Abstract
MITF, a gene that is mutated in familial melanoma and Waardenburg syndrome, encodes multiple isoforms expressed from alternative promoters that share common coding exons but have unique amino termini. It is not completely understood how these isoforms influence pigmentation in different tissues and how the expression of these independent isoforms of MITF is regulated. Here, we show that melanocytes express two isoforms of MITF, MITF-A and MITF-M. The expression of MITF-A is partially regulated by a newly identified retinoid enhancer element located upstream of the MITF-A promoter. Mitf-A knockout mice have only subtle changes in melanin accumulation in the hair and reduced Tyr expression in the eye. In contrast, Mitf-M-null mice have enlarged kidneys, lack neural crest-derived melanocytes in the skin, choroid, and iris stroma, yet maintain pigmentation within the retinal pigment epithelium and iris pigment epithelium of the eye. Taken together, these studies identify a critical role for MITF-M in melanocytes, a minor role for MITF-A in regulating pigmentation in the hair and Tyr expression in the eye, and a novel role for MITF-M in size control of the kidney.
Keywords: alternative promoters, choroid, CRISPR/CAS9, isoforms, melanogenesis, microphthalmia-associated transcription factor, MITF, retinal pigment epithelium, retinoids
1 ∣. INTRODUCTION
In the human genome, alternative promoters are present in 30%–50% of coding genes where they contribute to the diversity of the proteome and allow more sophisticated control of gene expression (Davuluri, Suzuki, Sugano, Plass, & Huang, 2008). Alternative promoters contain unique sequences to regulate the expression of distinct isoforms in response to environmental or developmental cues. Genes including EIF1AX and HBG1 utilize alternative promoters where one promoter lacks a TATA box for the expression of isoforms at the correct time during development (Davis & Schultz, 2000; Davuluri et al., 2008). In differentiation, alternative promoters allow for the expression of isoforms in tissue and cell-specific manners (Landry, Mager, & Wilhelm, 2003). Aberrant use of alternative promoters is associated with cancer. Transcripts generated through the TP53 P1 promoter produce p53 protein isoforms containing transactivation domains that allow p53 to regulate its tumor suppressor functions (Kim & An, 2016), while transcription from the P2 promoter produces p53 protein that lacks the transactivation domains (Kim & An, 2016), and promote breast cancer stem cell maintenance (Arsic et al., 2015).
Microphthalmia-associated transcription factor (MITF) is the master regulator of melanocyte cell identity, controlling the production of melanin that gives skin, hair, and eyes color (Vachtenheim & Ondrusov, 2013). Mutations in the human MITF locus are associated with increased risk for familial melanoma, Waardenburg syndrome, and Tietz syndrome (Bertolotto et al., 2011; Pingault et al., 2010). The MITF gene has nine distinct isoforms that are produced using alternative first coding exons (Bharti, Liu, Csermely, Bertuzzi, & Arnheiter, 2008). Some isoforms have ubiquitous expression, while others are more restricted to specific cell types and tissues (Amae et al., 1998). While the amino-terminus of MITF isoforms differ, all isoforms contain the basic helix–loop–helix leucine zipper that is required for DNA binding and dimer formation with other bHLH family transcription factors (Pogenberg et al., 2012). Mitf mutations in mice cause varied phenotypes with distinct features including premature graying, microphthalmia, and depigmentation (Levy, Khaled, & Fisher, 2006). Although mice with mutations in the promoter regions of Mitf have been identified (Steingrimsson, Copeland, & Jenkins, 2004), the mutant alleles involve the loss of multiple isoforms, so it is still unclear how specific isoforms of MITF regulate pigmentation. Established methods for studying the role of Mitf isoforms rely on (a) mutant alleles that affect multiple isoforms (Steingrímsson et al., 2004); (b) examination of the expression pattern of isoforms in target tissues of wild-type and Mitf mutant animals to infer their function during development (Bharti et al., 2008); (c) isoform-specific overexpression studies to determine the function of individual isoforms (Michael et al., 2018); or (d) isoform-specific knockouts as previously performed (Bharti et al., 2012; Phelep et al., 2017). More precise knockout models are needed to deconvolute the role of individual isoforms in tissue development and disease.
While published studies suggest that MITF-M is the main isoform expressed in melanocytes, the role of other MITF isoforms in melanocyte biology has not been studied. Here, we demonstrate that the human MITF-A isoform is expressed in melanocytes and its expression is regulated by retinoids. To distinguish the roles of Mitf-A and Mitf-M in pigment cell development and melanogenesis, we use CRISPR/Cas9 to generate Mitf-A and Mitf-M isoform-specific knockout mice. We observe that Mitf-A knockout mice have a subtle loss of pigmentation in the hair, while Mitf-M knockout mice lack melanocytes in the epidermis, hair follicle, iris, and choroid. Our study illustrates the utility of a CRISPR/Cas9-based approach to define specific roles for MITF isoforms in tissue development and pigmentation.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Mouse strains and genotyping
All animal experiments were approved by the UC Irvine Institutional Animal Care and Use Committee (IACUC) (AUP-17–230). ROSAmTmG/mTmG mice (Stock No: 007576), C57BL/6J (Stock No: 000664), and B6(Cg)-Tyrc-2J/J mice (Stock No: 000058) were obtained from The Jackson Laboratory. C57BL/6NTac zygotes for genome editing were obtained from Taconic Biosciences. C57BL/6N mice used for crb1 genotyping were obtained from the KOMP repository. Genotyping primers for Mitf mice were designed following the characterization of deletions. All other genotyping primers follow guidelines provided by The Jackson Laboratory. Genotyping primers are provided in Table S5 and additionally described in Appendix S1.
2.2 ∣. CRISPR/Cas9 genome editing
Mitf mutant mice were generated by pronuclear injection of CRISPR/Cas9 reagents into C57BL/6NTac zygotes. Guide RNAs (gRNAs) were designed using sgRNA Designer (Broad Institute, MIT) and GTScan (EMBL, Australia) algorithms. Guide-RNA templates were synthesized by PCR using oligos and GBLOCK oligos (IDT). In vitro-transcribed (IVT) gRNA was made using mMessage mMachine (Thermo Fisher) and purified by MEGAclear Kit (Thermo Fisher). Cas9 mRNA was made by IVT using the same reagents with px330 plasmid (Addgene plasmid #42230) (Cong, Ran, Cox, Lin, & Barretto, 2013). RNAs were injected at 20 ng/μl each. Resulting mice were screened by PCR and T7 endonuclease. T7 endo-positive alleles were amplified by nested PCR and Sanger-sequenced. CRISPR alleles were separated from hemizygous founders by outcrossing the mice to C57BL/6J mice (The Jackson Laboratory, Stock No: 000664). Lines were established from the F2 generation that was predicted to knock out expression of each specific Mitf transcript. All primers and gRNA used are listed in Table S6.
The Mitfem1Gane allele (MGI: 6273202) contains a 7 bp deletion in chromosome 6, spanning 97,807,194–97,807,200 in Ensembl assembly GRCm38.p6. The deletion in this allele results in a loss of the Mitf-A isoform, designated as Mitf variant 202 in Ensembl and NM_001113198 in NCBI. Homozygotes for the Mitfem1Gane are referred to as Mitf-A knockouts or nulls. The Mitfem2Gane allele (MGI: 6273203) contains an 18 bp deletion in chromosome 6, spanning 97,991,944–97,991,961 in Ensembl assembly GRCm38.p6. The deletion in the Mitfem2Gane allele results in a loss of the Mitf-M isoform through the loss of a splice site, and this is designated as Mitf variant 201 in Ensembl and NM_008601 in NCBI. Homozygotes for the Mitfem2Gane deletion are referred to as Mitf-M knockouts or nulls.
2.3 ∣. Single-cell isolation, sorting, and RNA-seq
Shaved and depilated whole back skin of P54 ROSAmTmG mice was dissociated into a single-cell suspension and incubated with Ghost Dye Red 780 (Tonbo Biosciences) to identify viable cells. Live GFP+ and tdTomato+ cells were sorted using a BD FACSAria Fusion Sorter, and RNA-seq was performed as described in Appendix S1. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar, Domrachev, & Lash, 2002) and are accessible through GEO Series accession number GSE138538 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138538).
2.4 ∣. Analysis of read junctions
BAM files generated in Tophat were visualized against the annotated mm10 transcriptome in IGV (version 2.4.8) (Robinson et al., 2011; Thorvaldsdóttir, Robinson, & Mesirov, 2013). After identifying read junctions present between exons 1A to 1B, 1B to 2A, and 1M to 2A, read junctions were counted between exons 1B and 2A as proxy for Mitf-A as no other isoforms containing exon 1B were found. Read junctions between exons 1M and 2A were counted as proxy for Mitf-M. The percentage of Mitf-A and Mitf-M transcripts was determined for GFP+ and tdTomato+ cells. To account for differential expression of Mitf between melanocytes and other skin cells, the percentage of each isoform was multiplied by the relative abundance of Mitf in the respective cell population. Pearson's chi-squared test performed in R (version 3.3.2) was used to determine whether the expression of individual isoforms was significantly different between samples.
