Summary
Drosophila larval musculature is a genetically and optically accessible system to study muscle development. Each larval muscle is a single fiber with conserved cytoarchitecture, including its sarcomere structure and composition. Here, we present a workflow for systematically analyzing muscle structure and function at discrete larval stages, as well as throughout the larval instars, using both newly developed and adapted methods.
For complete details on the use and execution of this protocol, please refer to Balakrishnan et al. (2020).
Subject areas: Genetics, Microscopy, Model organisms
Graphical abstract
Highlights
-
•
Detailed methods for preparation and analysis of Drosophila larval muscle structure
-
•
Methods for muscle structure and function analysis in the same larva over time
-
•
Steps for analyzing sarcomere size and organization
-
•
Pitfalls and troubleshooting approaches described
Drosophila larval musculature is a genetically and optically accessible system to study muscle development. Each larval muscle is a single fiber with conserved cytoarchitecture, including its sarcomere structure and composition. Here, we present a workflow for systematically analyzing muscle structure and function at discrete larval stages as well as throughout the larval instars, using both newly developed and adapted methods.
Before you begin
Larval muscle structure and function have been examined using numerous techniques at different instars. Previous techniques have examined muscle structure by larval dissection, fixation, and immunostaining, by heat-fixing larvae expressing fluorescent constructs, or by anesthetizing a larva at different instars to examine changes in muscle structure throughout development (Brent et al., 2009; Kakanj et al., 2020; Schnorrer et al., 2010; Choi et al., 2014; Fernandes and Schöck, 2014). Similarly, a low-cost technique to examine larval muscle function has been described previously (Brooks et al., 2016).
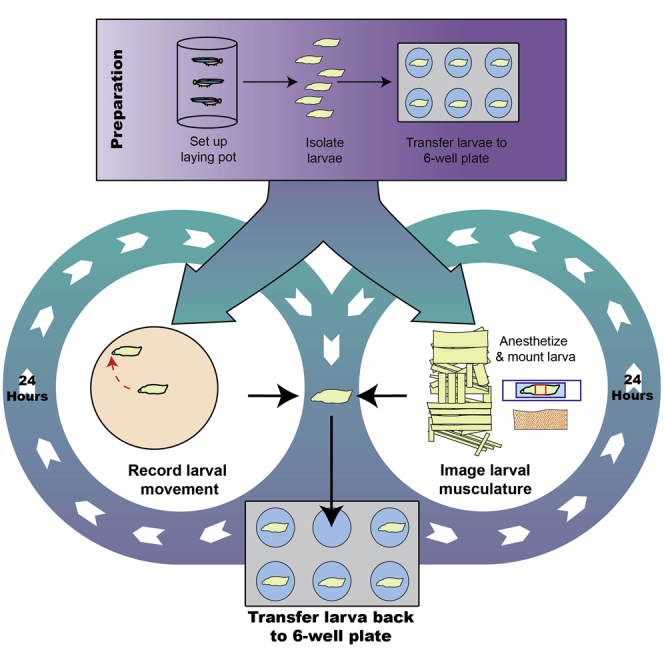
Here we describe a detailed workflow for systematically analyzing both muscle structure and function either at discrete larval stages or throughout the larval instars, using both newly developed and adapted methods. In comparison to previously published methods which enable visualization of muscle structure and function at discrete stages, our protocol can be used to observe changes in muscle structure or muscle function in the same larva throughout its development. We describe in detail the techniques used to isolate, store, and grow individual larva over time; likewise, we explain the methods used to quantify muscle and sarcomere structures such as muscle length, sarcomere width, and number from the embryonic period (when sarcomeres are established), as well as larval velocity, a proxy for muscle function, over time. We detail the procedures to collect and maintain the larva individually as they develop through the instars. This isolation of the individual larva is essential for monitoring changes in muscle structure and larval locomotion in a single larva throughout development. We recommend that the materials listed be acquired and that the following steps are followed.
Setting up laying pots
Timing: 3 h
-
1.Set up laying pots and embryo collection plates
-
a.Prepare Laying pot
-
i.Punch 100 mL plastic beaker (for 6 cm wide petri dishes) with holes to facilitate air exchange and prevent condensation. Holes are made using a hot syringe needle.
-
i.
-
b.Prepare Embryo collection plates (apple juice plates)
-
i.Microwave 1,500 mL of ddH2O, 50 g of granulated sugar, and 45 g of agar to form a homogenous solution. Add 500 mL of apple juice once the solution reaches 65°C, then add 40 mL of 10% Tegosept in 100% ethanol. Pour solution into 6 cm or 10 cm wide petri dishes.
-
i.
-
c.Prepare Yeast paste
-
i.Stir distilled water with dry baker’s yeast to make a paste. Store at 4°C.
-
i.
-
d.Spread a small quantity of yeast paste on a solidified apple juice plate.
-
a.
-
2.Collecting flies for laying pots
-
a.Raise flies of the desired genotypes in a light-dark incubator (12 h light:12 h dark) at 25°C.
-
b.Collect virgin females (♀) and males (♂) of the respective genotypes and store them separately.
-
c.Add flies to the laying pots and seal with embryo collection plate and rubber bands.Note: The number of male flies added is half the number of female virgins added, e.g., for approximately 60 virgin females, 30–35 male flies are added to a laying pot.
-
d.Place laying pots in a 25°C in a light-dark incubator, changing the agar plates every 24 h.Note: Prior to embryo collection, place laying pots at 25°C incubator for 2 days in order for the flies to acclimatize and mate, e.g., if planning on collecting embryos from Monday, set up laying pot in 25°C incubator on Friday evening changing plates every 24 h.
-
a.
Larval collection
Timing: 10 min
-
3.
Change the apple juice plates of your laying pots in ∼20 h from your last exchange.
Note: Changing the apple juice plates within ∼20 h of your last exchange only applies to the days you plan on collecting larvae for experiments. On all other days, changing plates every 24 h is sufficient.
-
4.
Examine the collection plate to ensure that there are no larvae present and then shift the plate with embryos to 25°C.
-
5.
