Summary
Fibroblast growth factor homologous factors (FHFs) are intracellular proteins which regulate voltage-gated sodium (Nav) channels in the brain and other tissues. FHF dysfunction has been linked to neurological disorders including epilepsy. Here, we describe two sibling pairs and three unrelated males who presented in infancy with intractable focal seizures and severe developmental delay. Whole-exome sequencing identified hemi- and heterozygous variants in the N-terminal domain of the A isoform of FHF2 (FHF2A). The X-linked FHF2 gene (also known as FGF13) has alternative first exons which produce multiple protein isoforms that differ in their N-terminal sequence. The variants were located at highly conserved residues in the FHF2A inactivation particle that competes with the intrinsic fast inactivation mechanism of Nav channels. Functional characterization of mutant FHF2A co-expressed with wild-type Nav1.6 (SCN8A) revealed that mutant FHF2A proteins lost the ability to induce rapid-onset, long-term blockade of the channel while retaining pro-excitatory properties. These gain-of-function effects are likely to increase neuronal excitability consistent with the epileptic potential of FHF2 variants. Our findings demonstrate that FHF2 variants are a cause of infantile-onset developmental and epileptic encephalopathy and underline the critical role of the FHF2A isoform in regulating Nav channel function.
Keywords: epilepsy, epileptic encephalopathy, FHF2, FGF13, voltage-gated sodium channel, developmental and epileptic encephalopathy, X linked, infantile onset
Main Text
Voltage-activated sodium channels (Nav) play an essential role in the generation and spread of action potentials in excitable tissues.1,2 Variants in Nav channels and their regulatory partners are a major cause of infantile-onset developmental and epileptic encephalopathies (DEEs).3,4 DEEs are typically associated with developmental delay or regression, treatment-resistant seizures, and electroencephalographic abnormalities.5,6 The developmental consequences of DEEs are due to frequent epileptiform activity in combination with the direct effects of the genetic variant.7,8
Fibroblast growth factor homologous factors (FHFs) are intracellular proteins that bind to the C-terminal domain of Nav channels to modulate their function and location.9, 10, 11, 12 FHFs were initially identified due to their homology with fibroblast growth factors (FGFs).13 However, FHFs are not secreted by cells and have only limited ability to activate FGF receptors.9,14,15 There are four FHF genes in mammals (often referred to by their FGF names): FHF1 (FGF12 [MIM: 601513]), FHF2 (FGF13 [MIM: 300070]), FHF3 (FGF11 [MIM: 601514]), and FHF4 (FGF14 [MIM: 601515]).9 The FHF genes have multiple transcription initiation sites. The alternative first exons produces multiple isoforms with variable N-terminal domains.16 The isoforms differ in their localization and ability to regulate Nav channels.9 FHF2, located at Xq26.3-q27.1, is highly expressed in the developing and adult brain.9,13 FHF2 is also expressed in endocrine tissues, ovaries, skeletal muscle, and the myocardium of the developing heart.17, 18, 19 FHF2 has been implicated in a variety of functions including microtubule stability, axonogenesis, neuronal migration,20 interneuron development,21 cardiac conduction,22 and thermogenesis.23 Here, we present evidence that variants in FHF2 cause a DEE.
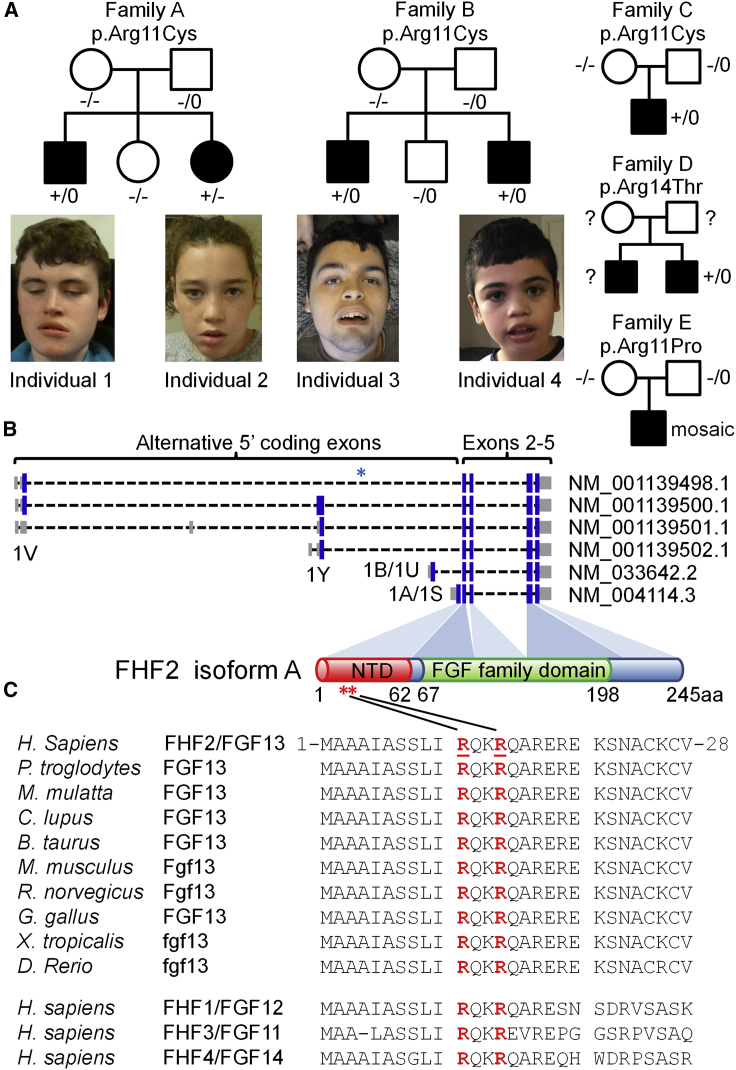
We identified seven individuals from five unrelated families who presented with severe infantile-onset seizures. Individuals 1 and 2 were a brother and sister from family A (Figure 1A) who had whole-exome sequencing (WES) as part of the Wales Epilepsy Research Network (WERN) family study. Individuals 3 and 4 were brothers from family B who had WES as part of the Deciphering Developmental Disorders (DDD) study.24 Individual 5 from family C had WES performed by GeneDx. Individuals 1 to 5 shared the same seemingly de novo missense variant in FHF2, chrX (GRCh38/hg38):g.138710973G>A, GenBank: NM_004114.5: c.31C>T (p.Arg11Cys). Individual 6 from family D also had WES as part of the DDD study. Individual 6 had a different nearby variant in FHF2, chrX (GRCh38/hg38): g.138710963C>G, GenBank: NM_004114.5: c.41G>C (p.Arg14Thr). Parental testing for this individual was not possible. Individual 7 from family E had trio whole-genome sequencing (WGS) performed by Cipher Gene. Individual 7 was mosaic (7 variant reads out of 17 total) for a different missense change at the Arg11 residue, chrX (GRCh38/hg38):g.138710972C>G, GenBank: NM_004114.5: c.32G>C (p.Arg11Pro). The mosaicism was confirmed by WES (14 variant reads out of 26 total) and Sanger sequencing. Identification of individuals 5 and 7 was achieved via GeneMatcher.25 Maternal mosaicism is likely to explain the sibling recurrence in families A and B. This was not detected by WES or Sanger sequencing. Families A and B and individual 6 also had WGS as part of the 100,000 Genomes Project.26 WGS confirmed the variants in the affected individuals. The p.Arg11Cys variant was not detected in the mother of family A but the mother of family B had 2 variant reads out of 40 total, consistent with low-level mosaicism. Individuals who had evaluation or analysis beyond routine clinical care were part of research studies approved by either the Research Ethics Committee for Wales (09/MRE09/51) or the Cambridge South Research Ethics Committee (10/H0305/83 and 14/EE/1112). The parents and carers of the individuals gave consent for their publication in this report.
