Abstract
Polycystic ovary syndrome (PCOS) is a common endocrine and metabolic disease in premenopausal women. Long non-coding RNAs (lncRNAs) constitute important factors in numerous biological processes. However, their roles in PCOS pathogenesis require further clarification. Our study aims to elucidate the roles of lncRNA lnc-CCNL1-3:1 (CCNL) in PCOS. CCNL expression in human luteinized granulosa cells (hLGCs) derived from women with and without PCOS was detected. The full length of CCNL was obtained by 5′ and 3′ rapid amplification of cDNA ends. CCNL roles in granulosa cell apoptosis, mitochondrial function, and glucose uptake were evaluated. The binding relationship between CCNL and forkhead box O1 (FOXO1) was determined by RPISeq, RNA immunoprecipitation, subcellular fractionation, and immunofluorescence. In KGN cells and hLGCs, CCNL overexpression upregulated FOXO1 expression, promoted cell apoptosis, reduced glucose transport capability, and impaired mitochondrial function, and these effects were partially abolished by silencing FOXO1. The interaction of CCNL with FOXO1 might prevents FOXO1 exclusion from the nucleus and subsequent degradation in the cytosol. We determined that CCNL serve as a facilitator in the processes of PCOS. CCNL might participate in PCOS pathologies such as follicular atresia and insulin resistance.
Keywords: LncRNA, Polycystic ovary syndrome, Granulosa cells, Follicular atresia, Insulin resistance
Graphical abstract
Polycystic ovary syndrome (PCOS), a common endocrine disease in premenopausal women, is associated with infertility and metabolic disorders such as obesity. Huang, Du, and colleagues reported that long non-coding RNA lnc-CCNL1-3:1 serves as a facilitator and mediator and is involved in the pathologies of PCOS, including follicular atresia and insulin resistance.
Introduction
Polycystic ovary syndrome (PCOS), one of the most common endocrine and metabolic disorders, has a prevalence of 6%–20% in women of reproductive age.1,2 PCOS is characterized by signs and symptoms of hyperandrogenemia, chronic oligo-ovulation/anovulation, and polycystic ovarian morphology on exclusion of other specific diagnoses.1 PCOS is commonly associated with reproductive co-morbidities such as female infertility, menstrual irregularity, ovulatory dysfunction, and pregnancy complications, along with metabolic disorders such as insulin resistance, metabolic syndrome, and type 2 diabetes, as well as psychological risk factors such as depression.2
Follicular atresia, a common process in female mammalian ovaries, constitutes the degeneration of follicles at any stage of growth and development.3 Follicles consist of granulosa cells, theca internal cells, and oocytes. The granulosa cell layers surround the oocytes, providing them with nutrients and regulators during oocyte development.2,4,5 Granulosa cell apoptosis is the major cause of follicular atresia, inducing DNA degradation and caspase activation.6,7 Women with PCOS exhibit more ripening and subsequent atretic follicles from an early stage,8 representing follicles that failed to develop into dominant follicles, along with follicular hypertrophy and degradation, concordant with the abnormal apoptosis and proliferation of granulosa cells in PCOS.1,2,9 Thus, granulosa cell apoptosis promotes abnormalities in PCOS folliculogenesis, which lead to anovulation and infertility.
Oxidative stress is closely associated with the levels of reactive oxygen species (ROS). In mitochondria, electron transfer between cytochrome c and p66Shc generates ROS,10 which regulate the ratio of B cell lymphoma-2 (Bcl-2)-associated X (Bax)/Bcl-2 expression, resulting in impaired mitochondrial membrane potential and caspase-induced apoptosis.11,12 Notably, mutations in different complexes of the electron transport chain led to the accumulation of ROS.11 In addition, patients with PCOS display features of mitochondrial impairment and oxidative stress as highlighted by elevated ROS production.13, 14, 15 Furthermore, oxidative stress-induced apoptosis has long been reported to play a vital role in follicular atresia. Specifically, increased ROS levels cause premature ovarian insufficiency and follicular atresia in the human ovary.16 Apoptosis and protein oxidation were also shown to be increased in sheep and mouse ovarian follicles during follicular atresia.17,18 These observations suggest that oxidative stress-induced granulosa cell apoptosis might contribute to the aberrant folliculogenesis observed in PCOS.
Insulin resistance, a clinical feature of PCOS, is defined as a decreased ability of insulin to mediate metabolic actions on glucose uptake, production, and lipolysis. Insulin resistance appears to constitute an important factor in the pathogenesis of PCOS.19 In women with PCOS, insulin resistance tends to worsen over time and is associated with the development of obesity and type 2 diabetes.20 In PCOS plus obesity, the capability of insulin-mediated glucose uptake of adipocytes is reduced, indicating a decrease in insulin sensitivity.21,22 Moreover, non-obese women with PCOS also suffer from metabolic disturbances and the risk of long-term metabolic complications.23 Thus, the associated metabolic disorders, obesity, and type 2 diabetes have recently become among the most important long-term concerns in PCOS and warrant increased attention.
Insulin resistance and compensatory hyperinsulinemia contribute to premature granulosa cell luteinization,24 leading to the arrest of cell proliferation and follicle growth in PCOS.25 Granulosa cells in the ovary are responsible for providing intermediates and energy substrates to the oocytes. Normal glucose metabolism in granulosa cells is essential for oocyte development, maturation, and protection.26 Previous studies have reported that human ovary tissues such as granulosa cells and endometrium from patients with PCOS showed reduced glucose uptake.27, 28, 29 The insulin resistance of granulosa cells may thus influence granulosa cell function, thereby impairing the development potential of the oocytes.30,31
Recently, the field of non-coding RNAs (ncRNAs) has markedly expanded, with the focus during the past decade on small ncRNAs such as microRNAs increasingly shifting to the analysis of long ncRNAs (lncRNAs), defined as ncRNAs with transcripts >200 nt.32 lncRNAs are emerging as vital regulators in abundant biological processes such as nuclear architecture, epigenetic modifiers, protein binding, and transcription in the nucleus, and molecular decay, translation, and post-translational modifications in the cytoplasm.32, 33, 34 lncRNAs are proposed as being involved in folliculogenesis, including cumulus expansion,35 luteinization,36,37 and oocyte development and maturation.38 In women with PCOS, lncRNAs have been found to regulate cell proliferation,39,40 apoptosis,41,42 endocrine function,40,42,43 and metabolism.44, 45, 46, 47, 48 Nevertheless, few studies have clarified the relationship between lncRNAs and PCOS. Thus, additional research related to the function of lncRNAs in the pathogenesis of PCOS is needed.
In this study, we focused on the potential roles and underlying mechanisms of lncRNAs in PCOS. This work derives from our previous microarray analysis of differentially expressed lncRNA profiles in human luteinized granulosa cells (hLGCs) in women with and without PCOS (GEO: GSE114419).39 Among these differentially expressed lncRNAs, intergenic lncRNA lnc-CCNL1-3:1 (CCNL), located on chromosome chr3:157131726–157132376 (Figure S1), is highly expressed in the ovary and liver (http://www.noncode.org/show_rna.php?id=NONHSAT092887&version=2&utd=1#), indicating a potential role in PCOS. Moreover, the PhyloCSF score suggested that CCNL is likely to constitute a ncRNA (Figure S2). In the present study, we found that CCNL was elevated in patients with PCOS and evaluated the effects of CCNL on PCOS pathogenesis using hLGCs and the human granulosa cell tumor-derived cell line, KGN. CCNL was found to promote granulosa cell apoptosis and suppress glucose uptake, partly contributing to the occurrence of follicular atresia and insulin resistance in PCOS. Our results regarding the function of dysregulated lncRNAs in PCOS provide new insight with respect to follicular atresia and insulin resistance in PCOS.
Results
lnc-CCNL1-3:1 expression is increased in granulosa cells of women with PCOS
The full length of CCNL was successfully obtained by 5′ and 3′ rapid amplification of cDNA ends (RACE) assays (Figure S3). We confirmed the full-length sequence of CCNL by running polymerase chain reaction (PCR) with three pairs of fragmented primers (Table S3) and compared our results with the CCNL sequence from the NONCODE database (Figure S4). Thus, the whole sequence of CCNL was validated and is provided in Table 1.
Table 1.
Full-length sequence of CCNL from RACE
| RNA sequence (lnc-CCNL1-3:1) |
|---|
| AGCTCTTCAGTGTTGTGCTCATTGGTTTCATCAAAGCAGGACAATACTTGGCACTAAGTCGACAAAATTCTCACTTCTGCAGTGGGTCTCTAGATGTCCATGACCCTGCCTTCTCAGGAAAAGAGGGAGGACATGCGGCTGGAGGGACACAGAGGGCAAGACTGCATCCCAGTTACAACACCAGGAAGCCACGTCTGAAAGGAAACTCCTCAGAAACCTGGGCATAGGCTATCGGGAAAGTAAAGAAGACATCTGGAGAGGCGAATATTCTGAAGGAATCTCTGCCAGAGGAGAAGGGAAAAGGAAAGCACTTATCCAGGTCAAAGGCATGAGTTTCCAGGAGATTATTCATCGTGACTAGAGGATGACTGTTGACTAGATTTATTCCACAGGTATGGAAGGAAAAGTCTTGGTACATTTTTTGCAGCATTTCCTCCTCTCCATATAGACTGTTCTCATTCTGCAGGGCTGGGGTTCACCCAGTATTCAACTCCAAGCCCTTCACATAGTAAAAGAAGTAAGGGGAGCCAGGCATGGTGGCTTACACCTGTAAACCCAGCACTTTGGGAGGGCAAAGCAGGCAGATTCATTGAGCTCAGGAGTTCGACCCCAACCTGGGCAACATGGTGAAACCCTGTCTCTACTAAAAATACAAAAATTAGCTGGGTGTGGTGGTGCATGCCTGTGGTCCCAGCTACTCTGGAGGCTGAGGTGGTAGGATTGCTTGAGCCCACATGTGTGAGGCTGCAGTGAGCTGTGATCACACCACTGCTCTCCAGCCTGGGTGACAGAGTGAGAGACCCTGTCCCCCCAACCACCCCCCCCAACAAAAAAAAAAAAAA |
RACE, rapid amplification of cDNA ends (5′ and 3′).