Examine collection plate after 2 h for any larvae that have hatched and isolate the newly hatched 1st instar larvae.
-
6.
Transfer the 1st instar larvae into fresh vials with food and incubate at 25°C in a light-dark incubator.
-
7.
Remove larvae at the desired instar stage (2nd, early 3rd or late 3rd instar) by floating them away from the food using 15% sucrose solution. To do this, pour the sucrose solution into the vial and gently disturb the food at the bottom of the vial using a spatula. Floating larvae can be collected and transferred using a paintbrush.
Note: Store 15% sucrose at 4°C to prevent contamination by microorganisms.
-
8.
Clean larvae gently by immersing and swishing them in a watch glass containing ddH2O. Transfer cleaned larvae into a fresh apple juice plate at room temperature.
Note: For further information and images on laying pot setup and larval collection please refer to Azevedo et al. (2016).
Setting up chambers/plates with food for larval isolation
Timing: 10 min
-
9.
Empty food from fresh vials (∼3–4) or a bottle into a beaker (Figure 1A).
-
10.
Heat food in a microwave in short bursts until food has completely melted (Figures 1B and 1C).
Note: Add ∼25 mL of water to food once melted to ensure that food does not dry out.
-
11.
Pipette ∼2.5 mL of food into each well of a 6-well plate (Figures 1D and 1E).
Note: Food in each well should sufficiently cover the bottom of the well and be approximately 0.25 cm thick.
-
12.
After food has cooled and solidified, transfer a larva into each well using a paintbrush.
-
13.
Seal plate using self-sealing plastic wrap and punch holes in the wrap over each well using a fine syringe needle (Figure 1F).
Note: Holes are punched to facilitate air exchange, around 10 holes per well is sufficient. If the holes are too large, larvae can crawl out of the holes.
-
14.
Incubate plate at 25°C in a light-dark incubator.
Note: To prevent food from drying out, place plates in a sealable container (either Pyrex/plastic) lined with wet paper towels. Refresh towels as they dry out.
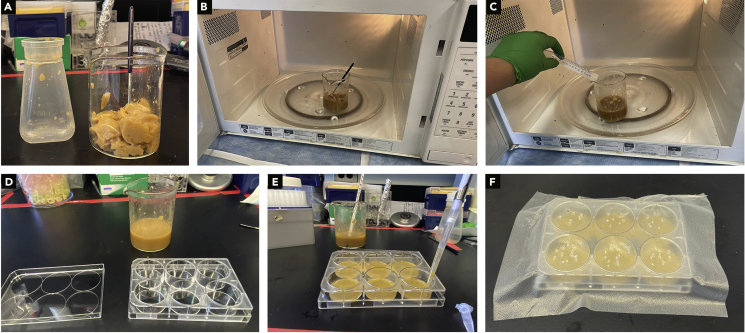
Figure 1.
Larval isolation plate setup
(A) Empty fly food from bottle into a glass beaker using spatula.
(B) Microwave until food completely melts.
(C) Add water to food.
(D) Mix food well.
(E) Pipette ~2.5 mL of food into each well of a 6-well plate.
(F) Once food solidifies transfer a larva into each well. Seal plate with plastic wrap and punch holes using syringe needle.
Anesthetizing larvae to visualize muscle structure at different larval stages
Timing: 10 min
-
15.
Set up laying pots of the desired genotypes.
-
16.
Set up larval collection plates for each of the genotypes.
-
17.
Fill wells of 6-well culture plate with Kwan Loong Medicated Oil, an anesthetic, and cover plate. Detailed steps on larval anesthetization and the subsequent imaging of their musculature are explained in the Step-by-step method details section.
Imaging platform to visualize larval movement
Timing: 5 min
-
18.Set up imaging platform.
-
a.Platform should be wide enough to accommodate a 10 cm apple juice plate at the bottom (Figure 2A). For dimensions of platform see Materials and equipment section (Figures 2A and 2B).
-
b.Ensure adequate number of 10 cm apple juice plates are at room temperature (1 plate per genotype/10 larvae). Detailed steps on larval locomotion analysis and its quantification are explained in the Step-by-step method details section.
-
a.
Note: We recommend collecting embryos and larvae from laying pots for no more than one week as fertility of flies decreases significantly after a week.
Note: While in our experiments we incubate embryos, larvae, and flies at 25°C, incubation at lower (i.e., 18°C) and higher temperatures (i.e., 28°C) can also be done. However, development is delayed (18°C) or accelerated (28°C) at these temperatures, and hence larval instars can progress slowly or quickly (Pulver and Berni, 2012).
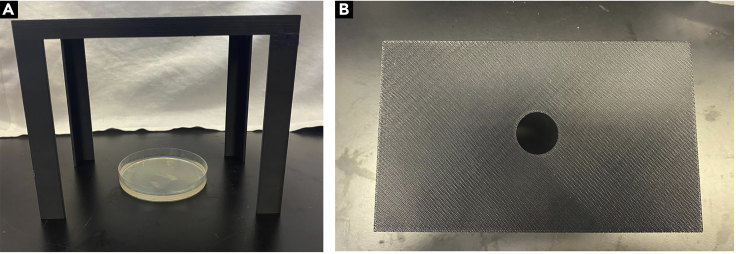
Figure 2.
Imaging platform setup and dimensions
(A) Lateral view image of the locomotion platform along with dimensions of platform legs and platform height.
