Abstract
The widespread nature of several viruses is greatly credited to their rapidly altering RNA genomes that enable the infection to persist despite challenges presented by host cells. Within the RNA genome of infections is RNA-dependent RNA polymerase (RdRp), which is an essential enzyme that helps in RNA synthesis by catalysing the RNA template-dependent development of phosphodiester bonds. Therefore, RdRp is an important therapeutic target in RNA virus-caused diseases, including SARS-CoV-2. In this review, we describe the promising RdRp inhibitors that have been launched or are currently in clinical studies for the treatment of RNA virus infections. Structurally, nucleoside inhibitors (NIs) bind to the RdRp protein at the enzyme active site, and nonnucleoside inhibitors (NNIs) bind to the RdRp protein at allosteric sites. By reviewing these inhibitors, more precise guidelines for the development of more promising anti-RNA virus drugs should be set, and due to the current health emergency, they will eventually be used for COVID-19 treatment.
Keywords: COVID-19, RNA virus, SARS-CoV-2, RNA-dependent RNA polymerase (RdRp), Nucleoside/non-nucleoside analogue inhibitor
Graphical abstract
Abbreviation index
- AEs
Adverse events
- AUC0-t
Area under the blood level-time curve
- BID
Bis in die
- C–H
Carbon-hydrogen bonds
- C-D
Carbon-deuterium bonds
- COVID-19
Coronavirus Disease 2019
- CTP
Cytidine triphosphate
- DENV
Dengue virus
- dNTP
Deoxyribonucleoside triphosphate
- DIEs
Deuterium isotope effects
- DAAs
Direct-acting antiviral agents
- dsRNA
Double-stranded RNA viruses
- GT
Genotype
- t1/2
Half-life
- HCV
Hepatitis virus C
- Cmax
Maximum plasma concentrations
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MHV
Mouse hepatitis virus
- NIAID
National Institute of Allergy and Infectious Diseases
- (−) ssRNA
Negative-sense ssRNA
- NNIs
Non-nucleoside inhibitors
- NNVs
Nonsegmented negative-sense RNA genome
- NS5B
Nonstructural 5B
- Nsp
Non-structural proteins
- NIs
Nucleoside inhibitors
- NTP
Nucleoside triphosphate
- NiRAN
Nucleotidyltransferase
- ORFs
Open reading frames
- F
Oral bioavailability
- Peg-IFNα
Pegylated interferon-α
- PSA
Polar surface area
- (+) ssRNA
Positive-sense ssRNA viruses
- QD
Quaque die
- FDA
The United States Food and Drug Administration
- RTC
Replicase/transcriptase complex
- RSV
Respiratory syncytial virus
- RBV
Ribavirin
- RdRp
RNA-dependent RNA polymerase
- sNSVs
Segmented negative strand RNA viruses
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- ssRNA
Single-stranded RNA
- SOC
Standard of care
- SAR
Structure-activity relationship
- TID
Ter in die
- TP
Triphosphate
- VEEV
Venezuelan equine encephalitis virus
- VSV
Vesicular stomatitis virus
- ZIKV
Zika virus
1. Introduction
1.1. RNA virus infections
Among the major groups of infections, RNA virus infections contribute considerably to the worldwide death and morbidity index related to viral infection. Chronic illness related to persistent RNA virus infections represents a crucial public health worry [1]. While human immunodeficiency virus-1 as well as hepatitis C virus (HCV) are perhaps the most popular instances of persistent RNA viruses that cause chronic disease, proof recommends that numerous other RNA viruses, consisting of re-emerging viruses such as Ebola virus and Zika virus, develop persistent infections [2]. Furthermore, swine and avian influenza infections, together with Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV), stand for substantial pandemic risks to the general populace [3].
Considering the high frequency and vast circulation of RNA viruses, their huge genetic diversity as well as the frequent recombination of their genomes and raising activity at the human-animal interface, these viruses are identified as a recurring hazard to human health and wellness [4]. This truth once again became noticeably obvious in late 2019 and very early 2020, when severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was found to be the root cause of a large and quickly spreading outbreak of lower respiratory tract infection and disease, consisting of potentially fatal pneumonia, in Wuhan, China [5,6]. This novel coronavirus-induced febrile respiratory system disease was formally named coronavirus disease 2019 (COVID-19) by the WHO. Presently, the COVID-19 pandemic is still spreading worldwide. Along with vaccinations, people are also hoping for particular medicines [7,8]. An increasing number of medication prospects have come to the attention of clinical scientists, and many clinical trials are being performed throughout the world. However, there is currently only one RNA-dependent RNA polymerase (RdRp) small molecule inhibitor, remdesivir, that has been approved by the United States Food and Drug Administration (FDA) for the treatment of COVID-19 [[9], [10], [11], [12]].
1.2. RNA virus and SARS-CoV-2
An RNA virus is a virus that utilizes RNA as its genetic material. Based on their genome and mode of replication, three distinct groups of RNA viruses have been classified: double-stranded RNA viruses (dsRNA), single-stranded RNA (ssRNA) and retroviruses [13,14]. dsRNA viruses contain one to a dozen different RNA molecules, each of which encodes one or more viral proteins [15]. The genome of positive-sense ssRNA viruses is directly used as mRNA, and the host ribosomes translate this genome into a single protein that is modified by host and viral proteins to form the various proteins needed for replication [[16], [17], [18]]. One of these includes RdRp, which copies the viral RNA to develop a double-stranded replicative type [19,20]. Consequently, this dsRNA routes the development of brand-new viral RNA. The genome of negative-sense ssRNA viruses must be copied by an RNA replicase to form positive-sense RNA [21,22]. The positive-sense RNA molecule then acts as viral mRNA, and this mRNA is translated into proteins by host ribosomes. In addition, retroviruses have a ssRNA genome but are generally not considered RNA viruses because they utilize DNA intermediates for replication [23]. The nucleic acid of a pathogenic virus, such as coronaviruses, hepacivirus, influenza viruses and respiratory syncytial virus (RSV), is typically ssRNA (Table 1 ) [24,25]. These RNA viruses can create lower respiratory system tract infections that cause bronchiolitis as well as pneumonia [26]. Young children, the elderly, and patients with compromised heart, lung, or immune systems are at the highest risk for significant disease related to these RNA virus-related breathing infections [27,28].
Table 1.
Highly pathogenic single-stranded RNA viruses.
| Demonstrative virus | Family | RNA |
|---|---|---|
| Norovirus | Caliciviridae | (+) ssRNA |
| SARS-CoV | Coronaviridae | (+) ssRNA |
| MERS-CoV | Coronaviridae | (+) ssRNA |
| SARS-CoV-2 | Coronaviridae | (+) ssRNA |
| Hepatitis C virus | Flaviviridae | (+) ssRNA |
| Dengue virus | Flaviviridae | (+) ssRNA |
| Zika virus | Flaviviridae | (+) ssRNA |
| Poliovirus | Picornaviridae | (+) ssRNA |
| Venezuelan equine encephalitis virus | Togaviridae | (+) ssRNA |
| Influenza A virus | Orthomyxoviridae | (−) ssRNA |
| Influenza B virus | Orthomyxoviridae | (−) ssRNA |
| Influenza C virus | Orthomyxoviridae | (−) ssRNA |
| Ebola virus | Filoviridae | (−) ssRNA |
| Marburgvirus | Filoviridae | (−) ssRNA |
| Vesicular stomatitis virus | Rhabdoviridae | (−) ssRNA |
| Respiratory syncytial virus | Paramyxoviridae | (−) ssRNA |
SARS-CoV: severe acute respiratory syndrome coronavirus; MERS-CoV: Middle East respiratory syndrome coronavirus; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; (+) ssRNA: positive-sense single-stranded RNA; (−) ssRNA: negative-sense single-stranded RNA.
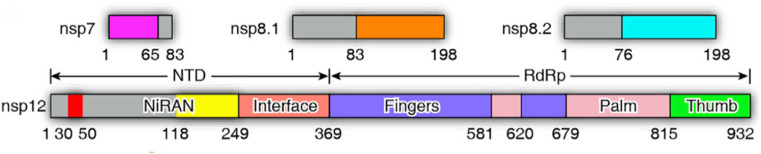
SARS-CoV-2 is a positive-sense ssRNA virus that can infect humans [29]. The infection largely spreads from human to human through close contact and by breathing droplets generated from sneezes or coughs. The 30-kb genome of SARS-CoV-2 contains 14 open reading frames (ORFs) that can encode at least 27 proteins [[30], [31], [32]]. The 3′ end of the genome encodes four structural proteins, namely, spike, envelope, membrane, and nucleocapsid proteins, and eight accessory proteins that disrupt the host’s inherent immune responses. The ORF1ab region at the 5′ end inscribes a polyprotein, which is hydrolysed into 16 nonstructural proteins (nsp 1-16) to create a replicase/transcriptase complex (RTC). The main RTC is RdRp (nsp12) (Fig. 1 ). In RNA viruses, such as SARS-CoV-2, RdRp creates the machinery needed for RNA synthesis and the organized replication and transcription of genomic RNA.
Fig. 1.
Domain organization of SARS-CoV-2 nsp 12 (RdRp). The interdomain borders are labelled with residue numbers. RdRp: RNA-dependent RNA polymerase.
1.3. RNA-dependent RNA polymerase
The genomic replication process of RNA infections is controlled by RdRp, which is inscribed by the virus itself [30,33]. After the virus attacks a host cell, the viral genomic RNA is directly utilized as a template, and the host cell protein synthesis system is utilized for the translation of RdRp. RdRp is consequently used to complete the transcriptional synthesis of negative-strand subgenomic RNA, the synthesis of different structural protein-related mRNAs, and the replication of viral genomic RNA. RdRp can properly and efficiently synthesize tens of thousands of nucleotides and thus facilitates all other biological activities that occur after the virus invades a host cell.
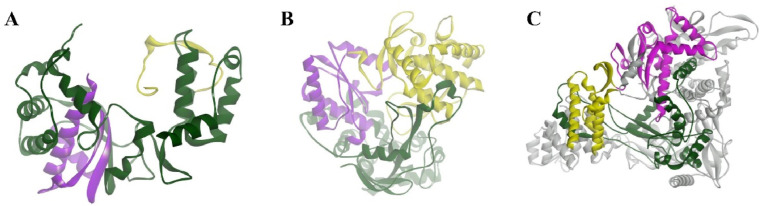
The structure of RdRp of positive-strand RNA viruses resembles that of a cupped right hand and includes fingers, palm and thumb subdomains that are largely associated with binding to the design template, polymerization, nucleoside triphosphate (NTP) access and associated features [[34], [35], [36]]. In addition to these three central subdomains, an N-terminal subdomain that bridges the fingers and thumb subdomains is located in all RdRps [37], and this subdomain serves as the active site of RdRp. The completely encircled active site cavity is responsible for considerable communication between the finger and thumb subdomains. The active site of RdRp is extremely well preserved. The finger subdomain plays a considerable role in establishing the geometry of the active site [38] by holding the template RNA in place and facilitating polymerization. The thumb subdomain harbours residues that are involved in packing against the template RNA and stabilizing the initiating NTP on the template [39]. This subdomain also facilitates the translocation of the template following polymerization by accommodating large conformational rearrangements. The thumb subdomain exhibits the greatest diversity among the identified RdRp and differences in size and complexity based on the mode of replication initiation. The palm subdomain translocates to the junction of the finger and thumb subdomains and houses many structurally conserved elements associated with catalysis [40]. The palm subdomain is involved in the selection of NTP over deoxyribonucleoside triphosphate (dNTP) and catalyses the phosphoryl transfer reaction by coordinating with two metal ions (Mg2+ and/or Mn2+). The best-known RdRps of positive-strand RNA viruses are the polioviral 3Dpol and hepatitis C virus nonstructural 5B (NS5B) proteins as well as the SARS-CoV-2 RdRp, which has recently attracted much attention.
The structure of the SARS-CoV-2 RdRp complex consists of a nsp 12 core catalytic unit, a nsp7-nsp8 (nsp8-1) heterodimer, and an additional nsp8 subunit (nsp8-2), and nsp12-nsp7-nsp8 is defined as the minimal core component for virus RNA replication [31]. The N-terminal portion of nsp12 contains a b-hairpin (V31–K50) and a nidovirus-specific extension domain (D60-R249). The b-hairpin is sandwiched by the palm subdomain in the RdRp core and nidovirus RdRp-associated nucleotidyltransferase (NiRAN), a configuration not observed in other coronavirus RdRp structures [41]. The C-terminal catalytic domain of nsp12 (A250-F369) connects to NiRAN through an interface subdomain. The C-terminal catalytic domain of nsp12 (S367–F920) adopts a canonical cupped right-handed configuration of all viral RdRp, composed of the finger, palm, and thumb subdomains. Catalytic metal ions are not observed in the absence of primer-template RNA and NTPs, although they are present in several structures of viral polymerases that synthesize RNA. The nsp7-nsp8 heterodimer binds above the thumb subdomain and stabilizes the thumb-finger interface. Nsp7 makes a major contribution to the binding of the heterodimer to nsp12, while nsp8 only contacts a few residues from nsp12. The other copy of nsp8 (nsp8-2) sits atop the finger subdomain and forms additional interactions with the interaction subdomain. In this structure, similar to other positive-strand RNA viruses RdRp, the template/primer entry channel, NTP entry channel, and nascent strand exit channel are all positively charged and converge in a central cavity, which is the active site of the SARS-CoV-2 RdRp that is formed by seven conserved catalytic motifs (A to G). In this central cavity, these RdRp motifs mediate template-guided RNA synthesis. Motif A (T611-M626) houses the catalytic motif DX2-4D, in which the first aspartic acid D618 is invariant in most viral polymerases. The flexible loop in Motif B (T680-T710) serves as a hinge to undergo conformational arrangement associated with template RNA and substrate binding. Motif C (F753–N767) contains the catalytic motif SDD, which is essential for binding the metal ion. Motif D contains residues L775-E796. Motif E contains residues H810–V820 and combines with the palm subdomain to support the primer chain. Motif F (K912-E921) interacts with the phosphate group of incoming NTP. The NTP entry channel is formed by a set of hydrophilic residues, including K545, R553 and R555 in motif F [42]. Motif G (K500–S518) interacts with the template strand. The RNA template strand enters the active site composed of motifs A and C through the groove sandwiched by motifs F and G. The product-template hybrid exits this active site through the RNA exit channel on the front side of the RdRp (Fig. 2 ).
Fig. 2.
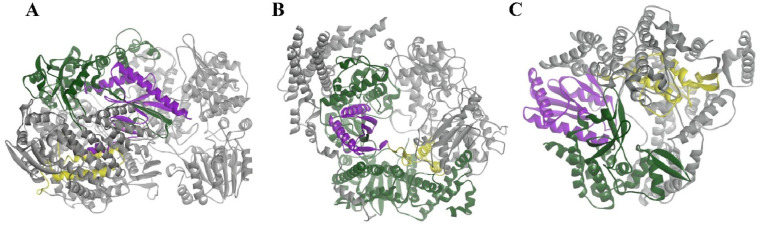
Structure of RdRps of positive-strand RNA viruses. A: poliovirus 3Dpol (1RDR); B: HCV NS5B polymerase (1NB4); and C: nsp 12-nsp7-nsp8 complexes of SARS-CoV-2 (7BW4). Nsp7 and nsp8 act as cofactors to promote the activity of RdRp (grey). The palm, finger and thumb subdomains are shown in purple, green and yellow, respectively. RdRp: RNA-dependent RNA polymerase.
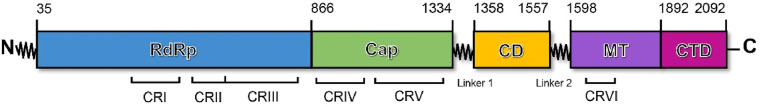
The polymerase of segmented negative-strand RNA viruses (sNSVs), including influenza virus, is composed of three polypeptides: PB1, PB2 and PA/P3. PB1 contains the polymerase active site, whereas PB2 and PA/P3 have cap-binding and endonuclease domains, respectively, needed for transcription initiation by cap snatching [[43], [44], [45]]. In addition, the catalytic core of nonsegmented negative-strand RNA viruses (NNVs), including vesicular stomatitis virus (VSV), RSV and Ebola virus, is the L protein, which catalyses RNA polymerization during both replication and transcription, cap addition, and cap methylation of nascent viral mRNAs [36,[46], [47], [48], [49]]. The RdRp domain is a functional domain of the L protein, and the L proteins of nonsegmented negative-sense single-stranded RNA viruses share six conserved regions and three functional domains (RdRp, capping, and cap methyltransferase) (Fig. 3, Fig. 4 ).
Fig. 3.
Domain organization of the L protein of vesicular stomatitis virus. The conserved regions within L proteins of nonsegmented negative-strand RNA viruses are labelled CR I–VI.
Fig. 4.
Structure of the RdRp domain of nonsegmented negative-strand RNA virus polymerases. A: Vesicular stomatitis virus L protein (5A22); B: respiratory syncytial virus protein L protein (6UEN); and C: rotavirus VP1 protein (2R7Q). The palm, finger and thumb subdomains are shown in purple, green and yellow, respectively. RdRp: RNA-dependent RNA polymerase.