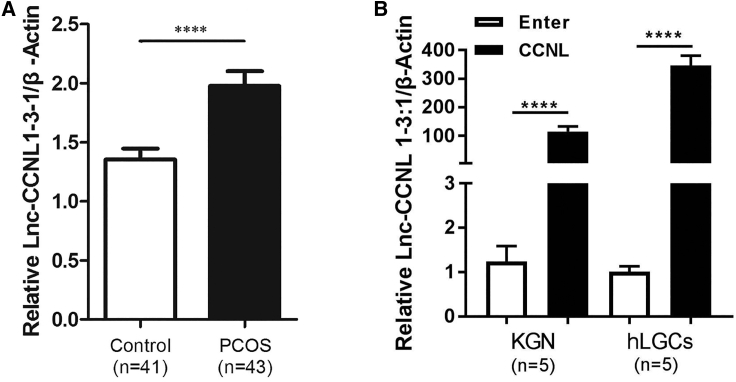
In our previous study,39 we found that the expression levels of CCNL were increased in patients with PCOS (p < 0.05) as revealed by microarray and quantitative real-time PCR analysis. To further validate these results, in the present study we analyzed the expression of CCNL by quantitative real-time PCR in a cohort of 43 women with PCOS and 41 controls (Figure 1A). The CCNL expression level was elevated in hLGCs of women with PCOS. The major anthropometric variables of the two groups are presented in Table 2. No significant difference was observed with regard to age and body mass index (BMI) between women with PCOS and the control group. Conversely, women with PCOS exhibited significantly higher serum levels of testosterone, luteinizing hormone, and anti-Müllerian hormone compared with those of the control group.
Figure 1.
lnc-CCNL1-3:1 is upregulated in granulosa cells of women with PCOS
(A) The expression level of CCNL in 43 women with PCOS and 41 healthy controls. The expression level was detected via quantitative real-time PCR. The first ΔCt normalization was to β-actin, and the second ΔCt normalization was to the control group. (B) Bar graph showing CCNL vector-mediated overexpression efficiency. KGN cells or hLGCs stably expressing empty vector (Enter) or CCNL overexpression vectors (CCNL) were cultured 72 h and then subjected to quantitative real-time PCR and normalized against β-actin. Error bars represent SDs of at least three independent experiments. ∗∗∗∗p < 0.0001 corresponds to two-tailed Student’s tests.
Table 2.
Relative clinical characteristics of PCOS and control groups
| Control (n = 41) | PCOS (n = 43) | p value | |
|---|---|---|---|
| Age (years) | 28.63 ± 2.79 | 27.81 ± 3.17 | 0.192 |
| BMI (kg/m2) | 23.89 ± 3.09 | 22.73 ± 3.34 | 0.095 |
| Basal FSH (IU/L) | 6.54 ± 1.38 | 6.19 ± 1.37 | 0.235 |
| Basal LH (IU/L) | 4.93 ± 1.80 | 7.86 ± 3.77 | <0.0001 |
| Basal T (ng/dL) | 19.71 ± 7.05 | 37.98 ± 19.28 | <0.0001 |
| AMH (ng/mL) | 4.52 ± 2.51 | 10.85 ± 5.89 | <0.0001 |
| Retrieved oocytes | 11.78 ± 3.65 | 17.17 ± 7.24 | <0.0001 |
All data are mean ± SD value. BMI, body mass index; FSH, follicle-stimulating hormone; LH, luteinizing hormone; T, testosterone; AMH, anti-Müllerian hormone. The Kolmogorov-Smirnov test was used to assess the distributions of relevant variables. Statistical comparisons between two variables (the PCOS group and the control group) were performed using the Student’s t test for normal distribution, whereas an unpaired test and the Mann-Whitney U test were used for continuous variables.
The abundance of CCNL in the granulosa cells of women with PCOS was significantly positively associated (R2 = 0.6044, p < 0.0001) with the concentration of testosterone in the serum (Figure S5A). To identify the potential for androgen-induced CCNL, we conducted quantitative real-time PCR on KGN cells that had been cultured in different concentration of double hydrogen testosterone (DHT) medium (0.1 or 10 mM) for 24 h (Figure S5B). We found that the expression level of CCNL in KGN cells was increased in the higher DHT group (10 mM).
Overexpressed lnc-CCNL1-3:1 promotes apoptosis of granulosa cells in PCOS
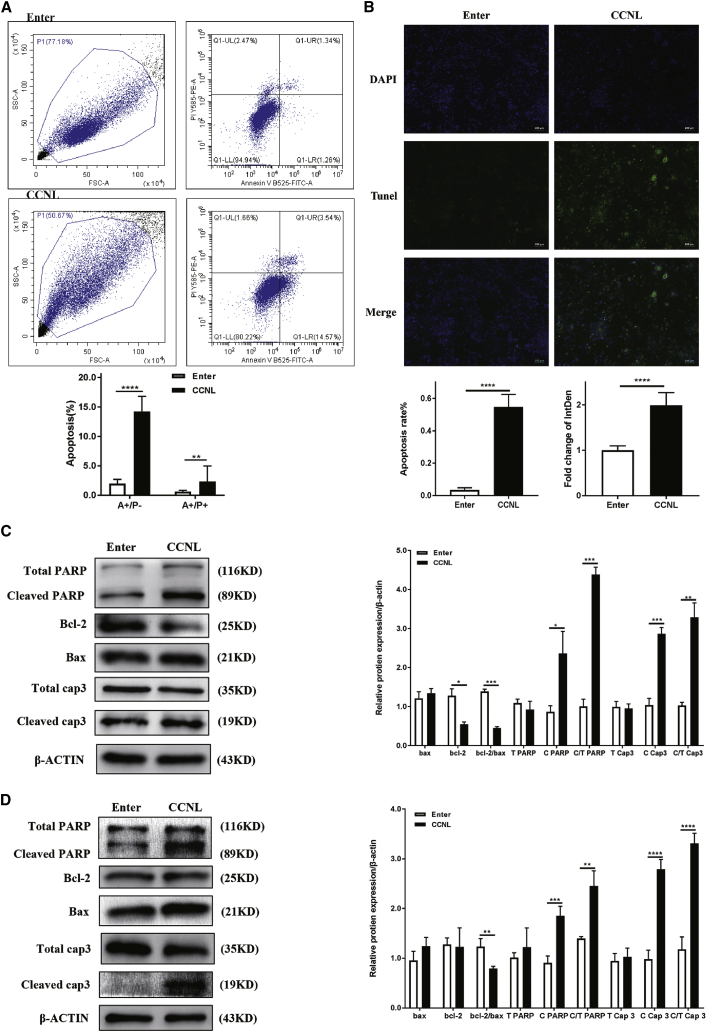
As CCNL was a previously unexplored lncRNA, its biological functions were unknown. To investigate the biological significance of CCNL in the development of PCOS, we generated KGN cells and hLGCs exhibiting overexpression of CCNL. CCNL vector-mediated overexpression efficiencies are shown in Figure 1B. KGN cells or hLGCs expressing empty vector (Enter) or CCNL-overexpression vector were cultured for 72 h and then subjected to quantitative real-time PCR. To evaluate the effects of CCNL on cell apoptosis, the Enter- and CCNL-transfected granulosa cells were subjected to annexin V/propidium iodide (PI) staining followed by flow cytometry to measure the percentages of annexin V-positive/PI-negative (A+/P−) and annexin V-positive/PI-positive (A+/P+) cells (Figure 2A). TUNEL (terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling) staining was also used to detect the apoptotic hLGCs (Figure 2B). Consistent with these results, western blot analyses of relative protein expression showed that overexpression of CCNL decreased the abundance of Bcl-2, whereas it increased the expression of Bax, cleaved poly(ADP-ribose) polymerase (PARP), and cleaved caspase-3 in both KGN cells and hLGCs (Figures 2C and 2D). The expression level of total PARP and total caspase-3 remained unchanged following transfection with CCNL. These data suggested that overexpressed CCNL promotes cell apoptosis in KGN cells and hLGCs.
Figure 2.
lnc-CCNL1-3:1 promoted granulosa cell apoptosis in vitro
(A and C) lnc-CCNL1-3:1 promoted cell apoptosis in KGN cells. (B and D) lnc-CCNL1-3:1 promoted cell apoptosis in hLGCs. (A) Enter- or CCNL-overexpression vector (CCNL)-transfected KGN cells were cultured in medium for 72 h and then subjected to annexin V/PI staining followed by fluorescence-activated cell sorting (FACS) analysis to measure the percentages of annexin V-positive/PI-negative (A+/P−) and annexin V-positive/PI-positive (A+/P+) cells or (C) to western blotting analysis to measure relative protein expression (cleaved/total PARP [C/T PARP], cleaved/total caspase-3 [C/T Cap3], Bcl-2, Bax). (B and D) Enter- or CCNL-transfected hLGCs were cultured in medium for 72 h before being subjected to (B) TUNEL (terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling) staining (green) to detect cell apoptosis and DAPI staining (blue) to detect the amount of cells (n = 3) or (D) to western blotting analysis to measure relative protein expression (C/T PARP, C/T Cap3, Bcl-2, Bax). Error bars represent SDs of at least three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 correspond to two-tailed Student’s tests.
