Abstract
Immunorelevant genes are among the most probable modulators of coronavirus disease 2019 (COVID-19) progression and prognosis. However, in the few months of the pandemic, data generated on host genetics has been scarce. The present study retrieved data sets of HLA-B alleles, KIR genes and functional single nucleotide polymorphisms (SNPs) in cytokines related to COVID-19 cytokine storm from two publicly available databases: Allele Frequency Net Database and Ensembl, and correlated these frequency data with Case Fatality Rate (CFR) and Daily Death Rates (DDR) across countries. Correlations of eight HLA-B alleles and polymorphisms in three cytokine genes (IL6, IL10, and IL12B) were observed and were mainly associated with DDR. Additionally, HLA-B correlations suggest that differences in allele affinities to SARS-CoV-2 peptides are also associated with DDR. These results may provide rationale for future host genetic marker surveys on COVID-19.
Keywords: HLA-B, KIR, Cytokine polymorphisms, COVID-19
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain which has caused the coronavirus disease 2019 (COVID-19) pandemic was originated in China in December 2019 and suddenly dispersed worldwide. The clinical manifestations vary from few or no symptoms to acute respiratory illness along with a wide spectrum of non-respiratory symptoms, including gastrointestinal, neurological, cardiovascular and ophthalmologic [1]. As of 10 November 2020, over 52 million cases were confirmed and more than 1.2 million deaths worldwide. Moreover, similarly to disease progression, lethality and mortality rates across different countries are also very heterogeneous, varying from 0 to 31% and from 0 to 48 daily deaths per 10 million inhabitants (rates estimated from WHO data as of 10 November 2020; https://covid19.who.int/). Interestingly, the fatality rate has been observed to be higher in Europe (1.8%) than in Asia (lower than 1.3%), but almost reaching 2.2% in Americas, where the European ancestry is high. Despite of the many intervening factors in these statistics, this trend should not be ignored, suggesting a genetic modulation of COVID-19 progression and outcomes.
Some preliminary evidences have pointed out to associations with ABO blood groups [2] and in silico predictions of some ACE2 genetic variants as predisposing/protective markers [3], reinforcing the genetic basis of COVID-19 heterogeneity. In this scenario, immunorelevant genes should be also highlighted as potential predictive markers, due to important immunological features of COVID-19, like the cytokine storm involving TNFa, IFNg, IL-6 and IL-10, CD4+/CD8+ lymphopenia [4] and development of Th17 responses [5]. Additional evidences may be inferred from studies with previous respiratory coronavirus epidemics, like SARS and Middle East respiratory syndrome (MERS), that showed association with human leukocyte antigen (HLA) alleles [6], [7] and the decrease of Natural Killer cells [8].
Hence, the present study retrieved worldwide allele frequencies, stored in the Allele Frequency Net Database (AFND) [9] and Ensembl database [10], for several immune-related genes including HLA-B, killer-cell immunoglobulin-like receptors (KIR) genes and genotypes, and functional polymorphisms related to cytokine expression, revealing correlation of HLA-B and cytokine polymorphisms with worldwide COVID-19 incidence and fatality rates. Additionally, HLA-B correlations seemed to be associated to differences in allele affinities to SARS-CoV-2 peptides. The present metanalysis may provide rationale for future host genetic marker surveys on COVID-19.
2. Material and methods
2.1. COVID-19 epidemiological statistics
Data on number of cases, daily deaths since the beginnings of the COVID-19 pandemic for all countries with genetic data available were retrieved from World Health Organization (https://covid19.who.int/), along with their respective inhabitant numbers. These data were used to estimate for each country: (i) the Case Fatality Rate (CFR), i.e. the number of deaths by COVID-19 divided by the number of confirmed cases and (ii) the Daily Death Rate (DDR), represented as the average number of deaths per day (since the first confirmed case) per ten million inhabitants. The data were obtained as of Nov 10, 2020. CFR and DDR estimates for all countries with genetic data are presented in Supplementary Data.
2.2. Choice of candidate markers
A total of 98 countries have any kind of genetic data available, about 50% of the countries in the world. We avoid polymorphisms with few population data and used SNPs, KIR and HLA data with available genetic data in at least 30 populations.
For multiple populations of a given country direct inspection for very discrepant frequencies was performed, if detected the cause was investigated and the more representative populations were chosen, likely these outlier populations were small aboriginal populations or immigrated populations. The preference was always the more urban populations because of their correspondence with DDR and CFR estimates that are more influenced by the great urban centers.
2.2.1. HLA-B
HLA genes have been postulated to be under pathogen-driven selection [11], revealing their importance in immune response to infections. Among class I HLA loci, HLA-B has shown the strongest selective signal [11]. Association studies showed HLA-B as a major locus associated with viruses and other infectious diseases [12]. Moreover, HLA-B-restricted viral derived epitopes have been reported to be targeted by CD8+ T cells more frequently than those presented by HLA-A [13], [14], [15]. Therefore, HLA-B becomes good candidate to be a marker of COVID-19 outcome.
Consulting AFND, data on 420 HLA-B alleles were obtained from 209 worldwide populations from 60 different countries. The allele frequencies of each country were obtained averaging the allele frequencies of the populations of each country they belong to.
HLA-B frequency data were retrieved only for populations that belong to the Gold Standard Dataset, described elsewhere [16]. This data set is composed of populations, where at least one HLA locus fulfill the criteria of high resolution allele genotyping (two fields), sample size larger than 50 and allele frequencies sum to 1 [16]. Supplementary Data presents HLA-B allele frequencies by country.
The entire FASTA-formatted proteome of a SARS-CoV-2 strain (Accession number: MN988713.1) was obtained from National Center of Biotechnology Information and used as input for predictions using NetMHCpan v4.0 online server [17]. Peptides with length 9mer were evaluated, because the preference of almost all HLA molecules for this length [17], and those assigned as strong binders (SB) to each HLA-B allele were recorded. The server’s default value of 0.5%-ranked as the threshold for SB was adopted. Running options included “Make BA predictions” (method that use binding affinity data for training model), “Sort by prediction score” (order from higher to lower scores) and “Save predictions to XLS file” (to save predictions in a Excel compatible file).
