Abstract
Diaporthe species have often been reported as important plant pathogens, saprobes and endophytes on a wide range of plant hosts. Although several Diaporthe species have been recorded, little is known about species able to infect forest trees in Jiangxi Province. Hence, extensive surveys were recently conducted in Jiangxi Province, China. A total of 24 isolates were identified and analysed using comparisons of DNA sequence data for the nuclear ribosomal internal transcribed spacer (ITS), calmodulin (cal), histone H3 (his3), partial translation elongation factor-1α (tef1) and β-tubulin (tub2) gene regions, as well as their morphological features. Results revealed five novel taxa, D. bauhiniae, D. ganzhouensis, D. schimae, D. verniciicola, D. xunwuensis spp. nov. and three known species, D. apiculatum, D. citri and D. multigutullata.
Keywords: DNA phylogeny, five new taxa, forest trees, systematics, taxonomy
Introduction
The genus Diaporthe Nitschke (Sordariomycetes, Diaporthales) represents a cosmopolitan group of fungi occupying diverse ecological behaviour as plant pathogens, endophytes and saprobes (Muralli et al. 2006; Rossman et al. 2007; Udayanga et al. 2014, 2015; Fan et al. 2015, 2018; Guarnaccia and Crous 2017; Guarnaccia et al. 2018; Yang et al. 2018, 2020; Manawasinghe et al. 2019; Marin-Felix et al. 2019). Diaporthe species are responsible for diseases on a wide range of plant hosts, including agricultural crops, forest trees and ornamentals, some of which are economically important. Several symptoms, such as root and fruit rots, dieback, stem cankers, leaf spots, leaf and pod blights and seed decay are caused by Diaporthe spp. (Uecker 1988; Rehner and Uecker 1994; Mostert et al. 2001; Santos et al. 2011; Thompson et al. 2011; Udayanga et al. 2011).
Diaporthe was historically considered as monophyletic, based on its typical sexual morph and Phomopsis asexual morph (Gomes et al. 2013). However, Gao et al. (2017) recently revealed its paraphyletic nature, showing that Mazzantia (Wehmeyer 1926), Ophiodiaporthe (Fu et al. 2013), Pustulomyces (Dai et al. 2014), Phaeocytostroma and Stenocarpella (Lamprecht et al. 2011) are embedded in Diaporthe s. lat. Furthermore, Senanayake et al. (2017) recently included additional two genera in Diaporthe s. lat., namely Paradiaporthe and Chiangraiomyces.
Species identification criteria in Diaporthe were originally based on host association, morphology and culture characteristics (Mostert et al. 2001; Santos and Phillips 2009; Udayanga et al. 2011), which led to the description of over 200 species (Hyde et al. 2020). Some species of Diaporthe were reported to colonise a single host plant, while other species were found to be associated with different host plants (Santos and Phillips 2009; Diogo et al. 2010; Santos et al. 2011; Gomes et al. 2013). In addition, considerable variability of the phenotypic characters was found to be present within a species (Rehner and Uecker 1994; Mostert et al. 2001; Santos et al. 2010; Udayanga et al. 2011). During the past decade, a polyphasic approach, based on multi-locus DNA data, morphology and ecology, has been employed for species boundaries in the genus Diaporthe (Crous et al. 2012; Huang et al. 2015; Gao et al. 2016, 2017; Guarnaccia and Crous 2017; Guarnaccia et al. 2018; Yang et al. 2018, 2020). The classification of Diaporthe has been progressing and the basis for the species identification is a combination of morphological, cultural, phytopathological and phylogenetical analyses (Gomes et al. 2013; Udayanga et al. 2014, 2015; Fan et al. 2015; Huang et al. 2015; Gao et al. 2016, 2017; Guarnaccia and Crous 2017; Guarnaccia et al. 2018; Yang et al. 2018, 2020; Manawasinghe et al. 2019).
In Jiangxi Province, China, some forest trees were observed to be infected with fungal pathogens that cause dieback and leaf spots. Cankered branches and leaves with typical Diaporthe fruiting bodies were also found in the area. However, we found that only limited research had been undertaken regarding the fungal pathogens isolated from forest trees in Jiangxi Province. Hence, the present study was conducted to identify Diaporthe species that cause dieback and leaf spots disease in the forest trees in Jiangxi Province through morphological and multi-locus phylogenetic analyses, based on modern taxonomic concepts.
Materials and methods
Isolates
Fresh specimens of Diaporthe were isolated from the collected branches and leaves of six host plants during the collection trips conducted in Jiangxi Province (Table 1). A total of 24 isolates were established by removing a mucoid conidia mass from conidiomata, spreading the suspension on the surface of 1.8% potato dextrose agar (PDA) and incubating at 25 °C for up to 24 h. A single germinating conidium was plated on to fresh PDA plates. Specimens were deposited at the Museum of the Beijing Forestry University (BJFC). Axenic cultures were maintained at the China Forestry Culture Collection Centre (CFCC).
Table 1.
Reference sequences included in molecular phylogenetic analyses of Diaporthe.
| Species | Isolate | Host | Location | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | cal | his3 | tef1 | tub2 | ||||
| D. acericola | MFLUCC 17-0956 | Acer negundo | Italy | KY964224 | KY964137 | NA | KY964180 | KY964074 |
| D. acerigena | CFCC 52554 | Acer tataricum | China | MH121489 | MH121413 | MH121449 | MH121531 | NA |
| D. acutispora | CGMCC 3.18285 | Coffea sp. | China | KX986764 | KX999274 | NA | KX999155 | KX999195 |
| D. alangii | CFCC 52556 | Alangium kurzii | China | MH121491 | MH121415 | MH121451 | MH121533 | MH121573 |
| D. alnea | CBS 146.46 | Alnus sp. | Netherlands | KC343008 | KC343250 | KC343492 | KC343734 | KC343976 |
| D. ampelina | STEU2660 | Vitis vinifera | France | AF230751 | AY745026 | NA | AY745056 | JX275452 |
| D. amygdali | CBS 126679 | Prunus dulcis | Portugal | KC343022 | KC343264 | KC343506 | AY343748 | KC343990 |
| D. angelicae | CBS 111592 | Heracleum sphondylium | Austria | KC343027 | KC343269 | KC343511 | KC343753 | KC343995 |
| D. apiculatum | CGMCC 3.17533 | Camellia sinensis | China | KP267896 | NA | NA | KP267970 | KP293476 |
| CFCC 53068 | Rhus chinensis | China | MK432651 | MK442973 | MK442998 | MK578127 | MK578054 | |
| CFCC 53069 | Rhus chinensis | China | MK432652 | MK442974 | MK442999 | MK578128 | MK578055 | |
| CFCC 53070 | Rhus chinensis | China | MK432653 | MK442975 | MK443000 | MK578129 | MK578056 | |
| D. arctii | CBS 139280 | Arctium lappa | Austria | KJ590736 | KJ612133 | KJ659218 | KJ590776 | KJ610891 |
| D. arecae | CBS 161.64 | Areca catechu | India | KC343032 | KC343274 | KC343516 | KC343758 | KC344000 |
| D. arengae | CBS 114979 | Arenga enngleri | Hong Kong | KC343034 | KC343276 | KC343518 | KC343760 | KC344002 |
| D. aseana | MFLUCC 12-0299a | Unknown dead leaf | Thailand | KT459414 | KT459464 | NA | KT459448 | KT459432 |
