Abstract
Background
Toxin-antitoxin (TA) systems, abundant in prokaryotes, are composed of a toxin gene and its cognate antitoxin. Several toxins are implied to affect the physiological state and stress tolerance of bacteria in a population. We previously identified a chromosomally encoded hok-sok type I TA system in Erwinia amylovora, the causative agent of fire blight disease on pome fruit trees. A high-level induction of the hok gene was lethal to E. amylovora cells through unknown mechanisms. The molecular targets or regulatory roles of Hok were unknown.
Results
Here, we examined the physiological and transcriptomic changes of Erwinia amylovora cells expressing hok at subtoxic levels that were confirmed to confer no cell death, and at toxic levels that resulted in killing of cells. In both conditions, hok caused membrane rupture and collapse of the proton motive force in a subpopulation of E. amylovora cells. We demonstrated that induction of hok resulted in upregulation of ATP biosynthesis genes, and caused leakage of ATP from cells only at toxic levels. We showed that overexpression of the phage shock protein gene pspA largely reversed the cell death phenotype caused by high levels of hok induction. We also showed that induction of hok at a subtoxic level rendered a greater proportion of stationary phase E. amylovora cells tolerant to the antibiotic streptomycin.
Conclusions
We characterized the molecular mechanism of toxicity by high-level of hok induction and demonstrated that low-level expression of hok primes the stress responses of E. amylovora against further membrane and antibiotic stressors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07376-w.
Keywords: Toxin:antitoxin, Fire blight, Phage shock protein, Transcriptome, Antibiotic tolerance
Background
Toxin-antitoxin (TA) systems are simple genetic loci that encode a stable proteinaceous toxin and an unstable counteracting antitoxin. TA systems are widely found throughout the chromosomes and plasmids of free-living prokaryotes [1]. In type I TA systems, the antitoxins are small RNAs that inhibit the translation of or facilitate the degradation of the transcript encoding the corresponding toxin (reviewed in [2, 3]). Type I toxins, such as Hok, HokB, and TisB, tend to be small (≤60 amino acids) hydrophobic proteins containing one transmembrane domain [4–6]. A high induction level of the toxin genes hok or tisB causes drastic cell death of E. coli cells, accompanied by collapse of the proton motive force (PMF) [7–9]. The gene products of both hokB and tisB form membrane pores in Escherichia coli [8, 10] and lead to leakage of cellular ATP during moderate [10] or high-level [7] induction of the toxin genes. The PMF, the proton gradient generated via oxidation of NADH and FADH2, is required to generate ATP through ATP synthase, as well as to power membrane-localized cell machinery, such as the flagellum [11, 12]. The hok/sok TA system in E. coli has been suggested as a target for killing host bacterial cells [13, 14]. Through sequestering the sRNA sok from interacting with hok mRNA by addition of anti-Sok peptide nucleic acid (PNA) oligomers [13] or doxycycline that inhibits RNase III degradation of the hok-sok dsRNA complex, hok mRNA is released and consequently causes cell death [14].
The molecular targets and regulatory roles of many TA systems are still enigmatic. Although inactivation of a single type I TA system does not frequently result in a phenotype [15], studies using low-level ectopic expression have revealed that a few membrane-associated TA systems can affect the physiological state and stress tolerance of bacteria in a population. In E. coli, expression of hokB or tisB at sub-toxic levels increased the proportion of persister cells with tolerance to multiple antibiotics, which was hypothesized to result from growth retardation following ATP leakage and the loss of the PMF [7, 8, 15–17]. Plasmid expression of the hok-sok locus also increased T4 bacteriophage exclusion in E. coli [18]. Interestingly, despite its role in compromising membrane integrity, moderate hokB expression was observed to increase metabolic activity in E. coli, determined via a fluorescent redox sensor [10].
Through transcriptomics and in vitro RNA degradation analyzes, Wang et al. demonstrated that the type V antitoxin GhoS cleaves the membrane-associated toxin ghoT mRNA [19]. However, the global transcriptional effects of a type I membrane-associated TA, to the best of our knowledge, have not been previously examined. It has been hypothesized that induction of hokB may activate phage shock protein (psp) genes, based on the protective effects of Psp proteins in mitigating various membrane stresses in E. coli [20, 21]. Though the effects vary in different bacteria, perturbation of the cell membrane seems to cause shared consequences in activating stress responses and downregulating genes that encode energy consuming machinery [22–26]. Addition of polymyxin, an antibiotic that causes formation of membrane pores and cell death in bacteria, caused increased expression of genes associated with vancomycin resistance and decreased expression of virulence factor-related genes in Staphylococcus aureus [22]; exposure of Klebsiella pneumoniae to 1-(1-Naphthylmethyl)-piperazine depolarized the membrane PMF yet upregulated many envelope stress response genes [26]. Still, it is not known whether endogenous pore-forming toxins also trigger stress response or influence the expression of virulence genes.
Recently, we identified a chromosomally encoded hok-sok type I TA system in Erwinia amylovora [27], a model enterobacterial plant-pathogenic bacterium that causes the destructive fire blight disease of pome fruit trees including apple (Malus sp.) and pear (Pyrus sp.) [28, 29]. Episomal overexpression of the hok gene caused massive killing of E. amylovora cells and arrested cell division after septa were formed [27]. We proposed that cell death due to hok induction at toxic levels in E. amylovora is likely to be associated with the disturbance of essential functions of the cell membrane. Although upregulation of toxin genes occurs under a variety of different stress conditions [30–34], natively expressed toxin genes are not known to be induced to cell-killing levels in any environmental context, to the best of our knowledge. Therefore, we hypothesized that hok might actually confer a selective advantage to E. amylovora at moderate (subtoxic) levels of induction, when no cell death is observed. In this study, we compared the transcriptome profiles of E. amylovora cultures expressing hok at toxic, subtoxic, and wild-type levels. We found that Hok plays important roles in activating ATP biosynthesis and priming the tolerance of E. amylovora cells against membrane and antibiotic damage.
Results
Moderate overexpression of hok does not suppress bacterial growth
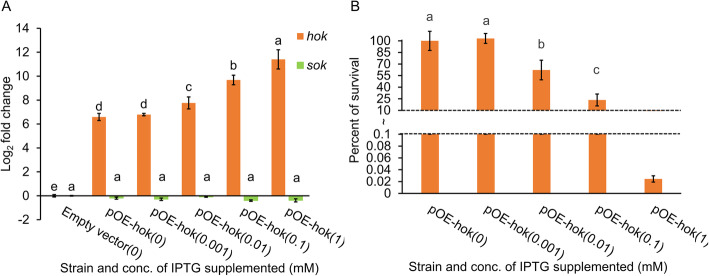
A hok overexpression construct, pOE-hok, was previously generated by cloning the E. amylovora Ea1189 hok gene into the lac promoter-containing plasmid pEVS143 [27]. The lac promoter allows low levels of transcription in the absence of the inducer isopropyl β-D-1-thiogalactopyranoside (IPTG) [35]. We did not observe any growth defect in E. amylovora Ea1189 cells transformed with pOE-hok (Fig. S1), suggesting that E. amylovora is able to tolerate leaky hok expression without inhibiting growth. Therefore, we hypothesized that Ea1189(pOE-hok) grown in the absence of IPTG induction may provide a useful system to identify the physiological roles of Hok separate from those caused by its toxicity. We used quantitative real-time PCR (qRT-PCR) to measure the expression levels of hok in Ea1189(pEVS143) and Ea1189(pOE-hok) without IPTG and in four progressively increasing doses of IPTG, and monitored the growth of the cultures in the same conditions. In the absence of IPTG, expression of hok was approximately 40-fold higher in Ea1189(pOE-hok) compared to Ea1189(pEVS143), and expression of hok increased by another ~ 130-fold when 1 mM IPTG was added to the Ea1189(pOE-hok) culture (Fig. 1a). The expression levels of the small RNA antitoxin sok remained almost unchanged in these conditions (Fig. 1a). Induction of hok did not result in cell death until expression reached about 60-fold induction or greater, induced by the addition of 0.01 mM IPTG (Fig. 1b). Henceforth, we will define hok expression from the lac promoter with 0.01 mM, 0.1 mM or 1 mM IPTG as the “toxic” expression conditions for this study, while expression from the lac promoter with 0.001 mM or no IPTG will be defined as the “subtoxic” expression conditions.