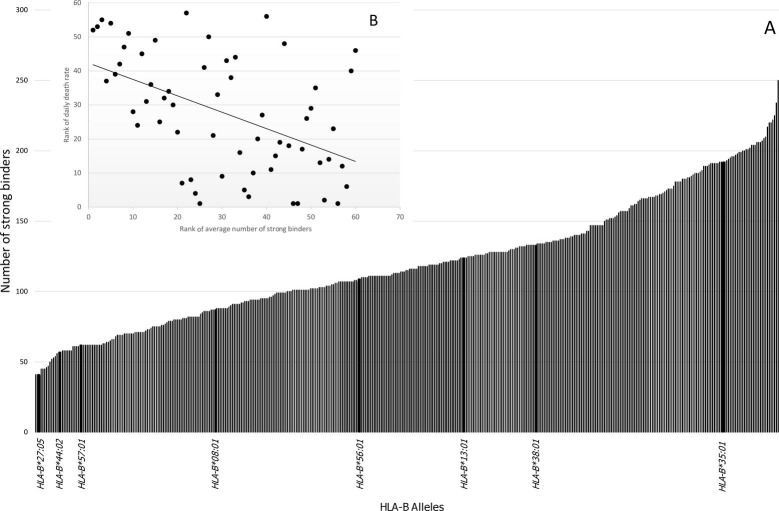
The total numbers of SB peptides for each allele were recorded and is showed in Fig. 1 .
Fig. 1.
[A]: Number of SARS-CoV-2-derived peptides that are strong binders (SB) to HLA-B alleles. The affinity was determined in silico by NetMHCpan 4.0 software. In the X-axis are presented the eight HLA-B alleles that correlated individually with Daily Death Rate (DDR) as showed in Table 1. [B]: Correlation between the average number of SB of each country (weighted by allele frequencies) and DDR (Spearman’s correlation coefficient = -0.49; p < 0.0001).
Additionally, for each country, the average number of SB was estimated, weighting the number of SB of the alleles observed in a given country by their respective frequencies. This average number of SB was correlated with CFR and DDR using Spearman’s correlation.
The HLA-B alleles with an average frequency of 1% or higher, considering all 60 countries, were then selected and their allele frequencies were correlated with CFR and DDR using Spearman’s correlation. Supplementary Data presents HLA-B allele frequencies by country. This approach allowed to pinpoint relevant alleles in terms of frequency and correlation with CFR and DDR.
Additionally, using HLA-B peptide sequence data obtained from IPD-IMGT/HLA database [18], the dimorphism at position −21 (M/T) were inferred for each HLA-B allele. This polymorphism in the gene segment encoding the leader peptide modulates whether NK cell regulation primarily relies on the KIRs or the NKG2A/CD94 receptor [19], [20]. Allele frequencies of HLA-B alleles were then pooled according their M or T dimorphism and correlated with DDR and CFR. The rationale of this approach are evidences that allele M is detrimental in HIV infection [19]
2.2.2. Cytokines
The cytokine storm observed in COVID-19 highlighted the relevance of inflammatory cytokines in disease pathogenesis. Consistently, high levels of IL-6, TNFa, IFNg, IL-1b, IL-2 and IL-10 have been reported [4], [5]. Their genes have mutations that affect the gene expression and may be associated with COVID-19 progression. The allele frequencies were obtained mainly from AFND (two IL1B polymorphisms have data only for less than four populations and were not considered) and complemented with data from Ensembl database release 100 [10]. The number of countries with data on cytokine polymorphisms frequencies varied from 16 to 54. The cytokine polymorphisms used, with their respective number of countries, are listed in Table 2 The allele frequencies were correlated (Spearman’s correlation) with CFR and DDR estimates. Supplementary Data presents cytokine polymorphism frequencies by country.
Table 2.
Correlation of CFR and DDR with cytokine polymorphisms.
| Marker | Number of countriesa | Average allele frequency (frequency range) | CFR |
DDR |
||
|---|---|---|---|---|---|---|
| rs | p | rs | p | |||
| IL6 −174C | 47 | 0.21 (0 – 0.5) | −0.099 | 0.51 | 0.54 | <0.0001 |
| IL6 + 565A | 19 | 0.26 (0–0.442) | 0.16 | 0.49 | 0.43 | 0.06 |
| TNFA −238A | 40 | 0.07 (0.007–0.225) | 0.31 | 0.04 | 0.41 | 0.0081 |
| TNFA −308A | 54 | 0.11 (0.017–0.267) | 0.32 | 0.04 | 0.35 | 0.0079 |
| TNFA −1031C | 17 | 0.21 (0.103–0.436) | −0.17 | 0.51 | −0.1 | 0.68 |
| TNFA −857 T | 21 | 0.21 (0.010–0.282) | −0.8 | 0.71 | 0.24 | 0.30 |
| TNFA −863A | 17 | 0.15 (0.065–0.331) | −0.09 | 0.73 | −0.32 | 0.20 |
| IFNG + 874 T | 31 | 0.34 (0.072–0.579) | −0.22 | 0.23 | 0.46 | 0.0092 |
| IFNG + 5644 T | 16 | 0.55 (0.225–0.584) | −0.04 | 0.87 | 0.28 | 0.28 |
| IL10 −1082G | 46 | 0.32 (0.024–0.549) | 0.11 | 0.46 | 0.58 | <0.0001 |
| IL10 −592C | 41 | 0.61 (0.258–0.796) | −0.02 | 0.89 | 0.60 | <0.0001 |
| IL10 −819C | 42 | 0.62 (0.249–0.800) | 0.03 | 0.87 | 0.59 | <0.0001 |
| IL1B + 3962 T | 31 | 0.18 (0.010–0.316) | −0.06 | 0.72 | 0.55 | 0.0013 |
| IL1B −511C | 34 | 0.45 (0.275–0.700) | −0.25 | 0.14 | 0.34 | 0.05 |
| IL2B −330G | 38 | 0.31 (0–0.567) | −0.2 | 0.21 | −0.05 | 0.78 |
| IL2B −166 T | 32 | 0.30 (0.053–0.640) | 0.13 | 0.45 | 0.15 | 0.38 |
| IL12B + 1188A | 32 | 0.68 (0.456–0.833) | −0.04 | 0.80 | 0.62 | <0.0001 |
Number of countries with cytokine data available. The correlations that remained significant after correction for multiple tests are highlighted in bold. CFR = case fatality rate; DDR = daily death rate; rs = Spearman's correlation coefficient; ns = not significant. Significant p value's after correction (pc): IL6 −174C, pc = 0.000064; IL10 −1082G, pc = 0.000064; IL10 −592C, pc = 0.00019; IL10 −819C, pc = 0.000032; IL12b + 1188A, pc = 0.00013.
2.2.3. Killer-cell immunoglobulin-like receptors (KIR)
KIR polymorphism relies most importantly on gene rearrangements that might delete one or more of the 15 genes of the KIR gene cluster, whose ligands are commonly HLA class I molecules. The presence or absence of these genes, as well as the KIR profiles, have been associated with some viral diseases [21]. In addition, a decay in Natural Killer (NK) cells number has been reported in SARS [8]. Together, these results encourage the investigation of KIR polymorphisms as marker for COVID-19.