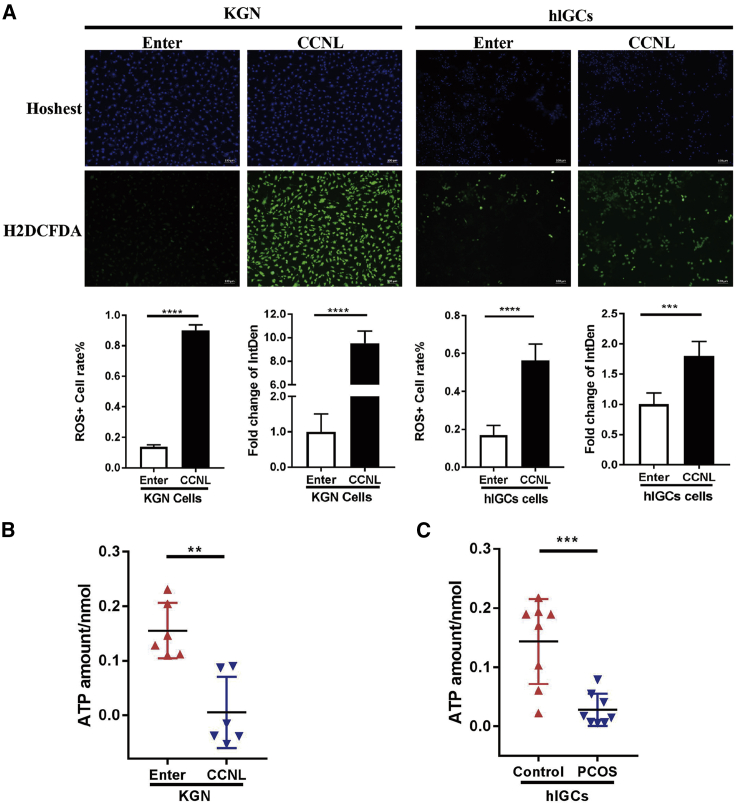
Overexpressed lnc-CCNL1-3:1 impairs mitochondrial function of granulosa cells in PCOS
As the hub proteins Bcl-2 and Bax are involved in the mitochondrial apoptotic pathway,12 their altered expression in CCNL-transfected granulosa cells inspired us to further investigate the potential role of CCNL in mitochondrial function. We compared the mitochondrial function in CCNL-overexpressed KGN cells and hLGCs with that of controls. The results showed that the ROS generation level in CCNL-overexpressed cells was significantly higher than that in the controls (Figure 3A), whereas the adenosine triphosphate (ATP) concentration was lower (Figure 3B). Because little is known regarding the ATP levels in PCOS, we also confirmed that the ATP concentration was lower in women with PCOS compared to that of controls (Figure 3C). Taken together, these results indicated that overexpressed CCNL impairs cell mitochondrial function.
Figure 3.
lnc-CCNL1-3:1 impaired cell mitochondrial function in vitro
(A) lnc-CCNL1-3:1 regulates cell reactive oxygen species (ROS) generation in KGN cells and hLGCs. Enter- or CCNL-transfected cells were cultured in medium for 72 h and then subjected to H2DCFDA staining (green) and Hoechst staining (blue). The level of intracellular ROS production was detected by fluorescence intensity of H2DCFDA and Hoechst 33342 to detect the amount of cells. (B) The concentration of intracellular ATP in Enter- or CCNL-transfected KGN cells. The level of ATP was detected via an Infinite M200 PRO microplate reader. (C) Concentration of intracellular ATP in granulosa cells of eight PCOS patients and eight healthy controls. Error bars represent SDs of at least three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 correspond to two-tailed Student’s tests.
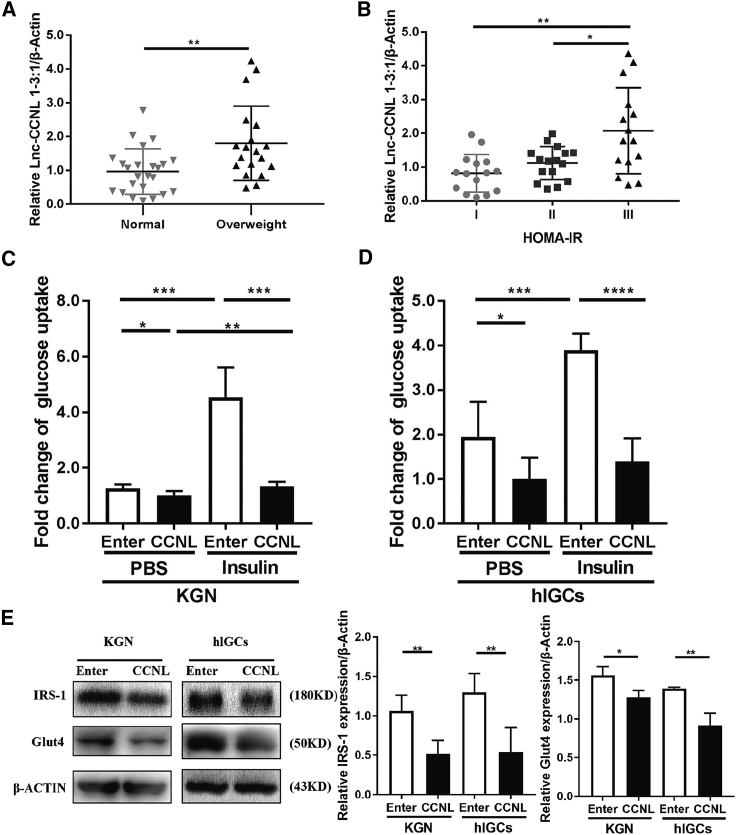
Overexpressed lnc-CCNL1-3:1 suppresses glucose uptake of granulosa cells in PCOS
As previously reported, mitochondrial function plays a vital role in insulin resistance.49 To verify whether CCNL is potentially involved in the etiology of PCOS with insulin resistance, we first analyzed relative CCNL expression in ovarian granulosa cells from a cohort of 43 women with PCOS. The relative clinical variables and endocrine parameters of the patients are presented in Table S5. The expression level of CCNL increased in patients with PCOS concomitant with elevated BMI (p = 0.0070) (Figure 4A). Ranges of homeostasis model assessment of insulin resistance (HOMA-IR) values in patients with PCOS were from 0.71 to 2.04 for tertile I, 2.04 to 3.89 for tertile II, and 3.89 to 20.73 for tertile III (Table S6). The ovarian granulosa cells of patients with PCOS exhibiting with the higher HOMA-IR values demonstrated higher expression levels of CCNL (p = 0.0784, tertile I versus II; p = 0.0017, tertile I versus III; p = 0.0267, tertile II versus III) (Figure 4B). We then ascertained whether CCNL affected cell glucose transport. The capabilities of glucose uptake at basal and insulin-stimulating levels were both impaired in CCNL-transfected granulosa cells compared to those of controls (Figures 4C and 4D). Detection of the expression of key proteins involved in glucose uptake (insulin receptor substrate 1 [IRS1] and glucose transporter 4 [GLUT4]) by western blot revealed that their abundance decreased when CCNL was overexpressed (Figure 4E).
Figure 4.
lnc-CCNL1-3:1 suppressed cell glucose uptake in vitro
(A) The expression level of CCNL was correlated with BMI in patients with PCOS (p = 0.0070). (B) The higher HOMA-IR values had a higher expression level of CCNL (p = 0.0784, tertile I versus II; p = 0.0017, tertile I versus III; p = 0.0267, tertile II versus III). (C and D) Basal and insulin-stimulated glucose uptake levels were reduced in CCNL-transfected KGN cells and hLGCs. The cells were serum starved for 12 h following insulin stimulation (100 nM, 20 min) or PBS as control before the detection. (E) Western blotting analysis to measure relative protein expression (GLUT4, IRS1) from KGN cells and hLGCs after transfection with vectors for 72 h. Error bars represent SDs of at least three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 correspond to two-tailed Student’s tests.
FOXO1 is involved in lnc-CCNL1-3:1-mediated regulation of granulosa cells in PCOS
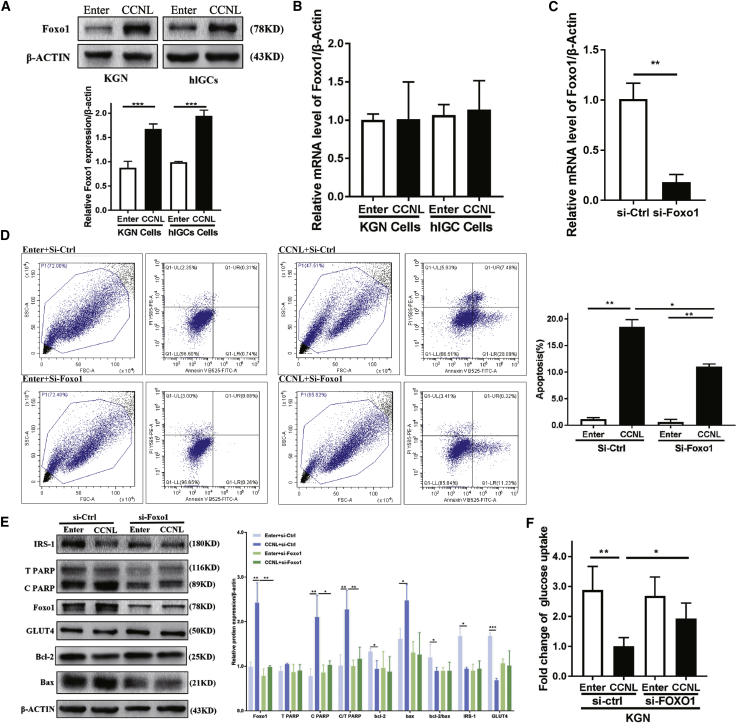
Forkhead box O1 (FOXO1) constitutes an important transcriptional factor and an RNA-binding protein (RBP) for inducing the expression of a series of genes involved in cell cycle arrest, apoptosis, insulin resistance, and oxidative stress.50, 51, 52, 53, 54 FOXO1 is highly expressed in the ovary, adipose tissues, fat, and endometrium (https://www.ncbi.nlm.nih.gov/gene/2308/?report=expression). Recent studies found that FOXO1 is involved in follicular development through the regulation of apoptosis and proliferation of granulosa cells.17,27 FOXO1 activity is regulated by subcellular localization and posttranslational modifications such as nuclear translocation and dephosphorylation.55 We found that CCNL regulated the expression of FOXO1. The overexpression of CCNL in KGN cells and hLGCs significantly increased the expression of FOXO1 at the protein level, whereas the mRNA level remained unchanged (Figures 5A and 5B). Moreover, CCNL and FOXO1 were predicted to exhibit high affinity as determined using the online algorithm RPISeq (http://pridb.gdcb.iastate.edu/RPISeq/index.html) (Table S7).
Figure 5.
