The stringent response is an essential mechanism of metabolic reprogramming during environmental stress that is mediated by the nucleotide alarmones guanosine tetraphosphate and pentaphosphate [(p)ppGpp]. In addition to physiological adaptations, (p)ppGpp also regulates virulence programs in pathogenic bacteria, including Salmonella enterica serovar Typhimurium. S. Typhimurium is a common cause of acute gastroenteritis, but it may also spread to systemic tissues, resulting in severe clinical outcomes.
KEYWORDS: Salmonella, complement, innate immunity, serum, virulence
ABSTRACT
The stringent response is an essential mechanism of metabolic reprogramming during environmental stress that is mediated by the nucleotide alarmones guanosine tetraphosphate and pentaphosphate [(p)ppGpp]. In addition to physiological adaptations, (p)ppGpp also regulates virulence programs in pathogenic bacteria, including Salmonella enterica serovar Typhimurium. S. Typhimurium is a common cause of acute gastroenteritis, but it may also spread to systemic tissues, resulting in severe clinical outcomes. During infection, S. Typhimurium encounters a broad repertoire of immune defenses that it must evade for successful host infection. Here, we examined the role of the stringent response in S. Typhimurium resistance to complement-mediated killing and found that the (p)ppGpp synthetase-hydrolase, SpoT, is required for bacterial survival in human serum. We identified the nucleotide hydrolase, PpnN, as a target of the stringent response that is required to promote bacterial fitness in serum. Using chromatography and mass spectrometry, we show that PpnN hydrolyzes purine and pyrimidine monophosphates to generate free nucleobases and ribose 5′-phosphate, and that this metabolic activity is required for conferring resistance to complement killing. In addition to PpnN, we show that (p)ppGpp is required for the biosynthesis of the very long and long O-antigen in the outer membrane, known to be important for complement resistance. Our results provide new insights into the role of the stringent response in mediating evasion of the innate immune system by pathogenic bacteria.
INTRODUCTION
Salmonella enterica is a bacterial pathogen with a broad host range for both animals and humans. Nontyphoidal strains of Salmonella, such as serovar Typhimurium, are acquired through contaminated food or water and cause gastrointestinal disease that is usually self-limiting (1). However, in developing countries, invasive, nontyphoidal serovars have been linked to severe clinical complications due to systemic bacteremia (2). One of the hallmarks of S. Typhimurium pathogenesis is its capacity to transition between the extracellular and intracellular environment due, in part, to the evolved ability of the bacteria to evade innate host defenses (3, 4).
A prominent arm of innate immunity that S. Typhimurium can encounter during infection is the complement system. Complement is composed of more than 30 proteins that are present in the circulation and at mucosal surfaces where frequent contact with microorganisms is made (5, 6). Activation of complement results in a proteolytic cascade that leads to the coating of target surfaces for phagocytic uptake or perturbation of bacterial membranes by the membrane attack complex (MAC) (6–8). Mechanisms of complement resistance by pathogenic bacteria have been described as proteolytic inactivation of complement proteins or the expression of cell surface structures to prevent complement deposition (9). For example, S. Typhimurium can express long variants of lipopolysaccharide (LPS) O-antigen, and this prevents the integration of the MAC onto the outer membrane (10, 11). Bacteria also undergo metabolic changes to ensure optimal use of resources upon exposure to serum (12–14). However, the role of these metabolic adaptations in promoting survival in serum is largely underexplored.
The stringent response is a mechanism of cellular reprogramming that allows bacteria to survive environmental stressors. It is characterized by the generation of the nucleotide alarmones ppGpp and pppGpp, collectively referred to as (p)ppGpp, and leads to rapid alterations in bacterial physiology (15, 16). The synthetase RelA produces (p)ppGpp during amino acid starvation through the sensing of uncharged tRNAs in the ribosomal A site (17–19). (p)ppGpp is also synthesized by SpoT in response to fatty acid, phosphate, iron, or carbon source limitation (20–22). In contrast to RelA, SpoT also has a hydrolase domain to balance cellular (p)ppGpp levels (23). Accumulation of (p)ppGpp inhibits the synthesis of ribosomal proteins, rRNA and tRNA, and activates stress-specific genes (15, 16). (p)ppGpp also regulates various metabolic pathways, including nucleotide biosynthesis, which occurs through allosteric regulation of enzymes such as PurF and PpnN in Escherichia coli (24–27). Bacterial pathogens often couple (p)ppGpp signaling to virulence gene expression (15, 16). In S. Typhimurium, (p)ppGpp helps activate genes required for invasion of the intestinal epithelium and intracellular survival (28–31). Thus, the stringent response mediates both physiological changes and expression of virulence factors in bacteria for successful host infection.
In this study, we examined the cross talk between the stringent response, metabolic reprogramming, and evasion of innate immunity. We found that SpoT is required for the survival of S. Typhimurium against human complement. Furthermore, we show that (p)ppGpp-mediated regulation of a nucleosidase called PpnN (formerly SL1344_2949) and biosynthesis of the LPS O-antigen promotes the survival of S. Typhimurium in serum. Our data provides insight into the role of nucleotide metabolism in bacterial resistance against complement.
RESULTS
The stringent response is required for complement resistance in S. Typhimurium.
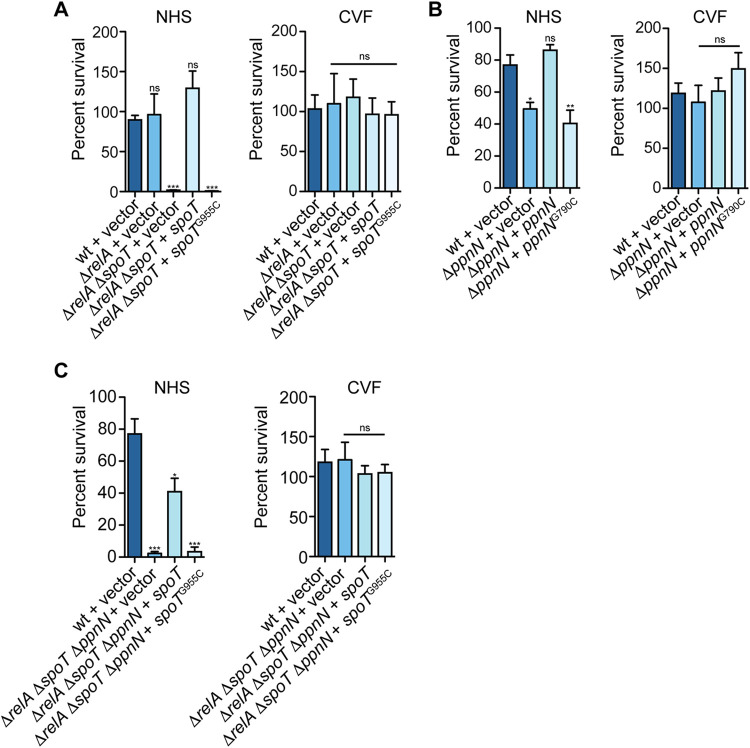
The stringent response is activated in response to environmental insults and is characterized by the production of (p)ppGpp. Mutants lacking the regulators, relA and spoT, are devoid of (p)ppGpp and have pleiotropic phenotypes, including the inability to grow in defined minimal media due to amino acid auxotrophies (32, 33). Consistent with this, we found that the growth of the ΔrelA ΔspoT mutant was severely attenuated in M9-glucose minimal medium compared to growth in nutrient-rich LB broth, whereas the ΔrelA mutant grew similarly to the wild type under both conditions (see Fig. S1 in the supplemental material). We were unable to generate a single ΔspoT mutant, because the (p)ppGpp hydrolase domain of SpoT is essential for bacterial viability in the presence of RelA (34). Given the role of the stringent response in enabling bacteria to adapt to changes in the environment, we next tested whether the stringent response was required for resistance against complement-mediated killing. We grew S. Typhimurium strains lacking relA or both relA and spoT in LB until stationary phase to mimic stringent response conditions and tested their survival in pooled human serum. The ΔrelA mutant survived similarly to the wild-type strain, whereas the ΔrelA ΔspoT mutant was highly susceptible to killing in normal human serum (Fig. 1A). Consistent with these data, complementation of the ΔrelA ΔspoT mutant with SpoT expressed in trans from its native promoter completely rescued bacterial viability to wild-type levels. In contrast, the double mutant remained highly susceptible to serum-mediated killing when it was complemented with spoTG955C, which encodes for a catalytically inactive SpoT mutant containing an E319Q point mutation that abrogates (p)ppGpp synthase activity (35). These data confirmed that the (p)ppGpp synthase activity of SpoT is important for S. Typhimurium survival in normal human serum. Moreover, we could recover bacterial survival of the wild-type and mutant strains following pretreatment of the serum with cobra venom factor (CVF), which depletes the serum of human C3 and prevents downstream activation of the membrane attack complex (Fig. 1A) (36). These data demonstrate that killing was due to the activation of the complement system (36, 37).