Figure 1.
Family pedigrees and locations of the FHF2A variants
(A) Pedigrees of families A–E with photographs of the affected individuals from families A and B. Genotypes are heterozygous mutant (+/−), homozygous wild type (−/−), hemizygous mutant (+/0), hemizygous wild type (−/0), or unknown (?).
(B) Exonic models of the FHF2 transcripts showing the alternative 5′ coding exons. The FHF2A protein isoform is derived from the GenBank: NM_004114.5 transcript which is initiated from an ATG start site in exon 1A (also known as exon 1S). The V, Y, U, and S nomenclature originates from Munoz-Sanjuan et al.27 The 1A/1S exon encodes the N-terminal domain (NTD) of FHF2A (amino acid residues 1 to 62). The shared C-terminal exons encode the core fibroblast growth factor (FGF) domain (amino acid residues 67 to 198). The blue asterisk indicates the location of the breakpoint of the translocation described by Puranam et al.28 The red asterisks indicate the location of the DEE-associated missense variants.
(C) The FHF2 (FGF13) substitutions are located at highly conserved residues in the N-terminal domain of the A isoform. Homology alignments for human FHF2 (GenBank: NP_004105.1, amino acid residues 1–28) and a range of orthologs and paralogs. Orthologs include chimpanzee (GenBank: XP_001138460.1), rhesus macaque (GenBank: NP_001252772.1), dog (GenBank: XP_549294.2), cow (GenBank: NP_001092362.1), mouse (GenBank: NP_034330.2), rat (GenBank: XP_006257654.1), chicken (GenBank: XP_015133693.1), western clawed frog (GenBank: XP_012824080.1), and zebrafish (GenBank: XP_005173268.1). Paralogs include FHF1/FGF12 (GenBank: NP_066360.1), FHF3/FGF11 (GenBank: NP_004103.1), and FHF4/FGF14 (GenBank: NP_004106.1). The positions of the Arg11 and Arg14 residues are underlined and in red.
The clinical features of the seven individuals are summarized in Table 1. Detailed case reports for the individuals are provided in the supplemental note. Shared features included global developmental delay and severe or profound intellectual disability. Individuals 2–4 and 6 were diagnosed with autism spectrum disorder. All seven individuals had treatment-resistant epilepsy. Five presented in the neonatal period with episodes of apnea and cyanosis. The predominant epilepsy phenotype was focal seizures often with secondary generalization. A range of other seizure types were reported including epileptic spasms, tonic seizures, gelastic seizures, absence seizures, drop attacks, and generalized tonic-clonic seizures. The focal seizures were associated with motor features (eye twitching or head deviation), apneas, and oroalimentary automatisms (chewing or repeated swallowing). Autonomic features included drooling, ictal vomiting, and skin flushing. EEGs found evidence of temporal lobe foci in some. Seizures were resistant to a wide range of anti-epileptic drugs (AED). Two individuals had vagus nerve stimulators implanted. Individual 2 underwent left anterior temporal lobectomy and partial amygdalohippocampectomy at 7 years of age. She was seizure free for 2 years; however, the focal seizures resumed and at last review (13 years of age) she was having fortnightly clusters despite three AEDs. Constipation was common in the group and one person (individual 1) had subtotal colectomy and ileostomy due to chronic constipation and recurrent severe abdominal pain. Physical examination of the individuals revealed normal or reduced muscle tone. There was no spasticity or hyperreflexia. Subtle dysmorphic features were noted but these were variable and may reflect the effects of AEDs and facial hypotonia. Photographs of individuals 1–4 are shown in Figure 1A. Brain MRI scans in infancy were normal. Brain MRI scans of individual 2 at 6 years of age and individual 7 at 3 years of age found evidence of cerebral atrophy. Individual 4 had a 3T MRI brain scan at 13 years of age which showed symmetrical T2 hyperintensity of the hippocampal body and head with loss of definition of the internal architecture.
Table 1.