Data for presence/absence frequencies of nine polymorphic KIR genes (framework genes were not included in the analysis) were retrieved from AFND for 70 countries. Framework genes are those present in almost all individuals.
Additionally, the KIR genotype AA frequencies for 64 countries were also retrieved from AFND. AA genotype was defined according AFND criteria that considers as AA genotypes where genes KIR2DL2, KIR2DL5, KIR3DS1, KIR2DS1, KIR2DS2, KIR2DS3 and KIR2DS5 are absent.
The list of polymorphic KIR genes are presented in Table 3 . These frequencies were correlated (Spearman’s correlation) with CFR and DDR estimates. Supplementary Data presents KIR gene and AA genotype frequencies by country.
Table 3.
Correlation of CFR and DDR with KIR polymorphisms.
| Marker | Number of countriesa | Average allele frequency (frequency range) | CFR |
DDR |
||
|---|---|---|---|---|---|---|
| rs | p | rs | p | |||
| Genotype AA | 64 | 0.29 (0.015 – 0.552) | −0.09 | 0.44 | −0.29 | 0.02 |
| KIR2DL2 | 69 | 0.51 (0.134 – 0.719) | −0.18 | 0.12 | 0.11 | 0.33 |
| KIR2DL3 | 69 | 0.87 (0.565 – 0.997) | 0.14 | 0.22 | −0.26 | 0.02 |
| KIR2DL5 | 58 | 0.55 (0.284 – 0.780) | −0.11 | 0.37 | 0.15 | 0.22 |
| KIR2DS1 | 68 | 0.37 (0.014 – 0.646) | 0.13 | 0.28 | 0.14 | 0.25 |
| KIR2DS2 | 68 | 0.49 (0.138 – 0.717) | −0.08 | 0.51 | 0.27 | 0.02 |
| KIR2DS3 | 65 | 0.30 (0.103 – 0.522) | 0.03 | 0.82 | 0.13 | 0.29 |
| KIR2DS5 | 64 | 0.34 (0.190 – 0.830) | 0.15 | 0.21 | 0.06 | 0.63 |
| KIR3DL1 | 69 | 0.93 (0.755 – 0.990) | −0.13 | 0.26 | −0.14 | 0.24 |
| KIR3DS1 | 69 | 0.35 (0.007 – 0.620) | 0.28 | 0.02 | 0.23 | 0.05 |
CFR = case fatality rate; DDR = daily death rate; rs = Spearman’s correlation coefficient; ns = not significant.
2.2.4. Statistical analysis
All polymorphisms frequency sets were tested for correlation with DDR and CFR, using the Spearman’s correlation test and p values were corrected for multiple tests using Bonferroni correction method. All tests were performed using the BioEstat 5.3 software [22].
Additionally, an ethnicity-based approach was carried out. Alleles that correlated significantly with DDR were evaluated in three major ethnicities, European (EUR), East Asian (EAS; includes North and South East Asia) and Sub-Saharan Africans (SSA). The regions each country belongs to, are given in Supplementary Table and follow AFND classification. Comparisons were made by Mann-Whitney test.
3. Results
3.1. Low affinity HLA-B alleles and COVID-19 outcomes
The number of strong binder peptides for each HLA-B allele is presented in Fig. 1A, as well as a correlation between the average number of SB of each country (weighted by allele frequencies) and DDR (Spearman's correlation coefficient = −0.49; p < 0.0001) (Fig. 1B). The correlation between the average number of SB of each country and CFR was also significant, however, this correlation was weaker (Spearman’s correlation coefficient = −0.30; p = 0.018).
The polymorphic HLA-B alleles, with average frequencies over 1%, were tested for correlation of their frequencies with CFR and DDR estimates (Table 1). This approach allowed to identify the putative alleles associated with COVID-19 outcome. CFR did not correlate with any HLA-B alleles. The allele frequencies correlated more strongly with DDR, where fifteen alleles showed significance at 0.05 level. However, after correction for multiple tests only eight remained significant, being only HLA-B*13:01 negatively correlated (protective). The significant HLA-B allele frequencies along with average DDR in each world region, defined by AFND dataset, is showed in Fig. 2 .
Table 1.
Correlation of CFR and DDR with HLA-B allele frequencies across 60 countries.
| Allele | Number of SBa | Average allele frequency | CFR |
DDR |
||
|---|---|---|---|---|---|---|
| rs | p | rs | P | |||
| −21 M | – | 0.165 | 0.03 | 0.78 | 0.37 | 0.0032 |
| −21 T | – | 0.590 | −0.02 | 0.84 | −0.15 | 0.23 |
| HLA-B*07:02 | 61 | 0.053 | 0.02 | 0.85 | 0.35 | 0.0053 |
| HLA-B*08:01 | 82 | 0.045 | 0.10 | 0.40 | 0.5 | <0.0001 |
| HLA-B*13:01 | 125 | 0.017 | −0.02 | 0.90 | −0.45 | 0.0003 |
| HLA-B*13:02 | 122 | 0.015 | −0.17 | 0.19 | 0.04 | 0.76 |
| HLA-B*14:02 | 167 | 0.017 | 0.04 | 0.76 | 0.37 | 0.0031 |
| HLA-B*15:01 | 128 | 0.032 | −0.01 | 0.91 | 0.22 | 0.09 |
