaudo M. 1982. Solid-state and solution conformation of scleroglucan. Carbohydrate Research 100, 117–130. [Google Scholar]
- Bluhm TL, Sarko A. 1977. The triple helical structure of lentinan, a linear β -(1→3)-d-glucan. Canadian Journal of Chemistry 55, 293–299. [Google Scholar]
- Bolouri Moghaddam MR, Van den Ende W. 2013. Sweet immunity in the plant circadian regulatory network. Journal of Experimental Botany 64, 1439–1449. [DOI] [PubMed] [Google Scholar]
- Bowman SM, Free SJ. 2006. The structure and synthesis of the fungal cell wall. Bioessays 28, 799–808. [DOI] [PubMed] [Google Scholar]
- Bozsoki Z, Cheng J, Feng F, Gysel K, Vinther M, Andersen KR, Oldroyd G, Blaise M, Radutoiu S, Stougaard J. 2017. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proceedings of the National Academy of Sciences, USA 114, E8118–E8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun SG, Meyer A, Holst O, Pühler A, Niehaus K. 2005. Characterization of the Xanthomonas campestris pv. campestris lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Molecular Plant-Microbe Interactions 18, 674–681. [DOI] [PubMed] [Google Scholar]
- Breedveld MW, Miller KJ. 1994. Cyclic beta-glucans of members of the family Rhizobiaceae. Microbiological Reviews 58, 145–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G. 2014. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonero ER, Sassaki GL, Gorin PA, Iacomini M. 2002. A (1→6)-linked beta-mannopyrananan, pseudonigeran, and a (1→4)-linked beta-xylan, isolated from the lichenised basidiomycete Dictyonema glabratum. FEMS Microbiology Letters 206, 175–178. [DOI] [PubMed] [Google Scholar]
- Carzaniga R, Bowyer P, O’Connell RJ. 2001. Production of extracellular matrices during development of infection structures by the downy mildew Peronospora parasitica. New Phytologist 149, 83–93. [DOI] [PubMed] [Google Scholar]
- Chen T, Nomura K, Wang X, et al. 2020. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JJ, Alba R, Côté F, Enkerli J, Hahn MG. 1993. Solubilization of functional plasma membrane-localized hepta-beta-glucoside elicitor-binding proteins from soybean. Plant Physiology 103, 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JJ, Birberg W, Fügedi P, Pilotti A, Garegg PJ, Hong N, Ogawa T, Hahn MG. 1991. Structure–activity relationships of oligo-beta-glucoside elicitors of phytoalexin accumulation in soybean. The Plant Cell 3, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheval C, Samwald S, Johnston MG, de Keijzer J, Breakspear A, Liu X, Bellandi A, Kadota Y, Zipfel C, Faulkner C. 2020. Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proceedings of the National Academy of Sciences, USA 117, 9621–9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley FG, Valent B. 1990. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Molecular Plant-Microbe Interactions 3, 135–143. [Google Scholar]
- Cobb BA, Kasper DL. 2005. Coming of age: carbohydrates and immunity. European Journal of Immunology 35, 352–356. [DOI] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BP. 2015. Understanding plant immunity as a surveillance system to detect invasion. Annual Review of Phytopathology 53, 541–563. [DOI] [PubMed] [Google Scholar]
- Cord-Landwehr S, Melcher RL, Kolkenbrock S, Moerschbacher BM. 2016. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Scientific Reports 6, 38018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado JE, Mneimneh S, Epstein SL, Qiu WG, Lipke PN. 2007. Conserved processes and lineage-specific proteins in fungal cell wall evolution. Eukaryotic Cell 6, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C. 2016. Regulation of pattern recognition receptor signalling in plants. Nature Reviews. Immunology 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Crespo-Rivas JC, Margaret I, Hidalgo A, et al. 2009. Sinorhizobium fredii HH103 cgs mutants are unable to nodulate determinate- and indeterminate nodule-forming legumes and overproduce an altered EPS. Molecular Plant-Microbe Interactions 22, 575–588. [DOI] [PubMed] [Google Scholar]
- Cunto-Amesty G, Dam TK, Luo P, Monzavi-Karbassi B, Brewer CF, Van Cott TC, Kieber-Emmons T. 2001. Directing the immune response to carbohydrate antigens. Journal of Biological Chemistry 276, 30490–30498. [DOI] [PubMed] [Google Scholar]
- Czaja LF, Hogekamp C, Lamm P, Maillet F, Martinez EA, Samain E, Dénarié J, Küster H, Hohnjec N. 2012. Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiology 159, 1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwish SF, Asfour HA. 2013. Investigation of biofilm forming ability in Staphylococci causing bovine mastitis using phenotypic and genotypic assays. TheScientificWorldJournal 2013, 378492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decote-Ricardo D, LaRocque-de-Freitas IF, Rocha JDB, Nascimento DO, Nunes MP, Morrot A, Freire-de-Lima L, Previato JO, Mendonça-Previato L, Freire-de-Lima CG. 2019. Immunomodulatory role of capsular polysaccharides constituents of Cryptococcus neoformans. Frontiers in Medicine 6, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deising H, Nicholson RL, Haug M, Howard RJ, Mendgen K. 1992. Adhesion pad formation and the involvement of cutinase and esterases in the attachment of uredospores to the host cuticle. The Plant Cell 4, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MH, Thomma BP. 2010. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955. [DOI] [PubMed] [Google Scholar]
