Abstract
Humans are a colonized with trillions of commensal microorganisms which exert a profound effect on normal host physiology and immune function through an abundance of genetic and metabolic by-products. Although the commensal microbiome has beneficial functions to host physiology, perturbations of the composition of the commensal microbiome or the homeostatic mucosal environment can lead to the induction of immune pathology and systemic inflammation. In the context of cancer progression or response to immune therapy, this inflammation can be detrimental, resulting in tumor growth and the promotion of immune suppression. On the other hand, significant associations have been identified whereby certain commensal microorganisms are able to enhance T cell function or are required for tumor control in cancer patients treated with certain immune therapies and chemotherapies. The focus of this chapter is to highlight the role of the commensal microbiome during tumor progression and in response to immune therapies.
Keywords: microbiome, commensal dysbiosis, cancer, immunotherapy, immunosuppression
Introduction
The cellular environment within solid tumors influences response to therapy and outcome in cancer. T cells are one of the only cell types capable of exerting spontaneous pressure against an evolving tumor, and frequencies of intratumoral T cells associate with significant survival benefit for multiple types of solid tumors (Zhang et al., 2003) (Brambilla et al., 2016; Erdag et al., 2012; Mahmoud et al., 2011). However, the tumor microenvironment (TME) diminishes anti-tumor T cell function through several mechanisms, including metabolic constraint of T cell function, recruiting suppressive myeloid cells into the TME, and establishing physical barriers that prohibit T cell entry into tumors. Thus, much of cancer research today focuses on enhancing T cell-mediated killing of tumors through immune therapy.
Immune therapy for cancer has been considered since 1891 when William Coley reported that administration of streptococcal organisms into sarcoma patients led to a reduction in tumor burden (Coley, 1891). While this particular treatment is not used today, newer forms of immune therapy are capable of achieving long-term disease-free survival for recurrent metastatic cancer, and in some cases, complete remission of disease. The primary goal of immune therapy is to re-invigorate T cell-mediated immune surveillance. However, significant variability in efficacy still exists for multiple cancer types. Increased T cell numbers and signatures of enhanced cytotoxic effector molecules, such as interferon (IFN)γ and granzyme B, correlate with increased response to immunotherapy and chemotherapy for multiple tumors (Melero et al., 2014; Sistigu et al., 2014; Stoll et al., 2014). On the other hand, studies have demonstrated that frequencies of myeloid cells (Kim et al., 2018; Krieg et al., 2018) or PD-1/PD-L1 expression within tumors (Daud et al., 2016) also associate with response to immune therapies. Although a multitude of tumor-associated biomarkers currently predict the ability of immune therapies to enhance T cell-mediated killing of established tumors, there is little consensus that any one particular mechanism associates with response across multiple tumor types. Moreover, recent studies have demonstrated that previously undefined host-intrinsic factors, such as the composition of the commensal microbiome, also have a significant influence on the efficacy of immune therapy.
The commensal microbiota is composed of bacteria, archaea, viruses, fungi and other eukaryotic species. Commensal microorganisms inhabit all mucosal barrier surfaces, with the most abundant population residing within the gut. The number of commensal microorganisms present in and on the human body is estimated to be approximately equal to the number of somatic cells within the host (Sender et al., 2016). In addition to their vast numbers, the microbiota also has incredible genetic diversity. For example, within the gut, approximately 1,000 species of bacteria have been identified that contain roughly 2,000 genes per microbial species (Gilbert et al., 2018). Therefore, compared to the estimated 20,000 protein-coding genes in humans, commensals in the gut have 2,000,000 genes (Turnbaugh et al., 2007). In essence, humans are a metaorganism – comprised of commensal microorganisms and human cells – where the genetic and metabolic products of the microbiota exert a profound influence upon normal host physiology, immune function, and pathology.
Although the commensal microbiome has beneficial functions, perturbations of the composition of the commensal microbiome or the homeostatic mucosal environment can lead to the induction of immune pathology and systemic inflammation. In the context of cancer progression or response to immune therapy, this inflammation can be detrimental, resulting in tumor growth and aiding in immune suppression. The microbiota and their microbial products have been implicated in the progression of inflammatory diseases, such as inflammatory bowel disease and colitis, as well as in many types of cancers, including colorectal, hepatocellular, and breast cancers. However, it is also clear that certain commensal microorganisms enhance T cell function and are required for tumor control in cancer patients treated with certain immune therapies. The focus of this chapter is to highlight the role of the commensal microbiome during tumor progression and in response to immune therapies.
Systemic outcomes associated with microbiome composition and function
Healthy individuals and microbial homeostasis
To appreciate the effects of the commensal microbiome on tumor progression and response to therapy, we will first discuss microbiome composition and function in healthy individuals. The microbiota of the intestinal tract is important in the development of the gastrointestinal immune system (Rhee et al., 2004). Commensal gut microbes have evolutionarily established and maintained a symbiotic relationship with their human hosts, specifically in the context of the human immune system (Silva et al., 2015). The microbiota provide a myriad of benefits to the host, such as aiding in the digestion of food, production of specific types of vitamins, protection from opportunistic pathogens, activation of different types of immune cells, and promotion of intestinal barrier function (Kamada et al., 2013).
The majority of microbiome colonization occurs in the short period of time after birth (Koenig et al., 2011). Human and mouse newborns initiate gut microbe colonization following exposure to maternal and environmental microorganisms, and the composition of the gut microbiome becomes well-established after weaning (Maynard et al., 2012; Weng and Walker, 2013). The influence of host genetics upon microbiome composition is suggested to contribute to microbiome composition. Although early studies suggested that monozygotic twins showed no more similarity in their gut microbiome composition than dizygotic twins (Ridaura et al., 2013; Turnbaugh et al., 2009; Turnbaugh et al., 2010), larger-scale studies of twin cohorts demonstrated that genetic differences have a significant impact on microbiome composition (Goodrich et al., 2016; Goodrich et al., 2014). Additional studies (Falony et al., 2016; Zhernakova et al., 2016) and re-analysis of the large twin cohort (Rothschild et al., 2018) have demonstrated that environmental factors such as diet, medication use, and lifestyle choices also significantly shape the composition of the microbiome. Genetic factors likely have an early influence on microbiome composition, whereas the effects of environmental factors may persist throughout the lifetime of the host. For example, it has recently been demonstrated that age-dependent Toll-like receptor (TLR) 5 expression within the intestinal epithelium negatively regulates the colonization of flagellated bacteria in neonatal mice, whereas the effects of TLR5 expression against flagellated commensals are reduced in adult mice (Fulde et al., 2018).
Early-life factors influencing the composition of commensal microorganisms also influence the early evolution of the immune system. For example, maternal acquisition of commensal-specific IgG through breast milk limits the development of T cell responses against commensal microorganisms (Koch et al., 2016). Mice that did not receive maternal IgG developed transient immune dysregulation and systemic inflammation, which eventually resolved in adult mice (Koch et al., 2016). The effect of the microbiome on immune function was observed decades ago in germ-free mice that were bred and maintained in the complete absence of microorganisms. These initial studies demonstrated that germ-free mice had defects in the development of multiple lymphoid organs (Bauer et al., 1963), defective myelopoiesis, and increased susceptibility to certain bacterial and viral pathogens due to reduced myeloid function (Abt et al., 2012a; Balmer et al., 2014; Clarke et al., 2010a; Ichinohe et al., 2011a; Khosravi et al., 2014).
Mechanistic studies demonstrated that commensal production of short-chain fatty acids, through fermentation of fiber, led to the development of mucosal-associated and peripheral Foxp3+ regulatory T cells (Arpaia et al., 2013; Furusawa et al., 2013; Lathrop et al., 2011). Additional studies have demonstrated that commensals are required for the development of IL-17-producing αβ T cells (Ivanov et al., 2009b; Shaw et al., 2012), γδ T cells (Duan et al., 2010) and invariant NKT cells (Olszak et al., 2012; Wei et al., 2010). Immune surveillance and sensitivity to antigens are also influenced by the composition of microbiome. Certain commensals also provide tonic stimulation to innate immune cells, through TLR or nucleotide-binding, oligomerization domain-containing protein (NOD) signaling pathways, resulting in decreased activation thresholds and increased anti-viral and bactericidal function (Abt et al., 2012b; Clarke et al., 2010b; Ichinohe et al., 2011b). Corresponding studies in humans have demonstrated that the composition of the gut microbiome attenuates the ability of immune cells to produce specific cytokines in response to pathogenic stimuli (Schirmer et al., 2016). For example, it was shown that degradation of tryptophan to tryptophol by commensal species is associated with reduced IFNγ production when PBMCs were stimulated with fungal pathogens (Schirmer et al., 2016). Additionally, palmitoleic acid synthesis reduced production of the proinflammatory cytokines IL-6, IL-1β, and tumor necrosis factor (TNF)α from monocytes (Schirmer et al., 2016). These effector cytokines are not only important for normal immune function and the maintenance of immune homeostasis, but have also been implicated in cancer progression or anti-tumor immune responses. Similarly, Bifidobacterium, a commensal determinant of immune responsiveness in melanoma patients treated with PD-1/PD-L1 blockade (Matson et al., 2018; Sivan et al., 2015), was linked with T cell production of IFNγ (Schirmer et al., 2016). These studies in healthy individuals offer insight into potential mechanisms whereby commensal microorganisms influence tumor-promoting inflammation and anti-tumor T cell immunity (Figure 1a).
Figure 1: Commensal microorganisms are crucial for the maintenance of immune homeostasis.
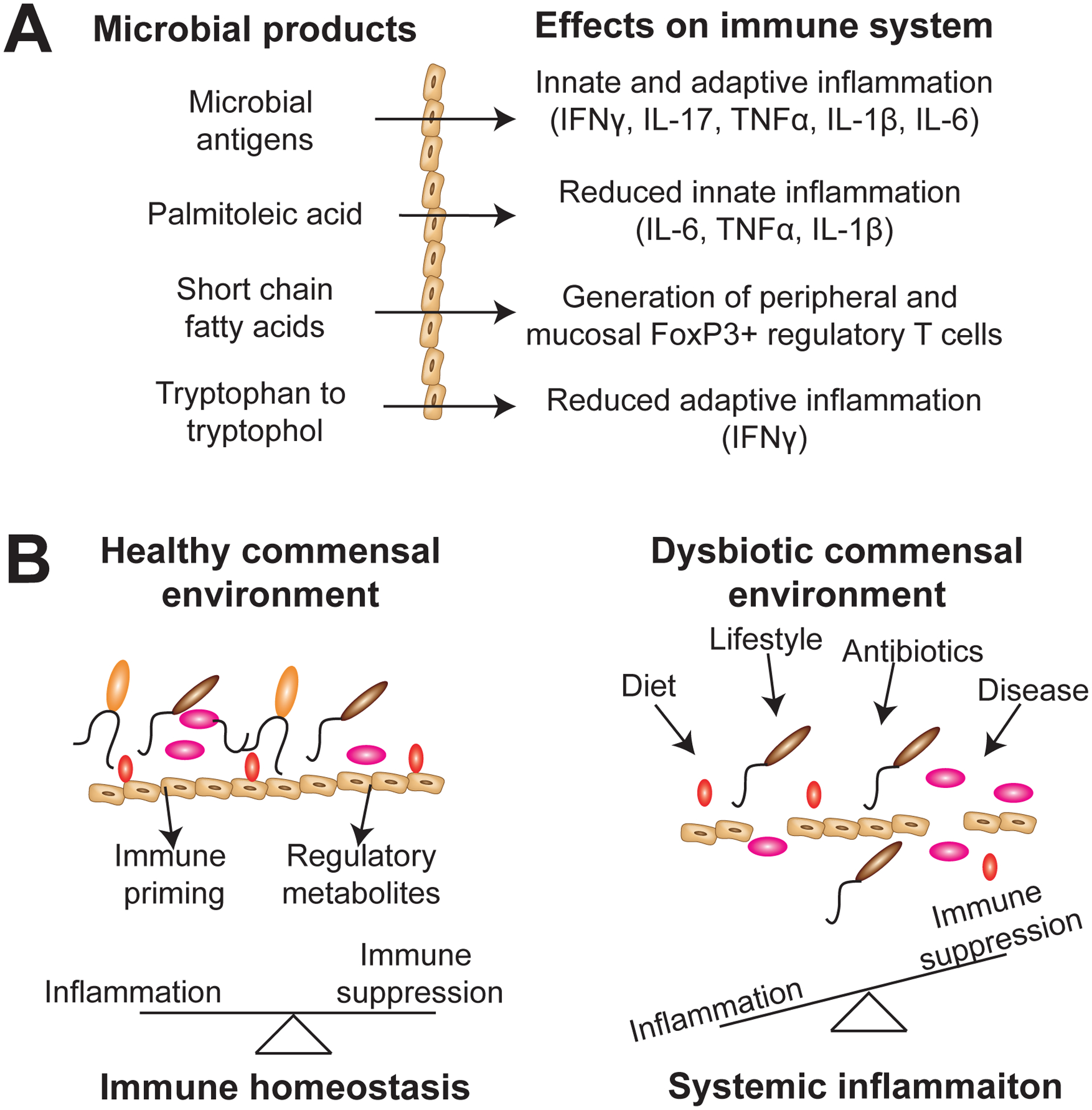
A) Microbial antigens activate microbe-specific T cells to produce IFNγ and/or IL-17 or induce myeloid cells through stimulation of TLR and NOD signaling pathways to produce inflammatory cytokines such as TNFα, IL-1β, and IL-6. Microbial metabolites such as palmitoleic acid, short chain fatty acids, and degradation of tryptophan to tryptophol result in reduced inflammation through modulation of innate-mediated inflammation, the generation of regulatory FoxP3+ T cells, or reducing IFNγ production from T cells. B) A balanced commensal microbiome results in balanced inflammatory and immune regulatory signals, which result in local and systemic immune homeostasis. However, factors such as poor diet, lifestyle factors, antibiotic usage, and disease can all negatively influence the balance of the commensal ecosystem resulting in dysbiosis. Intestinal leakage and translocation of commensals, enrichment for inflammatory commensals, and a loss of immune regulatory commensals results in local and systemic inflammation.