Human RNA-seq raw data files (SRP039354) were retrieved from the SRA database (Haltaufderhyde & Oancea, 2014) and aligned to the NCBI human reference genome GRCh38 using Tophat alignment software (version 2.0.12) (Trapnell, Pachter, & Salzberg, 2009). BAM files were visualized as described above. To delineate isoforms containing exon 1B, the proportion of read junctions from upstream exons (1A, 1J, 1C, 1E, and 1H) to 1B was multiplied by the proportion of read junctions to 2A containing 1B. Read junctions between 1M and 2A were still used as proxy for MITF-M.
2.5 ∣. Melanocyte isolation using CD117 microbeads
Neonatal mouse melanocytes were collected as previously described (Godwin et al., 2014; Liggins et al., 2018). Mice less than 3 days old were euthanized and sterilized. Skin was removed and cleaned of muscle. The epidermis was separated from the dermis following a 1-hr incubation in 5mg/ml trypsin (Sigma-Aldrich) at 37°C. The epidermis was chopped in 0.25% trypsin-EDTA solution (Gibco) and resuspended in RPMI with 5% FBS. The resuspended cells were filtered using a 100-μm cell strainer followed by a 40-μm cell strainer. Cells were sedimented and washed in PBS (pH 7.2) with 0.5% BSA (Fisher Scientific). The cell pellet was resuspended and incubated with CD117 MicroBeads (Miltenyi Biotec) following the manufacturers' protocol. CD117-positive and CD117-negative cells were then lysed for RNA extraction using RNAeasy Mini Kit (Qiagen), and cDNA was generated using a high-capacity RNA to cDNA Kit (Life Technologies).
2.6 ∣. Cell lines and cell culture
Detailed culturing methods for human MNT-1 melanoma cells, human deeply pigmented neonatal epidermal melanocytes, and HEK293T cells are provided in Appendix S1.
2.7 ∣. RNA isolation, reverse transcriptase-quantitative PCR
Human cell lines were harvested using Tri-Reagent solution (Ambion), and RNA was extracted using a Direct-zol RNA Miniprep Kit (Zymo Research). Complementary DNA (cDNA) was synthesized from total RNA using a high-capacity RNA to cDNA Kit (Life Technologies).
Enucleated eyes from adult mice were dissected to isolate the posterior chamber under a Leica DMC2900 stereo-microscope. The retina was carefully removed, and the cells of the choroid and RPE were scraped from the sclera and placed into RNAlater (Invitrogen). RNA was extracted using the RNAeasy Kit (Qiagen) and cDNA was synthesized utilizing Superscript II RT (Invitrogen) with random primers. All reverse transcriptase-quantitative PCR (RT-qPCR) primers used are listed in Table S4. For additional details, see Appendix S1.
2.8 ∣. Nanostring nCounter analysis on whole-mouse skin
RNA was isolated from mice at P60 following stimulation of 3rd Anagen as described (Paterson et al., 2015). A full description of RNA isolation and Nanostring analysis is described in Appendix S1. For RT-qPCR analysis on skin, cDNA was synthesized as described above for cells sorted with CD117 beads.
2.9 ∣. Identification of binding motifs and chromatin immunoprecipitation
Putative RXR/RAR binding sites in the MITF-A promoter were identified using MotifMap and the hg18 reference genome (Daily, Patel, Rigor, Xie, & Baldi, 2011; Xie, Rigor, & Baldi, 2009). Human NCBI accession numbers used were NM_00198159 (MITF-A) and NM_00248 (MITF-M) and corresponding mouse NCBI accession numbers are NM_002223198 (Mitf-A) and NM_008601 (Mitf-M). Conservation of the RXR/RAR binding site between human and mouse (mm10 reference genome) was determined by aligning 1,000 bp upstream of the transcription start site of MITF-A for the human and murine sequences in MegAlign (version 14.0.0) (DNASTAR). Two kilobases of DNA upstream of the transcription start site for each human promoter was designated as the “promoter region” and tiled using four 500 bp “tiles.” Primers for quantitative PCR targeted each of these four tiled regions are listed in Table S4 and described in Appendix S1.
2.10 ∣. Drug treatment and dual-luciferase reporter assay
HEK293T cells were transfected with 1 μg of a firefly luciferase reporter driven by human MITF-A full-length promoter (MITF-A EcoRI) or a truncated MITF-A promoter (MITF-A Smal). Both plasmids were a kind gift of Dr. Shigeki Shibahara as previously described (Udono et al., 2000). An additional construct with a mutagenized promoter (MITF-A Mut) was generated by site-specific mutagenesis (Agilent Technologies) as described in Appendix S1.
2.11 ∣. Melanin quantification
After shaving the dorsal hairs of mice at post-natal day 50 (P50), 1 mg of hair was dissolved overnight in 1 ml of 90% Soluene-350 (PerkinElmer) and 10% water at 65°C. Quadruplicate 150 μl aliquots for each mouse hair sample were analyzed for absorbance at 405 nm as previously described (Liggins et al., 2018). Melanin quantification of tail skin was performed on 1-cm sections of whole skin dissolved in 90% Soluene-350 as described above. To measure melanin in eyes of mice, the eyes were enucleated and dissected. The iris and the posterior chamber of the eye, following removal of retina, were separated and dissolved overnight in Soluene-350 as described above.
2.12 ∣. Histology
Eyes were collected from adult mice, and connective tissue around the eye was removed. The eyes were then fixed in 10% formalin, dehydrated in ethanol, and embedded in paraffin. Seven-micron-thick sections were dewaxed and rehydrated in ethanol and stained with hematoxylin and eosin to view general structure. Images at 20X were taken using a Nikon Eclipse E200. High magnification images of iris were taken with a 63X oil lens on a Zeica ApoTome2.
2.13 ∣. Western blots and immunoprecipitation
Total MITF levels were measured by generating protein lysates from both eyes of adult mice and subjecting them to immunoblotting with an MITF antibody as described in Appendix S1.
2.14 ∣. Immunofluorescence staining
Skin and eyes were collected at P56, fixed, and embedded in optimal cutting temperature (OCT) compound, sectioned, and probed with a CD117 antibodies to identify melanocytes (see Appendix S1 for more details).
2.15 ∣. Kidney characterization
Dissected kidneys were weighed, and one kidney per mouse was macerated to count glomeruli. Detailed methods are described in Appendix S1.
3 ∣. RESULTS
3.1 ∣. Multiple Mitf isoforms are expressed in melanocytes
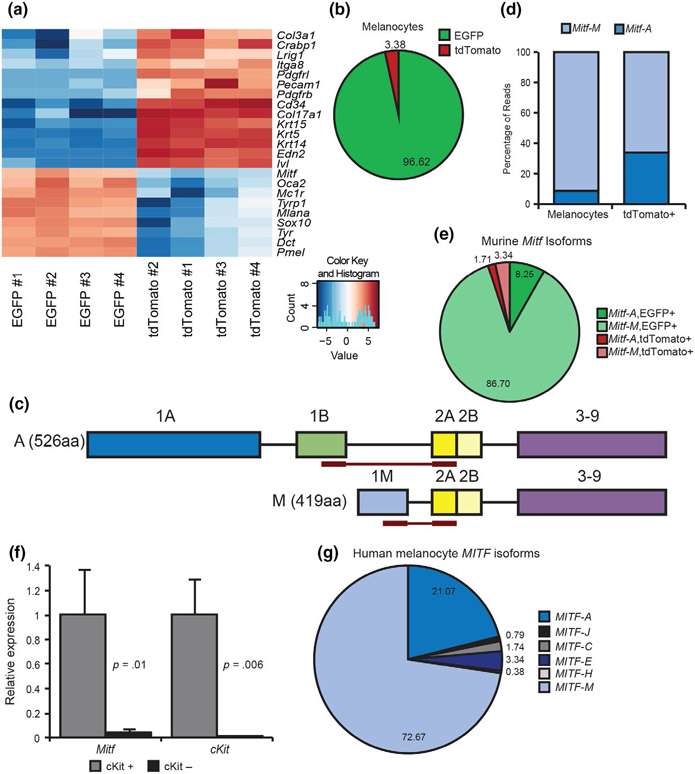
To identify which Mitf isoforms are expressed in melanocytes, we first crossed Tyr::CreERT2 mice (Bosenberg et al., 2006) with ROSAmTmG mice (Muzumdar, Tasic, Miyamichi, Li, & Luo, 2007). In Tyr::CreERT2, ROSAmTmG double heterozygous mice, injection of tamoxifen induces melanocytes to express EGFP, whereas all other cells in the epidermis express tdTomato (Figure S1a) (Liggins et al., 2018). Single-cell suspensions were generated from mouse skin of the indicated genotypes and sorted into tdTomato+ and EGFP+ populations for bulk RNA-seq. A PCA plot demonstrates that our sorting procedure could separate melanocytes from other skin cell types (Figure S1b). We next examined the expression of a panel of melanocyte, keratinocyte, fibroblast, and endothelial cell-specific transcripts in EGFP+ and tdTomato+ cells (Figure 1a, Table S1). Almost all (97%) of the transcripts encoding known melanogenesis regulators were expressed in the GFP+ sorted cells (Figure 1b). The remaining 3% of transcripts encoding melanocyte-specific markers were expressed in tdTomato+ cells, indicating that the induction of the Tyr::CreERT2 transgene was incomplete. In contrast, over 99% of Trp63 transcripts (a transcription factor that regulates keratinocyte development (Mills et al., 1999; Yang et al., 1999)) were found in the tdTomato+ populations (Figure S1c), indicating that our method could efficiently sort melanocytes from other skin cells.