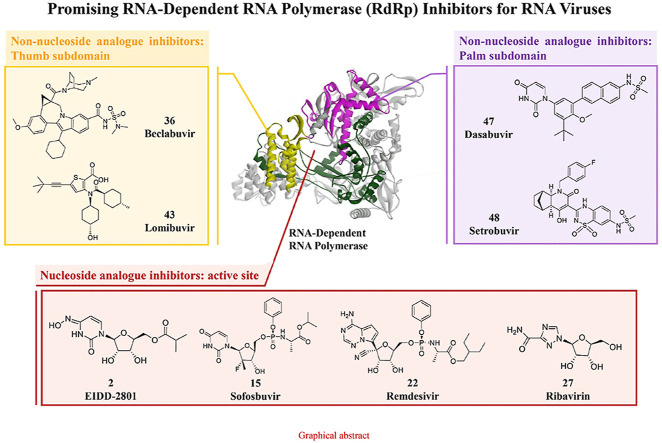
RdRp is an important therapeutic target because it plays a pivotal role in replication of the RNA genome and because the host lacks a functional equivalent to this protein. In addition, due to the absence of a counterpart to RdRp in mammalian cells, its inhibition is not expected to cause target-related side effects, and thus, RdRp is considered an attractive target in drug discovery and development. The development of effective RdRp inhibitors to block viral replication has long been a research topic in many scientific institutions and pharmaceutical companies. There are two known classes of RdRp inhibitors: nucleoside analogue inhibitors (NIs) and nonnucleoside analogue inhibitors (NNIs). The classes show differences in structure and the location that bind to RdRp: enzyme active site (Nis) and allosteric sites (NNIs). This review summarizes the promising RdRp inhibitors that have been launched or are currently in clinical studies for the treatment of RNA virus infections, including COVID-19.
2. Nucleoside inhibitors
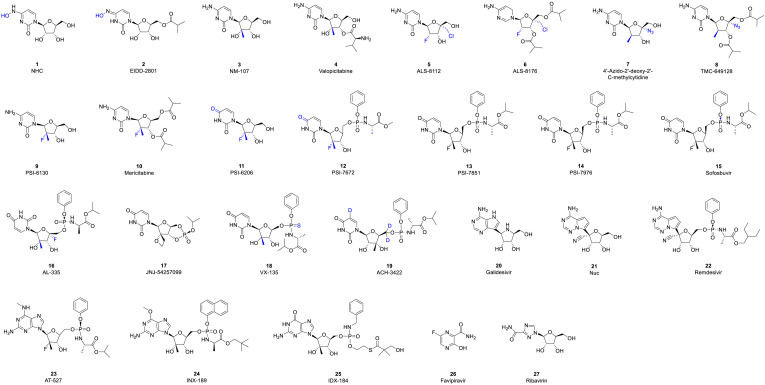
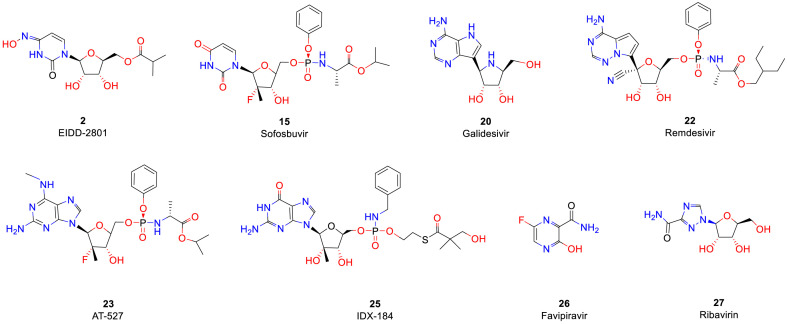
NIs terminate the RNA synthesis step, which is essential for RNA replication, through their incorporation by RdRp, which prevents incoming nucleotides from being added to the RNA chain [50]. It has been proposed that steric hindrance by nucleoside inhibitors, which contain a 3′-hydroxyl group, is responsible for the observed termination of chain elongation [51]. Due to this mechanism, NIs are sometimes called chain termination inhibitors [52]. NIs of RNA viruses, which have been developed as prodrugs, eventually become cleaved at their site of action in the liver by hepatic enzymes and undergo phosphorylation into a triphosphate form, which targets the polymerase at its highly conserved active site (Fig. 5 ). NIs of RdRp are classified into three major classes: pyrimidine nucleoside inhibitors, purine nucleoside inhibitors and miscellaneous nucleoside inhibitors (Table 2 ).
Fig. 5.
Structures of nucleoside analogue inhibitors.
Table 2.
RNA-dependent RNA polymerase nucleoside inhibitors.
| Number | Compound | CAS registry number | Classification | Condition and highest clinical phase |
|---|---|---|---|---|
| 1 | NHC | 3258-02-4 | Pyrimidine nucleoside | Biological test |
| 2 | EIDD-2801 | 2349386-89-4 | Pyrimidine nucleoside | Phase II - COVID-19- 2020 |
| 3 | NM-107 | 20724-73-6 | Pyrimidine nucleoside | Phase II - Hepatitis C - 2003 |
| 4 | Valopicitabine | 640281-90-9 | Pyrimidine nucleoside | Phase II - Hepatitis C - 2004 |
| 5 | ALS-8112 | 798009-58-2 | Pyrimidine nucleoside | Phase I - Infection, RSV -2014 |
| 6 | Lumicitabine | 1445385-02-3 | Pyrimidine nucleoside | Discontinued - Infection, RSV - 2019 Phase II - Infection, RSV -2014 |
| 7 | 4′-Azido-2′-deoxy-2′-C-methylcytidine | 1019639-20-3 | Pyrimidine nucleoside | Biological test |
| 8 | TMC-649128 | 1019639-33-8 | Pyrimidine nucleoside | Discontinued - Hepatitis C - 2011 Phase I - Hepatitis C - 2011 |
| 9 | PSI-6130 | 817204-33-4 | Pyrimidine nucleoside | Phase I - Hepatitis C - 2006 |
| 10 | Mericitabine | 940908-79-2 | Pyrimidine nucleoside | Phase II - Hepatitis C - 2011 |
| 11 | PSI-6206 | 1064684-44-1 | Pyrimidine nucleoside | Phase I - Hepatitis C - 2009 |
| 12 | PSI-7672 | 1015255-46-5 | Pyrimidine nucleoside | Biological test |
| 13 | PSI-7851 | 1064684-44-1 | Pyrimidine nucleoside | Biological test |
| 14 | PSI-7976 | 1190308-01-0 | Pyrimidine nucleoside | Biological test |
| 15 | Sofosbuvir | 1190307-88-0 | Pyrimidine nucleoside | Phase II - Porphyria cutanea tarda - 2017 Launched - Hepatitis C - 2014 Phase III - Hepatitis B - 2015 |
| 16 | AL-335 | 1613589-09-5 | Pyrimidine nucleoside | Phase II - Hepatitis C - 2015 |
| 17 | JNJ-54257099 | 1255860-33-3 | Pyrimidine nucleoside | Discontinued - Hepatitis C - 2015 Phase I - Hepatitis C - 2015 |
| 18 | VX-135 | 798007-79-1 | Pyrimidine nucleoside | Phase II - Hepatitis C - 2013 |
| 19 | ACH-3422 | 798779-31-4 | Pyrimidine nucleoside | Phase I - Hepatitis C - 2014 |
| 20 | Galidesivir | 249503-25-1 | Purine nucleoside | Phase I - COVID-19 -2020 Phase I - Viral haemorrhagic fever −2019 |
| 21 | Nuc | 1191237-69-0 | Purine nucleoside | Preclinical |
| 22 | Remdesivir | 1809249-37-3 | Purine nucleoside | Launched - COVID-19 -2020 Phase II/III - Ebola virus disease - 2018 |
| 23 | AT-527 | 2241337-84-6 | Purine nucleoside | Phase II - COVID-19 - 2020 Phase II - Hepatitis C −2019 |
| 24 | INX-189 | 1234490-83-5 | Purine nucleoside | Discontinued - Hepatitis C −2012 Phase II - Hepatitis C −2011 |
| 25 | IDX-184 | 1036915-08-8 | Purine nucleoside | Discontinued- Hepatitis C −2013 Phase II - Hepatitis C −2009 |
| 26 | Favipiravir | 259793-96-9 | Miscellaneous nucleoside | Phase III - COVID-19 - 2020 Launched - Influenza A - 2017 Launched - Influenza B - 2017 Phase II - Ebola virus disease - 2014 |
| 27 | Ribavirin | 36791-04-5 | Miscellaneous nucleoside | Phase I - COVID-19 - 2020 Phase I/II - Prostate cancer-2016 Phase II - Hepatitis E - 2012 Launched - Hepatitis C - 2001 Launched - Infection, RSV -1986 |
RSV: respiratory syncytial virus.
2.1. Pyrimidine nucleoside inhibitors
2.1.1. Cytosine analogue inhibitors
Cytidine analogues are metabolized to both cytidine and uridine triphosphates through the action of cytidine deaminase. 1 (NHC, EIDD-1931, β-D-N 4-hydroxycytidine) is an orally bioavailable ribonucleoside analogue with broad-spectrum antiviral activity against various unrelated RNA viruses, including SARS-CoV-2 (IC50 = 0.30 μM), MERS-CoV (EC50 = 0.56 μM), mouse hepatitis virus (MHV, EC50 = 0.17 μM) and venezuelan equine encephalitis virus (VEEV, EC50 = 1.00 μM), and increased potency against a coronavirus bearing resistance mutations to the nucleoside analogue inhibitor remdesivir [[53], [54], [55], [56], [57]]. 1 acts as a weak alternative substrate for cytidine triphosphate (CTP) to potentially modulate virus replication steps that are dependent on these structures, such as encapsidation, translation and replication [58]. 2 (EIDD-2801) is the 5′-isopropylester of 1 that exhibits broad activities against influenza viruses and multiple coronaviruses. In phase II clinical trials, Ridgeback Biotherapeutics is evaluating various candidates for the treatment of newly hospitalized adults with COVID-19 and symptomatic adult outpatients with COVID-19. Preclinical studies are also ongoing to determine its potential as a treatment for influenza and MERS-CoV infection.
The study of pyrimidine nucleoside inhibitors as potential antiviral drugs revealed that the unique substituents at the C2′ or C4′ position of the nucleoside exhibit obvious antiviral activity. The incorporation of 2′-C-modified monophosphates onto the 3′ termini of growing virus RNA strands promotes the termination of elongation due to steric hindrance between the incoming natural nucleotide and the unnatural 2′-C-group of the inhibitor [59,60]. 3 (NM-107) is a 2′-C-methylcytidine that was initially identified as a competitive inhibitor of NS5B polymerase, and the EC50 of 3 in wild-type replicon cells is 1.85 μM [61]. Upon phosphorylation into its 5-triphosphate form, this metabolite inhibits viral RNA chain elongation and viral RdRp activity, and these effects block the viral production of HCV RNA and thus viral replication. In addition to HCV, this compound inhibits the replication of a variety of other viruses, such as dengue virus (DENV) and norovirus [62,63]. 4 (valopicitabine, NM-283), which is the 3′-O-valinyl ester of 3, was synthesized to obtain a compound with improved oral bioavailability compared with that of its parent compound 2′-C-methylcytidine. Physicochemical, pharmacokinetic, and toxicokinetic studies have shown that 4 is an acid-stable prodrug of 2′-C-methylcytidine with excellent pharmacokinetic and toxicokinetic profiles [64]. 4 is currently being evaluated in phase II clinical trials for the treatment of chronic hepatitis virus C (HCV) infection.
Janssen screened a series of 4′-cytosine nucleoside analogues based on their pharmacodynamics and pharmacokinetic properties and found that 4′-chloromethyl-2′-deoxy-2′-fluorocytidine (5, ALS-8112) exhibited the most promising activity in the RSV replicon assay, with an EC50 of 0.15 μM [65]. 5 enters various types of epithelial cells in the respiratory tract and is subsequently phosphorylated to form an intracellular nucleoside triphosphate with a half-life (t1/2) of approximately 29 h [66]. The 5′-triphosphate of 5 is the active form of the drug and inhibits RSV polymerase with an IC50 of 0.02 μM, and no appreciable inhibition of human DNA and RNA polymerases was detected at a concentration of 100 μM [67]. 6 (lumicitabine, ALS-8176), the 3′,5′-di-O-isobutyryl prodrug of 5, is a first-in-class nucleoside RSV polymerase inhibitor that demonstrated excellent anti-RSV efficacy and safety in a phase II clinical trial for the treatment of RSV [68]. Moreover, a number of 2′-fluoro-4′-substituted cytidine nucleosides exhibited potent inhibition of the RSV replicon with a wide selectivity window in these studies, and their 5′-triphosphates effectively inhibited RSV polymerase with high selectivity with respect to the host polymerases [65]. These findings indicate that access to the F atom might allow the synthesis of first-in-class antiviral agents against RSV infection.
7 (4′-Azido-2′-deoxy-2′-C-methylcytidine) is a potent nucleoside inhibitor of the NS5B polymerase that displays an EC50 value of 1.2 μM and shows moderate in vivo bioavailability in rats (F = 14%) [69]. 8 (TMC-649128) is the di-isobutyryl ester of 7, and its pharmacokinetic properties in rats can potentially be improved by introducing prodrug esters. Isobutyryl ester 8 exhibits greatly improved mean maximum plasma concentrations (Cmax = 4.65 μM) and hence a larger area under the blood level-time curve (AUC0-t = 12.7 μM/h) and greater oral bioavailability (F = 65%).
Fluorinated nucleosides are well known for their antiviral and anticancer properties. Pharmasset screened a series of 2′-cytosine nucleoside analogues with obvious anti-HCV activity. β-D-2′-Deoxy-2′-fluoro-2′-C-methylcytidine (9, PSI-6130), in which the C2′ position of uridine is substituted by an F atom and methyl group, exerts a highly effective inhibitory effect on HCV replication [70,71]. Unfortunately, clinical phase I trials showed that 9 does not have good pharmacokinetic characteristics. The low oral bioavailability of 9 might be due to the presence of hydroxyl and amino groups, which are more polar, exhibit poor fat solubility, and cannot easily penetrate biofilms in its structure [72]. In addition, its amino group at position 4 is unstable under acidic conditions, and this amino group can be easily removed to generate carbonyl groups. To resolve the problem of bioavailability and metabolism, the 3′,5′-diisobutyrate prodrug 10 (mericitabine, RG-7128) was designed to promote the absorption and metabolism of the compound in the intestine [[73], [74], [75], [76]]. 10 is rapidly absorbed via the oral route and converted to 9, which is subsequently metabolized to its metabolite. The results also showed that 10 improves the pharmacokinetic parameters, and clinical trials have also shown that the drug exerts a certain effect. However, 10 is not the most ideal medicine due to its low general efficacy and short t1/2.
2.1.2. Uracil analogue inhibitors
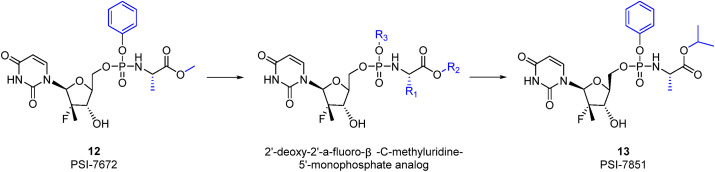
β-D-2′-deoxy-2′-fluoro-2′-C-methyluridine (11, PSI-6206) is a metabolite of 9 in vivo [77] that can significantly inhibit HCV replication. The patients with hepatitis C treated with 11 generally show good tolerance. However, the bioavailability of 11 is too low (25%) because 11 cannot be converted into 11-triphosphate (11-TP). However, observations of the metabolites of 9 revealed that monophosphate 9 can be further transformed into 11-TP, which has a long t1/2 and better activity than 9-TP [77,78]. This major discovery indicates that uridine monophosphate derivatives might be ideal direct-acting antiviral agents (DAAs). To ensure the safety and efficiency of the drug, the F atom and methyl group at the C2′ position are retained. Because compounds containing phosphoric acid groups are negatively charged, the related compounds cannot be easily absorbed by the human body. The prodrug design was finally adopted, and the first prodrug 12 (PSI-7672) was designed [77]. Since then, a large number of derivatives have been synthesized, and some of the related experience has been previously summarized [79]. The isomeric form of the amino acid is significant because the d-alanine derivative is inactive, which means that the natural l-amino acid is required for activity. Observations of the amino acid side chain (R1) have revealed that a small alkyl group is a viable substitution, but significant decreases in potency are observed with substitutions larger than ethyl. Methyl results in the greatest potency and is therefore a viable substitution. If the amino acid is alanine and the phosphate ester is a phenyl substituent, the groups at the carboxylic acid ester (R2) that provide the desired submicromolar activity are small alkyl groups and branched alkyl groups. However, cytotoxicity was observed with n-butyl, 2-butyl and n-pentyl esters. Phenyl and halogenated alkyl groups do not provide sufficient improvements in potency. The evaluation of the phosphoramidate ester substituent (R3) revealed that a derivative with phenyl as a substituent exhibits good potency and is not cytotoxic (Table 3 ).
Table 3.