- Denham ST, Verma S, Reynolds RC, Worne CL, Daugherty JM, Lane TE, Brown JCS. 2018. Regulated release of cryptococcal polysaccharide drives virulence and suppresses immune cell infiltration into the central nervous system. Infection and Immunity 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaki Y, Kouzai Y, Ninomiya Y, et al. 2018. OsCERK1 plays a crucial role in the lipopolysaccharide-induced immune response of rice. New Phytologist 217, 1042–1049. [DOI] [PubMed] [Google Scholar]
- D’Haeze W, Holsters M. 2004. Surface polysaccharides enable bacteria to evade plant immunity. Trends in Microbiology 12, 555–561. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo F, Speciale I, Silipo A, et al. 2020. Structure of the unusual Sinorhizobium fredii HH103 lipopolysaccharide and its role in symbiosis. Journal of Biological Chemistry 295, 10969–10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews. Genetics 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Doss RP 1999. Composition and enzymatic activity of the extracellular matrix secreted by germlings of Botrytis cinerea. Applied and Environmental Microbiology 65, 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie JA 2010. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiology Reviews 34, 150–170. [DOI] [PubMed] [Google Scholar]
- Durso L, Lehnen LP, Powell MJ. 1993. Characteristics of extracellular adhesions produced during Saprolegnia ferax secondary zoospore encystment and cystospore germination. Mycologia 85, 744–755. [Google Scholar]
- Dylan T, Nagpal P, Helinski DR, Ditta GS. 1990. Symbiotic pseudorevertants of Rhizobium meliloti ndv mutants. Journal of Bacteriology 172, 1409–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. 1996. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85, 673–681. [DOI] [PubMed] [Google Scholar]
- El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB. 2002. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytologist 156, 103–112. [Google Scholar]
- El Oirdi M, El Rahman TA, Rigano L, El Hadrami A, Rodriguez MC, Daayf F, Vojnov A, Bouarab K. 2011. Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. The Plant cell 23, 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L, Nicholson RL. 1997. Adhesion of spores and hyphae to plant surfaces. In: Carroll GC, Tudzynski P, eds. Plant relationships: Part A. Berlin, Heidelberg: Springer Berlin Heidelberg, 11–25. [Google Scholar]
- Erbs G, Newman MA. 2012. The role of lipopolysaccharide and peptidoglycan, two glycosylated bacterial microbe-associated molecular patterns (MAMPs), in plant innate immunity. Molecular Plant Pathology 13, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs G, Silipo A, Aslam S, et al. 2008. Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: structure and activity. Chemistry & Biology 15, 438–448. [DOI] [PubMed] [Google Scholar]
- Estrada-Garcia MT, Callow JA, Green JR. 1990. Monoclonal antibodies to the adhesive cell coat secreted by Pythium aphanidermatum zoospores recognise 200×103 Mr glycoproteins stored within large peripheral vesicles. Journal of Cell Science 95, 199–206. [Google Scholar]
- Evans NA, Hoyne PA, Stone BA. 1984. Characteristics and specificity of the interaction of a fluorochrome from aniline blue (sirofluor) with polysaccharides. Carbohydrate Polymers 4, 215–230. [Google Scholar]
- Fang A, Gao H, Zhang N, Zheng X, Qiu S, Li Y, Zhou S, Cui F, Sun W. 2019. A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice. Frontiers in Microbiology 10, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ. 2013. LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proceedings of the National Academy of Sciences, USA 110, 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Sun J, Radhakrishnan GV, et al. 2019. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nature Communications 10, 5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesel PH, Zuccaro A. 2016. Beta-glucan: crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genetics and Biology 90, 53–60. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. 2010. The biofilm matrix. Nature Reviews. Microbiology 8, 623–633. [DOI] [PubMed] [Google Scholar]
- Fliegmann J, Mithofer A, Wanner G, Ebel J. 2004. An ancient enzyme domain hidden in the putative beta-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. Journal of Biological Chemistry 279, 1132–1140. [DOI] [PubMed] [Google Scholar]
- Fontaine T, Simenel C, Dubreucq G, Adam O, Delepierre M, Lemoine J, Vorgias CE, Diaquin M, Latge JP. 2000. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. Journal of Biological Chemistry 275, 41528. [PubMed] [Google Scholar]
- Franken LE, Grünewald K, Boekema EJ, Stuart MCA. 2020. A technical introduction to transmission electron microscopy for soft-matter: imaging, possibilities, choices, and technical developments. Small 16, e1906198. [DOI] [PubMed] [Google Scholar]
- Fujikawa T, Sakaguchi A, Nishizawa Y, Kouzai Y, Minami E, Yano S, Koga H, Meshi T, Nishimura M. 2012. Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathogens 8, e1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius HJ 1997. Animal lectins. European Journal of Biochemistry 243, 543–576. [DOI] [PubMed] [Google Scholar]
- Gage DJ 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiology and Molecular Biology Reviews 68, 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci S, Maffei ME. 2017. DNA sensing across the tree of life. Trends in Immunology 38, 719–732. [DOI] [PubMed] [Google Scholar]
- Gao F, Zhang BS, Zhao JH, Huang JF, Jia PS, Wang S, Zhang J, Zhou JM, Guo HS. 2019. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nature Plants 5, 1167–1176. [DOI] [PubMed] [Google Scholar]