FIGURE 1.
Melanocytes express multiple isoforms of Mitf. Cells were collected from Tyr:CreERT2, ROSAmTmG mice at P54 and sorted into EGFP+ and tdTomato+ populations using FACS. (a) Heatmap of RNA-seq gene expression for melanocyte-specific genes and other skin cell markers in EGFP+ and tdTomato+ cells. Transcripts range from those with high abundance (dark red) to those with low abundance (dark blue). (b) Melanocyte-specific gene expression in EGFP+ and tdTomato+ cells. (c) Schematic illustrating read junctions for Mitf-A and Mitf-M. (d) Relative distribution of Mitf isoforms in mouse EGFP+ melanocytes and tdTomato+ skin cell subpopulations. n = 4, χ2 = 37,663, p < .0001. (e) Distribution of Mitf isoforms in melanocytes and other skin cell populations of the skin. (f) Relative Mitf expression of cKit-positive and cKit-negative cells from neonatal epidermis sorted on microbeads. (g) MITF isoform relative abundance in human melanocytes
To determine the isoforms of Mitf expressed in mouse skin, BAM files for MITF transcripts were visualized and unique read junctions were counted using IGV 2.3.77 (Robinson et al., 2011; Thorvaldsdóttir et al., 2013). Read junctions between exon 1A and 1B would correspond to Mitf-A. Notably, no other read junctions between exon 1B and other exons were observed in our dataset when aligned to the annotated mouse transcriptome. For simplicity, read junctions between exons 1B and 2A were counted as a proxy for Mitf-A, while read junctions between exons 1M and 2A were considered a proxy for Mitf-M (Figure 1c). In EGFP+ melanocytes, Mitf-A accounts for 9% of total Mitf, while Mitf-M accounts for the other 91% (Figure 1d). Of the Mitf expression in tdTomato+ skin cells, 66% was identified as Mitf-M (Figure 1d). To account for the differential expression of Mitf isoforms in EGFP+ and tdTomato+ cell populations, we calculated the relative abundance of these transcripts in melanocytes as compared to all other skin cells (Figure 1e). To validate RNA-seq findings, cKit+ cells (an established marker of melanocytes (Aoki et al., 2009)) were isolated from wild-type neonatal epidermis. RNA was isolated from cKit+ and cKit− cells and reverse-transcribed to quantify Mitf isoform expression. Mitf transcripts were abundant in cKit-expressing cells and largely absent in cKit-negative cells (Figure 1f), suggesting that the low levels of Mitf transcripts detected in tdTomato+ cells are a result of the presence of uninduced melanocytes in that sample.
To verify that both MITF-M and MITF-A are expressed in human melanocytes, we measured the relative abundance of MITF-M and MITF-A transcripts using isoform-specific RT-qPCR primers. In MNT-1-pigmented melanoma cells and deeply pigmented human melanocytes, both MITF-M and MITF-A are expressed at similar proportions as observed in the adult mouse (Figure S1d-e). Additionally, utilizing a published human epidermal melanocyte RNA-seq dataset (Haltaufderhyde & Oancea, 2014), we determined the relative isoform abundance of MITF isoforms by counting read junctions as proxy for individual isoforms. Multiple isoforms containing exon 1B were identified, so read junctions between exons 1B and 2 were taken as proxy for all exon 1B containing isoforms (Figure S1f). Relative abundance for each individual isoform containing exon 1B was proportioned by counting read junctions from exons 1A, 1J, 1C, 1E, and 1H to exon 1B (Figure S1f). Read junctions between exons 1M and 2 were counted as proxy for MITF-M (Figure S1f). In human melanocytes, 73% of MITF was MITF-M and the next most abundant isoform was MITF-A, which accounted for 21% (Figure 1g). The other isoforms present accounted for less than 6% of total MITF expression (Figure 1g). These results confirm that MITF-M is the primary isoform expressed in both mouse and human melanocytes, and MITF-A is the second most abundant isoform expressed in melanocytes.
3.2 ∣. The MITF-A promoter contains a RXR/RAR binding site
To determine how MITF-A is differentially regulated in melanocytes, we searched for unique binding motifs in the alternative promoters for human MITF-A and MITF-M. Utilizing MotifMap (Daily et al., 2011; Xie et al., 2009), we verified the presence of known sites that mediate MITF-M transcription including CREB and PAX (Table S2). We also identified a putative RXR/RAR DR5 binding site 714 base pairs (bp) upstream of the transcription start for MITF-A (Table S3). While the promoter regions, including the RXR/RAR binding site, for the A isoform, are conserved in the mouse 740 bp upstream of the transcription start site (Figure S2), the expression of MITF-A is more robust in human melanocytes (Figure 1, Figure S1). We previously showed that 9-cis retinoic acid upregulates MITF and TYR expression in cultured melanocytes, stimulating pigment production in melanocyte and melanoma cell lines (Paterson, Ho, Kapadia, & Ganesan, 2013). Both the retinoic acid receptors (RARs) and retinoid x receptors (RXRs) form dimers that are activated in response to retinoids (Allenby et al., 1993). Additionally, RXRα is expressed in melanocytes (Reichrath et al., 1995) and a RXRα hypomorphic (I273N) mouse mutant exhibits premature graying (Du et al., 2005).
Having established that MITF-A expression levels were higher in human melanocytes compared with mouse, we next sought to validate the binding of RXR and RAR to the MITF-A human promoter. For ChIP analysis, we designed tiled primers for 2kb upstream of transcription start sites for both MITF-A and MITF-M (Figure 2a). The putative RXR/RAR DR5 binding site is located in tile A3. In the human MNT-1 melanoma cell line, we found that pulldown with RARα significantly enriched for tiles A3 and A4 (Figure 2b) and RXRα enriched for tile A4 (Figure 2c). Using deeply pigmented (DP) human melanocytes, tile A4 was significantly enriched with RARα, while tiles A2 and A3 were approaching significance (Figure 2d). For RXRα in DP melanocytes, only tile A3 was approaching significance (Figure 2e). Consistent with our MotifMap findings, RARα and RXRα did not bind to the MITF-M tiles (Figure 2b-e).
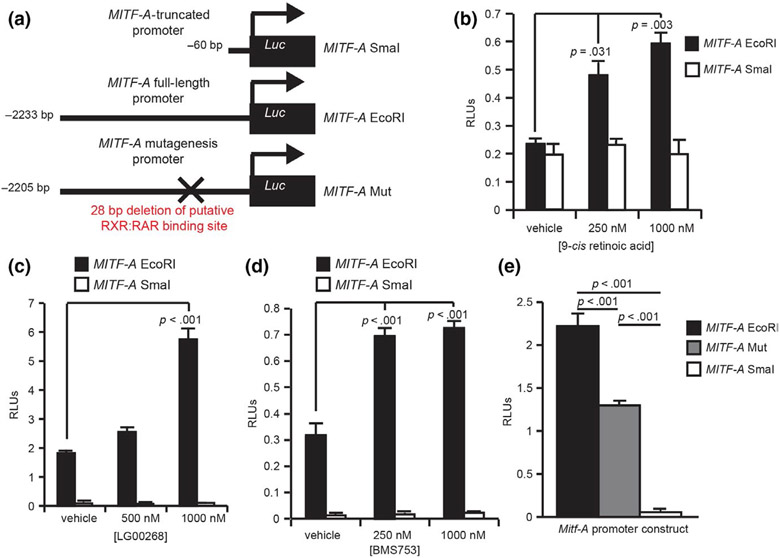
FIGURE 2.