Structure–activity relationship of the 2′-deoxy-2′-a-fluoro-β-C-methyluridine-5′-monophosphate analogue.
| R1 | R2 | R3 | EC90 | Inhibition of cellular rRNA replication at 50 μM (%) |
|---|---|---|---|---|
| Methyl | Isopropyl | Phenyl | 0.52 | 25.9 |
| Methyl | Methyl | Phenyl | 1.62 | 0.0 |
| Methyl | c-Hexyl | Phenyl | 0.25 | 61.1 |
| Methyl | Ethyl | 4-F- Phenyl | 0.76 | 55.3 |
| Methyl | Isopropyl | 4-F- Phenyl | 0.77 | 0.0 |
| Methyl | Isopropyl | 4-Cl- Phenyl | 0.42 | 0.0 |
| Methyl | c-Hexyl | 4-Cl- Phenyl | 0.04 | 52.1 |
It was finally determined that 13 (PSI-7851) is an ideal DAA with a favourable pharmacokinetic profile for inhibiting HCV [77,78,80]. 13 contains a chiral phosphorous atom and is therefore a mixture of two diastereomers, S p diastereoisomer 14 (PSI-7976) and R p diastereoisomer 15 (sofosbuvir, PSI-7977) [81]. The activity of 15 is significantly better than that of 14, and this difference might be due to the different binding orientations of 14 and 15 to the enzyme, which are productive and nonproductive, respectively. 15 might preferentially bind in the nonproductive orientation and form a dead-end complex to exert a significant antiviral effect [82]. Compound 15 is an approved NS5B polymerase inhibitor (EC90 = 0.42 μM) and exerts pangenotypic antiviral effects against HCV genotypes (GTs) 1–6. The potent antiviral activities of 15 are higher than 90% [83]. In addition, 15 has the ability to suppress different families of viruses, including Zika virus (ZIKV), DENV and chikungunya virus [[84], [85], [86]]. Moreover, 15 exhibits a rapid response, and over 0.8 and 2 days, this compound exerts estimated potencies of 99 and 99.9%, respectively [87]. The bioavailability of 15 is high, and maximum plasma concentrations (Cmax) were detected 0.5–2 h after oral administration [88]. 15 has been considered a potential effective drug for inhibiting SARS-CoV-2 infection since the emergence of the COVID-19 pandemic. The administration of 15 and daclatasvir in combination with standard of care (SOC) for the treatment of patients with COVID-19 resulted in better 14-day recovery rates and a shorter hospital stay. The patients in the therapy group experienced a shorter duration of hospital stay and a shorter median time to discharge than the control group (6 vs 8 days and 6 vs 11 days, respectively). Recently, Pinar Mesci et al. [89] reported that 15 can potentially be used to alleviate COVID-19-related neurological symptoms.
16 (AL-335) is a type of uridine analogue with 4′-fluoro-2′-C-substituted sugar moieties [90]. 16-TP exhibits potent inhibition of NS5B polymerase with IC50 values as low as 27 nM. In an HCV subgenomic replicon assay, the phosphoramidate prodrug of 16 demonstrated very potent activity with EC50 values as low as 20 nM [91]. The administration of 16 in combination with simeprevir and odalasvir has been evaluated in human phase II clinical trials and has shown promising efficacy and safety results. 16 is well tolerated when administered as single and multiple doses and exhibits an acceptable pharmacokinetic profile [92]. 17 (JNJ-54257099) is a cyclic phosphate ester derivative belonging to the class of 2′-deoxy-2′-spirooxetane uridine nucleotide prodrugs [93]. This compound profoundly dose-dependently decreases HCV RNA levels in mouse models of HCV GT 1a and 3a infections. 17 was terminated following completion of phase I clinical studies conducted by Janssen Pharmaceutical. This is because the clinical antiviral activity of 17 in patients infected with HCV GT 1 was insufficient to justify further clinical studies. Compound 18 (VX-135) exhibited pronounced antiviral activity against GTs 1–6 (EC50 values between 12 and 390 nM) in vitro [94] and has been evaluated in phase II clinical studies for the treatment of hepatitis C. The phase I study evaluated the pharmacokinetics, safety and antiviral activity of 18 in 48 healthy controls and 30 patients with HCV GT 1 infection. The most common adverse events were headache and diarrhoea (two subjects each), and no severe adverse events were recorded. Compound 18 demonstrated potent antiviral activity with a 4.5 log10 decrease in HCV RNA over 7 days at a dose of 200 mg quaque die (QD) in patients infected with chronic hepatitis C [95].
2.1.3. Thymine analogue inhibitors
19 (ACH-3422) is an NS5B polymerase inhibitor that displays pangenotypic activity and a high in vitro barrier to resistance. 19 is designed to introduce three deuteriums on the side chains of pyrimidine and ribose groups. The incorporation of deuterium into pharmacologically active agents according to the principle of deuterium isotope effects (DIEs) offers potential benefits, such as improved exposure profiles and decreased production of toxic metabolites that could yield improvements in efficacy, tolerability, or safety [96]. Therefore, 19 is well tolerated and induces no serious adverse events in healthy volunteers and hepatitis C patients [97]. Among active patients, increasing doses of 19 resulted in increased viral decline. In the proof of concept group administered 700 mg of the antiviral, mean decreases in the maximum viral load of 3.4 log10, 4.2 log10, and 4.6 log10 were obtained after 7, 10, and 14 days of treatment, respectively. Three of six patients (50%) achieved viral clearance after the administration of 700 mg for 14 days.
2.2. Purine nucleoside inhibitors
2.2.1. Adenine analogue inhibitors
20 (galidesivir, BCX4430) is an adenosine nucleoside analogue developed by BioCryst Pharmaceuticals. This compound was originally intended as a treatment for HCV but was subsequently developed as a potential treatment for deadly filovirus infections [98,99]. Studies have shown that 20 protects against both Ebola and Marburg viruses in both rodents and monkeys [99]. This compound also shows broad-spectrum antiviral effectiveness against a range of other RNA virus families, including SARS-CoV and MERS-CoV [100]. 20 can bind SARS-CoV-2 RdRp, with a binding energy of −7.0 kcal/mol [101]. In April 2020, BioCryst opened enrolment into a randomized, double-blind, placebo-controlled clinical trial aiming to assess the safety, clinical impact and antiviral effects of 20 in patients with COVID-19.
21 (Nuc, GS-441524) is a 1′–CN–modified adenosine C-nucleoside analogue that exhibits antiviral activity against a variety of RNA viruses [102]. Structurally, the 1′-CN group provides potency and selectivity for viral RdRp. A study conducted in 2019 revealed that 21 can potentially be used for the treatment of feline infectious peritonitis caused by a coronavirus [103]. 21 is synthesized into 21-TP through intracellular metabolism, and 21-TP can interfere with the activity of viral RdRp. However, the kinetics of the monophosphorylation of 21 are slow, and the use of a parent nucleoside modified with monophosphate might greatly increase the intracellular NTP concentration [102]. Compound 22 (remdesivir, GS-5734) is the Sp isomer of the 2-ethylbutyl l-alanine phosphoramidate prodrug and effectively bypasses the rate-limiting step of 21 monomer phosphorylation [104]. Compound 22 is activated more rapidly than 21 in human cells infected with SARS-CoV and MERS-CoV, and multiple uses of this compound have been explored with the aim of helping to address urgent and unmet medical needs around the world, including Ebola disease, SARS, MERS and most recently COVID-19 [105]. In Vero E6 cells, 22 effectively blocks SARS-CoV-2 infection at low concentrations (EC50 = 0.77 μM) and exhibits low cytotoxicity (CC50 > 100 μM). In addition, the EC90 value of 22 against SARS-CoV-2 in Vero E6 cells is 1.76 μM [10]. A study using the rhesus macaque model of SARS-CoV-2 infection revealed that therapeutic treatment with 22 initiated early during infection results in a clear clinical benefit in SARS-CoV-2-infected rhesus macaques [106,107]. NIAID reported that remdesivir was superior to placebo in shortening the time to recovery in adults who were hospitalized with COVID-19 and had evidence of lower respiratory tract infection in phase III clinical trials [108]. Recently, the FDA approved 22 for the treatment of patients with COVID-19 requiring hospitalization. To date, 22 is the first and only FDA-approved treatment for COVID-19 in the United States. In addition, some researchers have argued for the direct administration of 21 as a COVID-19 treatment because 21 exhibits either similar to or more potency than 22 against SARS-CoV-2 in cell culture [109].
23 (AT-527) is a novel modified guanosine nucleotide prodrug inhibitor of the NS5B polymerase that belongs to the same category as 22. Compound 23 exhibits higher in vitro antiviral activity than compound 15. The free base of 23 had an EC95 value of 25 nM and thus exhibited 10-fold higher potency than 15 in Huh-7 cells bearing the HCV GT 1b replicon [110]. The antiviral activity and safety of 23 has been demonstrated in phase II clinical studies of hepatitis C patients. The mean maximum reductions observed in noncirrhotic subjects with HCV GT 1b, noncirrhotic subjects with HCV GT 3, and subjects with compensated cirrhosis after 7 days of treatment were 4.4, 4.5, and 4.6 log10 IU/mL, respectively [111]. A phase II clinical trial was established to evaluate the safety and efficacy of 23 for the treatment of adult patients hospitalized with moderate COVID-19 disease. 24 (INX-189), the phosphoramidate nucleoside analogue prodrug of 2′-C-methylguanosine, is a potent HCV replication inhibitor (EC50 = 35 nM) that is currently being investigated in a phase II clinical trial by Bristol-Myers Squibb for the oral treatment of hepatitis C virus infections [112].
2.2.2. Guanine analogue inhibitors
25 (IDX-184), a highly potent inhibitor of HCV replication in vitro, was designed to achieve enhanced targeting to the liver through monophosphorylation and reduce the exposure of other tissues to the drug. Compound 25 is preferentially cleaved by hepatic enzymes to form TP, and 25-TP potently inhibits NS5B polymerase (IC50 = 0.31 mM, Ki = 52.3 nM) but does not inhibit human polymerases a, b or g (IC50 > 50 mM) [113]. The administration of 25 at single and multiple doses of up to 100 mg/day for three days revealed that the compound is safe and well tolerated in both healthy volunteers and treatment-naïve HCV GT 1-infected subjects, respectively. 25 in combination with pegylated interferon-α (Peg-IFN) and ribavirin (RBV) was generally safe and well tolerated in HCV GT-1-infected subjects, and its lowest dose of 50 mg QD resulted in marked viral load reductions compared with those obtained with Peg-IFN/RBV alone [114].
2.3. Miscellaneous nucleoside inhibitors
26 (favipiravir, T-705) is a broad-spectrum anti-RNA virus drug that was approved in 2014 for the oral treatment of influenza A and for the treatment of influenza B infection. Studies have shown that 26 also exhibits good antiviral effects against a variety of RNA viruses, such as Ebola virus and rabies virus, in addition to influenza viruses [[115], [116], [117]]. Wang et al. [10] showed that 26 can effectively reduce SARS-CoV-2 infection in vitro (EC50 = 61.88 μM, CC50 > 400 μM, SI > 6.46). Ongoing clinical trials have shown that 26 can accelerate the recovery of COVID-19 patients, as demonstrated by a median cure time of 2.5–9 days, whereas that obtained with the control group is 11 days (8–13 days). Compared with the control group, the patients with nonsevere new coronavirus belonging to the 26 group exhibited a shorter virus clearance time, and chest CT also showed significant improvement. In addition, the patients in the 26 treatment group experienced fewer adverse reactions and better tolerance [118]. The results of the clinical trial conducted in Wuhan suggest that among patients with common COVID-19, the 7-day clinical recovery rate of the patients in the 26 treatment group was 71.43%, which was significantly higher than the rate of 55.68% obtained with the control group; in addition, treatment with 26 significantly shortened the times to fever and cough relief in the patients with hypertension/diabetes [119]. Phase III clinical trials of 26 for the treatment of hospitalized patients with SARS-CoV-2 infection are outgoing. Future large-scale clinical trials will help verify the effectiveness and safety of 26 as a drug for the treatment of COVID-19.
27 (ribavirin, ICN-1229) can directly induce antiviral activity against a number of RNA viruses by increasing the mutation frequency in the genomes of several RNA viruses [120]. This compound is primarily indicated for the treatment of hepatitis C and viral haemorrhagic fevers. 27-TP also exhibits an inhibitory action on viral mRNA guanylyltransferase and mRNA 2′-O-methyltransferase of DENV [121] and was used as a therapeutic drug in the SARS outbreak in 2003 [122]. Tong et al. compared 27 and supportive therapies for patients with severe COVID-19 and found that ribavirin therapy is not associated with an improved negative conversion time in the SARS-CoV-2 test or with an improved mortality rate in patients with severe COVID-19 [123]. In addition, a clinical trial studied the efficacy and safety of the combination of interferon beta-1b, lopinavir-ritonavir, and 27 for the treatment of patients with COVID-19. The results showed that early triple antiviral therapy was safe and superior to lopinavir-ritonavir alone in alleviating symptoms and shortening the durations of viral shedding and hospital stay in patients with mild symptoms, and the side effects were mild and controllable [124]. However, the results from the trial need to be further verified by an expanded double-blind trial. It is worth noting that the safety of ribavirin has been controversial. The adverse reactions that have been reported include teratogenicity and haemolytic anaemia [125].
3. Non-nucleoside inhibitors
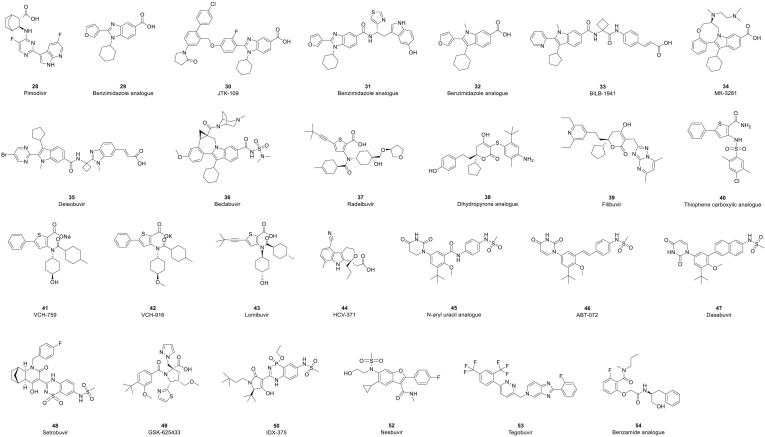
The structures of NNIs are diverse. Most NNIs change the spatial conformation of RdRp by binding to allosteric sites on the surface of the enzyme and thereby inhibit its activity and the replication of RNA viruses (Fig. 6 ). 28 (pimodivir, JNJ-63623872) is an NNI of the PB2 domain of the RdRp of influenza A virus [126]. Phase I and II clinical trials have shown that 28 has the potential to not only reduce the viral load but also have a clinical impact on patients [127,128]. However, due to the unsatisfactory results of phase III clinical trials, the clinical study of 28 has been terminated. In addition, clinical studies of allosteric site inhibitors have mainly focused on anti-HCV infection. Five different allosteric binding sites for NNIs in HCV NS5B polymerase have been discovered [129,130]. Two of these sites are located in the thumb subdomain of the polymerase, and the other three are located in the palm subdomain. The NNIs of HCV can be divided into five different classes according to the location of their respective binding sites (referred to as thumbs I and II and palms I, II and III) (Table 4 ).
Fig. 6.
Structures of nonnucleoside analogue inhibitors.
Table 4.
RNA-dependent RNA polymerase nonnucleoside inhibitors.
| Number | Compound | CAS registry number | Allosteric sites | Condition and highest clinical phase |
|---|---|---|---|---|
| 28 | Pimodivir | 1629869-44-8 | PB2 | Discontinued - Influenza A - 2020 Phase III - Influenza A - 2018 |
| 29 | Benzimidazole analogue | / | Thumb Ⅰ | Biological test |
| 30 | JTK-109 | 480462-62-2 | Thumb Ⅰ | Discontinued - Hepatitis C - 2003 Phase I - Hepatitis C – 2002 |
| 31 | Benzimidazole analogue | / | Thumb Ⅰ | Biological test |
| 32 | Benzimidazole analogue | / | Thumb Ⅰ | Biological test |
| 33 | BILB-1941 | 494856-61-0 | Thumb Ⅰ | Discontinued - Hepatitis C - 2014 Phase I/II - Hepatitis C - 2014 |
| 34 | MK-3281 | 886043-45-4 | Thumb Ⅰ | Discontinued - Hepatitis C - 2007 Phase I - Hepatitis C - 2007 |
| 35 | Deleobuvir | 1221574-24-8 | Thumb Ⅰ | Discontinued - Hepatitis C - 2014 Phase III - Hepatitis C - 2013 |
| 36 | Beclabuvir | 958002-33-0 | Thumb Ⅰ | Launched - Hepatitis C - 2017 |
| 37 | Radalbuvir | 1314795-11-3 | Thumb II | Phase II - Hepatitis C - 2011 |
| 38 | Dihydropyrone analogue | / | Thumb II | Biological test |
| 39 | Filibuvir | 877130-28-4 | Thumb II | Discontinued - Hepatitis C - 2013 Phase II - Hepatitis C – 2009 |
| 40 | Thiophene carboxylic analogue | / | Thumb II | Biological test |
| 41 | VCH-759 | 713139-25-4 | Thumb II | Phase I/II - Hepatitis C - 2006 |
| 42 | VCH-916 | 1200133-34-1 | Thumb II | Phase I - Hepatitis C - 2008 |
| 43 | Lomibuvir | 1026785-55-6 | Thumb II | Licenced - Infections - 2016 Discontinued- Hepatitis C −2010 Phase II - Hepatitis C - 2010 |
| 44 | HCV-371 | 675184-27-7 | Thumb II | Discontinued - Hepatitis C - 2003 Phase I - Hepatitis C - 2002 |
| 45 | N-aryl uracil analogue | / | Palm Ⅰ | Biological test |
| 46 | ABT-072 | 1132936-00-5 | Palm Ⅰ | Phase II - Hepatitis C - 2009 |
| 47 | Dasabuvir | 1132935-63-7 | Palm Ⅰ | Launched - Hepatitis C - 2015 |
| 48 | Setrobuvir | 1071517-39-9 | Palm Ⅰ | Phase II - Hepatitis C - 2009 |
| 49 | GSK-625433 | 885264-71-1 | Palm Ⅰ | Discontinued - Hepatitis C - 2009 Phase I - Hepatitis C - 2007 |
| 50 | IDX-375 | 1256735-81-5 | Palm Ⅰ | Phase II - Hepatitis C - 2009 |
| 51 | CC-31244 | Undisclosed structure | Palm Ⅰ | Phase II - Hepatitis C - 2019 |
| 52 | Nesbuvir | 1132935-63-7 | Palm II | Phase II - Hepatitis C - 2008 |
| 53 | Tegobuvir | 1000787-75-6 | Palm II | Phase II - Hepatitis C - 2008 |
| 54 | Benzamide analogue | / | Palm Ⅲ | Biological test |
3.1. Thumb Ⅰ inhibitors
Thumb I inhibitors are mainly benzimidazole and indole compounds, and the compounds under clinical studies all show excellent anti-HCV activity. These types of compounds bind to thumb I mainly through hydrophobic interactions and salt bridge/hydrogen bonds between the ester group or carbonyl group of the compound and the guanidine group of the amino acid residue R503.
