Significance
Due to the difficulty in their biochemical characterization, until recently intrinsically disordered proteins have gone largely unannotated. However, an appreciation for their biological significance is emerging, as they are now known to facilitate liquid–liquid phase separation (LLPS), which can produce membrane-lacking organelles that enable the spatial regulation of important cellular processes. This report characterizes the Ligand of Ate1 (Liat1) as an intrinsically disordered protein that participates in LLPS in the nucleolus, a prominent membrane-lacking organelle specialized in ribosome biogenesis. Furthermore, it shows that the lysyl-hydroxylase activity of Jumonji Domain Containing 6 modifies Liat1 and inhibits its nucleolar targeting and potential functions.
Keywords: intrinsically disordered protein, liquid–liquid phase separation, nucleolus, Jmjd6, Liat1
Abstract
The Ligand of Ate1 (Liat1) is a protein of unknown function that was originally discovered through its interaction with arginyl-tRNA protein transferase 1 (Ate1), a component of the Arg/N-degron pathway of protein degradation. Here, we characterized the functional domains of mouse Liat1 and found that its N-terminal half comprises an intrinsically disordered region (IDR) that facilitates its liquid–liquid phase separation (LLPS) in the nucleolus. Using bimolecular fluorescence complementation and immunocytochemistry, we found that Liat1 is targeted to the nucleolus by a low-complexity poly-K region within its IDR. We also found that the lysyl-hydroxylase activity of Jumonji Domain Containing 6 (Jmjd6) modifies Liat1, in a manner that requires the Liat1 poly-K region, and inhibits its nucleolar targeting and potential functions. In sum, this study reveals that Liat1 participates in nucleolar LLPS regulated by Jmjd6.
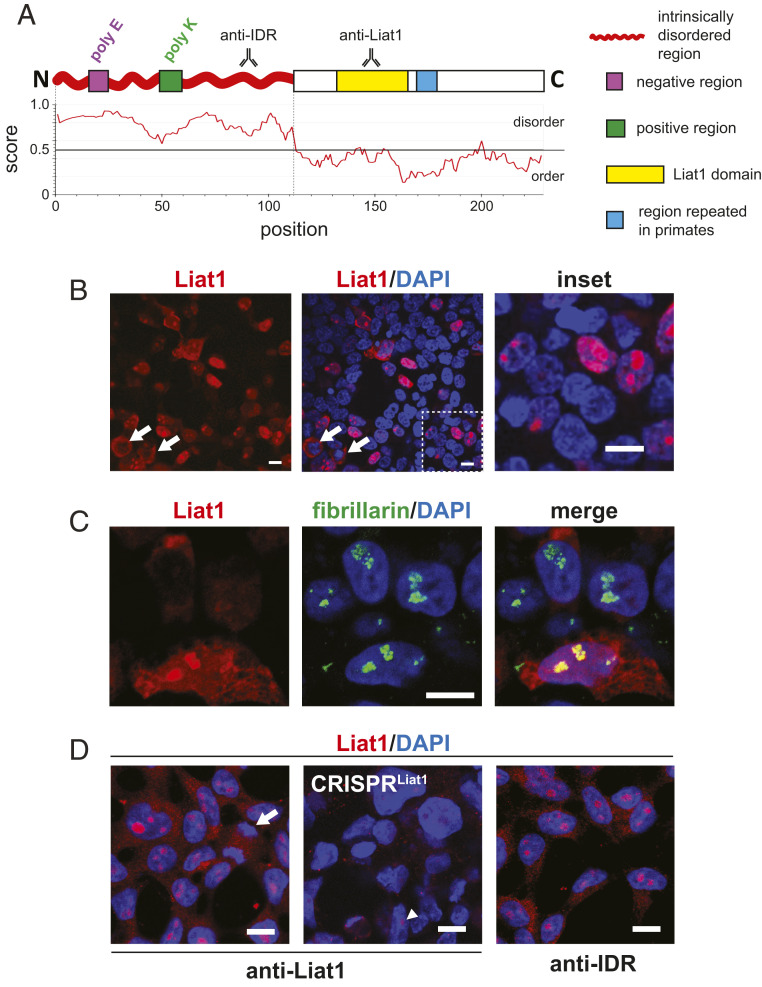
The Ligand of Ate1 (Liat1) was a previously uncharacterized protein that was identified through its interaction with specific isoforms of arginyl-tRNA protein transferase 1 (Ate1), a component of the Arg/N-degron pathway of protein degradation (1). Although the function of Liat1 is still unknown, its amino acid sequence contains several regions of interest that are conserved among metazoans, which provide clues to its function (Fig. 1A). Two regions of low sequence complexity are located within the N-terminal portion of Liat1, a negatively charged polyglutamic acid (poly-E) region and a positively charged polylysine (poly-K) region. The “Liat1 domain,” internally located within the primary sequence, is an ∼30-residue region of particularly high conservation among vertebrates and invertebrate Liat1 species. This domain is required for Ate1 binding (1). Finally, C-terminal to the Liat1 domain is a 10-amino acid motif of unknown function that is tandemly repeated (between 1 and >20 times) in various primate Liat1 species (Fig. 1A).
Fig. 1.
Conserved features of Liat1 and its subcellular localization. (A) Schematic of the mouse Liat1 protein showing evolutionarily conserved features (indicated in Right). Shown are the relative locations of anti-IDR and anti-Liat1 antibody recognition. Also shown is an IUPred2A plot revealing that the N-terminal half of Liat1 comprises an IDR (41). (B) Anti-Liat1 immunocytochemistry of HEK293T cells transfected with plasmids expressing mouse 3xHALiat1. (C) Colocalization of Liat1 with fibrillarin, a marker of the DFC of the nucleolus. (D) Anti-Liat1 immunocytochemistry of nontransfected HEK293T cells (Left). Also probed was a mixed population of cells transfected with CRISPRLIAT1-B, targeting the portion of LIAT1 encoding the repeated motif. The arrowhead indicates Liat1-containing nucleoli that could be seen in some cells (Center). Anti-IDR immunocytochemistry of nontransfected HEK293T cells (Right). White arrows, mitotic cells. (Scale bars: 10 µm.)
Our previous work revealed that Liat1 is multivalent and interacts with a number of structurally and functionally unrelated proteins (1). In addition to Ate1, Liat1 was shown to interact with the Jumonji Domain Containing 6 (Jmjd6) protein, a member of the Jumonji C domain-containing family of Fe+2- and 2-oxoglutarate (2OG)-dependent oxygenases (1, 2). Jmjd6 is a bifunctional arginine demethylase and lysyl-hydroxylase (3–5). Several studies have shown that Jmjd6 plays a role in messenger RNA (mRNA) splicing, particularly through lysine C-5 hydroxylation of the U2 auxiliary factor 65-kDa subunit (U2AF65), a principal splicing regulator that couples RNA splicing with DNA transcription (6, 7). Jmjd6 was also shown to increase productive transcription by facilitating the release of paused RNA polymerase II during the initiation and elongation steps of transcription (8). Jmjd6 is known to enhance tumorigenesis, particularly in breast cancers (9–12), and to promote colon carcinogenesis by negatively regulating the p53 tumor suppressor through hydroxylation (13, 14). In addition to intermolecular hydroxylation, Jmjd6 hydroxylates its own N terminus, resulting in its self-association to form oligomers that are resistant to denaturing polyacrylamide gel electrophoresis (PAGE) (15–19).
Liquid–liquid phase separation (LLPS) has emerged in recent years as a mechanism governing biomolecular condensation. This process is capable of producing membrane-lacking organelles, which enable the temporal and spatial regulation of important cellular processes such as RNA metabolism, genome organization, DNA repair, and transcription regulation (20). For example, the nucleolus contains three immiscible, liquid phases that allow distinct steps of ribosome biogenesis to be carried out efficiently in an “assembly line” fashion starting in the fibrillar center, progressing through the dense fibrillar component (DFC), and concluding in the granular component (GC) (21). Other examples of biomolecular condensates include Cajal bodies, paraspeckles, nuclear speckles, and histone locus bodies in the nucleus (22, 23) as well as stress granules and processing bodies in the cytoplasm (24). Signifying its importance to cell biology, several studies have shown that defects in phase separation contribute to human diseases including cancer and neurodegeneration (25–27). A driving force in LLPS is multivalent protein–protein and protein–RNA interactions involving proteins with intrinsically disordered regions (IDRs). These domains are rich in polar amino acids such as Gly, Ser, Pro, and Gln and charged amino acids such as Glu, Lys, and Arg, and they often contain regions of low sequence complexity (25). While it is clear that IDR-containing proteins play a role in distinguishing between liquid phase states, how cells regulate phase separation in a spatially and temporally appropriate fashion is only beginning to be understood.
Here, we report that the N-terminal half of mouse Liat1 comprises an IDR and participates in biomolecular condensation. Using a combination of bimolecular fluorescence complementation (BiFC), yeast two-hybrid analysis, and site-directed mutagenesis, we report that Liat1 undergoes LLPS and is targeted to the nucleolus by a low-complexity poly-K region within its IDR. We also found that the lysyl-hydroxylase activity of Jmjd6 modifies Liat1, in a manner that requires the poly-K region, and inhibits its nucleolar targeting. This study reveals that Liat1 participates in nucleolar LLPS regulated by Jmjd6.
Results
Liat1 Is Targeted to the Nucleolus.
