Abstract
Spread of antibiotic resistance via mobile genetic elements associates with transfer of genes providing resistance against multiple antibiotics. Use of various comparative genomics analysis techniques enables to find intrinsic and acquired genes associated with phenotypic antimicrobial resistance (AMR) in Campylobacter jejuni genome sequences with exceptionally high-level multidrug resistance. In this study, we used whole genome sequences of seven C. jejuni to identify isolate-specific genomic features associated with resistance and virulence determinants and their role in multidrug resistance (MDR). All isolates were phenotypically highly resistant to tetracycline, ciprofloxacin, and ceftriaxone (MIC range from 64 to ≥256 µg/mL). Besides, two C. jejuni isolates were resistant to gentamicin, and one was resistant to erythromycin. The extensive drug-resistance profiles were confirmed for the two C. jejuni isolates assigned to ST-4447 (CC179). The most occurring genetic antimicrobial-resistance determinants were tetO, beta-lactamase, and multidrug efflux pumps. In this study, mobile genetic elements (MGEs) were detected in genomic islands carrying genes that confer resistance to MDR, underline their importance for disseminating antibiotic resistance in C. jejuni. The genomic approach showed a diverse distribution of virulence markers, including both plasmids and phage sequences that serve as horizontal gene transfer tools. The study findings describe in silico prediction of AMR and virulence genetics determinants combined with phenotypic AMR detection in multidrug-resistant C. jejuni isolates from Lithuania.
Keywords: Campylobacter jejuni, multidrug resistance, mobile genetic elements, antibiotic-resistance genes, horizontal gene transfer, genomic islands, whole genome sequence
1. Introduction
Campylobacteriosis is one of the most common foodborne bacterial diseases worldwide [1]. The reported confirmed cases of human campylobacteriosis reached 246,571 in 2018 and remained the most frequently reported foodborne illness in the EU with notification rate of 64.1 per 100,000 population. [2]. Campylobacter jejuni is an important zoonotic pathogen causing significant infections, especially in young children, geriatric and immunocompromised patients [3,4]. Bacterial antibiotic resistance, especially multidrug resistance (MDR) and extensive drug resistance (XDR), has become a global emerging threat to public health systems [5,6]. Multidrug-resistance bacteria are frequently detected in humans and animals from developed and developing countries and pose a serious threat to human health [7].
Bacteria can acquire antimicrobial resistance through two leading pathways: chromosomal mutation and the acquisition of mobile genetic elements (MGEs) by horizontal gene transfer (HGT). Horizontal gene transfer of mobile genetic elements allows bacteria to exchange the genetic materials among pathogenic and non-pathogenic bacteria from different environments [8,9]. The HGT partly causes an increase in the adaptability of bacteria to environmental changes [10]. The transposition of MGEs can radically alter genome structure and genome sequence of bacteria, as antibiotic resistance is often spread via mobile genetic elements, and carry resistance against multiple antibiotics. Such bacteria with acquired AMR can spread and be transmitted to another environment, as resistant bacteria transfer from wild birds and cattle host to humans [11,12]. A better understanding of antibiotic resistance genes’ (ARGs’) circulation within bacterial species in different host niches and the mobility of these genes between different hosts could be important for identifying and analyzing multidrug resistance [13].
The study aimed to search the possible links among the phenotypic multidrug antimicrobial resistance and whole genome sequencing data (WGS) of C. jejuni isolates from different sources (cattle and wild bird feces).
2. Materials and Methods
2.1. Study Isolates
In total, seven C. jejuni isolates from bacterial culture collection of the Department of Food Safety and Quality of Lithuanian University of Health Sciences were tested in this study. These isolates were previously characterized by Multi Locus Sequence Typing (MLST) and assigned to CC179 and CC21 clonal complexes, with wide spread in Lithuania [14,15]. The isolates were stored at −80 °C in brain heart infusion broth (BHI) (Oxoid, Basingstoke, UK) with 30% glycerol (Stanlab, Lublin, Poland). The isolates’ recovery were performed by plating the stocks on Blood agar base No. 2 (Oxoid, Basingstoke, UK) supplemented with 5% defibrinated horse blood (E&O Laboratories Limited, Scotland, UK) and further incubation under microaerophilic conditions (5% oxygen, 10% carbon dioxide, and 85% nitrogen) at 42 °C for 48 h.