FIG 1.
Stringent response is required for complement resistance in S. Typhimurium. (A) ΔrelA ΔspoT mutant is susceptible to killing in pooled normal human serum (NHS) and complementation with spoT, or inhibition of complement with cobra venom factor (CVF) is sufficient to rescue bacterial survival. (B) ΔppnN mutant is susceptible to killing in pooled NHS. Complementation with ppnN or inhibition of complement with CVF is sufficient to rescue bacterial viability, whereas complementation with ppnNG790C does not recover the survival of the bacteria. (C) ΔrelA ΔspoT ppnN::cat mutant complemented with spoT or spoTG955C is susceptible to killing in pooled NHS, and inhibition of complement with CVF is sufficient to rescue bacterial viability. Data are the means ± standard errors of the means (SEM; error bars) from at least three independent experiments. Strains are carrying the empty pGEN-MCS vector control unless otherwise specified. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
It is well established that (p)ppGpp signaling results in global regulatory changes in bacteria, including nucleotide metabolism (15). Recent work in Escherichia coli has shown that (p)ppGpp can allosterically activate the nucleotide hydrolase, PpnN, to degrade nucleotide 5′-monophosphates to the corresponding free bases and ribose 5′-phosphate (25, 26, 38, 39). PpnN is part of the Lonely Guy (LOG) protein family (Pfam PF03641), which are mainly single-domain phosphohydrolases that share a conserved “PGGxGTxxE” motif that is important for the biosynthesis of cytokinins in plant growth and development (39–43) (Fig. S2A). Furthermore, an amino acid sequence alignment between PpnN from E. coli and SL1344_2949 (PpnN hereafter) in S. Typhimurium showed that these proteins share 94% pairwise identity in amino acids (Fig. S2B). We also found that S. Typhimurium ppnN is coregulated with genes, such as pagP and pgtE, that are involved in bacterial resistance against innate immunity (4, 44–46). To study the role of nucleotide metabolism in promoting serum resistance, we deleted ppnN in S. Typhimurium and tested the viability of the mutant in human serum. Although the ΔppnN mutant grew similarly to the wild-type strain in both LB and M9-glucose medium (Fig. S1), it showed a significant ∼30% reduction in viability upon exposure to human serum compared to wild-type S. Typhimurium. Expression of PpnN in trans or pretreatment of the serum with CVF restored bacterial survival back to wild-type levels, indicating a direct connection of this phenotype to complement (Fig. 1B). To determine whether the PGGxGTxxE motif was required for PpnN-mediated complement resistance in S. Typhimurium, we generated a ppnNG790C variant using site-directed mutagenesis, which mutates residue E264 to glutamine. A ΔppnN mutant expressing PpnNE264Q remained susceptible to killing in human serum, confirming that the function of PpnN is required for complement resistance (Fig. 1B).
PpnN is regulated in a (p)ppGpp-dependent manner.
Based on our findings that the ΔrelA ΔspoT and ΔppnN mutants were compromised for serum survival, we hypothesized that (p)ppGpp-mediated regulation of PpnN is required for complement resistance. To test this, we generated a deletion of ppnN in the ΔrelA ΔspoT background and tested the survival of the ΔrelA ΔspoT ppnN::cat mutant in human serum. Consistent with our previous data with the ΔrelA ΔspoT mutant, the ΔrelA ΔspoT ppnN::cat mutant was highly susceptible to complement killing (Fig. 1A and C). Complementation of ΔrelA ΔspoT ppnN::cat with spoT partially recovered S. Typhimurium survival in human serum compared to the wild-type strain, whereas expression of spoT fully restored the survival of the ΔrelA ΔspoT double mutant (Fig. 1A and C). Similar to the ΔrelA ΔspoT mutant, expression of SpoTE319Q was unable to rescue the viability of the ΔrelA ΔspoT ppnN::cat mutant (Fig. 1A and C). In contrast, pretreatment of the serum with CVF rescued the survival of both mutants to wild-type levels (Fig. 1A and C). Taken together, these data support that (p)ppGpp production by SpoT acts through ppnN and other unknown factor(s) to confer resistance to complement killing by S. Typhimurium.
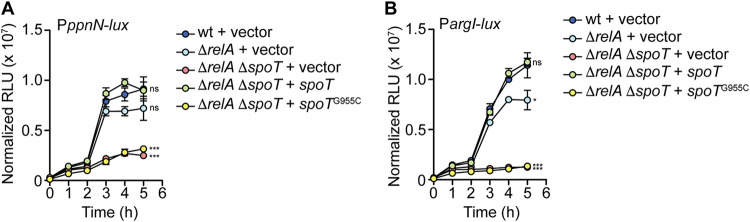
Previously it was shown that PpnN in E. coli K-12 is allosterically regulated by (p)ppGpp (25, 26, 39). This prompted us to investigate the regulation of PpnN by (p)ppGpp in S. Typhimurium. First, we tested whether the activation of the ppnN promoter was dependent on (p)ppGpp. ppnN was activated to similar levels in wild-type S. Typhimurium and the ΔrelA mutant, but its expression was significantly reduced in the ΔrelA ΔspoT mutant that is defective for (p)ppGpp synthesis (Fig. 2A). Complementation with spoT was sufficient to recover activation of ppnN, whereas complementation with spoTG955C was unable to restore activity of the ppnN promoter. Notably, the activation of the ppnN promoter between the wild type and the ΔrelA ΔspoT mutant was significantly different at 5 h of growth in LB when the bacteria are in stationary phase and (p)ppGpp levels increase (47, 48). As a control, we also tested the expression of a known (p)ppGpp-regulated target, argI, which is involved in l-arginine biosynthesis (49). The argI promoter was partially repressed in the relA mutant compared to the wild type, which is consistent with the role of RelA in responding to amino acid stress during bacterial growth in stationary phase (Fig. 2B) (47). Furthermore, argI promoter activity was completely abolished in the ΔrelA ΔspoT mutant, and complementation with spoT, but not spoTG955C, was sufficient to restore argI activation to wild-type levels (Fig. 2B). The amino acid sequence alignment of PpnN in E. coli and S. Typhimurium also showed that the (p)ppGpp binding residues, R68, R70, K73, R341, and Y347, are conserved between the two homologues, strongly suggesting that (p)ppGpp regulates PpnN in S. Typhimurium post-translationally (Fig. S2B) (25).
FIG 2.
PpnN is regulated in a (p)ppGpp-dependent manner. (A and B) Transcriptional reporter of the full-length ppnN promoter (A) and argI promoter (B), a canonical gene target regulated during the stringent response, showed (p)ppGpp-dependent up-regulation. RLU, relative light units. Data are the means ± SEM (error bars) from three independent experiments. Strains are carrying the empty pWSK129 vector control unless otherwise specified. ns, not significant; *, P < 0.05; ***, P < 0.0001.
PpnN is a pyrimidine/purine nucleosidase.