| D. bauhiniae | CFCC 53071 | Bauhinia purpurea | China | MK432648 | MK442970 | MK442995 | MK578124 | MK578051 |
| CFCC 53072 | China | MK432649 | MK442971 | MK442996 | MK578125 | MK578052 | ||
| CFCC 53073 | China | MK432650 | MK442972 | MK442997 | MK578126 | MK578053 | ||
| D. beilharziae | BRIP 54792 | Indigofera australis | Australia | JX862529 | NA | NA | JX862535 | KF170921 |
| D. betulicola | CFCC 51128 | Betula albo-sinensis | China | KX024653 | KX024659 | KX024661 | KX024655 | KX024657 |
| D. biconispora | CGMCC 3.17252 | Citrus grandis | China | KJ490597 | KJ490539 | KJ490539 | KJ490476 | KJ490418 |
| D. biguttulata | CGMCC 3.17248 | Citrus limon | China | KJ490582 | NA | KJ490524 | KJ490461 | KJ490403 |
| CFCC 52584 | Juglans regia | China | MH121519 | MH121437 | MH121477 | MH121561 | MH121598 | |
| D. bohemiae | CPC 28222 | Vitis vinifera | Czech Republic | MG281015 | MG281710 | MG281361 | MG281536 | MG281188 |
| D. brasiliensis | CBS 133183 | Aspidosperma tomentosum | Brazil | KC343042 | KC343284 | KC343526 | KC343768 | KC344010 |
| D. caatingaensis | CBS 141542 | Tacinga inamoena | Brazil | KY085927 | NA | NA | KY115603 | KY115600 |
| D. caryae | CFCC 52563 | Carya illinoensis | China | MH121498 | MH121422 | MH121458 | MH121540 | MH121580 |
| D. celeris | CPC 28262 | Vitis vinifera | Czech Republic | MG281017 | MG281712 | MG281363 | MG281538 | MG281190 |
| D. celastrina | CBS 139.27 | Celastrus sp. | USA | KC343047 | KC343289 | KC343531 | KC343773 | KC344015 |
| D. cercidis | CFCC 52565 | Cercis chinensis | China | MH121500 | MH121424 | MH121460 | MH121542 | MH121582 |
| D. charlesworthii | BRIP 54884m | Rapistrum rugostrum | Australia | KJ197288 | NA | NA | KJ197250 | KJ197268 |
| D. cinnamomi | CFCC 52569 | Cinnamomum sp. | China | MH121504 | NA | MH121464 | MH121546 | MH121586 |
| D. citri | AR 3405 | Citrus sp. | USA | KC843311 | KC843157 | NA | KC843071 | KC843187 |
| CFCC 53079 | Citrus sinensis | China | MK573940 | MK574579 | MK574595 | MK574615 | MK574635 | |
| CFCC 53080 | Citrus sinensis | China | MK573941 | MK574580 | MK574596 | MK574616 | MK574636 | |
| CFCC 53081 | Citrus sinensis | China | MK573942 | MK574581 | MK574597 | MK574617 | MK574637 | |
| CFCC 53082 | Citrus sinensis | China | MK573943 | MK574582 | MK574598 | MK574618 | MK574638 | |
| D. citriasiana | CGMCC 3.15224 | Citrus unshiu | China | JQ954645 | KC357491 | KJ490515 | JQ954663 | KC357459 |
| D. citrichinensis | CGMCC 3.15225 | Citrus sp. | China | JQ954648 | KC357494 | NA | JQ954666 | NA |
| D. collariana | MFLU 17-2770 | Magnolia champaca | Thailand | MG806115 | MG783042 | NA | MG783040 | MG783041 |
| D. conica | CFCC 52571 | Alangium chinense | China | MH121506 | MH121428 | MH121466 | MH121548 | MH121588 |
| D. cucurbitae | CBS 136.25 | Arctium sp. | Unknown | KC343031 | KC343273 | KC343515 | KC343757 | KC343999 |
| D. cuppatea | CBS 117499 | Aspalathus linearis | South Africa | KC343057 | KC343299 | KC343541 | KC343783 | KC344025 |
| D. discoidispora | ZJUD89 | Citrus unshiu | China | KJ490624 | NA | KJ490566 | KJ490503 | KJ490445 |
| D. endophytica | CBS 133811 | Schinus terebinthifolius | Brazil | KC343065 | KC343307 | KC343549 | KC343791 | KC343065 |
| D. eres | AR5193 | Ulmus sp. | Germany | KJ210529 | KJ434999 | KJ420850 | KJ210550 | KJ420799 |
| D. fraxini-angustifoliae | BRIP 54781 | Fraxinus angustifolia | Australia | JX862528 | NA | NA | JX862534 | KF170920 |
| D. fraxinicola | CFCC 52582 | Fraxinus chinensis | China | MH121517 | MH121435 | NA | MH121559 | NA |
| D. fructicola | MAFF 246408 | Passiflora edulis × P. edulis f. flavicarpa | Japan | LC342734 | LC342738 | LC342737 | LC342735 | LC342736 |
| D. fukushii | MAFF 625034 | Pyrus pyrifolia | Japan | JQ807469 | NA | NA | JQ807418 | NA |
| D. fusicola | CGMCC 3.17087 | Lithocarpus glabra | China | KF576281 | KF576233 | NA | KF576256 | KF576305 |
| D. ganjae | CBS 180.91 | Cannabis sativa | USA | KC343112 | KC343354 | KC343596 | KC343838 | KC344080 |
| D. ganzhouensis | CFCC 53087 | Unknown dead wood | China | MK432665 | MK442985 | MK443010 | MK578139 | MK578065 |
| CFCC 53088 | Unknown dead wood | China | MK432666 | MK442986 | MK443011 | MK578140 | MK578066 | |
| D. garethjonesii | MFLUCC 12-0542a | Unknown dead leaf | Thailand | KT459423 | KT459470 | NA | KT459457 | KT459441 |
| D. guangxiensis | JZB320094 | Vitis vinifera | China | MK335772 | MK736727 | NA | MK523566 | MK500168 |
| D. gulyae | BRIP 54025 | Helianthus annuus | Australia | JF431299 | NA | NA | KJ197271 | JN645803 |
| D. helicis | AR5211 | Hedera helix | France | KJ210538 | KJ435043 | KJ420875 | KJ210559 | KJ420828 |
| D. heterophyllae | CBS 143769 | Acacia heterohpylla | France | MG600222 | MG600218 | MG600220 | MG600224 | MG600226 |
| D. hispaniae | CPC 30321 | Vitis vinifera | Spain | MG281123 | MG281820 | MG281471 | MG281644 | MG281296 |
| D. hubeiensis | JZB320123 | Vitis vinifera | China | MK335809 | MK500235 | NA | MK523570 | MK500148 |
| D. incompleta | CGMCC 3.18288 | Camellia sinensis | China | KX986794 | KX999289 | KX999265 | KX999186 | KX999226 |
| D. infecunda | CBS 133812 | Schinus terebinthifolius | Brazil | KC343126 | KC343368 | KC343610 | KC343852 | KC344094 |
| D. juglandicola | CFCC 51134 | Juglans mandshurica | China | KU985101 | KX024616 | KX024622 | KX024628 | KX024634 |
| D. kadsurae | CFCC 52586 | Kadsura longipedunculata | China | MH121521 | MH121439 | MH121479 | MH121563 | MH121600 |
| D. kochmanii | BRIP 54033 | Helianthus annuus | Australia | JF431295 | NA | NA | JN645809 | NA |
| D. kongii | BRIP 54031 | Portulaca grandiflora | Australia | JF431301 | NA | NA | JN645797 | KJ197272 |
| D. litchicola | BRIP 54900 | Litchi chinensis | Australia | JX862533 | NA | NA | JX862539 | KF170925 |
| D. lithocarpus | CGMCC 3.15175 | Lithocarpus glabra | China | KC153104 | KF576235 | NA | KC153095 | KF576311 |
| D. lonicerae | MFLUCC 17-0963 | Lonicera sp. | Italy | KY964190 | KY964116 | NA | KY964146 | KY964073 |
| D. lusitanicae | CBS 123212 | Foeniculum vulgare | Portugal | KC343136 | KC343378 | KC343620 | KC343862 | KC344104 |
| D. masirevicii | BRIP 57892a | Helianthus annuus | Australia | KJ197277 | NA | NA | KJ197239 | KJ197257 |
| D. middletonii | BRIP 54884e | Rapistrum rugostrum | Australia | KJ197286 | NA | NA | KJ197248 | KJ197266 |
| D. miriciae | BRIP 54736j | Helianthus annuus | Australia | KJ197282 | NA | NA | KJ197244 | KJ197262 |
| D. momicola | MFLUCC 16-0113 | Prunus persica | China | KU557563 | KU557611 | NA | KU557631 | KU55758 |
| D. multigutullata | ZJUD98 | Citrus grandis | China | KJ490633 | NA | KJ490575 | KJ490512 | KJ490454 |
| D. multigutullata | CFCC 53095 | Citrus maxima | China | MK432645 | MK442967 | MK442992 | MK578121 | MK578048 |
| CFCC 53096 | Citrus maxima | China | MK432646 | MK442968 | MK442993 | MK578122 | MK578049 | |
| CFCC 53097 | Citrus maxima | China | MK432647 | MK442969 | MK442994 | MK578123 | MK578050 | |