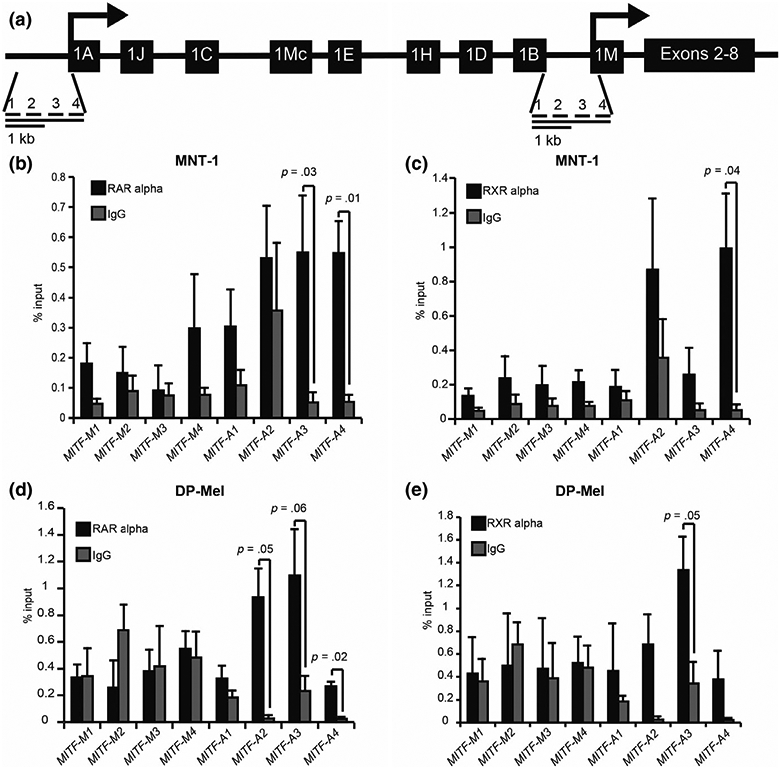
RXR/RAR binds upstream of the human MITF-A promoter. (a) Tiled primers for ChIP analysis were designed to target the promoter region 2 kb upstream of the transcription start site for both MITF-A and MITF-M isoforms. (b–e) Enrichment of both (b, d) RARα and (c, e) RXRα in MITF promoter regions was investigated using ChIP RT-qPCR analysis in (b-c) MNT-1 cells and (d-e) darkly pigmented human melanocytes (DP-Mel), n = 3
3.3 ∣. Retinoids stimulate MITF-A promoter
Having demonstrated binding of RXR/RAR to a region within the MITF-A promoter, we next investigated whether RXR and RAR ligands could induce MITF-A promoter activity. To test the activity of the human MITF-A promoter, we used MITF-A promoter luciferase plasmid constructs containing either full-length or truncated MITF-A promoter (Figure 3a). Luciferase experiments were conducted in HEK293T cells. The full-length MITF-A promoter construct had activity similar to the truncated construct in serum-starved, vehicle (ethanol)-treated cells (Figure 3b). By contrast, 9-cis retinoic acid induced luciferase activity of the full-length MITF-A promoter construct, but failed to induce luciferase activity of the truncated MITF-A construct (Figure 3b). In the presence of DMSO, the vehicle for some retinoid receptor ligands, the full-length construct had a higher baseline activation compared with the truncated promoter (Figure 3c-e). Stimulation of the RXR subunit alone using the pan-RXR agonist LG100268 increased the activation of the luciferase reporter above baseline (Figure 3c), verifying that RXR can induce the luciferase activity of this reporter. Likewise, increasing concentrations of BMS753, a potent RARα agonist, stimulated the activity of the full-length MITF-A promoter construct (Figure 3d). Taken together, these results suggest that RXR/RAR heterodimers can regulate the expression of MITF-A. This is consistent with previous findings that 9-c/s retinoic acid can regulate MITF expression (Paterson et al., 2013).
FIGURE 3.
Retinoids stimulate the human MITF-A promoter through RXR/RAR binding site. (a) Schematic showing the size of MITF-A promoter upstream of the luciferase gene in the pGL3-luciferase plasmid constructs. (b-d) HEK293T cells were transfected with MITF-A promoter constructs and co-transfected with a Renilla luciferase reporter as an internal control. The cells were treated with the indicated dose of (b) 9-cis retinoic acid; (c) LG100268, a RXR agonist; and (d) BMS753, a RARα-specific agonist. (e) HEK293T cells were transfected with promoter constructs including a mutant MITF-A promoter lacking the putative RXR/RAR binding site. n = 3
Having verified that the MITF-A promoter responds to retinoids, RXR, and RAR agonists, we next tested whether RXR/RAR regulates MITF-A expression via the newly identified RXR/RAR binding site. Using site-directed mutagenesis, we deleted 28 bp encompassing the 17 bp motif identified through MotifMap to generate a mutant promoter construct (Figure 3a). Baseline luciferase activity from the mutant construct was significantly reduced compared with the full-length promoter construct, although the mutant retained higher levels of luciferase expression when compared to the truncated promoter (Figure 3e). These results can be explained by the presence of other enhancer elements in the promoter that are known to activate MITF-A expression. Taken together, these studies indicate that RXR/RAR activation can induce the expression of MITF-A.
3.4 ∣. Mitf isoform-specific mutant mice
While expression studies have defined Mitf-M as the melanocyte-specific isoform and Mitf-A as a regulator of eye development (Reinisalo, Putula, Mannermaa, Urtti, & Honkakoski, 2012), the specific function of each isoform during development is unclear. Established mouse Mitf mutant alleles have loss of expression or function of multiple isoforms (Steingrímsson, 2008; Steingrímsson et al., 2003, 2004), making it difficult to determine the distinct roles of Mitf isoforms in tissue development. Because each isoform has its own unique first exon, we sought to generate mutant mice specifically lacking a single isoform. We designed guide RNAs for CRISPR/Cas9 gene editing for exons 1A and 1M of Mitf. Utilizing this technique, we generated a 7 bp deletion at the beginning of the coding region of exon 1A that produces a frameshift mutation that causes multiple premature stop codons (Figure 4a, Figure S3a,b). This allele was designated Mitfem1Gane, but for clarity, homozygous mice will hereafter be referred to as Mitf-A knockout or null mice. Similarly, we also generated an 18 bp deletion spanning the splice site for exon 1M of Mitf (Figure 4a, S3c) designated Mitfem2Gane. Mice homozygous for the Mitfem2Gane allele, with targeted deletion of the Mitf-M isoform, will hereafter be referred to as Mitf-M knockout or null.
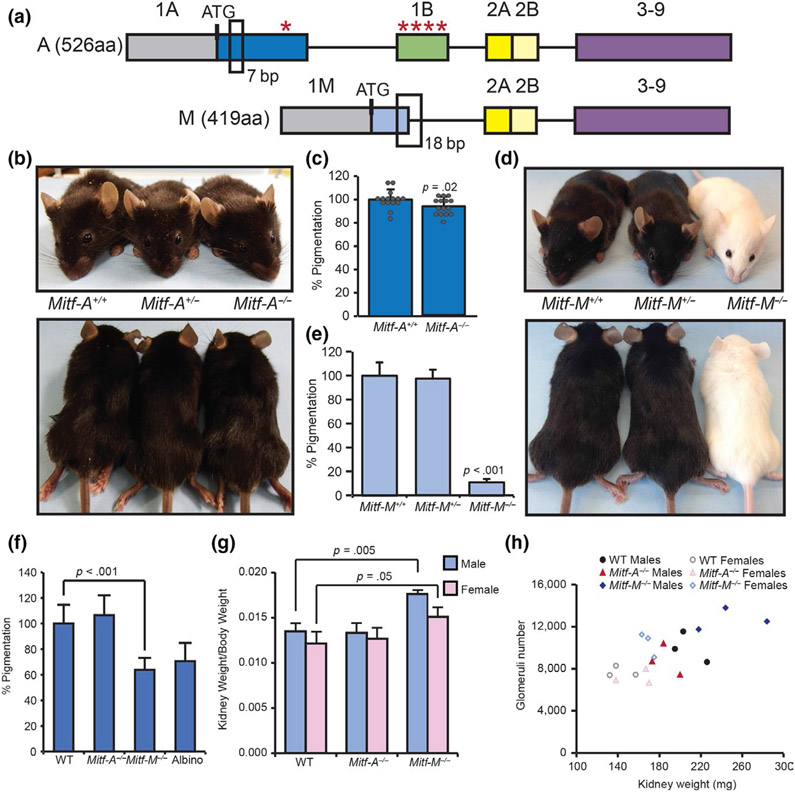
FIGURE 4.
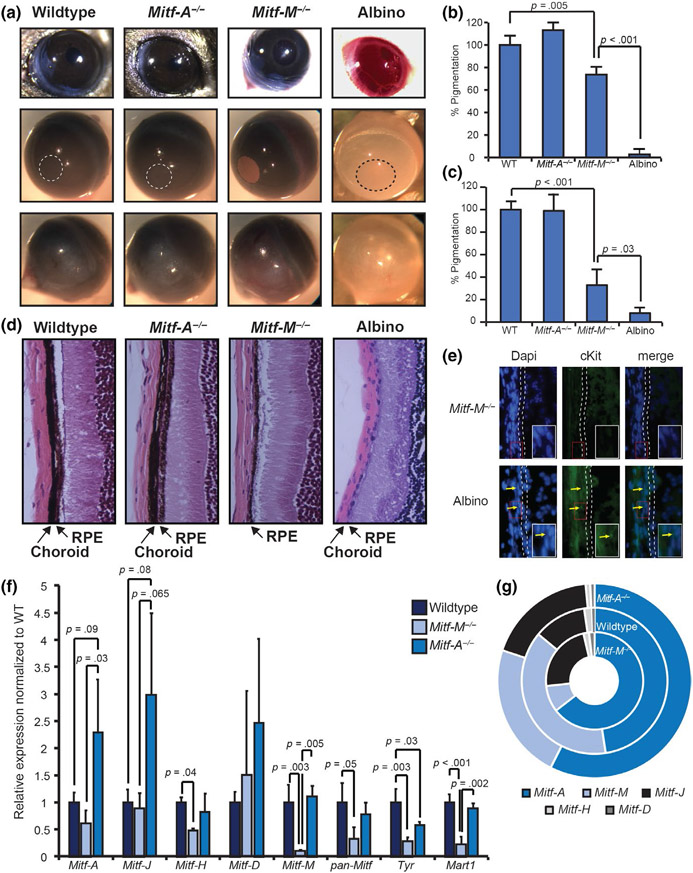
Mitf-A and Mitf-M isoform-specific mutant mice generated using CRISPR/Cas9 gene targeting platform. (a) Schematic of 7 bp deletion in exon 1A for Mitf-A knockout mice and 18 bp deletion in exon 1M and intron for Mitf-M knockout mice. Untranslated regions of exons 1A and 1M are gray, while coding regions are blue. Red asterisks denote premature stop codons in first 2 exons caused by 7 bp deletion. (b) Wild-type and Mitf-A isoform-specific knockout heterozygous and homozygous mice imaged at p50. (c) Melanin quantification of hair collected at p50 normalized to wild-type mice. n = 15. (d) Comparison of wild-type and Mitf-M isoform-specific knockout heterozygous and homozygous mice imaged at p50. (e) Melanin quantification of hair collected at p50 normalized to wild-type mice. n = 10. (f) Melanin quantification on tail skin of wild-type, Mitf-A−/−, Mitf-M−/−, and B6 albino mice. (g) Quantification of total kidney mass to body mass for males and females of indicated genotypes, n = 3. (h) Scatterplot of glomeruli number and kidney weight for individual male (filled shapes) and female (open shapes) of wild-type (circles), Mitf-A−/− (triangles), and Mitf-M−/− (diamonds) mice
The loss of specific isoforms of Mitf produced distinct pigment phenotypes. While mice lacking Mitf-A have no visible defect on a black (non-agouti) coat-color background (Figure 4b, Figure S4a-b, S5a-b), quantitative analysis of the hair detected a 7% reduction in melanin accumulation of Mitf-A knockout mice compared with wild-type black mice (Figure 4c). In contrast, mutation of Mitf-M results in a loss of melanin in the hair and the skin of the ears and tail similar to the phenotype seen in albino mice (Figure 4d-f, Figure S4c-d, S5c-d). Despite this dramatic loss of coat color, Mitf-M knockout mice are distinct from albino mice since their eyes contain some pigment and appear dark (Figure 4d, Figure S5c). Other studies have shown that mice heterozygous for mutations in the Mitf promoter region do not have appreciable differences in coat color (Steingrímsson et al., 2003). Similarly, the loss of one copy of Mitf-M does not result in an appreciable change in pigmentation as mice appear black (Figure 4d). Moreover, there were no quantifiable differences in pigmentation in these animals, as they accumulate similar amounts of melanin in their hair as wild-type mice (Figure 4e).
