Abstract
In order to control the COVID-19 pandemic caused by SARS-CoV-2 infection, serious progress has been made to identify infected patients and to detect patients with a positive immune response against the virus. Currently, attempts to generate a vaccine against the coronavirus are ongoing. To understand SARS-CoV-2 immunoreactivity, we compared the IgG antibody response against SARS-CoV-2 in infected versus control patients by dot blot using recombinant viral particle proteins: N (Nucleocapsid), M (Membrane) and S (Spike). In addition, we used different protein fragments of the N and S protein to map immune epitopes. Most of the COVID-19 patients presented a specific immune response against the full length and fragments of the N protein and, to lesser extent, against a fragment containing amino acids 300–685 of the S protein. In contrast, immunoreactivity against other S protein fragments or the M protein was low. This response is specific for COVID-19 patients as very few of the control patients displayed immunoreactivity, likely reflecting an immune response against other coronaviruses. Altogether, our results may help develop method(s) for measuring COVID-19 antibody response, selectivity of methods detecting such SARS-CoV-2 antibodies and vaccine development.
Keywords: Nucleocapsid protein, Membrane protein, Spike protein, SARS-CoV-2, COVID-19, Immunotest
Abbreviations: COVID-19 Coronavirus disease 2019, CTD C-terminal Domain; E protein Envelope protein, LKR Serine-rich linker region; M protein Membrane protein, N protein Nucleocapsid protein; NTD N-terminal Domain, RBD Receptor Binding Domain; SARS-CoV-2 Severe acute respiratory syndrome corona virus 2, S protein Spike protein
1. Introduction
Severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) infection causes the coronavirus disease 2019 (COVID-19) [1]. COVID-19 has spread quickly over the world and is responsible for more than a million deaths despite severe containment measures in many countries (https://covid19.who.int). To limit the spread of the disease, efforts are undertaken by the scientific community to detect patients that are undergoing or have passed the infection [2]. Also, many research centers and pharmaceutical companies are working on the development for an effective vaccine that protects against COVID-19 [3,4].
Upon infection, the immune system recognizes and tries to neutralize the SARS-CoV-2 virus particle. Here we investigated which protein(s) and/or protein fragments from the SARS-CoV-2 virus particle induce a humoral immune response in patients. We believe our work contributes to an accurate method to detect patients that suffer COVID-19 but also gives information on what are the most immunoreactive parts of the virus, information that might contribute to developing an effective vaccine.
2. Materials and methods
2.1. Cloning and purification of SARS-CoV-2 viral particle proteins and fragments
Full length N and M proteins from SARS-CoV-2 and fragments containing amino acids 1–350, 300–685, 500–700, 686–811 from the Spike (S protein) and 1–200, 100–300 and 200–419 from the N protein were expressed in purified in E. Coli. For this, the corresponding cDNAs obtained from N.J. Krogan (San Francisco, CA, USA) [5] were subcloned into pET30a (Novagen) vector and proteins were expressed in E. coli. Expressed proteins were purified after obtaining an insoluble fraction that was solubilized with urea buffer (Urea 7 M, 400 mM NaCl, 50 mM Tris pH 8) and using Ni-NTA (Qiagen) following the manufacturer’s instructions. Eluted proteins were quantified using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). SARS-CoV-2 N protein cDNA was subcloned into peGFP-N1 (Clontech) to generate a GFP-SARS-CoV-2 N fusion. 293T cells were transfected with GFP-tagged SARS-CoV-2 N protein and extracts were generated by urea-SDS buffer as described before [6].
2.2. Antibody generation
The recombinant His-tagged full length SARS-CoV-2 N protein was used to immunize a rabbit. Serum was obtained after five immunizations and used in experiments of Fig. S1. The anti-GFP antibody was described before [7].