| D. musigena | CBS 129519 | Musa sp. | Australia | KC343143 | KC343385 | KC343627 | KC343869 | KC344111 |
| D. neilliae | CBS 144.27 | Spiraea sp. | USA | KC343144 | KC343386 | KC343628 | KC343870 | KC344112 |
| D. neoarctii | CBS 109490 | Ambrosia trifida | USA | KC343145 | KC343387 | KC343629 | KC343871 | KC344113 |
| D. oraccinii | CGMCC 3.17531 | Camellia sinensis | China | KP267863 | NA | KP293517 | KP267937 | KP293443 |
| D. ovoicicola | CGMCC 3.17093 | Citrus sp. | China | KF576265 | KF576223 | NA | KF576240 | KF576289 |
| D. pandanicola | MFLU 18-0006 | Pandanus sp. | Thailand | MG646974 | NA | NA | NA | MG646930 |
| D. pascoei | BRIP 54847 | Persea americana | Australia | JX862532 | NA | NA | JX862538 | KF170924 |
| D. passifloricola | CBS 141329 | Passiflora foetida | Malaysia | KX228292 | NA | KX228367 | NA | KX228387 |
| D. penetriteum | CGMCC 3.17532 | Camellia sinensis | China | KP714505 | NA | KP714493 | KP714517 | KP714529 |
| D. perjuncta | CBS 109745 | Ulmus glabra | Austria | KC343172 | KC343414 | KC343656 | KC343898 | KC344140 |
| D. perseae | CBS 151.73 | Persea gratissima | Netherlands | KC343173 | KC343415 | KC343657 | KC343899 | KC344141 |
| D. pescicola | MFLUCC 16-0105 | Prunus persica | China | KU557555 | KU557603 | NA | KU557623 | KU557579 |
| D. podocarpi-macrophylli | CGMCC 3.18281 | Podocarpus macrophyllus | China | KX986774 | KX999278 | KX999246 | KX999167 | KX999207 |
| D. pseudomangiferae | CBS 101339 | Mangifera indica | Dominican Republic | KC343181 | KC343423 | KC343665 | KC343907 | KC344149 |
| D. pseudophoenicicola | CBS 462.69 | Phoenix dactylifera | Spain | KC343184 | KC343426 | KC343668 | KC343910 | KC344152 |
| D. psoraleae-pinnatae | CBS 136413 | Psoralea pinnata | South Africa | KF777159 | NA | NA | NA | KF777252 |
| D. pterocarpicola | MFLUCC 10-0580a | Pterocarpus indicus | Thailand | JQ619887 | JX197433 | NA | JX275403 | JX275441 |
| D. pulla | CBS 338.89 | Hedera helix | Yugoslavia | KC343152 | KC343394 | KC343636 | KC343878 | KC344120 |
| D. pyracanthae | CAA483 | Pyracantha coccinea | Portugal | KY435635 | KY435656 | KY435645 | KY435625 | KY435666 |
| D. racemosae | CBS 143770 | Euclea racemosa | South Africa | MG600223 | MG600219 | MG600221 | MG600225 | MG600227 |
| D. rostrata | CFCC 50062 | Juglans mandshurica | China | KP208847 | KP208849 | KP208851 | KP208853 | KP208855 |
| D. sackstonii | BRIP 54669b | Helianthus annuus | Australia | KJ197287 | NA | NA | KJ197249 | KJ197267 |
| D. sambucusii | CFCC 51986 | Sambucus williamsii | China | KY852495 | KY852499 | KY852503 | KY852507 | KY852511 |
| D. schimae | CFCC 53103 | Schima superba | China | MK432640 | MK442962 | MK442987 | MK578116 | MK578043 |
| CFCC 53104 | Schima superba | China | MK432641 | MK442963 | MK442988 | MK578117 | MK578044 | |
| CFCC 53105 | Schima superba | China | MK432642 | MK442964 | MK442989 | MK578118 | MK578045 | |
| D. schini | CBS 133181 | Schinus terebinthifolius | Brazil | KC343191 | KC343433 | KC343675 | KC343917 | KC344159 |
| D. schisandrae | CFCC 51988 | Schisandra chinensis | China | KY852497 | KY852501 | KY852505 | KY852509 | KY852513 |
| D. schoeni | MFLU 15-1279 | Schoenus nigricans | Italy | KY964226 | KY964139 | NA | KY964182 | KY964109 |
| D. sennae | CFCC 51636 | Senna bicapsularis | China | KY203724 | KY228875 | NA | KY228885 | KY228891 |
| D. serafiniae | BRIP 55665a | Helianthus annuus | Australia | KJ197274 | NA | NA | KJ197236 | KJ197254 |
| D. siamensis | MFLUCC 10-573a | Dasymaschalon sp. | Thailand | JQ619879 | NA | NA | JX275393 | JX275429 |
| D. sojae | FAU635 | Glycine max | USA | KJ590719 | KJ612116 | KJ659208 | KJ590762 | KJ610875 |
| D. sterilis | CBS 136969 | Vaccinium corymbosum | Italy | KJ160579 | KJ160548 | MF418350 | KJ160611 | KJ160528 |
| D. subclavata | ICMP20663 | Citrus unshiu | China | KJ490587 | NA | KJ490529 | KJ490466 | KJ490408 |
| D. subellipicola | MFLU 17-1197 | Dead wood | China | MG746632 | NA | NA | MG746633 | MG746634 |
| D. subordinaria | CBS 464.90 | Plantago lanceolata | New Zealand | KC343214 | KC343456 | KC343698 | KC343940 | KC344182 |
| D. taoicola | MFLUCC 16-0117 | Prunus persica | China | KU557567 | NA | NA | KU557635 | KU557591 |
| D. tectonae | MFLUCC 12-0777 | Tectona grandis | China | KU712430 | KU749345 | NA | KU749359 | KU743977 |
| D. tectonendophytica | MFLUCC 13-0471 | Tectona grandis | China | KU712439 | KU749354 | NA | KU749367 | KU749354 |
| D. tectonigena | MFLUCC 12-0767 | Tectona grandis | China | KU712429 | KU749358 | NA | KU749371 | KU743976 |
| D. terebinthifolii | CBS 133180 | Schinus terebinthifolius | Brazil | KC343216 | KC343458 | KC343700 | KC343942 | KC344184 |
| D. ternstroemia | CGMCC 3.15183 | Ternstroemia gymnanthera | China | KC153098 | NA | NA | KC153089 | NA |
| D. thunbergii | MFLUCC 10-576a | Thunbergia laurifolia | Thailand | JQ619893 | JX197440 | NA | JX275409 | JX275449 |
| D. tibetensis | CFCC 51999 | Juglandis regia | China | MF279843 | MF279888 | MF279828 | MF279858 | MF279873 |
| D. tulliensis | BRIP 62248a | Theobroma cacao | Australia | KR936130 | NA | NA | KR936133 | KR936132 |
| D. ukurunduensis | CFCC 52592 | Acer ukurunduense | China | MH121527 | MH121445 | MH121485 | MH121569 | NA |
| D. unshiuensis | CGMCC 3.17569 | Citrus unshiu | China | KJ490587 | NA | KJ490529 | KJ490408 | KJ490466 |
| CFCC 52594 | Carya illinoensis | China | MH121529 | MH121447 | MH121487 | MH121571 | MH121606 | |
| D. undulata | CGMCC 3.18293 | Leaf of unknown host | China-Laos border | KX986798 | NA | KX999269 | KX999190 | KX999230 |
| D. vawdreyi | BRIP 57887a | Psidium guajava | Australia | KR936126 | NA | NA | KR936129 | KR936128 |
| D. verniciicola | CFCC 53109 | Vernicia montana | China | MK573944 | MK574583 | MK574599 | MK574619 | MK574639 |
| CFCC 53110 | Vernicia montana | China | MK573945 | MK574584 | MK574600 | MK574620 | MK574640 | |
| CFCC 53111 | Vernicia montana | China | MK573946 | MK574585 | MK574601 | MK574621 | MK574641 | |
| CFCC 53112 | Vernicia montana | China | MK573947 | MK574586 | MK574602 | MK574622 | MK574642 | |
| D. viniferae | JZB320071 | Vitis vinifera | China | MK341551 | MK500107 | MK500119 | MK500112 | |
| D. virgiliae | CMW40748 | Virgilia oroboides | South Africa | KP247566 | NA | NA | NA | KP247575 |
| D. xishuangbanica | CGMCC 3.18282 | Camellia sinensis | China | KX986783 | NA | KX999255 | KX999175 | KX999216 |
| D. xunwuensis | CFCC 53085 | Unknown dead wood | China | MK432663 | MK442983 | MK443008 | MK578137 | MK578063 |
| CFCC 53086 | Unknown dead wood | China | MK432664 | MK442984 | MK443009 | MK578138 | MK578064 | |
| D. yunnanensis | CGMCC 3.18289 | Coffea sp. | China | KX986796 | KX999290 | KX999267 | KX999188 | KX999228 |
| Diaporthella corylina | CBS 121124 | Corylus sp. | China | KC343004 | KC343246 | KC343488 | KC343730 | KC343972 |
Newly sequenced material is indicated in bold type. NA, not applicable.