FOXO1 was involved in CCNL regulation of granulosa cells
(A) The protein level of FOXO1 was upregulated in CCNL-transfected KGN cells and hLGCs. Enter- or CCNL-transfected cells were cultured 72 h then subjected to western blotting. (B) The mRNA level of FOXO1 remained unchanged via quantitative real-time PCR. (C–F) CCNL-induced cell apoptosis and glucose uptake are partially attenuated by knocking down FOXO1 in granulosa cells. (C) Bar graph showing FOXO1-siRNA-mediated knockdown efficiency by quantitative real-time PCR analysis. KGN cells stably transfecting si-Ctrl or si-FOXO1 were cultured 72 h and then subjected to quantitative real-time PCR and normalized against β-actin. (E) Knocking down FOXO1 in granulosa cells can partially rescue the CCNL-induced relative protein expression. Enter- or CCNL-transfected cells were treated with si-Ctrl or si-FOXO1 for 72 h and then subjected to western blotting analysis to measure relative protein expression (C/T PARP, Bcl-2, Bax, IRS1, GLUT4). (D and F) Knocking down FOXO1 in granulosa cells can partially rescue the CCNL effect on cell apoptosis and glucose transport. Enter- or CCNL-transfected cells were treated with si-Ctrl or si-FOXO1 for 72 h and then subjected to annexin V/PI staining followed by FACS analysis or glucose uptake assay. Error bars represent SDs of at least three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 correspond to two-tailed Student’s tests.
Therefore, we conducted series of rescue experiments to determine whether FOXO1 is involved in CCNL-induced cell apoptosis. KGN cells overexpressing CCNL followed by FOXO1 knockdown were cultured for 72 h and then subjected to quantitative real-time PCR and western blot. The efficiency of the FOXO1 small interfering RNA (siRNA)-mediated knockdown is shown in Figure 5C. Western blot analysis showed that the effects of overexpressed CCNL on relative Bcl-2, Bax, cleaved PARP, IRS1, and GLUT4 protein abundance were rescued by the siRNA-mediated knockdown of FOXO1 (Figure 5E). Moreover, the effects of CCNL overexpression on cell apoptosis (Figure 2) and glucose transport (Figure 4) were attenuated by FOXO1 knockdown (Figures 5D and 5F). These results suggested that CCNL promotes granulosa cell apoptosis and glucose uptake at least partly through an increase in FOXO1 expression.
lnc-CCNL1-3:1 interaction with FOXO1 enhances its nuclear location and expression
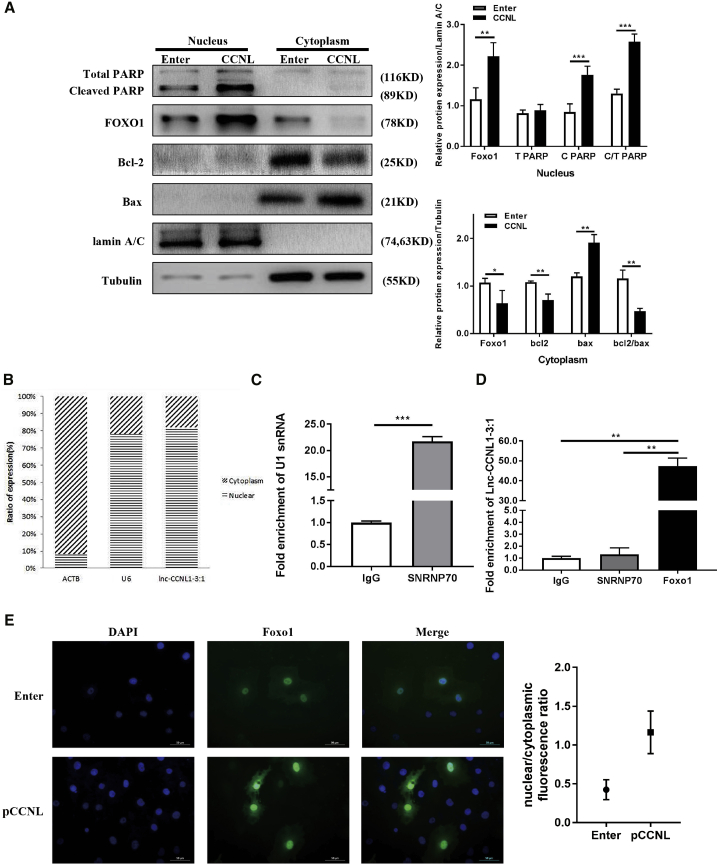
To explore the molecular mechanism underlying the regulation of FOXO1 by CCNL, we first detected the subcellular localization of CCNL in granulosa cells. The sequence-based tool iLoc-LncRNA predicted that the subcellular location of CCNL is nucleolus, nucleus, nucleoplasm (probability score, 0.694883) (http://lin-group.cn/server/iLoc-LncRNA/pre.php). RNA from nuclear and cytoplasmic fractions was reverse transcribed and detected via quantitative real-time PCR, revealing that CCNL was predominantly distributed in the nucleus (Figure 6B). ACTB and U6 were used as cytoplasm and nucleus controls, respectively.
Figure 6.
lnc-CCNL1-3:1 interaction with FOXO1 by enhancing its nuclear location
(A) Nuclear and cytoplasmic fractions of Enter- or CCNL-transfected cells were subjected to western blotting analysis to measure relative protein expression. The cytoplasmic tubulin and nuclear lamin A/C were used as controls. (B) Nuclear and cytoplasmic fractions of granulosa cells were subjected to quantitative real-time PCR to detect the intracellular localization of CCNL. The cytoplasmic ACTB and nuclear U6 were used as controls. (C) The efficiency of RNA immunoprecipitation (RIP) is shown as quantitative real-time PCR of U1 snRNA. (D) Enrichment of CCNL in granulosa cells was measured by quantitative real-time PCR from RIP with FOXO1 antibody or immunoglobulin G (IgG). IgG immunoprecipitation (IP) was used as a negative control. (E) Cells were incubated with CCNL (pCCNL) or Enter for 72 h. Subcellular localization of FOXO1 was visualized using anti-FOXO1 (green), and the nuclei were counterstained with DAPI (blue). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 correspond to two-tailed Student’s tests.
Next, we identified the intracellular localization of related proteins (Figure 6A). The cytoplasmic and nuclear proteins were separated and subjected to western blot analysis. FOXO1 protein tends to re-translocate in the nucleus after CCNL overexpression. Moreover, the abundance of FOXO1 increased in the nucleus but decreased in the cytoplasm in overexpressed CCNL cells (Figure 6A). Cleaved PARPs were primarily located in the nucleus, and its expression levels were also elevated following CCNL overexpression. In comparison, Bcl-2/Bax were distributed in the cytoplasm, and this localization decreased/increased, respectively, following CCNL overexpression. Lamin A/C and tubulin were used as nucleus and cytoplasm controls.
Notably, CCNL was shown to possess the ability to bind to FOXO1 (Table S7). The full-length sequence of CCNL was validated via a RACE assay (Table 1; Figure S3). Khail et al.56 reported that approximately 20% of lncRNAs bind to PRC2, which includes EZH2. We therefore used the EZH2-HOTAIR interaction pair as a positive control (interaction probability, 0.75). Using RPISeq, we found that the CCNL-FOXO1 interaction pair had a higher score (interaction probability, 0.75) (Table S7) than that of CCNL-EZH2 (interaction probability, 0.65) (Table S8). Therefore, we hypothesized that CCNL may interact with FOXO1 in granulosa cells. An RNA immunoprecipitation (RIP) assay to verify this hypothesis revealed that enrichment of CCNL in the precipitates of FOXO1 substantially increased compared with that of the immunoglobulin G (IgG) control (Figure 6D). The efficiency of RIP results is shown in Figure 6C using SNRNP70 as a positive control. Immunofluorescence staining further illustrated that CCNL-transfected cells exhibited higher FOXO1 expression in the nucleus (Figure 6E).
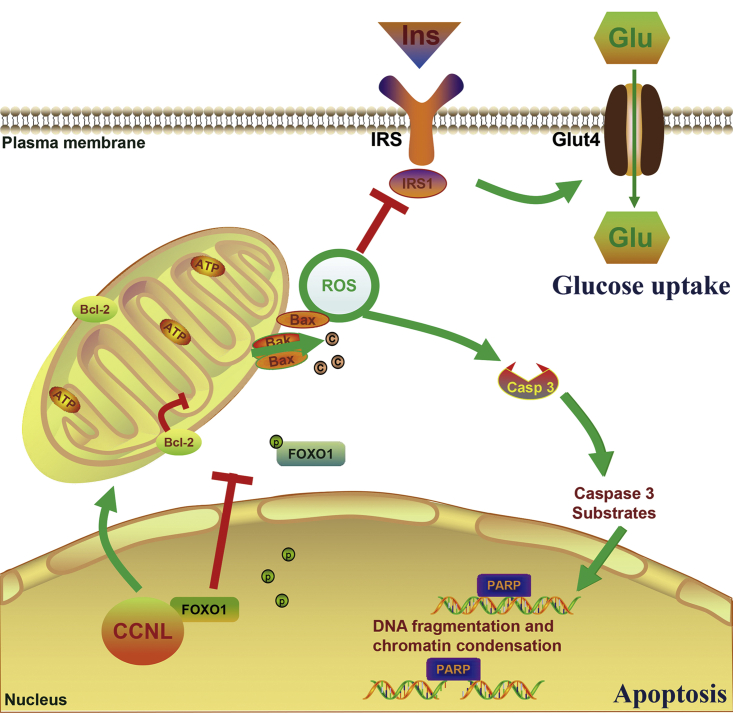
These results suggested that CCNL may interact with FOXO1 and regulate its activity. Hence, we elucidated that CCNL induces cell apoptosis and reduces glucose uptake in granulosa cells partly by enhancing FOXO1 nuclear location and expression (Figure 7).
Figure 7.
Schematic diagram for lnc-CCNL1-3:1 function in women with PCOS
Upregulation of CCNL interacts with the transcription factor FOXO1, impairs the mitochondria function, promotes cell apoptosis, and reduces glucose uptake in women with PCOS.