Previously, we reported that Liat1 is expressed in a broad array of mouse tissues, ranging from high in the spleen and thymus to nondetectable in neuronal tissues (1). Multiple Liat1 isoforms could be detected in cells of human origin (e.g., HEK293T and HeLa) at least in part due to variations in the number of tandem repeats in human Liat1 (1). Despite this assessment, the function of Liat1 remains unknown, and its subcellular localization has not been explored. To determine Liat1 subcellular localization, we transfected HEK293T cells with plasmids encoding the mouse Liat1 cDNA and detected its expression immunocytologically using a polyclonal anti-Liat1 antibody. This antibody was raised against the highly conserved Liat1 domain and is capable of recognizing both recombinant and endogenous mouse and human Liat1 proteins (1). Transfected cells were easily discernable due to their high reactivity to the anti-Liat1 antibody. Whereas Liat1 was detected in the cytoplasm and the nucleoplasm of transfected cells, a significant portion was detected as intense punctate structures within the nucleus (Fig. 1B). Since transfection was carried out with asynchronous cultures, some cells were undergoing mitosis, which could be identified using 4′,6-diamidino-2-phenylindole (DAPI) staining. Liat1 was detected in the cytoplasm of these cells presumably due to breakdown of the nuclear membrane (Fig. 1B, white arrows).
Punctate nuclear staining can originate from a number of molecularly and functionally distinct membrane-lacking organelles, collectively referred to as nuclear bodies. To identify Liat1-containing nuclear structures, we examined its colocalization with fibrillarin, a constituent of the nucleolar DFC (28), and with the Ser/Arg-rich splicing factor SC35 (SC-35), a constituent of nuclear speckles (29). Whereas Liat1 did not colocalize with SC-35, a significant portion of nuclear Liat1 colocalized with fibrillarin (Fig. 1C). To ensure that the nuclear punctate staining was not an artifact of Liat1 overexpression, we examined endogenous Liat1 in nontransfected cells. Similar to exogenously expressed Liat1, endogenous Liat1 was detected in both the cytosol and in punctate nuclear structures with the anti-Liat1 antibody (Fig. 1D). Validating these results, Liat1-containing nuclear structures were largely absent in a mixed population of cells treated with CRISPR-Cas9 targeting the human LIAT1 gene (Fig. 1D). In further corroboration, identical Liat1 immunostaining was obtained using an independently derived antibody (anti-IDR) raised against a conserved portion of the N-terminal IDR of mouse Liat1. As such, there is no overlap between the anti-IDR and the anti-Liat1 antibody-binding site. Collectively, these data indicate that Liat1 can shuttle between the cytoplasm and nucleoplasm and that a significant portion is localized to the nucleolus.
Liat1 Is Detected as a High-Molecular-Weight Protein in the Nucleolus.
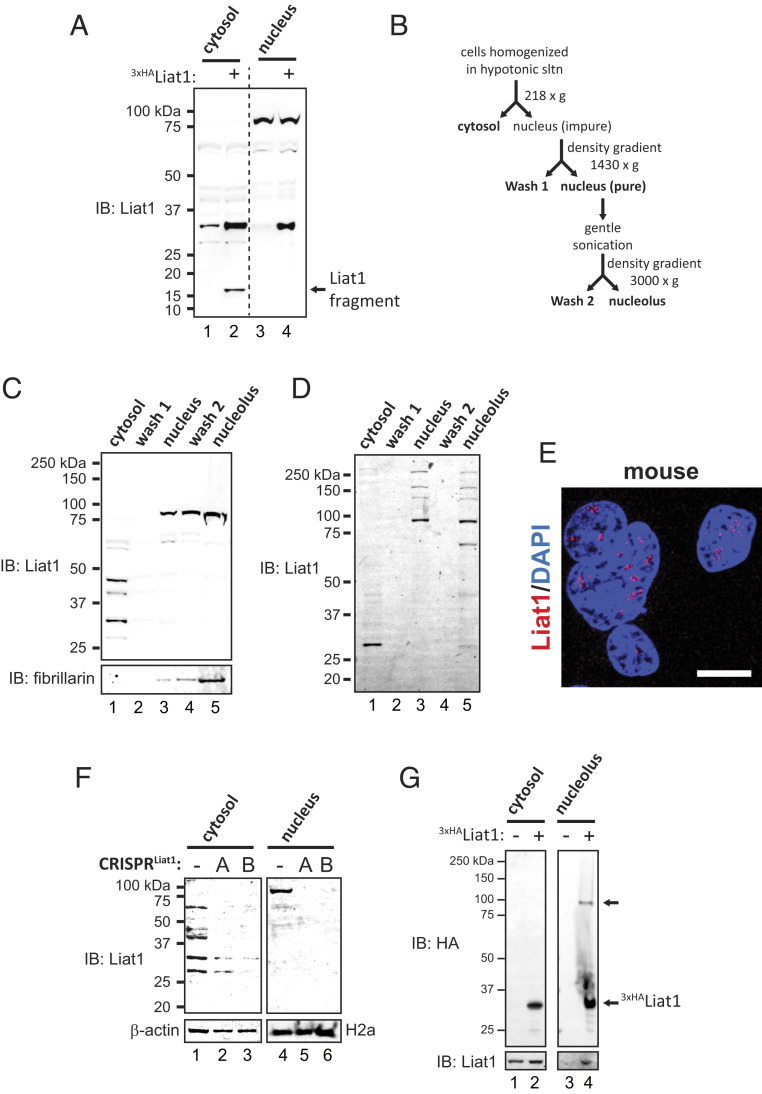
To examine Liat1 subcellular expression biochemically, we carried out immunoblotting using the anti-Liat1 antibody with cytosolic and nuclear fractions of transfected and nontransfected HEK293T cells. Mouse Liat1 bearing three N-terminal antibody epitopes derived from the human influenza hemagglutinin (HA) protein (3xHALiat1) migrates in denaturing PAGE similar to the endogenous human Liat1 with a single repeat. Consistent with immunocytochemistry, 3xHALiat1 was detected in both the cytosol and the nucleus (Fig. 2A). A portion of cytosolic 3xHALiat1 was cleaved suggesting that “excess” Liat1 is processed by an unknown cytosolic protease. Importantly, we detected an ∼80-kDa anti-Liat1 reactive protein in the nuclear fraction of both transfected and nontransfected cells (Fig. 2A). To examine nucleolar Liat1, we used the method of Lam and Lamond (30) to isolate nucleolar fractions (Fig. 2B) (31). Using this method, a number of Liat1 species were detected in the cytosol, but the ∼80-kDa species was restricted to the nucleus and the nucleolus of HEK293T cells (Fig. 2C).
Fig. 2.
Liat1 forms a high-molecular-weight species in the nucleolus. (A) Anti-Liat1 immunoblot of the cytosolic and nuclear fractions showing endogenous Liat1 (lanes 1 and 3) and exogenously expressed 3xHALiat1 (lanes 2 and 4) in HEK293T cells. Note that an ∼16-kDa Liat1 proteolytic fragment was also produced in conditions of excess cytosolic Liat1. (B) Schematic showing procedure for the isolation of nucleoli using the Lam and Lamond (30) methods. (C, Upper) Anti-Liat1 immunoblot of HEK293T cells fractionated by the method shown in B. (C, Lower) Antifibrillarin immunoblot. (D) Same as in C except using mouse NIH 3T3 cells. (E) Anti-Liat1 immunocytochemistry of nontransfected mouse primary embryonic fibroblasts. (Scale bar: 10 µm.) (F) CRISPR-Cas9 knockdown of endogenous Liat1. HEK293T cells were transfected with CRISPRLIAT1-A, targeting the portion of human LIAT1 encoding the Liat1 domain, or CRISPRLIAT1-B, targeting the portion of LIAT1 encoding the repeated motif. Anti-Liat1 immunoblot of cytosolic and nuclear fractions to detect endogenous Liat1 species (Upper). Anti–β-actin and anti-Histone H2a of the cytosolic and nuclear fractions, respectively (Lower). (G) Anti-HA immunoblot of the cytosolic and nucleolar fractions of HEK293T cells expressing 3xHALiat1 (Upper). Anti-Liat1 immunoblot (Lower).IB: immunoblot using the indicated antibodies.
Since primates contain additional Liat1 isoforms not apparent in nonprimates, we also examined fractions from mouse embryonic fibroblasts. In mouse cells, Liat1 was detected as a single species in the cytosol (Fig. 2D, lane 1) and formed several higher-molecular-mass species in nucleolus, the most predominant being ∼80 kDa, which could also be detected in the nucleus (Fig. 2D, lanes 3 and 5). Similar to its immunostaining in human cells (Fig. 1D), endogenous Liat1 was detected as punctate staining resembling nucleoli in mouse cells (Fig. 2E).
Although searches of the human expressed sequence tags (EST) database using the basic local alignment search tool reveal additional Liat1 isoforms due to variations in the number of the repeats, none contain an open reading frame predicted to produce an ∼80-kDa species. Therefore, this protein either is an unrelated polypeptide that cross-reacts with the anti-Liat1 antibody or results from Liat1 posttranslational modifications that are resistant to the reducing conditions. To determine if it is encoded by the human LIAT1 gene (C17orf97), we treated cells with CRISPR-Cas9 independently targeting to two separate regions of the endogenous LIAT1 gene in hopes of ablating its protein product. CRISPRLIAT1-A targets the region of LIAT1 encoding the highly conserved Liat1 domain common to all known Liat1 isoforms. CRISPRLIAT1-B targets the region encoding the repeat region. Notably, CRISPRLIAT1-B targets C17orf97 in multiple locations (e.g., locus NM_001013672 would be targeted in 12 locations) and likely has a higher efficiency of LIAT1 ablation. Following CRISPR-Cas9 treatment, cytosolic anti-Liat1 reactive bands were reduced to ∼10% and nearly undetectable in mixed cell populations selected for CRISPRLIAT1-A or CRISPRLIAT1-B, respectively (Fig. 2F, compare lanes 1 through 3), and the nuclear ∼80-kDa species was reduced to undetectable levels with either CRISPRLIAT1-A or CRISPRLIAT1-B (Fig. 2F, compare lanes 4 through 6). This indicates that the ∼80-kDa anti-Liat1 reactive band is indeed encoded by the LIAT1 gene.
