Abstract
Paternal obesity is known to have a negative impact on the male’s reproductive health as well as the health of his offspring. Although epigenetic mechanisms have been implicated in the non-genetic transmission of acquired traits, the effect of paternal obesity on gene expression in the preimplantation embryo has not been fully studied. To this end, we investigated whether paternal obesity is associated with gene expression changes in eight-cell stage embryos fathered by males on a high-fat diet. We used single embryo RNA-seq to compare the gene expression profile of embryos generated by males on a high fat (HFD) versus control (CD) diet. This analysis revealed significant upregulation of the Samd4b and Gata6 gene in embryos in response to a paternal HFD. Furthermore, we could show a significant increase in expression of both Gata6 and Samd4b during differentiation of stromal vascular cells into mature adipocytes. These findings suggest that paternal obesity may induce changes in the male germ cells which are associated with the gene expression changes in the resulting preimplantation embryos.
Subject terms: Development, Gene expression, Gene regulation, Obesity, Epigenetic memory
Introduction
Global obesity rates have more than doubled over the past three decades1. Despite increasing recognition of the problem, the prevalence of obesity is rising in most countries worldwide. Obesity is a significant risk factor and contributor to morbidity from several diseases, including diabetes, cardiovascular diseases, and cancer2. The predicted heritability rate of obesity in humans is in the range of 40–75%3–5, nevertheless, genetic studies using whole-genome sequencing as well as genome-wide association studies (GWAS) could explain < 30% of the genetic link to high BMI levels6. Apart from genetics, epigenetic modifications are currently primary targets when searching for factors that increase the risk of obesity7. Recently, several studies in humans and functional work in animal models proposed a role for epigenetic modifications in shaping obesity risk8–10. Epigenetic information in form of histone modifications and DNA methylation plays an important role in regulating gene expression11. There is increasing evidence for the role of epigenetic marks, like DNA methylation, histone modification, and chromatin remodeling in transmitting parental effects to the next generation(s)12,13. Furthermore, recent reports showed a role for sperm-borne RNA (mRNA and small non-coding RNAs) as a carrier of epigenetic information14. As such, transgenerational epigenetic inheritance may contribute to the epidemic increase of metabolic diseases including diabetes and obesity in only one generation. Several reports linked paternal obesity with impaired fertility and altered sperm parameters including declined motility and sperm count, as well as abnormal morphology15,16. Furthermore, such defects can have a negative impact on preimplantation embryo development and physiology17. Paternal-induced obesity in mice causes adverse effects at early stages of embryo development, which results in delayed fetal development as well as reduced placenta size and smaller progeny18. Multiple studies in rodents have shown that paternal high-fat diet can cause transmission of adverse metabolic effects to the F1 generation possibly via epigenetic germline inheritance19–21. Interestingly, bariatric surgery was reported to reverse the diet-induced epigenetic effects on human spermatozoa22. This clearly indicates that the sperm epigenome is malleable and sensitive to dietary influences. It is generally assumed that epigenetic alterations which are transmitted to the offspring can cause adverse health effects in later life.
In this study, we aimed to determine gene expression changes of post-fertilization embryos of males on a high-fat diet in a murine model of diet-induced obesity. We focused on eight-cell embryos since blastomeres are morphologically identical and symmetrically distributed at this stage in addition to zygotic genome activation being dramatically upregulated23. Single-embryo RNA-seq (seRNA-seq) was applied to compare 8-cell embryos fathered by males on a high fat (HFD) versus control (CD) diet. Single cell RNA-seq allows the assessment of biological and molecular diversity across single embryos which is not possible to resolve when applying bulk approaches24.
