Key Points
Question
Is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) present in the human cornea?
Findings
In this case series carried out in a tertiary care facility, SARS-CoV-2 genomic RNA was detected in the cornea of 6 of 11 eyes (55%) of patients with viremic coronavirus disease 2019; subgenomic SARS-CoV-2 RNA was present in 4 of these 6 eyes (67%). Infectivity or presence of viral structural proteins could not be confirmed in any eye.
Meaning
This report demonstrates the presence of viral genomic and subgenomic RNA of SARS-CoV-2 in the human cornea.
Abstract
Importance
Current recommendations are to avoid tissue for corneal transplant from donors with coronavirus disease 2019 (COVID-19) or those who were recently exposed to COVID-19 owing to the lack of knowledge about the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in corneal tissues. Evidence of SARS-CoV-2 in corneal tissue would seem to have clinical relevance for corneal transplant.
Objectives
To investigate the presence of viral SARS-CoV-2 RNA in corneal discs of deceased patients with confirmed COVID-19 and assess viral genomic and subgenomic RNA load, possible infectivity, and histologic abnormalities.
Design, Setting, and Participants
A case series was conducted of 11 deceased patients with COVID-19 who underwent autopsy between March 20 and May 14, 2020. Eleven corneal discs (1 corneal disc per patient) were harvested for molecular detection of viral genomic and subgenomic RNA, virus isolation, and immunohistochemistry. The SARS-CoV-2 RNA loads were compared with RNA loads in the conjunctival and throat swab samples and aqueous humor, vitreous humor, and blood samples.
Main Outcomes and Measures
Evidence of SARS-CoV-2 RNA in human corneas.
Results
This study comprised 11 patients (6 women [55%]; mean [SD] age, 68.5 [18.8] years). In 6 of 11 eyes (55%), SARS-CoV-2 genomic RNA was detected in the cornea; subgenomic RNA was present in 4 of these 6 eyes (67%). Infectivity or the presence of viral structural proteins could not be confirmed in any eye. However, patients whose corneal disc was positive for SARS-CoV-2 RNA also had positive results for SARS-CoV-2 RNA in 4 of 6 conjunctival swab samples, 1 of 3 aqueous humor samples, 3 of 5 vitreous humor samples, and 4 of 5 blood samples. Overall, conjunctival swab samples had positive results for SARS-CoV-2 RNA in 5 of 11 cases. Postmortem SARS-CoV-2 viremia was detected in 5 of 9 patients.
Conclusions and Relevance
Viral genomic and subgenomic RNA of SARS-CoV-2 was detected in the cornea of patients with COVID-19 viremia. The risk of COVID-19 infection via corneal transplant is low even in donors with SARS-CoV-2 viremia, but further research is necessary to assess the rate of SARS-CoV-2 transmission via corneal transplant.
This case series investigates the presence of viral severe acute respiratory syndrome coronavirus 2 RNA in corneal discs of deceased patients with confirmed coronavirus disease 2019 and assesses viral genomic and subgenomic RNA load, possible infectivity, and histologic abnormalities.
Introduction
The highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters the target cells by binding to its main receptor, angiotensin-converting enzyme 2 (ACE2),1 while also being dependent on type II transmembrane serine protease (TMPRSS2).2 Because both ACE2 and TMPRSS2 are present in the limbal superficial cells and the superficial conjunctiva, susceptibility to SARS-CoV-2 infection can be suspected.3 In contrast, the human corneal stroma and endothelium do not express ACE2 nor TMPRSS2.3 However, only limited knowledge about SARS-CoV-2 in corneal tissues is available because only 1 publication, to our knowledge, has reported on the absence of the virus in 5 patients.4
The cornea is the most frequently transplanted tissue worldwide—in 2012, approximately 185 000 corneas were transplanted worldwide.5 However, corneal transplants have a limited shelf life of approximately 30 days if stored in organ culture and approximately 14 days if stored in preservation medium at 4 °C, implicating planning and economic challenges for corneal transplant in particular. Currently, the recommendation from the Global Alliance of Eye Bank Associations is to avoid tissue from donors infected with coronavirus disease 2019 (COVID-19) or who have recently been exposed to COVID-19.6 The risk of transmission is considered low, but owing to the lack of knowledge about SARS-CoV-2 in corneal tissues, precautionary measures have been taken.
