Summary
Plants that successfully acclimate to stress can resume growth under stressful conditions. The grass Brachypodium distachyon can grow a cold‐adaptive morphology during cold acclimation. Studies on transcriptional memory (TM) have revealed that plants can be primed for stress by adjusting their transcriptional responses, but the function of TM in stress acclimation is not well understood. We investigated the function of TM during cold acclimation in B. distachyon.
Quantitative polymerase chain reaction (qPCR), RNA‐seq and chromatin immunoprecipitation qPCR analyses were performed on plants exposed to repeated episodes of cold to characterize the presence and stability of TM during the stress and growth responses of cold acclimation.
Transcriptional memory mainly dampened stress responses as growth resumed and as B. distachyon became habituated to cold stress. Although permanent on vernalization gene VRN1, TMs were short‐term and reversible on cold‐stress genes. Growing under cold conditions also coincided with the acquisition of new and targeted cold‐induced transcriptional responses.
Overall, TM provided plasticity to cold stress responses during cold acclimation in B. distachyon, leading to stress habituation, acquired stress responses, and resumed growth. Our study shows that chromatin‐associated TMs are involved in tuning plant responses to environmental change and, as such, regulate both stress and developmental components that characterize cold‐climate adaptation in B. distachyon.
Keywords: Brachypodium distachyon, chromatin, cold acclimation, growth, phenotypic plasticity, stress, transcriptional memory
Introduction
Understanding the mechanisms of phenotypic plasticity in plants is crucial to building a sustainable agriculture, especially under the current world‐wide climate and environmental crises. Human activities have caused abrupt environmental changes that will affect the global climate well beyond this century, imposing stress of increasing intensity upon plants and crops (Gray & Brady, 2016; USGCRP, 2017; Bathiany et al., 2018). Environmental disruptions can be widely problematic; for example, cold spells and late frosts impair crop production in a wide variety of environments, from temperate Canada to sub‐tropical India (Aggarwal, 2008; Kutcher et al., 2010). Finding ways to employ or enhance the acclimation mechanisms of plants can contribute to building more resilient food production systems.
Stressful conditions generally trigger responses that alleviate the negative effects of stress on plant growth and development. Depending on the severity of the stress and the adaptability of the species, plants either die, enter a state of dormancy until conditions improve, or acclimate and continue growing (Ciarmiello et al., 2011). In the literature, acclimation is usually confined to reversible physiological changes, although stress conditions can irreversibly influence plant structure (Liu & Su, 2016; Klem et al., 2019; Mayer et al., 2020). Recently, stress responses were found to be particularly plastic, improving over multiple exposures (Ding et al., 2012; Li et al., 2019; Zuther et al., 2019; Mayer et al., 2020). These modified responses are influenced by stress exposures during which plants build ‘experience’ through stress memories (Crisp et al., 2016; Yeung et al., 2018).
Stress responses are often linked to extensive changes in gene expression (Ingram & Bartels, 1996; Thomashow, 1999; Zhang et al., 2006). Studies have hence reported cases of stress memories linked to transcription, called transcriptional memories (TMs). Genes that show TM typically display different transcriptional responses when the same stimulus is applied repeatedly (Avramova, 2015; Lämke & Bäurle, 2017). Transcriptional memory events were associated with specific epigenetic remodelers and to changes in chromatin marks left by stress exposure, which can, at least in part, explain the encoding of stress memories (Ding et al., 2012; Lamke et al., 2016). The di‐ and tri‐methylation of histone 3 lysine 4 (H3K4me2/3) were identified as markers of TM in various plant abiotic stress contexts, including heat, cold, drought and salt stress (Liu et al., 2014, 2018; Shen et al., 2014; Feng et al., 2016; Lamke et al., 2016; Zeng et al., 2019), while DNA methylation is involved in stress‐responses, TM and adaptation to environmental stress (Verhoeven et al., 2010; Jiang et al., 2014; Mayer et al., 2015; Sanchez & Paszkowski, 2014; Wibowo et al., 2016). By changing the regulation of gene expression, mechanisms of TM contribute to the plasticity of stress responses. As plants can progress from ‘shock‐like’ stress responses to resuming growth in stressful conditions, TM mechanisms may hence confer plasticity to such responses, leading to successful acclimation and adapted morphology. Although stress‐response mechanisms, in general terms, have been well studied, the function of TMs in this stress‐to‐growth requires further investigation.
The traits that characterize cold adaptation in the model cereal Brachypodium distachyon, namely cold acclimation and vernalization, provide a useful system for the study of the interaction between stress and growth, and the function of TM in acclimation. Recently established as a model for cold‐induced responses in temperate cereals, this undomesticated grass responds to cold by physiologically acclimating, gaining flowering competence through vernalization, and growing a cold‐hardy morphology (Li et al., 2012; Colton‐Gagnon et al., 2014; Ream et al., 2014; Ryu et al., 2014; Mayer et al., 2020). The acquisition of freezing tolerance, aimed at maximizing survival to winter, encompasses physiological changes associated with cold acclimation and morphological plasticity, leading to a freezing tolerant structure (Chouard, 1960; Thomashow, 1999; Körner, 2016). In addition, timely flowering, crucial in temperate climates, is ensured by a process known as vernalization. In temperate grasses, overwintering provides flowering competence by activating the transcription factor VERNALIZATION1 (VRN1) (Danyluk et al., 2003; Oliver et al., 2009; Woods et al., 2017; Mayer et al., 2020). Under cold conditions, the chromatin state of VRN1 becomes gradually depleted of the silencing epigenetic mark histone 3 lysine 27 trimethylation (H3K27me3), and this change remains after cold exposure (Oliver et al., 2009, 2013; Chen & Dubcovsky, 2012; Woods et al., 2017; Huan et al., 2018). Hence, vernalization ensures timely flowering over long‐term cold exposure through an epigenetically regulated permanent TM. Although well studied in vernalization, little is known about the function of TM in cold acclimation and morphological responses to cold. Recent work in Arabidopsis has demonstrated the existence of cold‐stress memories affecting freezing tolerance, and transcriptomic and metabolomic responses to chilling, and has highlighted the involvement of chromatin modifications in cold memory (Zuther et al., 2019; Vyse et al., 2020), though other studies have also described cold memory and changes to chromatin states associated with cold exposure other than vernalization (Zhu et al., 2008; Kwon et al., 2009; Mayer et al., 2015; Bittner et al., 2020). Unlike in Arabidopsis, however, vernalization and the expression of freezing tolerance encompassing cold acclimation and morphology are interconnected in B. distachyon, notably through VRN1 which, other than regulating vernalization, influences cold acclimation and the acquisition of a winter‐hardy morphology (Bond et al., 2011; Mayer et al., 2020). Hence, studying the function of TM when growth is resumed in cold conditions in B. distachyon can provide valuable insights into the mechanisms that regulate phenotypic plasticity during stress acclimation in plants.
Materials and Methods
Plant growth and cold treatments
Bd21‐3 seeds were planted in 3 × 3 inch 0.5 l pots containing 160 g of G2 Agromix (Fafard et Frères Ltd, Saint‐Remi, QC, Canada), grown in an environmental growth chamber (Conviron, Winnipeg, MB, Canada) under nonstress control conditions (under a 16 h : 8 h, light : dark photoperiod at 22°C; 150 μmol m−2 s−1 photosynthetically active radiation (PAR) intensity) for 14 d, then transferred to 4°C or diurnal‐freezing conditions in an LT‐36VL growth chamber (Percival Scientific, Perry, IA, USA). The diurnal‐freezing treatment is characterized by air temperature cycles reaching −1°C at night and 22°C during the day, as described in Fig. 3(a) and as explained in more detail in Fig. 1(d) and (e) of Mayer et al. (2020). Plants were kept equally watered throughout.
Fig. 3.
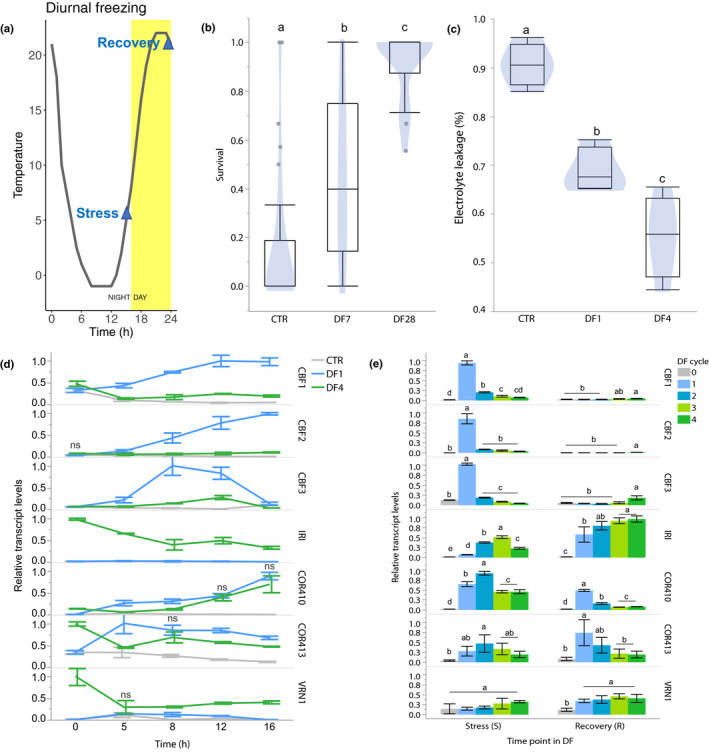
Cycles of diurnal freezing gradually increase freezing tolerance and change transcriptional responses to cold in Brachypodium distachyon. (a) One cycle of diurnal freezing, with time points S (stress) and R (recovery). (b) Freezing tolerance of plants exposed to 7 and 28 cycles of diurnal freezing (DF; DF7 and DF28), measured in whole‐plant freeze tests performed three times independently on 27 plants per temperature plateau between −8 and −12°C. (c) Damage induced by freezing of plants previously exposed to 1 or 4 cycles of DF (DF1 and DF4) measured by electrolyte leakage. (d) Relative transcript levels of CBFs, IRI, COR410, COR413 and VRN1 measured at 0, 5, 8, 12 and 16 of exposure to 1 or 4 cycles of DF (DF1 and DF4) relative to control (CTR). (e) Relative transcript level of COR genes in response to 1, 2, 3 and 4 cycles of DF (DF1–4) samples at S (S1–4) and R (R1–4) time points. Different letters indicate statistical difference; ns, no significant difference; P < 0.05. Error bars represent SD between three biological replicates.
Fig. 1.
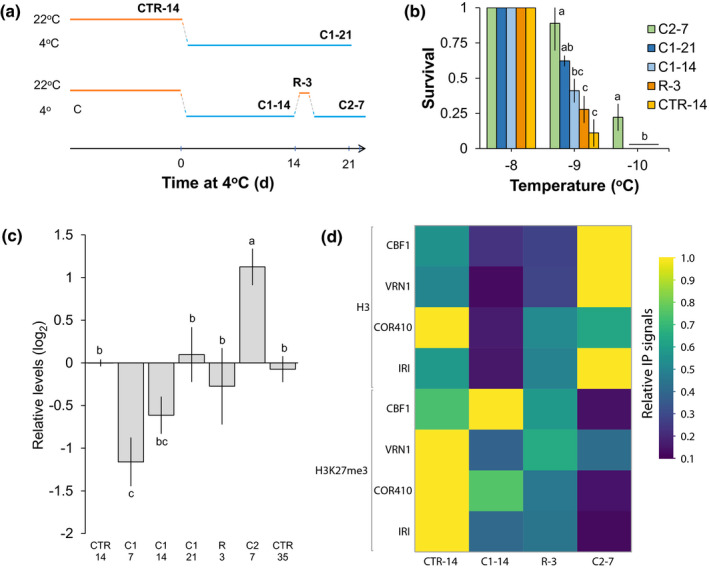
Repeated exposure to chilling leads to higher freezing tolerance, and to global and gene‐specific epigenetic changes. (a) Experimental design where one period of recovery at 22°C was inserted during cold acclimation under constant‐chilling (CC) at 4°C. C1, first exposure to chilling for 14 or 21 d (C1‐14 and C1‐21); C2‐7, plants re‐exposed to chilling for 7 d after recovery R‐3; CTR‐14, plants grown under control conditions at 22°C; R‐3, plants recovered from chilling for 3 d. (b) Survival rate under freezing conditions of CTR‐14, C1‐14, C1‐21, R‐3, and C2‐7 Brachypodium distachyon plants measured by whole‐plant freeze tests. (c) Relative global DNA‐methylation. C1, first exposure to chilling for 7, 14 or 21 d (C1‐7, C1‐14, C1‐21); C2‐7, second exposure to chilling for 7 d; CTR‐14, B. distachyon plants grown under control conditions at 22°C for 14 d; CTR‐35, B. distachyon plants grown under control conditions at 22°C for 35 d; R‐3, recovery from chilling at 22°C for 3 d. (d) Levels of histone 3 (H3) relative to input levels and of H3K27me3 relative to H3 of cold‐regulated genes C‐REPEAT BINDING FACTOR1 (CBF1), VERNALIZATION1 (VRN1), COLD‐REGULATED410 (COR410) and ICE RECRYSTALLIZATION INHIBITOR (IRI) at CTR‐14, C1‐14, R‐3 and C2‐7. Different letters indicate statistical difference; P < 0.05; error bars represent SD between three biological replicates.