Commensal dysbiosis and immune pathology
Changes within the composition and/or the functional attributes of the microbiome associate with several diseases, including cancer. Commensal dysbiosis, the shift from commensal homeostasis to an environment in which the balance of symbionts and pathobionts has been perturbed or negatively altered, has adverse effects on the host. This is in part due to the fact that the beneficial capabilities of many resident, symbiotic microbes are lost (Silva et al., 2015). Many of these benefits are involved in maintaining or aiding effective immune responses within the gut and systemic periphery. Commensal microorganisms have dynamic control on immune homeostasis through induction of inflammatory and/or regulatory immune responses at mucosal surfaces. For example, segmented filamentous bacteria (SFB), which are more abundant in the small intestines of mice from Taconic Biosciences, are required for the robust generation of mucosal-associated Th17 T cells (Ivanov et al., 2009a). This occurs through presentation of SFB-associated antigens to CD4 T cells, resulting in polarization of mucosal-associated Th17 T cells (Goto et al., 2014; Lecuyer et al., 2014). Conversely, polysaccharide A from the symbiont Bacteroides fragilis promotes the differentiation of Foxp3+ regulatory T cells which restrain pathogenic Th17 responses at mucosal surfaces, promoting mucosal tolerance (Round et al., 2011). These studies underscore the dynamic relationship between inflammatory and regulatory commensals, highlighting a scenario where commensal dysbiosis can result in mucosal or systemic pathology through the loss of immune homeostasis (Figure 1b) (Silva et al., 2015).
Dysbiotic shifts within the microbiome that enable the outgrowth of pathogenic or pathogen-associated commensals are frequently related to specific diseases, including cancer. In addition to the gut, dysbiosis of the microbiota can also occur within the reproductive tract, skin, lungs, and other mucosal surfaces where commensals reside. Fusobacterium has been associated with oral infections (Griffen et al., 2012; Moore and Moore, 1994), adverse pregnancy outcomes (Dixon et al., 1994; Han et al., 2010), and gastrointestinal disorders (Kostic et al., 2013; McCoy et al., 2013). Within the intestines, colorectal cancer has been associated with increased prevalence of Fusobacterium, Porphyromonas, Peptostreptococcus, Parvimonas, and Enterobacter genera (Lozupone et al., 2013). It was reported that skin colonization of Staphylococcus epidermidis, a commensal capable of producing 6-N-hydroxyaminopurine, inhibited the proliferation of melanoma tumor cell lines and reduced tumor growth in mouse models of melanoma (Nakatsuji et al., 2018). Although skin and tumor samples from melanoma patients were not assessed, this study demonstrated the potential protective effects that local commensal microorganisms may have in preventing oncogenesis.
Patients with breast cancer have also been reported to have a distinct breast tissue microbiome. Differences were demonstrated within tissues adjacent to breast malignancies when compared to normal tissue within the same individual (Xuan et al., 2014a), or when compared to tissues taken from women with benign disease (Hieken et al., 2016). As reviewed by Fernandez et al, multiple follow-up studies in women with breast cancer have validated that breast cancer patients harbor a distinct microbiome within the mammary tissue and distally within the gut (Fernandez et al., 2018). Significant alterations in the oral microbiome have been reported in pancreatic cancer, where patients with cancer have significantly reduced levels of Neisseria elongata and Streptococcus mitis (Farrell et al., 2012). Although the significance of these findings has yet to be experimentally validated, these reports provide comprehensive evidence that multiple cancers are associated with changes to the local and systemic microbiome. Further investigation is required to elucidate whether these changes promote tumorigenesis or occur in response to cancer. Clarifying the role for specific tissue- and gut-specific commensal microorganisms in the prevention or progression of these cancers could pave the way towards targeting the microbiome for therapeutics, as discussed below.
Alterations in the microbial composition of the female reproductive tract associate with reproductive disorders such as endometritis and adenomyosis (Chen et al., 2017). Increased levels of proinflammatory cytokines such as IFNγ, IL-1β, TNF-α, and granulocyte macrophage-colony stimulating factor (GM-CSF) also associate with dysbiosis within the reproductive tract (Anderson et al., 2011). One study that evaluated the composition of bacterial species within the cervical microenvironment found that dysbiosis associates with alterations in the cervical mucosal barrier, increasing the levels of proinflammatory cytokines and proteins associated with proteolysis and increased cell death within the cervical microenvironment (Borgdorff et al., 2016). Similarly, changes in lung microbial diversity and composition in patients with asthma associate with increased airway hyperresponsiveness (Huang et al., 2011) and increased inflammation (Simpson et al., 2016). Although these studies did not evaluate the effects of local dysbiosis on cancer occurrence or outcome, these results suggest that changes to the microbiome composition affect local tissue immunity and pathology.
Commensal dysbiosis can occur in response to antibiotic usage, changes in diet, obesity, and genetic polymorphisms (Buchta Rosean and Rutkowski, 2017). Commensal dysbiosis is associated with reduced gene richness and microbial diversity, as has been shown in obese individuals (Le Chatelier et al., 2013). It was demonstrated that in obesity, microbial gene signatures are altered that associate with mucus degradation, oxidative stress management, and decreased production of immune regulatory molecules, while frequencies of inflammatory bacterial species are increased. These associations link the dysbiotic reduction of gene richness to increased inflammation and mucosal damage (Le Chatelier et al., 2013). Thus, dysbiosis driven by alterations in host diet can ultimately lead to both intestinal and systemic inflammation through disruption of homeostasis within the intestinal microenvironment.
Intestinal barrier damage and translocation of microbial products
One consequence of commensal dysbiosis is the negative impact it can have on maintaining the integrity of the intestinal barrier. When this barrier is compromised, bacteria can translocate from the intestinal lumen to peripheral, systemic sites (Hand and Belkaid, 2010). Bacterial translocation activates pattern recognition receptors on cells, most notably the membrane-bound TLRs and the cytoplasmic NOD-like receptors. Many of the receptors that fall under these two categories have been implicated in tumor initiation and tumor proliferation (Schwabe and Jobin, 2013) due to their ability to promote inflammation. Indeed, translocation of commensal bacteria due to reduced intestinal barrier function has been associated with both autoimmune diseases and cancer (Tlaskalova-Hogenova et al., 2011).
Under homeostatic conditions within the intestinal lumen, commensal microbes and their by-products do not induce immune pathology. However, some bacterial components or by-products such as lipopolysaccharide (LPS), peptidoglycan, and flagellin, when present systemically, illicit systemic inflammation. Additionally, some bacterial components have been implicated in the initiation and/or progression of multiple types of cancers (Buchta Rosean and Rutkowski, 2017). A well-studied example is LPS, a cell wall component of Gram-negative bacteria. LPS has been shown to increase both tumor angiogenesis and metastasis through stimulation of endothelial cells to produce vascular endothelial growth factor (VEGF) (Pollet et al., 2003). VEGF increases the permeability of host vasculature, which aids in tumor cell dissemination and eventual metastatic disease (Harmey et al., 2002). In macrophages and other immune cells, LPS stimulation enhances production of proinflammatory cytokines such as IL-6, TNFα, and IL-8 – all of which have been implicated in tumor progression and immune regulation. Thus, it is likely that continued exposure of bacterial LPS and the subsequent chronic inflammation associated with systemic LPS signaling can negatively impact the host during cancer progression.
Flagellin, a protein that forms the flagellum of many motile flagellated bacteria, has been categorized as having both beneficial and harmful effects on the host immune system depending upon the concentration, location, and targets throughout the host. Immune and epithelial cell inflammatory cytokine production is activated through recognition of flagellin via TLR5. Translocation of flagellated bacteria or shed flagellum from the gut into circulation has been implicated in inducing systemic immune responses (Grimes et al., 2016; Svard et al., 2015). It has been shown that sustained or out of context TLR5-mediated inflammation promotes lung inflammation (Liaudet et al., 2003) in addition to inducing cardiac (Rolli et al., 2010) and liver pathology (Xiao et al., 2015). Thus, there is precedence to suggest that in certain inflammatory contexts, TLR5 engagement by translocated flagellin can lead to excessive inflammation. Supporting this, it was previously demonstrated that IL-6-mediated inflammation during ovarian cancer and in soft tissue sarcomas occurs in response to TLR5-mediated recognition of commensal microbes, resulting in sustained T cell dysfunction and enhanced tumor growth (Rutkowski et al., 2015). TLR5−/− mice or antibiotic-sterilized wild-type mice had reduced IL-6 signaling, functional anti-tumor T cells, and reduced tumor burden. These findings suggest that interactions with the host microbiome, through TLR5 signaling, regulate the pro-tumorigenic, IL-6-mediated inflammatory axis in certain types of cancer.
Peptidoglycan, a cell-wall component in many species of bacteria, has been shown to prime the innate immune system systemically when it is recognized by NOD1. NOD1 is a pattern recognition receptor that affects the functioning of neutrophils, which play an important role in innate immunity and in the killing of extracellular pathogens. Peptidoglycan shed from commensal bacteria is excreted or transported out of the gut into the systemic periphery, where it is recognized by NOD1 and prompts neutrophil killing under basal conditions (Clarke et al., 2010a) while also enabling the survival of circulatory phagocytic cells during homeostasis (Hergott et al., 2016). Systemic levels of peptidoglycan in the absence of infection were shown to prime the immune system to promote a more rapid response to extracellular pathogens and/or infections. Dysbiosis can thus present a consequence whereby the levels of bacteria that produce peptidoglycan are reduced, resulting in decreased priming of neutrophils and phagocytic cells during tumor progression.
Role of microbiota in cancer initiation and progression
Through influence on host inflammatory and pathogenic responses, gut dysbiosis greatly impacts the initiation and progression of multiple types of cancer (Sheflin et al., 2014). It has been shown that certain microbes within the gut can induce inflammation, enhance cellular proliferation, and produce by-products that impact the host immune response. When imbalanced, it is these traits that can subsequently lead to the promotion of tumorigenesis and cancer progression (Zou et al., 2018).
The impact of the microbiome on cancer progression in colorectal cancer (CRC) is well-studied. In CRC, dysbiosis results in a decrease in regulatory commensal species, leading to increased inflammation. More specifically, this inflammation arises due to the production of secondary bile acids, such as deoxycholic acid (DCA), from Clostridium species (Yoshimoto et al., 2013). DCA also has pro-inflammatory roles in other cancer types, as it promotes the development of hepatocellular carcinoma (HCC) in obese mice (Yoshimoto et al., 2013).