Considering the possible implications of SARS-CoV-2 for corneal transplants, detailed knowledge on virus tropism, particularly with regard to corneal tissue, is of high clinical relevance. We therefore prospectively analyzed corneal tissue from 11 deceased patients with COVID-19 to investigate the presence of SARS-CoV-2 genomic and subgenomic RNA (sgRNA) by reverse transcriptase–polymerase chain reaction (RT-PCR). For correlation of the results, conjunctival and throat swab samples, blood samples, aqueous humor (AH) samples, and vitreous humor (VH) samples were examined. To assess potential infectivity, virus isolation was attempted. Immunohistochemistry was performed on SARS-CoV-2 and leukocyte common antigen (LCA) from the cornea to characterize a possible SARS-CoV-2 infection of the anterior segment of the eye.
Methods
Study Design
Approximately 170 autopsies of deceased patients with COVID-19 were performed between March 20 and May 14, 2020, at the Institute of Forensic Medicine at the University Medical Center Hamburg-Eppendorf in Germany. Autopsies were allowed according to § 25 of the German Infection Protection Law and other relevant legal regulations.7 Institutional review board approval from the independent ethics committee of Hamburg University was obtained. Informed consent was waived by the German government according to § 25 of the German Infection Protection Law. Individual case presentations are protected by privacy safeguard (part of § 27 of the federal Data Protection Act). The study complied with the tenets of the Declaration of Helsinki.8
All deceased patients were screened for SARS-CoV-2 viral RNA using a throat smear followed by immediate RT-PCR. Primarily patients with a high viral load of more than 104 copies per cell (ie, copies per milliliter) in the throat swab were selected, but there was no specific minimum viral load requested for inclusion. Corneal discs, AH samples, VH samples, conjunctival smears, and venous blood samples were obtained. Clinical records were checked for preexisting medical conditions and antemortem diagnostic findings.
Corneal Preparation and Aqueous Fluid Sample
Until autopsy, the bodies were stored at 4 °C. Prior to autopsy, a conjunctival smear was obtained from 11 deceased patients with COVID-19. The conjunctiva was separated from the limbus, and the globe was enucleated. After enucleation, AH was obtained through the clear cornea using a 27-gauge needle (Becton, Dickinson and Co), if possible, and the anterior chamber was reestablished using Balanced Salt Solution (Deltamedica GmbH). For 4 of 11 patients, an AH sample could not be obtained because of an insufficient amount of liquid for further processing. For corneal discs and VH, the eyes were dissected at the equator of the globe. Consequently, the vitreous automatically detached from the retina as the anterior part of the eye was removed. A native sample of the VH was obtained. Corneal discs without adjacent sclera or conjunctiva were dissected and cut in half. Two samples of corneal discs were obtained—one native and the other fixed in buffered 4% formaldehyde.
Molecular Diagnostic Procedure
All tissue samples were homogenized by using ceramic beads (Precellys lysing kit; Bertin Instruments).9 Automated extraction of nucleic acids was performed on a MagNA Pure 96 system (Roche) according to manufacturer recommendations. A Light Cycler 480 II instrument (Roche) was used to perform SARS-CoV-2 RT-PCR as described previously10 using the 1-step RNA control kit (Roche). SARS-CoV-2 RNA in cell culture supernatant was detected by RT-PCR as described previously.11 SARS-CoV-2 sgRNA in homogenized tissue samples was quantified by RT-PCR as published previously12 using SARS-CoV-2 (isolate HH-113)–infected Vero cells (CRL-1586; ATCC) as a control.
Cell Culture and Virus Isolation
Vero cells were cultivated under standard culture conditions in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum; 1% penicillin-streptomycin; 1% l-glutamine, 200mM; 1% sodium pyruvate; and 1% nonessential amino acids (all Gibco; Thermo Fisher Scientific) under standard culture conditions. For virus isolation attempts, cells were seeded to 80% to 90% confluency into 24-well plates (TPP Techno Plastic Products AG). A total of 250 μL of the homogenized tissue solution was used for infection after 1 hour of absorption at 37 °C and was added to each well, cells were washed once with phosphate-buffered saline, and 1 mL of fresh cell culture medium was added. Cells were monitored daily for cytopathic effect. Absence of virus growth was confirmed by quantitative RT-PCR.11
Immunohistochemistry
Formalin-fixed, paraffin embedded tissue samples from corneas were processed and stained with hematoxylin-eosin following standard laboratory procedures. Furthermore, immunohistochemistry with an antibody against the LCA CD45 (1:200; clone M742; Dako) was performed on a Ventana benchmark XT autostainer following the manufacturer’s recommendations. For immunohistochemistry of SARS-CoV-2 spike protein, antibody (1:300; GTX632604; clone 1A9; GeneTex) was validated on SARS-CoV-2–infected (Hamburg isolate13) and SARS-CoV-2–noninfected Vero cells that were processed to formalin-fixed paraffin, embedded blocks. Slides were examined by experienced morphologists (M.G. and S.K.), and selected slides were electronically scanned at high magnification (×40) as high-resolution images (1900 × 1200 pixels) using a NanoZoomer 2.0-HT scanner (Hamamatsu Ltd).