2.2. Antimicrobial Susceptibility Testing
All isolates were tested for antimicrobial susceptibility to erythromycin, tetracycline, gentamicin, ciprofloxacin, and ceftriaxone (all Sigma-Aldrich, Saint Louis, MO, USA). The minimum inhibitory concentration (MIC) was determined by the agar dilution method performed according to recommendations by the Clinical and Laboratory Standards Institute (CLSI) guidelines [16]. Isolates were cultured on Mueller-Hinton agar (Liofilchem, Italy) plates with dilutions ranging from 0.25 to 256 mg/mL for all antimicrobials. For each individual C. jejuni isolate, 5 µL of approximately 1 × 107 CFU/mL (OD600 = 0.1) bacterial suspension dissolved in phosphate-buffered saline (PBS) (E&O Laboratories Limited, Scotland, UK) were spotted onto Mueller-Hinton agar plates containing the corresponding antimicrobial agent concentration. The inoculated plates were incubated under microaerophilic conditions at 42 °C temperature for 24 h. The determination of MIC for each isolate was performed in triplicate. The MIC values were defined as the lowest concentration that produces complete inhibition of C. jejuni growth. For quality control, the reference isolate C. jejuni ATCC 33,560 was included. These breakpoints defined by the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) for antimicrobial susceptibility were used: for erythromycin ≥8 mg/mL, for tetracycline ≥2 mg/mL, for ciprofloxacin ≥1 mg/mL, for gentamicin ≥4 mg/mL, and for ceftriaxone ≥8 mg/mL. Isolates showing resistance to three or more antimicrobials were considered as multidrug-resistant.
2.3. Whole Genome Sequencing
DNA extraction was performed using the PureLink Genomic DNA Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and finally eluted in 50 µL of sterile Mili-Q water. The concentration and integrity of DNA were quantified by Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) with the double-stranded DNA (dsDNA) assay HS kit (lifetechnologies, Eugene, OR, USA) and 1% agarose gel, respectively.
According to the manufacturer’s instructions, DNA libraries were prepared using the Nextera XT library preparation kit (Illumina, San Diego, CA, USA). The sequencing was carried out at the NGS-MiSeq core facility of the University of Copenhagen using an Illumina MiSeq platform (Illumina, San Diego, CA, USA) with 250 bp paired-end read format and aiming to obtain an average genome depth of 50X. CLC Genomics Workbench version 6.5.1 was used for the adapter and quality trimming of the raw reads. Sequence reads were de novo assembled into contigs using SPAdes v.3.10 assembler [17]. The quality of the assembly was evaluated with QUAST v.2.3 (3). The subsystems’ annotation was obtained using the SEED-based automated annotation system after the data were uploaded to RAST (Rapid Annotation using Subsystem Technology) [18,19] genome server. Ribosomal multilocus sequence typing (rMLST) employing 53 genes encoding the bacterial ribosome protein subunits (rps genes) was performed using the Genome Comparator module of the BIGSdb platform on the PubMLST website [20]. BlastKOALA (https://www.kegg.jp/blastkoala/) [21] was used to perform KO (KEGG Orthology) assignments to characterize individual gene functions and reconstruct KEGG pathways, BRITE hierarchies, and KEGG modules. The presence of potential genes encoding antibiotic resistance was checked using the NCBI AMRFinder v.3.1.1 tool and the ResFinder v.3.0 and PointFinder v.3.1.0 (https://cge.cbs.dtu.dk/services/ResFinder/) [22] databases using thresholds of 90% identity and 60% gene coverage. Besides, a resistome prediction which uses BLAST algorithms to search of the AMR genes and SNPs was performed with Resistance Gene Identifier (RGI v.4.2.2) [23] with a previous coding sequence of the genome submission to the Comprehensive Antimicrobial Resistance Database (https://card.mcmaster.ca/analyze/rgi). Detection of antibiotic-resistance genes, mobile genetic elements, and mutation were performed through alignment to perform multiple alignments of the query with reference sequence downloaded from the NCBI reference (RefSeq) database using the CLUSTAW alignment tool https://www.genome.jp/tools-bin/clustalw [24]. CRISPR finder (https://crispr.i2bc.paris-saclay.fr/Server/) [25] and PathogenFinder 1.1 (https://cge.cbs.dtu.dk/services/PathogenFinder/) [26] databases of the Center for Genomic Epidemiology were used for the potential prediction of pathogenicity. A genome BLAST atlas was generated using isolates of C. jejuni comparison against the reference genome NCTC11168 (AL111168.1) using Gview (https://server.gview.ca/) [27]. For the dataset, alignment was used with BLASTn analysis with an e-value of 1 × 10−10, coverage 100%, and identity 80%. The IslandViewer version 4 server (https://www.pathogenomics.sfu.ca/islandviewer/) [28] was used to predict the putative genomic islands (GIs).