To determine biochemically if exogenously expressed Liat1 is targeted to the nucleolus and could assemble into an ∼80-kDa species, we examined 3xHALiat1 expression in the cytosol and nucleolar fractions of transfected cells using an anti-HA antibody. Consistently, 3xHALiat1 was detected as an ∼32-kDa species in both the cytosol and the nucleolus (Fig. 2G, lanes 2 and 4). Remarkably, an anti-HA reactive ∼80-kDa species was also detected in the nucleolus (Fig. 2F, lane 4). Collectively, these data indicate that Liat1 undergoes posttranslational modification in the nucleolus (and possibly in the nucleus) to form an ∼80-kDa species that is resistant to denaturing conditions.
Liat1 Self-Associates.
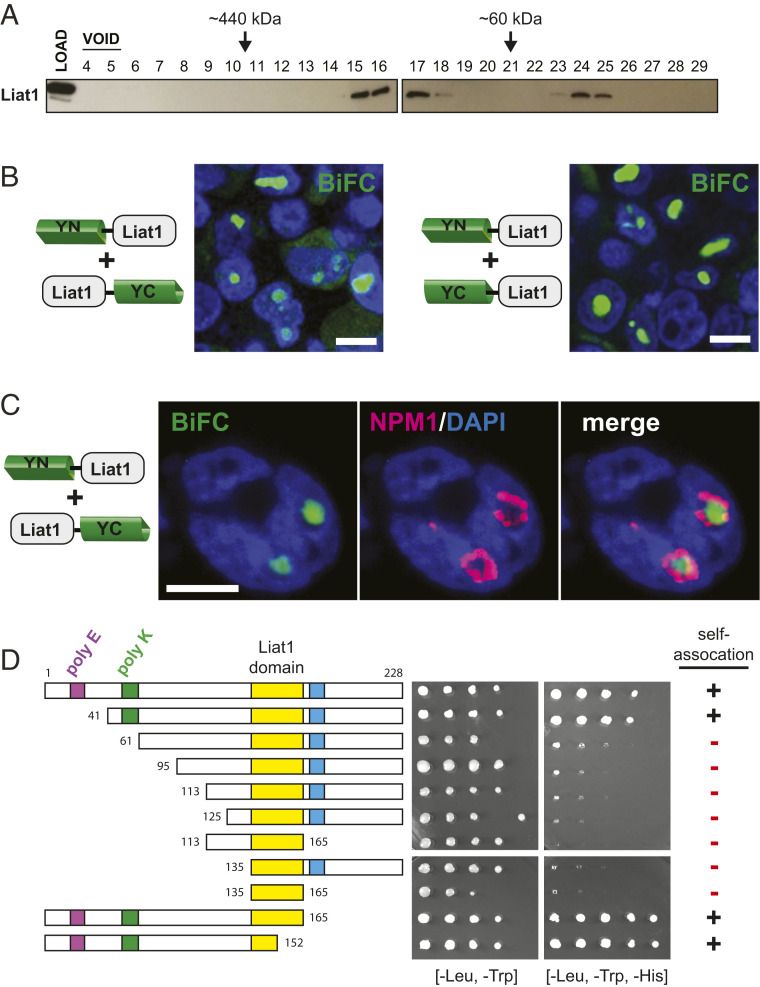
Size exclusion–fast protein liquid chromatography fractionation of a mouse testis extract in physiological (nondenaturing) conditions revealed that endogenous Liat1 partitioned into two distinct fractions: one <60 kDa and another between 60 and 440 kDa (Fig. 3A). A common signature of proteins that participate in nuclear bodies is their ability to self-associate (32–36). Therefore, to determine if Liat1 self-associates, we used BiFC, which is based on the reconstitution of fluorescence when two complementary, nonfluorescent fragments of the yellow fluorescent protein (YFP) are joined through a pair of interacting proteins (37, 38). For this, we generated four fusion constructs: 1) YN-Liat1, YFP amino acids 1 to 154 fused to the N terminus of Liat1; 2) YC-Liat1, YFP amino acids 154 to 239 fused to the N terminus of Liat1; 3) Liat1-YN, YFP amino acids 1 to 154 fused to the C terminus of Liat1; and 4) Liat1-YC, YFP amino acids 154 to 239 fused to the C terminus of Liat1. BiFC was carried out in HEK293T cells by transfecting each construct alone or in combination (SI Appendix). Fluorescence complementation by coexpression of YN-Liat1 with Liat1-YC or YN-Liat1 with YC-Liat1 was detected as diffuse fluorescence in the cytosol and large punctate structures in the nucleus (Fig. 3B). This indicates that Liat1 self-associates, which may at least partially explain distinct Liat1 fractions in size exclusion chromatography.
Fig. 3.
Liat1 self-associates and is targeted to the nucleolus. (A) Anti-Liat1 immunoblot of mouse testis cytosol fractionated by size exclusion chromatography. (B) BiFC between YN-Liat1 and Liat1-YC (Left) and between YN-Liat1 and YC-Liat1 (Right). YN indicates the N-terminal half of YFP; YC indicates the C-terminal half of YFP. (Scale bars: 10 µm.) (C) Colocalization of NPM1 and BiFC of YN-Liat1 and Liat1-YC. (Scale bar: 10 µm.) (D) A yeast two-hybrid assay was performed in S. cerevisiae by cotransforming Liat1-DBD and full-length Liat1-AD or the indicated Liat1 truncations fused to the AD. Interactions were detected as growth on SC medium lacking Leu, Trp, and His.
To determine if the punctate Liat1–BiFC nuclear structures colocalize with the nucleolus, we coupled BiFC with immunostaining to Nucleophosmin (NPM1), a constituent of the nucleolar GC. Interestingly, the Liat1 BiFC signal was enclosed within NPM1, suggesting that Liat1 is a component of a more interior nucleolar compartment (Fig. 3C), consistent with its colocalization with fibrillarin (Fig. 1C). Alternatively, it is possible that the expression of BiFC fusions and/or experimental conditions of BiFC induce nucleolar stress, resulting in the appearance of nucleolar NPM1 “necklaces.” Such structures have been detected during conditions that allow ribosomal DNA transcription but impair ribosomal RNA processing (39, 40). In either case, these results indicate that Liat1 self-associates and participates in LLPS at the nucleolus.
To determine the regions of mouse Liat1 required for its self-association, we carried out a yeast two-hybrid analysis of full-length Ligand of Ate1 fused to the Gal4 DNA-binding domain (Liat1-DBD) and truncated forms of Ligand of Ate1 fused to the Gal4 activation domain (Liat1-AD). Consistent with our BiFC studies, full-length Liat1-AD was able to associate with full-length Liat1-DBD such that it could activate grow on histidine (HIS)-lacking media (Fig. 3D). N-terminal truncations resulting in the loss of the poly-E region (41 to 228) and C-terminal truncations resulting in the loss of the repeated motif or a portion of the Liat1 domain (1 to 165 and 1 to 152) maintained self-association with Liat1-DBD, indicating that these regions are not required for Liat1 self-association (Fig. 3D). In contrast, N-terminal truncations that removed the poly-K region resulted in a loss of Liat1 self-association as shown by a lack of growth on HIS-lacking media. These results indicate that the Liat1 poly-K region, between amino acids 41 and 61, is required for its self-association.
The N-Terminal Half of Liat1 Is an IDR That Participates in LLPS.
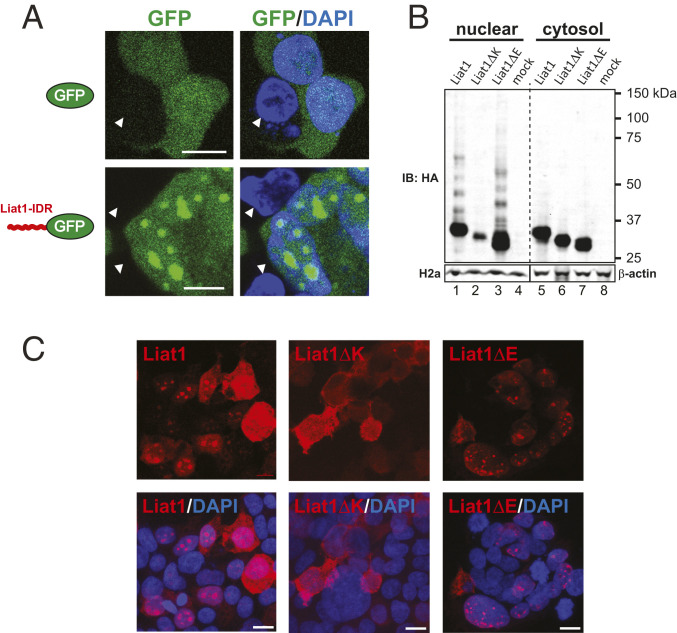
Membrane-lacking organelles such as the nucleolus are formed through biomolecular condensation facilitated by LLPS of its constituents. A driving force in LLPS is multivalent protein–protein and protein–RNA interactions involving proteins with IDRs. Since we found that Liat1 self-associates extensively at the nucleolus, we used the intrinsically unstructured protein predictor, IUPred2A (https://iupred2a.elte.hu/) to ask if Liat1 contains an IDR (41). IUPred2A predicted that amino acids 1 to 112 of mouse Liat1 comprise an IDR, whereas the C-terminal half is structured (Fig. 1A). To determine if the Liat1 IDR can drive LLPS, we fused it to the enhanced green fluorescence protein (eGFP; IDR-GFP) and compared its subcellular localization in cells with that of eGFP. In contrast to the diffuse GFP signal detected throughout the cytosol and the nucleus of transfected cells, a significant amount of IDR-GFP was detected in the nucleus as large punctate structures resembling nucleoli (Fig. 4A). This indicates that the disordered N-terminal half of Liat1 participates in LLPS.