- Gauthier A, Trouvelot S, Kelloniemi J, et al. 2014. The sulfated laminarin triggers a stress transcriptome before priming the SA- and ROS-dependent defenses during grapevine’s induced resistance against Plasmopara viticola. PLoS One 9, e88145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay-Fraret J, Ardissone S, Kambara K, Broughton WJ, Deakin WJ, Le Quéré A. 2012. Cyclic-β -glucans of Rhizobium (Sinorhizobium) sp. strain NGR234 are required for hypo-osmotic adaptation, motility, and efficient symbiosis with host plants. FEMS Microbiology Letters 333, 28–36. [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, et al. 2013. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytologist 198, 190–202. [DOI] [PubMed] [Google Scholar]
- Genre A, Russo G. 2016. Does a common pathway transduce symbiotic signals in plant–microbe interactions? Frontiers in Plant Science 7, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan I, Steinberg G, Gurr S. 2017. The role of the fungal cell wall in the infection of plants. Trends in Microbiology 25, 957–967. [DOI] [PubMed] [Google Scholar]
- Girardin A, Wang T, Ding Y, et al. 2019. LCO receptors involved in arbuscular mycorrhiza are functional for rhizobia perception in legumes. Current Biology 29, 4249–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43, 205–227. [DOI] [PubMed] [Google Scholar]
- González-Hourcade M, Del Campo EM, Braga MR, Salgado A, Casano LM. 2020. Disentangling the role of extracellular polysaccharides in desiccation tolerance in lichen-forming microalgae. First evidence of sulfated polysaccharides and ancient sulfotransferase genes. Environmental Microbiology 22, 3096–3111. [DOI] [PubMed] [Google Scholar]
- Gow NAR, Latge JP, Munro CA. 2017. The fungal cell wall: structure, biosynthesis, and function. Microbiology Spectrum 5, doi:10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PubMed] [Google Scholar]
- Gravelat FN, Beauvais A, Liu H, et al. 2013. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β -glucan from the immune system. PLoS Pathogens 9, e1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube M, Cardinale M, de Castro JV Jr, Müller H, Berg G. 2009. Species-specific structural and functional diversity of bacterial communities in lichen symbioses. The ISME Journal 3, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Guberman M, Seeberger PH. 2019. Automated glycan assembly: a perspective. Journal of the American Chemical Society 141, 5581–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Hardham AR. 1988. Secretion of adhesive material during encystment of Phytophthora cinnamomi zoospores, characterized by immunogold labelling with monoclonal antibodies to components of peripheral vesicles. Journal of Cell Science 90, 225–235. [Google Scholar]
- Guennoc CM, Rose C, Labbe J, Deveau A. 2018. Bacterial biofilm formation on the hyphae of ectomycorrhizal fungi: a widespread ability under controls? FEMS Microbiology Ecology 94, fiy093. [DOI] [PubMed] [Google Scholar]
- Gust AA, Biswas R, Lenz HD, et al. 2007. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. Journal of Biological Chemistry 282, 32338–32348. [DOI] [PubMed] [Google Scholar]
- Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. 2017. Interplay between innate immunity and the plant microbiota. Annual Review of Phytopathology 55, 565–589. [DOI] [PubMed] [Google Scholar]
- Ham K-S, Wu S-C, Darvill AG, Albersheim P. 1997. Fungal pathogens secrete an inhibitor protein that distinguishes isoforms of plant pathogenesis-related endo-β -1,3-glucanases. The Plant Journal 11, 169–179. [Google Scholar]
- Hansel CS, Holme MN, Gopal S, Stevens MM. 2020. Advances in high-resolution microscopy for the study of intracellular interactions with biomaterials. Biomaterials 226, 119406. [DOI] [PubMed] [Google Scholar]
- Hawkins JP, Geddes BA, Oresnik IJ. 2017. Succinoglycan production contributes to acidic pH tolerance in Sinorhizobium meliloti Rm1021. Molecular Plant-Microbe Interactions 30, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Hayafune M, Berisio R, Marchetti R, et al. 2014. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proceedings of the National Academy of Sciences, USA 111, E404–E413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhang C, Dai H, et al. 2019. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Molecular Plant 12, 1561–1576. [DOI] [PubMed] [Google Scholar]
- Hodkinson BP, Lutzoni F. 2009. A microbiotic survey of lichen-associated bacteria reveals a new lineage from the Rhizobiales. Symbiosis 49, 163–180. [Google Scholar]
- Iizasa S, Iizasa E, Matsuzaki S, Tanaka H, Kodama Y, Watanabe K, Nagano Y. 2016. Arabidopsis LBP/BPI related-1 and -2 bind to LPS directly and regulate PR1 expression. Scientific Reports 6, 27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa S, Iizasa E, Watanabe K, Nagano Y. 2017. Transcriptome analysis reveals key roles of AtLBR-2 in LPS-induced defense responses in plants. BMC Genomics 18, 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irieda H, Inoue Y, Mori M, et al. 2019. Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proceedings of the National Academy of Sciences, USA 116, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jashni MK, Dols IH, Iida Y, Boeren S, Beenen HG, Mehrabi R, Collemare J, de Wit PJ. 2015. Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin-binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Molecular Plant-Microbe Interactions 28, 996–1008. [DOI] [PubMed] [Google Scholar]