Additionally, Fusobacterium nucleatum induces inflammation and proliferation in CRC through invasion of cancer cells and activation of β-catenin signaling through FadA binding to E-cadherin (Rubinstein et al., 2013). Inflammation in this context results in increased production of nitric oxide synthase, increasing the number of facultative anaerobic bacteria present in the gut and further promoting dysbiosis (Winter et al., 2013). Pro-inflammatory responses driven by the host immune system can also disrupt the intestinal barrier, which can allow for the translocation of different strains of gut bacteria to distal sites (Brenchley and Douek, 2012). This often intensifies the overall inflammatory response that is observed in the host, as bacteria and their by-products become systemically distributed (Sheflin et al., 2014). In the case of CRC, it has been demonstrated that barrier disruption and microbial invasion of adenomas leads to IL-23 production by tumor-associated myeloid cells, inducing pro-tumorigenic, IL-17-producing Th17 cells (Grivennikov et al., 2012). Further mechanistic insight into the role of the microbiome during liver cancer confirmed that secondary bile acids increase liver cancer metastasis and reduce anti-tumor immune surveillance through inhibition of CXCL16, resulting in reduced accumulation of NKT cells into the tumor microenvironment (Ma et al., 2018). Interestingly, primary bile acids had an opposing effect on CXCL16 expression, and conversion of primary bile acids into secondary bile acids by Clostridium species associated with higher incidence of liver tumor progression and reduced immune surveillance (Ma et al., 2018).
Multiple studies have begun to demonstrate an association between commensal dysbiosis and cancer. However, no study has determined whether pre-existing dysbiosis influences cancer development and progression, or if the presence of cancer and cancer-associated inflammation contributes to commensal dysbiosis. Although this “chicken and the egg” relationship between dysbiosis and cancer remains undefined, differences in microbial composition between cancer patients and healthy controls exist. In CRC, microbial composition associated with inflammatory gene expression patterns as compared with healthy controls, with CRC patients having elevated expression of CXCL1, IL-17a and IL-23 (Flemer et al., 2017). In breast cancer, dysbiosis has been demonstrated in both the gut as well as the breast tissue in cancer patients as compared to healthy controls. One study found a significant reduction in the overall number of bacteria and an alteration in the proportions of specific bacterial species during dysbiosis, such as Sphingomonas yanoikuyae, in breast tumors when compared to healthy tissue (Xuan et al., 2014b). There was also a decrease in antibacterial responses within the tumor-adjacent tissue, as marked by a reduction in the expression of genes associated with microbial sensors and receptors (Xuan et al., 2014b). Although the mechanistic relevance of microbiome changes in tumor-bearing individuals has yet to be elucidated, these studies provide evidence that cross-talk between tumors and the microbiota occurs.
Dysbiosis within the female reproductive tract has also been implicated in the severity and progression of endometrial cancer, and vaginal microbial compositional changes have been observed in women with cervical cancer (Kyrgiou et al., 2017; Walther-Antonio et al., 2016). A recent study that evaluated ovarian cancer tumor samples using a pathogen array and next-generation sequencing identified a unique microbial signature associated with ovarian cancer compared to matched and non-matched ovarian control samples (Banerjee et al., 2017). Mechanistically, it was shown that interactions with gut commensal microorganisms, through TLR5 signaling, amplified IL-6-mediated tumor inflammation, myeloid-induced immune dysfunction, and the promotion of ovarian tumor growth (Rutkowski et al., 2015). Thus, these studies suggest that both local and systemic commensal microorganisms influence ovarian tumor progression.
Cancer therapies impacted by the microbiome
Microbes are beginning to be appreciated for their roles in efficacy of cancer treatment. Although the majority of papers on this subject have been published in the last decade, microbes and microbial products have been used as cancer therapeutics for over a hundred years. As previously mentioned, in 1891, Coley successfully used Streptococcus erysipelas and Bacillus prodigiosus toxins as a treatment for sarcoma (Coley, 1910). In addition to its use as a tuberculosis vaccine, Bacillus Calmette-Guerin, a live attenuated strain of Mycobacterium bovis, has been used to treat bladder cancer for almost forty years (Redelman-Sidi et al., 2014). More recently, several studies have shown impaired anti-tumor responses in antibiotic-treated or germ-free mice, demonstrating a beneficial role for the commensal microbiome in regulating protective responses to cancer therapy. In this section, we will review the current literature on the types of cancer therapies that are known to be impacted by microbes, the mechanisms by which the microbiome can impact cancer therapy, and potential microbial-driven therapeutics that could enhance the efficacy of cancer treatment (Figure 2).
Figure 2: Microbiota-mediated regulation of anti-cancer therapies and immune therapy.
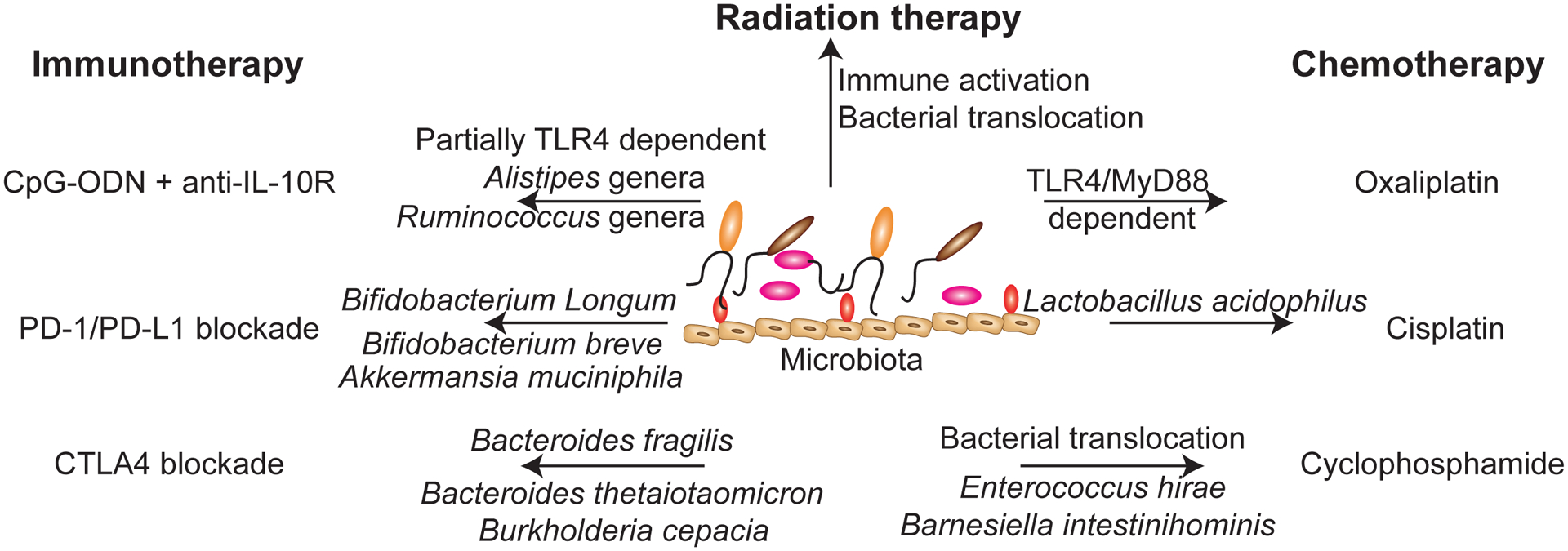
Recent human and mouse studies have identified commensal microorganisms or microbiota-dependent mechanisms that enhance the efficacy of immune therapies, radiation therapy, and chemotherapy. Systemic delivery of the TLR9 agonist, CpG Oligodeoxynucleotide (ODN) in combination with IL-10 blockade is partially TLR4 dependent. Alistipes and Ruminococcus genera were found to be overrepresented in treatment-responsive mice, whereas mono-colonization with A. shahii was sufficient to restore response to this therapy in mice that were treated with antibiotics. For PD-1/PD-L1 blockade, studies have demonstrated that mono-colonization with Bifidobacterium longum, B. breve, and Akkermansia muciniphila were sufficient to enhance the efficacy of this therapy in tumor-bearing mice. For blockade of CTLA4 signaling, gavage of antibiotic-treated mice with Bacteroides fragilis, B. thetaiotamicron, and Burkholderia cepacian were sufficient to restore efficacy of this therapy in antibiotic-treated or germ-free mice. Radiation therapy on the other hand can alter microbiome composition and associate with gut toxicity, commensal translocation, and pathologic inflammation. However, total body irradiation is sufficient to improve the efficacy of adoptive T cell transfer due to the translocation of gram-negative species into the mesenteric lymph nodes and TLR4 signaling. For chemotherapies, oxaliplatin-induced cell death is dependent upon TLR4 signaling whereas the anti-tumor responses are MyD88 dependent. Recolonization of antibiotic-treated mice with Lactobacillus acidophilus is sufficient to restore the efficacy of cisplatin in tumor-bearing antibiotic-treated mice. The beneficial effects of cyclophosphamide require the translocation of gram-positive bacteria from the gut into secondary lymphoid organs, where commensals such as Enterococcus hirae act to prime T cells in a MyD88-dependent manner. Additionally, gram-negative Barnesiella intestinihominis associates with enhanced cyclophosphamide-mediated anti-tumor immunity. Mono-colonization with either microorganism is able to partially restore the immunogenic effects of cyclophosphamide in antibiotic-treated mice.
Chemotherapies
Platinum compounds
Platinum-based compounds such as oxaliplatin and cisplatin exert their cytotoxic effects through the formation of DNA-platinum adducts that result in DNA damage and the upregulation of pathways that promote apoptosis (Siddik, 2003). Additionally, oxaliplatin induces immunogenic cell death, promoting anti-tumor immune responses by activating antigen-presenting cells and their cognate T cells in a TLR4-dependent mechanism (Tesniere et al., 2010). A murine EL4 lymphoma model was used to investigate the impact of the microbiota on the efficacy of platinum compounds (Iida et al., 2013).
The anti-tumor effects of oxaliplatin and cisplatin were significantly reduced in microbiota-deficient mice, both in germ-free animals and in mice that had been treated with broad-spectrum antibiotics to deplete commensal microbes. Although DNA-platinum adducts were formed at comparable levels in both experimental groups, antibiotic-treated mice had a significant reduction in gene expression modification and cytotoxicity compared with control animals. Additionally, tumor-infiltrating myeloid cells from antibiotic-treated animals showed markedly diminished production of reactive oxygen species (ROS) (Iida et al., 2013): a crucial contributing factor to the efficacy of platinum compound therapy (Santoro et al., 2016). Oxaliplatin was also largely ineffective in GR-1+ myeloid cell-depleted mice and in mice deficient for the NADPH oxidase NOX2, demonstrating the previously unknown importance of myeloid-derived ROS in the anti-tumor effects of platinum compound therapy (Iida et al., 2013).
The authors hypothesized that commensal microbes prime myeloid cells for ROS production, presumably through ligation of pattern recognition receptors. Indeed, signaling through MyD88, an adaptor protein crucial for the activity of most pattern recognition receptors, was found to be necessary for the reduction in tumor burden in response to oxaliplatin (Iida et al., 2013). In a similar study, recolonizing antibiotic-treated mice with Lactobacillus acidophilus, a bacterial species commonly used in probiotics, restored the anti-tumor effects of cisplatin in a murine lung cancer model (Gui et al., 2015), further demonstrating the importance of commensal microbes in determining efficacy of this therapy.
Cyclophosphamide
The DNA-alkylating agent cyclophosphamide has been in use as an anti-cancer therapy for almost sixty years. At high doses, cyclophosphamide has immunosuppressive properties and thus is also used to treat certain autoimmune disorders. However, at low doses, it acts to promote anti-tumor immunity through depletion of regulatory T cells and activation of effector T cells (Scurr et al., 2017). Additionally, like oxaliplatin, cyclophosphamide induces immunogenic cell death, further promoting anti-tumor immune responses (Kroemer et al., 2013). Viaud et al demonstrated that the anti-cancer effects of cyclophosphamide are largely dependent on microbial translocation from the gut (Viaud et al., 2013).