Measures of freezing tolerance
For whole‐plant freeze tests, plants were subjected to gradually decreasing temperatures from −1°C to −12°C at a rate of 1°C h–1 in a LT‐36VL chamber (Percival Scientific). Pots containing nine plants each were watered to soil saturation and placed randomly in the chamber. Three randomly selected pots were removed hourly as the temperature decreased from −8°C to −12°C, left to thaw at 4°C in the dark for 24 h, then switched to 22°C for an additional 24 h before being transferred to control conditions; percent survival was determined after 1 wk of recovery. This experiment was repeated three times. Electrolyte leakage for leaf tissue was measured as described in a study by Lee & Zhu (2010) for five leaf replicates, and was performed three times.
RNA extraction and quantitative reverse transcription polymerase chain reaction (RT‐qPCR)
For each biological triplicate, a pool of aerial tissue from three plants was collected, flash‐frozen in liquid nitrogen, and extracted using an EZ‐10 RNA kit (cat. no. BS82314; Bio Basic, New York, NY, USA) following the manufacturer's protocol. Reverse‐transcriptase cDNA was obtained using iScript (cat. no. 1725037; Bio‐Rad) and was RT‐qPCR performed using Green‐2‐Go (cat. no. QPCR004; Bio Basic) and CFX Connect Real Time (BioRad), following the manufacturer's protocol. Relative transcript levels were determined by ΔΔCT using the UBC18 gene as a reference for three biologically independent replicates (Hong et al., 2008; Mayer et al., 2020). The cold‐responsive genes selected have been studied previously and exhibit different functions during cold acclimation in B. distachyon. These include transcription factors C‐REPEAT BINDING FACTOR1, 2 and 3 (CBF1, CBF2, CBF3) (Colton‐Gagnon et al., 2014; Ryu et al., 2014), which likely induce the expression of the following structural genes: membrane‐protein COLD‐REGULATED413, dehydrin COLD‐REGULATED410 (Colton‐Gagnon et al., 2014; Mayer et al., 2020), and possibly the anti‐freeze protein ICE RECRYSTALLIZATION INHIBITOR (IRI) (Bredow et al., 2016; Mayer et al., 2020). The transcription factor VERNALIZATION1 (VRN1) is involved in vernalization but was recently shown to play an important function in cold acclimation and freezing tolerance (Ream et al., 2014; Mayer et al., 2020). Primer sequences can be found in Supporting Information Table S1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed for three biological replicates as previously described (Mayer et al., 2015), using antibodies anti‐Histone H3 (cat. no. ab1791; Abcam, Cambridge, UK), anti‐Histone H3 tri‐methyl‐K27 (cat. no. ab6002; Abcam), anti‐Histone H3 di‐methyl‐K4 (cat. no. ab11946; Abcam) and anti‐Histone H3 tri‐methyl‐K4 (cat. no. ab8580; Abcam). Immunoprecipitated samples were analyzed using qPCR and their percent input or percent H3 values were ascertained.
Global DNA methylation assay
Global DNA methylation was performed using an Imprint Methylated‐DNA Quantification Kit (Sigma‐Aldrich) following the manufacturer's protocol. Genomic DNA was extracted using phenol‐chloroform for two independent biological replicates, each consisting of a pool of aerial tissue from three plants. Each replicate was measured in technical quadruplicate using a Microplate reader (Bio‐Rad).
Sample preparation for RNA‐sequencing
For the diurnal‐freezing priming experiment, three plants for each treatment were collected 10 min before the light phase of the photoperiod. For the primed response to chilling experiment, three plants for each treatment were collected 3 h after the plants were moved to the chilling environment. After collection, plant tissue was flash‐frozen and stored at −80°C until extraction. This experiment was performed twice. RNA was extracted using the RNeasy Kit (Qiagen), following the manufacturer's protocol. Two libraries, composed of three biological replicates each, were built using NEBNext Multiplex Oligos for Illumina (cat. no. E7600S; New England Biolabs, Ipswich, MA, USA) and sequenced using a HiSeq 4000 (diurnal‐freezing priming experiment; Illumina, San Diego, CA, USA) and a NovaSeq 6000 (primed response to chilling; Illumina) at Le Centre d’Expertise et de Services Genome, Québec.
Transcriptome analysis
Using galaxy, FASTQ reads (available online at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA629906), were analysed with fastqc and processed with trimmomatic to remove adapters (Andrews, 2010; Bolger et al., 2014; Afgan et al., 2018). Using rna star, trimmed reads were mapped to Bd21v.3.1 obtained via Phytozome (Brachypodium distachyon Bd21v.3.1 DOE‐JGI, http://phytozome.jgi.doe.gov), and reads were counted using featurecounts (Vogel, 2010; Dobin et al., 2013; Liao et al., 2014). Using deseq2 with cold/priming and replicates as factors, fold‐change was calculated for S1/S0, S4/S0, S4/S1, and naïve and primed responses to chilling by comparing cold‐treated plants with their respective non‐stressed controls (Anders & Huber, 2010). Genes displaying significant differential expression (P‐adj < 0.05) and absolute fold change > 2 (FC > 2) in both replicates were selected for further analysis.
Gene ontology (GO) enrichment analysis
Gene ontology analyses were performed using agrigo v.2.0 (Tian et al., 2017) with a false discovery rate (FDR) adjusted p‐value of 0.05 for genes with FC > 2, goseq for genes with FC > 4 (Young et al., 2010), and revigo (Supek et al., 2011) with associated over‐represented P‐adj values. The Oryza sativa GO term size database was used, along with simrel for semantic similarity (Supek et al., 2011). The annotated B. distachyon gene list was obtained from Phytozome. Gene ontology terms shown in Fig. 4(e) (see below) were obtained by summarizing agrigo terms with revigo. Treemaps were visualized using the package treemap in R (R Core Team, 2013).
Fig. 4.
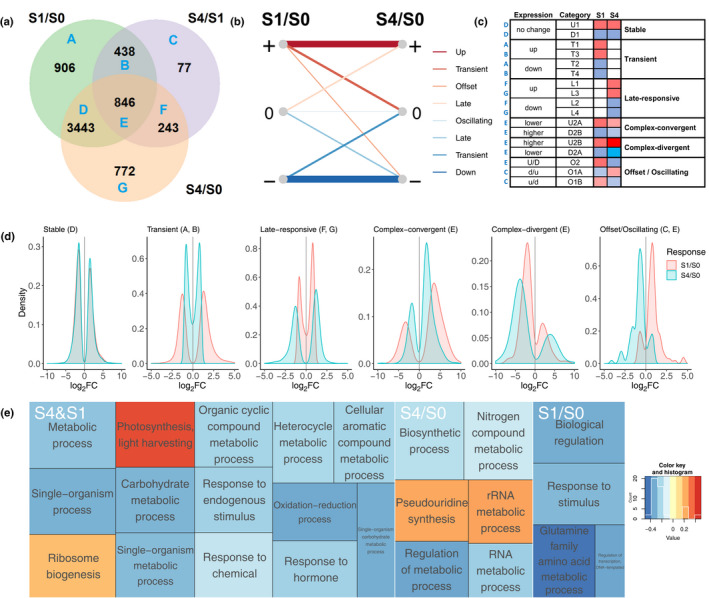
Transcriptional responses of Brachypodium distachyon evolve through repeated cycles of diurnal freezing following six profiles. (a) Venn diagrams of significantly differentially‐expressed genes in S1 (sampled during a first cycle of diurnal freezing) compared to S0 (control conditions), labelled S1/S0; in S4 (sampled during a fourth cycle of diurnal freezing) compared to S1, labelled S4/S1; and in S4 compared to S0, labelled S4/S0. (b) Sign of differential expression of genes responsive to diurnal freezing in S1/S0 and S4/S0. +, upregulated; 0, nonresponsive; −, downregulated. Lines are proportional to the number of genes following each trend. (c) Genes responsive to diurnal freezing divided into 17 categories according to their distribution between S1/S0, S4/S1 and S4/S0, as illustrated in (a), and their upregulation or downregulation in S1 and S4 responses (S1/S0 and S4/S0) as illustrated in (b). (d) Six profiles that describe the transcriptional behavior of genes under diurnal freezing. (e) Significantly over‐represented GO terms that are unique to the S1 response (S1/S0), unique to the S4 response (S4/S0), or shared by both (S4&S1). Colours depict the transcriptional change of genes associated with each GO term between S1 and S4 (S4/S1 – FC negative to positive, blue to red).
Transcription factors and transcriptional regulator analysis
Transcription factors were identified using itak (Zheng et al., 2016) and visualized with heatmap.2 in R (R Core Team, 2013).
Statistical analyses
All experimental data, except for RNA‐seq results, were analyzed by one‐way ANOVA followed by Tukey's honest significance (HSD) test using jmp (SAS Institute, Cary, NC, USA). Statistical significance was determined with P < 0.05.
Results
Intermittent exposure to chilling resulted in a higher survival rate than continuous treatment
To determine whether B. distachyon can acquire cold‐stress memories, we exposed plants to a cold acclimation treatment interrupted by recovery in control conditions. By performing whole‐plant freeze tests, we measured survival under freezing conditions for plants that were exposed to chilling continuously for 14 or 21 d, or intermittently for 21 d (first for 14 d, then for 7 d separated by a 3‐d recovery; Fig. 1a). Plants exposed to chilling for a total of 21 d were most tolerant to freezing. However, those that were subjected to an intermittent treatment survived better than those exposed to continuous chilling (Fig. 1b). Interestingly, the second 7‐d exposure induced an increase in the 50% lethal temperature (LT50) value from c. 25% to c. 90% at − 9°C (re‐acclimated compared to recovery plants), compared to an increase from c. 15% to c. 40% after the first 14‐d exposure (acclimated compared to nonacclimated plants). Therefore, these results indicate that B. distachyon acclimated to cold faster and more efficiently during the second exposure to chilling.
Repeated chilling led to global and gene specific chromatin changes
As chilling induces both cold acclimation and vernalization in B. distachyon, we investigated the chromatin response during both continuous and intermittent chilling treatments. We first measured the levels of global DNA methylation, which decreased significantly after 7 d of chilling but gradually increased back to control levels after 14 and 21 d of continuous exposure (Fig. 1c). Moving plants to recovery after 14 d in chilling did not alter the global levels of DNA methylation, while subsequent re‐exposure induced a hypermethylation significantly higher than the continuous 21‐d treatment (Fig. 1c). Therefore, continuous exposure to chilling led to a transient hypomethylation, while a second exposure to chilling led to global DNA hypermethylation.