Clinical and molecular findings in the individuals with N-terminal FHF2A variants
| Individual | 1 (family A) | 2 (family A) | 3 (family B) | 4 (family B) | 5 (family C) | 6 (family D) | 7 (family E) |
|---|---|---|---|---|---|---|---|
| Age | 15 y | 13 y | 19 y | 12 y | 2 y 3 m | 5 y | 5y 8 m |
| Sex | male | female | male | male | male | male | male |
| Variant | c.31C>T (p.Arg11Cys) | c.31C>T (p.Arg11Cys) | c.31C>T (p.Arg11Cys) | c.31C>T (p.Arg11Cys) | c.31C>T (p.Arg11Cys) | c.41G>C (p.Arg14Thr) | c.32G>C (p.Arg11Pro) |
| Inheritance | maternal gonadal mosaicisma | maternal gonadal mosaicisma | maternal somatic mosaicism | maternal somatic mosaicism | de novo | unknown | mosaic |
| OFC | −1.4 SD at 13 y 10 m | +0.8 SD at 6 y 9 m | −0.2 SD at 19 y | −1.4 SD at 12 y 5 m | −1.1 SD at 22 m | −2.5 SD at 5 y | −2.6 SD at 3 y 2 m |
| ID/DD | profound | severe | severe | severe | profound | severe | severe |
| Initial concerns | 11 d, apneas, repetitive swallowing, head deviation, eye twitching | 11 d, apneas, lip smacking, repetitive swallowing, eye deviation, facial twitching | 4 w, apneas, stiffness | 6 m, head and eye deviation, twitching | 5 d, head and eye deviation, blinking, repetitive swallowing | 1 d, apnea, cyanosis | 6 m, focal sz |
| Epilepsy | focal sz at 2 m. generalized sz from 7 m | focal sz from 11 m. rarely generalized | focal sz at 2 m. flexor spasms from 6 m | focal sz at 6 m. generalized sz from 2.5 y, episodes of NCSE, tonic sz and vomiting currently | focal dyscognitive seizures | yes | focal sz at 6 m. also spasms, myoclonic sz, GTCS |
| EEG | multiple epileptogenic temporal foci, EIMFS considered | ictal EEG at 6 y 8 m, left fronto-temporal focus | hypsarrhythmia at 6 m. later EEG suggestive of LGS | hypsarrhythmia at 2.5 y, EEG at 11 y suggestive of LGS | multiple epileptogenic temporal foci, suggestive of LGS | n/k | atypical hypsarrhythmia and intermittent burst suppression at 23 m |
| Neurology | hypotonia, no hyperreflexia | normal tone and reflexes | low axial tone, mild limb hypertonia, no hyperreflexia or tremor | broad-based, unsteady gait, mildly increased limb tone | hypotonia, periodic abnormal posturing | n/k | limb hypertonia, positive Babinski sign and ankle clonus |
| Other features | constipation, abdominal pain, subtotal colectomy and ileostomy | left anterior temporal lobectomy and partial amygdalo-hippocampectomy | severe scoliosis | antenatal renal pelvic dilatation, recurrent UTI, nephrectomy | hypothyroidism | atrial septal defect | regression at 14 m |
Variants based on transcript GenBank: NM_004114.5. Age in y(ears), m(onths), w(eeks), or d(ays); AED, antiepileptic drugs; ASD, autism spectrum disorder; EEG, electroencephalogram; EIMFS, epilepsy of infancy with migrating focal seizures; GTCS, generalized tonic-clonic seizures; ID/DD, intellectual disability or developmental delay; LGS, Lennox-Gastaut syndrome; NCSE, nonconvulsive status epilepticus; n/k, not known; SD, standard deviations; sz, seizures; UTI, urinary tract infection.
Maternal gonadal mosaicism is presumed in family A.
The three variants p.Arg11Cys, p.Arg11Pro, and p.Arg14Thr identified in the affected individuals are not present in population databases such as gnomAD.29 The variants were predicted to be deleterious by a range of in silico prediction programs (Table S1).30 The variants are located in exon 1A of FHF2. This exon encodes the N-terminal domain specific to the A isoform of the FHF2 protein (FHF2A) (Figure 1B). The 1A exon is highly conserved across species and across all four FHF genes (Figure 1C). The Arg11 and Arg14 residues are completely conserved in all sequences. Overall, FHF2 is moderately intolerant to variation (gnomAD missense z-scores range from 1.59 to 2.11 for the different isoforms). However, the N-terminal of the A isoform (ENST00000315930.6) is particularly constrained with no gnomAD missense variants in the first 21 residues.29 In contrast, there are 5 gnomAD synonymous variants in the same region and 7–10 missense variants in the first 21 residues of the other FHF2 isoforms.
The FHF2 exon 1A missense variants are at residues known to contribute to long-term Nav channel block.31 Nav channels cycle between closed (or resting), open, and inactivated states (Figure 2A).32 Membrane depolarization from resting potential to the voltage threshold for Nav channel activation triggers the shift from a closed to an open state. The channel is open for around a millisecond before inactivation occurs. The majority of channels are inactivated by a mechanism intrinsic to the Nav channel but around one third are inactivated by an “inactivation particle” contained by the N-terminal domain of A-type FHF proteins.33 Channels can recover (or reprime) to a closed state upon hyperpolarization of the membrane. Recovery from the intrinsic Nav inactivation mechanism is complete within 10 ms while recovery from FHF-mediated inactivation takes hundreds of milliseconds. Channels may also proceed directly from a closed to an inactivated state (“steady-state” inactivation). A-type FHF isoforms have a mixture of inhibitory and pro-excitatory effects on the inactivation mechanisms of Nav channels. Repeated cycles of channel opening promote accumulation of channels in FHF-mediated long-term block. This contributes to a run-down in Nav channel availability immediately following a cluster of action potentials.33,34 Through this inhibitory mechanism, A-type FHF isoforms can induce spike-frequency adaptation, also termed accommodation.31 One potential function of accommodation is to prevent persistent excitation of neural networks and seizures.35,36 In contrast, A-type FHF isoforms also induce a depolarizing shift in the voltage dependence of Nav channels for steady-state inactivation.33,34,37,38 This is pro-excitatory because it limits steady-state inactivation of channels at resting potential. Another pro-excitatory effect of A-type FHF isoforms is to slow the intrinsic fast inactivation of channels during membrane depolarization and action potential initiation.10,34
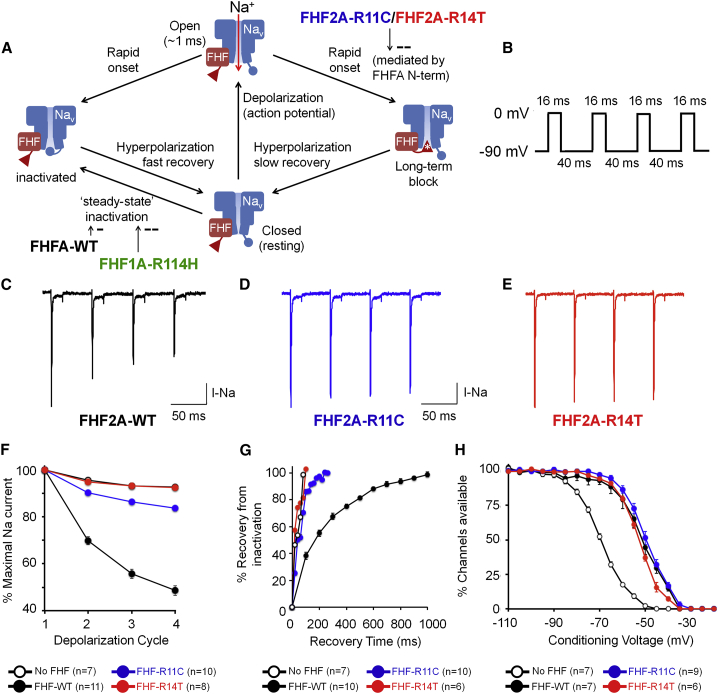
Figure 2.