In a mouse model of MCA205 sarcoma, cyclophosphamide treatment resulted in significant damage to the gut mucosa that disrupted gut barrier integrity, allowing for translocation of bacteria from the lumen of the intestine into mesenteric lymph nodes and the spleen. Additionally, cyclophosphamide altered the composition of the commensal bacteria in the small intestine, resulting in dysbiosis (Viaud et al., 2013). Specifically, cyclophosphamide treatment resulted in a decrease in both the Firmicutes phylum and species of lactobacilli and enterococci as well as a significant enrichment in species of Gram-positive bacteria, including Enterococcus hirae (Daillere et al., 2016; Viaud et al., 2013). These Gram-postive bacteria translocated into mesenteric lymph nodes where they primed both pro-inflammatory “pathogenic” Th17 (pTh17) and memory Th1 cells in a MyD88-dependent manner with subsequent activation of tumor-specific CD8+ T cells. Additionally, translocation of E. hirae resulted in a reduction in immunosuppressive regulatory T cells and γδ T cells within the tumor (Viaud et al., 2013).
A follow-up study from the same group identified the importance of an additional bacterial strain, as cyclophosphamide treatment resulted in an overrepresentation of the Gram-negative species Barnesiella intestinihominis within the colon of tumor-bearing animals. High levels of B. intestinihominis correlated with enhanced systemic Th1 cells, cytotoxic CD8+ T cells, and tumor-infiltrating γδ T cells that produced IFNγ while also correlating with reduced regulatory T cells in the tumor microenvironment. This study also demonstrated that both E. hirae and B. intestinihominis are regulated by intestinal NOD2 receptors, preventing their egress from the gut under homeostatic conditions (Daillere et al., 2016). The cumulative effects of the presence of these bacteria promote a proinflammatory environment within the tumor, driving anti-tumor immune responses.
As expected, cyclophosphamide-mediated anti-tumor responses were blunted in both germ-free and antibiotic-treated mice due to reduced proinflammatory pTh17 responses and enhanced tumor burden in models of MCA205 sarcoma and P815 mastocytoma. Interestingly, treatment with broad-spectrum antibiotics or colistin (specifically targeting Gram-negative bacteria) or vancomycin (specifically targeting Gram-positive bacteria) alone were all sufficient to blunt responses to cyclophosphamide, demonstrating that multiple bacterial species promote response to this treatment. Adoptive transfer of pTh17 cells into antibiotic-treated mice was sufficient to rescue cyclophosphamide efficacy (Viaud et al., 2013). Additionally, recolonization of antibiotic-treated mice with E. hirae or B. intestinihominis partially restored the effects of cyclophosphamide, resulting in decreased MCA205 tumor burden and enhanced cytotoxic CD8+ T cell responses, respectively. These findings are clinically relevant, as T cell-mediated immune responses against E. hirae and B. intestinihominis are positive predictors of progression-free survival in chemotherapy-treated patients with ovarian and lung cancers (Daillere et al., 2016).
Radiation
Many cancer patients receive radiation therapy, either as a primary treatment (targeted radiation) to reduce localized tumor burden or as a tool (total-body irradiation) to lymphodeplete patients prior to hematopoietic stem cell transplantation or adoptive T cell transfer therapy. Radiation is genotoxic, resulting in tumor cell death. Additionally, radiation can kill additional non-targeted cells through the bystander effect, in part due to activation of the immune system by radiation-induced inflammation (Demaria and Formenti, 2012; Vacchelli et al., 2013). These, off-target effects, such as damage to the intestinal mucosa, can result in substantial toxicity, and the microbiota is known to be involved in this process.
Radiation can alter the composition of the microbiome, with one group describing decreased total gut bacteria with specific decreases in Enterobacteriaceae and Lactobacillus strains after irradiation of the ileum in mice (Johnson et al., 2004). Similarly, a study in patients with gynecological cancers found that radiation treatment significantly reduced diversity within the gut, with decreases in Firmicutes and enrichment for Fusobacterium (Nam et al., 2013). Another group demonstrated a local enrichment in Proteobacteria and decreases in Firmicutes and Bacteroidetes species after radiation treatment in mice, resulting in increased production of proinflammatory cytokines within the gut. Additionally, transfer of post-radiation microbiota into germ-free animals enhanced their susceptibility to mucosal inflammation after radiation treatment, demonstrating the impact of the microbiome on radiation-related gut toxicity (Gerassy-Vainberg et al., 2018). Pre-existing alterations in microbial composition can also predict susceptibility to gut toxicity after radiation treatment, with decreased levels of diversity and species richness correlating with enhanced mucosal inflammation in two small cohorts of patients undergoing pelvic radiotherapy (Manichanh et al., 2008; Wang et al., 2015).
Germ-free mice are resistant to radiation-induced gut toxicity, with fewer apoptotic intestinal epithelial cells and reduced infiltration of lymphocytes as compared with specific pathogen-free mice. Germ-free mice survive substantially longer than conventionally-raised mice after radiation therapy, with higher doses of radiation needed to result in mortality. Angiopoietin-like 4, also known as Fiaf, is secreted by intestinal epithelial cells and promotes endothelial survival and integrity (Kim et al., 2000). Expression of Fiaf is regulated by the presence of bacteria, with certain species promoting or suppressing protein expression. Germ-free mice lacking Fiaf were susceptible to gut toxicity after radiation, demonstrating its ability to suppress mucosal inflammation after radiotherapy (Crawford and Gordon, 2005). Similarly, mice lacking TLR3 were resistant to gut toxicity after ionizing radiation therapy, with enhanced survival and decreased apoptosis of intestinal crypts after treatment (Takemura et al., 2014). In mice with a replete microbiome, the enhanced intestinal permeability caused by radiation treatment results in bacterial translocation from the lumen of the intestine into draining lymph nodes, resulting in significant mucosal inflammation, toxicity, and immune activation against bacterial species (Barker et al., 2015; Paulos et al., 2007).
However, this immune activation can be beneficial, as in the case of radiation preceding adoptive T cell transfer. Adoptive transfer of cytotoxic, tumor-specific CD8+ T cells can be an effective therapy to dramatically reduce tumor burden. In order for these transferred cells to survive, an immunological niche must be created through lymphodepletion of the host by chemotherapy and/or radiation. Total-body irradiation improves the efficacy of adoptive T cell transfer in both humans and mice as a result of bacterial translocation of Gram-negative species into draining mesenteric lymph nodes, resulting in activation of innate immune cells through a TLR4-dependent mechanism. Paulos et al showed that in mice, treatment with broad-spectrum antibiotics or genetic deletion of TLR4 were both sufficient to ameliorate the anti-tumor promoting effects of total-body irradiation prior to T cell transfer, demonstrating the importance of commensal bacteria in promoting response to this therapy (Paulos et al., 2007). This phenomenon has also been observed in humans, as patients with metastatic melanoma responded better to transfer of tumor-infiltrating lymphocytes when chemotherapeutic lymphodepletion was combined with total-body irradiation (Dudley et al., 2008). With the advent of chimeric antigen receptor (CAR)-T cell therapy, use of lymphodepleting radiation in combination with chemotherapy may be effective at enhancing anti-tumor immune responses in these patients following T cell transfer (reviewed in (Flynn et al., 2017)).
Hematopoietic stem cell transplant
In certain cancers, primarily hematologic malignancies, transfer of hematopoietic stem cells from a genetically-similar donor can be curative when other treatment options fail. However, decreased diversity of gut microbiota is associated with reduced survival and increased incidence of graft versus host disease (GVHD) in patients receiving allogeneic hematopoietic stem cells transplantation (HSCT) (Holler et al., 2014; Jenq et al., 2012; Shono et al., 2016; Taur et al., 2014). Importantly, HSCT recipients are frequently pre-treated with broad-spectrum antibiotics, immunosuppressants, and total-body irradiation: all of which are known to impact the composition of the gut microbiota and result in dysbiosis.
Taur et al stratified HSCT recipients into groups of high, intermediate, or low gut bacterial diversity. Low diversity, a hallmark of gut commensal dysbiosis, associated with a significant reduction in 3-year survival as compared to the group with high microbial diversity. Furthermore, the microbial composition after engraftment significantly differed between subjects who lived or died after transplant, with survivors showing decreases in Proteobacteria and enrichment in Actinobacteria (Taur et al., 2014). A follow-up study from the same group found similar results, with enrichment for the Blautia genus associated with reduced risk for GVHD and enhanced survival (Jenq et al., 2015).
Additional studies also demonstrated a loss of bacterial diversity in HSCT recipients, with some groups identifying enrichments in Enterococci (Heimesaat et al., 2010; Holler et al., 2014; Taur et al., 2012), Lactobacilli (Jenq et al., 2012), Streptococci (Taur et al., 2012), and/or Proteobacteria (Taur et al., 2012) after treatment. Lactobacilli (Jenq et al., 2012) and Bacteroidetes (Biagi et al., 2015) were found to be protective against GVHD, while enrichments in Enterococci associated with the development of both GVHD (Heimesaat et al., 2010; Holler et al., 2014) and bacteremia (Taur et al., 2012), presumably through a TLR9-dependent mechanism (Heimesaat et al., 2010). The species of predominating bacteria post-transplant greatly depended on the classes of antibiotics used pre- and post-HSCT, which may inform future clinical decisions for patients receiving HSCT (Shono et al., 2016; Taur et al., 2012).
Immunotherapy
CpG-ODN and anti-IL-10R
Unmethylated 5’-cytosine-phosphate-guanosine-3’ (CpG) sites are rare in vertebral DNA, but are abundant in bacterial DNA and are recognized by the pattern recognition receptor TLR9. Although expression levels differ between mice and humans (Rehli, 2002), TLR9 is primarily expressed on antigen-presenting cells, including myeloid cells and B cells, and its ligation induces expression of pro-inflammatory cytokines and a robust anti-microbial immune response. Synthetic CpG oligodeoxynucleotides (ODN) mimic microbial DNA and are currently being examined for use as adjuvants for vaccines against cancer and infectious pathogens (Scheiermann and Klinman, 2014). CpG-ODN have shown limited efficacy as a monotherapy, but they enhance anti-tumor immune responses when used in combination with other therapeutics. Iida et al examined the impact of the microbiome on combined immunotherapy using intratumoral CpG-ODN and systemic blocking antibodies against the IL-10 receptor (CpG/aIL-10R).
CpG/aIL-10R treatment effectively controls the growth of subcutaneous MC38 colon cancer in mice by inducing tumor necrosis factor (TNF)-dependent hemorrhagic necrosis. The TNF is produced in large part by formerly immunosuppressive tumor-infiltrating myeloid cells that have been reprogrammed to a more pro-inflammatory phenotype (Guiducci et al., 2005). However, this treatment is rendered ineffective in germ-free and antibiotic-treated mice. In these mice with a deficient microbiome, tumor-infiltrating myeloid cells showed diminished inflammatory cytokine production, including production of TNF, and thus the anti-tumor adaptive immune response and resulting hemorrhagic tumor necrosis were blunted. The efficacy of treatment did not rely solely on the adaptive immune system, however, as CpG/aIL-10R therapy was similarly ineffective in antibiotic-treated Rag1−/− mice, while vehicle-treated Rag1−/− mice exhibited partial responses to therapy. However, TNF−/− mice were resistant to treatment with CpG/aIL-10R whether the animals had been treated with antibiotics or not, demonstrating the necessity of this cytokine for efficacy of CpG/aIL-10R treatment (Iida et al., 2013).
Oral gavage with the bacterial component LPS, the ligand for TLR4, partially rescued the diminished anti-tumor response to CpG/aIL-10R, including the decreased TNF production from myeloid cells, seen in antibiotic-treated mice. Thus, bacteria or bacterial products prime tumor-associated myeloid cells to respond to TLR9 stimulation with CpG-ODN, at least in part through ligation of TLR4. The authors went on to identify bacterial genera that correlated with Tnf expression in the tumor. Both the Gram-negative (LPS-containing) Alistipes and Gram-positive Ruminococcus genera were overrepresented in the gut in treatment-responsive mice and positively correlated with Tnf expression, suggesting that additional TLR4-independent mechanisms exist to prime myeloid cells for response to CpG/aIL-10R. Conversely, an overrepresentation of Gram-positive Lactobacillus species negatively correlated with Tnf expression. When antibiotic-treated mice were mono-colonized with Alistipes shahii, tumor-infiltrating myeloid cells produced more TNF, and CpG/aIL-10R was more effective at reducing MC38 tumor burden. However, reconstitution of intact mice with Lactobacillus fermentum had the opposite effect, reducing the myeloid-driven TNF response within the tumor. Thus, although commensal microbes are necessary to prime myeloid cells for response to CpG/aIL-10R, different bacterial species can have opposing effects on the immune system.