The initially discovered benzimidazole 29 has weak inhibitory activity (IC50 = 1.6 μM) against HCV GT 1 NS5B polymerase, and its EC50 value is higher than 10 μM in a cell-based model for subgenomic replicon [131]. The structure has been optimized to improve the activity of benzimidazole compounds. JTK-109 (30) is a superior compound that was obtained by modifying position 2 of benzimidazole. 30 inhibits HCV GT 1NS5B polymerase with an IC50 value of 0.022 μM and an EC50 value of 0.62 μM [132]. In addition, the carboxyl group at position 5 of benzimidazole can be modified to enhance the antiviral activity of the compound. A longer side chain was linked by an amide bond to obtain 31, which exerts an inhibitory effect on NS5B polymerase GT 1 polymerase (IC50 = 0.3 μM) with an EC50 value of 1.7 μM [133]. To further improve the activity of the compounds on enzymes and cells, the structure of these compounds was further modified by replacing the benzimidazole ring with the indole ring, which yielded 32. The IC50 value of 32 in inhibiting HCV GT 1b NS5B polymerase is 0.016 μM, and the EC50 value is 4.2 μM [134]. Compared with 29, the activity of 32 at the enzyme level was increased 100-fold, and the activity in the cell model was also improved. The structure of 32 was modified using the same strategy as that used to obtain 29, which yielded 33 (BILB-1941). The IC50 value of 33 in inhibiting HCV GT 1b NS5B polymerase is 0.045 μM, and the EC50 value is 0.084 μM [131]. Compound 33 was administered via a single oral dose in a clinical phase I trial, which revealed that this compound has antiviral activity against HCV GT 1. Adverse events (AEs) were mainly related to the gastrointestinal tract (most frequent diarrhoea), and the frequency increased with increasing dose [135]. However, at high doses (450 mg), all five actively treated patients were unable to tolerate the compound due to gastrointestinal reactions, and clinical studies have been discontinued [136].
Whether the compound has benzimidazole or indole as its core, the dihedral angle between the core and the 2-position aryl group exerts a greater impact on the activity of the compound during the process of structural modification [137]. Structure-activity relationship (SAR) studies involving an indolo-benzoxazocine scaffold led to the identification of 34 (MK-3281), an inhibitor that exhibits good potency in the HCV subgenomic replication assay and attractive molecular properties suitable for a clinical candidate [138]. Compound 34 inhibited HCV NS5B polymerase with an IC50 value of 0.006 μM, and an EC50 value of 0.038 μM was obtained in the replicator model (GT 1b). The compound caused a consistent decrease in viremia in vivo, as demonstrated with the chimaeric mouse and chimpanzee model of HCV infection [139]. In a 7-day clinical trial of 34 monotherapies, patients with HCV GT 1b infection showed the greatest decrease in viral load and no viral load rebound, whereas patients with HCV GT 1a infection exhibited a small decrease in viral load, and their viral load appeared to rebound. One patient developed severe myoclonus side effects, and clinical trials of the compound were terminated. The EC50 values of 35 (deleobuvir, BI 207127) in the cell-based HCV GT 1b and GT 1a subgenomic replicons are 23 nM and 11 nM, respectively [140]. In a clinical trial of 35 combined with SOC for the treatment of HCV infection, the patients showed a significant reduction in viral load [141]. However, the use of 35 for the treatment of hepatitis C has been discontinued by Boehringer Ingelheim due to weak market competition. 36 (beclabuvir, BMS-791325) is a thumb Ⅰ-NS5B polymerase ligand [142]. In cell culture, 36 inhibits the replication of HCV subgenomic replicons representing HCV GTs 1a and 1b at EC50 values of 3 nM and 6 nM, respectively, and similar values (3–18 nM) were obtained for GTs 3a, 4a, and 5a [143]. The oral bioavailability of 36 is 66%, and its volume of distribution is 2.7 L/kg; following intravenous administration, a plasma clearance of 3.5 mL/min/kg and an estimated plasma t1/2 of 8.3 h were obtained in a 24-h rat pharmacokinetic study [144]. 36 represents a valid drug that exhibits a good tolerability and safety profile, and together with other antivirals, this compound exhibits optimal efficacy against HCV in compensated phases of the diseases, as demonstrated in clinical studies [145,146]. At present, 36 in combination with asunaprevir and daclatasvir has been launched in Japan for the treatment of hepatitis C.
3.2. Thumb Ⅱ inhibitors
The thumb II site is a spatially distinct allosteric site on the polymerase situated at the base of the thumb subdomain at a distance of 30 Å from the enzyme active site. Inhibitors acting on the thumb II site are mainly dihydropyrones, thiophene carboxylic acids and pyranoindole compounds. The lipophilic substituents of these compounds occupy the shallow grooves formed by the amino acid residues Leu 419, Tyr477 and Trp 528 in the thumb Ⅱ site. The acidic groups of the compounds generate hydrogen bonds with the backbone amide bonds of the amino acid residues Ser 476 and Tyr477 [147,148].
37 (radalbuvir, GS-9669) is an inhibitor of NS5B polymerase that is currently in phase II clinical trials for the oral treatment of patients with HCV infection. In replicon cell lines, compound 37 exerts a high antiviral effect against HCV GT 1a (EC50 = 11 nM), HCV GT 1b (EC50 = 2.8 nM) and HCV GT 5a (EC50 = 8 nM) [149]. Due to its synergistic or additive effects with other antivirals and the lack of cross-resistance, this compound might be an important component of interferon-free combinations for the treatment of HCV infection [150].
38 is a seed compound identified by high-throughput screening. The compound has an IC50 value of 0.93 μM for HCV GT 1b NS5B polymerase inhibition, and an EC50 value of 48 μM was obtained in the replicon model [148]. To optimize the structure of the compound, one strategy is to introduce an aromatic group that can interact with the residue and undergo π-π stacking interactions, and another strategy is to replace the sulfur atom connecting the dihydropyrone and the aromatic group on the right with a carbon atom. These strategies aim to improve the membrane permeability and pharmacokinetic properties of the compound and yield highly active compound 39 (filibuvir, PF-868554) [151,152]. Compound 39 exhibits an IC50 value of 0.01 μM for HCV GT 1b NS5B polymerase inhibition and an EC50 value of 0.59 μM in the replicon model. In the phase I trial of patients with HCV GT 1 infection, the single administration of 39 resulted in decreases in HCV RNA. In a phase II clinical trial, the combination of 39 and Peg-IFN/RBV increased the rapid viral response rate and was well tolerated. However, compared with the development of Gilead Sciences’ hepatitis C drug, Pfizer has been far behind, and continued development of 39 cannot take the lead in the market; thus, Pfizer terminated its clinical research of 39.
40 was the first discovered anti-HCV inhibitor of thiophene-2-carboxylic acid acting on the thumb Ⅱ site, and its IC50 value for HCV GT 1b NS5B polymerase inhibition is 14 μM [153]. 41 (VCH-759), 42 (VCH-916) and 43 (lomibuvir, VCH-222) were obtained through the structural modification of 40. These three compounds have all entered clinical trials. Compound 41 inhibits GT1a NS5B polymerase and GT1b NS5B polymerase with IC50 values of 0.41 and 0.38 μM, respectively [154]. In a phase I clinical trial of 41, the average viral load of HCV type 1 patients who were treated with 800 mg TID was evaluated 10 days later, and the largest decrease in volume reached 2.5 log10 IU/mL [155]. Although more patients had diarrhoea, no serious adverse reactions were observed. The EC50 value of 42 for HCV GT 1 in the replicon model was found to be equal to 0.1 μM [156]. In a clinical phase I trial, the compound rapidly reduced the viral load of HCV GT 1 patients after 3 days of treatment, and the maximum reduction in viral load obtained with these treatments was 1.5 log10 IU/mL [157]. The compound was well tolerated in healthy volunteers administered a single oral dose of 1500 mg and in patients with HCV orally administered 750 mg BID, and the maximum average viral load observed 3 days after the treatment was decreased by 3.7 log10 IU/mL. The phase II clinical trial of 43 in combination with telaprevir for the treatment of patients with HCV GT 1 was terminated [158,159].
Compound 44 (HCV-371) is a type of pyranoindole HCV thumb Ⅱ inhibitor that has a structure that differs from that of the two abovementioned types of compounds [[160], [161], [162]]. For 90% HCV GTs 1a and 1b, the IC50 value was 0.3–1.4 μM, the IC50 value for HCV GT3a inhibition was 1.8 μM, and the EC50 values for HCV GTs 1a and 1b in the replicon model were 6.1 and 4.8 μM, respectively. The compound was well tolerated in clinical phase I trials, but due to a lack of significant antiviral activity, clinical trials of the compound have been terminated.
3.3. Palm Ⅰ inhibitors
The palm I binding site is located between the active site and the palm II site and contains a deep hydrophobic pocket. The inhibitors that bind to this site mainly include N-aryl uracil analogues, benzothiadiazines and acyl pyrrolidines.
A series of N-aryl uracil analogues was reported as a novel structural class of NS5B polymerase NNIs [163]. Compound 45 is a potent inhibitor of GTs 1a (EC50 = 51 nM) and 1b (EC50 = 19 nM) NS5B polymerase [164]. Replicon activity was maintained when the assay was conducted in the presence of 40% human plasma [EC50 = 61 nM (1a), EC50 = 22 nM (1b)]. However, 45 exhibited poor pharmacokinetic properties in rats, with high plasma clearance and poor oral bioavailability (F = 1.4%). The physical properties of this compound that can be associated with poor oral exposure include low aqueous solubility and poor membrane permeability. The solubility problem is likely related to the high melting point of the compound. Based on its excellent antiviral activity profile, 46 (ABT-072) was developed. The replacement of the amide linkage in 45 with a trans-olefin yielded a compound with improved permeability and solubility and markedly better pharmacokinetic properties in preclinical species. The replacement of the dihydrouracil in 45 with an N-linked uracil provided better potency in the HCV GT 1 replicon assay. Compound 46 is a potent inhibitor of HCV GT 1 replicons, with EC50 values of 1 nM and 0.3 nM against HCV GT 1a and GT 1b, respectively. The results from phase Ⅰ clinical studies supported the once-daily oral dosing of HCV-infected patients with 46 [163,165]. A phase II clinical study that combined 46 with the HCV protease inhibitor ABT-450 revealed a sustained virologic response at 24 weeks after dosing (SVR24) in 10 of 11 patients who received treatment [166]. 47 (dasabuvir, ABT-333) is also an N-linked uracil derivative that was identified via throughput screening of the aryl dihydrouracil fragment [167]. This compound does not exhibit stereoisomerism, is thermodynamically stable and shows aqueous solubility, dissolution, and Caco-2 permeability. The IC50 for clinical isolates ranges between 2.2 and 10.7 nM for HCV GTs 1a and 1b [168]. In 2016, 47 in combination with ombitasvir/paritaprevir/ritonavir was launched for the treatment of chronic hepatitis C infection. However, 47 has limitations in terms of its limited genotypic coverage, and its administration to patients with advanced cirrhosis is difficult. This compound also represents a large pill burden when added to combination therapy [169].
Compounds with benzothiadiazine as the basic skeleton show strong activity at the enzyme level and in the cell model, and their physical and chemical properties are not ideal due to their special compound structure (intramolecular hydrogen bonds bring the aromatic ring close to the same plane), which results in poor pharmacokinetic properties in the body [170,171]. Such structures are optimized by introducing carbon-containing branches, reducing the number of aromatic rings, and reducing the polar surface area (PSA) of the molecule. 48 (setrobuvir, ANA598) is a benzothiadiazine analogue with a reduced aromatic ring that is currently in phase II clinical trials. In the replicon model, the EC50 values of 48 in inhibiting HCV GTs 1a and 1b NS5B polymerases were found to be equal to 0.05 and 0.003 μM, respectively [172]. In a phase I clinical trial, BID treatment with 48 at doses of 200 mg, 400 mg, and 800 mg for 4 days decreased the viral load by 2.4, 2.3 and 2.9 log10 IU/ml, respectively. In an earlier phase II study, 48 was administered in combination with Peg-IFN/RBV to naïve HCV GT 1-infected patients for 12 weeks, and the combination exhibited potent antiviral activity and good safety and tolerability [173].
49 (GSK-625433) is a representative acyl pyrrolidine HCV polymerase inhibitor with IC50 values of 1.1 (1a) and 0.028 (1b) μM and EC50 values of 0.29 (1a) and 0.003 (1b) μM in the replicon model [174]. 50 (IDX-375) demonstrates low nanomolar potency in vitro (EC50 of 2.3 nM) in the HCV GT 1b replicon with a selectivity index of 43,000 [175]. 50 is well absorbed and well tolerated by all healthy male volunteers included in the phase Ⅰ study. A single-day 200-mg BID dose resulted in exposure-related HCV activity with maximal 0.5 to 1.1 log10 reductions in plasma HCV RNA levels [176]. 51 (CC-31244, undisclosed structure) is a pangenotypic inhibitor of NS5B polymerase (GTs 1–5) that was designed for the treatment of hepatitis C infection. Compound 51 shows no significant cytotoxicity, CYP450 inhibition, or off-target or drug-drug interactions [177]. In the phase Ⅰ study, a rapid and marked decline in HCV RNA levels, slow viral rebound after treatment, and no viral breakthrough during treatment were observed in the patients, which indicates that this compound is highly favourable compared with the currently approved NNIs [178].
3.4. Palm Ⅱ inhibitors
The palm II site is mainly composed of a large hydrophobic pocket in the palm area, and the RdRp inhibitors that bind to this site include benzofurans. These palm II site inhibitors are different from other nonnucleoside inhibitors in that they exhibit potent activity against HCV GTs 1 to 4 and NS5B polymerase.
52 (nesbuvir, HCV-796) is the first palm II-NNI inhibitor to enter phase II clinical trials. In hepatoma cells containing an HCV GT 1b replicon, the IC50 value of 52 was found to be 9 nM [179]. In a phase I clinical trial, the greatest decreases in the average viral load, which reached 1.4 log IU/mL, were observed with the 1000 mg BID and 1500 mg BID treatments [180]. The initial results of phase II clinical trials showed that the combined treatment of 52 and PEG-IFNα can increase the therapeutic effect and reduce the occurrence of mutant strains. However, elevated liver enzyme levels were observed in some patients administered the combination treatment for at least 8 weeks [181]. The efficacy and safety of 52 for the treatment of hepatitis C via intravenous injection are currently being evaluated. 53 (tegobuvir, GS-9190) is currently in a phase II clinical trial for the treatment of HCV infection [182]. Its EC50 values were lower than 16 nM against HCV GT 1 and higher than 100 nM for other GTs [183]. In the clinical studies of 53 for the treatment of HCV GT 1-infected patients (doses of 40 mg and 120 mg BID), the viral load decreased by 1.4 and 1.7 log IU/ml after 8 days, respectively.
3.5. Palm Ⅲ inhibitors
Cliff C. Cheng et al. [184] described a novel series of NS5B polymerase inhibitors based on the 2-oxy-6-fluoro-N-((S)-1-hydroxy-3-phenylpropan-2-yl)-benzamide-based scaffold. Among them, the first palm III inhibitor identified was 54, and its IC50 value related to the inhibition of HCV GT 1b NS5B polymerase was 0.8 μM. The X-ray crystal structure of a complex with 54 has provided structural insights into the mechanism of inhibition and aided the rationalization of the structure-activity relationships. Compound 54 binds within the active site cavity of NS5B polymerase near the top of the palm subdomain, and the binding site of this compound is a new binding site, which has been denoted the palm III site.
4. Discussion and perspectives
Currently, only 22 has been approved in the United States as the first COVID-19 treatment drug, the specific drug for the treatment of COVID-19 remains scarce, and the rapid identification of an effective strategy for the treatment of COVID-19 is currently a major challenge facing researchers. In terms of development time, research progress, safety and effectiveness, small-molecule drugs are the best choice compared with other therapies, such as monoclonal antibodies, oligonucleotide-based therapies, plasma therapies, and peptide therapies. However, the medicinal chemistry of SARS-CoV-2 infection remains in its infancy, and target-specific lead molecules remain to be identified. The existing antiviral drugs have established safety characteristics and effectiveness against related coronaviruses. The reuse of existing small molecule antiviral drugs is an important and promising strategy for addressing the COVID-19 epidemic. If the existing drugs can be repurposed for the treatment of SARS-CoV-2 infection, preclinical research (such as animal experiments and pharmacological research) and early clinical research can be bypassed, and the drugs can directly enter phase Ⅱ or Ⅲ clinical trials.