Fig. 1.
Induction of hok and its effect on cell survival in E. amylovora. a Expression levels of hok induced with four progressive doses (0.001 mM, 0.01 mM, 0.1 mM, and 1 mM) of isopropyl β-D-1-thiogalactopyranoside (IPTG) or with water. b The effect hok induction on survival rate of E. amylovora. The concentrations of IPTG supplemented are indicated in parentheses. After IPTG or water addition, cultures were incubated at 28 °C with 200 rpm shaking for 1 h. Expression levels of hok were measured using quantitative real-time PCR (qRT-PCR), and fold changes were calculated using the 2-ΔΔCT formula. The recA gene was used as an endogenous control. Survival rate was determined as the ratio of colony forming units (CFU)/ml in Ea1189(pOE-hok) after and before the addition of IPTG. Results represent the means of three replications, and error bars indicate the standard deviation. Different letters indicate significant differences (P < 0.05) using Tukey’s HSD (honestly significant difference) test. The experiments were conducted three times with similar results
Induction of hok causes PMF collapse and membrane rupture
Membrane-associated type I toxins of E. coli, including HokB and TisB, form membrane pores [8, 10, 19], and cause collapse of the PMF [9, 10, 17]. We therefore wondered if the transmembrane domain-containing E. amylovora Hok, sharing 48 and 14% amino acid identity to HokB and TisB, respectively, also causes membrane depolarization and rupture. To assess this possibility, we measured membrane potential using DiBAC4 [3] (bis-(1,3-dibutylbarbituric acid) trimethine oxonol), a membrane potential-sensitive fluorescent dye. Fluorescence level negatively correlates to membrane potential, meaning that higher fluorescence indicates a greater level of PMF collapse. Carbonyl cyanide-m-chlorophenylhydrazone (CCCP), a protonophore that uncouples the PMF, was used as a positive control for the DiBAC4 [3] staining (Fig. S2). Propidium iodide (PI) was used as an indicator of membrane rupture, which binds to nucleic acid and generates fluorescence in membrane integrity compromised cells. Ethanol disturbs the physical structure of cell membranes and was used as a positive control for the PI staining (Fig. S2). Fluorescence was measured in single cells using a flow cytometer. We found that induction of hok to subtoxic levels caused membrane depolarization and rupture in a subpopulation of cells, though many cells remained unchanged in their membrane states (Fig. 2a). More drastic membrane depolarization and rupture was observed when hok was induced to toxic levels (Fig. 2a). At the highest level of hok induction, almost the entire population was shifted to the membrane depolarization state, with varied levels of membrane rupture. We next asked whether mannitol, a bacterial metabolite that feeds into glycolysis and was shown to stimulate the PMF in E. coli [36], was able to restore the collapsed PMF and rupture of cell membrane due to the toxicity of Hok in E. amylovora. In cells expressing hok with 0.1 mM IPTG induction, mannitol partially relieved the membrane stress (Fig. 2a). Similarly, addition of mannitol significantly alleviated the inhibitory effect of bacterial growth during 0.01 or 0.1 mM induction of hok (Fig. 2b). However, when 1 mM IPTG was supplemented, the protective effect of mannitol was not observed in any of these phenotypes (Fig. 2a and Fig. 2b). Arabinose, which does not contribute to the PMF [36], was used a negative control for the assays (Fig. S3).
Fig. 2.
hok induction disturbs essential membrane functions of E. amylovora. Effect of hok induction on the proton motive force (PMF) and membrane integrity without (panel labelled as “None”) or with the addition of 10 mM mannitol (panel labelled as “Mannitol”) immediately before IPTG supplementation (a), and effect of mannitol in reversing the toxicity of Hok (b). E. amylovora cultures grown overnight for 20 h in LB broth were washed twice and diluted to OD600 = 0.2 in fresh LB broth. The concentrations of IPTG supplemented are indicated in parentheses. After incubation at 28 °C with 200 rpm shaking for 1 h, the PMF of cultures was examined using bis-(1,3-dibutylbarbituric acid) trimethine oxonol (DiBAC4 [3]), and membrane integrity was determined via propidium iodide (PI). Fluorescence was measured using a BD LSR II flow cytometer. Ten thousand events were examined with a 488 nm laser and a 530/30 emission filter (DiBAC4 [3]) staining and a 561 nm laser and a 620/15 emission filter (PI). Subsequent analyses were conducted on Flowing Software 2.5.1 and R v3.4.0. Increased fluorescence after treatment with DiBAC4 [3] or PI indicates greater collapse of the PMF or compromised membrane integrity, respectively. To test the effect of bacterial metabolites on the toxicity of Hok, 10 mM mannitol or 10 mM arabinose was added to the cultures (OD600 = 0.2) immediately before IPTG was added, and the OD600 was measured 4 h after the incubation using a Tecan spectrophotometer. Bacterial growth was monitored by measuring OD600 of the cultures using a Tecan spectrophotometer. Results represent the means of three biological replicates and error bars indicate the standard deviation. Different letters indicate significant differences (P < 0.05) using Student’s t-test. The assays were done three times with similar results
Transcriptomic analysis reveals that hok overexpression affects genes involved in stress responses and energy generation/consumption
While overexpression of E. amylovora hok causes extreme disturbance of essential membrane functions, it is not clear how the membrane disruption capacity of these toxins may affect bacterial physiology when hok is expressed in subtoxic or native expression conditions. To distinguish potential downstream effects of E. amylovora Hok from those resulting from toxicity, we compared the transcriptomes of E. amylovora cultures expressing hok at wild-type levels (i.e., wild-type strains carrying the empty vector) with cultures expressing hok at subtoxic (IPTG untreated) and toxic (1 mM IPTG treated) levels. Expression of each gene was quantified as counts per million reads (CPM), and differentially-expressed genes (DEGs) were defined as those having greater than 2-fold change of CPM values and less than 0.05 of the corresponding false discovery rate (FDR) values (Fig. S4 and Table S1).
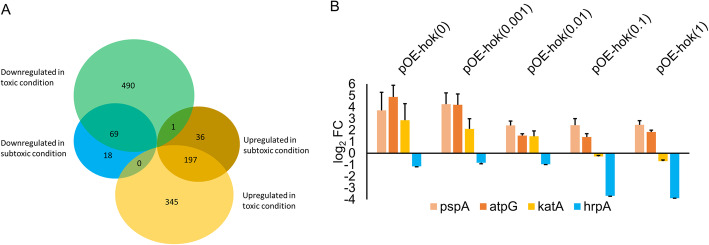
Compared with Ea1189(pEVS143), which was also untreated with IPTG, 321 DEGs were identified in IPTG-untreated Ea1189(pOE-hok), of which 234 had increased expression and 87 had decreased expression (Fig. 3a). After 1 mM IPTG treatment of Ea1189(pOE-hok), a much larger set of 541 and 560 genes were up- and down-regulated, respectively (Fig. 3a). Approximately 83% of the DEGs identified in the subtoxic condition were differentially expressed in the same direction and to a greater extent in the toxic condition. Expression of representative genes in Ea1189(pOE-hok) in subtoxic and toxic conditions was validated through qRT-PCR (Fig. 3b). The housekeeping gene recA was used as an endogenous control, that had negligible differences in expression among E. amylovora cultures expressing wild-type, subtoxic, or toxic levels of hok in our transcriptomic analysis. Based on the read count, the ratio of hok to sok was approximately 18 in the wild-type condition, that increased to ~ 200 in the subtoxic condition and ~ 6000 in the toxic condition (Fig. S5). Gene ontology (GO) enrichment analysis of the DEGs further revealed that hok exerts substantial effects in the essential metabolism of E. amylovora (Fig. 4 and Table S2). Oxidative phosphorylation-related genes (GO:0006119), that include NADH-coenzyme Q oxidoreductase (complex I), Succinate-Q oxidoreductase (complex II), Cytochrome c oxidase (complex IV) and F1Fo-ATPase (complex V), were enriched among the higher expressed genes in both toxic and subtoxic conditions. Specifically, in the toxic condition, higher expressed genes were also significantly associated with the “tricarboxylic acid cycle” GO term (GO:0006099).