- Javvadi S, Pandey SS, Mishra A, Pradhan BB, Chatterjee S. 2018. Bacterial cyclic β -(1,2)-glucans sequester iron to protect against iron-induced toxicity. EMBO Reports 19, 172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian J, Liang X. 2019. One small RNA of Fusarium graminearum targets and silences CEBiP gene in common wheat. Microorganisms 7, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Thürich J, Petutschnig EK, et al. 2018. A poly(A) ribonuclease controls the cellotriose-based interaction between Piriformospora indica and its host Arabidopsis. Plant Physiology 176, 2496–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. 2008. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proceedings of the National Academy of Sciences, USA 105, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N. 2006. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proceedings of the National Academy of Sciences, USA 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagawa M, Satoh T, Ikeda A, Adachi Y, Ohno N, Yamaguchi Y. 2011. Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor. Journal of Biological Chemistry 286, 29158–29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Kirui A, Muszyński A, Widanage MCD, Chen A, Azadi P, Wang P, Mentink-Vigier F, Wang T. 2018. Molecular architecture of fungal cell walls revealed by solid-state NMR. Nature Communications 9, 2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, et al. 2015. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312. [DOI] [PubMed] [Google Scholar]
- Kawaharada Y, Nielsen MW, Kelly S, et al. 2017. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nature Communications 8, 14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen NT, Yoshikawa M. 1983. beta-1,3-Endoglucanase from soybean releases elicitor-active carbohydrates from fungus cell walls. Plant Physiology 71, 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW. 2013. Conditional requirement for exopolysaccharide in the Mesorhizobium–Lotus symbiosis. Molecular Plant-Microbe Interactions 26, 319–329. [DOI] [PubMed] [Google Scholar]
- Klarzynski O, Plesse B, Joubert JM, Yvin JC, Kopp M, Kloareg B, Fritig B. 2000. Linear beta-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiology 124, 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink A, Rovenich H, Shi-Kunne X, et al. 2017. Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Molecular Plant Pathology 18, 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CM. 2008. Cross talk in defense signaling. Plant Physiology 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzai Y, Mochizuki S, Nakajima K, et al. 2014. Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Molecular Plant-Microbe Interactions 27, 975–982. [DOI] [PubMed] [Google Scholar]
- Kutschera A, Dawid C, Gisch N, et al. 2019. Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364, 178–181. [DOI] [PubMed] [Google Scholar]
- Laus MC, van Brussel AA, Kijne JW. 2005. Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Molecular Plant-Microbe Interactions 18, 533–538. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Geller AM, Bamford NC, et al. 2016. Deacetylation of fungal exopolysaccharide mediates adhesion and biofilm formation. mBio 7, e00252–e00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Liu H, Barker BM, et al. 2015. The fungal exopolysaccharide galactosaminogalactan mediates virulence by enhancing resistance to neutrophil extracellular traps. PLoS Pathogens 11, e1005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Sheppard DC. 2016. Recent advances in the understanding of the Aspergillus fumigatus cell wall. Journal of Microbiology 54, 232–242. [DOI] [PubMed] [Google Scholar]
- Lehman AP, Long SR. 2013. Exopolysaccharides from Sinorhizobium meliloti can protect against H2O2-dependent damage. Journal of Bacteriology 195, 5362–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Signer ER, Walker GC. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proceedings of the National Academy of Sciences, USA 82, 6231–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Luo H, Meng J, Sun W, Wang X, Lu S, Peng Y, Zhou L. 2014. Effects of oligosaccharides from endophytic Fusarium oxysporum Dzf17 on activities of defense-related enzymes in Dioscorea zingiberensis suspension cell and seedling cultures. Electronic Journal of Biotechnology 17, 156–161. [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang Ch, Qiu J, Stacey G. 2013. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341, 1384–1387. [DOI] [PubMed] [Google Scholar]
- Liang Y, Tóth K, Cao Y, Tanaka K, Espinoza C, Stacey G. 2014. Lipochitooligosaccharide recognition: an ancient story. New Phytologist 204, 289–296. [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. 2003. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. [DOI] [PubMed] [Google Scholar]
- Limpens E, van Zeijl A, Geurts R. 2015. Lipochitooligosaccharides modulate plant host immunity to enable endosymbioses. Annual Review of Phytopathology 53, 311–334. [DOI] [PubMed] [Google Scholar]
- Lin S, Yang L, Chen G, Li B, Chen D, Li L, Xu Z. 2017. Pathogenic features and characteristics of food borne pathogens biofilm: biomass, viability and matrix. Microbial Pathogenesis 111, 285–291. [DOI] [PubMed] [Google Scholar]