(B) Top view image of the locomotion platform along with dimensions of platform surface used to hold smartphone used to acquire larval crawling movies.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Active dry yeast | Fleischmann's | Cat# 2192 |
| Agar | Moorehead and Company | Cat# 700B |
| Tegosept (hydrobenzoic acid methyl ester) | Sigma | Cat# H5501 |
| Sucrose | Fisher Scientific | Cat# BP220-1 |
| Apple juice | Monarch | Cat# 515815 |
| Ethanol | Fisher Scientific | Cat# 04-355-223 |
| Glycerol | Fisher Scientific | Cat# BP2291 |
| Kwan looong medicated oil | Kwan looong | http://www.kwanloongoil.com/ |
| 10× PBS | Fisher Scientific | Cat# BP3994 |
| Bleach, concentrated Clorox (30966) | Owens and Minor | Cat# 5406 |
| Halocarbon oil 700 | Sigma | Cat# H8898 |
| Experimental models: organisms/strains | ||
| D. melanogaster: Mhc-Gal4 | Bloomington Drosophila Stock Center | BDSC# 67044 |
| D. melanogaster: UAS-2x-GFP | Bloomington Drosophila Stock Center | BDSC # 6874 |
| D. melanogaster: Zasp66::GFP (ZCL0663) | Bloomington Drosophila Stock Center | BDSC #6824 |
| D. melanogaster:α-actinin::GFP | Bloomington Drosophila Stock Center | BDSC #51573 |
| D. melanogaster: UAS-moesin::mCherry | (Millard and Martin, 2008) | N/A |
| Software and algorithms | ||
| ImageJ/Fiji | NIH | https://fiji.sc ; RRID: SCR_002285 |
| GraphPad Prism | GraphPad |
http://www.graphpad.com; RRID:SCR_002798 |
| Photoshop | Adobe | https://www.adobe.com/products/photoshop.html; RRID:SCR_014199 |
| Illustrator | Adobe | http://www.adobe.com/products/illustrator.html; RRID:SCR_010279 |
| Excel | Microsoft | https://www.microsoft.com/en-gb/; RRID:SCR_016137 |
| MPEG Streamclip | Squared 5 | http://www.squared5.com/ |
| Other | ||
| 6-well cell culture plates | Thermo Scientific | Cat# 130184 |
| Precision Glide Sub-Q Needle 26G × 5/8 inch | Becton, Dickinson and Company | Cat# 305115 |
| 6 cm petri dish (60 mm × 15 mm) | Fisher Scientific | Cat# 08-757-100B |
| 10 cm petri dish (100 mm × 15 mm) | Fisher Scientific | Cat# FB0875712 |
| Fly vials | Crystalgen | Cat# 23-5056 |
| Fly bottles | Crystalgen | Cat# 5065 |
| Spoonula | Fisher Scientific | Cat# S35888 |
| Paintbrush size 0 | Blick | Cat# 05565-1000 |
| Beaker (150 mL glass) | Fisher Scientific | Cat# 02-255-25A |
| Beaker (100 mL plastic) | Globe Scientific | Cat# 3641 |
| Forceps (#5 Dumont) | Fine Science Tools | Cat# 11251-20 |
| Microscope glass slide (3 × 1 × 1 mm) | Fisher Scientific | Cat# 12-544-3 |
| Coverslip (22 mm × 40 mm) | Fisher Scientific | Cat# 12-544-BP |
| Plastic wrap (Press ’n Seal) | Glad | N/A |
Materials and equipment
Imaging platform
The imaging platform was 3D printed to the dimensions as indicated (Figures 2A and 2B).
Step-by-step method details
Analyzing muscle function throughout larval development
Timing: 30–45 min/day ×4 days
The steps below describe how to setup and record larval movement for muscle function analysis. Refer to steps 15–18 in the Before you begin section before starting.
-
1.
Isolate 1st instar larvae of your desired genotypes from your vial/plate. Clean larvae gently by immersing and swishing them in a watch glass containing ddH2O and transfer to a clean apple juice plate.
-
2.
Set up imaging platform as shown (Figure 3A).
-
3.
Place a fresh 10 cm wide apple juice agar plate below the imaging platform (Figure 3A).
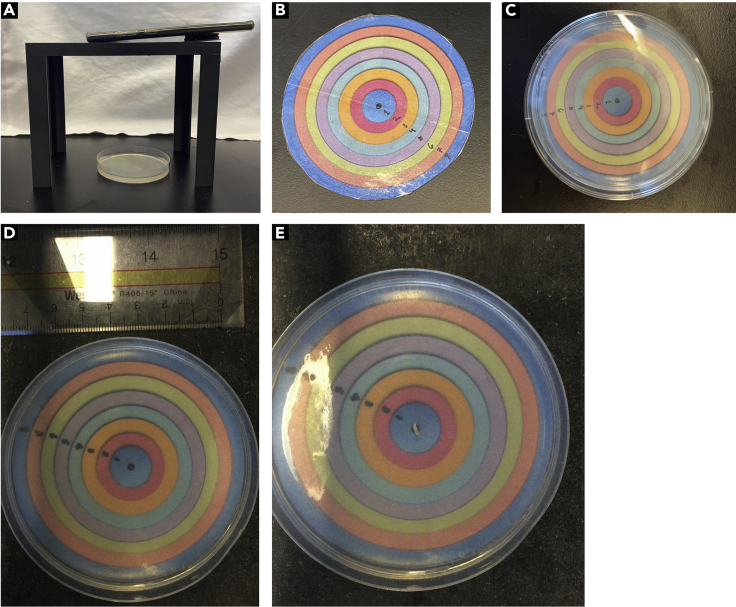
Optional: An additional aid to help record larval crawling data is a dartboard paper, a circular piece of paper the same size as agar plate divided into concentric circles. Each circle in the dartboard is numbered, recording how far a larva moves with respect to the dartboard can prevent further confusion when handling large numbers of larvae (Figures 3B and 3C).
-
4.
Place a ruler/measuring tape alongside the 10 cm plate. Acquire an image of the agar plate with ruler beside it (Figure 3D).
Note: The ruler or measuring tape is required to set scale and measure distance traveled by the larvae in standard units (cm, mm, etc.)
-
5.
Place larva from the first well of genotype of interest at the center of apple juice agar plate (Figure 3E).
Note: It is not required to provide an attractant for the larvae, the larvae crawl both in the presence and absence of an attractant with no significant difference in velocities observed between the two groups.
-
6.
Start recording larval movement using a smartphone or camera for 45 s.
Note: Choose a time period for tracking larval movement, e.g., 30, 45, 60 s, etc., based on your preferences. On selection of a time period, keep the time constant throughout all tracking experiments. We do not recommend choosing a time under 30 s as larvae often need time to acclimatize and begin moving. Allowing the larvae 2 s to acclimatize before recording their movement is beneficial.