Morphological observation
Agar plugs (6 mm diam.) were taken from the edge of actively-growing cultures on PDA and transferred on to the centre of 9 cm diam. Petri dishes containing 2% tap water agar, supplemented with sterile pine needles (PNA; Smith et al. 1996) and potato dextrose agar (PDA) and incubated at 25 °C under a 12 h near-ultraviolet light/12 h dark cycle to induce sporulation, as described in recent studies (Gomes et al. 2013; Lombard et al. 2014). Colony characters and pigment production on PNA and PDA were noted in the 10-day culture. Colony features were rated according to the colour charts of Rayner (1970). Cultures were examined periodically for the development of conidiomata. The microscopic examination was based on the morphological features of conidiomata obtained from the fungal growth, mounted in clear lactic acid. At least 30 conidia were measured to calculate the mean size/length. Micro-morphological observations were done at 1000× magnification using a Leica compound microscope (DM 2500) with interference contrast (DIC) optics. Descriptions, nomenclature and illustrations of taxonomic novelties were deposited at MycoBank (www.MycoBank.org).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from colonies grown on cellophane-covered PDA, using a CTAB (cetyltrimethylammonium bromide) method (Doyle and Doyle 1990). DNA was estimated by electrophoresis in 1% agarose gel and the yield was measured using the NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA), following the user manual (Desjardins et al. 2009). The PCR amplifications were performed in the DNA Engine Peltier Thermal Cycler (PTC-200; Bio-Rad Laboratories, Hercules, CA, USA). The primer set ITS1/ITS4 (White et al. 1990) was used to amplify the ITS region. The primer pair CAL228F/CAL737R (Carbone and Kohn 1999) was used to amplify the calmodulin gene (cal) and the primer pair CYLH4F (Crous et al. 2004) and H3-1b (Glass and Donaldson 1995) were used to amplify part of the histone H3 (his3) gene. The primer pair EF1-728F/EF1-986R (Carbone and Kohn 1999) was used to amplify a partial fragment of the translation elongation factor 1-α gene (tef1). The primer sets T1 (O’Donnell and Cigelnik 1997) and Bt2b (Glass and Donaldson 1995) were used to amplify the beta-tubulin gene (tub2); the additional combination of Bt2a/Bt2b (Glass and Donaldson 1995) was used in case of amplification failure of the T1/Bt2b primer pair. The PCR amplifications of the genomic DNA with the phylogenetic markers were done using the same primer pairs and conditions as in Yang et al. (2018). The PCR products were assayed via electrophoresis in 2% agarose gels, while the DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyser with a BigDye Terminater Kit v.3.1 (Inv-itrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
Phylogenetic analyses
The quality of the amplified nucleotide sequences was checked and combined using SeqMan v.7.1.0 and reference sequences were retrieved from the National Center for Biotechnology Information (NCBI), based on recent publications on the genus Diaporthe (Guarnaccia et al. 2018; Yang et al. 2018, 2020). Sequences were aligned using MAFFT v. 6 (Katoh and Toh 2010) and corrected manually using Bioedit 7.0.9.0 (Hall 1999). The best-fit nucleotide substitution models for each gene were selected using jModelTest v. 2.1.7 (Darriba et al. 2012) under the Akaike Information Criterion.
The phylogenetic analyses of the combined gene regions were performed using Maximum Likelihood (ML) and Bayesian Inference (BI) methods. ML was conducted using PhyML v. 3.0 (Guindon et al. 2010), with 1000 bootstrap replicates while BI was performed using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v. 3.0 (Ronquist et al. 2003). Two MCMC chains, started from random trees for 1,000,000 generations and trees, were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25% of trees were discarded as burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BPP) were estimated in the remaining 7500 trees. Phylogenetic trees were viewed with FigTree v.1.3.1 (Rambaut and Drummond 2010) and processed by Adobe Illustrator CS5. Sequence alignment and phylogenetic trees were deposited in TreeBASE (submission ID: S25213). The nucleotide sequence data of the new taxa were deposited in GenBank (Table 1).
Results
The phylogenetic position of the 24 isolates of Diaporthe was determined by the phylogenetic analysis of the combined ITS, cal, his3, tef1 and tub2 sequences data. Reference sequences of the representative species used in the analysis were selected from Yang et al. (2018) and supplemented with sequences from GenBank. The ITS, cal, his3, tef1tub2 and combined data matrices contained 522, 541, 529, 520, 535 and 2 659 characters with gaps, respectively. The alignment comprised of 142 strains together with Diaporthella corylina (culture CBS 121124) which was selected as the outgroup. The best nucleotide substitution model used for the analysis of ITS, his3 and tub2 was TrN+I+G, while HKY+I+G was used for cal and tef1. The topologies resulting from ML and BI analyses of the concatenated dataset were congruent (Fig. 1) and the sequences from the 24 Diaporthe isolates formed eight distinct clades as shown in Fig. 1, representing five undescribed species and three known species.
Figure 1.
Phylogram of Diaporthe from a Maximum Likelihood analysis based on combined ITS, cal, his3, tef1 and tub2. Values above the branches indicate Maximum Likelihood bootstrap (left, ML BP ≥ 50%) and Bayesian probabilities (right, BI PP ≥ 0.90). The tree is rooted with Diaporthella corylina. Strains in current study are in blue font and the ex-type cultures are in bold font.
Figure 11.
Continued.
Taxonomy
Diaporthe apiculatum
Y.H. Gao & L. Cai, in Gao, Liu & Cai, Syst. Biodiv. 14: 106. 2016.
6FAC7E1B-E3D9-59EA-A41B-9780AD378F43
Figure 2.
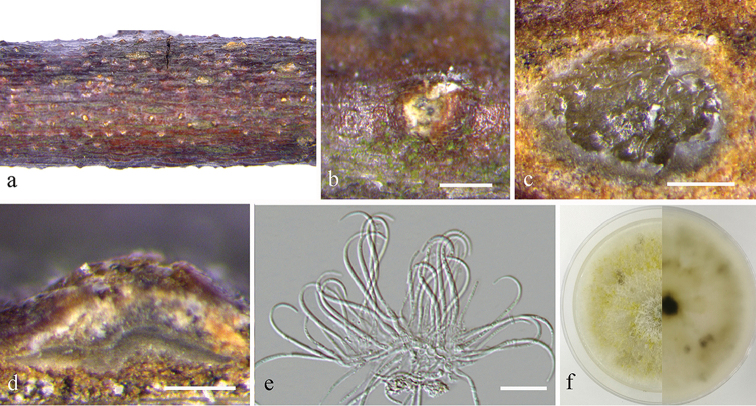
Diaporthe apiculatum on Rhus chinensis (BJFC-S1680) a, b habit of conidiomata in wood c transverse section of conidiomata d longitudinal section through conidiomata e conidiogenous cells attached with beta conidia f the colony on PDA. Scale bars: 200 μm (b–d); 10 μm (e).
Description.
Conidiomata pycnidial, discoid, immersed in bark, scattered, slightly erumpent through bark surface, with a solitary undivided locule. Ectostromatic disc yellowish to grey, one ostiole per disc, (300–)305–357(–368) μm diam. Ostiole medium black, up to level of disc. Locule undivided, (338–)357–450(–464) μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical, hyaline, densely aggregated, phiailidic, unbranched, straight or slightly curved. Beta conidia hyaline, aseptate, filiform, hamate, eguttulate, base subtruncate, tapering towards one apex, (26.5–)30–39.5(–43) × 1.5–2 µm. Alpha conidia not observed.
Culture characters.
Colony originally flat with white fluffy aerial mycelium, becoming yellowish to pale green mycelium with age, marginal area irregular, conidiomata absent.