Previous studies have shown that overexpression of Mitf-A in kidneys can increase the ratio of kidney to body mass and increase the number of glomeruli, while the deletion of Mitf-A and elements upstream of the Mitf-A promoter results in experimental mice with kidneys that have less glomeruli (Phelep et al., 2017). Histological analysis of kidneys from our Mitf-A null, 4-month-old mice showed no gross morphological changes as determined by two separate pathologists (Figure S6a-d). Additionally, in our mice, the loss of Mitf-A did not significantly alter the ratio of kidney to body mass (Figure 4f, Figure S6e) or the number of glomeruli in the kidney (Figure 4g, Figure S5e). The melanocyte-specific isoform Mitf-M was also detected in the kidneys (Figure S6f-g) and loss did significantly increase the ratio of kidney to body mass in both males and females (Figure 4f). However, while there was a trend toward increased glomeruli in Mitf-M knockout mice, the difference was not significant (Figure 4g, Figure S6e). In Mitf-M knockout mice, there was no significant change in relative isoform abundance with the exception of Mitf-A and total Mitf (Figure S6f); however, there is a trend toward enrichment of the Mitf-C isoform with the loss of Mitf-M in the kidney (Figure S6g).
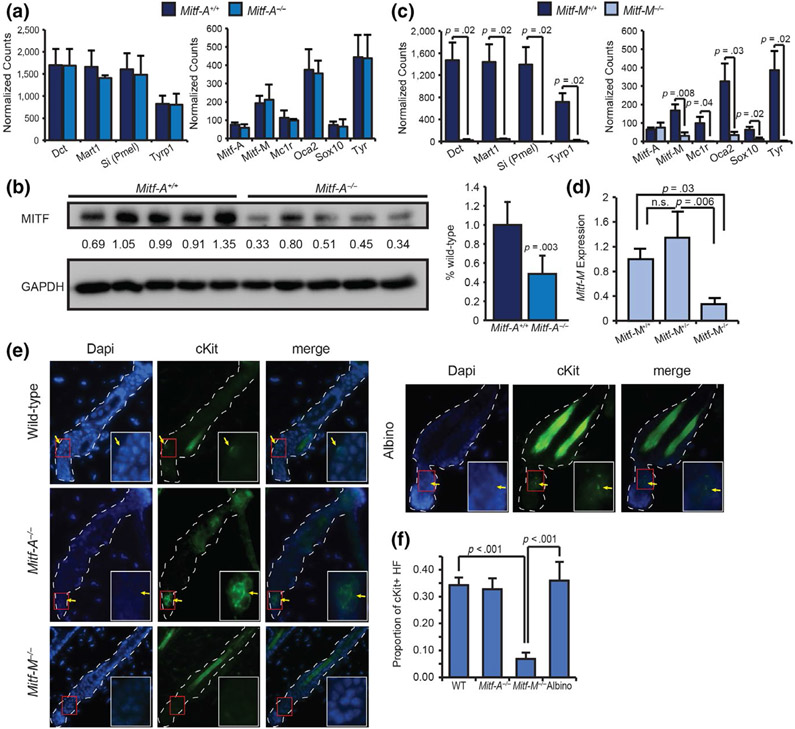
To more closely examine the expression of melanocyte isoforms of Mitf in the skin, we examined melanocyte-specific gene expression in whole back skin from wild-type and isoform-specific knockout mice using Nanostring. Mitf-A knockout mice had a similar gene expression profile compared with wild-type mice (Figure 5a). While the 7 bp deletion causes a frameshift (Figure S3b), it did not alter the stability of the mRNA transcript, and Mitf-A expression levels were unchanged in knockout mice. To verify that the Mitf-A knockout mice lack functional MITF-A protein, we collected protein lysate from whole eyes of wild-type and Mitf-A knockout mice, as Mitf-A is the predominant isoform in the eye. Mitf-A knockout mice had ~50% reduced levels of total MITF in the eye (p-value = .003; Figure 5b). Consistent with the visual loss of pigmentation, Mitf-M knockout mice had decreased the expression of all melanocyte-specific genes in the skin without notable changes in the expression of Mitf-A (Figure 5c). Mice heterozygous for the Mitf-M mutation showed no significant change in Mitf-M expression using RT-qPCR (Figure 5d), suggesting that the wild-type allele is upregulated in Mitf-M heterozygotes. RT-qPCR was not sensitive enough to detect the different isoforms of Mitf expressed in the skin. This type of the analysis would likely require isolation of melanocytes from the skin to generate enough Mitf transcripts that could be reproducibly measured. We did note that the downregulation of melanogenesis markers in Mitf-M knockout mice coincides with the loss of cKit-positive melanocytes in the hair follicle (Figure 5e-f). This loss of cKit-positive cells was observed in Mitf-M knockout mice, but not in Mitf-A null or albino mice, suggesting that the pigment phenotype observed in these mice is secondary to the loss of melanocytes.
FIGURE 5.
Mitf isoform-specific knockout mice have distinct skin gene expression phenotypes. Skin was collected at P60 for RNA extraction after stimulating the hair cycle at P50. Purified RNA was subjected to Nanostring analysis on melanocyte-specific genes. Data shown are mean normalized counts of mRNA for each gene in (a) Mitf-A knockout mice and (c) Mitf-M knockout mice compared with wild-type littermates. Wild-type, Mitf-M−/− n = 3; Mitf-A−/− n = 2. (b) Western blot for pan-Mitf on whole-eye protein lysates from Mitf-A+/+ and Mitf-A−/− mice. (d) Relative expression of Mitf-M in Mitf-M wild-type, heterozygous, and knockout mice. (e) cKit staining on indicated mouse skin collected at P56 with cKit-positive melanocytes indicated by arrows and bounded by the red box, inset. (f) Quantification of cKit-positive hair follicles (HF), n = 3
During normal development, MITF controls the migration of melanoblasts to target tissues including the epidermal basal membrane, the hair follicle, the iris stroma, and the choroid of the eye (Vachtenheim & Ondrusov, 2013). Choroidal and iris stromal melanocytes and the pigmented epithelia of the iris (IPE) and retina (RPE) are thought to rely on MITF to induce melanin production. Unlike the neural crest-derived melanocytes, the cells of the pigmented epithelium are derived from the optic cup and express multiple isoforms of Mitf (Bharti, Nguyen, Skuntz, Bertuzzi, & Arnheiter, 2006). Because these distinct pigment layers in the eye rely on different isoforms of Mitf, we investigated whether Mitf isoform-specific knockout mice had any eye phenotype. Live animal images taken of mice suggest a subtle loss of pigment in the iris of Mitf-M knockout mice (Figure 6a (top), Figure S7a-d). There was a significant reduction (~35%) in pigmentation in the iris of Mitf-M knockout mice (Figure 6b) when melanin absorbance of the isolated iris was quantified. In contrast, Mitf-A knockout mice had no significant change in iris pigmentation (Figure 6b). Moreover, the difference in iris pigmentation was somewhat apparent when hematoxylin and eosin (H&E)-stained sections were visualized at 20× magnification (Figure S8a-d), and even more apparent when these sections were visualized at 63× magnification (Figure S8e-h). Upon enucleation of the eyes, it became apparent that the loss of Mitf-M, but not Mitf-A, affected pigment accumulation in the posterior of the eye (Figure 6a, Figure S7e-h), a result that was even more evident after removal of the retina from the posterior segment. Overall, the posterior segment of the eye of Mitf-M knockout mice had 30% of the pigment present in wild-type eyes as measured by relative melanin absorbance (Figure 6c). Mitf-M knockout mice did have significantly more pigment in their eyes when compared to B6 albino mice (Figure 6b-c), which are known to lack pigment in both melanocytes and the retinal pigment epithelium. H&E staining of wild-type and Mitf-A knockout mice revealed pigmentation in both the choroid and RPE; however, the loss of MITF-M results in the loss of choroidal pigmentation while the RPE appears normally pigmented (Figure 6d, Figure S8i-I). Choroidal cKit-positive melanocytes are also absent in Mitf-M knockout mice (Figure 6e). While histology reveals no gross morphological defects in the retina, we next sought to ensure any phenotypes observed in the Mitf knockout mice are not due to changes in retinal degeneration caused by the presence of the Crb1 rd8 mutation from the C57BL/6N line. Since the Mitf-A and Mitf-M lines are on a mixed 6N and 6J background, 50 mice of each line were genotyped for the Crb1 wild-type and rd8 alleles. Primers were validated with C57BL/6J mice, which lack the rd8 mutation, and C57BL/6N, which are homozygous for the rd8 mutation. The Mitf-M mice tested in these studies were all either heterozygous or lack the rd8 allele altogether. Their eye phenotypes were compared against the eyes of wild-type mice, including those from the C57BL/6N line, which are known to be homozygous for rd8. Given these results, it is highly unlikely that the phenotypes observed are related to rd8, as none of the Mitf-M mice examined were rd8 homozygotes while, in contrast, the control mice examined were rd8 homozygotes (Figure S7i-j). Using RT-qPCR, we compared the expression of isoforms in the combined choroid and RPE of the mice. The mutation in Mitf-M resulted in the expected downregulation of the Mitf-M transcript as well as total Mitf, Tyr, and Mart1 (Figure 6f). In Mitf-A knockout mice, there was an increase in the mutated Mitf-A transcript that results in premature stop codons. The mutation was verified using Sanger sequencing of wild-type and knockout Mitf-A transcripts (Figure S9). Interestingly, more Mitf-J transcripts also accumulated in Mitf-A knockout eyes when compared to the choroid and RPE of wild-type mice. These changes in specific transcripts are highlighted by the change in relative abundance of Mitf isoforms in the eye (Figure 6g). Finally, we also observed decreased accumulation of Tyr transcript in Mitf-A knockout eyes (Figure 6f), consistent with other studies that indicate that MITF-A regulates TYR expression in the eye (Amae et al., 1998). Together, these studies show that the deletion of Mitf-A or Mitf-M has distinct effects on the development of the skin, eyes, and kidneys.