Functional characterization of the variants in the FHF2A N-terminal domain
(A) Scheme of voltage-dependent sodium (Nav) channel state transitions as modulated by A-type fibroblast growth factor homologous factors (FHFs), as described previously.31,33 Strong rapid depolarization results in transient opening of channels and sodium influx (vertical red arrow). Within milliseconds, channels intrinsically inactivate (leftward descending arrow). When channels are physically associated with A-type FHF, the N terminus (red triangle) competes with the intrinsic Nav mechanism (blue circle) to rapidly induce long-term inactivation (rightward descending arrow). Recovery to the closed state upon repolarization requires hundreds of milliseconds. Long-term inactivation of channels is impaired in presence of the FHF2AR11C or FHF2AR14T (white asterisk). A-type FHFs raise the voltage dependence of steady-state inactivation, preserving more channels in the closed (resting) state. The pro-excitatory DEE-associated FHF1AR114H mutant protein further raises the voltage dependence of steady-state inactivation.4
(B) Voltage-clamp protocol for accumulating long-term inactivation used in (C)–(E). The 40 ms intervals at −90 mV allow for full recovery from intrinsic fast inactivation, but only partial recovery from long-term inactivation, which has a far slower recovery rate.
(C–E) Representative sodium current traces in Neuro2A cells expressing Nav1.6 together with either (C) FHF2AWT, (D) FHF2AR11C, or (E) FHF2AR14T. The variants impair channel long-term inactivation.
(F) Analysis of long-term inactivation induced by wild-type and mutant FHF2A proteins. n represents the number of transfected cells recorded for a given experimental protocol. Data points represent the mean ± standard error (error bars) from the n recorded cells. The impaired long-term inactivation of FHF2AR11C and FHF2AR14T in comparison to FHF2AWT upon depolarizations 2,3,4 is highly significant (p < 10−7).
(G) Analysis of Nav1.6 recovery from long-term inactivation at −90 mV. Nav1.6 recovers faster in cells expressing FHF2AR11C or FHF2AR14T compared to cells expressing FHF2AWT.
(H) FHF2AR11C and FHF2AR14T retain the ability to induce 17–19 mV depolarizing shifts in the voltage dependence of Nav1.6 steady-state inactivation.
To assess the functional effects of FHF2A p.Arg11Cys and p.Arg14Thr on neuronal sodium channel gating, we measured sodium currents during voltage clamp protocols in FHF-negative Neuro2A cells co-transfected with constructs expressing Nav1.6 (SCN8A) and either wild-type FHF2A (FHF2AWT) or a mutated version (FHF2AR11C or FHF2AR14T). Detailed descriptions of the plasmids, mutagenesis, transfection of Neuro2A cells and measurement of sodium currents are provided in the supplemental material and methods. All procedures parallel those we have described previously.31,33 Patched cells were depolarized to 0 mV for four 16 ms intervals separated by 40 ms recovery periods at −90 mV to allow for sodium channel recovery from intrinsic fast inactivation (Figure 2B). In cells expressing FHF2AWT, transient Nav1.6 sodium current amplitude diminishes with each depolarization cycle (Figures 2C and 2F), reflecting accumulating long-term inactivation of Nav1.6. By contrast, the extent of long-term channel inactivation is greatly reduced in cells expressing FHF2AR11C (Figures 2D and 2F), while the sodium currents during four depolarization cycles in cells expressing FHF2AR14T are indistinguishable from those in cells without FHF2 (Figures 2E and 2F), demonstrating that FHF2AR14T is incapable of promoting Nav1.6 long-term inactivation.
In a second experiment to assess the effects of the FHF2A variants on long-term channel inactivation, cells were subjected to multiple rapid depolarization cycles followed by a −90 mV recovery period of varying duration (20 ms to 1 s) before a subsequent test depolarization. In cells expressing FHF2AWT, near-full recovery of Nav1.6 from long-term inactivation required 1 s, while in cells expressing FHF2AR11C channels fully recovered by 200 ms (Figure 2G). Cells expressing FHF2AR14T again show no evidence of Nav1.6 long-term inactivation, as channels fully recover as quickly as in cells lacking FHF (Figure 2G).
Although impaired in their ability to promote Nav1.6 long-term inactivation, FHF2AR11C and FHF2AR14T are expressed at comparable levels to FHF2AWT in transfected cells and the mutant isoforms associate with Nav1.6 as effectively as does FHF2AWT, as demonstrated by co-immunoprecipitation (Figure S1). This indicates the variants do not disrupt binding of FHF2 to the C-terminal cytoplasmic tail of Nav channels. This binding is mediated by the conserved β-trefoil core domain shared by all FHF isoforms.16,39 In contrast, residues 2–21 of A-type FHFs contain the inactivation particle that competes with the intrinsic fast inactivation mechanism of Nav channels following membrane depolarization.33 The reduced ability of mutant FHF2A to promote Nav1.6 long-term inactivation indicates the N-terminal variants impair function of this inactivation particle. Mutagenesis of FHF2A to replace Arg11 or Arg14 with glutamine prevented binding of an antibody specific to the N-terminal of A-type FHFs.31 This suggests that both residues are prominent surface features. Both FHF2AR11Q and FHF2AR14Q showed impaired accumulation of long-term inactivation and hastened recovery of Nav channels while having little effect on the ability of FHF2A to modulate steady-state inactivation. These findings are consistent with our results for the two DEE-associated variants which suggest the effects are due to loss of the cationic arginine side chains rather than specific effects of the substituted side chains.