2.3. Dot blot analysis
Different amounts of purified and quantified proteins were spotted on a nitrocellulose membrane (Amersham Protran Premium 0.45 μm NC, GE Healthcare) in different quantities (2.5, 10 and 40 μg of protein). Membranes were dried, blocked with TBS+0.1% Tween 20 (TBS-T) with 5% non-fat dry milk for 1 h at RT and subsequently incubated overnight with TBS-T+5% milk containing a 1:500 dilution of each patient serum or 1:8000 in the case of the control rabbit anti-N protein. After washing 3 times for 10 min with TBS-T, strips were incubated with a rabbit anti-Human IgG (H + L) secondary antibody conjugated to HRP (Thermo Fisher) for 1 h at RT for patient serum, and with goat anti-rabbit HRP (Jackson Immunoresearch) in the case of the rabbit anti-N protein. After washing as before, the strips were incubated for 5 min with the superSignal™ West Pico PLUS chemiluminescent substrate (Thermo Fisher) and the images were obtained using the ImageQuant LAS 4000 mini. For quantification 60 s exposures were selected for all strips.
2.4. Quantification of dot blots from patients
The quantification analysis was performed based on the dot blot results using the ImageQuant TL 1D v8.1 software following analysis performed by integrating the levels of pixels (volume) surrounded by a circular selection. Density of immunoreactive dots is reported as arbitrary units (a.u.).
The data are expressed as mean (bar) with the 25th −75th percentile range (box) and the 10th – 90th percentile range (whiskers). A two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparisons test was applied for statistical significance determination (∗∗∗∗P < 0.0001, ∗∗P < 0.01, ∗P < 0.05).
2.5. Patient sera
Banked sera collected for other diagnostic purposes in the Department of Microbiology at the Hospital Universitario de Canarias during February 2019 was used as negative controls. For positive controls, we used sera collected from COVID-19 patients with clinical and microbiological (nasopharyngeal swab tests, positive by FDA-approved RT-PCR (Seegene, Roche)) evidence of SARS-CoV-2 infection. The sera were collected 14–66 days after appearance of first symptoms. This project was approved by the ethical committee of the Hospital Universitario de Canarias, which included the informed consent from the subjects involved in the study (Comité de Ética de la Investigación con medicamentos del Complejo Hospitalario Universitario de Canarias, #CHUC_2020_45).
3. Results
3.1. Purification of SARS-CoV-2 full length proteins and fragments
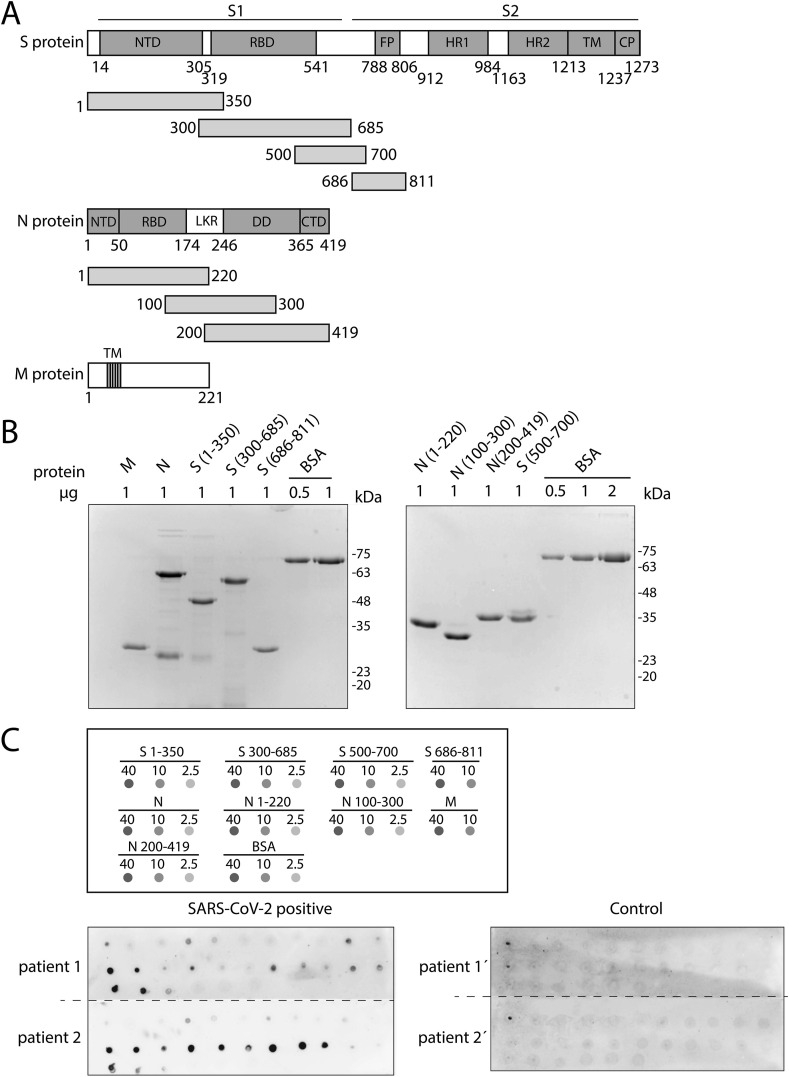
The viral particle of SARS-CoV-2 contains the viral positive-sense, single-stranded RNA genome that is surrounded by a lipid bilayer that contains the spike (S), nucleocapsid (N), membrane (M) and envelope (E) proteins [8]. The S protein is a large protein (1273 amino acids) that is processed into two subunits, S1 (amino acids 14 to 685) and S2 (amino acids 686 to 1273). S1 is responsible of the binding to the ACE2 receptor in the host cell and S2 subunit facilitates membrane fusion. Different subdomains are described in the S1 and S2 subunits, including a Receptor Binding Domain (RBD) comprising amino acids 319–541, that binds to the human ACE2 receptor with high affinity [9] (Fig. 1 A). The N protein is the most