-
7.
Stop recording larval movement once 45 s is over or when the larva reaches the edge of the plate, whichever comes first.
-
8.
Remove larva from plate using paintbrush and transfer back to well 1. Transfer larva from well 2 to the plate and repeat steps 5–7 until all larvae of the genotype/plate have been recorded.
Note: In our experiments we tracked 8–10 larvae per genotype.
-
9.
Change apple juice plate before tracking larvae from the next genotype.
Note: As larvae crawl on the apple juice, plate they leave their tracks on the plate. Over time, subsequent larvae follow the same tracks skewing the results. Track a maximum of 8 larvae on a single plate.
-
10.
Repeat steps 4–8 till all larvae from all genotypes/plates are tracked. Place plates with larvae back at 25°C.
-
11.
After 24 h, isolate 2nd instar larvae and repeat steps 1–10. Repeat the process every 24 h till larvae reach late 3rd instar stage.
Note: Day 1, 1st instar; day 2, 2nd instar; day 3, early 3rd instar; day 4, late 3rd instar.
Figure 3.
Assessing muscle function through larval locomotion
(A) Set up imaging platform, agar plate, and smartphone as indicated.
(B) Image of optional dartboard paper used for recording final larval movement.
(C) Setup of dartboard paper placed under agar plate.
(D) Image of agar plate and ruler beside it to set scale during the experiments.
(E) Place larva at the center of the plate/dart board paper and record movement.
For more details on how temperature can influence development, please refer to Figure 4A.
Note: While currently there is no evidence to suggest that larval movement is affected by circadian rhythms, ensure that larval tracking occurs during the light cycle of the larva (Sawin et al., 1994). It is recommended to track the larvae at the same time every day.
-
12.
Once late 3rd instar larvae have been tracked, dissect, fix, stain, and mount each larva individually as described in Azevedo et al. (2016). Examine muscle structure and count number of muscles with structural defects.
-
13.
Quantify muscle structure defects by scoring the number of defective muscles present in the larval abdominal hemisegments A2-A7 (Figures 4B–4D). The affected muscles were divided into different classes/groups based on severity of defects. The percentages of muscles affected were plotted as graphs, and statistical significance was calculated using two-way ANOVA.
-
14.
The above experiment was repeated again using a different group of larvae. Quantification of muscle function and larval tracking analysis are explained in detail in the Quantification and statistical analysis section.
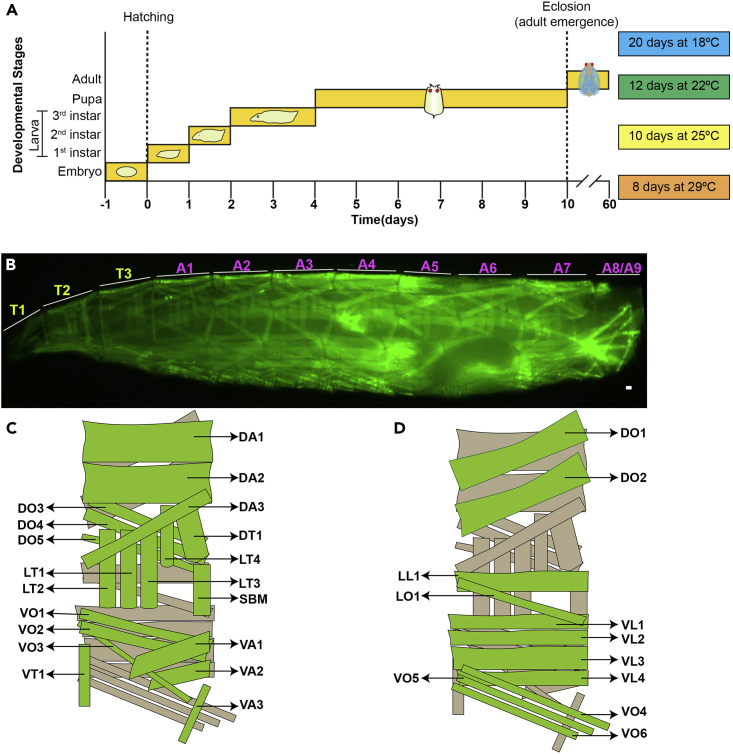
Figure 4.
Drosophila development and larval musculature
(A) Illustration depicting the influence of temperature on Drosophila development.
(B) Image of a heat-fixed 3rd instar larva expressing Tropomyosin-GFP with the different thoracic and abdominal hemisegments labeled. Scale bar, 100 μm.
(C) Schematic diagram depicting muscles from abdominal hemisegments (A1–A5) of the larva when viewed externally.
(D) Schematic diagram depicting muscles from abdominal hemisegments (A1–A5) of the larva when viewed internally.
Muscle names are as follows: muscle position (D, dorsal; V, ventral; L, lateral) followed by orientation (A, acute; L, longitudinal; O, oblique; T, transverse); SBM, segment border muscle.
Anesthetizing larvae to visualize muscle structure at different larval stages
Timing: 30–45 min/day × 4 days
The steps below describe how to anesthetize a larva and image its musculature throughout larval development.
-
15.
Isolate 1st instar larvae of desired genotype and clean larvae gently by immersing and swishing them in a watch glass containing ddH2O.
-
16.
Transfer and place larva on the lid of 6-well plate containing Kwan Loong Medicated Oil , such that the larva is placed directly over the well containing the oil.
Note: Ensure that the larva does not dry out on the lid. Adding droplets of water to the larva using a paintbrush can help prevent larva from drying out.
-
17.
Incubate larva in the chamber until it is anesthetized, i.e., stops moving when disturbed with a paintbrush.
Note: Incubation times of larvae greatly vary, so need optimization. Here, we present sample incubation times experienced with a larva; e.g., 1st instar, 3–5 min; 2nd instar, 5–8 min; early 3rd instar, 10–12 min; late 3rd instar, 15 min.
-
18.
Transfer anesthetized larvae to a microscope slide containing 50% glycerol and mount with a cover slip.
-
19.