Fig. 3.
Comparative transcriptomic analysis of E. amylovora cells expressing hok at wild-type, subtoxic and toxic levels, respectively. a Venn diagram of the differentially-expressed genes (DEGs) in E. amylovora cells expressing hok at subtoxic or toxic level. b Expression of representative DEGs in subtoxic or toxic condition examined using qRT-PCR. Fold changes were calculated using the 2-ΔΔCT formula. The recA gene was used as an endogenous control. The error bars indicate standard deviation. The concentrations of IPTG supplemented are indicated in parentheses
Fig. 4.
Overrepresented Gene Ontology (GO) terms enriched in the GO enrichment analysis with a cutoff FDR of 0.01. Scale bar indicates the color key of log2 fold-change values
Several genes with demonstrated importance to bacterial plant pathogenesis were negatively affected by elevated hok expression. Specifically, hrpA and flhD, encoding a T3SS protein and a flagellar transcriptional activator, respectively, decreased in expression at both levels of hok induction. In toxic but not subtoxic conditions, down-regulated genes were primarily comprised of flagellar genes and “protein secretion” (GO:0009306) genes, which included type II secretion system (T2SS) and type III secretion system (T3SS)-related genes.
Induction of hok also activated multiple genes involved in stress responses. Several genes with known roles in antibiotic persistence and other stress responses, i.e. groS, groL, dnaK, dnaJ, skp, surA, sucB and lon [37–42], were consistently more highly expressed in both hok induction conditions. Also upregulated were genes in the “response to virus” ontology (GO:0009615), including genes encoding phage shock proteins, i.e. pspABCD, and CRISPR-associated proteins. The catalase gene katA showed increased expression in the subtoxic condition, consistent with our previous observation that catalase activity is significantly compromised in a hok-sok deletion mutant [27]. The stress-induced ATP-dependent chaperone gene clpB was also more highly expressed in the subtoxic but not the toxic condition. Together, these results show that different hok expression levels exert diverse and overlapping effects on the E. amylovora transcriptome, enhancing expression of metabolic and stress-related traits while suppressing genes required for infection.
hok positively affects ATP biosynthesis
Membrane-associated type I toxins have been shown to cause leakage of cellular ATP as indicated by either decrease level of intracellular ATP or increase level of extracellular ATP [7, 10, 19]. In this study, we found that genes associated with oxidative phosphorylation, the process of ATP generation through electron transfer, were higher expressed in the subtoxic condition and were higher expressed to a greater extent in the toxic condition (Fig. 5). We hypothesized that the upregulation of ATP biogenesis-related genes could be part of a response to compensate for the possible leakage of intracellular ATP through increased ATP synthesis in Ea1189(pOE-hok) cultures in both subtoxic and toxic conditions. To determine whether ATP leakage was occurring, we performed simultaneous measurements of both the intracellular and the extracellular levels of ATP in both subtoxic and toxic conditions. When induced with 0.1 or 1 mM IPTG, conditions causing more than 70% dieoff (Fig. 1b), E. amylovora Hok caused dramatic leakage of ATP from the cells, indicated by the decreased level of intracellular ATP and increased level of extracellular ATP (Fig. 5 and Fig. S6). In contrast, a significant increase in intracellular ATP was measured after induction with 0.01 mM or less IPTG (Fig. 5 and Fig. S6), expression conditions that were associated with minimal or no cell death of E. amylovora (Fig. 1b). No ATP leakage was observed in these subtoxic conditions.
Fig. 5.
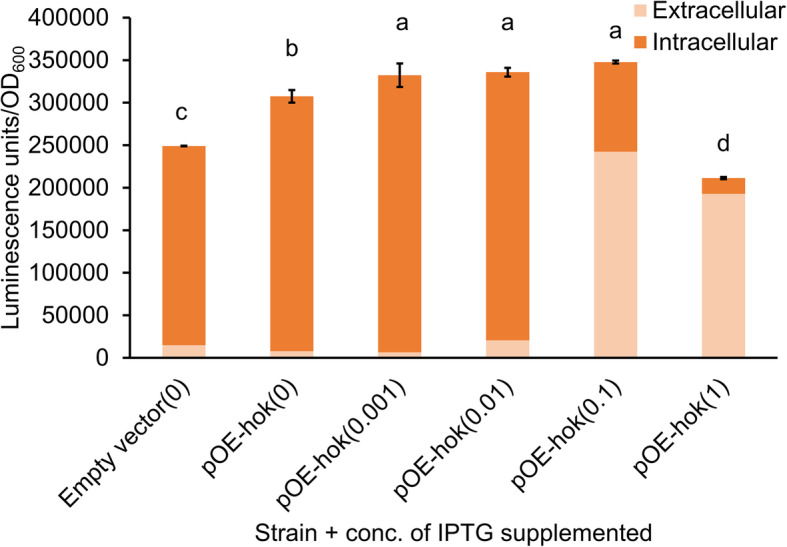
Effect of hok induction on ATP biosynthesis in E. amylovora. Both extracellular and intracellular levels of ATP were simultaneously quantified using a luciferase reporter system. Results represent the means of three biological replications and error bars indicate the standard deviation. Different letters indicate significant differences (P < 0.05) using Tukey’s HSD test. The assays were done twice with similar results
Combining intercellular and extracellular ATP measurements allowed us to assess the total ATP concentration under each expression condition. In the absence of IPTG, total ATP was greater in Ea1189(pOE-hok) cultures than Ea1189(pEVS143). Total ATP in Ea1189(pOE-hok) increased with IPTG addition at concentrations up to 0.1 mM (Fig. 5). At the highest concentration of IPTG tested, 1 mM, the total ATP in Ea1189(pOE-hok) cultures started to decrease compared with lower levels of inducer, likely due to the massive kill-off of ATP-generating cells at this induction level. Taken together, our results suggest that hok positively affects the biosynthesis of ATP, and leakage of ATP only occurs when hok was induced at toxic levels.
Overexpression of the ATP synthase gene atpB is toxic to E. coli cells; it allows leakage of protons through the F0 sector of F1Fo-ATPase [43–46]. Given that hok positively affects ATP synthase gene expression and ATP biosynthesis in subtoxic conditions, we wondered if the toxicity of Hok was increased by the upregulation in ATP synthase genes. To test this hypothesis, we generated ATP synthase gene deletion mutants, Ea1189ΔatpB and Ea1189ΔatpBEFHAGDC. The growth of Ea1189ΔatpB and Ea1189ΔatpBEFHAGDC mutants was severely reduced, as overnight cultures only reached OD600 ≈ 0.3 compared with OD600 ≈ 1.5 in the wild-type Ea1189 strain (data not shown). pOE-hok was transformed into the ATP synthase mutants to generate Ea1189ΔatpB(pOE-hok) and Ea1189ΔatpBEFHAGDC(pOE-hok), respectively. Hok expression was induced in the wild-type and ATPase mutant backgrounds with 1 mM IPTG, and survival rates were measured. Hok killing efficiency was not changed between the wild-type and the mutants (Fig. S7), suggesting that the toxicity of Hok is not affected by the increased expression of ATP biosynthesis genes.