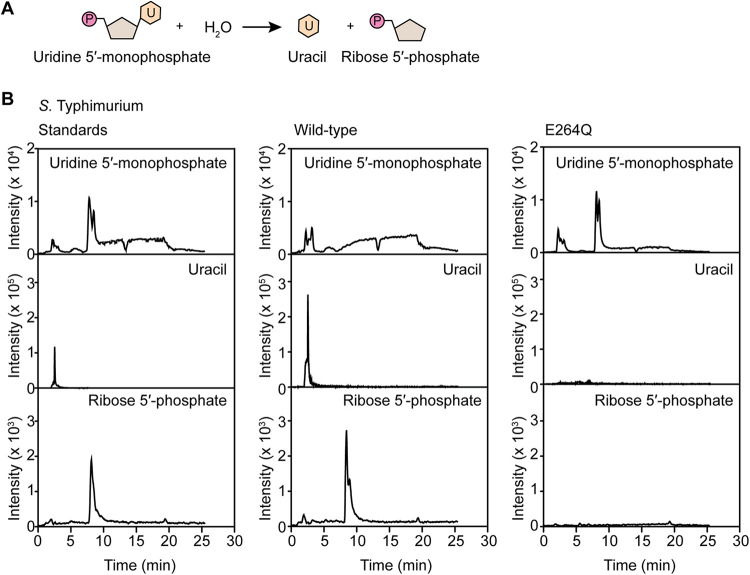
Our data indicated that PpnN promotes the survival of S. Typhimurium in human serum. In E. coli, PpnN is a cytoplasmic hydrolase that degrades purine and pyrimidine nucleotide 5′-monophosphates to the corresponding free bases and ribose 5′-phosphate (25, 26, 38, 39). To compare PpnN in S. Typhimurium to its E. coli homologue, we tested their enzymatic activity by purifying PpnN from S. Typhimurium and E. coli using affinity chromatography and incubated it with the nucleotide substrate, uridine 5′-monophosphate (UMP) (38). Hydrophilic interaction chromatography combined with mass spectrometry (HILIC-MS) was used to monitor the presence of the substrate and the potential products. Similar to the E. coli homologue, PpnN from S. Typhimurium degraded UMP to uracil and ribose 5′-phosphate (Fig. 3A and B and Fig. S3). As a control, we also purified and incubated a catalytic inactive variant of PpnN containing the E264Q point mutation with UMP, and, as expected, the function of the protein was abrogated (Fig. 3B). To determine the nucleotide specificity of PpnN, we also tested its ability to hydrolyze purines such as guanosine 5′-monophosphate (GMP). Similar to UMP, PpnN hydrolyzed GMP to guanine and ribose 5′-phosphate (Fig. S4). UMP and GMP were used as representative substrates, as the detection of the reaction standards was optimal using HILIC-MS. Together, these data confirm that PpnN in S. Typhimurium is a cytosolic nucleotide 5′-phosphate nucleosidase with broad substrate specificity.
FIG 3.
PpnN from S. Typhimurium strain SL1344 is a pyrimidine/purine nucleosidase. (A and B) PpnN hydrolyzes uridine 5′-monophosphate (UMP) to uracil and ribose 5′-phosphate. Introduction of an E264Q catalytic site mutation in PpnN abrogates protein function. Data are representative of two replicates.
PpnN does not confer complement resistance by contributing to cell wall biogenesis.
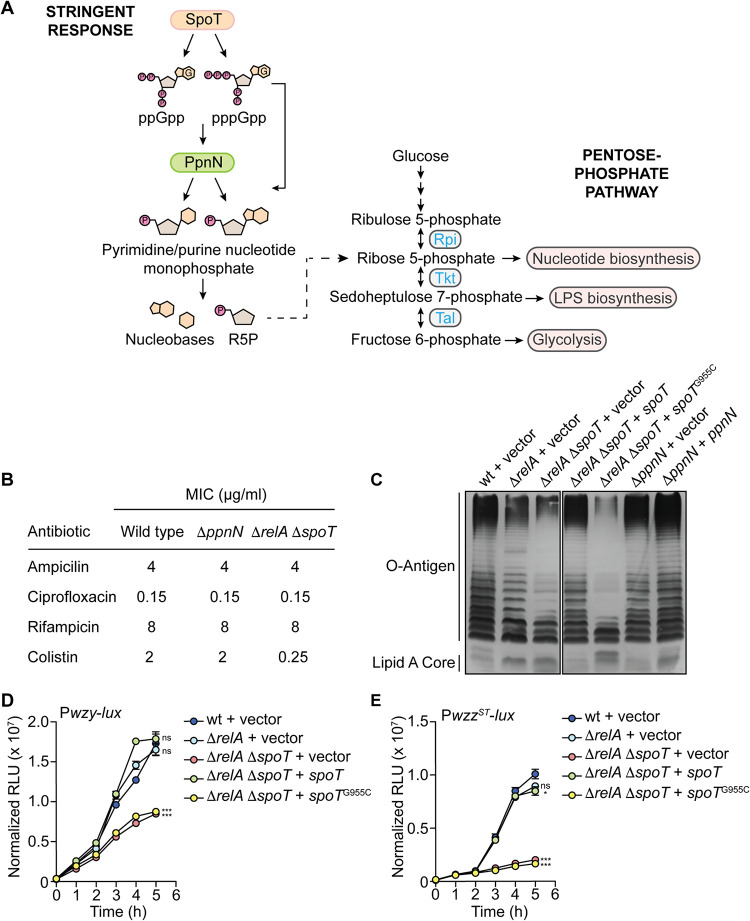
The cytosolic localization and function of PpnN suggested that it mediates complement resistance through its metabolic activity. A common metabolite that PpnN produces from the hydrolysis of purine and pyrimidine monophosphates is ribose 5′-phosphate. We hypothesized that ribose 5′-phosphate could be entering carbon metabolism and conferring complement resistance by contributing to LPS biosynthesis (Fig. 4A) (50, 51). To investigate whether PpnN contributes to maintaining the integrity of the cell wall, we determined the MIC of ampicillin and colistin, which are antibiotics that target peptidoglycan biosynthesis and LPS, respectively, for wild-type S. Typhimurium and the ΔppnN and ΔrelA ΔspoT mutants (52). As negative controls, we also tested rifampin and ciprofloxacin, which affect the synthesis of RNA and DNA, respectively (8, 53). The ΔppnN mutant did not display an increase in susceptibility to any of these antibiotics, whereas the ΔrelA ΔspoT mutant showed an 8-fold increase in susceptibility to colistin (Fig. 4B). These data suggested that the lack of (p)ppGpp production compromises the ability of the bacteria to synthesize the full-length variants of LPS. To investigate this possibility, we extracted whole LPS from wild-type and mutant strains of S. Typhimurium and used gel electrophoresis to examine the O-antigen polymers produced by each strain. The ΔrelA ΔspoT mutant showed a decreased production of the very long and long O-antigen relative to the wild-type strain, and expression of SpoT in trans completely restored its production (Fig. 4C). However, complementation of the ΔrelA ΔspoT mutant with SpoTE319Q was unable to restore the O-antigen, demonstrating that the (p)ppGpp synthetase activity of SpoT is required for regulating its biosynthesis (Fig. 4C). We next engineered bioluminescence reporters to confirm the regulatory input of (p)ppGpp in the production of the LPS O-antigen. Using lux-transcriptional fusions for wzy and wzzST, which encode the O-antigen polymerase and the long O-antigen chain length regulator, respectively, we found that both genes are expressed in a (p)ppGpp-dependent manner (Fig. 4D and E) (54). In contrast to the ΔrelA ΔspoT mutant, the O-antigen profile of the ΔppnN mutant was similar to wild type (Fig. 4C). Consistent with our previous data, we also found that the outer membrane protein composition of the ΔppnN mutant was similar to that of the wild-type strain, whereas the ΔrelA ΔspoT mutant showed significant differences (Fig. S5). These findings demonstrate that PpnN promotes the survival of S. Typhimurium in human serum in a manner independent of cell wall integrity and suggest that the regulation of LPS O-antigen biosynthesis by (p)ppGpp is another mechanism contributing to complement resistance during the stringent response.
FIG 4.
PpnN does not confer complement resistance by contributing to cell wall biogenesis. (A) Model showing (p)ppGpp-mediated regulation of ppnN. PpnN produces free nucleobases and ribose 5′-phosphate (R5P), the latter of which could enter the pentose phosphate pathway and regulate carbon metabolism. (B) MIC of ampicillin, ciprofloxacin, rifampin, and colistin for wild-type S. Typhimurium strain SL1344 and ΔppnN and ΔrelA ΔspoT mutants. The ΔppnN mutant does not show increased susceptibility to any of the tested antibiotics relative to wild-type S. Typhimurium, whereas the ΔrelA ΔspoT mutant is 8-fold more susceptible to colistin. Data are representative of three replicates. (C) LPS analysis of the ΔrelA ΔspoT and ΔppnN mutants compared to wild-type S. Typhimurium. The ΔrelA ΔspoT mutant expresses significantly lower levels of very long and long O-antigen, whereas the LPS O-antigen of the ΔppnN mutant is similar to that of the wild-type strain. Data are representative of two replicates. Strains carry the empty pGEN-MCS vector control unless otherwise specified. Transcriptional reporter of the full-length wzy promoter (D) and wzzST promoter (E) showed (p)ppGpp-dependent up-regulation. Data are the means ± SEM (error bars) from three independent experiments. Strains are carrying the empty pWSK129 vector control unless otherwise specified. ns, not significant; *, P < 0.05; ***, P < 0.0001.
DISCUSSION
Here, we showed that the stringent response is required for S. Typhimurium survival against the complement system, a component of innate immunity that bacteria encounter during host infection. By using bioinformatics to guide mechanistic predictions, we identified the nucleotide monophosphate nucleosidase, PpnN, as being important for complement resistance through the SpoT-dependent second messenger, (p)ppGpp.