Consequently, we tested the ability of mutant FHF2A proteins to modulate the voltage dependence of Nav1.6 steady-state inactivation. As shown in Figure 2H, FHF2AR11C and FHF2AR14T raised the V1/2 for channel steady-state inactivation by 17 mV and 19 mV, respectively, which was statistically indistinguishable from the 18 mV V1/2 elevation of Nav1.6 inactivation mediated by FHF2AWT. Therefore, the DEE-associated variants preserve this pro-excitatory property of FHF2A while suppressing the sodium channel long-term inactivation mechanism by which FHF2A can attenuate excitation.
The FHF2 exon 1A missense variants join a growing number of isoform-specific variants that have been reported in epileptic encephalopathies.40, 41, 42 Around 30% of neonatal epilepsy genes have brain-expressed alternative coding regions, suggesting that isoform-specific variants are relatively common.43 However, variants in alternative transcripts are prone to being missed or misinterpreted due to problems with panel designs and genome annotation.44 This may have contributed to FHF2-DEE not being recognized until now.
Deleterious variation in FHF genes has previously been associated with neurological disorders. In 2015, a balanced reciprocal translocation, t(X; 14) (q27; q21), disrupting FHF2 was reported segregating in a family (two brothers and their mother) with genetic epilepsy and febrile seizures plus (GEFS+ [MIM: 604233]).28 One of the brothers and other members of their extended family had cognitive impairment. The translocation breakpoint occurred after two of the alternative first coding exons of FHF2 (exons 1V and 1Y, using the notation from Munoz-Sanjuan et al.27) but before the first coding exons of the two main isoforms, exon 1A (also known as 1S, employed by FHF2A) and exon 1B (also known as 1U, used by FHF2B). The predicted consequence was to disrupt the minor isoforms of FHF2 which use the more 5′ exons. There was a subsequent report of a de novo missense variant in FHF2 (GenBank: NM_004114.5: c.638C>T [p.Thr213Met]) in one individual with mild febrile seizures, normal development, and facial edema.45 However, the significance of this variant was uncertain.
Gain-of-function variants in FHF1 (FGF12) have recently been identified in individuals with DEE (MIM: 617166).4,46,47 There are strong similarities between FHF1- and FHF2-DEE. FHF1-DEE (familial early-onset epileptic encephalopathy with progressive cerebral atrophy) presents with seizures in early infancy and severe developmental delay. The epileptic phenotypes of FHF1 and FHF2 disease are similar. FHF1-DEE is typically associated with focal epilepsy or a combined generalized and focal epilepsy.48 Tonic seizures with autonomic signs are common in both FHF1- and FHF2-DEE. Other seizure types reported in both disorders include generalized tonic-clonic seizures, epileptic spasms, and absence and myoclonic seizures. The epilepsy in FHF1-DEE is often intractable but responsiveness to phenytoin (a sodium channel blocker) has been noted.47, 48, 49 Individuals 3 and 4 in our series gained at least transient benefit from phenytoin. The other subjects have either not been given phenytoin or their response is unknown.
Individuals with FHF1-DEE have been noted to have progressive cerebellar atrophy, potentially as a result of excitotoxic damage.4 Loss-of-function variants in FHF4 (FGF14) have also been associated with cerebellar dysfunction (and occasionally cerebellar atrophy) as the cause of spinocerebellar ataxia type 27 (MIM: 609307).50,51 In contrast, structural and functional abnormalities of the cerebellum were not observed in FHF2-DEE. Different gene expression patterns may explain this difference as FHF1 and FHF4 are both highly expressed in the cerebellum while FHF2 is not.9,18 An additional difference between FHF1- and FHF2-DEE is that the recurrent FHF1-DEE variant is present in both the A (GenBank: NM_021032.4: c.341G>A [p.Arg114His]) and B (GenBank: NM_004113.6: c.155G>A [p.Arg52His]) isoforms. This residue is located in the B4-B5 loop of the core FHF domain that contributes to the highly conserved structural interface between FHF proteins and the cytoplasmic tails of Nav channels.4 Substitution of FHF1A Arg114/FHF1B Arg52 to other amino acids increased the depolarizing shift in the voltage dependence of steady-state inactivation of Nav1.6 (Figure 2A). The shift of this voltage dependence (which increases the availability of resting channel) was greatest for the mutant form of the A isoform. In contrast, FHF2-DEE variants do not alter steady-state inactivation; instead, they disrupt the ability of the N-terminal inactivation particle to cause long-term inhibition of Nav channels.
We recognize the need for caution in interpreting the significance of the variant in female individual 2 when FHF2 is X linked and the other individuals are male. However, individual 2’s epilepsy phenotype was strikingly similar to her brother’s and no other cause for her DEE was identified. X chromosome inactivation assays found no significant skewing in her blood (65:35 at AR [MIM: 313700] and 55:45 at ZNF261 [MIM: 300061]), suggesting the variant allele is active in a moderate proportion of her somatic tissues (although this may not reflect the inactivation pattern in her brain). Consistent with being heterozygous, individual 2 had a milder phenotype than her hemizygous brother and the other males. A difference in severity was also present in the GEFS+ family with the balanced translocation described by Puranam et al.28 The mother with the translocation had febrile seizures in infancy but was neurologically normal and seizure free in adulthood. In contrast, her sons had febrile seizures and temporal lobe epilepsy and one had cognitive impairment. A sex difference is recapitulated by the Fhf2 knockout animal model. Female heterozygous knockout mice are viable but have increased susceptibility to hyperthermia-induced seizures.28 The brains of female heterozygous knockout mice demonstrate mosaic expression of wild-type and null Fhf2 alleles (C.M. and M.G., unpublished data). There is evidence for reduced embryonic survival of male mice, although this may be dependent on strain background.20,22,28 Surviving male knockout mice experience progressive conduction failure in response to higher body temperatures. No cardiac conduction abnormalities were observed in our series. However, loss-of-function partial or whole gene deletions are likely to have different effects from gain-of-function missense variants.