Expression of pspA is induced in known PMF dissipation conditions and relieves the toxicity of Hok
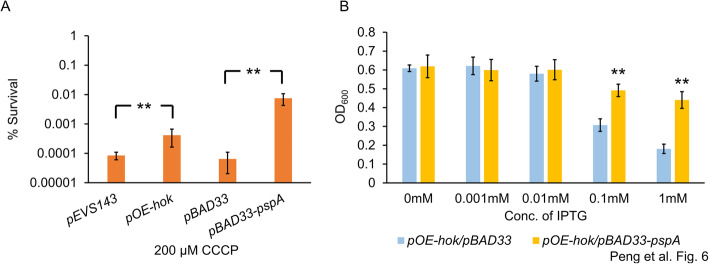
Our transcriptome results indicated that psp genes were upregulated in both expression conditions. The psp genes are induced on exposure to conditions that dissipate the PMF, such as bacteriophage infection, alkaline pH, and addition of uncoupling agents, in both Gram-negative and -positive bacteria (reviewed in [47]). The protective roles of PspA in managing membrane stresses have been validated in E. coli and Salmonella enterica serovar Typhimurium [48–50]. As the functions of psp genes have not been previously investigated in E. amylovora, we constructed a transcriptional fusion of the promoter region of the pspABCD operon to a green fluorescence protein (gfp) reporter. As expected, the promoter activity of the pspABCD operon was significantly increased in E. amylovora cells after exposure to bacteriophage, and was increased to a lesser extent in the presence of CCCP, ethanol, or Triton X-100 (Fig. S8). To examine the possible protective role of pspA under the condition of membrane stress in E. amylovora, we generated the pspA-overexpression construct, pBAD33-pspA, through cloning the pspA gene into the pBAD33 plasmid, containing the arabinose-inducible PBAD promoter. Compared with Ea1189(pBAD33), Ea1189(pBAD33-pspA) cultures were ~ 100 times more tolerant to CCCP (Fig. 6a). Interestingly, without supplementing any IPTG, Ea1189(pOE-hok) cultures survived at significantly higher rates than Ea1189(pEVS143) (Fig. 6a), suggesting that induction of hok at subtoxic levels protect E. amylovora cells from further membrane damage by activating the expression of pspA. Interestingly, pspA overexpression significantly alleviated the toxicity due to high levels of hok induction (Fig. 6b), further validating the defensive role of pspA in response to membrane stress in E. amylovora.
Fig. 6.
Effect of pspA in managing membrane stresses in E. amylovora. a Carbonyl cyanide-m-chlorophenylhydrazone (CCCP) was added to 200 μM to E. amylovora cultures and incubated for 5 h. Survival rate was determined as the ratio of CFU/ml after the treatment to that before the treatment. Arabinose at 10 mM, but no IPTG, was supplemented to the cultures. b Overexpression of pspA largely reverses the killing of E. amylovora cells expressing hok at toxic levels. Growth of cultures was determined by measuring OD600 using a Tecan spectrophotometer. Arabinose at 10 mM was supplemented to the cultures. Results represent the means of four biological replications and error bars indicate the standard deviation. Asterisk symbols indicate significant differences (P < 0.05 using Student’s t-test). The assays were done three times with similar results
Subtoxic expression of hok increases tolerance of stationary-phase E. amylovora cells to the aminoglycoside antibiotic streptomycin
Transcriptome results showed that hok expression upregulated several genes previously associated with antibiotic persistence, so we next asked whether hok has a role in antibiotic tolerance during stationary phase. Without addition of IPTG, stationary phase E. amylovora cultures expressing hok had 10 times the number of survivors to streptomycin exposure than the vector control strain (Fig. 7). concentration that is routinely used for management of fire blight and screening of streptomycin-resistant E. amylovora isolates [51–53]. Of note, we did not observe altered tolerance of E. amylovora cultures overexpressing pspA, suggesting that hok does not affect antibiotic tolerance through overproduction of PspA.
Fig. 7.
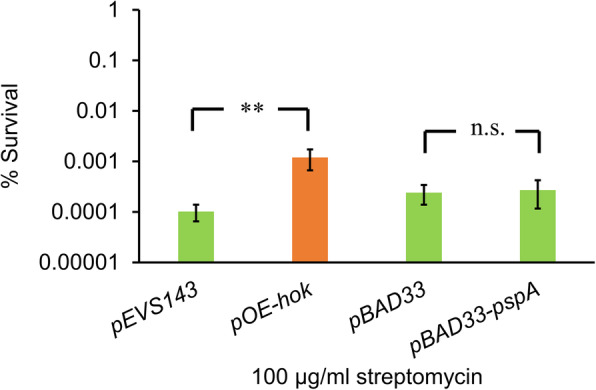
Subtoxic level of hok increases tolerance of stationary-phase E. amylovora cells to streptomycin. Single colonies of E. amylovora cultures were grown in LB broth amended with selective antibiotics for 20 h to reach stationary phase. Cultures were washed twice using fresh LB broth before streptomycin was added to 100 μg/ml; the culture was subsequently incubated at 28 °C with shaking for 5 h. Survival rate was determined as the ratio of CFU/ml after the treatment to that before the treatment. Arabinose at 10 mM and no IPTG was supplemented into the cultures. Results represent the means of four biological replications and error bars indicate the standard deviation. Asterisk symbols indicate significant differences (P < 0.05 using Student’s t-test). The assays were done three times with similar results
Discussion
In this study, by taking advantage of the “leaky” expression of the lac promoter, we showed that induction of the type I toxin hok disturbs essential functions of the cell membrane in both subtoxic and toxic conditions. In an effort to understand the physiological roles of hok in subtoxic and native expression conditions, we examined the transcriptomic changes in E. amylovora cells expressing hok at subtoxic and toxic levels. We demonstrated that subtoxic expression of hok in E. amylovora stimulates ATP biogenesis, activates pspA expression to protect cells from further membrane damage, and renders stationary phase cell cultures more tolerant to streptomycin.
Consistent with observations in E. coli expressing tisB at a toxic level or hokB at a subtoxic level, induction of E. amylovora hok at both subtoxic and toxic levels caused collapse of the PMF and membrane rupture in a subpopulation of the cultures. Interestingly, although induction of hok with 1 mM IPTG killed more than 97% of E. amylovora cells, a significant subpopulation of cells retained low levels of membrane rupture under the same conditions. Therefore, the culturability of E. amylovora cells expressing a toxic level of hok is not entirely correlated with the level of membrane rupture. Addition of mannitol alleviated the toxicity of Hok at lower induction levels, indicating that collapse of the PMF indeed contributes to the toxicity of Hok. However, mannitol did not reduce toxicity when Hok was expressed to the highest levels. Mannitol provides the PMF through glycolysis, and the massive killing due to high-level hok induction may severely impair the central metabolism machineries of E. amylovora.
Our transcriptomic profiling experiments demonstrate that induction of hok positively affects the expression of all the F1Fo-ATPase genes. F1Fo-ATPase, consist of a proton-conducting structure and a catalytic portion, exhibits a central role in energy transduction in bacteria. In line with the transcriptomic changes, quantification of intracellular and extracellular ATP showed that induction of hok positively affects ATP biogenesis, though leakage of ATP from cells was observed in toxic conditions. Although leakage of cellular ATP has also been suggested in a few other membrane-associated toxins at toxic conditions in E. coli, only intracellular [7, 54] or extracellular ATP [10] alone was measured.
Though a subtoxic level of hok triggered primarily higher expression of genes in E. amylovora, a greater proportion of DEGs were negatively affected in cells expressing toxic levels of hok, including genes of flagellum, type II secretion system, and type III secretion system. A previous study of a Lon protease mutant in E. amylovora showed that Lon negatively affects the expression of pathogenesis-related genes hrpA and flhD [55]. In this study, lon expression was increased, while hrpA and flhD expression was reduced, by more than two-fold in both hok expression conditions. This suggests that induction of hok inactivates these energy-consuming cell machineries that are important for the pathogenesis of E. amylovora, and this favors energy conservation during the stress.
The transcriptome analysis in this study showed that hok triggers higher expression of psp genes in E. amylovora. Though not previously studied in E. amylovora, Psp proteins and homologs are widely found in bacteria, archaea, and plants [47]. Bacterial Psp proteins are activated in response to phage, extreme temperature, ethanol, mislocalization of outer membrane secretins, and other events that cause membrane dissipation [23, 30, 31, 56–61]. We demonstrated that the promoter activity of E. amylovora pspA was induced upon exposure to several membrane stress-inducing substances, and that overexpression of pspA increased tolerance to Hok. These results show that in addition to protection from external stresses, PspA can serve a protective function from the effects of bacterially produced stresses, such as toxin-antitoxin systems.
We found that subtoxic hok induction increased tolerance of stationary cultures of E. amylovora to the aminoglycoside antibiotic streptomycin. This is consistent with a previous report that low-level expression of the hok homolog hokB confers increased antibiotic tolerance to E. coli [10]. Although energy state is an important factor for persister development in different bacteria [34, 62], we did not observe any leakage of ATP from E. amylovora cells expressing subtoxic levels of hok. Instead, ATP levels were significantly higher in these conditions. Given that several other genes associated with persistence were also upregulated during hok overexpression [37–40, 63], it is possible that hok affects antibiotic tolerance in E. amylovora through induction of stress response genes that help cells to mitigate antibiotic stressors. While the importance of type II TA systems in antibiotic survival is still being debated, this study adds to the body of evidence that Type I systems may play such a role.