Our findings share some parallels with SpoT in Helicobacter pylori and Borrelia burgdorferi, where SpoT is required for optimal growth in the presence of serum (55). An H. pylori ΔspoT mutant exhibits relaxed growth over wild-type bacteria in serum-free medium, suggesting that SpoT is able to sense serum starvation. There have also been other studies relating the stringent responses to bacterial survival either in whole blood or in serum (56–59). For example, a (p)ppGpp0 mutant in Enterococcus faecalis is attenuated in serum due to dysregulated metal homeostasis. Supplementation of the serum with iron or manganese was able to restore bacterial viability (59). In Salmonella Typhi, (p)ppGpp regulates the expression of the (Vi) capsular polysaccharide, which prevents complement deposition and formation of the MAC on the cell surface (58, 60). In contrast to S. Typhi, S. Typhimurium does not express a capsule, which suggests that (p)ppGpp mediates complement resistance through a different mechanism.
The role of a LOG protein in mammalian pathogens has only been investigated in Mycobacterium tuberculosis. In M. tuberculosis, the LOG protein Rv1205 produces cytokinins that are maintained at basal levels by a proteasome system. The function of the cytokinins is unclear, but it has been shown that proteasome-deficient M. tuberculosis is susceptible to killing due to synergy between host nitric oxide and cytokinins that accumulate (41, 61). To our knowledge, our study is the first to report that a LOG-like protein aids in resistance to complement killing. Previously it was shown that the biosynthesis of purines and pyrimidines is needed for bacterial growth in serum (62). However, this study used heat-inactivated serum where complement was presumably inactive, suggesting that nucleotides were not serving to resist the bactericidal activity of complement. Several biochemical screens have identified that nucleotide metabolism is a key output of the stringent response (24, 25). For example, (p)ppGpp inhibits the purine biosynthesis enzymes Gpt, Hpt, GuaB, PurA, and PurF (24, 63). It has also been shown that (p)ppGpp enhances the activity of PpnN in E. coli to degrade nucleotides (25, 26, 38, 39). Deletion of ppnN in S. Typhimurium did not affect the integrity of the cell wall, suggesting that PpnN promotes complement resistance through its metabolic roles in the cytoplasm. It is possible that the generation of nucleotide precursors by PpnN allows for the reallocation of resources to other pathways relevant for survival in serum. In addition, PpnN may facilitate the transition from serum to more favorable conditions by increasing the cellular pool of nucleotide metabolites (26, 64, 65). This is consistent with the finding that a ΔppnN mutant is outcompeted by wild-type E. coli when the bacteria transition between nutrient-rich and -poor media (26). Although the mechanism of PpnN-mediated complement resistance is not fully understood, it is unlikely that it interacts directly with complement proteins, as it is not secreted (66). Exploring the contributions of PpnN to metabolism and how this influences complement resistance in S. Typhimurium will be the focus of future research. A functional genomics approach, such as RNA sequencing of wild-type S. Typhimurium and the ΔppnN mutant exposed to human serum, may reveal insights into the potential pathways affected by PpnN. This may be followed by the systematic deletion of metabolic enzymes in a ΔppnN mutant to test which pathways enhance complement resistance.
Our data suggest that there are other genetic targets in addition to ppnN that contribute to (p)ppGpp-mediated resistance to complement killing. Another possible mechanism is through the regulation of cell wall biogenesis genes by the alternative sigma factor, RpoS, and (p)ppGpp during bacterial growth in stationary phase (33, 47, 67). In particular, bacteria exhibit an increase in LPS, cross-linking of outer membrane lipoproteins, and an increase in thickness of the peptidoglycan layer under nutrient-limited conditions (10, 47, 68). This is supported by our findings that an S. Typhimurium ΔrelA ΔspoT mutant displays significantly lower levels of the very long and long O-antigen and increased susceptibility to complement. The decreased production of O-antigen by the ΔrelA ΔspoT mutant was corroborated by lower expression levels of wzy and wzzST. Our LPS silver staining also suggests that wzzfepE from the O-antigen biosynthesis pathway is regulated in a (p)ppGpp-dependent manner (54). We and others have also shown that the LPS O-antigen of S. Typhimurium is involved in colistin resistance (52). Furthermore, RNA sequencing of S. Typhimurium grown to stationary phase in LB showed that genes such as pagC and pgtE are activated by (p)ppGpp (30). PagC and PgtE are outer membrane proteins that have been directly implicated in complement resistance in Salmonella (45, 46, 69). Together, these data suggest that the stringent response coordinates resistance to complement in S. Typhimurium by remodeling the cell membrane and reprogramming metabolism.
In summary, our findings highlight the role of nucleotide metabolism and the biosynthesis of the LPS O-antigen during the stringent response in mediating evasion of innate immunity by pathogenic bacteria. (p)ppGpp signaling is an essential mechanism of the bacterial stress response that involves the regulation of virulence gene expression for successful host infection. Identifying the genetic factors that allow bacteria to evade the immune system will reveal new therapeutic targets that could be integral in informing drug discovery efforts.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Typhimurium (SL1344) and isogenic derivatives were used in the study and are listed in Table 1. Bacteria were grown in LB or M9 minimal medium supplemented with 1% glucose and 0.135 mM l-histidine. Antibiotics were added where appropriate (streptomycin, 100 μg/ml; kanamycin, 50 μg/ml; ampicillin, 200 μg/ml; chloramphenicol, 34 μg/ml), and strains were grown at 37°C with shaking.
TABLE 1.
Bacterial strains used in the study
| Strain | Source or reference |
|---|---|
| Wild-type S. Typhimurium strain SL1344 | Coombes et al. (79) |
| ΔppnN S. Typhimurium strain SL1344 | This study |
| ppnN::aph S. Typhimurium strain 14028s | Porwollik et al. (70) |
| ΔrelA S. Typhimurium strain SL1344 | This study |
| ΔrelA ΔspoT S. Typhimurium strain SL1344 | This study |
| ΔrelA ΔspoT ppnN::cat S. Typhimurium strain SL1344 | This study |
| Wild-type E. coli strain K-12 | Baba et al. (80) |
| BL21 (DE3) E. coli | Agilent Technologies |
| TOP10 E. coli | Invitrogen |
Cloning and mutant generation.
ppnN::aph from the Salmonella single-gene deletion (SGD) library was transduced to S. Typhimurium strain SL1344 as previously described, with modifications (70, 71). The donor mutant strain was grown overnight in LB supplemented with 0.5× E-Salts and 0.2% d-glucose and mixed with ∼1.5 × 105 PFU of P22 HT phage at 37°C. Next, 1 ml of the donor strain was pelleted and the supernatant was mixed with 100 μl of chloroform to lyse any remaining live bacteria. Two microliters of the donor lysate was then mixed with 200 μl of a wild-type S. Typhimurium strain SL1344 overnight culture and incubated at 37°C for 1 h before plating on LB supplemented with 50 μg/ml kanamycin. Colonies were restreaked onto green indicator agar to screen for the absence of phage. To ensure that the colonies were free of phage lysogen, white colonies were streaked across a line of P22 H5 phage on green agar and incubated overnight at 37°C. Successful transduction of the mutant was verified by PCR. The aph cassette was removed using pCP20, resulting in ΔppnN::FRT in S. Typhimurium strain SL1344.
Lambda Red recombination was used to generate an in-frame, marked mutant of S. Typhimurium relA::cat (72). Wild-type S. Typhimurium carrying pKD46 was transformed with linear PCR products amplified using Phire Hot Start II DNA polymerase (Thermo Fisher) and primers (Sigma-Aldrich) containing gene-specific regions of homology and flanking the cat cassette carried by pKD3. Transformants were selected on LB agar supplemented with chloramphenicol (34 μg/ml), and knockouts were verified by PCR. The cat cassette was removed using pCP20, resulting in the ΔrelA::FRT strain, and confirmed by PCR. Lambda Red treatment was repeated in the ΔrelA::FRT background to generate the ΔrelA::FRT ΔspoT::FRT double mutant and in the ΔrelA::FRT ΔspoT::FRT double mutant background to generate the ΔrelA::FRT ΔspoT::FRT ppnN::cat triple mutant.