2.4. Data Availability
All genomes were submitted to NCBI under the following Accession Numbers: SAMN08794492; SAMN08794493; SAMN08803042; SAMN08803043; SAMN08803044; SAMN08803060; SAMN08803062 (BioProject PRJNA445645).
3. Results and Discussion
3.1. Phenotypic Antimicrobial Resistance Determination
All seven C. jejuni isolates were resistant to three or more antibiotics, and were identified as multidrug-resistant (MDR). MIC data are provided in Table 1. All isolates were phenotypically highly resistant to tetracycline, ciprofloxacin, and ceftriaxone (MIC range 64 ≥ 256 µg/mL). Besides, two C. jejuni isolates were resistant to gentamicin, and one was resistant to erythromycin. The extensive drug-resistance profiles were confirmed for the two C. jejuni isolates assigned to ST-4447 (CC179) (Table 2).
Table 1.
Minimum inhibitory concentration among Campylobacter jejuni isolates from cattle and wild birds.
| Antimicrobial Agent | |||||
|---|---|---|---|---|---|
| Isolate | TET | CIP | GEN | AXO | ERY |
| MIC, µg/mL | |||||
| CCm26 | 128 | 128 | 2 | 128 | 4 |
| CCm31 | >256 | 64 | 0.5 | 128 | 0.5 |
| CCm32 | 128 | 256 | 8 | 64 | 8 |
| CCm33 | >256 | 128 | 4 | 128 | 0.25 |
| CCm35 | 128 | 128 | 0.5 | 128 | 0.5 |
| CCm36 | 256 | 128 | 0.25 | 256 | 0.5 |
| CCm37 | 64 | 64 | 0.25 | 64 | 0.5 |
| Breakpoint | ≥2 | ≥1 | ≥4 | ≥8 | ≥8 |
TET, tetracycline; ERY, erythromycin; CIP, ciprofloxacin; GEN, gentamicin; AXO, ceftriaxone; MIC, minimum inhibitory concentration.
Table 2.
Phenotypic antimicrobial resistance profiles of C. jejuni isolates from cattle and wild birds.
| Isolate | Source | Clonal Complex | Sequence Type | Antimicrobial Resistance Profile |
|---|---|---|---|---|
| CCm26 | Wild bird | CC179 | ST-6424 | TET + CIP + AXO |
| CCm31 | Wild bird | CC179 | ST-4447 | TET + CIP + AXO |
| CCm32 | Wild bird | CC179 | ST-4447 | TET + CIP + AXO + GEN + ERY * |
| CCm33 | Wild bird | CC179 | ST-4447 | TET + CIP + AXO + GEN * |
| CCm35 | Cattle | CC21 | ST-21 | TET + CIP + AXO |
| CCm36 | Cattle | CC21 | ST-21 | TET + CIP + AXO |
| CCm37 | Cattle | CC21 | ST-21 | TET + CIP + AXO |
TET, tetracycline; ERY, erythromycin; CIP, ciprofloxacin; GEN, gentamicin; AXO, ceftriaxone; CC, clonal complex; ST, sequence type; * extensive drug resistant (XRD) isolates.
3.2. Genomics
All isolates with high-level multidrug resistance were further characterized by whole genome sequencing to identify transferable genes encoding antimicrobial resistance.
The general genome stats suggest that genome sizes range from 1.68 to 1.70 Mbp, with the average 30% G + C content, well within the range of available genome sequences. The summary of the genomes’ sequencing assignment is listed in Table 3.
Table 3.
List of genomic data of C. jejuni isolates.