To our knowledge, this is the first study of the transcriptomic response to a type I membrane-disrupting toxin. Overall, the response of E. amylovora to hok expression appears highly congruent with bacterial responses to other PMF-dissipating stresses. Alkaline conditions, in which the PMF is dissipated to maintain the inverted pH gradient, results in upregulation of ATP synthases and downregulation of chemotaxis genes in E. coli [64]. The membrane-targeting antibiotic polymyxin strongly upregulates metabolic pathways while repressing key virulence factor genes in Staphylococcus aureus [22]. Exposure to the membrane destabilizer 1-(1-naphthylmethyl)-piperazine causes upregulation of many stress response genes, including dnaJ, dnaK, and pspABCD [26]. Although it is still unclear whether Hok exhibits any direct regulatory roles, the overlapping downstream transcriptional effects due to hok induction and other membrane perturbing agents suggest that formation of membrane pore is at least one of the main reasons for the transcriptomic changes observed during hok induction.
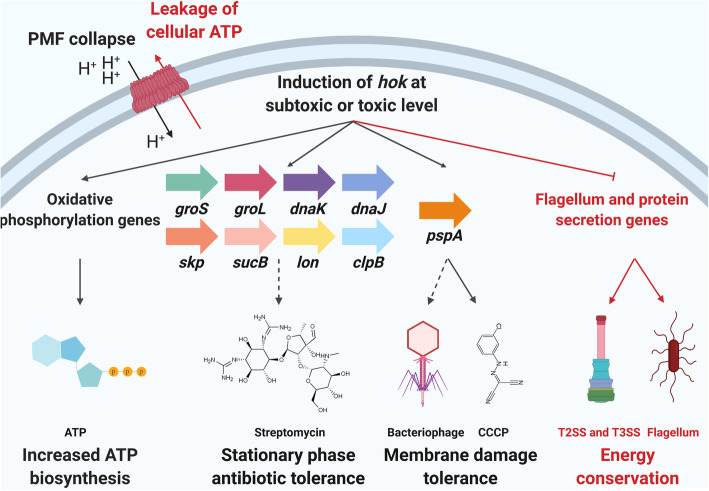
A working model summarizing the effects of hok induced at subtoxic or toxic levels in E. amylovora is shown in Fig. 8. The shared phenotypes of membrane-associated type I TA systems that have been investigated in this study and previous studies are compared (Table S3).
Fig. 8.
Working model of the effects of hok induction at subtoxic or toxic level. Induction of hok at a subtoxic level causes dissipation of the PMF, increased ATP biosynthesis, increased tolerance to antibiotics and further membrane stress (black arrows and letters). Induction of hok at a toxic level leads to leakage of cellular ATP and inactivation of energy-consuming cell machineries including T2SS, T3SS, and flagellum (red arrows and letters)
Conclusions
By examining the physiological and transcriptomic changes of Erwinia amylovora cells expressing the type I toxin/antitoxin system toxin gene hok at subtoxic or toxic levels, we demonstrated that low-level expression of hok, while not affecting bacterial culturability, triggers expression of the ATP synthase genes and overproduction of cellular ATP. Low-level expression of hok also activates multiple genes associated with stress response, and triggers expression of the phage shock protein gene pspA, which then functions to protect E. amylovora cells from further membrane stressors. This study contributes to the idea that stress management is an important selective advantage of TA systems when these systems are under low level expression conditions. Although the E. coli Hok has been implied as a target for killing host bacterial cells [13, 14], the significant transcriptomic and physiological changes during E. amylovora hok overexpression presented in this study suggest that additional considerations are requisite in applying this toxin to fire blight disease management.
Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. amylovora strains were routinely grown at 28 °C in Luria-Bertani (LB) agar or broth. Unless stated otherwise, the following antibiotics were supplemented in the medium at the concentrations indicated: ampicillin (Ap; 100 μg/ml), chloramphenicol (Cm; 15 μg/ml), or kanamycin (Km; 25 μg/ml). For tolerance screening, streptomycin was applied at 100 μg/ml.
Table 1.
Bacteria strains or plasmids used in this study and their relevant characteristics
| Strain or plasmid | Genotype | Reference | |
|---|---|---|---|
| Strains | Ea1189 | Wild type | [65] |
| Ea1189ΔatpB | atpB deletion mutant | This study | |
| Ea1189ΔatpBC | Deletion of the chromosomal region that spans the atpB, atpE, atpF, atpH, atpA, atpG, atpD, and atpC genes | This study | |
| Plasmids | pKD3 | Contains FRT-flanked Cmr cassette sites; R6K ori; Cmr | [66] |
| pKD46 | Contains lambda Red recombinase induced by L-Arabinose; R101 ori; Apr | [66] | |
| pEVS143 | Broad-host-range cloning vector; IPTG inducible Cmr; pES213 ori; Kmr | [67] | |
| pOE-hok | pEVS143 cmR::hok; overexpression vector; Kmr | [27] | |
| pBAD33 | Broad-host-range cloning vector; arabinose inducible Cmr; pACYC18 ori | [68] | |
| pBAD33-pspA | pBAD33 cmR::pspA; overexpression vector; Cmr | This study | |
| pPROBE-NT | Broad-host-range promoter-probe vector; pBBR1 ori; Kmr | [69] | |
| pPROBE-pspA | pPROBE-NT::pspA; native promoter of pspA in pPROBE-NT; Kmr | This study | |
DNA manipulations
The atpB gene or the chromosomal region that spanned the entire ATP synthase gene cluster (atpB, atpE, atpF, atpH, atpA, atpG, atpD, and atpC) was deleted from E. amylovora Ea1189 using the λ Red recombinase system [65, 66] to generate Ea1189ΔatpB and Ea1189ΔatpBC, respectively. The pBAD33-pspA and pPROBE-NT:pspA constructs were generated by cloning the coding region of pspA or the 500 bp upstream the coding region into pBAD33 and pPROBE-NT, respectively, using the FastCloning approach [70]. The oligonucleotide primers used in this study are listed in Table 2. E. coli and E. amylovora cells were transformed routinely using heat shock and electroporation approaches, respectively.
Table 2.