For the cloning of spoT, primers EC215F (GTA TCA TAT GGC ACG CGT AAC TGT TCA GGA CGC TG) and EC215R (AGT CCC ATG GCT AGT TTC GGT TAC GGG TGA CTT TA) or EC224F (GTA TGA GCT CGC ACG CGT AAC TGT TCA GGA CGC TG) and EC224R (AGT CGC GGC CGC CTA GTT TCG GTT ACG GGT GAC TTT A) were used to PCR amplify the coding sequence of spoT and 291 bp upstream of the start codon. PCR-amplified products were then cloned into pGEN-MCS or pWSK129 after digestion with NdeI/NcoI (Thermo Fisher) or SacI/NotI (Thermo Fisher), respectively. For the cloning of ppnN, the coding sequence and 800 bp upstream of the start codon of ppnN in SL1344 was PCR amplified using primers EC100F (ATG CGA ATT CGG ATA TCT GGA CGT TGT ATG AAC TT) and EC100R (GTA TCA TAT GTT AAG CGC AGA TCT CGT AAC AGG GG) and then cloned as an EcoRI/NdeI (Thermo Fisher) DNA product into pGEN-MCS. pGEN-MCS-ppnNG790C (encoding PpnNE264Q) and pGEN-MCS-spoTG955C (encoding SpoTE319Q) were generated using the Q5 site-directed mutagenesis kit (NEB) and primers EC186F (TAC GGC GGA ACA GCT GCT TTA TTT GCT G) and EC186R (CCC ACG CCG CCC GGG AAG ATG ATG ATA C) or EC320F (GGC GTT CCT GTT CAA GTC CAG ATC CGT A) and EC320R (GGC GTT CCT GTT CAA GTC CAG ATC CGT A), respectively. pWSK129-spoTG955C was cloned by PCR amplifying EC224F (GTA TGA GCT CGC ACG CGT AAC TGT TCA GGA CGC TG)/EC325R (ACG GAT CTG GAC TTG AAC AGG AAC G) and EC326F (GGC GTT CCT GTT CAA GTC CAG ATC C)/EC224R (AGT CGC GGC CGC CTA GTT TCG GTT ACG GGT GAC TTT A) and then performing splicing by overlap extension (SOE) PCR using EC224F and EC224R. The product of the SOE PCR was digested with SacI/NotI (Thermo Fisher) and cloned into pWSK129.
Bioluminescence reporters were generated using primers EC97F (ATG CGG ATC CGG ATA TCT GGA CGT TGT ATG AAC TT) and EC97R (GCC ATA CGT AGT AAA CTC CTT ATG GGA CGC AAC AC) to PCR amplify 800 bp upstream of the start codon of ppnN, EC188F (ATG CGG ATC CCA ATG GTG GCT TTC GCC AGG) and EC188R (GCC ATA CGT AAT AGA GCC TTT AGA AAA AAT GCT TA) to amplify 1,000 bp upstream of the start codon of wzy, EC212F (AGT CGG ATC CGT GAC GCA CGC CGT CGT CAT) and EC212R (CGG CTA CGT AAC TTC CCT CAC ATG GCT TAG GCC TC) to PCR amplify 579 bp upstream of the start codon of argI, and EC321F (ATG CGG ATC CGT GAT CAG CAT CAA CCC CGC) and EC321R (GCC ATA CGT AAG ATA CCC TAA CTA AAA AAA GGA TG) to PCR amplify 1,000 bp upstream of the start codon of wzzST. PCR-amplified products were then cloned into pGEN-luxCDABE after digestion with SnaBI/BamHI (Thermo Fisher) (73).
All plasmid constructs were sequence verified by Sanger sequencing (GENEWIZ) and then transformed by electroporation (Bio-Rad) into the appropriate strain backgrounds for downstream experiments. Plasmids used in the study are listed in Table 2, and primers (Sigma-Aldrich) are listed in Table 3.
TABLE 2.
Plasmids used in the study
| Plasmida | Description | Source or reference |
|---|---|---|
| pGEN-MCS | Low-copy-no. cloning vector | Lane et al. (73) |
| pGEN-luxCDABE | Lux transcriptional reporter plasmid | Lane et al. (73) |
| pWSK129 | Low-copy-no. cloning vector | Wang and Kushner (81) |
| pEXT20 | Expression vector | Dykxhoorn et al. (82) |
| pET-24a | Expression vector | Novagen |
| pKD3 | Template plasmid for Lambda Red recombination | Datsenko and Wanner (72) |
| pKD46 | Lambda Red recombinase expression plasmid | Datsenko and Wanner (72) |
| pCP20 | Lambda Red flippase expression plasmid | Datsenko and Wanner (72) |
| pGEN-MCS-ppnNST | ppnNST with 800 bp upstream of coding sequence cloned into pGEN-MCS for complementation experiments | This study |
| pGEN-MCS-ppnNST(G790C) | ppnNST containing G790C point mutation with 800 bp upstream of coding sequence cloned into pGEN-MCS for complementation experiments | This study |
| pGEN-MCS-spoT | spoT with 291 bp upstream of coding sequence cloned into pGEN-MCS for complementation experiments | This study |
| pGEN-MCS-spoTG955C | spoT containing G955C point mutation with 291 bp upstream of coding sequence cloned into pGEN-MCS for complementation experiments | This study |
| pWSK129-spoT | spoT with 291 bp upstream of coding sequence cloned into pGEN-MCS for complementation experiments | This study |
| pWSK129-spoTG955C | spoT containing G955C point mutation with 291 bp upstream of coding sequence cloned into pWSK129 for complementation experiments | This study |
| pEXT20-ppnNST-6HIS | PpnNST expression plasmid, C-terminal His6 tagged | This study |
| pEXT20-ppnNEC-6HIS | PpnNEC expression plasmid, C-terminal His6 tagged | This study |
| pET-24a-ppnNST | PpnNST expression plasmid, C-terminal His6 tagged | This study |
| pET-24a-ppnNST(G790C) | PpnNST expression plasmid, C-terminal His6 tagged containing G790C point mutation in ppnNST coding sequence | This study |
| pGEN-luxCDABE-ppnNST | Lux transcriptional reporter for ppnNST promoter | This study |
| pGEN-luxCDABE-argI | Lux transcriptional reporter for argI promoter | This study |
| pGEN-luxCDABE-wzy | Lux transcriptional reporter for wzy promoter | This study |
| pGEN-luxCDABE-wzzST | Lux transcriptional reporter for wzzST promoter | This study |
ST and EC denote ppnN from S. Typhimurium strain SL1344 and E. coli strain K-12, respectively.
TABLE 3.
Primers used in the study
| Primer | Genea | Direction | Destination | Sequence (5′–3′) |
|---|---|---|---|---|
| EC85 | ppnNST | F | pET24-a | CGAACATATGTTGATTACACATATTAGCCCGCTTG |
| EC85 | ppnNST | R | pET24-a | GTATCTCGAGAGCGCAGATCTCGTAACAGGGGATG |
| EC96 | ppnNST | F | pEXT20 | CGAAGGATCCTTGATTACACATATTAGCCCGCTTG |
| EC98 | ppnNST | R | pEXT20 | GTATAAGCTTTTAATGGTGGTGGTGATGATGAGCG CAGATCTCGTAACAGGGG |
| EC97 | ppnNST | F | pGEN-luxCDABE | ATGCGGATCCGGATATCTGGACGTTGTATGAACTT |
| EC97 | ppnNST | R | pGEN-luxCDABE | GCCATACGTAGTAAACTCCTTATGGGACGCAACAC |
| EC100 | ppnNST | F | pGEN-MCS | ATGCGAATTCGGATATCTGGACGTTGTATGAACTT |
| EC100 | ppnNST | R | pGEN-MCS | GTATCATATGTTAAGCGCAGATCTCGTAACAGGGG |
| EC167 | ppnNEC | F | pEXT20 | CGAAGAATTCTTGATTACACATATTAGCCCGCTTG |
| EC167 | ppnNEC | R | pEXT20 | GTATGGTACCTTAATGGTGGTGGTGATGATGCGTGCAGATTTCGTAGCAAGGG |
| EC186 | ppnNST | F | ppnNST | TACGGCGGAACAGCTGCTTTATTTGCTG |
| EC186 | ppnNST | R | ppnNST | CCCACGCCGCCCGGGAAGATGATGATAC |
| EC188 | wzy | F | pGEN-luxCDABE | ATGCGGATCCCAATGGTGGCTTTCGCCAGG |
| EC188 | wzy | R | pGEN-luxCDABE | GCCATACGTAATAGAGCCTTTAGAAAAAATGCTTA |
| EC200 | relA | F | ΔrelA | CGCATGTAATGATTACCGGCTTACCGACTTCGGTAGGCCTGGTCCCTTAAGGAGAGGACGATGGTCGCGGTAAGAAGTGCACATATTAATGTGTAGGCTGGAGCTGCTTCG |
| EC200 | relA | R | ΔrelA | GTTGCTAATGCGGCTTTGCTGAACGAGTAGCAAAGCCGCTACATGATTACTGTCTGGGGTTTACCCCCCGTGCAGTCGCCGTGCATCAATCATATGAATATCCTCCTTAG |
| EC204 | spoT | F | ΔspoT | GAATTACAAGCCGTTACCGCTATTGCTGAAGGTCGTCGTTAATCACAAAGCGGGTCGCCCTTGTATCTGTTTGAAAGCCTGAATCAACTGGTGTAGGCTGGAGCTGCTTCG |
| EC204 | spoT | R | ΔspoT | TCAGGCTGACGCCTGGCGAGCATTTCGCATATACGCGCATAACGTTTTGGATTCATAGCGCTAGTTTCGGTTACGGGTGACTTTAATGACCATATGAATATCCTCCTTA |
| EC212 | argI | F | pGEN-luxCDABE | AGTCGGATCCGTGACGCACGCCGTCGTCAT |
| EC212 | argI | R | pGEN-luxCDABE | CGGCTACGTAACTTCCCTCACATGGCTTAGGCCTC |