| CCm26 | CCm31 | CCm32 | CCm33 | CCm35 | CCm36 | CCm37 | |
|---|---|---|---|---|---|---|---|
| Metrics of sequence data | |||||||
| Coverage (x) | 160 | 120 | 180 | 400 | 220 | 180 | 160 |
| Genomic data report | |||||||
| Size (bp) | 1682833 | 1685191 | 1685033 | 1721979 | 1701391 | 1705164 | 1705114 |
| No. of contigs | 31 | 59 | 25 | 29 | 23 | 24 | 21 |
| GC (%) | 30.3 | 30.4 | 30.4 | 30.3 | 30.3 | 30.3 | 30.3 |
| N50 | 122854 | 62137 | 158860 | 184236 | 116706 | 122859 | 135281 |
| L50 | 4 | 9 | 4 | 4 | 6 | 6 | 5 |
| L75 | 9 | 17 | 8 | 7 | 10 | 9 | 8 |
| Genes | 1768 | 1772 | 1770 | 1814 | 1781 | 1781 | 1779 |
| CDs | 1725 | 1729 | 1727 | 1771 | 1737 | 1738 | 1736 |
| Subsystems | |||||||
| rRNAs | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| tRNAs | 40 | 40 | 40 | 40 | 41 | 40 | 40 |
3.3. Whole Genome Sequence-Based Genotypic Predictions of Antibiotic-Resistance Genes
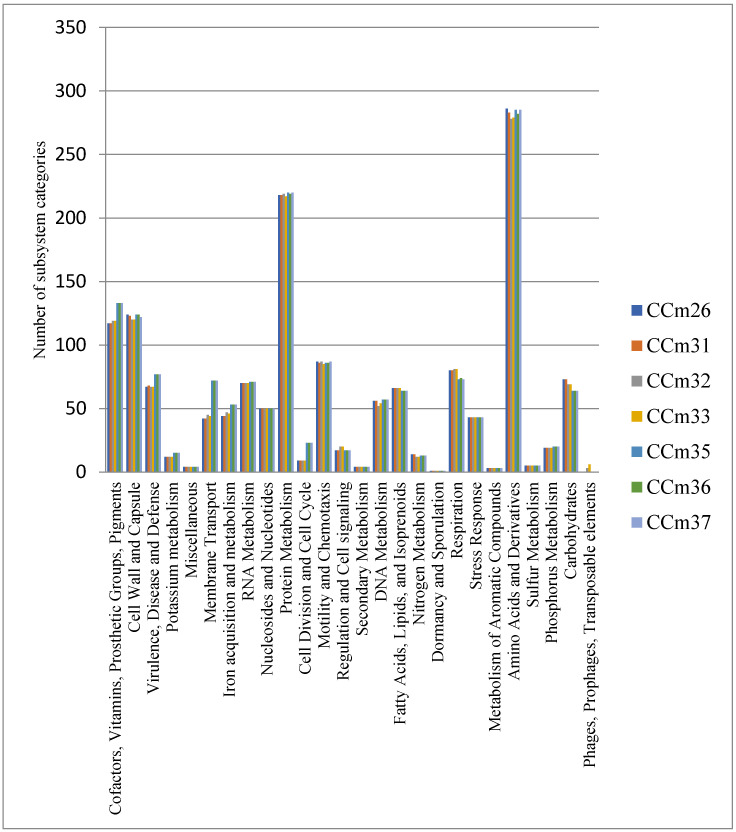
The genomes of C. jejuni isolates were clustered into orthologue groups and annotated in RAST with the aim to identify traits involved in antimicrobial resistance and survival. Based on RAST analysis, 77 genes in three cattle, in each genome assigned to ST-21 (CC21), were annotated in association with virulence, disease, and defense. The analysis revealed the presence of virulence marker genes associated with adhesion (cadF and pEB1), invasion (yidC and yidD), and cytotoxin production (cdtA, cdtB, and cdtC). Nine protein-coding genes in genomes of CCm32 and CCm33 isolates were identified in phages, prophages, and transposable elements’ category including phages’ proteins involved in phage replication process, phage tail, and phage capsid proteins. Figure 1 shows the diagram of the genes associated with the functional categories of examined isolates.
Figure 1.
Subsystem category distribution of seven C. jejuni isolates.
The WGS data of examined isolates were mapped based on intrinsic and acquired genes known to be associated with phenotypic AMR. The genomes were also manually searched for genes known to being involved in AMR and virulence. The isolates CCm31 and CCm32 assigned to CC179 harbored cobalt-zinc-cadmium resistance determinants composed of czc, chr, ncc, and mer genes responsible for resistance to Zn, Cr, Ni and Hg, respectively. In three isolates, CCm35, CCm36, and CCm37, all assigned to CC21 (ST-21), platinum drug resistance ctpA, and a cationic antimicrobial peptide (CAMP) system genes cluster were identified (Table 4). The ctpA gene encodes the C-terminal processing protease for the photosystem’s D1 protein II reaction center complex related to virulence and cytotoxicity against host cells [29,30].
Table 4.
Genetic determinants associated of virulence and resistance markers found in C. jejuni. czc, cobalt zinc cadmium resistance system; ctpA, platinum resistance; AMPs, antimicrobial peptides; CAMP, cationic antimicrobial peptide; T4S, Type IV secretion system; flgE, flagellar motility; trg, chemotaxis; bdlA, biofilm formation; +, positive; −, negative; +/−, uncomplete system.