RdRp is one of the most important viral proteins of RNA viruses for RNA synthesis and has been proposed as a valuable target for the development of antiviral therapeutics. Considering the similarity of the key drug-binding pockets between SARS-CoV-2, SARS-CoV, and MERS-CoV RdRps, repurposing known RdRp inhibitors for SARS-CoV-2 remains a promising strategy [185]. W. Yin et al. reported the complex structure of 22 inhibiting SARS-CoV-2 RdRp, which provided insights into the mechanism of viral RNA replication and a rational template for drug design to combat viral infection [186]. In this study, the complex structure revealed that the partial double-stranded RNA template was inserted into the central channel of RdRp, where 22 was covalently incorporated into the primer strand at the first replicated base pair and terminated chain elongation. At present, the candidate drugs that have shown obvious anti-SARS-CoV-2 at the cellular level or in clinical trials are 2, 15, 20, 22, 23, 25, 26 and 27 (Fig. 7 ). Like 22, these nucleotide analogues can converge into a central cavity of viral RdRp and inhibit viral RdRp through nonobligate RNA chain termination, a mechanism that requires conversion of the parent compound to the triphosphate active form. 2, 20, 22, 26 and 27 retain the entire ribose group, so they may be able to form a stable hydrogen bond network similar to natural substrates [31]. In addition, the unmodified side chain hydroxyl groups on the 20 and 27 nucleosides can also form hydrogen bonds. In particular, 2 has been shown to be 3 to 10 times as potent as 22 in blocking SARS-CoV-2 replication [54]. The N4 hydroxyl group off the cytidine ring forms an extra hydrogen bond with the side chain of K545, and the cytidine base also forms an extra hydrogen bond with the guanine base from the template strand. These two extra hydrogen bonds may explain the apparent higher potency of 2 in inhibiting SARS-CoV-2 replication [186]. However, 15 only formed 7H-bonds and two hydrophobic contacts with the SARS-CoV-2 RdRp residues. This is because fluorine substitution occurs on the ribose group of 15, so they cannot form a hydrogen bond network, but this is necessary to keep the incoming natural nucleotides stable. The same phenomenon occurs when compound 23 is combined with SARS-CoV-2 RdRp. The guanosine triphosphate derivative 25 formed 10H-bonds with the SARS-CoV-2 RdRp residues and two metal interactions with the active site residue of RdRp [101]. Moreover, among all intracellular NTPs, cytidine triphosphate is found at a lower intracellular concentration than other NTPs. Therefore, pyrimidine nucleoside inhibitors such as compound 2 are more likely to be developed into antiviral drugs.
Fig. 7.
Structure of the RdRp inhibitor repurposing for the COVID-19 pandemic.
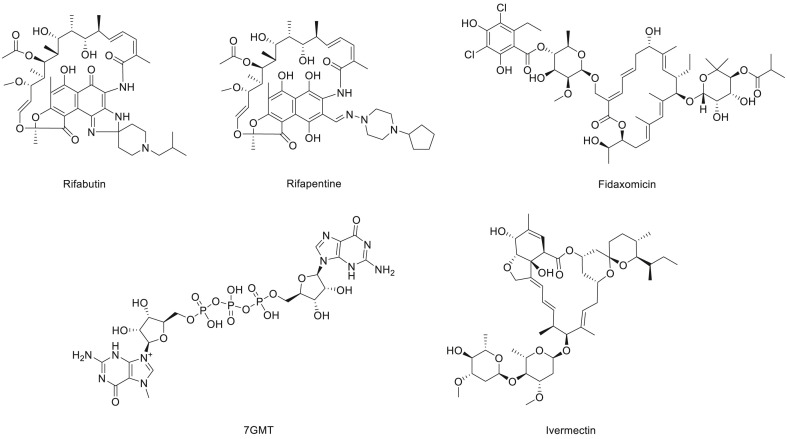
In addition, a variety of antiviral drugs were suggested as lead candidates against COVID-19 through homologue model-based virtual screening and molecular docking, including candidate drugs that have been applied in other diseases (Fig. 8 ). M.S.A. Parvez et al. [187] revealed that antibacterial drugs, including rifabutin, rifapentine, fidaxomicin, 7-methyl-guanosine-5′-triphosphate-5′-guanosine and ivermectin, have a potential inhibitory interaction with RdRp of SARS-CoV-2 and could be effective drugs for COVID-19. The drug surface hotspot study revealed that the molecular binding sites of all the compounds had a similar pattern. The vast number of noncovalent interactions between these screened compounds and RdRp suggests that the protein-inhibitor complexes are very stable. Considering that patients with COVID-19 may have combined bacterial or fungal infections, the proportion of antibiotics used in clinical treatment is relatively high [188]. If these antibacterial drugs that have inhibitory effects on RdRp can show a significant decrease in viral load in in vitro/vivo studies, they would be good therapies and play a dual antiviral/antibacterial role.
Fig. 8.
Structure of the antibacterial drugs that have potential inhibitory interactions with RdRp of SARS-CoV-2.
For the more effective screening of candidate drugs, we compared the advantages and disadvantages of NIs and NNIs. NIs targeting the active site of virus RdRps and NNIs targeting allosteric sites have different biochemical properties. NIs mostly exhibit spectral antiviral activity. Many anti-HCV NIs are active against multiple HCV GTs, which indicates that the catalytic active sites bound by such inhibitors are relatively highly conserved. However, the problem faced in the development of NIs is the high concentration of intracellular natural NTP. For triphosphorylated NIs to compete with high concentrations of cellular NTP to exert their antiviral activity, the dose of the drug needs to be increased, which increases the risk of drug toxicity. In addition, NNIs acting on allosteric sites exert antiviral activity by affecting the binding of the catalytically active site of RdRp to the substrate. Such inhibitors do not need to undergo metabolic activation and do not need to compete with intracellular NTP. Combined with the structural diversity of such inhibitors, these compounds appear to be better antiviral drugs than nucleoside analogues. However, the structural variability and nonconservation of adjacent allosteric sites cause the RNA virus to rapidly develop resistance to allosteric site inhibitors.
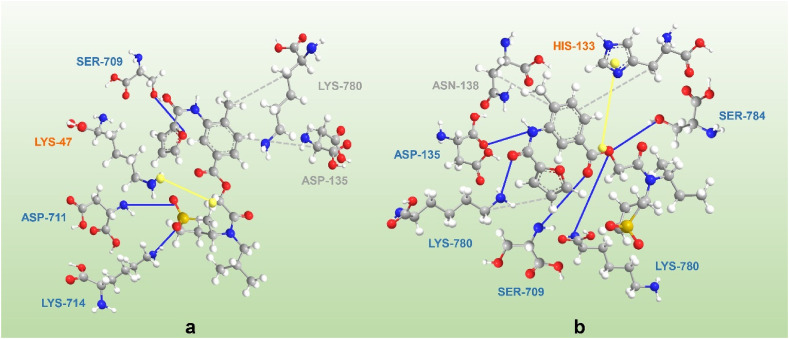
As part of ongoing global efforts to prevent the spread of SARS-COV-2 and treat the resulting infection, the use of approved drugs for nonapproved uses can alleviate urgent needs. However, to prevent viruses with similar genomic and pathological characteristics from returning a few years later, more specific, safe and effective drugs need to be developed. 2, the isopropyl ester prodrug of N4-hydroxycytidine, has been approved for use in clinical trials for the treatment of patients with COVID-19 and has shown positive effects. Since deuterium atoms are twice as heavy as hydrogen atoms, the vibrational zero-point energy of carbon-deuterium bonds (C-D) is lower than that of carbon-hydrogen bonds (C–H), and C-D is more stable than C–H [189]. Wen et al. replaced the hydrogen in the active molecular group with isotope deuterium to close the metabolic site and prolong the t1/2 of the drug, which further improves the metabolism of remdesivir in vivo and expands the scope of the treatment window and thereby reduces the therapeutic dose [190]. Additionally, the pharmacophore model is a ligand-based drug design tool that starts from the structure of known active compounds to find common pharmacodynamic feature information, thereby guiding the rational design or virtual screening of new compounds. M.S.A. Parvez et al. [187] designed a pharmacophore using 22, which was used further for screening the ZINC database. Molecular docking analysis revealed that two compounds (ZINC09128258 and ZINC09883305) with pharmacophore features that interact effectively with RdRp of SARS-CoV-2, indicating their potential as effective inhibitors of the enzyme (Fig. 9 ). In addition, in recent years, the research and development of RdRp inhibitors for RNA viruses has continued to be hot. Wang et al. reported the synthesis and biological evaluation of a series of 2′,3′- and 2′,4′-substituted guanosine nucleotide analogues as HCV NS5B polymerase inhibitors [191]. 6′-Fluorinated aristeromycins were designed as dual-target antiviral compounds for the development of broad-spectrum antiviral agents that target RNA viruses [192]. These new compounds may also be a hope against the COVID-19 epidemic.
Fig. 9.
The interaction of SRAS-CoV-2 RdRp with (a) ZINC09128258 and (b) ZINC09883305. Solid blue lines represent H-bonds, while hydrophobic interactions are grey dashed lines. In addition, π-cation stacking is represented by yellow spheres connected by dashed lines.
Undeniably, the development of efficient anti-SARS-CoV-2 drugs over a short time is associated with considerable obstacles and unidentified threats. Nonetheless, efforts to establish antiviral medicines to fight the novel coronavirus are urgently needed. Scientists from clinical research organizations and pharmaceutical companies along with front-line doctors should enhance their cooperation to jointly advertise pharmaceutical, scientific and preclinical tests of appropriate anti-coronavirus medications.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81602967 and 81803784), the China Postdoctoral Science Foundation (Grant Nos. 2016 M592898XB and 2019 M663921XB), the Basic Research Program of Natural Science of Shaanxi Province (Grant Nos. 2019JQ-779, 2020CGXNG-044), the Basic Research Plan of the Education Department of Shaanxi Province (Grant No. 19JC006), the Key Scientific Research Group of Shaanxi Province (2020TD-009), and the Youth Innovation Team of Shaanxi Universities.
References
- 1.McCarthy M.K., Morrison T.E. Persistent RNA virus infections: do PAMPS drive chronic disease? Current Opinion in Virology. 2017;23:8–15. doi: 10.1016/j.coviro.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddharta A., Pfaender S., Vielle N.J., Dijkman R., Friesland M., Becker B., Yang J., Engelmann M., Todt D., Windisch M.P., Brill F.H., Steinmann J., Steinmann J., Becker S., Alves M.P., Pietschmann T., Eickmann M., Thiel V., Steinmann E. Virucidal activity of world health organization-recommended formulations against enveloped viruses, including Zika, ebola, and emerging coronaviruses. JID (J. Infect. Dis.) 2017;215:902–906. doi: 10.1093/infdis/jix046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrasco-Hernandez R., Jácome R., López Vidal Y., Ponce de León S. Are RNA viruses candidate agents for the next global pandemic? A Review, ILAR Journal. 2017;58:343–358. doi: 10.1093/ilar/ilx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reperant L.A., Osterhaus A. AIDS, avian flu, SARS, MERS, ebola, Zika… what next? Vaccine. 2017;35:4470–4474. doi: 10.1016/j.vaccine.2017.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acter T., Uddin N., Das J., Akhter A., Choudhury T.R., Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: a global health emergency. Sci. Total Environ. 2020;730:138996. doi: 10.1016/j.scitotenv.2020.138996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D., Chen B., Zhang Z., Guan W., Ling Z., Jiang R., Hu T., Ding Y., Lin L., Gan Q., Luo L., Tang X., Liu J. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imag. 2020;47:1275–1280. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghaebi M., Osali A., Valizadeh H., Roshangar L., Ahmadi M. Vaccine development and therapeutic design for 2019-nCoV/SARS-CoV-2: challenges and chances. J. Cell. Physiol. 2020;235:9098–9109. doi: 10.1002/jcp.29771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause P., Fleming T.R., Longini I., Henao-Restrepo A.M., Peto R., Dean N., Halloran M., Huang Y., Fleming T., Gilbert P. COVID-19 vaccine trials should seek worthwhile efficacy. Lancet. 2020;396:741–743. doi: 10.1016/S0140-6736(20)31821-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 10.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X.B., Yu Y., Xu J.Q., Shu H.Q., Xia J.A., Liu H., Wu Y.R., Zhang L., Yu Z., Fang M.H., Yu T., Wang Y.X., Pan S.W., Zou X.J., Yuan S.Y., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respiratory Medicine. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baltimore D. Evolution of RNA viruses. Ann. N. Y. Acad. Sci. 1980;354:492–497. doi: 10.1111/j.1749-6632.1980.tb27988.x. [DOI] [PubMed] [Google Scholar]
- 14.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui Y., Zhang Y., Zhou K., Sun J., Zhou Z.H. Conservative transcription in three steps visualized in a double-stranded RNA virus. Nat. Struct. Mol. Biol. 2019;26:1023–1034. doi: 10.1038/s41594-019-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahlquist P., Noueiry A.O., Lee W.M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comas-Garcia M. Packaging of genomic RNA in positive-sense single-stranded RNA viruses: a complex story. Viruses-Basel. 2019;11:253. doi: 10.3390/v11030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thébaud G., Chadoeuf J., Morelli M.J., McCauley J.W., Haydon D.T. The relationship between mutation frequency and replication strategy in positive-sense single-stranded RNA viruses. Proceedings: Biol. Sci. 2010;277:809–817. doi: 10.1098/rspb.2009.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maida Y., Yasukawa M., Furuuchi M., Lassmann T., Possemato R., Okamoto N., Kasim V., Hayashizaki Y., Hahn W.C., Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkataraman S., Prasad B., Selvarajan R. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses-Basel. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamin M., Yabukarski F. Nonsegmented negative-sense RNA viruses-structural data bring new insights into nucleocapsid assembly. Adv. Virus Res. 2017;97:143–185. doi: 10.1016/bs.aivir.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Luo M., Terrell J.R., McManus S.A. Nucleocapsid structure of negative strand RNA virus. Viruses. 2020;12:835. doi: 10.3390/v12080835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller A.D. Retroviral Vectors in Gene Therapy. 2006. [DOI] [Google Scholar]
- 24.Parvez M.K., Parveen S. Evolution and emergence of pathogenic viruses: past, present, and future. Intervirology. 2017;60:1–7. doi: 10.1159/000478729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawood F.S., Iuliano A.D., Reed C., Meltzer M.I., Shay D.K., Cheng P.-Y., Bandaranayake D., Breiman R.F., Brooks W.A., Buchy P. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect. Dis. 2012;12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 26.Poltronieri P., Sun B., Mallardo M. RNA viruses: RNA roles in pathogenesis, coreplication and viral load. Curr. Genom. 2015;16:327–335. doi: 10.2174/1389202916666150707160613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeler S.P., Agapov E.V., Hinojosa M.E., Letvin A.N., Wu K., Holtzman M.J. Influenza A virus infection causes chronic lung disease linked to sites of active viral RNA remnants. J. Immunol. 2018;201:2354–2368. doi: 10.4049/jimmunol.1800671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitko V., Barik S. Respiratory viral diseases: access to RNA interference therapy. Drug Discov. Today Ther. Strat. 2007;4:273–276. doi: 10.1016/j.ddstr.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buonaguro L., Tagliamonte M., Tornesello M.L., Buonaguro F.M. SARS-CoV-2 RNA polymerase as target for antiviral therapy. J. Transl. Med. 2020;18:185. doi: 10.1186/s12967-020-02355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Y., Zhao Q., Liu H., Cao W., Xie X., Zhang D., Wang Y., Jian Y. The development of Coronavirus 3C-Like protease (3CL(pro)) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206:112711. doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White K.A., Enjuanes L., Berkhout B. RNA virus replication, transcription and recombination. RNA Biol. 2011;8:182–183. doi: 10.4161/rna.8.2.15663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Farias S.T., Dos Santos Junior A.P., Rêgo T.G., José M.V. Origin and evolution of RNA-dependent RNA polymerase. Front. Genet. 2017;8:125. doi: 10.3389/fgene.2017.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkataraman S., Prasad B.V., Selvarajan R. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen J.L., Long A.M., Schultz S.C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 37.Xiao M., Li H., Wang Y., Wang X., Wang W., Peng J., Chen J., Li B. Characterization of the N-terminal domain of classical swine fever virus RNA-dependent RNA polymerase. J. Gen. Virol. 2006;87:347–356. doi: 10.1099/vir.0.81385-0. [DOI] [PubMed] [Google Scholar]