Results
HFD effects on weight gain and fertilization rate
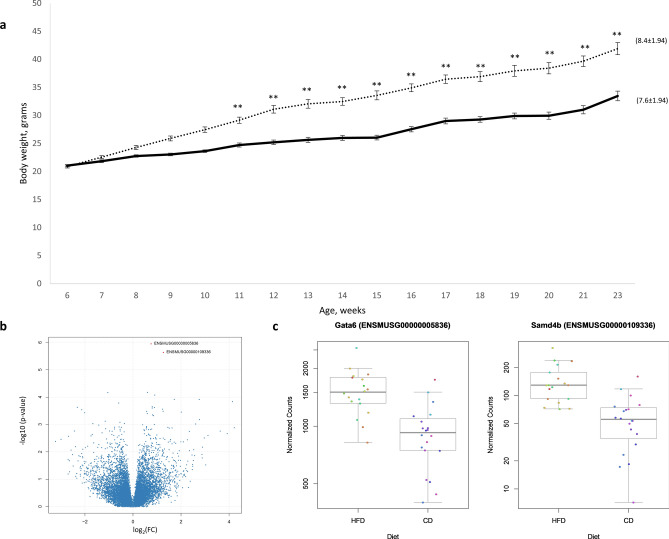
The body weight of males fed a HFD was recorded weekly and compared to males fed a CD. The starting body weight at 6 weeks of age was similar between the HFD (20.9 ± 0.268) and the CD group (21.0 ± 0.202). After 2 weeks, the mean recorded body-weights of HFD males (24.3 ± 0.382) was significantly higher compared to CD males (22.8 ± 0.218, p = 0.003; paired t-test). This significant increase in HFD male body-weights was observed every week including at the final week of weight measurements (22–23 weeks of age) prior to mating (HFD = 41.9 ± 1.077, CD = 33.5 ± 0.858, p < 0.001) (Fig. 1A). Following fertilization, we identified a total of 17 females with eight-cell stage embryos out of 29 cycles with plugs detected (17/29 = 58.6%) in females fertilized by CD males (N = 10). On average, 7.6 ± 1.94 eight-cell stage embryos were obtained per mating. On the other hand, 33 out of 38 females (86.8%) mated with HFD males produced embryos with an average of 8.4 ± 1.94 eight-cell embryos per mating. The number of recovered embryos was not significantly different (p = 0.20; paired t-test) when comparing both groups.
Figure 1.
(a) Average body weight for male mice fed a high fat diet (HFD) or a control diet (CD) from 6 to 23 weeks of age. Mean body weight ± standard error of the mean (SEM). p-values < 0.05 were considered statistically significant, ** for p < 0.01. Solid line = CD (n = 10). Dashed line = HFD (n = 10). Data in brackets indicates the average number of 8-cell embryos recovered for each group (ns). (b) Volcano plot showing log-fold change and −log10 (p-value) for the comparison HFD vs CD embryos. (c) Gata6 and Samd4b expression in 8-cell embryos fathered by males on a high fat diet and control diet. Each dot represents the normalized gene count for each embryo and dot colour indicates “father of origin”.
Single embryo expression analysis
We compared the gene expression profile of 8-cell stage embryos fathered by males on a high fat (HFD) versus control (CD) diet. In total, we performed single-embryo RNA sequencing (seRNA-seq) on 38 embryos including 20 CD and 18 HFD embryos with the majority of studied embryos recovered after fertilizing a different female mouse. In the HFD cohort, the analyzed embryos were conceived by 8 males while in the CD group 7 males were used to conceive the studied embryos. Following seRNA-seq, on average 64.2% of the reads could be uniquely mapped to the mouse genome with the majority of transcripts in protein coding regions. We performed a comparative analysis to identify candidate genes associated with altered expression in HFD vs CD embryos. This revealed GATA binding protein 6 (Gata6) and the sterile alpha motif domain containing 4B (Samd4b) as differentially expressed between two groups after multiple testing correction (Fig. 1B,C, Table 1).
Table 1.
Top 20 differentially expressed genes when comparing 8-cell stage embryos fathered by males on a high fat vs control diet. A positive log2 fold change indicates upregulation in HFD embryos whereas a negative log2 fold change denotes increased expression in control diet embryos.