As a depletion of the silencing mark H3K27me3 occurs during vernalization in B. distachyon, which is a long‐term epigenetic response, we investigated H3K27me3 levels on the gene VERNALIZATION1 (VRN1), in acclimated, recovered and re‐acclimated plants. To determine whether this mark is also involved in cold acclimation, we measured H3K27me3 levels on the transcription factor C‐REPEAT BINDING FACTOR1 (CBF1), the dehydrin COR410, and the anti‐freeze protein ICE RECRYSTALLIZATION INHIBITOR (IRI), which are involved in cold acclimation in B. distachyon (Colton‐Gagnon et al., 2014; Ryu et al., 2014; Bredow et al., 2016; Mayer et al., 2020). After 14 d of chilling, nucleosome levels significantly decreased on VRN1, CBF1, COR410 and IRI, while levels of H3K27me3 generally decreased, suggesting that these genes were activated upon cold exposure (Fig. 1d; Table S2). Interestingly, 7 d of re‐acclimation led to even lower levels of H3K27me3, while nucleosome levels increased on the four genes. Therefore, cold acclimation led to fewer nucleosomes and lower levels of H3K27me3, while reacclimation led to denser chromatin, suggesting that B. distachyon had a different chromatin response upon re‐exposure to chilling.
Cold acclimation induces the formation of TMs
The transcriptional response of cold acclimation typically occurs, unlike vernalization, during the first few h of cold exposure. Hence, to determine whether cold acclimation led to the formation of TMs, we measured the transcript levels of CBF1, COR410, IRI and VRN1 at 1, 3, 6 and 24 h of exposure, during re‐exposure to chilling, and in recovery. The transcript levels of all four genes were measurably higher at least at the first h of recovery compared to nonacclimated controls (Fig. 2a). Interestingly, transcript levels of IRI increased within the first 6 h of recovery, while those of VRN1, known to be regulated by a permanent TM, remained elevated throughout recovery. Re‐exposure to chilling led to a lower transcriptional response at all time points in CBF1 but only at the later ones in COR410 and IRI, while VRN1 showed higher activation at all time points (Fig. 2a). Therefore, cold exposure induced the formation of TMs that affected the transcript levels of IRI during recovery, of CBF1 upon re‐exposure and of VRN1 throughout.
Fig. 2.
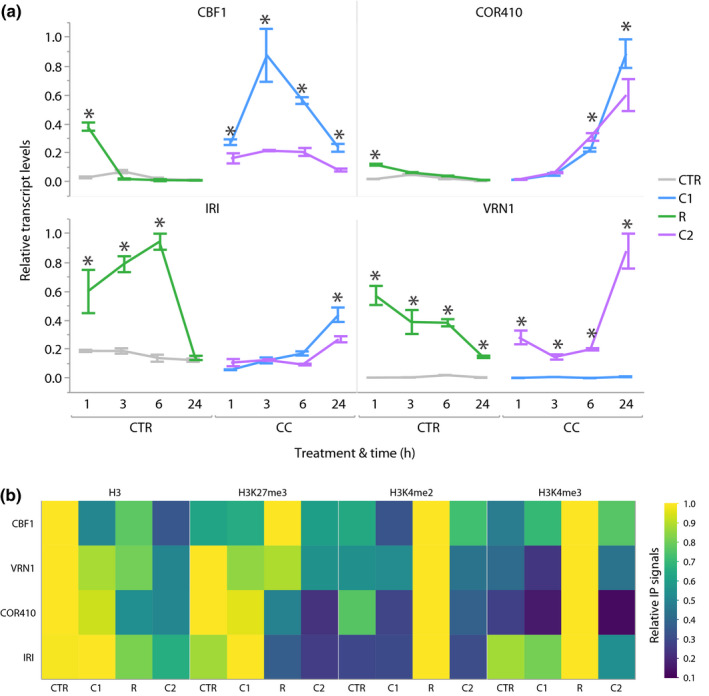
Repeated chilling influences transcriptional and chromatin responses to cold in Brachypodium distachyon. (a) Transcript levels of COR genes CBF1, VRN1, COR410 and IRI at the first 1, 3, 6 and 24 h of exposure to a first cold treatment (C1), during recovery from chilling (R) and to a second cold treatment (C2) relative to CTR, C1‐14 and R‐3. (b) Levels of histone H3 relative to input levels, and of H3K27me3, H3K4me2 and H3K4me3 relative to H3 on CBF1, VRN1, COR410 and IRI at 3 h into C1, R and C2 compared to control CTR. Asterisks indicate statistical difference (*, P < 0.05); error bars represent SD between three biological replicates.
As chilling led to the formation of TMs affecting CBF1, COR410, IRI and VRN1, we investigated whether these were connected, at their gene loci, to nucleosome occupancy, to levels of H3K27me3 and to the levels of chromatin marks involved in stress‐induced TMs, namely H3K4me2 and H3K4me3. These were measured during the early transcriptional response to cold acclimation at 3 h into the first and second exposures, and into recovery. The nucleosome and H3K27me3 levels measured under chilling were lower in plants exposed for a second time, indicating that these adopted a looser chromatin structure (Fig. 2b; Table S2). Moreover, the levels of H3K4me2 and H3K4me3 peaked at recovery, suggesting that these were deposited after the first episode of cold, and dropped to lower levels upon re‐exposure, which indicates that they are involved in the acquired transcriptional responses of CBF1, VRN1 IRI and COR410 (Fig. 2b). Therefore, the TMs observed for CBF1, COR410, IRI and VRN1 occurred along with different epigenetic signatures at their gene loci.
Longer exposures to diurnal freezing translate into higher freezing tolerance
We previously showed that chilling induces artificial responses and limits cold acclimation in B. distachyon partly through a high expression of VRN1, which inhibits the transcription of CBFs (Mayer et al., 2020). Hence, the high VRN1 expression induced during the first chilling exposure has likely interfered with the transcriptional responses of re‐exposed plants. To address this limitation, we investigated cold acclimation under diurnal freezing, which models more closely the signals inducing cold acclimation and vernalization in the native range of B. distachyon (Mayer et al., 2020). Diurnal freezing is characterized by repeated 24‐h temperature cycles that fluctuate between day (22°C) and sub‐zero night temperatures (−1°C) under a 16 h : 8 h, light : dark photoperiod, and are therefore characterized by daily cycles of low‐temperature stress and recovery (Fig. 3a). Whole‐plant freeze tests performed on plants exposed to 7 and 28 cycles of diurnal freezing indicate that the treatment induces a gradually higher tolerance (Fig. 3b). In addition, while whole‐plant freeze‐tests also indicate this trend in plants exposed to 1 and 4 cycles of diurnal freezing (Fig. S1), electrolyte leakage assays show that plants exposed to 4 cycles were less damaged by freezing than plants exposed to only 1 cycle of diurnal freezing, which were both less damaged than nontreated control plants (Fig. 3b). Therefore, the freezing tolerance of B. distachyon increased over repeated cycles of diurnal freezing, indicating that these gradually prime B. distachyon to survive freezing (Fig. 3b,c).
Transcriptional responses evolve over cycles of diurnal freezing
To determine whether cold‐induced transcriptional responses change over time in diurnal freezing, we measured the transcript levels of CBF1, COR410, IRI and VRN1. In addition, to deepen our analysis, the study of additional genes involved in cold acclimation in the species, namely the transcription factors CBF2 and CBF3 and the structural gene COR413, were also included. The transcript levels of the seven genes were measured at 0, 5, 8, 12 and 16 h of exposure to low‐temperature conditions during a first and a fourth cycle of diurnal freezing (Fig. 3a,d). Except for IRI, all transcripts accumulated within 16 h of exposure to the first cycle. During the fourth cycle, transcript levels were higher for IRI and VRN1, and lower for CBFs compared to the first, indicating that repeated cycles of diurnal freezing led to the establishment of TMs on these genes.
To investigate the changing transcript levels under diurnal freezing, we selected two time points: a stress time point which showed the highest difference in transcript levels from cycle 1 to cycle 4 (time point = 16 in Fig. 3d), and a recovery time point (time point = 0 or 24 in Fig. 3d). Transcript levels of CBFs decreased sharply after cycle 1 at the stress time point, but slowly increased from cycle 1 to 4 at recovery (Fig. 3d). The transcript levels of IRI gradually increased from cycle 1 to 4, especially at recovery, while the transcriptional responses of COR410 and COR413 remained fairly similar at stress time points, but decreased at recovery (Fig. 3d). VRN1 transcripts gradually increased from cycle 1 to 4 at both stress and recovery time points (Fig. 3d). Therefore, all genes showed altered transcriptional responses to cycles of diurnal freezing. CBFs and COR410–413 showed clear signs of TM that decreased their expression during stress and recovery, respectively, while IRI showed a TM that increased its transcriptional response at recovery.
Six profiles describe the evolution of transcriptional responses to diurnal freezing
To characterize the transcriptional change induced by diurnal freezing, we compared the transcriptomes of plants exposed to the stress time point of cycle 1 (S1), or of cycle 4 (S4) of diurnal freezing, to those of nontreated controls (S0; Fig. 3a). A total of 6725 genes showed an FC > 2 when comparing at least one condition to another: cycle 1 of diurnal freezing compared to control (S1 compared to S0; S1/S0), cycle 4 of diurnal freezing compared to control (S4 compared to S0; S4/S0), or cycle 4 of diurnal freezing compared to cycle 1 (S4 compared to S1; S4/S1), which can be summarized using a Venn diagram, as shown in Fig. 4(a). We further classified these genes depending on whether they were upregulated, downregulated or nonresponsive in response to cold under diurnal freezing conditions at S1 and S4 (S1/S0 and S4/S0; Fig. 4b). Combining the categories of the Venn diagram and the genes’ responses in S1 and S4, we obtained a total of 17 distinct categories, which describe in detail the transcript level outcomes in plants responding to diurnal freezing (Table 1; Fig. 4c). These could be further regrouped into six expression profiles (Fig. 4c,d): stable genes showed no change in response to diurnal freezing, transient genes responded only in S1/S0, while late‐responsive genes responded only in S4/S0. Complex‐convergent and complex‐divergent genes responded in both S1/S0 and S4/S0, but complex‐convergent had lower expression in S4/S0 compared to S1/S0, and complex‐divergent had increased expression between S4/S0 and S1/S0. Offset/oscillating moved from upregulated to downregulated (offset with FC > 2, oscillating with FC < 2) or from downregulated to upregulated (oscillating FC < 2) between S1/S0 and S4/S0. Hence, these six expression profiles describe the main outcomes of transcriptional change between the first and the fourth cycles of diurnal freezing (Table 1; Fig 4c,d).
Table 1.