| HLA-B*15:02 | 195 | 0.019 | 0.17 | 0.19 | −0.25 | 0.06 |
| HLA-B*15:03 | 180 | 0.015 | 0.04 | 0.76 | 0.09 | 0.50 |
| HLA-B*18:01 | 101 | 0.037 | −0.22 | 0.08 | 0.14 | 0.27 |
| HLA-B*27:05 | 41 | 0.015 | 0.12 | 0.36 | 0.53 | <0.0001 |
| HLA-B*35:01 | 191 | 0.051 | 0.09 | 0.46 | 0.42 | <0.0001 |
| HLA-B*35:03 | 147 | 0.016 | −0.11 | 0.40 | 0.41 | 0.0011 |
| HLA-B*35:05 | 184 | 0.015 | 0.15 | 0.24 | −0.15 | 0.24 |
| HLA-B*38:01 | 134 | 0.012 | −0.06 | 0.64 | 0.51 | <0.0001 |
| HLA-B*39:01 | 157 | 0.015 | 0.04 | 0.78 | 0.04 | 0.76 |
| HLA-B*40:01 | 71 | 0.038 | 0.01 | 0.91 | −0.4 | 0.06 |
| HLA-B*40:02 | 70 | 0.03 | −0.04 | 0.75 | 0.03 | 0.83 |
| HLA-B*40:06 | 91 | 0.01 | −0.03 | 0.82 | −0.10 | 0.42 |
| HLA-B*42:01 | 80 | 0.011 | −0.03 | 0.80 | 0.008 | 0.95 |
| HLA-B*44:02 | 58 | 0.027 | 0.03 | 0.79 | 0.45 | 0.0003 |
| HLA-B*44:03 | 62 | 0.033 | 0.05 | 0.70 | 0.18 | 0.15 |
| HLA-B*45:01 | 95 | 0.013 | 0.06 | 0.64 | 0.08 | 0.53 |
| HLA-B*46:01 | 225 | 0.012 | −0.06 | 0.61 | −0.37 | 0.0034 |
| HLA-B*48:01 | 119 | 0.012 | 0.08 | 0.56 | −0.16 | 0.22 |
| HLA-B*49:01 | 77 | 0.013 | −0.05 | 0.69 | 0.20 | 0.12 |
| HLA-B*50:01 | 103 | 0.018 | −0.15 | 0.24 | 0.27 | 0.03 |
| HLA-B*51:01 | 111 | 0.059 | −0.08 | 0.50 | 0.38 | 0.0022 |
| HLA-B*52:01 | 147 | 0.022 | −0.04 | 0.74 | 0.13 | 0.30 |
| HLA-B*53:01 | 108 | 0.023 | −0.07 | 0.56 | 0.04 | 0.71 |
| HLA-B*56:01 | 113 | 0.015 | −0.08 | 0.50 | −0.38 | 0.0027 |
| HLA-B*57:01 | 71 | 0.016 | 0.09 | 0.48 | 0.48 | <0.0001 |
| HLA-B*58:01 | 87 | 0.028 | −0.07 | 0.57 | −0.34 | 0.0062 |
The correlations that remained significant after correction for multiple tests are highlighted in bold. CFR = case fatality rate; DDR = daily death rate; rs = Spearman’s correlation coefficient; ns = not significant. aThe number of strong binders is the number of peptides of the SARS-CoV-2 proteome that binds strongly each HLA-B allele. Significant p values after correction (pc): HLA-B*08:01, pc = 0.00016; HLA-B*13:01, pc = 0.0096; HLA-B*27:05, pc = 0.0000000032; HLA-B*35:01, pc = 0.0022; HLA-B*35:03, pc = 0.03; HLA-B*38:01, pc = 0.00022; HLA-B*44:02, pc = 0.0096; HLA-B*57:01, pc = 0.00013.
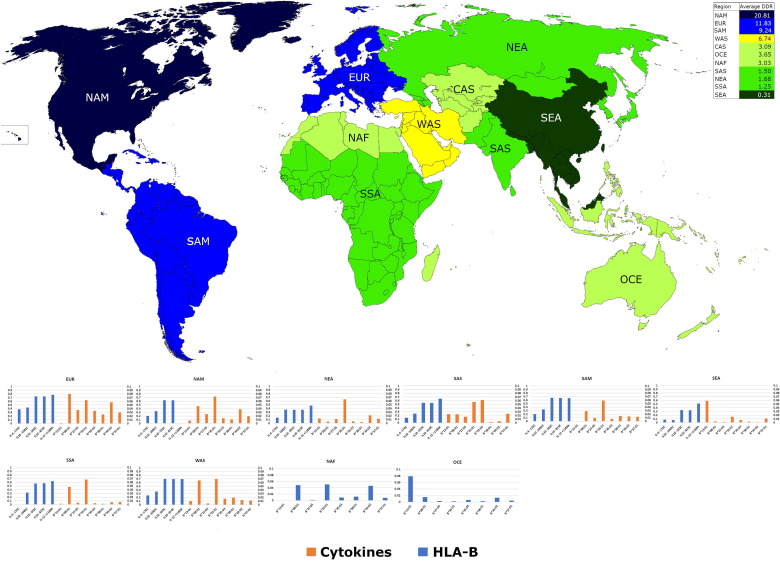
Fig. 2.
World heatmap showing average Daily Death Rate (DDR) in world regions (according to AFND) and their respective HLA-B alleles frequency distributions in associated graphs. Only the HLA-B alleles and cytokine SNPs that correlated significantly with DDR, after correction for multiple tests, were presented. NAM = North America; EUR = Europe; SAM = South and Central America; WAS = West Asia; CAS = Central Asia; OCE = Oceania; NAF = North Africa; SAS = South Asia; NEA = North-East Asia; SSA = Sub-Saharan Africa; SEA = South-East Asia. Australia and Oceania AFND regions were joined under OCE designation. CAS has no genetic data available, OCE and NAF have only HLA-B genetic data.
The frequencies of HLA-B −21 M allele correlated positively with DDR (Table 1). The spearman's correlation coefficient for HLA-B −21 M allele (rs) was 0.37, however it loses significance after correction for multiple tests (Table 1).
3.2. Influence of KIR and cytokine polymorphisms in COVID-19 outcomes
The frequencies of KIR genes KIR2DL3, KIR2DS2 and genotype AA showed significant correlation with DDR and KIR3DS1 with CFR, but these significances were lost after correlation for multiple tests. However, some significant correlations were observed for cytokine polymorphisms (Table 2). Two cytokine polymorphisms correlated with CFR but none remained significant after correction for multiple tests. Otherwise, the correlations with DDR were stronger and from nine correlations observed, five remained significant after correction for multiple tests (Table 3). The significant cytokine allele frequencies along with average DDR in each world region, defined by AFND dataset, can be seen in Fig. 2. This trend followed the same from HLA-B, with DDR showing the stronger correlation signals.
3.3. Ethnicity analysis
The Table 4 clearly shows a higher DDR in European that is associated with the higher average frequencies of predisposing HLA-B and cytokine alleles. Moreover, East Asians and the Sub-Saharan have the lowest DDR estimate and the highest frequencies of protective HLA-B*13:01 (protective allele).
Table 4.