Results
Patient Cohort
Of the 11 patients analyzed (6 women [55%]; mean [SD] age, 68.5 [18.8] years), 10 (91%) died of COVID-19 (Table). One patient (9%) died of hemorrhagic shock, which is likely to be unrelated to COVID-19 and was defined as a non–COVID-19 death. The mean (SD) postmortem interval was 2.7 (1.7) days. The Table lists patient characteristics and risk factors for severe conditions. No preexisting ocular conditions associated with COVID-19 were listed in the medical record of any patient.
Table. Characteristics of Patients, Cause of Death, Place of Death, and Risk Factors for Severe Condition.
| Characteristic | Patients, No. (%) (N = 11) |
|---|---|
| Patient characteristics | |
| Female | 6 (55) |
| Age, mean (SD), y | 68.5 (18.8) |
| Postmortem interval, mean (SD), d | 2.7 (1.7) |
| Invasive ventilation | 6 (55) |
| BMI, mean (SD) | 28 (8) |
| Died of COVID-19 | 10 (91) |
| Risk factors, mean (SD), No. | 2.4 (1.1) |
| Cause of death, No. of patientsa | |
| Pneumonia | 10 |
| Sepsis | 2 |
| Hemorrhagic shock | 1 |
| Place of death | |
| Intensive care unit | 6 (55) |
| Normal ward | 2 (18) |
| Emergency unit | 1 (9) |
| Retirement home | 1 (9) |
| Own home | 1 (9) |
| Comorbiditya | |
| Liver | 1 (9) |
| Chronic heart disease | 7 (64) |
| Lung | 2 (18) |
| Neurologic (dementia or stroke) | 1 (9) |
| Kidney | 3 (27) |
| Oncologic | 4 (36) |
| Endocrine | 3 (27) |
| Immunologic | 1 (9) |
| Condition after embolism | 1 (9) |
| Other | 3 (27) |
Abbreviations: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); COVID-19, coronavirus disease 2019.
Multiple inclusion of 1 patient in the various categories possible.
SARS-CoV-2 RNA in Corneal Tissue
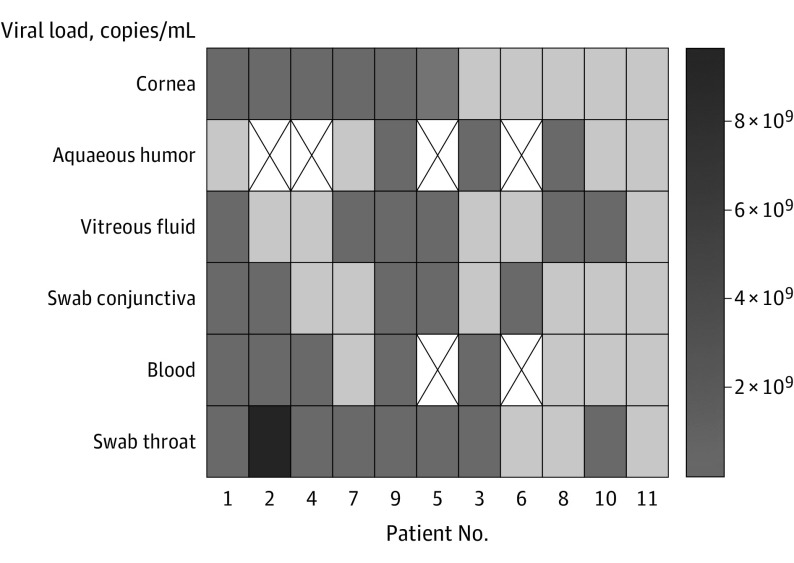
In 6 of 11 corneal disc samples (55%), SARS-CoV-2 RNA was detected (Figure). In 4 of the 6 corneal discs (67%) with SARS-CoV-2 RNA, sgRNA was detected. Virus isolation failed in all corneal disc samples. Patients with SARS-CoV-2 RNA detected in the corneal disc also had positive results for SARS-CoV-2 RNA in 4 of 6 conjunctival swab samples, 1 of 3 AH samples, 3 of 5 VH samples, and 4 of 5 blood samples (Figure).