Distribution of diurnal‐freezing responsive genes of Brachypodium distachyon according to fold change value for S1 and S4 responses, and according to the change in expression between S1 and S4.
| Venn diagram groups | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | ||||||||
| S1/S0 | FC > 2 | FC > 2 | FC > 2 | FC > 2 | ||||||||||
| S4/S0 | FC > 2 | FC > 2 | FC > 2 | FC > 2 | ||||||||||
| S1/S4 | FC > 2 | FC > 2 | FC > 2 | FC > 2 | ||||||||||
| Trend relative to control | Transient | Oscillating | Stable | Complex | Late responsive | |||||||||
| S1 | S4 | S4–S1 | A | B* | C | D | E* | F | G | Total | % | Group | Transcriptional profiles | |
| UP 2851 (42%) | UP 2063 (31%) | + | 72 (1%) | 72 | (1%) | U2B | Complex‐divergent | 72 | ||||||
| = | 1600 (24%) | 1600 | (23%) | U1 | Stable | 1600 | ||||||||
| − | 391 (6%) | 391 | (5%) | U2A | Complex‐convergent | 391 | ||||||||
| = | − | 444 (7%) | 314 (5%) | 758 | (11%) | T1–T3 | Transient | 444–314 | ||||||
| DOWN | − | 30 (0.4%) | 30 | (0.4%) | O2 | Offset | 30 | |||||||
| = 1092 (16%) | UP | + | 69 (1%) | 461 (7%) | 530 | (7%) | L1–L3 | Late response | 69–461 | |||||
| = | + | 16 (0.2%) | 16 | (0.2%) | O1A | Oscillating | 16 | |||||||
| − | 61 (0.9%) | 61 | (0.9%) | O1B | Oscillating | 61 | ||||||||
| DOWN | − | 174 (2%) | 311 (5%) | 485 | (7%) | L2–L4 | Late response | 174–311 | ||||||
| DOWN 2782 (41%) | = | + | 462 (7%) | 124 (2%) | 586 | (8%) | T2–T4 | Transient | 462–124 | |||||
| DOWN (33%) | + | 163 (2%) | 163 | (2%) | D2B | Complex‐convergent | 163 | |||||||
| = | 1843 (27%) | 1843 | (27%) | D1 | Stable | 1843 | ||||||||
| − | 190 (3%) | 190 | (2%) | D2A | Complex‐divergent | 190 | ||||||||
| Total | 906 (13%) | 438 (7%) | 77 (1%) | 3443 (51%) | 846 (13%) | 243 (4%) | 772 (11%) | 6725 | ||||||
The progression from S1 to S4 responses suggests a transition from stress to growth in diurnal freezing
The response to cycle 1 of diurnal freezing (S1 response) and cycle 4 of diurnal freezing (S4 response) shared respectively 61% and 65% of their transcriptomes (the same transcripts in the same amount, corresponding to stable genes). In addition, 15% and 16% of their transcriptomes contained the same transcripts, but in different amounts (corresponding to complex‐convergent, complex‐divergent and offset/oscillating genes), while unique transcripts corresponded to 24% and 19% of their transcriptomes respectively (transient or late‐responsive genes; Table 2). To characterize the progression from S1 to S4, enriched gene ontology terms shared between, and unique to, S1 and S4 were visualized along with the average S4/S1 fold‐change of associated genes (Figs. 4e, S2, S3). Photosynthesis and ribosome biogenesis genes were mostly upregulated, while oxidation/reduction and metabolism genes were mostly downregulated between S1 and S4 (Fig. 4e). Further analysis revealed that transcription factor activity and responses related to oxygen and heme binding were significantly enriched in S1 and S4 responses (Table 3). Interestingly, iron ion binding, response to auxin, photosynthesis and sequence‐specific DNA binding were enriched in S1, but not in S4, indicating that S1 partly included transient responses. Specifically, the absence of the term photosynthesis in S4 and the upregulation of associated genes between S1 and S4 suggest that the normal expression of photosynthesis was restored in S4 (Fig. 4e; Table 3). Hence, the response to diurnal freezing was plastic, and clearly changed between S1 and S4. Priest et al. (2014) described 22 distinct gene modules that characterize the plasticity of the abiotic stress response in B. distachyon. A comparison between the diurnal‐freezing responsive genes and these modules supports the idea that S1 was associated with transiently expressed transcription factors and S4 with growth‐related responses and restored photosynthesis (Fig. S4). Therefore, repeated cycles of diurnal freezing led to extensive reorganization of cold‐induced responses from S1 to S4, indicating that B. distachyon exhibited plastic responses during cold acclimation.
Table 2.
Distribution of common and unique genes between S1 and S4 responses in Brachypodium distachyon.
| S1 | S4 | |
|---|---|---|
| Responsive | 5633 (84%) | 5304 (79%) |
| Up | 2851 | 2756 |
| Down | 2782 | 2548 |
| Shared | 4289 (76%) | 4289 (81%) |
| Similar levels | 3443 (61%) | (65%) |
| Different levels | 846 (15%) | (16%) |
| Unique | 1344 (24%) | 1015 (19%) |
| Unresponsive | 1092 (16%) | 1421 (21%) |
| Total | 6725 |
Table 3.
Gene ontology enrichment analysis in diurnal freezing responsive genes (DFRG) and their distribution in S1 and S4 responses in of Brachypodium distachyon using GOseq.
| GO term | DFRG | S1 | S4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ontol. | DE | Tot | p.adj.or | p.adj.ur | DE | Tot | p.adj.or | p.adj.ur | DE | Tot | p.adj.or | p.adj.ur | ||
| S1&S4 | Transcription factor activity, sequence‐specific DNA binding | MF | 164 | 364 | 8.01E‐08 | 1 | 81 | 364 | 2.99E‐11 | 1 | 61 | 364 | 4.26E‐06 | 1 |
| S1&S4 | Regulation of transcription, DNA‐templated | BP | 284 | 707 | 8.01E‐08 | 1 | 109 | 707 | 1.17E‐06 | 1 | 89 | 707 | 7.43E‐05 | 1 |
| S1&S4 | Oxidoreductase activity, acting on paired donors…* | MF | 71 | 150 | 0.00095 | 1 | 31 | 150 | 0.00064 | 1 | 25 | 150 | 0.01005 | 1 |
| S1&S4 | Heme binding | MF | 108 | 249 | 0.00046 | 1 | 43 | 249 | 0.00419 | 1 | 37 | 249 | 0.01005 | 1 |
| S1 | Iron ion binding | MF | 76 | 180 | 0.02655 | 1 | 33 | 180 | 0.0041 | 1 | 26 | 180 | 0.06271 | 1 |
| S1 | Response to auxin | BP | 22 | 34 | 0.00521 | 1 | 12 | 34 | 0.03424 | 1 | 10 | 34 | 0.13828 | 1 |
| S1 | Photosynthesis, light harvesting | BP | 15 | 18 | 0.00095 | 1 | 12 | 18 | 5.85E‐06 | 1 | 7 | 18 | 0.06271 | 1 |
| S1 | Sequence‐specific DNA binding | MF | 103 | 215 | 2.86E‐06 | 1 | 41 | 215 | 0.00127 | 1 | 30 | 215 | 0.11307 | 1 |
| DFRG | Calmodulin binding | MF | 18 | 26 | 0.00488 | 1 | 3 | 26 | 1 | 1 | 2 | 26 | 1 | 1 |
| DFRG | Response to stress | BP | 35 | 67 | 0.01228 | 1 | 15 | 67 | 0.12457 | 1 | 13 | 67 | 0.17717 | 1 |
| DFRG | Oxidation‐reduction process | BP | 291 | 812 | 0.00654 | 1 | 94 | 812 | 0.23589 | 1 | 81 | 812 | 0.18156 | 1 |
| S1 | Carboxylic acid metabolic process | BP | 13 | 25 | 0.72934 | 1 | 9 | 25 | 0.02881 | 1 | 7 | 25 | 0.17717 | 1 |
| S4 | Trehalose biosynthetic process | BP | 12 | 17 | 0.05087 | 1 | 4 | 17 | 1 | 1 | 8 | 17 | 0.00071 | 1 |
| S1&S4 | Structural constituent of ribosome | MF | 49 | 247 | 1 | 0.0299 | 0 | 247 | 1 | 0 | 1 | 247 | 1 | 0 |
| S1&S4 | GTP binding | MF | 24 | 155 | 1 | 0.0082 | 2 | 155 | 1 | 0.0179 | 0 | 155 | 1 | 0.00177 |
| S1&S4 | Ribosome | CC | 47 | 239 | 1 | 0.0299 | 0 | 239 | 1 | 0 | 1 | 239 | 1 | 2.52E‐07 |
| S1&S4 | Translation | BP | 51 | 256 | 1 | 0.0299 | 0 | 256 | 1 | 0 | 1 | 256 | 1 | 0 |
p.adj.or adjusted P‐value for over‐representation.
p.adj.ur adjusted P‐value for under‐representation.
…with incorporation or reduction of molecular oxygen.
Transcriptional memories regulate transient stress responses
To characterize the transcriptional regulation that accompanied the response plasticity in diurnal freezing, we investigated the distribution of transcription factors within the six expression profiles identified in Fig. 4(c,d). Transient and complex‐convergent expression profiles were especially enriched in transcription factors, while the stable profile regrouped the most (Table 4). Interestingly, there were twice as many transcription factors in the transient than there were in the late‐responsive expression profile. Hence, S1 regrouped more transcription factors both in terms of proportion and absolute number compared to S4. Moreover, transient transcription factors function mostly in stress responses, late‐responsive in growth and chromatin remodeling, and complex‐convergent in both stress and development‐related factors (Table 4). Hence, as suggested by the previous gene ontology analyses, the response to diurnal freezing generally progressed from the expression of stress‐related to growth‐related transcriptional responses.
Table 4.
Distribution of transcription factors in the six expression profiles of diurnal‐freezing responsive genes in Brachypodium distachyon.
| Number of transcription factors and regulators | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DFRG profile | % | Category | % | S1 | % | S4 | % | Representative | General function | References | ||
| Transient | 128 | 10 | T1 | 41 | 9 | 128 | 26 | 0 | C2H2 | Stress response | Wang et al. (2019) | |
| T2 | 41 | 9 | E2F‐DP | Cell cycle, cell growth | Ramirez‐Parra et al. (2018) | |||||||
| T3 | 36 | 11 | GeBP | Cytokinin signalling | Chevalier et al. (2008) | |||||||
| T4 | 10 | 8 | HMG | Stress response, growth | Kwak et al. (2007) | |||||||
| IWS1 | Brassinosteroid‐responsive | Belkhadir & Jaillais (2015) | ||||||||||
| Li et al. (2010) | ||||||||||||
| MBF1 | Stress response, development | Jaimes‐Miranda & Chávez Montes (2020) | ||||||||||
| Tify | Stress response, development | Zhang et al. (2015) | ||||||||||
| Stable | 278 | 8 | U1 | 126 | 8 | 361 | 73 | 361 | 83 | |||
| D1 | 152 | 8 | ||||||||||
| Complex‐convergent | 61 | 11 | D2B | 16 | 10 | AP2/ERF | Stress response, hormone and development | Xie et al. (2019) | ||||
| U2A | 45 | 12 | LDB | Plant growth and development | Fan et al. (2012) | |||||||
| WRKY | Stress response | Phukan et al. (2016) | ||||||||||
| Complex‐divergent | 22 | 8 | D2A | 16 | 8 | |||||||
| U2B | 6 | 8 | ||||||||||
| Late response | 64 | 6 | L1 | 13 | 19 | 0 | 64 | 15 | Alfin‐like | Chromatin modifier | Sanchez & Zhou (2011) | |
| L2 | 13 | 7 | BES1 | Plant growth from multiple signals | Li et al. (2018) | |||||||
| L3 | 18 | 4 | PHD | Chromatin modifier | Sanchez & Zhou (2012) | |||||||
| L4 | 20 | 6 | SNF2 | Chromatin remodelling, may modulate stress‐growth | Mlynárová et al. (2007) | |||||||
| Offset/Oscillating | 7 | 7 | O2 | 2 | 7 | 7 | 1 | 7 | 2 | |||
| O1A | 3 | 19 | ||||||||||
| O1B | 4 | 7 | ||||||||||
| Total DFRG | 542 | 8 | ||||||||||
Genes responsive in S1 and which showed significant changes in expression between S4 and S1 (i.e. transient T3 and T4 and complex genes), were regulated by TMs (Fig. 4a,d; Table 1). Hence, memory genes responded differently between the first and the fourth cycle of diurnal freezing, which was validated by RT‐qPCR analysis (Fig. S5). Accounting for 19% of the diurnal‐freezing responsive genes, memory genes were enriched in transcription factors generally involved in the stress response. For example, most members of the AP2/ERF family were regulated by TMs (Fig. S6). This analysis indicated that CBF1 and CBF2 were complex‐convergent, IRI and CBF3 were complex‐divergent and COR410 and COR413 were stable genes. VRN1 behaved like complex‐divergent genes (but with lower fold‐change). Hence, RNA‐seq data support our previous results that CBFs, IRI and VRN1 displayed TM.