- 38.Hernández S., Figueroa D., Correa S., Díaz A., Aguayo D., Villanueva R.A. Phosphorylation at the N-terminal finger subdomain of a viral RNA-dependent RNA polymerase. Biochem. Biophys. Res. Commun. 2015;466:21–27. doi: 10.1016/j.bbrc.2015.08.082. [DOI] [PubMed] [Google Scholar]
- 39.te Velthuis A.J. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 2014;71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrer-Orta C., Ferrero D., Verdaguer N. RNA-dependent RNA polymerases of picornaviruses: from the structure to regulatory mechanisms. Viruses. 2015;7:4438–4460. doi: 10.3390/v7082829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., Liu H.T., Wu C.Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92:693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukarska M., Fournier G., Pflug A., Resa-Infante P., Reich S., Naffakh N., Cusack S. Structural basis of an essential interaction between influenza polymerase and Pol II CTD. Nature. 2017;541:117–121. doi: 10.1038/nature20594. [DOI] [PubMed] [Google Scholar]
- 44.Hengrung N., El Omari K., Serna Martin I., Vreede F.T., Cusack S., Rambo R.P., Vonrhein C., Bricogne G., Stuart D.I., Grimes J.M., Fodor E. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature. 2015;527:114–117. doi: 10.1038/nature15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S., Toyoda T. Molecular mechanisms of transcription and replication of the influenza A virus genome. Front. Biol. 2011;6:446–461. [Google Scholar]
- 46.Cao D., Gao Y., Roesler C., Rice S., D’Cunha P., Zhuang L., Slack J., Domke M., Antonova A., Romanelli S., Keating S., Forero G., Juneja P., Liang B. Cryo-EM structure of the respiratory syncytial virus RNA polymerase. Nat. Commun. 2020;11:368. doi: 10.1038/s41467-019-14246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang B., Li Z., Jenni S., Rahmeh A.A., Morin B.M., Grant T., Grigorieff N., Harrison S.C., Whelan S.P.J. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tchesnokov E.P., Raeisimakiani P., Ngure M., Marchant D., Götte M. Recombinant RNA-dependent RNA polymerase complex of Ebola virus. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-22328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma P.L., Nurpeisov V., Hernandez-Santiago B., Beltran T., Schinazi R.F. Nucleoside inhibitors of human immunodeficiency virus type 1 reverse transcriptase. Curr. Top. Med. Chem. 2004;4:895–919. doi: 10.2174/1568026043388484. [DOI] [PubMed] [Google Scholar]
- 51.Deval J. Antimicrobial strategies: inhibition of viral polymerases by 3’-hydroxyl nucleosides. Drugs. 2009;69:151–166. doi: 10.2165/00003495-200969020-00002. [DOI] [PubMed] [Google Scholar]
- 52.Deval J., Powdrill M.H., D’Abramo C.M., Cellai L., Götte M. Pyrophosphorolytic excision of nonobligate chain terminators by hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 2007;51:2920–2928. doi: 10.1128/AAC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urakova N., Kuznetsova V., Crossman D.K., Sokratian A., Guthrie D.B., Kolykhalov A.A., Lockwood M.A., Natchus M.G., Crowley M.R., Painter G.R., Frolova E.I., Frolov I. β-d-N (4)-hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018;92 doi: 10.1128/JVI.01965-17. e01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X.T., Andres E.L., Bluemling G.R., Lockwood M.A., Sheahan T.P., Sims A.C., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G.R., Baric R.S., Denison M.R. Small-molecule antiviral beta-D-N-4-Hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93:14. doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urakova N., Kuznetsova V., Crossman D.K., Sokratian A., Guthrie D.B., Kolykhalov A.A., Lockwood M.A., Natchus M.G., Crowley M.R., Painter G.R., Frolova E.I., Frolov I. beta-D-N-4-Hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018;92:22. doi: 10.1128/JVI.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheahan T.P., Sims A.C., Zhou S.T., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X.T., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:15. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynard O., Nguyen X.-N., Alazard-Dany N., Barateau V., Cimarelli A., Volchkov V.E. Identification of a new ribonucleoside inhibitor of ebola virus replication. Viruses. 2015;7:6233–6240. doi: 10.3390/v7122934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dulin D., Arnold J.J., van Laar T., Oh H.-S., Lee C., Perkins A.L., Harki D.A., Depken M., Cameron C.E., Dekker N.H. Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Rep. 2017;21:1063–1076. doi: 10.1016/j.celrep.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi K.H. Springer; 2012. Viral Polymerases, Viral Molecular Machines; pp. 267–304. [Google Scholar]
- 61.Qu C., Xu L., Yin Y., Peppelenbosch M.P., Pan Q., Wang W. Nucleoside analogue 2’-C-methylcytidine inhibits hepatitis E virus replication but antagonizes ribavirin. Arch. Virol. 2017;162:2989–2996. doi: 10.1007/s00705-017-3444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee J.C., Tseng C.K., Wu Y.H., Kaushik-Basu N., Lin C.K., Chen W.C., Wu H.N. Characterization of the activity of 2’-C-methylcytidine against dengue virus replication. Antivir. Res. 2015;116:1–9. doi: 10.1016/j.antiviral.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 63.Rocha-Pereira J., Jochmans D., Dallmeier K., Leyssen P., Cunha R., Costa I., Nascimento M.S., Neyts J. Inhibition of norovirus replication by the nucleoside analogue 2’-C-methylcytidine. Biochem. Biophys. Res. Commun. 2012;427:796–800. doi: 10.1016/j.bbrc.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Pierra C., Amador A., Benzaria S., Cretton-Scott E., D’Amours M., Mao J., Mathieu S., Moussa A., Bridges E.G., Standring D.N., Sommadossi J.P., Storer R., Gosselin G. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2’-C-methylcytidine. J. Med. Chem. 2006;49:6614–6620. doi: 10.1021/jm0603623. [DOI] [PubMed] [Google Scholar]
- 65.Deval J., Hong J., Wang G., Taylor J., Smith L.K., Fung A., Stevens S.K., Liu H., Jin Z., Dyatkina N. Molecular basis for the selective inhibition of respiratory syncytial virus RNA polymerase by 2’-fluoro-4’-chloromethyl-cytidine triphosphate. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deval J., Fung A., Stevens S.K., Jordan P.C., Gromova T., Taylor J.S., Hong J., Meng J., Wang G., Dyatkina N. Biochemical effect of resistance mutations against synergistic inhibitors of RSV RNA polymerase. PloS One. 2016;11 doi: 10.1371/journal.pone.0154097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang G., Deval J., Hong J., Dyatkina N., Prhavc M., Taylor J., Fung A., Jin Z., Stevens S.K., Serebryany V., Liu J., Zhang Q., Tam Y., Chanda S.M., Smith D.B., Symons J.A., Blatt L.M., Beigelman L. Discovery of 4’-chloromethyl-2’-deoxy-3’,5’-di-O-isobutyryl-2’-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J. Med. Chem. 2015;58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- 68.Wang G., Deval J., Hong J., Dyatkina N., Prhavc M., Taylor J., Fung A., Jin Z., Stevens S.K., Serebryany V. Discovery of 4′-chloromethyl-2′-deoxy-3′, 5′-di-O-isobutyryl-2′-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J. Med. Chem. 2015;58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- 69.Nilsson M., Kalayanov G., Winqvist A., Pinho P., Sund C., Zhou X.-X., Wähling H., Belfrage A.-K., Pelcman M., Agback T. Discovery of 4′-azido-2′-deoxy-2′-C-methyl cytidine and prodrugs thereof: a potent inhibitor of Hepatitis C virus replication. Bioorg. Med. Chem. Lett. 2012;22:3265–3268. doi: 10.1016/j.bmcl.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 70.Clark J.L., Mason J.C., Hollecker L., Stuyver L.J., Tharnish P.M., McBrayer T.R., Otto M.J., Furman P.A., Schinazi R.F., Watanabe K.A. Synthesis and antiviral activity of 2′-deoxy-2′-fluoro-2′-C-methyl purine nucleosides as inhibitors of hepatitis C virus RNA replication. Bioorg. Med. Chem. Lett. 2006;16:1712–1715. doi: 10.1016/j.bmcl.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Murakami E., Bao H., Ramesh M., McBrayer T.R., Whitaker T., Steuer H.M.M., Schinazi R.F., Stuyver L.J., Obikhod A., Otto M.J. Mechanism of activation of β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase. Antimicrob. Agents Chemother. 2007;51:503–509. doi: 10.1128/AAC.00400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee C.H., Lee Y.J., Kim J.H., Lim J.H., Kim J.-H., Han W., Lee S.-H., Noh G.-J., Lee S.-W. Inhibition of hepatitis C virus (HCV) replication by specific RNA aptamers against HCV NS5B RNA replicase. J. Virol. 2013;87:7064–7074. doi: 10.1128/JVI.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wedemeyer H., Forns X., Hézode C., Lee S.S., Scalori A., Voulgari A., Le Pogam S., Nájera I., Thommes J.A. Mericitabine and either Boceprevir or Telaprevir in combination with peginterferon alfa-2a plus Ribavirin for patients with chronic hepatitis C genotype 1 infection and prior null response: the randomized DYNAMO 1 and DYNAMO 2 studies. PloS One. 2016;11 doi: 10.1371/journal.pone.0145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guedj J., Dahari H., Shudo E., Smith P., Perelson A.S. Hepatitis C viral kinetics with the nucleoside polymerase inhibitor mericitabine (RG7128) Hepatology. 2012;55:1030–1037. doi: 10.1002/hep.24788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gane E.J., Roberts S.K., Stedman C.A., Angus P.W., Ritchie B., Elston R., Ipe D., Morcos P.N., Baher L., Najera I. Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1): a randomised, double-blind, placebo-controlled, dose-escalation trial. Lancet. 2010;376:1467–1475. doi: 10.1016/S0140-6736(10)61384-0. [DOI] [PubMed] [Google Scholar]
- 76.Le Pogam S., Seshaadri A., Ewing A., Kang H., Kosaka A., Yan J.-M., Berrey M., Symonds B., De La Rosa A., Cammack N. RG7128 alone or in combination with pegylated interferon-α2a and ribavirin prevents hepatitis C virus (HCV) replication and selection of resistant variants in HCV-infected patients. J. Infect. Dis. 2010;202:1510–1519. doi: 10.1086/656774. [DOI] [PubMed] [Google Scholar]
- 77.Murakami E., Niu C., Bao H., Micolochick Steuer H.M., Whitaker T., Nachman T., Sofia M.A., Wang P., Otto M.J., Furman P.A. The mechanism of action of beta-D-2’-deoxy-2’-fluoro-2’-C-methylcytidine involves a second metabolic pathway leading to beta-D-2’-deoxy-2’-fluoro-2’-C-methyluridine 5’-triphosphate, a potent inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 2008;52:458–464. doi: 10.1128/AAC.01184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma H., Jiang W.R., Robledo N., Leveque V., Ali S., Lara-Jaime T., Masjedizadeh M., Smith D.B., Cammack N., Klumpp K., Symons J. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-D-2’-Deoxy-2’-fluoro-2’-C-methylcytidine (PSI-6130) and identification of a novel active 5’-triphosphate species. J. Biol. Chem. 2007;282:29812–29820. doi: 10.1074/jbc.M705274200. [DOI] [PubMed] [Google Scholar]
- 79.Sofia M.J., Bao D., Chang W., Du J., Nagarathnam D., Rachakonda S., Reddy P.G., Ross B.S., Wang P., Zhang H.-R. Discovery of a β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 2010;53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 80.Denning J., Cornpropst M., Flach S.D., Berrey M.M., Symonds W.T. Pharmacokinetics, safety, and tolerability of GS-9851, a nucleotide analog polymerase inhibitor for hepatitis C virus, following single ascending doses in healthy subjects. Antimicrob. Agents Chemother. 2013;57:1201–1208. doi: 10.1128/AAC.01262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murakami E., Tolstykh T., Bao H., Niu C., Steuer H.M.M., Bao D., Chang W., Espiritu C., Bansal S., Lam A.M. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 2010;285:34337–34347. doi: 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirby B.J., Symonds W.T., Kearney B.P., Mathias A.A. Pharmacokinetic, pharmacodynamic, and drug-interaction profile of the hepatitis C virus NS5B polymerase inhibitor sofosbuvir. Clin. Pharmacokinet. 2015;54:677–690. doi: 10.1007/s40262-015-0261-7. [DOI] [PubMed] [Google Scholar]
- 83.Gentile I., Maraolo A.E., Buonomo A.R., Zappulo E., Borgia G. The discovery of sofosbuvir: a revolution for therapy of chronic hepatitis C. Expet Opin. Drug Discov. 2015;10:1363–1377. doi: 10.1517/17460441.2015.1094051. [DOI] [PubMed] [Google Scholar]
- 84.Ferreira A.C., Zaverucha-do-Valle C., Reis P.A., Barbosa-Lima G., Vieira Y.R., Mattos M., Silva P.P., Sacramento C., de Castro Faria Neto H.C., Campanati L., Tanuri A., Brüning K., Bozza F.A., Bozza P.T., Souza T.M.L. Sofosbuvir protects Zika virus-infected mice from mortality, preventing short- and long-term sequelae. Sci. Rep. 2017;7:9409. doi: 10.1038/s41598-017-09797-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Onorati M., Li Z., Liu F., Sousa A.M.M., Nakagawa N., Li M., Dell’Anno M.T., Gulden F.O., Pochareddy S., Tebbenkamp A.T.N., Han W., Pletikos M., Gao T., Zhu Y., Bichsel C., Varela L., Szigeti-Buck K., Lisgo S., Zhang Y., Testen A., Gao X.B., Mlakar J., Popovic M., Flamand M., Strittmatter S.M., Kaczmarek L.K., Anton E.S., Horvath T.L., Lindenbach B.D., Sestan N. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 2016;16:2576–2592. doi: 10.1016/j.celrep.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu H.T., Colby-Germinario S.P., Hassounah S.A., Fogarty C., Osman N., Palanisamy N., Han Y., Oliveira M., Quan Y., Wainberg M.A. Evaluation of Sofosbuvir (β-D-2’-deoxy-2’-α-fluoro-2’-β-C-methyluridine) as an inhibitor of Dengue virus replication<sup/>. Sci. Rep. 2017;7:6345. doi: 10.1038/s41598-017-06612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guedj J., Pang P.S., Denning J., Rodriguez-Torres M., Lawitz E., Symonds W., Perelson A.S. Analysis of hepatitis C viral kinetics during administration of two nucleotide analogues: sofosbuvir (GS-7977) and GS-0938. Antivir. Ther. 2014;19:211–220. doi: 10.3851/IMP2733. [DOI] [PubMed] [Google Scholar]
- 88.Kirby B.J., Symonds W.T., Kearney B.P., Mathias A.A. Pharmacokinetic, pharmacodynamic, and drug-interaction profile of the hepatitis C virus NS5B polymerase inhibitor sofosbuvir. Clin. Pharmacokinet. 2015;54:677–690. doi: 10.1007/s40262-015-0261-7. [DOI] [PubMed] [Google Scholar]
- 89.Mesci P., Macia A., Saleh A., Martin-Sancho L., Yin X., Snethlage C., Avansini S., Chanda S., Muotri A. bioRxiv; 2020. Sofosbuvir Protects Human Brain Organoids against SARS-CoV-2. Published online 31 May 2020. [DOI] [Google Scholar]
- 90.Wang G., Dyatkina N., Prhavc M., Williams C., Serebryany V., Hu Y., Huang Y., Wan J., Wu X., Deval J. Synthesis and anti-HCV activities of 4′-fluoro-2′-substituted uridine triphosphates and nucleotide prodrugs: discovery of 4′-fluoro-2′-C-methyluridine 5′-phosphoramidate prodrug (AL-335) for the treatment of hepatitis C infection. J. Med. Chem. 2019;62:4555–4570. doi: 10.1021/acs.jmedchem.9b00143. [DOI] [PubMed] [Google Scholar]
- 91.McClure M.W., Berliba E., Tsertsvadze T., Streinu-Cercel A., Vijgen L., Astruc B., Patat A., Westland C., Chanda S., Zhang Q. Safety, tolerability, and pharmacokinetics of AL-335 in healthy volunteers and hepatitis C virus-infected subjects. PloS One. 2018;13 doi: 10.1371/journal.pone.0204974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeuzem S., Bourgeois S., Greenbloom S., Buti M., Aghemo A., Lampertico P., Janczewska E., Lim S.G., Moreno C., Buggisch P. JNJ-4178 (AL-335, odalasvir, and simeprevir) for 6 or 8 Weeks in hepatitis C virus-infected patients without cirrhosis: omega-1. Hepatology. 2019;69:2349–2363. doi: 10.1002/hep.30527. [DOI] [PubMed] [Google Scholar]
- 93.Jonckers T.H., Tahri A., Vijgen L., Berke J.M., Lachau-Durand S., Stoops B., Snoeys J., Leclercq L., Tambuyzer L., Lin T.I., Simmen K., Raboisson P. Discovery of 1-((2R,4aR,6R,7R,7aR)-2-Isopropoxy-2-oxidodihydro-4H,6H-spiro[furo[3,2-d][1,3,2]dioxaphosphinine-7,2’-oxetan]-6-yl)pyrimidine-2,4(1H,3H)-dione (JNJ-54257099), a 3’-5’-cyclic phosphate ester prodrug of 2’-deoxy-2’-spirooxetane uridine triphosphate useful for HCV inhibition. J. Med. Chem. 2016;59:5790–5798. doi: 10.1021/acs.jmedchem.6b00382. [DOI] [PubMed] [Google Scholar]