Checkpoint blockade
The discovery of immune checkpoints revolutionized approaches to cancer therapy, with Drs. James Allison and Tasuku Honjo recently awarded the 2018 Nobel Prize in Physiology or Medicine for their pioneering work using antibodies against cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) and programmed death 1 (PD-1), respectively. The CTLA-4 and PD-1 signaling pathways function to inhibit effector T cell function and proliferation. Antibodies directed against these proteins block interactions with the cognate ligand or receptor, releasing the inhibition of tumor-specific T cells and allowing for reinvigoration of the immune response (Sharma and Allison, 2015). Additionally, anti-CTLA-4 may exert its anti-tumor effects, at least in part, through macrophage-mediated depletion of immunosuppressive regulatory T cells in the tumor microenvironment, as this cell type characteristically expresses high levels of CTLA-4 (Simpson et al., 2013).
Monoclonal antibodies against CTLA-4 and PD-1 or its ligand PD-L1 have shown remarkable efficacy in a subset of patients with many types of cancer including advanced melanoma (Brahmer et al., 2012; Weber et al., 2008), renal cell carcinoma (Yang et al., 2007), and non-small cell lung cancer (Lynch et al., 2012; Topalian et al., 2012), with additional types of cancer currently being tested in clinical trials. However, there is a substantial variability in responsiveness to checkpoint blockade therapy (Robert et al., 2014), and identifying factors that influence responsiveness is of high importance. Two landmark papers in Science provided the first evidence that alterations in commensal gut bacteria could influence responses to checkpoint blockade through CTLA-4 and PD-1 (Sivan et al., 2015; Vetizou et al., 2015).
CTLA-4:
Vetizou et al reported that while CTLA-4 blockade was an effective therapeutic in mouse models of sarcoma, melanoma, and colon cancer, tumors did not respond to this treatment in germ-free mice or in mice that had been previously treated with antibiotics. Following anti-CTLA-4 therapy in animals with an intact microbiome, mucosal damage and subsequent shifts in the composition of the gut microbiota were noted, with decreases in Bacteroidales and Burkholderiales and relative enrichment in species of Clostridiales. However, despite this overall decrease in Bacteroidales, anti-CTLA-4 treatment resulted in enrichment of specific species of Bacteroides. When antibiotic-treated or germ-free animals were orally recolonized with strains of Bacteroides fragilis, Bacteroides thetaiotaomicron, or Burkholderia cepacia, the anti-tumor effects of anti-CTLA-4 treatment were restored, while reconstitution with control bacteria, such as Escherichia coli, had no effect. Interestingly, responses to anti-CTLA-4 in antibiotic-treated mice varied depending on the type of antibiotic used for pre-treatment, with animals treated with vancomycin having increased efficacy of CTLA-4 blockade as vancomycin specifically targets Gram-positive bacteria, allowing for retention and outgrowth of Gram-negative species such as Bacteroides and Burkholderia.
The presence of these species, in particular B. fragilis, promoted Th1 responses in secondary lymphoid structures and activated and matured tumor-infiltrating dendritic cells, likely through inducing production of IL-12. Responses to anti-CTLA-4 could also be partially restored through adoptive transfer of B. fragilis-specific CD4+ T cells into antibiotic-treated or germ-free mice. Neutralization of IL-12 partially abrogated treatment efficacy in the context of B. fragilis, demonstrating the importance of this cytokine to anti-CTLA-4-mediated anti-tumor responses. The crucial bacterial component driving these anti-tumor responses was found to be the extracellular polysaccharides of species of Bacteroides. Interestingly, it appears that these polysaccharides do not exert their immunostimulatory effects solely through ligation of their classical receptors TLR2 or TLR4, as mice deficient in these receptors had similar responses to anti-CTLA-4 as those seen in control animals. These polysaccharides are sufficient to restore responsiveness to treatment, however, as immunization with B. fragilis polysaccharides enhanced responses to anti-CTLA-4 in mice previously treated with antibiotics.
The authors observed similar effects in melanoma patients treated with anti-CTLA-4, with treated patients having enhanced ex vivo T cell responses against species of Bacteroides as compared to untreated patients or healthy controls. Additionally, patients associated into distinct clusters, or enterotypes, of gut bacterial composition: enterotype A had significant enrichment of Prevotella species, while enterotypes B and C were predominately controlled by species of Bacteroides. In some patients, anti-CTLA-4 treatment shifted gut microbiota composition from that of enterotype B at pre-treatment to enterotype C following treatment. Performing fecal microbiota transfer from patients with these enterotypes into tumor-bearing germ-free mice demonstrated that only transfer of enterotype C was capable of promoting responsiveness to anti-CTLA-4 treatment, with concurrent engraftment of B. thetaiotaomicron and/or B. fragilis observed in these animals (Vetizou et al., 2015). Thus, anti-CTLA-4 therapy can result in alterations to the composition of gut bacteria that affect responsiveness to treatment in the host.
PD-1/PD-L1:
Similar studies using PD-1/PD-L1 blockade showed differences in tumor outgrowth in mice purchased from two different vendors. Although these animals were of the same strain and genetically near-identical, mice from different vendors are known to have differences in the composition of their commensal microbiome (Ericsson et al., 2015). While mice sourced from Jackson Laboratories showed moderate outgrowth of B16.SIY melanoma and sensitivity to treatment with PD-L1 blockade, mice from Taconic Biosciences had greatly enhanced tumor growth kinetics and blunted responses to anti-PD-L1. This enhanced tumor growth was diminished after co-housing with or fecal transfer from Jackson mice, although co-housing and fecal transfer from Taconic animals did not enhance tumor growth in Jackson mice. Sivan et al showed that the efficacy of anti-PD-L1 blockade in Jackson mice was a result of enriched Bifidobacterium species in the gut, specifically B. longum and B. breve. These Bifidobacterium served to activate and mature dendritic cells that promoted the expansion of tumor-specific CD8+ T cells and their subsequent accumulation into tumor lesions. Oral gavage of a probiotic cocktail including B. breve and B. longum was sufficient to slow tumor growth and enhance the efficacy of PD-L1 blockade in Taconic mice (Sivan et al., 2015).
Three follow-up studies confirmed these findings and expanded into clinical studies that examined the impact of the microbiome on PD-1/PD-L1 blockade in human patients. Gopalakrishnan et al and Matson et al sequenced the fecal microbiota of metastatic melanoma patients undergoing PD-1 and PD-L1 blockade, respectively. In Gopalakrishnan’s study, responding patients, with longer progression-free survival after treatment, had significantly greater alpha diversity of bacterial species within the gut, with specific enrichment of the Ruminococcaceae family, Clostridiales order, and Faecalibacterium genus. In contrast, patients that did not respond to treatment had lower bacterial diversity with enrichment for the Bacteroidales order. The gut microbial composition of responding patients was associated with enhanced systemic and intratumoral T cell responses and upregulation of pathways involved with antigen processing and presentation. These responses were transferrable, as fecal transplantation from responders and non-responders into tumor-bearing germ-free mice transferred responsiveness or non-responsiveness to anti-PD-L1 therapy, respectively. Mice that received responder fecal transplants had increased numbers of CD8+ T cells that correlated with decreased BRAFV600E/PTEN−/− melanoma burden after treatment when compared with mice that received a fecal transplant from non-responders (Gopalakrishnan et al., 2018).
Conversely, Matson et al identified different bacterial species as being critical for response to therapy in their patients with advanced melanoma, with Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium, among others, found to be enriched in the feces of patients that responded to anti-PD-L1. Although the sample size was small, all patients that carried the species Akkermansia muciniphila responded to treatment, in agreement with findings from Routy et al, discussed below. Despite these differences in bacterial strains identified to be important for response, transfer of responder vs. non-responder feces into tumor-bearing germ-free mice was similarly capable of transferring responsiveness to treatment, as animals that received fecal transplants from responding patients had a reduction in B16.SIY tumor burden after PD-L1 blockade. These responding animals had increased tumor-specific CD8+ T cells in the tumor microenvironment whereas numbers of immunosuppressive Foxp3+ regulatory T cells remained unchanged, allowing for a more effective anti-tumor immune response (Matson et al., 2018).
Similar findings were reported by Routy et al in patients with advanced urothelial carcinoma, non-small cell lung cancer, and renal cell carcinoma. Patients who had been treated with antibiotics within several months before, during, or after treatment with PD-1/PD-L1 blockade had shorter progression-free survival and lower overall survival rates compared with patients who had not received antibiotics. After sequencing fecal samples from these patients, the genera Akkermansia and Alistipes were enriched and the bacterial species A. muciniphila specifically was found to be highly represented in patients that responded to checkpoint blockade. Ex vivo T cell recall responses to A. muciniphila also correlated with enhanced survival in patients undergoing treatment. Similar to what was found by Gopalakrishnan et al and Matson et al, when fecal transplantation was performed using fecal samples from treatment responders or non-responders into tumor-bearing germ-free mice, mice that received fecal transplants from responding patients were then also able to respond to PD-1 blockade while mice receiving transplants from non-responding patients remained resistant to treatment. However, anti-PD-1 responses could be restored in animals receiving non-responder transplants with concurrent oral administration of A. muciniphila. These animals showed increased tumor-infiltrating immune cells that were promoted by CCR9+CXCR3+CD4+ T cells within the tumor microenvironment after PD-1 blockade. This study was the first to demonstrate that in humans, antibiotic-induced dysbiosis can negatively affect responses to checkpoint blockade therapy (Routy et al., 2018). As antibiotics are so frequently prescribed to cancer patients, these findings have tremendous clinical relevance.
Potential for microbial-driven therapeutics to enhance cancer treatment efficacy
Pre- pro- and postbiotics
In recent years, the use of pre- pro- and postbiotics has gained traction as a method of altering the bacterial species within one’s individual microbiome. The most commonly recognized member of this trio is probiotics: live bacteria that can be orally administered with the intention of shifting the composition of the gut microbiota (Fuller, 1991). Prebiotics also aim to shift microbiome composition through expansion of selected beneficial types of bacteria. Prebiotics, or food components that cannot be digested by the human body, are selectively fermented by specific types of bacteria, promoting their health and expansion (Collins and Gibson, 1999). Postbiotics are the most recent addition to the group, and are defined as bacterial products or metabolites that have beneficial activity within the human host (Klemashevich et al., 2014). As the previously-discussed studies have identified a multitude of different bacterial species as being associated with or necessary for response to various cancer treatments, it has become clear that microbial-driven responses to cancer therapies are likely a direct result of bacterial community composition and interaction, not specific differences on the species level. However, use of both pre- and probiotics have shown some efficacy in preventing infection and ameliorating cancer treatment-related side effects in a number of studies.
In addition to the previously mentioned studies that tested administration of various bacterial species (particularly Bifidobacterium) on enhancing responses to cancer therapy, several additional studies have examined the impact of various strains of Lactobacillus in cancer treatment in mice. L. rhamnosus given both prior to and following HSCT improved overall survival and reduced the incidence of GVHD following transplant (Gerbitz et al., 2004). As previously mentioned, L. acidophilus treatment was also associated with reduced tumor burden and longer survival in a mouse model of lung cancer, enhancing responses to cisplatin (Gui et al., 2015). Additionally, Yeung et al demonstrated that administration of L. casei or L. acidophilus in combination with Bifidobacterium bifidum was sufficient to attenuate toxicity associated with fluorouracil (5-FU) chemotherapy. Mice treated with either probiotic culture had reduced diarrhea, intestinal damage, and release of proinflammatory cytokines as compared with mice treated with saline (Yeung et al., 2015). However, other reports have shown no benefit of probiotics in this model, with the caveat that different probiotic strains were tested in these studies (Maioli et al., 2014; Mauger et al., 2007; Prisciandaro et al., 2012). Despite the efficacy of Lactobacillus strains in these studies, L. fermentum associated with decreased responses to CpG-ODN treatment, demonstrating the importance of identifying potentially beneficial bacterial strains in the context of a specific treatment (Iida et al., 2013).