- Liu B, Li JF, Ao Y, et al. 2012. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. The Plant Cell 24, 3406–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Liu Z, Song C, et al. 2012. Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Liu X, Grabherr HM, Willmann R, et al. 2014. Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. eLife 3, e01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Baena FJ, Ruiz-Sainz JE, Rodriguez-Carvajal MA, Vinardell JM. 2016. Bacterial molecular signals in the Sinorhizobium fredii–soybean symbiosis. International Journal of Molecular Sciences 17, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Löhrer M, Hempel M, Mathea S, Schliebner I, Menzel M, Kiesow A, Schaffrath U, Deising HB, Horbach R. 2014. Melanin is not required for turgor generation but enhances cell-wall rigidity in appressoria of the corn pathogen Colletotrichum graminicola. Molecular Plant-Microbe Interactions 27, 315–327. [DOI] [PubMed] [Google Scholar]
- Madala NE, Molinaro A, Dubery IA. 2012. Distinct carbohydrate and lipid-based molecular patterns within lipopolysaccharides from Burkholderia cepacia contribute to defense-associated differential gene expression in Arabidopsis thaliana. Innate Immunity 18, 140–154. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, et al. 2003. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Banerjee D. 2013. Fungal exopolysaccharide: production, composition and applications. Microbiology Insights 6, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F, Fournier J, Mendis HC, Tadege M, Wen J, Ratet P, Mysore KS, Gough C, Jones KM. 2020. Sinorhizobium meliloti succinylated high-molecular-weight succinoglycan and the Medicago truncatula LysM receptor-like kinase MtLYK10 participate independently in symbiotic infection. The Plant Journal 102, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, et al. 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. [DOI] [PubMed] [Google Scholar]
- Margaret I, Crespo-Rivas JC, Acosta-Jurado S, et al. 2012. Sinorhizobium fredii HH103 rkp-3 genes are required for K-antigen polysaccharide biosynthesis, affect lipopolysaccharide structure and are essential for infection of legumes forming determinate nodules. Molecular Plant-Microbe Interactions 25, 825–838. [DOI] [PubMed] [Google Scholar]
- Marshall R, Kombrink A, Motteram J, Loza-Reyes E, Lucas J, Hammond-Kosack KE, Thomma BP, Rudd JJ. 2011. Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiology 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, Lopez-Ribot JL, Oliveira R. 2010. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 169, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, et al. 2016. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nature Microbiology 1, 16043. [DOI] [PubMed] [Google Scholar]
- Mélida H, Sandoval-Sierra JV, Diéguez-Uribeondo J, Bulone V. 2013. Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryotic Cell 12, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida H, Sopeña-Torres S, Bacete L, Garrido-Arandia M, Jordá L, López G, Muñoz-Barrios A, Pacios LF, Molina A. 2018. Non-branched β -1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. The Plant Journal 93, 34–49. [DOI] [PubMed] [Google Scholar]
- Ménard R, Alban S, de Ruffray P, Jamois F, Franz G, Fritig B, Yvin JC, Kauffmann S. 2004. Beta-1,3 glucan sulfate, but not beta-1,3 glucan, induces the salicylic acid signaling pathway in tobacco and Arabidopsis. The Plant Cell 16, 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendis HC, Madzima TF, Queiroux C, Jones KM. 2016. Function of succinoglycan polysaccharide in Sinorhizobium meliloti host plant invasion depends on succinylation, not molecular weight. mBio 7, e00606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlak TA, Kombrink A, Shinya T, et al. 2012. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. The Plant Cell 24, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KJ, Hadley JA, Gustine DL. 1994. Cyclic [beta]-1,6-1,3-glucans of Bradyrhizobium japonicum USDA 110 elicit isoflavonoid production in the soybean (Glycine max) host. Plant Physiology 104, 917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milling A, Babujee L, Allen C. 2011. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS One 6, e15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KF, Zarnowski R, Sanchez H, Edward JA, Reinicke EL, Nett JE, Mitchell AP, Andes DR. 2015. Community participation in biofilm matrix assembly and function. Proceedings of the National Academy of Sciences, USA 112, 4092–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Bhagwat AA, Feger M, Ebel J. 1996. Suppression of fungal β -glucan-induced plant defence in soybean (Glycine max L) by cyclic 1,3-1,6-β -glucans from the symbiont Bradyrhizobium japonicum. Planta 199, 270–275. [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. 2007. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proceedings of the National Academy of Sciences, USA 104, 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloshok TD, Leinhos GME, Staples RC, Hoch HC. 1993. The autogenic extracellular environment of Uromyces appendiculatus urediospore germlings. Mycologia 85, 392–400. [Google Scholar]
- Money PN 2008. Insights on the mechanics of hyphal growth. Fungal Biology Reviews 22, 71–76. [Google Scholar]
- Muszyński A, Heiss C, Hjuler CT, Sullivan JT, Kelly SJ, Thygesen MB, Stougaard J, Azadi P, Carlson RW, Ronson CW. 2016. Structures of exopolysaccharides involved in receptor-mediated perception of Mesorhizobium loti by Lotus japonicus. Journal of Biological Chemistry 291, 20946–20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi I, Kimura K, Suzuki T, Ishikawa M, Banno I, Sakane T, Harada T. 1976. Demonstration of curdlan-type polysaccharide and some other beta-1,3-glucan in microorganisms with aniline blue. Journal of General and Applied Microbiology 22, 1–11. [Google Scholar]