- 94.Gentile I., Coppola N., Buonomo A.R., Zappulo E., Borgia G. Investigational nucleoside and nucleotide polymerase inhibitors and their use in treating hepatitis C virus. Expet Opin. Invest. Drugs. 2014;23:1211–1223. doi: 10.1517/13543784.2014.921680. [DOI] [PubMed] [Google Scholar]
- 95.Marcellin P., Popa S., Streinu-Cercel A., Boyer N., Dospinoiu D., Patat A., Ghicavii N., Garg V., Kauffman R., Koziel M. ALS-2200, a novel once-daily nucleotide hcv polymerase inhibitor, demonstrated potent antiviral activity in treatment-nave patients with compensated cirrhosis or genotype 2-4 chronic hepatitis c. J. Hepatol. 2013;58:S355. [Google Scholar]
- 96.Foster A.B. Deuterium isotope effects in studies of drug metabolism. Trends Pharmacol. Sci. 1984;5:524–527. [Google Scholar]
- 97.Gane E., Schwabe C., Stedman C., Weilert F., Stuart K., Cheng W., Yang J., Robison H., Hui J., Lahey J. LP27: ACH-3422, A novel nucleotide prodrug inhibitor of HCV NS5B polymerase. J. Hepatol. 2015;62:S277. [Google Scholar]
- 98.Kamat S.S., Burgos E.S., Raushel F.M. Potent inhibition of the C–P lyase nucleosidase PhnI by immucillin-A triphosphate. Biochemistry. 2013;52:7366–7368. doi: 10.1021/bi4013287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G., Honnold S., Bantia S., Kotian P., Chen X., Taubenheim B.R., Welch L.S., Minning D.M., Babu Y.S., Sheridan W.P., Bavari S. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Westover J.B., Mathis A., Taylor R., Wandersee L., Bailey K.W., Sefing E.J., Hickerson B.T., Jung K.H., Sheridan W.P., Gowen B.B. Galidesivir limits Rift Valley fever virus infection and disease in Syrian golden hamsters. Antivir. Res. 2018;156:38–45. doi: 10.1016/j.antiviral.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pedersen N.C., Perron M., Bannasch M., Montgomery E., Murakami E., Liepnieks M., Liu H. Efficacy and safety of the nucleoside analog GS-441524 for treatment of cats with naturally occurring feline infectious peritonitis. J. Feline Med. Surg. 2019;21:271–281. doi: 10.1177/1098612X19825701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murphy B.G., Perron M., Murakami E., Bauer K., Park Y., Eckstrand C., Liepnieks M., Pedersen N.C. The nucleoside analog GS-441524 strongly inhibits feline infectious peritonitis (FIP) virus in tissue culture and experimental cat infection studies. Vet. Microbiol. 2018;219:226–233. doi: 10.1016/j.vetmic.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alanazi A.S., James E., Mehellou Y. The ProTide prodrug technology: where next? ACS Med. Chem. Lett. 2019;10:2–5. doi: 10.1021/acsmedchemlett.8b00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liang C., Tian L., Liu Y., Hui N., Qiao G., Li H., Shi Z., Tang Y., Zhang D., Xie X., Zhao X. A promising antiviral candidate drug for the COVID-19 pandemic: a mini-review of remdesivir. Eur. J. Med. Chem. 2020;201:112527. doi: 10.1016/j.ejmech.2020.112527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Pérez-Pérez L., Okumura A., Lovaglio J., Hanley P.W., Saturday G., Bosio C.M., Anzick S., Barbian K., Cihlar T., Martens C., Scott D.P., Munster V.J., de Wit E. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tempestilli M., Caputi P., Avataneo V., Notari S., Forini O., Scorzolini L., Marchioni L., Ascoli Bartoli T., Castilletti C., Lalle E., Capobianchi M.R., Nicastri E., D’Avolio A., Ippolito G., Agrati C., Group C.I.S. Pharmacokinetics of remdesivir and GS-441524 in two critically ill patients who recovered from COVID-19. J. Antimicrob. Chemother. 2020;75:2977–2980. doi: 10.1093/jac/dkaa239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yan V.C., Muller F.L. Advantages of the parent nucleoside GS-441524 over remdesivir for covid-19 treatment. ACS Med. Chem. Lett. 2020;11:1361–1366. doi: 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Good S.S., Moussa A., Zhou X.J., Pietropaolo K., Sommadossi J.P. Preclinical evaluation of AT-527, a novel guanosine nucleotide prodrug with potent, pan-genotypic activity against hepatitis C virus. PloS One. 2020;15 doi: 10.1371/journal.pone.0227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berliba E., Bogus M., Vanhoutte F., Berghmans P.J., Good S.S., Moussa A., Pietropaolo K., Murphy R.L., Zhou X.J., Sommadossi J.P. vol. 63. 2019. Safety, pharmacokinetics, and antiviral activity of AT-527, a novel purine nucleotide prodrug; p. 12. (Hepatitis C Virus-Infected Subjects with or without Cirrhosis, Antimicrobial Agents and Chemotherapy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coats S.J., Garnier-Amblard E.C., Amblard F., Ehteshami M., Amiralaei S., Zhang H., Zhou L., Boucle S.R., Lu X., Bondada L., Shelton J.R., Li H., Liu P., Li C., Cho J.H., Chavre S.N., Zhou S., Mathew J., Schinazi R.F. Chutes and ladders in hepatitis C nucleoside drug development. Antivir. Res. 2014;102:119–147. doi: 10.1016/j.antiviral.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Standring D.N., Lanford R., Cretton-Scott E., Licklider L., Larsson M., Pierra C., Gosselin G., Perigaud C., Surleraux D., Mayes B., Moussa A., Selden J. Potent antiviral activity of second generation nucleoside inhibitors, IDX102 and IDX184, in HCV-infected chimpanzees. J. Hepatol. 2008;48:S30. [Google Scholar]
- 114.Lalezari J., Poordad F., Mehra P., Nguyen T., Dejesus E., Godofsky E., Patrick G.D., Chen J., Pietropaolo K., Zhou X.J., Sullivan-Bolyai J.Z., Mayers D., Grp I.-C.I. Antiviral activity, pharmacokinetics and safety of IDX184 in combination with pegylated interferon (PEGIFN) and ribavirin (RBV) in treatment - naive HCV genotype 1- infected subjects. J. Hepatol. 2010;52:S469. [Google Scholar]
- 115.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Madelain V., Oestereich L., Graw F., Nguyen T.H., de Lamballerie X., Mentré F., Günther S., Guedj J. Ebola virus dynamics in mice treated with favipiravir. Antivir. Res. 2015;123:70–77. doi: 10.1016/j.antiviral.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 118.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering (Beijing) 2020;6:1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Du Y.X., Chen X.P. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020;108:242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 120.Nyström K., Waldenström J., Tang K.-W., Lagging M. Ribavirin: pharmacology, multiple modes of action and possible future perspectives. Future Virol. 2019;14:153–160. [Google Scholar]
- 121.Chang J., Schul W., Butters T.D., Yip A., Liu B., Goh A., Lakshminarayana S.B., Alonzi D., Reinkensmeier G., Pan X., Qu X., Weidner J.M., Wang L., Yu W., Borune N., Kinch M.A., Rayahin J.E., Moriarty R., Xu X., Shi P.Y., Guo J.T., Block T.M. Combination of α-glucosidase inhibitor and ribavirin for the treatment of dengue virus infection in vitro and in vivo. Antivir. Res. 2011;89:26–34. doi: 10.1016/j.antiviral.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van Vonderen M.G., Bos J.C., Prins J.M., Wertheim-van Dillen P., Speelman P. Ribavirin in the treatment of severe acute respiratory syndrome (SARS) Neth. J. Med. 2003;61:238–241. [PubMed] [Google Scholar]
- 123.Tong S., Su Y., Yu Y., Wu C., Chen J., Wang S., Jiang J. Ribavirin therapy for severe COVID-19: a retrospective cohort study. Int. J. Antimicrob. Agents. 2020;56:106114. doi: 10.1016/j.ijantimicag.2020.106114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hung I.F., Lung K.C., Tso E.Y., Liu R., Chung T.W., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., Shum H.P., Chan V., Wu A.K., Sin K.M., Leung W.S., Law W.L., Lung D.C., Sin S., Yeung P., Yip C.C., Zhang R.R., Fung A.Y., Yan E.Y., Leung K.H., Ip J.D., Chu A.W., Chan W.M., Ng A.C., Lee R., Fung K., Yeung A., Wu T.C., Chan J.W., Yan W.W., Chan W.M., Chan J.F., Lie A.K., Tsang O.T., Cheng V.C., Que T.L., Lau C.S., Chan K.H., To K.K., Yuen K.Y. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chiou H.E., Liu C.L., Buttrey M.J., Kuo H.P., Liu H.W., Kuo H.T., Lu Y.T. Adverse effects of ribavirin and outcome in severe acute respiratory syndrome: experience in two medical centers. Chest. 2005;128:263–272. doi: 10.1378/chest.128.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Trevejo J.M., Asmal M., Vingerhoets J., Polo R., Robertson S., Jiang Y., Kieffer T.L., Leopold L. Pimodivir treatment in adult volunteers experimentally inoculated with live influenza virus: a Phase IIa, randomized, double-blind, placebo-controlled study. Antivir. Ther. 2018;23:335–344. doi: 10.3851/IMP3212. [DOI] [PubMed] [Google Scholar]
- 127.Finberg R.W., Lanno R., Anderson D., Fleischhackl R., van Duijnhoven W., Kauffman R.S., Kosoglou T., Vingerhoets J., Leopold L. Phase 2b study of pimodivir (JNJ-63623872) as monotherapy or in combination with oseltamivir for treatment of acute uncomplicated seasonal influenza A: TOPAZ trial. JID (J. Infect. Dis.) 2019;219:1026–1034. doi: 10.1093/infdis/jiy547. [DOI] [PubMed] [Google Scholar]
- 128.Beigel J.H., Nam H.H., Adams P.L., Krafft A., Ince W.L., El-Kamary S.S., Sims A.C. Advances in respiratory virus therapeutics - a meeting report from the 6th isirv Antiviral Group conference. Antivir. Res. 2019;167:45–67. doi: 10.1016/j.antiviral.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Le Pogam S., Kang H., Harris S.F., Leveque V., Giannetti A.M., Ali S., Jiang W.R., Rajyaguru S., Tavares G., Oshiro C., Hendricks T., Klumpp K., Symons J., Browner M.F., Cammack N., Nájera I. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 2006;80:6146–6154. doi: 10.1128/JVI.02628-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Beaulieu P.L., Bös M., Bousquet Y., Fazal G., Gauthier J., Gillard J., Goulet S., LaPlante S., Poupart M.A., Lefebvre S., McKercher G., Pellerin C., Austel V., Kukolj G. Non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase: discovery and preliminary SAR of benzimidazole derivatives. Bioorg. Med. Chem. Lett. 2004;14:119–124. doi: 10.1016/j.bmcl.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 131.McKercher G., Beaulieu P.L., Lamarre D., LaPlante S., Lefebvre S., Pellerin C., Thauvette L., Kukolj G. Specific inhibitors of HCV polymerase identified using an NS5B with lower affinity for template/primer substrate. Nucleic Acids Res. 2004;32:422–431. doi: 10.1093/nar/gkh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hirashima S., Suzuki T., Ishida T., Noji S., Yata S., Ando I., Komatsu M., Ikeda S., Hashimoto H. Benzimidazole derivatives bearing substituted biphenyls as hepatitis C virus NS5B RNA-dependent RNA polymerase inhibitors: structure-activity relationship studies and identification of a potent and highly selective inhibitor JTK-109. J. Med. Chem. 2006;49:4721–4736. doi: 10.1021/jm060269e. [DOI] [PubMed] [Google Scholar]
- 133.Beaulieu P.L., Bousquet Y., Gauthier J., Gillard J., Marquis M., McKercher G., Pellerin C., Valois S., Kukolj G. Non-nucleoside benzimidazole-based allosteric inhibitors of the hepatitis C virus NS5B polymerase: inhibition of subgenomic hepatitis C virus RNA replicons in Huh-7 cells. J. Med. Chem. 2004;47:6884–6892. doi: 10.1021/jm040134d. [DOI] [PubMed] [Google Scholar]
- 134.Beaulieu P.L., Gillard J., Bykowski D., Brochu C., Dansereau N., Duceppe J.S., Haché B., Jakalian A., Lagacé L., LaPlante S., McKercher G., Moreau E., Perreault S., Stammers T., Thauvette L., Warrington J., Kukolj G. Improved replicon cellular activity of non-nucleoside allosteric inhibitors of HCV NS5B polymerase: from benzimidazole to indole scaffolds. Bioorg. Med. Chem. Lett. 2006;16:4987–4993. doi: 10.1016/j.bmcl.2006.07.074. [DOI] [PubMed] [Google Scholar]
- 135.Larrey D., Benhamou Y., Lohse A.W., Trepo C., Moelleken C., Bronowicki J.P., Arasteh K., Bourliere M., Heim M., Enriquez J., Erhardt A., Zarski J.P., Wiest R., Gerlach T., Wedemeyer H., Berg T., Stern J., Wu K., Abdallah N., Nehmiz G., Boecher W., Steffgen J. SAFETY, pharmacokinetics and antiviral effect OF BI 207127, a novel HCV RNA polymerase inhibitor, after 5 days oral treatment IN patients with chronic hepatitis C. J. Hepatol. 2009;50:S383–S384. [Google Scholar]
- 136.Snape M.D., Kelly D.F., Salt P., Green S., Snowden C., Diggle L., Borkowski A., Yu L.M., Moxon E.R., Pollard A.J. Serogroup C meningococcal glycoconjugate vaccine in adolescents: persistence of bactericidal antibodies and kinetics of the immune response to a booster vaccine more than 3 years after immunization. Clin. Infect. Dis. 2006;43:1387–1394. doi: 10.1086/508776. [DOI] [PubMed] [Google Scholar]
- 137.Stansfield I., Ercolani C., Mackay A., Conte I., Pompei M., Koch U., Gennari N., Giuliano C., Rowley M., Narjes F. Tetracyclic indole inhibitors of hepatitis C virus NS5B-polymerase. Bioorg. Med. Chem. Lett. 2009;19:627–632. doi: 10.1016/j.bmcl.2008.12.068. [DOI] [PubMed] [Google Scholar]
- 138.Narjes F., Crescenzi B., Ferrara M., Habermann J., Colarusso S., Ferreira Mdel R., Stansfield I., Mackay A.C., Conte I., Ercolani C., Zaramella S., Palumbi M.C., Meuleman P., Leroux-Roels G., Giuliano C., Fiore F., Di Marco S., Baiocco P., Koch U., Migliaccio G., Altamura S., Laufer R., De Francesco R., Rowley M. Discovery of (7R)-14-cyclohexyl-7-{[2-(dimethylamino)ethyl](methyl) amino}-7,8-dihydro-6H-indolo[1,2-e][1,5]benzoxazocine-11-carboxylic acid (MK-3281), a potent and orally bioavailable finger-loop inhibitor of the hepatitis C virus NS5B polymerase. J. Med. Chem. 2011;54:289–301. doi: 10.1021/jm1013105. [DOI] [PubMed] [Google Scholar]
- 139.Brainard D., Anderson M., Petry A., Van Dyck K., De Lepeleire I., Sneddon K., Cummings C., Nachbar R., Barnard R., Sun P. Safety and antiviral activity of NS5B polymerase inhibitor MK-3281. Treatment-Naïve Genotype A. 2009;1:1568. [Google Scholar]
- 140.Zeuzem S., Soriano V., Asselah T., Gane E.J., Bronowicki J.-P., Angus P., Lohse A.W., Stickel F., Müllhaupt B., Roberts S. Efficacy and safety of faldaprevir, deleobuvir, and ribavirin in treatment-naive patients with chronic hepatitis C virus infection and advanced liver fibrosis or cirrhosis. Antimicrob. Agents Chemother. 2015;59:1282–1291. doi: 10.1128/AAC.04383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Love R.A., Parge H.E., Yu X., Hickey M.J., Diehl W., Gao J., Wriggers H., Ekker A., Wang L., Thomson J.A., Dragovich P.S., Fuhrman S.A. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 2003;77:7575–7581. doi: 10.1128/JVI.77.13.7575-7581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kukolj G., McGibbon G.A., McKercher G., Marquis M., Lefèbvre S., Thauvette L., Gauthier J., Goulet S., Poupart M.A., Beaulieu P.L. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J. Biol. Chem. 2005;280:39260–39267. doi: 10.1074/jbc.M506407200. [DOI] [PubMed] [Google Scholar]
- 143.Lemm J.A., Liu M., Gentles R.G., Ding M., Voss S., Pelosi L.A., Wang Y.K., Rigat K.L., Mosure K.W., Bender J.A., Knipe J.O., Colonno R., Meanwell N.A., Kadow J.F., Santone K.S., Roberts S.B., Gao M. Preclinical characterization of BMS-791325, an allosteric inhibitor of hepatitis C Virus NS5B polymerase. Antimicrob. Agents Chemother. 2014;58:3485–3495. doi: 10.1128/AAC.02495-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gentile I., Zappulo E., Buonomo A.R., Maraolo A.E., Borgia G. Beclabuvir for the treatment of hepatitis C. Expet Opin. Invest. Drugs. 2015;24:1111–1121. doi: 10.1517/13543784.2015.1059820. [DOI] [PubMed] [Google Scholar]
- 145.Poordad F., Sievert W., Mollison L., Bennett M., Tse E., Bräu N., Levin J., Sepe T., Lee S.S., Angus P., Conway B., Pol S., Boyer N., Bronowicki J.P., Jacobson I., Muir A.J., Reddy K.R., Tam E., Ortiz-Lasanta G., de Lédinghen V., Sulkowski M., Boparai N., McPhee F., Hughes E., Swenson E.S., Yin P.D. Fixed-dose combination therapy with daclatasvir, asunaprevir, and beclabuvir for noncirrhotic patients with HCV genotype 1 infection. JAMA, J. Am. Med. Assoc. 2015;313:1728–1735. doi: 10.1001/jama.2015.3860. [DOI] [PubMed] [Google Scholar]