Studies in humans have also focused on probiotic treatment with strains of Lactobacillus and Bifidobacterium. Similar to what was shown in mice, L. rhamnosus reduced intestinal toxicity after 5-FU treatment, with subjects experiencing reduced diarrhea and abdominal discomfort (Osterlund et al., 2007). Lactobacillus administration also reduced intestinal symptoms in the context of pelvic cancers treated with radiation (Delia et al., 2007), and head and neck squamous cell carcinoma treated with radiation and cisplatin (Sharma et al., 2012). Combining Lactobacillus and Bifidobacterium species as a probiotic treatment has also shown efficacy at reducing side effects in cancer treatment. Treatment with both L. acidophilus and B. bifidum reduced diarrhea in subjects undergoing pelvic radiotherapy (Chitapanarux et al., 2010). This combination treatment was also effective at reducing chemotherapy-related side effects, with treatment with L. casei and B. breve (in combination with galacto-oligosaccharide prebiotics) associating with reduced diarrhea, lymphopenia, and febrile neutropenia in patients with esophageal cancer undergoing chemotherapy (Motoori et al., 2017). Similarly, colorectal cancer patients undergoing chemotherapy had reductions in diarrhea incidence after administration of a cocktail containing ten different species of Lactobacillus and Bifidobacterium (Mego et al., 2015). Current clinical trials are focusing on the efficacy of probiotic treatment at promoting anti-cancer responses and reducing side effects in the treatment of cancers including colorectal cancer (NCT03742596, NCT03705442), breast cancer (NCT03358511), kidney cancer (NCT02944617), gynecologic cancers (NCT02351089), and non-small cell lung cancer (NCT03642548).
Fecal microbiota transplantation
To avoid the confounding issues involved with transferring specific bacterial species as probiotics, some groups have examined fecal microbiota transplantation (FMT) as a potential therapeutic to shift the composition of the gut microbiome. Transplanting fecal microbiota from a healthy subject into a diseased subject can alter the composition of commensal microbes in the transplant recipient (Khoruts et al., 2010). These transplants have been effective at treating Clostridium difficile infections, presumably by increasing both the diversity of the gut microbiome and competition for resources within the gastrointestinal tract (Kassam et al., 2013; van Nood et al., 2013). It stands to reason that this therapy could be similarly effective in cancer patients undergoing therapies such as checkpoint blockade, ionizing radiation, or HSCT, where previously-discussed studies have shown the impact of bacterial composition and diversity on responses to treatment. Indeed, results from initial studies of FMT for treatment of GVHD following HSCT were promising, despite the small sample size, with most subjects showing complete response (Kakihana et al., 2016). Additionally, several ongoing clinical trials are examining the effects of FMT on enhancing anti-tumor responses in patients with advanced melanoma (NCT03341143, NCT03353402), avoiding gut toxicity in patients with acute myeloid leukemia (NCT02928523), and preventing or treating infection and GVHD in patients undergoing HSCT (NCT03678493, NCT03720392, NCT03214289).
However, FMT can have wide-ranging and unpredictable effects, commonly resulting in mild side effects including fever, diarrhea, and vomiting (van Nood et al., 2013), with potentially serious side effects including gastrointestinal bleeding or perforation (Kelly et al., 2014). Additionally, bacterial transfer is sufficient to drive increases in systemic inflammatory markers that can have deleterious effects (Angelberger et al., 2013; Vermeire et al., 2016). Moreover, due to the immunosuppressed status of many cancer patients, donors need to be screened for the presence of pathogens, including viral and fungal pathogens, that could result in infection after transplant (Quera et al., 2014; Schwartz et al., 2013). Further complicating matters, administration of FMT is not currently approved by the FDA. The FDA is now evaluating methods of regulation for these transplants, including designating feces as a medication that requires approval as an Investigational New Drug (IND) (Smith et al., 2014). As this review has demonstrated, microbiota composition is exceptionally personalized, with many individual-to-individual differences in species number, richness, and localization within the gastrointestinal tract. Thus, FMT treatment outcomes for cancer patients are likely to be both unpredictable and inconsistent. With murine data demonstrating that fecal transfer also results in transfer of pathological conditions, including obesity, anxiety behaviors, and allergic skin disease, use of FMT to enhance responses to cancer therapy should be approached with caution (Bercik et al., 2011; De Palma et al., 2017; Ellekilde et al., 2014; Plantamura et al., 2018; Ridaura et al., 2013).
Bacterial engineering
While probiotics and FMT aim to promote anti-cancer responses by reconfiguring the gut microbiome, bacteria can also be an effective cancer therapeutic outside of their role as a commensal microorganism. One area of interest lies in engineering bacteria for effective targeting of cancer tissue and delivery of therapeutic cargo, effectively transforming bacteria into anti-cancer factories. Bacteria can easily be stably transfected with vectors encoding many products, including RNAi (Xiang et al., 2006; Zhang et al., 2007), cytokines (Hu et al., 2009; Loeffler et al., 2007, 2008; Yoon et al., 2007), toxins (Nguyen et al., 2010), antiangiogenic factors (Jia et al., 2005; Li et al., 2003), and antibodies (Massa et al., 2013). One benefit to using bacteria in this way is their quick replication rate, providing amplification of the transgene within the target microenvironment (Dang et al., 2001; Zhao et al., 2005). Anaerobic bacteria are particularly well-suited to invade hypoxic tumor microenvironments. Bacteria such as Shigella and Listeria monocytogenes can enter into the cytoplasm of mammalian host cells to deliver their engineered payloads, and some bacterial species also possess secretion systems, which can deliver a therapeutic product into a target cell without the need for the bacteria itself to enter the cell (Blanco-Toribio et al., 2010).
However, this therapeutic strategy is not without risks. A fine balance must be struck between the number of introduced bacteria that are necessary to elicit a therapeutic effect and the number at which the bacteria overwhelm the host immune system, particularly in the context of host immunosuppression which is common in cancer patients. A broader overview of this subject is well-reviewed in (Forbes, 2010).
Concluding Statements
The cooperation between the host and the commensal microbiome influence health and disease. Commensal microorganisms are central to the maintenance of immune homeostasis and tolerance, while providing signals to the immune system that enable the elimination of invading pathogens. When the equilibrium between the host and its commensal population is disrupted, due to disease or life-style related factors, commensal dysbiosis and related pathologies occur. Although this field is nascent, several studies have helped to uncover the importance of commensal microorganisms in maintaining immune homeostasis, host physiology, metabolism, cancer progression, and response to immune therapies. Understanding the implications of commensal dysbiosis in the context of cancer progression will pave the way towards harnessing the beneficial effects of specific microorganisms to aid in cancer treatment and eventually prevention. Importantly, these studies may also uncover commensal-associated biomarkers, in the context of specific microbial signatures or metabolic byproducts, that aid in the diagnosis and treatment of multiple types of cancers. Undoubtedly, more research is required to elucidate methods to manipulate the microbiome in order to achieve better patient outcomes.
REFERENCES
- Abt M, Osborne L, Monticelli L, Doering T, Alenghat T, Sonnenberg G, Paley M, Antenus M, Williams K, Erikson J, et al. (2012a). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. (2012b). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BL, Cu-Uvin S, Raker CA, Fitzsimmons C, and Hillier SL (2011). Subtle perturbations of genital microflora alter mucosal immunity among low-risk pregnant women. Acta Obstet Gynecol Scand 90, 510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, Novacek G, Trauner M, Loy A, and Berry D (2013). Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. The American journal of gastroenterology 108, 1620–1630. [DOI] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, Deroos P, Liu H, Cross J, Pfeffer K, Coffer P, et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer ML, Schürch CM, and Saito Y (2014). Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. The Journal of …. [DOI] [PubMed]
- Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, Coukos G, and Robertson ES (2017). The ovarian cancer oncobiome. Oncotarget 8, 36225–36245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker HE, Paget JT, Khan AA, and Harrington KJ (2015). The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nature reviews Cancer 15, 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Horowitz RE, Levenson SM, and Popper H (1963). The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol 42, 471–483. [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, et al. (2011). The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141, 599–609, 609.e591–593. [DOI] [PubMed] [Google Scholar]
- Biagi E, Zama D, Nastasi C, Consolandi C, Fiori J, Rampelli S, Turroni S, Centanni M, Severgnini M, Peano C, et al. (2015). Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone marrow transplantation 50, 992–998. [DOI] [PubMed] [Google Scholar]
- Blanco-Toribio A, Muyldermans S, Frankel G, and Fernandez LA (2010). Direct injection of functional single-domain antibodies from E. coli into human cells. PloS one 5, e15227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgdorff H, Gautam R, Armstrong SD, Xia D, Ndayisaba GF, van Teijlingen NH, Geijtenbeek TB, Wastling JM, and van de Wijgert JH (2016). Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal Immunol 9, 621–633. [DOI] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. (2012). Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. The New England journal of medicine 366, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla E, Le Teuff G, Marguet S, Lantuejoul S, Dunant A, Graziano S, Pirker R, Douillard JY, Le Chevalier T, Filipits M, et al. (2016). Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J Clin Oncol 34, 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, and Douek DC (2012). Microbial translocation across the GI tract. Annual review of immunology 30, 149–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchta Rosean CM, and Rutkowski MR (2017). The influence of the commensal microbiota on distal tumor-promoting inflammation. Seminars in immunology 32, 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, Li F, Yu X, Feng Q, Wang Z, et al. (2017). The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun 8, 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitapanarux I, Chitapanarux T, Traisathit P, Kudumpee S, Tharavichitkul E, and Lorvidhaya V (2010). Randomized controlled trial of live lactobacillus acidophilus plus bifidobacterium bifidum in prophylaxis of diarrhea during radiotherapy in cervical cancer patients Radiation oncology (London, England: ) 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, and Weiser JN (2010a). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature medicine 16, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, and Weiser JN (2010b). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature medicine 16, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley WB (1891). II. Contribution to the Knowledge of Sarcoma. Ann Surg 14, 199–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley WB (1910). The Treatment of Inoperable Sarcoma by Bacterial Toxins (the Mixed Toxins of the Streptococcus erysipelas and the Bacillus prodigiosus). Proceedings of the Royal Society of Medicine 3, 1–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, and Gibson GR (1999). Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. The American journal of clinical nutrition 69, 1052s–1057s. [DOI] [PubMed] [Google Scholar]
- Crawford PA, and Gordon JI (2005). Microbial regulation of intestinal radiosensitivity. Proceedings of the National Academy of Sciences of the United States of America 102, 13254–13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daillere R, Vetizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP, et al. (2016). Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 45, 931–943. [DOI] [PubMed] [Google Scholar]
- Dang LH, Bettegowda C, Huso DL, Kinzler KW, and Vogelstein B (2001). Combination bacteriolytic therapy for the treatment of experimental tumors. Proceedings of the National Academy of Sciences of the United States of America 98, 15155–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daud AI, Loo K, Pauli ML, Sanchez-Rodriguez R, Sandoval PM, Taravati K, Tsai K, Nosrati A, Nardo L, Alvarado MD, et al. (2016). Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 126, 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G, Lynch MD, Lu J, Dang VT, Deng Y, Jury J, Umeh G, Miranda PM, Pigrau Pastor M, Sidani S, et al. (2017). Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Science translational medicine 9. [DOI] [PubMed] [Google Scholar]
- Delia P, Sansotta G, Donato V, Frosina P, Messina G, De Renzis C, and Famularo G (2007). Use of probiotics for prevention of radiation-induced diarrhea. World journal of gastroenterology 13, 912–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria S, and Formenti SC (2012). Radiation as an immunological adjuvant: current evidence on dose and fractionation. Frontiers in oncology 2, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon NG, Ebright D, Defrancesco MA, and Hawkins RE (1994). Orogenital contact: a cause of chorioamnionitis? Obstetrics and gynecology 84, 654–655. [PubMed] [Google Scholar]