Specimens examined.
China. Jiangxi Province: Ganzhou City, Fengshan Forest Park, on branches of Rhus chinensis, 25°45'12"N, 115°00'41"E, 23 Jul 2018, Q. Yang, Y. Liu, Y.M. Liang & C.M. Tian (BJFC-S1680; living culture: CFCC 53068, CFCC 53069 and CFCC 53070).
Notes.
Diaporthe apiculatum was originally described as an endophyte from healthy leaves of Camellia sinensis in Jiangxi Province, China (Gao et al. 2015). In the present study, three isolates (CFCC 53068, CFCC 53069 and CFCC 53070) from symptomatic branches of Rhus chinensis were found congruent with D. apiculatum, based on DNA sequence and morphological data (Fig. 1). The clade was, therefore, confirmed to be D. apiculatum and was found to be both an endophyte and a pathogen.
Diaporthe bauhiniae
C.M. Tian & Q. Yang sp. nov.
13521E6A-F2E0-57FC-91D1-D6461FF110AC
829519
Figure 3.
Diaporthe bauhiniae on Bauhinia purpurea (BJFC-S1621) a habit of conidiomata in wood b transverse section of conidiomata c longitudinal section through conidiomata d the colony on PDAe conidiogenous cells attached with alpha conidia f Alpha conidia g Beta conidia. Scale bars: 100 μm (b, c); 10 μm (e–h).
Diagnosis.
Distinguished from the phylogenetically closely-related species D. psoraleae-pinnatae in alpha and beta conidia.
Etymology.
Named after Bauhinia, the host genus where the fungus was isolated.
Description.
Conidiomata pycnidial, immersed in bark, scattered, slightly erumpent through bark surface, nearly flat, discoid, with a solitary undivided locule. Ectostromatic disc grey to brown, one ostiole per disc. Locule circular, undivided, (180–)200–290(–300) μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, cylindrical, unbranched, straight, tapering towards the apex. Alpha conidia hyaline, aseptate, ellipsoidal to fusiform, biguttulate to multi-guttulate, (7.5–)9–13(–14) × (1.5–)2–2.5(–3) μm. Beta conidia hyaline, aseptate, filiform, straight to sinuous, eguttulate, (25–)28.5–40(–43) × 1 µm.
Culture characters.
Colony at first white, becoming wine-red in the centre with age. Aerial mycelium white, dense, fluffy, conidiomata absent.
Specimens examined.
China. Jiangxi Province: Ganzhou City, on branches of Bauhinia purpurea, 25°52'21"N, 114°56'44"E, 11 May 2018, Q. Yang, Y. Liu & Y.M. Liang (holotype BJFC-S1621; ex-type living culture: CFCC 53071; living culture: CFCC 53072 and CFCC 53073).
Notes.
Three isolates representing D. bauhiniae cluster in a well-supported clade and appear most closely related to D. psoraleae-pinnatae. Diaporthe bauhiniae can be distinguished from D. psoraleae-pinnatae, based on ITS and tub2 (38/458 in ITS and 11/418 in tub2). Morphologically, D. bauhiniae differs from D. psoraleae-pinnatae in having narrower alpha conidia (2–2.5 vs. 2.5–3 μm) and the beta conidia of D. psoraleae-pinnatae were not observed (Crous et al. 2013).
Diaporthe citri
(H.S. Fawc.) F.A. Wolf, J. Agric. Res., Washington 33(7): 625, 1926.
6F1FD04B-82E2-56E3-AC79-428FF8679E08
Figure 4.
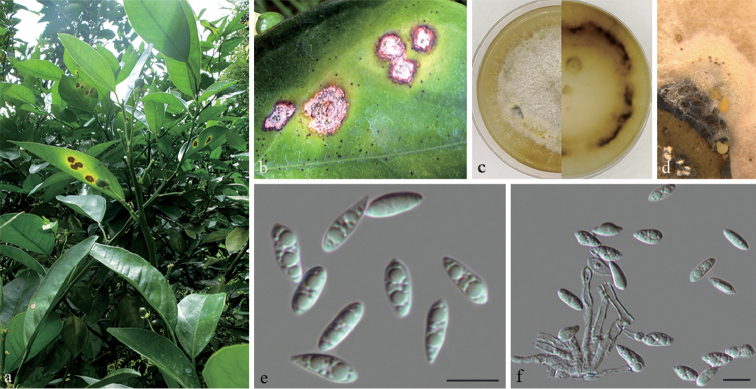
Diaporthe citri on Citrus sinensis (BJFC-S1658) a, b symptoms on leaves of host plant c culture on PDA (30d) d conidiomata e alpha conidia f conidiophores and alpha conidia. Scale bars: 10 μm (e, f).
Description.
Leaf spots subcircular to irregular, pale brown, with dark brown at margin. Pycnidia solitary, scattered on the leaf surface. Pycnidial conidiomata in culture, globose, erumpent, single or clustered in groups of 3–5 pycnidia, coated with hyphae, cream to yellowish translucent conidial droplets exuded from ostioles. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, unbranched, septate, straight, slightly tapering towards the apex, 14.5–25 × 2–3 μm. Alpha conidia hyaline, aseptate, rounded at one end, apex at the other end, usually with two large guttulate, (9.5–)10.5–12 × 3.5–4.5 μm. Beta conidia not observed.
Culture characters.
Colony originally flat with white fluffy aerial mycelium, becoming greyish mycelium with age, with yellowish-cream conidial drops exuding from the ostioles.
Specimens examined.
China. Jiangxi Province: Ganzhou City, on leaves of Citrus sinensis, 24°59'44"N, 115°31'01"E, 13 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1658; living culture: CFCC 53079 and CFCC 53080); 24°59'45"N, 115°31'02"E, 13 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1659; living culture: CFCC 53081 and CFCC 53082).
Notes.
Diaporthe citri is a widely distributed species in citrus-growing regions. In the present study, four isolates (CFCC 53079, CFCC 53080, CFCC 53081 and CFCC 53082) from symptomatic leaves of Citrus sinensis were congruent with D. citri, based on DNA sequence and morphological data (Fig. 1). The clade was, therefore, confirmed to be D. citri.
Diaporthe ganzhouensis
C.M. Tian & Q. Yang sp. nov.
90B3BE1F-2BF5-5865-BCDC-C58684BB6AEE
829522
Figure 5.
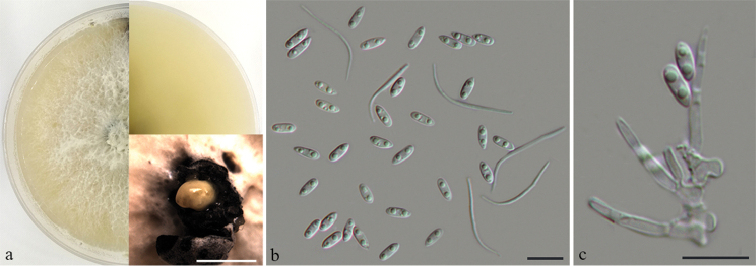
Diaporthe ganzhouensis on unknown host (BJFC-S1678) a the colony on PDA and conidiomata b alpha and beta conidia c conidiogenous cells and alpha conidia. Scale bars: 10 μm (b, c).
Diagnosis.
Distinguished from the phylogenetically closely-related species D. vawdreyi in having longer conidiophores and wider alpha conidia.
Etymology.
Named after Ganzhou City where the species was first collected.
Description.
On PDA: Conidiomata pycnidial, subglobose, solitary, deeply embedded in the medium, erumpent, dark brown to black. Pale yellow conidial drops exuding from ostioles. Conidiophores (12–)15.5–21 × 1.5–2 μm, cylindrical, hyaline, phiailidic, branched, straight or slightly curved. Alpha conidia 6.5–8.5(–9) × 2–2.5(–3) μm, aseptate, hyaline, ellipsoidal to fusiform, rounded at one end, slightly apex at the other end, biguttulate. Beta conidia hyaline, aseptate, filiform, sinuous at one end, eguttulate, (21.5–)25.5–31(–33) × 1 µm.
Culture characters.
Colony at first white, becoming yellowish with age. Aerial mycelium white, dense, fluffy, with visible solitary conidiomata at maturity.
Specimens examined.
China. Jiangxi Province: Ganzhou City, unknown dead wood, 25°45'17"N, 115°00'41"E, 23 Jul 2018, Q. Yang, Y. Liu, Y.M. Liang & C.M. Tian (holotype BJFC-C004; ex-type culture: CFCC 53087; living culture: CFCC 53088).