| Ensembl | Symbol | Mean Count Normalized | log2 fold change | p-value | Adjusted p-value |
|---|---|---|---|---|---|
| ENSMUSG00000005836 | Gata6 | 1213.41 | 0.76 | 1.11E−06 | 0.0154 |
| ENSMUSG00000109336 | Samd4b | 99.73 | 1.28 | 2.28E−06 | 0.0158 |
| ENSMUSG00000027782 | Kpna4 | 491.38 | 0.61 | 6.58E−05 | 0.2301 |
| ENSMUSG00000030965 | Fam175b | 323.94 | − 1.03 | 6.67E−05 | 0.2301 |
| ENSMUSG00000024068 | Spast | 205.78 | 0.92 | 8.30E−05 | 0.2301 |
| ENSMUSG00000004988 | Fxyd4 | 128.86 | 1.73 | 0.00011928 | 0.2369 |
| ENSMUSG00000028222 | Calb1 | 109.21 | 2.77 | 0.00011964 | 0.2369 |
| ENSMUSG00000030711 | Sult1a1 | 5.71 | 4.15 | 0.00014511 | 0.2493 |
| ENSMUSG00000031504 | Rab20 | 2918.93 | − 0.58 | 0.00016182 | 0.2493 |
| ENSMUSG00000063410 | Stk24 | 310.58 | 0.81 | 0.00022196 | 0.2628 |
| ENSMUSG00000105238 | 111.2 | − 2.31 | 0.00023341 | 0.2628 | |
| ENSMUSG00000030970 | Ctbp2 | 176.96 | 0.9 | 0.0002375 | 0.2628 |
| ENSMUSG00000036002 | Fam214b | 168.8 | 0.81 | 0.00024648 | 0.2628 |
| ENSMUSG00000047843 | Bri3 | 27.94 | 1.24 | 0.00027214 | 0.2636 |
| ENSMUSG00000080076 | 147.27 | − 2.12 | 0.0002852 | 0.2636 | |
| ENSMUSG00000022528 | Hes1 | 186.48 | 1.26 | 0.00032274 | 0.2669 |
| ENSMUSG00000036698 | Ago2 | 1020.2 | 0.58 | 0.0003273 | 0.2669 |
| ENSMUSG00000096056 | 12.05 | 2.02 | 0.00034708 | 0.2673 | |
| ENSMUSG00000078878 | Gm14432 | 6.01 | − 2 | 0.00036631 | 0.2673 |
| ENSMUSG00000062691 | Cebpzos | 812.74 | 0.6 | 0.00039294 | 0.2724 |
| ENSMUSG00000032870 | Smap2 | 303.81 | 0.83 | 0.0004404 | 0.2907 |
Significant genes following multiple testing correction are highlighted in bold.
Both Gata6 and Samd4b showed an increased expression in HFD embryos with a log2-fold-change (HFD/CD) of 0.7 (adjusted p-value = 0.0154) and 1.02 (adjusted p-value = 0.0158), respectively. The top 20 significant genes also included Kpna4, Fam175b, Spast, Fxyd4, Calb1, Sult1a1, Rab20, Stk24, Ctbp2, Fam214b, Bri3, Hes1, Ago2, Gm14432, Cebpzos, and Smap2, however, none of these genes had a p-value < 0.05 after multiple testing adjustment (Table 1).
Expression analysis in mouse adipocyte-like cells during differentiation
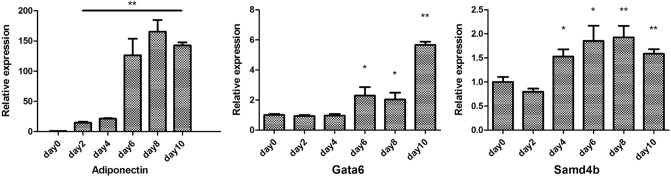
An increase in differentiated adipocytes is crucial for adipose tissue accumulation in obesity. Therefore, we hypothesized that Gata6 and Samd4b are upregulated during adipocyte differentiation. We used stromal vascular cells isolated from the subcutaneous white adipose tissue of C57/B16 mice and differentiated them into adipocyte-like cells. Adiponectin gene expression was included as a positive control to monitor differentiation. Significances were tested using a two-tailed student’s t-test for independent groups (N = 6 for each condition) after comparing each day of differentiation to day 0. We observed a significant increase in Gata6 expression from day 6 (p-value = 0.049) onwards including upregulation at day 8 (p-value = 0.045) and day 10 (p-value = 2.259e−09) of adipocyte differentiation (Fig. 2). Furthermore, we observed a significant upregulation in Samd4b expression from day 4 (p-value = 0.015) until differentiation day 8 (p-value = 0.005) and 10 (p-value = 0.002) (Fig. 2).
Figure 2.