Oligonucleotide primers used in this study
| Primer Name | Sequence (5′-3′) | Reference |
|---|---|---|
| Primers used for making knockout mutants | ||
| atpBC_F | ATGGCTGCAGGAGAAATCTCTACGCCGCAAGAGTACATAGGTCATCATCTGTGTAGGCTGGAGCTGCTTC | This study |
| atpBC_R | TTACATCGCGTTTTTGGTCAACTCGATCACGCGCAGTTTGGCGATCGCTTCATATGAATATCCTCCTTA | This study |
| atpB_R | TCAATGTTCTTCAGATGCCATCGACAGATAGACAATCGTTAAGACCATGACATATGAATATCCTCCTTA | This study |
| Primers used for the overexpression construct of pspA | ||
| pBAD_FC_F | GGCTGTTTTGGCGGATGAGA | This study |
| pBAD_FC_R | AATTCGCTAGCCCAAAAAAACGG | This study |
| pspA_FC_F | CCGTTTTTTTGGGCTAGCGAATTATGGGTATTTTTTCACGTTTTGCCG | This study |
| pspA_FC_R | TCTCATCCGCCAAAACAGCCTTATTCACCGATACGCCGGTT | This study |
| Primers used for confirming knockout of genes | ||
| atpBC_CF | CCGGCTGTAATTAACAACAAAG | This study |
| atpBC_CR | TTTCCTGACTGGCCTTCT | This study |
| atpB_CR | CCATGTACAGCAGATCCATATT | This study |
| Primers used for the transcriptional fusion construct of pspA | ||
| pspA_tsc_F | CGACCTGAATGGAAGCCGGCGCCAGTTCGTCGAGAAACAAC | This study |
| pspA_tsc_R | GAGCTCGGTACCCGGGGATCCTCAATCAAATTCCTCATCAGTCTGG | This study |
| pPROBE-NT_tsc_F | GAGGATCCCCGGGTACCGAGCTC | [27] |
| pPROBE-NT_tsc_R | GCCGGCTTCCATTCAGGTCG | [27] |
| Primers used for qRT-PCR | ||
| qhok_F | TGGTGCGTACTTATAGTGTGTG | This study |
| qhok_R | CCGGATTCGTAAGCCATGAA | This study |
| qpspA_F | GACCTGATTGCTGCTTTGC | This study |
| qpspA_R | GGTTTCAGCCAGTTTACTTTCC | This study |
| qatpG_F | GTCGGCTATCTGGTCGTATCT | This study |
| qatpG_R | GCCTTTATCAGCCCAGGATTT | This study |
| qkatA_F | CGCGACTGGGTGTTAACTATAA | This study |
| qkatA_R | TGGATCAGAGGCAAGATCAATAC | This study |
| qhrpA_F | AGCACTTCAGCATCCAAGAC | This study |
| qhrpA_R | CGAGTTCTGCGTATCCATCTTC | This study |
Growth arrest assay
E. amylovora cultures grown overnight for 20 h in LB broth were washed twice and diluted to OD600 = 0.2 in fresh LB broth. Serially diluted IPTG (1 mM, 0.1 mM, 0.01 mM, 0.001 mM) or H2O was supplemented, and the cultures were incubated at 28 °C with 200 rpm shaking for 1 h. Survival rate was determined by the ratio of colony forming units (CFUs) per ml calculated through dilution plating in Ea1189(pOE-hok) to that in Ea1189(pEVS143). To test the effect of bacterial metabolites on the toxicity of Hok, cultures were prepared the same as the growth arrest assay, except that 10 mM mannitol or 10 mM arabinose was added to the cultures right before IPTG was added, and the OD600 was measured 4 h after the incubation using a spectrophotometer plate reader (Tecan; Männedorf, Switzerland).
Measurement of membrane polarity and membrane integrity through flow cytometry
Changes in membrane polarity and membrane integrity was measured using bis-(1,3-dibutylbarbituric acid) trimethine oxonol (DiBAC4 [3]; Thermo Fisher Scientific; Waltham, MA) and propidium iodide (PI; Thermo Fisher Scientific), respectively, following a published protocol [71] with minor modifications. DiBAC4 [3] is a slow-response potential-sensitive dye that enters depolarized cells and exhibits fluorescence (excitation/emission = 490 nm/516 nm) after binding to membrane proteins. Propidium iodide exhibits fluorescence (excitation/emission, 535 nm/617 nm) after binding to DNA of membrane integrity-compromised cells. Briefly, E. amylovora cultures grown for 20 h were washed twice with fresh LB and adjusted to OD600 = 0.5. Mannitol or arabinose at 10 mM or H2O was supplemented followed by addition of serially diluted IPTG (1 mM, 0.1 mM, 0.01 mM, 0.001 mM) or H2O of the same volume for 1 h at 28 °C with shaking. Cells were washed twice with 0.5× Phosphate-Buffered Saline (PBS) to the original volume, and 250 nM DiBAC4 [3] or 20 μg/ml PI was added. The mixtures were incubated in a 28 °C incubator for 1 h without shaking. Fluorescence was measured using a BD LSR II flow cytometer (BD Biosciences; Franklin Lakes, NJ) equipped with a 488 nm laser and a 530/30 emission filter for DiBAC4 [3] staining and a 561 nm laser and a 620/15 emission filter for PI staining. A minimum of 10,000 events were collected for each sample. Subsequent analyses were conducted using Flowing Software 2.5.1 and R v3.4.0.
Total RNA extraction
Cultures of Ea1189(pEVS143) and Ea1189(pOE-hok) grown for 20 h were washed twice and diluted to OD600 = 0.2 in fresh LB broth. H2O or 1 mM IPTG was supplemented to Ea1189(pOE-hok) cultures, representing induction of hok at subtoxic and toxic level, respectively. Ea1189(pEVS143) cultures supplemented with H2O of the same volume represented cells expressing wild-type level of hok. Each treatment had three biological replicates. Bacterial cells were harvested 1 h after the addition of IPTG or H2O. Total RNA extraction was modified from a previous reported method [72]. Briefly, cell pellets of E. amylovora cultures were resuspended in 200 μl of 0.1% N-lauroyl sarcosine sodium salt followed by centrifugation at 13,000×g for 1 min. The cell pellets were resuspended in 100 μl acetate/SDS solution (1% SDS in 10 mM EDTA and 50 mM sodium acetate, pH 5.1) and incubated for 5 min in boiling water. After centrifugation, RNA was extracted from the supernatants using the RNA Clean & Concentrator-25 kit (Zymo Research; Irvine, CA) following manufacturer’s instructions. Residual DNA contamination was removed by TURBO DNA-free Kit (Thermo Fisher Scientific) following the manufacturer’s instructions.
Library preparation and sequencing
The quality of RNA was analyzed on a 2100 Bioanalyzer (Agilent Technologies; Santa Clara, CA). The integrity of RNA was examined via electrophoresis on a 1% agarose gel. RNA libraries were prepared by Illumina TruSeq Stranded Total RNA Library Prep Kit (bacteria) (Illumina; San Diego, CA). Sequencing was conducted with the 50 bp single-end format on the Illumina HiSeq 4000 platform at the Michigan State University Research Technology Support Facility.
Data analysis
Adaptor sequences and low-quality reads were filtered through Trimmomatic v0.32 with following parameters: ILLUMINACLIP:$ADAPTOR:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:30 [73]. The remaining reads were mapped to the genome of E. amylovora ATCC 49964 using Bowtie 2 v2.3.4.3 [74]. Counts of reads for each annotated gene were obtained using HTSeq v0.6.1 [75]. TMM-normalized CPM were called via edgeR [76]. DEGs were defined as greater than 2-fold change of CPM value and less than 0.05 of the corresponding FDR value. GO enrichment analysis was conducted on AgriGO v2.0 [77], with a cutoff FDR of 0.01. Redundant GO terms were filtered through REVIGO using the default setting [78].
qRT-PCR
Cultures were grown in the same condition as the RNA-seq experiment and total RNA from three biological replicates was extracted and pooled together. cDNA was synthesized from 1000 ng total RNA using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific) following manufacturer’s instructions. Gene expression levels were quantified via qRT-PCR using a StepOne Plus Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) under the routine condition [79] with three replications. The housekeeping gene recA was used as an endogenous reference gene [79]. Fold changes of gene expression were quantified via the 2-ΔΔCT method.
Quantification of intracellular and extracellular ATP
BacTiter-Glo™ reagent (Promega; Madison, WI) was prepared following the manufacturer’s instructions. The BacTiter-Glo™ reagent supports bacterial lysis, ATP extraction, and ATP quantification based on the luminescent signal from a thermostable luciferase. E. amylovora cultures were grown and treated in the same condition as that of the growth arrest assay in this study. To measure intracellular ATP levels, cultures were washed twice and resuspended in 0.5× PBS of the same volume to remove any extracellular ATP. One hundred μl of the culture suspension was mixed with equal volume of the BacTiter-Glo™ reagent prepared in a 96-well plate. The mixtures were incubated at room temperature for 5 min before luminescence was measured. Extracellular ATP levels were measured following the same procedure for intracellular ATP measurement, except that supernatants of the cultures from the first centrifugation were used for quantification. Total ATP was defined as the sum up of the intracellular and the extracellular levels of ATP.
Promoter activity of pspABCD operon and CCCP tolerance assay
Promoter activity of pspABCD was monitored in E. amylovora Ea1189 cultures containing the transcriptional fusion construct pPROBE-NT:pspA. The overnight cultures of Ea1189(pPROBE-NT:pspA) were washed and resuspended in LB broth to a final OD600 of 1.0. The cultures were incubated with approximately 108 plaque forming units of bacteriophage ΦEa31–3 [80], 40 μM CCCP, 5% ethanol, or 0.1% Triton X-100 for 5 h. The Gfp fluorescence was measured on a spectrophotometer plate reader (Tecan). To measure tolerance of E. amylovora cultures during membrane stress, 200 μM CCCP was supplemented to the washed E. amylovora cultures at OD600 = 1.0 followed by incubation for 5 h. Survival rate was determined as the ratio of CFU/ml after the treatment to that before the treatment.