Notes.
Diaporthe ganzhouensis comprises the isolates CFCC 53087 and CFCC 53088, revealed to be closely related to D. vawdreyi in the combined phylogenetic tree (Fig. 1). Diaporthe ganzhouensis can be distinguished, based on ITS, tef1-α and tub2 loci from D. vawdreyi (6/456 in ITS, 63/357 in tef1-α and 40/469 in tub2). Diaporthe ganzhouensis differs morphologically from D. vawdreyi in having longer conidiopores (15.5–21 vs. 6–15 μm) and wider alpha conidia (2–2.5 vs. 1.5–2 μm) (Crous et al. 2015).
Diaporthe multiguttulata
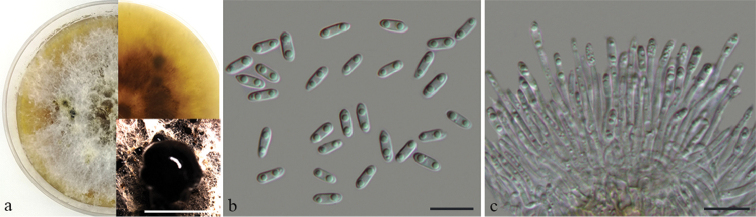
F. Huang, K.D. Hyde & Hong Y. Li, in Huang et al., Fungal Biology 119(5): 343. 2015.
FF202454-C6E7-5DAF-AFA6-F783DE9AA27A
Figure 6.
Diaporthe multiguttulata on Citrus maxima (BJFC-S1614) a, b habit of conidiomata on twig c conidiomata on PDAd transverse section through conidiomata e longitudinal section through conidiomata f conidiogenous cells attached with alpha conidia g alpha conidia h the colony on PDA. Scale bars: 200 μm (b, d, e); 10 μm (f, g).
Description.
Conidiomata pycnidial, 692–750(–800) μm diam., solitary and with single necks erumpent through host bark. Tissue around neck is cylindrical. Locule circular, undivided, 450–565(–600) μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells unbranched, straight or slightly curved, apical or base sometimes swelling, (8.5–)9–10.5(–11) × 1.5–2 μm. Alpha conidia hyaline, aseptate, ellipsoidal, biguttulate or with one large guttulate, rounded at one end, slightly apex at the other end, occasionally submedian constriction, (7.5–)8–9(–10.5) × 4–5(–5.5) μm. Beta conidia not observed.
Culture characters.
Colony originally flat with white felty aerial mycelium, becoming pale green mycelium with age, margin area irregularly, with visible solitary conidiomata at maturity.
Specimens examined.
China. Jiangxi Province: Ganzhou City, on branches of Citrus maxima, 25°51'28"N, 114°55'19"E, 11 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1614; living culture: CFCC 53095, CFCC 53096 and CFCC 53097).
Notes.
Diaporthe multiguttulata was originally described as an endophyte from a healthy branch of Citrus grandis in Fujian Province, China (Huang et al. 2015). In the present study, three isolates (CFCC 53095, CFCC 53096 and CFCC 53097) from symptomatic branches of Citrus maxima were congruent with D. multigutullata, based on DNA sequence data and confirmed from the morphological analysis (Fig. 1). The clade, therefore, was verified as D. multigutullata which could exist both as an endophyte and a pathogen.
Diaporthe schimae
C.M. Tian & Q. Yang sp. nov.
B56097C9-3E21-5805-B9F2-80D8060CA4A3
829526
Figure 7.
Diaporthe schimae on Schima superba (BJFC-S1661) a symptoms on leaves of host plant b the colony on PDAc conidiomata on PDAd conidiophores cells attached with beta conidia e Alpha conidia. Scale bars: 10 μm (d, e).
Diagnosis.
Distinguished from the phylogenetically closely-related species D. sennae in having larger alpha conidia and longer beta conidia.
Etymology.
Named after the host genus Schima on which the fungus was isolated.
Description.
Leaf spots subcircular to irregular, pale brown, with dark brown at margin. Pycnidia solitary, scattered on the leaf surface. Pycnidial conidiomata in culture, globose, (150–)173–357(–373) µm in its widest diam., erumpent, single or clustered in groups of 3–5 pycnidia, coated with hyphae, cream to yellowish translucent conidial droplets exuded from ostioles. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, unbranched, septate, straight, slightly tapering towards the apex. Alpha conidia scarce, hyaline, aseptate, ellipsoidal to spindle-shaped, four small guttulate, (7.5–)8–8.5(–9) × 2.5–3 μm. Beta conidia abundant, hyaline, aseptate, filiform, straight to sinuous at one end, eguttulate, (25–)27.5–38.5(–40.5) × 1–1.5 µm.
Culture characters.
Colony entirely white, with fluffy aerial mycelium, concentric zonation, margin fimbricate, reverse slightly yellowish.
Specimens examined.
China. Jiangxi Province: Ganzhou City, Fengshan Forest Park, on leaves of Schima superba, 25°44'22"N, 114°59'40"E, 15 May 2018, Q. Yang, Y. Liu & Y.M. Liang (holotype BJFC-S1661; ex-type culture: CFCC 53103); 24°40'51"N, 115°34'36"E, 15 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1662; living culture: CFCC 53104); 24°40'52"N, 115°34'54"E, 15 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1663; living culture: CFCC 53105).
Notes.
Diaporthe schimae occurs in an independent clade (Fig. 1) and was revealed to be phylogenetically distinct from D. sennae. Diaporhe schimae can be distinguished with D. sennae by 41 nucleotides in concatenated alignment, in which three were distinct in the ITS region, 20 in the tef1-α region and 18 in the tub2 region. Diaporthe schimae differs morphologically from D. sennae in having larger alpha conidia and longer beta conidia (8–8.5 × 2.5–3 vs. 5.5–6.3 × 1.5–1.7 µm in alpha conidia; 27.5–38.5 vs. 18.4–20 µm in beta conidia) (Yang et al. 2017a).
Diaporthe verniciicola
C.M. Tian & Q. Yang sp. nov.
F4EAE0F5-46B8-58C0-B937-A95F82DCDF39
832921
Figure 8.
Diaporthe verniciicola on Vernicia montana (BJFC-S1622) a, b habit of conidiomata on twig c transverse section through conidiomata d longitudinal section through conidiomata e alpha conidia f conidiophores g culture on PDA (30d). Scale bars: 500 μm (b); 200 μm (c); 10 μm (e, f).
Diagnosis.
Distinguished from the phylogenetically closely-related species D. rostrata in having smaller alpha conidia; and from D. juglandicola in having wider alpha conidia.
Etymology.
Named after the host genus Vernicia on which the fungus was isolated.
Description.
Conidiomata pycnidial, 825–1050 × 445–500 μm diam., solitary and with single necks erumpent through host bark. Tissue around neck is conical. Locule circular, undivided, 400–665 μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells unbranched, straight or sinuous, 14.5–21.5 × 1–1.5 μm. Alpha conidia hyaline, aseptate, ellipsoidal to fusiform, with 1–2-guttulate, 7–8.5 × 3–3.5 μm. Beta conidia not observed.
Culture characters.
Colony white to yellowish, with dense and felted mycelium in the centre, lacking aerial mycelium, conidiomata absent.
Specimens examined.
China. Jiangxi Province: Ganzhou City, on branches of Vernicia montana, 24°40'51"N, 115°34'52"E, 12 May 2018, Q. Yang, Y. Liu & Y.M. Liang (holotype BJFC-S1622; ex-type culture: CFCC 53109); 24°40'52"N, 115°34'50"E, 12 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1623; living culture: CFCC 53110); 24°45'14"N, 115°34'00"E, 12 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1624; living culture: CFCC 53111); 25°44'15"N, 114°59'32"E, 15 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1624; living culture: CFCC 53112).
Notes.
Two isolates of D. verniciicola clustered in a well-supported clade (ML/BI = 100/1) and appeared closely related to D. rostrata and D. juglandicola (Fig. 1). Morphologically, D. verniciicola is similar to D. rostrata characterised by conidiomata with single necks erumpent through the host bark. However, the new taxon can be distinguished from D. rostrata in having smaller alpha conidia (7–8.5 × 3–3.5 vs. 8.5–11.5 × 4–5 μm) (Fan et al. 2015) and D. verniciicola differs from D. juglandicola in having wider alpha conidia (3–3.5 vs. 2.5–3 μm) (Yang et al. 2017b). This is the first discovery of a Diaporthe species isolated from infected branches or twigs on Vernicia montana and was confirmed as a new species, based on phylogeny and morphology.