Discussion
In the present study, we revealed that CCNL, an intergenic lncRNA, was upregulated in PCOS and likely contributed to the associated follicular atresia and insulin resistance in this condition. Our results further indicated that alternation of CCNL expression may participate in the PCOS pathological process.
lncRNAs are emerging as important regulatory elements that control cell growth, development, and metabolic function in human disease.34,57,58 Accordingly, an increasing number of studies have demonstrated that lncRNAs are involved in the development of PCOS as well. In particular, three general aspects of lncRNA function in PCOS have emerged. The first relates to cell growth, development, and apoptosis. LINC-01572:2839 and lncRNA HCG2640 were found to influence cell proliferation and cell-cycle progression in the granulosa cells of PCOS. Han et al.41 indicated that lncRNA-LET inhibits cell viability and migration whereas it induces apoptosis. lncRNA SRA (steroid receptor RNA activator)42 has also been reported as a regulator of cell growth and apoptosis in mouse granulosa cells. The second aspect deals with steroid genesis, secretion, and receptor function. lncRNA HCG2640 and lncRNA SRA42 affect steroidogenesis, as well as estradiol and progesterone secretion in granulosa cells. CTBP1-AS43 positively associates with androgen receptor activity and serum testosterone level in PCOS. Finally, the third aspect is related to metabolic processes. lncRNA SRA positively associates with obesity in women with PCOS.45 Moreover, Jiao et al.44 conducted RNA sequencing analysis in women with or without PCOS, identifying differentially expressed lncRNAs that were associated with the metabolic process in the follicular fluid of mature follicles. Butler et al.46 found eight differentially expressed lncRNAs in PCOS with significant insulin resistance and hyperandrogenemia. An insulin resistance-associated lncRNA-mRNA network of PCOS was also constructed using starBase and GEO, allowing the identification of hub lncRNAs.47 Among these, lncRNA RP11-151A6.4 was highly expressed in PCOS and positively correlated with higher BMI, hyperinsulinemia, and higher HOMA-IR values. Conversely, lncRNA GAS548 was negatively associated with HOMA-IR. In turn, the expression of H19 was positively associated with fasting plasma glucose levels.59 Our results are in accordance with these prior studies. Specifically, we presented the CCNL as being related to PCOS pathogenesis and evaluated its potential function, demonstrating that CCNL promotes apoptosis, regulates mitochondrial function, and suppresses glucose uptake in granulosa cells.
Multiple studies have reported that increased apoptosis in the ovarian granulosa cells of PCOS leads to the induction of premature arrest of follicle growth.9,60, 61, 62, 63 Our results illustrated that upregulated CCNL induced apoptosis in both hLGCs and KGN cells as detected by annexin V/PI and TUNEL staining and the alteration of related protein levels (e.g., cleaved/total PARP, cleaved/total caspase-3, Bcl-2, and Bax). Caspase-3, a member of the caspase family of 13 aspartate-specific cysteine proteases, is mainly responsible for the cleavage of PARP during cell death.64 PARP cleavage catalyzed by caspase-3 is generally involved in the cellular DNA damage response, required for cell apoptosis,65 as PARP contributes to programmed cell death by depleting the cell of nicotinamide adenine dinucleotide (NAD) and ATP.66 The anti-apoptotic Bcl-2 and pro-apoptotic Bax proteins exhibit the ability to regulate mitochondrial outer membrane permeabilization, which impacts mitochondrial redox metabolism.49 The alteration of Bcl-2/Bax protein levels is thus indicative of mitochondria-dependent apoptosis and ROS production. Notably, oxidative stress can trigger apoptosis in granulosa cells and is thought to constitute a major cause of follicular atresia.17 We found that overexpression of CCNL increased the level of ROS generation, decreased the ATP concentration, and concomitantly impacted mitochondrial function. Thus, we demonstrated that dysregulation of CCNL impaired mitochondria, leading to the stimulation of apoptosis in granulosa cells.
Substantial evidence indicates the significant role of insulin in women with PCOS. For example, insulin resistance and compensatory hyperinsulinemia in PCOS contribute to hyperandrogenemia and ovulatory disturbances.67 In PCOS, a post-binding defect is observed in receptor signaling, likely owing to inactivated IRS1 that selectively affects metabolic action in classic insulin target tissues and in the ovary.68 We found that CCNL expression levels were also increased in patients with PCOS exhibiting higher BMI and higher HOMA-IR values. However, we note that the relationship between CCNL expression and relative endocrine parameters in the normal population remain to be further evaluated owing to the lack of clinical data. Our data also highlighted that CCNL impaired glycometabolism in granulosa cells as evaluated by glucose uptake capability and relative key protein expression (e.g., IRS1, GLUT4). Insulin regulates blood glucose by binding to the insulin receptor, then stimulating IRS1 and downstream proteins. Glucose uptake is activated via the canonical IRS-mediated GLUT4 vesicle translocation to the membrane.69 In the present study, we found that increased CCNL led to decreased glucose uptake along with IRS1 and GLUT4 abundance. Moreover, previous studies have reported that patients with PCOS display features of mitochondrial impairment and oxidative stress, and that these can aggravate insulin resistance,13,14 which is consistent with the identification of chronic oxidative stress and associated inflammation as risk factors that contribute to insulin resistance.26,70 Consequently, we suggest that upregulated CCNL affects the glucose uptake ability and oxidative stress levels in PCOS granulosa cells, contributing in part to tissue-specific insulin resistance in the ovary. In addition, CCNL is highly expressed not only in the ovary but also in the liver, which constitutes an important insulin target organ, and it is responsible for hepatic insulin resistance. However, owing to the lack of human hepatic tissue sources, we were unable to determine the CCNL expression and effects on glucose uptake in the liver. Future work may thus include exploring the function of CCNL in the liver.
Investigation of the underlying mechanisms of CCNL-mediated regulation of the PCOS-related phenotypes revealed that the transcription factor FOXO1 was highly expressed in KGN cells and hLGCs. FOXO1 protein levels were elevated whereas mRNA levels remained unchanged. The luciferase reporter assays also echo that CCNL has no effects on transcription activity of FOXO1 (Figure S10). FOXO1, a member of the forkhead transcription factor family, is highly expressed in the ovary, adipose tissues, fat, and endometrium (https://www.ncbi.nlm.nih.gov/gene/2308/?report=expression). FOXO family members regulate various processes, including apoptosis, cell cycle, DNA repair, ROS, glucose metabolism, mitophagy, and mitochondrial metabolism.50, 51, 52 Activated FOXO1 promotes apoptosis both in a mitochondria-dependent and mitochondria-independent manner by regulating various pro-apoptotic proteins, including Fas ligand and Bcl-2 family members, such as Bim, bNIP3, and Bcl-xL.30,50 The members of the FOXO family of transcription factors function in the regulation of stress-inducible genes, such as GADD45, resulting in the arrest of cell growth in the G2 phase of the cell cycle. The oxidative stress increases the expression of GADD45 by a FOXO-dependent, but p53-independent, pathway.31,71 We also detected the expression of some classic FOXO1 target genes in overexpressed CCNL cells (Figure S8). The FOXO1 target genes, such as Bim, TRAIL, and GADD45α, were evaluated under CCNL overexpression while some of cell cycle markers remained unchanged (Figure S6). We suggest that upregulated CCNL might thereby participate in the FOXO1 selective activation of downstream signal. Notably, recent studies highlight a potential role for FOXO1 in follicular development through the regulation of apoptosis and proliferation of granulosa cells.17,27 In addition, previous studies illustrated that FOXO1 mediates the metabolic consequences of insulin resistance.28,29,72,73 Specifically, FOXO1 serves as the major intracellular transcription factor of insulin action and contributes to the regulation of glucose metabolism, including gluconeogenesis and glycolysis.74, 75, 76, 77
FOXO1 proteins function primarily as transcription factors by binding as monomers to their target gene sequences in the nucleus. FOXO1 activity is regulated by numerous post-translational modifications, including phosphorylation, ubiquitylation, and acetylation.55 Following such modifications, FOXO1 tends to be excluded from the nucleus and subsequently degraded in the cytoplasm, which inhibits FOXO1-dependent transcription. The abundance of phosphorylated FOXO1 (p-FOXO1) was decreased in CCNL-overexpressed KGN cells and hLGCs (Figure S7). Moreover, FOXO1 was reported to have binding ability toward ncRNAs, thus exerting certain biological functions.53,54 Consistent with the ability of endonuclear lncRNAs to interact with RBPs and regulate the transcriptional activity of their target genes in an epigenetic manner or through the direct stabilization of the mRNAs and proteins,33,34 we found that CCNL could interact with FOXO1 at the protein level owing to its high affinity. Thus, we propose that CCNL binds to FOXO1 to increase the levels of activated FOXO1 in the nucleus, as verified through RIP, immunofluorescence staining, and western blotting. Specifically, CCNL may interact with FOXO1 and potentially regulate its activity by preventing FOXO1 exclusion from the nucleus and subsequent degradation in the cytosol. Moreover, rescue experiments revealed that CCNL affects cell apoptosis, glucose uptake ability, and the levels of related protein expression (e.g., cleaved/total PARP, cleaved/total caspase-3, Bcl-2, Bax, IRS1, and GLUT4) were diminished upon FOXO1 knockdown, further supporting that FOXO1 is involved in the CCNL-mediated regulation of granulosa cells. This finding thus indicates a new interaction mode for lncRNAs with proteins. To further explore CCNL-protein interaction and its function in PCOS, in vitro or in vivo approaches such as proximity proteomics78 should be considered for future work determining the relationship between CCNL and protein expression in patients with PCOS.
In addition, we found that the abundance of CCNL in the granulosa cells of women with PCOS was positively associated with the concentration of testosterone in the serum, whereas no correlation existed between other PCOS clinical characteristics and CCNL expression. Evaluation in KGN cells cultured in DHT medium indicated that excess androgen induces CCNL expression (Figure S5). However, owing to the unavailability of sufficient human theca cells, the main source of androgen in the ovary, we were unable to directly determine the relationship between CCNL levels and androgen excess in the patients.
To date, the etiology of PCOS remains largely unknown. However, increasing evidence has revealed that PCOS might constitute a multigenic disorder with environmental influences, including diet and lifestyle.1,2 These environmental factors, which are strongly influenced by race and/or ethnicity, might be responsible for the differences in PCOS-related genes among populations of diverse origins.1 As most patients recruited in this study were from an Asian population of ethnic Han ethnicity, the conclusions should be carefully applied to other population-based research.
In conclusion, our findings showed that CCNL was highly expressed in the granulosa cells of women with PCOS. Overexpressed CCNL interacts with the transcription factor FOXO1 to undermine mitochondria function, ultimately promoting apoptosis and insulin resistance in PCOS (Figure 7). These findings thus identify lnc-CCNL1-3:1 as a potential therapeutic target for PCOS and provide a new perspective regarding the molecular mechanisms underlying the pathogenesis of this disorder.