- Nars A, Lafitte C, Chabaud M, et al. 2013. Aphanomyces euteiches cell wall fractions containing novel glucan-chitosaccharides induce defense genes and nuclear calcium oscillations in the plant host Medicago truncatula. PLoS One 8, e75039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann TA 2011. Modification of recombinant maize ChitA chitinase by fungal chitinase-modifying proteins. Molecular Plant Pathology 12, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Daniels MJ, Dow JM. 1995. Lipopolysaccharide from Xanthomonas campestris induces defense-related gene expression in Brassica campestris. Molecular Plant-Microbe Interactions 8, 778–780. [DOI] [PubMed] [Google Scholar]
- Nicholson RL, Moraes WBC. 1980. Survival of Colletotrichum graminicola: importance of the spore matrix. Phytopathology 70, 225–261. [Google Scholar]
- Nizam S, Qiang X, Wawra S, Nostadt R, Getzke F, Schwanke F, Dreyer I, Langen G, Zuccaro A. 2019. Serendipita indica E5'NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonization. EMBO Reports 20, e47430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaka Y, Mimori K, Takeo K, Kitamura S, Takeuchi Y, Yamaoka N, Yoshikawa M. 1995. A structural model for the mechanisms of elicitor release from fungal cell walls by plant beta-1,3-endoglucanase. Plant Physiology 109, 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ökmen B, Kemmerich B, Hilbig D, Wemhöner R, Aschenbroich J, Perrar A, Huesgen PF, Schipper K, Doehlemann G. 2018. Dual function of a secreted fungalysin metalloprotease in Ustilago maydis. New Phytologist 220, 249–261. [DOI] [PubMed] [Google Scholar]
- Olafsdottir ES, Ingólfsdottir K. 2001. Polysaccharides from lichens: structural characteristics and biological activity. Planta Medica 67, 199–208. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews. Microbiology 11, 252–263. [DOI] [PubMed] [Google Scholar]
- Oliveira-Garcia E, Deising HB. 2013. Infection structure-specific expression of β -1,3-glucan synthase is essential for pathogenicity of Colletotrichum graminicola and evasion of β -glucan-triggered immunity in maize. The Plant Cell 25, 2356–2378. [Google Scholar]
- Oliveira-Garcia E, Deising HB. 2016. Attenuation of PAMP-triggered immunity in maize requires down-regulation of the key β -1,6-glucan synthesis genes KRE5 and KRE6 in biotrophic hyphae of Colletotrichum graminicola. The Plant Journal 87, 355–375. [DOI] [PubMed] [Google Scholar]
- Osumi M 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29, 207–233. [DOI] [PubMed] [Google Scholar]
- Pérez-Mendoza D, Rodríguez-Carvajal MÁ, Romero-Jiménez L, Farias Gde A, Lloret J, Gallegos MT, Sanjuán J. 2015. Novel mixed-linkage β -glucan activated by c-di-GMP in Sinorhizobium meliloti. Proceedings of the National Academy of Sciences, USA 112, E757–E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino F, Sasaki I, Turbic A, Wilson SM, Bacic A, Hrmova M, Fincher GB. 2009. Hyphal cell walls from the plant pathogen Rhynchosporium secalis contain (1,3/1,6)-beta-d-glucans, galacto- and rhamnomannans, (1,3;1,4)-beta-d-glucans and chitin. The FEBS Journal 276, 3698–3709. [DOI] [PubMed] [Google Scholar]
- Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. 2010. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. Journal of Biological Chemistry 285, 28902–28911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast I, Lünemann JD. 2014. Fc glycan-modulated immunoglobulin G effector functions. Journal of Clinical Immunology 34 Suppl 1, S51–S55. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, et al. 2003. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. [DOI] [PubMed] [Google Scholar]
- Ranf S 2016. Immune sensing of lipopolysaccharide in plants and animals: same but different. PLoS Pathogens 12, e1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Gisch N, Schäffer M, et al. 2015. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nature Immunology 16, 426–433. [DOI] [PubMed] [Google Scholar]
- Rapicavoli JN, Blanco-Ulate B, Muszyński A, Figueroa-Balderas R, Morales-Cruz A, Azadi P, Dobruchowska JM, Castro C, Cantu D, Roper MC. 2018. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nature Communications 9, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhardt C, Joubert LM, Clemons KV, Stevens DA, Cegelski L. 2019. Integration of electron microscopy and solid-state NMR analysis for new views and compositional parameters of Aspergillus fumigatus biofilms. Medical Mycology 57, S239–S244. [DOI] [PubMed] [Google Scholar]
- Reis RA, Tischer CA, Gorin PA, Iacomini M. 2002. A new pullulan and a branched (1→3)-, (1→6)-linked beta-glucan from the lichenised ascomycete Teloschistes flavicans. FEMS Microbiology Letters 210, 1–5. [DOI] [PubMed] [Google Scholar]
- Rigano LA, Payette C, Brouillard G, et al. 2007. Bacterial cyclic beta-(1,2)-glucan acts in systemic suppression of plant immune responses. The Plant Cell 19, 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudi LV, González JE. 2009. The low-molecular-weight fraction of exopolysaccharide II from Sinorhizobium meliloti is a crucial determinant of biofilm formation. Journal of Bacteriology 191, 7216–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocafort M, Fudal I, Mesarich CH. 2020. Apoplastic effector proteins of plant-associated fungi and oomycetes. Current Opinion in Plant Biology 56, 9–19. [DOI] [PubMed] [Google Scholar]