Average frequencies of HLA-B and cytokine alleles that correlated with DDR in three major well defined ethnic regions.
| Ethnicity | EUR | EAS | SSA |
|---|---|---|---|
| DDR | 12.6 | 0.90 | 1.95 |
| Alleles | |||
| IL6 −174C | 0.383 | 0.103c | 0.003c |
| IL10 −1082G | 0.434 | 0.168c | 0.321c |
| IL10 −592C | 0.731 | 0.344c | 0.567c |
| IL10 −819C | 0.731 | 0.341c | 0.573c |
| IL12B + 1188A | 0.775 | 0.502c | 0.632c |
| B*08:01 | 0.080 | 0.003c | 0.043b |
| B*13:01 | 0.001 | 0.045c | 0.002ª |
| B*27:05 | 0.036 | 0.004c | 0.004c |
| B*35:01 | 0.063 | 0.030c | 0.073ª |
| B*35:03 | 0.034 | 0.006c | 0.003c |
| B*38:01 | 0.024 | 0.002c | 0.002c |
| B*44:02 | 0.057 | 0.007c | 0.006c |
| B*57:01 | 0.030 | 0.011c | 0.007c |
| Sum of Predisposing HLA-B Alleles | 0.324 | 0.063c | 0.138a |
| HLA-B −21 allele M | 0.238 | 0.080c | 0.178b |
| HLA-B −21 allele T | 0.623 | 0.670a | 0.553b |
DDR = daily Death Rate, EUR = Europe, EAS = East Asia, SSA = Sub-Saharan Africa, in bold HLA-B alleles that correlated negatively with DDR (protective). Mann-Whitney tests for comparison between European and the other ethnicities: a = p not significant; b = p < 0.05 but lost significance after correction for multiple tests; c = p < 0.05 and remain significant after correction for multiples tests.
4. Discussion
4.1. Mortality rates of COVID-19
The results showed that HLA-B and cytokine polymorphisms correlations are stronger and more extensive with DDR than CFR. The CFR is obtained by dividing the number of deaths by the number of confirmed cases. The denominator is known to be compromised by low testing coverage, asymptomatic or mild cases [23] that depends on socio-economic and political management factors. Moreover, CFR’s denominator is composed by both active and recovered cases, being the proportion of active cases that would evolve to death not yet detectable.
The testing, in most countries, have been preferentially applied to severe/critical cases and to clear suspect death by COVID-19. Hence, differences in testing coverage would impact less in the number of deaths than in the number of cases, because the resources in countries with limited testing capacity would be redirected to resolve the cause of deaths. This scenario favors epidemiological statistics like DDR that considers country’s population as denominator and take in account the time. Thus, DDR, while an average daily incidence, seems to be more suitable to COVID-19 that is still ongoing.
The results showed consistently stronger correlation signals with DDR than with CFR, suggesting that DDR is a more accurate to represent the death rates of COVID-19.
4.2. HLA-B
The strong correlation of the average number of SB with DDR (Fig. 1B) suggests a functional role of peptide affinity with COVID-19 prognosis, in agreement with a previous study that, using a similar approach, associated fewer predicted binding peptides with COVID-19 vulnerability [24].
The first association studies with SARS-CoV suggested that HLA-B*07:03 [25] and HLA-B*46:01 [6] would be predisposing alleles. However, these associations could not be confirmed afterward [26], [27], [28]. An important point is that all studies were carried out with SARS-CoV patients and restricted to Chinese/Taiwanese samples. Thus, extrapolation to other ethnicities must be carefully considered.
In this scenario, the pooled frequencies of all seven alleles that correlated positively with DDR are lower in SEA (0.1), NEA (0.13) and SSA (0.14) than in those with predominantly European ancestry (EUR, 0.32 and NAM 0.23), where the DDR are higher. These data corroborate the best performance of East Asia countries against COVID-19, if using as criteria the death rates.
The MHC region is known by the strong linkage disequilibrium between alleles from different loci. Although HLA-B displays the strongest selective signal [11] and associations with infectious diseases [12] the linkage of HLA-B alleles with other HLA alleles should be considered, in special with HLA-C alleles because of the close distance between both loci. Moreover, HLA-C alleles are ligands to KIRs that regulate NK cell responses to viral infection [21]. In this context, along with evidences that HLA-B molecules affinity to SARS-CoV-2 epitopes play a role in the infection, future studies should consider additional HLA loci and haplotypes and their functional relationship with KIR.
4.3. KIR and cytokine polymorphisms
The literature reported that the Natural Killer cell counts decrease [8] in SARS, suggesting that low NK cell activity would be relevant in SARS, being interesting to highlight that, although the lack of significance after correction for multiple tests, the trend for positive correlation of inhibitory genotype AA and KIR2DL3 with DDR and negative correlation of stimulatory KIR2DS2 with DDR could be observed. Additionally, HLA-B*27:05, *38:01, *44:02 and *57:01 are Bw4 and KIR3DL1 ligands while HLA-B*08:01, *35:01 and *35:03 are the Bw6 and the most frequent alleles. Thus, HLA-B ligands also do not show a clear pattern. NK cells also respond to several cytokines like IL-12 and IL-2 and produce many others like IFNg, TNFa and IL-6 [29], being all these cytokines augmented in COVID-19 cytokine storm. Additionally, TNFA is close to HLA-B and -C genes and the determination of HLA-B, -C and TNFA SNP haplotypes would be relevant in the determination of possible association of these haplotypes with COVID-19.
Despite of the absence of clear correlation with KIR and their ligands, polymorphisms at other NK receptors, like NKG2D, and their ligands may still be evaluated as candidate marker, once they have been also associated with infectious and inflammatory diseases [30]. In this context the correlation with HLA-B signal peptide allele M suggests an effect similar to observe for HIV, where allele M compromise antiviral NK-mediated activity [19].
Cytokine polymorphisms showed stronger correlation with DDR. A clear trend showing that alleles that augment the gene expression correlate consistently with a poor COVID-19 prognosis (Table 5 ). Considering that all these cytokines display elevated levels in the COVID-19 cytokine storm the correlations may directly reflect their functional role.
Table 5.
Cytokine polymorphisms found to be correlated significantly with COVID-19 Daily Death Rate and their respective functional effects and associations with comorbidities that give poor prognosis in COVID-19.
| Cytokine polymorphisms | Functional effect | Association comorbities |
|---|---|---|
| IL6 −174C | Increase IL6 expression [38] | Hypertension [39], cardiovascular diseases [40], diabetes [41], obesity [42] |
| IL10 −1082G/ IL10 −592C/ IL10 −819C | Linked in a haplotype that increases IL-10 expression[43] | Diabetes [44], cardiovascular diseases [45] |
| IL12B + 1188A | Controversy [46] | Diabetes [47], asthma [48] |
Alternatively, underlying medical conditions with poor prognosis to COVID-19, like cardiovascular disease, diabetes, chronic respiratory disease, hypertension, and cancer [31], have been also reported as associated to these polymorphisms (Table 5). Hence, the correlations could represent, indirectly, correlation of the prevalence of these comorbidities with cytokine polymorphisms.