Tolerance of stationary phase cultures to streptomycin
Single colonies of E. amylovora cultures were in grown in LB broth amended with selective antibiotics for 20 h to reach stationary phase. Cultures were washed twice by fresh LB broth before subjecting to 100 μg/ml streptomycin. After incubation at 28 °C with shaking for 5 h, cultures were washed once and serially diluted in 0.5× PBS buffer, and plated in LB plates without any antibiotics. Survival rate was determined as the ratio of CFU/ml after the treatment to that before the treatment.
Supplementary Information
Acknowledgments
This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program Grant no. 2015-67013-23068 from the USDA National Institute of Food and Agriculture and by Michigan State University AgBioResearch. We thank Antonet Svircev, Agriculture and Agri-Food Canada, for the gift of E. amylovora bacteriophage ΦEa31-3.
Abbreviations
- TA
Toxin-antitoxin
- PMF
Proton motive force
- PNA
Peptide nucleic acid
- psp
Phage shock protein
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- qRT-PCR
Quantitative real-time PCR
- bis-(1,3-dibutylbarbituric acid) trimethine oxonol
DiBAC4 (3)
- CCCP
Carbonyl cyanide-m-chlorophenylhydrazone
- CPM
Counts per million reads
- DEGs
Differentially-expressed genes
- FDR
False discovery rate
- T2SS
Type II secretion system
- T3SS
Type III secretion system
- gfp
Green fluorescence protein
- LB
Luria-Bertani
- PI
Propidium iodide
- PBS
Phosphate-Buffered Saline
- GO
Gene Ontology
- CFU
Colony forming unit
- Ap
Ampicillin
- Cm
Chloramphenicol
- Km
Kanamycin
Authors’ contributions
JP and GWS designed the study and experiments, JP conducted the experiments, and collected and analyzed the data, JP, LRT, and GWS interpreted the results and wrote the manuscript. The author(s) read and approved the final manuscript.
Funding
This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program Grant no. 2015–67013-23068 from the USDA National Institute of Food and Agriculture and by Michigan State University AgBioResearch. The funders had no role in experimental design, data collection, data analysis, data interpretation or writing of the manuscript.
Availability of data and materials
The reference genome sequence of E. amylovora was obtained from GenBank with the accession number of FN666575. The datasets generated and analyzed during the current study are available in the National Center for Biotechnology Information Sequence Read Archive repository (https://www.ncbi.nlm.nih.gov/sra) with the following accession number: SRR10489885-SRR10489893. The E. amylovora strains and mutants analyzed in this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests in this section.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Unterholzner SJ, Poppenberger B, Rozhon W. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elem. 2013;3(5):e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brielle R, Pinel-Marie ML, Felden B. Linking bacterial type I toxins with their actions. Curr Opin Microbiol. 2016;30:114–121. doi: 10.1016/j.mib.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012;9(12):1488–1490. doi: 10.4161/rna.23045. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32(5):1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 5.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38(11):3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thisted T, Gerdes K. Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J Mol Biol. 1992;223(1):41–54. doi: 10.1016/0022-2836(92)90714-U. [DOI] [PubMed] [Google Scholar]
- 7.Unoson C, Wagner EG. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol Microbiol. 2008;70(1):258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]
- 8.Gurnev PA, Ortenberg R, Dörr T, Lewis K, Bezrukov SM. Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 2012;586(16):2529–2534. doi: 10.1016/j.febslet.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes K, Bech FW, Jørgensen ST, Løbner-Olesen A, Rasmussen PB, Atlung T, et al. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilmaerts D, Bayoumi M, Dewachter L, Knapen W, Mika JT, Hofkens J, et al. The persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. mBio. 2018;9(4):e00744–e00718. doi: 10.1128/mBio.00744-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogawa S, Lee TM. The relation between the internal phosphorylation potential and the proton motive force in mitochondria during ATP synthesis and hydrolysis. J Biol Chem. 1984;259(16):10004–10011. doi: 10.1016/S0021-9258(18)90918-X. [DOI] [PubMed] [Google Scholar]
- 12.Streif S, Staudinger WF, Marwan W, Oesterhelt D. Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. J Mol Biol. 2008;384(1):1–8. doi: 10.1016/j.jmb.2008.08.057. [DOI] [PubMed] [Google Scholar]
- 13.Faridani OR, Nikravesh A, Pandey DP, Gerdes K, Good L. Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Res. 2006;34(20):5915–5922. doi: 10.1093/nar/gkl750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chukwudi CU, Good L. The hok/sok toxin/antitoxin locus enhances bacterial susceptibility to doxycycline. bioRxiv. 2020;2020.02.13:948752. [Google Scholar]
- 15.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell. 2015;59(1):9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Edelmann D, Berghoff BA. Type I toxin-dependent generation of superoxide affects the persister life cycle of Escherichia coli. Sci Rep. 2019;9(1):14256. doi: 10.1038/s41598-019-50668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dörr T, Vulić M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8(2):e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178(7):2044–2050. doi: 10.1128/JB.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Lord DM, Cheng H-Y, Osbourne DO, Hong SH, Sanchez-Torres V, et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012;8(10):855–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jovanovic G, Lloyd LJ, Stumpf MP, Mayhew AJ, Buck M. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem. 2006;281(30):21147–21161. doi: 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes K. Hypothesis: type I toxin-antitoxin genes enter the persistence field-a feedback mechanism explaining membrane homoeostasis. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2016;371(1707):20160189. doi: 10.1098/rstb.2016.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, Cheah S-E, Roberts KD, Nation RL, Thompson PE, Velkov T, et al. Transcriptomic analysis of the activity of a novel polymyxin against Staphylococcus aureus. mSphere. 2016;1(4):e00119–e00116. doi: 10.1128/mSphere.00119-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fallico V, Ross RP, Fitzgerald GF, McAuliffe O. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, D-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J Virol. 2011;85(22):12032–12042. doi: 10.1128/JVI.00275-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urfer M, Bogdanovic J, Lo Monte F, Moehle K, Zerbe K, Omasits U, et al. A Peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli. J Biol Chem. 2016;291(4):1921–1932. doi: 10.1074/jbc.M115.691725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han ML, Zhu Y, Creek DJ, Lin YW, Gutu AD, Hertzog P, et al. Comparative metabolomics and transcriptomics reveal multiple pathways associated with polymyxin killing in Pseudomonas aeruginosa. mSystems. 2019;4:1. doi: 10.1128/mSystems.00149-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anes J, Sivasankaran SK, Muthappa DM, Fanning S, Srikumar S. Exposure to sub-inhibitory concentrations of the chemosensitizer 1-(1-naphthylmethyl)-piperazine creates membrane destabilization in multi-drug resistant Klebsiella pneumoniae. Front Microbiol. 2019;10:92. doi: 10.3389/fmicb.2019.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng J, Triplett LR, Schachterle JK, Sundin GW. Chromosomally encoded hok-sok toxin-antitoxin system in the fire blight pathogen Erwinia amylovora: identification and functional characterization. Appl Environ Microbiol. 2019;85:e00724–e00719. doi: 10.1128/AEM.00724-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatterjee A. Fire blight: The disease and its causative agent, Erwinia amylovora. Edited by J.L. Vanneste. European Journal of Plant Pathology. 2001;107(5):569. doi: 10.1023/A:1011254217275. [DOI] [Google Scholar]
- 29.Malnoy M, Martens S, Norelli JL, Barny MA, Sundin GW, Smits TH, et al. Fire blight: applied genomic insights of the pathogen and host. Annu Rev Phytopathol. 2012;50:475–494. doi: 10.1146/annurev-phyto-081211-172931. [DOI] [PubMed] [Google Scholar]