abundant viral protein and binds to the viral RNA forming the core of a ribonucleoprotein complex [10]. The Nucleocapsid protein contains an N-terminal Domain (NTD) that functions in the genomic RNA binding, which is connected by a central serine-rich linker region (LKR) to the and C-terminal Domain (CTD), involved in dimerization and also with RNA-binding activity [8,11] (Fig. 1A). The E protein is a membrane protein of only 75 amino acids and contains a hydrophobic region. Together with the M protein of 221 amino acids, containing 3 transmembrane domains, the E protein plays a role in viral assembly [12] (Fig. 1A).
Fig. 1.
Detection of antibodies against SARS-CoV-2 by dot plot. (A) Schematic representation of the different domains in S, N and M proteins of SARS-CoV-2 and the fragments used in this work. For the S protein, the following domains/motifs are indicated: N-terminal domain (NTD), receptor binding domain (RBD), fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), transmembrane domain (TM) and cytoplasmic domain (CP). The protein is proteolytically cleaved into two subunits: S1 (amino acids 14–685) and S2 (amino acids 686–1472). N protein-indicated domains are the N-terminal domain (NTD), RNA binding domain (RBD), dimerization domain (DD), a C-terminal domain (CTD) and a linker region (LKR). The 3 transmembrane domains (TM) are shown for the M protein. (B) Coomassie gel staining of all purified proteins used in this study. BSA was used as a control. (C) Top panel displays a diagram of the dot blot design, with the proteins spotted and the amount of protein used (in ng). Bottom panel represents two representative blots developed with sera from SARS-CoV-2 positive (left) and control (right) patients, respectively.
In order to study the patient immune response to the SARS-CoV-2 viral particle, we set out to investigate the antibody response to the different virus proteins in COVID-19 patients’ sera. For this, different SARS-CoV-2 proteins/protein fragments were cloned into a plasmid to produce his-tagged proteins, that were subsequently expressed in E. Coli, purified and quantified (Fig. 1A). Due to technical difficulties, we could not purify the E protein or a fragment containing the C-terminal part of the S2 subunits, but we successfully purified 4 different overlapping regions of the S protein, 3 regions of the N protein together with the full length M and N proteins (Fig. 1A and B).
3.2. Dot blot assay detects immunoglobulins against SARS-CoV-2 virions proteins
An immunoglobulin G (IgG) antibody response against the full length S protein was observed in COVID-19 patients, from 15 days after appearance of the first symptoms [13]. To study the possible differential response to the different structural proteins of the virus, we set up a simple assay to detect specific IgGs. First preliminary experiments were performed with an anti-N protein antibody that we raised in rabbit using the full length N protein as immunogen. As shown in Supplementary Fig. S1A, the antibody very specifically (at dilutions up to 1:8000) recognized the overexpressed N protein fused to GFP by Western blot, but also worked in a dot blot assay recognizing small amounts of purified recombinant N protein spotted on a nitrocellulose membrane (Supplementary Fig. S1B).