Diaporthe xunwuensis
C.M. Tian & Q. Yang sp. nov.
6EF44D7F-DB53-53B3-99E0-B6C9247F20A0
829521
Figure 9.
Diaporthe xunwuensis on unknown host (BJFC-S1679) a the colony on PDA and conidiomata b alpha conidia c conidiogenous cells attached with alpha conidia. Scale bars: 10 μm (a–c).
Diagnosis.
Distinguished from the phylogenetically closely-related species D. oraccinii in having longer conidiophores and larger alpha conidia.
Etymology.
Named after the county (Xunwu) where the species was first collected.
Description.
On PDA: Conidiomata pycnidial, globose, solitary or aggregated, deeply embedded in the medium, erumpent, dark brown to black. Hyaline conidial drops exuding from ostioles. Conidiophores (18.5–)21.5–30(–32.5) × 1–1.5(–2) μm, cylindrical, hyaline, phiailidic, unbranched, straight to sinuous. Alpha conidia (6.5–)7–8.5 × 2–3 μm, aseptate, hyaline, ellipsoidal to fusiform, rounded at one end, slightly apex at the other end, usually with 2-guttulate. Beta conidia not observed.
Culture characters.
Colony at first white, becoming dark brown in the centre with age. Aerial mycelium white, dense, fluffy, with black conidial drops exuding from the ostioles.
Specimens examined.
China. Jiangxi Province: Ganzhou City, unknown dead wood, 25°45'17"N, 115°00'41"E, 23 Jul 2018, Q. Yang, Y. Liu, Y.M. Liang & C.M. Tian (holotype BJFC-C003; ex-type culture: CFCC 53085; living culture: CFCC 53086).
Notes.
Two isolates representing D. xunwuensis clustered in a well-supported clade and appear most closely related to D. oraccinii. Diaporthe xunwuensis can be distinguished from D. oraccinii, based on ITS, his3 and tef1-α loci (5/471 in ITS, 5/432 in his3 and 5/325 in tef1-α). Morphologically, D. xunwuensis differs from D. oraccinii in having longer conidiopores (21.5–30 vs. 10.5–22.5 μm) and larger alpha conidia (7–8.5 × 2–3 vs. 5.5–7.5 × 0.5–2 μm) (Gao et al. 2016).
Discussion
The current study described eight Diaporthe species from 24 strains, based on a large set of freshly-collected specimens. It includes five new species and three known species, which were sampled from six host genera distributed in Jiangxi Province of China (Table 1). In this study, 142 reference sequences (including outgroup) were selected, based on BLAST searches of NCBIs GenBank nucleotide database and included in the phylogenetic analyses (Table 1). Phylogenetic analyses, based on five combined loci (ITS, cal, his3, tef1 and tub2), as well as morphological characters, revealed the diversity of Diaporthe species in Jiangxi Province, mainly focusing on diebacks from major ecological or economic forest trees.
The identification and characterisation of novel taxa and new host records indicate the high potential of Diaporthe to evolve rapidly. In the present study, five species were first reported in China as pathogens. Amongst these species, D. bauhiniae was characterised by having longer alpha conidia (9–13 × 2–2.5 μm). Diaporthe ganzhouensis and D. xunwuensis were isolated from unknown dead wood, but D. ganzhouensis can be distinguish from D. xunwuenesis in having beta conidia and was supported by analysis of the sequence data. Diaporthe schimae was identified as the most widespread species from isolates collected in Jiangxi Province. Diaporthe verniciicola have conidiomata with single necks erumpent through the host bark. Furthermore, two new host records were described, D. apiculatum from Rhus chinensis and D. multiguttulata from Citrus maxima.
Recent plant pathological studies have revealed that several Diaporthe species cause disease, particularly to important plant hosts on a wide range of economically-significant agricultural crops, such as blueberries, citrus, grapes, oaks, sunflowers, soybeans, tea plants, tropical fruits, vegetables and various trees (van Rensburg et al. 2006; Santos and Phillips 2009; Santos et al. 2011; Thompson et al. 2011; Grasso et al. 2012; Lombard et al. 2014; Huang et al. 2015; Udayanga et al. 2015; Gao et al. 2016; Guarnaccia et al. 2018; Yang et al. 2020). For example, research conducted by Huang et al. (2015) revealed seven endophytic Diaporthe species on Citrus; Gao et al. (2016) demonstrated that Diaporthe isolates associated with Camellia spp. could be assigned to seven species and two species complexes; Guarnaccia et al. (2018) explored the occurrence, diversity and pathogenicity of Diaporthe species associated with Vitis vinifera and revealed four new Diaporthe species; Yang et al. (2018) provided the first molecular phylogenetic framework of Diaporthe diversity associated with dieback diseases in China. Following the adoption of DNA sequence-based methods, Diaporthe taxonomy is actively changing, with numerous species being described each year.
The present study is the first evaluation of Diaporthe species, associated with dieback diseases in Jiangxi Province using the combined morphology and molecular data and provided useful information for evaluating the pathogenicity of various species. Multiple strains from different locations should also be subjected to multi-locus phylogenetic analysis to determine intraspecific variation and redefine species boundaries. The descriptions and molecular data of Diaporthe species, provided in this study, represent a resource for plant pathologists, plant quarantine officials and taxonomists for identification of Diaporthe.
Supplementary Material
Acknowledgements
This study is financed by the National Natural Science Foundation of China (Project No.: 31670647). We are grateful to Chungen Piao and Minwei Guo (China Forestry Culture Collection Center (CFCC), Chinese Academy of Forestry, Beijing) for support of strain preservation during this study.
Citation
Yang Q, Jiang N, Tian C-M (2021) New species and records of Diaporthe from Jiangxi Province, China. MycoKeys 77: 41–64. https://doi.org/10.3897/mycokeys.77.59999
Funding Statement
the National Natural Science Foundation of China (Project No.: 31670647); the Research Foundation of Education Bureau of Hunan Province, China (Project No.: 19B608) and the introduction of talent research start-up fund project of CSUFT (Project No.: 2019YJ025).