Expression of gata6 and samd4b during the differentiation of stromal vascular cells (SVC) into mature adipocytes. The results are presented as mean values ± standard error of the mean (SEM). p-values < 0.05 were considered statistically significant and indicated with a * for p < 0.05 and ** for p < 0.01).
Discussion
Parental obesity has been shown to have a negative impact on fertility and embryo development25,26. Several epidemiological studies reported that paternal and/or maternal obesity can affect multiple generations and cause adverse health conditions in the non-affected offspring27,28. The mechanism behind the non-genetic inheritance of acquired traits remains unclear; however several reports have shed light on epigenetic factors such as DNA methylation, histone modifications, or small RNAs29. In this study, we aimed to elucidate the effect of paternal obesity on gene expression in 8-cell embryos fathered by HFD males to determine whether paternal obesity has possible consequences on embryonic gene expression and development. Gata6 and Samd4b were the only differentially expressed genes in HFD embryos following multiple testing corrections. Samd4b is a mammalian homolog of the Drosophila Smaug, which represses mRNA translation of developmental regulators in early fly embryos30,31. The second mammalian Smaug homolog is Samd4, which has been previously shown to play a role in body weight regulation where a missense mutation in this gene prevented diet-induced obesity in C57BL/6J mice32. SAMD4B is a widely expressed gene in human adult and embryonic tissues and mediates AP-1-, p53-and p21 signaling activity33. Our analysis of Samd4b expression during adipocyte differentiation revealed a significant increase in expression similar to the GATA Binding Protein 6 (Gata6) gene. Gata6 is a member of the GATA family of zinc-finger transcription factors and plays an important role during vertebrate development34. GATA6 mutations have been previously linked to diabetes and pancreatic agenesis and it was shown to have a role in endoderm formation, pancreas development, and β-like cell functionality35,36. Obesity is a metabolic disease that is characterized by excessive ectopic fat accumulation. Adipocytes respond to energy surplus by a rapid increase in their size (hypertrophy) and also in their number (hyperplasia)37. Retrospective analysis of human cohorts subjected to malnutrition during prenatal and early postnatal life provide strong evidence to support intergenerational effects38. Here, we showed that Gata6 and Samd4b are upregulated during adipocyte differentiation, which supports a role for these genes in predisposing offspring of obese fathers to diet-induced obesity in later life. However, additional experiments are needed to functionally validate the role of Gata6 and Samd4b in non-genetic transmission of paternal obesity.
Recent reports have identified several epigenetic mechanisms that might explain the non-genetic transmission of obesity39,40. This includes small RNA molecules where Chen et al. reported that injecting sperm transfer RNA (tRNA) fragments from obese male mice in control oocytes induces metabolic disorders in the progeny41. Similarly, a high fat diet in utero induced DNA methylation alterations at the rDNA locus correlating with growth restriction42. Stable DNA methylation changes in sperm ribosomal DNA were also shown to occur in response to a protein restricted diet during intrauterine development42. Recently, we have shown a hypermethylation of the rDNA locus in aging germ cells across several mammalian species43. Therefore, epigenetic modifications of germ cells might be a possible mechanism via which a paternal high fat diet might influence the regulation of Gata6 and Samd4b, in embryos.
Here, we provide evidence that paternal high fat diet is associated with upregulation of Gata6 and Samd4b in eight-cell stage embryos. We propose that an epigenetic mechanism might be responsible for the non-genetic transmission of paternal diet-induced obesity. Future research is warranted to determine how molecular changes in response to a high-fat diet are transmitted via male germ cells and their effect on embryo development as well as the health of the offspring.
Materials and methods
Housing and dietary intervention
Housing and embryo collection of the mice were performed as described in Mitchell et al.44. Male C57/B16 mice (Janvier Labs, France) were assigned to either a high fat (N = 10) or a control diet (N = 10) for a total of 15 weeks to generate an obese (HFD) and a lean (CD) mouse cohort (ssniff Spezialdiaeten GmbH, Soest, Germany; E15721-34 (HFD) and customised adjusted E15720-04 (CD)). Diets were individually formulated to match the CD and HFD used previously (Mitchell et al.44), with some modifications to suit the locally available raw materials (e.g. corn starch in place of wheat starch). The HFD provided 22% fat (0.15% cholesterol), 19% protein and 49.5% carbohydrate, and the CD provided 6% fat, Protein 19% protein and 64.7% carbohydrate. Each male mouse was housed individually and food was available ad libitum. The body weight was recorded weekly.