Fig. 4.
The IDR of Liat1 facilitates LLPS and contains a poly-K nucleolar targeting signal. (A) Direct fluorescence of eGFP (GFP; Upper) and of eGFP bearing an N-terminal fusion of the Liat1 IDR (Lower). Arrowheads indicate nontranfected cells. (Scale bars: 10 µm.) (B) Anti-HA immunoblot of the nuclear and cytosolic fractions of HEK293T cells expressing wild-type 3xHALiat1 or Liat1 lacking the poly-E region (Liat1ΔE) or the poly-K region (Liat1ΔK; Upper). Anti-Histone H2a and anti–β-actin of the nuclear and cytosolic fractions, respectively (Lower). IB: immunoblot using the indicated antibody. (C) Anti-Liat1 immunocytochemistry of cells expressing wild-type Liat1, Liat1ΔK, or Liat1ΔE. (Scale bars: 15 µm.)
IDRs are often rich in polar amino acids such as Gly as well as charged amino acids such as Glu, Lys, and Arg (25). Whereas the amino acid composition of the C-terminal half of mouse Liat1 largely reflects the amino acid composition of proteins across different taxa (42), the amino acid composition of N-terminal disordered half of mouse Liat1 contained approximately twice the content of Gly as well as Glu and Lys contributed by the low-complexity poly-E and poly-K regions, respectively. Since lysine has been shown to drive biomolecular condensation (43), we sought to determine if the low-complexity regions within the Liat1 IDR play a role in its nucleolar targeting by comparing the subcellular localization of 3xHALiat1 (Liat1) with Liat1 lacking the poly-E region (Liat1ΔE) or the poly-K region (Liat1ΔK). Biochemical fractionation of HEK293T cells expressing 3xHALiat1 was consistent with immunocytochemistry data such that Liat1 was distributed similarly between the nuclear and cytosolic fraction (Fig. 4B, lanes 1 and 5). Liat1ΔE was also localized in the cytosol and slightly enriched in the nuclear fraction (Fig. 4B, lanes 3 and 7). In contrast, Liat1ΔK was largely restricted to the cytosol (Fig. 4B, lanes 2 and 6). Interestingly, both Liat1 and Liat1ΔE formed higher-molecular-weight, SDS/PAGE-resistant species in the nuclear fraction. In contrast, higher-molecular-weight species of Liat1ΔK were not detected (Fig. 4B). We also examined the subcellular localization of these proteins using immunocytochemistry. Consistent with the biochemical fractionation, wild-type Liat1 and Liat1ΔE were detected in the cytoplasm, nucleoplasm, and in the nucleolus (Fig. 4C). In contrast, Liat1ΔK was largely restricted to the cytoplasm, and no punctate nuclear structures were detected (Fig. 4C). Together, these data indicate that the low-complexity poly-K region within the N-terminal IDR of Liat1 forms a nucleolar targeting signal that is required for its import into the nucleus and nucleolar targeting.
Liat1 Interacts with Jmjd6, a Lysyl-Hydroxylase.
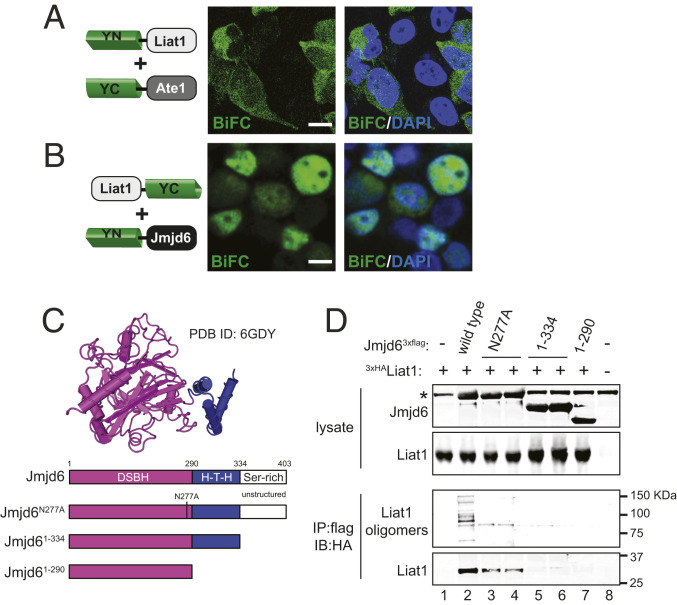
Previously, Liat1 was shown to interact with Jmjd6 (1, 2). To determine the subcellular location of this interaction, we carried out BiFC between Liat1 and Jmjd6 and compared it with BiFC between Liat1 and another interacting partner, ATE11A7A (1). In contrast to BiFC reconstituted in the cytoplasm by YN-Liat1 and YC-Ate1 (Fig. 5A), BiFC between Liat1-YC and YN-Jmjd6 was detected in the cytoplasm and especially in the nucleus (Fig. 5B). Interestingly, whereas BiFC from Liat1 self-association produced intense punctate nuclear structures corresponding to the nucleolus (Fig. 3C), BiFC between Liat1 and Jmjd6 was detected in the entire nucleus and contained circular voids indicative of the absence of BiFC in the nucleolus (Fig. 5B). These coincided with voids in DAPI staining that are due to the nucleolus, which has an approximately threefold lower DNA content relative to the surrounding nucleoplasm (44).
Fig. 5.
Liat1 interacts with Jmjd6. (A) BiFC between YN-Liat1 and YC-Ate1. (B) BiFC between Liat1-YC and YN-Jmjd6. Right contains DAPI. (Scale bars: 10 µm.) (C, Upper) Structure of Jmjd6 residues 1 to 334 (Protein Data Bank [PDB] ID code 6GDY). Not shown is the unstructured C-terminal serine-rich domain. (C, Lower) Schematic of wild-type Jmjd6 and its mutants used in this study. DSBH, double-stranded β-helix fold; H-T-H, helix–turn–helix. (D) Anti-flag coimmunoprecipitation of 3xHALiat1 with wild-type Jmjd63xflag or its indicated mutants. Note that higher-molecular-weight Liat1 species are detected in the presence of wild-type Liat1 but not with Jmjd6N277A despite its ability to interact with Liat1. *Antibody heavy chain. IP: immunoprecipitation using the indicated antibody. IB: immunoblot using the indicated antibody.
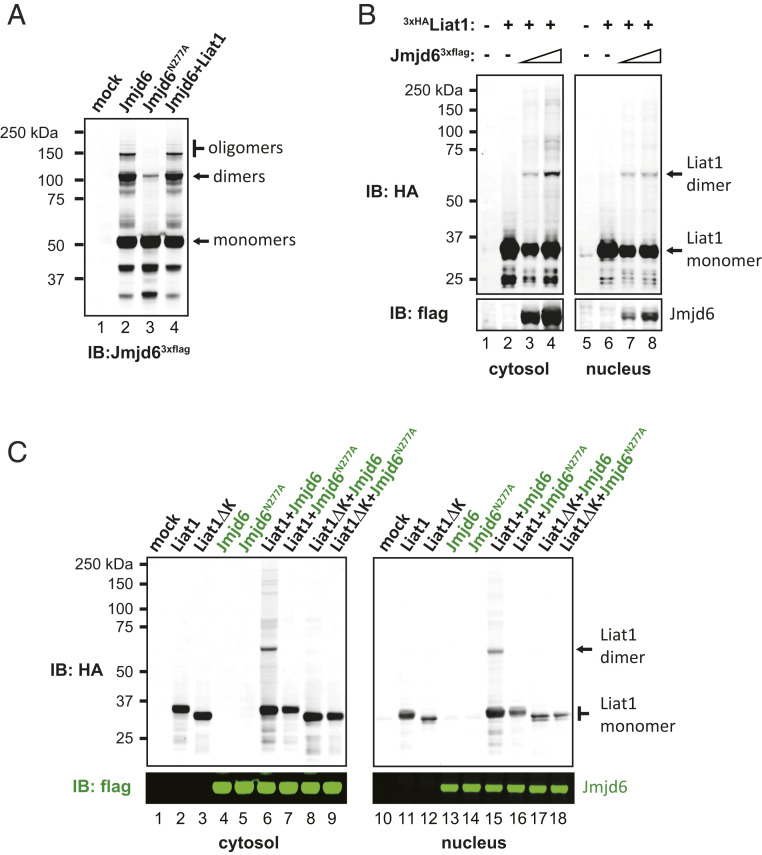
Jmjd6 consists of three domains. The N-terminal 290 amino acids form a large domain containing a typical cupin or double-stranded β-helix fold, characteristic of 2OG-dependent oxygenases. It is connected to a helix–turn–helix-like motif (residues 291 to 334) through an inflexible linker thought to restrict movement of the two domains (4, 14, 15). The C-terminal domain (residues 335 to 403) is serine rich and unstructured but important for the oligomerization and localization of Jmjd6 (4, 15, 45, 46) (Fig. 5C). To determine which domain is important for its interaction with Liat1, we created Jmjd6 truncations lacking the C terminus (Jmjd61-334) or lacking both the C terminus and helix–turn–helix-like motif (Jmjd61-290) (Fig. 5C). In contrast to wild-type Jmjd6, Liat1 was not coimmunoprecipitated by either Jmjd61-334 or Jmjd61-290 (Fig. 5D, compare lane 2 with lanes 5 to 7). We also asked if the hydroxylase activity of Jmjd6 was required for its interaction with Liat1. Previous studies have shown that substitution of metal-binding residues does not always block hydroxylase activity (14). Therefore, we mutated Asn277, a critical 2OG-binding residue (4), to Ala (Jmjd6N277A) in efforts to abrogate hydroxylase activity without disrupting Jmjd6 structure (Fig. 5C). Similar to wild-type Jmjd6, Liat1 was able to interact with Jmjd6N277A (Fig. 5D, lanes 3 and 4), suggesting that structure needed for Liat1 binding was not disrupted by this mutation. That Jmjd6N277A was unable to form oligomers resistant to denaturing PAGE indicated that it lacked lysyl-hydroxylase activity (Fig. 6A, compare lanes 2 and 3). Together, these results indicate that the disordered C terminus of Jmjd6 is required for its interaction with Liat1 and that its lysyl-hydroxylase activity is dispensable. Interestingly, we detected several anti-HA reactive Liat1 oligomers that also coimmunoprecipitated with wild-type Jmjd6, but not with Jmjd6N277A (Fig. 5D, compare lane 2 with lanes 3 and 4). This indicates that Liat1 is posttranslationally modified by Jmjd6 lysyl-hydroxylase activity.