- Romero-Contreras YJ, Ramírez-Valdespino CA, Guzmán-Guzmán P, Macías-Segoviano JI, Villagómez-Castro JC, Olmedo-Monfil V. 2019. Tal6 from Trichoderma atroviride is a LysM effector involved in mycoparasitism and plant association. Frontiers in Microbiology 10, 2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Ham KS, Darvill AG, Albersheim P. 2002. Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. The Plant Cell 14, 1329–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovenich H, Zuccaro A, Thomma BP. 2016. Convergent evolution of filamentous microbes towards evasion of glycan-triggered immunity. New Phytologist 212, 896–901. [DOI] [PubMed] [Google Scholar]
- Rydahl MG, Krac Un SK, Fangel JU, et al. 2017. Development of novel monoclonal antibodies against starch and ulvan—implications for antibody production against polysaccharides with limited immunogenicity. Scientific Reports 7, 9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S. 2018. Pattern recognition receptors and signaling in plant–microbe interactions. The Plant Journal 93, 592–613. [DOI] [PubMed] [Google Scholar]
- Samalova M, Mélida H, Vilaplana F, Bulone V, Soanes DM, Talbot NJ, Gurr SJ. 2017. The beta-1,3-glucanosyltransferases (Gels) affect the structure of the rice blast fungal cell wall during appressorium-mediated plant infection. Cell Microbiology 19, e12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vallet A, Mesters JR, Thomma BP. 2015. The battle for chitin recognition in plant–microbe interactions. FEMS Microbiology Reviews 39, 171–183. [DOI] [PubMed] [Google Scholar]
- Scarpari M, Reverberi M, Parroni A, et al. 2017. Tramesan, a novel polysaccharide from Trametes versicolor. Structural characterization and biological effects. PLoS One 12, e0171412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz AM, Harrison MJ. 2014. Signaling events during initiation of arbuscular mycorrhizal symbiosis. Journal of Integrative Plant Biology 56, 250–261. [DOI] [PubMed] [Google Scholar]
- Sharp JK, Albersheim P, Ossowski P, Pilotti A, Garegg P, Lindberg B. 1984a Comparison of the structures and elicitor activities of a synthetic and a mycelial-wall-derived hexa(beta-d-glucopyranosyl)-d-glucitol. Journal of Biological Chemistry 259, 11341–11345. [PubMed] [Google Scholar]
- Sharp JK, Valent B, Albersheim P. 1984b Purification and partial characterization of a beta-glucan fragment that elicits phytoalexin accumulation in soybean. Journal of Biological Chemistry 259, 11312–11320. [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, et al. 2010. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. The Plant Journal 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockman GD, Barrett JF. 1983. Structure, function, and assembly of cell walls of gram-positive bacteria. Annual Review of Microbiology 37, 501–527. [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG. 2009. A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiology 151, 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harbor Perspectives in Biology 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A, Erbs G, Shinya T, Dow JM, Parrilli M, Lanzetta R, Shibuya N, Newman MA, Molinaro A. 2010. Glyco-conjugates as elicitors or suppressors of plant innate immunity. Glycobiology 20, 406–419. [DOI] [PubMed] [Google Scholar]
- Silipo A, Molinaro A, Sturiale L, Dow JM, Erbs G, Lanzetta R, Newman MA, Parrilli M. 2005. The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. Journal of Biological Chemistry 280, 33660–33668. [DOI] [PubMed] [Google Scholar]
- Souza CA, Li S, Lin AZ, Boutrot F, Grossmann G, Zipfel C, Somerville SC. 2017. Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiology 173, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spribille T, Tagirdzhanova G, Goyette S, Tuovinen V, Case R, Zandberg WF. 2020. 3D biofilms: in search of the polysaccharides holding together lichen symbioses. FEMS Microbiology Letters 367, fnaa023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Miller JB, Granqvist E, et al. 2015. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. The Plant Cell 27, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathogens 8, e1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara H, Hacquard S, Kombrink A, et al. 2016. Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytologist 211, 1323–1337. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Ochiai M, Horiuchi M, Kumeta H, Ogura K, Ashida M, Inagaki F. 2009. Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition. Proceedings of the National Academy of Sciences, USA 106, 11679–11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira PJP, Colaianni NR, Fitzpatrick CR, Dangl JL. 2019. Beyond pathogens: microbiota interactions with the plant immune system. Current Opinion in Microbiology 49, 7–17. [DOI] [PubMed] [Google Scholar]
- Umemoto N, Kakitani M, Iwamatsu A, Yoshikawa M, Yamaoka N, Ishida I. 1997. The structure and function of a soybean beta-glucan-elicitor-binding protein. Proceedings of the National Academy of Sciences, USA 94, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R, Andersen TG, Marhavý P, Geldner N. 2018. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. The Plant Journal 93, 399–412. [DOI] [PubMed] [Google Scholar]