- 146.Tatum H., Thuluvath P.J., Lawitz E., Martorell C., DeMicco M., Cohen S., Rustgi V., Ravendhran N., Ghalib R., Hanson J., Zamparo J., Zhao J., Cooney E., Treitel M., Hughes E. A randomized, placebo-controlled study of the NS5B inhibitor beclabuvir with peginterferon/ribavirin for HCV genotype 1. J. Viral Hepat. 2015;22:658–664. doi: 10.1111/jvh.12372. [DOI] [PubMed] [Google Scholar]
- 147.Wang M., Ng K.K., Cherney M.M., Chan L., Yannopoulos C.G., Bedard J., Morin N., Nguyen-Ba N., Alaoui-Ismaili M.H., Bethell R.C., James M.N. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 2003;278:9489–9495. doi: 10.1074/jbc.M209397200. [DOI] [PubMed] [Google Scholar]
- 148.Love R.A., Maegley K.A., Yu X., Ferre R.A., Lingardo L.K., Diehl W., Parge H.E., Dragovich P.S., Fuhrman S.A. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure. 2004;12:1533–1544. doi: 10.1016/j.str.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 149.Fenaux M., Eng S., Leavitt S.A., Lee Y.J., Mabery E.M., Tian Y., Byun D., Canales E., Clarke M.O., Doerffler E., Lazerwith S.E., Lew W., Liu Q., Mertzman M., Morganelli P., Xu L., Ye H., Zhang J., Matles M., Murray B.P., Mwangi J., Zhang J., Hashash A., Krawczyk S.H., Bidgood A.M., Appleby T.C., Watkins W.J. Preclinical characterization of GS-9669, a thumb site II inhibitor of the hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 2013;57:804–810. doi: 10.1128/AAC.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gentile I., Buonomo A.R., Zappulo E., Coppola N., Borgia G. GS-9669: a novel non-nucleoside inhibitor of viral polymerase for the treatment of hepatitis C virus infection. Expert Rev. Anti-infect. Ther. 2014;12:1179–1186. doi: 10.1586/14787210.2014.945432. [DOI] [PubMed] [Google Scholar]
- 151.Shi S.T., Herlihy K.J., Graham J.P., Nonomiya J., Rahavendran S.V., Skor H., Irvine R., Binford S., Tatlock J., Li H., Gonzalez J., Linton A., Patick A.K., Lewis C. Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 2009;53:2544–2552. doi: 10.1128/AAC.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li H., Tatlock J., Linton A., Gonzalez J., Jewell T., Patel L., Ludlum S., Drowns M., Rahavendran S.V., Skor H. Discovery of (R)-6-cyclopentyl-6-(2-(2, 6-diethylpyridin-4-yl) ethyl)-3-((5, 7-dimethyl-[1, 2, 4] triazolo [1, 5-a] pyrimidin-2-yl) methyl)-4-hydroxy-5, 6-dihydropyran-2-one (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor. J. Med. Chem. 2009;52:1255–1258. doi: 10.1021/jm8014537. [DOI] [PubMed] [Google Scholar]
- 153.Chan L., Das S.K., Reddy T.J., Poisson C., Proulx M., Pereira O., Courchesne M., Roy C., Wang W., Siddiqui A., Yannopoulos C.G., Nguyen-Ba N., Labrecque D., Bethell R., Hamel M., Courtemanche-Asselin P., L’Heureux L., David M., Nicolas O., Brunette S., Bilimoria D., Bédard J. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 1: Sulfonamides. Bioorg. Med. Chem. Lett. 2004;14:793–796. doi: 10.1016/j.bmcl.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 154.Hassan G.S., Georgey H.H., Mohammed E.Z., Omar F.A. Anti-hepatitis-C virus activity and QSAR study of certain thiazolidinone and thiazolotriazine derivatives as potential NS5B polymerase inhibitors. Eur. J. Med. Chem. 2019;184:111747. doi: 10.1016/j.ejmech.2019.111747. [DOI] [PubMed] [Google Scholar]
- 155.Cooper C., Lawitz E.J., Ghali P., Rodriguez-Torres M., Anderson F.H., Lee S.S., Bédard J., Chauret N., Thibert R., Boivin I., Nicolas O., Proulx L. Evaluation of VCH-759 monotherapy in hepatitis C infection. J. Hepatol. 2009;51:39–46. doi: 10.1016/j.jhep.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 156.Proulx L., Bourgault B., Chauret N., Larouche R., Tanguay M., Thibert R. Results of a safety, tolerability and pharmacokinetic phase I study of VCH-916, a novel polymerase inhibitor for HCV, following single ascending doses in healthy volunteers. J. Hepatol. 2008;48:S320–S321. [Google Scholar]
- 157.Yi G., Deval J., Fan B., Cai H., Soulard C., Ranjith-Kumar C.T., Smith D.B., Blatt L., Beigelman L., Kao C.C. Biochemical study of the comparative inhibition of hepatitis C virus RNA polymerase by VX-222 and filibuvir. Antimicrob. Agents Chemother. 2012;56:830–837. doi: 10.1128/AAC.05438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bedard J., Nicolas O., Bilimoria D., L’Heureux L., Fex P., David M., Chan L. 935 identification and characterization of VCH-222, a novel potent and selective non-nucleoside HCV polymerase inhibitor. J. Hepatol. 2009;50:S340. [Google Scholar]
- 159.Cooper C., Larouche R., Bourgault B., Chauret N., Proulx L. Safety, toperability and pharmacokinetics of the HCV polymerase inhibitor VCH-222 following single dose administration in healthy volunteers and antiviral activity in HCV-infected individuals. J. Hepatol. 2009;50:S342. [Google Scholar]
- 160.Gopalsamy A., Lim K., Ciszewski G., Park K., Ellingboe J.W., Bloom J., Insaf S., Upeslacis J., Mansour T.S., Krishnamurthy G., Damarla M., Pyatski Y., Ho D., Howe A.Y., Orlowski M., Feld B., O’Connell J. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J. Med. Chem. 2004;47:6603–6608. doi: 10.1021/jm0401255. [DOI] [PubMed] [Google Scholar]
- 161.Gopalsamy A., Shi M., Ciszewski G., Park K., Ellingboe J.W., Orlowski M., Feld B., Howe A.Y. Design and synthesis of 2,3,4,9-tetrahydro-1H-carbazole and 1,2,3,4-tetrahydro-cyclopenta[b]indole derivatives as non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA polymerase. Bioorg. Med. Chem. Lett. 2006;16:2532–2534. doi: 10.1016/j.bmcl.2006.01.105. [DOI] [PubMed] [Google Scholar]
- 162.Gopalsamy A., Aplasca A., Ciszewski G., Park K., Ellingboe J.W., Orlowski M., Feld B., Howe A.Y. Design and synthesis of 3,4-dihydro-1H-[1]-benzothieno[2,3-c]pyran and 3,4-dihydro-1H-pyrano[3,4-b]benzofuran derivatives as non-nucleoside inhibitors of HCV NS5B RNA dependent RNA polymerase. Bioorg. Med. Chem. Lett. 2006;16:457–460. doi: 10.1016/j.bmcl.2005.08.114. [DOI] [PubMed] [Google Scholar]
- 163.Schoenfeld R.C., Bourdet D.L., Brameld K.A., Chin E., de Vicente J., Fung A., Harris S.F., Lee E.K., Le Pogam S., Leveque V., Li J., Lui A.S., Najera I., Rajyaguru S., Sangi M., Steiner S., Talamas F.X., Taygerly J.P., Zhao J. Discovery of a novel series of potent non-nucleoside inhibitors of hepatitis C virus NS5B. J. Med. Chem. 2013;56:8163–8182. doi: 10.1021/jm401266k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Randolph J.T., Krueger A.C., Donner P.L., Pratt J.K., Liu D., Motter C.E., Rockway T.W., Tufano M.D., Wagner R., Lim H.B., Beyer J.M., Mondal R., Panchal N.S., Colletti L., Liu Y., Koev G., Kati W.M., Hernandez L.E., Beno D.W.A., Longenecker K.L., Stewart K.D., Dumas E.O., Molla A., Maring C.J. Synthesis and biological characterization of aryl uracil inhibitors of hepatitis C virus NS5B polymerase: discovery of ABT-072, a trans-stilbene analog with good oral bioavailability. J. Med. Chem. 2018;61:1153–1163. doi: 10.1021/acs.jmedchem.7b01630. [DOI] [PubMed] [Google Scholar]
- 165.Randolph J.T., Krueger A.C., Donner P.L., Pratt J.K., Liu D., Motter C.E., Rockway T.W., Tufano M.D., Wagner R., Lim H.B. Synthesis and biological characterization of aryl uracil inhibitors of hepatitis C virus NS5B polymerase: discovery of ABT-072, a trans-Stilbene analog with good oral bioavailability. J. Med. Chem. 2018;61:1153–1163. doi: 10.1021/acs.jmedchem.7b01630. [DOI] [PubMed] [Google Scholar]
- 166.Lawitz E., Poordad F., Kowdley K.V., Cohen D.E., Podsadecki T., Siggelkow S., Larsen L., Menon R., Koev G., Tripathi R. A phase 2a trial of 12-week interferon-free therapy with two direct-acting antivirals (ABT-450/r, ABT-072) and ribavirin in IL28B C/C patients with chronic hepatitis C genotype 1. J. Hepatol. 2013;59:18–23. doi: 10.1016/j.jhep.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 167.Liu Y., Lim B.H., Jiang W.W., Flentge C.A., Hutchinson D.K., Madigan D.L., Randolph J.T., Wagner R., Maring C.J., Kati W.M., Molla A. Identification of aryl dihydrouracil derivatives as palm initiation site inhibitors of HCV NS5B polymerase. Bioorg. Med. Chem. Lett. 2012;22:3747–3750. doi: 10.1016/j.bmcl.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 168.Kati W., Koev G., Irvin M., Beyer J., Liu Y., Krishnan P., Reisch T., Mondal R., Wagner R., Molla A., Maring C., Collins C. In vitro activity and resistance profile of dasabuvir, a nonnucleoside hepatitis C virus polymerase inhibitor. Antimicrob. Agents Chemother. 2015;59:1505–1511. doi: 10.1128/AAC.04619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.El Kassas M., Elbaz T., Hafez E., Wifi M.N., Esmat G. Discovery and preclinical development of dasabuvir for the treatment of hepatitis C infection. Expet Opin. Drug Discov. 2017;12:635–642. doi: 10.1080/17460441.2017.1322955. [DOI] [PubMed] [Google Scholar]
- 170.Shaw A.N., Tedesco R., Bambal R., Chai D., Concha N.O., Darcy M.G., Dhanak D., Duffy K.J., Fitch D.M., Gates A., Johnston V.K., Keenan R.M., Lin-Goerke J., Liu N., Sarisky R.T., Wiggall K.J., Zimmerman M.N. Substituted benzothiadizine inhibitors of Hepatitis C virus polymerase. Bioorg. Med. Chem. Lett. 2009;19:4350–4353. doi: 10.1016/j.bmcl.2009.05.091. [DOI] [PubMed] [Google Scholar]
- 171.Tedesco R., Shaw A.N., Bambal R., Chai D., Concha N.O., Darcy M.G., Dhanak D., Fitch D.M., Gates A., Gerhardt W.G., Halegoua D.L., Han C., Hofmann G.A., Johnston V.K., Kaura A.C., Liu N., Keenan R.M., Lin-Goerke J., Sarisky R.T., Wiggall K.J., Zimmerman M.N., Duffy K.J. 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem. 2006;49:971–983. doi: 10.1021/jm050855s. [DOI] [PubMed] [Google Scholar]
- 172.Lawitz E., Rodriguez-Torres M., DeMicco M., Nguyen T., Godofsky E., Appleman J., Rahimy M., Crowley C., Freddo J. 1055 antiviral activity OF ANA598, a potent NON-nucleoside polymerase inhibitor, IN chronic hepatitis C patients. J. Hepatol. 2009;50:S384. [Google Scholar]
- 173.I. Gaultier, D. Cohen, E. Dumas, L. Larsen, T. Podsadecki, B. Bernstein, 12-week efficacy and safety of ABT-072 or ABT-333 with pegylated interferon + ribavirin, following 3-day monotherapy in genotype 1 HCV infected treatment-naive subjects, 21st Conference of the Asian Pacific Association for the Study of the Liver (APASL), 17-20 February 2011, Thailand.
- 174.Gray F., Amphlett E., Bright H., Chambers L., Cheasty A., Fenwick R., Haigh D., Hartley D., Howes P., Jarvest R. GSK625433; a novel and highly potent inhibitor of the HCV NS5B polymerase. J. Hepatol. 2007;46:S225. [Google Scholar]
- 175.Standring D.N., Bilello J.P., Dousson C.B., Griffon J.F., LaColla M., Lallos L., Liuzzi M., Loi A.G., McCarville J., Paparin J.L., Pierra C., Roland A., Seifer M., Surleraux D. In vitro activity and pharmacological properties of IDX375, a novel HCV non-nucleoside inhibitor. Hepatology. 2008;48:1166A–1167A. [Google Scholar]
- 176.de Bruijne J., van de Wetering de Rooij J., van Vliet A.A., Zhou X.J., Temam M.F., Molles J., Chen J., Pietropaolo K., Sullivan-Bólyai J.Z., Mayers D., Reesink H.W. First-in-human study of the pharmacokinetics and antiviral activity of IDX375, a novel nonnucleoside hepatitis C virus polymerase inhibitor. Antimicrob. Agents Chemother. 2012;56:4525–4528. doi: 10.1128/AAC.00451-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jacobson I., Feese M., Xiao H., Uher L., Lin B., Sanchez E., Tomkiewicz R., Whitaker T., McBrayer T., Pascual L. CC-31244, A novel, pan-genotypic, potent NS5B non-nucleoside polymerase inhibitor for the treatment of chronic hepatitis C. J. Hepatol. 2016;64:S421–S422. [Google Scholar]
- 178.Lee S., Pascual L., Pattassery J., Sulkowski M. Phase 1 study assessing the safety, pharmacokinetics, and antiviral activity of CC-31244, a pan-genotypic, potent non-nucleoside NS5B polymerase inhibitor for the treatment of hepatitis C virus infection. J. Hepatol. 2017;66:S720. [Google Scholar]
- 179.Howe A.Y., Cheng H., Johann S., Mullen S., Chunduru S.K., Young D.C., Bard J., Chopra R., Krishnamurthy G., Mansour T., O’Connell J. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor. HCV-796, Antimicrobial Agents and Chemotherapy. 2008;52:3327–3338. doi: 10.1128/AAC.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kneteman N.M., Howe A.Y., Gao T., Lewis J., Pevear D., Lund G., Douglas D., Mercer D.F., Tyrrell D.L., Immermann F., Chaudhary I., Speth J., Villano S.A., O’Connell J., Collett M. HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology. 2009;49:745–752. doi: 10.1002/hep.22717. [DOI] [PubMed] [Google Scholar]
- 181.Feldstein A., Kleiner D., Kravetz D., Buck M. Severe hepatocellular injury with apoptosis induced by a hepatitis C polymerase inhibitor. J. Clin. Gastroenterol. 2009;43:374–381. doi: 10.1097/MCG.0b013e318178d91f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bavisotto L., Wang C.C., Jacobson I.M., Marcellin P., Zeuzem S., Lawitz E.J., Lunde M., Sereni P., O’Brien C., Oldach D.W., Rhodes G. Antiviral, pharmacokinetic and safety data for GS-9190, a non-nucleoside HCVNS5B polymerase inhibitor, in a phase-1 trial in HCV genotype 1 infected subjects. Hepatology. 2007;46:255A. [Google Scholar]
- 183.Wong K.A., Xu S., Martin R., Miller M.D., Mo H. Tegobuvir (GS-9190) potency against HCV chimeric replicons derived from consensus NS5B sequences from genotypes 2b, 3a, 4a, 5a, and 6a. Virology. 2012;429:57–62. doi: 10.1016/j.virol.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 184.Cheng C.C., Shipps G.W., Jr., Yang Z., Kawahata N., Lesburg C.A., Duca J.S., Bandouveres J., Bracken J.D., Jiang C.K., Agrawal S., Ferrari E., Huang H.C. Inhibitors of hepatitis C virus polymerase: synthesis and characterization of novel 2-oxy-6-fluoro-N-((S)-1-hydroxy-3-phenylpropan-2-yl)-benzamides. Bioorg. Med. Chem. Lett. 2010;20:2119–2124. doi: 10.1016/j.bmcl.2010.02.054. [DOI] [PubMed] [Google Scholar]
- 185.Jácome R., Campillo-Balderas J.A., Ponce de León S., Becerra A., Lazcano A. Sofosbuvir as a potential alternative to treat the SARS-CoV-2 epidemic. Sci. Rep. 2020;10:9294. doi: 10.1038/s41598-020-66440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Parvez M.S.A., Karim M.A., Hasan M., Jaman J., Karim Z., Tahsin T., Hasan M.N., Hosen M.J. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int. J. Biol. Macromol. 2020;163:1787–1797. doi: 10.1016/j.ijbiomac.2020.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hughes S., Troise O., Donaldson H., Mughal N., Moore L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020;26:1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.McKenzie R.H., Bekker C., Athokpam B., Ramesh S.G. Effect of quantum nuclear motion on hydrogen bonding. J. Chem. Phys. 2014;140:174508. doi: 10.1063/1.4873352. [DOI] [PubMed] [Google Scholar]
- 190.Wen W.D., Chen Q., Shi W.Q., Deng C., Shi Y., Tang W., Chen K., Wu X., Dai M.X., Tang Y.J. 5 June 2020. Deuterium A Nucleoside Analogue and Preparation Method and Use Thereof. CN111233929A. [Google Scholar]
- 191.Wang G., Dyatkina N., Prhavc M., Williams C., Serebryany V., Hu Y., Huang Y., Wu X., Chen T., Huang W., Rajwanshi V.K., Deval J., Fung A., Jin Z., Stoycheva A., Shaw K., Gupta K., Tam Y., Jekle A., Smith D.B., Beigelman L. Synthesis and anti-HCV activity of sugar-modified guanosine analogues: discovery of AL-611 as an HCV NS5B polymerase inhibitor for the treatment of chronic hepatitis C. J. Med. Chem. 2020;63:10380–10395. doi: 10.1021/acs.jmedchem.0c00935. [DOI] [PubMed] [Google Scholar]
- 192.Yoon J.-s., Kim G., Jarhad D.B., Kim H.-R., Shin Y.-S., Qu S., Sahu P.K., Kim H.O., Lee H.W., Wang S.B., Kong Y.J., Chang T.-S., Ogando N.S., Kovacikova K., Snijder E.J., Posthuma C.C., van Hemert M.J., Jeong L.S. Design, synthesis, and anti-RNA virus activity of 6′-fluorinated-aristeromycin analogues. J. Med. Chem. 2019;62:6346–6362. doi: 10.1021/acs.jmedchem.9b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]