- Duan J, Chung H, Troy E, and Kasper DL (2010). Microbial Colonization Drives Expansion of IL-1 Receptor 1-Expressing and IL-17-Producing γ/δ T Cells. Cell Host & Microbe 7, 140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley ME, Yang JC, Sherry R, Hughes MS, Royal R, Kammula U, Robbins PF, Huang J, Citrin DE, Leitman SF, et al. (2008). Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 26, 5233–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellekilde M, Selfjord E, Larsen CS, Jakesevic M, Rune I, Tranberg B, Vogensen FK, Nielsen DS, Bahl MI, Licht TR, et al. (2014). Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Scientific reports 4, 5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, Patterson JW, and Slingluff CL Jr. (2012). Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res 72, 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, McIntosh M, and Franklin CL (2015). Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PloS one 10, e0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder M, Valles-Colomer M, Vandeputte D, et al. (2016). Population-level analysis of gut microbiome variation. Science 352, 560–564. [DOI] [PubMed] [Google Scholar]
- Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, and Wong DT (2012). Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MF, Reina-Perez I, Astorga JM, Rodriguez-Carrillo A, Plaza-Diaz J, and Fontana L (2018). Breast Cancer and Its Relationship with the Microbiota. Int J Environ Res Public Health 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, O’Riordain M, Shanahan F, and O’Toole PW (2017). Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 66, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JP, O’Hara MH, and Gandhi SJ (2017). Preclinical rationale for combining radiation therapy and immunotherapy beyond checkpoint inhibitors (i.e., CART). Translational lung cancer research 6, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes NS (2010). Engineering the perfect (bacterial) cancer therapy. Nature reviews Cancer 10, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, Basic M, Klopfleisch R, Rosenstiel P, Bleich A, et al. (2018). Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature 560, 489–493. [DOI] [PubMed] [Google Scholar]
- Fuller R (1991). Probiotics in human medicine. Gut 32, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo T, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. [DOI] [PubMed] [Google Scholar]
- Gerassy-Vainberg S, Blatt A, Danin-Poleg Y, Gershovich K, Sabo E, Nevelsky A, Daniel S, Dahan A, Ziv O, Dheer R, et al. (2018). Radiation induces proinflammatory dysbiosis: transmission of inflammatory susceptibility by host cytokine induction. Gut 67, 97–107. [DOI] [PubMed] [Google Scholar]
- Gerbitz A, Schultz M, Wilke A, Linde HJ, Scholmerich J, Andreesen R, and Holler E (2004). Probiotic effects on experimental graft-versus-host disease: let them eat yogurt. Blood 103, 4365–4367. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, and Knight R (2018). Current understanding of the human microbiome. Nat Med 24, 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, and Ley RE (2016). Genetic determinants of the gut microbiome in UK twins. Cell host & microbe 19, 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. (2014). Human genetics shape the gut microbiome. Cell 159, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients Science (New York, NY: ) 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, and Ivanov II (2014). Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, and Leys EJ (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME journal 6, 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes L, Doyle A, Miller AL, Pyles RB, Olah G, Szabo C, Hoskins S, and Eaves-Pyles T (2016). Intraluminal Flagellin Differentially Contributes to Gut Dysbiosis and Systemic Inflammation following Burn Injury. PloS one 11, e0166770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S, Wang K, Mucida D, Stewart C, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher C, Hung K, et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui QF, Lu HF, Zhang CX, Xu ZR, and Yang YH (2015). Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genetics and molecular research : GMR 14, 5642–5651. [DOI] [PubMed] [Google Scholar]
- Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, and Colombo MP (2005). Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer research 65, 3437–3446. [DOI] [PubMed] [Google Scholar]
- Han YW, Fardini Y, Chen C, Iacampo KG, Peraino VA, Shamonki JM, and Redline RW (2010). Term stillbirth caused by oral Fusobacterium nucleatum. Obstetrics and gynecology 115, 442–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand T, and Belkaid Y (2010). Microbial control of regulatory and effector T cell responses in the gut. Current opinion in immunology 22, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmey JH, Bucana CD, Lu W, Byrne AM, McDonnell S, Lynch C, Bouchier-Hayes D, and Dong Z (2002). Lipopolysaccharide-induced metastatic growth is associated with increased angiogenesis, vascular permeability and tumor cell invasion. International journal of cancer 101, 415–422. [DOI] [PubMed] [Google Scholar]
- Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, Loddenkemper C, Steinhoff U, Tchaptchet S, Thiel E, Freudenberg MA, et al. (2010). MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut 59, 1079–1087. [DOI] [PubMed] [Google Scholar]
- Hergott CB, Roche AM, Tamashiro E, Clarke TB, Bailey AG, Laughlin A, Bushman FD, and Weiser JN (2016). Peptidoglycan from the gut microbiota governs the lifespan of circulating phagocytes at homeostasis. Blood 127, 2460–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, Xiao J, Radisky DC, Knutson KL, Kalari KR, et al. (2016). The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci Rep 6, 30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, et al. (2014). Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 20, 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Kou L, Li C, Zhu LP, Fan YR, Wu ZW, Wang JJ, and Xu GX (2009). Bifidobacterium longum as a delivery system of TRAIL and endostatin cooperates with chemotherapeutic drugs to inhibit hypoxic tumor growth. Cancer gene therapy 16, 655–663. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, et al. (2011). Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol 127, 372–381 e371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, and Iwasaki A (2011a). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America 108, 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, and Iwasaki A (2011b). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America 108, 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. (2013). Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, and Cell S-T (2009a). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I, Atarashi K, Manel N, Brodie E, Shima T, Karaoz U, Wei D, Goldfarb K, Santee C, Lynch S, et al. (2009b). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, et al. (2015). Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 21, 1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, Liu C, West ML, Singer NV, Equinda MJ, et al. (2012). Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. The Journal of experimental medicine 209, 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia LJ, Xu HM, Ma DY, Hu QG, Huang XF, Jiang WH, Li SF, Jia KZ, Huang QL, and Hua ZC (2005). Enhanced therapeutic effect by combination of tumor-targeting Salmonella and endostatin in murine melanoma model. Cancer biology & therapy 4, 840–845. [DOI] [PubMed] [Google Scholar]
- Johnson LB, Riaz AA, Adawi D, Wittgren L, Back S, Thornberg C, Osman N, Gadaleanu V, Thorlacius H, and Jeppsson B (2004). Radiation enteropathy and leucocyte-endothelial cell reactions in a refined small bowel model. BMC surgery 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakihana K, Fujioka Y, Suda W, Najima Y, Kuwata G, Sasajima S, Mimura I, Morita H, Sugiyama D, Nishikawa H, et al. (2016). Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood 128, 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, and Nunez G (2013). Role of the gut microbiota in immunity and inflammatory disease. Nature reviews Immunology 13, 321–335. [DOI] [PubMed] [Google Scholar]
- Kassam Z, Lee CH, Yuan Y, and Hunt RH (2013). Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. The American journal of gastroenterology 108, 500–508. [DOI] [PubMed] [Google Scholar]
- Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, Aroniadis O, Barto A, Borody T, Giovanelli A, et al. (2014). Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. The American journal of gastroenterology 109, 1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, and Sadowsky MJ (2010). Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. Journal of clinical gastroenterology 44, 354–360. [DOI] [PubMed] [Google Scholar]
- Khosravi A, Yáñez A, Price JG, Chow A, and Merad M (2014). Gut microbiota promote hematopoiesis to control bacterial infection. Cell host & …. [DOI] [PMC free article] [PubMed]
- Kim HR, Park SM, Seo SU, Jung I, Yoon HI, Gabrilovich DI, Cho BC, Seong SY, Ha SJ, and Youn JI (2018). The Ratio of Peripheral Regulatory T Cells to Lox-1(+) PMN-MDSC Predicts the Early Response to Anti-PD-1 Therapy in Non-Small Cell Lung Cancer Patients. Am J Respir Crit Care Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Kim HG, Kim H, Kim HH, Park SK, Uhm CS, Lee ZH, and Koh GY (2000). Hepatic expression, synthesis and secretion of a novel fibrinogen/angiopoietin-related protein that prevents endothelial-cell apoptosis. The Biochemical journal 346 Pt 3, 603–610. [PMC free article] [PubMed] [Google Scholar]
- Klemashevich C, Wu C, Howsmon D, Alaniz RC, Lee K, and Jayaraman A (2014). Rational identification of diet-derived postbiotics for improving intestinal microbiota function. Current opinion in biotechnology 26, 85–90. [DOI] [PubMed] [Google Scholar]
- Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, and Barton GM (2016). Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell 165, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, and Ley RE (2011). Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl 1, 4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, Dummer R, Robinson MD, Levesque MP, and Becher B (2018). High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med 24, 144–153. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Kepp O, and Zitvogel L (2013). Immunogenic cell death in cancer therapy. Annual review of immunology 31, 51–72. [DOI] [PubMed] [Google Scholar]
- Kyrgiou M, Mitra A, and Moscicki AB (2017). Does the vaginal microbiota play a role in the development of cervical cancer? Translational research : the journal of laboratory and clinical medicine 179, 168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop S, Bloom S, Rao S, Nutsch K, Lio C-W, Santacruz N, Peterson D, Stappenbeck T, and Hsieh C-S (2011). Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, et al. (2014). Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620. [DOI] [PubMed] [Google Scholar]
- Li X, Fu GF, Fan YR, Liu WH, Liu XJ, Wang JJ, and Xu GX (2003). Bifidobacterium adolescentis as a delivery system of endostatin for cancer gene therapy: selective inhibitor of angiogenesis and hypoxic tumor growth. Cancer gene therapy 10, 105–111. [DOI] [PubMed] [Google Scholar]
- Liaudet L, Szabo C, Evgenov OV, Murthy KG, Pacher P, Virag L, Mabley JG, Marton A, Soriano FG, Kirov MY, et al. (2003). Flagellin from gram-negative bacteria is a potent mediator of acute pulmonary inflammation in sepsis. Shock 19, 131–137. [DOI] [PubMed] [Google Scholar]
- Loeffler M, Le’Negrate G, Krajewska M, and Reed JC (2007). Attenuated Salmonella engineered to produce human cytokine LIGHT inhibit tumor growth. Proceedings of the National Academy of Sciences of the United States of America 104, 12879–12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler M, Le’Negrate G, Krajewska M, and Reed JC (2008). IL-18-producing Salmonella inhibit tumor growth. Cancer gene therapy 15, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, Jansson JK, Gordon JI, and Knight R (2013). Meta-analyses of studies of the human microbiota. Genome research 23, 1704–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bondarenko I, Luft A, Serwatowski P, Barlesi F, Chacko R, Sebastian M, Neal J, Lu H, Cuillerot JM, et al. (2012). Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter phase II study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 30, 2046–2054. [DOI] [PubMed] [Google Scholar]
- Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, et al. (2018). Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, and Green AR (2011). Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 29, 1949–1955. [DOI] [PubMed] [Google Scholar]
- Maioli TU, de Melo Silva B, Dias MN, Paiva NC, Cardoso VN, Fernandes SO, Carneiro CM, Dos Santos Martins F, and de Vasconcelos Generoso S (2014). Pretreatment with Saccharomyces boulardii does not prevent the experimental mucositis in Swiss mice. Journal of negative results in biomedicine 13, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Varela E, Martinez C, Antolin M, Llopis M, Dore J, Giralt J, Guarner F, and Malagelada JR (2008). The gut microbiota predispose to the pathophysiology of acute postradiotherapy diarrhea. The American journal of gastroenterology 103, 1754–1761. [DOI] [PubMed] [Google Scholar]
- Massa PE, Paniccia A, Monegal A, de Marco A, and Rescigno M (2013). Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. Blood 122, 705–714. [DOI] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger CA, Butler RN, Geier MS, Tooley KL, and Howarth GS (2007). Probiotic effects on 5-fluorouracil-induced mucositis assessed by the sucrose breath test in rats. Digestive diseases and sciences 52, 612–619. [DOI] [PubMed] [Google Scholar]
- Maynard CL, Elson CO, Hatton RD, and Weaver CT (2012). Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, and Keku TO (2013). Fusobacterium is associated with colorectal adenomas. PloS one 8, e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mego M, Chovanec J, Vochyanova-Andrezalova I, Konkolovsky P, Mikulova M, Reckova M, Miskovska V, Bystricky B, Beniak J, Medvecova L, et al. (2015). Prevention of irinotecan induced diarrhea by probiotics: A randomized double blind, placebo controlled pilot study. Complementary therapies in medicine 23, 356–362. [DOI] [PubMed] [Google Scholar]
- Melero I, Rouzaut A, Motz GT, and Coukos G (2014). T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov 4, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, and Moore LV (1994). The bacteria of periodontal diseases. Periodontology 2000 5, 66–77. [DOI] [PubMed] [Google Scholar]