Materials and methods
Ethics statement
The study was approved by the ART Ethnics Committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University (no. 2017041411), and informed consent was obtained from all participants.
Human specimens
Ovarian granulosa cell samples were collected from patients who underwent in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) treatment at the Center for Reproductive Medicine, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University from March 2016 to June 2019. The diagnosis of PCOS was based on the Revised Rotterdam Diagnostic Criteria, which require the presence of at least two of the following criteria for a PCOS diagnosis: (1) oligo-ovulation and/or anovulation; (2) clinical and/or biochemical signs of hyperandrogenism; and (3) polycystic ovaries.79 Diagnoses of PCOS were made after exclusion of other etiologies for hyperandrogenemia and ovulatory dysfunction such as congenital adrenal hyperplasia, Cushing syndrome, androgen-secreting tumors, thyroid disease, 21-hydroxylase deficiency, and hyperprolactinemia. The inclusion criteria for control women were as follows: regular menstrual cycles (25–35 days), no endocrine abnormalities, and normal ovarian morphology confirmed by ultrasound; total testosterone was evaluated to exclude hyperandrogenism. The exclusion criteria for both groups were as follows: previous history of gynecological surgery, such as oophorocystectomy and unilateral ovariectomy; abnormal chromosome karyotypes, steroid use or illegal drug use in past 3 months, recurrent abortion and pregnancy loss, with other reproductive disorders, such as adenomyosis, endometriosis, or uterine fibroids, with other underlying diseases or complications that are not consistent with pregnancy, such as diabetes, systemic lupus erythematosus, or anti-cardiolipin antibody syndrome. Anthropometric variables, such as age, BMI, and selected endocrine and biochemical parameters, were recorded (Table 2). Peripheral blood samples were collected from all subjects during days 2–4 of spontaneous cycles or after cessation of bleeding after a 12-h overnight fast. Follicle-stimulating hormone, luteinizing hormone, testosterone, and anti-Müllerian hormone levels were determined using either chemiluminescent assay (Roche Diagnostics, Indianapolis, IN, USA) or enzyme-linked immunosorbent assay (Kangrun, Guangzhou, China) kits. The World Health Organization cutoff points for normal (BMI, 18.5–23.9 kg/m2) and overweight (BMI, ³24 kg/m2) for Asians were utilized.80 The HOMA-IR index was calculated according to the previously published formula.81
Isolation and culture of ovarian granulosa and KGN cells
The ovarian stimulation for patients undergoing IVF/ICSI treatment consisted of the gonadotropin-releasing hormone (GnRH) antagonist or agonist protocol. Human chorionic gonadotropin (hCG; Lvzhu, Zhuhai, China) was administered to trigger ovulation after adequate follicle development was detected via both ovarian ultrasound and a serum estradiol assay. Ultrasound-guided oocyte retrieval was performed after 34–36 h. hLGCs were retrieved from the follicular fluid as previously described.39 The follicular fluid was pooled and centrifuged at 2,500 rpm (628.875 × g) for 10 min. Granulosa cells were purified by Ficoll-Paque density centrifugation, then the pellets were re-suspended in phosphate-buffered saline (PBS) and dispersed in 0.1% hyaluronidase (Sigma, St. Louis, MO, USA) at 37°C for 10 min. The isolated hLGCs were stored at −80°C or used after 3 days in culture.
The human granulosa cell tumor-derived cell line, KGN, was a generous gift from Shandong University. All the hLGCs and KGN cells were cultured in Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 (DMEM/F12) (Gibco, Grand Island, NY, USA) containing 10% charcoal-stripped fetal bovine serum (Biological Industries, Cromwell, CT, USA) and 1% penicillin-streptomycin-neomycin (Gibco). Cells were cultured at 37°C in a humidified atmosphere containing 95% air and 5% CO2. KGN cells were passaged every 3 days.
Cell transfection and treatment
The cells were transfected with the constructed lnc-CCNL1-3:1 overexpression plasmid (GV144-CCNL) and control plasmid (GV144 vector) (Sangon, Shanghai, China; Genechem, Shanghai, China). The full length of the major transcript of CCNL was synthesized into the pBluescript II SK+ and then cut and synthesized into transient vector GV144 (Figure S9) (vector backbone: https://www.genechem.com.cn/index/supports/zaiti_info.html?id=9).
KGN cells and hLGCs (2 × 105) were seeded into six-well plates, cultured for 1 day, and then transfected with plasmid DNA and/or siRNAs (siFOXO1) (GenePharma, Shanghai, China) using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. After transfection, the cells were incubated for 72 h prior to further treatment. siRNA (Sigma) sequences are shown in Table S1.
RNA extraction and quantitative real-time PCR
Total RNA from patient ovarian granulosa cells was extracted using TRIzol (Invitrogen) and reverse transcribed into cDNA (PrimeScript RT reagent kit, TaKaRa, Dalian, China). Target gene expression was detected with the use of quantitative real-time PCR. The relative expression of RNA was calculated using the formula 2−ΔΔCt. β-Actin was used as an internal control for the quantification of target genes, using ΔCt normalization against β-actin. Primers used in this study are shown in Table S2.
RNA ligase-mediated RACE (RLM-RACE)
Total RNA from KGN cells (2 × 106 to 2 × 107) was extracted via a TRIzol Plus RNA purification kit (Invitrogen; 12183-555) and reverse transcribed into cDNA. According to the manufacturer’s instructions, 5′ RACE and 3′ RACE were performed using a GeneRacer kit (Invitrogen; L1500-01). The gene-specific primers used for PCR are provided in Table S3.
Immunoblotting assay
Cells were harvested and lysed in ice-cold radioimmunoprecipitation assay lysis buffer (Sangon) containing a protease inhibitor cocktail (Sangon) and phosphatase inhibitor (Sangon). The protein was extracted from cells and quantified as previously described82 using 10%–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Relative band density was detected using Gel-Pro Analyzer (Media Cybernetics, Rockville, MD, USA) and normalized to β-actin (Proteintech, Wuhan, Hubei, China). Specifically, the relative cleaved target protein amount was calculated by the ratios of cleaved target protein to total protein. The antibodies used in the western blot analysis are summarized in Table S4.
Subcellular fractionation
Cells were collected using trypsin and washed twice with PBS. Cell pellets were processed using the cytoplasmic and nuclear RNA purification kit (Sangon). Cytoplasmic and nuclear fractions were split for RNA extraction and quantitative real-time PCR or protein extraction and western blotting. Tubulin and lamin A/C were used as markers of the cytoplasm and nucleus in western blotting. β-Actin and nuclear U6 were used as markers of the cytoplasm and nucleus in quantitative real-time PCR.
Flow cytometry analysis
To measure apoptosis, the cells were stained using the annexin V kit according to the manufacturer’s instructions (BD Biosciences, San Jose, CA, USA). Cells were harvested by trypsinization 48 h after transfection with siRNAs and/or plasmid DNA. Cells were washed once with PBS and then 1 × 106 to 1 × 107 cells were resuspended in 100 μL of 1× binding buffer. Fluorescein isothiocyanate (FITC)-labeled annexin V and propidium iodide (PI) were added to samples and incubated in the dark for 15 min at room temperature (18°C–22°C). Subsequently, cells were subjected to fluorescence-activated cell sorting analysis. The results were analyzed using CytExpert 2.0 (Beckman Coulter, Miami, FL, USA).
TUNEL assay
Cells were fixed with 4% paraformaldehyde for 30 min and washed twice with PBS. Then, the TUNEL assay was performed using an in situ cell detection kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions with a fluorescence microscope (Carl Zeiss, Oberkochen, Germany). ImageJ software (Rawak Software, Stuttgart, Germany) was used to quantify fluorescence intensity.
Determination of ROS levels
Cellular ROS levels were assessed using 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA) (Life Technology/Thermo Fisher Scientific, Waltham, MA, USA). ROS generation was induced by treatment with 20 mM H2O2 for 5 min, and then the cells were incubated with 30 μM H2DCFDA in PBS at 37°C for 30 min. Cells were washed three times in H2DCFDA-free PBS and images were captured on a fluorescence microscope (Carl Zeiss). ImageJ software was used to quantify fluorescence intensity.
ATP measurement
Cellular ATP concentration was measured using an ATP assay kit following the manufacturer’s protocol (BioVision, Milpitas, CA, USA).
Glucose uptake assay
A glucose uptake assay was performed in KGN cells and hLGCs following serum starvation for 12 h and insulin stimulation (Sigma-Aldrich, 100 nM, 20 min) using the Glucose Uptake-Glo assay kit J1342 (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
RIP assay
The RIP assay was performed using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Burlington, MA, USA; 17-701) according to the manufacturer’s instructions. Briefly, cells were lysed and incubated with magnetic beads conjugated with human anti-FOXO1 antibody (Cell Signaling Technology, Danvers, MA, USA) or anti-rabbit IgG (Millipore; 03-110). The immunoprecipitated RNA was detected by quantitative real-time PCR. Relative quantities of gene expression level were normalized. The relative quantities of RIP samples were normalized by the respective individual inputs.
Immunofluorescence experiment
The stable CCNL- or Enter-transfected KGN cells were seeded on four-well glass slides (Millipore). After washing in PBS three times, the cells were fixed with 4% paraformaldehyde for 20 min, permeabilized using 0.5% Triton X-100 in PBS for 15 min, and blocked with goat serum for 1 h at room temperature. The slides were then incubated with anti-FOXO1 antibody (Cell Signaling Technology; 1:100) for 24 h at 4°C. The cells were incubated with Alexa Fluor 488 (green)-conjugated goat anti-rabbit IgG (Invitrogen; 1:100) for 1 h in the dark. After nuclear staining with DAPI (Invitrogen) for 1 min, the slides were washed with PBS three times and observed under a microscope in the dark.