Noteworthy, all cytokines and HLA-B alleles positively correlated with DDR reached their highest frequencies in Europe and lowest frequencies in Sub-Saharan and East Asia. The populations in these regions belongs mainly to major ethnicities and reinforces the relevance of genetic backgrounds in COVID-19 context.
4.4. Studies on host genetics
There are a few studies that associates COVID-19 with genetics factors, mostly related to the immune response, like the HLA Class I genes [32]. ABO blood group and single nucleotide polymorphisms (SNP) in 3p21.31 and 9q34.2 [33]; 19p13.3, 12q24.13 and 21q22.1 [34] regions were also recently reported as associated in genome wide association studies (GWAS). Ongoing GWAS initiatives may reveals additional key genetic markers [35].
Moreover, an exome approach identified polymorphisms at immune related genes in regions 4q35.1, 11q13.2, 19p13.3, 12q14.2, 19q13.33, 11p15.5 and 21q22.11 [36], where three of them at chromosomes 12, 19 and 21 are coincident with previous GWAS studies.
Hence, the trend towards association with immunorrelevant genes corroborates indirectly the results suggesting that immunological status would be a major player in COVID-19 context.
5. Concluding remarks
The present study provided preliminary evidence that death by COVID-19 could be associated with immunogenetic markers (HLA-B, IL6, IL10 and IL12B). However, these results should be considered for future evaluation and need to be corroborated in more structured protocols. Noteworthy, the design of further studies must consider poor prognosis underlying disorders, due to their association with these immunogenetic markers. Additionally, careful sampling strategies should be employed avoiding stratification [37], because these polymorphisms have a wide range of variability associated with ethnicity. Finally, additive effects also should be considered and approached in the evaluation of these markers.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The present work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brasil (CNPq) fellowship to Eduardo José Melo dos Santos under grant number 401235/2020-3 and was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES) under grant number 88887.198075/2018-00 fellowship to MML. Federal University of Pará provided logistics for the completion of this study.
Author Contributions Statement
All authors were actively involved in the surveys, AFND development, analysis, interpreting of results and writing of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.humimm.2021.01.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Vetter P., Vu D.L., L’Huillier A.G., Schibler M., Kaiser L., Jacquerioz F. Clinical features of covid-19. BMJ. 2020;369:1–2. doi: 10.1136/bmj.m1470. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J., Yang Y., Huang H.-P., Li D., Gu D.-F., Lu X.-F., Zhang Z., Liu L., Liu T., Liu Y.-K., He Y.-J., Sun B., Wei M.-L., Yang G.-Y., Wang X., Zhang L., Zhou X.-Y., Xing M.-Z., Wang P.G. Relationship between the ABO Blood Group and the COVID-19 Susceptibility. MedRxiv. 2020;2020(03) doi: 10.1101/2020.03.11.20031096. [DOI] [Google Scholar]
- 3.Stawiski E.W., Diwanji D., Suryamohan K., Gupta R., Fellouse F.A., Sathirapongsasuti J.F., Liu J., Jiang Y.-P., Ratan A., Mis M., Santhosh D., Somasekar S., Mohan S., Phalke S., Kuriakose B., Antony A., Junutula J.R., Schuster S.C., Jura N., Seshagiri S. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. BioRxiv. 2020:24. doi: 10.1101/2020.04.07.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen S.F., Ho Y.-C. SARS-CoV-2: A Storm is Raging. J. Clin. Invest. 2020 doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu D., Yang X.O. TH17 Responses in Cytokine Storm of COVID-19: An Emerging Target of JAK2 Inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020:17–19. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin M., Tseng H.K., Trejaut J.A., Lee H.L., Loo J.H., Chu C.C., Chen P.J., Su Y.W., Lim K.H., Tsai Z.U., Lin R.Y., Lin R.S., Huang C.H. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;4:1–7. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajeer A.H., Balkhy H., Johani S., Yousef M.Z., Arabi Y. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome - coronavirus infection. Ann. Thorac. Med. 2016:211–213. doi: 10.4103/1817-1737.185756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C., Xia C.Q. The Involvement of Natural Killer Cells in the Pathogenesis of Severe Acute Respiratory Syndrome. Am. J. Clin. Pathol. 2004;121:507–511. doi: 10.1309/WPK7Y2XKNF4CBF3R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Galarza F.F., Takeshita L.Y.C., Santos E.J.M., Kempson F., Maia M.H.T., Da Silva A.L.S., Teles E Silva A.L., Ghattaoraya G.S., Alfirevic A., Jones A.R., Middleton D. Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(2015):D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A.D. Yates, P. Achuthan, W. Akanni, J. Allen, J. Allen, J. Alvarez-Jarreta, M.R. Amode, I.M. Armean, A.G. Azov, R. Bennett, J. Bhai, K. Billis, S. Boddu, J.C. Marugán, C. Cummins, C. Davidson, K. Dodiya, R. Fatima, A. Gall, C.G. Giron, L. Gil, T. Grego, L. Haggerty, E. Haskell, T. Hourlier, O.G. Izuogu, S.H. Janacek, T. Juettemann, M. Kay, I. Lavidas, T. Le, D. Lemos, J.G. Martinez, T. Maurel, M. McDowall, A. McMahon, S. Mohanan, B. Moore, M. Nuhn, D.N. Oheh, A. Parker, A. Parton, M. Patricio, M.P. Sakthivel, A.I. Abdul Salam, B.M. Schmitt, H. Schuilenburg, D. Sheppard, M. Sycheva, M. Szuba, K. Taylor, A. Thormann, G. Threadgold, A. Vullo, B. Walts, A. Winterbottom, A. Zadissa, M. Chakiachvili, B. Flint, A. Frankish, S.E. Hunt, G. IIsley, M. Kostadima, N. Langridge, J.E. Loveland, F.J. Martin, J. Morales, J.M. Mudge, M. Muffato, E. Perry, M. Ruffier, S.J. Trevanion, F. Cunningham, K.L. Howe, D.R. Zerbino, P. Flicek, Ensembl 2020, Nucleic Acids Res. 48 (2020) D682–D688. 10.1093/nar/gkz966.