FHF2 was historically considered a candidate gene for Börjeson-Forssman-Lehmann syndrome (BFLS [MIM: 301900]), an X-linked intellectual disability syndrome associated with dysmorphism, epilepsy, obesity, and hypogonadism.20,52 An individual with BFLS-like features (and neonatal-onset seizures) had a maternally inherited Xq26q28 duplication.52 The breakpoint was mapped to a ∼400 kb interval in Xq26.3, a region which contains FHF2. BFLS was subsequently mapped to variants in PHF6 (MIM: 300414), a gene nearby in Xq26.2. The clinical significance of copy-number variation involving FHF2 remains uncertain. However, metrics of dosage sensitivity (haploinsufficiency score 3.17, top 5th percentile)53 and mutational constraint (probability of being loss-of-function intolerant 0.97),29 and evidence from human and animal studies20,22,28 suggests they are likely to have clinically significant effects. Notably, duplications of FHF1 have been reported in individuals with DEE.49,54
A-type FHFs, including FHF2A, and slow-activating/deactivating voltage-gated potassium channels (Kv7.2 and Kv7.3) both contribute to attenuation of excitatory drive by either run-down of the sodium conductance or build-up of the potassium conductance, respectively.55,56 Consequently, both of these processes contribute to spike frequency adaptation (accommodation), as documented in CA1 hippocampal pyramidal neurons.31,57,58 Variants in the genes encoding Kv7.2 or Kv7.3 (KCNQ2 [MIM: 602235] and KCNQ3 [MIM: 602232]) cause early-onset epilepsy phenotypes including DEEs.55,56 Our new findings regarding FHF2A thereby suggest that disturbed spike frequency adaptation is an important mechanism in epileptogenesis.
In conclusion, we have identified hemi- and heterozygous missense variants in the N-terminal of the A isoform of FHF2 as a cause of infantile-onset DEE. Our functional results demonstrate that the variants lead to gain of function because they disrupt the ability of FHF2A to cause long-term inactivation of Nav channels while preserving pro-excitatory properties of FHF2A. Our study provides evidence for the critical role of the FHF2A isoform in regulating Nav channel function and neuronal excitability.
Data and Code Availability
DDD exome data are available through the European Genome-phenome Archive (Study ID EGAS00001000775). Researchers must complete a Data Access Form detailing their research aims and be approved by the DDD data access committee. Whole-genome sequence data generated by the 100,000 Genomes Project is available to researchers at institutions who join the Genomics England Clinical Interpretation Partnership. All other exome data are available from the authors upon reasonable request and with the permission of the families.
Declaration of Interests
X.W. is an employee of Cipher Gene Ltd.
Acknowledgments
We thank the individuals and their families for participating in this study. We thank Dr. David Bunyan of the Wessex Regional Genetics Laboratory for performing the X-chromosome inactivation analysis. A.E.F., M.I.R., and S.-K.C. were supported by WERN, BRAIN Unit, and Wales Gene Park. WERN was funded by The National Institute of Social Care and Health Research. BRAIN Unit and Wales Gene Park are funded by Health and Care Research Wales. C.M. and M.G. were funded in part by grant R01HL142498 from the National Heart Lung and Blood Institute at the National Institutes of Health. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003). This study makes use of DECIPHER, which is funded by Wellcome. See Deciphering Developmental Disorders Study24 or DDD website (see web resources) for full acknowledgment. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support.
Published: November 26, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.10.017.
Web Resources
DDD study, https://www.ddduk.org
DECIPHER, https://decipher.sanger.ac.uk
Ensembl, https://www.ensembl.org
European Genome-phenome Archive (EGA), https://www.ebi.ac.uk/ega
GenBank, https://www.ncbi.nlm.nih.gov/genbank
GeneMatcher, https://genematcher.org
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org
Genomics England, https://www.genomicsengland.co.uk
Human Protein Atlas, https://www.proteinatlas.org
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org
Supplemental information
References
- 1.Kaplan D.I., Isom L.L., Petrou S. Role of Sodium Channels in Epilepsy. Cold Spring Harb. Perspect. Med. 2016;6:6. doi: 10.1101/cshperspect.a022814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catterall W.A. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J. Physiol. 2012;590:2577–2589. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McTague A., Howell K.B., Cross J.H., Kurian M.A., Scheffer I.E. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15:304–316. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]
- 4.Siekierska A., Isrie M., Liu Y., Scheldeman C., Vanthillo N., Lagae L., de Witte P.A.M., Van Esch H., Goldfarb M., Buyse G.M. Gain-of-function FHF1 mutation causes early-onset epileptic encephalopathy with cerebellar atrophy. Neurology. 2016;86:2162–2170. doi: 10.1212/WNL.0000000000002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D., Nassif C., Diallo O., Monlong J., Cadieux-Dion M., Deciphering Developmental Disorders Study High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EuroEPINOMICS-RES Consortium. Epilepsy Phenome/Genome Project. Epi4K Consortium De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheffer I.E., Berkovic S., Capovilla G., Connolly M.B., French J., Guilhoto L., Hirsch E., Jain S., Mathern G.W., Moshé S.L. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheffer I.E., Liao J. Deciphering the concepts behind “Epileptic encephalopathy” and “Developmental and epileptic encephalopathy”. Eur. J. Paediatr. Neurol. 2020;24:11–14. doi: 10.1016/j.ejpn.2019.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16:215–220. doi: 10.1016/j.cytogfr.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldfarb M., Schoorlemmer J., Williams A., Diwakar S., Wang Q., Huang X., Giza J., Tchetchik D., Kelley K., Vega A. Fibroblast growth factor homologous factors control neuronal excitability through modulation of voltage-gated sodium channels. Neuron. 2007;55:449–463. doi: 10.1016/j.neuron.