FIGURE 6.
Mitf isoform-specific knockout mice have distinct eye phenotypes. (a) Representative images of whole eyes of indicated mice before (top) and following enucleation of iris with pupil indicated (middle) and posterior surface of the eye (bottom). (b) Melanin quantification on dissected iris of indicated mice, n = 3. (c) Pigment assay on combined RPE, choroid, and sclera of wild-type, Mitf-A−/−, Mitf-M−/−, and Albino mice, n = 4. (d) H&E sections from paraffin-embedded eyes with choroid and RPE indicated by arrows. (e) cKit staining of choroid, with RPE highlighted and cKit-positive melanocytes indicated with arrows. The inset area is highlighted by the red box. (f) Relative expression of Mitf isoforms in the isolated RPE and choroid of wild-type and Mitf isoform-specific mutant mice, n = 3. (g) Further comparison of Mitf isoform abundance in the RPE and choroid of wild-type and Mitf knockout mice
4 ∣. DISCUSSION
The use of alternative promoters by a gene enables more sophisticated control of the gene's expression. Such elaboration helps facilitate development and tissue specification, and is also associated with unique disease states (Davuluri et al., 2008). In the eye, mouse Mitf isoforms are known to be differentially regulated during the formation of the RPE (Bharti et al., 2008). We show here that the expression of MITF-A, the second most abundant isoform in human melanocytes (Figure 1), is regulated by retinoids, transcriptional activators that have a prominent role in eye development (McBee, Palczewski, Baehr, & Pepperberg, 2001). Retinoids can regulate alternative promoters. The Stra6 gene contains two alternative promoters, with the intronic promoter containing a retinoic acid response element (RARE). Under excessive vitamin A levels, there is increased expression of Stra6S from the intronic promoter (Laursen, Kashyap, Scandura, & Gudas, 2015). Similarly, it is conceivable that levels of retinoids may dictate MITF isoform expression within the RPE, as retinoids are known regulators of eye development.
Our group has shown that 9-cis retinoic acid and retinal stimulate melanogenesis in pigmented cell lines (Paterson et al., 2013), while others have shown that retinal increases the sensitivity of melanocytes to UVA-induced pigment production (Wicks, Chan, Najera, Ciriello, & Oancea, 2011). Based on the finding that 9-cis retinoic acid led to an upregulation in MITF expression (Paterson et al., 2013), we identified a RARE binding motif in the human MITF promoter upstream of the 1A exon. Apart from the MITF-M promoter, the regulators of other isoform-specific promoters are poorly understood (Steingrímsson, 2008). While it is possible that the RARE facilitates chromatin looping to regulate distant genes or additional MITF isoforms, we have shown this binding motif contributes to human MITF-A promoter activity (Figure 3). Consistent with this notion, retinoid receptors have also been linked to pigmentation. A germline mutation of RXRα in the RXRaPke mouse results in premature graying starting when mice reach 5 weeks (Du et al., 2005). RXRaPke mice also experience alopecia and are hairless at 4 months (Du et al., 2005), so the isolated effect of RXRα on melanocytes and pigmentation is still unknown. Conditional deletion of RXRα in keratinocytes causes a dilute coat color (Li et al., 2001), suggesting that RXRα may regulate pigmentation in a melanocyte cell-autonomous and non-autonomous manner. Taken together, these results indicate that the deletion of MITF-A in melanocytes, like the RXRα knockout, only partially affects hair follicle pigmentation.
In humans, mutations in MITF have been linked to Waardenburg syndrome, type 2A and Tietz syndrome, and disorders characterized by pigmentary defects including a white forelock and hearing loss (Pingault et al., 2010; Wildhardt et al., 2013). These mutations occur throughout the MITF gene, but are most common in the DNA-binding domains (Grill et al., 2013) with some of the mutations recapitulated in mice (Pingault et al., 2010). Similarly, mutations in humans associated with familial melanoma and predisposition to renal cell carcinoma (Bertolotto et al., 2011; Grill et al., 2013) are also within the DNA-binding domains. The characterized mouse mutations in Mitf only partially recapitulate pigmentary disorders, but cause a wide range of phenotypes including pigmentation defects, microphthalmia, hearing loss, changes in bone density, and deficiency in immune cells (Steingrímsson et al., 2004). The majority of mouse mutations occur in the DNA-binding and dimerization domains that can alter all isoforms of MITF, which explains the wide range of tissues affected (Steingrímsson et al., 2004). Even mutations in the promoter regions have variable phenotypes. Both the Mitfmi-vga9, a MITF null, and Mitfmi-rw mice, which has a deletion of multiple unique first exons and promoter regions, have pigmentation and eye defects found only in homozygous mice (Hodgkinson et al., 1993; Watanabe et al., 2002). To distinguish the role of different isoforms, we generated Mitf-A and Mitf-M isoform-specific knockout mice. These unique specific isoform knockout reagents can be used to better delineate the function of individual isoforms, as described in this manuscript.
While Mitf-A was first thought to play an important role in the development of the RPE (Amae et al., 1998), similar to the loss of Mitf-D and Mitf-H in the Mitfmi-rw mouse (Bharti et al., 2008), we found that Mitf-A is not required for the development of the RPE (Figure 6). These results suggest that though Mitf-A plays a role in pigmentation of the hair and eye (Figures 4-6), it is redundant in melanocytes and both the IPE and RPE cells. Lack of a phenotype in the RPE may be a result of compensation by additional Mitf isoforms, as was shown with the loss of Mitf-D (Bharti et al., 2012). Since Mitf-A is expressed in multiple tissues (Amae et al., 1998), the A isoform may have more specific roles in other organs such as the kidney, where the overexpression of Mitf-A caused an increase in size and nephron number while the deletion of the promoter and Mitf-A decreases the glomeruli number (Phelep et al., 2017). In our study, we found no significant change in kidney mass in our Mitf-A knockout mice (Figure 4, Figure S5). This difference in phenotype may be secondary to the deletion of almost 6,000 bp that includes exon 1A and the Mitf-A promoter in Ref. (Phelep et al., 2017) as compared to the more specific deletion within the 1A exon presented in this study. Since the Mitf isoforms can be regulated by distant regulatory elements, large changes in the promoter regions may modulate the expression of multiple isoforms, as observed in mice with the Mitfvga9 allele. In contrast, exon-specific deletions could affect promoter regulation in cases where Mitf isoforms themselves differentially activate Mitf promoters. Future studies will address the broad impacts of loss of individual Mitf isoforms on the expression of other Mitf isoforms and Mitf target genes at different times during development.