The TMs of genes involved in cold acclimation are mostly reversible
Plants exposed to diurnal freezing became increasingly tolerant to freezing and gained TMs. Hence, B. distachyon became primed to respond to freezing, with the response to cycle 1 of diurnal freezing considered naïve, and the response to cycle 4 considered primed. To determine the stability of TMs on genes involved in cold acclimation, primed plants (exposed to four cycles of diurnal freezing) were exposed to an extended recovery (lag phase) for 1, 3, 6 or 9 d at 22°C, then to a trigger cycle of diurnal freezing (Fig. 5a). Measured at the stress time point of diurnal freezing, the response of COR410 and COR413 was naïve throughout, while the VRN1 response remained primed after all lag times (Fig. 5b). Moreover, the transcriptional responses of CBF1, CBF2, CBF3 and IRI gradually returned to their naïve states after 3–6 d of lag. Therefore, the TM was stable for VRN1 but reversible for CBF1, CBF2, CBF3 and IRI. Interestingly, all seven genes responded differently to priming at the recovery time point (Fig. 5b). For instance, the transcriptional responses of CBF1, CBF2, CBF3 and IRI were naïve at all stages, hence showing no signs of memory at recovery, while VRN1, COR410 and COR413 showed reversible memories. Overall, diurnal freezing induced TMs in primed plants that were mostly reversible after 3–6 d under control conditions.
Fig. 5.
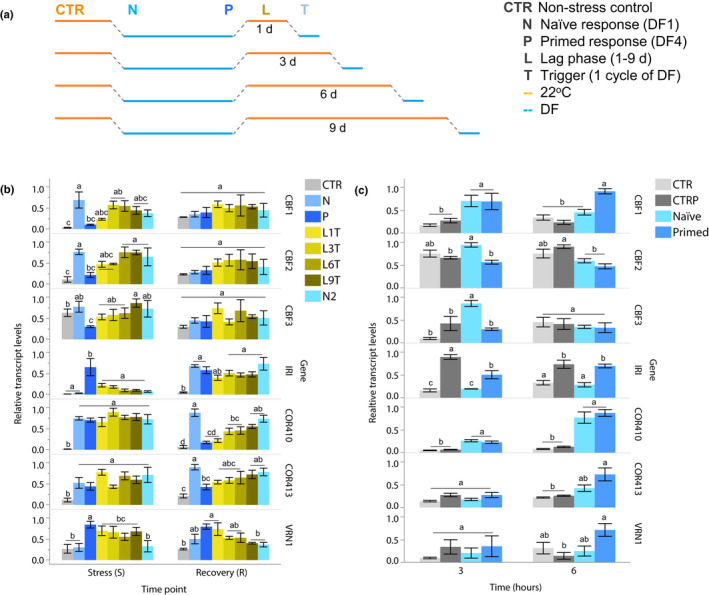
Priming induced by diurnal freezing leads to reversible transcriptional memories on cold‐responsive genes in Brachypodium distachyon. (a) Diagram of the experimental design, showing the naïve (N) and primed (P) responses, lag phase (L), and a trigger cycle (T). (b) Naïve (N), primed (P), trigger responses after 1, 3, 6 and 9 d of lag (L1‐9T), and naïve age control (N2). (c) Transcriptional levels of cold‐responsive genes in response to chilling in primed and naïve plants compared to primed control (CTRP) and naïve control (CTR). Different letters indicate statistical difference; P < 0.05; error bars represent SD between three biological replicates.
Primed plants respond less intensely to chilling and retained the responses acquired in diurnal freezing
Genes involved in cold acclimation demonstrated different types of TMs at the stress and recovery time points of diurnal freezing, which could indicate that temperature cycles influenced their expression, for example inducing a change in circadian regulation. To address this possibility and confirm the presence of TM, we applied cold at an unexpected time (during the middle of the day as opposed to dusk/night as in diurnal freezing) and measured the transcriptional response of primed plants exposed to chilling after one lag‐cycle (when memories had not reverted to the naïve state) compared to naïve plants exposed to chilling, and to the nonstressed primed and naïve control plants (Fig. 5c). In primed plants, CBF1 showed a more sustained response at 6 h, while CBF2 and CBF3 responded with lower levels at 3 h compared to naïve plants (Fig. 5c). Moreover, IRI levels were higher in primed plants both in chilling and control conditions, confirming that IRI acquired a strong TM, while COR410 and COR413 showed no difference in expression after priming. As expected, VRN1 transcripts accumulated to higher levels in primed plants, which have likely undergone early vernalization (Fig. 5b). Therefore, primed plants exhibited TMs formed under diurnal freezing conditions when exposed to chilling.
Furthermore, we compared the transcriptome of naïve and primed plants at 3 h of exposure to chilling to nonstressed controls. The primed response consisted of approximately half the number of responsive genes observed for the naïve response (Fig. 6a; Table 5). 412 genes showed no memory, while 942 were altered by TMs – a result that was validated by RT‐qPCR analysis (Table 5; Fig. S7). All six expression profiles identified in diurnal freezing were represented in the naïve and primed responses to chilling. The primed response had a 3‐fold depletion in complex‐convergent genes and a 2.3‐fold enrichment in late‐responsive genes compared to the naïve response, indicating that the responses acquired in diurnal freezing were retained in primed plants (Fig. 6a; Table 5). Diurnal freezing led to TMs on all naïve‐specific genes (unique to the naïve response), and on genes that showed a different response in naïve and primed plants. Over 70% of the naïve response consisted of memory genes.
Fig. 6.
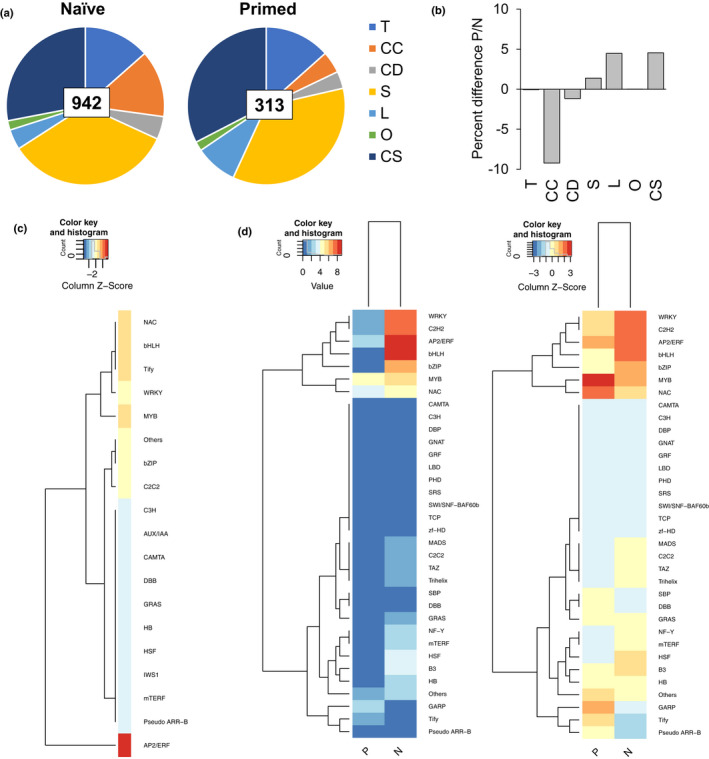
Primed Brachypodium distachyon acquired an attenuated response to chilling. (a) Proportion of chilling‐responsive genes unique to naïve (942 genes) or primed responses (313 genes) that belong to the expression profiles transient (T), complex‐convergent (CC), complex‐divergent (CD), stable (S), late‐response (L) and offset/oscillating (O) or not found to be responsive under diurnal‐freezing (chilling‐specific; CS). (b) Difference between the distribution of genes in diurnal‐freezing expression profiles and the chilling‐specific category in naïve (N) or in primed response (P). (c) Transcription factors and transcriptional regulators that are responsive in both N and P. (d) Number of transcription factors and transcriptional regulators found in genes unique to N and P in absolute value (left) and relative to the total number of genes in P and N (right).
Table 5.
Transcriptional behavior of chilling‐responsive genes that fit into diurnal‐freezing expression profiles and that show transcriptional memory in Brachypodium distachyon.
| N & P same response (FC < 2) | N & P different response (FC > 2) | Unique to N –nonresponsive in P | Response in P only | Total | % | |
|---|---|---|---|---|---|---|
| DF‐responsive profiles | ||||||
| Transient | 76 | 7 | 127 | 42 | 252 | 15 |
| Complex‐convergent | 107 | 16 | 129 | 14 | 266 | 15 |
| Complex‐divergent | 9 | 3 | 44 | 11 | 67 | 4 |
| Stable | 131 | 17 | 321 | 111 | 580 | 34 |
| Late response | 17 | 1 | 39 | 27 | 84 | 5 |
| Offset/Oscillating | 7 | 0 | 18 | 6 | 32 | 2 |
| Chilling‐specific | 64 | 6 | 264 | 102 | 436 | 25 |
| Total | 412 | 50 | 942 | 313 | 1717 | 100 |
| % Total | 24 | 3 | 55 | 18 | ||
| % Naive | 29 | 4 | 67 | – | ||
| % Primed | 53 | 6 | – | 40 | ||
| DF‐responsive | ||||||
| Memory (M) | 174 (50%) | 27 (59%) | 249 (37%) | 44 (21%) | 494 | 30 |
| No memory (NM) | 173 (50%) | 18 (41%) | 429 (63%) | 167 (79%) | 787 | 50 |
| Ratio M/NM | 1 | 1.5 | 0.6 | 0.3 | 0.6 | |
| TF/TR | 72 | 6 | 96 | 33 | ||
| In response to CC | Stable | Memory, complex | Memory, silenced | Acquired response | ||
| Genes showing memory overall | 174 | 50 | 942 | 44 | 1210 | 70 |
The primed response to chilling is depleted in transcription factors and enriched in structural genes
Naïve‐specific genes were enriched in nucleic acid binding and sequence‐specific DNA binding transcription factor activity whereas primed‐specific genes were enriched in metabolic, oxido‐reduction, oxoacid, organic acid and cellular processes (Table S3). Although the number of primed‐specific genes was three times lower than the number of naïve‐specific genes, the primed‐specific genes were associated with four times the number of enriched gene ontology terms, and hence were connected to a wider diversity of functions, related to plant metabolism and cellular processes, as opposed to naïve‐specific genes, whose function is mostly related to transcriptional regulation. Among the genes that were differently expressed between the naïve and primed responses, 66% were dampened by diurnal freezing (Table S4). Of these genes, which became hyposensitive to cold, many were stress‐response transcription factors (Tables S4, S5). Indeed, the primed response contained fewer stress‐response transcription factors of the families AP2/ERF, bHLH, WRKY and C2C2 (Figs 6c,d, S8). Overall, the primed response to chilling was mostly depleted in stress‐response transcription factors and contained a higher proportion of responsive structural genes.
Repeated priming in diurnal freezing led to the formation of similar TMs but a different chromatin response
To investigate whether a second episode of priming could reinforce TMs and induce chromatin changes, we compared the transcriptional response of genes involved in cold acclimation and their associated levels of H3, H3K27me3, H3K4me2, H3K4me3, and the levels of global DNA methylation, to two episodes of priming in diurnal freezing separated by 3‐d recovery. The changes in the transcriptional responses to diurnal freezing were similar for both priming episodes (Fig. 7a). However, the first and second episodes of priming led to distinct chromatin signatures. Genes became generally depleted in H3 and H3K27me3 after the first priming episode, and this effect remained at recovery, along with increased levels of H3K4me3. During the second episode of priming, genes also became generally depleted in H3K4me3, but enriched in H3K4me2 (Figs S9, 7b). Global DNA methylation increased at the first priming episode, but decreased over recovery and during the second episode of priming, indicating contrasting chromatin responses to diurnal freezing (Fig. 7c). Overall, the chromatin composition of genes involved in cold acclimation evolved in response to repeated episodes of priming in diurnal freezing without affecting the formation of TMs, suggesting that chromatin composition can evolve separately from transcriptional responses.
Fig. 7.