References
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in fila-mentous ascomycetes. Mycologia 3: 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Summerell BW, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GESTJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ. (2012) Fungal Planet description sheets: 107–127. Persoonia 28: 138–182. 10.3767/003158512X652633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, Cheewangkoon R, van der Bank M, Swart WJ, Stchigel AM, Cano-Lira JF, Roux J, Madrid H, Damm U, Wood AR, Shuttleworth LA, Hodges CS, Munster M, de Jesús Yáñez-Morales M, Zúñiga-Estrada L, Cruywagen EM, De Hoog GS, Silvera C, Najafzadeh J, Davison EM, Davison PJN, Barrett MD, Barrett RL, Manamgoda DS, Minnis AM, Kleczewski NM, Flory SL, Castlebury LA, Clay K, Hyde KD, Maússe-Sitoe SND, Shuaifei C, Lechat C, Hairaud M, Lesage-Meessen L, Pawłowska J, Wilk M, Śliwińska-Wyrzychowska A, Mętrak M, Wrzosek M, Pavlic-Zupanc D, Maleme HM, Slippers B, Mac Cormack WP, Archuby DI, Grünwald NJ, Tellería MT, Dueñas M, Martín MP, Marincowitz S, de Beer ZW, Perez CA, Gené J, Marin-Felix Y, Groenewald JZ. (2013) Fungal Planet description sheets: 154–213. Persoonia 31: 188–296. 10.3767/003158513X675925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane MS, Tan YP, Altés A, Barasubiye T, Barnes CW, Blanchette RA, Boertmann D, Bogo A, Carlavilla JR, Cheewangkoon R, Daniel R, de Beer ZW, Yáñez-Morales M de Jesús, Duong TA, Fernández-Vicente J, Geering ADW, Guest DI, Held BW, Heykoop M, Hubka V, Ismail AM, Kajale SC, Khemmuk W, Kolařík M, Kurli R, Lebeuf R, Lévesque CA, Lombard L, Magista D, Manjón JL, Marincowitz S, Mohedano JM, Nováková A, Oberlies NH, Otto EC, Paguigan ND, Pascoe IG, Pérez-Butrón JL, Perrone G, Rahi P, Raja HA, Rintoul T, Sanhueza RMV, Scarlett K, Shouche YS, Shuttleworth LA, Taylor PWJ, Thorn RG, Vawdrey LL, Solano-Vidal R, Voitk A, Wong PTW, Wood AR, Zamora JC, Groenewald JZ. (2015) Fungal Planet description sheets: 371–399. Persoonia 35: 264–327. 10.3767/003158515X690269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DQ, Wijayawardene NN, Bhat DJ, Chukeatirote E, Bahkali AH, Zhao R-L, Xu J-C, Hyde KD. (2014) Pustulomyces gen. nov. accommodated in Diaporthaceae, Diaporthales, as revealed by morphology and molecular analyses. Cryptogamie, Mycologie 35: 63–72. 10.7872/crym.v35.iss1.2014.63 [DOI] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: e772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed]
- Desjardins P, Hansen JB, Allen M. (2009) Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. Journal of Visualized Experiments: JoVE 33: 1–3. 10.3791/1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo E, Santos JM, Phillips AJ. (2010) Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115. 10.1007/s13225-010-0057-x [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. 10.2307/2419362 [DOI] [Google Scholar]
- Fan XL, Hyde KD, Udayanga D, Wu XY, Tian CM. (2015) Diaporthe rostrata, a novel ascomycete from Juglans mandshurica associated with walnut dieback. Mycological Progress 14: 1–8. 10.1007/s11557-015-1104-5 [DOI] [Google Scholar]
- Fan XL, Yang Q, Bezerra JDP, Alvarez LV, Tian CM. (2018) Diaporthe from walnut tree (Juglans regia) in China, with insight of Diaporthe eres complex. Mycological Progress 1–13. 10.1007/s11557-018-1395-4 [DOI]
- Fu CH, Hsieh HM, Chen CY, Chang TT, Huang YM, Ju YM. (2013) Ophiodiaporthe cyatheae gen. et sp. nov., a diaporthalean pathogen causing a devastating wilt disease of Cyathea lepifera in Taiwan. Mycologia 105: 861–872. 10.3852/12-346 [DOI] [PubMed] [Google Scholar]
- Gao YH, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14: 102–117. 10.1080/14772000.2015.1101027 [DOI] [Google Scholar]
- Gao YH, Liu F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA Fungus 8: 153–187. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YH, Su YY, Sun W, Cai L. (2015) Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biology 115: 295–309. 10.1016/j.funbio.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. 10.1128/AEM.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1–41. 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed]
- Grasso FM, Marini M, Vitale A, Firrao G, Granata G. (2012) Canker and dieback on Platanus × acerifolia caused by Diaporthe scabra. Forest Pathology 42: 510–513. 10.1111/j.1439-0329.2012.00785.x [DOI] [Google Scholar]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8: 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Woodhall J, Armengol J, Cinelli T, Eichmeier A, Ezra D, Fontaine F, Gramaje D, Gutierrez-Aguirregabiria A, Kaliterna J, Kiss L, Larignon P, Luque J, Mugnai L, Naor V, Raposo R, Sándor E, Váczy KZ, Crous PW. (2018) Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40: 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hall T. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, Fu Y, Hyde KD, Li HY. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119: 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Jayarama Bhat D, Gareth Jones EB, Liu N-G, Abeywickrama PD, Mapook A, Wei D, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao D-F, Li J, Samarakoon MC, Chaiwan N, Chuan-Gen Lin, Phutthacharoen K, Zhang S-N, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang H-B, Yang J, Zeng M, Huanraluek N, Liu J-K, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang S-K, Thiyagaraja V, Lu Y-Z, Jayawardena RS, Dong W, Yang E-F, Singh SK, Singh MS, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga LRM, Gibertoni BT, Pfliegler WP, Horváth E, Imre A, Alves LA, da Silva Santos CA, Tiago VP, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu J, Sheng J. (2020) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht SC, Crous PW, Groenewald JZ, Tewoldemedhin YT, Marasas WFO. (2011) Diaporthaceae associated with root and crown rot of maize. IMA Fungus 2: 13–24. 10.5598/imafungus.2011.02.01.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van Leeuwen GCM, Guarnaccia V, Polizzi G, Van Rijswick PC, Karin Rosendahl KC, Gabler J, Crous PW. (2014) Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53: 287–299. 10.14601/Phytopathol_Mediterr-14034 [DOI] [Google Scholar]
- Manawasinghe IS, Dissanayake A, Liu M, Liu M, Wanasinghe DN, Xu J, Zhao W, Zhang W, Zhou Y, Hyde KD, Brooks S, Yan J. (2019) High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Frontiers in Microbiology 10: e1936. 10.3389/fmicb.2019.01936 [DOI] [PMC free article] [PubMed]
- Marin-Felix Y, Hernández-Restrepo M, Wingfield M J, Akulov A, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F, Lombard L, Luangsa-ard J, Marincowitz S, Moslemi A, Mostert L, Quaedvlieg W, Schumacher RK, Spies CFJ, Thangavel R, Taylor PWJ, Wilson AM, Wingfield BD, Wood AR, Crous PW. (2019) Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology 92: 47–133. 10.1016/j.simyco.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Kang JC, Phillips AJ. (2001) Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93: 146–167. 10.1080/00275514.2001.12061286 [DOI] [Google Scholar]
- Muralli TS, Suryanarayanan TS, Geeta R. (2006) Endophytic Phomopsis species: host range and implications for diversity estimates. Canadian Journal of Microbiology 52: 673–680. 10.1139/w06-020 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. (2010) FigTree v.1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh.
- Rayner RW. (1970) A mycological colour chart. Commonwealth Mycological Institute, Kew, 34 pp. [Google Scholar]
- Rehner SA, Uecker FA. (1994) Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Canadian Journal of Botany 72: 1666–1674. 10.1139/b94-204 [DOI] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48: 135–144. 10.1007/S10267-007-0347-7 [DOI] [Google Scholar]
- Santos JM, Correia VG, Phillips AJL. (2010) Primers for mating-type diagnosis in Diaporthe and Phomopsis, their use in teleomorph induction in vitro and biological species definition. Fungal Biology 114: 255–270. 10.1016/j.funbio.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Diversity 34: 111–125. [Google Scholar]
- Santos JM, Vrandečić K, Ćosić J, Duvnjak T, Phillips AJL. (2011) Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27: 9–19. 10.3767/003158511X603719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ, Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Phillips AJL, Bhat JD, Perera RH, Li QR, Li WJ, Tangthirasunun N, Norphanphoun C, Karunarathna SC, Camporesi E, Manawasighe IS, Al-Sadi AM, Hyde KD. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86: 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Wingfeld MJ, Coutinho TA, Crous PW. (1996) Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. 10.1016/S0254-6299(15)30596-2 [DOI]
- Thompson SM, Tan YP, Young AJ, Neate SM, Aitken EAB, Shivas RG. (2011) Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27: 80–89. 10.3767/003158511X617110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014) Insights into the genus Diaporthe: phylogenetic species delimitation in the D. eres species complex. Fungal Diversity 67: 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthe sojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119: 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Bahkali AH, Hyde KD. (2011) The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50: 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Uecker FA. (1988) A world list of Phomopsis names with notes on nomenclature, morphology and biology. Mycological Memoirs 13: 1–231. [Google Scholar]
- van Rensburg JCJ, Lamprecht SC, Groenewald JZ, Castlebury LA, Crous PW. (2006) Characterization of Phomopsis spp. associated with die-back of rooibos (Aspalathus linearis) in South Africa. Studies in Mycology 55: 65–74. 10.3114/sim.55.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer LE. (1926) A biologic and phylogenetic study of stromatic Sphaeriales. American Journal of Botany 13: 575–645. 10.1002/j.1537-2197.1926.tb05903.x [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017b) Diaporthe juglandicola sp. nov. (Diaporthales, Ascomycetes), evidenced by morphological characters and phylogenetic analysis. Mycosphere 8: 817–826. 10.5943/mycosphere/8/5/3 [DOI] [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, Tian CM. (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017a) Diaporthe species occurring on Senna bicapsularis in southern China, with descriptions of two new species. Phytotaxa 302: 145–155. 10.11646/phytotaxa.302.2.4 [DOI] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2020) Three new Diaporthe species from Shaanxi Province, China. Mycokeys 67: 1–18. 10.3897/mycokeys.67.49483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.