Figure. Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) RNA.
SARS-CoV-2 RNA loads in eye samples, blood samples, and throat swab samples are displayed for each patient. The darkness of the fields indicates the viral load; darker color indicates higher viral load (see color bar on the right side), and white boxes with an X indicate missing values.
Overall, SARS-CoV-2 RNA was detected in 5 of 11 conjunctival swab samples, 3 of 7 AH samples, and 6 of 11 VH samples (Figure). Postmortem SARS-CoV-2 viremia was detected in 5 of 9 patients (Figure).
Histologic and Immunohistochemistry Findings
The eFigure in the Supplement displays a representative image of corneal disc samples that had negative and positive PCR results for SARS-CoV-2. The hematoxylin-eosin staining showed a preserved corneal structure in samples that had negative and positive PCR results for SARS-CoV-2 (eFigure, A, B, C, G, H, and I in the Supplement). SARS-CoV-2 spike protein–positive cells could not be detected by immunohistochemistry regardless of a positive or negative PCR result for SARS-CoV-2 PCR. Staining on LCA showed some LCA-positive cells in corneal discs. There was no difference between the samples that had negative PCR results for SARS-CoV-2 and those that had positive PCR results (eFigure, E, F, K, and L in the Supplement).
Discussion
The main receptor ACE2 and the cofactor TMPRSS2 required for efficient viral entry1,14 are expressed in some anatomical parts of the eye, including the basal corneal epithelium, limbal niche, corneal wing cells, transit amplifying cells, limbal superficial cells, corneal epithelial superficial cells, and limbal stem cells. This expression suggests that SARS-CoV-2 infection of eye tissues—especially at the limbus and the cornea—is possible.
In this study, we provide evidence of SARS-CoV-2 in the human cornea because we detected SARS-CoV-2 RNA in the corneal discs of 6 of 11 (55%) deceased patients with COVID-19. In the patient cohort, we also obtained a rather high rate of 5 of 11 positive PCR results (46%) in conjunctival swab samples. These findings are in contrast to previous studies that reported on SARS-CoV-2 detection in conjunctival swab samples,15,16,17,18,19 with a low rate of positive PCR results ranging from 1.4% (1 of 72)20 to 4.5% (3 of 67).21
Most patients (4 of 5) with conjunctival swab samples with a positive PCR result for SARS-CoV-2 had SARS-CoV-2 viremia. Moreover, for 4 of 6 patients (67%) with positive PCR results for SARS-CoV-2 in the corneal disc, SARS-CoV-2 could be detected in blood samples. The rate of viremia in the patient cohort is high compared with other studies that reported a positive RT-PCR result of SARS-CoV-2 in 1% of the blood samples (3 of 307)22 or 8% of the blood samples (1 of 12).23 Because our study analyzed postmortem viremia in deceased patients with COVID-19, the observed high viremia rate of 56% might be explained by the severity of disease and high viral loads induced by patient selection, especially because patients with a high viral load in throat swab samples were selected. This finding characterizes our patient cohort as a high-risk group, with a high rate of positive conjunctival swab samples and viremia compared with other studies. In addition, patients with high viral RNA loads in nasopharyngeal swab samples and viremia were more likely to show ocular involvement with positive PCR results in ocular samples of corneal discs, conjunctival swab samples, and AH samples. Consequently, the possibility of contamination has to be considered as an explanation for the positive SARS-CoV-2 detection in corneal discs.
During corneal disc preparation, the limbal vascular arcade might be cut or partly included in the corneal disc sample, although sclera and conjunctiva were cautiously avoided. Because isolation of the virus failed in all patients and viral loads in the cornea were low compared with viral loads in the blood (cornea: median, 7.14 × 102 copies/mL [range, ≤2.89 × 105 copies/mL]; blood: median, 4.95 × 103 copies/mL [range, ≤2.68 × 107 copies/mL]), contamination of the corneal disc through blood is possible.