Luciferase reporter assay
pcDNA3.1-FOXO1 or pcDNA3.1-FOXO1-mut was co-transfected with pcDNA3.1 or pcDNA3.1-CCNL into 293T cells (psb66 vector) (Sangon) (Figure S9). 48 h after transfection, luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega) in accordance with the manufacturer’s protocol. Relative firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis and reproducibility
Most experiments were repeated more than three times to be eligible for the indicated statistical analyses. No estimation of group variation was performed prior to experiments. All calculations were performed using the SPSS statistical software package (version 20.0) (IBM, Armonk, NY, USA). Results are expressed as the means ± standard deviation (SD). The Kolmogorov-Smirnov test was used to assess the distributions of relevant variables. Statistical comparisons between two variables (the PCOS group and the control group) were performed using the Student’s t test for normal distribution, whereas an unpaired test and the Mann-Whitney U test were used for continuous variables. The Spearman’s correlation and linear regression analyses were used to examine the association between gene expression levels. Data were considered statistically significant for p <0.05.
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper. This work was supported in part by grants from the National Key Research and Development Program of China (nos. 2017YFC1001002 and 2018YFC1003202), the National Natural Science Foundation (nos. 81971343, 81671414, and 81671413), the National Institutes of Health (project no. 1R01HD085527), and by the Shanghai Commission of Science and Technology (nos. 19410760300 and 17DZ2271100).
Author contributions
Guarantor of integrity of the entire study: Z.J.C. and Y.D. Study design: Y.D., J.H., and J.Z. Experimental studies: J.H., J.Z., and X.G. Manuscript editing: J.H., X.G., W.C., S.L., and Y.D. All authors contributed to manuscript revision and read and approved the submitted version.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.12.008.
Supplemental information
References
- 1.Escobar-Morreale H.F. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat. Rev. Endocrinol. 2018;14:270–284. doi: 10.1038/nrendo.2018.24. [DOI] [PubMed] [Google Scholar]
- 2.Azziz R. PCOS in 2015: new insights into the genetics of polycystic ovary syndrome. Nat. Rev. Endocrinol. 2016;12:183. doi: 10.1038/nrendo.2016.9. [DOI] [PubMed] [Google Scholar]
- 3.Baerwald A.R., Adams G.P., Pierson R.A. Ovarian antral folliculogenesis during the human menstrual cycle: a review. Hum. Reprod. Update. 2012;18:73–91. doi: 10.1093/humupd/dmr039. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda F., Inoue N., Manabe N., Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012;58:44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- 5.Munakata Y., Kawahara-Miki R., Shiratsuki S., Tasaki H., Itami N., Shirasuna K., Kuwayama T., Iwata H. Gene expression patterns in granulosa cells and oocytes at various stages of follicle development as well as in in vitro grown oocyte-and-granulosa cell complexes. J. Reprod. Dev. 2016;62:359–366. doi: 10.1262/jrd.2016-022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J.Y., Cheung C.K., Wang Y., Tsang B.K. Regulation of cell death and cell survival gene expression during ovarian follicular development and atresia. Front. Biosci. 2003;8:d222–d237. doi: 10.2741/949. [DOI] [PubMed] [Google Scholar]
- 7.Asselin E., Xiao C.W., Wang Y.F., Tsang B.K. Mammalian follicular development and atresia: role of apoptosis. Biol. Signals Recept. 2000;9:87–95. doi: 10.1159/000014627. [DOI] [PubMed] [Google Scholar]
- 8.Hughesdon P.E. Morphology and morphogenesis of the Stein-Leventhal ovary and of so-called “hyperthecosis”. Obstet. Gynecol. Surv. 1982;37:59–77. doi: 10.1097/00006254-198202000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Das M., Djahanbakhch O., Hacihanefioglu B., Saridogan E., Ikram M., Ghali L., Raveendran M., Storey A. Granulosa cell survival and proliferation are altered in polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2008;93:881–887. doi: 10.1210/jc.2007-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giorgio M., Migliaccio E., Orsini F., Paolucci D., Moroni M., Contursi C., Pelliccia G., Luzi L., Minucci S., Marcaccio M. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Liu X., Bhalla K., Kim C.N., Ibrado A.M., Cai J., Peng T.I., Jones D.P., Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 13.González F., Considine R.V., Abdelhadi O.A., Acton A.J. Oxidative stress in response to saturated fat ingestion is linked to insulin resistance and hyperandrogenism in polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2019;104:5360–5371. doi: 10.1210/jc.2019-00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bañuls C., Rovira-Llopis S., Martinez de Marañon A., Veses S., Jover A., Gomez M., Rocha M., Hernandez-Mijares A., Victor V.M. Metabolic syndrome enhances endoplasmic reticulum, oxidative stress and leukocyte-endothelium interactions in PCOS. Metabolism. 2017;71:153–162. doi: 10.1016/j.metabol.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Zhao W., Xu H., Hu M., Guo X., Jia W., Liu G., Li J., Cui P., Lager S. Hyperandrogenism and insulin resistance-induced fetal loss: evidence for placental mitochondrial abnormalities and elevated reactive oxygen species production in pregnant rats that mimic the clinical features of polycystic ovary syndrome. J. Physiol. 2019;597:3927–3950. doi: 10.1113/JP277879. [DOI] [PubMed] [Google Scholar]
- 16.Kumar M., Pathak D., Kriplani A., Ammini A.C., Talwar P., Dada R. Nucleotide variations in mitochondrial DNA and supra-physiological ROS levels in cytogenetically normal cases of premature ovarian insufficiency. Arch. Gynecol. Obstet. 2010;282:695–705. doi: 10.1007/s00404-010-1623-x. [DOI] [PubMed] [Google Scholar]
- 17.Shen M., Lin F., Zhang J., Tang Y., Chen W.K., Liu H. Involvement of the up-regulated FoxO1 expression in follicular granulosa cell apoptosis induced by oxidative stress. J. Biol. Chem. 2012;287:25727–25740. doi: 10.1074/jbc.M112.349902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega-Camarillo C., González-González A., Vergara-Onofre M., González-Padilla E., Avalos-Rodríguez A., Gutiérrez-Rodríguez M.E., Arriaga-Pizano L., Cruz M., Baiza-Gutman L.A., Díaz-Flores M. Changes in the glucose-6-phosphate dehydrogenase activity in granulosa cells during follicular atresia in ewes. Reproduction. 2009;137:979–986. doi: 10.1530/REP-08-0463. [DOI] [PubMed] [Google Scholar]
- 19.Diamanti-Kandarakis E., Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr. Rev. 2012;33:981–1030. doi: 10.1210/er.2011-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasquali R., Gambineri A. Glucose intolerance states in women with the polycystic ovary syndrome. J. Endocrinol. Invest. 2013;36:648–653. doi: 10.1007/BF03346757. [DOI] [PubMed] [Google Scholar]
- 21.Ciaraldi T.P., Aroda V., Mudaliar S., Chang R.J., Henry R.R. Polycystic ovary syndrome is associated with tissue-specific differences in insulin resistance. J. Clin. Endocrinol. Metab. 2009;94:157–163. doi: 10.1210/jc.2008-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciaraldi T.P., el-Roeiy A., Madar Z., Reichart D., Olefsky J.M., Yen S.S. Cellular mechanisms of insulin resistance in polycystic ovarian syndrome. J. Clin. Endocrinol. Metab. 1992;75:577–583. doi: 10.1210/jcem.75.2.1322430. [DOI] [PubMed] [Google Scholar]
- 23.Zhu S., Zhang B., Jiang X., Li Z., Zhao S., Cui L., Chen Z.J. Metabolic disturbances in non-obese women with polycystic ovary syndrome: a systematic review and meta-analysis. Fertil. Steril. 2019;111:168–177. doi: 10.1016/j.fertnstert.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Franks S., Gilling-Smith C., Watson H., Willis D. Insulin action in the normal and polycystic ovary. Endocrinol. Metab. Clin. North Am. 1999;28:361–378. doi: 10.1016/s0889-8529(05)70074-8. [DOI] [PubMed] [Google Scholar]
- 25.Dumesic D.A., Abbott D.H. Implications of polycystic ovary syndrome on oocyte development. Semin. Reprod. Med. 2008;26:53–61. doi: 10.1055/s-2007-992925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruegsegger G.N., Creo A.L., Cortes T.M., Dasari S., Nair K.S. Altered mitochondrial function in insulin-deficient and insulin-resistant states. J. Clin. Invest. 2018;128:3671–3681. doi: 10.1172/JCI120843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen M., Liu Z., Li B., Teng Y., Zhang J., Tang Y., Sun S.C., Liu H. Involvement of FoxO1 in the effects of follicle-stimulating hormone on inhibition of apoptosis in mouse granulosa cells. Cell Death Dis. 2014;5:e1475. doi: 10.1038/cddis.2014.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puigserver P., Rhee J., Donovan J., Walkey C.J., Yoon J.C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B.M. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 29.O-Sullivan I., Zhang W., Wasserman D.H., Liew C.W., Liu J., Paik J., DePinho R.A., Stolz D.B., Kahn C.R., Schwartz M.W., Unterman T.G. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat. Commun. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marfè G., Tafani M., Fiorito F., Pagnini U., Iovane G., De Martino L. Involvement of FOXO transcription factors, TRAIL-FasL/Fas, and sirtuin proteins family in canine coronavirus type II-induced apoptosis. PLoS ONE. 2011;6:e27313. doi: 10.1371/journal.pone.0027313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran H., Brunet A., Grenier J.M., Datta S.R., Fornace A.J., Jr., DiStefano P.S., Chiang L.W., Greenberg M.E. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 32.Jandura A., Krause H.M. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33:665–676. doi: 10.1016/j.tig.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 34.Yao R.W., Wang Y., Chen L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019;21:542–551. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 35.Yerushalmi G.M., Salmon-Divon M., Yung Y., Maman E., Kedem A., Ophir L., Elemento O., Coticchio G., Dal Canto M., Mignini Renzinu M. Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol. Hum. Reprod. 2014;20:719–735. doi: 10.1093/molehr/gau031. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., Fujita Y., Fujimori T., Standaert L., Marine J.C., Hirose T. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsubara S., Kurihara M., Kimura A.P. A long non-coding RNA transcribed from conserved non-coding sequences contributes to the mouse prolyl oligopeptidase gene activation. J. Biochem. 2014;155:243–256. doi: 10.1093/jb/mvt113. [DOI] [PubMed] [Google Scholar]