Record orientation of mounted larva (e.g., dorsal side up or down, head and mouth left or right) and identify the hemisegments of the larval musculature to be imaged. Image hemisegment(s) and record hemisegment numbers.
Note: While imaging, take care not to squish or squeeze the larva, as it can lead to larval death.
-
20.
After imaging, carefully remove coverslip from larva, remove larva, and wash gently with 1× PBS, and then with ddH2O.
-
21.
Transfer larva to clean apple juice plate until it awakens. Once larva begins moving transfer larva to the first well of larval collection plate.
-
22.
Isolate second larva and repeat steps 15–21, until all larvae of desired genotypes have been anesthetized and imaged.
-
23.
Move larval collection plate at 25°C in light-dark incubator.
-
24.
After 24 h isolate 2nd instar larvae and repeat steps 15–23. Repeat the process every 24 h till larvae reach late 3rd instar stage.
Note: When imaging the same larva at successive instars, ensure that the larva is mounted in the same orientation as done for the 1st instar larva. Image the same hemisegments imaged in the 1st instar larva.
Note: Day 1, 1st instar; day 2, 2nd instar; day 3, early 3rd instar; day 4, late 3rd instar.
Visualizing and quantifying sarcomere structure and size from the late embryonic stage throughout the larval stages using heat fixation
Timing: 30 min/day ×4 days
These steps describe how to analyze and image muscle structure analysis at embryonic and larval stages.
-
25.
Set up laying pots of appropriate genotype containing fluorescent reporter(s) (e.g., Zasp-GFP) at 25°C.
-
26.
Label and change apple juice plates on laying pots within 20 h, keeping the old ones for embryo and larval collection.
-
27.
Isolate 1st instar larvae and distribute larvae into 3 different vials and an apple juice plate.
Note: We isolated and transferred ∼10 larvae into each vial and apple juice plate.
-
28.Use the remaining embryos on the plate for collection.
-
a.Pour enough water to cover embryos on plate and remove dead flies using paintbrush.
-
b.Gently disturb the embryos on the plate using a paintbrush and then transfer them to a collection basket.
-
c.Place collection baskets in a plate with 50% bleach and dechorionate embryos for 4 min at room temperature.
-
d.Rinse the embryos in the collection basket with distilled water repeatedly to remove any traces of bleach from the embryos.
-
a.
-
29.Heat Fixation
-
a.Embryos: Dip collection baskets with embryos into a water bath at 65°C for 1–2 s, ensuring that embryos are submerged in water.
-
b.Larvae: Pick up larvae of any developmental stage by the tail or head using forceps and submerge into a water bath at 65°C for 1–2 s.
-
a.
-
30.Mounting
-
a.Embryos: Transfer embryos from the collection basket to a microscope slide using a paintbrush and then spread on the slide. Mount in either halocarbon oil or 50% glycerol, and cover with a coverslip.
-
b.Larvae: Transfer larvae onto a microscope slide containing halocarbon oil or 50% glycerol. Orient larvae such that their dorsal sides (containing trachea) are either parallel or perpendicular to the long axis of the slide, and cover with a cover slip.
-
a.
-
31.
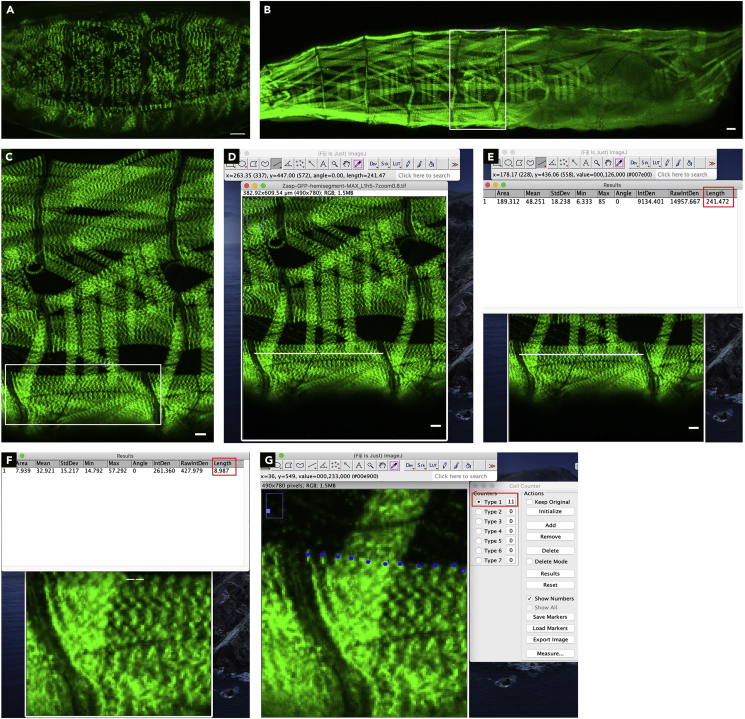
Image the embryos or larvae immediately using a microscope (Figures 5A and 5B).
Note: Both embryos and larvae were imaged on an inverted LSM 700 laser-scanning confocal microscope using either the 10×/0.30 NA or 20×/0.8 NA air objectives.
-
32.Further analysis of muscle structure such as muscle length, sarcomere size, and number can be performed upon image acquisition.
-
a.Muscle length
-
i.Open image of muscle in Fiji (Figure 5C).
-
ii.Using Line tool draw a line from one end of the muscle to other (Figure 5D).Note: Sometimes it is difficult to determine muscle ends from a single fluorescent reporter line. Using an additional marker such as moesin-mCherry (Millard and Martin, 2008) aids in visualization of the muscle ends.
-
iii.To register each measurement, select: Analyze> Measure (a new window will pop up when this is assessed for the first time) (Figure 5E).
-
iv.Copy all measurements into Excel, and plot graphs using wither Excel or GraphPad Prism. Statistical significance of muscle length between genotypes at different stages were calculated using multiple t tests.
-
i.
-
b.Sarcomere width
-
i.Open image of muscle in Fiji.Note: To measure sarcomere width or number, it is preferable to use a fluorescent Z-disc marker such as Zasp-GFP or α-actinin-GFP (Clark and Kadrmas, 2013).