To our knowledge, thus far there is no evidence of SARS-CoV-2 detection in the AH of patients with COVID-19, but ACE2 expression has been detected in AH.24 This finding supports our findings of SARS-CoV-2 genomic RNA in 3 of 7 AH samples. In corneal disc samples positive for SARS-CoV-2, AH was positive in only 1 case. Contamination of the corneal sample through AH is conceivable because the AH is in direct anatomical contact with the corneal endothelium, but owing to the low number, contamination is rather unlikely. In addition, contamination of the AH is unlikely because AH samples were obtained through the clear cornea and VH samples were obtained before the eyes were dissected. Consequently, the relatively high rate of VH samples positive for SARS-CoV-2 might be explained by the severe and finally fatal course of COVID-19 in our severely infected patient cohort.
We could not detect viral protein in corneal cells by immunohistology; isolation of infectious virus also failed. Owing to comparably low RNA load in the cornea, this result was not surprising because recent studies show that SARS-Cov-2 isolation is unlikely from samples with low RNA loads or high cycle threshold values.25,26 Staining of viral protein by immunohistochemistry in formalin-fixed tissue is less sensitive compared with diagnostic RT-PCR because no amplification of target structures is involved. To prove the presence of virus in tissue, a large amount of protein needs to be accumulated. Moreover, infected cells might be rare in the corneal disc. Thus, although we could not detect the presence of SARS-CoV-2 infectious particles in any of the samples analyzed, our findings do not prove their absence.
Recently, Bayyoud et al4 investigated 10 globes from 5 patients and reported on the absence of SARS-CoV-2 RNA in the cornea, AH, and conjunctiva. These findings do not necessarily contradict our results because, in their study, viral load in the blood was not examined and no patient selection was performed. In our study, two-thirds of the PCR-positive corneal discs were obtained from patients with viremia, which is uncommon in patients with COVID-19.
In general, the transmission of donor diseases to recipients is rare during a corneal transplant. Several viral diseases, including HIV type 1 and 2, hepatitis B and C, rabies, and West Nile virus, are contraindications for cornea donations, whereas other viral diseases, such as influenza, are not contraindications for cornea donations.27 In routine diagnostics, blood samples are obtained 24 hours post mortem and screened for infectious diseases. Owing to the lack of knowledge about SARS-CoV-2 in corneal tissue and its possible transmission to the graft recipient, corneal transplants from donors infected with COVID-19 or recently exposed to COVID-19 are avoided.6
According to the distribution of the viral entry receptors and the lack of ACE2 and TMPRSS2 in the corneal endothelium, a Descemet membrane endothelial keratoplasty is likely to be safer than a perforating keratoplasty. In addition, less tissue and, consequently, less viral RNA would be transplanted via a contaminated graft.
SARS-CoV-2 RNA can be detected in the corneas of patients with COVID-19 viremia. Hence, our findings might be explained by contamination through blood or AH. The probable absence of an actively replicating virus is further underlined by the missing histopathologic changes in analyzed tissue samples and by the failure of virus isolation in all samples, including some with higher RNA loads.
During coronavirus replication, the infected cells accumulate sgRNA.28 As the amount of sgRNA generally correlates to viral replication activity, the absence of sgRNA is highly indicative for the absence of replication. However, the molecular detection of sgRNA cannot necessarily prove active replication. Detected sgRNA might originate from leakage from dead cells in different tissues; moreover, it cannot be excluded that sgRNA is circulating within immune cells and/or in the blood.
Limitations
This study has some limitations, including the small sample size and the broad inclusion criteria. The small sample size and prioritization of patients with viremia, which is generally a rare condition in patients with COVID-19, poses the risk of selection bias. The percentage of positive results in corneal tissue would most likely be lower if more patients with a lower viral load had been included in this case series.
Owing to the limited number of available cases, patients with or without viremia were included in the study. Consequently, the investigated cohort is not homogenous concerning infectious conditions at the time of death. Nevertheless, because SARS-CoV-2 RNA could be detected in the corneal tissue of patients without viremia, these limitations do not seem to change the results of corneal transplant and eye banking. Further evaluation with a much larger sample size is needed to adjust the risk of viral transmission in patients without viremia, although possible transmission can never be excluded.