The loss of Mitf-M results in the loss of melanocytes in the hair, skin, iris stroma, and choroid, but not the cells of the IPE and RPE (Figures 4-6). This phenotype is similar to the phenotype of Mitfmi-bw mice, where a LINE1 element is inserted between the common exons 3 and 4 (Hozumi et al., 2012; Yajima et al., 1999). While the Mitfmi-bw mice have the loss of Mitf-M expression in the skin and eye, the LINE insertion also results in reduced expression and alternatively spliced transcripts of Mitf-A and Mitf-H (Yajima et al., 1999). During embryonic development, aberrant Mitf-M transcripts were also detected (Takeda et al., 2014), indicating the Mitfmi-bw allele is not a clean knockout of the M isoform. Mitf-M heterozygotes have no pigment defects (Figure 4d-e), which suggests that one copy of Mitf-M is sufficient to maintain Mitf-M expression levels (Figure 5d). In this study, we have generated a specific Mitf-M knockout mouse and demonstrated that the loss of MITF-M results in the loss of iris stromal and choroidal melanocytes. Our findings also demonstrate that while Mitf-M may be expressed in the adult RPE (Maruotti, Thein, Zack, & Esumi, 2012) and can rescue the loss of pigmentation in Mitfvga9 mice (Michael et al., 2018), it is not required for pigmentation of the RPE. Additionally, the loss of Mitf-M increased the size of the kidneys (Figure 4), indicating that the loss of a single isoform affects distinct tissues differently, further highlighting the utility of the isoform-specific knockout reagents generated in this study.
Recent studies in the eye have identified the Crb1 mutant rd8 allele present in the C57BL/6N mice as a cause of retinal degeneration (Mattapallil et al., 2012; Moore et al., 2018). While the rd8 allele is present in both of the knockout lines from the original C57BL/6NTac embryos, both lines were outcrossed to C57BL/6J to isolate individual CRISPR alleles. In the 50 mice from the Mitf-M line studied here, all were either wild-type or heterozygous for the rd8 allele at the Crb1 locus (Figure S7j). Published studies have shown that rd8 heterozygous mice show no signs of retinal degeneration as compared to rd8 homozygous mice (Luhmann et al., 2015). In this study, eye phenotype comparisons were made between Mitf-M knockout mice and C57BL/6N mice, which are homozygous for the rd8 allele. While we cannot exclude that the rd8 allele can influence the RPE phenotypes observed here, it is unlikely that our observed results are caused by an rd8 mutation. Moreover, Mitf-M knockout mice not only have a pigment phenotype in the choroid but also in the iris stroma, further evidence that the phenotype observed here is independent of rd8 mutation.
In summary, this study demonstrates that differential regulation of MITF isoforms plays a critical role in tissue development. MITF-A is regulated by retinoids, signaling molecules that are crucial in eye development, yet Mitf-A knockout mice have no eye phenotype although they have reduced MITF protein levels. These findings suggest that low levels of MITF are likely required for RPE pigmentation and Mitf isoforms play redundant roles in this process. In contrast, Mitf-M deletion results in the loss of melanocytes in both the skin and choroid, indicating that this isoform is necessary for melanocyte development in these tissues. This work provides a first glimpse at how two distinct Mitf isoforms regulate three distinct tissues and is another example of how protein isoforms can be differentially regulated in tissues, resulting in distinct developmental phenotypes that influence human disease.
Supplementary Material
Significance.
Mutations in MITF cause hypopigmentation and increase the risk for familial melanoma in humans. There are multiple different isoforms of MITF, and it is currently unclear how these different isoforms contribute to pigment cell development in the eye, hair, and skin. In this study, we identified two isoforms of MITF that are present in melanocytes of the skin and use genome editing technology to block the expression of one or the other of these isoforms in mice. Taken together, our work illustrates how protein isoforms can differentially contribute to biological phenotypes, findings that have relevance to human disease.
ACKNOWLEDGEMENTS
We thank Kai-Xuan Shi and Shuling Wang of the UCI Transgenic Mouse Facility (TMF) for pronuclear injection and molecular analysis, respectively, for the production of Mitf-A and Mitf-M CRISPR knockout mice. We thank Dr. Shigeki Shibahara for his kind gift of the MITF-A luciferase plasmid constructs and Feng Zhang for his kind gift of the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid. We thank Sahil Telang and Madeline McCanne for assistance with luciferase assays and Yumay Chen for examining kidney histology. We thank Amber Habowski, Chi-Fen Chen, Jessica Shiu, and Klemens Hertel for advice on experimental design and editing the manuscript. We thank the UCI Genomics High Throughput Facility (GHTF) for their help with Nanostring nCounter and RNA sequencing. We thank the Institute for Immunology Flow Cytometry Core Facility (FCCF) for assistance with sorting cells using FACS. This work was supported by grants from the National Institutes of Health (R01AR063116, R01CA151513, and U54-CA217378) to AKG. The research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health Award Number T32CA009054-37 and an Anti-Cancer Challenge research grant from the University of California, Irvine Chao Family Comprehensive Cancer Center to JLF. The TMF, GHTF, and FCCF are Shared Resources funded in part by the Chao Family Comprehensive Cancer Center Support Grant (P30CA062203) from the National Cancer Institute and NIH Shared Instrumentation Grants (1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding information
National Center for Research Resources, Grant/Award Number: 1S10RR025496-01; NIH Office of the Director, Grant/Award Number: 1S10OD010794-01 and 1S10OD021718-01; National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Number: R01AR063116; Chao Family Comprehensive Cancer Center; National Cancer Institute, Grant/Award Number: P30CA062203, R01CA151513, T32CA009054-37 and U54-CA217378
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, … Chambon P (1993). Retinoic acid receptors and retinoid X receptors: Interactions with endogenous retinoic acids. Proceedings of the National Academy of Sciences, 90(1), 30–34. 10.1073/pnas.90.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amae S, Fuse N, Yasumoto K-I, Sato S, Yajima I, Yamamoto H, … Shibahara S (1998). Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochemical and Biophysical Research Communications, 247(3), 710–715. 10.1006/bbrc.1998.8838 [DOI] [PubMed] [Google Scholar]
- Aoki H, Yamada Y, Hara A, Kunisada T, Kunisada T, Pavan WJ, & Arnheiter H (2009). Two distinct types of mouse melanocyte: differential signaling requirement for the maintenance of non-cutaneous and dermal versus epidermal melanocytes. Development (Cambridge, England), 136(15), 2511–2521. 10.1242/dev.037168 [DOI] [PubMed] [Google Scholar]
- Arsic N, Gadea G, Lagerqvist EL, Busson M, Cahuzac N, Brock C, … Roux P (2015). The p53 isoform Δ133p53β promotes cancer stem cell potential. Stem Cell Reports, 4(4), 531–540. 10.1016/j.stemcr.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, … Bressac-de Paillerets B (2011). A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature, 480(7375), 94–98. 10.1038/nature10539 [DOI] [PubMed] [Google Scholar]
- Bharti K, Gasper M, Ou J, Brucato M, Clore-Gronenborn K, Pickel J, & Arnheiter H (2012). A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genetics, 8(7), e1002757 10.1371/journal.pgen.1002757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, Liu W, Csermely T, Bertuzzi S, & Arnheiter H (2008). Alternative promoter use in eye development: The complex role and regulation of the transcription factor MITF. Development (Cambridge, England), 135(6), 1169–1178. 10.1242/dev.014142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, Nguyen MTT, Skuntz S, Bertuzzi S, & Arnheiter H (2006). The other pigment cell: Specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Research, 19(5), 380–394. 10.1111/j.1600-0749.2006.00318.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, Chin L (2006). Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis, 44(5), 262–267. 10.1002/dvg.20205 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, & Barretto R (2013). Multiplex genome engineering using CRISPR/Cas systems. Multiplex Genome Engineering Using CRISPR/Cas Systems, 339(6121), 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily K, Patel VR, Rigor P, Xie X, & Baldi P (2011). MotifMap: Integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics, 12(1), 495 10.1186/1471-2105-12-495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W, & Schultz RM(2000). Developmental change in TATA-box utilization during preimplantation mouse development. Developmental Biology, 218(2), 275–283. 10.1006/DBIO.1999.9486 [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Plass C, & Huang T-H-M (2008). The functional consequences of alternative promoter use in mammalian genomes. Trends in Genetics, 24(4), 167–177. 10.1016/j.tig.2008.01.008 [DOI] [PubMed] [Google Scholar]
- Du X, Tabeta K, Mann N, Crozat K, Mudd S, & Beutler B (2005). An essential role for Rxr alpha in the development of Th2 responses. European Journal of Immunology, 35(12), 3414–3423. 10.1002/eji.200535366 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, & Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30(1), 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin LS, Castle JT, Kohli JS, Goff PS, Cairney CJ, Keith WN, … Bennett DC (2014). Isolation, culture, and transfection of melanocytes In Current protocols in cell biology (pp. 1.8.1–1.8.20). Hoboken, NJ, USA: John Wiley & Sons, Inc; 10.1002/0471143030.cb0108s63 [DOI] [PubMed] [Google Scholar]