-
ii.Using a line tool draw a line from one Z-disc to the next Z-disc. Register each measurement by Select: Analyze> Measure. Repeat the step by drawing a line from each Z-disc to its neighbor throughout the length of the muscle (Figure 5F).
-
iii.Copy all measurement values into an Excel sheet. Calculate average sarcomere size by averaging the measurements of sarcomere size from a single muscle. Repeat the above steps for subsequent muscles.Note: Alternatively, another option is to draw a line from one end of the muscle to the other. Register the measurement by selecting Analyze > Measure. Count the number of Z discs, i.e., sarcomere number. To calculate average sarcomere size in the muscle, use the following formula: Muscle length/Sarcomere number.
-
iv.In our experiments, we calculated average sarcomere sizes for at least 10 different muscles from 5 different larvae.
-
i.
-
c.Sarcomere number
-
i.Open image of muscle in Fiji.
-
ii.Open the Cell counter plugin by selecting Plugins> Cell Counter (a new window will pop up when this plugin is selected). Select initialize option followed by Type 1 option in the pop-up window.
-
iii.Begin clicking all the Z lines within the muscle. Once all the Z lines are counted, the number next to the Type 1 option in the pop-up window indicates the number of sarcomeres.
-
iv.Select Type 2 option and then quantify the sarcomere number in the next muscle (Figure 5G). Once all muscles in an image are quantified, open new image and repeat above steps.
-
v.Copy all measurement values into an Excel sheet. Calculate average sarcomere number by averaging the number of sarcomeres from different muscles. Graphs were plotted in Excel or in GraphPad Prism.
-
vi.In our experiments, we calculated sarcomere numbers from at least 10 different muscles from 5 different larvae for each genotype. Statistical significance of sarcomere number between genotypes and/or at different stages were calculated using either Student’s t test or multiple t tests.
-
i.
-
a.
Figure 5.
Visualization and quantification of muscle and sarcomere structure
(A) Image of sarcomeres and muscle structure in a heat-fixed stage 17 embryo expressing Zasp-GFP (green). Scale bar, 20 μm.
(B) Image of heat-fixed 2nd instar larva expressing Zasp-GFP. Zasp-GFP marks sarcomeres. White rectangle highlights a single hemisegments in the larva. Scale bar, 20 μm.
(C) High-magnification image of a single hemisegments from a heat-fixed 3rd instar larva opened in Fiji for further analysis. White box highlights 2 ventral longitudinal (VL) muscles, VL1 and VL2, used for muscle and sarcomere structure analysis. Scale bar, 25 μm.
(D) Using line tool in Fiji. White line is drawn from one end of VL muscle to the other. Scale bar, 25 μm.
(E) Measuring length of the line using Measure function under Analyze tool, generates information about line draw in a new window. Red box highlights the length of the line, i.e., muscle length which is then plotted. Scale bar, 25 μm.
(F) Image displaying the results of measuring sarcomere length using the line tool. Red box highlights the length of the line, i.e., sarcomere length which is then plotted.
(G) Image of a larval muscle segment expressing Zasp-GFP to count number of sarcomeres using Cell counter plugin. Red box highlights the number sarcomeres counted, i.e., number of blue dots on the muscle, in a segment of the muscle.
Expected outcomes
This protocol describes how Drosophila larval muscle function and structure can be quantified and analyzed, respectively. Quantification of larval crawling velocity by tracking larval movement serves as a proxy for muscle function and can help understand how mutations in genes involved in muscle development and function can affect muscle contraction, thereby impairing larval movement. In control larvae, larval locomotion velocity increases as the larvae progress through the instars [Figure 5I, (Balakrishnan et al., 2020)]. This increase in locomotion velocity throughout larval stages is consistent with an increase in muscle size that occurs during the instars (Balakrishnan et al., 2020, Demontis and Perrimon, 2009, Bawa et al., 2020). Mutations in genes affecting larval locomotion thus far have predominantly decreased locomotion, with very few mutations identified that increase locomotion of the larva (Camuglia et al., 2018). Measuring larval velocity throughout the larval instars can identify when the first defects in muscle function are observed, and if these defects coincide with the defects observed in muscle structure.
Examining muscle structure in control larvae highlights that both sarcomere and muscle structure, which are established in the late embryonic stages, are maintained throughout the larval stages with an increase in sarcomere number as muscles grow (Balakrishnan et al., 2020). In control muscles, sarcomere size remains the same as muscles increase in size (Balakrishnan et al., 2020). However, muscles harboring mutations often show defects in sarcomere/muscle structure, which are observed as early as the embryonic stage or 1st and 2nd larval instars (Balakrishnan et al., 2020). While muscle defects observed in the embryonic musculature are indicative of perturbations in muscle formation, defects during the 1st or 2nd instar stages are suggestive of defects in muscle growth and/or maintenance (Balakrishnan et al., 2020).
Quantification and statistical analysis
Analyzing muscle function throughout larval development
-
1.