Fig. 6.
Liat1 is modified by Jmjd6 hydroxylase activity. (A) Anti-Flag immunoblot of Jmjd63xflag or Jmjd6N277A in the presence or absence of 3xHALiat1. (B) Anti-HA immunoblot of 3xHALiat1 in the cytosol and nuclear fractions in the absence or presence of increasing amounts of Jmjd63xflag (Upper). Anti-flag immunoblots indicated relative expression of Jmjd63xflag (Lower). (C) Anti-HA immunoblot of 3xHALiat1 and 3xHALiat1ΔK in the cytosol and nuclear fractions in the presence of wild-type Jmjd6 or Jmjd6N277A (Upper). Anti-flag immunoblots indicated relative expression of Jmjd63xflag (Lower). IB: immunoblot using the indicated antibodies.
Liat1 Is Modified by Jmjd6 Lysyl-Hydroxylase Activity.
To explore Liat1 modifications by Jmjd6 in cells, we examined 3xHALiat1 expression alone or in the presence of Jmjd63xflag by anti-HA immunoblotting of cytosolic and nuclear fractions. In the absence of Jmjd63xflag, 3xHALiat1 was detected as a single protein of the expected size in the cytosol and in the nucleus using an anti-HA antibody. In the presence of increasing levels of Jmjd63xflag, higher-molecular-mass Liat1 species were increasingly detected, most notably ∼60 kDa, the size of an Liat1 dimer (Fig. 6B). To determine if Jmjd6 lysyl-hydroxylase activity is necessary for Liat1 dimer formation and if the poly-K region of Liat1 is required, we examined 3xHALiat1 and Liat1ΔK expression in the presence and absence of Jmjd63xflag and Jmjd6N277A. Consistently, Liat1 modifications, particularly Liat1 dimers, were detected in the cytosolic and nuclear fractions in the presence of wild-type Jmjd6 (Fig. 6C, lanes 6 and 15). Remarkably, modifications were not detected in the presence of Jmjd6N277A despite its ability to interact with Liat1 (Fig. 6C, compare lane 6 with lane 7 and compare lane 15 with lane 16). Furthermore, modifications of Liat1ΔK were absent even in the presence of wild-type Jmjd6 (Fig. 6C, lanes 8 and 17). Collectively, these data indicate that Liat1 is modified by Jmjd6 lysyl-hydroxylase activity and forms Jmjd6-induced dimers in a manner that requires the poly-K region within the Liat1 IDR.
Jmjd6 Inhibits Liat1 Targeting to the Nucleolus.
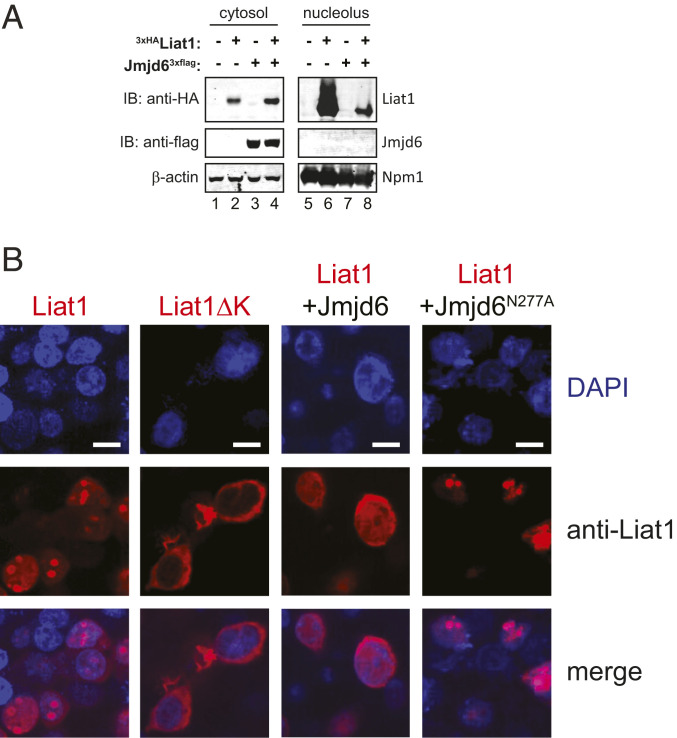
To determine if Jmjd6 influences Liat1 nucleolar targeting, we expressed 3xHALiat1 in the presence and absence of Jmjd63xflag and examined their levels in cytosolic and nucleolar fractions. In the absence of exogenously added Jmjd63xflag, 3xHALiat1 was detected in the cytosol and to a large degree, in the nucleolus (Fig. 7A, lanes 2 and 6). In the presence of exogenously added Jmjd63xflag, 3xHALiat1 levels were dramatically reduced in the nucleolar fraction (Fig. 7A, compare lanes 6 and 8).
Fig. 7.
Jmjd6 lysyl-hydroxylase activity inhibits the nucleolar localization of Liat1. (A) Anti-HA and anti-Flag immunoblots of the cytosol and nucleolar fractions isolated from cells expressing the indicated proteins (Top and Middle). Anti–β-actin and anti-NPM1 to indicate cytosol and nucleolar fractions, respectively (Bottom). IB: immunoblot using the indicated antibodies. (B) Anti-Liat1 immunocytochemistry showing subcellular localization of wild-type Liat1 or Liat1ΔK in the presence or absence of wild-type Jmjd6 or Jmjd6N277A. (Scale bars: 10 µm.)
To examine Jmjd6 effects on Liat1 nucleolar targeting using immunocytochemistry, we compared subcellular location of 3xHALiat1 and Liat1ΔK and in the presence of Jmjd63xflag and Jmjd6N277A. Similar to data in Fig. 4C, Liat1 was detected in the nucleolus, whereas Liat1ΔK was excluded from the nucleus and nucleolus (Fig. 7B). In the presence of Jmjd6, Liat1 was detected in the cytoplasm and nucleoplasm but was absent from the nucleolus (Fig. 7B). In contrast, Jmjd6N277A was unable to inhibit the nucleolar targeting of Liat1 (Fig. 7B). These data indicate that the lysyl-hydroxylase activity of Jmjd6 inhibits the LLPS of Liat1 at the nucleolus.
Discussion
In this study, we found that the N-terminal half of Liat1 comprises an IDR that facilitates LLPS. We also found that the Liat1 IDR harbors a low-complexity poly-K region that functions as a nucleolar targeting signal and is regulated by Jmjd6 lysyl-hydroxylase activity. Similar to other IDR-containing proteins, Liat1 self-associates to form oligomers and higher-molecular-weight species that are resistant to denaturing conditions. Of note, Liat1 is detected as an ∼80-kDa denaturing PAGE-resistant species in the nucleolus and in the nucleoplasm. A number of independent lines of evidence indicate that this species is a product of the LIAT1 gene. First, this species was detected in both human cells and in mouse cells using the same antibody (Fig. 2 C and D). Second, the ∼80-kDa species was not detected in cells treated with either of two independent CRISPR-Cas9 constructs targeting the LIAT1 gene (Fig. 2F). Third, the ∼80-kDa species was detected using an anti-HA antibody in nucleolar fractions of cells expressing 3xHALiat1 (Fig. 2G). That 3xHALiat1 generates the ∼80-kDa band indicates that it is a posttranslationally modified species rather than an alternatively spliced product. This species appears to form in the nucleolus but is also detected in the nucleoplasm, consistent with the dynamic nature of the nucleolus and its highly mobile constituents. More work is needed to understand how this species is formed as well as its function.
Our previous work revealed that Liat1 is a multivalent protein that interacts with a variety of structurally and seemingly functionally unrelated proteins including Jmjd6 (1). Here, we show that Liat1 is posttranslationally modified by Jmjd6 in a manner that requires its lysyl-hydroxylase activity (Fig. 6). Interestingly, the poly-K region of Liat1, required for its nucleolar targeting, is necessary for this modification. This suggests that Jmjd6 hydroxylates one or more of the conserved lysine residues clustered within the poly-K region. Studies are underway to examine Liat1 hydroxylation in vitro. Although the Jmjd6-dependent modification of Liat1 appears as a smear extending to ∼250 kDa in denaturing PAGE, the predominant Jmjd6-modified Liat1 species is an ∼60-kDa band, consistent with an Liat1 dimer (Fig. 6 B and C). Several studies have shown that Jmjd6 hydroxylates its own N terminus, causing its self-association and oligomerization (15–19). In this study, we also observed the formation of dimers and oligomers with wild-type Jmjd6 but not with Jmjd6N277A, which lacks hydroxylase activity (Fig. 6A). Therefore, a similar mechanism may account for Jmjd6-induced oligomerization of Liat1. That Liat1ΔK was unable to dimerize, even in the presence of excess Jmjd6 (Fig. 6C), is consistent with our yeast two-hybrid data indicating that the poly-K region is required for Liat1 self-association (Fig. 3D).