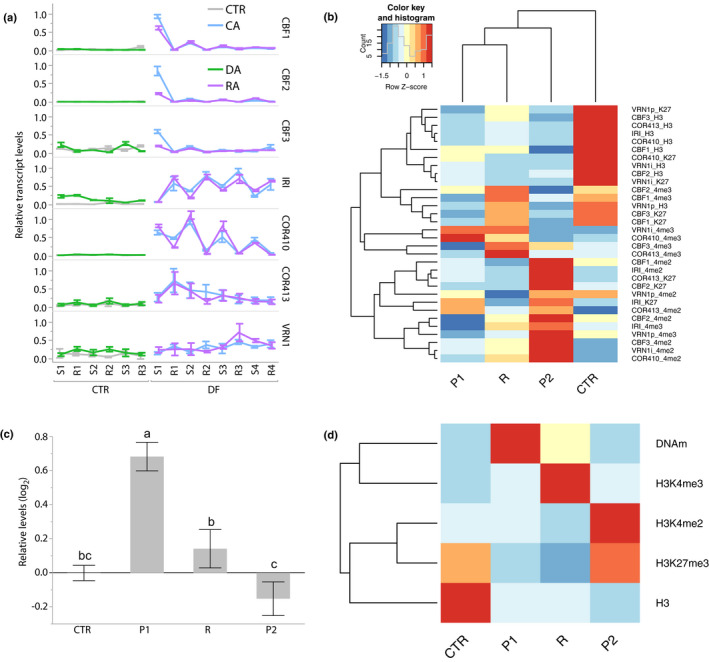
Repetitive priming under diurnal freezing (DF) induced similar effects on transcription but distinct chromatin responses in Brachypodium distachyon. (a) Relative transcript levels during a first (P1) and second (P2) episode of priming under DF separated by a 3‐d recovery from DF (R) in COR genes CBFs, IRI, COR410–413 and VRN1. (b) Relative levels of epigenetic marks H3, H3K27me3 (k27), H3K4me2 (4me2) and H3K4me3 (4me3) in CTR, P1, R and P2. (c) Relative global levels of DNA methylation in CTR, P1, R and P2. (d) Summary of the epigenetic change on COR genes and global DNA methylation levels during repeated priming. Different letters indicate statistical difference; P < 0.05; error bars represent SD between three biological replicates.
Discussion
Cold acclimation in B. distachyon is characterized by a transition from stress to growth responses
Brachypodium distachyon gradually acquired higher freezing‐tolerance under diurnal‐freezing conditions (Fig. 3b,c). This treatment first induced transcriptional responses typical of cold acclimation, including the activation of the transcription factors, C‐repeat binding factors (CBFs), and structural genes IRI, COR410 and COR413 (Fig. 3d). Over repetitions of diurnal‐freezing cycles, the transcriptional responses associated with cold acclimation were dampened, while the initial downregulation of photosynthesis, which is a typical sign of stress, returned to control levels (Figs 3d–e, 4e, 6; Tables 3, 4). As plants grown under diurnal‐freezing conditions eventually produce biomass equivalent to nonstressed control plants, B. distachyon could hence successfully acclimate to the treatment (Crisp et al., 2016; Mayer et al., 2020). Interestingly, almost 40% of the cold‐response transcriptome changed at the fourth cycle of diurnal freezing, where initial stress response genes were generally replaced by others linked to primary metabolic processes, and cellular growth and development (Figs 4, S2–3; Table 4). Furthermore, responsive genes were enriched in iron binding, heme binding and oxygen signaling genes. Plant hemoglobins are important for normal plant growth (Hebelstrup et al., 2006), and by regulating the interplay between oxygen and nitric oxide, they mediate stress and growth responses to environmental changes (Gupta et al., 2011; Simontacchi et al., 2015). Therefore, these results indicate that B. distachyon readjusted stress responses, growth and cellular homeostasis over time under diurnal‐freezing conditions (Table 3). As this treatment was designed to reproduce the natural signals of cold acclimation in B. distachyon (Mayer et al., 2020), the stress‐to‐growth transition indicates that morphology may be an extension of cold acclimation in the species.
The transiently high expression of CBFs, important regulators of cold acclimation in several plant species, is likely important for morphological cold acclimation in B. distachyon. Well studied in cold acclimation, CBFs are known regulators of structural cold‐regulated genes’ expression (Jaglo‐Ottensen et al., 1998). In response to repeated chilling and diurnal‐freezing, the initial transcriptional responses of CBFs are dampened (Figs 2a, 3d, 5b). C‐repeat binding factors induce the expression of COR genes by binding to the C‐repeat motif in their promoter (Thomashow, 1999), but this has yet to be determined experimentally in B. distachyon. Here, results indicate that the maintenance of COR410/413 expression post‐cold is correlated to the TM status of CBFs, suggesting the existence of a CBF/COR regulatory link in B. distachyon (Figs 5b,c, S10). Furthermore, CBF1, CBF2 and CBF3 have redundant and essential cold‐acclimating functions in Arabidopsis, confer freezing tolerance in many plant species, and are involved in other processes, such as seedling and chloroplast development (Gilmour et al., 2004; Savitch et al., 2005; Benedict et al., 2006; Jia et al., 2016; Zhao et al., 2016). Their regulation receives inputs from the circadian clock, light, and temperature for an appropriate response (Nakamichi et al., 2009; Jiang et al., 2017; Liu et al., 2017). The constitutive overexpression of CBFs usually provides high freezing tolerance but severely limits plant growth and delays development in many plant species, notably through the accumulation of DELLA proteins (Achard et al., 2008; Jeknic et al., 2014; Wisniewski et al., 2015). Therefore, the high expression of CBFs under diurnal‐freezing conditions may be transient to allow B. distachyon to grow.
Cold acclimation in B. distachyon relies on a habituation response mediated by TMs
Cold acclimation and the increase in freezing tolerance coincided with the establishment of TMs. These mainly dampened initial cold‐stress transcriptional responses (Figs 2a, 3d,e). Approximately 20% of diurnal‐freezing responsive genes showed strong TMs, with 75% of these leading to downregulation (Table 1). Moreover, over 67% of naïve‐response genes became nonresponsive under chilling in primed plants, revealing that most TMs were silencing/downregulating memories (Table 5). Generally, studies report memory genes that show stronger and faster responses (i.e. showing hyperactivation) under repeated exposure to a priming stimulus (Ding et al., 2012; Lamke et al., 2016; D'Urso & Brickner, 2017; Liu et al., 2018), although different types of memory have also been described (Ding et al., 2013). A recent review stated that plant memory genes fit into one of two classes, showing either a sustained expression during stress recovery or hyperactivation upon re‐exposure (Bäurle & Trindade, 2020). According to the definition that memory genes show an altered response to a given repeated stimulus (Avramova, 2015; Lämke & Bäurle, 2017), the present study reports TM events that mainly lead, instead, to hypoactivation.
The decrease in cold‐stress responses suggests that B. distachyon habituated to cold stress under diurnal‐freezing conditions (Figs 2a, 3d,e, 6). ‘Habituation’ is a reversible response that becomes weaker over time (Rankin et al., 2009; Gagliano et al., 2018), and it has previously been used to describe plant responses to mechanical stimuli (Gagliano et al., 2014), cell cultures growing autonomously from hormones and growth factors (Christou, 1988; Pischke et al., 2006) or from toxins and growth inhibitors (Brochu et al., 2010; Mélida et al., 2010), and more recently, along with the term ‘sensitization’, to describe TMs in the context of the abiotic stress response (Liu et al., 2014; Liu & Avramova, 2016; Csermely et al., 2020). In general, the explanations of habituation responses relate to energy balance, suggesting that organisms mitigate the energy cost of constantly responding to a given stimulus (Duijn, 2017). In our system, B. distachyon shows reversible hypoactivation of cold‐regulated genes and can fully acclimate by resuming growth under diurnal‐freezing conditions (Figs 3d,e, 5b; Mayer et al., 2020). Hence, habituation likely coincided, along with the return of growth‐associated gene expression, with a diversion of the energy spent on initial stress responses towards the acquisition of a freezing‐tolerant plant morphology.
The regulation of phenotypic plasticity
The phenotype acquired under diurnal‐freezing conditions is the result of dynamic signals that, over time, led to converging stress and growth responses. Recovery from stress is a critical period during which stress memories can either be discarded, encoded for future responses, or reinforced to maintain the expression stress‐activated genes (e.g. VRN1; Crisp et al., 2016). In Arabidopsis, recovery from cold exposure, or deacclimation, induces rapid hormonal, structural, metabolic and transcriptomic changes that relate to growth and development, and are likely to kickstart growth in spring (Zuther et al., 2015; Pagter et al., 2017). The higher temperatures of diurnal‐freezing cycles allow B. distachyon to grow and gain high freezing tolerance, unlike constant low temperature which can also induce chilling stress in the species (Fig. 3b; Mayer et al., 2020). Hence, the alternation of low/high temperature signals experienced during diurnal‐freezing conditions, or stress/recovery, appears to favour an equilibrium between stress response and growth, which could explain the ‘carried‐over’ expression of growth‐associated genes during cold exposure, and the dampening of acute cold‐stress responses over time (Figs 4e, S2–S4, S6; Table 3). Similarly, transcripts of IRI, CBFs and VRN1 gradually accumulate at recovery time points under diurnal freezing, hence gaining higher basal expression outside of cold exposure (Fig. 3e). The gradual loss of TM of VRN1 and the rapid loss associated with IRI at recovery time points also indicate that the extended expression of cold‐activated genes into the high temperatures of diurnal freezing is an acquired response to the treatment (Fig. 5a). The diurnal‐freezing phenotype therefore implies a gradual convergence of cold‐stress and growth responses. It is important to consider that, although this treatment models seasonal cues, the plasticity displayed by B. distachyon in response to diurnal freezing can result from both cold‐adaptive plasticity traits and conditions of the treatment itself (Fig. 4; Tables 1, 5). Although the experiments of this study do not allow for differentiation between these factors, it is probably a combination of the two. For instance, plants exposed to diurnal freezing become fully vernalized (Mayer et al., 2020), and the establishment of similar TMs was observed in repeated chilling and diurnal freezing conditions, although these induce contrasting responses in B. distachyon (Figs 2a, 3d; Mayer et al., 2020), which supports the idea that the shaping of cold responses via TMs likely regulates cold acclimation in B. distachyon.
Cold acclimation and vernalization are responses that ensure persistence in temperate climates. The expression of these two adaptive plasticity responses is linked in temperate grasses, notably through the expression of VRN1 (Galiba et al., 2009; Dhillon et al., 2010; Deng et al., 2015; Mayer et al., 2020). This study shows that the regulatory mechanism of vernalization, namely the epigenetic regulation of VRN1’s TM, may also regulate cold acclimation in the temperate grass B. distachyon (Figs 2a, 3d,e, 5b,c). Our results show that repeated cold exposure affected the levels of global DNA methylation and of H3K27me3, H3K4me2 and H3K4me3 at the loci of genes involved in cold acclimation in B. distachyon (Figs 1c, 2b, 7b,c). In Arabidopsis, H3K4me3 and H3K4me2 mark hyperactive memory genes during heat stress, and mediate tolerance to salt and salt‐stress priming responses (Shen et al., 2014; Feng et al., 2016; Lamke et al., 2016). While H3K4me2 is usually associated with gene repression (Lämke & Bäurle, 2017; Liu et al., 2019), H3K4me3 is deposited on active cold‐regulated genes in potato exposed to cold conditions, along with the repressive mark H3K27me3 (Zeng et al., 2019). However, studies show that levels of H3K27me3 and H3K4me3 tend to be negatively correlated to one another, especially in TMs linked to vernalization in Arabidopsis, barley and B. distachyon – this is also supported by our results (Figs 7, S11; Finnegan et al., 2005; Oliver et al., 2009; Zhang et al., 2009; Huan et al., 2018). Our results suggest that depletions of H3 and H3K27me3 were linked to TMs established under diurnal‐freezing conditions, that H3K4me3 participated in their maintenance during recovery and the enrichment in H3K4me2 occurred in a second response to diurnal freezing (Figs 7d, S9). It is intriguing that this change in epigenetic signature occurred while the progression of transcriptional responses to diurnal freezing did not change during the second exposure (Fig. 7). Extended recovery from diurnal freezing hence induced the onset of different chromatin marks, also leading to a slight decrease in global DNA methylation upon re‐exposure, rather than a hypermethylation as observed during the first priming episode (Fig. 7c). Because cold acclimation extends to morphology in B. distachyon, and because vernalization occurs simultaneously, the chromatin responses observed during diurnal freezing can be related to stress memories as well as development (e.g. linked to vernalization and growth stages), demonstrating complex links between chromatin marks, and stress and development responses in B. distachyon.