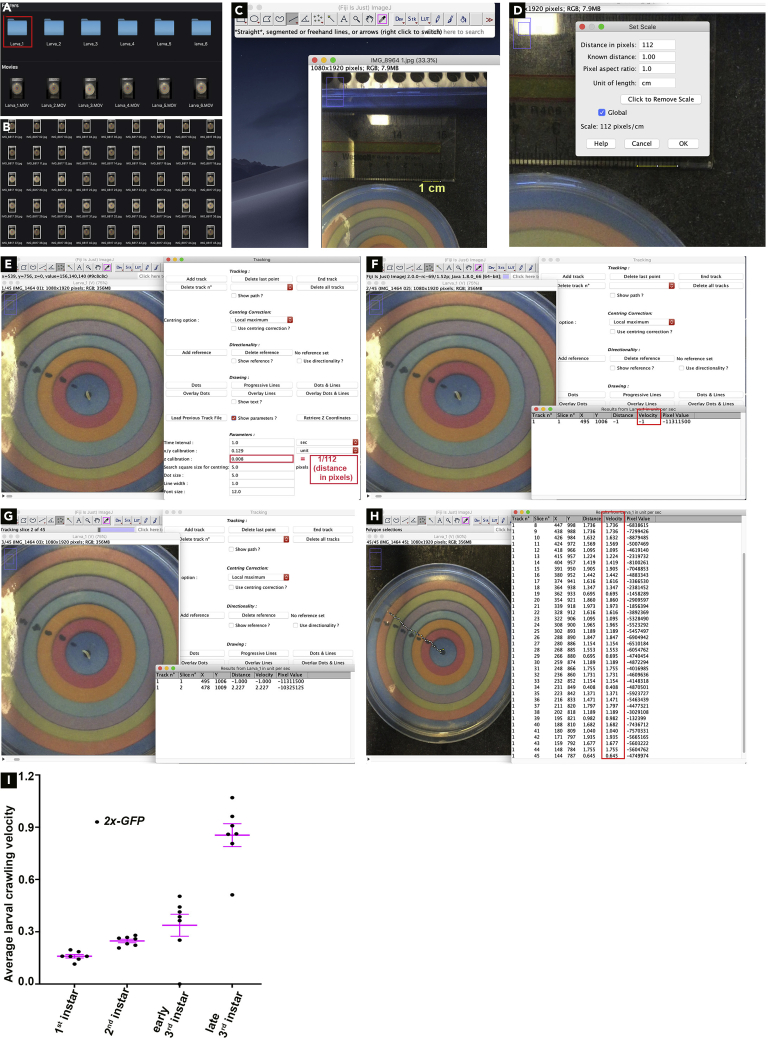
Convert video files to image sequence format (jpeg) using a video converting software such as MPEG Streamclip. Ensure that all jpeg files from the same video file are saved in one folder (Figures 6A and 6B).
Note: When converting from video to image sequence, format select output as 1 frame/s. This will result in every folder (1 per larva) containing files of each frame, e.g., 45 jpeg files if larva was imaged for 45 s.
-
2.
To set scale, open image of apple juice plate with ruler beside it by dragging it into the Fiji toolbar. Select line tool and draw line along ruler for a distance of 1 cm. Select: Analyze>Set Scale. In the dialog box set known distance to 1.00 and Unit of length as cm. Note the distance in pixels, e.g., 112. Select the Global option. The scale is indicated at the bottom of the box, e.g., 112 pixel/cm. Accept all changes (Figures 6C and 6D).
-
3.
To begin manual larval tracking, Select: Plugins > Tracking > Manual Tracking. In the parameters section of the dialog box Time interval is 1 s and z calibration is 1/112 (distance in pixels) (Figure 6E).
-
4.
Open folder containing jpeg images of larval crawling by dragging it into the Fiji toolbar and check the use Virtual Stack option in the dialog box. Once the larval sequence loads, select the Add Track option in the manual tracking dialog box.
-
5.
To begin tracking, select Add Track in the manual tracking window. Then identify the mouth parts of the larva (black structures) and click on it. A dialog box will appear that displays the X and Y coordinates along with velocity and distance traveled at that time point. The image also automatically moves to the next frame (Figure 6F).
-
6.
In the subsequent frames, repeatedly click on the mouth parts of the larva again till the last frame. The last frame will display the track/path taken by the larva along with a dialog box containing all the measurements (Figure 6G).
-
7.
Copy all measurements and paste them in an Excel sheet. Calculate average velocity (cm/s) by averaging velocity values from Slice 2-45. For velocity in mm/s multiple average velocity(cm/s) × 10 (Figure 6H).
Note: The first velocity measurement is excluded from the quantification as the measurement is negative and is used as the starting point of the larval analysis.
-
8.
Repeat the above steps for the remaining larvae and the other genotypes.
-
9.
Average velocities (mm/s) were plotted using GraphPad Prism. Statistical significance between genotypes at different larval stages were calculated using multiple t tests (Figure 6I).
Figure 6.
Quantification of muscle function through larval locomotion analysis
(A) List of folders and larval crawling videos from a genotype. Red box highlights a folder containing all files of a single larva.
(B) List of images (.jpeg) obtained from converting a larval crawling video into an image sequence format. Each .jpeg file is a single second of the larval crawling video.
(C) Set scale for the experiment by drawing a line of 1 cm on ruler.
(D) Scale is set by equating distance in pixels of the line to its metric length of 1 cm.
(E) Manual Tracking option under the Tracking plugin is selected, z calibration is entered, and track is added.
(F) Dialog box displaying results after tracking the first second of larval crawling. Note that the velocity is −1.00 as this is the reference point of the larva and this value is discarded during average velocity calculation (highlighted by red box) .
(G) Dialog box displaying results from tracking first 2 s of larval crawling.
(H) Dialog box displaying results from tracking the 45 s of larval crawling, along with the path taken by the larva. Red box highlights the velocity column that is used to average the velocity of the larva that is then plotted.
(I) Crawling velocities of larvae at different larval stages. p values by multiple t tests. Mhc-Gal4 is used to drive expression of UAS-2x-GFP in the muscles. Error bars, mean ±SEM.
Limitations
Muscle function can be affected by defects in the musculature or in the nervous system, particularly in the motor neurons that innervate the musculature. Hence, while assessing zygotic mutants for muscle function using the larval crawling assay, it is important to confirm any observed changes in muscle function with muscle structure analysis. It is also important to follow up any observed differences in muscle function in zygotic mutants with mutants where the gene is specifically knocked down in the muscle.
Examination of muscle structure through heat fixation is limited by the availability of fluorescent transgenes. To analyze muscle structure in detail, larval dissection, followed by fixation and staining of musculature in larval fillet should be performed [as described in (Azevedo et al., 2016, Brent et al., 2009)]. Dissection is technically challenging in the 1st and 2nd instar larvae.
Troubleshooting
Problem 1
Larval development extends for over 4 days
Potential solution
Handling the larvae every day, as in the larval locomotion or anesthetization protocols, inflicts stress on the larvae and thereby delays their development, i.e., pupariation. Collecting age matched larvae of the same genotype that are not handled everyday can help by size matching experimental larvae to determine the stage.