| EC215 | spoT | F | pGEN-MCS | GTATCATATGGCACGCGTAACTGTTCAGGACGCTG |
| EC215 | spoT | R | pGEN-MCS | AGTCCCATGGCTAGTTTCGGTTACGGGTGACTTTA |
| EC224 | spoT | F | pWSK129 | GTATGAGCTCGCACGCGTAACTGTTCAGGACGCTG |
| EC224 | spoT | R | pWSK129 | AGTCGCGGCCGCCTAGTTTCGGTTACGGGTGACTTTA |
| EC245 | ppnNST | F | ΔppnNST | ATCAGCCAGGGCTATTGTAATCAACAGGGAATGGCGTGTTGCGTCCCATAAGGAGTTTACTTGATTACACATATTAGCCCGCTTGGCTCAGTGTAGGCTGGAGCTGCTTCG |
| EC245 | ppnNST | R | ΔppnNST | ATACCGCAATGAAAGGAATGGGAGAAGCGCCCGGCTGCTGGCGGCAACCGGGCATAAGCGTTAAGCGCAGATCTCGTAACAGGGGATGTACATATGAATATCCTCCTTAG |
| EC320 | spoT | F | pGEN-MCS | GGCGTTCCTGTTCAAGTCCAGATCCGTA |
| EC320 | spoT | R | pGEN-MCS | GTGCGGGCCGATCATTGAGGTGTGCAAA |
| EC321 | wzzST | F | pGEN-luxCDABE | ATGCGGATCCGTGATCAGCATCAACCCCGC |
| EC321 | wzzST | R | pGEN-luxCDABE | GCCATACGTAAGATACCCTAACTAAAAAAAGGATG |
| EC325 | spoT | R | pWSK129 | ACGGATCTGGACTTGAACAGGAACG |
| EC326 | spoT | F | pWSK129 | GGCGTTCCTGTTCAAGTCCAGATCC |
ST and EC denote ppnN from S. Typhimurium strain SL1344 and E. coli strain K-12, respectively.
Bacterial growth curves.
Bacteria grown overnight in LB medium were harvested and normalized to an optical density at 600 nm (OD600) of 0.5. Cells were washed and resuspended in phosphate-buffered saline (PBS) and diluted to an OD600 of 0.05 in fresh LB or M9 minimal medium supplemented with 1% glucose and 0.135 mM l-histidine. Bacteria were then grown in 96-well flat, clear-bottom polystyrene plates (Corning) at 37°C with shaking, and the A600 was measured every 1 h using the BioTek Epoch 2 plate reader.
Serum bactericidal assay.
Bacteria grown overnight in LB medium were harvested and normalized to an OD600 of 0.5. Cells were washed and resuspended in PBS and diluted (1:10) further for the assay. The equivalent OD600 of 0.005 of each strain was incubated in 90% pooled normal human serum (Innovative Research) at 37°C. Serum treated with 5 U/ml cobra venom factor (Quidel) for 30 min at 37°C was used as a negative control for each strain. The number of viable bacteria was determined by plating on LB agar supplemented with 200 μg/ml of ampicillin to select for strains carrying the pGEN-MCS empty vector control or complementation plasmid, and percent survival was calculated as the number of CFU/ml at 30 min relative to 0 min.
Bioluminescence reporter assay.
S. Typhimurium strains containing lux transcriptional fusions were subcultured (1:50) and grown to mid-exponential phase (OD600 of 0.4 to 0.5) in LB and then subcultured (1:50) again into LB in black 96-well flat, clear-bottom polystyrene plates (Corning). Plates were incubated at 37°C with shaking, and luminescence and A600 were measured every 1 h up to 5 h using the PerkinElmer Plate Reader. Luminescence was normalized to A600.
Protein purification.
BL21(DE3) E. coli harboring pET-24a-ppnN-6HIS or pET-24a-ppnNG790C-6HIS was subcultured (1:50) into LB broth at 37°C with shaking and grown until an OD600 of 0.2. Cells were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) until the culture reached mid-logarithmic phase (OD600 of 0.5), after which the culture was left to incubate overnight at 18°C. Cells were pelleted by centrifugation for 10 min at 5,000 × g and resuspended in lysis buffer (100 mM Tris-HCl, 5 mM MgCl2, 10 mM imidazole, 1 mM 2-mercaptoethanol, 300 mM NaCl, 4 mM phenylmethanesulfonyl fluoride, pH 7.5). Cells were lysed using a Continuous Cell Disruptor (Constant Systems Ltd.) operated at 20,000 lb/in2 and then centrifuged at 30,000 × g for 30 min to pellet cellular debris. Lysates were applied to a nickel-nitrilotriacetic acid affinity column and washed with a gradient buffer (100 mM Tris-HCl, 5 mM MgCl2, 500 mM NaCl, pH 7.5) containing 20 mM, 40 mM, and 60 mM imidazole. Proteins were eluted in 100 mM Tris-HCl, 5 mM MgCl2, 500 mM NaCl, 500 mM imidazole, pH 7.5. Eluted fractions were run on a 12% SDS-PAGE gel and stained with Coomassie brilliant blue to verify isolation of the target protein. In a second purification step, elution fractions were combined and applied to a HiLoad 16/60 Superdex 200-pg filtration column (GE Healthcare). Proteins were eluted in 1-ml fractions in buffer containing 10 mM Tris-HCl, 1 mM MgCl2, pH 7.5. Fractions containing the target proteins (as determined by SDS-PAGE) were pooled and stored at −80°C with 20% glycerol.
Enzymatic assay.
Purified proteins (50 μg/ml) were incubated with 0.2 mM of substrate at 37°C in buffer (10 mM Tris-HCl, 1 mM MgCl2, pH 7.5) for 1.5 h. At 0 h and 1.5 h, 150 μl of each reaction mixture was inactivated with 150 μl of methanol cooled at −80°C. For negative-control reactions, a catalytic inactive protein variant was used or wild-type protein was heat inactivated at 95°C for 60 min. Relative abundance of reactant compounds and products was measured using hydrophilic chromatography and mass spectrometry (HILIC-MS) on an LTQ Orbitrap XL (Thermo Fisher).
Antibiotic susceptibility testing.
MIC determinations of ampicillin, colistin, rifampin, and ciprofloxacin (BioShop Canada) were performed using broth microdilution in 96-well flat, clear-bottom polystyrene plates (Corning) (74). Antibiotics were 2-fold serially diluted in LB medium from 512 μg/ml down to ~1e-3 μg/ml. Bacterial cultures grown overnight were diluted to 106 CFU/ml in fresh LB medium and then diluted further to 105 CFU/ml in the microplates containing the antibiotics. Plates were incubated at 37°C in sealed plastic bags. The A600 was read after overnight incubation using a PerkinElmer plate reader.
LPS analysis.
Bacteria grown overnight in LB medium were harvested and normalized to an OD600 of 3.0. Two milliliters of each strain under investigation was pelleted by centrifugation at 16,000 × g for 2 min, and the LPS was extracted as previously described (75), with some modifications. Cells were resuspended in 200 μl of TRIzol reagent (Thermo Fisher) and then incubated for 10 min at room temperature. Next, 20 μl of chloroform was added for every milliliter of culture. The resulting TRIzol-chloroform mixture was vortexed vigorously and incubated at room temperature for an additional 10 min, followed by centrifugation at 16,000 × g for 10 min to separate the aqueous and organic phases. The aqueous phase was transferred to a new microcentrifuge tube, and 100 μl of distilled water was added to the organic phase, vortexed briefly, incubated for an additional 10 min, and centrifuged again for 10 min at 16,000 × g to create phase separation. Two additional water extractions were performed to ensure complete removal of the LPS. The combined aqueous phases were then dried at 45°C for ∼2 h using a Vacufuge (Eppendorf). Dried pellets were resuspended in 500 μl of 0.375 M MgCl2 dissolved in 95% ethanol that had been cooled at −20°C and then centrifuged for 15 min at 16,000 × g (76). The final pellets were normalized by weight/volume (grams per microliter) and resuspended in distilled water. LPS was run on 16% SDS-PAGE gels and stained with silver nitrate as previously described (77) and imaged using the ChemiDoc MP imaging system (Bio-Rad).