After preliminary tests to determine the optimal amounts of recombinant proteins spotted, dot blot assays with the 4 fragments of the S protein, 3 of the N protein, together with the full length M and N proteins were performed (see schedule in Fig. 1C, top). These dot blots were probed with the sera of patients from our hospital that were diagnosed as COVID-19 positive by RT-PCR tests. We also included sera of control patients collected before the pandemic, in February 2019.
Fig. 1C (bottom) shows a representative example from a COVID-19 patient and control patient against all structural proteins/fragments used in this study. Supplementary Figs. S2 and S3 show all the dot blots obtained for all COVID-19 and control patients, respectively. The results demonstrate an IgG antibody response in almost all COVID-19 patients, whereas the general response in control patients was low, as expected.
3.3. N protein is the most immunogenic of the structural proteins
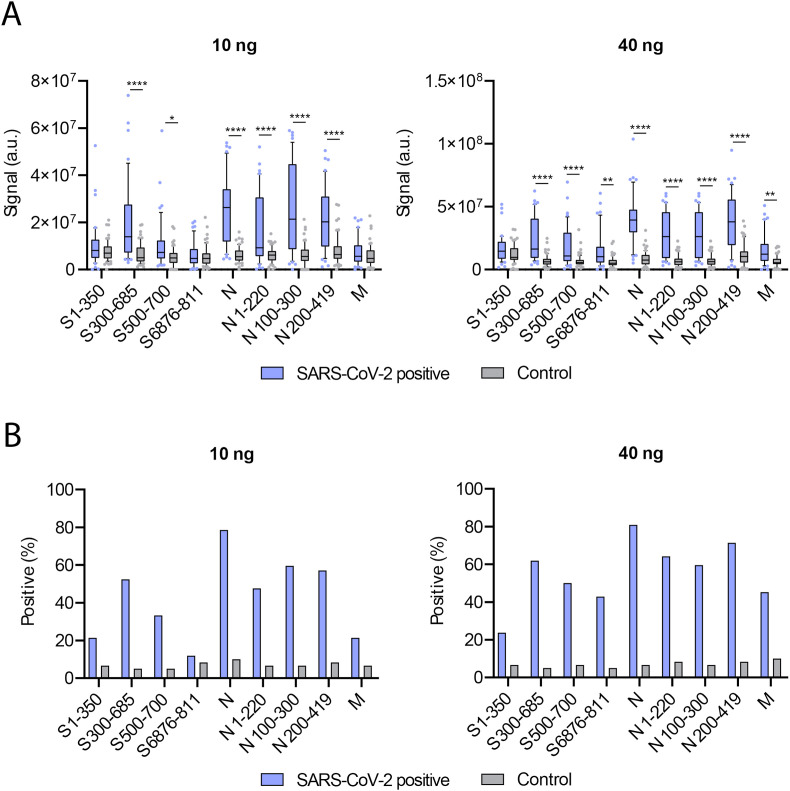
Next, all dot blots were quantified, and the data were analyzed. As shown in Fig. 2 A, a clear difference in the IgG immunoreactivity was observed, both for 10 ng and 40 ng of antigens in the dot blot, between COVID-19 patient sera and controls. In the SARS-CoV-2 positive patients, a strong IgG response to the full length N protein and all the N protein fragments was observed, as compared to control patients. Although not as striking, a significant difference between control and SARS-CoV-2 positive patients was also observed with the fragment containing amino acids 300–685 of the S protein. In contrast, only minor differences in immunoreactivity to all the other S protein fragments and the M protein were seen.
Fig. 2.