Embryo collection
A total of 120 female C57/ B16 mice at an age of 42 + days were housed for ten days and fed a CD prior to mating. After 8 weeks, each week one male and a new naturally cycling female mouse were housed individually to achieve pregnancy. The observation date of the mating plug was assigned as “day 1–9” and based on this date embryos were isolated at the 8-cell stage. The females were euthanized by cervical dislocation and the 8-cell stage embryos were recovered directly from the females, without in vitro culture. The recovered embryos were then separately transferred to 0.2 ml PCR tubes containing ~ 9 µl PBS, snap frozen in liquid nitrogen, and stored at − 80 °C.
mRNA profiling of 8-cell stage embryos using SMART technology
The “SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing” (Takara/Clontech Laboratories) was used for preparing full length cDNA libraries. This kit is based on the SMART technology, which utilizes the template switching activity of the reverse transcriptase and captures specifically poly-adenylated RNA. Briefly, a total of 38 single 8-cell embryos each in 9 µl PBS were directly taken as input material. Manufacturer`s instructions were followed and 18 cycles were used for cDNA amplification. Library preparation for sequencing on the Nextseq 500 platform (Illumina) was performed using the Nextera XT DNA Library Preparation Kit (Illumina) and 150 pg of amplified cDNA was used as input volume as recommended in the sample preparation guide. The rest of the protocol was followed according to manufacturer`s instructions. Quantitative and qualitative assessment of the library was implemented using a Bioanalyzer High Sensitivity DNA chip. Finally, 2 × 76 paired end sequencing of 38 (18 HFD vs. 20 CD) libraries in parallel was performed on the NextSeq 500 platform (Illumina) using the NextSeq 500/550 High Output v2 Kit (150 cycles) (Illumina).
Primary adipocyte isolation and differentiation
Stromal vascular cells (SVC) from subcutaneous white adipose tissue (sWAT) were isolated and differentiated into mature adipocytes as described in El-Merahbi et al.45. Briefly, sWAT of 8 weeks old mice were collected on a 10 cm Petri dish, washed with PBS, and cleaned from lymph-nodes and visible blood vessels. Tissues were then minced and further digested with 2 mg/ml collagenase D (Roche). After hemolysis, digested tissue was filtered through a 40-µm mesh, washed in PBS by centrifugation, and cultured in DMEM/F-12 containing 10% FBS, 1% SP, 1% non-essential amino acids (NEAA) and 1% Penicillin–Streptomycin (P/S). Two days post-confluence, adipocytes differentiation was induced by adding a cocktail including 0.2 µM indomethacin, 1 μM dexamethasone, 0.5 mM IBMX, and 1.5 μg/ml insulin for the first 4 days, followed by insulin treatment for additional 4 days to achieve adipocyte’s maturation.
RNA isolation and RT-qPCR
Total RNA was extracted from stromal vascular cells in triplicates throughout defined time points (stages) of differentiation using Qiazol manufacturer instructions. cDNA was then synthesized by First Strand cDNA Synthesis Kit (Thermo Fischer Scientific) according to the manufacturer’s protocol. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using the Power SYBR green PCR master mix (Thermo Fischer Scientific) on a QuantStudio 5 Real-Time PCR System (Thermo Fischer Scientific). Expression of all genes was obtained in technical duplicates and normalized to the Rpl13a housekeeping gene.
Bioinformatic data analysis
Following quality control of sequenced libraries and adapter removal with Cutadapt46 all reads have been aligned to the mouse genome (GRCm38, including ERCC Sequences) using HISAT2 (version 2.0.5)47. Subsequently, the mapped reads have been assigned to genes and counted using 'featureCounts' as implemented in the Rsubread package (version 1.20.6)48. Genes which have been detected in less than 50% of samples have been removed from further analysis. Differential expression of genes between the high fat diet and control diet group has been analyzed using a generalized linear model as implemented in the DESeq2 (version 1.10.1)49. Gene-wise p-values have been multiple testing corrected using the Benjamini and Hochberg method50 and adjusted p-values < 0.05 have been considered significant. All statistical analyses have been performed using R (version 3.2.2) including packages from the Bioconductor project51.