Outer membrane profiling.
The outer membrane of different S. Typhimurium strains was isolated as previously described (78), with some modifications. Bacterial cell pellets were harvested from overnight cultures and resuspended in 50 mM Tris-HCl (pH 8.0), 50 mM MgCl2, 150 mM NaCl, and then lysed by sonication. Lysates were centrifuged at 8,000 × g for 10 min and then filtered through 0.2-μm low-protein-binding filters to remove insoluble debris. Total membranes were harvested from cell-free lysates by ultracentrifugation at 100,000 × g for 1 h. To isolate the outer membrane, total membrane pellets were solubilized in the buffer described above supplemented with 1.5% Triton X-100 and EDTA-free protease inhibitor cocktail (Roche Applied Science) for 24 h at 4°C. Outer membranes were pelleted by ultracentrifugation at 100,000 × g for 1 h and then resuspended in sterile 1× PBS buffer. Outer membrane preparations were normalized by protein content following quantification using the 2-D Quant kit (GE Healthcare) and then separated on 10% SDS-PAGE gels. Coomassie brilliant blue R-250 dye (BioShop) was used to stain the gels, followed by imaging using the ChemiDoc MP imaging system (Bio-Rad).
Statistical analysis.
Data were analyzed using GraphPad Prism 5.0a software (GraphPad Inc., San Diego, CA) using one-way analysis of variance. P values of <0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicola Henriquez at the Centre for Microbial Chemical Biology for technical assistance with the HILIC-MS experiments.
This work was supported by a grant to B.K.C. from the Canadian Institutes of Health Research (CIHR) (FRN 156361), a grant to Y.E.Z. from the Novo Nordisk Foundation (NNF19OC0058331), and grants to V.H.B. from Consejo Nacional de Ciencia y Tecnología (254531)-Mexico and Dirección General de Asuntos del Personal Académico de la UNAM (IN202418)-Mexico. B.K.C. holds the Canada Research Chair in Infectious Disease Pathogenesis. N.Y.E.C. is a recipient of an Ontario Graduate Scholarship. W.E. was supported by a CIHR Postdoctoral Fellowship.
We have no conflicts of interest with the contents of this article to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.LaRock DL, Chaudhary A, Miller SI. 2015. Salmonellae interactions with host processes. Nat Rev Microbiol 13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez-Morales D, Banda MM, Chau NYE, Salgado H, Martinez-Flores I, Ibarra JA, Ilyas B, Coombes BK, Bustamante VH. 2017. The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PLoS Pathog 13:e1006497. doi: 10.1371/journal.ppat.1006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilyas B, Mulder DT, Little DJ, Elhenawy W, Banda MM, Perez-Morales D, Tsai CN, Chau NYE, Bustamante VH, Coombes BK. 2018. Regulatory evolution drives evasion of host inflammasomes by Salmonella Typhimurium. Cell Rep 25:825–832. doi: 10.1016/j.celrep.2018.09.078. [DOI] [PubMed] [Google Scholar]
- 5.Sorbara MT, Foerster EG, Tsalikis J, Abdel-Nour M, Mangiapane J, Sirluck-Schroeder I, Tattoli I, van Dalen R, Isenman DE, Rohde JR, Girardin SE, Philpott DJ. 2018. Complement C3 drives autophagy-dependent restriction of cyto-invasive bacteria. Cell Host Microbe 23:644–652. doi: 10.1016/j.chom.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Ricklin D, Hajishengallis G, Yang K, Lambris JD. 2010. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heesterbeek DA, Bardoel BW, Parsons ES, Bennett I, Ruyken M, Doorduijn DJ, Gorham RD Jr, Berends ET, Pyne AL, Hoogenboom BW, Rooijakkers SH. 2019. Bacterial killing by complement requires membrane attack complex formation via surface-bound C5 convertases. EMBO J 38:e99852. doi: 10.15252/embj.201899852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heesterbeek DAC, Martin NI, Velthuizen A, Duijst M, Ruyken M, Wubbolts R, Rooijakkers SHM, Bardoel BW. 2019. Complement-dependent outer membrane perturbation sensitizes Gram-negative bacteria to Gram-positive specific antibiotics. Sci Rep 9:3074. doi: 10.1038/s41598-019-43208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambris JD, Ricklin D, Geisbrecht BV. 2008. Complement evasion by human pathogens. Nat Rev Microbiol 6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bravo D, Silva C, Carter JA, Hoare A, Alvarez SA, Blondel CJ, Zaldivar M, Valvano MA, Contreras I. 2008. Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J Med Microbiol 57:938–946. doi: 10.1099/jmm.0.47848-0. [DOI] [PubMed] [Google Scholar]
- 11.Maldonado RF, Sa-Correia I, Valvano MA. 2016. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev 40:480–493. doi: 10.1093/femsre/fuw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huja S, Oren Y, Biran D, Meyer S, Dobrindt U, Bernhard J, Becher D, Hecker M, Sorek R, Ron EZ. 2014. Fur is the master regulator of the extraintestinal pathogenic Escherichia coli response to serum. mBio 5:e01460-14. doi: 10.1128/mBio.01460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng ZX, Gong QY, Wang Z, Chen ZG, Ye JZ, Li J, Wang J, Yang MJ, Ling XP, Peng B. 2017. Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front Immunol 8:1706. doi: 10.3389/fimmu.2017.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng ZX, Guo C, Chen ZG, Yang TC, Zhang JY, Wang J, Zhu JX, Li D, Zhang TT, Li H, Peng B, Peng XX. 2019. Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat Commun 10:3325. doi: 10.1038/s41467-019-11129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 17.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci U S A 70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown A, Fernandez IS, Gordiyenko Y, Ramakrishnan V. 2016. Ribosome-dependent activation of stringent control. Nature 534:277–280. doi: 10.1038/nature17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winther KS, Roghanian M, Gerdes K. 2018. Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-site. Mol Cell 70:95–105. doi: 10.1016/j.molcel.2018.02.033. [DOI] [PubMed] [Google Scholar]
- 20.Battesti A, Bouveret E. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol 62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee JW, Park YH, Seok YJ. 2018. Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc Natl Acad Sci U S A 115:E6845–E6854. doi: 10.1073/pnas.1722514115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinella D, Albrecht C, Cashel M, D'Ari R. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 23.Sarubbi E, Rudd KE, Xiao H, Ikehara K, Kalman M, Cashel M. 1989. Characterization of the spoT gene of Escherichia coli. J Biol Chem 264:15074–15082. [PubMed] [Google Scholar]
- 24.Wang B, Dai P, Ding D, Del Rosario A, Grant RA, Pentelute BL, Laub MT. 2019. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat Chem Biol 15:141–150. doi: 10.1038/s41589-018-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YE, Zbornikova E, Rejman D, Gerdes K. 2018. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio 9:e02188-17. doi: 10.1128/mBio.02188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YE, Bærentsen RL, Fuhrer T, Sauer U, Gerdes K, Brodersen DE. 2019. ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Mol Cell 74:1239–1249. doi: 10.1016/j.molcel.2019.03.035. [DOI] [PubMed] [Google Scholar]
- 27.Wang B, Grant RA, Laub MT. 2020. ppGpp coordinates nucleotide and amino-acid synthesis in E. coli during starvation. Mol Cell 80:29–42. doi: 10.1016/j.molcel.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson A, Rolfe MD, Lucchini S, Schwerk P, Hinton JC, Tedin K. 2006. The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J Biol Chem 281:30112–30121. doi: 10.1074/jbc.M605616200. [DOI] [PubMed] [Google Scholar]
- 29.Tapscott T, Kim JS, Crawford MA, Fitzsimmons L, Liu L, Jones-Carson J, Vazquez-Torres A. 2018. Guanosine tetraphosphate relieves the negative regulation of Salmonella pathogenicity island-2 gene transcription exerted by the AT-rich ssrA discriminator region. Sci Rep 8:9465. doi: 10.1038/s41598-018-27780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandran VK, Shearer N, Thompson A. 2014. The primary transcriptome of Salmonella enterica serovar Typhimurium and its dependence on ppGpp during late stationary phase. PLoS One 9:e92690. doi: 10.1371/journal.pone.0092690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzsimmons LF, Liu L, Kant S, Kim JS, Till JK, Jones-Carson J, Porwollik S, McClelland M, Vazquez-Torres A. 2020. SpoT induces intracellular Salmonella virulence programs in the phagosome. mBio 11:e03397-19. doi: 10.1128/mBio.03397-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 33.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentry DR, Cashel M. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol 19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 35.Schakermann M, Langklotz S, Narberhaus F. 2013. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J Bacteriol 195:1912–1919. doi: 10.1128/JB.02134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alper CA, Balavitch D. 1976. Cobra venom factor: evidence for its being altered cobra C3 (the third component of complement). Science 191:1275–1276. doi: 10.1126/science.56780. [DOI] [PubMed] [Google Scholar]