Conclusion
This study is the first to report that reversible TMs mediate the progressive return of plant growth following initial stress responses. By regulating transcriptional habituation, TMs provide plasticity to B. distachyon's stress responses, to develop a freezing tolerant morphology during cold acclimation. Hence, chromatin‐associated TMs are involved in regulating both stress and developmental components of cold‐climate adaptive plasticity in B. distachyon.
Author contributions
BFM and J‐BC designed the research, BFM performed the experiments and analyses, BFM and J‐BC wrote the manuscript.
Supporting information
Fig. S1 Whole‐plant freeze tests performed on plants exposed to one and four cycles of diurnal freezing, compared to non‐acclimated plants.
Fig. S2 Significantly enriched gene ontology (GO) terms in the 17 categories regrouped into 6 expression profiles identified in diurnal‐freezing responsive genes.
Fig. S3 Transcript levels of genes whose expression changed under diurnal‐freezing conditions (S4/S1) that are associated with significantly enriched GO terms.
Fig. S4 Distribution and differential expression of diurnal‐freezing responsive genes in abiotic stress response modules.
Fig. S5 Quantitative reverse transcription polymerase chain reaction (RT‐qPCR) validation of RNA‐seq analysis of plants exposed to diurnal freezing.
Fig. S6 Families of transcription factors in the six expression profiles identified under diurnal freezing.
Fig. S7 RT‐qPCR validation of RNA‐seq analysis of primed plants exposed to chilling.
Fig. S8 Brachypodium distachyon gene modules identified as being associated with abiotic stress responses and their distribution in chilling‐responsive genes.
Fig. S9 Chromatin marks at the loci of genes involved in cold acclimation in response to repeated priming under diurnal freezing.
Fig. S10 Transcript levels of CBF1–3 at stress (S) and COR410/413 at recovery (R) are positively correlated.
Fig. S11 Correlation R 2 adj between epigenetic marks at COR gene loci.
Table S1 Primers used in this study.
Table S2 Chromatin immunoprecipitation (ChIP)‐qPCR signals and statistical difference.
Table S3 Gene ontology analysis of chilling‐responsive genes.
Table S4 Chilling‐responsive genes common to naïve and primed responses that show transcriptional memory and their categorization as diurnal‐freezing responsive genes.
Table S5 Annotated chilling‐responsive memory genes common to naïve and primed responses.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was funded by Natural Sciences and Engineering Research Council of Canada grant RGPIN‐2020‐05243 to J‐BC. BFM was supported by the Vanier Canada Graduate Scholarship. The authors also acknowledge support from Centre SEVE.
References
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. 2008. The cold‐inducible CBF1 factor‐dependent signaling pathway modulates the accumulation of the growth‐repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA et al 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research 46: W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal P. 2008. Global climate change and Indian agriculture: impacts, adaptation and mitigation. Indian Journal of Agricultural Sciences 78: 911. [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data . [WWW document] URL http://www.bioinformatics.babraham.ac.uk/projects/fastqc [accessed 22 March 2020].
- Avramova Z. 2015. Transcriptional 'memory' of a stress: transient chromatin and memory (epigenetic) marks at stress‐response genes. The Plant Journal 83: 149–159. [DOI] [PubMed] [Google Scholar]
- Bathiany S, Scheffer M, van Nes EH, Williamson MS, Lenton TM. 2018. Abrupt climate change in an oscillating world. Scientific Reports 8: 5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I, Trindade I. 2020. Chromatin regulation of somatic abiotic stress memory. Journal of Experimental Botany 71: 5269–5279. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y. 2015. The molecular circuitry of brassinosteroid signaling. New Phytologist 206: 522–540. [DOI] [PubMed] [Google Scholar]
- Benedict C, Skinner JS, Meng R, Chang Y, Bhalerao R, Huner NP, Finn CE, Chen TH, Hurry V. 2006. The CBF1‐dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant, Cell & Environment 29: 1259–1272. [DOI] [PubMed] [Google Scholar]
- Bittner A, van Buer J, Baier M. 2020. Cold priming uncouples light‐ and cold‐regulation of gene expression in Arabidopsis thaliana . BMC Plant Biology 20: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DM, Dennis ES, Finnegan EJ. 2011. The low temperature response pathways for cold acclimation and vernalization are independent. Plant, Cell & Environment 34: 1737–1748. [DOI] [PubMed] [Google Scholar]
- Bredow M, Vanderbeld B, Walker VK. 2016. Knockdown of ice‐binding proteins in Brachypodium distachyon demonstrates their role in freeze protection. PLoS ONE 11: e0167941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochu V, Girard‐Martel M, Duval I, Lerat S, Grondin G, Domingue O, Beaulieu C, Beaudoin N. 2010. Habituation to thaxtomin A in hybrid poplar cell suspensions provides enhanced and durable resistance to inhibitors of cellulose synthesis. BMC Plant Biology 10: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J. 2012. Wheat TILLING mutants show that the vernalization gene VRN1 down‐regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genetics 8: e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Perazza D, Laporte F, Le Hénanff G, Hornitschek P, Bonneville J‐M, Herzog M, Vachon G. 2008. GeBP and GeBP‐like proteins are noncanonical leucine‐zipper transcription factors that regulate cytokinin response in Arabidopsis. Plant Physiology 146: 1142–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard P. 1960. Vernalization and its relations to dormancy. Annual Review of Plant Physiology 11: 191–238. [Google Scholar]
- Christou P. 1988. Habituation in in vitro soybean cultures. Plant Physiology 87: 809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarmiello LF, Woodrow P, Fuggi A, Pontecorvo G, Carillo P. 2011. Plant genes for abiotic stress, abiotic stress in plants – mechanisms and adaptations. Rijeka, Croatia: IntechOpen. [WWW document] URL https://www.intechopen.com/books/abiotic‐stress‐in‐plants‐mechanisms‐and‐adaptations/plant‐genes‐for‐abiotic‐stress [accessed 12 April 2020].
- Colton‐Gagnon K, Ali‐Benali MA, Mayer BF, Dionne R, Bertrand A, Do Carmo S, Charron JB. 2014. Comparative analysis of the cold acclimation and freezing tolerance capacities of seven diploid Brachypodium distachyon accessions. Annals of Botany 113: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ. 2016. Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Science Advances 2: e1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Kunsic N, Mendik P, Kerestély M, Faragó T, Veres DV, Tompa P. 2020. Learning of signaling networks: molecular mechanisms. Trends in Biochemical Sciences 45: 284–294. [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. 2003. TaVRT‐1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiology 132: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, Trevaskis B. 2015. Direct links between the vernalization response and other key traits of cereal crops. Nature Communications 6: 1–8. [DOI] [PubMed] [Google Scholar]
- Dhillon T, Pearce SP, Stockinger EJ, Distelfeld A, Li C, Knox AK, Vashegyi I, Vágújfalvi A, Galiba G, Dubcovsky J. 2010. Regulation of freezing tolerance and flowering in temperate cereals: the VRN‐1 connection. Plant Physiology 153: 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z. 2012. Multiple exposures to drought “train” transcriptional responses in Arabidopsis . Nature Communications 3: 1–9. [DOI] [PubMed] [Google Scholar]
- Ding Y, Liu N, Virlouvet L, Riethoven J‐J, Fromm M, Avramova Z. 2013. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana . BMC Plant Biology 13: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijn MV. 2017. Phylogenetic origins of biological cognition: convergent patterns in the early evolution of learning. Interface Focus 7: 20160158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Urso A, Brickner JH. 2017. Epigenetic transcriptional memory. Current Genetics 63: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y. 2012. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Research 22: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XJ, Li JR, Qi SL, Lin QF, Jin JB, Hua XJ. 2016. Light affects salt stress‐induced transcriptional memory of P5CS1 in Arabidopsis . Proceedings of the National Academy of Sciences, USA 113: E8335–E8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA, Jaligot E, Sheldon CC, James Peacock W, Dennis ES. 2005. The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant Journal 44: 420–432. [DOI] [PubMed] [Google Scholar]
- Gagliano M, Abramson CI, Depczynski M. 2018. Plants learn and remember: lets get used to it. Oecologia 186: 29–31. [DOI] [PubMed] [Google Scholar]
- Gagliano M, Renton M, Depczynski M, Mancuso S. 2014. Experience teaches plants to learn faster and forget slower in environments where it matters. Oecologia 175: 63–72. [DOI] [PubMed] [Google Scholar]
- Galiba G, Vágújfalvi A, Li C, Soltész A, Dubcovsky J. 2009. Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Science 176: 12–19. [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. 2004. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Molecular Biology 54: 767–781. [DOI] [PubMed] [Google Scholar]
- Gray SB, Brady SM. 2016. Plant developmental responses to climate change. Developmental Biology 419: 64–77. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Hebelstrup KH, Mur LAJ, Igamberdiev AU. 2011. Plant hemoglobins: Important players at the crossroads between oxygen and nitric oxide. FEBS Letters 585: 3843–3849. [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, Hunt P, Dennis E, Jensen SB, Jensen EØ. 2006. Hemoglobin is essential for normal growth of Arabidopsis organs. Physiologia Plantarum 127: 157–166. [Google Scholar]
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM. 2008. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real‐time PCR. BMC Plant Biology 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan Q, Mao Z, Chong K, Zhang J. 2018. Global analysis of H3K4me3/H3K27me3 in Brachypodium distachyon reveals VRN3 as critical epigenetic regulation point in vernalization and provides insights into epigenetic memory. New Phytologist 219: 1373–1387. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. 1996. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47: 377–403. [DOI] [PubMed] [Google Scholar]
- Jaglo‐Ottensen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. 1998. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. [DOI] [PubMed] [Google Scholar]
- Jaimes‐Miranda F, Chávez Montes RA. 2020. The plant MBF1 protein family: a bridge between stress and transcription. Journal of Experimental Botany 71: 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeknic Z, Pillman KA, Dhillon T, Skinner JS, Veisz O, Cuesta‐Marcos A, Hayes PM, Jacobs AK, Chen TH, Stockinger EJ. 2014. Hv‐CBF2A overexpression in barley accelerates COR gene transcript accumulation and acquisition of freezing tolerance during cold acclimation. Plant Molecular Biology 84: 67–82. [DOI] [PubMed] [Google Scholar]
- Jia Y, Ding Y, Shi Y, Zhang X, Gong Z, Yang S. 2016. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytologist 212: 345–353. [DOI] [PubMed] [Google Scholar]
- Jiang B, Shi Y, Zhang X, Xin X, Qi L, Guo H, Li J, Yang S. 2017. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA 114: E6695–E6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Mithani A, Belfield EJ, Mott R, Hurst LD, Harberd NP. 2014. Environmentally responsive genome‐wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Research 24: 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem K, Gargallo‐Garriga A, Rattanapichai W, Oravec M, Holub P, Veselá B, Sardans J, Peñuelas J, Urban O. 2019. Distinct morphological, physiological, and biochemical responses to light quality in barley leaves and roots. Frontiers in Plant Science 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C. 2016. Plant adaptation to cold climates. F1000Research 5: F1000 Faculty Rev‐2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutcher HR, Warland JS, Brandt SA. 2010. Temperature and precipitation effects on canola yields in Saskatchewan, Canada. Agricultural and Forest Meteorology 150: 161–165. [Google Scholar]
- Kwak KJ, Kim JY, Kim YO, Kang H. 2007. Characterization of transgenic Arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress. Plant and Cell Physiology 48: 221–231. [DOI] [PubMed] [Google Scholar]
- Kwon CS, Lee D, Choi G, Chung WI. 2009. Histone occupancy‐dependent and ‐independent removal of H3K27 trimethylation at cold‐responsive genes in Arabidopsis . Plant Journal 60: 112–121. [DOI] [PubMed] [Google Scholar]
- Lämke J, Bäurle I. 2017. Epigenetic and chromatin‐based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biology 18: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamke J, Brzezinka K, Altmann S, Baurle I. 2016. A hit‐and‐run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO Journal 35: 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B‐h, Zhu J‐K. 2010. Phenotypic analysis of Arabidopsis mutants: electrolyte leakage after freezing stress. Cold Spring Harbor Protocols 2010: pdb.prot4970. [DOI] [PubMed] [Google Scholar]