- 30.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186(24):8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen-Dalsgaard M, Jørgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol. 2010;75(2):333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5(12):e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K. ATP-dependent persister formation in Escherichia coli. mBio. 2017;8:1. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oehler S, Eismann ER, Krämer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9(4):973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473(7346):216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma C, Sim S, Shi W, Du L, Xing D, Zhang Y. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol Lett. 2010;303(1):33–40. doi: 10.1111/j.1574-6968.2009.01857.x. [DOI] [PubMed] [Google Scholar]
- 38.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51(6):1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 39.Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology (Reading, England) 2007;153(Pt 9):3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 40.Hansen S, Lewis K, Vulić M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008;52(8):2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goltermann L, Good L, Bentin T. Chaperonins fight aminoglycoside-induced protein misfolding and promote short-term tolerance in Escherichia coli. J Biol Chem. 2013;288(15):10483–10489. doi: 10.1074/jbc.M112.420380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costello SM, Plummer AM, Fleming PJ, Fleming KG. Dynamic periplasmic chaperone reservoir facilitates biogenesis of outer membrane proteins. Proc Natl Acad Sci. 2016;113(33):E4794–EE800. doi: 10.1073/pnas.1601002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Meyenburg K, Jørgensen BB, Michelsen O, Sørensen L, McCarthy JE. Proton conduction by subunit a of the membrane-bound ATP synthase of Escherichia coli revealed after induced overproduction. EMBO J. 1985;4(9):2357–2363. doi: 10.1002/j.1460-2075.1985.tb03939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Venkat S, Wilson J, McGuire P, Chang AL, Gan Q, et al. Genome-wide quantification of the effect of gene overexpression on Escherichia coli growth. Genes. 2018;9:8. doi: 10.3390/genes9080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arechaga I, Miroux B, Runswick MJ, Walker JE. Over-expression of Escherichia coli F1Fo-ATPase subunit a is inhibited by instability of the uncB gene transcript. FEBS Lett. 2003;547(1–3):97–100. doi: 10.1016/S0014-5793(03)00677-X. [DOI] [PubMed] [Google Scholar]
- 46.Na YA, Lee JY, Bang WJ, Lee HJ, Choi SI, Kwon SK, et al. Growth retardation of Escherichia coli by artificial increase of intracellular ATP. J Ind Microbiol Biotechnol. 2015;42(6):915–924. doi: 10.1007/s10295-015-1609-6. [DOI] [PubMed] [Google Scholar]
- 47.Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, et al. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev. 2010;34(5):797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 48.Kleerebezem M, Crielaard W, Tommassen J. Involvement of stress protein PspA (phage shock protein a) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 1996;15(1):162–171. doi: 10.1002/j.1460-2075.1996.tb00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein a (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol. 2007;66(1):100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- 50.Becker LA, Bang IS, Crouch ML, Fang FC. Compensatory role of PspA, a member of the phage shock protein operon, in rpoE mutant Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005;56(4):1004–1016. doi: 10.1111/j.1365-2958.2005.04604.x. [DOI] [PubMed] [Google Scholar]
- 51.McGhee GC, Guasco J, Bellomo LM, Blumer-Schuette SE, Shane WW, Irish-Brown A, et al. Genetic analysis of streptomycin-resistant (SmR) strains of Erwinia amylovora suggests that dissemination of two genotypes is responsible for the current distribution of SmRE. amylovora in Michigan. Phytopathology. 2010;101(2):182–191. doi: 10.1094/PHYTO-04-10-0127. [DOI] [PubMed] [Google Scholar]
- 52.Förster H, McGhee GC, Sundin GW, Adaskaveg JE. Characterization of streptomycin resistance in isolates of Erwinia amylovora in California. Phytopathology. 2015;105(10):1302–10. doi: 10.1094/PHYTO-03-15-0078-R. [DOI] [PubMed] [Google Scholar]
- 53.Sundin GW, Wang N. Antibiotic resistance in plant-pathogenic bacteria. Annu Rev Phytopathol. 2018;56(1):161–180. doi: 10.1146/annurev-phyto-080417-045946. [DOI] [PubMed] [Google Scholar]
- 54.Cheng HY, Soo VW, Islam S, McAnulty MJ, Benedik MJ, Wood TK. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ Microbiol. 2014;16(6):1741–1754. doi: 10.1111/1462-2920.12373. [DOI] [PubMed] [Google Scholar]
- 55.Lee JH, Ancona V, Zhao Y. Lon protease modulates virulence traits in Erwinia amylovora by direct monitoring of major regulators and indirectly through the Rcs and Gac-Csr regulatory systems. Mol Plant Pathol. 2018;19(4):827–840. doi: 10.1111/mpp.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivastava D, Moumene A, Flores-Kim J, Darwin AJ. Psp stress response proteins form a complex with mislocalized secretins in the Yersinia enterocolitica Cytoplasmic Membrane. mBio. 2017;8(5):e01088–e01017. doi: 10.1128/mBio.01088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brissette JL, Russel M, Weiner L, Model P. Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990;87(3):862–866. doi: 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darwin AJ. The phage-shock-protein response. Mol Microbiol. 2005;57(3):621–628. doi: 10.1111/j.1365-2958.2005.04694.x. [DOI] [PubMed] [Google Scholar]
- 59.Model P, Jovanovic G, Dworkin J. The Escherichia coli phage-shock-protein (psp) operon. Mol Microbiol. 1997;24(2):255–261. doi: 10.1046/j.1365-2958.1997.3481712.x. [DOI] [PubMed] [Google Scholar]
- 60.Flores-Kim J, Darwin AJ. The phage shock protein response. Annu Rev Microbiol. 2016;70:83–101. doi: 10.1146/annurev-micro-102215-095359. [DOI] [PubMed] [Google Scholar]
- 61.Armstrong RM, Adams KL, Zilisch JE, Bretl DJ, Sato H, Anderson DM, et al. Rv2744c is a PspA ortholog that regulates lipid droplet homeostasis and nonreplicating persistence in Mycobacterium tuberculosis. J Bacteriol. 2016;198(11):1645–1661. doi: 10.1128/JB.01001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 63.Wu N, He L, Cui P, Wang W, Yuan Y, Liu S, et al. Ranking of persister genes in the same Escherichia coli genetic background demonstrates varying importance of individual persister genes in tolerance to different antibiotics. Front Microbiol. 2015;6:1003. doi: 10.3389/fmicb.2015.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187(1):304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edmunds AC, Castiblanco LF, Sundin GW, Waters CM. Cyclic Di-GMP modulates the disease progression of Erwinia amylovora. J Bacteriol. 2013;195(10):2155–2165. doi: 10.1128/JB.02068-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72(1):802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/JB.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller WG, Leveau JH, Lindow SE. Improved gfp and inaZ broad-host-range promoter-probe vectors. Molecular plant-microbe interactions : MPMI. 2000;13(11):1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 70.Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clementi EA, Marks LR, Roche-Håkansson H, Håkansson AP. Monitoring changes in membrane polarity, membrane integrity, and intracellular ion concentrations in Streptococcus pneumoniae using fluorescent dyes. JoVE. 2014;84:e51008. doi: 10.3791/51008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivas R, Vizcaíno N, Buey RM, Mateos PF, Martínez-Molina E, Velázquez E. An effective, rapid and simple method for total RNA extraction from bacteria and yeast. J Microbiol Methods 2001;47(1):59–63. [DOI] [PubMed]
- 73.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed]
- 74.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England). 2015;31(2):166–9. [DOI] [PMC free article] [PubMed]
- 76.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics (Oxford, England). 2008;9(2):321–32. [DOI] [PubMed]
- 77.Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45(W1):W122–W1w9. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeng Q, McNally RR, Sundin GW. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J Bacteriol. 2013;195(8):1706–1717. doi: 10.1128/JB.02056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gill JJ, Svircev AM, Smith R, Castle AJ. Bacteriophages of Erwinia amylovora. Appl Environ Microbiol. 2003;69(4):2133–2138. doi: 10.1128/AEM.69.4.2133-2138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The reference genome sequence of E. amylovora was obtained from GenBank with the accession number of FN666575. The datasets generated and analyzed during the current study are available in the National Center for Biotechnology Information Sequence Read Archive repository (https://www.ncbi.nlm.nih.gov/sra) with the following accession number: SRR10489885-SRR10489893. The E. amylovora strains and mutants analyzed in this study are available from the corresponding author upon reasonable request.