| C. jejuni | Virulence Markers | Resistance Markers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heavy Metal Resistance | AMPs Sensing System | Invasion | Multidrug Efflux Pupms | Tetracycline | β-Lactams | ||||||||
| czc | ctpA | CAMP | T4S | flgE, trg, bdlA | cmeABC | pmrA | tetO | tetM | blaOXA-448 | blaOXA-61 | blaOXA-451 | blaOXA-133 | |
| CCm26 | − | − | − | − | +++ | + | + | + | − | + | − | − | − |
| CCm31 | + | − | − | − | +++ | − | − | − | − | + | − | − | − |
| CCm32 | + | − | − | − | +++ | − | − | − | − | + | − | − | − |
| CCm33 | − | − | − | +/− | +++ | − | − | − | − | + | − | − | − |
| CCm35 | − | + | + | − | +++ | − | − | + | − | − | + | − | + |
| CCm36 | − | + | + | +/− | +++ | + | − | + | + | − | + | − | + |
| CCm37 | − | + | + | +/− | +++ | + | + | + | + | − | + | + | + |
Type IV secretion system (T4S) genes virB2, virB4, virB8 and virB9 were identified in genomes of C. jejuni CCm33, CCm36, CCm37 isolates. The operon of the cmeABC multidrug efflux pump, consisting of cmeA, cmeB, and cmeC genes were identified in the genomes of three isolates (Table 4). The cmeABC, a resistance-nodulation-division (RND) type of efflux pump, contributes significantly to both intrinsic and acquired resistance to various antimicrobials in C. jejuni [31].
Additionally, the pmrA efflux pump, which belongs to the resistance-nodulation-division family of transporters and contributes to multidrug resistance of antimicrobials, was found in two C. jejuni isolates. The tetO gene, which codes the resistance to tetracycline, was detected in all C. jejuni isolates from cattle and in one isolate from a wild bird. Another tetracycline resistance gene tetM was found in the same two isolates, which had a tetO gene. The β-lactamase resistance gene blaOXA-448 was identified in all C. jejuni isolates assigned to clonal complex CC179; however, blaOXA-61 was identified in all isolates assigned to clonal complex CC21. Among the resistant isolates, several genes coding the virulence factors were found. The chemotaxis and flagellar motility genes, trg, flgE, and biofilm dispersion bdlA gene with increased adherent properties required for biofilm formation, were identified. These genes were identified in all C. jejuni isolates, which shows their widespread dissemination throughout C. jejuni genomes.
3.4. Point Mutation
Nucleotide and amino acid changes of C. jejuni genomic sequences are shown in Table 5. All CIP-resistant isolates harbored gyrA point mutation T86I. The G→A transversion in the rpsL gene was detected in two ST-4447 isolates. The six different point mutations, including deletion Lys→del in the L22 ribosomal protein gene rpIV, were observed in ST-6424. The amino acid changes in the cmeR gene, including D121N and E159K, were identified in two ST-21 isolates. Fourteen non-synonymous mutations were detected in the 23S rRNA gene.
Table 5.
Nucleotide and amino acid changes of C. jejuni genomic sequences.
| L22 | cmeR | gyrA | rpsL | 23S rRNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | Nucleotide Change | Amino Acid Change | Mutation | Nucleotide Change | Amino Acid Change | Mutation | Nucleotide Change | Amino Acid Change | Mutation | Nucleotide Change | Amino Acid Change | Mutation | Nucleotide Change |
| I165V 1 | ATT→GTT | Ile→Val | G144D 1,2,3,4,5 | GGT→GAT | Gly→Asp | R285K 1,2,3,4 | AGG→AAG | Arg→Lys | A119T 3,4 | GCT→ACT | Ala→Thr | 287G > A 5,6,7 | G→A |
| S109A 1 | TCT→GCT | Ser→Ala | S207G 1,2,3,4 | AGC→GGC | Ser→Gly | A312T 1,2,3,4 | GCT→ACT | Ala→Thr | 296C > G 5,6,7 | C→G | |||
| T119A 1 | ACT→GCT | Thr→Ala | D121N 6,7 | GAC→AAC | Asp→Asn | A664V 1,2,3,4 | GCC→GTC | Ala→Val | 298G > A 5,6,7 | G→A | |||
| T120P 1 | ACA→CCA | Thr→Pro | E159K 6,7 | GAA→AAA | Glu→Lys | T665S 1,2,3,4 | ACT→AGT | Thr→Ser | 327G > A 5,6,7 | G→A | |||
| V137A 1 | GTG→GCG | Val→Ala | T804A 1,2,3,4 | ACA→GCA | Thr→Ala | 364G > C 5,6,7 | G→C | ||||||
| K123→del 1 | AAA→del | Lys→del | T86I 1,2,3,4,5,6,7 | ACA→ATA | Trr→Ile | 554A > C 5,6,7 | A→C | ||||||
| 571T > G 5,6,7 | T→G | ||||||||||||
| 1027A > G 5,6,7 | A→G | ||||||||||||
| 1485C > T 4 | C→T | ||||||||||||
| 1735T > C 4 | T→C | ||||||||||||
| 1739T > C 4 | T→C | ||||||||||||
| 1752T > C 4 | T→C | ||||||||||||
| 1759A > G 4 | A→G | ||||||||||||
| 1761G > A 4 | G→A | ||||||||||||
Superscript numbers indicate the C. jejuni isolates harboring specific nucleotide and amino acid changes: 1 CCm26; 2 CCm31; 3 CCm32; 4 CCm33; 5 CCm35; 6 CCm36; 7 CCm37.