- Vadassery J, Ranf S, Drzewiecki C, Mithöfer A, Mazars C, Scheel D, Lee J, Oelmüller R. 2009. A cell wall extract from the endophytic fungus Piriformospora indica promotes growth of Arabidopsis seedlings and induces intracellular calcium elevation in roots. The Plant Journal 59, 193–206. [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, de Wit PJ. 2006. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Molecular Plant-Microbe Interactions 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- van Overbeek LS, Saikkonen K. 2016. Impact of bacterial–fungal interactions on the colonization of the endosphere. Trends in Plant Science 21, 230–242. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiology Reviews 32, 149–167. [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. 2008. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. The Plant Cell 20, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Liang Z, Kang BH. 2019. Electron tomography of plant organelles and the outlook for correlative microscopic approaches. New Phytologist 223, 1756–1761. [DOI] [PubMed] [Google Scholar]
- Wanke A, Rovenich H, Schwanke F, Velte S, Becker S, Hehemann JH, Wawra S, Zuccaro A. 2020. Plant species-specific recognition of long and short β -1,3-linked glucans is mediated by different receptor systems. The Plant Journal 102, 1142–1156. [DOI] [PubMed] [Google Scholar]
- Wawra S, Fesel P, Widmer H, et al. 2016. The fungal-specific β -glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nature Communications 7, 13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawra S, Fesel P, Widmer H, Neumann U, Lahrmann U, Becker S, Hehemann JH, Langen G, Zuccaro A. 2019. FGB1 and WSC3 are in planta-induced β -glucan-binding fungal lectins with different functions. New Phytologist 222, 1493–1506. [DOI] [PubMed] [Google Scholar]
- Weishaupt MW, Hahm HS, Geissner A, Seeberger PH. 2017. Automated glycan assembly of branched beta-(1,3)-glucans to identify antibody epitopes. Chemical Communications 53, 3591–3594. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annual Review of Biochemistry 83, 99–128. [DOI] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, et al. 2011. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proceedings of the National Academy of Sciences, USA 108, 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse E, Braet F, Duimel H, et al. 2010. Fixation methods for electron microscopy of human and other liver. World Journal of Gastroenterology 16, 2851–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JEMM, Gysel K, Birkefeldt TG, et al. 2020. Structural signatures in EPR3 define a unique class of plant carbohydrate receptors. Nature Communications 11, 3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY. 2016. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wang J, Zhao J, Xu J, Sun S, Zhang H, Wu J, Tang C, Kang Z, Wang X. 2020. A polysaccharide deacetylase from Puccinia striiformis f. sp. tritici is an important pathogenicity gene that suppresses plant immunity. Plant Biotechnology Journal 18, 1830–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue DX, Li CL, Xie ZP, Staehelin C. 2019. LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. Journal of Experimental Botany 70, 5507–5516. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Yamada A, Hong N, Ogawa T, Ishii T, Shibuya N. 2000. Differences in the recognition of glucan elicitor signals between rice and soybean: beta-glucan fragments from the rice blast disease fungus Pyricularia oryzae that elicit phytoalexin biosynthesis in suspension-cultured rice cells. The Plant Cell 12, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS 1995. A conformational model for cyclic beta-(1→2)-linked glucans based on NMR analysis of the beta-glucans produced by Xanthomonas campestris. Carbohydrate Research 278, 205–225. [DOI] [PubMed] [Google Scholar]
- Yun MH, Torres PS, El Oirdi M, Rigano LA, Gonzalez-Lamothe R, Marano MR, Castagnaro AP, Dankert MA, Bouarab K, Vojnov AA. 2006. Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiology 141, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Chrisman CJ, Castelli MV, Frases S, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2008. Capsule enlargement in Cryptococcus neoformans confers resistance to oxidative stress suggesting a mechanism for intracellular survival. Cellular Microbiology 10, 2043–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J. 2004. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proceedings of the National Academy of Sciences, USA 101, 15811–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeković DB, Kwiatkowski S, Vrvić MM, Jakovljević D, Moran CA. 2005. Natural and modified (1→3)-beta-d-glucans in health promotion and disease alleviation. Critical Reviews in Biotechnology 25, 205–230. [DOI] [PubMed] [Google Scholar]
- Zeng T, Rodriguez-Moreno L, Mansurkhodzaev A, et al. 2020. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytologist 225, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgadzaj R, James EK, Kelly S, Kawaharada Y, de Jonge N, Jensen DB, Madsen LH, Radutoiu S. 2015. A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genetics 11, e1005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wang Y, Wang W, Jia J, Li Y, Wang Q, Wu Y, Tang J. 2009. Generation of monoclonal antibodies against highly conserved antigens. PLoS One 4, e6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Oldroyd GE. 2017. Plant signalling in symbiosis and immunity. Nature 543, 328–336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the content of this study are available within the review and within its supplementary data published online.