- 11.Prugnolle F., Manica A., Charpentier M., Guégan J.F., Guernier V., Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 2005;15:1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 12.Blackwell J.M., Jamieson S.E., Burgner D. HLA and infectious diseases. Clin. Microbiol. Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacey S.F., Villacres M.C., La Rosa C., Wang Z., Longmate J., Martinez J., Brewer J.C., Mekhoubad S., Maas R., Leedom J.M., Forman S.J., Zaia J.A., Diamond D.J. Relative dominance of HLA-B*07 restricted CD8+ T-lymphocyte immune responses to human cytomegalovirus pp65 in persons sharing HLA-A*02 and HLA-B*07 Alleles. Hum. Immunol. 2003;64:440–452. doi: 10.1016/S0198-8859(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 14.Boon A.C.M., de Mutsert G., Fouchier R.A.M., Sintnicolaas K., Osterhaus A.D.M.E., Rimmelzwaan G.F. Preferential HLA Usage in the Influenza Virus-Specific CTL Response. J. Immunol. 2004;172:4435–4443. doi: 10.4049/jimmunol.172.7.4435. [DOI] [PubMed] [Google Scholar]
- 15.Kiepiela P., Leslie A.J., Honeyborne I., Ramduth D., Thobakgale C., Chetty S., Rathnavalu P., Moore C., Pfafferott K.J., Hilton L., Zimbwa P., Moore S., Allen T., Brander C., Addo M.M., Altfeld M., James I., Mallal S., Bunce M., Barber L.D., Szinger J., Day C., Klenerman P., Mullins J., Korber B., Coovadia H.M., Walker B.D., Goulder P.J.R. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–774. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 16.dos Santos E.J.M., McCabe A., Gonzalez-Galarza F.F., Jones A.R., Middleton D. Allele Frequencies Net Database: Improvements for storage of individual genotypes and analysis of existing data. Hum. Immunol. 2015 doi: 10.1016/j.humimm.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: Improved Peptide–MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merino A.M., Sabbaj S., Easlick J., Goepfert P., Kaslow R.A., Tang J. Dimorphic HLA-B signal peptides differentially influence HLA-E- and natural killer cell-mediated cytolysis of HIV-1-infected target cells. Clin. Exp. Immunol. 2013;174:414–423. doi: 10.1111/cei.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallner A., Bernson E., Hussein B.A., Sander F.E., Brune M., Aurelius J., Martner A., Hellstrand K., Thorén F.B. The HLA-B 221 dimorphism impacts on NK cell education and clinical outcome of immunotherapy in acute myeloid leukemia. Blood. 2019;133:1479–1488. doi: 10.1182/blood-2018-09-874990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni S., Martin M.P., Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayres M., Ayres M., Jr, Ayres D., Santos A. Manual Biostat, 5a Edição. Belém. 2007 [Google Scholar]
- 23.Rajgor D.D., Lee M.H., Archuleta S., Bagdasarian N., Quek S.C. The many estimates of the COVID-19 case fatality rate. Lancet. Infect. Dis. 2020;3099:30244. doi: 10.1016/S1473-3099(20)30244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., Thompson R.F. Human leukocyte antigen susceptibility map for SARS-CoV-2. J. Virol. 2020 doi: 10.1128/jvi.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng M.H.L., Lau K., Li L., Cheng S., Chan W.Y., Hui P.K., Zee B., Leung C., Sung J.J.Y. Association of Human-Leukocyte-Antigen Class I (B*0703) and Class II (DRB1*0301) Genotypes with Susceptibility and Resistance to the Development of Severe Acute Respiratory Syndrome. J. Infect. Dis. 2004;190:515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan F.F., Velickovic Z., Ashton L.J., Dyer W.B., Geczy A.F., Dunckley H., Lynch G.W., Sullivan J.S. Influence of HLA gene polymorphisms on susceptibility and outcome post infection with the SARS-CoV virus. Virol. Sin. 2014;29:128–130. doi: 10.1007/s12250-014-3398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong P., Zeng X., Song M.S., Jia S.W., Zhong M.H., Xiao L.L., Lan W., Cai C., Wu X.W., Gong F.L., Wang W. Lack of association between HLA-A, -B and -DRB1 alleles and the development of SARS: A cohort of 95 SARS-recovered individuals in a population of Guangdong, southern China. Int. J. Immunogenet. 2008;35:69–74. doi: 10.1111/j.1744-313X.2007.00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng M.H.L., Cheng S.H., Lau K.M., Leung G.M., Khoo U.S., Zee B.C.W., Sung J.J.Y. Immunogenetics in SARS: A casecontrol study. Hong Kong Med. J. 2010;16:29–33. [PubMed] [Google Scholar]
- 29.Semino C., Rubartelli A. NK cell-derived cytokines and delivery: NK cell synapses. Elsevier Ltd. 2010 doi: 10.1016/B978-0-12-370454-2.00013-2. [DOI] [Google Scholar]
- 30.Zuo J., Mohammed F., Moss P. The biological influence and clinical relevance of polymorphism within the NKG2D ligands. Front. Immunol. 2018;9:1–8. doi: 10.3389/fimmu.2018.01820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, Epidemiology, Pathogenesis, and Control of COVID-19. Viruses. 2020;12:1–17. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Zhang W., Zhang J., He J., Zhu F. Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease-2019 (COVID-19) Hla. 2020;96:194–196. doi: 10.1111/tan.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D. Ellinghaus, F. Degenhardt, L. Bujanda, M. Buti, A. Albillos, P. Invernizzi, J. Fernández, D. Prati, G. Baselli, R. Asselta, M.M. Grimsrud, C. Milani, F. Aziz, J. Kässens, S. May, M. Wendorff, L. Wienbrandt, F. Uellendahl-Werth, T. Zheng, X. Yi, R. de Pablo, A.G. Chercoles, A. Palom, A.-E. Garcia-Fernandez, F. Rodriguez-Frias, A. Zanella, A. Bandera, A. Protti, A. Aghemo, A. Lleo, A. Biondi, A. Caballero-Garralda, A. Gori, A. Tanck, A. Carreras Nolla, A. Latiano, A.L. Fracanzani, A. Peschuck, A. Julià, A. Pesenti, A. Voza, D. Jiménez, B. Mateos, B. Nafria Jimenez, C. Quereda, C. Paccapelo, C. Gassner, C. Angelini, C. Cea, A. Solier, D. Pestaña, E. Muñiz-Diaz, E. Sandoval, E.M. Paraboschi, E. Navas, F. García Sánchez, F. Ceriotti, F. Martinelli-Boneschi, F. Peyvandi, F. Blasi, L. Téllez, A. Blanco-Grau, G. Hemmrich-Stanisak, G. Grasselli, G. Costantino, G. Cardamone, G. Foti, S. Aneli, H. Kurihara, H. ElAbd, I. My, I. Galván-Femenia, J. Martín, J. Erdmann, J. Ferrusquía-Acosta, K. Garcia-Etxebarria, L. Izquierdo-Sanchez, L.R. Bettini, L. Sumoy, L. Terranova, L. Moreira, L. Santoro, L. Scudeller, F. Mesonero, L. Roade, M.C. Rühlemann, M. Schaefer, M. Carrabba, M. Riveiro-Barciela, M.E. Figuera Basso, M.G. Valsecchi, M. Hernandez-Tejero, M. Acosta-Herrera, M. D’Angiò, M. Baldini, M. Cazzaniga, M. Schulzky, M. Cecconi, M. Wittig, M. Ciccarelli, M. Rodríguez-Gandía, M. Bocciolone, M. Miozzo, N. Montano, N. Braun, N. Sacchi, N. Martínez, O. Özer, O. Palmieri, P. Faverio, P. Preatoni, P. Bonfanti, P. Omodei, P. Tentorio, P. Castro, P.M. Rodrigues, A. Blandino Ortiz, R. de Cid, R. Ferrer, R. Gualtierotti, R. Nieto, S. Goerg, S. Badalamenti, S. Marsal, G. Matullo, S. Pelusi, S. Juzenas, S. Aliberti, V. Monzani, V. Moreno, T. Wesse, T.L. Lenz, T. Pumarola, V. Rimoldi, S. Bosari, W. Albrecht, W. Peter, M. Romero-Gómez, M. D’Amato, S. Duga, J.M. Banales, J.R. Hov, T. Folseraas, L. Valenti, A. Franke, T.H. Karlsen, Genomewide Association Study of Severe Covid-19 with Respiratory Failure, N. Engl. J. Med. (2020) NEJMoa2020283. 10.1056/NEJMoa2020283.