3.5. Mobile Genetic Elements: Genomic Islands, Prophages and Plasmids
The different genomic approaches analysis revealed the prevalence of MGEs, including plasmids, pathogenicity islands, and bacteriophages in C. jejuni isolates. Most of the AMR and virulence factors were distributed within genomic island (GI) regions. Analysis of ARGs’ composition based on GI revealed 18 GIs with the size ranging in length from 5.61 kb to 58.83 kb. The largest GI was detected in the CCm36 (58.83 kb), containing gene tetO encoding tetracycline resistance, trg gene encoding chemotaxis, virB2 gene, putative DNA-invertase pinR, and ansZ coding L-asparaginase 2. In total, six GI (size 5.73 to 45.9 kb) were uniquely found in one of the ST-4447 (CCm33) isolate, which contains prophage CPS-53 integrase intS, flgE encoding flagellar motility, virB2, virB4, virB8, and virB9 genes. GIs play crucial roles in microbial genome evolution and adaptation of microbes to environments as part of a flexible gene pool [32]. Whole genome sequencing also revealed a 131.1 kb phage harbored by one isolate (C. jejuni CCm33) with high homology (identity 98%; e value 1.37 × 10−118) to Campylobacter phage PC5 (KX229736.1). Prophages are genomes of temperate phages that have infected a susceptible host bacterium, where they either integrate into the chromosome or exist as circular or linear plasmids [33].
The plasmid of 20,765 bp was identified in two C. jejuni isolates assigned to ST-21 (CC21) with high homology of repUS59 (pSSG1) plasmid sequence (FR824044) in the NCBI repository database. Furthermore, the nucleotide sequence comparison of genomic island in CCm37 showed presence of pTet plasmids with the tetO gene, L-asparginase 2, flagellin A and various homologous hypothetical genes. These acquired elements expand the genetic flexibility of pathogens. Plasmids and bacteriophages contribute to the C. jejuni evolution via adaptation and survivability with the novel functions integrated into the chromosome [34,35].
3.6. BLAST Identification and Diverse Genomic Locations of the C. jejuni
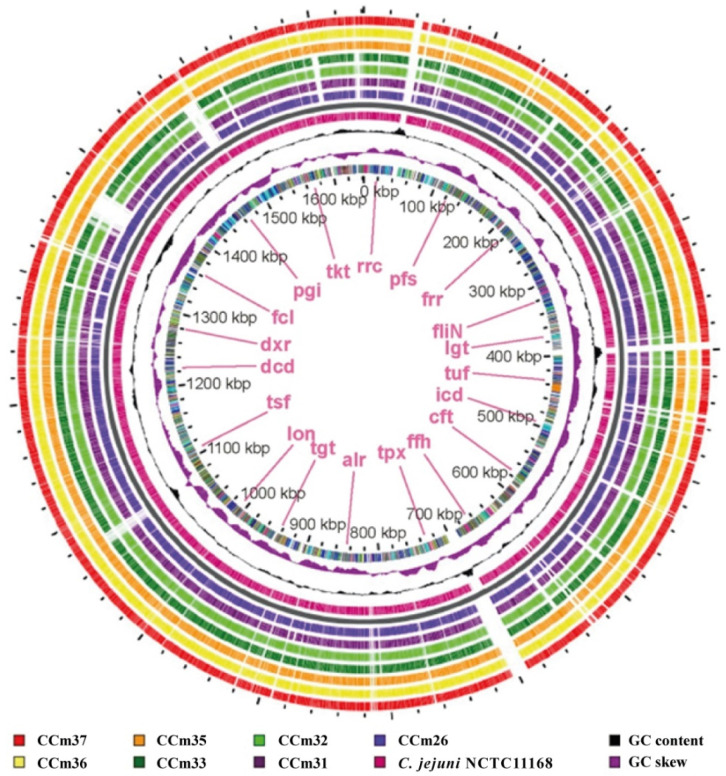
BLAST genomic atlas provides detailed valuable genomic insights that support genome-wide gene characterization of C. jejuni isolates and reveals how similar any genome is to the reference genome (NCTC 11168). Analysis of the G + C content distribution showed localized peak deviations from the genome’s average GC content. At positions from 71,887 to 50,973 bp, the chemotactic transducer gene pctC was detected in CCm33, CCm36, and CCm37 isolates. This gene consists of 2102 bp and encodes a transmembrane signaling receptor activity, which is to enhance the ability of signal transduction [36]. Genomic comparison with the reference C. jejuni NCTC11168 isolate genome revealed that the ssa1 gene encoding serotype 1-specific antigen (identity 100%) was found in all ST-21 C. jejuni isolates. This serotype-specific antigen has been shown to be involved in the endopeptidase activity regulation complex of Neisseria meningitides M0579 and Pasteurella haemolytica but is not present in C. jejuni NCTC 11168. The multidrug resistance gene MdtG (identity 97.88%) was observed in all ST-4447 isolates.