- Li C, Rudi H, Stockinger EJ, Cheng H, Cao M, Fox SE, Mockler TC, Westereng B, Fjellheim S, Rognli OA et al 2012. Comparative analyses reveal potential uses of Brachypodium distachyon as a model for cold stress responses in temperate grasses. BMC Plant Biology 12: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye H, Guo H, Yin Y. 2010. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proceedings of the National Academy of Sciences, USA 107: 3918–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Yang H, Wang L, Liu H, Huo H, Zhang C, Liu A, Zhu A, Hu J, Lin Y et al 2019. Physiological and transcriptome analyses reveal short‐term responses and formation of memory under drought stress in rice. Frontiers in Genetics 10: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q‐F, Lu J, Yu J‐W, Zhang C‐Q, He J‐X, Liu Q‐Q. 2018. The brassinosteroid‐regulated transcription factors BZR1/BES1 function as a coordinator in multisignal‐regulated plant growth. Biochimica et Biophysica Acta (BBA)‐Gene Regulatory Mechanisms 1861: 561–571. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Liu H‐c, Lämke J, Lin S‐y, Hung M‐J, Liu K‐M, Charng Y‐y, Bäurle I. 2018. Distinct heat shock factors and chromatin modifications mediate the organ‐autonomous transcriptional memory of heat stress. The Plant Journal 95: 401–413. [DOI] [PubMed] [Google Scholar]
- Liu N, Avramova Z. 2016. Molecular mechanism of the priming by jasmonic acid of specific dehydration stress response genes in Arabidopsis. Epigenetics Chromatin 9: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Fromm M, Avramova Z. 2014. H3K27me3 and H3K4me3 chromatin environment at super‐induced dehydration stress memory genes of Arabidopsis thaliana . Molecular Plant 7: 502–513. [DOI] [PubMed] [Google Scholar]
- Liu W, Su J. 2016. Effects of light acclimation on shoot morphology, structure, and biomass allocation of two Taxus species in southwestern China. Scientific Reports 6: 35384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu K, Yin L, Yu Y, Qi J, Shen W‐H, Zhu J, Zhang Y, Dong A. 2019. H3K4me2 functions as a repressive epigenetic mark in plants. Epigenetics & Chromatin 12: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S. 2017. Plasma membrane CRPK1‐mediated phosphorylation of 14‐3‐3 proteins induces their nuclear import to fine‐tune CBF signaling during cold response. Molecular Cell 66: 117–128. [DOI] [PubMed] [Google Scholar]
- Mayer BF, Ali‐Benali MA, Demone J, Bertrand A, Charron JB. 2015. Cold acclimation induces distinctive changes in the chromatin state and transcript levels of COR genes in Cannabis sativa varieties with contrasting cold acclimation capacities. Physiologia Plantarum 155: 281–295. [DOI] [PubMed] [Google Scholar]
- Mayer BF, Bertrand A, Charron J‐B. 2020. Treatment analogous to seasonal change demonstrates the integration of cold responses in Brachypodium distachyon . Plant Physiology 182: 1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida H, Encina A, Álvarez J, Acebes JL, Caparrós‐Ruiz D. 2010. Unraveling the biochemical and molecular networks involved in maize cell habituation to the cellulose biosynthesis inhibitor dichlobenil. Molecular Plant 3: 842–853. [DOI] [PubMed] [Google Scholar]
- Mlynárová L, Nap JP, Bisseling T. 2007. The SWI/SNF chromatin‐remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. The Plant Journal 51: 874–885. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. 2009. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant and Cell Physiology 50: 447–462. [DOI] [PubMed] [Google Scholar]
- Oliver SN, Deng W, Casao MC, Trevaskis B. 2013. Low temperatures induce rapid changes in chromatin state and transcript levels of the cereal VERNALIZATION1 gene. Journal of Experimental Botany 64: 2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B. 2009. Vernalization‐induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proceedings of the National Academy of Sciences, USA 106: 8386–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagter M, Alpers J, Erban A, Kopka J, Zuther E, Hincha DK. 2017. Rapid transcriptional and metabolic regulation of the deacclimation process in cold acclimated Arabidopsis thaliana . BMC Genomics 18: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan UJ, Jeena GS, Shukla RK. 2016. WRKY transcription factors: molecular regulation and stress responses in plants. Frontiers in Plant Science 7: 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke MS, Huttlin EL, Hegeman AD, Sussman MR. 2006. A transcriptome‐based characterization of habituation in plant tissue culture. Plant Physiology 140: 1255–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest HD, Fox SE, Rowley ER, Murray JR, Michael TP, Mockler TC. 2014. Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS ONE 9: e87499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2013. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ramirez‐Parra E, del Pozo JC, Desvoyes B, de la Paz Sanchez M, Gutierrez C. 2018. E2F–DP transcription factors. Annual Plant Reviews online 138–163. [Google Scholar]
- Rankin CH, Abrams T, Barry RJ, Bhatnagar S, Clayton DF, Colombo J, Coppola G, Geyer MA, Glanzman DL, Marsland S et al 2009. Habituation revisited: an updated and revised description of the behavioral characteristics of habituation. Neurobiology of Learning and Memory 92: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream TS, Woods DP, Schwartz CJ, Sanabria CP, Mahoy JA, Walters EM, Kaeppler HF, Amasino RM. 2014. Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiology 164: 694–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JY, Hong SY, Jo SH, Woo JC, Lee S, Park CM. 2014. Molecular and functional characterization of cold‐responsive C‐repeat binding factors from Brachypodium distachyon . BMC Plant Biology 14: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DH, Paszkowski J. 2014. Heat‐induced release of epigenetic silencing reveals the concealed role of an imprinted plant gene. PLoS Genetics 10: e1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou M‐M. 2011. The PHD finger: a versatile epigenome reader. Trends in Biochemical Sciences 36: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitch LV, Allard G, Seki M, Robert LS, Tinker NA, Huner NPA, Shinozaki K, Singh J. 2005. The effect of overexpression of two brassica CBF/DREB1‐like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant and Cell Physiology 46: 1525–1539. [DOI] [PubMed] [Google Scholar]
- Shen Y, CondeeSilva N, Audonnet L, Servet C, Wei W, Zhou D‐X. 2014. Over‐expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis . Frontiers in Plant Science 5: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simontacchi M, Galatro A, Ramos‐Artuso F, Santa‐María GE. 2015. Plant survival in a changing environment: the role of nitric oxide in plant responses to abiotic stress. Frontiers in Plant Science 6: 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE: 0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Research 45: 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Global Change Research Program (USGCRP) . 2017. In: Wuebbles DJ, Fahey DW, Hibbard KA, Dokken DJ, Stewart BC, Maycock TK, eds. Climate science special report: Fourth national climate assessment, Volume 1. Washington, DC, USA: U.S. Global Change Research Program. doi: 10.7930/J0J964J6. [DOI] [Google Scholar]
- Verhoeven KJ, Jansen JJ, van Dijk PJ, Biere A. 2010. Stress‐induced DNA methylation changes and their heritability in asexual dandelions. New Phytologist 185: 1108–1118. [DOI] [PubMed] [Google Scholar]
- Vogel JP. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Vyse K, Faivre L, Romich M, Pagter M, Schubert D, Hincha DK, Zuther E. 2020. Transcriptional and post‐transcriptional regulation and transcriptional memory of chromatin regulators in response to low temperature. Frontiers in Plant Science 11: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Ding Y, Cai C, Chen Z, Zhu C. 2019. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiologia Plantarum 165: 690–700. [DOI] [PubMed] [Google Scholar]
- Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J, Papareddy R, Putra H, Kageyama J, Becker J et al 2016. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5: e13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M, Norelli J, Artlip T. 2015. Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Frontiers in Plant Science 6: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DP, Ream TS, Bouché F, Lee J, Thrower N, Wilkerson C, Amasino RM. 2017. Establishment of a vernalization requirement in Brachypodium distachyon requires REPRESSOR OF VERNALIZATION1 . Proceedings of the National Academy of Sciences, USA 114: 6623–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Nolan TM, Jiang H, Yin Y. 2019. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis . Frontiers in Plant Science 10: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung E, van Veen H, Vashisht D, Sobral Paiva AL, Hummel M, Rankenberg T, Steffens B, Steffen‐Heins A, Sauter M, de Vries M et al 2018. A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 15: E6085–E6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA‐seq: accounting for selection bias. Genome Biology 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Zhang W, Marand AP, Zhu B, Buell CR, Jiang J. 2019. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biology 20: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, You J, Chan Z. 2015. Identification and characterization of TIFY family genes in Brachypodium distachyon . Journal of Plant Research 128: 995–1005. [DOI] [PubMed] [Google Scholar]
- Zhang W, Culley DE, Hogan M, Vitiritti L, Brockman FJ. 2006. Oxidative stress and heat‐shock responses in Desulfovibrio vulgaris by genome‐wide transcriptomic analysis. Antonie van Leeuwenhoek 90: 41–55. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. 2009. Genome‐wide analysis of mono‐, di‐ and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana . Genome Biology 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu JK. 2016. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiology 171: 2744–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ et al 2016. iTAK: a program for genome‐wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Molecular Plant 9: 1667–1670. [DOI] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ et al 2008. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proceedings of the National Academy of Sciences, USA 105: 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E, Juszczak I, Ping Lee Y, Baier M, Hincha DK. 2015. Time‐dependent deacclimation after cold acclimation in Arabidopsis thaliana accessions. Scientific Reports 5: 12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E, Schaarschmidt S, Fischer A, Erban A, Pagter M, Mubeen U, Giavalisco P, Kopka J, Sprenger H, Hincha DK. 2019. Molecular signatures associated with increased freezing tolerance due to low temperature memory in Arabidopsis . Plant, Cell & Environment 42: 854–873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Whole‐plant freeze tests performed on plants exposed to one and four cycles of diurnal freezing, compared to non‐acclimated plants.
Fig. S2 Significantly enriched gene ontology (GO) terms in the 17 categories regrouped into 6 expression profiles identified in diurnal‐freezing responsive genes.
Fig. S3 Transcript levels of genes whose expression changed under diurnal‐freezing conditions (S4/S1) that are associated with significantly enriched GO terms.
Fig. S4 Distribution and differential expression of diurnal‐freezing responsive genes in abiotic stress response modules.
Fig. S5 Quantitative reverse transcription polymerase chain reaction (RT‐qPCR) validation of RNA‐seq analysis of plants exposed to diurnal freezing.
Fig. S6 Families of transcription factors in the six expression profiles identified under diurnal freezing.
Fig. S7 RT‐qPCR validation of RNA‐seq analysis of primed plants exposed to chilling.
Fig. S8 Brachypodium distachyon gene modules identified as being associated with abiotic stress responses and their distribution in chilling‐responsive genes.
Fig. S9 Chromatin marks at the loci of genes involved in cold acclimation in response to repeated priming under diurnal freezing.
Fig. S10 Transcript levels of CBF1–3 at stress (S) and COR410/413 at recovery (R) are positively correlated.
Fig. S11 Correlation R 2 adj between epigenetic marks at COR gene loci.
Table S1 Primers used in this study.
Table S2 Chromatin immunoprecipitation (ChIP)‐qPCR signals and statistical difference.
Table S3 Gene ontology analysis of chilling‐responsive genes.
Table S4 Chilling‐responsive genes common to naïve and primed responses that show transcriptional memory and their categorization as diurnal‐freezing responsive genes.
Table S5 Annotated chilling‐responsive memory genes common to naïve and primed responses.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