Problem 2
Mutants show delayed development.
Potential solution
Some mutants are developmentally delayed, which can be further exacerbated when anesthetizing and imaging the larvae daily. Under these circumstances it is important to stage the larvae based on the differentiation of the mouth hooks or anterior and posterior spiracles (Okada, 1963; Schubiger et al., 1998; Vaufrey et al., 2018). For example, with respect to staging larvae based on spiracles, 1st instar larvae have white posterior spiracles, 2nd instar larvae have orange posterior spiracles and apparition of club shaped anterior spiracles, early 3rd instar have branched anterior spiracles, late 3rd instar larvae have everted anterior spiracles, and wandering 3rd instar larvae are found out of the food (Park et al., 2002).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mary Baylies (bayliesm@mskcc.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This protocol did not generate new data or code.
Acknowledgments
We would like to thank the Baylies lab members for helpful discussions. This work was supported by the United States National Institute of Arthritis, Musculoskeletal and Skin Diseases (R21AR067361, R01AR108981) and the National Institute of General Medicine (R01GM121971) to M.K.B.
Author contributions
M.B. and M.K.B. wrote the manuscript with input from W.J.S. W.J.S. took images for all the protocols and helped in protocol optimization.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Mridula Balakrishnan, Email: mridula.balakrishnan@mpi-bn.mpg.de.
Mary K. Baylies, Email: bayliesm@mskcc.org.
References
- Azevedo M., Schulman V.K., Folker E., Balakrishnan M., Baylies M. Imaging approaches to investigate myonuclear positioning in Drosophila. Methods Mol. Biol. 2016;1411:291–312. doi: 10.1007/978-1-4939-3530-7_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan M., Yu S.F., Chin S.M., Soffar D.B., Windner S.E., Goode B.L., Baylies M.K. Cofilin loss in Drosophila muscles contributes to muscle weakness through defective sarcomerogenesis during muscle growth. Cell Rep. 2020;32:107893. doi: 10.1016/j.celrep.2020.107893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa S., Brooks D.S., Neville K.E., Tipping M., Sagar M.A., Kollhoff J.A., Chawla G., Geisbrecht B.V., Tennessen J.M., Eliceiri K.W., Geisbrecht E.R. Drosophila TRIM32 cooperates with glycolytic enzymes to promote cell growth. eLife. 2020;9:e52358. doi: 10.7554/eLife.52358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent J.R., Werner K.M., McCabe B.D. Drosophila larval NMJ dissection. J. Vis. Exp. 2009:1107. doi: 10.3791/1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.S., Vishal K., Kawakami J., Bouyain S., Geisbrecht E.R. Optimization of wrMTrck to monitor Drosophila larval locomotor activity. J. Insect Physiol. 2016;93-94:11–17. doi: 10.1016/j.jinsphys.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camuglia J.M., Mandigo T.R., Moschella R., Mark J., Hudson C.H., Sheen D., Folker E.S. An RNAi based screen in Drosophila larvae identifies fascin as a regulator of myoblast fusion and myotendinous junction structure. Skelet. Muscle. 2018;8:12. doi: 10.1186/s13395-018-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B.J., Imlach W.L., Jiao W., Wolfram V., Wu Y., Grbic M., Cela C., Baines R.A., Nitabach M.N., McCabe B.D. Miniature neurotransmission regulates Drosophila synaptic structural maturation. Neuron. 2014;82:618–634. doi: 10.1016/j.neuron.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.A., Kadrmas J.L. Drosophila melanogaster muscle LIM protein and alpha-actinin function together to stabilize muscle cytoarchitecture: a potential role for Mlp84B in actin-crosslinking. Cytoskeleton (Hoboken) 2013;70:304–316. doi: 10.1002/cm.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F., Perrimon N. Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development. 2009;136:983–993. doi: 10.1242/dev.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes I., Schöck F. The nebulin repeat protein Lasp regulates I-band architecture and filament spacing in myofibrils. J. Cell Biol. 2014;206:559–572. doi: 10.1083/jcb.201401094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakanj P., Eming S.A., Partridge L., Leptin M. Long-term in vivo imaging of Drosophila larvae. Nat. Protoc. 2020;15:1158–1187. doi: 10.1038/s41596-019-0282-z. [DOI] [PubMed] [Google Scholar]
- Millard T.H., Martin P. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development. 2008;135:621–626. doi: 10.1242/dev.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T. Caenogenetic differentiation of mouth hooks in drosophilid larvae. Evolution. 1963;17:84–98. [Google Scholar]
- Park Y., Filippov V., Gill S.S., Adams M.E. Deletion of the ecdysis-triggering hormone gene leads to lethal ecdysis deficiency. Development. 2002;129:493–503. doi: 10.1242/dev.129.2.493. [DOI] [PubMed] [Google Scholar]
- Pulver S.R., Berni J. The fundamentals of flying: simple and inexpensive strategies for employing Drosophila genetics in neuroscience teaching laboratories. J. Undergrad. Neurosci. Educ. 2012;11:A139–A148. [PMC free article] [PubMed] [Google Scholar]
- Sawin E.P., Dowse H.B., Hamblen-Coyle M.J., Hall J.C., Sokolowski M.B. A lack of locomotor activity rhythms in Drosophila melanogaster larvae (Diptera: Drosophilidae) J. Insect Behav. 1994;7:249–262. [Google Scholar]
- Schnorrer F., Schönbauer C., Langer C.C., Dietzl G., Novatchkova M., Schernhuber K., Fellner M., Azaryan A., Radolf M., Stark A., Keleman K., Dickson B.J. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- Schubiger M., Wade A.A., Carney G.E., Truman J.W., Bender M. Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development. 1998;125:2053–2062. doi: 10.1242/dev.125.11.2053. [DOI] [PubMed] [Google Scholar]
- Vaufrey L., Balducci C., Lafont R., Prigent C., Le Bras S. Size matters! Aurora A controls Drosophila larval development. Dev. Biol. 2018;440:88–98. doi: 10.1016/j.ydbio.2018.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol did not generate new data or code.