Dot blot analysis reveals N protein as the best candidate for antibody reactivity against SARS-CoV-2. (A) Quantification of all dot blots for SARS-CoV-2 positive (blue) and control (grey) patients for 10 ng (left) and 40 ng (right) of the indicated proteins. Density of immunoreactive dots is reported as arbitrary units (a.u.). Data are expressed as mean (bar) with the 25th −75th percentile range (box) and the 10th – 90th percentile range (whiskers). Two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparisons test was applied for statistical significance determination. (∗∗∗∗P < 0.0001, ∗∗P < 0.01, ∗P < 0.05). (B) The percentage of positive individuals within the SARS-CoV-2-positive and the control groups for each protein (fragment) from (A) was determined by setting the signal corresponding to the 90th percentile of the control group as a threshold. Data from both 10 ng (left) and 40 ng (right) were used. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Finally, the dot blot quantifications of Fig. 2A were used to examine the sensitivity of our assay in detecting positive and negative patients, upon analysis of the different proteins. A signal above the 90th percentile of the control group was scored as positive. The results of these analyses are shown in Fig. 2B. Importantly, we observed the highest predictive value with the full length N protein: around 80% of the positive patients were detected, with less than 10% of false positives. Using all three fragments of the N protein resulted in a slightly lower predictive value. These results suggest that although most patients generate a strong IgG antibody response to the N protein, this response does not seem to be triggered to a particular region of the protein. Furthermore, using the S protein 300–685 fragment also resulted in a good sensitivity detecting around 55–60% of positive patients, depending on the amount of protein tested. Studying the IgG antibody response to the rest of the fragments of the S protein and the full length M protein showed to be less effective for detecting positive COVID-19 patients.
4. Discussion
The new SARS-CoV-2 coronavirus has quickly spread over the world. Due to its potent capacity for infection and the fact it can cause severe morbidity and mortality, the virus became a serious health threat that led to economic and social disruptions. Here we compared the humoral immune response against different fragments or full length proteins of SARS-CoV-2 in serum from verified COVID-19 patients to that of control patients.
Our observations demonstrate that dot blot analysis is a relatively simple and efficient technique to examine the humoral immune response to SARS-CoV-2. Analyzing the response to the different proteins of SARS-CoV-2 showed that the N protein is the most immunogenic among all analyzed proteins. While a strong IgG antibody response to the N protein was also observed by others [11], the immunogenic regions were not mapped nor was its relative immunogenicity compared to other viral proteins. Here we show that multiple regions of the N protein elicit an immune response in patient serum that is relatively more potent than that elicited by other viral proteins. Interestingly, also the N protein of the related SARS-CoV was shown to be highly immunogenic [14]. In contrast, even though the overall IgG response to the SARS-CoV-2 S protein was lower, possibly due to the glycosylation status of this protein [15], the majority of the COVID-19 patients did trigger a response to a region in the S1 subunit of the protein, containing the RBD.
Despite the clear overall IgG response to the N-protein in the majority of the patients, not all patients showed this reaction. One explanation could be that the adaptive immune response of these patients was still early and IgG titers were still very low. Alternatively, innate immunity could have played a role in defense against the SARS-CoV-2 infection in these patients [16,17]. In addition, a few control patients displayed an IgG response to the SARS-CoV-2 N proteins, suggesting that there might be some reactivity with other coronaviruses.
Antibody testing is not only important to identify those who have gained immunity but also in understanding the prevalence of the virus in the community. In addition, we believe that this information could help in the development of a safe vaccine, as we identified the 300–685 region as the most immunogenic among the S protein, together with the full length N protein.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We thank the Krogan laboratory for providing the cDNAs that encode for the S, N and M proteins of SARS-CoV-2. This work was supported by the Fundación Canaria Instituto de Investigación Sanitaria de Canarias (FIISC) [PIFIISC20/47].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.01.073.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., et al. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 4.Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 5.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-Castro A.J., Freire R. Rad9B responds to nucleolar stress through ATR and JNK signalling, and delays the G1-S transition. J. Cell Sci. 2012;125:1152–1164. doi: 10.1242/jcs.091124. [DOI] [PubMed] [Google Scholar]
- 7.Refolio E., Cavero S., Marcon E., Freire R., San-Segundo P.A. The Ddc2/ATRIP checkpoint protein monitors meiotic recombination intermediates. J. Cell Sci. 2011;124:2488–2500. doi: 10.1242/jcs.081711. [DOI] [PubMed] [Google Scholar]
- 8.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51:482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., et al. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 11.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16 doi: 10.1186/s12985-019-1182-0. 69–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 14.Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah V.K., Firmal P., Alam A., Ganguly D., Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connell P., Aldhamen Y.A. Systemic innate and adaptive immune responses to SARS-CoV-2 as it relates to other coronaviruses. Hum. Vaccines Immunother. 2020;138:1–12. doi: 10.1080/21645515.2020.1802974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.