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pablo J.L., Wang C., Presby M.M., Pitt G.S. Polarized localization of voltage-gated Na+ channels is regulated by concerted FGF13 and FGF14 action. Proc. Natl. Acad. Sci. USA. 2016;113:E2665–E2674. doi: 10.1073/pnas.1521194113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wittmack E.K., Rush A.M., Craner M.J., Goldfarb M., Waxman S.G., Dib-Hajj S.D. Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J. Neurosci. 2004;24:6765–6775. doi: 10.1523/JNEUROSCI.1628-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smallwood P.M., Munoz-Sanjuan I., Tong P., Macke J.P., Hendry S.H., Gilbert D.J., Copeland N.G., Jenkins N.A., Nathans J. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc. Natl. Acad. Sci. USA. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen S.K., Garbi M., Zampieri N., Eliseenkova A.V., Ornitz D.M., Goldfarb M., Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J. Biol. Chem. 2003;278:34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- 15.Sochacka M., Opalinski L., Szymczyk J., Zimoch M.B., Czyrek A., Krowarsch D., Otlewski J., Zakrzewska M. FHF1 is a bona fide fibroblast growth factor that activates cellular signaling in FGFR-dependent manner. Cell Commun. Signal. 2020;18:69. doi: 10.1186/s12964-020-00573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C., Chung B.C., Yan H., Lee S.-Y., Pitt G.S. Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure. 2012;20:1167–1176. doi: 10.1016/j.str.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartung H., Feldman B., Lovec H., Coulier F., Birnbaum D., Goldfarb M. Murine FGF-12 and FGF-13: expression in embryonic nervous system, connective tissue and heart. Mech. Dev. 1997;64:31–39. doi: 10.1016/s0925-4773(97)00042-7. [DOI] [PubMed] [Google Scholar]
- 18.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Li S., Tao T., Li X., Zhu Q., Liao Y., Ma J., Sun Y., Liu W. Intrafollicular fibroblast growth factor 13 in polycystic ovary syndrome: relationship with androgen levels and oocyte developmental competence. J. Ovarian Res. 2018;11:87. doi: 10.1186/s13048-018-0455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q.-F., Yang L., Li S., Wang Q., Yuan X.-B., Gao X., Bao L., Zhang X. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell. 2012;149:1549–1564. doi: 10.1016/j.cell.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 21.Favuzzi E., Deogracias R., Marques-Smith A., Maeso P., Jezequel J., Exposito-Alonso D., Balia M., Kroon T., Hinojosa A.J.F., F Maraver E., Rico B. Distinct molecular programs regulate synapse specificity in cortical inhibitory circuits. Science. 2019;363:413–417. doi: 10.1126/science.aau8977. [DOI] [PubMed] [Google Scholar]
- 22.Park D.S., Shekhar A., Marra C., Lin X., Vasquez C., Solinas S., Kelley K., Morley G., Goldfarb M., Fishman G.I. Fhf2 gene deletion causes temperature-sensitive cardiac conduction failure. Nat. Commun. 2016;7:12966. doi: 10.1038/ncomms12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinden D.S., Holman C.D., Bare C.J., Sun X., Gade A.R., Cohen D.E., Pitt G.S. Knockout of the X-linked Fgf13 in the hypothalamic paraventricular nucleus impairs sympathetic output to brown fat and causes obesity. FASEB J. 2019;33:11579–11594. doi: 10.1096/fj.201901178R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomics England (2019). The National Genomics Research and Healthcare Knowledgebase v5. 10.6084/m9.figshare.4530893.v5 [DOI]
- 27.Munoz-Sanjuan I., Smallwood P.M., Nathans J. Isoform diversity among fibroblast growth factor homologous factors is generated by alternative promoter usage and differential splicing. J. Biol. Chem. 2000;275:2589–2597. doi: 10.1074/jbc.275.4.2589. [DOI] [PubMed] [Google Scholar]
- 28.Puranam R.S., He X.P., Yao L., Le T., Jang W., Rehder C.W., Lewis D.V., McNamara J.O. Disruption of Fgf13 causes synaptic excitatory-inhibitory imbalance and genetic epilepsy and febrile seizures plus. J. Neurosci. 2015;35:8866–8881. doi: 10.1523/JNEUROSCI.3470-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Shi L., Zhang K., Zhang Y., Hu S., Zhao T., Teng H., Li X., Jiang Y., Ji L., Sun Z. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018;46(D1):D1039–D1048. doi: 10.1093/nar/gkx1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatesan K., Liu Y., Goldfarb M. Fast-onset long-term open-state block of sodium channels by A-type FHFs mediates classical spike accommodation in hippocampal pyramidal neurons. J. Neurosci. 2014;34:16126–16139. doi: 10.1523/JNEUROSCI.1271-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagal S.K., Marron B.E., Owen R.M., Storer R.I., Swain N.A. Voltage gated sodium channels as drug discovery targets. Channels (Austin) 2015;9:360–366. doi: 10.1080/19336950.2015.1079674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dover K., Solinas S., D’Angelo E., Goldfarb M. Long-term inactivation particle for voltage-gated sodium channels. J. Physiol. 2010;588:3695–3711. doi: 10.1113/jphysiol.2010.192559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rush A.M., Wittmack E.K., Tyrrell L., Black J.A., Dib-Hajj S.D., Waxman S.G. Differential modulation of sodium channel Na(v)1.6 by two members of the fibroblast growth factor homologous factor 2 subfamily. Eur. J. Neurosci. 2006;23:2551–2562. doi: 10.1111/j.1460-9568.2006.04789.x. [DOI] [PubMed] [Google Scholar]
- 35.Peters H.C., Hu H., Pongs O., Storm J.F., Isbrandt D. Conditional transgenic suppression of M channels in mouse brain reveals functions in neuronal excitability, resonance and behavior. Nat. Neurosci. 2005;8:51–60. doi: 10.1038/nn1375. [DOI] [PubMed] [Google Scholar]