Our data indicate that interactions between Liat1 and specific binding partners occur in distinct subcellular locations. For example, in BiFC experiments, the Liat1–Ate1 interaction was restricted to the cytoplasm, whereas the Liat1–Jmjd6 interaction occurred in the cytoplasm as well as in the nucleoplasm. In the absence of Jmjd6, Liat1 is targeted to the nucleolus. Nucleolar targeting of Liat1 was shown using two independently derived antibodies with nonoverlapping Liat1 epitopes. Three independent lines of evidence suggest that the lysyl-hydroxylase activity of Jmjd6 regulates Liat1 nucleolar targeting. First, BiFC between Liat1 and Jmjd6 was robust in the nucleus but appeared to be restricted from the nucleolus (Fig. 5B). Second, anti-HA immunoblot analysis of normalized cytosolic and nucleolar fractions revealed a significant reduction in nucleolar 3xHALiat1 in the presence of Jmjd63xflag (Fig. 7A). Third, immunocytochemistry revealed that wild-type Jmjd6, but not the hydroxylase-deficient mutant Jmjd6N277A, inhibited the nucleolar targeting of 3xHALiat1 (Fig. 7B). Although it is possible that Jmjd6 binding simply prevents Liat1 entry into the nucleolus, this model is unlikely since Jmjd6N277A is unable to inhibit Liat1 nucleolar targeting (Fig. 7B) despite its ability to interact with Liat1 (Fig. 5D). A more likely model is that Jmjd6-dependent hydroxylation of the Liat1 poly-K region prevents its targeting and function in the nucleolus. Since LiatΔK was largely restricted to the cytoplasm (Figs. 4C and 7B), modifications to the poly-K region may also result in increased cytosolic Liat1 levels. Our data show that the Liat1–Ate1 interaction is restricted to the cytosol (Fig. 5A) and that when overexpressed, a portion of Liat1 undergoes limited proteolytic digestion in the cytoplasm (Fig. 2A). As such, one possibility is that Ate1 facilitates the Arg/N-degron–mediated degradation of Liat1 fragments generated during conditions of excess cytosolic Liat1 or perhaps during mitosis. This remains to be tested.
It is unclear how (or if) Jmjd6-induced dimer formation inhibits nucleolar targeting, especially in light of our BiFC data revealing that Liat1 self-association is linked to nucleolar targeting (Fig. 3C). Depending upon cellular conditions and available binding partners, IDR-containing proteins such as Liat1 may adopt distinct conformations to facilitate different activities (i.e., Jmjd6-induced Liat1 dimerization may have a distinct conformation from those formed in its absence). Self-interactions of the nucleolar protein NPM1 were shown to control its valency and switching between different LLPS mechanisms to influence ribosome biogenesis (36). A similar mechanism may exist for Liat1 within the crowded protein and nucleic acid milieu of the nucleolus. Lastly, dimers were only a fraction of detected Jmjd6-induced Liat1 modifications. As such, they may represent an early or intermediate product in Liat1 modification (e.g., oligomerization). Further work is needed to understand Jmjd6-dependent modifications of Liat1 and their biological significance.
Although Liat1 does not influence the oligomerization of Jmjd6 (Fig. 6A), it may influence Jmjd6 function. Jmjd6 functions include the regulation of alternative mRNA splicing, particularly through lysine hydroxylation of U2AF65 (6, 7). In addition, Jmjd6 functions in DNA transcription through its recruitment to antipause enhancers where it demethylates H4R3me2s and the 7SK small nuclear RNA. This releases the positive transcription elongation factor b (P-TEFb) from the 7SK small nuclear ribonucleoprotein (snRNP) complex and stimulates transcription elongation (8). Suggesting its participation, Liat1 was previously shown to interact with La ribonucleoprotein 7, a component of the 7SK snRNP complex (1). Studies to examine the role of Liat1 in mRNA splicing and DNA transcription are underway.
While this study firmly establishes Liat1 targeting to the nucleolus, it does not establish a role for Liat1 in ribosome biogenesis. Liat1 lacks canonical DNA- or RNA-binding motifs. However, it was previously shown to interact with the ribosomal small subunit proteins S14 and S19, suggesting a direct role in ribosome biogenesis (1). Alternatively, since Liat1 is multivalent, disordered, and participates in LLPS, it may play a scaffolding role in nucleolar organization, particularly during stress. Nucleoli undergo reorganization during cellular stress conditions including DNA damage, temperature change, reactive oxygen species, osmotic stress, nutrient stress, or viral infection (39, 47). They also act as a “detention center” to sequester and inhibit specific enzymes during stress (48). For example, the nucleolus regulates p53 export and degradation and can suppress p53 activation in response to oncogenic stress (40, 49, 50). Here, we show that Liat1 participates in nucleolar LLPS in a manner controlled by Jmjd6 lysyl-hydroxylase activity. Since Jmjd6 requires Fe+2, 2OG, and O2 as cofactors (51), we predict that Liat1 nucleolar targeting is regulated in an O2-dependent manner. Such a mechanism would serve as an oxygen sensor allowing cells to tailor nucleolar reorganization and/or function according to available oxygen levels.
Materials and Methods
Cell Culture.
Cells were cultured at 37 °C in Dulbecco’s modified Eagle media (Fisher Scientific), which was supplemented with 10% fetal bovine serum (Gemini BioProducts), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Gemini BioProducts) in the presence of 5% CO2. Cells were transfected using BioT transfection reagent (BioLand Scientific) according to the manufacturer’s instructions. For isolation of nucleoli from transfected cells, cells were transfected using the calcium phosphate method (52). Briefly, plasmids were mixed with CaCl2 (250 mM final concentration) and added into 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) buffered saline to form calcium phosphate–DNA coprecipitates.
Antibody to the Liat1 IDR.
Rabbits were immunized with the peptide KNQPLSSSFHDILNPC, corresponding to mouse Liat1 sequence, to generate antisera using standard methods (Abgent). Total IgG was isolated by affinity chromatography using GammaBind G-Sepharose (GE Healthcare). Pooled IgG fractions were dialyzed against phosphate buffered saline (PBS; 0.15 M NaCl, 50 mM potassium phosphate [pH 7.5]) containing 10% glycerol and then incubated with Affi-Gel-10 beads conjugated to purified mouse Liat1 for 2 h at 4 °C. After washing in PBS, anti-IDR IgG was eluted with 0.2 M glycine (pH 2.8) into tubes containing one-third of elution volume of 1 M K2HPO4 (pH 8.5). The resulting fractions were pooled, dialyzed against PBS (0.15 M NaCl, 50 mM potassium phosphate [pH 7.5]) containing 10% glycerol, and stored at −20 °C.
Immunocytochemistry.
Cells were grown on chamber slides, fixed with 4% formaldehyde for 15 min at room temperature (RT), and permeabilized with 0.5% Triton X-100 for 15 min. Fixed cells were then blocked in 10% goat serum (Sigma-Aldrich) for 1 h. Cells were then incubated with primary antibody for 3 h at 37 °C followed by washing three times in PBS containing 0.1% Tween for 5 min each. Cells were then incubated with secondary antibodies diluted 1:500 in 5% goat serum for 1 h at RT and washed three times in PBS containing 0.1% Tween for 5 min each. Slides were washed once in PBS and mounted in Vectashield containing DAPI (Vector Laboratories). Primary antibodies used for immunocytochemistry were C17ORF97 (HPA023583; Sigma) at 1:500, anti-IDR at 1:500, Fibrillarin (sc-374022; Santa Cruz) at 1:500, NPM1/B23 (sc-47725; Santa Cruz) at 1:250, and anti-GFP antibody (ab13970; abcam) at 1:2,000. Secondary antibodies used were goat anti-rabbit IgG, Alexa Fluor 546 (A-11010; ThermoFisher Scientific), goat anti-mouse IgG, Alexa Fluor 488 (A-11001; ThermoFisher Scientific), goat anti-rabbit IgG, Alexa Fluor 488 (A-11008; ThermoFisher Scientific), and goat anti-mouse IgG, Cy3.5 (ab6946; abcam). Slides were imaged using a Nikon A1 confocal microscope, and images were analyzed using Nikon Instrument Service (NIS) elements AR 3.2 imaging software. Imaging of nontransfected cells was carried out using a Zeiss LSM900 with Airyscan 2 and images were processed using Zen3.1 software.
BiFC Assay.
HEK293T cells at ∼40 to 50% confluency were transfected with BiFC plasmids as indicated in figures and incubated at 37 °C and 5% CO2 for 48 h followed by an additional 24 h at 30 °C and 5% CO2 prior to fixation. Cells were then washed in PBS, fixed with 4% formaldehyde for 15 min at RT, washed again in PBS, and mounted in Vectashield containing DAPI (Vector Laboratories). Fluorescence reconstitution was detected and imaged using a Nikon A1 confocal microscope, and images were analyzed using NIS elements AR 3.2 software.
Plasmid Construction.