- 38.Xu X.F., Li J., Cao Y.X., Chen D.W., Zhang Z.G., He X.J., Ji D.M., Chen B.L. Differential expression of long noncoding RNAs in human cumulus cells related to embryo developmental potential: a microarray analysis. Reprod. Sci. 2015;22:672–678. doi: 10.1177/1933719114561562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J., Xu J., Wang W., Zhao H., Liu H., Liu X., Liu J., Sun Y., Dunaif A., Du Y., Chen Z.J. Long non-coding RNA LINC-01572:28 inhibits granulosa cell growth via a decrease in p27 (Kip1) degradation in patients with polycystic ovary syndrome. EBioMedicine. 2018;36:526–538. doi: 10.1016/j.ebiom.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y.D., Li Y., Feng S.X., Ye D.S., Chen X., Zhou X.Y., Chen S.L. Long noncoding RNAs: potential regulators involved in the pathogenesis of polycystic ovary syndrome. Endocrinology. 2017;158:3890–3899. doi: 10.1210/en.2017-00605. [DOI] [PubMed] [Google Scholar]
- 41.Han Q., Zhang W., Meng J., Ma L., Li A. lncRNA-LET inhibits cell viability, migration and EMT while induces apoptosis by up-regulation of TIMP2 in human granulosa-like tumor cell line KGN. Biomed. Pharmacother. 2018;100:250–256. doi: 10.1016/j.biopha.2018.01.162. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Wang H., Zhou D., Shuang T., Zhao H., Chen B. Up-regulation of long noncoding RNA SRA promotes cell growth, inhibits cell apoptosis, and induces secretion of estradiol and progesterone in ovarian granular cells of mice. Med. Sci. Monit. 2018;24:2384–2390. doi: 10.12659/MSM.907138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z., Hao C., Song D., Zhang N., Bao H., Qu Q. Androgen receptor coregulator CTBP1-AS is associated with polycystic ovary syndrome in Chinese women: a preliminary study. Reprod. Sci. 2015;22:829–837. doi: 10.1177/1933719114565037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiao J., Shi B., Wang T., Fang Y., Cao T., Zhou Y., Wang X., Li D. Characterization of long non-coding RNA and messenger RNA profiles in follicular fluid from mature and immature ovarian follicles of healthy women and women with polycystic ovary syndrome. Hum. Reprod. 2018;33:1735–1748. doi: 10.1093/humrep/dey255. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z., Hao C., Huang X., Zhang N., Bao H., Qu Q. Peripheral blood leukocyte expression level of lncRNA steroid receptor RNA activator (SRA) and its association with polycystic ovary syndrome: a case control study. Gynecol. Endocrinol. 2015;31:363–368. doi: 10.3109/09513590.2014.999763. [DOI] [PubMed] [Google Scholar]
- 46.Butler A.E., Hayat S., Dargham S.R., Malek J.A., Abdulla S.A., Mohamoud Y.A., Suhre K., Sathyapalan T., Atkin S.L. Alterations in long noncoding RNAs in women with and without polycystic ovarian syndrome. Clin. Endocrinol. (Oxf.) 2019;91:793–797. doi: 10.1111/cen.14087. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J., Huang J., Geng X., Chu W., Li S., Chen Z.J., Du Y. Polycystic ovary syndrome: novel and hub lncRNAs in the insulin resistance-associated lncRNA-mRNA network. Front. Genet. 2019;10:772. doi: 10.3389/fgene.2019.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin H., Xing W., Li Y., Xie Y., Tang X., Zhang Q. Downregulation of serum long noncoding RNA GAS5 may contribute to insulin resistance in PCOS patients. Gynecol. Endocrinol. 2018;34:784–788. doi: 10.1080/09513590.2018.1459548. [DOI] [PubMed] [Google Scholar]
- 49.Chong S.J., Low I.C., Pervaiz S. Mitochondrial ROS and involvement of Bcl-2 as a mitochondrial ROS regulator. Mitochondrion. 2014;19(Pt A):39–48. doi: 10.1016/j.mito.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 50.Fu Z., Tindall D.J. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilhelm K., Happel K., Eelen G., Schoors S., Oellerich M.F., Lim R., Zimmermann B., Aspalter I.M., Franco C.A., Boettger T. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature. 2016;529:216–220. doi: 10.1038/nature16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bella L., Zona S., Nestal de Moraes G., Lam E.W. FOXM1: a key oncofoetal transcription factor in health and disease. Semin. Cancer Biol. 2014;29:32–39. doi: 10.1016/j.semcancer.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 53.Yan C., Li J., Feng S., Li Y., Tan L. Long noncoding RNA Gomafu upregulates Foxo1 expression to promote hepatic insulin resistance by sponging miR-139-5p. Cell Death Dis. 2018;9:289. doi: 10.1038/s41419-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Z., Gong Z., Sun X. lncRNA DANCR involved osteolysis after total hip arthroplasty by regulating FOXO1 expression to inhibit osteoblast differentiation. J. Biomed. Sci. 2018;25:4. doi: 10.1186/s12929-018-0406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H., Tindall D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 56.Muppirala U.K., Honavar V.G., Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics. 2011;12:489. doi: 10.1186/1471-2105-12-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goyal N., Kesharwani D., Datta M. Lnc-ing non-coding RNAs with metabolism and diabetes: roles of lncRNAs. Cell. Mol. Life Sci. 2018;75:1827–1837. doi: 10.1007/s00018-018-2760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kornfeld J.W., Brüning J.C. Regulation of metabolism by long, non-coding RNAs. Front. Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin L., Huang C.C., Yan X.M., Wang Y., Li Z.Y., Wei X.C. Long non-coding RNA H19 is associated with polycystic ovary syndrome in Chinese women: a preliminary study. Endocr. J. 2019;66:587–595. doi: 10.1507/endocrj.EJ19-0004. [DOI] [PubMed] [Google Scholar]
- 60.Ambekar A.S., Kelkar D.S., Pinto S.M., Sharma R., Hinduja I., Zaveri K., Pandey A., Prasad T.S., Gowda H., Mukherjee S. Proteomics of follicular fluid from women with polycystic ovary syndrome suggests molecular defects in follicular development. J. Clin. Endocrinol. Metab. 2015;100:744–753. doi: 10.1210/jc.2014-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding L., Gao F., Zhang M., Yan W., Tang R., Zhang C., Chen Z.J. Higher PDCD4 expression is associated with obesity, insulin resistance, lipid metabolism disorders, and granulosa cell apoptosis in polycystic ovary syndrome. Fertil. Steril. 2016;105:1330–1337.e3. doi: 10.1016/j.fertnstert.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 62.Zheng Q., Li Y., Zhang D., Cui X., Dai K., Yang Y., Liu S., Tan J., Yan Q. ANP promotes proliferation and inhibits apoptosis of ovarian granulosa cells by NPRA/PGRMC1/EGFR complex and improves ovary functions of PCOS rats. Cell Death Dis. 2017;8:e3145. doi: 10.1038/cddis.2017.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y., Tao M., Wei M., Du S., Wang H., Wang X. Mesenchymal stem cells derived exosomal miR-323-3p promotes proliferation and inhibits apoptosis of cumulus cells in polycystic ovary syndrome (PCOS) Artif. Cells Nanomed. Biotechnol. 2019;47:3804–3813. doi: 10.1080/21691401.2019.1669619. [DOI] [PubMed] [Google Scholar]
- 64.Slee E.A., Adrain C., Martin S.J. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J. Biol. Chem. 2001;276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 65.Boulares A.H., Yakovlev A.G., Ivanova V., Stoica B.A., Wang G., Iyer S., Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J. Biol. Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 66.Pascal J.M. The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst.) 2018;71:177–182. doi: 10.1016/j.dnarep.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cassar S., Misso M.L., Hopkins W.G., Shaw C.S., Teede H.J., Stepto N.K. Insulin resistance in polycystic ovary syndrome: a systematic review and meta-analysis of euglycaemic-hyperinsulinaemic clamp studies. Hum. Reprod. 2016;31:2619–2631. doi: 10.1093/humrep/dew243. [DOI] [PubMed] [Google Scholar]
- 68.Diamanti-Kandarakis E., Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr. Rev. 2012;33:981–1030. doi: 10.1210/er.2011-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samuel V.T., Shulman G.I. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J. Clin. Invest. 2016;126:12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newsholme P., Cruzat V.F., Keane K.N., Carlessi R., de Bittencourt P.I., Jr. Molecular mechanisms of ROS production and oxidative stress in diabetes. Biochem. J. 2016;473:4527–4550. doi: 10.1042/BCJ20160503C. [DOI] [PubMed] [Google Scholar]
- 71.Furukawa-Hibi Y., Yoshida-Araki K., Ohta T., Ikeda K., Motoyama N. FOXO forkhead transcription factors induce G2-M checkpoint in response to oxidative stress. J. Biol. Chem. 2002;277:26729–26732. doi: 10.1074/jbc.C200256200. [DOI] [PubMed] [Google Scholar]
- 72.Matsumoto M., Han S., Kitamura T., Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J. Clin. Invest. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong X.C., Copps K.D., Guo S., Li Y., Kollipara R., DePinho R.A., White M.F. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8:65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang W., Patil S., Chauhan B., Guo S., Powell D.R., Le J., Klotsas A., Matika R., Xiao X., Franks R. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 75.Rui L. Energy metabolism in the liver. Compr. Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hall R.K., Yamasaki T., Kucera T., Waltner-Law M., O’Brien R., Granner D.K. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 2000;275:30169–30175. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- 77.Schmoll D., Walker K.S., Alessi D.R., Grempler R., Burchell A., Guo S., Walther R., Unterman T.G. Regulation of glucose-6-phosphatase gene expression by protein kinase Bα and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J. Biol. Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 78.Ramanathan M., Porter D.F., Khavari P.A. Methods to study RNA-protein interactions. Nat. Methods. 2019;16:225–234. doi: 10.1038/s41592-019-0330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rotterdam E.A.P., Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil. Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 80.Expert Consultation, W.H.O. WHO Expert Consultation Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363:157–163. doi: 10.1016/S0140-6736(03)15268-3. [DOI] [PubMed] [Google Scholar]
- 81.Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 82.Zhu Q., Zuo R., He Y., Wang Y., Chen Z.J., Sun Y., Sun K. Local regeneration of cortisol by 11β-HSD1 contributes to insulin resistance of the granulosa cells in PCOS. J. Clin. Endocrinol. Metab. 2016;101:2168–2177. doi: 10.1210/jc.2015-3899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.