- 36.Buchin A., Kerr C.C., Huberfeld G., Miles R., Gutkin B. Adaptation and Inhibition Control Pathological Synchronization in a Model of Focal Epileptic Seizure. eNeuro. 2018;5:5. doi: 10.1523/ENEURO.0019-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laezza F., Lampert A., Kozel M.A., Gerber B.R., Rush A.M., Nerbonne J.M., Waxman S.G., Dib-Hajj S.D., Ornitz D.M. FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol. Cell. Neurosci. 2009;42:90–101. doi: 10.1016/j.mcn.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou J.-Y., Laezza F., Gerber B.R., Xiao M., Yamada K.A., Hartmann H., Craig A.M., Nerbonne J.M., Ornitz D.M. Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J. Physiol. 2005;569:179–193. doi: 10.1113/jphysiol.2005.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goetz R., Dover K., Laezza F., Shtraizent N., Huang X., Tchetchik D., Eliseenkova A.V., Xu C.-F., Neubert T.A., Ornitz D.M. Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J. Biol. Chem. 2009;284:17883–17896. doi: 10.1074/jbc.M109.001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvill G.L., Engel K.L., Ramamurthy A., Cochran J.N., Roovers J., Stamberger H., Lim N., Schneider A.L., Hollingsworth G., Holder D.H., EuroEPINOMICS Rare Epilepsy Syndrome, Myoclonic-Astatic Epilepsy, and Dravet Working Group Aberrant Inclusion of a Poison Exon Causes Dravet Syndrome and Related SCN1A-Associated Genetic Epilepsies. Am. J. Hum. Genet. 2018;103:1022–1029. doi: 10.1016/j.ajhg.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epilepsy Genetics Initiative De novo variants in the alternative exon 5 of SCN8A cause epileptic encephalopathy. Genet. Med. 2018;20:275–281. doi: 10.1038/gim.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perenthaler E., Nikoncuk A., Yousefi S., Berdowski W.M., Alsagob M., Capo I., van der Linde H.C., van den Berg P., Jacobs E.H., Putar D. Loss of UGP2 in brain leads to a severe epileptic encephalopathy, emphasizing that bi-allelic isoform-specific start-loss mutations of essential genes can cause genetic diseases. Acta Neuropathol. 2020;139:415–442. doi: 10.1007/s00401-019-02109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bodian D.L., Kothiyal P., Hauser N.S. Pitfalls of clinical exome and gene panel testing: alternative transcripts. Genet. Med. 2019;21:1240–1245. doi: 10.1038/s41436-018-0319-7. [DOI] [PubMed] [Google Scholar]
- 44.Schoch K., Tan Q.K.-G., Stong N., Deak K.L., McConkie-Rosell A., McDonald M.T., Goldstein D.B., Jiang Y.H., Shashi V., Undiagnosed Diseases Network Alternative transcripts in variant interpretation: the potential for missed diagnoses and misdiagnoses. Genet. Med. 2020;22:1269–1275. doi: 10.1038/s41436-020-0781-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rigbye K.A., van Hasselt P.M., Burgess R., Damiano J.A., Mullen S.A., Petrovski S., Puranam R.S., van Gassen K.L.I., Gecz J., Scheffer I.E. Is FGF13 a major contributor to genetic epilepsy with febrile seizures plus? Epilepsy Res. 2016;128:48–51. doi: 10.1016/j.eplepsyres.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Al-Mehmadi S., Splitt M., Ramesh V., DeBrosse S., Dessoffy K., Xia F., Yang Y., Rosenfeld J.A., Cossette P., Michaud J.L., For DDD Study group∗; For CENet Study group‡ FHF1 (FGF12) epileptic encephalopathy. Neurol. Genet. 2016;2:e115. doi: 10.1212/NXG.0000000000000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paprocka J., Jezela-Stanek A., Koppolu A., Rydzanicz M., Kosińska J., Stawiński P., Płoski R. FGF12p.Gly112Ser variant as a cause of phenytoin/phenobarbital responsive epilepsy. Clin. Genet. 2019;96:274–275. doi: 10.1111/cge.13592. [DOI] [PubMed] [Google Scholar]
- 48.Trivisano M., Ferretti A., Bebin E., Huh L., Lesca G., Siekierska A., Takeguchi R., Carneiro M., De Palma L., Guella I. Defining the phenotype of FHF1 developmental and epileptic encephalopathy. Epilepsia. 2020;61:e71–e78. doi: 10.1111/epi.16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi R.-M., Kobayashi T., Kikuchi A., Sato R., Uematsu M., An K., Kure S. Phenytoin-responsive epileptic encephalopathy with a tandem duplication involving FGF12. Neurol. Genet. 2017;3:e133. doi: 10.1212/NXG.0000000000000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miura S., Kosaka K., Fujioka R., Uchiyama Y., Shimojo T., Morikawa T., Irie A., Taniwaki T., Shibata H. Spinocerebellar ataxia 27 with a novel nonsense variant (Lys177X) in FGF14. Eur. J. Med. Genet. 2019;62:172–176. doi: 10.1016/j.ejmg.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 51.van Swieten J.C., Brusse E., de Graaf B.M., Krieger E., van de Graaf R., de Koning I., Maat-Kievit A., Leegwater P., Dooijes D., Oostra B.A., Heutink P. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia [corrected] Am. J. Hum. Genet. 2003;72:191–199. doi: 10.1086/345488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gecz J., Baker E., Donnelly A., Ming J.E., McDonald-McGinn D.M., Spinner N.B., Zackai E.H., Sutherland G.R., Mulley J.C. Fibroblast growth factor homologous factor 2 (FHF2): gene structure, expression and mapping to the Börjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum. Genet. 1999;104:56–63. doi: 10.1007/s004390050910. [DOI] [PubMed] [Google Scholar]
- 53.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oda Y., Uchiyama Y., Motomura A., Fujita A., Azuma Y., Harita Y., Mizuguchi T., Yanagi K., Ogata H., Hata K. Entire FGF12 duplication by complex chromosomal rearrangements associated with West syndrome. J. Hum. Genet. 2019;64:1005–1014. doi: 10.1038/s10038-019-0641-1. [DOI] [PubMed] [Google Scholar]
- 55.Villa C., Combi R. Potassium Channels and Human Epileptic Phenotypes: An Updated Overview. Front. Cell. Neurosci. 2016;10:81. doi: 10.3389/fncel.2016.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miceli F., Soldovieri M.V., Ambrosino P., De Maria M., Migliore M., Migliore R., Taglialatela M. Early-onset epileptic encephalopathy caused by gain-of-function mutations in the voltage sensor of Kv7.2 and Kv7.3 potassium channel subunits. J. Neurosci. 2015;35:3782–3793. doi: 10.1523/JNEUROSCI.4423-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu H., Vervaeke K., Storm J.F. M-channels (Kv7/KCNQ channels) that regulate synaptic integration, excitability, and spike pattern of CA1 pyramidal cells are located in the perisomatic region. J. Neurosci. 2007;27:1853–1867. doi: 10.1523/JNEUROSCI.4463-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hönigsperger C., Marosi M., Murphy R., Storm J.F. Dorsoventral differences in Kv7/M-current and its impact on resonance, temporal summation and excitability in rat hippocampal pyramidal cells. J. Physiol. 2015;593:1551–1580. doi: 10.1113/jphysiol.2014.280826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DDD exome data are available through the European Genome-phenome Archive (Study ID EGAS00001000775). Researchers must complete a Data Access Form detailing their research aims and be approved by the DDD data access committee. Whole-genome sequence data generated by the 100,000 Genomes Project is available to researchers at institutions who join the Genomics England Clinical Interpretation Partnership. All other exome data are available from the authors upon reasonable request and with the permission of the families.