SI Appendix, Tables S1 and S2 have comprehensive lists of plasmids and oligonucleotides, respectively, used in this study. For constructs expressing Liat1ΔE, Liat1ΔK, Jmjd6N277A, Jmjd6(1-334), and Jmjd6(1-290), the Q5 Site-Directed Mutagenesis kit (New England Biolabs) was used according to the manufacturer's protocol. Briefly, for pAA01, the region of Liat1 cDNA encoding the poly-E was removed from pCB179 using primers AA01F and AA02R. For pAA02, the region of Liat1 cDNA encoding poly-K was removed from pCB179 using primers AA03F and AA04R. For pCB500, the Asn227Ala mutation was made by altering the Jmjd6 cDNA in pCB458 using CB475F and CB476R. For pCB501, the region encoding Jmjd6 C-terminal amino acids 335 to 403 was removed from pCB458 using CB477F and CB478R. For pCB502, the region encoding Jmjd6 C-terminal amino acids 291 to 403 was removed from pCB458 using CB479F and CB480R. PCR was carried out for 25 cycles, and 1 μL of the resulting reaction mix was then used in a kinase–ligase–DpnI (KLD) reaction for 5 min at room temperature to obtain ligation and template removal. NEB5α competent cells (New England Biolabs) were transformed with 5 μL of the KLD reaction mixture and selected on Luria-Bertani (LB) plates supplemented with ampicillin (100 μg/mL).
For pAA03, cDNA encoding the N-terminal IDR of Liat1 was amplified from pCB179 such that it had a 5′ NheI site and 3′ KpnI site using primers CB200F and AA05R. The PCR product and pEGFP-N1 were digested with Nhe1 and Kpn1 and ligated using the Quick Ligation Kit (New England Biolabs). DH5α cells (New England Biolabs) were transformed with pAA03 and selected by growth on LB plates supplemented with 100 µg/mL Kanamycin.
To construct parental BiFC plasmids, the N-terminal half of YFP complementary DNA (cDNA) was PCR amplified from pEYFP-N1 using CB443F and CB444R to generate plasmids for N-terminal fusions and CB447F and CB448R to generate C-terminal fusions. The C-terminal half of YFP cDNA was PCR amplified using CB445F and CB446R for N-terminal fusions and CB449F and CB450R for C-terminal fusions. Amplified cDNA fragments for N-terminal fusions were digested with HindIII and KpnI; those for C-terminal fusions were digested with XbaI and ApaI and cloned into pcDNA3.0 to produce pCB481, pCB482 with the C-terminal, and pCB483 and pCB484 with an N-terminal linker (5′CGATCGATTGCTACA3′). The Ate1 BiFC constructs, pCB486 and pCB487, were made by PCR amplifying Ate11A7A cDNA from ATE1-3 in pcDNA3.1 using CB451F and CB452R, and the resulting fragment was cloned into EcoRI- and XbaI-digested pCB481 and pCB482, respectively. pCB488 and pCB489 were made by PCR amplifying Ate11A7A cDNA from pCB409 using CB453F and CB454R, and the resulting fragment was cloned into HindIII- and XhoI-digested pCB483 and pCB484, respectively. The Liat1 BiFC constructs, pCB490 and pCB491, were made by PCR amplifying Liat1 cDNA from pCB179 using CB455F and CB456R, and the resulting fragment was cloned into EcoRI- and XbaI-digested pCB481 and pCB482, respectively. pCB492 and pCB493 were made by PCR amplifying Liat1 cDNA from pCB179 with CB457F and CB458R, and the resulting fragment was cloned into HindIII- and XhoI-digested pCB483 and pCB484, respectively. The Jmjd6 BiFC constructs, pCB494 and pCB495, were made by PCR amplifying Jmjd6 cDNA from pCB458 using CB459F and CB460R, and the resulting fragment was cloned into KpnI- and XbaI-digested pCB481 and pCB482, respectively. pCB496 and pCB497 were made by PCR amplifying Jmjd6 cDNA from pCB458 using CB461F and CB462R, and the resulting fragment was cloned into HindIII- and XbaI-digested pCB483 and pCB484, respectively.
CRISPRLiat1 constructs pAA05 (CRISPRLIAT1-A) and pAA06 (CRISPRLIAT1-B) were constructed by the ligation of a phosphorylated double-strand oligomer (made by the denaturation and renaturation of oligonucleotides JD10 and JD11 for pAA05 and JD12 and JD13 for pAA06) into BsmBI-digested LentiCRISPv2, a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA (Addgene plasmid # 52961; http://n2t.net/addgene:52961; RRID:Addgene_52961).
IUPred2A.
We used IUPred2A to predict the IDRs in Liat1 (41, 53). IUPred2A is a web interface that predicts the ordered and disordered regions in a protein depending on the redox state of the environment. The amino acid sequence of mouse Liat1 from the National Center for Biotechnology Information (accession no. NP 941039) was used to predict the IDRs in the amino acid sequence of Liat1.
Cell Lysis and Fractionation.
Cells were scraped in PBS and centrifuged at 3,500 rpm for 5 min, washed with PBS and centrifuged at 3,500 rpm for 5 min. Cells were lysed in tissue lysis buffer (TLB; 50 mM Hepes, 10% glycerol, 0.05% Nonidet P-40, 150 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) containing complete protease-inhibitor mixture (Roche) followed by freeze–thawing in liquid nitrogen. The lysate was centrifuged at 13,000 rpm for 15 min at 4 °C. The supernatant collected was considered the cytosolic fraction. The pellet was washed in TLB and dissolved in nuclear extract buffer (420 mM NaCl, 20 mM Hepes [pH 7.5], 0.2 mM ethylenediaminetetraacetic acid [EDTA], 0.5 mM DTT, 1.5 mM MgCl2, 0.5 mM PMSF, 25% glycerol) containing complete protease-inhibitor mixture (Roche). The pellet was sonicated with 10 bursts using a sonicator with a microtip probe. The lysate is rotated for 20 min at 4 °C and then centrifuged at 13,000 rpm for 10 min at 4 °C. The supernatant collected was labeled nuclear fraction.
Experiments utilizing mouse tissues were approved by the Texas Woman’s University Institutional Animal Care and Use Committee and were conducted in strict compliance with the Animal Welfare Act, implementing Animal Welfare Regulations, and the principles of the Guide for the Care and Use of Laboratory Animals (54). For gel filtration of mouse testis fractions, mouse testis (450 mg/mL buffer) was homogenized in Tris-magnesium chloride-sucrose-DTT (TMSD) buffer (250 mM sucrose, 10 mM Tris⋅HCl [pH 7.5], 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF) containing “complete protease-inhibitor mixture” (Roche), using FastPrep-24 and Lysing Matrix D (MP Biomedicals), with three runs at 6.5 m/s for 20 s each and with 5-min incubations on ice between runs. The homogenate was centrifuged at 900 × g for 15 min at 4 °C. The supernatant (cytosol) was dialyzed using a “Slide-A-lyzer” with a molecular mass cutoff of 10 kDa (Pierce) for 4 h in Buffer G (400 mM Hepes [pH 7.9], 150 mM NaCl, 0.1 mM DTT, 10% glycerol) at 4 °C and filtered through a 0.45-µm filter. Five milligrams of total protein (at 5 mg/mL) was loaded onto a Superdex 200 HiLoad 16/60 (GE Life Sciences) and run in Buffer G at 0.5 mL/min collecting 2-mL fractions.
For experiments analyzing the nucleolar fraction, nucleoli were isolated according to the method described by Lam and Lamond (30). Briefly, cells were grown in ten 10-cm plates to ∼80 to 90% confluency and harvested by trypsinization. Cells were then washed three times in cold PBS and centrifuged at 1,000 rpm at 4 °C. Cell pellets were then resuspended in 5 mL of Buffer A (10 mM Hepes [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, protease inhibitor tablets), incubated on ice for 5 min, and lysed using a precooled Dounce tissue homogenizer. The lysate was then centrifuged at 1,000 rpm for 5 min at 4 °C. The supernatant was stored as the cytosolic fraction. The pellet was resuspended in 3 mL of solution 1 (0.25 M sucrose, 10 mM MgCl2 supplemented with protease inhibitor tablet) and layered over 3 mL of solution 2 (0.35 M sucrose, 0.5 mM MgCl2 supplemented with protease inhibitor tablet). After centrifugation at 2,500 rpm for 5 min at 4 °C, the pellet was resuspended in 3 mL of solution 2, sonicated using a microtip probe, layered on top of 3 mL of solution 3 (0.88 M sucrose, 0.5 mM MgCl2) supplemented with complete protease-inhibitor mixture (Roche), and centrifuged at 3,500 rpm for 10 min at 4 °C. The supernatant was stored as nuclear fraction. The pellet was washed in 0.5 mL of solution 2 and centrifuged at 2,500 rpm for 5 min at 4 °C. The remaining pellet (pure nucleoli) was resuspended in 300 µL of solution 2 and stored at −80 °C.
Immunoprecipitation and Immunoblotting.