- Motoori M, Yano M, Miyata H, Sugimura K, Saito T, Omori T, Fujiwara Y, Miyoshi N, Akita H, Gotoh K, et al. (2017). Randomized study of the effect of synbiotics during neoadjuvant chemotherapy on adverse events in esophageal cancer patients Clinical nutrition (Edinburgh, Scotland: ) 36, 93–99. [DOI] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Butcher AM, Trzoss LL, Nam SJ, Shirakawa KT, Zhou W, Oh J, Otto M, Fenical W, et al. (2018). A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci Adv 4, eaao4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YD, Kim HJ, Seo JG, Kang SW, and Bae JW (2013). Impact of pelvic radiotherapy on gut microbiota of gynecological cancer patients revealed by massive pyrosequencing. PloS one 8, e82659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VH, Kim HS, Ha JM, Hong Y, Choy HE, and Min JJ (2010). Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer research 70, 18–23. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera M, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. (2012). Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science 336, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, Kouri M, Elomaa I, and Joensuu H (2007). Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. British journal of cancer 97, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, Palmer DC, Boni A, Muranski P, Yu Z, et al. (2007). Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. The Journal of clinical investigation 117, 2197–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantamura E, Dzutsev A, Chamaillard M, Djebali S, Moudombi L, Boucinha L, Grau M, Macari C, Bauche D, Dumitrescu O, et al. (2018). MAVS deficiency induces gut dysbiotic microbiota conferring a proallergic phenotype. Proceedings of the National Academy of Sciences of the United States of America 115, 10404–10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollet I, Opina CJ, Zimmerman C, Leong KG, Wong F, and Karsan A (2003). Bacterial lipopolysaccharide directly induces angiogenesis through TRAF6-mediated activation of NF-kappaB and c-Jun N-terminal kinase. Blood 102, 1740–1742. [DOI] [PubMed] [Google Scholar]
- Prisciandaro LD, Geier MS, Chua AE, Butler RN, Cummins AG, Sander GR, and Howarth GS (2012). Probiotic factors partially prevent changes to caspases 3 and 7 activation and transepithelial electrical resistance in a model of 5-fluorouracil-induced epithelial cell damage. Supportive care in cancer : official journal of the Multinational Association of Supportive Care in Cancer 20, 3205–3210. [DOI] [PubMed] [Google Scholar]
- Quera R, Espinoza R, Estay C, and Rivera D (2014). Bacteremia as an adverse event of fecal microbiota transplantation in a patient with Crohn’s disease and recurrent Clostridium difficile infection. Journal of Crohn’s & colitis 8, 252–253. [DOI] [PubMed] [Google Scholar]
- Redelman-Sidi G, Glickman MS, and Bochner BH (2014). The mechanism of action of BCG therapy for bladder cancer--a current perspective. Nature reviews Urology 11, 153–162. [DOI] [PubMed] [Google Scholar]
- Rehli M (2002). Of mice and men: species variations of Toll-like receptor expression. Trends in immunology 23, 375–378. [DOI] [PubMed] [Google Scholar]
- Rhee KJ, Sethupathi P, Driks A, Lanning DK, and Knight KL (2004). Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. Journal of immunology (Baltimore, Md : 1950) 172, 1118–1124. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Ribas A, Wolchok JD, Hodi FS, Hamid O, Kefford R, Weber JS, Joshua AM, Hwu WJ, Gangadhar TC, et al. (2014). Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial Lancet (London, England: ) 384, 1109–1117. [DOI] [PubMed] [Google Scholar]
- Rolli J, Rosenblatt-Velin N, Li J, Loukili N, Levrand S, Pacher P, Waeber B, Feihl F, Ruchat P, and Liaudet L (2010). Bacterial flagellin triggers cardiac innate immune responses and acute contractile dysfunction. PLoS One 5, e12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. [DOI] [PubMed] [Google Scholar]
- Round JL, Lee MS, Li J, Tran G, Jabri B, Chatila TA, and Mazmanian SK (2011). The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science, 1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors Science (New York, NY: ) 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Rubinstein M, Wang X, Liu W, Hao Y, Cai G, and Han YW (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/ -catenin signaling via its FadA adhesin. Cell host & microbe 14, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski MR, Stephen TL, Svoronos N, Allegrezza MJ, Tesone AJ, Perales-Puchalt A, Brencicova E, Escovar-Fadul X, Nguyen JM, Cadungog MG, et al. (2015). Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell 27, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro V, Jia R, Thompson H, Nijhuis A, Jeffery R, Kiakos K, Silver AR, Hartley JA, and Hochhauser D (2016). Role of Reactive Oxygen Species in the Abrogation of Oxaliplatin Activity by Cetuximab in Colorectal Cancer. Journal of the National Cancer Institute 108, djv394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann J, and Klinman DM (2014). Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine 32, 6377–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A, et al. (2016). Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167, 1125–1930850304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe RF, and Jobin C (2013). The microbiome and cancer. Nature reviews Cancer 13, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Gluck M, and Koon S (2013). Norovirus gastroenteritis after fecal microbiota transplantation for treatment of Clostridium difficile infection despite asymptomatic donors and lack of sick contacts. The American journal of gastroenterology 108, 1367. [DOI] [PubMed] [Google Scholar]
- Scurr M, Pembroke T, Bloom A, Roberts D, Thomson A, Smart K, Bridgeman H, Adams R, Brewster A, Jones R, et al. (2017). Low-Dose Cyclophosphamide Induces Antitumor T-Cell Responses, which Associate with Survival in Metastatic Colorectal Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 6771–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, and Milo R (2016). Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS biology 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Rath GK, Chaudhary SP, Thakar A, Mohanti BK, and Bahadur S (2012). Lactobacillus brevis CD2 lozenges reduce radiation- and chemotherapy-induced mucositis in patients with head and neck cancer: a randomized double-blind placebo-controlled study. European journal of cancer (Oxford, England : 1990) 48, 875–881. [DOI] [PubMed] [Google Scholar]
- Sharma P, and Allison JP (2015). The future of immune checkpoint therapy Science (New York, NY: ) 348, 56–61. [DOI] [PubMed] [Google Scholar]
- Shaw MH, Kamada N, Kim Y-GG, and Núñez G (2012). Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. The Journal of experimental medicine 209, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheflin AM, Whitney AK, and Weir TL (2014). Cancer-promoting effects of microbial dysbiosis. Current oncology reports 16, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, Slingerland AE, Smith OM, Young LF, Gupta J, et al. (2016). Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Science translational medicine 8, 339ra371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddik ZH (2003). Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22, 7265–7279. [DOI] [PubMed] [Google Scholar]
- Silva MJ, Carneiro MB, dos Anjos Pultz B, Pereira Silva D, Lopes ME, and dos Santos LM (2015). The multifaceted role of commensal microbiota in homeostasis and gastrointestinal diseases. J Immunol Res 2015, 321241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JL, Daly J, Baines KJ, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, Hugenholtz P, Willner D, et al. (2016). Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J 47, 792–800. [DOI] [PubMed] [Google Scholar]
- Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD, et al. (2013). Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. The Journal of experimental medicine 210, 1695–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remédios C, et al. (2014). Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nature medicine 20, 1301–1309. [DOI] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MB, Kelly C, and Alm EJ (2014). Policy: How to regulate faecal transplants. Nature 506, 290–291. [DOI] [PubMed] [Google Scholar]
- Stoll G, Enot D, Mlecnik B, Galon J, Zitvogel L, and Kroemer G (2014). Immune-related gene signatures predict the outcome of neoadjuvant chemotherapy. Oncoimmunology 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svard J, Paquin-Proulx D, Buggert M, Noyan K, Barqasho B, Sonnerborg A, and Nowak P (2015). Role of translocated bacterial flagellin in monocyte activation among individuals with chronic HIV-1 infection Clinical immunology (Orlando, Fla: ) 161, 180–189. [DOI] [PubMed] [Google Scholar]
- Takemura N, Kawasaki T, Kunisawa J, Sato S, Lamichhane A, Kobiyama K, Aoshi T, Ito J, Mizuguchi K, Karuppuchamy T, et al. (2014). Blockade of TLR3 protects mice from lethal radiation-induced gastrointestinal syndrome. Nature communications 5, 3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, et al. (2014). The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 124, 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, et al. (2012). Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 55, 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesniere A, Schlemmer F, Boige V, Kepp O, Martins I, Ghiringhelli F, Aymeric L, Michaud M, Apetoh L, Barault L, et al. (2010). Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 29, 482–491. [DOI] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H, Stepankova R, Kozakova H, Hudcovic T, Vannucci L, Tuckova L, Rossmann P, Hrncir T, Kverka M, Zakostelska Z, et al. (2011). The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol 8, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine 366, 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, and Gordon JI (2007). The human microbiome project. Nature 449, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, et al. (2010). Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proceedings of the National Academy of Sciences of the United States of America 107, 7503–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacchelli E, Vitale I, Tartour E, Eggermont A, Sautes-Fridman C, Galon J, Zitvogel L, Kroemer G, and Galluzzi L (2013). Trial Watch: Anticancer radioimmunotherapy. Oncoimmunology 2, e25595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. The New England journal of medicine 368, 407–415. [DOI] [PubMed] [Google Scholar]
- Vermeire S, Joossens M, Verbeke K, Wang J, Machiels K, Sabino J, Ferrante M, Van Assche G, Rutgeerts P, and Raes J (2016). Donor Species Richness Determines Faecal Microbiota Transplantation Success in Inflammatory Bowel Disease. Journal of Crohn’s & colitis 10, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. (2013). The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide Science (New York, NY: ) 342, 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther-Antonio MR, Chen J, Multinu F, Hokenstad A, Distad TJ, Cheek EH, Keeney GL, Creedon DJ, Nelson H, Mariani A, et al. (2016). Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med 8, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Ling Z, Yang Z, Kiela PR, Wang T, Wang C, Cao L, Geng F, Shen M, Ran X, et al. (2015). Gut microbial dysbiosis may predict diarrhea and fatigue in patients undergoing pelvic cancer radiotherapy: a pilot study. PloS one 10, e0126312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JS, O’Day S, Urba W, Powderly J, Nichol G, Yellin M, Snively J, and Hersh E (2008). Phase I/II study of ipilimumab for patients with metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 26, 5950–5956. [DOI] [PubMed] [Google Scholar]
- Wei B, Wingender G, Fujiwara D, Chen D, McPherson M, Brewer S, Borneman J, Kronenberg M, and Braun J (2010). Commensal Microbiota and CD8+ T Cells Shape the Formation of Invariant NKT Cells. The Journal of Immunology 184, 1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M, and Walker WA (2013). The role of gut microbiota in programming the immune phenotype. Journal of developmental origins of health and disease 4, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut Science (New York, NY: ) 339, 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Fruehauf J, and Li CJ (2006). Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nature biotechnology 24, 697–702. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Liu F, Yang J, Zhong M, Zhang E, Li Y, Zhou D, Cao Y, Li W, Yu J, et al. (2015). Over-activation of TLR5 signaling by high-dose flagellin induces liver injury in mice. Cell Mol Immunol 12, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan C, Shamonki JM, Chung A, and DiNome ML (2014a). Microbial dysbiosis is associated with human breast cancer. PloS one. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, and Lee DJ (2014b). Microbial dysbiosis is associated with human breast cancer. PLoS One 9, e83744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Hughes M, Kammula U, Royal R, Sherry RM, Topalian SL, Suri KB, Levy C, Allen T, Mavroukakis S, et al. (2007). Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. Journal of immunotherapy (Hagerstown, Md : 1997) 30, 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung CY, Chan WT, Jiang CB, Cheng ML, Liu CY, Chang SW, Chiang Chiau JS, and Lee HC (2015). Amelioration of Chemotherapy-Induced Intestinal Mucositis by Orally Administered Probiotics in a Mouse Model. PloS one 10, e0138746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon WS, Choi WC, Sin JI, and Park YK (2007). Antitumor therapeutic effects of Salmonella typhimurium containing Flt3 Ligand expression plasmids in melanoma-bearing mouse. Biotechnology letters 29, 511–516. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. (2013). Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101. [DOI] [PubMed] [Google Scholar]
- Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, et al. (2003). Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 348, 203–213. [DOI] [PubMed] [Google Scholar]
- Zhang L, Gao L, Zhao L, Guo B, Ji K, Tian Y, Wang J, Yu H, Hu J, Kalvakolanu DV, et al. (2007). Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer research 67, 5859–5864. [DOI] [PubMed] [Google Scholar]
- Zhao M, Yang M, Li XM, Jiang P, Baranov E, Li S, Xu M, Penman S, and Hoffman RM (2005). Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proceedings of the National Academy of Sciences of the United States of America 102, 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder M, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila A, Falony G, Vieira-Silva S, et al. (2016). Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Fang L, and Lee MH (2018). Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterology report 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]