- 34.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A., Rawlik K., Pasko D., Walker S., Richmond A., Head Fourman M., Russell C.D., Law A., Furniss J., Gountouna E., Wrobel N., Moutsianas L., Wang B., Meynert A., Yang Z., Zhai R., Zheng C., Griffiths F., Oosthuyzen W., Grimes G., Shih B., Keating S., Zechner M., Haley C., Porteous D.J., Knight J., Summers C., Shankar-Hari M., Klenerman P., Rowan K., Murphy L., Ponting C.P., Tenesa A., Caulfield M., Scott R. Genetic mechanisms of critical illness in Covid-19. MedRxiv. 2020;17:25. [Google Scholar]
- 35.Ovsyannikova I.G., Haralambieva I.H., Crooke S.N., Poland G.A., Kennedy R.B. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol. Rev. 2020;296:205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.-H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.-L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.-C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.-E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Gallego C.R., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., Snow A.L., Dalgard C.L., Milner J., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R., Zhang S.-Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.-L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science (80-) 2020;4570:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J.N. Hellwege, J.M. Keaton, A. Giri, X. Gao, D.R. Velez Edwards, T.L. Edwards, Population Stratification in Genetic Association Studies, Curr. Protoc. Hum. Genet. 95 (2017) 1.22.1-1.22.23. 10.1002/cphg.48 [DOI] [PMC free article] [PubMed]
- 38.Nokso-Koivisto J., Patel J.A., Chonmaitree T. IL-6 -174 C/C Genotype Is Not Conclusively a Low IL-6 Production Phenotype. J. Infect. Dis. 2011;203:1876–1878. doi: 10.1093/infdis/jir181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeng J.R., Wang J.H., Liu W.S., Chen S.P., Chen M.Y.C., Wu M.H., Hsu W.L., Lin S.Z. Association of interleukin-6 gene G-174C polymorphism and plasma plasminogen activator inhibitor-1 level in Chinese patients with and without hypertension. Am. J. Hypertens. 2005;18:517–522. doi: 10.1016/j.amjhyper.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 40.Antonicelli R., Olivieri F., Bonafè M., Cavallone L., Spazzafumo L., Marchegiani F., Cardelli M., Recanatini A., Testarmata P., Boemi M., Parati G., Franceschi C. The interleukin-6 -174 G>C promoter polymorphism is associated with a higher risk of death after an acute coronary syndrome in male elderly patients. Int. J. Cardiol. 2005;103:266–271. doi: 10.1016/j.ijcard.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 41.Bouhaha R., Baroudi T., Ennafaa H., Vaillant E., Abid H., Sassi R., Vatin V., Froguel P., el Gaaied A.B., Meyre D., Vaxillaire M. Study of TNFα-308G/A and IL6 -174G/C polymorphisms in type 2 diabetes and obesity risk in the Tunisian population. Clin. Biochem. 2010;43:549–552. doi: 10.1016/j.clinbiochem.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Goyenechea E., Parra D., Martínez J.A. Impact of interleukin 6–174G>C polymorphism on obesity-related metabolic disorders in people with excess in body weight. Metabolism. 2007;56:1643–1648. doi: 10.1016/j.metabol.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Kremer K.N., Kumar A., Hedink K.E. Haplotype-independent co-stimulation of IL-10 secretion by SDF-1/CXCL12 proceeds via AP-1 binding to the human IL-10 promoter. J Immunol. 2007;178:1581–1588. doi: 10.4049/jimmunol.178.3.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai H., Jing D., Guo A., Yin S. Association between interleukin 10 gene polymorphisms and risk of type 2 diabetes mellitus in a Chinese population. J. Int. Med. Res. 2014;42:702–710. doi: 10.1177/0300060513505813. [DOI] [PubMed] [Google Scholar]
- 45.Cruz M., Fragoso J.M., Alvarez-León E., Escobedo-de-la-Peña J., Valladares A., Juárez-Cedillo T., Pérez-Méndez O., Vargas-Alarcón G. The TGF-B1 and IL-10 gene polymorphisms are associated with risk of developing silent myocardial ischemia in the diabetic patients. Immunol. Lett. 2013;156:18–22. doi: 10.1016/j.imlet.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Eskandari-Nasab E., Moghadampour M., Asadi-Saghandi A., Kharazi-nejad E., Rezaeifar A., Pourmasoumi H. Levels of Interleukin-(IL)-12p40 are Markedly increased in Brucellosis among patients with specific IL-12B genotypes. Scand. J. Immunol. 2013;78:85–91. doi: 10.1111/sji.12054. [DOI] [PubMed] [Google Scholar]
- 47.Eirís N., González-Lara L., Santos-Juanes J., Queiro R., Coto E., Coto-Segura P. Genetic variation at IL12B, IL23R and IL23A is associated with psoriasis severity, psoriatic arthritis and type 2 diabetes mellitus. J. Dermatol. Sci. 2014;75:167–172. doi: 10.1016/j.jdermsci.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Chen T., Liang W., Gao L., Wang Y., Liu Y., Zhang L., Zhang L. Association of single nucleotide polymorphisms in interleukin 12 (IL-12A and -B) with asthma in a Chinese population. Hum. Immunol. 2011;72:603–606. doi: 10.1016/j.humimm.2011.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.