Ethical approval
Animal experimental procedures were performed according to the National Research Council's publication Guide for Care and Use of Laboratory Animals, and were approved by the Animal Care Committee of the University of Erlangen-Nürnberg and the Government of Mittelfranken, Germany (54-2532.1-37/12). The study was carried out in compliance with the ARRIVE guidelines.
Acknowledgements
The authors would like to thank Dr Dieter Engelkamp and Martina Doehler from the Biotechnisches Entwicklungslabor at the Friedrich-Alexander University, Erlangen for their technical assistance with the mouse experiment.
Author contributions
L.B., R.E.M., A.E.S., S.N., M.M., and N.E.H. performed the main experiments. M.D. and T.M. performed bioinformatic analyses. A.E.S., G.S., J.V., M.M., T.H., and N.E.H. designed research. T.H. and N.E.H wrote the manuscript. M.D., A.E.S., G.S., J.V., M.M., T.H. and N.E.H. critically reviewed, discussed, and edited the manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by the Qatar National Library.
Data availability
The datasets generated during and/or analysed during the current study are available in SRA under accession number: SRP293075.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stevens GA, et al. National, regional, and global trends in adult overweight and obesity prevalences. Popul. Health Metr. 2012 doi: 10.1186/1478-7954-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pi-Sunyer X. The medical risks of obesity. Postgrad. Med. 2009;121:21–33. doi: 10.3810/pgm.2009.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wardle J, Carnell S, Haworth CMA, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am. J. Clin. Nutr. 2008;87:398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 4.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 5.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256:51–54. doi: 10.1001/jama.1986.03380010055024. [DOI] [PubMed] [Google Scholar]
- 6.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–U401. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Hajj N, Schneider E, Lehnen H, Haaf T. Epigenetics and life-long consequences of an adverse nutritional and diabetic intrauterine environment. Reproduction. 2014;148:R111–R120. doi: 10.1530/Rep-14-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalgaard K, et al. Trim28 haploinsufficiency triggers Bi-stable epigenetic obesity. Cell. 2016;164:353–364. doi: 10.1016/j.cell.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dick KJ, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 10.Pfeiffer S, et al. Hypoxia-inducible factor gene expression and methylation in adipose tissue is related to adipose tissue dysfunction. Sci. Rep. 2016 doi: 10.1038/srep27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Rando OJ. Intergenerational transfer of epigenetic information in sperm. Csh Perspect Med. 2016 doi: 10.1101/cshperspect.a022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atsem S, et al. Paternal age effects on sperm FOXK1 and KCNA7 methylation and transmission into the next generation. Hum. Mol. Genet. 2016;25:4996–5005. doi: 10.1093/hmg/ddw328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma U, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakos HW, Mitchell M, Setchell BP, Lane M. The effect of paternal diet-induced obesity on sperm function and fertilization in a mouse model. Int. J. Androl. 2011;34:402–410. doi: 10.1111/j.1365-2605.2010.01092.x. [DOI] [PubMed] [Google Scholar]
- 16.Fullston T, et al. Paternal obesity affects male offspring's health and sperm function. Aust. Nz. J. Obstet. Gyn. 2010;50:22–22. [Google Scholar]
- 17.Binder NK, Mitchell M, Gardner DK. Parental diet-induced obesity leads to retarded early mouse embryo development and altered carbohydrate utilisation by the blastocyst. Reprod. Fertil. Dev. 2012;24:804–812. doi: 10.1071/Rd11256. [DOI] [PubMed] [Google Scholar]
- 18.Binder NK, Hannan NJ, Gardner DK. Paternal diet-induced obesity retards early mouse embryo development, mitochondrial activity and pregnancy health. PLoS ONE. 2012 doi: 10.1371/journal.pone.0052304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huypens P, et al. Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat. Genet. 2016;48:497. doi: 10.1038/ng.3527. [DOI] [PubMed] [Google Scholar]
- 20.Ng SF, et al. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell M, et al. Gene expression and epigenetic aberrations in F1-placentas fathered by obese males. Mol. Reprod. Dev. 2017;84:316–328. doi: 10.1002/mrd.22784. [DOI] [PubMed] [Google Scholar]