Moreover, the BLAST search of C. jejuni ST-21 isolates revealed a novel acquired complex that harbors the β-lactamase resistance gene blaOXA-133 (100% identity) belonging to the class-D β-lactamase family, yafP gene, and ykkC multidrug resistance gene in the region from 273,300 to 282,500 bases (Figure 2). The changes, most often associated with MGEs that have been acquired by HGT, can initiate a rapid mechanism for acquiring genes and confer novel function [37].
Figure 2.
Circular genomic atlas of 7 C. jejuni isolates in comparison to the reference C. jejuni NCTC11168 genome. The circle is divided into arcs representing the genomes as labeled. The black histogram represents the G + C content, and the purple-green histogram represents the G + C deviation.
In conclusion, this knowledge provides insights on the distribution and genetic content of MGEs in multidrug Campylobacter jejuni isolates. Mobile genetic elements are principally important to facilitate horizontal genetic exchange and, therefore, induce the acquisition and spread of resistance genes. This may allow an assessment of how genes carried by mobile genetic elements can contribute to traits that are responsible for antimicrobial resistance and virulence. These findings highlighted the role of resistance determinants in the epidemiology of MGEs in C. jejuni genome sequences and indicate the potential for bacteria’s genomic plasticity. In this context, a better understanding of the acquisition of resistance determinants is critical to understand the capacity of resistance genes to spread in C. jejuni population.
Author Contributions
M.M. supervised the work; J.A. and E.K. performed the bioinformatics analysis; J.A. wrote the manuscript with input from all authors; M.M., E.K., A.G. and A.N. contributed to interpreting the results. All authors contributed to manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oyarzabal O.A., Carrillo C.D. Chapter 4—Isolation, identification, and typing of Campylobacter strains from food samples. In: Klein G., editor. Campylobacter. Academic Press; Cambridge, MA, USA: 2017. pp. 61–83. [Google Scholar]
- 2.European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC) The European Union One Health 2018 Zoonoses Report. EFSA J. 2019;17:e05926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakran W., Hexner-Erlichman Z., Spiegel R., Batheesh H., Halevy R., Koren A. Campylobacter gastroenteritis in children in north-eastern Israel comparison with other common pathogens. Sci. Rep. 2020;10:5823. doi: 10.1038/s41598-020-62744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abebe E., Gugsa G., Ahmed M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop. Med. 2020;2020:e4674235. doi: 10.1155/2020/4674235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 6.Van Duin D., Paterson D. Multidrug Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016;30:377–390. doi: 10.1016/j.idc.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle M.E. Multidrug-resistant pathogens in the food supply. Foodborne Pathog. Dis. 2015;12:261–279. doi: 10.1089/fpd.2014.1865. [DOI] [PubMed] [Google Scholar]
- 8.Bello-López J.M., Cabrero-Martínez O.A., Ibáñez-Cervantes G., Hernández-Cortez C., Pelcastre-Rodríguez L.I., Gonzalez-Avila L.U., Castro-Escarpulli G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms. 2019;7:363. doi: 10.3390/microorganisms7090363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K., Kim D.-W., Lee D.-H., Kim Y.-S., Bu J.-H., Cha J.-H., Thawng C.N., Hwang E.-M., Seong H.J., Sul W.J., et al. Mobile resistome of human gut and pathogen drives anthropogenic bloom of antibiotic resistance. Microbiome. 2020;8:2. doi: 10.1186/s40168-019-0774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emamalipour M., Seidi K., Zununi Vahed S., Jahanban-Esfahlan A., Jaymand M., Majdi H., Amoozgar Z., Chitkushev L.T., Javaheri T., Jahanban-Esfahlan R., et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front. Cell Dev. Biol. 2020;8:229. doi: 10.3389/fcell.2020.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waddell S., Barnstedt O., Treiber C. Chapter Four—Neural Transposition in the Drosophila Brain: Is It All Bad News? In: Yamamoto D., editor. Advances in Genetics; Epigenetic Shaping of Sociosexual Interactions. Volume 86. Academic Press; Cambridge, MA, USA: 2014. pp. 65–92. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y.-M., Holmes E.C., Chen X., Tian J.-H., Lin X.-D., Qin X.-C., Gao W.-H., Liu J., Wu Z., Zhang Y. Diverse and abundant resistome in terrestrial and aquatic vertebrates revealed by transcriptional analysis. Sci. Rep. 2020;10:18870. doi: 10.1038/s41598-020-75904-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramonaite S., Kudirkiene E., Tamuleviciene E., Leviniene G., Malakauskas A., Gölz G., Alter T., Malakauskas M. Prevalence and genotypes of Campylobacter jejuni from urban environmental sources in comparison with clinical isolates from children. Pt 9J. Med. Microbiol. 2014;63:1205–1213. doi: 10.1099/jmm.0.072892-0. [DOI] [PubMed] [Google Scholar]