- 37.Berends ET, Mohan S, Miellet WR, Ruyken M, Rooijakkers SH. 2015. Contribution of the complement membrane attack complex to the bactericidal activity of human serum. Mol Immunol 65:328–335. doi: 10.1016/j.molimm.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Sévin DC, Fuhrer T, Zamboni N, Sauer U. 2017. Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli. Nat Methods 14:187–194. doi: 10.1038/nmeth.4103. [DOI] [PubMed] [Google Scholar]
- 39.Bærentsen RL, Brodersen DE, Zhang YE. 2019. Evolution of the bacterial nucleosidase PpnN and its relation to the stringent response. Microb Cell 6:450–453. doi: 10.15698/mic2019.09.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 41.Naseem M, Sarukhanyan E, Dandekar T. 2015. LONELY-GUY knocks every door: crosskingdom microbial pathogenesis. Trends Plant Sci 20:781–783. doi: 10.1016/j.tplants.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo H, Kim S, Sagong HY, Son HF, Jin KS, Kim IK, Kim KJ. 2016. Structural basis for cytokinin production by LOG from Corynebacterium glutamicum. Sci Rep 6:31390. doi: 10.1038/srep31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189–198. doi: 10.1016/S0092-8674(00)81750-X. [DOI] [PubMed] [Google Scholar]
- 45.Ramu P, Tanskanen R, Holmberg M, Lahteenmaki K, Korhonen TK, Meri S. 2007. The surface protease PgtE of Salmonella enterica affects complement activity by proteolytically cleaving C3b, C4b and C5. FEBS Lett 581:1716–1720. doi: 10.1016/j.febslet.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 46.Hammarlöf DL, Kröger C, Owen SV, Canals R, Lacharme-Lora L, Wenner N, Schager AE, Wells TJ, Henderson IR, Wigley P, Hokamp K, Feasey NA, Gordon MA, Hinton JCD. 2018. Role of a single noncoding nucleotide in the evolution of an epidemic African clade of Salmonella. Proc Natl Acad Sci U S A 115:E2614–E2623. doi: 10.1073/pnas.1714718115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro Llorens JM, Tormo A, Martinez-Garcia E. 2010. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev 34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 48.Potrykus K, Murphy H, Philippe N, Cashel M. 2011. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol 13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci U S A 102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw JA, Henard CA, Liu L, Dieckman LM, Vazquez-Torres A, Bourret TJ. 2018. Salmonella enterica serovar Typhimurium has three transketolase enzymes contributing to the pentose phosphate pathway. J Biol Chem 293:11271–11282. doi: 10.1074/jbc.RA118.003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murray GL, Attridge SR, Morona R. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol 188:2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ricci V, Zhang D, Teale C, Piddock LJV. 2020. The O-antigen epitope governs susceptibility to colistin in Salmonella enterica. mBio 11:e02831-19. doi: 10.1128/mBio.02831-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaara M, Vaara T. 1983. Sensitization of Gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature 303:526–528. doi: 10.1038/303526a0. [DOI] [PubMed] [Google Scholar]
- 54.Larue K, Kimber MS, Ford R, Whitfield C. 2009. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J Biol Chem 284:7395–7403. doi: 10.1074/jbc.M809068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou YN, Coleman WG Jr, Yang Z, Yang Y, Hodgson N, Chen F, Jin DJ. 2008. Regulation of cell growth during serum starvation and bacterial survival in macrophages by the bifunctional enzyme SpoT in Helicobacter pylori. J Bacteriol 190:8025–8032. doi: 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hooven TA, Catomeris AJ, Bonakdar M, Tallon LJ, Santana-Cruz I, Ott S, Daugherty SC, Tettelin H, Ratner AJ. 2017. The Streptococcus agalactiae stringent response enhances virulence and persistence in human blood. Infect Immun 86:e00612-17. doi: 10.1128/IAI.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez-Larrayoz AF, Elhosseiny NM, Chevrette MG, Fu Y, Giunta P, Spallanzani RG, Ravi K, Pier GB, Lory S, Maira-Litran T. 2017. Complexity of complement resistance factors expressed by Acinetobacter baumannii needed for survival in human serum. J Immunol 199:2803–2814. doi: 10.4049/jimmunol.1700877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dasgupta S, Das S, Biswas A, Bhadra RK, Das S. 2019. Small alarmones (p)ppGpp regulate virulence associated traits and pathogenesis of Salmonella enterica serovar Typhi. Cell Microbiol 21:e13034. doi: 10.1111/cmi.13034. [DOI] [PubMed] [Google Scholar]
- 59.Colomer-Winter C, Gaca AO, Lemos JA. 2017. Association of metal homeostasis and (p)ppGpp regulation in the pathophysiology of Enterococcus faecalis. Infect Immun 85:e00260-17. doi: 10.1128/IAI.00260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson RP, Winter SE, Spees AM, Winter MG, Nishimori JH, Sanchez JF, Nuccio SP, Crawford RW, Tukel C, Baumler AJ. 2011. The Vi capsular polysaccharide prevents complement receptor 3-mediated clearance of Salmonella enterica serotype Typhi. Infect Immun 79:830–837. doi: 10.1128/IAI.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samanovic MI, Tu S, Novak O, Iyer LM, McAllister FE, Aravind L, Gygi SP, Hubbard SR, Strnad M, Darwin KH. 2015. Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol Cell 57:984–994. doi: 10.1016/j.molcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA. 2008. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog 4:e37. doi: 10.1371/journal.ppat.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hochstadt-Ozer J, Cashel M. 1972. The regulation of purine utilization in bacteria. V. Inhibition of purine phosphoribosyltransferase activities and purine uptake in isolated membrane vesicles by guanosine tetraphosphate. J Biol Chem 247:7067–7072. [PubMed] [Google Scholar]
- 64.Rinas U, Hellmuth K, Kang R, Seeger A, Schlieker H. 1995. Entry of Escherichia coli into stationary phase is indicated by endogenous and exogenous accumulation of nucleobases. Appl Environ Microbiol 61:4147–4151. doi: 10.1128/AEM.61.12.4147-4151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Link H, Fuhrer T, Gerosa L, Zamboni N, Sauer U. 2015. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat Methods 12:1091–1097. doi: 10.1038/nmeth.3584. [DOI] [PubMed] [Google Scholar]
- 66.Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. 2008. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanchez-Vazquez P, Dewey CN, Kitten N, Ross W, Gourse RL. 2019. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc Natl Acad Sci U S A 116:8310–8319. doi: 10.1073/pnas.1819682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. 2009. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nishio M, Okada N, Miki T, Haneda T, Danbara H. 2005. Identification of the outer-membrane protein PagC required for the serum resistance phenotype in Salmonella enterica serovar. Microbiology 151:863–873. doi: 10.1099/mic.0.27654-0. [DOI] [PubMed] [Google Scholar]
- 70.Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang HJ, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One 9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 72.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 75.Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651–656. doi: 10.1039/b000368i. [DOI] [PubMed] [Google Scholar]
- 76.Darveau RP, Hancock RE. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol 155:831–838. doi: 10.1128/JB.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsai CM, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem 119:115–119. doi: 10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- 78.Page WJ, Taylor DE. 1988. Comparison of methods used to separate the inner and outer membranes of cell envelopes of Campylobacter spp. J Gen Microbiol 134:2925–2932. doi: 10.1099/00221287-134-11-2925. [DOI] [PubMed] [Google Scholar]
- 79.Coombes BK, Brown NF, Kujat-Choy S, Vallance BA, Finlay BB. 2003. SseA is required for translocation of Salmonella pathogenicity island-2 effectors into host cells. Microbes Infect 5:561–570. doi: 10.1016/S1286-4579(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 80.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 82.Dykxhoorn DM, St Pierre R, Linn T. 1996. A set of compatible tac promoter expression vectors. Gene 177:133–136. doi: 10.1016/0378-1119(96)00289-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.