Protein concentrations were determined using Bio-Rad Protein Assay (BioRad) according to the manufacturer’s instruction and normalized for immunoprecipitation reactions. Protein G magnetic beads (Bio-Rad) were incubated with 0.5 µg anti-FLAG M2 antibody (Sigma-Aldrich) per sample and incubated for 30 min at 4 °C with rotation. The antibody–bead mixture was added to the normalized lysates and rotated for 3 h at 4 °C. Nonbound material was removed using a magnet, and beads were washed three times in TLB and once in PBS; then, bound material was eluted in 2× Laemmli buffer. Samples (normalized lysates or immunoprecipitated samples) were heated to 95 °C for 5 min, loaded onto 4 to 12% NuPage Bis-Tris gels (Invitrogen), and electrophoresed in 3-(N-morpholino)propanesulfonic acid (Mops) buffer at 70 V. Separated proteins were then transferred to a 0.4-µm polyvinylidene difluoride membrane (BioRad) in Towbin buffer (25 mM Tris, 192 mM glycine, 20% methanol) overnight at 4 °C. Membranes were blocked with 5% milk in PBS containing 0.01% Tween-20 for 1 h at room temperature and incubated in primary antibodies at 1:1,000 concentration for 3 h at RT followed by three 5-min washes in PBS containing 0.01% Tween. Membranes were then incubated in secondary antibodies at 1:7,000 concentration for 1 h at RT followed by three 5-min washes in PBS containing 0.01% Tween followed by a final wash in PBS. The following primary antibodies were used for immunoblotting: HA (H3663; Sigma), Histone H2A.X (sc-517336; Santa Cruz), β-actin (sc-47778; Santa Cruz), C17ORF97 (HPA-023583; Sigma), Fibrillarin (374022; Santa Cruz), FLAG (F1804; Sigma), and NPM1/B23 (sc-47725; Santa Cruz). The following secondary antibodies were used: goat anti-mouse Immunoglobulin G (IgG), Dy light 680 (35519; ThermoFisher Scientific), goat anti-mouse IgG, Dy light 800 (SA5-10176; ThermoFisher Scientific), goat anti-rabbit IgG, Dy light 680 (35569; ThermoFisher Scientific), goat anti-rabbit IgG, and Dy light 800 (SA5-10036; ThermoFisher Scientific). Blots were imaged using a Licor Odyssey CLx system.
CRISPR-Cas9 Knockdown of Liat1 in Cells.
HEK293T cells were transfected with either pAA05 (CRISPRLIAT1-A) or pAA06 (CRISPRLIAT1-B) encoding mammalian codon-optimized Cas9 enzyme derived from Streptococcus pyogenes and a chimeric guide RNA targeted to the human LIAT1 gene (C17orf97) region encoding the Liat1 domain (pAA05) or the region repeated in primates (pAA06). To obtain a population of cells within which endogenous Liat1 was reduced, transfected cells were selected and grown in 3 µg/mL puromycin. LIAT1-targeted cells were then lysed and immunoblotted for endogenous Liat1 expression.
Yeast Two-Hybrid Assay.
Yeast two-hybrid assays were performed in the yeast strain Saccharomyces cerevisiae AH109 (MATα trp1-901 leu2-3, 112 ura3-52 his3-200 Gal4Δgal80, LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ, MEL1). Culturing was carried out in synthetic complete (SC) media containing 2% dextrose and 0.67% yeast nitrogen base supplemented with ammonium sulfate and amino acids. AH109 was cotransformed using the Lithium Acetate method with Gal4DBD-Liat1 (pCB239) and Gal4AD-Liat1 (pCB432) or the Liat1 mutants: pCB531, pCB433, pCB439, pCB434, pCER020, pCER021, pCB435, pCB436, pCB437, and pCER022. Cotransformants were selected by growth on SC medium lacking Leu and Trp and then tested for growth on SC medium lacking Leu, Trp, and His to determine the interaction between the Gal4DBD-Liat1 and Gal4AD-Liat1 mutants.
Supplementary Material
Acknowledgments
This work was supported by the Texas Woman’s University (TWU) Student Research Grant Program (A.A.), a TWU Quality Enhancement Program grant (to J.D.), a TWU Research Enhancement Program grant (to C.S.B.), and National Institute of Neurological Disorders and Stroke of the NIH Grant R15NS095317 (to C.S.B.). We thank Hope Parkin for her technical assistance and other members of the laboratory of C.S.B. for their help and advice.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015887118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Brower C. S., et al. , Liat1, an arginyltransferase-binding protein whose evolution among primates involved changes in the numbers of its 10-residue repeats. Proc. Natl. Acad. Sci. U.S.A. 111, E4936–E4945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weimann M., et al. , A Y2H-seq approach defines the human protein methyltransferase interactome. Nat. Methods 10, 339–342 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Heim A., et al. , Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine-serine-rich (RS) domains of SR- and SR-like proteins. Nucleic Acids Res. 42, 7833–7850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong X., et al. , Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 107, 14568–14572 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwok J., O’Shea M., Hume D. A., Lengeling A., Jmjd6, a JmjC dioxygenase with many interaction partners and pleiotropic functions. Front. Genet. 8, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webby C. J., et al. , Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Yi J., et al. , JMJD6 and U2AF65 co-regulate alternative splicing in both JMJD6 enzymatic activity dependent and independent manner. Nucleic Acids Res. 45, 3503–3518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W., et al. , Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155, 1581–1595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulard C., et al. , Role of JMJD6 in breast tumourigenesis. PLoS One 10, e0126181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aprelikova O., et al. , The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin. Epigenetics 8, 38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y. F., et al. , JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 14, R85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao W. W., et al. , JMJD6 licenses ERα-dependent enhancer and coding gene activation by modulating the recruitment of the CARM1/MED12 co-activator complex. Mol. Cell 70, 340–357.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F., et al. , JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 12, e1001819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam M. S., et al. , Biochemical and structural investigations clarify the substrate selectivity of the 2-oxoglutarate oxygenase JMJD6. J. Biol. Chem. 294, 11637–11652 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantri M., et al. , Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6. J. Mol. Biol. 401, 211–222 (2010). [PubMed] [Google Scholar]
- 16.Mantri M., et al. , The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. ChemBioChem 12, 531–534 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Mantri M., et al. , Self-hydroxylation of the splicing factor lysyl hydroxylase, JMJD6. MedChemComm 3, 80–85 (2012). [Google Scholar]
- 18.Han G., et al. , The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J. Cell. Biochem. 113, 1663–1670 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Mantri M., Zhang Z., McDonough M. A., Schofield C. J., Autocatalysed oxidative modifications to 2-oxoglutarate dependent oxygenases. FEBS J. 279, 1563–1575 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Sawyer I. A., Dundr M., “Nuclear bodies” in Nuclear Architecture and Dynamics, Lavelle C., Victor J.-M., Eds. (Academic Press, Boston, MA, 2018), pp. 235–256. [Google Scholar]
- 21.Feric M., et al. , Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawyer I. A., Sturgill D., Dundr M., Membraneless nuclear organelles and the search for phases within phases. Wiley Interdiscip. Rev. RNA 10, e1514 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Sleeman J. E., Trinkle-Mulcahy L., Nuclear bodies: New insights into assembly/dynamics and disease relevance. Curr. Opin. Cell Biol. 28, 76–83 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Decker C. J., Parker R., P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4, a012286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Alberti S., Dormann D., Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Molliex A., et al. , Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochs R. L., Lischwe M. A., Spohn W. H., Busch H., Fibrillarin: A new protein of the nucleolus identified by autoimmune sera. Biol. Cell 54, 123–133 (1985). [DOI] [PubMed] [Google Scholar]
- 29.Hall L. L., Smith K. P., Byron M., Lawrence J. B., Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288, 664–675 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam Y. W., Lamond A. I., “Isolation of nucleoli” in Cell Biology, Celis J. E., Ed. (Academic Press, Burlington, MA, ed. 3, 2006), chap. 15, pp. 103–107. [Google Scholar]
- 31.Li Z. F., Lam Y. W., A new rapid method for isolating nucleoli. Methods Mol. Biol. 1228, 35–42 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Hebert M. D., Matera A. G., Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11, 4159–4171 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T., Boisvert F. M., Bazett-Jones D. P., Richard S., A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell 10, 3015–3033 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis J. D., Tollervey D., Like attracts like: Getting RNA processing together in the nucleus. Science 288, 1385–1389 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Terzo E. A., et al. , Distinct self-interaction domains promote Multi Sex Combs accumulation in and formation of the Drosophila histone locus body. Mol. Biol. Cell 26, 1559–1574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitrea D. M., et al. , Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 9, 842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerppola T. K., Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerppola T. K., Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7, 449–456 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang K., Yang J., Yi J., Nucleolar stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2, 125–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latonen L., Phase-to-phase with nucleoli—stress responses, protein aggregation and novel roles of RNA. Front. Cell. Neurosci. 13, 151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dosztányi Z., Prediction of protein disorder based on IUPred. Protein Sci. 27, 331–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogatyreva N. S., Finkelstein A. V., Galzitskaya O. V., Trend of amino acid composition of proteins of different taxa. J. Bioinform. Comput. Biol. 4, 597–608 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Ukmar-Godec T., et al. , Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat. Commun. 10, 2909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pontvianne F., et al. , Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 16, 1574–1587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui P., Qin B., Liu N., Pan G., Pei D., Nuclear localization of the phosphatidylserine receptor protein via multiple nuclear localization signals. Exp. Cell Res. 293, 154–163 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Wolf A., et al. , The polyserine domain of the lysyl-5 hydroxylase Jmjd6 mediates subnuclear localization. Biochem. J. 453, 357–370 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I., The nucleolus under stress. Mol. Cell 40, 216–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Audas T. E., Jacob M. D., Lee S., The nucleolar detention pathway: A cellular strategy for regulating molecular networks. Cell Cycle 11, 2059–2062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyd M. T., Vlatkovic N., Rubbi C. P., The nucleolus directly regulates p53 export and degradation. J. Cell Biol. 194, 689–703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woods S. J., Hannan K. M., Pearson R. B., Hannan R. D., The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim. Biophys. Acta 1849, 821–829 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Boeckel J. N., et al. , Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl. Acad. Sci. U.S.A. 108, 3276–3281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kingston R. E., Chen C.A., Okayama H., Calcium phosphate transfection. Curr. Protoc. Immunol. 31, 10.13.1–10.13.9 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Erdős G., Dosztányi Z., Analyzing protein disorder with IUPred2A. Curr. Protoc. Bioinformatics 70, e99 (2020). [DOI] [PubMed] [Google Scholar]
- 54.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.