- Grill C, Bergsteinsdóttir K, Ögmundsdóttir MH, Pogenberg V, Schepsky A, Wilmanns M, … Steingrímsson E (2013). MITF mutations associated with pigment deficiency syndromes and melanoma have different effects on protein function. Human Molecular Genetics, 22(21), 4357–4367. 10.1093/hmg/ddt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltaufderhyde KD, & Oancea E (2014). Genome-wide transcriptome analysis of human epidermal melanocytes. Genomics, 104(6), 482–489. 10.1016/j.ygeno.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, & Arnheiter H (1993). Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell, 74(2), 395–404. 10.1016/0092-8674(93)90429-T [DOI] [PubMed] [Google Scholar]
- Hozumi H, Takeda K, Yoshida-Amano Y, Takemoto Y, Kusumi R, Fukuzaki-Dohi U, … Shibahara S (2012). Impaired development of melanoblasts in the black-eyed white Mitfmi-bw mouse, a model for auditory-pigmentary disorders. Genes to Cells, 17(6), 494–508. 10.1111/j.1365-2443.2012.01603.x [DOI] [PubMed] [Google Scholar]
- Kim S, & An SSA (2016). Role of p53 isoforms and aggregations in cancer. Medicine, 95(26), e3993 10.1097/MD.0000000000003993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J-R, Mager DL, & Wilhelm BT (2003). Complex controls: The role of alternative promoters in mammalian genomes. Trends in Genetics, 19(11), 640–648. 10.1016/j.tig.2003.09.014 [DOI] [PubMed] [Google Scholar]
- Laursen KB, Kashyap V, Scandura J, & Gudas LJ (2015). An alternative retinoic acid-responsive Stra6 promoter regulated in response to retinol deficiency. The Journal of Biological Chemistry, 290(7), 4356–4366. 10.1074/jbc.M114.613968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, & Fisher DE (2006). MITF: Master regulator of melanocyte development and melanoma oncogene. Trends in Molecular Medicine, 12(9), 406–414. 10.1016/j.molmed.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Li M, Chiba H, Warot X, Messaddeq N, Gérard C, Chambon P, & Metzger D (2001). RXR-alpha ablation in skin keratinocytes results in alopecia and epidermal alterations. Development (Cambridge, England), 128(5), 675–688. [DOI] [PubMed] [Google Scholar]
- Liggins MC, Flesher JL, Jahid S, Vasudeva P, Eby V, Takasuga S, … Ganesan AK (2018). PIKfyve regulates melanosome biogenesis. PLoS Genetics, 14(3), 10.1371/journal.pgen.1007290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhmann UFO, Carvalho LS, Holthaus S-M-K, Cowing JA, Greenaway S, Chu CJ, … Ali RR (2015). The severity of retinal pathology in homozygous Crb1rd8/rd8 mice is dependent on additional genetic factors. Human Molecular Genetics, 24(1), 128–141. 10.1093/hmg/ddu424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruotti J, Thein T, Zack DJ, & Esumi N (2012). MITF-M, a ‘melanocyte-specific’ isoform, is expressed in the adult retinal pigment epithelium. Pigment Cell & Melanoma Research, 25(5), 641–644. 10.1111/j.1755-148X.2012.01033.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil MJ, Wawrousek EF, Chan C-C, Zhao H, Roychoudhury J, Ferguson TA, & Caspi RR (2012). The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Investigative Opthalmology & Visual Science, 53(6), 2921 10.1167/iovs.12-9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBee JK, Palczewski K, Baehr W, & Pepperberg DR (2001). Confronting complexity: The interlink of phototransduction and retinoid metabolism in the vertebrate retina. Progress in Retinal and Eye Research, 20(4), 469–529. 10.1016/S1350-9462(01)00002-7 [DOI] [PubMed] [Google Scholar]
- Michael HT, Graff-Cherry C, Chin S, Rauck C, Habtemichael AD, Bunda P, … Day C-P (2018). Partial rescue of ocular pigment cells and structure by inducible ectopic expression of Mitf-M in MITF-deficient mice. Investigative Opthalmology & Visual Science, 59(15), 6067 10.1167/iovs.18-25186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang X-J, Vogel H, Roop DR, & Bradley A (1999). p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature, 398(6729), 708–713. 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Moore BA, Roux MJ, Sebbag L, Cooper A, Edwards SG, Leonard BC, … Moshiri A (2018). A population study of common ocular abnormalities in C57BL/6N rd8 Mice. Investigative Opthalmology & Visual Science, 59(6), 2252 10.1167/iovs.17-23513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, & Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis, 45(9), 593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Paterson EK, Fielder TJ, MacGregor GR, Ito S, Wakamatsu K, Gillen DL, … Ganesan AK (2015). Tyrosinase depletion prevents the maturation of melanosomes in the mouse hair follicle. PLoS ONE, 10(11), e0143702 10.1371/journal.pone.0143702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson EK, Ho H, Kapadia R, & Ganesan AK (2013). 9-cis retinoic acid is the ALDH1A1 product that stimulates melanogenesis. Experimental Dermatology, 22, 202–209. 10.1111/exd.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelep A, Laouari D, Bharti K, Burtin M, Tammaccaro S, Garbay S, … Terzi F (2017). MITF – A controls branching morphogenesis and nephron endowment. PLOS Genetics, 13(12), e1007093 10.1371/journal.pgen.1007093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, & Bondurand N (2010). Review and update of mutations causing Waardenburg syndrome. Human Mutation, 31(4), 391–406. 10.1002/humu.21211 [DOI] [PubMed] [Google Scholar]
- Pogenberg V, Ogmundsdóttir MH, Bergsteinsdóttir K, Schepsky A, Phung B, Deineko V, … Wilmanns M (2012). Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes & Development, 26(23), 2647–2658. 10.1101/gad.198192.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichrath J, Münssinger T, Kerber A, Rochette-Egly C, Chambon P, Bahmer FA, & Baum HP (1995). In situ detection of retinoid-X receptor expression in normal and psoriatic human skin. The British Journal of Dermatology, 133(2), 168–175. 10.1111/j.1365-2133.1995.tb02612.x [DOI] [PubMed] [Google Scholar]
- Reinisalo M, Putula J, Mannermaa E, Urtti A, & Honkakoski P (2012). Regulation of the human tyrosinase gene in retinal pigment epithelium cells: The significance of transcription factor orthodenticle homeobox 2 and its polymorphic binding site. Molecular Vision, 18, 38–54. [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, & Mesirov JP (2011). Integrative genomics viewer. Nature Biotechnology, 29(1), 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrímsson E (2008). All for one, one for all: Alternative promoters and Mitf. Pigment Cell & Melanoma Research, 21(4), 412–414. 10.1111/j.l755-148X.2008.00473.x [DOI] [PubMed] [Google Scholar]
- Steingrímsson E, Arnheiter H, Hallsson JH, Lamoreux ML, Copeland NG, Jenkins NA, … Shibahara S (2003). Interallelic complementation at the mouse Mitf locus. Genetics, 163(1), 267–276. 10.1007/bf03023302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrímsson E, Copeland NG, & Jenkins NA (2004). Melanocytes and the Microphthalmia transcription factor network. Annual Review of Genetics, 38(1), 365–411. 10.1146/annurev.genet.38.072902.092717 [DOI] [PubMed] [Google Scholar]
- Takeda K, Hozumi H, Nakai K, Yoshizawa M, Satoh H, Yamamoto H, & Shibahara S (2014). Insertion of long interspersed element-1 in the Mitf gene is associated with altered neurobehavior of the black-eyed white Mitfmi-bwmouse. Genes to Cells, 19(2), 126–140. 10.1111/gtc.12117 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, & Mesirov JP (2013). Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Briefings in Bioinformatics, 14(2), 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, & Salzberg SL (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics, 25(9), 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udono T, Yasumoto KI, Takeda K, Amae S, Watanabe KI, Saito H, … Shibahara S (2000). Structural organization of the human microphthalmia-associated transcription factor gene containing four alternative promoters. Biochimica Et Biophysica Acta-Gene Structure and Expression, 1491, 205–219. 10.1016/S0167-4781(00)00051-8 [DOI] [PubMed] [Google Scholar]
- Vachtenheim J, & Ondrusov L (2013). MITF: A critical transcription factor in melanoma transcriptional regulatory network In Recent Advances in the Biology, Therapy and Management of Melanoma. InTech; 10.5772/55191 [DOI] [Google Scholar]
- Watanabe K, Takeda K, Yasumoto K, Udono T, Saito H, Ikeda K, … Shibahara S (2002). Identification of a distal enhancer for the melanocyte-specific promoter of the MITF gene. Pigment Cell Research, 15(3), 201–211. 10.1034/j.1600-0749.2002.01080.x [DOI] [PubMed] [Google Scholar]
- Wicks NL, Chan JW, Najera JA, Ciriello JM, & Oancea E (2011). UVA phototransduction drives early melanin synthesis in human melanocytes. Current Biology: CB, 21(22), 1906–1911. 10.1016/j.cub.2011.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildhardt G, Zirn B, Graul-Neumann LM, Wechtenbruch J, Suckfüll M, Buske A, … Steinberger D (2013). Spectrum of novel mutations found in Waardenburg syndrome types 1 and 2: Implications for molecular genetic diagnostics. British Medical Journal Open, 3(3), e001917 10.1136/bmjopen-2012-001917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Rigor P, & Baldi P (2009). MotifMap: A human genome-wide map of candidate regulatory motif sites. Bioinformatics, 25(2), 167–174. 10.1093/bioinformatics/btn605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima I, Sato S, Kimura T, Yasumoto K-I, Shibahara S, Goding CR, & Yamamoto H (1999). An L1 element intronic insertion in the black-eyed white (Mitfmi-bw) gene: The loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Human Molecular Genetics, 8(8), 1431–1441. 10.1093/hmg/8.8.1431 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, … McKeon F (1999). p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature, 398(6729), 714–718. 10.1038/19539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.