Conclusions
The potential of SARS-CoV-2 infection via a corneal transplant is low, but further research is warranted to assess the rate of SARS-CoV-2 transmission. The low RNA loads in corneal samples suggest a low risk of infection through a corneal transplant, even in a high-risk cohort of patients with viremia. Nevertheless, infection via a contaminated corneal graft cannot be fully excluded.
eFigure. Histology and Immunohistochemistry—Cornea
References
- 1.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127-20. doi: 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-280. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sungnak W, Huang N, Bécavin C, et al. ; HCA Lung Biological Network . SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681-687. doi: 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayyoud T, Iftner A, Iftner T, et al. Absence of severe acute respiratory syndrome-coronavirus-2 RNA in human corneal tissues. Cornea. Published online June 29, 2020. doi: 10.1097/ICO.0000000000002479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gain P, Jullienne R, He Z, et al. Global survey of corneal transplantation and eye banking. JAMA Ophthalmol. 2016;134(2):167-173. doi: 10.1001/jamaophthalmol.2015.4776 [DOI] [PubMed] [Google Scholar]
- 6.Global Alliance of Eye Bank Associations. ALERT UP-DATE: coronavirus (COVID-2019) and ocular tissue donation. Updated November 12, 2020. Accessed May 15, 2020. https://www.gaeba.org/2020/alert-coronavirus-2019-ncov-and-ocular-tissue-donation/
- 7.Püschel K, Heinemann A, Dietz E, Hellwinkel O, Henners D, Fitzek A. New developments and possibilities in the field of post-mortem medicine mortui vivos docent. Rechtsmedizin. 2020;30:425-429. doi: 10.1007/s00194-020-00402-3 [DOI] [Google Scholar]
- 8.World Medical Association . World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 9.Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383(6):590-592. doi: 10.1056/NEJMc2011400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3). doi: 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25(9). doi: 10.2807/1560-7917.ES.2020.25.9.2000152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465-469. doi: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 13.Pfefferle S, Huang J, Nörz D, et al. Complete genome sequence of a SARS-CoV-2 strain isolated in northern Germany. Microbiol Resour Announc. 2020;9(23):e00520-20. doi: 10.1128/MRA.00520-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562-569. doi: 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P, Duan F, Luo C, et al. Characteristics of ocular findings of patients with coronavirus disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 2020;138(5):575-578. doi: 10.1001/jamaophthalmol.2020.1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Liu M, Zhang Z, et al. Ocular manifestations of a hospitalised patient with confirmed 2019 novel coronavirus disease. Br J Ophthalmol. 2020;104(6):748-751. doi: 10.1136/bjophthalmol-2020-316304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiello F, Gallo Afflitto G, Mancino R, et al. Coronavirus disease 2019 (SARS-CoV-2) and colonization of ocular tissues and secretions: a systematic review. Eye (Lond). 2020;34(7):1206-1211. doi: 10.1038/s41433-020-0926-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun CB, Wang YY, Liu GH, Liu Z. Role of the eye in transmitting human coronavirus: what we know and what we do not know. Front Public Health. 2020;8:155. doi: 10.3389/fpubh.2020.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia J, Tong J, Liu M, Shen Y, Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J Med Virol. 2020;92(6):589-594. doi: 10.1002/jmv.25725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Chen X, Chen L, et al. The evidence of SARS-CoV-2 infection on ocular surface. Ocul Surf. 2020;18(3):360-362. doi: 10.1016/j.jtos.2020.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Zeng Y, Tong Y, Chen C. Ophthalmologic evidence against the interpersonal transmission of 2019 novel coronavirus through conjunctiva. Posted February 12, 2020. medRxiv 20021956. doi: 10.1101/2020.02.11.20021956 [DOI]
- 22.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843-1844. doi: 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young BE, Ong SWX, Kalimuddin S, et al. ; Singapore 2019 Novel Coronavirus Outbreak Research Team . Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488-1494. doi: 10.1001/jama.2020.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holappa M, Valjakka J, Vaajanen A. Angiotensin(1-7) and ACE2, “the hot spots” of renin-angiotensin system, detected in the human aqueous humor. Open Ophthalmol J. 2015;9:28-32. doi: 10.2174/1874364101509010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang CG, Lee KM, Hsiao MJ, et al. Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J Clin Microbiol. 2020;58(8):e01068-20. doi: 10.1128/JCM.01068-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059-1061. doi: 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desautels JD, Moshirfar M, Martheswaran T, Shmunes KM, Ronquillo YC. Risks posed to corneal transplant recipients by COVID-19-affected donors. Ophthalmol Ther. 2020;9(3):371-379. doi: 10.1007/s40123-020-00254-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1-23. doi: 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure. Histology and Immunohistochemistry—Cornea