- 22.Donkin I, et al. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab. 2016;23:369–378. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Jukam D, Shariati SAM, Skotheim JM. Zygotic genome activation in vertebrates. Dev. Cell. 2017;42:316–332. doi: 10.1016/j.devcel.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell JM, Lane M, Owens JA, Bakos HW. Paternal obesity negatively affects male fertility and assisted reproduction outcomes: a systematic review and meta-analysis. Reprod. Biomed. Online. 2015;31:593–604. doi: 10.1016/j.rbmo.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Lane M, Zander-Fox DL, Robker RL, McPherson NO. Peri-conception parental obesity, reproductive health, and transgenerational impacts. Trends Endocrinol. Metab. 2015;26:84–90. doi: 10.1016/j.tem.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Bygren LO, et al. Change in paternal grandmothers' early food supply influenced cardiovascular mortality of the female grandchildren. Bmc Genet. 2014 doi: 10.1186/1471-2156-15-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period. Eur. J. Hum. Genet. 2002;10:682–688. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- 29.Sharma U, Rando OJ. Metabolic inputs into the epigenome. Cell Metab. 2017;25:544–558. doi: 10.1016/j.cmet.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Jeske M, Moritz B, Anders A, Wahle E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J. 2011;30:90–103. doi: 10.1038/emboj.2010.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smibert CA, Wilson JE, Kerr K, Macdonald PM. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, et al. Mutation of mouse Samd4 causes leanness, myopathy, uncoupled mitochondrial respiration, and dysregulated mTORC1 signaling. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7367–7372. doi: 10.1073/pnas.1406511111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo N, et al. SAMD4B, a novel SAM-containing protein, inhibits AP-1-, p53- and p21-mediated transcriptional activity. BMB Rep. 2010;43:355–361. doi: 10.5483/bmbrep.2010.43.5.355. [DOI] [PubMed] [Google Scholar]
- 34.Charron F, Nemer M. GATA transcription factors and cardiac development. Semin. Cell Dev. Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- 35.De Franco E, et al. GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes. 2013;62:993–997. doi: 10.2337/db12-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiyaboonchai A, et al. GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic beta cells. Stem Cell Rep. 2017;8:589–604. doi: 10.1016/j.stemcr.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlman I, et al. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. Int. J. Obesity. 2015;39:910–919. doi: 10.1038/ijo.2015.31. [DOI] [PubMed] [Google Scholar]
- 38.Sales VM, Ferguson-Smith AC, Patti M-E. Epigenetic mechanisms of transmission of metabolic disease across generations. Cell Metab. 2017;25:559–571. doi: 10.1016/j.cmet.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potabattula R, et al. Allele-specific methylation of imprinted genes in fetal cord blood is influenced by cis-acting genetic variants and parental factors. Epigenomics-Uk. 2018;10:1315–1326. doi: 10.2217/epi-2018-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potabattula R, et al. Male obesity effects on sperm and next-generation cord blood DNA methylation. PLoS ONE. 2019;14:e0218615. doi: 10.1371/journal.pone.0218615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 42.Holland ML, et al. Early-life nutrition modulates the epigenetic state of specific rDNA genetic variants in mice. Science. 2016;353:495–498. doi: 10.1126/science.aaf7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potabattula R, et al. Increasing methylation of sperm rDNA and other repetitive elements in the aging male mammalian germline. Aging Cell. 2020 doi: 10.1111/acel.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell M, Bakos HW, Lane M. Paternal diet-induced obesity impairs embryo development and implantation in the mouse. Fertil. Steril. 2011;95:1349–1353. doi: 10.1016/j.fertnstert.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 45.El-Merahbi R, et al. The adrenergic-induced ERK3 pathway drives lipolysis and suppresses energy dissipation. Genes Dev. 2020;34:495–510. doi: 10.1101/gad.333617.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;2011:17. doi: 10.14806/ej.17.1.200pp.10-12. [DOI] [Google Scholar]
- 47.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995;57:289–300. [Google Scholar]
- 51.Huber W, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in SRA under accession number: SRP293075.