- 15.Ramonaite S., Tamuleviciene E., Alter T., Kasnauskyte N., Malakauskas M. MLST genotypes of Campylobacter jejuni isolated from broiler products, dairy cattle and human campylobacteriosis cases in Lithuania. BMC Infect. Dis. 2017;17:430. doi: 10.1186/s12879-017-2535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel and Clinical and Laboratory Standards Institute—2017—Performance Standards for Antimicrobial Susceptibility Testing. [(accessed on 1 October 2020)]; Available online: https://clsi.org/media/1469/m100s27_sample.pdf.
- 17.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolley K.A., Bliss C.M., Bennett J.S., Bratcher H.B., Brehony C., Colles F.M., Wimalarathna H., Harrison O.B., Sheppard S.K., Cody A.J., et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Pt 4Microbiology. 2012;158:1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.-L.V., Cheng A.A., Liu S., et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J.D., Gibson T.J., Higgins D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003;1:2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 25.Grissa I., Vergnaud G., Pourcel C. CRISPRcompar: A website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2008;36:W145–W148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosentino S., Voldby Larsen M., Møller Aarestrup F., Lund O. PathogenFinder—Distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE. 2013;8:e77302. doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petkau A., Stuart-Edwards M., Stothard P., Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertelli C., Laird M.R., Williams K.P., Simon Fraser University Research Computing Group. Lau B.Y., Hoad G., Winsor G.L., Brinkman F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anbudurai P.R., Mor T.S., Ohad I., Shestakov S.V., Pakrasi H.B. The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. USA. 1994;91:8082–8086. doi: 10.1073/pnas.91.17.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inagaki N., Yamamoto Y., Mori H., Satoh K. Carboxyl-terminal processing protease for the D1 precursor protein: Cloning and sequencing of the spinach cDNA. Plant Mol. Biol. 1996;30:39–50. doi: 10.1007/BF00017801. [DOI] [PubMed] [Google Scholar]
- 31.Su C.-C., Yin L., Kumar N., Dai L., Radhakrishnan A., Bolla J.R., Lei H.-T., Chou T.-H., Delmar J.A., Rajashankar K.R., et al. Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump. Nat. Commun. 2017;8:171. doi: 10.1038/s41467-017-00217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hacker J., Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2001;2:5–81. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uelze L., Grützke J., Borowiak M., Hammerl J.A., Juraschek K., Deneke C., Tausch S.H., Malorny B. Typing methods based on whole genome sequencing data. One Health Outlook. 2020;2:3. doi: 10.1186/s42522-020-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobrindt U., Hacker J. The Comprehensive Sourcebook of Bacterial Protein Toxins. Elsevier Ltd.; Amsterdam, The Netherlands: 2006. Mobile genetic elements and pathogenicity islands encoding bacterial toxins; pp. 44–63. [Google Scholar]
- 35.Oliveira P.H., Touchon M., Cury J., Rocha E.P.C. The chromosomal organization of horizontal gene transfer in bacteria. Nat. Commun. 2017;8:841. doi: 10.1038/s41467-017-00808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parales R.E., Ditty J.L. Chemotaxis. In: Timmis K.N., editor. Handbook of Hydrocarbon and Lipid Microbiology. Springer; Berlin/Heidelberg, Germany: 2010. pp. 1529–1543. [DOI] [Google Scholar]
- 37.Sheppard S.K., Guttman D.S., Fitzgerald J.R. Population genomics of bacterial host adaptation. Nat. Rev. Genet. 2018;19:549–565. doi: 10.1038/s41576-018-0032-z. [DOI] [PubMed] [Google Scholar]